Abstract
CRISPR–Cas systems provide prokaryotes with adaptive defense against bacteriophage infections. Given an enormous variety of strategies used by phages to overcome their hosts, one can expect that the efficiency of protective action of CRISPR–Cas systems against different viruses should vary. Here, we created a collection of Escherichia coli strains with type I-E CRISPR–Cas system targeting various positions in the genomes of bacteriophages λ, T5, T7, T4 and R1-37 and investigated the ability of these strains to resist the infection and acquire additional CRISPR spacers from the infecting phage. We find that the efficiency of CRISPR–Cas targeting by the host is determined by phage life style, the positions of the targeted protospacer within the genome, and the state of phage DNA. The results also suggest that during infection by lytic phages that are susceptible to CRISPR interference, CRISPR–Cas does not act as a true immunity system that saves the infected cell but rather enforces an abortive infection pathway leading to infected cell death with no phage progeny release.
INTRODUCTION
Bacteriophages are the most abundant and diverse biological objects on the planet (1). They are constantly forced to improve and diversify the mechanisms and strategies of infection while their prokaryotic hosts develop increasingly sophisticated mechanisms to protect themselves. CRISPR–Cas systems represent one class of such host defense mechanisms. They provide prokaryotes with adaptive immunity against bacteriophages by targeting their DNA and/or RNA. CRISPR–Cas systems comprise Clusters of Regularly Interspaced Short Palindromic Repeats and CRISPR associated (cas) genes. Between the arrays of CRISPR repeats there are spacers, DNA sequences that have been acquired from foreign DNA and stored as ‘memories’ of prior infections (2,3). CRISPR arrays are transcribed to generate short protective CRISPR RNAs (crRNAs) each containing individual spacer sequence flanked by fragments of repeats (4). During CRISPR interference, effector complexes composed of Cas proteins with individual crRNAs recognize complementary sequences in foreign DNA or RNA (referred to as ‘protospacers’). Upon target protospacer recognition the effector complex either itself destroys the target by endonucleolytic cleavage (5,6) or recruits dedicated executor Cas endonucleases to degrade the target DNA (7–9). In some CRISPR–Cas systems, target recognition activates non-specific degradation of non-target nucleic acids (6,10,11), a strategy that does not benefit individual infected cell but should be advantageous to the population by limiting the spread of infection. In most CRISPR–Cas systems interference requires, in addition to a perfect or almost perfect match between crRNA spacer and target protospacer, an additional protospacer adjacent motif (PAM) that makes it possible to distinguish between the non-self target and the fully matching spacer in CRISPR array (12).
CRISPR interference has been shown to be very efficient in protecting cells from transformation or conjugation with plasmids (13) and infection with phages containing protospacers matching crRNA spacers (14). The protection against phage infection can only occur when a spacer matching a protospacer in the phage genome is present in the array. The acquisition of such spacer must happen at an earlier point in the course of infection of an initially ‘naïve’ cell. There is evidence that spacer acquisition occurs most efficiently during the infection by non-virulent defective viral particles (15), which should simplify the task of acquisition of protective spacers and passing them along to progeny cells.
In response to CRISPR interference by the host, phages with mutations that introduce mismatches between protospacer and crRNA spacer or inactivate PAM are selected (14,16). Such ‘escape’ mutations render CRISPR interference ineffective. Using Escherichia coli cells chronically infected with the M13 bacteriophage we earlier showed that residual interaction of type I-E CRISPR–Cas effector complex with partially matched protospacer triggers efficient acquisition of additional spacers (17) in a process named ‘primed CRISPR adaptation’ (17,18). Primed adaptation leads to preferential acquisition of additional spacers from viral protospacers with canonical PAM allowing the cell to restore interference against escape phage (17).
Some phages have been shown to counter the protective CRISPR interference of their hosts with anti-CRISPR proteins that bind to and inactivate either the effector complex or executor nuclease (19,20). Considering the commonality of CRISPR–Cas systems in prokaryotes it is somewhat surprising that viral anti-CRISPR systems are rare. Many phages have evolved elaborate strategies that allow them to take over the host macromolecular synthesis early in infection, essentially converting the infected cell into an assembly line for phage progeny production. For some lytic phages, the earliest stage of infection, injection of phage DNA into the host cell, proceeds in an orderly manner, with host-takeover genes injected and expressed first, followed by injection of the rest of the viral DNA into a pre-conditioned cell (21,22). Such conditioning involves, for example, degradation of host DNA to nucleotides, which are then used to synthesize viral progeny DNA (21). Host take-over strategies used by a phage may severely limit the efficiency of CRISPR–Cas response at either the adaptation or interference stages even in the absence of specialized anti-CRISPR proteins. However, no systematic study in this regard was undertaken yet. In this work, we investigated the infection of E. coli cells containing CRISPR arrays with spacers targeting different locations in the genomes of classical bacteriophages λ, T5, T7 and T4 and a giant phage R1–37. Our results reveal complex dynamics of CRISPR–Cas system interactions with genetic parasites and show that phages can limit the efficiency of CRISPR–Cas immunity by strategies other than dedicated anti-CRISPR proteins.
MATERIALS AND METHODS
Plasmid and strain construction
All strains (Supplementary Table S1) are based on KD263 (23) or KD349 (an F-derivative of KD263) that were engineered with a Red recombinase-based procedure (24). Both strains contain a g8 spacer targeting a protospacer in the M13 genome. The same protospacer is present in plasmids with cloned fragments of lytic phage genomes. The cas3 gene expression is driven by the lac promoter in both strains. The remaining cas genes are transcribed from the araB8 promoter.
The pG8mut, a pT7Blue-based plasmid carrying a 209-bp M13 fragment with the g8 protospacer (genome positions 1311–1519) with an escape mutation C1T at the first position of the protospacer is described in (25,26) and was used to clone fragments of phage DNA. Phage DNA was purified from lysed cultures by a protocol described in (27).
To construct pG8mut-based plasmids containing λ, T5 or T7 fragments, the plasmid was treated with NdeI and ligated with phage DNA digested with MseI. Plasmids containing phage DNA inserts of desired size (2–4 kbp) were selected for further use. To construct pG8mut-based plasmids containing fragments of T4 or R1–37 DNA 2–4 kb regions of phage genomes were amplified using appropriate primers and individually cloned in pG8mut. The sequences of primers are listed in Supplementary Table S2.
Construction of KD263 or KD349 derivatives harboring phage targeting CRISPR spacers using plasmids containing cloned phage DNA fragments was performed as described in (25) and as summarized in Supplementary Figure S1. We use the following nomenclature to refer to strains carrying spacers targeting phage protospacers: ‘phage name—temporal class of the gene carrying targeted protospacer, protospacer #—protospacer orientation’. For example, ‘T7-M3-F’ refers, depending on the context, to a strain or a protospacer from phage T7 middle (M) gene in the forward orientation, i.e. the sequence of the spacer in crRNA or the protospacer in phage genome matches the genomic sequence deposited in the Genbank. ‘R’ indicates reverse orientation when crRNA spacer is complementary to the GenBank sequence. To distinguish spacers in different strains and targeted protospacers, they are numbered (in the example above we refer to strain or protospacer number 3 of the four distinct strains targeting middle T7 genes that are present in our collection, see Supplementary Table S1). All phage targeting strains are listed in Supplementary Table S1.
CRISPR interference assays
To assay for plasmid interference, cells were grown in LB medium at 37°C in the presence or in the absence of 1 mM arabinose and 1 mM IPTG until the culture OD600 reached 0.6. Electrocompetent cells were prepared using a standard protocol (28) and transformed with 5 ng of plasmids containing protospacers. After 1-h 37°C outgrowth in 500 μl LB without antibiotic, 100 μl aliquots of serial dilutions of transformation mixtures were plated onto LB agar plates containing 100 μg/ml ampicillin. Plates were incubated at 37°C overnight. For each plasmid, transformation was repeated three times. Efficiency of transformation was determined as a ratio of transformants formed by induced and uninduced cells.
