Abstract
Co-expression of physically linked genes occurs surprisingly frequently in eukaryotes. Such chromosomal clustering may confer a selective advantage as it enables coordinated gene regulation at the chromatin level. We studied the chromosomal organization of genes involved in male reproductive development in Arabidopsis thaliana. We developed an in-silico tool to identify physical clusters of co-regulated genes from gene expression data. We identified 17 clusters (96 genes) involved in stamen development and acting downstream of the transcriptional activator MS1 (MALE STERILITY 1), which contains a PHD domain associated with chromatin re-organization. The clusters exhibited little gene homology or promoter element similarity, and largely overlapped with reported repressive histone marks. Experiments on a subset of the clusters suggested a link between expression activation and chromatin conformation: qRT-PCR and mRNA in situ hybridization showed that the clustered genes were up-regulated within 48 h after MS1 induction; out of 14 chromatin-remodeling mutants studied, expression of clustered genes was consistently down-regulated only in hta9/hta11, previously associated with metabolic cluster activation; DNA fluorescence in situ hybridization confirmed that transcriptional activation of the clustered genes was correlated with open chromatin conformation. Stamen development thus appears to involve transcriptional activation of physically clustered genes through chromatin de-condensation.
INTRODUCTION
During the past decade, chromosomal clusters of functionally related but non-homologous co-expressed genes have been identified in the genomes of plants, animals, and fungi (see e.g. (1) and references within). Furthermore, a study by Al-Shahrour et al. (2) claimed that many eukaryotic genes are organized in functional neighborhoods that are evolutionarily conserved (2). Unlike prokaryotic operons, the genes in these eukaryotic clusters are, with few known exceptions, not transcribed in a single transcriptional unit but are organized in physical clusters, i.e. their chromosomal locations are close to each other, and the clustered genes are co-expressed (1). These observations prompt questions as to how physical clustering of co-expressed genes arises and what are the functional advantages of such clusters. In order to start tackling such intriguing but large-scale questions, we have developed a bioinformatics platform to efficiently collect relevant insights from global transcriptome data.
In plants, co-expression of physically linked genes that have arisen through serial tandem duplication events occurs more frequently than expected by random chance (3). Several gene clusters of distinct secondary metabolite pathways have been identified, for example for the syntheses of cyclic hydroxamic acids in maize (Zea mays) (4) and the triterpenes thalianol and marneral in A. thaliana (5,6). Several studies report clustering tendencies also among co-expressed genes not belonging to the same metabolic pathway, indicating that clustering of genes may play a role during the execution of integrative molecular pathways, such as developmental programs (7–9).
Co-regulation of physically linked genes may be mediated by common regulatory elements or by a shared chromatin environment due to localized changes in chromatin structure (1). Cell and organ identity-specific gene expression profiles are thought to be maintained via chromatin-level regulation (9), and physical clustering would facilitate stable co-regulation of genes via chromatin re-organization. In fact, cell-type specific chromatin de-condensation has been associated with expression of the avenacin metabolism gene cluster in oat (10). Recently, it was also demonstrated that metabolic clustered pathways are enriched in histone 3 lysine trimethylation (H3K27me3) chromatin signatures and histone 2 variant H2A.Z, associated with cluster repression and activation, respectively (11,12). This indicates that chromatin level regulation drives the co-expression of physically clustered genes.
Genome-wide surveys of co-regulated gene clusters provide clues to the regulatory modes deployed in various tissues and developmental stages. Clusters can be identified in a number of ways in silico, using genetic distance (13); based on gene IDs (14); and sequential gene processing with large windows of physical distance (15). To investigate the functional or evolutionary significance of the clusters, additional characteristics of the clusters need to be examined. Previous studies have included analysis of duplication or homology between clustered genes (16), synteny together with recorded gene expression values (17), or known transcription factor binding motifs (15).
Here, we addressed whether physical clustering occurred in genes acting in development and differentiation of A. thaliana stamens, and whether co-regulation of the clustered genes was associated with changes in chromatin state. To this end, we developed a new bioinformatics platform to detect physical gene clustering among a proposed set of genes involved in a specific developmental pathway, in our case identified by genome-wide expression analyses. We refer to these genes as GOIs, genes of interest. The clusters were constructed based on the genomic coordinates of the GOIs, and the statistical significance of the set of identified clusters was calculated through simulations. To determine the influence of non-chromatin-level mechanisms for gene co-expression in the clusters, we augmented our clustering analysis with assessment of (i) gene duplications, through homology detection, and (ii) promoter element similarities, through detection of known regulatory motifs combined with unbiased identification of overrepresented DNA oligomers.
We used this platform to analyse physical clustering of co-expressed genes during the development of male reproductive organs, the stamens, in A.thaliana. The stamen is a complex organ with many specialized cell types, including male gametophytes, and its development involves multiple rounds of fate specification; therefore, it is particularly well suited for studies of sequential developmental pathways. Stamen identity is specified by a set of homeotic proteins that belong to the MADS-box family of transcription factors (for review see (18)). To assess clustering among genes active during stamen development we reanalyzed global expression data of spatial gene expression in A. thaliana flowers and focused on genes down-regulated in inflorescences of the floral homeotic mutants apetala3, pistillata and agamous (19). The combined set of genes downregulated in those mutants represent genes specifically or preferentially expressed in stamens. A multitude of processes, ranging from hormone signalling to boundary formation, are regulated by the homeotic factors responsible for stamen identity, often through direct or indirect regulation of other transcriptional regulators (18).
Another attractive feature of stamen development is that there are many mutants in which the differentiation pathways of unique cell types are impaired. In order to focus on a specific stage of stamen development and to provide a more direct link between coordinated activation of clustered stamen enriched genes and chromatin de-condensation, we also applied the same in silico characterization to genes regulated by the transcriptional activator MALE STERILITY1 (MS1) (20). MS1 is necessary for pollen coat formation and the protein contains a plant homeo-domain (PHD)-finger domain (21–23). The PHD-finger domain has been linked to control of chromatin structure (21–24) mediated through protein-protein interactions (25). MS1 acts downstream of the homeotic genes during stamen development, and is specifically active in the tapetum cells that are required for pollen maturation from the late tetrad stage until the free microspore stage (23), which corresponds approximately to floral stage 10 (26).
We combined the analyses of the two datasets to identify overlapping clusters containing genes that were enriched during stamen organogenesis and also expressed during the specific process of pollen maturation in response to chromatin-level transcriptional regulation. We surveyed the expression of genes in the overlapping clusters in 57 datasets from A. thaliana representing different tissue types at specific developmental stages and compared the expression with the occurrence of a repressive histone mark, H3K27me3. For a subset of the overlapping clusters, we assayed gene expression in 14 A. thaliana chromatin remodelling mutant lines. We experimentally verified the relationship between active expression of clustered genes and chromatin de-condensation using DNA FISH in combination with in situ individual cluster monitoring of chromatin state using structured illumination (SIM) super-resolution microscopy.
MATERIALS AND METHODS
Availability of data and materials
Scripts and code developed to analyse chromosomal clustering are deposited at https://github.com/b97jre/ClusterAnalysisTools. Material and methods are described below and in Supplemental Text S1, Methods and Supplemental Data S1.
Expression data, H3K27me3 data and genome annotation
All genome sequences and gene annotations were downloaded from http://www.phytozome.net/. The following versions of sequences and annotation for each species were used: A. thaliana (TAIR10) (27,28), A.lyrata (v. 1.0) (29), C. rubella (v. 1.0) (30), E. salsugineum (v 1.0) (31) and B. rapa (v 1.1). In A. thaliana, there were 35 186 gene models, corresponding to 27 416 different gene loci (chloroplast and mitochondrial encoded genes excluded).
The Wellmer et al. (19) oligonucleotide microarray platform layout (GPL1077; oligos are from the Operon Arabidopsis Genome Oligo Set Version 1.0, based on TAIR4) was extracted from the GEO database (http://www.ncbi.nlm.nih.gov/geo/) (19). We remapped the array data from TAIR4 to TAIR10 using Blast (Supplemental Text S1 Methods and Supplemental Data S2). For each gene locus, we picked the gene model with the lowest present ‘extension’ number to represent the gene locus. For instance, for the gene locus AT1G07700 we picked AT1G07700.1. For the cluster formation algorithms, we used the gene locus information directly from TAIR10 annotation.
Normalised expression data from the A. thaliana microarray Atlas of Development (AtGenExpress Development) were used to examine the expression pattern of the SEG-MS1 genes in 57 distinct tissues. Meristem, leaf and seedling samples were used to compare expression levels with the accumulation of a repressive histone mark, H3K27me3. Physical clustering of genes marked with H3K27me3 in vegetative tissues was performed as described in Supplemental Text S1 Methods.
