Abstract
Eukaryotic gene expression is regulated by enhancer–promoter interactions but the molecular mechanisms that govern specificity have remained elusive. Genome-wide studies utilizing STARR-seq identified two enhancer classes in Drosophila that interact with different core promoters: housekeeping enhancers (hkCP) and developmental enhancers (dCP). We hypothesized that the two enhancer classes are occupied by distinct architectural proteins, affecting their enhancer–promoter contacts. By evaluating ChIP-seq occupancy of architectural proteins, typical enhancer-associated proteins, and histone modifications, we determine that both enhancer classes are enriched for RNA Polymerase II, CBP, and architectural proteins but there are also distinctions. hkCP enhancers contain H3K4me3 and exclusively bind Cap-H2, Chromator, DREF and Z4, whereas dCP enhancers contain H3K4me1 and are more enriched for Rad21 and Fs(1)h-L. Additionally, we map the interactions of each enhancer class utilizing a Hi-C dataset with <1 kb resolution. Results suggest that hkCP enhancers are more likely to form multi-TSS interaction networks and be associated with topologically associating domain (TAD) borders, while dCP enhancers are more often bound to one or two TSSs and are enriched at chromatin loop anchors. The data support a model suggesting that the unique architectural protein occupancy within enhancers is one contributor to enhancer–promoter interaction specificity.
INTRODUCTION
Eukaryotic gene expression is regulated by a complex interplay of different regulatory elements. Genes contain core promoters that are bound by general transcription factors (GTFs) and RNA Polymerase II to form the pre-initiation complex adjacent to the transcription start site (TSS) (1,2). In addition, promoter proximal regulatory elements, typically located ∼100–200 bp upstream of the core promoter, bind transcription factors and promote the expression of the neighboring genes by enhancing the recruitment of the GTFs to core promoters or improving the recruitment of distal regulatory elements to promoters (3,4). Distal regulatory elements, commonly referred to as enhancers, are often many kilobases away from TSSs and interact with promoters to stimulate transcriptional output (1). The molecular mechanisms determining which combination of regulatory elements interact with a given promoter have remained somewhat elusive because comprehensive identification of promoter–enhancer interactions has proven technically challenging.
In recent years, whole genome sequencing technologies have significantly enhanced the mapping of enhancer–promoter interactions. Chromatin Interaction Analysis Paired-End Tag (ChIA-PET) analysis, a method that identifies a subset of the chromatin interactions mediated by a specific protein, for RNA Polymerase II led to the discovery of many promoter–enhancer as well as promoter–promoter interactions in five different human cell types (5). In addition, Capture Hi-C, a modified Hi-C technique that enriches for contacts occurring at genomic loci of interest, has been utilized to map the loci that interact with the ∼22 000 promoters in mouse and human cells (6–8). These comprehensive analyses of enhancer–promoter interactions have demonstrated that a given promoter often associates with multiple regulatory elements, which is supported by 4C-seq studies of 92 enhancers in flies (9). Complementary studies utilizing an approach called self-transcribing active regulatory region sequencing (STARR-seq) has also improved the genome-wide detection of enhancers in Drosophila and human cells (10–12). Of particular interest, a recent study utilizing STARR-seq with two different core promoters identified two distinct enhancer classes in Drosophila. One class is promoter-proximal and interacts specifically with a housekeeping core promoter, whereas the second class is located distal to promoters and interacts with a developmentally regulated core promoter (11). Notably, previous studies have demonstrated that housekeeping genes are active, while developmental enhancers tend to be silent in any particular Drosophila cell line, suggesting that a portion of the developmental enhancers identified by STARR-seq may be active in one cell line but inactive in another. The increase in comprehensive identification of enhancers and potential identification of additional subclasses will likely be instrumental in elucidating the molecular mechanisms that regulate enhancer–promoter specificity.
A number of potential molecular mechanisms have been described to explain the observed specificity between enhancers and promoters, including an intrinsic compatibility between promoter and enhancer sequences, and the 3D chromatin architecture surrounding a locus (13). These mechanisms are not mutually exclusive and both likely contribute to the establishment of enhancer–promoter specificity. There are many examples of individual promoter–enhancer studies demonstrating that the motifs present within a core promoter influence promoter–enhancer compatibility, a conclusion that has now been supported on a genome-wide scale by STARR-seq (11,14–20). In addition, the strongest evidence that 3D chromatin architecture regulates enhancer–promoter contacts comes from an analysis of phenotypes resulting from altered Topologically Associating Domains (TADs), which represent regions of highly interacting chromatin and compartmentalization within individual chromosomes (21–24). Genomic deletions and inversions that alter the location of a TAD border result in ectopic interactions between the EPHA4 enhancer and three neighboring genes, ultimately generating malformed limb phenotypes and implicating 3D chromatin architecture as an important contributor to enhancer–promoter interactions (25). Notably, the EPHA4 enhancer does not interact with all the genes in the novel TAD generated by the genomic rearrangement, further supporting the notion that a combination of intrinsic compatibility and chromatin architecture regulate enhancer–promoter interactions (25).
The proteins that regulate chromatin architecture and enhancer–promoter specific interactions are still poorly understood, but there is growing evidence that a family of architectural proteins mediate these contacts. In Drosophila, architectural proteins can be divided into two groups: DNA-binding proteins (CTCF, SuHw, BEAF-32, DREF, TFIIIC, Z4, Elba, ZIPIC, Ibf1 and Ibf2) and accessory proteins that form complexes with their DNA-binding counterparts (CP190, Mod(mdg4), Rad21 (a component of the cohesin complex), Cap-H2 (a component of the condensin complex), Fs(1)h-L, L3mbt and Chromator) (26–30). Although some of these architectural proteins have been experimentally shown to mediate or support chromatin interactions, others, including Z4, Fs(1)h-L and DREF, have been proposed to function as architectural proteins only based on their presence at genomic sites bound by functionally characterized architectural proteins (31). Architectural protein genomic occupancy is highly correlated with regulatory elements (31). Furthermore, ChIA-PET analysis demonstrated that cohesin and CTCF are present at the anchors of genome-wide enhancer–promoter interactions in mammals (32,33). Functional evidence that architectural proteins contribute to enhancer interactions was demonstrated by CRISPR-mediated deletion or inversion of genomic CTCF motifs, resulting in altered interactions between neighboring genomic loci (32,34,35). In addition to mediating individual chromatin interactions, architectural proteins have also been implicated in regulating TAD structure, and their location in the genome is highly correlated with TAD borders in mammals and flies (22–24,36). Furthermore, depletion of either CTCF or Rad21 results in a loss of intra-TAD interactions and an increase of inter-TAD interactions, indicative that these architectural proteins are required to maintain TADs (37–40). Altogether, these studies have led to a model suggesting that architectural proteins are key regulators of chromatin interactions and architecture and thus, potential mediators of enhancer–promoter specificity.
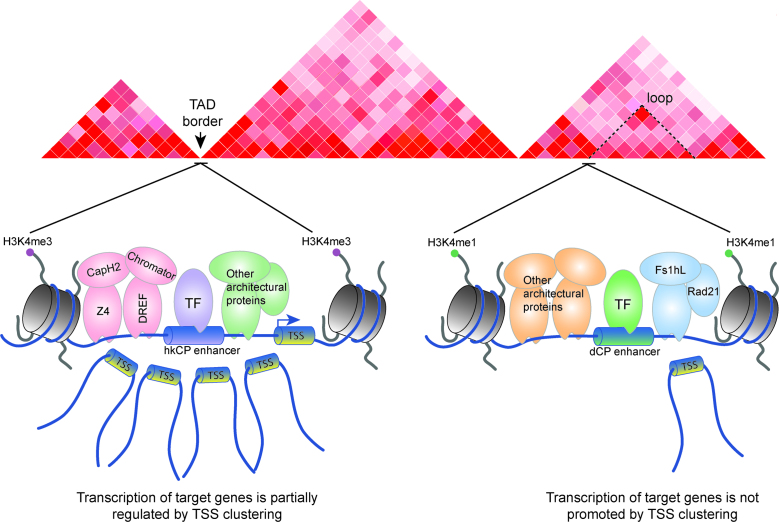
Here, we utilize the two distinct enhancer classes identified by STARR-seq in Drosophila S2 cells, the housekeeping core promoter interacting enhancers (hkCP enhancers) and the developmental core promoter interacting enhancers (dCP enhancers), as a model to further investigate the role of architectural proteins in the regulation of enhancer–promoter interactions. Zabidi et al. demonstrated that mutation of the DRE motif, which likely recruits DREF or BEAF-32, is essential for hkCP enhancer core promoter function, suggesting that architectural protein occupancy is a key contributor to enhancer–promoter specificity (11,41). Because the STARR-seq enhancers were discovered with an ectopic assay, we utilized histone modification and architectural protein ChIP-seq analyses of active enhancers to demonstrate that the two enhancer classes have distinct protein occupancy profiles. Notably, only dCPs contain the classical enhancer modification H3K4me1, while hkCP enhancers are enriched for H3K4me3. Both enhancer classes are occupied by many architectural proteins but enrichment of subcomplexes of architectural proteins within each class is observed. CAP-H2, Chromator, DREF, and Z4 are almost exclusively associated with hkCP enhancers, while Rad21 and Fs(1)h-L occupancy is more enriched in dCP enhancers compared to hkCP enhancers but not exclusive to dCP enhancers. Using high resolution Hi-C, we show that hkCP and dCP enhancers make distinct types of long-range chromatin interactions and are associated with TAD borders or chromatin loop anchors, respectively. Altogether, the results suggest that differential architectural protein occupancy contributes to distinct enhancer identity, ultimately affecting the interactions and architecture generated by these regulatory elements.
MATERIALS AND METHODS
Cell culture
Drosophila Kc167 cells were cultured at 25°C in Hyclone SFX insect cell culture media (GE Healthcare). Asynchronously growing cells were harvested and utilized for ChIP-seq and Hi-C experiments.
Antibodies
The following antibodies were generous gifts from the following sources: anti-Pita and anti-ZIPIC from Pavel Georgiev (Russian Academy of Sciences), anti-Ibf1 and anti-Ibf2 from M. Lluisa Espinás (Institute of Molecular Biology of Barcelona), and anti-GAF from Carl Wu (Janelia Research Campus). anti-Nup98 polyclonal antibodies were generated by immunizing rabbits with full length Nup98 (Pocono Rabbit Farm and Laboratory, Canadensis, PA). The following antibodies were obtained from commercial sources: anti-H3K27me3 (Millipore Cat# 07-449) and anti-H3 (Abcam ab1791).
