Abstract
In situ fabricated nucleic acids microarrays are versatile and very high-throughput platforms for aptamer optimization and discovery, but the chemical space that can be probed against a given target has largely been confined to DNA, while RNA and non-natural nucleic acid microarrays are still an essentially uncharted territory. 2΄-Fluoroarabinonucleic acid (2΄F-ANA) is a prime candidate for such use in microarrays. Indeed, 2΄F-ANA chemistry is readily amenable to photolithographic microarray synthesis and its potential in high affinity aptamers has been recently discovered. We thus synthesized the first microarrays containing 2΄F-ANA and 2΄F-ANA/DNA chimeric sequences to fully map the binding affinity landscape of the TBA1 thrombin-binding G-quadruplex aptamer containing all 32 768 possible DNA-to-2΄F-ANA mutations. The resulting microarray was screened against thrombin to identify a series of promising 2΄F-ANA-modified aptamer candidates with Kds significantly lower than that of the unmodified control and which were found to adopt highly stable, antiparallel-folded G-quadruplex structures. The solution structure of the TBA1 aptamer modified with 2΄F-ANA at position T3 shows that fluorine substitution preorganizes the dinucleotide loop into the proper conformation for interaction with thrombin. Overall, our work strengthens the potential of 2΄F-ANA in aptamer research and further expands non-genomic applications of nucleic acids microarrays.
INTRODUCTION
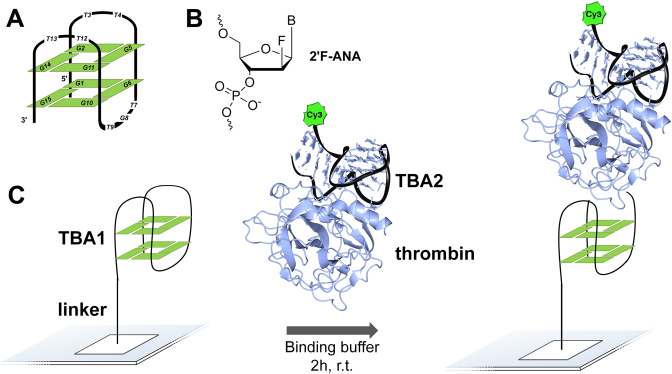
G-quadruplexes are naturally-occurring nucleic acid motifs consisting of stacked guanine tetrads stabilized by monovalent cations. Found in telomeres and oncogene promoter regions, they are believed to play multiple roles in the maintenance and the regulation of gene expression (1,2). G-quadruplex inhibition of telomerase has sparked strong interest as a target in anticancer drug discovery (3,4). The protein-binding spectrum of G-quadruplexes also extends to thrombin, a central enzyme in the process of blood coagulation, which certain quadruplex topologies have been found to bind to and inactivate (5,6). One sequence in particular, the 15mer 5΄-GGTTGGTGTGGTTGG (referred to as TBA1) folds into a unimolecular, chair-like conformation featuring two stacked G-tetrads. Each tetrad is composed of syn- and anti-oriented guanines in an alternating fashion. The strands are aligned in an antiparallel manner and connected by three loops, two of which are TT dimers that contact the fibrinogen binding site of thrombin (Figure 1A), as shown in the crystal structure of the TBA1·thrombin complex (7) and whose Kd was reported at 75–100 nM (8). TBA2, a 29mer showing structural features from both duplex and quadruplex conformations, binds to thrombin with even higher affinity (Kd = 0.5 nM (8)) and, relevantly, to a different epitope than TBA1 (9). As sequences whose structural, biophysical and binding properties are well characterized, TBA1 and TBA2 have become model aptamers and, as such, have been widely investigated and subjected to a plethora of chemical modifications (10,11), for the most part in TBA1, however, with little success in improving the thrombin-binding properties of the original oligomer.
Figure 1.
(A) Schematic representation of TBA1 in a chair-like, antiparallel G-quadruplex conformation. (B) Chemical structure of 2΄-Fluoroarabinonucleic acid (2΄F-ANA). (C) Overview of the ‘sandwich’ assay on folded TBA1 arrays using thrombin and a second aptamer, TBA2, labelled with Cy3.
One modification that stands out is 2΄-deoxy-2΄-fluoroarabinonucleic acid (2΄F-ANA), a well-studied derivative of arabinose with DNA-like character (12,13), with particular relevance in oligonucleotide-based therapeutic strategies (14–16) and in the study of non-canonical structures, particularly G-quadruplexes (17–19). The introduction of 2΄F-araG nucleotides at anti-oriented dG positions in TBA1 was found to increase the stability of the resulting quadruplex while keeping an antiparallel chair-like folding pattern. Furthermore, replacing dT units with 2΄F-araT has, in a few select cases, improved the binding affinity towards thrombin (19). These encouraging observations prompted us to consider the systematic substitution of all deoxynucleotides in TBA1 with 2΄F-aranucleotides and to examine the effect of each modification on the thrombin-binding ability of TBA1. The number of all possible DNA-to-2΄F-ANA permutations in TBA1 amounts to 215 (32 768) unique sequences; while certainly a formidable endeavour by standard solid-phase synthesis, this number is easily within the reach of in situ photolithography microarray synthesis.
Maskless photolithography is an in situ array fabrication technique that provides a very high number of unique, spatially-defined sequences (‘features’), up to >2 million per microarray (20,21). Microarray oligonucleotides are fabricated using a slightly modified version of the familiar phosphoramidite synthesis cycle. The main difference is that an array of computer-controlled micromirrors within an optical imaging system delivers UV light to defined positions on the surface, triggering photodeprotection of the terminal 5΄ hydroxyl groups, allowing coupling reactions to proceed at selected locations in the next cycle. The overwhelming majority of oligonucleotide arrays are used as platforms to interrogate the relative abundance of DNA or RNA sequences in gene expression experiments (22). However, the technology is well suited to reach beyond genomics applications and has emerged as a very high throughput technique for aptamer study and discovery (23), allowing for the resolution of binding motifs and the evaluation of binding affinity as well. Common study targets on microarray platforms include DNA- and RNA-binding proteins (24–26), and thrombin in particular (27,28), but also other classes of DNA ligands (29).
We present herein the first in situ synthesis of 2΄F-ANA and 2΄F-ANA/DNA chimeric microarrays, and their use in fully mapping the binding affinity landscape of the DNA to 2΄F-ANA permutation space of the TBA1 thrombin-binding aptamer. A fluorescently labelled TBA2 DNA aptamer is used in the experiments to perform ‘sandwich’-like thrombin-binding assays (30–36). Our method not only provides a complete picture of the binding landscape of 2΄F-ANA-containing TBA1, but also demonstrates the potential of highly complex microarrays of natural and chemically modified nucleic acids in the exploration and discovery of novel binding patterns and affinities.