Selection of stamen-enriched genes (SEG set) and MS1 mutant genes (MS1 set) in TAIR10
Wellmer et al. (19), using the microarray platform GPL1077, identified 1162 stamen-enriched genes (SEGs) in TAIR4. 1106 of them could be re-annotated, as described above, to TAIR10 (19). Genes up- or down-regulated in ms1 mutants were collected from Alves-Ferreira et al. (20). Using the Wellmer et al. identified in total 1914 gene loci with an affected expression pattern in ms1 mutants compared to wild-type (GPL1077). 1854 of them could be uniquely re-annotated, as described above, to TAIR10 (19). Out of these genes, 1615 had at least a 2-fold change in expression level (up or down) coupled with a Benjamini–Hochberg adjusted P-value ≤0.05, in any out of seven samples collected at seven different A. thaliana anther developmental stages. 1095 of these were down-regulated in at least one of the seven samples (939 were down-regulated in all seven samples), while 520 were up-regulated (or neutral) in all seven samples.
Testing the distribution of differentially expressed genes on the chromosomes
To test if the distribution of distances (measured in bases) between genes of interest (GOIs) on the chromosomes, Ditrue, differed from the overall gene distribution, Dibackground, on the chromosomes, we used the two-sample Wilcoxon test (also called the Mann–Whitney test). The Wilcoxon test is a non-parametric test with the null hypothesis that the two samples of observations are from the same distribution. It does not require or assume that data follow a normal distribution or that sample sizes are equal. The density functions of the SEG distances were clearly non-normal (Supplemental Figure S1). We used the two-tail P-value generated from this test. Each chromosome Ci has ni GOI. To get Ditrue on each chromosome Ci we calculated the distances between adjacent GOIs along the chromosome. The distance between a pair of GOIs was calculated as the difference between the rightmost coordinate of the most 5΄ gene and the leftmost coordinate of the most 3΄ gene. The distance was only calculated if the GOIs were on the same chromosome. To get Dibackground distribution on each chromosome Ci we randomly chose ni genes on chromosome Ci and calculated the distances between those genes. This was done 10 000 times to generate a background distribution (Table 1).
Table 1. Median values of intergenic distances for true and randomized GOIs.
| Median values of intergenic distances for true and randomised GOIs | ||||||
|---|---|---|---|---|---|---|
| SEG | SEG | SEG | MS1 | MS1 | MS1 | |
| Chr | True GOIs | Randomized GOIs | P-value | True GOIs | Randomized GOIs | P-value |
| 1 | 53859.5 | 65815 | 0.00486 | 49623.5 | 60880 | 0.00774 |
| 2 | 68514.5 | 74790 | 0.03891 | 62540.5 | 72001 | 0.06423 |
| 3 | 44507 | 56098 | 0.00152 | 57261 | 67632 | 0.01647 |
| 4 | 63472 | 75674 | 0.57229 | 63557 | 71321 | 0.38548 |
| 5 | 51902 | 67863 | 0.00021 | 47005 | 69771 | 0.00002 |
Chr, chromosome. SEG, stamen-enriched gene data set. MS1, MS1-regulated gene data set. GOI, gene of interest. Distances are measured in bases. P-values are from the two-sample Wilcoxon test comparing true to randomized values. See main text for details.
Identification of physical clusters of genes on the chromosomes
To identify GOIs that were located close to each other on the chromosomes we created a program that search for physical clusters of GOIs. A physical cluster was defined as a set of N GOIs where the distance between two adjacent GOI was at most L bases (Figure 1A) on the chromosome. A cluster must also contain at least hn groups of homologous genes (how homologous genes were identified, see next section). The distance between a pair of GOIs was calculated as the difference between the rightmost coordinate of the most 5΄ gene and the leftmost coordinate of the most 3΄ gene. The distance was only calculated if the GOIs were on the same chromosome. A cluster was formed or expanded regardless of the orientation (forward/reverse strand) of the GOIs or of the number of non-GOIs in between the GOIs. To test if the number of physical clusters identified from a set of n GOIs, and given L,N and hn is more than what we expect to see by chance, we investigated whether the number of clusters that was found in the true data set, Ctrue, was different from the null distribution Crandom. To identify the null distribution, Crandom, we sampled the process of identifying clusters on n randomly chosen genes with the same L, N and hn parameters M times. We defined the P-value of the true datasets as the number of times Crandom ≥ Ctrue divided by the total sample size M. If Ctrue > Crandom for all M then the P-value was less than 1/M. See Figure 1B and Supplemental Figure S2 for results on different parameter settings. The P-values are in Supplemental Table S1.
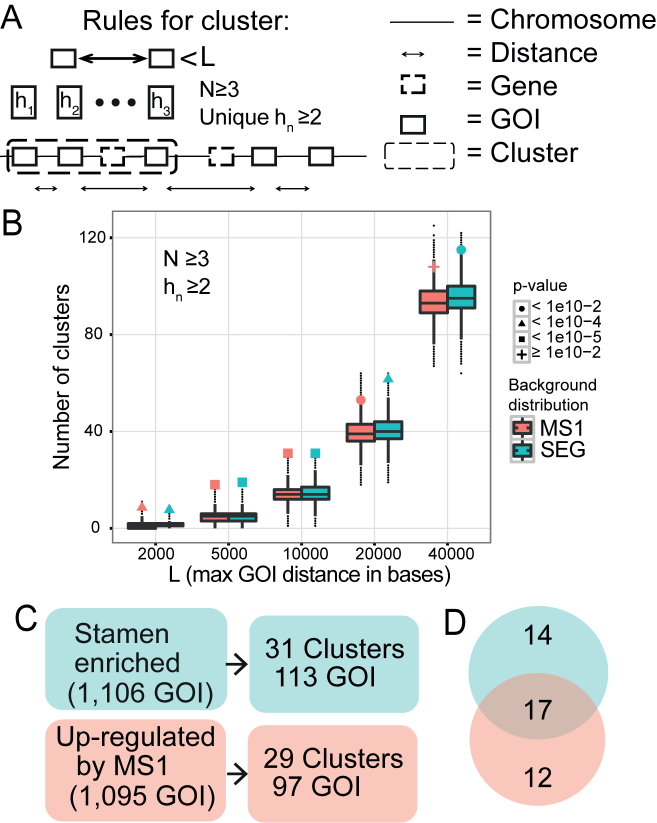
Figure 1.
Identification of co-expressed physical clusters in stamen development. (A) Schematic view of the physical clustering analysis tool developed to identify clusters of co-regulated genes. To be assigned as a cluster at least N GOIs (genes of interest) with a maximal distance L bases between adjacent GOIs has to be identified, and with at least hn groups of homologous genes. The distance L is measured between the gene ends closest to each other. (B) Number of clusters detected in the SEG (green) and MS1 (red) datasets, as a function of L. The background distributions, obtained from simulations, are shown as a boxplot. The actual number of clusters found is marked with a filled dot, triangle, square, or plus sign, depending on the P-value assigned by the corresponding background distribution. N was set to three and hn (number of homologous groups) was at least two. (C) The number of clusters found in the two datasets when N ≥ 3, hn ≥ 2, and L ≤ 10 000 bases. (D) Venn diagram showing the number of clusters that were unique and in common for the two datasets. The intersection is the 17 SEG-MS1 clusters studied in depth.
Identification of homologous genes
To identify genes on the chromosomes that have been recently duplicated we used OrthoMCL v.1.4, which identifies clusters of homologous genes across and within species (35), based on amino acid sequence similarities detected using Blast. To allow maximum sequence diversity within each cluster we used the lowest recommended settings (–inflation 1.0) on the coding genes of Arabidopsis thaliana (TAIR10) and a set of related species: Arabidopsis lyrata (v. 1.0), Capsella rubella (v. 1.0), E. salsugineum (v 1.0; previously known as Thellungiella halophila), and Brassica rapa (v 1.1). Any A. thaliana genes that were placed in the same cluster by OrthoMCL were considered as homologs. For genes with >1 variant, it sufficed if one of the variants was homologous in order to consider the gene homologous.
Promoter analysis: promoter regions
We searched for regulatory motifs in regions surrounding the transcription start site (TSS) of each of the genes in A. thaliana (TAIR10 annotation). For each gene, we collected 1000 bases upstream of its transcription start site, the 5΄UTR regions of 5΄ exon(s), and the first intron. We allowed overlap with possible open reading frames (ORFs), both upstream of the TSS (i.e. overlap with ORFs of other genes) and within the 5΄UTR of the gene itself (uORFs). This constitutes our set of promoter regions.
Atted-II motifs and promoter similarity score definition
The 304 heptamers predicted to be cis-elements in Atted-II (36) were extracted (http://atted.jp/browsing/browsing_cis_hc.html) and matched, including reverse complement, using exact string matching to the promoter regions of all genes. Atted-II contains regulatory motifs compiled taking known co-expression into account, and is hence a suitable database in order to identify similarities between promoter regions of co-expressed genes. The pairwise similarity of all promoter regions was calculated: for each pairwise comparison a promoter similarity score was calculated as the percentage of shared motifs of all motifs in the pairwise comparison, i.e. 100 × {intersection of motifs in the promoter regions of the gene pair}/{union of motifs in the promoter regions of the gene pair}. This has a maximum value of 100, which would correspond to identical motif sets, while 0 means no identical motifs.