ChIP-seq data generation and processing
Previously reported ChIP-seq data for S2 and Kc167 cells was obtained from the Gene Expression Omnibus (GEO) database. Raw ChIP data for Kc167 cells was obtained from the following sources: BEAF-32 (GSE 30740, GSE63518), CAP-H2 (GSE 54529, GSE63518), CBP (GSE 63518), CP190 (GSE 3074, GSE54529, GSE63518), Chromator (GSE 54529, GSE63518), CTCF (GSE30740), DREF (GSE63518, GSE39664), Fs(1)h-L (GSE63518, GSE42086), GAF (GSE 54529 and data from this study), H3K4me1 (GSE 36374, GSE63518), H3K4me3 (GSE63518), H3K27ac (GSE36374), H3K27me3 (37444 and data from this study), IgG (GSE63518), L3mbt (GSE36393, GSE63518), Mod(mdg4) (GSE36393), Rad21 (GSE54529, GSE63518), RNA Polymerase II (GSE63518), SuHw (GSE30740), ttk (GSE34698), TFIIIC (GSE63518,GSE54529) and Z4 (GSE63518). In addition, raw ChIP data for S2 cells was obtained from the following sources: CP190, CTCF, Mod(mdg4) and SuHw (GSE41354), H3K4me1 and H3K4me3 (GSE41440), Ibf1 and Ibf2 (GSE47559), Input (GSE41440, GSE41354, GSE54337), Pita and ZIPIC (GSE54337). For proteins with multiple replicates, the reads from each replicate were combined prior to genomic mapping.
New ChIP experiments were performed in Kc167 cells for GAF, H3, H3K27me3, Ibf1, Ibf2, Pita, Nup98 and ZIPIC as described previously with minor modifications (28). More precisely, antibody-bound protein complexes were isolated with Protein A or Protein G Dynabeads (ThermoFisher Scientific) and library generation was completed utilizing the KAPA SYBR FAST qPCR Master Mix (Kapa Biosystems) and size selected with Agencourt AMPure XP beads (Beckman Coulter Inc). ChIP-seq libraries were sequenced on an Illumina HiSeq 2500 at the HudsonAlpha Genomic Services Laboratory.
Sequences were mapped to the dm6 reference genome using bowtie version 1.0.0, using default settings with the addition of the (-m 1) parameter, which removes any sequences that align to more than one genomic site (42). PCR duplicates were removed with samtools version 0.1.18 (43). Peak calling was conducted utilizing MACS(v1.4.2), requiring peaks to have a P-value of 1e−10 (44). Peaks were called with an equal number of reads for the protein of interest and the IgG ChIP or input control for Kc167 and S2 cells, respectively. An equal number of reads were utilized for each cell type when conducting comparative analyses between S2 and Kc167 cells. The full MACS peak was utilized when analyzing histone modifications. The summit of the peak ±200 bp was utilized as the protein peak for all other proteins analyzed.
Hi-C
Hi-C libraries of two biological replicates were generated utilizing the recently published in situ Hi-C methodology with minor modifications (45). The genomes were digested with either DpnII (NEB) or HinfI (NEB) and 5΄ overhangs were filled in with Biotin-16-dUTP (Jena Bioscience). HiC libraries were sequenced at the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) sequencing facility and the HudsonAlpha Genomic Services Laboratory. Paired reads were mapped to the genome, processed and matrix resolution was calculated as described previously (45). To call significant interactions, contact matrixes were first normalized by ICE and processed by Fit-Hi-C at 1 kb resolution for all interaction distances between 3 kb and 1 Mb (46,47). Interaction calls from Fit-Hi-C were further filtered by their probability of occurring in a random generated list of interactions. Using this secondary filtering step, we obtained lists of interactions below a secondary false discovery rate (FDR < 0.001). To ensure Fit-Hi-C calls were accurate across replicates and/or experimental conditions, KR normalized reads (and further normalized by total sequencing depth) from HinfI and DpnII were plotted and the spearman correlation was calculated.
STARR-seq enhancers
Genome coordinates for enhancers defined by STARR-seq were downloaded from GSE57876 and lifted over to dm6 utilizing the FlyBase Coordinates Converter. Enhancer summits ±250 bp from the hkCP and dCP STARR-seq peaks were intersected, and any enhancer with at least a 1 bp overlap with the other enhancer class was denoted a BothCP enhancer. All other enhancers were considered either unique hkCP or dCP enhancers. Enhancer strength was obtained from the original publication (11). Raw data was also downloaded and processed as described for ChIP-seq to obtain genome-browser compatible wiggle files. TSS-proximal enhancers were defined as any enhancer summit that overlapped a TSS ± 250 bp. Any enhancer with a summit >250 bp from a TSS was named TSS-distal. Enhancers were additionally classified as active or inactive. An enhancer was denoted as active if the enhancer peak (summit ± 250 bp) overlapped an H3K27ac peak by at least 1 bp. Enhancers not meeting these criteria were defined as inactive.
Bioinformatics
General bioinformatics methods
Profile plots and heatmaps were generated using ngs.plot (48). All genomic intersections were conducted with bedtools v.2.25.0 (49). Boxplots and T-tests were conducted in R Studio. Visualization of the Hi-C data was conducted using Juicebox (45). Genomic alignment for STARR-seq and ChIP-seq reads were visualized with the IGV genome browser (50,51).
Enrichment scores
The enrichment of architectural proteins or enhancers to various genomic elements (TAD borders, lamin associated domains (LADs), and chromatin loop anchors) were conducted with bedtools. The expected overlap was generated using 1000 permutations of randomized genomic elements and the median value from the 1000 permutations was reported as the expected value. Enrichment values are reported as the log2(Observed/Expected). LAD coordinates were obtained from van Bemmel et al. and converted to dm6 utilizing the FlyBase Coordinates Converter (52).
Force-directed interaction layouts
Interaction networks were visualized by Cytoscape, with interaction anchors represented as nodes (53). Distances between nodes were computed from Fit-Hi-C significance scores using a prefuse force-directed layout.
Generation of high occupancy APBS list
APBS occupancy was assessed by expanding peak summits by 200 bp on both sides. Expanded regions overlapping each other by more than one architectural protein were merged and the midpoint taken as the new summit and expanded ±200 bp. Reads from each ChIP-seq dataset were counted for each expanded region, normalized by RPM, and calculated as a fold change over IgG. Protein occupancy was deemed positive if there was at least a three-fold change over IgG.
GROseq
GROseq from S2 cells was downloaded from GSE23543 and GSE42117 and mapped to the dm6 reference genome using bowtie2 (54,55). Transcriptional output was determined by using bedtools to count all of the GROseq reads within the coding region of the gene and normalized by gene size.
TAD calling
TADs were called using the directionality index and hidden Markov model as described (22). In lieu of discrete bins for directionality, we used fragment based resolution intensities over a 2 kb sliding window with a step size of 200 bp.
Chromatin loop calling
Loops were called in a similar manner to Rao et al. with a few modifications (45). To call spots at higher resolution, each bin within a normalized 2 kb matrix was examined and was required to pass several filters. (i) We examined only bins greater than 6 kb apart. (ii) Focal points (2 kb bins) were required to have a higher signal intensity than each point for two bins up, down, left, and right as well as each of the bins at the corners. (iii) The median of the central cross (center spot plus one bin up, down, left and right), had to be higher than the median of the corners. (iv) To obtain an estimation of local signal variance, the focal point was shifted 7 bins from the original center and signal intensity information was gathered from bins two up, down, left, right as well as the corners. This focal point shift was performed several times to obtain estimations for the vertical, horizontal, top left, top right, bottom right, and bottom left regions. Each shifted focal region was then tested against the original focal region by a two-sample Kolmogorov–Smirnov test. This produced twenty P-values describing enrichment of the central region. Only spots with a median P-value score <0.05 were kept. If spots were too close to the diagonal (within 26 kb of each other) the most extreme left, bottom, and bottom left regions were omitted from statistical analysis. (v) As an additional filter, the distance normalized mean of the center region had to be greater than the means of each neighboring region. These five modifications permitted high-confidence loop calls at 2 kb resolution.
Identification of structural variants in the Kc167 cell line genome
Structural variants were called using BWA and BreakDancer on paired-end IgG ChIP-seq reads. Variants above an 80% confidence interval were kept and called. Fit-Hi-C interaction distances were corrected to the actual distance considering structural variation. Fit-Hi-C calls that fell below 4 kb and those where read counts were less than the cutoff of unaffected pairs at the same distance category were considered to be a result of structural variation.
RESULTS
Enhancer classes have distinct H3K4 modifications
We conducted our studies in Kc167 cells because of the extensive characterization of architectural proteins and chromatin interactions in this cell line. Drosophila enhancers were defined using STARR-seq in S2 cells. Kc167 and S2 cells are derived from plasmatocytes, have similar transcriptional profiles (56), and we have previously shown that enhancers defined by STARR-seq are conserved between the two cell lines (40). Throughout this study, we analyzed the enhancers previously shown to activate an ectopically express the hkCP core promoter (4137) or the dCP core promoter (3586), not including the ∼1800 enhancers that activated both the dCP and hkCP core promoters. To reduce any bias caused by protein recruitment related to the core promoter within the predominantly promoter-proximal hkCP enhancers, we will distinguish between TSS proximal and distal enhancers for each enhancer class. The distribution of distances between each enhancer class and the closest TSS is shown in Supplementary Figure S1. Proximal enhancers are defined as those with a summit within 250 bp of a TSS, while distal enhancers have a summit at least 251 bp away from a TSS. Using these definitions, we subdivided the hkCP enhancers into 2944 proximal enhancers and 1193 distal enhancers, and the dCP enhancers into 222 proximal enhancers and 3364 distal enhancers. As a control, we also analyzed randomized genomic regions that are either proximal or distal to TSSs.
Because enhancers can be defined by epigenetic marks, we investigated the presence of histone modifications and enhancer-associated proteins at different enhancer classes identified by STARR-seq. We compared the occupancy of known enhancer-resident proteins in each enhancer class utilizing ChIP-seq data from Kc167 cells. First, we analyzed RNA Polymerase II and CBP, both of which are present in active enhancers (57). As expected, both proteins are enriched in each enhancer class compared to randomized controls. RNA Polymerase II and CBP are more enriched in proximal hkCP enhancers compared to proximal dCP enhancers but are observed at comparable levels when the distal hkCP and dCP enhancers were analyzed (Supplementary Figure S2A). Thus, we conclude that RNA Polymerase II and CBP protein occupancy does not distinguish the two enhancer classes.