To determine interference with phage infection, aliquots of induced and uninduced cultures grown as described above were combined with serial dilutions of phage lysates, combined with soft LB agar and overlaid on LB plates with or without 1 mM arabinose and 1 mM IPTG. Each experiment was repeated at least three times. After drying, plates were incubated at 37°C overnight. The efficiency of plaquing was determined as a ratio of phage plaques formed by induced and uninduced cells.
Monitoring phage infection
The growth of induced and uninduced cultures with or without the phage was continuously monitored in EnSpire Multimode Plate Reader (PerkinElmer). At least three growth curves were determined for each strain at every condition. Induced (1 mM arabinose and 1 mM IPTG) and uninduced cells were grown in LB at 37°C to OD600 ≈ 0.4 and infected with phages at various multiplicities. Cultures growth at 37°C was continued for 2–3 h with intensive aeration.
To determine phage burst size, infections were carried out essentially as described in (29). Cultures grown to OD600 0.6 were infected at MOI of 0.1. After 3–7 min at 37°C to allow phage adsorption, cells were separated from unbound phage by centrifugation, resuspended in fresh medium with or without inducers and incubation at 37°C was continued. At various times, aliquots were taken and phage titer was determined.
DNA extraction
Induced or uninduced cultures were grown to OD600 of 0.6 and infected at MOI of 2. Cells were collected shortly before lysis and DNA was isolated by phenol-chloroform method from cell pellet as described (27). The DNA was next digested with restriction endonucleases recognizing multiple sites in phage DNA and analyzed by agarose electrophoresis.
CRISPR adaptation assay
Induced cells cultures (OD600 = 0.6) were infected at the MOI of 0.001–1.0 with wild-type or escape phages and grown for 16 h at 37°C. Culture aliquots were withdrawn and cells were subjected to PCR with primers annealing to the CRISPR array leader sequence and to M13 g8 spacer. Amplification reaction products were analyzed as described in (23).
High throughput sequencing and data analysis
PCR-products corresponding to expanded CRISPR array were subjected to high-throughput sequencing with MiSeq Illumina system. The resulting data were analyzed using ShortRead (30) and BioStrings (31) Bioconductor packages. Sequences located between two CRISPR repeats were considered as spacers. They were mapped on phage genomes with no mismatches allowed. R scripts were used for statistical analysis and Circos (32) was used for graphical representation of the data.
To sequence T7 phage genome in infected cells, total DNA was extracted from infected cells (27) several minutes before expected lysis time and treated with restriction endonucleases BamHI, EcoRV, HindIII, PstI (do not recognize T7 DNA) to destroy host DNA. Samples were loaded on an 0.8% agarose gel, resolved by electrophoresis and a high-molecular weight band of phage genomic DNA was extracted from gel with GeneJET Extraction Kit (ThermoFisher Scientific) and sequenced with MiSeq Illumina system. Reads were trimmed and mapped on the phage genome with no mismatches allowed.
Microscopy
All microscopy experiments were performed using Zeiss AxioImager.Z1 upright microscope equipped with a custom made incubation system to maintain cells at 37°C. Semrock mCherry-40LP filter set was used for propidium iodide fluorescence detection. Images were collected using Cascade II 1024 back-illuminated EM-CCD camera (Photometrics). Autofocusing, multichannel and multifield time-lapse acquisition were implemented using MicroManager (33) with custom scripts. Image analyses were performed using Fiji (ImageJ) (34,35).
Microscope chambers were prepared using previously described procedure (36) to provide time-lapse imaging. 1.5% agarose (Helicon) diluted in LB (Amresco) was used as a medium to monitor bacterial growth during microscopy. One microscope chamber allowed simultaneous cultivation and monitoring of cells with and without cas gene induction by using two or more separate agarose pads with and without inducers. Cells were mixed with phage and propidium iodide (Molecular Probes), which was added to the final concentration of 20 μM and placed in microscope chamber (0.5 μl on each pad). When cells were absorbed onto agarose pads the chamber was sealed and installed in the microscope. Typically 4–10 fields of view were visualized in a single experiment. Each field was imaged every 5–10 min during 1–4 h in transmitted light channel and propidium iodide fluorescence channel.
RESULTS
Generation of E. coli cells carrying CRISPR spacers targeting different phages
The primed adaptation phenomenon can be harnessed for facile construction of strains targeting various regions of phage and/or plasmid DNA (Supplementary Figure S1). The procedure involves creation of a plasmid containing a protospacer partially matching a CRISPR spacer of parental strain and a fragment of DNA that needs to be targeted (25). Escherichia coli cells carrying inducible cas genes and a miniature CRISPR array containing a single g8 spacer (23) were transformed with pT7Blue-based plasmids containing a protospacer corresponding to g8 spacer but bearing a mismatch at position +1 and cloned fragments of different genomic regions of E. coli bacteriophages λ, T5, T7, T4. Plasmids containing cloned fragments of R1–37, a giant phage infecting Yersinia pseudotuberculosis (37), were also constructed. Following transformation and induction of primed adaptation, clones that have lost the plasmid and expanded their CRISPR arrays by acquiring spacers from DNA of interest were identified. Since cloned phage DNA fragments were between 2 and 4 kb in length, which is comparable with ∼3 kb length of the pT7Blue vector, after growth at conditions of cas genes expression approximately half of ampicillin-sensitive clones with expanded CRISPR arrays acquired spacers from phage DNA inserts. Strains harboring phage-specific spacers in our collection are listed in Supplementary Table S1.
Bacteriophage λ
Fifteen E. coli strains targeting λ early or late genes (Supplementary Table S1) were tested for ability to be infected with λvir, a phage λ mutant that is only capable of lytic cycle. The number of plaques on induced (CRISPR–Cas system ON) and uninduced (CRISPR–Cas system OFF) cell lawns was recorded and efficiency of plaquing (EOP) was calculated as the ratio of the two numbers. All strains strongly (10−5–10−9) interfered with λvir infection in the plaque-forming assay (Supplementary Table S1 and Figure 1B). For seven strains, no phage plaques were observed, so the calculated EOP of 10−9 presents a lower estimate of CRISPR interference efficiency. Phages that formed plaques on remaining strains contained escape mutations which included point substitutions in the PAM or the seed of targeted protospacer; a 15-bp deletion in the protospacer (the affected protospacer is in an intergenic region of the phage); a double mutation outside the seed region (positions +20/+22 and +25/+28) but with intact PAM/seed segments, or single nucleotide deletions in positions 23 and 20 (Supplementary Table S1).
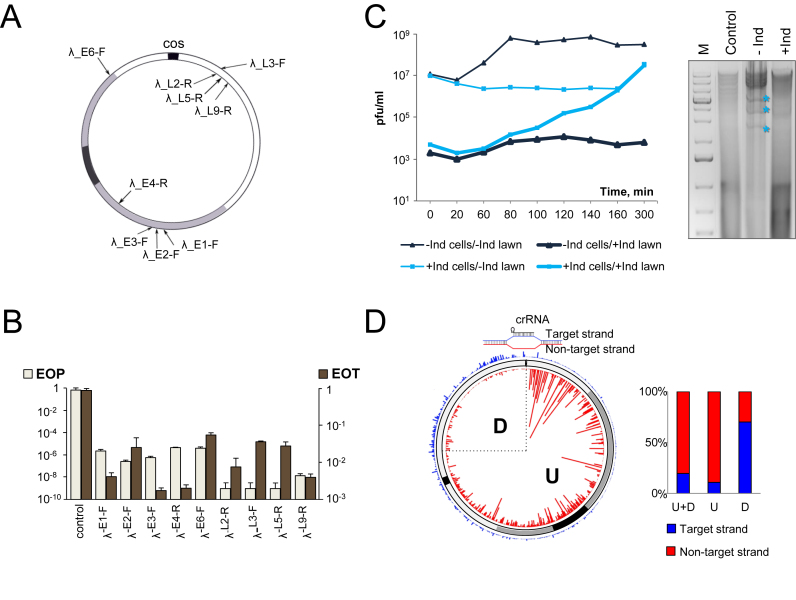
Figure 1.
