Abstract
Enhancers are intergenic DNA elements that regulate the transcription of target genes in response to signaling pathways by interacting with promoters over large genomic distances. Recent studies have revealed that enhancers are bi-directionally transcribed into enhancer RNAs (eRNAs). Using single-molecule fluorescence in situ hybridization (smFISH), we investigated the eRNA-mediated regulation of transcription during estrogen induction in MCF-7 cells. We demonstrate that eRNAs are localized exclusively in the nucleus and are induced with similar kinetics as target mRNAs. However, eRNAs are mostly nascent at enhancers and their steady-state levels remain lower than those of their cognate mRNAs. Surprisingly, at the single-allele level, eRNAs are rarely co-expressed with their target loci, demonstrating that active gene transcription does not require the continuous transcription of eRNAs or their accumulation at enhancers. When co-expressed, sub-diffraction distance measurements between nascent mRNA and eRNA signals reveal that co-transcription of eRNAs and mRNAs rarely occurs within closed enhancer–promoter loops. Lastly, basal eRNA transcription at enhancers, but not E2-induced transcription, is maintained upon depletion of MLL1 and ERα, suggesting some degree of chromatin accessibility prior to signal-dependent activation of transcription. Together, our findings suggest that eRNA accumulation at enhancer–promoter loops is not required to sustain target gene transcription.
INTRODUCTION
Enhancers are intergenic DNA elements that regulate the transcription of target genes by interacting with promoters over large genomic distances (1–3). They contain binding sites for transcription factors, which promote RNA polymerase II (RNAPII) recruitment and transcription activation (4). Enhancers carry unique epigenetic marks that distinguish them from promoters, including monomethylated lysine 4 of Histone H3 (H3K4me1) and acetylated lysine 27 of Histone H3 (H3K27ac) (5,6). In addition, these regulatory elements have an open chromatin conformation, which increases accessibility to transcription factors and to RNAPII (7). Several genome-wide studies have shown that enhancers are transcribed into long noncoding RNAs (ncRNAs) in a tissue-specific manner in response to extracellular signals. For example, stimulation of cortical neurons via membrane depolarization was shown to induce the recruitment of RNAPII to enhancers and the initiation of bi-directional transcription of noncoding RNAs, termed enhancer RNAs (eRNAs) (8). Importantly, eRNA expression positively correlated with increased mRNA expression from proximal target genes, suggesting that eRNA transcription marks active enhancers. Although genome-wide studies in other cell types have also established a correlation between eRNA and target mRNA expression, whether there is a unified mechanism by which eRNAs regulate their target genes remains unclear (8–17).
Since long ncRNAs are more functionally diverse than other classes of ncRNAs, such as microRNAs, rRNAs and tRNAs, they are thought to be under lower selective constraints (18). Although enhancers possess conserved transcription factor binding sites, enhancer-derived transcripts often lack conserved motifs or secondary structures that could provide a hint for a unified mechanism of action (7). Similarly, little is known about eRNA biogenesis. Most eRNAs are capped, unspliced and non-polyadenylated with a median length of ∼350 nucleotides (19). Recent studies showed that the RNAPII-associated integrator complex mediates transcription termination at enhancers, and that 3΄ cleavage of eRNAs by the integrator complex is required for their function (20). Interestingly, several experimentally validated eRNAs were detected as distinct bands in northern blot analyses, further corroborating the idea that eRNA transcription is terminated in a uniform manner (16,21,22). In addition, ChromRNA-seq analyses have suggested that eRNAs are enriched in the chromatin fraction (20). Collectively, these observations indicate that eRNAs act as processed transcripts in cis on their target loci. Supporting this idea, tethering of eRNAs to luciferase reporter plasmids enhanced promoter activity (15,16). However, it cannot be excluded that enhancer transcription rather than the eRNA transcript per se is required for enhancer function.
While the features that constitute a functional eRNA molecule are currently unknown, several studies have implicated eRNAs in different steps of transcription regulation, including the modulation of chromatin accessibility at promoters and the release of the negative elongation factor NELF (11,23,24). In particular, eRNAs have been suggested to mediate looping interactions between enhancers and promoters of target genes by facilitating the recruitment of specific factors known to stabilize long-range chromatin interactions, including cohesin and the mediator complex (15,21,25). However, treatment with flavopiridol to block eRNA transcription did not affect chromatin looping at the P2RY2 and GREB1 loci upon E2 stimulation of MCF-7 cells (14). Furthermore, eRNA inhibition affected neither enhancer-specific epigenetic modifications nor the recruitment of transcriptional regulators, suggesting that eRNA synthesis is not required for the assembly of enhancer–promoter complexes (14). Interestingly, a recent study has shown that eRNAs bind to the transcription factor Ying Yang 1 (YY1), and that this interaction can enhance the recruitment of this transcription factor to enhancers (26). Whether transcription factor trapping by eRNAs is a widespread mechanism of gene expression regulation remains to be investigated. Therefore, while several different studies suggest that eRNAs are likely to be implicated in regulating target gene transcription, the mechanistic details of eRNA function remain unclear.
One difficulty in studying induced transcriptional responses is that different cells and even different alleles within a cell may not show the same induction kinetics upon stimulation, due to the stochastic nature of many cellular processes (27,28). Different studies have shown that eRNA transcription precedes the peak of mRNA transcription (9,11). In addition, transcription often occurs in short bursts, where individual alleles switch between active and inactive states (27,29–31). Observing such behavior, therefore, requires monitoring the transcriptional response at the single-cell and single-allele level. Most previous studies investigating eRNA function used ensemble measurements of cell populations, and were thus unable to provide spatial or allele-specific information regarding eRNA and target mRNA transcription (14,15). In particular, whether eRNAs are required for every round of transcription initiation, and whether they modulate transcription bursts is still unknown.
To investigate the role of eRNAs in transcription, we characterized the spatiotemporal expression of these noncoding transcripts in individual cells using single-molecule resolution fluorescence in situ hybridization (smFISH). Specifically, we used ERα-positive MCF-7 breast cancer cells as a model system to study eRNA-mediated transcriptional programs during estrogen induction. Estrogen binds directly to the nuclear receptor ERα, which is recruited to estrogen response elements (EREs) as a homodimer and regulates target gene transcription by recruiting transcriptional cofactors (32,33). ERα binding to chromatin is facilitated by the pioneer factor FOXA1, which itself is recruited at H3K4me1 marks deposited by the histone methyltransferase MLL1 on the enhancer regions of target genes (34,35). While the mechanism whereby MLL1 recognizes its target loci is presently unknown, roles for the CpG Binding protein CGBP, the transcription factor YY1, and the core subunit of the SWI–SNF complex hSNF5 have been suggested (36–38).