Simulation of promoter regions for Atted-II motif analysis
First, we simulated 17 random clusters with in total 96 genes. This procedure was iterated 10 000 times: For each iteration, we started by choosing 17 genes at random (without replacement) and used these as seeds for clusters. For each randomly chosen gene, a cluster size (in number of genes) was randomly chosen without replacement from the list of 17 actual cluster sizes (sizes including non-GOIs as well as GOIs). The randomly chosen gene is then assigned that number of nearby downstream genes (as defined by accession numbers), thus forming a cluster. In the end, a set of 17 randomly placed clusters with the same sizes as the actual clusters were generated for each iteration. P-values were derived from comparing the average promoter score of each actual cluster (for local promoter similarity analysis) or the average promoter score within and between all clusters (for global promoter similarity analysis) to the corresponding empirical distribution of averaged promoter scores from the simulation, i.e., without assuming a particular distribution.
Next, we restricted the analysis to GOIs only. Similar to above, we simulated 17 random clusters with in total 96 genes and for each cluster we picked the number of GOIs that corresponded to the number of GOIs in the actual clusters. For each cluster, the two genes at the ends were assigned as GOIs, as our definition of a cluster requires any cluster to start with a GOI and end with a GOI. Remaining GOIs were chosen randomly from the remaining genes within the clusters. Thus, in the end, a set of 17 randomly placed clusters with the same sizes and number of GOIs as the actual clusters were generated for each iteration (procedure iterated 10 000 times, as above). P-values were calculated as above.
Detecting overrepresented k-mers in promoter regions with Rsat
We used the Rsat tool (http://rsat.ulb.ac.be/rsat/) to detect overrepresented k-mers, and followed the protocol in (37) with A. thaliana as background model organism. Rsat searched for overrepresented k-mers (k ≥ 6) in the promoter regions and was thus a complement to the Atted-II based method that searched for presence of a set of predefined motifs. We used Rsat both applied to each cluster individually (local promoter similarity), and applied to all clusters taken together (global promoter similarity). Rsat calculates the expected significance of each possible k-mer and reports those k-mers that meet an E-value threshold (that corresponds to the number of instances of the k-mer that would be expected by random chance given the background model). We discarded isolated motifs with high E-value, as suggested by the Rsat tutorial at the web site.
Global promoter similarity analysis
In the Atted-II global promoter similarity analysis, the pairwise promoter similarity scores were averaged between each combination of pairs among the 96 genes in the 17 clusters (i.e. 96 × 95/2 comparisons), or, among the 68 GOIs in the 17 clusters (i.e. 68 × 67/2 comparisons), thus yielding a collective average promoter similarity score for the set of clusters (one using all clustered genes, one using all clustered GOIs). The promoter similarity scores of the simulated clusters were averaged in the same way. Thus, all Atted-II global promoter similarity results pertain to scores that have been averaged within as well as between the promoter regions of all clusters. In the Rsat global promoter similarity analysis, Rsat was, in a single run, provided as input the promoter regions of all 96 genes, and a background model, which was the upstreams regions of all A. thaliana genes (Supplemental Table S2).
Local promoter similarity analysis
In the Atted-II local promoter similarity analysis, the pairwise promoter similarity scores were averaged only within each cluster (and not between clusters), thus creating an average promoter similarity score for each cluster. This was done both for all genes in a cluster, and for all GOIs in a cluster. For instance cluster #1 contains five genes: the average promoter similarity score for cluster #1 (16.72) is based on 5 × 4/2 comparisons. But out of the five genes in cluster #1, only three are GOIs: the average promoter similarity score for cluster #1 based on the GOIs only (10.46) is calculated from 3 × 2/2 comparisons. The promoter similarity scores of the simulated clusters were averaged in the same way. In the Rsat local promoter similarity analysis, Rsat was run once for each cluster (i.e., 17 times), and was provided as input the promoter regions for the genes in one cluster at a time. The background model was the upstreams regions of all A. thaliana genes (Supplemental Table S3).
Gene ontology (GO) term analysis
To identify common characteristics of the clustered genes we identified enriched GO terms using topGO v.2.22.0 (38). The gene to GO term gene association file (gene_association.tair.gz) with the submission date 11 February 2015 was downloaded from the Gene Ontology Consortium webpage (39). Significantly enriched GO terms were identified using Fisher's exact test comparing the number of GO terms in the selected genes compared to a background set of genes. P-values were adjusted for multiple testing using the Benjamini–Hochberg approach (40). Three different comparisons were performed: the selected genes versus (i) all genes present on the array, (ii) the union of non-clustered MS1 and SEG GOIs, (iii) the intersection of non-clustered MS1 and SEG GOIs.
Plant material
The A. thaliana lines and the growth conditions used in the present study are described in Supplemental Text S1, Methods.
Quantitative real-time PCR (qRT-PCR)
To assay dynamics of transcriptional responses after DEX induction, whole inflorescences with flower buds at stages 1–13 were sampled from wild type, ms1-1, and MS1-GR plants. DEX or Mock treatment was performed once, and inflorescences were collected 0, 4, 12, 24 and 48 h after the induction. Buds from at least six independent plants were pooled and snap frozen in liquid nitrogen. To assay transcription of clustered genes at different floral stages, buds were examined under a stereomicroscope and classified according to stages specified by (41). Floral buds at stages 1–9, 10, 11 and 12 were separately collected at 48 h after induction, each sample consisting of buds from at least 10 plants. Similarly, floral buds at floral stage 11 were collected from wild type and chromatin-remodeling mutant plants. For details on bud collection, RNA extraction, cDNA synthesis, and subsequent qRT-PCR analysis see Supplemental Text S1, Methods.
RNA and DNA in situ hybridization (mRNA in situ/ DNA FISH)
Inflorescences with stages 1–12 flower buds were collected 48 h after DEX or MOCK treatment. Templates for mRNA in situ probes were generated using primers listed in Supplemental Table S4. Manufacturing of probes, tissue fixation and wax embedding, and mRNA in situ hybridization experiments were performed as described previously (42). DNA FISH experiments were performed essentially as described in (10). The details of DNA FISH probe making, labeling procedures, image acquisition and analysis are described in Supplemental Text S1, Methods.
RESULTS
Genes involved in stamen development form physical clusters in Arabidopsis thaliana
In order to study physical clustering tendencies among the genes preferentially active during stamen development, we compiled 1106 genes from global gene expression data (19) that comprise genes active during all stages of stamen development (SEG data set). To enable our analysis to target a specific stage of stamen development, we also compiled 1095 genes that were down-regulated in the male sterility1-1 (ms1-1) mutant compared to wild-type (20), i.e. genes up-regulated by MS1, which is a known transcriptional activator (MS1 data set). The overlap of the two sets was 686 genes. All expression data were remapped to the TAIR10 genome annotation.
We used two distinct approaches to evaluate physical clustering tendencies of SEGs and MS1-regulated genes (Materials and Methods). First, we examined whether the inter-genic distances between neighboring GOIs (in our case the genes in either the SEG or the MS1 dataset) were different from the ones between randomly chosen genes. Genes in both the SEG and MS1 datasets were closer than expected to each other on chromosomes 1 and 5 at a statistically significant level (Bonferroni corrected P-value ≤0.05 from two-sample Mann–Whitney test), and also on chromosome 3 for SEG (Table 1; Supplemental Figure S1). Second, we clustered the genes transitively; any two GOIs within a certain genomic distance L of each other were placed into a putative cluster. The cluster was iteratively expanded with the closest GOI if it was contained within L. We tested different settings for the maximum distance between GOIs, L = [2, 5, 10, 20, 40 kb], the required number of GOIs in a cluster, N = [2,3], and how many unique homologous groups should be present in a cluster, hn = [2,3], where hn ≤ N. This approach was applied separately to the SEG and MS1 sets and we observed significantly more clusters than expected by random chance for both sets, for several parameter combinations (Supplemental Figure S2). We chose a highly significant setting (L = 10 kb, N = 3, and hn ≥ 2; P < 10−5) that both allowed for extending clusters over genes potentially missing from the expression array and assured that a cluster was not solely formed by a single set of duplicated genes (since hn > 1) (Figure 1A and B).
We identified 31 SEG clusters containing 113 GOIs (10.3% of all SEG GOIs) and 29 MS1 clusters containing 97 GOIs (8.9% of all MS1 GOIs) (Figure 1C). Shared between these two sets were 17 clusters that we call the ‘SEG-MS1 clusters’ or ‘shared clusters’ (Table 2). The SEG-MS1 clusters contained 96 genes, whereof 68 GOIs (64 SEG GOIs and 63 MS1 GOIs) (Figure 1D, Supplemental Table S5 and Figure S3). The average cluster size was 5.6 genes or 4.0 GOIs.