The effect of CRISPR–Cas targeting on λvir infection. (A) The circular genome of bacteriophage λ formed after annealing at the cos sites is shown. The region containing immediate early genes is colored black; the region containing delayed early genes is gray, late gene region is white. Black arrows indicate the positions of protospacers targeted by crRNA spacers from different strains. Protospacers located at different strands of the genome are shown by arrows located inside and outside the circle representing the phage genome. (B) The efficiency of plaque formation (EOP) by λvir and efficiency of transformation (EOT) by cognate protospacer-containing plasmids into indicated strains. Mean values and standard deviations from three independent experiments with each strain are presented. EOP values for strains targeting additional protospacers can be found in Supplementary Table S1. (C) On the left, dynamics of plaque forming units (pfu) during the infection of induced (‘+Ind’, blue lines) or uninduced (‘–Ind’, black lines) λ-L9-R culture is shown. Phage from each infection was plated on lawns of uninduced (thin lines) and induced (thick lines) λ-L9-R cells to determine the total number of infectious phages and the number of escape phages, respectively. On the right, an agarose gel showing the products of BspHI digestion of DNA prepared from induced (‘+Ind’) and uninduced (‘–Ind’) λ-L9-R cells infected with λvir and collected 60 min post-infection is presented. ‘M’ is a molecular weight marker lane. Lane labeled ‘Control’ shows DNA from uninfected cells. Asterisks indicate restriction fragments of phage DNA. (D) Graphical representation of HTS analysis of spacers acquired during the infection of λ-L9-R cells by a λvir G-1T escape mutant. The position of the priming protospacer is indicated at the top and a structure of the R-loop formed by crRNA at this protospacer is shown. Acquired spacers are mapped on the circular viral genome. Red and blue lines show spacers matching, respectively, non-target strand and target strands. Line heights indicate relative frequency of reads corresponding to different spacers. Rectangular bars show the proportion of spacers matching the non-target (red) and target (blue) strands for the entire genome (‘U+D’); in the downstream (‘D’) quarter of the viral genome, and in the remaining upstream part (‘U’).
The efficiency of CRISPR interference could be affected by spacer-protospacer sequences. To account for such effects, competent cells from induced and uninduced strains targeting phage protospacers shown in Figure 1A were prepared, transformed with plasmids containing matching phage protospacers and efficiency of transformation (EOT) was determined by calculating the ratio of numbers of transformants formed by induced and uninduced cells. EOT values for tested λ targeting strains varied by as much as 2–3 orders of magnitude (Figure 1B), demonstrating that the sequences of crRNA spacer and cognate protospacer indeed strongly affect CRISPR interference. There was no correlation between the EOP and EOT values (Figure 1B).
λvir infected liquid cultures lyzed poorly at our conditions even when infected at high multiplicity of infection (MOI) since phage receptor requires glucose for maximal expression (38), while cas gene expression requires arabinose and the absence of glucose. Therefore, it was impossible to monitor the effect of CRISPR interference at the growth of infected λ targeting strains. The total amount of infectious phage particles in infected liquid cultures was determined by counting plaques formed on lawns of uninduced cells. The amount of escape phages in infected cultures was determined by plating on induced cell lawns. A representative result is shown in Figure 1C. In the absence of cas gene induction the phage titer in infected cultures increased about 100-fold ∼80 min post-infection (thin black line). In the presence of active CRISPR–Cas system (thin blue line) the increase in phage titer was delayed to ∼160 min. The late increase in phage titer was due to accumulation of escape phages, which were rare (10−4) at the start of infection but completely overtook the population at late stages (Figure 1C, left, thick blue line). There was no such increase in escape mutants in the course of infection of uninduced cells (thick black line).
To monitor accumulation of phage DNA in infected cells, total DNA was extracted from infected cells, digested with restriction endonuclease BspHI and examined by agarose electrophoresis. A representative experiment showing accumulation of λ DNA in the presence and in the absence of cas gene expression inducers is shown on the right of Figure 1C. Distinct bands corresponding to fragments of λ genome were clearly visible on the smeary background of host DNA in the absence of cas genes expression. In contrast, only cellular DNA was present when cas genes were induced. Thus, CRISPR interference prevents accumulation of replicated λvir DNA in infected cells.
CRISPR array expansion was detected in the course of infection of λ targeting cells with escape phages. Several bacterial clones that survived the infection of λ-L9-R strain with the G-1T escape phage carrying a G to T substitution in the PAM of the L9-R protospacer and harbored an expanded CRISPR array were isolated. These clones were resistant to both wild-type and G-1T phage infection at conditions of cas genes induction. Thus, newly acquired λ-derived spacers are competent for interference. PCR amplified material corresponding to expanded λ-L9-R array from infected culture was subjected to Illumina high-throughput sequencing (HTS) and spacers were extracted from sequencing reads and mapped on the circular phage genome (Figure 1D). Ninety two percent of new spacers were acquired from protospacers associated with canonical AAG PAM and there was an overall 79% bias for selection of spacers from the non-targeted strand (Supplementary Table S3). These values are typical for primed adaptation (39). However, the extent and even the direction of strand bias was not uniform throughout the viral genome (Figure 1D, and Supplementary Table S3). 89% of spacers acquired from the upstream (clockwise in Figure 1D) three quarters of the λ genome matched protospacers in non-targeted strand. In the downstream (counterclockwise) quarter of phage genome the direction of strand bias was reversed and most (∼70%) of spacers matched protospacer in the opposite, targeted strand (Figure 1D). Efficiency of spacer selection decreased as the distance from the priming protospacer increased and overall spacer acquisition was most efficient close to and upstream of the priming protospacer (the bias towards non-targeted strand was also the strongest in this area, approaching 99%).
Bacteriophage T5
Bacteriophage T5 is a strictly lytic virus with a linear ∼121 300 bp genome containing terminal 10 160 bp direct repeats (Figure 2A). The proximal repeat carries pre-early genes that are injected in the host cell at the first stage of infection followed by a pause to allow the synthesis of pre-early proteins that inactivate several essential cellular processes and destroy host DNA (21,22). The rest of phage DNA from which early and late genes are expressed is next injected into the infected cell. Strains targeting unique protospacers in pre-early, early and late T5 genes are listed in Supplementary Table S1 and some are shown in Figure 2A. In the plaque formation assay, induced strains carrying spacers targeting late or early T5 genes were infected by the phage as efficiently as uninduced or the parental non-targeting strain (Figure 2B, right and Supplementary Table S1). The morphology of phage plaques was also unaffected (data not shown). In contrast, all strains targeting pre-early genes exhibited some level of interference with the infection (Figure 2B, left and Supplementary Table S1). The EOP values ranged from 10−1 for T5-PE3-F to <10−8 for T5-PE9-R strain (Figure 2B, left; in the latter case, no phage plaques was observed, so the interference efficiency value given is a lower estimate). Phages were recovered from several individual randomly picked plaques formed on lawns of induced strains targeting pre-early region and analyzed further. Phages recovered from strains that gave low (10−1–10−2) levels of interference continued to be restricted at the same level upon reinfection and when sequenced through the protospacer region were found to contain a wild-type sequence. Phages recovered from strains that efficiently interfered with infection (10−4–10−6) demonstrated an EOP of one when replated on the same strain expressing cas genes and therefore behaved as escape mutants. Indeed, such phage contained mutations in the PAM or seed (positions +1 to +8) segments of targeted protospacers (Supplementary Table S1).