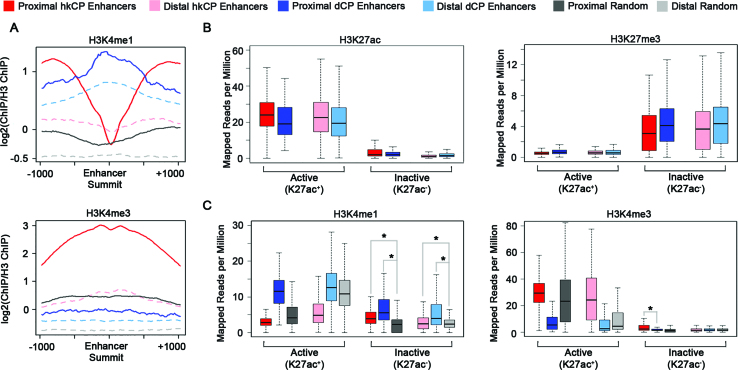
Next we analyzed the levels of H3K4me1, a classical enhancer modification, and H3K4me3, a classical promoter modification, in the two enhancer classes (58). Both distal and proximal dCP enhancers exhibit high levels of H3K4me1 and low levels of H3K4me3 compared to randomized controls, albeit to a lesser degree in the distal enhancers (Figure 1A). It is unclear why the developmental enhancers in our study exhibit a strong enrichment in H3K4me1 compared to the developmental enhancers shown to be devoid of chromatin modifications in a recent study (59). In contrast, hkCP enhancers are enriched for high levels of H3K4me3 compared to the randomized controls and depleted for H3K4me1 around the enhancer summit (Figure 1A). The high levels of H3K4me3 are consistent with hkCP enhancers being predominantly promoter-proximal regulatory elements, but surprisingly, even distal hkCP enhancers are enriched for H3K4me3 (Figure 1A). A similar pattern of H3K4 methylation was observed when utilizing ChIP-seq data from S2 cells, underscoring the similarities between these two cell types (Supplementary Figure S2B). High transcriptional activity levels could explain the H3K4me3 enrichment in TSS-distal hkCP enhancers, but we cannot discount the possibility that TSSs greater than 250 bp away from the distal hkCP enhancers are present nearby and at least partially affecting the H3K4me3 levels (60,61). Because enhancers were defined utilizing STARR-seq, an assay in which the enhancer regions are located on a plasmid, it is possible that a subset of the enhancers identified are normally inactive at their endogenous location due to the chromatin environment. Thus, we sought to analyze the potentially active and inactive enhancers separately by analyzing the levels of H3K27ac and H3K27me3, as was shown previously (10). We defined active enhancers as any enhancer overlapping an H3K27ac peak by at least one base pair and all non-overlapping enhancers were classified as inactive. The ‘inactive’ enhancers that have more H3K27ac reads than ‘active’ enhancers likely occur in regions of the genome that contain relatively high levels of H3K27ac but were not called as H3K27ac peaks by MACS. These regions would not have resulted in an ‘active’ enhancer call based on our method. The numbers of each enhancer class fitting this definition are listed in Table 1. Utilizing this approach, we observe a strong enrichment of H3K27me3, a histone modification typically associated with inactive transcription, in the inactive but not the active TSS-proximal and TSS-distal enhancers for both classes (Figure 1B). As a control, H3K27ac enrichment for active and inactive enhancers is also shown (Figure 1B). This approach permits successful differentiation between the active and inactive enhancer groups defined using STARR-seq and helps reduce any bias generated by the ectopic discovery of the STARR-seq enhancers.
Figure 1.
dCP and hkCP Enhancers Have Distinct H3K4 Methylation. (A) Profile plots depicting ChIP-seq read density for H3K4me1 and H3K4me3 at enhancer summits ±1 kb. Y-axis depicts log2(ChIP Reads/H3 Reads). (B and C) Boxplots depicting the mapped reads per million for H3K27ac, H3K27me3, H3K4me1 and H3K4me3 within the 501 bp enhancer categories. Four separate randomized data categories are shown: TSS-proximal and overlapping an H3K27ac peak, TSS-proximal and not overlapping an H3K27ac peak, TSS-distal and overlapping an H3K27ac peak, and TSS-distal and not overlapping an H3K27ac peak. ChIP-seq data shown is from Kc167 cells. Asterisks denote a P value < 0.009 (Student's t-test).
Table 1. Number of enhancers in each activity category.
| Total | Active (K27ac+) | Inactive (K27ac−) | |
|---|---|---|---|
| hkCP Enhancers | 4137 | 3343 | 794 |
| dCP Enhancers | 3586 | 1465 | 2121 |
| TSS Proximal hkCP Enhancers | 2944 | 2807 | 137 |
| TSS Distal hkCP Enhancers | 1193 | 536 | 657 |
| TSS Proximal dCP Enhancers | 222 | 110 | 112 |
| TSS Distal dCP Enhancers | 3364 | 1355 | 2009 |
Next, we reassessed the H3K4 methylation status taking enhancer activity into account. Consistent with our previous analysis, active dCP enhancers show an enrichment for H3K4me1 while active hkCP enhancers are enriched for H3K4me3 (Figure 1C). Notably, inactive dCP enhancers still have a strong H3K4me1 enrichment, consistent with previous reports that H3K4me1 is a marker of enhancer identity, independent of activity (57). Interestingly, inactive hkCP enhancers are also enriched for H3K4me1 compared to randomized controls (P value 1.869e−10 for proximal and 0.008736 for distal hkCP enhancers), suggesting that inactive hkCP enhancers may be denoted with an H3K4me1 modification state. Inactive proximal but not inactive distal hkCP enhancers are enriched in H3K4me3 (P values: 5.184e−7 for proximal; 0.6495 for distal), but it is unclear if this is simply a consequence of TSS proximity. Although we do not see any particular distance bias associated with distal enhancers (Supplementary Figure S1), we wanted to rule out any TSS specific effects for the H3K4me3 presence on distal active hkCP enhancers. We therefore examined active enhancers that are at least 3 kb away from any annotated TSS and still see H3K4me3 enrichment on hkCP enhancers (Supplementary Figure S2C). Altogether, our results suggest that both enhancer classes contain CBP and RNA Polymerase II but dCP enhancers are enriched for H3K4me1 modification, while active hkCP enhancers are associated with H3K4me3.
Architectural proteins bind both enhancer classes but unique architectural protein subcomplexes are observed in each class
Because the two enhancer classes interact with different core promoters and have unique epigenetic marks, we hypothesized that each enhancer class is also occupied by distinct subfamilies of architectural proteins to regulate their interactions. Since hkCP and dCP enhancers were defined using STARR-seq in S2 cells and the genomic occupancy of the entire architectural protein family has been characterized in Kc167 cells, we first validated that the architectural protein occupancy is consistent between the two plasmatocyte cell lines S2 and Kc167 (Supplementary Figure S3). We analyzed the distribution of architectural proteins CP190, CTCF, Ibf1, Ibf2, Mod(mdg4), Pita, SuHw and ZIPIC for which ChIP-seq data is available in both cell types and called peaks in S2 cells using an equivalent number of reads in the input control. We aligned an equal number of ChIP-seq reads from both cell types to the architectural protein peaks defined in S2 cells. Strikingly, all eight architectural proteins exhibit an enrichment of reads in Kc167 cells at the architectural protein peaks defined in S2 cells (Supplementary Figure S3). Our observations indicate that the architectural protein occupancy is highly conserved between these two cell types.
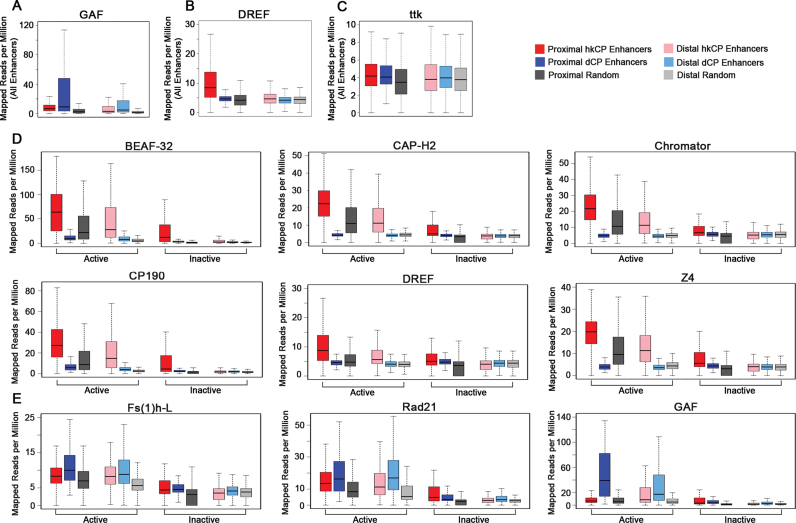
Next, we utilized ChIP-seq data from Kc167 cells for the architectural protein family and evaluated the occupancy of each in the two enhancer classes, focusing on the active enhancers to reduce any bias generated by the ectopic assay utilized to discover the STARR-seq enhancers. As a positive control, we assessed the occupancy of DREF and GAF (also known as Trl) because the protein levels and motifs of each protein were shown to be enriched in hkCP and dCP enhancers, respectively (11). Consistent with previous results, we observed a specific enrichment for DREF in hkCP enhancers and GAF in dCP enhancers compared to randomized controls, with the proximal enhancers showing a more pronounced signal than distal enhancers (Figure 2A and B). As a negative control, we analyzed the occupancy of ttk, a transcription factor known to repress GAGA-mediated activation, which would not be expected to be present at active enhancers (62). hkCP and dCP enhancers do not exhibit an enrichment for ttk compared to the randomized controls (Figure 2C). When analyzing the occupancy of all the architectural proteins in the enhancers, three clear patterns of enrichment were observed. First, the majority of architectural proteins (CTCF, L3mbt, Mod(mdg4), Ibf1, Ibf2, Nup98, SuHw, TFIIIC, Pita and ZIPIC) exhibit an enrichment in both hkCP and dCP enhancers compared to controls, with a more pronounced enrichment observed when comparing hkCP enhancer to dCP enhancer enrichment or when comparing TSS proximal to TSS distal enhancer enrichment (Supplementary Figure S4A). Consistent with architectural proteins mediating contacts between active enhancers and promoters, a similar enrichment for these ten architectural proteins is observed when analyzing only active enhancers (Supplementary Figure S4B). The second group of architectural proteins (BEAF-32, Cap-H2, Chromator, CP190, DREF and Z4) shows a very strong enrichment in active hkCP enhancers and is nearly depleted in active dCP enhancers compared to randomized controls (Figure 2D). Notably, the enrichment of BEAF-32, Cap-H2, Chromator, CP190, DREF and Z4 is strong in both proximal and distal enhancers and is not observed when inactive enhancers are analyzed (Figure 2D). It is unclear if the small enrichment of these six proteins in inactive proximal hkCP enhancers is biologically significant. Consistent with these observations, CP190 and BEAF-32 were shown to bind near housekeeping gene promoter regions previously (63–65). The final group of architectural proteins (Fs(1)h-L and Rad21) is enriched in both active enhancer classes compared to randomized controls, but shows a stronger enrichment in dCP enhancers compared to hkCP enhancers (Figure 2E). Fs(1)h-L and Rad21 occupancy is only observed in active but not inactive enhancers and is analogous to the transcription factor GAF occupancy, which is known to specifically bind dCP enhancers (Figure 2E) (11). Notably, many hkCP enhancers also have Fs(1)h-L and Rad21 occupancy, so the biological significance of the increased occupancy observed in dCP enhancers remains to be determined. Overall, our observations demonstrate that the majority of architectural proteins are present in both enhancer classes but that BEAF-32, Cap-H2, Chromator, CP190, DREF and Z4 are preferentially associated with the often promoter-associated hkCP enhancers, while Fs(1)h-L and Rad21 are enriched to higher levels in dCP than hkCP enhancers.