MATERIALS AND METHODS
Monomer synthesis
All solvents and reagents were purchased from Sigma-Aldrich and used without further purification. THF was dried using an MB-SPS-800 solvent dryer (MBraun) and pyridine was dried over 4 Å activated molecular sieves. Thin layer chromatography was performed on EM Science Kieselgel 60 F254 (1 mm) plates and flash chromatography on Silicycle 40–63 μm (230–400 mesh) silica gel. 1H-NMR, 13C-NMR, 19F-NMR and 31P-NMR spectra were recorded on a Varian 500 MHz spectrophotometer with chemical shift values reported in ppm. 1H-NMR and 13C-NMR spectra were referenced to residual solvent. 31P-NMR were measured from 85% H3PO4 as an external standard.
Microarray design
The sequences to be synthesized on each array are listed in a text file, which is then uploaded into a MatLab (MathWorks) program, which generates the virtual masks and coupling order. Sequences were distributed randomly throughout the synthesis area to reduce spatial artefacts. Each microarray was designed with positive and negative control sequences and includes empty features to serve as a background reference. All possible DNA to 2΄F-ANA permutations of TBA1 were generated using Excel with a built-in ‘Mix and Match’ macro. The negative control in TBA1 arrays has the sequence 5΄-AATGGTAGTTTAAAT-3΄. An array layout with ∼191 000 features was chosen, allowing for the synthesis of 5–6 replicates of each permutation. For the design of the microarrays containing the 178 best aptamer candidates, a four-part array layout was selected. This layout results in the synthesis of four small identical sub-arrays, within the footprint of a full array, each containing 2340 features. These sub-arrays can then be independently exposed to thrombin. Each sub-array contained 13 replicates of each of the 178 best aptamers.
Microarray synthesis and deprotection
The fabrication of microarrays followed the protocols for maskless array synthesis as already described elsewhere (37,38). In the dual array synthesis system, the bottom slide was first drilled with a 0.9 mm diamond bit, washed and rinsed in an ultrasonic bath (39). Both top and bottom slides (Schott Nexterion Glass D) were then functionalized using N-(3-triethoxysilylpropyl)-4-hydroxybutyramide (Gelest SIT8189.5, 10g) in 500 ml of a 95:5 (v/v) solution of EtOH/H2O containing 1 ml of acetic acid for 4h at room temperature under gentle agitation. After washing in EtOH/H2O 95:5 +0.2% AcOH (2 × 500 ml, 20 min each), the slides were dried and cured in a preheated vacuum oven at 120°C overnight, after which they were stored in a desiccator cabinet until use.
The maskless array synthesis consists of an optical imaging system that uses a digital micromirror device (DMD, 1024 × 768 micromirrors) to deliver patterned 365 nm ultraviolet light onto the surface of the silanized slides, where synthesis takes place. A high-power 365 nm UV-LED (Nichia NVSU333A) serves as a light source (40). The layout of the array is determined by the pattern of UV irradiation, which triggers the selective removal of the photolabile 2-(2-nitrophenyl)-propyloxycarbonyl (NPPOC) 5΄-OH protecting group. Light exposure is performed with a radiant energy density of 6 J/cm2, with typical exposure times of 60 s at an irradiance of 100 mW/cm2. Reagent delivery and exposure to UV light are synchronized and controlled by a computer coupled to an Expedite 8909 synthesizer (PerSeptive Biosystems), which controls chemistry delivery to the array synthesis chamber. The cycle-based phosphoramidite chemistry is, other than NPPOC removal under UV irradiation, similar to that of standard solid-phase oligonucleotide synthesis. 5΄-NPPOC dT/2΄F-araT, dG (N2-isopropylphenoxyacetyl, iPrPac) and 2΄F-araG (N2-iPrPac) phosphoramidites were used. For the coupling step, amidites were prepared as 0.03 M solutions in dry acetonitrile and the coupling step was set to 15 s for DNA and 10 min for 2΄F-ANA. After synthesis, microarrays were deprotected at r.t. for 2 h in a 1:1 mixture of ethylenediamine (EDA)/EtOH (50 ml in staining glass jar), washed twice with deionized water then dried by blowing a stream of argon and stored in a desiccator cabinet.
Microarray hybridization
Complementary high pressure (or high performance) liquid chromatography-purified 5΄-Cy3-labelled oligonucleotides (rA18 for poly-dT arrays and r(AC)6 for (TG)6 arrays) were obtained from IDT or Eurogentec. Deprotected microarrays were hybridized in a self-adhesive chamber (Grace Bio-Labs SA200) filled with a 300 μl solution of 10 nM 5΄-Cy3 labelled complementary strand in hybridization buffer (0.1 M 2-(N-morpholino)ethanesulfonic acid, 0.9 M NaCl, 20 mM ethylenediaminetetraacetic acid, 0.01% Tween20, 0.05% bovine serum albumine (BSA)) at 4°C for poly-dT arrays and at room temperature in a hybridization oven (Boekel Scientific) at a slow rotation rate for (TG)6 arrays. After 2 h, the chamber was removed and the arrays were washed first in Non-Stringent Wash buffer (SSPE; 0.9 M NaCl, 0.06 M phosphate, 6 mM ethylenediaminetetraacetic acid, 0.01% Tween20) for 2 min, in Stringent Wash Buffer (100 mM 2-(N-morpholino)ethanesulfonic acid, 0.1 M NaCl, 0.01% Tween20) for 1 min and then briefly dipped in Final Wash Buffer (0.1X sodium saline citrate). The microarrays were then dried on a microarray centrifuge and scanned at 5 μm resolution with an excitation wavelength of 532 nm in a microarray scanner (GenePix 4100A, Molecular Devices). Fluorescence intensities are reported as arbitrary units (a.u.). The scanned images were processed using NimbleScan (Roche NimbleGen) and the extracted data were analysed using Excel.
Measure of coupling efficiency
The coupling efficiency of 2΄F-araT and 2΄F-araG was measured using the method of terminal labelling. Oligomers of 2΄F-araT or 2΄F-araG of various lengths (1 to 12mers) were fabricated on the same microarray and labelled with a terminal 5΄-Cy3. A capping step followed each coupling of 2΄F-araT or 2΄F-araG and was performed by coupling DMTr-dT phosphoramidite (0.03 M) for 60 s. The terminal labelling consisted in two consecutive coupling events of 300 s with Cy3 phosphoramidite at a 0.05 M concentration. After synthesis, the arrays were washed in acetonitrile at room temperature for 2 h, dried then scanned and the data extracted using NimbleScan. The decrease in fluorescence intensity as the length of the oligomer increases reflects the coupling efficiency of the 2΄F-ANA monomers and follows the mathematical model of an exponential decay. The extracted data were first normalized and then plotted in SigmaPlot 11.0 (Systat Software) and the resulting curve was fit to the model curve 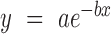 where y is the fluorescence intensity, x the number of couplings, a is the maximum intensity and b corresponds to the fractional stepwise coupling yield.
where y is the fluorescence intensity, x the number of couplings, a is the maximum intensity and b corresponds to the fractional stepwise coupling yield.