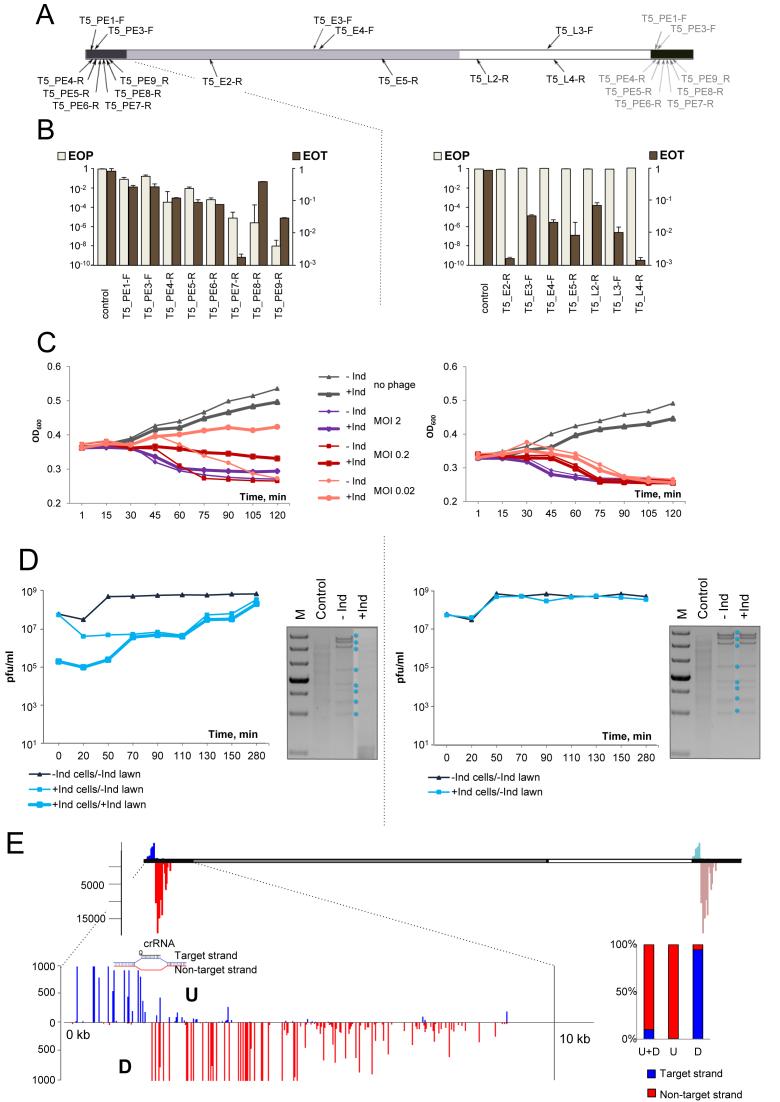
Figure 2.
The effect of CRISPR–Cas targeting on T5 infection. (A) The linear genome of bacteriophage T5 and positions of protospacers targeted by crRNA spacers of strains from our collection are schematically shown. Pre-early genes are colored black, the region containing early genes is gray, late gene region is white. The T5 genome is terminally redundant with the entire pre-early area present at either end of the genome. Spacers targeting pre-early region thus have two matching sites. Protospacers at the right end are shown as grey arrows. Protospacers originating from different strands of the genome are shown above or below the genome scheme. (B) EOP by T5 and EOT by cognate protospacer-containing plasmids into indicated strains. Results obtained with pre-early targeting strains are shown on the left. Results obtained with early and late targeting strains are on the right. Mean values and standard deviations from three independent experiments with each strain are presented. EOP values for strains targeting additional protospacers can be found in Supplementary Table S1. (C) Growth curves of induced (‘+Ind’) and uninduced (‘–Ind’) cultures of T5-PE-6-R (left) and T5-E4-F (right) strains infected with T5 at indicated MOIs. (D) Phage progeny production during the infection of T5-PE-6-R (left) or T5-E4-F (right) cultures. See Figure 1C legend for details. At the right of each diagram with growth curves, agarose gels showing the products of SalI and MluI digestion of DNA prepared from cells from an uninfected culture, or induced and uninduced infected (MOI = 2) cultures collected 45 min post-infection are presented. Phage DNA restriction fragments are marked by asterisks. (E) Graphical representation of HTS results of spacers acquired by the T5-PE-6-R cultures infected with the A2G escape T5 phage. Red vertical lines indicate spacers matching the non-target strand, blue lines—spacers matching the target strand. Line heights indicate relative frequency of reads corresponding to each acquired spacer. Scale bar at the left shows numbers of Illumina reads. The pre-early region at the left end of the genome is expanded below to show mapping results at a larger scale. The position of the priming protospacer is indicated. A vertical black line shows the position until which phage DNA is initially inserted into the infected cell (21,22). Rectangular bars show the proportion of spacers matching non-target (red) and target (blue) strands upstream (‘U’) and downstream (‘D’) of the priming protospacer and the total strand bias of acquired spacers (‘U+D’).
Plasmid transformation experiments indicated that CRISPR–Cas was able to recognize and interfere with target protospacers from pre-early, early, and late T5 genes (Figure 2B), suggesting that genomic location is responsible for lack of protection of cells with early and late-gene targeting spacers from phage infection.
Cultures of cells targeting early or late genes behaved indistinguishably from uninduced cultures and were lysed by the phage at all MOIs tested (Figure 2C, right). Induced cultures targeting pre-early genes infected at low MOI of ∼0.02 were able to partially withstand the infection, with cell density rising slightly over the course of the experiment (Figure 2C, left, thick pink line). At the intermediate MOI of ∼0.2 the optical density of induced cultures declined slowly (Figure 2C, left, thick red line). Induced cultures infected at the MOI of ∼2 lysed as fast as the uninduced control (Figure 2C compare thick and thin purple lines).
In single-burst experiments, no difference in phage progeny yield or eclipse time was observed between induced and uninduced cells targeting early or late T5 genes (Figure 2D, right). For pre-early targeting cells the increase in phage titer in infected cultures was delayed and was due to accumulation of escape phages (Figure 2D, left).
Total DNA was extracted from infected cells and digested with SalI and MluI restriction endonucleases. No changes in the amounts or profile of phage DNA restriction fragments was observed when DNA purified from cells bearing spacers against early or late T5 genes was compared with DNA prepared from uninduced infected cells and host DNA was degraded (Figure 2D, right). In the case of pre-early targeting no viral DNA was present. Host DNA was also absent, apparently degraded by phage pre-early proteins (Figure 2D, left).
Individual T5 phage isolates that behaved as escape mutants were tested for ability to cause primed adaptation during infection of corresponding targeting cells. CRISPR array expansion was observed only after infection of induced pre-early targeting cells. Several bacterial clones that survived the infection of the T5-PE6-R culture with an A2G escape phage bearing an A to G substitution in the second position of the targeted protospacer and expanded their CRISPR array were isolated. All clones were resistant to both the wild-type and the A2G phage at conditions of cas genes induction. Thus, newly acquired T5-derived spacers are competent for interference.