Table 2. Characteristics of the 17 SEG-MS1 clusters.
| Characteristics of the 17 SEG-MS1 clusters | |||||||
|---|---|---|---|---|---|---|---|
| Cluster | Gene loci (TAIR10) | Number of genes | Number of GOIs | Length (kb) | Atted-II P-value | Rsat k-mer motifs | Orthologous genes in cluster |
| #1 | At1g04880-920 | 5 | 3 | 19.8 | 0.946 | 0 | - |
| #2 | At1g06250-280 | 4 | 3 | 7.7 | 0.370 | 0 | - |
| #3 | At1g20120-150 | 6 | 3 | 15 | 0.977 | 0 | Yes |
| #4 | At1g22100-150 | 6 | 4 | 24 | 0.544 | 1 | - |
| #5 | At1g23510-690 | 17 | 8 | 37.3 | 0.431 | 0 | Yes |
| #6 | At1g51240-260 | 3 | 3 | 5.1 | 0.895 | 0 | Yes |
| #7 | At1g75910-940 | 4 | 3 | 12.9 | 0.388 | 0 | - |
| #8 | At2g47030-050 | 3 | 3 | 8.4 | 0.061* | 0 | Yes |
| #9 | At3g01230-270 | 5 | 4 | 8.2 | 0.598 | 0 | - |
| #10 | At3g07820-850 | 4 | 4 | 11 | 0.316 | 0 | Yes |
| #11 | At3g13220-229 | 8 | 3 | 22 | 0.952 | 0 | - |
| #12 | At3g26860-880 | 3 | 3 | 5.2 | 0.301 | 1 | - |
| #13 | At3g28780-840 | 6 | 6 | 34 | 0.079* | 0 | Yes |
| #14 | At5g07410-430 | 3 | 3 | 8.4 | 0.330 | 0 | - |
| #15 | At5g07490-560 | 8 | 7 | 21 | 0.330 | 1 | - |
| #16 | At5g45810-840 | 4 | 3 | 13 | 0.381 | 0 | - |
| #17 | At5g46940-700 | 7 | 5 | 12.1 | 0.440 | 0 | Yes |
Loci, refers to the A. thaliana accession numbers. Number of genes, number of genes in total in each cluster. Number of GOIs, number of genes-of-interest in each cluster. Length, the physical genomic distance from one end of the cluster to the other end of the cluster, measured in kilobases (kb). Atted-II P-value, the probability of finding a an average pairwise promoter similarity score equal to or larger than the observed one in the cluster (P-values are estimated from simulations), using Atted-II regulatory motifs, and where values marked with (*) indicate nominally significant at alpha <0.1. Rsat k-mer motifs, the number of overrepresented k-mer motifs present in the promoter regions of all GOIs in the cluster. Orthologous genes in cluster, whether the OrthoMCL-based gene orthology analysis revealed that orthologous genes were present in the cluster (see Supplemental Figure S3 for details). GOI, gene of interest.
We tested for overrepresentation of functional gene ontology (GO) categories in the 68 GOIs in the SEG-MS1 clusters compared to three different background models (Methods). Sixteen out of 558 GO terms were overrepresented in at least one comparison at adjusted P-value < 0.005 (Supplemental Table S6 and Figure S4). The two most enriched biological processes were sexual reproduction and lipid storage. Sexual reproduction (GO:0019953) was present in 31 GOIs, of which eight were in clusters, and lipid storage (GO:0019915) showed up in 17 GOIs of which six were in clusters, with adjusted P-values <0.001 in both cases. Thus, reproductive function annotation was overrepresented in the clustered GOIs as compared to the set of non-clustered GOIs. All GO-terms for the clustered genes are presented in Supplemental Data S3. Further, we investigated the gene expression levels of all A. thaliana genes in 57 other tissues (32). We observed that most genes in the 17 SEG-MS1 clusters were expressed at a low level in all tissues or developmental stages not associated with sexual reproduction, e.g. shoot apex and root, while in comparison they were highly expressed in most samples associated with reproduction, e.g. stamens in floral stage 12 (Supplemental Figure S5). This supports the importance of the clustered genes in sexual reproduction.
The SEG-, MS1- and shared clusters were unevenly distributed among the chromosomes, with chromosomes 1, 3 and 5 having the most clusters and chromosome 4 the fewest. Chromosome 4 lacked shared clusters altogether (Figure 2A; Supplemental Figure S6). This was in accordance with the inter-genic distance analysis. We discovered no specific bias regarding the position of the clusters on the chromosomes, except that centromere regions were devoid of clusters.
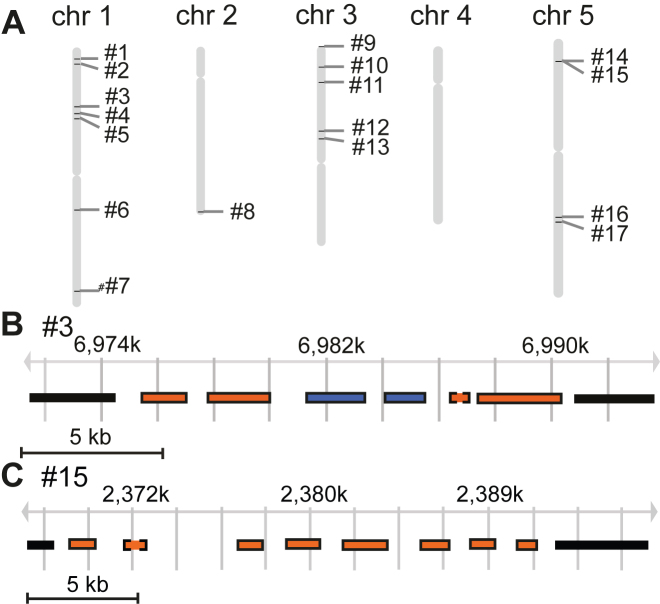
Figure 2.
Chromosomal position and gene composition of the clusters. (A) The position of the clusters on the A. thaliana chromosomes. (B) and (C) show a close-up of cluster #3 (loci At1g20120-150) and cluster #15 (loci At5g07490-560), respectively. (Supplemental Figure S3 shows close-ups of all 17 clusters). The chromosomal region of each cluster is indicated by position coordinates (in kilobases). Each gene in the clusters is shown as a rectangle. Blue color indicates that the gene has a homolog within the cluster, whereas orange indicates lack of homolog within the cluster. Surrounding solid line: GOI; Surrounding dashed line: non-GOI. Graphs are adopted from Gbrowse using TAIR10 as the data source.
Gene duplication does not explain co-regulation of the clustered genes in Arabidopsis thaliana
Co-expression of physically linked genes could be due to gene duplication events. Duplicated genes are by definition homologous, and we used the Blast-based tool OrthoMCL (35) to identify potentially duplicated genes based on the similarities of their protein sequences (Supplemental Data S1). OrthoMCL is suitable since it resolves ambiguous results, e.g. how to form groups of orthologs when not all pairwise comparisons meet the chosen threshold. For reference, the Blast pairwise alignment scores for genes in clusters are provided in Supplemental Data S4.
Six out of the 17 SEG-MS1 clusters contained homologous genes (Table 2; Supplemental Figure S3), but our requirement that clusters must contain genes from at least two groups of homologs (hn ≥ 2) assured that no cluster contained only homologous genes. We noted that the cluster enrichment signal was significant for various parameter settings (Figure 1B; Supplemental Figure S2). Clusters #6 and #10 were examples with all but one of their genes assigned as homologs. Cluster #3 (Figure 2B) contained some homologous genes (AT1G20132 and AT1G20135) and the gene AT1G20150 was homologous with AT1G20160, which is immediately adjacent to, yet outside of, the cluster. Cluster #15 (Figure 2C) was composed of all unrelated genes. We conclude that the co-regulation of the clustered genes cannot be explained exclusively by gene duplications.
Common promoter motifs do not underlie co-regulation of the clustered genes in Arabidopsis thaliana
The promoters of the clustered genes could provide information whether the genes were co-regulated because of similar or identical regulatory motif patterns. We assessed the promoter regions of the genes in the clusters in two distinct ways: mapping the presence of a set of 304 known cis-regulatory motifs (Atted-II) (36), and detecting overrepresented k-mers (Rsat) (37). Pairwise promoter similarity scores were calculated based on the presence of the Atted-II cis-regulatory motifs.
First, we analysed all 96 genes in the 17 clusters collectively, as well as the subset of 68 GOIs. The promoter score averaged over all pairwise comparisons within and between clusters was not significantly higher than expected by chance, P-value = 0.12 and 0.09 for all genes and all GOIs, respectively, from simulations. Still, six motifs were found in all clusters (albeit not in the promoter regions of all genes, nor in the promoter regions of all GOIs): four 7-mers from the Atted-II analysis and two significant 6-mers (E-value ≤ 0.1) from the Rsat analysis (Supplemental Table S2).