Figure 2.
dCP and hkCP Enhancers Contain Distinct Subcomplexes of Architectural Proteins. Boxplots depicting the mapped reads per million from Kc167 cells for (A) GAF, (B) DREF, (C) ttk within each 501 bp enhancer class. Boxplots depicting the mapped reads per million for the architectural proteins in active and inactive 501 bp enhancer classes including the architectural proteins (D) unique to hkCP enhancers (BEAF-32, Cap-H2, Chromator, CP190, DREF, Z4) or (E) enriched in dCP enhancers compared to hkCP enhancers (Fs(1)h-L and Rad21). GAF is shown as a control.
Most architectural protein sites are associated with enhancers or promoters
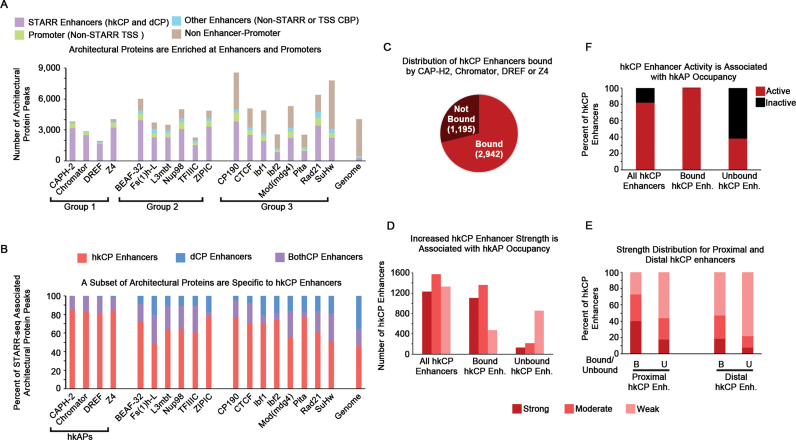
To evaluate the functional significance of unique architectural protein occupancy in each enhancer class, we determined the number of architectural protein sites that can be accounted for by enhancers and promoters. We utilized ChIP-seq data to call peaks using an equal number of reads for the architectural protein ChIP and an IgG control. To obtain a comprehensive list of architectural protein sites likely contributing to enhancer–promoter interactions, we intersected architectural protein peaks with multiple classifications of enhancers or promoters: STARR-seq enhancers, Promoters (TSSs not associated with STARR-seq enhancers) and Other Enhancers (CBP sites not associated with STARR-seq or TSSs). Notably, all of the architectural proteins are enriched at sites of enhancers and promoters, but three distinct groups are detected (Figure 3A). More than 93% of the Cap-H2, Chromator, DREF and Z4 peaks (Group 1) are detected at either enhancers or promoters, whereas ∼80–87% of BEAF-32, Fs(1)h-L, L3mbt, Nup98, TFIIIC and ZIPIC peaks (Group 2) are enhancer or promoter associated. Thus, it is interesting to speculate that the predominant function of Group 1 and Group 2 architectural proteins is to regulate 3D enhancer–promoter interactions. In contrast, at least 25% of the peaks for the third subclass of architectural proteins, including CP190, CTCF, Ibf1, Ibf2, Mod(mdg4), Pita, Rad21 and SuHw (Group 3), are independent of enhancers or promoters, indicative that Group 3 architectural proteins likely have at least one other, non-enhancer–promoter function in the cell (Figure 3A). Notably, the enhancer–promoter independent sites of nearly all architectural proteins are enriched at TAD borders, with the non-enhancer protein Group 3 architectural protein sites showing small enrichments for lamin associated domains (LADs), possibly indicative that these architectural protein sites contribute to chromatin organization in inactive genomic regions (Supplementary Figure S5). However, the functional significance of the non-enhancer–promoter sites remains to be empirically examined. Overall, these findings suggest that the majority of architectural protein sites genome-wide can be explained by enhancer–promoter interactions, but only the architectural proteins in Group 1 (Cap-H2, Chromator, DREF and Z4) are almost exclusively associated with enhancers and promoters.
Figure 3.
Architectural Proteins Predominantly Occupy Enhancers/Promoters, with Cap-H2, Chromator, DREF, and Z4 Distinguishing hkCP Enhancers. (A) 401 bp architectural protein peaks were overlapped with either 501 bp STARR-seq enhancers, single bp TSSs, or 401 bp CBP peaks to determine the enhancer–promoter association of each architectural protein. A single base pair overlap was considered a positive association. Groups 1 and 2 proteins are predominantly explained by enhancers and promoters, whereas Group 3 proteins have at least 25% of their sites unexplained by enhancers or promoters. (B) The fraction of STARR-seq enhancer classes that are bound by each architectural protein are shown. BothCP enhancers denote enhancer elements that activate both core promoter elements (hkCP and dCPs). Only Cap-H2, Chromator, DREF and Z4 are unique to hkCP enhancers and are denoted as housekeeping architectural proteins (hkAPs). (C) Pie chart depicting the number of hkCP enhancers bound or unbound by at least one of the hkAPs. (D) The enhancer strength distribution for all hkCP enhancers, hkCP enhancers bound by the hkAPs, and hkCP enhancers not bound by the hkAPs. (E) The distribution of enhancer strength as described in D, except TSS-proximal and –distal hkCP enhancers are analyzed separately. (F) The distribution of enhancer activity for all hkCP enhancers, hkCP enhancers bound by the hkAPs, and hkCP enhancers not bound by the hkAPs. ChIP-seq data shown is derived from Kc167 cells.
Cap-H2, chromator, DREF and Z4 are markers of hkCP enhancer identity
Because the Group 1 architectural proteins are also highly associated with hkCP enhancers (Figures 2D and 3A), we assessed the distribution of all the architectural protein sites within the STARR-seq enhancers using a more quantitative approach. For this analysis, we included the STARR-seq enhancers that were shown to interact with both core promoters (denoted BothCP enhancers) in addition to the class-specific enhancers. Consistent with our previous analysis, more than 80% of the architectural protein peaks associated with STARR-seq enhancers for Cap-H2, Chromator, DREF and Z4 are hkCP enhancers. Furthermore, if hkCP enhancers and BothkCP enhancers are taken into account, <1% of the STARR-seq associated peaks for the Cap-H2, Chromator, DREF or Z4 overlap dCP-specific enhancers (Figure 3B). The architectural proteins in Groups 2 and 3, including BEAF-32, CP190, Rad21 and Fs(1)h-L, also showed an enrichment for hkCP and BothCP enhancers but exhibited a 7–25% of the STARR-seq peaks overlapping dCP enhancers (Figure 3B). Altogether, these data contibute to a model suggesting that although there are distinct architectural protein subcomplexes enriched in the different enhancer classes, only the occupancy of Cap-H2, Chromator, DREF or Z4 is specific to hkCP enhancers.
To investigate whether Cap-H2, Chromator, DREF or Z4 genomic occupancy is representative of hkCP enhancer identity, we determined the percentage of hkCP enhancers that correspond to a peak in at least one of the architectural proteins. Strikingly, nearly 75% of all identified hkCP enhancers were bound by Cap-H2, Chromator, DREF or Z4 (Figure 3C). Furthermore hkCP enhancers bound by Cap-H2, Chromator, DREF or Z4 correspond to strong and moderate enhancers, while the unbound enhancers correspond to weak hkCP enhancers (Figure 3D). The association of hkCP enhancer strength and occupancy by Cap-H2, Chromator, DREF or Z4 was most prevalent in hkCP enhancers proximal to TSSs but was consistent in distal hkCP enhancers as well (Figure 3E). Finally, we determined that nearly all of the inactive hkCP enhancers are not bound by Cap-H2, Chromator, DREF or Z4 (Figure 3F). Thus, we speculate that the strongly associated architectural proteins Cap-H2, Chromator, DREF and Z4 at least partially contribute to mediating hkCP enhancer interactions.
hkCP and dCP enhancers are involved in distinct genomic interactions
Because of the differential architectural protein occupancy in enhancers, we sought to determine if the endogenous interactions mediated by hkCP and dCP enhancers are distinct. We generated two novel Hi-C genomic libraries digested with HinfI from a single biological replicate and a new library digested with DpnII and combined these data with the four biological replicates of Hi-C genomic libraries generated by DpnII that we published previously (40). Utilizing this method, we obtained nearly 1 billion (983 799 884) uniquely mapped read pairs to the dm6 reference genome. After removal of reads between adjacent fragments, we obtained 605 million usable reads (Supplementary Table S1) for identifying chromatin interactions, resulting in a ‘matrix resolution’ or the smallest locus where 80% of the loci have >1000 contacts (45) of <1 kb. This resolution allows the mapping of interactions between 1 kb bins ranging from 3 kb to 1 Mb in distance. Processed reads were normalized using the iterative correction and eigenvector (ICE) decomposition method, which assumes that the total number of contacts for all genomic loci should be the same, and then interactions were identified by Fit-Hi-C (46,47). In total, 1 258 936 significant chromatin interactions ranging from 3 kb to 1 Mb were identified utilizing a 0.001 FDR cutoff (Supplementary Table S2).