Thrombin-binding assays and identification of aptamer candidates
To record and measure the binding of thrombin to G-quadruplex chips, a ‘sandwich’ assay was employed whereby a secondary, Cy3-labelled G-quadruplex aptamer, referred to as TBA2 (5΄-AGT CCG TGG TAG GGC AGG TTG GGG TGA CT-3΄) binds to the heparin binding site of thrombin with very high affinity (Kd = 0.5 nM (8)). A 10 μM stock solution of Cy3-TBA2 (Eurogentec) in 100 mM KCl was first heated up to 90°C for 10 min and slowly cooled down to r.t. overnight to achieve correct folding of the Cy3-TBA2 aptamer. All 32 768 sequences in the TBA1 arrays were also folded in binding buffer (20 mM Tris, 5 mM KCl, 140 mM NaCl, 1 mM MgCl2 pH 7.5 supplemented with 0.5% BSA and 0.01% Tween20) for 2 h at r.t. Meanwhile, the Cy3-TBA2·thrombin complex was assembled by mixing human thrombin (Sigma-Aldrich, T1063, 200 nM) and Cy3-TBA2 (400 nM) in binding buffer and left to bind at r.t. overnight. The Cy3-TBA2·thrombin complex solution was then transferred to a self-adhesive chamber over the TBA1 array and the resulting mix was allowed to bind for 2 h at r.t. in a hybridization oven at a slow rotation rate. The chamber was then removed and the array was briefly washed three times in phosphate buffered saline buffer (10 mM Na2HPO4, 1.8 mM KH2PO4, 140 mM NaCl, 0.01% Tween20), dried by blowing a stream of argon and scanned at 2.5 μm resolution using an excitation wavelength of 532 nm in a microarray scanner (GenePix 4400A, Molecular Devices). Scanned images were processed using NimbleScan and Excel. The aptamer candidates were selected after a binding assay at an initial thrombin concentration of 200 nM, significantly above the Kd of the unmodified TBA1. The recorded fluorescence intensities were ordered from lowest to highest (unmodified TBA1 = 640 a.u.) and a cut-off was set at 1000, above which any sequence was surmised to bind to thrombin with higher affinity than the control TBA1. Partial analysis of the data based on the type, the number or the location of 2΄F-ANA modifications was done in Excel by applying various filters. To calculate the number of r combinations of 2΄F-ANA modifications in a sequence of length n, the following equation was used: 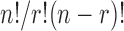 . To graphically represent the proportion of DNA or 2΄F-ANA nucleotide found at each position of the 15mer in a set of sequences, sequence logos can be created from this set and displayed in the form of a consensus sequence. Sequence logos were generated by WebLogo (weblogo.berkeley.edu (41)) using the following descriptors: dT = T; 2΄F-araT = U; dG = G and 2΄F-araG = A. Letters U and A were then replaced with fT and fG, respectively.
. To graphically represent the proportion of DNA or 2΄F-ANA nucleotide found at each position of the 15mer in a set of sequences, sequence logos can be created from this set and displayed in the form of a consensus sequence. Sequence logos were generated by WebLogo (weblogo.berkeley.edu (41)) using the following descriptors: dT = T; 2΄F-araT = U; dG = G and 2΄F-araG = A. Letters U and A were then replaced with fT and fG, respectively.
Thrombin-binding assays and determination of binding constants
Thrombin-binding assays were repeated at various thrombin concentrations (5, 10, 20, 50, 100 and 200 nM and a corresponding 2X excess of Cy3-TBA, i.e. 10, 20, 100, 200 and 400 nM, respectively) for TBA1 arrays containing all aptamer candidates. All experiments were performed in triplicate as described above. The recorded fluorescence values for each sequence was corrected for background, normalized and plotted as a function of thrombin concentration in SigmaPlot. The dissociation constant was then extracted by fitting a four-parameter logistic curve 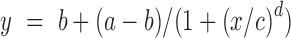 to the experimental curve where y is the fluorescence intensity, b the maximum fluorescence, a the minimum fluorescence, x the thrombin concentration, c the concentration yielding a fluorescence of 0.5 and d the slope factor.
to the experimental curve where y is the fluorescence intensity, b the maximum fluorescence, a the minimum fluorescence, x the thrombin concentration, c the concentration yielding a fluorescence of 0.5 and d the slope factor.
Oligonucleotide synthesis and purification
Oligonucleotide synthesis was performed on an ABI 3400 DNA synthesizer from Applied Biosystems at 1 μmol scale on Unylinker (ChemGenes) CPG solid support. Thymidine (dT) and deoxyguanosine (ibu) (dG) phosphoramidites were used at 0.1 M concentration in acetonitrile, and coupled for 110 s and 300 s, respectively. 2΄F-araT and 2΄F-araG were used at 0.11 M and 0.13 M concentration respectively and coupled for 600 s. After completion of the synthesis, CPG was transferred to a 1.5 ml screw-cap eppendorf. One millilitre aqueous ammonium hydroxide was added and the eppendorf was placed on a shaker at room temperature for 48 h or at 55°C for 16 h. The deprotection solution was centrifuged and decanted from the CPG. Samples were vented for 60 min, chilled in dry ice and evaporated to dryness. Crude oligonucleotides were purified by preparative denaturing polyacrylamide gel electrophoresis using 24% polyacrylamide gels and 0.5x TBE running buffer. Bands were extracted and left overnight on the shaker in autoclaved DEPC-treated Millipore water. Samples were then filtered, lyophilized to dryness and desalted with NAP-25 Sephadex columns according to the manufacturer's protocol. The extinction coefficient of the 2΄F-araT and 2΄F-araG was similar to that of the unmodified sequence. Masses were verified by ESI-MS.
Thermal melting temperature experiments
UV thermal denaturation data were obtained on a Varian CARY 100 UV-visible spectrophotometer equipped with a Peltier temperature controller. Thrombin-binding aptamers were dissolved in 10 mM Tris, pH 6.8 with and without 25 mM KCl at a final concentration of 8 μM. Concentrations were determined after quantitating the samples by UV absorbance at λ = 260 nm. Samples were annealed in Tm buffer at 90°C for 15 min, then cooled slowly to room temperature, and stored at 5°C at least 12 h before the measurements were performed. Denaturation curves were acquired at 295 nm at a rate of 0.5°C/min. Samples were kept under a nitrogen flow at temperatures below 12°C to avoid condensation. The dissociation temperatures were calculated as the midpoint of the transition (Tm) values using the first derivatives of the experimental data. Experiments were performed in triplicate.
Circular dichroism
Circular dichroism (CD) studies were performed at 15°C on a JASCO J-810 spectropolarimeter using a 1 mm path length cuvette. Temperature was maintained using the instrument's Peltier unit. Spectra were recorded from 350 to 230 nm at a scan rate of 100 nm·min−1 and a response time of 2.0 s with three acquisitions recorded for each spectrum. The spectra were normalized by subtraction of the background scan with buffer. Data were smoothed using the means-movement function within the JASCO graphing software. Thrombin-binding aptamers were annealed in 10 mM Tris, pH 6.8 with and without 25 mM KCl at a final concentration of 8 μM.