Next, we analysed each cluster individually, with the aim to reveal local promoter features that could explain simultaneous regulation of all genes within a particular cluster. We used, again, the Atted-II set of regulatory motifs and calculated the average pairwise promoter similarity score, but now one individual average for each of the 17 clusters, analysing both all genes and restricting to the GOIs. Including all genes in the analysis, only the two highest-scoring clusters, #8 and #13, showed nominal P-value <0.1, indicating similar promoter regions within each of the two clusters, but they were not significant at α = 0.1 after multiple testing correction. Restricting the analysis to GOIs changed the promoter score for some clusters, but did not change the statistical significance (Table 2). Rsat analysis identified three clusters that each contained an overrepresented k-mer in all the promoter regions of all genes in the cluster (E-value ≤ 0.1): ATAGAG in cluster #4, GCTGGTAC in cluster #12 and CATGCA in cluster #15. Restricting the analysis to only GOIs returned the same three clusters and k-mers (Table 2; Supplemental Table S3). Additional details from the results of the promoter motif analyses are available in Supplemental Text S1, Materials and Methods, and Results.
In summary, we grouped the 17 clusters into four classes according to their homology and promoter region properties, Supplemental Table S7. We conclude that the co-regulation of the clustered genes is not fully explained by shared promoter elements among genes or GOIs.
Genes with H3K27me3 marks are lowly expressed in vegetative tissues and form clusters that overlap with SEG-MS1 clusters
We applied our clustering algorithm to genes with H3K27me3 marks (a repressive histone mark) in three A. thaliana tissues: rosette leaves, shoot apical meristem, and seedlings (data from (33,34)). We observed 773 (3622 genes) [14], 681 (3146 genes) [14], and 434 (1768 genes) [11] clusters of genes marked as tri-methylated, respectively, where the numbers in square brackets give the overlap between tri-methylated clusters and the SEG-MS1 clusters. Eleven of the 17 SEG-MS1 clusters were present in all three H3K27me3 cluster sets, and only two (#1 and #12) did not show any H3K27me3 clustering tendencies in these tissues.
Next, we investigated the gene expression levels of all A. thaliana genes in the three tissues (data from (32)). The genes were divided into four classes: present only in SEG-MS1 clusters; present only in H3K27me3 clusters; present in both cluster types; present in neither cluster type (Supplemental Figure S7). Genes present in both cluster types showed the lowest expression levels, while genes found in neither cluster showed the highest expression levels. SEG-MS1 clusters were significantly enriched for H3K27me3-clustered genes, Fisher's exact test P < 2.2E–16 (contrasting the fraction of clustered H3K27me3 genes in SEG-MS1 clusters versus the fraction outside of SEG-MS1 clusters).
MS1 dependent activation of the clustered genes
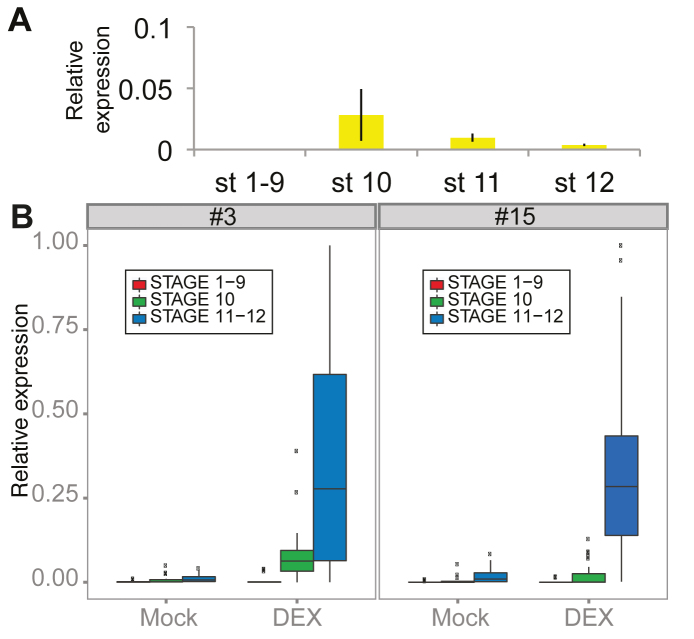
To experimentally verify that genes in the chromosomal clusters are activated in an MS1 dependent manner we used transgenic ms1-1 plants harbouring an inducible construct in which MS1 is fused to the rat glucocorticoid receptor (GR), (pMS1::MS1-GR) (43). In our experimental conditions, MS1 expression was initiated in stage 10 flowers with subsiding expression in stages 11 and 12 (Figure 3A; stages according to Smyth et al. (41)). Screening for MS1 dependent activation of clustered genes demonstrated that genes in clusters #3, #7 and #15 were up-regulated in samples of whole inflorescences within 48 h after dexamethasone (DEX) induction (Supplemental Figure S8).
Figure 3.
Expression levels in floral buds of different developmental stages. (A) Relative expression levels of MS1 in floral buds of stages 1–9, 10, 11 and 12 assayed using qRT-PCR. (B) Box-plots showing the relative expression levels of all GOIs present in clusters #3 and #15. Expression levels were assayed 48 h after DEX or Mock treatment. Error bars in (A) denote standard error of three biological replicates. Expression of each gene was normalised against the value of two reference genes, ß-tub and UBQ5.
To examine if the transcription of the clustered genes was confined to a specific developmental stage, cDNA derived from floral buds of stages 1–9, 10, 11 and 12 were used as the templates in qRT-PCR experiments. Transcription of all GOIs present in clusters #3 and #15 were assayed 48 h after DEX or Mock (negative control) treatment (Figure 3B; Supplemental Figures S9 and S10). Transcript levels of the clustered genes were very low in stages 1–9, before MS1 became active, very low or low at the onset of MS1 transcription in stage 10 buds, and clearly elevated in stage 11 and 12 flower buds (P < 0.01) in both clusters. Hence, transcript levels of the clustered genes were elevated subsequent to MS1 induction. A comparison of transcript levels between DEX and Mock treated samples did not show any significant difference in stages 1–9 (cluster #3, P = 0.95; cluster #15, P = 0.41), whereas statistically significant differences were detected in stages 11 and 12 (P < 0.01). At the onset of MS1 transcription (in stage 10), a significant expression difference was detected in cluster #15 (P ≤ 0.01), but not in cluster #3 (P = 0.14). This indicated that the majority of the genes present in cluster #3 and cluster #15 were activated in late stages relative to MS1 activation, but did not exclude occasional low-level expression of individual GOIs in stage 10 buds.
In order to estimate if the spatial expression pattern of the clustered genes coincides with that of MS1, mRNA in situ hybridization experiments were performed on selected cluster genes. For the assayed genes, signal was detected specifically in the tapetal cells of anthers in late stage 11 flowers (Supplemental Figure S11), indicating that DEX induction of the MS1-GR construct led to a tapetum specific activation of clustered genes within 48 h.
Activation of clustered gene expression is correlated with chromatin de-condensation
To assess the potential role of chromatin level regulation of the clustered genes we first assayed (with quantitative RT-PCR) the expression levels of representative genes from clusters #3 and #15 in stage 11 flowers in 14 different A. thaliana chromatin remodeling mutant lines (Supplemental Figure S12A and Supplemental Table S8). Expression was consistently down-regulated in the hta9/hta11 double mutant, which is mutated in two of the three H2A.Z genes (44), which was also found to be associated with metabolic cluster activation (11,12), but not in any other mutant. Thus, we next investigated the hta9/hta11mutant in more depth: we assayed the expression of all GOIs as well as the flanking genes of these clusters (Supplemental Figure S12B). The clustered GOIs were down-regulated (P < 0.0005), while genes flanking the clusters were not significantly down-regulated (P = 0.2). These results indicate that chromatin-mediated transcriptional regulation of the clustered genes likely take place.