Here we monitored the expression and localization of eRNAs derived from the enhancers of the FOXC1 and P2RY2 loci, as well as the expression of their cognate mRNAs over a time course of estrogen induction in individual MCF-7 cells by smFISH. In the uninduced state, FOXC1 and P2RY2 eRNAs were expressed at low levels, independently of MLL1 and ERα. Estrogen treatment induced the transcription of eRNAs and target mRNAs with similar kinetics, and required both chromatin modification by MLL1 and ERα recruitment. However, co-expression of eRNAs and mRNAs at individual alleles was infrequent, and did not correlate with bursting mRNA transcription. Furthermore, distance measurements between eRNA and nascent mRNA signals at co-expressing alleles revealed infrequent co-localization within closed enhancer–promoter loops. Taken together, our data suggest that ongoing eRNA transcription is neither required to stabilize chromatin loops, nor to sustain continuous transcription from target alleles.
MATERIALS AND METHODS
Reagents
17β-estradiol (E2) (E1024) and the antiestrogens (Z)-4-hydroxytamoxifen (H7904) and ICI 182 780 (I4409) were purchased from Sigma. Cells were pretreated with 100 nM antiestrogens for 3 h prior to E2 induction (5 nM) for 40 min in the absence of the antiestrogens.
MCF-7 cell culture
The MCF-7 cell line was maintained in α-minimal Eagle's medium (α-MEM) (Wisent, St-Bruno, QC, Canada) supplemented with 10% fetal bovine serum (FBS) (Invitrogen, 10437028) as described previously (39). The cells were attached to poly-l-lysine (Sigma, P8920) coated coverslips in regular media. Three days prior to induction, the cells were washed twice with PBS, and the media was changed to phenol red–free Dulbecco's modified Eagle's medium (DMEM) (Wisent), supplemented with 10% charcoal-treated FBS (Invitrogen, 12676029), 1% sodium pyruvate and 1% l-glutamine (Wisent). The cells were induced with 17β-estradiol (E2) at 100 nM final concentration (except for the ERα inhibition experiments, where the E2 concentration was lowered to 5 nM) for the indicated lengths of time. Validated MLL1 and non-specific (NS) siRNAs were transfected at 50 nM final concentration using Lipofectamine RNAiMax (Invitrogen, 13778100) according to the manufacturer's instructions. Twenty-four hours after transfection, the media was replaced with supplemented phenol red-free DMEM. Estrogen treatments were initiated 48 h later (a total of 72 h post-siRNA transfection). All experiments represent two to three independent biological replicates.
smRNA FISH
Custom DNA probe sets were designed using Stellaris® Probe Designer, synthesized by Biosearch Technologies containing a 3΄ amine reactive group, and labeled with Cy5 (GEPA25001), Cy3 (GEPA23001) or Cy3.5 (GEPA23501) from Sigma or their DyLight (Thermo Scientific) equivalents DyLight 650 (62266), DyLight 550 (62263), DyLight 594 (46413). Probe sequences are shown in Supplementary Table S1. For smRNA FISH, the cells were briefly washed with 1× PBS, fixed with 4% paraformaldehyde (pH 7.2) for 10 min at room temperature, washed three times with 1× PBS, and stored overnight in 70% ethanol at −20°C. Prior to hybridization, the cells were air-dried and rehydrated in 10% formamide/2× SSC for 10 min at room temperature. The cells were hybridized with 10–20 ng of each probe plus 40 ug ssDNA/tRNA mix resuspended in the hybridization solution (10% dextran sulfate/10% formamide/2× SSC/2 mM VRC/0.1 mg/ml BSA) for 3 h in the dark at 37°C. Two post-hybridization washes were carried out at 37°C with 10% formamide/2× SSC for 30 min each. Samples were then rinsed with 1× PBS and mounted with ProLong Gold antifade reagent with DAPI (P36935, Invitrogen). Images were acquired with a 63× NA 1.4 oil objective on a Zeiss Axioimager Z2 equipped with an AxioCam mRm CCD camera and the following filter sets: Zeiss 488050-9901-000 (Cy5), Chroma SP102 v1 (Cy3), Chroma SP103 v2 (Cy3.5), Zeiss 488049-9901-000 (DAPI).
Protease and RNaseA treatments
For protease treatment, prior to smFISH probe hybridization, fixed cells were rehydrated in 1× PBS and treated with 1 mg/ml pepsin in 10 mM HCl, at 37°C, for 1 min. Enzyme activity was inhibited with 0.1 M Glycine in 1× PBS, pH 7.2 for 10 min. For RNAse treatment, fixed cells were treated with 0.1 mg/ml RNaseA for 1 h at 37°C. The cells were washed three times with 1× PBS, pre-incubated in 10% formamide/2× SSC, and hybridized with the indicated probes as described above.
Immunofluorescence
Cells stored in 70% ethanol were air-dried and rinsed with 1× PBS for 5 min. This was followed by permeabilization with 0.5% Triton x-100/1× PBS for 10 min at RT. Cells were then washed three times with 1× PBS before proceeding with the smFISH protocol as described above. After smFISH hybridization, the cells were washed three times with 1× PBS and blocked with 4% BSA (Ambion/Life Technologies molecular biology grade BSA Catalog # AM2616) in 1× PBS for 10 min at RT. Cells were then incubated with the rabbit polyclonal anti-MLL-C (EMD Millipore, ABE240) antibody (diluted 1:100 in 1% BSA/1× PBS) for 1 h at RT. After the primary antibody incubation, cells were washed three times with 1× PBS for 5 min. This was followed by incubation with the secondary anti-rabbit-Alexa 488 antibody (diluted 1:500 in 1% BSA/1× PBS) for 1 h at RT in the dark. Cells were then washed three times with 1× PBS for 5 min and mounted on slides with Prolong Gold antifade reagent containing DAPI.
Image processing and spot detection
For image analysis, 3D datasets were reduced to 2D data using maximum projections in Fiji. Spot detection was done by 2D Gaussian fitting as described previously (29,40). To correct for pixel shifts between channels, TetraSpec beads (Invitrogen T-7279) were imaged in all the channels and their position was determined by 2D Gaussian fitting. Relative pixel shifts were used to align channels after image acquisition and spot detection using a custom script in MATLAB R2013a (8.1.0.604) (The Mathworks, Inc.). The mRNA channel was used as a reference to correct eRNA positions relative to their target mRNAs.
RNA quantification and distance measurements
Nuclear masks were created in Fiji after manual segmentation of DAPI stained nuclei. Assignment of eRNA and mRNA signals within the nuclear masks was done using custom scripts in MATLAB. Detection of mRNA transcription sites was done in two steps: (i) computing the single RNA intensity Is and (ii) locating transcription sites by searching for nuclear spots with at least n* Is intensity, where n=1.5. To determine the intensity of a single mRNA, mRNA spots were clustered into different classes using ck-means, based on intensity. For FOXCI mRNA signals, the single mRNA intensity was calculated by computing the mean intensity of spots outside the nuclear boundary that belong to the low intensity group (with cytoplasmic mRNAs corresponding to single mRNAs). For P2RY2 intron signals, the single RNA intensity was calculated by computing the mean intensity of the nuclear intron spots in the low intensity group. To measure the distance between mRNA transcription sites and eRNAs, we searched for co-localizations occurring within a 400 nm radius around each mRNA transcription site. When eRNAs were detected within that window, we measured the 2D Euclidian distance between the centroids of the eRNAs and mRNAs, as calculated above. Distances between active transcription sites were measured using Volocity 6.0 (PerkinElmer).