NMR spectroscopy and structural modelling
Samples of TBA1 and its 2΄F-araT3 modification were suspended in 200 μl of either D2O or H2O/D2O 9:1 in 10 mM potassium phosphate, 25 mM KCl, pH 7 (DNA concentration ∼1.0 mM). NMR spectra were acquired in a Bruker Avance spectrometer operating at 600 MHz, and processed with Topspin software. The NOESY spectra were acquired with mixing times of 100 and 250 ms, and the TOCSY spectra were recorded with standard MLEV 17 spin lock sequence, and 80 ms mixing time. NOESY spectra were recorded at 5°C. The spectral analysis program Sparky (42) was used for semiautomatic assignment of the NOESY cross-peaks and quantitative evaluation of the NOE intensities. Almost complete assignment of the NMR resonances could be carried out from TOCSY and NOESY experiments on the basis of the strong similarity with the NMR spectra of unmodified TBA (Table S1, Supplementary Data) (43). Interproton distance constraints were set according to NOE intensities. A list of distance constraints involving lateral loops protons is shown in Table S2 (Supplementary Data).
Structural models of 2΄F-araT3-TBA were built from the coordinates of unmodified TBA as reported in the literature (43). T3 was mutated to 2΄F-araT3, preserving the geometry of the original coordinates. The structures were first minimized in vacuo, and then immersed in a water box of around 4000 water molecules. The whole system was then submitted to the standard equilibration protocol used in our group (44) and to a run of 1 ns of constrained molecular dynamics calculation with the molecular dynamics package AMBER (45). The BSC1 force-field (46) was used to describe the DNA, and the TIP3P model was used to simulate water molecules. Force field parameters for 2΄F-araT residues were taken from previous work (47). The last 25 ps of each trajectory were averaged and then minimized. The analysis of the final structures was carried out with the program MOLMOL (48). The coordinates of the structure have been deposited to the Protein Data Bank (code 5MJX).
RESULTS
Synthesis and incorporation of NPPOC-protected 2΄F-araT and 2΄F-araG phosphoramidites into microarrays
Synthesis of NPPOC-protected 2΄F-araT and 2΄F-araG phosphoramidites
The synthesis of the photolabile 2΄F-ANA phosphoramidites took first a common route leading to the 3΄,5΄-OH 2΄F-araT and 2΄F-araG nucleosides (49). The exocyclic amine of the guanine base was then protected with a 4-isopropylphenoxyacetyl (iPrPac), as in the standard NPPOC deoxyguanosine, using iPrPac chloride (Scheme S1, Supplementary Data). The photosensitive protecting group was attached to the 5΄ position by reacting the free nucleosides with NPPOC chloride at 0°C, minimizing the formation of a 3΄-NPPOC side-product. A final phosphitylation step, employing CEP-Cl and DIPEA, afforded the corresponding phosphoramidites 6 and 7 in very high yields.
Incorporation into microarrays and evaluation of the coupling efficiency
DNA and fully-modified 2΄F-ANA oligomers of various lengths (1 to 12mers) were synthesized in parallel on microarrays using amidites 6 and 7 and each sequence was terminated with a Cy3 dye. The coupling efficiency can be extracted from the curve plotting the decrease in Cy3 intensity against the increase in sequence length. While a 1 min coupling time for 2΄F-araT and 2 min for 2΄F-araG performed poorly and only achieved an 80.5% and 92% stepwise yield, respectively (Figure S1, Supplementary Data), extending it to 10 min proved sufficient to reach near quantitative coupling (>99.9%), thus showing that multiple incorporations of photolabile 2΄F-ANA phosphoramidites are feasible.
Hybridization of 2΄F-ANA-containing sequence models on microarrays
To verify the integrity of 2΄F-ANA oligonucleotides fabricated on microarrays, we proceeded with the hybridization of sequence models to their complementary, dye-labelled strands. First, polymers of 2΄F-araT of various lengths (1 to 18 mers) were synthesized, deprotected and hybridized to 5΄-Cy3-rA18 at 4°C for 2 h. The recorded fluorescence intensities for poly-2΄F-araT:rA18 double strands even exceed those of the corresponding DNA-only sequences (Figure S2, Supplementary Data), suggesting more stable 2΄F-araT:rA duplexes than dT:rA; this observation is consistent with a previous study on similar duplexes in solution (12). Next, 2΄F-araG was incorporated in a 12mer composed of alternating dTs and dGs. Scanning after hybridization revealed a fluorescence emission for the (dT/2΄F-araG)6:r(AC)6 duplex of comparable intensity to that of the d(TG)6:r(AC)6 duplex (Figure S3, Supplementary Data), indicating correct removal of the base protecting group in 2΄F-araG, similar duplex stability and further validating the correct structures of 2΄F-ANA-containing oligonucleotides on arrays.
Synthesis of all DNA/2΄F-ANA permutations in a G-quadruplex on microarrays
Synthesis and thrombin-binding experiments
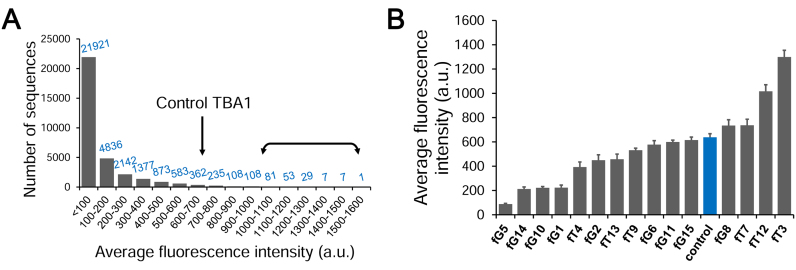
Having shown that functional arrays can be produced with 2΄F-ANA chemistry, we proceeded with the synthesis of G-quadruplexes on the high-density microarray platform. We opted for a layout with 1 91 000 features in order to span all 32 768 permutations in the 15mer TBA1 (215) on the same array while including several replicates, negative controls and empty features to be used as background reference. A spacer of ten thymidine nucleotides is fabricated first to distance the folded nucleic acid structure from the glass surface and ensure an optimal sensitivity and availability for binding (30,50). After synthesis and deprotection, folding of the quadruplexes was carried out in a potassium buffer. To bind and detect thrombin on arrays, we adopted the idea of a ‘sandwich’ assay that avoids the labelling of thrombin (Figure 1); specifically, a preformed complex of a secondary aptamer, TBA2, bound to a different, distal epitope of thrombin carries a Cy3 dye at its 5΄-end. The tightly bound complex was added over the folded quadruplex array, washed off after 2 h and the resulting TBA1·thrombin·TBA2 ternary complex was scanned. Our initial attempt looked at the response for a thrombin concentration of 200 nM, high enough to ensure binding to the unmodified control TBA1. The emitted fluorescence signals are displayed in Figure 2A, ordered from lowest to highest. A large majority of 2΄F-ANA permutations (65% of all 32 768 sequences) give rise to very low signals, close to background values, indicating very poor binding to thrombin. The fully 2΄F-ANA-modified TBA1 also performs poorly, as was already noted previously (19). Interestingly however, the signals for a significant number of sequences (1015 sequences, 3% of the total amount of permutations) exceed that of the control TBA1, indicating stronger binding affinity towards thrombin. A similar assay on blank surfaces yielded no recordable fluorescence, meaning that the Cy3-TBA2·thrombin complex does not bind non-specifically to glass. Likewise, the addition of Cy3-TBA2 alone in binding buffer over G-quadruplex arrays did not produce any signal, confirming that fluorescence emission solely arises from the interaction of the labelled complex and the folded structures. Finally, the removal of BSA, a protein commonly used to reduce non-specific binding of the target protein to the glass surface, had no effect on the relative fluorescence intensities between strong and weak thrombin binders, suggesting that sequences with higher affinity towards thrombin do not, simultaneously, display stronger affinity to BSA as well.