To assess the validity of the Fit-Hi-C significant interaction calls, we conducted two control analyses. First we determined that the interactions called between the biological replicates generated by either DpnII or HinfI digestion are highly correlated (Spearman's Correlation R = 0.90, Supplementary Figure S6). Second, we assessed how structural variants such as inversions, translocations or deletions, might lead to false significant interactions called by Fit-Hi-C. We performed this analysis using the variant caller BreakDancer at a relatively low confidence interval (80%) to obtain as many likely structural variants as possible. We adjusted Fit-Hi-C distances by the actual distance after accounting for structural variants between or overlapping significant interactions. We then determined how many Fit-Hi-C interactions were possibly due to differences between the reference genome and that of Kc167 cells by filtering out interactions either falling <4 kb in distance or below a minimum score threshold for interactions at the same distance (see Materials and Methods). We found that structural variants have minimal impact on the significant interaction calls (1.13% of interactions) and that hkCP and dCP enhancer interactions are affected equally (0.92% and 0.95%, respectively) (Supplementary Table S3). Altogether, these analyses suggest that the interaction calling method generates robust pair-wise interactions in Kc167 cells.
To gain insight into potentially unique interactions mediated by each enhancer class, we analyzed the enhancer–TSS and enhancer-enhancer contacts for each. When evaluating enhancer contacts, only interaction bins containing a single enhancer type were utilized. We omitted 94 interaction anchors due to the presence of hkCP and dCP enhancers, or a BothCP enhancer in combination with either unique enhancer class. Genome-wide, dCP and hkCP enhancers are involved in a similar number of significant interactions (hkCP = 43 079 and dCP = 38 279, which equates to 10.4 and 10.7 interactions per enhancer). All interactions deemed significant occurred between 3 kb and 1 Mb distances, so interactions between TSS-proximal hkCP enhancers and their neighboring genes were omitted from this analysis. Approximately 21.2% of hkCP enhancer interactions and 11.6% of dCP enhancer interactions involve a TSS on the other interaction anchor, which could suggest that hkCP enhancers are more predominantly engaged with genes than dCP enhancers (Figure 4A). Both hkCP and dCP enhancers preferentially interact with their own enhancer class, with hkCP enhancers interacting with themselves more frequently than dCP enhancers do (9.4% and 5.2% respectively) (Figure 4B). From this analysis, it is unclear if the increased promoter–promoter or promoter–enhancer interactions of hkCP enhancers are indicative of multi-TSS–enhancer interactions or rather a higher proportion of this enhancer class interacting with another regulatory element.
Figure 4.
hkCP Enhancers Mediate More Multi-TSS Clustered Interactions than dCP Enhancers to Increase Transcriptional Output. (A) Bargraph representing the percent of enhancer interactions containing an enhancer in one anchor with a TSS on the opposite anchor. (B) Bargraph representing the percent of enhancer interactions containing an enhancer in one anchor with an enhancer on the opposite anchor. (C) Cytoscape force directed layout depicting the significant interactions on Chromosome 2L. Interaction anchors are represented as dots (anchors containing hkCP enhancers are in red, dCP enhancers are in blue, TSSs are in green and all other anchors are in gray). The distance between dots is representative of interaction frequency. (D) Boxplot representing the distance distribution of interactions between enhancers and TSSs. (E and F) Line graphs highlighting the percent of enhancers interacting with varying numbers of TSSs. (G) Boxplot demonstrating the distribution of TSSs bound per enhancer for all active and inactive enhancer classifications.(H) Boxplot depicting the contact strength (q value is determined by Fit-Hi-C) of active and inactive enhancer–TSS interactions. All chromatin interaction data shown is from Kc167 cells. (I and J) Boxplots depicting the transcriptional output (GRO-seq reads/kb) for the genes interacting with each enhancer class. P values calculated by the Student's t-test. The GRO-seq data shown is derived from S2 cells.
To qualitatively evaluate how hkCP and dCP enhancers may cluster with TSSs, we generated a visual representation of the genome-wide chromatin interactions (Figure 4C). The significant interactions from chromosome 2L are depicted as a force directed layout. The interaction anchors are represented as dots and the frequency of the interactions is denoted by the distance between the anchors. Interaction anchors containing TSSs, hkCP enhancers, and dCP enhancers are highlighted, while all others are shown in gray. TSS-containing anchors are often found together, suggesting that promoter clusters are readily detectable in our data (Figure 4C). Of particular interest, hkCP enhancers are often found clustered with multiple TSSs and are frequently bound to other hkCP enhancers. dCP enhancers, on the other hand, are bound to individual TSSs as well as in multi-TSS interactions but are predominantly isolated away from other dCP enhancers (Figure 4C). This observation led us to hypothesize that hkCP enhancers may be more actively engaged in large promoter clusters than dCP enhancers.
To assess a potential distinction in promoter cluster formation between the two enhancer classes, we quantitatively measured the number of TSS interactions occurring per enhancer. First, we evaluated the distance between interacting TSSs and enhancers, observing that the majority of both hkCP and dCP enhancer–TSS interactions occur within the median TAD size of 32.5 kDa (Figure 4D). Next, we quantified the number of TSSs bound per enhancer more specifically. A higher percentage of hkCP enhancers are engaged with 3, 4, 5 or >6 TSSs compared to dCP enhancers (Figure 4E and G), suggesting that hkCP enhancers may regulate the formation of large gene clusters and dCP enhancers are involved in more specific point to point enhancer–promoter contacts. Because significant enhancer–promoter contacts were defined as occurring between 3 kb and 1 Mb, any enhancer–promoter interactions occurring within 3 kb, such as hkCP enhancer interactions with their neighboring promoters, will not be included in this analysis. Notably, the distinction between class-specific-TSS associations is due to active enhancer contacts, as inactive hkCP and dCP enhancers interact with similar numbers of TSSs (Figure 4F and G). Consistent with a previous report, inactive and active dCPs interact with a similar distribution of TSSs (9). However, the contact strength is higher between TSSs and active enhancers than inactive enhancers, suggesting that the TSS–enhancer interactions mediated by active enhancers either occur more frequently within the cellular population or are more stably associated than the interactions of inactive enhancers (Figure 4H). Altogether, these observations suggest that hkCP enhancers are more likely to interact with higher numbers of TSSs compared to dCP enhancers, supporting a model suggesting that hkCP enhancers regulate multigene interactions while dCP enhancer interactions mediate single enhancer–promoter associations.
Finally, we evaluated the transcriptional output of the genes interacting with each enhancer class by utilizing GROseq data derived from S2 cells. As expected, the genes interacting with housekeeping enhancers were expressed at higher levels than the genes interacting with dCP enhancers (P value < 0.0005 for active enhancers) (Figure 4I). The genes bound by inactive enhancers had significantly lower GROseq reads, consistent with the notion that a TSS–enhancer contact is necessary but not sufficient for transcriptional activation (Figure 4I) (66). Next, we evaluated the effect of promoter clustering on the expression of genes interacting with either an hkCP or a dCP enhancer (Figure 4J). Strikingly, genes bound by hkCP enhancers show a dose-dependent expression pattern, with an increase in gene expression correlating with the rise in the number of TSSs bound per enhancer (P value < 0.02 for expression mediated by enhancers bound to 1 or 6 TSSs) (Figure 4J). These observations suggest that the activation of a given housekeeping gene is at least partially related to the number of promoters interacting with it, but additional studies are required to prove a causal relationship. This pattern was not observed when comparing dCP enhancers (P value < 0.5 for expression mediated by enhancers bound to 1 or 6 TSSs). Altogether, these data support a model suggesting that hkCP enhancers mediate multi-TSS enhancer contacts to promote robust expression of housekeeping genes, while dCP enhancers make specific point to point contacts critical to drive expression of tightly regulated genes. Because the hkCP enhancers are more predominantly TSS-proximal compared to the dCP enhancers, this analysis cannot rule out that both the hkCP enhancer and its neighboring TSS are not contributing to these 3 kb–1 Mb interactions.
hkCP and dCP enhancers are associated with distinct forms of chromatin architecture
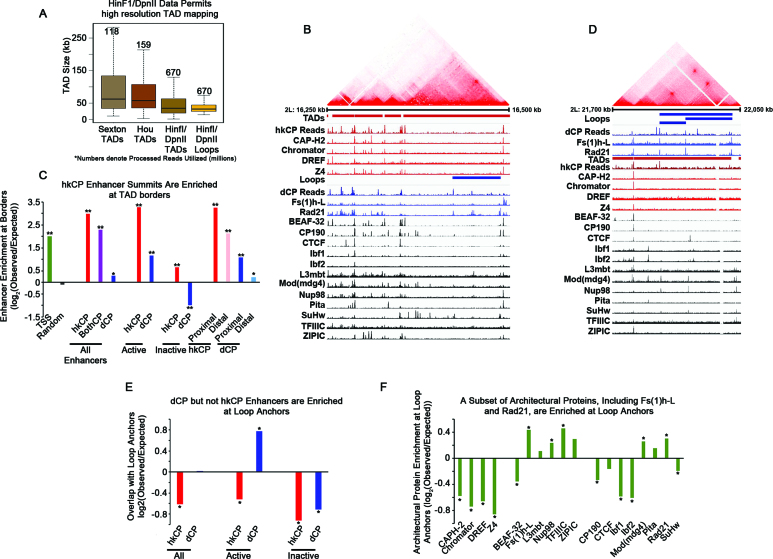
Utilizing the high resolution Hi-C dataset, we sought to define TADs as well as the more recently described chromatin loops (21,22,45). Chromatin loops are regions of enriched chromatin interactions within TADs and may be analogous to subTADs, which exhibit cell type specificity and have been suggested to be enhancer–promoter interactions that change during differentiation (45,67). In Hi-C heatmaps, a TAD is visualized as a triangle of strong interactions between neighboring loci, while a chromatin loop appears as a strong site of interaction at the top point of a triangle that lacks strong interactions within itself (45). We mapped TADs utilizing a Hidden Markov model based on the directionality index described previously (22), resulting in the identification of 2543 TADs (Supplementary Table S4). The median TAD size was 34 500 bp (average 49 008), with a maximum size of 376 000 bp, which is smaller than the previously reported TAD sizes in Drosophila (Figure 5A) (23,24). It is likely that the observed decrease in TAD size can be attributed to higher resolution data and more accurate TAD calls as the number of reads in our data exceed 4-fold the previously published work (Figure 5A). Consistent with previous reports, the TAD borders are highly associated with architectural proteins, with each architectural protein showing an enrichment at TAD borders (Supplementary Figure S7A, 1000 permutations P < 0.001) and high occupancy architectural protein binding sites (APBSs) showing the strongest association with TAD borders (Supplementary Figure S7B and C; Supplementary Table S5) (36,40). In addition, we observed chromatin loops (45) when visualizing our data. We identified the chromatin loops computationally using HiCCUPS (45) with a few modifications to permit loop calls at higher resolution. We identified genomic sites where interactions exhibit higher frequencies compared to the intervening genomic sequences between them, resulting in the identification of 458 chromatin loops with 2 kb resolution of loop anchors (Supplementary Figure S8 and Supplementary Table S6). The loops are comparable in size to TADs (median 32 000 bp and mean of 41 150 bp), but overall, have a smaller size distribution than the TADs (Figure 5A).