RTq-PCR
Total RNA was harvested in TRIzol Reagent (ThermoFisher Scientific, 15596026) and cDNA synthesis was carried out with random hexamers using SuperScript II Reverse Transcriptase (ThermoFisher Scientific, 18064014) according to the manufacturer's instructions. The RTq-PCR assay was performed in duplicate on a LightCycler 480 System (Roche Life Science) using SYBR Green. The ΔCT (delta threshold cycle) method was used for quantification and transcript levels were normalized to GAPDH. RT-qPCR primers are listed in Supplementary Table S2.
Chromatin Immunoprecipitation
For ChIP, the cells were seeded at a density of 1.66 × 106 cells per 10 cm dish in phenol red-free DMEM media supplemented with 10% charcoal-stripped serum and transfected 24 h after seeding with 50 nM siRNAs using siLentFect Lipid Reagent (BioRad, 170-3361) according to manufacturer's instructions. The next day, media was replaced with supplemented phenol red-free DMEM. The cells were induced for 40 min with 100 nM estradiol 72 h post-transfection and harvested for ChIP. Briefly, the cells were crosslinked with 1% formaldehyde for 10 min at room temperature. The crosslinking reaction was quenched with 0.15 M glycine for 5 min and the cells were washed twice with ice-cold PBS and harvested. The cell pellets were resuspended in lysis buffer (5 mM PIPES pH 8, 85 mM KCl, 0.5% NP40), supplemented with a mix of protease and phosphatase inhibitors. Following centrifugation, nuclei were resuspended in nuclear lysis buffer (50 mM Tris pH 8.1, 10 mM EDTA pH 8, 1% SDS), supplemented with a mix of protease and phosphatase inhibitors, sonicated with a Bioruptor (Diagenode) at medium power for four rounds of 8 min with 30 s intervals between pulses. The DNA was quantified and 50 ug of each sample was diluted 20 times in dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris pH 8.1, 167 mM NaCl), and incubated overnight at 4°C with 4 ug of one of the following antibodies: ERα (Santa Cruz, sc-543), H3K4me (abcam, ab8895), rabbit IgG (Jackson ImmunoResearch, 011-000-003). The complexes were immunoprecipitated for 2 h at 4°C with 40 ul of a 1:1 mix of Dynabeads A and G (Life technologies). The beads were washed twice with dialysis buffer (2 mM EDTA pH 8, 50 mM Tris pH 8.1, 0.2% Sarkosyl) for 15 min with rotation at room temperature and four times with wash buffer (0.5 M LiCl, 1% NP40, 1% sodium deoxycholate, 33.2 mM Tris pH 8.1). The crosslinking was reversed and the DNA was eluted by heating the beads for 30 min at 65°C in elution buffer (50 mM NaHCO3, 1% SDS). Proteins were digested with Proteinase K (ThermoScientific, EO0491) and the DNA was purified on EZ-10 columns (BioBasic). The abundance of the immunoprecipitated DNA fragments was quantified by real time qPCR on a LightCycler 480 System (Roche Life Science) using SYBR Green. ChIP results were analyzed by the Percent Input Method. The knockdown efficiency was assessed by western blotting with the MLL-C antibody (Millipore, ABE240), using β-actin (Sigma, A5441) as a loading control. The qPCR primers and siRNAs used in the ChIP experiments are listed in Supplementary Table S2.
Statistical analysis
K-sample permutation tests were used to analyze the number of nascent transcripts per allele over the time course of E2 induction. The Fisher's exact test was used to compare the proportions of eRNAs that were co-localized and non co-localized with active TSs. P values were corrected for multiple comparisons using the Holm–Bonferroni method, and a threshold of P <0.05 was used throughout.
RESULTS
eRNAs and target mRNAs are co-induced upon E2 treatment in MCF-7 cells
To examine the role of eRNAs in modulating transcription at the single-cell level, we examined their spatial expression patterns in MCF-7 breast cancer cells over a time course of 17β-estradiol (E2) induction using smFISH. This technique uniquely allows the simultaneous localization of eRNAs and target mRNAs in individual cells and at individual alleles with single-molecule resolution (29,41). Specifically, we examined the E2-induced FOXC1 and P2RY2 loci as model genes. Genome-wide studies have shown previously that the FOXC1 and P2RY2 enhancers are transcribed into eRNAs in a hormone-dependent manner (14,15). The FOXC1 gene encodes a single exon of 3.4 kb and its expression is regulated by a single E2-responsive enhancer, located 26 kb downstream of its transcription termination site. The P2RY2 gene is 18 kb-long and its expression is regulated by an E2 responsive enhancer located 22 kb upstream of its transcription start site (TSS). The FOXC1 enhancer is bi-directionally transcribed, producing sense and antisense eRNAs (Figure 1A). We designed oligonucleotide probes against both target mRNAs and eRNAs (based on GRO-seq data published by Li et al., 2013), which we hybridized to paraformaldehyde-fixed MCF-7 cells at different time points of E2 induction. Addition of E2 results in a transient transcriptional response that peaks at 40 min (13); we therefore analyzed eRNA and mRNA expression at 0, 20, 40 and 60 min after E2 treatment. As shown in Figure 1B, FOXC1 and P2RY2 mRNAs were expressed at a low level in uninduced cells. Upon E2 induction, bright spots representing active sites of transcription, consisting of multiple nascent mRNAs, were detected within the nuclei of single cells. Since MCF-7 cells are triploid for the FOXC1 and P2RY2 genes, three to six transcription sites were observed in interphase and dividing cells, respectively.
Figure 1.
eRNAs are low-abundance nuclear transcripts that are induced with similar kinetics as their target genes. (A) Diagram of the genomic organization of the FOXC1 and P2RY2 loci. (B) smFISH showing the expression and localization of FOXC1 (left) and P2RY2 (right) eRNAs and mRNAs before and after 40 min of E2 induction in MCF-7 cells. Arrows indicate different eRNA and transcription site configurations. (C) Frequency distributions of FOXC1 (left panels) and P2RY2 (right panels) eRNA and active mRNA transcription sites during E2 induction. Representative data of three independent experiments; n = 100 cells per time point.