Figure 2.
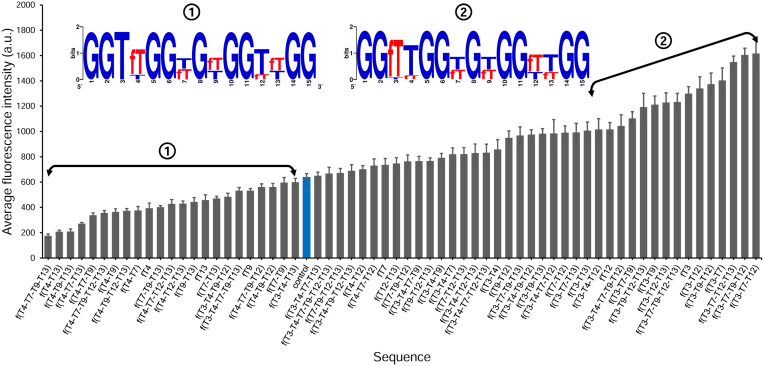
Recorded fluorescence intensities at a thrombin concentration ([Thr]) of 200 nM. (A) Number of sequences found per subrange (100 a.u.) of fluorescence. Background fluorescence is 47. The arrow denotes the subrange where the control sequence is found (fluorescence intensity of ∼640). The double arrow marks the range where high-binding sequences were identified and used for the measurement of binding affinity (178 sequences). (B) Average fluorescence intensity for each single-modified TBA1 sequence. The unmodified control is coloured blue. The error bars represent the standard error of the mean of the fluorescence signal.
Sequences containing single and double-2΄F-ANA modifications
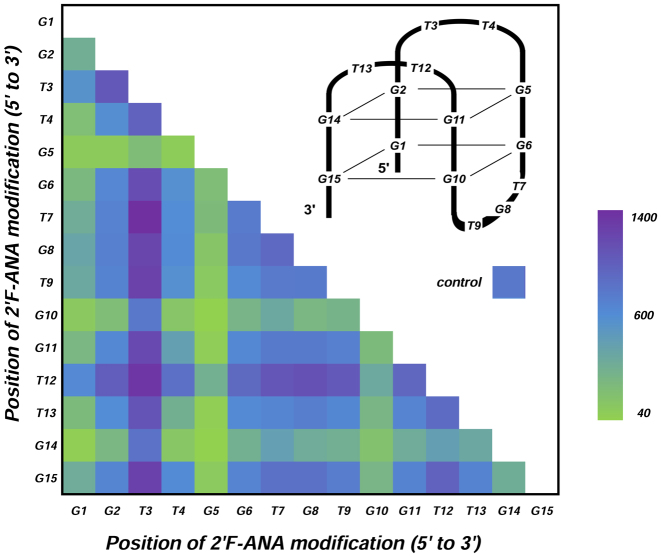
Having access to the entire chemical binding landscape of DNA to 2΄F-ANA permutations in TBA1, we reasoned that focusing on specific subsets of permutations may return relevant information about the position-dependence of 2΄F-ANA modifications on thrombin binding. Thus, we first turned our attention to sequences containing a single 2΄F-ANA modification (15 occurrences), for which the binding signals are shown in Figure 2B. The lowest intensities occur where 2΄F-ANA was incorporated at G1, G5, G10 or G14. At these positions the guanine base is usually found adopting a syn conformation, which is known to result in a steric clash between the 2΄-β-fluorine and the syn-oriented nucleobase (19,51). While most single-modified TBA1 yield binding signals comparable to that of the DNA control, two combinations stand out and give relatively high signals, one where 2΄F-ANA is at position T3, and the other at T12. Both positions are in the small TT loops directly contacting thrombin. Surprisingly, replacing the other two loop-dT units—namely T4 and T13—with 2΄F-ANA, reduces the binding affinity. We then turned to studying the influence of an additional 2΄F-ANA incorporation on the thrombin-binding affinity of singly-modified 2΄F-ANA sequences. To do so, we mapped out the fluorescence signals of all 105 possible sequences mutated with two 2΄F-ANA units. A few trends emerge from the diagram shown in Figure 3. The largest fluorescence counts are almost exclusively populated with sequences containing a 2΄F-ANA at position T3, and sequences with an additional 2΄β fluorine at positions T7 or T12 form the strongest combinations. However, a combination of 2΄F-ANA at T3 and G5 dramatically decreases binding, indicating that the conformational changes provoked by a steric clash between fluorine and a syn-oriented guanine cannot be compensated for by the simple introduction of a second 2΄F-ANA at a favourable position. Indeed, almost any combination featuring 2΄F-araG5, G10 or G14 fails to yield significant binding signals.
Figure 3.
Average fluorescence intensities for all possible sequences (105) containing two 2΄F-ANA modifications. The diagram plots the position of a first 2΄F-ANA unit against the position of the second 2΄F-ANA unit. The recorded intensities are arranged as a coloured heatmap, green corresponds to lowest signals and purple to the highest. Colour for the control TBA1 is given as reference.
Multiple incorporations of 2΄F-ANA in TBA1
The systematic analysis of all combinations of three or more 2΄F-ANA modifications in the TBA1 sequence indicates that the vast majority are poor binders (Figure S4, Supplementary Data). For five incorporations of 2΄F-ANA nucleotides, already 92% of the 3003 possible sequences yield weaker signals than that of the original model. Nonetheless, we identified a combination of three 2΄F-ANA substitutions (T3, T7 and T12) whose signal significantly exceeds that of any single or double-modified sequence, hinting at an additive effect of 2΄F-araT on binding affinity.