PCR amplified fragments corresponding to expanded CRISPR arrays in the T5-PE6-R culture infected with the A2G phage were subjected to Illumina sequencing. The results of mapping of acquired spacers are presented in Figure 2E. Almost all (99.7%) spacers were acquired from protospacers associated with canonical AAG PAM (Supplementary Table S3) and an overall 89% bias towards acquisition of spacers from the non-targeted strand was observed (Figure 2E, Supplementary Table S3). More than 99% of spacers matching protospacers located upstream of the priming site had the same orientation as the priming protospacer. Conversely, 95% of spacers that originated downstream had the opposite orientation. The efficiency of spacer acquisition declined as the distance from the priming site increased in both upstream and downstream directions, resulting in a gradient of protospacer use.
Bacteriophage T7
The linear genome of bacteriophage T7 gradually enters host cell starting with the left (early) end in a process that is coupled to transcription of early T7 genes by the host RNA polymerase (21). Our collection contained 11 strains targeting four protospacers in T7 early genes, four in middle genes and three in late genes (Figure 3A, Supplementary Table S1). All strains interfered with T7 infection, decreasing EOP between one to at least nine orders of magnitude (Figure 3B). Each strain also interfered with plasmid transformation (Figure 3B). The EOT and EOP efficiencies for different spacers appeared to be correlated, suggesting a lack of significant contribution of genomic context to CRISPR interference against phage T7 infection.
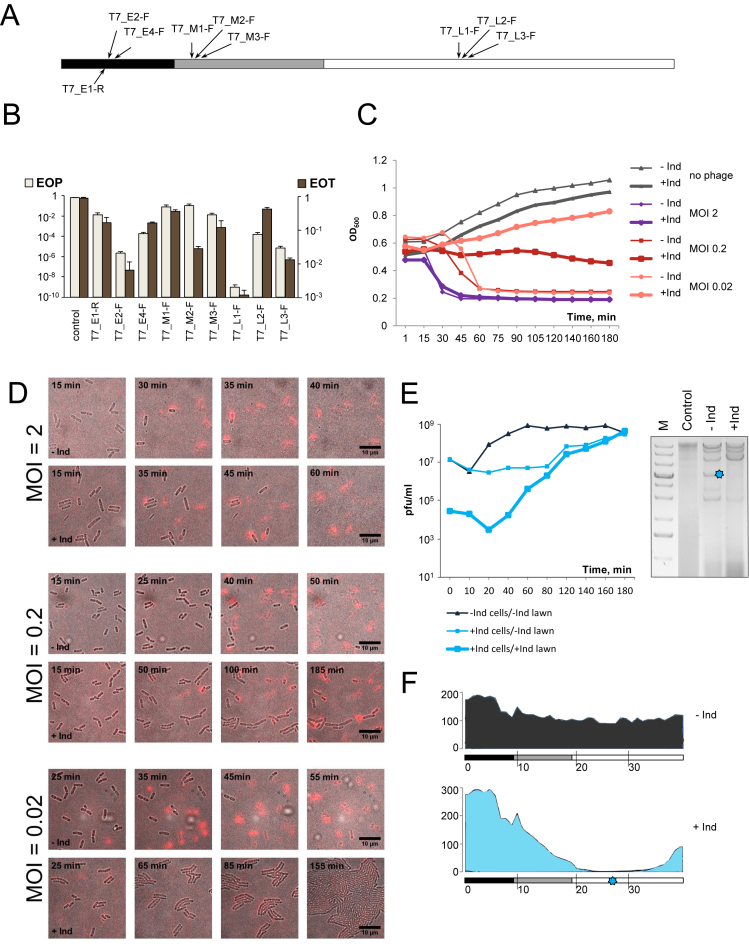
Figure 3.
The effect of CRISPR–Cas targeting on T7 infection. (A) The linear genome of bacteriophage T7 and positions of protospacers targeted by crRNA spacers of strains from our collection are schematically shown. The early genes region is colored black, the region containing middle genes is gray, late gene region is white. An area that contains both early and middle genes is striped. (B) EOP by T7 and EOT by cognate protospacer-containing plasmids into indicated strains. Mean values and standard deviations from three independent experiments with each strain are shown. EOP values for strains targeting additional protospacers can be found in Supplementary Table S1. (C) Growth curves of induced and uninduced cultures of T7-E-2-F infected with T7 at indicated MOIs. (D) Images of induced and uninduced T7-E2-F cultures infected with T7 at different MOIs. Images were taken at times indicated. Depolarized or lyzed cells appear red in the images. For each MOI, images in the top row show the progress of infection in the absence of cas gene expression; images at the lower row show infection of induced cells. (E) On the left, phage progeny production during the infection of induced and uninduced T7-L2-F cells. See Figure 1C legend for details. On the right, an agarose gel showing the products of SspI digestion of DNA prepared from uninfected, or infected (MOI = 2) induced and uninduced T7-L2-F cells collected 18 min post-infection. (F) Graphical representation of mapping of Illumina reads from total DNA prepared from T7-infected uninduced (top) or induced (bottom) T7-L2-F cells onto the T7 genome. Scale bars at the left show numbers of Illumina reads. The position of the protospacer targeted by T7-L2-F crRNA is shown at the bottom with a blue asterisk.
Escape mutants were recovered for 8 strains targeting all temporal classes of T7 genes. In addition to point escape mutants changing the PAM of the protospacer seed, a deletion removing 59 nucleotides from early gene 0.7 was obtained. Gp0.7 codes for a protein kinase that is dispensable for growth at laboratory conditions (40) and so the deletion did not interfere with phage viability. Another escape mutant contained a GGT insertion between positions 24 and 25 of protospacer T7-E4-F (Supplementary Table S1). This protospacer is part of gene 1, which codes for T7 RNA polymerase. The insertion occurred between glycine-coding codons GGT and GGC. The mutation may have arisen due to slippage during replication of a G-rich stretch of phage genome, resulting in insertion of an additional glycine residue in phage RNA polymerase.
When growth of cultures infected at different MOIs was monitored, similar results were obtained with cells targeting any one of the three gene expression classes. Representative results obtained with an early gene targeting T7-E2-F strain are shown Figure 3C. In the absence of cas gene induction cells lysis occurred shortly after the addition of the phage (MOI of 2) or after a delay (MOIs of 0.2 and 0.002) needed for progeny phage to appear, infect, and lyse remaining uninfected cells. At high MOI, induced cultures lysed as efficiently as uninduced control cultures, indicating that phage was able to overrun the defensive action of CRISPR–Cas. Induced cultures infected at the MOI of 0.2 maintained constant OD600 at first and then gradually started to decline. Induced cultures infected at the MOI of 0.02 withstood the infection and grew almost as well as the uninfected control.
Cells in infected T7-E2-F cultures were monitored by live microscopy in the presence of propidium iodide, which selectively stains cells with depolarized membrane. Membrane depolarization occurs shortly before cell lysis by the phage (21,41). The results showed that all cells in cultures infected at the MOI of 2 were depolarized and lyzed irrespective of whether cas gene expression was induced or not (Figure 3D). At the intermediate MOI of 0.2 cells in uninduced cultures lysed completely, while in induced cultures a dynamic behavior including lysis of some cells and continued growth of others was observed during the course of experiment. At the MOI of 0.02 some cells lysed between 20 and 60 min, but later no lysis was observed and the culture grew as well as the uninfected control.
In single burst experiments, progeny phage appearance during infection of induced T7 targeting cells was delayed and was due to accumulation of escape phages (Figure 3E, left).