Analysis of the bi-directionally transcribed FOXC1 eRNAs showed similar induction kinetics as the target mRNAs, but different localization patterns. In uninduced cells, FOXC1 eRNAs were detected in the nucleus and were expressed at low levels (Figure 1B). Even though 38.2% of the cells expressed antisense and 23.5% expressed sense FOXC1 eRNAs, only a single eRNA spot was detected in the majority of these cells (Figure 1C). Upon E2 induction, eRNA and target mRNA expression increased in parallel, with 62.5% of the cells expressing antisense and 55.8% expressing sense FOXC1 eRNAs at 40 min of treatment. Notably, the fraction of cells expressing more than one eRNA increased three-fold compared to the uninduced state. Importantly, the number of eRNA spots rarely exceeded the number of alleles in either interphase or dividing cells (Figure 1C). A similar pattern of expression was observed for the P2RY2 sense eRNA and its cognate mRNA.
Since enhancer elements recruit many proteins, including transcriptional co-activators and RNAPII, eRNAs could be incorporated into large complexes and become inaccessible to the RNA FISH probes. To address this point, we carried out protease treatment prior to probe hybridization as described previously by Buxbaum et al., but found similar levels of eRNA expression in the presence and absence of protease digestion (42). Specifically, we observed comparable numbers of FOXC1 AS eRNAs per cell with or without pepsin treatment (Supplementary Figure S1A). Likewise, we found that the intensities of single FOXC1 AS eRNA spots did not change, suggesting that probe accessibility to the target is not obscured by eRNA incorporation into protein complexes (Supplementary Figure S1B). In addition, quantification of eRNA and mRNA fold induction levels in response to estrogen treatment by RT-qPCR was consistent with smFISH measurements, implying that eRNA and mRNA detection by smFISH is efficient (Supplementary Figure S2). Furthermore, in the presence of RNAse A, neither FOXC1 mRNAs nor AS eRNAs were detected, validating that the signals observed by smFISH are specific to RNA (Supplementary Figure S3).
In summary, our time course analysis revealed that eRNA transcripts are nuclear and induced with similar kinetics as the target mRNA. Combined with previous findings showing that eRNAs have a short half-life of about 7 min, and act in cis, these observations provide evidence that most eRNAs detected by smFISH are nascent and are mostly restricted to the enhancer from which they are transcribed (11,24).
Induction of eRNA and target mRNA transcription requires ERα and MLL1
As shown in Figure 1, low levels of eRNA expression were detectable at estrogen-responsive enhancers even in uninduced cells. This could be due to hormone-independent activation of ERα or to continuous basal transcription of the enhancer prior to ERα recruitment. In the latter case, low levels of eRNA transcription could be implicated in chromatin remodeling, thereby facilitating ERα accessibility once the cells are exposed to estrogen. To distinguish between these possibilities, we pre-treated the cells prior to the addition of E2 with either Tamoxifen, a drug that blocks estrogen receptor function by inhibiting E2 binding, or with ICI 182 780 (ICI), which induces ERα degradation (43–45). Both drugs effectively abolished the induction of FOXC1 and P2RY2 mRNA, as shown by the lack of bursting transcription upon E2 treatment following the addition of either antiestrogen (Figure 2A and 2B, and Supplementary Figure S4A and S4B). Similarly, eRNA induction was inhibited by both Tamoxifen and ICI (Figure 2A and 2B, and Supplementary Figure S4A and S4B). However, basal eRNA expression was unaffected by ICI 182 780 treatment, suggesting that the low level of eRNA transcription observed at these enhancers in uninduced cells is independent of ERα. This observation further raises the possibility that low-level eRNA transcription primes enhancers to respond rapidly to environmental stimuli.
Figure 2.
Induction of FOXC1 eRNA and mRNA transcription requires ERα. (A) FOXC1 eRNA and mRNA expression in the presence or absence of a 3 h pre-treatment with Tamoxifen (100 nM) or ICI (100 nM) followed by E2 induction (5 nM) for 40 min. (B) Quantification of data from (A). Frequency distributions of FOXC1 antisense eRNAs and nascent FOXC1 transcripts, representative of two independent experiments; n = 70–200 cells for each condition.
Since ERα inhibition did not attenuate the basal level of eRNA expression, we investigated whether upstream factors are required to activate eRNA transcription. As noted above, the histone methyltransferase MLL1 deposits the H3K4me1 mark on enhancer regions, which recruits the pioneering factor FOXA1, and subsequently, ERα (34). Thus, we examined whether siRNA-mediated MLL1 knockdown affects basal eRNA transcription. We found similar basal levels of P2RY2 eRNA transcription in the absence of E2 in MLL1-depleted cells and in the non-specific siRNA control (Figure 3A and 3B). Therefore, low basal eRNA transcription may occur prior to chromatin remodeling and participate in the recruitment of MLL1 (46,47).
Figure 3.
MLL1 is required for E2-induced eRNA transcription. (A) smFISH analysis for P2RY2 sense eRNA and mRNA combined with MLL1 immunofluorescence in MLL1-depleted or non-specific siRNA-treated cells before and after E2 induction. (B) Frequency distribution of P2RY2 sense eRNAs and mRNA in MLL1-depleted or non-specific siRNA-treated cells before and after E2 induction. Data are representative of four independent experiments; n = 160–200 cells for each condition. (C) ChIP analysis for H3K4me1 (left) and ERα (right) presence at the P2RY2 enhancer in MLL1-depleted or non-specific siRNA-treated cells in the presence or absence of E2 treatment; n = 2 independent experiments; error bars indicate SD.
In contrast to the basal level of eRNA transcription, P2RY2 eRNA induction was significantly reduced upon E2 treatment in MLL1-depleted cells compared to the non-specific siRNA control (Figure 3B), and paralleled a decrease in the number of transcriptionally active P2RY2 loci (Figure 3B). Consistent with these observations, ChIP analysis revealed a ten-fold decrease in ERα binding to the P2RY2 and FOXC1 enhancers upon E2 induction in MLL1-depleted cells compared to the control (Figure 3C, Supplementary Figure S5A). Furthermore, MLL1 knockdown resulted in a two-fold decrease in the level of H3K4me1 at these regulatory elements both in the presence and in the absence of E2 induction (Figure 3C, Supplementary Figure S5A). Taken together, these findings corroborate the RNA FISH results showing that MLL1 depletion inhibits both eRNA and target mRNA induction (Figure 3A and 3B), and suggest that MLL1-dependent deposition of H3K4me1 and the recruitment of ERα are required for the induction of eRNA transcription.