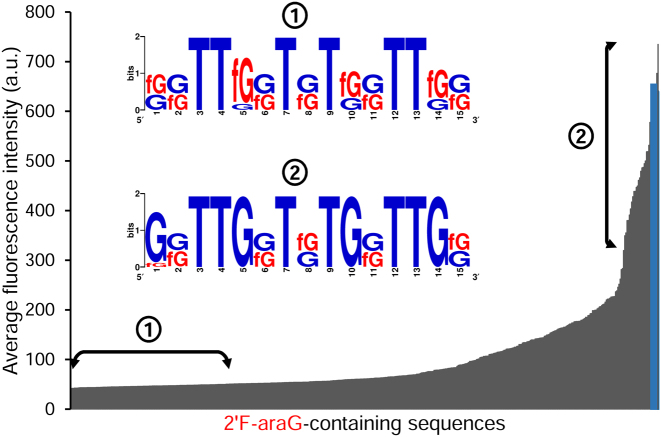
These results prompted us to examine permutations composed of only 2΄F-araG or 2΄F-araT (512 and 64 sequences, respectively). Figure 4 displays the fluorescence intensities of all dG-to-2΄F-araG mutations which, almost invariably, remain below that of the DNA control. Two exceptions, both featuring a 2΄F-araG at position G8, the only guanine residue not involved in the formation of a G-quartet, marginally stand above the DNA control. In order to identify permutation motifs that adversely or favourably affect the binding properties of TBA1 and determine which positions are frequently populated with 2΄F-araG in the low and high fluorescence region of Figure 4, we selected 2΄F-araG-containing sequences leading to very poor or better binding signals and generated a consensus sequence. As shown in Figure 4, very low signals are mainly populated with sequences containing 2΄F-araG at G1, G5, G10 and/or G14, and amongst those, combinations with a 2΄F-araG at G5 dominate, suggesting that introducing 2΄F-ANA at this position destines any mutated candidate to be a weak thrombin binder. At the other end of the fluorescence spectrum in Figure 4, sequences whose binding signals are comparable to that of the control preferentially contain 2΄F-araG at G8 and/or G15, which is in line with our interpretation on single-modified TBA1s. The replacement of dT with 2΄F-araT yielded a very different picture (Figure 5). Indeed, the majority of 2΄F-araT combinations appear to bind thrombin with higher affinity than the unmodified TBA1 and in the corresponding consensus sequence, a 2΄F-araT at T3 is almost always present, clearly identifying it as a key residue for improving the binding affinity of the original TBA1 structure. Conversely, modifying position T4 with 2΄F-araT negatively affects TBA1 and holds true regardless of the number and the location of additional 2΄F-araT units. Thus, like G5, the position T4 does not seem to tolerate 2΄F-ANA chemistry.
Figure 4.
Average fluorescence intensities of all possible sequence permutations containing 2΄F-araG (29, 512 sequences) at a [Thr] of 200 nM. The control (unmodified) TBA1 is shown in blue. The left double arrow marks the region of poor binders used to extract the sequence logo shown in the inset (from 40 to 50 a.u). The right double arrow marks the region of better binders used to extract the sequence logo shown in the inset (from 250 to 740 a.u). Sequence names are omitted for clarity.
Figure 5.
Average fluorescence intensities of all possible sequence permutations containing 2΄F-araT (26, 64 sequences) at a [Thr] of 200 nM. The control (unmodified) TBA1 is shown in blue. The left double arrow marks the region of poor-to-average binders used to extract the first sequence logo shown in the inset. On the right side of the diagram, the second double arrow marks the region of high thrombin binders used to extract the associated sequence logo shown in the inset. The error bars represent the standard error of the mean of the fluorescence signal.
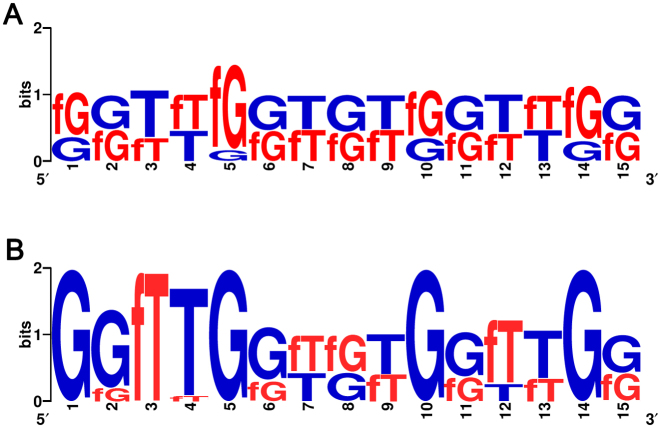
To address whether the recognized consensus and motifs transpose well to a mixed 2΄F-araT/G population, we obtained sequence logos from regions of low and high fluorescence emission in the entire permutation library (Figure 6). Again, the extremely weak binding signals (<10 a.u. above background) largely stem from sequences containing a 2΄F-araG at position G5, which confirms that mutating G5 to 2΄F-ANA eliminates significant thrombin binding, even if accompanied with other favourable 2΄F-ANA substitutions. Indeed, at the opposite end of the fluorescence emission spectrum, G5 appears only in DNA form and, in addition, a consensus sequence emerges in strong binding candidates wherein the syn-oriented G1, G5, G10 and G14 remain unmodified while position T3 is exclusively found modified with 2΄F-araT.
Figure 6.
Sequence logos computed by feeding (A) sequences yielding the lowest fluorescent counts (<50 a.u.) of the entire library and (B) sequences yielding the highest fluorescence counts (>1000 a.u.) at a [Thr] of 200 nM. G and T = dG and dT, respectively. fG and fT = 2΄F-araG and 2΄F-araT, respectively.