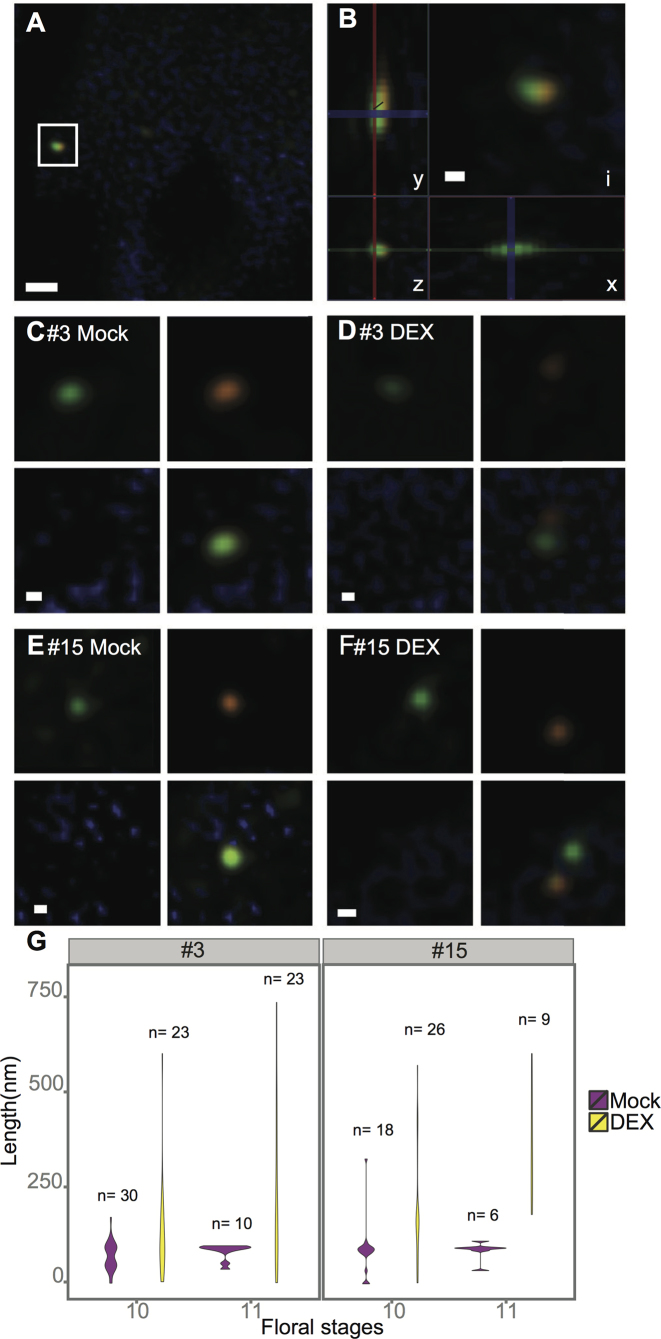
The timing of MS1 dependent activation of the clustered genes provides an opportunity to examine if transcription, at least in part, is mediated by changes in chromatin structure, manifested as a de-condensation of the chromatin region spanning the clustered genes. To address this possibility, DNA fluorescence in situ hybridization (FISH) was performed, combined with analysis with structured illumination super-resolution microscopy (SIM). SIM microscopy allows resolution below 100 nm, thus enabling in situ single cell monitoring of the shift in chromatin state from closed to open upon MS1 induction (45). DNA FISH experiments were performed for clusters #3 and #15. Cluster #3 harbours six genes and covers 15.8 kb, while cluster #15 harbours eight genes and covers 21 kb.
To detect the borders of the clusters, biotin-labelled (orange) and DIG-labelled (green) probes against the flanking regions of either side of the clusters were generated and hybridized in pairs. The hybridization signals of each probe pair frequently comprised more than one pair of fluorescent foci, probably reflecting hybridization to the clusters on sister chromosomes. We then examined the distance between the green and orange signals, i.e. the region encompassing the cluster itself. To do this, we measured the Euclidean distance between the fluorescent DIG-labelled and biotin-labeled foci in tapetum cells of buds in floral stages 10 and 11 (Figure 4A–F).
Figure 4.
Chromosomal de-condensation estimated by DNA FISH. Biotin- (orange) or DIG- (green) labelled probes targeted against the two flanking regions of either cluster #3 or #15 were used. (A) A tapetum cell with a pair of green and orange signals, marked by a square (cluster #15, DEX). (B) The inset (i) from micrograph (A) and distance measurement along the x-, y- and z-axis. (C–F) Micrographs showing examples of signals collected from: tapetum cells of MOCK- (C, E) and DEX- (D, F) treated samples, using the probes targeted against cluster #3 (C, D) or cluster #15 (E, F). (G) Violin plots showing the frequency distribution of the distance between Biotin- and DIG-labelled probes directed against the flanking regions of cluster #3 (left) and cluster #15 (right). Mock: negative control; DEX: dexamethasone treated samples. Size bars: 500 nm (B) and 100 nm (C to G).
The floral tissues used were the transverse sections of either Mock- or DEX-treated MS1-GR inflorescences fixed 48 hours after the induction. Statistically significant differences in the distribution of length measurements were detected both in stage 10 (P < 0.05 and < 0.001 for cluster #3 and #15, respectively; Kolmogorov–Smirnov test) and in stage 11 (P < 0.05 and < 0.01 for cluster #3 and #15, respectively) (Figure 4G). Corresponding differences in length distributions between Mock- and DEX-treated samples were not found in negative control cells outside of the tapetum layer (Supplemental Figure S10). On average, we detected a 70-fold chromatin compaction in Mock-treated samples, whereas DEX-treated samples displayed only a 32-fold (stage 10) and 22-fold (stage 11) compaction, compared with naked B-DNA (Supplemental Table S9).
The distances between cluster borders revealed a significant de-condensation of the chromosomal regions spanning clusters #3 and #15 at the onset of MS1-activated transcription: De-condensation had already occurred in stage 10 flowers, corresponding to the floral stage when transcription was initiated, but was even more pronounced in stage 11 flowers, where a majority of the genes in the clusters were transcribed at elevated levels. Thus, we have demonstrated that chromatin de-condensation coincided with MS1 dependent transcriptional activation of the genes present in cluster #3 and cluster #15, as judged by stage specific qRT-PCR and mRNA in situ hybridiszation experiments.
DISCUSSION
Physical clustering of co-expressed genes occurs among genes involved in biosynthesis pathways of secondary metabolites in plants (6). The current work addresses if physical clustering also occurs in genes acting in development and differentiation of A. thaliana stamens; in other words if the execution of a developmental program involves activation of specific sets of physically linked genes. To address this question we have analysed clustering tendencies among genes present in genome-wide gene expression datasets representing two different aspects of stamen development (19,20). One dataset represents the genes active during the entire course of stamen development (i.e., genes down-regulated in the homeotic apetala3 mutant), and the other dataset includes the genes active during later stages of stamen development, when pollen is maturing (i.e., genes down-regulated in the ms1 mutant). Both datasets contained significantly more clusters than expected by chance. All the 17 shared clusters contained genes originating from at least two different homologous groups showing that the clustering of co-regulated genes is not only due to recent gene duplication events on the chromosomes (Table 2). Even though the number of clusters we find depends on the clustering parameters, our finding of a statistically significant number of clusters was robust against changes in the parameters (Figure 1; Supplemental Figure S2).
We did not find any clusters with both gene homology and significant promoter similarities, although clusters #8 and #13 both contained homologs and were nominally significant in the promoter similarity score analysis. There might be a selection against promoter similarity connected to homology—if two genes are both homologous (and hence, possibly very similar in sequence, structure, and function) and have very similar promoter regions (and hence, regulated in a similar fashion), this would entail a redundancy that there might not be a selective pressure to maintain. We used OrthoMCL, which identifies homologous genes by comparing amino acid sequences within and between species. This means that genes that were duplicated before speciation can be separated if there are more similar sequences between species. Other metrics for assigning homology, e.g. max Blast bit score over a limited portion of the sequence (as was used in (16)) could be more permissive in the homology assignment but with an increased risk of identifying non-homologous genes with common domains. In our data set, cluster #7 would also be considered as containing homologs if using the bit score. In the promoter score analysis, we used a similarity score reminiscent of a score used previously for time-series gene expression data (46). It could in a future study be augmented by, e.g., including motif position in the score.
The presence of similar regulatory motifs in promoter regions of co-regulated genes is indicative of transcriptional regulation by a common transcription factor. The promoter analysis did not identify a common regulatory motif for all genes in all clusters. However, six distinct regulatory motifs were present in the promoter region of at least one gene in each cluster. One example was the E-box motif, CAGCTG, which is recognized by transcription factors in the ‘basic helix-loop-helix’ domain protein superfamily (bHLH) (47). This motif was present in 70 of the 96 clustered genes, but not all 70 genes were part of the up-regulated set. In cluster #15, we observed overrepresentation of the motif CATGCA, which has previously been linked to transcriptional regulation due to association with chromatin remodeling factors that repress transcription (48). Thus, the promoter analysis does not support the idea that a single common transcription factor is responsible for the up-regulation of the clustered genes, but also does not exclude a possible interaction between a transcriptional regulator and the entire chromosomal region of a cluster. Additionally, we did not find any evident role for chromatin insulator proteins in the clustering (Supplemental Text S1, Materials and Methods, and Results).
Genome wide localization of two MADS-box transcription factors AP3 and PI binding sites have been analysed in synchronized floral buds at early floral stages using Chromatin Immuno-precipitation followed by massively parallel sequencing (49). Individual genes in three of the 17 clusters studied here are indeed bound by the AP3/PI heterodimer (clusters #1, #6 and #15). It has been proposed that MADS-box transcription factors may act as pioneer factors by accessing closed chromatin and directly or indirectly trigger changes in chromatin accessibility (50), possibly through interaction with chromatin remodeling factors. Our findings that AP3-regulated genes (SEGs) did form a significant amount of clusters without clear homology or common regulatory motif support involvement of chromatin modification in gene regulation by the homeotic proteins. It is tempting also to speculate that interactions could occur between AP3/PI and the PHD-finger protein MS1, since PHD-finger proteins are suggested to regulate transcription via the modification of chromatin (51). Further MS1-studies analogous to the experiments outlined by Wuest et al. (49) may require single cell analyses, since MS1 activity is restricted to relatively late stages of floral development and to a small number of cells over a short period of time.