eRNA- mRNA co-expression is infrequent and is not required to maintain transcription
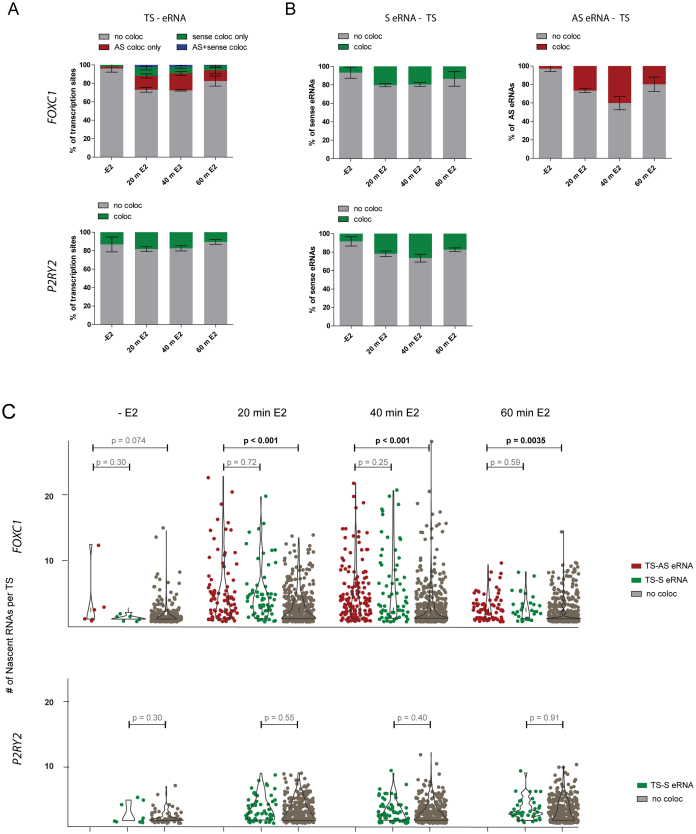
Previous studies have suggested that eRNAs facilitate transcription by stabilizing enhancer-promoter interactions (15,21,25). These interactions could persist for the duration of a single initiation event or for the entire length of the transcriptional response. If eRNA transcription or accumulation at a specific allele were required for enhancer–promoter communication or for maintaining transcription, frequent co-localization of eRNAs at active alleles should be observed. To test this hypothesis, we determined whether the simultaneous induction of eRNAs and target mRNAs is coordinated at individual alleles, and measured the frequency of eRNA and nascent mRNA signal co-localization. To identify FOXC1 TSs, we determined the position and intensity of mRNA signals, and clustered them into low and high intensity groups, with the low intensity spots representing single mRNAs. Nuclear spots of >1.5 times the intensity of the mean value of single mRNAs were scored as transcription sites, assuming that actively transcribed loci contain multiple nascent mRNAs due to transcription bursting (27,29,30,48). Since the P2RY2 gene contains several short exons at the 5΄ end that are too short for smFISH probe design, we used the first intron region to detect the nascent mRNA, and applied a similar spot-clustering algorithm to locate the transcription sites (Supplementary Figure S6A). We then measured the frequency of co-localization of actively transcribing alleles with eRNAs (Figure 4A) and vice versa (Figure 4B). Specifically, co-localization was defined by the presence of an eRNA within a 400 nm radius from the center of the TS signal. This cutoff was selected to include open enhancer–promoter configurations, but to exclude signals from neighboring alleles, which are separated by >1 um, as determined from distance measurements between active P2RY2 transcription sites (Supplementary Figure S6B).
Figure 4.
Simultaneous expression of eRNAs and mRNAs is infrequent in single cells and is not required to maintain transcription. (A) Frequency of co-localization between FOXC1 (top panel) and P2RY2 (bottom panel) mRNA transcription sites and their cognate eRNAs using the transcription site as a reference. eRNA-mRNA transcription site co-localization was scored within a 400 nm radius. (B) Frequency of co-localization of eRNAs with mRNA transcription sites using the eRNA signal as a reference. Panels A and B indicate the mean and SD from three independent experiments. (C) Quantification of the relationship between eRNA-transcription site co-localization and the RNAPII density on individual alleles. Outlines represent the relative density of data points. Data was pooled from three independent experiments; active transcription sites from n = 300 cells per time point were analyzed.
As shown in Figure 4A, FOXC1 eRNA–TS co-localization was infrequent before E2 treatment, with only 4% of transcription sites co-localizing with an eRNA. FOXC1 eRNA–TS co-localization increased to 27% at 40 min of E2 treatment. While E2 induction significantly increased the proportion of FOXC1 eRNA-TS co-localization (Fisher's exact test; P < 0.0001), the majority of transcription sites did not have an associated eRNA. As noted in Figure 1C, the fraction of cells expressing more than one eRNA increased three-fold at 40 min of E2 treatment compared to the uninduced state. Additionally, there was a five-fold increase in the fraction of cells with more than one active transcription site at 40 min of E2 treatment compared to the uninduced state. Despite this increase in the numbers of eRNAs and transcription sites detected per cell, the majority of TSs did not co-localize with eRNAs, indicating that mRNA transcription does not require the simultaneous expression of eRNAs. Triple co-localization of FOXC1 mRNA, antisense and sense eRNA was present at only 2% of active transcription sites at 40 min of E2 induction, further suggesting that simultaneous transcription of sense and antisense eRNAs is rare at active TSs.
Although the low frequency of eRNA–mRNA co-expression at individual alleles suggests that eRNAs are not required for sustained transcription, eRNAs could influence the magnitude of the transcription output. To determine whether eRNAs modulate transcription strength, we correlated the presence or absence of eRNAs at the TS with the RNAPII density on the target gene. As shown in Figure 4C, in uninduced cells, basal FOXC1 expression averaged two to three transcripts per TS and occurred primarily from alleles that had no associated eRNAs. During peak induction, both FOXC1 and P2RY2 showed strong bursting transcription, independent of the presence of eRNAs; however, for the FOXC1 gene, there was a small but significant increase in the number of nascent RNAs per transcription site at active alleles that co-localized with eRNAs compared to those that did not (Figure 4C). Furthermore, comparisons of mRNA signal intensities at transcription sites before and after E2 treatment showed that the RNAPII density along the gene increased over time (Supplementary Figure S7). In contrast, although the proportion of cells transcribing eRNAs increased with induction, the eRNA signal intensities did not change (Supplementary Figure S7). This observation suggests that compared to mRNA, eRNA transcription initiation occurs at a lower frequency, and that eRNAs do not accumulate at enhancers in high numbers.
mRNA–eRNA co-transcription rarely occurs within a closed enhancer–promoter loop conformation
Chromosome conformation capture (3C) analyses conducted in the presence or absence of enhancer-derived transcripts proposed that eRNAs mediate enhancer–promoter interactions (15,21,25). Such interactions are thought to be maintained by large protein complexes, containing cohesin, the mediator complex and other co-factors, and are thought to occur within 10–100 nm (49,50). Although these distances are below the 200 nm resolution limit of conventional light microscopy, signals within this range can be localized at higher spatial resolution by 2D Gaussian fitting (40). Using the coordinates of eRNA and mRNA transcription site signals as markers of enhancers and genes, respectively, we assessed whether transcribed enhancers interact with active genes. To achieve this, we first determined the minimal distance at which we could resolve co-localizing signals using a P2RY2 intron probe labeled with two different colors (Figure 5A). After pixel shift correction, signals identifying the same intron were co-localized within 63 nm (Figure 5A). We then measured pairwise eRNA–nascent mRNA distances among co-expressing alleles at 40 min of E2 induction. At this time point, 29% of antisense and 19% of sense FOXC1 eRNA–mRNA co-expressing alleles showed a separation of 100 nm or less (Figure 5B). These data show that the small percentage of active transcription sites that co-express eRNAs are infrequently transcribed within a closed enhancer–promoter loop. Furthermore, these observations suggest that looping interactions are either transient or that eRNAs are preferentially transcribed before enhancer–promoter loops are established (Figure 5C).