Figure 5.
hkCP enhancers are associated with TAD borders, while dCP Enhancers are Enriched in Chromatin Loop Anchors. (A) Boxplot depicting the size distribution of TADs or chromatin loops from various publications. Numbers denote the millions of Hi-C reads obtained in each study (23,24). (B) Hi-C data, from Kc167 cells, visualized using Juicebox and aligned with ChIP-seq and STARR-seq reads visualized with the IGV genome browser. Architectural proteins associated with hkCP enhancers are shown in red, those enriched in dCP enhancers are shown in blue, and proteins associated with both enhancer classes are shown in black. (C) Barplot depicting the enrichment of the various classes of enhancers and TAD borders. Overlaps were conducted with enhancer summits and TAD borders ±500 bp (* denotes P-value < 0.05 and ** denotes P-value < 0.005). (D) Hi-C data visualized using Juicebox and aligned with ChIP-seq and STARR-seq reads visualized with the IGV genome browser. Architectural proteins associated with hkCP enhancers are shown in red, those enriched in dCP enhancers are shown in blue, and proteins associated with both enhancer classes are shown in black. (E) Barplot depicting the enrichment of the various classes of enhancers and the 2 kb anchors of chromatin loops. Overlaps were conducted with enhancer summits and the 2 kb anchors (* denotes P-value < 0.02). (F) Barplot depicting the enrichment of the individual architectural proteins and the 2 kb anchors of chromatin loops. Overlaps were conducted with architectural protein peak summits and the 2 kb anchors (* denotes P-value < 0.042).
Because TAD borders are associated with high occupancy APBSs, we hypothesized that hkCP enhancers are also likely associated with TAD borders, which was also reported in a recent study (68). We utilized Juicebox (45) to visualize Hi-C data and further investigate the possible association between TAD borders, STARR-seq reads for each enhancer class, and ChIP-seq data for architectural proteins (Figure 5B). Consistent with previous reports, the vast majority of architectural proteins have peaks at TAD borders (Figure 5B) (36). Strikingly, TAD borders are associated with hkCP enhancer reads, as well as strong ChIP-seq peaks for the architectural proteins associated with these enhancers, CAP-H2, Chromator, DREF and Z4 (Figure 5B). The dCP STARR-seq reads also show some TAD border association, but predominantly at sites where there is also a signal for hkCP enhancers, indicative that these enhancers would be classified as BothCP enhancers (Figure 5B). To further assess a preference for hkCP enhancers compared to dCP enhancers at TAD borders, we calculated the enrichment of each enhancer class at TAD borders. We find that hkCP and BothCP enhancers but not dCP enhancers are strongly enriched at TAD borders (±500 bp) (Figure 5C). Active, TSS-proximal, and TSS-distal hkCP enhancers all exhibit a higher enrichment at TAD borders than TSSs alone. However, we cannot discount that the enrichment of housekeeping genes at TAD borders at least partially contributes to the observed hkCP enhancer enrichment (22,68). Surprisingly, active but not inactive dCP enhancers are enriched at TAD borders. This is likely due to the high gene density at TAD borders, since a similar enrichment was observed for proximal but not distal dCP enhancers, suggesting that dCP enhancers are preferentially localized inside TADs (Figure 5C). We then explored the association of high occupancy APBSs with hkCP enhancers (36). As expected, hkCP enhancers are predominantly associated with high occupancy APBSs, whereas dCP enhancers lack a strong enrichment (Supplementary Figure S7D and E). Altogether, these observations indicate that active and inactive hkCP enhancers are associated with TAD borders, while only active dCP enhancers show enrichment near TAD borders.
Finally, we investigated a potential correlation between chromatin loops and the different enhancer classes. Consistent with chromatin loops being distinct from TADs, the majority of architectural proteins are not present at the loop anchors, indicative that loop anchors are not high occupancy APBSs (Figure 5D). The anchors of chromatin loops are often associated with strong dCP STARR-seq reads in the absence of hkCP STARR-seq reads, suggesting that unique dCP enhancers occur within loop anchors (Figure 5D). Furthermore, the architectural proteins that are most enriched in dCP enhancers, Fs(1)h-L and Rad21, exhibit strong peaks at loop anchors, whereas the hkCP enhancer-associated architectural proteins (CAP-H2, Chromator, DREF and Z4) are depleted at loop anchors (Figure 5D). Altogether, these qualitative observations suggest that dCP enhancers may be associated with chromatin loop architecture. To confirm this conclusion, we measured the enrichment of each enhancer class in the chromatin loop anchors. We find that active dCP enhancers show a strong enrichment for loop anchors, while inactive dCP enhancers as well as active and inactive hkCP enhancers are depleted (Figure 5E). Furthermore, a quantitative enrichment analysis of each architectural protein demonstrates that the dCP-enriched architectural proteins are enriched in loop anchors while the hkCP-associated proteins are depleted (Figure 5F). In addition to Fs(1)h-L and Rad21, the architectural proteins Nup98, TFIIIC, and Mod(mdg4) are also significantly enriched on loop anchors (1000 permutations P value < 0.042) (Figure 5F). In conclusion, these data suggest that dCP enhancers are more likely to be associated with chromatin loop architecture than the hkCP enhancers, which is consistent with a model suggesting that each enhancer class contributes to different forms of chromatin organization.
DISCUSSION
In this study, we characterize the protein occupancy, chromatin interactions and architecture profiles for the two enhancer classes found in Drosophila (11). We demonstrate that each enhancer class has distinct H3K4 methylation states, is bound by both common and distinct architectural proteins, and is involved in distinct types of chromatin interactions (Figure 6). First, we establish that hkCP enhancers exclusively bind CAP-H2, Chromator, DREF and Z4, while dCP enhancers do not and are preferentially enriched for but not exclusively bound by Fs(1)h-L and Rad21. In addition, hkCP enhancers are more likely than dCP enhancers to associate with multiple TSSs, which promotes a higher transcriptional output (Figure 6). Finally, we show that hkCP enhancers preferentially associate with TAD borders, whereas dCP enhancers are enriched at chromatin loop anchors present inside TADs. Interestingly, enhancers activated by both core promoters exhibite more hkCP enhancer like characteristics, indicating that the bothCP enhancers may represent an intermediate among the distinctive hkCP and dCP enhancers. Altogether, our results provide strong correlative evidence, supporting a model suggesting that architectural proteins are critical regulators of enhancer–promoter interaction specificity and that the interactions between enhancers and promoters significantly contribute to the generation of 3D chromatin architecture.
Figure 6.
hkCP and dCP enhancers bind unique architectural protein subcomplexes and mediate unique chromatin interactions. Cartoon summarizing the main findings presented in this study. hkCP enhancers are associated with H3K4me3, are bound by architectural proteins including CAP-H2, Chromator, DREF and Z4, are more likely to generate multi-TSS chromatin interactions to promote robust transcription, and are often found at TAD borders. In contrast, dCP enhancers are associated with H3K4me1, are bound by architectural proteins excluding CAP-H2, Chromator, DREF and Z4, are more likely to generate single TSS contacts, and are enriched at chromatin loop anchors.
The importance of architectural proteins in regulating enhancer–promoter interactions in Drosophila is supported by the observation that the vast majority of architectural protein sites present in the genome correspond to enhancers and promoters. Historically, architectural proteins were identified as insulators, which were functionally demonstrated to block enhancer–promoter interactions (31,69). The insulator function of architectural proteins correlates with their enrichment at TAD borders. However, several lines of evidence, including ChIA-PET analysis of CTCF- and cohesin-mediated interactions in mammals, suggest that these architectural proteins help mediate long range contacts among regulatory sequences (32,33). In Drosophila we observe that nearly all of the Group 1 and Group 2 architectural protein sites are associated with enhancers or promoters defined by STARR-seq, TSSs or CBP peaks, suggesting that architectural proteins help mediate enhancer–promoter interactions (69). Notably, Group 3 architectural proteins include the classic insulator proteins CTCF, CP190, Mod(mdg4) and SuHw, and at least 25% of their peaks cannot be explained by enhancers or promoters. It is interesting to speculate that the non-enhancer–promoter sites may be involved in more classical insulator functions or contributing to the chromatin architecture of inactive regions of the genome.
The conclusion that architectural proteins are critical regulators of the specificity between enhancers and promoters is supported by two main lines of evidence. First, our results demonstrate a strong correlation between each enhancer class and distinct architectural protein subcomplexes. Functional evidence supporting this conclusion comes from mutational analyses of the DRE motif in the distinct enhancer classes, which likely recruits DREF and the other hkCP enhancer associated architectural proteins (41). Zabidi et al. demonstrated that the tandem DRE motif alone was sufficient to enhance expression of the housekeeping core promoter and that mutation of DRE motifs within an hkCP enhancer reduced its promoter interactions in a luciferase assay (11). Furthermore, addition of a DRE motif to a dCP enhancer changed its promoter specificity (11). Because DREF and potentially BEAF-32 bind to the DRE motif, these results strongly support a model suggesting that the differential occupancy of Cap-H2, Chromator, DREF and Z4 in the two enhancer classes is a critical regulator of their specific interactions with the core promoter types. However, our data cannot discount the notion that unique transcription factor binding at proximal TSSs also contribute to the specificity of enhancer–promoter interactions. Although hkCP enhancer identity is most highly correlated with CAP-H2, Chromator, DREF and Z4 localization, these four architectural proteins are not found in isolation within hkCP enhancers. BEAF-32 and CP190 are also strongly enriched in hkCP enhancers, which are also associated with high occupancy APBSs and TAD borders. Thus, the full architectural protein complement at hkCP enhancers is far more complex than the four hkCP-specific architectural proteins. In addition, we did not detect any architectural proteins that are truly unique to dCP enhancers. Because dCP enhancers exhibit higher cell type specificity, we cannot discount that there are additional dCP enhancers present in the Drosophila genome that were not identified by STARR-seq and thus, excluded from this analysis (11). From our studies, it is unclear if the enrichment of Fs(1)h-L and Rad21, particularly because Fs(1)h-L and Rad21 are present in hkCP enhancers at lower levels, or the absence of BEAF-32, CAP-H2, Chromator, CP190, DREF and Z4 truly distinguishes the architectural protein complexes found at dCP enhancers. In the future, careful biochemical analyses will be required to gain a comprehensive understanding of the complete organization of architectural protein subcomplexes associated with each enhancer class.