Phage DNA accumulated in infected cells that grew without induction of cas genes as judged by the appearance of restriction fragments of expected size (Figure 3E, right). Restriction fragments corresponding to T7 genomic DNA were also observed in induced cells. However, the pattern of these fragments was different and some fragments were missing (Figure 3F). To better understand the state of T7 DNA accumulating at conditions of CRISPR interference DNA from induced and uninduced cultures was subjected to HTS (Figure 3F and Supplementary Figure S2). Mapping of reads from uninduced cultures revealed uniform coverage over the entire length of the T7 genome. In contrast, there was a dramatic decrease in coverage for DNA prepared from targeting cells. The area of decreased coverage was extensive and centered at the targeted protospacer (Figure 3F and Supplementary Figure S2). We conclude that at conditions of CRISPR interference large portions of the T7 genome are lost, preventing production of progeny phage.
The primed adaptation experiment was attempted with every T7 targeting strain from our collection with wild-type and appropriate escape phages. In no case could we detect CRISPR array expansion in infected cultures.
Phages with modified genomes: T4 and R1-37
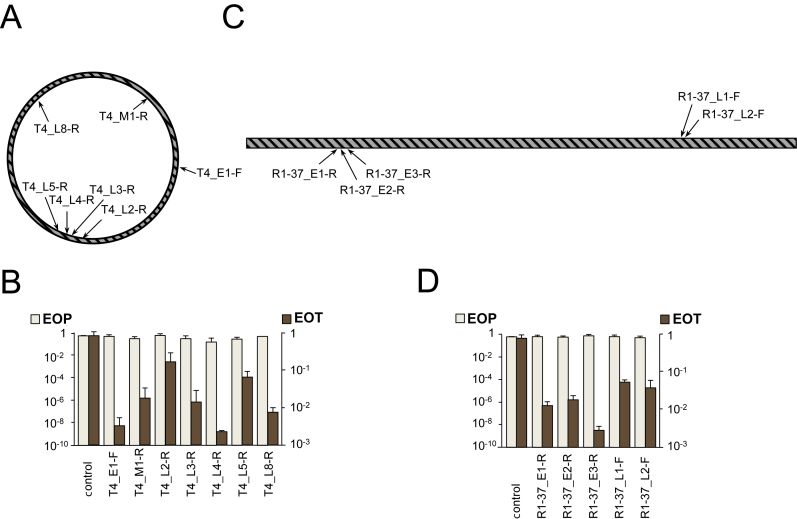
Bacteriophage T4 contains hyrdoxymethylcytosine instead of cytosine in its large (170 000 kb) circularly permuted genome. The unusual nucleotide is additionally modified by glycosylation (21,42). Bacteriophage R1–37 contains uracil instead of thymine in its 260 000 kb genome (37). Both phages have genes belonging to different expression classes scattered throughout their genomes (Figure 4A and C, respectively). Escherichia coli is naturally resistant to R1–37, however, the phage can infect E. coli carrying a plasmid expressing its receptor (35). Accordingly, a plasmid expressing the R1–37 receptor was introduced in strains containing R1–37-specific spacers. When T4 and R1–37 targeting strains were tested for ability to interfere with cognate phage infection, no resistance was observed (Figure 4B and D). In contrast, when these cells were transformed by plasmids carrying protospacers, a decrease of transformation efficiency by one to three orders of magnitude was observed. Neither phage was able to induce CRISPR array expansion in infected induced targeting cultures.
Figure 4.
The effect of CRISPR–Cas targeting on T4 and R1–37 infections. (A) The circular genome of bacteriophage T4 is shown. Genes belonging to different expression classes are scattered throughout the genome. Arrows indicate the positions of protospacers targeted by crRNA spacers of strains from our collection. (B) EOP by T4 and EOT by cognate protospacer-containing plasmids into indicated strains. Mean values and standard deviations from three independent experiments with each strain are shown. EOP values for strains targeting additional protospacers can be found in Supplementary Table S1. (C) The linear genome of bacteriophage R1–37 and positions of protospacers targeted by crRNA spacers of strains from our collection. Genes belonging to different expression classes are scattered throughout the genome. (D) EOP by R1–37 and EOT by cognate protospacer-containing plasmids into indicated strains. Mean values and standard deviations from three independent experiments with each strain are shown. EOP values for strains targeting additional protospacers can be found in Supplementary Table S1.
DISCUSSION
The goal of this work was to assess how different dsDNA phages are affected by the action of the type I-E CRISPR–Cas system of E. coli. Whenever phage DNA can be effectively targeted by crRNA-effector complex (λvir, T7, pre-early region of T5), a delay in the appearance of phage progeny is observed in infected cultures. Eventually, phage titers recover, and this recovery is due to accumulation of escape mutants, which must either preexist in phage population used for infection, or are rapidly selected in the course of infection. The results thus show that CRISPR–Cas interference provides a strong selective pressure on diverse lytic phages, which, however, are able to rapidly overcome it. CRISPR–Cas systems can thus be regarded as a potent factor in generating phage variety. If effector complex targets phage protospacers where no escape mutants can be selected, no phage progeny is observed. Thus, within the bacterial population there shall be a strong selection of cells that acquired spacers targeting phage sequences that cannot be easily mutated, to allow long-lasting protection.
From the ‘point of view’ of a clonal bacterial culture, the outcome of infections by ‘professional’ lytic viruses T5 and T7 at conditions of countering CRISPR defense very much depends on the multiplicity of infection. When MOI is higher than one, all infected cells succumb to infection even though no progeny phage is released. When multiplicity of infection is significantly less than one, most cells remain uninfected, while infected cells die from the infection. However, since no or very little phage progeny is released, infected cultures continue to grow and the infection process dies off unless an escape phage appears. Therefore, it appears that in the cases studied here the CRISPR immunity functions similar to abortive infection mechanisms (43,44), not curing individual infected cells but preventing the spread of the virus through the population.
Targeting of phage λ or pre-early region of T5 DNA prevents accumulation of viral DNA in infected cells, presumably by destroying templates for replication of circular λ DNA or preventing the entry of stage II early and late T5 genes by the cleavage of pre-early region, respectively. This, however, does not rescue T5-infected cells, which die because of cytotoxic pre-early proteins (22). In the case of T7, replication of phage DNA occurs even at conditions of ongoing CRISPR interference. However, extensive regions in both directions from the targeted protospacer are destroyed, presumably due to the function of Cas3 nuclease-helicase (8). T7 is nevertheless able to replicate parts of its DNA at these conditions because of the active recombination of phage genomes during replication (45,46).
CRISPR interference is ineffective against early and late T5 genes, which are inserted in the cells after the point when cellular DNA is degraded. Our experiments do not directly address the question whether effector complexes that pre-exist in infected cells are present in infected cells, but even if they do, they clearly cannot prevent phage progeny production. The strategy used by T5 and its relatives thus appears to be an effective anti-CRISPR mechanism, with less than 10% of the genome being susceptible to CRISPR interference and only for a short period of time.
Since most cells infected by phages used here die even in the presence of CRISPR–Cas targeting a phage, only low levels of primed adaptation are observed. The situation contrasts with that seen with the M13 phage infections, where infected cells continue to grow and very high levels of spacer acquisition in infected cultures are observed (17). Analysis of spacers acquired during primed adaptation by cells targeting pre-early region of T5 reveals a gradient of spacer acquisition efficiency and a switch in the direction of strand bias of acquired spacers upstream and downstream of the priming site. Most acquired spacers match protospacers in the non-targeted strand upstream of the priming protospacer and target strand protospacers in the downstream direction. Since there are ∼10 times more spacers acquired from the upstream region, there is a ∼90% bias towards the non-targeted strand when all spacers are considered.