Measurement of binding affinity
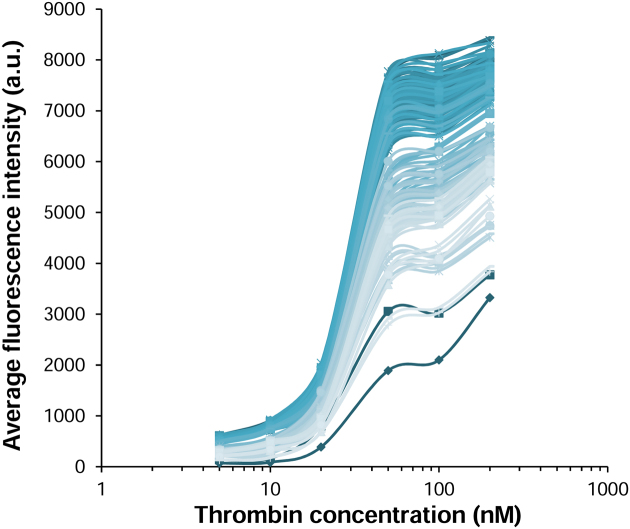
There is good evidence that the intensity of fluorescence emission on microarrays qualitatively reflects binding affinity (24,52,53). To confirm that a strong signal correlates with a strong binding, we wished to provide a quantitative measure of the stability of the TBA1·[thrombin·Cy3-TBA2] complex. To do so, we scanned a quadruplex microarray under a range of thrombin concentrations spanning two orders of magnitude. The lower limit was set at 5 nM so as to ensure the formation of the first Cy3-TBA2·thrombin complex and the higher limit at 200 nM, as the non-specific binding of the preformed fluorescent thrombin·Cy3-TBA2 to the glass surface becomes significant at higher concentrations and reduces data reliability. We selected the 178 sequences yielding the highest signals (>1000 a.u.) and, together with positive and negative controls, synthesized microarrays containing all the best aptamer candidates. The recorded fluorescence intensities were plotted as a function of the various amounts of thrombin to which the folded aptamer arrays were exposed and the results are shown in Figure 7. While the curves marginally differ from one sequence to another, the maxima of fluorescence when close to the saturation regime show some disparity. The curves yielding the highest fluorescence counts, corresponding to sequences 1 and 2 in Table 1, were fit to a sigmoidal model in order to determine binding constants, and found to be 28 nM in both cases. In addition, they share a common pattern of chemical modifications; with T3, G8 and T12 identified as 2΄F-ANA, in line with the consensus sequence presented in Figure 6B. The unmodified TBA1 was found to bind thrombin with a Kd of 93 nM, when it was estimated at 210 nM in our previous study (19). Despite the broad spectrum of maximal fluorescence intensities, the variation in Kd among all aptamer candidates only spans 6 nM (28–34 nM), indicating that there are 178 combinations of 2΄F-ANA TBA1 aptamers able to bind thrombin with a ∼3-fold improvement over the original quadruplex structure. Surprisingly, a single incorporation of 2΄F-ANA at position T3 (sequence 5) is able to bring the dissociation constant down to 31 nM, clearly designating 2΄F-araT3 as the main contributor to the increased binding affinity. The two combinations previously identified as good binders using nitrocellulose filter binding assays, sequences 3 and 4 (19), are not members of this subgroup, yet their binding constants were measured on arrays and found to closely match those obtained by standard methods (39 nM versus 58 nM for 3, 41 nM versus 40 nM for 4). Two 2΄F-araT-containing sequences, 15 and 16, were found to bind thrombin with Kds of 85 and 117 nM, respectively, apparently stronger binders than originally measured (Kds of 300 and 250 nM, respectively), yet in the same range as the reported binding constant of the unmodified TBA1 in each experimental setup. This allows us to consider that Kd values for 2΄F-ANA aptamers on microarray surfaces reflect, qualitatively, those calculated previously (19).
Figure 7.
Binding curves of all 178 G-quadruplex aptamer candidates. Average fluorescence intensities are plotted as a function of thrombin concentration. Error bars are omitted for clarity.
Table 1. Dissociation constants, melting temperatures and folding alignment for 2΄F-ANA-rich aptamers identified with microarrays or with standard solid-phase synthesis (19). T or G = 2΄F-araT or 2΄F-araG.
| Code | Sequence (5΄ to 3΄) | K d (nM) | T m (°C) with KCla | T m (°C) w/o KCl | Folding alignment |
|---|---|---|---|---|---|
| control | GGTTGGTGTGGTTGG | 93 ± 23 (210)c | 46.9 ± 0.2 | 15.8 ± 0.2 | antiparallelb |
| 1 | GGTTGGTGTGGTTGG | 28 ± 1.6 | n.d. | n.d. | n.d. |
| 2 | GGTTGGTGTGGTTGG | 28 ± 1.2 | n.d. | n.d. | n.d. |
| 3 | GGTTGGTGTGGTTGG | 39 ± 10 (58)c | 51.0c | n.d. | antiparallel |
| 4 | GGTTGGTGTGGTTGG | 41 ± 3.7 (40)c | 50.6c | n.d. | antiparallel |
| 5 | GGTTGGTGTGGTTGG | 31 ± 2.4 | 54.1 ± 0.5 | 24.0 ± 0.1 | antiparallel |
| 6 | GGTTGGTGTGGTTGG | 29 ± 2.7 | 66.3 ± 0.2 | 36.8 ± 0.2 | antiparallel |
| 7 | GGTTGGTGTGGTTGG | 31 ± 4 | 68.1 ± 0.5 | 41.0 ± 0.1 | antiparallel |
| 8 | GGTTGGTGTGGTTGG | 31 ± 3.4 | 66.8 ± 0.2 | 38.3 ± 0.3 | antiparallel |
| 9 | GGTTGGTGTGGTTGG | 31 ± 3.4 | 66.9 ± 0.5 | 38.4 ± 0.2 | antiparallel |
| 10 | GGTTGGTGTGGTTGG | 45 ± 14 | 67.7 ± 0.2 | 38.7 ± 0.2 | antiparallel |
| 11 | GGTTGGTGTGGTTGG | 53 ± 16 | 68.1 ± 0.1 | 39.1 ± 0.3 | antiparallel |
| 12 | GGTTGGTGTGGTTGG | n.d. | 51.7 ± 0.2 | 22.6 ± 0.3 | antiparallel |
| 13 | GGTTGGTGTGGTTGG | n.d. | 47.6 ± 0.1 | 14.6 ± 0.5 | antiparallel |
| 14 | GGTTGGTGTGGTTGG | n.d. | 47.7 ± 0.2 | 15.0 ± 0.3 | antiparallel |
| 15 | GGTTGGTGTGGTTGG | 85 ± 21 (300)c | 57.1c | n.d. | antiparallelc |
| 16 | GGTTGGTGTGGTTGG | 117 ± 44 (250)c | 51.2c | n.d. | antiparallelc |
aConditions: 8 μM thrombin-binding aptamers in 10 mM Tris, pH 6.8 with or without 25 mM KCl.
bAs determined by CD spectroscopy. Conditions: 8 μM thrombin-binding aptamers in 10 mM Tris, 25 mM KCl, pH 6.8.
cValues and measurements performed in (19).
n.d. not determined.