hkCP enhancers are associated with multi-TSS chromatin interactions and TAD borders. The promoter-clustering by hkCP enhancers results in a dose-dependent increase in transcriptional output for the interacting genes. Thus, one likely molecular mechanism by which hkCP enhancers promote robust transcriptional activation is by increasing the local concentration of RNA Polymerase II and general transcription factors (GTFs) by bringing multiple TSSs into close proximity. It is interesting that the hkCP enhancers, which form promoter clusters, are associated with TAD borders. We speculate that the hkCP enhancer interactions involve inter-TAD contacts within the A-type compartment, indicative of the formation of transcription factories (70). From our analysis, it is unclear if the hkCP enhancers alone are sufficient for the formation of the 3D interactions or the neighboring TSSs and their associated transcription factors are also contributing to these contacts. We hypothesize that the genes recruited to the factories contain the housekeeping promoter motifs (DRE, Ohler 1, Ohler 6 and TCT) and that the hkCP enhancer residents Cap-H2, Chromator, DREF and Z4, are critical to the formation of these 3D contacts.
dCP enhancers are more likely to be present within TADs and are enriched on the subTAD-like chromatin loop anchors. dCP enhancers do not form promoter clusters, but are more likely to interact with individual TSSs. One possible explanation for this observation is that the genes interacting with dCP enhancers require the binding of sequence-specific transcription factors, and increasing the concentration of GTFs and RNA polymerase II is not an effective mechanism to promote transcriptional output. The chromatin loop association is consistent with dCP enhancers forming a strong contact with a single TSS. However, we acknowledge that dCP enhancers are likely one of multiple molecular mechanisms contributing to chromatin loop formation. Surprisingly, the chromatin loops that we observe in Drosophila are distinct from the chromatin loops described in humans (45). Rao et al. recently reported approximately 10 000 chromatin loops in the genome of GM12878 lymphoblastoid cells (45), but we only detected 458 chromatin loops in Drosophila utilizing a similar method. The reason why there are so few chromatin loops in Drosophila compared to humans is unclear. It is possible that chromatin loops represent a more precise level of architecture within TADs between specific enhancers and promoters in mammals, but because TADs are significantly smaller in flies (median size 32.5 kb compared to 880 kb in mice (22)), the chromatin loops are not as prominent or easily detected in the Drosophila genome. Notably, it appears that the chromatin loops are generated by different architectural proteins in the two species. The chromatin loops in humans are anchored by convergent CTCF motifs, while the results presented here demonstrate that the chromatin loop anchors in Drosophila are depleted of CTCF (35,45). Because the chromatin loops in Drosophila show a strong enrichment for Fs(1)h-L, a Brd4 homolog, and the architectural proteins Rad21, Nup98, TFIIIC and Mod(mdg4), it is possible that a combination of transcription and architectural proteins is required for chromatin loop formation in flies, which may be different from mammals (71). Altogether, it is clear that dCP enhancers are involved in individual contacts with TSSs and are likely one mechanism by which chromatin loops form in Drosophila.
Surprisingly, only ∼20% and ∼12.5% of all hkCP enhancer and ∼7.5% and ∼8.5% of dCP enhancer interactions involve a TSS or enhancer on the opposite anchor, respectively. The biological significance of the enhancer to non-TSS association is unclear. One possible explanation is that our current methods for identifying statistically significant interactions are not sufficiently robust and that many of the enhancer to non-TSS interactions are not representative of biologically significant contacts. However, we cannot discount the possibility that the non-TSS interactions mediated by enhancers are real and the biological significance of these contacts remains to be determined. Throughout our analysis, we compared the patterns of TSS interactions with each enhancer class instead of drawing conclusions about the absolute number of TSSs bound per enhancer, minimizing the impact of any non-specific interactions within the data. Additional molecular studies for the various type of enhancer interactions (enhancer to promoter, enhancer to non-TSS, etc.) will be required to evaluate the various biological contributions of each.
In this study, we find that the functional differences between enhancers that activate housekeeping versus developmental genes are reflected in their chromatin and architectural protein composition, and in the type of interactions they mediate. hkCP enhancers are marked by H3K4me3, associate with TAD borders, and mediate large TSS-clustered interactions to promote robust transcription. This class of enhancers contain the architectural proteins CAP-H2, Chromator, DREF and Z4. In contrast, dCP enhancers are marked by H3K4me1, associate with chromatin loop anchors and are more commonly associated with single TSS-contacts. dCP enhancers are depleted of the hkCP-specific architectural proteins and show an enrichment for Fs(1)h-L and Rad21. The results support a model suggesting that the unique occupancy of architectural proteins in the distinct enhancer classes are key contributors to the types of interactions that enhancers can mediate genome-wide, ultimately affecting enhancer–promoter specificity and 3D chromatin organization. In the future, further characterization of the broadly defined housekeeping and developmental enhancers into smaller subclasses may yield additional levels of regulation and formation of unique architectural protein and transcription factor protein complexes as key mediators of long range chromatin contacts.
ACCESSION NUMBERS
All ChIP-seq and Hi-C data generated for this publication are publicly available from GEO (Gene Expression Omnibus) under accession number GSE80702.
Supplementary Material
ACKNOWLEDGEMENTS
We appreciate all of the helpful comments and discussions provided by the members of the Corces lab. We would also like to thank Drs Carl Wu, Pavel Georgiev and Lluisa Espinás for their generous contributions of antibodies. We thank the HudsonAlpha Institute for Biotechnology and the NIDDK sequencing facility for performing the Illumina sequencing for this project. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Present addresses:
Ge Li, Division of Digestive Diseases, Emory University, 1670 Clairmont Rd, Decatur, GA 30033, USA.
Caelin Cubeñas-Potts, Meningitis and Vaccine Preventable Diseases Branch, Division of Bacterial Diseases, National Center for Immunization and Respiratory Diseases, Centers for Disease Control and Prevention, 1600 Clifton Rd., Atlanta, GA 30333, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
U.S. Public Health Service Award R01 GM035463 (to V.G.C.); Intramural Research Program of the NIDDK (to E.L.) from the National Institutes of Health and Ruth L. Kirschstein National Research Service Award F32 GM113570 (to M.J.R.). Funding for open access charge: U.S. Public Health Service Award [R01 GM035463].
Conflict of interest statement. None declared.
REFERENCES
- 1.Vernimmen D., Bickmore W.A.. The hierarchy of transcriptional activation: from enhancer to promoter. Trends Genet. 2015; 31:696–708. [DOI] [PubMed] [Google Scholar]
- 2.Lagha M., Bothma J.P., Esposito E., Ng S., Stefanik L., Tsui C., Johnston J., Chen K., Gilmour D.S., Zeitlinger J. et al. Paused Pol II coordinates tissue morphogenesis in the Drosophila embryo. Cell. 2013; 153:976–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim T.K., Shiekhattar R.. Architectural and functional commonalities between enhancers and promoters. Cell. 2015; 162:948–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akbari O.S., Bae E., Johnsen H., Villaluz A., Wong D., Drewell R.A.. A novel promoter-tethering element regulates enhancer-driven gene expression at the bithorax complex in the Drosophila embryo. Development. 2008; 135:123–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li G., Ruan X., Auerbach R.K., Sandhu K.S., Zheng M., Wang P., Poh H.M., Goh Y., Lim J., Zhang J. et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012; 148:84–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mifsud B., Tavares-Cadete F., Young A.N., Sugar R., Schoenfelder S., Ferreira L., Wingett S.W., Andrews S., Grey W., Ewels P.A. et al. Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat. Genet. 2015; 47:598–606. [DOI] [PubMed] [Google Scholar]
- 7.Schoenfelder S., Sugar R., Dimond A., Javierre B.M., Armstrong H., Mifsud B., Dimitrova E., Matheson L., Tavares-Cadete F., Furlan-Magaril M. et al. Polycomb repressive complex PRC1 spatially constrains the mouse embryonic stem cell genome. Nat. Genet. 2015; 47:1179–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sahlen P., Abdullayev I., Ramskold D., Matskova L., Rilakovic N., Lotstedt B., Albert T.J., Lundeberg J., Sandberg R.. Genome-wide mapping of promoter-anchored interactions with close to single-enhancer resolution. Genome Biol. 2015; 16:156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghavi-Helm Y., Klein F.A., Pakozdi T., Ciglar L., Noordermeer D., Huber W., Furlong E.E.. Enhancer loops appear stable during development and are associated with paused polymerase. Nature. 2014; 512:96–100. [DOI] [PubMed] [Google Scholar]
- 10.Arnold C.D., Gerlach D., Stelzer C., Boryn L.M., Rath M., Stark A.. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science. 2013; 339:1074–1077. [DOI] [PubMed] [Google Scholar]
- 11.Zabidi M.A., Arnold C.D., Schernhuber K., Pagani M., Rath M., Frank O., Stark A.. Enhancer-core-promoter specificity separates developmental and housekeeping gene regulation. Nature. 2015; 518:556–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vanhille L., Griffon A., Maqbool M.A., Zacarias-Cabeza J., Dao L.T., Fernandez N., Ballester B., Andrau J.C., Spicuglia S.. High-throughput and quantitative assessment of enhancer activity in mammals by CapStarr-seq. Nat. Commun. 2015; 6:6905. [DOI] [PubMed] [Google Scholar]
- 13.van Arensbergen J., van Steensel B., Bussemaker H.J.. In search of the determinants of enhancer–promoter interaction specificity. Trends Cell Biol. 2014; 24:695–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharpe J., Nonchev S., Gould A., Whiting J., Krumlauf R.. Selectivity, sharing and competitive interactions in the regulation of Hoxb genes. EMBO J. 1998; 17:1788–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Merli C., Bergstrom D.E., Cygan J.A., Blackman R.K.. Promoter specificity mediates the independent regulation of neighboring genes. Genes Dev. 1996; 10:1260–1270. [DOI] [PubMed] [Google Scholar]
- 16.Li X., Noll M.. Compatibility between enhancers and promoters determines the transcriptional specificity of gooseberry and gooseberry neuro in the Drosophila embryo. EMBO J. 1994; 13:400–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ohtsuki S., Levine M., Cai H.N.. Different core promoters possess distinct regulatory activities in the Drosophila embryo. Genes Dev. 1998; 12:547–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Butler J.E., Kadonaga J.T.. enhancer–promoter specificity mediated by DPE or TATA core promoter motifs. Genes Dev. 2001; 15:2515–2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaertner B., Johnston J., Chen K., Wallaschek N., Paulson A., Garruss A.S., Gaudenz K., De Kumar B., Krumlauf R., Zeitlinger J.. Poised RNA polymerase II changes over developmental time and prepares genes for future expression. Cell Rep. 2012; 2:1670–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hendrix D.A., Hong J.W., Zeitlinger J., Rokhsar D.S., Levine M.S.. Promoter elements associated with RNA Pol II stalling in the Drosophila embryo. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:7762–7767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nora E.P., Lajoie B.R., Schulz E.G., Giorgetti L., Okamoto I., Servant N., Piolot T., van Berkum N.L., Meisig J., Sedat J. et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012; 485:381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dixon J.R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J.S., Ren B.. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012; 485:376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sexton T., Yaffe E., Kenigsberg E., Bantignies F., Leblanc B., Hoichman M., Parrinello H., Tanay A., Cavalli G.. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012; 148:458–472. [DOI] [PubMed] [Google Scholar]