Figure 5.
eRNA-mRNA co-expressing alleles are infrequently found in a closed enhancer–promoter loop configuration. (A) Determination of the upper limit of co-localization detection (63 nm) using smFISH probes spanning the P2RY2 5΄ intron labeled in two different colors and 2D Gaussian fitting. (B) Frequency distribution plots displaying the distances between mRNA transcription sites and nascent eRNAs within a 400 nm radius (red) overlaid with the localization precision plot (green) shown in (A). The data represent the 40 min E2 induction point combined from three independent experiments. (C) Cartoon illustrating the spatial organization of nascent eRNAs relative to the transcription site. Three different scenarios of eRNA and target mRNA co-expression are observed at single alleles. (i) Simultaneous eRNA and mRNA transcription consistent with a closed enhancer–promoter loop conformation; least frequent. (ii) Simultaneous eRNA and mRNA transcription in an open enhancer-promoter loop conformation, and (iii) mRNA transcription from alleles that are not co-expressed with an eRNA; most prevalent. Factors involved in enhancer-promoter communication are shown schematically.
DISCUSSION
Our study reveals important insights into eRNA expression that are unique to the use of single-cell approaches, and complements previous investigations of eRNA function that have largely relied on ensemble measurements. Population-level studies are limited in their ability to reveal the differential behavior of cells within a population, and cannot distinguish whether individual alleles show alternate transcriptional activity. Therefore, such studies typically report average expression profiles that are limited in their scope to accurately describe what occurs in individual cells and at individual alleles. Transcriptional responses induced by external stimuli have been shown to be particularly noisy, as gene activation depends on a cascade of events, such as signal sensing, transcription factor binding and chromatin remodeling (27,29). Our analysis of E2-mediated transcription kinetics, previously described as a strong and synchronous transcriptional response, showed highly variable induction in individual cells, both for mRNA and eRNA expression. Even at the peak of E2 induction (40 min), one third of all the cells did not exhibit strong FOXC1 and P2RY2 mRNA transcription, suggesting that only a fraction of the cells respond to the stimulus at any given time. Furthermore, individual active alleles showed variable transcriptional strength, as measured by the number of nascent mRNAs, consistent with a bursting expression pattern.
Interestingly, although the eRNA and mRNA transcription frequencies increase over the time course of E2 induction, we did not observe an increase in eRNA signal intensity at individual alleles as observed for mRNAs (Supplementary Figure S7). Typically, an increase in transcription initiation frequency results in the association of multiple nascent RNAs with a locus. The ability to detect nascent mRNAs by smFISH depends not only on the initiation frequency, but also on the length of the gene, on the elongation velocity, and on the kinetics of release of an RNA from chromatin. Although eRNA transcription units are shorter than those of mRNAs, strong transcriptional bursting at enhancers should result in an increase in eRNA signal intensity, which was not observed over the entire time course examined. Furthermore, if eRNAs were infrequently initiated and released shortly after transcription termination, smFISH probes would hybridize to partially transcribed eRNAs, yielding signals of variable intensities. Northern blot analyses have demonstrated that these transcripts are detected as bands of discrete size, which correlate well with the uniform eRNA intensities measured by smFISH and imply that most eRNAs are present as fully synthesized transcripts of defined length (16,21,22). Additionally, fractionation experiments revealed that eRNAs are mainly chromatin-associated, consistent with our findings that the number of eRNA signals rarely exceed the number of alleles in individual nuclei. Together, these lines of evidence indicate that eRNA transcription is infrequent and that eRNA release from chromatin is slow.
Various functions have been ascribed to eRNAs during the transcription cycle. For instance, eRNAs are thought to facilitate the transition of paused RNAPII into productive elongation by mediating the release of the negative elongation factor NELF (24). If eRNAs were required for RNAPII pause release at target alleles, we would predict to observe comparable frequencies of eRNA and mRNA expression, since every cycle of mRNA transcription would require an eRNA to act as a decoy for NELF. The low transcription frequency of eRNAs observed at the FOXC1 and P2RY2 enhancers, however, does not favor such a model, at least for the genes investigated here.
Alternatively, several studies have proposed a role for eRNAs in facilitating or stabilizing enhancer-promoter loops through interactions with cohesin and the mediator complex (15,21,25). Specifically, knockdown of different eRNAs were shown to reduce enhancer–promoter contacts in 3C assays. One limitation of 3C analysis is the inability to determine whether there is heterogeneity in enhancer–promoter interactions at individual alleles within a population. DNA FISH studies have reported both transient and stable interactions between regulatory regions and promoters, depending on the system investigated (50,51). In favor of stable looping, FRAP analysis of cohesin sub-complexes in G1 measured chromatin-bound residence times of ∼24 min (52). Although it is not clear whether cohesin-stabilized enhancers–promoter interactions have a similar duration, this data suggest that such interactions could persist for many minutes (52). Our measurements of eRNA and mRNA co-expression at individual alleles revealed two interesting features: (i) that co-expressing alleles are rare, and (ii) that co-expression infrequently occurs in a closed enhancer–promoter configuration. One interpretation of this observation is that enhancer–promoter interactions are dynamic. Recent studies show compartmentalization of genes and their regulatory regions in topologically associating domains (TADs), which position enhancers and promoters in proximity. Thus, TADs are thought to facilitate contacts between regulatory regions and target gene promoters, and to circumvent the need for stable loops (49,50). Alternatively, the infrequent co-expression of eRNAs and mRNAs in a closed-loop configuration could indicate that eRNA transcription precedes looping or that eRNA transcription is mostly inhibited in the closed loop configuration (Figure 5C). Positioning of ERα, co-activators, the mediator complex and the basal transcription machinery in a closed loop configuration could indeed facilitate mRNA over eRNA transcription. Therefore, while the accumulation of eRNAs at active enhancers is unlikely to be required to stabilize enhancer–promoter loops, eRNA transcription may, nonetheless, initiate this communication, which promotes target gene activation.