For genes in metabolic clusters, histone 2 variant H2A.Z has been associated with activation, and histone 3 lysine tri-methylation (H3K27me3) with repression (11,12). The expression of clustered genes was suppressed in stage 11 flowers of the H2A.Z-deficient hta9/hta11 mutant (Supplemental Figure S12A). Further, H3K27me3 marks on clusters were enriched in tissues where the clustered genes were silent (Supplemental Figure S7). These observations indicate chromatin-mediated transcriptional regulation of the clustered genes. To test the hypothesis of whether MS1 induction leads to loosening in chromatin compaction, which could make the promoters of the clustered genes accessible for the transcriptional regulators, we analyzed chromosome de-condensation in single cells using DNA FISH in combination with super-resolution SIM microscopy. The estimated de-condensation lengths of clusters #3 and #15 in DEX treated tapetum cells were in agreement with the differences in number of kb each cluster is spanning. The fold compaction compared with naked B-DNA was similar for both clusters (22-fold or 20-fold compaction in stage 11, respectively) and also in agreement with estimates of chromatin compaction in other species, e.g. the Sad1–Sad2 locus in diploid oat (10). In line with this notion, the analysis of stage specific mRNA expression levels of the clustered genes demonstrated that the genes in the clusters express at their highest in floral stages 11 and 12. However, we cannot exclude that individual genes in the clusters initiate transcription at an earlier stage, i.e. that chromatin de-condensation in fact coincides with initiation of transcription of distinct clustered genes.
Clustering of genes involved in similar cellular or developmental processes is likely to confer two selective advantages: co-inheritance and co-regulation (52). Previous studies of physical clustering tended to focus on either possibility, but here we studied both. While tandem repeats of duplicated genes may constitute a source of genetic material available for sub- and neo-functionalization, clustering of unrelated genes may bestow a selective advantage provided that the genes contribute to the similar pathways and that they are inherited as a single unit. One example of this is the pollination syndrome in petunia where traits adapted to attract specific pollinator guilds (i.e. floral scent, color and morphology) have become clustered and are thus inherited as a monogenic trait (53). This tight genetic linkage of independent regulators facilitates co-segregation of co-adaptive variation and limits the production of unfit recombinant forms. In addition, physical clustering may be a consequence of recruitment of new gene function via expansion of chromatin-level regulated regions (54). Evolutionary studies of gene clusters would shed light on the selective forces that promote the formation of chromosomal gene clusters.
Identification of physical gene clusters by combining global gene expression data with chromosomal gene distances, together with gene homology and promoter element assessments, provides a useful tool for investigating co-regulated physical gene clusters. The clustering analysis platform is applicable to any eukaryotic expression data sets, provided there is a stable genomic sequence and gene annotation, as well as a set of related annotated species (for homology analysis) and, ideally, a set of defined regulatory motifs. Our findings indicate that chromosomal clusters of co-expressed genes are important for different stages of stamen development. By focusing on genes acting downstream of MS1, we are likely to identify clusters harboring genes important for tapetum function and pollen development. Lipid storage and sexual reproduction are enriched biological processes among the clustered genes (Supplemental Table S6): Cluster #11 contains genes involved in transport of pollen wall precursors from the tapetum to the developing pollen grain. Cluster #8, #10 and #17 harbor pectin-modifying enzymes. Pectin is a major constituent of the innermost (intine) layer of the pollen wall, which is required for structural integrity of pollen grains and pollen germination. Cluster #3, #7 and #15 harbor genes involved in formation of trypine, the outermost layer of the pollen wall, sometimes referred to as the pollen coat. Loss of pollen coat lipids and proteins results in defective or delayed pollen hydration on the stigma surface (54). Cluster #6 and #12 harbor genes with similarity to proteins that regulate the rejection or acceptance of non-self-/self-pollen in Papaver rhoeas (55). Hence, several clusters contain genes with implicated functions in pollen stigma interactions.
Transcription of the clustered genes is associated with MS1 activation and shifts in chromatin de-condensation in individual tapetum cells. This suggests that male reproductive development in A. thaliana is dependent on physical gene clusters at least in part regulated at the chromatin level. Further studies of other organ types and related species will show whether cell and organ differentiation generally involve chromatin-level co-regulation of functionally related genes through physical clustering.
Supplementary Material
ACKNOWLEDGEMENTS
Cajsa Lithell (SLU) is acknowledged for providing help with layout and illustrations, John Tim Fisher (University of Edinburgh) for assisting the cleaning of the code.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
UK Biotechnology and Biological Sciences Research Council [ISP grant BB/ J004588/1 to P.S.]; EMBO and University of Edinburgh [LT Fellowship and Mary Kinross Seedcorn Award to N.N.]; Carl Tryggers Foundation [CTS 08:376, 09:371 to J.S.]; Formas [2013-650 to J.S. and O.E.]; Swedish Research Council [80358701, 80358702, 8035870 to O.E]. Funding for open access charge: KTH Royal Institute of Technology.
Conflict of interest statement. None declared.
REFERENCES
- 1. Osbourn A.E., Field B.. Operons. Cell. Mol. Life Sci.: CMLS. 2009; 66:3755–3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Al-Shahrour F., Minguez P., Marques-Bonet T., Gazave E., Navarro A., Dopazo J.. Selection upon genome architecture: conservation of functional neighborhoods with changing genes. PLoS Comput. Biol. 2010; 6:e1000953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Williams E.J., Bowles D.J.. Coexpression of neighboring genes in the genome of Arabidopsis thaliana. Genome Res. 2004; 14:1060–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Frey M., Chomet P., Glawischnig E., Stettner C., Grun S., Winklmair A., Eisenreich W., Bacher A., Meeley R.B., Briggs S.P. et al. Analysis of a chemical plant defense mechanism in grasses. Science. 1997; 277:696–699. [DOI] [PubMed] [Google Scholar]
- 5. Field B., Fiston-Lavier A.S., Kemen A., Geisler K., Quesneville H., Osbourn A.E.. Formation of plant metabolic gene clusters within dynamic chromosomal regions. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:16116–16121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Field B., Osbourn A.E.. Metabolic diversification–independent assembly of operon-like gene clusters in different plants. Science. 2008; 320:543–547. [DOI] [PubMed] [Google Scholar]
- 7. Chen W.H., de Meaux J., Lercher M.J.. Co-expression of neighbouring genes in Arabidopsis: separating chromatin effects from direct interactions. BMC Genomics. 2010; 11:178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wada M., Takahashi H., Altaf-Ul-Amin M., Nakamura K., Hirai M.Y., Ohta D., Kanaya S.. Prediction of operon-like gene clusters in the Arabidopsis thaliana genome based on co-expression analysis of neighboring genes. Gene. 2012; 503:56–64. [DOI] [PubMed] [Google Scholar]
- 9. Zhan S., Horrocks J., Lukens L.N.. Islands of co-expressed neighbouring genes in Arabidopsis thaliana suggest higher-order chromosome domains. Plant J. 2006; 45:347–357. [DOI] [PubMed] [Google Scholar]
- 10. Wegel E., Koumproglou R., Shaw P., Osbourn A.. Cell type-specific chromatin decondensation of a metabolic gene cluster in oats. Plant Cell. 2009; 21:3926–3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nutzmann H.W., Osbourn A.. Regulation of metabolic gene clusters in Arabidopsis thaliana. New Phytologist. 2015; 205:503–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yu N., Nutzmann H.W., MacDonald J.T., Moore B., Field B., Berriri S., Trick M., Rosser S.J., Kumar S.V., Freemont P.S. et al. Delineation of metabolic gene clusters in plant genomes by chromatin signatures. Nucleic Acids Res. 2016; 44:2255–2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Miller M.A., Cutter A.D., Yamamoto I., Ward S., Greenstein D.. Clustered organization of reproductive genes in the C. elegans genome. Curr. Biol.: CB. 2004; 14:1284–1290. [DOI] [PubMed] [Google Scholar]
- 14. Pignatelli M., Serras F., Moya A., Guigo R., Corominas M.. CROC: finding chromosomal clusters in eukaryotic genomes. Bioinformatics. 2009; 25:1552–1553. [DOI] [PubMed] [Google Scholar]
- 15. Dottorini T., Palladino P., Senin N., Persampieri T., Spaccapelo R., Crisanti A.. CluGene: a bioinformatics framework for the identification of co-localized, co-expressed and co-regulated genes aimed at the investigation of transcriptional regulatory networks from high-throughput expression data. PLoS One. 2013; 8:e66196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Alexeyenko A., Millar A.H., Whelan J., Sonnhammer E.L.. Chromosomal clustering of nuclear genes encoding mitochondrial and chloroplast proteins in Arabidopsis. Trends Genet.: TIG. 2006; 22:589–593. [DOI] [PubMed] [Google Scholar]