Spacers acquired from the circular λ genome exhibit an 80% overall bias towards the non-targeted strand. In contrast to situation with T5, a pronounced gradient of spacer acquisition efficiency is observed only upstream of the priming site and the strand bias disappears as the distance from the priming site is increased in the upstream direction (Figure 1D). The reason for this behavior is likely an ‘overlap’ of spacers acquired from different strands of the circular genome coupled with the overall lower efficiency of spacer selection downstream of the priming site. Previous studies of primed adaptation in E. coli using plasmids (18,26,37) or M13 infected phage (17) revealed a strand bias but no gradient of spacer acquisition efficiency as a function of distance from the priming site. The lack of observable gradients is likely caused by the smaller sizes of these circular genomes, which results in extensive overlaps of upstream and downstream spacer selection gradients and insufficient depth of sequencing of newly acquired spacers. Indeed, analysis of spacers acquired during the M13 phage infection presented in Supplementary Figure S3 reveals gradients of spacer acquisition efficiency and change in the direction of strand bias that were undetected previously (17).
The difference in the overall efficiency of spacer acquisition upstream and downstream of priming sites revealed in our work is consistent with the preferential cleavage by the E. coli Cas3 of the non-targeted strand in the R-loop complex and its 3΄-5΄ helicase activity (47). One can envision that Cas3 processively unwinds the priming protospacer and, after the initial cleavage of its single-stranded non-target strand, moves upstream (in the 3΄ to 5΄ direction with respect to the non-target strand) generating substrates that are channeled for integration into CRISPR array by Cas1 and Cas2. The strand bias in spacer acquisition would be consistent with recent in vitro data showing that Cas3 generated fragments that fuel primed adaption are partially single-stranded (48). The gradient in spacer acquisition efficiency must be related to either the processivity of Cas3 nuclease or the life times of Cas3 generated DNA fragments. Acquisition of spacers downstream from the priming site must be caused by Cas3 induced cleavage of the target strand followed by its subsequent degradation. In vitro, target strand cleavage proceeds with lower efficiency (47), which should explain smaller number of spacers acquired in the downstream direction.
Gradients of spacer acquisition efficiency were observed during primed adaptation by the I-F and I-B CRISPR–Cas systems (49–52). Spacer acquisition efficiency gradients observed in this work are extensive and much longer than that reported for type I-B system (49) but comparable to what was observed in the case of I-F system (51). Interestingly, the length of the gradient is also different between λ and T5, indicating that phage genome structure/replication affects adaptation. In the case of T5, the gradient may be also physically limited by the two-stage viral DNA injection mechanism. Since no early or late DNA is present in the cell during the first stage of injection, spacers cannot be acquired from these areas. In the upstream direction, the length of the gradient is limited by the end of the linear genome. Another possibility is that the length of the time window when adaptation can happen in infected cell differs for different phages (however, it should be much shorter during the T5 infection compared to λ infection).
We were unable to detect primed adaptation during T7 infection. Given our success with T5 and λ, the result may indicate that T7 has a mechanism to inhibit primed adaptation machinery. This putative mechanism must be targeting the adaptation proteins only, since CRISPR interference against T7 is highly efficient. Such a system could be biologically advantageous for the phage, given the ease with which escape phage mutants, that promote primed adaptation, appear.
The results with the two phages with modified DNA studied here, T4 and R1-37, show that the type I-E CRISPR–Cas system of E. coli is totally ineffective against these viruses. The result may indicate that DNA modifications provide protection against a type I system effector complex. It was recently reported that T4 can be successfully targeted by a type II Cas9 effector (53), which would mean that these systems are more robust. Alternatively, given the complexity of T4 and R1–37, it is entirely possible that the lack of CRISPR protection is due to other, yet unknown phage mechanisms that are unrelated to DNA modification but specifically inactivate type I CRISPR–Cas systems.
Overall, our results establish certain commonalities but also reveal differences that may reflect specific viral adaptations used to overcome the CRISPR–Cas defense. The outcome of infections in the presence of functional CRISPR–Cas system targeting viral DNA is determined not by the intrinsic efficiency of target protospacer recognition by the effector complex but by the phage development strategy, genomic positions of targeted protospacers, and, possibly, DNA modification state. These factors can allow phages to overcome or at least limit the damaging effects of the CRISPR–Cas systems even in the absence of dedicated anti-CRISPR proteins.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NIH [R01 grant GM10407]; Russian Science Foundation [14-14-00988]; Ministry of Education and Science of the Russian Federation [14.B25.31.0004 to K.S.]; Russian Foundation for Basic Research of Russian Federation [16-34-01176 mol_a grant to A.S.]. Funding for open access charge: Skolkovo Institute of Science and Technology.
Conflict of interest statement. None declared.
REFERENCES
- 1. Bergh O., Borsheim K. Y., Bratbak G., Heldal M.. High abundance of viruses found in aquatic environments. Nature. 1989; 340:467–468. [DOI] [PubMed] [Google Scholar]
- 2. Mojica F.J., Díez-Villaseñor C., García-Martínez J., Soria E.. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 2005; 60:174–182. [DOI] [PubMed] [Google Scholar]
- 3. Bolotin A., Quinquis B., Sorokin A., Ehrlich S.D.. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005; 151:2551–2561. [DOI] [PubMed] [Google Scholar]
- 4. Brouns S.J., Jore M.M., Lundgren M., Westra E.R., Slijkhuis R.J., Snijders A.P., Dickman M.J., Makarova K.S., Koonin E.V., van der Oost J.. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008; 321:960–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E.. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012; 337:816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Estrella M.A., Kuo F.T., Bailey S.. RNA-activated DNA cleavage by the Type III-B CRISPR–Cas effector complex. Genes Dev. 2016; 30:460–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sinkunas T., Gasiunas G., Fremaux C., Barrangou R., Horvath P., Siksnys V.. Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR/Cas immune system. EMBO J. 2011; 30:1335–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Westra E.R., Van Erp P.B., Künne T., Wong S.P., Staals R.H., Seegers C.L., Bollen S., Jore M.M., Semenova E., Severinov K. et al. CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol. Cell. 2012; 46:595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mulepati S., Bailey S.. In vitro reconstitution of an Escherichia coli RNA-guided immune system reveals unidirectional, ATP-dependent degradation of DNA target. J. Biol. Chem. 2013; 288:22184–22192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Abudayyeh O.O., Gootenberg J.S., Konermann S., Joung J., Slaymaker I.M., Cox D.B.T., Shmakov S., Makarova K.S., Semenova E., Minakhin L. et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016; 353:aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Elmore J.R., Sheppard N.F., Ramia N., Deighan T., Li H., Terns R.M., Terns M.P.. Bipartite recognition of target RNAs activates DNA cleavage by the Type III-B CRISPR–Cas system. Genes Dev. 2016; 28:2432–2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Westra E.R., Semenova E., Datsenko K.A., Jackson R.N., Wiedenheft B., Severinov K., Brouns S.J.. Type I-E CRISPR-cas systems discriminate target from non-target DNA through base pairing-independent PAM recognition. PLoS Genet. 2013; 9:e1003742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Marraffini L.A., Sontheimer E.J.. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008; 322:1843–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Semenova E., Jore M.M., Datsenko K.A., Semenova A., Westra E.R., Wanner B., van der Oost J., Brouns S.J., Severinov K.. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:10098–10103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hynes A.P., Villion M., Moineau S.. Adaptation in bacterial CRISPR–Cas immunity can be driven by defective phages. Nat. Commun. 2014; 5:4399. [DOI] [PubMed] [Google Scholar]