Lastly, our data show that eRNA transcription at the FOXC1 and P2RY2 enhancers occurs at a basal level even in the absence of MLL1 and ERα. An additional role of eRNAs in mediating enhancer–promoter communication suggests that the process of transcription at enhancers modifies chromatin, thereby facilitating the binding of transcription factors and co-activators (46). Such low-level transcription at enhancers could maintain this regulatory region in an open chromatin state, and allow cells to respond rapidly to E2 stimulation. In support of this idea, we found that knockdown of MLL1 reduced both the level of H3K4me1 and the binding of ERα to the FOXC1 and P2RY2 enhancers. Enhancer-mediated gene activation may be characterized by a feed-forward loop, whereby basal eRNA transcription facilitates the recruitment of TFs and co-regulators, which then further remodel chromatin and increase the frequency of eRNA and mRNA transcription initiation. Understanding how eRNAs and TFs act either jointly or independently to stimulate transcription will require targeted deletion of different enhancer regions to alter eRNA structure or abolish eRNA expression, while maintaining the binding of ERα.
In conclusion, our smFISH analysis of eRNA and mRNA expression patterns over a time course of E2 induction showed that the majority of eRNAs are not co-localized with active mRNA transcription sites, and vice versa, implying that eRNA and mRNA transcription is rarely coupled on individual alleles. The lack of co-localization of eRNAs with a TS may reflect roles at an early stage of transcription initiation, when the enhancer is primed for activating target gene expression. This observation is consistent with previous studies showing that the onset of eRNA transcription precedes target gene activation (9,11). Reciprocally, the absence of eRNAs at TSs indicates that eRNAs are not required to sustain bursting transcription. Furthermore, the lack of accumulation of eRNAs at enhancers upon E2 induction suggests that transcription of these noncoding RNAs is initiated at low frequency, and that eRNAs act transiently during the early stages of activation, possibly to alter the local chromatin environment, facilitate transcription factor access or to initiate enhancer–promoter communication. Notably, alleles that co-express eRNAs and mRNAs show a broad distribution of enhancer–promoter configurations, in which eRNAs are rarely transcribed within distances compatible with direct interactions. This suggests that while eRNAs may initiate enhancer–promoter communication, their transcription is mostly repressed once looping interactions are established. Elucidating the dynamics of enhancer–promoter interactions in the presence or absence of eRNAs will require live cell analyses at high spatial and temporal resolution.
Supplementary Material
ACKNOWLEDGEMENTS
We thank F. Robert, M. Oeffinger, N. Francis, D. Scott, P. Raymond and P. Bensidoun for helpful discussions. We are grateful to J. Kulpa and S. Haidar for technical advice and to W. Li and Q. Ma for providing eRNA sequences.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Bourse FESP from Université de Montréal (to S.R.); Canadian Imperial Bank of Commerce Breast Cancer Chair at Université de Montréal (to S.M.); Canadian Institute for Health Research [MOP-BMB-232642, Project Grant-366682 to D.Z., MOP-288154, Project Grant-125863 to S.M.]; Canadian Foundation for Innovation (to D.Z.). Funding for open access charge: Canadian Institute for Health Research.
Conflict of interest statement. None declared.
REFERENCES
- 1. Bulger M., Groudine M.. Enhancers: the abundance and function of regulatory sequences beyond promoters. Dev. Biol. 2010; 339:250–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Deng W., Rupon J.W., Krivega I., Breda L., Motta I., Jahn K.S., Reik A., Gregory P.D., Rivella S., Dean A. et al. Reactivation of developmentally silenced globin genes by forced chromatin looping. Cell. 2014; 158:849–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Deng W., Lee J., Wang H., Miller J., Reik A., Gregory P.D., Dean A., Blobel G.A.. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell. 2012; 149:1233–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Marsman J., Horsfield J.A.. Long distance relationships: enhancer–promoter communication and dynamic gene transcription. Biochim. Biophys. Acta. 2012; 1819:1217–1227. [DOI] [PubMed] [Google Scholar]
- 5. Heintzman N.D., Stuart R.K., Hon G., Fu Y., Ching C.W., Hawkins R.D., Barrera L.O., Van Calcar S., Qu C., Ching K.A. et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007; 39:311–318. [DOI] [PubMed] [Google Scholar]
- 6. Rada-Iglesias A., Bajpai R., Swigut T., Brugmann S.A., Flynn R.A., Wysocka J.. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011; 470:279–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Natoli G., Andrau J.C.. Noncoding transcription at enhancers: general principles and functional models. Annu. Rev. Genet. 2012; 46:1–19. [DOI] [PubMed] [Google Scholar]
- 8. Kim T.K., Hemberg M., Gray J.M., Costa A.M., Bear D.M., Wu J., Harmin D.A., Laptewicz M., Barbara-Haley K., Kuersten S. et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010; 465:182–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Arner E., Daub C.O., Vitting-Seerup K., Andersson R., Lilje B., Drablos F., Lennartsson A., Ronnerblad M., Hrydziuszko O., Vitezic M. et al. Gene regulation. Transcribed enhancers lead waves of coordinated transcription in transitioning mammalian cells. Science. 2015; 347:1010–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Orom U.A., Shiekhattar R.. Long noncoding RNAs usher in a new era in the biology of enhancers. Cell. 2013; 154:1190–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. De Santa F., Barozzi I., Mietton F., Ghisletti S., Polletti S., Tusi B.K., Muller H., Ragoussis J., Wei C.L., Natoli G.. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 2010; 8:e1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang D., Garcia-Bassets I., Benner C., Li W., Su X., Zhou Y., Qiu J., Liu W., Kaikkonen M.U., Ohgi K.A. et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011; 474:390–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hah N., Danko C.G., Core L., Waterfall J.J., Siepel A., Lis J.T., Kraus W.L.. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell. 2011; 145:622–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hah N., Murakami S., Nagari A., Danko C.G., Kraus W.L.. Enhancer transcripts mark active estrogen receptor binding sites. Genome Res. 2013; 23:1210–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li W., Notani D., Ma Q., Tanasa B., Nunez E., Chen A.Y., Merkurjev D., Zhang J., Ohgi K., Song X. et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013; 498:516–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Melo C.A., Drost J., Wijchers P.J., van de Werken H., de Wit E., Oude Vrielink J.A., Elkon R., Melo S.A., Leveille N., Kalluri R. et al. eRNAs are required for p53-dependent enhancer activity and gene transcription. Mol. cell. 2013; 49:524–535. [DOI] [PubMed] [Google Scholar]