- 24.Hou C., Li L., Qin Z.S., Corces V.G.. Gene density, transcription, and insulators contribute to the partition of the Drosophila genome into physical domains. Mol. Cell. 2012; 48:471–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lupianez D.G., Kraft K., Heinrich V., Krawitz P., Brancati F., Klopocki E., Horn D., Kayserili H., Opitz J.M., Laxova R. et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell. 2015; 161:1012–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Labrador M., Mongelard F., Plata-Rengifo P., Baxter E.M., Corces V.G., Gerasimova T.I.. Protein encoding by both DNA strands. Nature. 2001; 409:1000. [DOI] [PubMed] [Google Scholar]
- 27.Wood A.M., Van Bortle K., Ramos E., Takenaka N., Rohrbaugh M., Jones B.C., Jones K.C., Corces V.G.. Regulation of chromatin organization and inducible gene expression by a Drosophila insulator. Mol. Cell. 2011; 44:29–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Bortle K., Ramos E., Takenaka N., Yang J., Wahi J.E., Corces V.G.. Drosophila CTCF tandemly aligns with other insulator proteins at the borders of H3K27me3 domains. Genome Res. 2012; 22:2176–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cuartero S., Fresan U., Reina O., Planet E., Espinas M.L.. Ibf1 and Ibf2 are novel CP190-interacting proteins required for insulator function. EMBO J. 2014; 33:637–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zolotarev N., Fedotova A., Kyrchanova O., Bonchuk A., Penin A.A., Lando A.S., Eliseeva I.A., Kulakovskiy I.V., Maksimenko O., Georgiev P.. Architectural proteins Pita, Zw5,and ZIPIC contain homodimerization domain and support specific long-range interactions in Drosophila. Nucleic Acids Res. 2016; 44:7228–7241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cubenas-Potts C., Corces V.G.. Architectural proteins, transcription, and the three-dimensional organization of the genome. FEBS Lett. 2015; 589:2923–2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dowen J.M., Fan Z.P., Hnisz D., Ren G., Abraham B.J., Zhang L.N., Weintraub A.S., Schuijers J., Lee T.I., Zhao K. et al. Control of cell identity genes occurs in insulated neighborhoods in Mammalian chromosomes. Cell. 2014; 159:374–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Handoko L., Xu H., Li G., Ngan C.Y., Chew E., Schnapp M., Lee C.W., Ye C., Ping J.L., Mulawadi F. et al. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat. Genet. 2011; 43:630–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Narendra V., Rocha P.P., An D., Raviram R., Skok J.A., Mazzoni E.O., Reinberg D.. Transcription. CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science. 2015; 347:1017–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo Y., Xu Q., Canzio D., Shou J., Li J., Gorkin D.U., Jung I., Wu H., Zhai Y., Tang Y. et al. CRISPR Inversion of CTCF Sites Alters Genome Topology and Enhancer/Promoter Function. Cell. 2015; 162:900–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Bortle K., Nichols M.H., Li L., Ong C.T., Takenaka N., Qin Z.S., Corces V.G.. Insulator function and topological domain border strength scale with architectural protein occupancy. Genome Biol. 2014; 15:R82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sofueva S., Yaffe E., Chan W.C., Georgopoulou D., Vietri Rudan M., Mira-Bontenbal H., Pollard S.M., Schroth G.P., Tanay A., Hadjur S.. Cohesin-mediated interactions organize chromosomal domain architecture. EMBO J. 2013; 32:3119–3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zuin J., Dixon J.R., van der Reijden M.I., Ye Z., Kolovos P., Brouwer R.W., van de Corput M.P., van de Werken H.J., Knoch T.A., van I.W.F. et al. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:996–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seitan V.C., Faure A.J., Zhan Y., McCord R.P., Lajoie B.R., Ing-Simmons E., Lenhard B., Giorgetti L., Heard E., Fisher A.G. et al. Cohesin-based chromatin interactions enable regulated gene expression within preexisting architectural compartments. Genome Res. 2013; 23:2066–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li L., Lyu X., Hou C., Takenaka N., Nguyen H.Q., Ong C.T., Cubenas-Potts C., Hu M., Lei E.P., Bosco G. et al. Widespread rearrangement of 3D chromatin organization underlies polycomb-mediated stress-induced silencing. Mol. Cell. 2015; 58:216–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gurudatta B.V., Yang J., Van Bortle K., Donlin-Asp P.G., Corces V.G.. Dynamic changes in the genomic localization of DNA replication-related element binding factor during the cell cycle. Cell Cycle. 2013; 12:1605–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Langmead B. Aligning short sequencing reads with Bowtie. Curr. Protoc. Bioinformatics. 2010; doi:10.1002/0471250953.bi1107s32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., Genome Project Data Processing S.. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rao S.S., Huntley M.H., Durand N.C., Stamenova E.K., Bochkov I.D., Robinson J.T., Sanborn A.L., Machol I., Omer A.D., Lander E.S. et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014; 159:1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Imakaev M., Fudenberg G., McCord R.P., Naumova N., Goloborodko A., Lajoie B.R., Dekker J., Mirny L.A.. Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nat. Methods. 2012; 9:999–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ay F., Bailey T.L., Noble W.S.. Statistical confidence estimation for Hi-C data reveals regulatory chromatin contacts. Genome Res. 2014; 24:999–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shen L., Shao N., Liu X., Nestler E.. ngs.plot: Quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics. 2014; 15:284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robinson J.T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P.. Integrative genomics viewer. Nat. Biotechnol. 2011; 29:24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thorvaldsdottir H., Robinson J.T., Mesirov J.P.. Integrative Genomics Viewer (IGV): high-pserformance genomics data visualization and exploration. Brief Bioinform. 2013; 14:178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Bemmel J.G., Pagie L., Braunschweig U., Brugman W., Meuleman W., Kerkhoven R.M., van Steensel B.. The insulator protein SU(HW) fine-tunes nuclear lamina interactions of the Drosophila genome. PLoS One. 2010; 5:e15013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T.. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003; 13:2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Core L.J., Waterfall J.J., Gilchrist D.A., Fargo D.C., Kwak H., Adelman K., Lis J.T.. Defining the status of RNA polymerase at promoters. Cell Rep. 2012; 2:1025–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kwak H., Fuda N.J., Core L.J., Lis J.T.. Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science. 2013; 339:950–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chopra V.S., Kong N., Levine M.. Transcriptional repression via antilooping in the Drosophila embryo. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:9460–9464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zentner G.E., Scacheri P.C.. The chromatin fingerprint of gene enhancer elements. J. Biol. Chem. 2012; 287:30888–30896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Heintzman N.D., Hon G.C., Hawkins R.D., Kheradpour P., Stark A., Harp L.F., Ye Z., Lee L.K., Stuart R.K., Ching C.W. et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009; 459:108–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Perez-Lluch S., Blanco E., Tilgner H., Curado J., Ruiz-Romero M., Corominas M., Guigo R.. Absence of canonical marks of active chromatin in developmentally regulated genes. Nat. Genet. 2015; 47:1158–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pekowska A., Benoukraf T., Zacarias-Cabeza J., Belhocine M., Koch F., Holota H., Imbert J., Andrau J.C., Ferrier P., Spicuglia S.. H3K4 tri-methylation provides an epigenetic signature of active enhancers. EMBO J. 2011; 30:4198–4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shen H., Xu W., Guo R., Rong B., Gu L., Wang Z., He C., Zheng L., Hu X., Hu Z. et al. Suppression of enhancer overactivation by a RACK7-histone demethylase complex. Cell. 2016; 165:331–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pagans S., Ortiz-Lombardia M., Espinas M.L., Bernues J., Azorin F.. The Drosophila transcription factor tramtrack (TTK) interacts with Trithorax-like (GAGA) and represses GAGA-mediated activation. Nucleic Acids Res. 2002; 30:4406–4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bartkuhn M., Straub T., Herold M., Herrmann M., Rathke C., Saumweber H., Gilfillan G.D., Becker P.B., Renkawitz R.. Active promoters and insulators are marked by the centrosomal protein 190. EMBO J. 2009; 28:877–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jiang N., Emberly E., Cuvier O., Hart C.M.. Genome-wide mapping of boundary element-associated factor (BEAF) binding sites in Drosophila melanogaster links BEAF to transcription. Mol. Cell. Biol. 2009; 29:3556–3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Emberly E., Blattes R., Schuettengruber B., Hennion M., Jiang N., Hart C.M., Kas E., Cuvier O.. BEAF regulates cell-cycle genes through the controlled deposition of H3K9 methylation marks into its conserved dual-core binding sites. PLoS Biol. 2008; 6:2896–2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Deng W., Lee J., Wang H., Miller J., Reik A., Gregory P.D., Dean A., Blobel G.A.. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell. 2012; 149:1233–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Phillips-Cremins J.E., Sauria M.E., Sanyal A., Gerasimova T.I., Lajoie B.R., Bell J.S., Ong C.T., Hookway T.A., Guo C., Sun Y. et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell. 2013; 153:1281–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ulianov S.V., Khrameeva E.E., Gavrilov A.A., Flyamer I.M., Kos P., Mikhaleva E.A., Penin A.A., Logacheva M.D., Imakaev M.V., Chertovich A. et al. Active chromatin and transcription play a key role in chromosome partitioning into topologically associating domains. Genome Res. 2016; 26:70–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Phillips-Cremins J.E., Corces V.G.. Chromatin insulators: linking genome organization to cellular function. Mol. Cell. 2013; 50:461–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lieberman-Aiden E., van Berkum N.L., Williams L., Imakaev M., Ragoczy T., Telling A., Amit I., Lajoie B.R., Sabo P.J., Dorschner M.O. et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009; 326:289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kellner W.A., Van Bortle K., Li L., Ramos E., Takenaka N., Corces V.G.. Distinct isoforms of the Drosophila Brd4 homologue are present at enhancers, promoters and insulator sites. Nucleic Acids Res. 2013; 41:9274–9283. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.