- 16. Deveau H, Garneau J.E., Moineau S.. CRISPR/Cas system and its role in phage-bacteria interactions. Annu. Rev. Microbiol. 2010; 64:475–493. [DOI] [PubMed] [Google Scholar]
- 17. Datsenko K.A., Pougach K., Tikhonov A., Wanner B.L., Severinov K., Semenova E.. Molecular memory of prior infections activates the CRISPR–Cas adaptive bacterial immunity system. Nat. Commun. 2012; 3:945. [DOI] [PubMed] [Google Scholar]
- 18. Swarts D.C., Mosterd C., van Passel M.W., Brouns S.J.. CRISPR interference directs strand specific spacer acquisition. PLoS One. 2012; 7:e35888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bondy-Denomy J., Pawluk A., Maxwell K.L., Davidson A.R.. Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature. 2013; 493:429–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pawluk A., Bondy-Denomy J., Cheung V.H., Karen L.M., Davidson A.. A new group of phage anti-CRISPR genes inhibits the type I-E CRISPR–Cas system of Pseudomonas aeruginosa. MBio. 2014; 5:e00896–e00814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Calendar R., Abedon S.T.. The Bacteriophages. 2006; 2st ednNY: Oxford University Press. [Google Scholar]
- 22. Davison J. Pre-early functions of bacteriophage T5 and its relatives. Bacteriophage. 2015; 5:e1086500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shmakov S., Savitskaya E., Semenova E., Datsenko K.A., Severinov K.. Pervasive generation of oppositely-oriented spacers during CRISPR adaptation. Nucleic Acids Res. 2014; 42:5907–5916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Datsenko K.A., Wanner B.L.. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Strotksaya A., Semenova E., Savitskaya E., Severinov K.. Rapid multiplex creation of Escherichia coli strains capable of interfering with phage infection through CRISPR. Methods Mol. Biol. 2015; 1311:147–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Semenova E., Savitskaya E., Musharova O., Strotskaya A., Vorontsova D., Datsenko K.A., Logacheva M.D., Severinov K.. Highly efficient primed spacer acquisition from targets destroyed by the Escherichia coli type I-E CRISPR–Cas interfering complex. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:7626–7631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pickard D.J. Preparation of bacteriophage lysates and pure DNA. Methods Mol. Biol. 2009; 502:3–9. [DOI] [PubMed] [Google Scholar]
- 28. Miller E.M., Nickoloff J.A.. Escherichia coli electrotransformation. Methods Mol. Biol. 1995; 47:105. [DOI] [PubMed] [Google Scholar]
- 29. Hyman P, Abedon S.T.. Practical methods for determining phage growth parameters. Methods Mol. Biol. 2009; 501:175–202. [DOI] [PubMed] [Google Scholar]
- 30. Morgan M., Anders S., Lawrence M., Aboyoun P., Pagès H., Gentleman R.. ShortRead: a bioconductor package for input, quality assessment and exploration of high-throughput sequence data. Bioinformatics. 2009; 25:2607–2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pages H., Aboyoun P., Gentleman R., DebRoy S.. Biostrings: String objects representing biological sequences, and matching algorithms. R package version 2.24.1. 2012. [Google Scholar]
- 32. Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A.. Circos: an information aesthetic for comparative genomics. Genome Res. 2009; 19:1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Edelstein A., Stuurman N., Amodaj N., Hoover K.H., Vale R.D.. Computer control of microscopes using microManager. Curr. Protoc. Mol. Biol. 2010; doi:10.1002/0471142727.mb1420s92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schneider C.A., Rasband W.S., Eliceiri K.W.. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012; 9:671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012; 9:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Morozova N., Sabantsev A., Bogdanova E., Fedorova Y., Maikova A., Vedyaykin A., Rodic A., Djordjevic M., Khodorkovskii M., Severinov K. et al. Temporal dynamics of methyltransferase and restriction endonuclease accumulation in individual cells after introducing a restriction-modification system. Nucleic Acids Res. 2016; 44:790–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Skurnik M., Hyytiäinen H.J., Happonen L.J., Kiljunen S., Datta N., Mattinen L., Williamson K., Kristo P., Szeliga M., Kalin-Mänttäri L. et al. Characterization of the genome, proteome, and structure of Yersiniophage phiR1-37. Virol. 2012; 86:12625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schwartz M. The adsorption of coliphage lambda to its host: effect of variations in the surface density of receptor and in phage-receptor affinity. J. Mol. Biol. 1976; 103:521–536. [DOI] [PubMed] [Google Scholar]
- 39. Savitskaya E., Semenova E., Dedkov V., Metlitskaya A., Severinov K.. High-throughput analysis of type I-E CRISPR–Cas spacer acquisition in E. coli. RNA Biol. 2013; 10:716–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nguyen H.M., Kang C.. Lysis delay and burst shrinkage of coliphage T7 by deletion of terminator Tφ reversed by deletion of early genes. J Virol. 2014; 88:2107–2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Young R. Bacteriophage lysis: mechanism and regulation. Microbiol. Rev. 1992; 56:430–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Miller E. S., Kutter E., Mosig G., Arisaka F., Kunisawa T., Ruger W.. Bacteriophage T4 genome. Microbiol. Mol. Biol. Rev. 2003; 67:86–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chopin M.C., Chopin A., Bidnenko E.. Phage abortive infection in lactococci: variations on a theme. Curr. Opin. Microbiol. 2005; 8:473–479. [DOI] [PubMed] [Google Scholar]
- 44. Labrie S.J., Samson J.E., Moineau S.. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 2010; 8:317–327. [DOI] [PubMed] [Google Scholar]
- 45. Nossal N.G., Richardson C.C., Kong D.. Role of the bacteriophage T7 and T4 single-stranded DNA-binding proteins in the formation of joint molecules and DNA helicase-catalyzed polar branch migration. J. Biol. Chem. 1997; 272:8380–8387. [DOI] [PubMed] [Google Scholar]
- 46. Sun B, Pandey M., Inman T., Yang Y., Kashlev M., Patel S.S., Wang M.D.. T7 replisome directly overcomes DNA damage. Nat. Commun. 2015; 6:10260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mulepati S., Bailey S.. In vitro reconstitution of an Escherichia coli RNA-guided immune system reveals unidirectional, ATP-dependent degradation of DNA target. J. Biol. Chem. 2013; 288:22184–22192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kunne T., Kieper S.N., Bannenberg J.W., Vogel A.I., Miellet W.R., Klein M., Depken M., Suarez-Diez M., Brouns S.J.. Cas3-derived target DNA degradation fragmnents fuel primed CRISPR adaptation. Mol. Cell. 2016; 63:852–864. [DOI] [PubMed] [Google Scholar]
- 49. Li M., Wang R., Zhao D., Xiang H.. Adaptation of the Haloarcula hispanica CRISPR–Cas system to a purified virus strictly requires a priming process. Nucleic Acids Res. 2014; 42:2483–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Richter C., Dy R.L., McKenzie R.E., Watson B.N., Taylor C., Chang J.T., McNeil M.B., Staals R.H., Fineran P.C.. Priming in the Type I-F CRISPR–Cas system triggers strand-independent spacer acquisition, bi-directionally from the primed protospacer. Nucleic Acids Res. 2014; 42:8516–8526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Vorontsova D., Datsenko K.A., Medvedeva S., Bondy-Denomy J., Savitskaya E.E., Pougach K., Logacheva M., Wiedenheft B., Davidson A.R., Severinov K. et al. Foreign DNA acquisition by the I-F CRISPR–Cas system requires all components of the interference machinery. Nucleic Acids Res. 2015; 43:10848–10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Westra E.R., van Houte S., Oyesiku-Blakemore S., Makin B., Broniewski J.M., Best A., Bondy-Denomy J., Davidson A., Boots M., Buckling A.. Parasite exposure drives selective evolution of constitutive versus inducible defense. Curr. Biol. 2015; 25:1043–1049. [DOI] [PubMed] [Google Scholar]
- 53. Yaung S.J., Esvelt K.M., Church G.M.. CRISPR/Cas9-mediated phage resistance is not impeded by the DNA modifications of phage T4. PLoS One. 2014; 9:e98811. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.