- 17. Lam M.T., Cho H., Lesch H.P., Gosselin D., Heinz S., Tanaka-Oishi Y., Benner C., Kaikkonen M.U., Kim A.S., Kosaka M. et al. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature. 2013; 498:511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Johnsson P., Lipovich L., Grander D., Morris K.V.. Evolutionary conservation of long non-coding RNAs; sequence, structure, function. Biochim. Biophys. Acta. 2014; 1840:1063–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Andersson R., Gebhard C., Miguel-Escalada I., Hoof I., Bornholdt J., Boyd M., Chen Y., Zhao X., Schmidl C., Suzuki T. et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014; 507:455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lai F., Gardini A., Zhang A., Shiekhattar R.. Integrator mediates the biogenesis of enhancer RNAs. Nature. 2015; 525:399–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hsieh C.L., Fei T., Chen Y., Li T., Gao Y., Wang X., Sun T., Sweeney C.J., Lee G.S., Chen S. et al. Enhancer RNAs participate in androgen receptor-driven looping that selectively enhances gene activation. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:7319–7324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pnueli L., Rudnizky S., Yosefzon Y., Melamed P.. RNA transcribed from a distal enhancer is required for activating the chromatin at the promoter of the gonadotropin alpha-subunit gene. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:4369–4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mousavi K., Zare H., Dell'orso S., Grontved L., Gutierrez-Cruz G., Derfoul A., Hager G.L., Sartorelli V.. eRNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Mol. Cell. 2013; 51:606–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schaukowitch K., Joo J.Y., Liu X., Watts J.K., Martinez C., Kim T.K.. Enhancer RNA facilitates NELF release from immediate early genes. Mol. Cell. 2014; 56:29–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lai F., Orom U.A., Cesaroni M., Beringer M., Taatjes D.J., Blobel G.A., Shiekhattar R.. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature. 2013; 494:497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sigova A.A., Abraham B.J., Ji X., Molinie B., Hannett N.M., Guo Y.E., Jangi M., Giallourakis C.C., Sharp P.A., Young R.A.. Transcription factor trapping by RNA in gene regulatory elements. Science. 2015; 350:978–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Raj A., Peskin C.S., Tranchina D., Vargas D.Y., Tyagi S.. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 2006; 4:e309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sanchez A., Golding I.. Genetic determinants and cellular constraints in noisy gene expression. Science. 2013; 342:1188–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zenklusen D., Larson D.R., Singer R.H.. Single-RNA counting reveals alternative modes of gene expression in yeast. Nat. Struct. Mol. Biol. 2008; 15:1263–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chubb J.R., Trcek T., Shenoy S.M., Singer R.H.. Transcriptional pulsing of a developmental gene. Curr. Biol.: CB. 2006; 16:1018–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Suter D.M., Molina N., Gatfield D., Schneider K., Schibler U., Naef F.. Mammalian genes are transcribed with widely different bursting kinetics. Science. 2011; 332:472–474. [DOI] [PubMed] [Google Scholar]
- 32. Evans R.M., Mangelsdorf D.J.. Nuclear receptors, RXR, and the big bang. Cell. 2014; 157:255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sanchez R., Nguyen D., Rocha W., White J.H., Mader S.. Diversity in the mechanisms of gene regulation by estrogen receptors. Bioessays. 2002; 24:244–254. [DOI] [PubMed] [Google Scholar]
- 34. Jeong K.W., Andreu-Vieyra C., You J.S., Jones P.A., Stallcup M.R.. Establishment of active chromatin structure at enhancer elements by mixed-lineage leukemia 1 to initiate estrogen-dependent gene expression. Nucleic Acids Res. 2014; 42:2245–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lupien M., Eeckhoute J., Meyer C.A., Wang Q., Zhang Y., Li W., Carroll J.S., Liu X.S., Brown M.. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008; 132:958–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ansari K.I., Mishra B.P., Mandal S.S.. Human CpG binding protein interacts with MLL1, MLL2 and hSet1 and regulates Hox gene expression. Biochim. Biophys. Acta. 2008; 1779:66–73. [DOI] [PubMed] [Google Scholar]
- 37. Rezai-Zadeh N., Zhang X., Namour F., Fejer G., Wen Y.D., Yao Y.L., Gyory I., Wright K., Seto E.. Targeted recruitment of a histone H4-specific methyltransferase by the transcription factor YY1. Genes Dev. 2003; 17:1019–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rozenblatt-Rosen O., Rozovskaia T., Burakov D., Sedkov Y., Tillib S., Blechman J., Nakamura T., Croce C.M., Mazo A., Canaani E.. The C-terminal SET domains of ALL-1 and TRITHORAX interact with the INI1 and SNR1 proteins, components of the SWI/SNF complex. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:4152–4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bourdeau V., Deschenes J., Laperriere D., Aid M., White J.H., Mader S.. Mechanisms of primary and secondary estrogen target gene regulation in breast cancer cells. Nucleic Acids Res. 2008; 36:76–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Thompson R.E., Larson D.R., Webb W.W.. Precise nanometer localization analysis for individual fluorescent probes. Biophys. J. 2002; 82:2775–2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Raj A., van den Bogaard P., Rifkin S.A., van Oudenaarden A., Tyagi S.. Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods. 2008; 5:877–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Buxbaum A.R., Wu B., Singer R.H.. Single beta-actin mRNA detection in neurons reveals a mechanism for regulating its translatability. Science. 2014; 343:419–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Deschenes J., Bourdeau V., White J.H., Mader S.. Regulation of GREB1 transcription by estrogen receptor alpha through a multipartite enhancer spread over 20 kb of upstream flanking sequences. J. Biol. Chem. 2007; 282:17335–17339. [DOI] [PubMed] [Google Scholar]
- 44. Liu Z., Merkurjev D., Yang F., Li W., Oh S., Friedman M.J., Song X., Zhang F., Ma Q., Ohgi K.A. et al. Enhancer activation requires trans-recruitment of a mega transcription factor complex. Cell. 2014; 159:358–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shang Y., Hu X., DiRenzo J., Lazar M.A., Brown M.. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000; 103:843–852. [DOI] [PubMed] [Google Scholar]
- 46. Kaikkonen M.U., Spann N.J., Heinz S., Romanoski C.E., Allison K.A., Stender J.D., Chun H.B., Tough D.F., Prinjha R.K., Benner C. et al. Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Mol. Cell. 2013; 51:310–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Milne T.A., Dou Y., Martin M.E., Brock H.W., Roeder R.G., Hess J.L.. MLL associates specifically with a subset of transcriptionally active target genes. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:14765–14770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bahar Halpern K., Tanami S., Landen S., Chapal M., Szlak L., Hutzler A., Nizhberg A., Itzkovitz S.. Bursty gene expression in the intact mammalian liver. Mol. Cell. 2015; 58:147–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dekker J., Marti-Renom M.A., Mirny L.A.. Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat. Rev. Genet. 2013; 14:390–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Giorgetti L., Galupa R., Nora E.P., Piolot T., Lam F., Dekker J., Tiana G., Heard E.. Predictive polymer modeling reveals coupled fluctuations in chromosome conformation and transcription. Cell. 2014; 157:950–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ghavi-Helm Y., Klein F.A., Pakozdi T., Ciglar L., Noordermeer D., Huber W., Furlong E.E.. Enhancer loops appear stable during development and are associated with paused polymerase. Nature. 2014; 512:96–100. [DOI] [PubMed] [Google Scholar]
- 52. Gerlich D., Koch B., Dupeux F., Peters J.M., Ellenberg J.. Live-cell imaging reveals a stable cohesin-chromatin interaction after but not before DNA replication. Curr. Biol.: CB. 2006; 16:1571–1578. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.