- 17. Lemay D.G., Martin W.F., Hinrichs A.S., Rijnkels M., German J.B., Korf I., Pollard K.S.. G-NEST: a gene neighborhood scoring tool to identify co-conserved, co-expressed genes. BMC Bioinformatics. 2012; 13:253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. O'Maoileidigh D.S., Graciet E., Wellmer F.. Gene networks controlling Arabidopsis thaliana flower development. New Phytologist. 2014; 201:16–30. [DOI] [PubMed] [Google Scholar]
- 19. Wellmer F., Riechmann J.L., Alves-Ferreira M., Meyerowitz E.M.. Genome-wide analysis of spatial gene expression in Arabidopsis flowers. Plant Cell. 2004; 16:1314–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Alves-Ferreira M., Wellmer F., Banhara A., Kumar V., Riechmann J.L., Meyerowitz E.M.. Global expression profiling applied to the analysis of Arabidopsis stamen development. Plant Physiol. 2007; 145:747–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ito T., Nagata N., Yoshiba Y., Ohme-Takagi M., Ma H., Shinozaki K.. Arabidopsis MALE STERILITY1 encodes a PHD-type transcription factor and regulates pollen and tapetum development. Plant Cell. 2007; 19:3549–3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wilson Z.A., Morroll S.M., Dawson J., Swarup R., Tighe P.J.. The Arabidopsis MALE STERILITY1 (MS1) gene is a transcriptional regulator of male gametogenesis, with homology to the PHD-finger family of transcription factors. Plant J. 2001; 28:27–39. [DOI] [PubMed] [Google Scholar]
- 23. Yang C., Vizcay-Barrena G., Conner K., Wilson Z.A.. MALE STERILITY1 is required for tapetal development and pollen wall biosynthesis. Plant Cell. 2007; 19:3530–3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wysocka J., Swigut T., Xiao H., Milne T.A., Kwon S.Y., Landry J., Kauer M., Tackett A.J., Chait B.T., Badenhorst P. et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006; 442:86–90. [DOI] [PubMed] [Google Scholar]
- 25. Merkel D.J., Wells S.B., Hilburn B.C., Elazzouzi F., Perez-Alvarado G.C., Lee B.M.. The C-terminal region of cytoplasmic polyadenylation element binding protein is a ZZ domain with potential for protein-protein interactions. J. Mol. Biol. 2013; 425:2015–2026. [DOI] [PubMed] [Google Scholar]
- 26. Sanders P.M., Bui A.Q., Weterings K., McIntire K.N., Hsu Y.C., Lee P.Y., Truong M.T., Beals T.P., Goldberg R.B.. Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex Plant Reprod. 1999; 11:297–322. [Google Scholar]
- 27. Lamesch P., Berardini T.Z., Li D., Swarbreck D., Wilks C., Sasidharan R., Muller R., Dreher K., Alexander D.L., Garcia-Hernandez M. et al. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 2012; 40:D1202–D1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Swarbreck D., Wilks C., Lamesch P., Berardini T.Z., Garcia-Hernandez M., Foerster H., Li D., Meyer T., Muller R., Ploetz L. et al. The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res. 2008; 36:D1009–D1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hu T.T., Pattyn P., Bakker E.G., Cao J., Cheng J.F., Clark R.M., Fahlgren N., Fawcett J.A., Grimwood J., Gundlach H. et al. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat. Genet. 2011; 43:476–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Slotte T., Hazzouri K.M., Agren J.A., Koenig D., Maumus F., Guo Y.L., Steige K., Platts A.E., Escobar J.S., Newman L.K. et al. The Capsella rubella genome and the genomic consequences of rapid mating system evolution. Nat. Genet. 2013; 45:831–835. [DOI] [PubMed] [Google Scholar]
- 31. Yang R., Jarvis D.E., Chen H., Beilstein M.A., Grimwood J., Jenkins J., Shu S., Prochnik S., Xin M., Ma C. et al. The reference genome of the halophytic plant Eutrema salsugineum. Front. Plant Sci. 2013; 4:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schmid M., Davison T.S., Henz S.R., Pape U.J., Demar M., Vingron M., Scholkopf B., Weigel D., Lohmann J.U.. A gene expression map of Arabidopsis thaliana development. Nat. Genet. 2005; 37:501–506. [DOI] [PubMed] [Google Scholar]
- 33. Lafos M., Kroll P., Hohenstatt M.L., Thorpe F.L., Clarenz O., Schubert D.. Dynamic regulation of H3K27 trimethylation during Arabidopsis differentiation. PLoS Genet. 2011; 7:e1002040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang X., Clarenz O., Cokus S., Bernatavichute Y.V., Pellegrini M., Goodrich J., Jacobsen S.E.. Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol. 2007; 5:e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li L., Stoeckert C.J. Jr, Roos D.S.. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003; 13:2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Obayashi T., Kinoshita K., Nakai K., Shibaoka M., Hayashi S., Saeki M., Shibata D., Saito K., Ohta H.. ATTED-II: a database of co-expressed genes and cis elements for identifying co-regulated gene groups in Arabidopsis. Nucleic Acids Res. 2007; 35:D863–D869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Defrance M., Janky R., Sand O., van Helden J.. Using RSAT oligo-analysis and dyad-analysis tools to discover regulatory signals in nucleic sequences. Nat. Protoc. 2008; 3:1589–1603. [DOI] [PubMed] [Google Scholar]
- 38. Alexa A., Rahnenfuhrer J.. R package version 2.22.0. 2010; Vol. R package version 2.22.0. [Google Scholar]
- 39. Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T. et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000; 25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Benjamini Y., Hochberg Y.. Controlling the false discovery rate—a practical and powerful approach to multiple testing. J. R. Stat. Soc. B Met. 1995; 57:289–300. [Google Scholar]
- 41. Smyth D.R., Bowman J.L., Meyerowitz E.M.. Early flower development in Arabidopsis. Plant Cell. 1990; 2:755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Karlgren A., Carlsson J., Gyllenstrand N., Lagercrantz U., Sundstrom J.F.. Non-radioactive in situ hybridization protocol applicable for Norway spruce and a range of plant species. J. Visual. Exp.: JoVE. 2009; 26, doi:10.3791/1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Aoyama T., Chua N.H.. A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 1997; 11:605–612. [DOI] [PubMed] [Google Scholar]
- 44. Coleman-Derr D., Zilberman D.. Deposition of histone variant H2A.Z within gene bodies regulates responsive genes. PLoS Genet. 2012; 8:e1002988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Markaki Y., Smeets D., Cremer M., Schermelleh L.. Fluorescence in situ hybridization applications for super-resolution 3D structured illumination microscopy. Methods Mol. Biol. 2013; 950:43–64. [DOI] [PubMed] [Google Scholar]
- 46. Nilsson R., Bajic V.B., Suzuki H., di Bernardo D., Bjorkegren J., Katayama S., Reid J.F., Sweet M.J., Gariboldi M., Carninci P. et al. Transcriptional network dynamics in macrophage activation. Genomics. 2006; 88:133–142. [DOI] [PubMed] [Google Scholar]
- 47. Toledo-Ortiz G., Huq E., Quail P.H.. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell. 2003; 15:1749–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Suzuki M., Wang H.H., McCarty D.R.. Repression of the LEAFY COTYLEDON 1/B3 regulatory network in plant embryo development by VP1/ABSCISIC ACID INSENSITIVE 3-LIKE B3 genes. Plant Physiol. 2007; 143:902–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wuest S.E., O'Maoileidigh D.S., Rae L., Kwasniewska K., Raganelli A., Hanczaryk K., Lohan A.J., Loftus B., Graciet E., Wellmer F.. Molecular basis for the specification of floral organs by APETALA3 and PISTILLATA. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:13452–13457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pajoro A., Madrigal P., Muino J.M., Matus J.T., Jin J., Mecchia M.A., Debernardi J.M., Palatnik J.F., Balazadeh S., Arif M. et al. Dynamics of chromatin accessibility and gene regulation by MADS-domain transcription factors in flower development. Genome Biol. 2014; 15:R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Aasland R., Gibson T.J., Stewart A.F.. The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci. 1995; 20:56–59. [DOI] [PubMed] [Google Scholar]
- 52. Field B., Osbourn A.. Order in the playground: Formation of plant gene clusters in dynamic chromosomal regions. Mobile Genet. Elem. 2012; 2:46–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hermann K., Klahre U., Moser M., Sheehan H., Mandel T., Kuhlemeier C.. Tight genetic linkage of prezygotic barrier loci creates a multifunctional speciation island in Petunia. Curr. Biol.: CB. 2013; 23:873–877. [DOI] [PubMed] [Google Scholar]
- 54. Updegraff E.P., Zhao F., Preuss D.. The extracellular lipase EXL4 is required for efficient hydration of Arabidopsis pollen. Sex Plant Reprod. 2009; 22:197–204. [DOI] [PubMed] [Google Scholar]
- 55. Foote H.C., Ride J.P., Franklin-Tong V.E., Walker E.A., Lawrence M.J., Franklin F.C.. Cloning and expression of a distinctive class of self-incompatibility (S) gene from Papaver rhoeas L. Proc. Natl. Acad. Sci. U.S.A. 1994; 91:2265–2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Scripts and code developed to analyse chromosomal clustering are deposited at https://github.com/b97jre/ClusterAnalysisTools. Material and methods are described below and in Supplemental Text S1, Methods and Supplemental Data S1.