Stability of single-modified and 2΄F-ANA-rich TBA1 quadruplexes
UV and CD spectroscopy
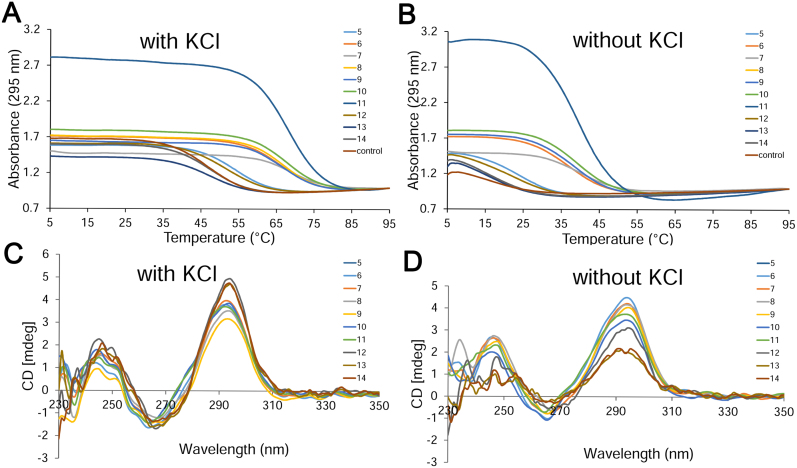
An additional parameter to be taken into consideration when modifying the structure of the TBA1 aptamer is the stability of the folding arrangement, which can be assessed using spectroscopic techniques. The effect of the introduction of 2΄F-ANA in TBA1 was already partially examined (19), and a high-2΄F-ANA content generally translated into an increase of the melting temperature of the quadruplex, which still retained an antiparallel chair-like conformation, as evidenced by CD spectroscopy. Only the presence of 2΄F-ANA on syn-oriented guanine positions was found to compromise the stability of the TBA1 aptamer. The thermal stability of single-modified sequences was not reported. We therefore wished to complete this study by measuring the stability of sequences containing a single 2΄F-araT unit at positions which were previously found to be key residues for thrombin binding (T3, T4, T12 and T13). We thus prepared sequences 5, 12, 13 and 14 (modified with 2΄F-ANA at position T3, T12, T13 and T4, respectively) by solid-phase synthesis and investigated their behaviour in potassium-supplemented buffer. The profile of all Tm curves (Figure 8A) is consistent with G-quadruplex formation, with a decrease in absorbance at 295 nm upon melting of the conformer (54). The presence of 2΄F-ANA at position T3 (sequence 5) not only drastically strengthens binding (Table 1), but also leads to a striking increase in thermal stability: a +7°C increment compared to the control (54°C versus 47°C, Table 1). Upon modification of position T12 (sequence 12), a ΔTm of +5°C was observed, in agreement with the binding assays whereby position T12 appeared to be the next best choice for 2΄F-ANA incorporation. Conversely, 2΄F-ANA-modified sequences at positions T13 or T4 (13 and 14, respectively) were found to be as stable as the unmodified control, clearly showing that the thymidines in the 2-nt loops are structurally distinct.
Figure 8.
(A and B) UV-melting profiles of quadruplexes 5–14 as well as the unmodified control measured at 295 nm in 10 mM Tris, pH 6.8 (A) with and (B) without 25 mM KCl at a final concentration of 8 μM. (C and D) CD spectra of quadruplexes 5–14 recorded at 15°C in 10 mM Tris, pH 6.8 (C) with and (D) without 25 mM KCl at a final concentration of 8 μM.
In parallel, we sought to identify high thrombin-binding sequences featuring a large number of 2΄F-ANA units and evaluate their biophysical properties. Amongst the pre-selected 178 candidates mentioned previously, sequences 6–9 include eight 2΄F-ANA residues and bind thrombin with Kds of ∼30 nM. Further down the catalogue of permutations, we found that up to 10 2΄F-ANA mutations can be introduced in the 15mer (sequences 10 and 11) while still maintaining a good thrombin binding ability (Kds of 45 and 53 nM, respectively). Oligonucleotides 6–11 were therefore synthesized on solid-phase synthesis. All 2΄F-ANA aptamers were found to be more stable than the control TBA1, with Tms ranging from 66 to 68°C; an overall Tm increase of 20°C compared to the control (47°C, Table 1). These sequences are more stable than any 2΄F-ANA quadruplex studied previously (19).
We then looked at the CD spectra of sequences 5–14, which displayed a strong positive absorption band at 295 nm and a weaker, negative absorption band at 260 nm (Figure 8C); characteristic features of an antiparallel folding pattern wherein the guanine bases are oriented in an alternating syn-anti fashion in the G-quartet (55). TBA1 also adopts this conformation and exhibits a similar CD spectrum (19). We then performed UV and CD spectroscopy on 5–14 in the absence of K+ and, interestingly for sequences 5–12, noticed a hypochromic transition of high amplitude (Figure 8B), suggesting the formation of a G-quadruplex in those oligonucleotides even in the absence of potassium ions. The formation of a G-tetrad under potassium-free conditions has been observed independently, in particular for the native TBA1 (56), although usually yielding fragile structures. Indeed, we obtained a denaturation curve with very low amplitude for the control TBA1 in the absence of K+ and measured a Tm of 15.8°C, in agreement with the literature (57). Similarly, sequences 13 and 14 showed a weak hypochromic transition, corresponding to Tms slightly below that of the original structure (14.6 and 15°C). Aptamers 6–11, on the other hand, formed stable structures up to 36–41°C in the absence of potassium, a Tm difference of 20–25°C compared to the control, and are only 5–10°C less stable than the unmodified TBA1 when studied in a potassium-rich environment (Tm of 47°C). A G-quadruplex modified only at position T3 (sequence 5) exhibits a Tm of 24°C, well above that of the control TBA1 (ΔTm of +8°C), while a ΔTm of +6.5°C was measured for sequence 12 modified at position T12. In addition, CD spectra recorded without potassium (Figure 8D) show that all aptamers (5 to 12) maintain an antiparallel folding (type II spectrum) with a positive band at 295 nm and a negative band at 260 nm, and the same ellipticities as those recorded in the presence of potassium. The CD spectra of sequences 13 and 14 display somewhat weaker ellipticities, in line with their relative instability under potassium-depleted conditions. Thus, our study reveals that not only are the 2΄F-ANA aptamers 5 to 11 better thrombin binders than the original model, they are considerably more stable and do not seem to require potassium to be able to fold into their distinct chair-like conformation.
Solution structure of TBA1 modified with 2΄F-araT3
Another striking feature of the presence of 2΄F-ANA in the TBA1 aptamer unveils upon modification of T3, as it leads to both increased thrombin binding and thermal stability. We thus sought to shed some light onto the source of stabilization and improved affinity using NMR. We first recorded NMR spectra of aptamer 5 under a range of increasing temperatures and clearly confirm its higher stability relative to unmodified TBA1 (Figure S7, Supplementary Data). The complete assignment of the NMR spectra was carried out and shows that the proton chemical shifts are almost identical to the unmodified TBA with the exception of some thymine protons in the lateral loops (Figures S8, S9 and Table S1, Supplementary Data) including not only the modified residue T3, but also T4 and, to a lesser extent, T13. Since chemical shifts are extremely sensitive to structural changes, we can safely conclude that the structures of aptamer 5 and the unmodified TBA1 only differ in the lateral loops region, where a large number of NOEs involving these residues were detected (Table S2, Supplementary Data for a complete list). Based on these NOEs, a model structure of aptamer 5 was calculated using restrained molecular dynamics methods. It was found that, in contrast to the solution structure of the unmodified TBA1 in which T3 and T12 are mainly observed in a disordered arrangement (43,58), the resulting structure of aptamer 5 exhibits very well-defined loops (Figure 9B), both at T3 and T12 positions. In particular, the conformation of 2΄F-araT3 in the solution structure resembles that of T3 in the crystallographic structure of the TBA1-alpha thrombin complex (7) (Figure 9C). Thus, these results indicate that 2΄F-ANA substitution at residue T3 comes with better structured loops, both in terms of rigidity and compactness, seemingly adopting the appropriate conformation for thrombin recognition. Of interest is the fact that the structural organization of T3 upon mutation with 2΄F-ANA affects position T12 as well. Given the symmetrical position of T3 and T12 with respect to the overall quadruplex structure, it is expected that the substitution of T12 with 2΄F-ANA would produce similar changes.
Figure 9.
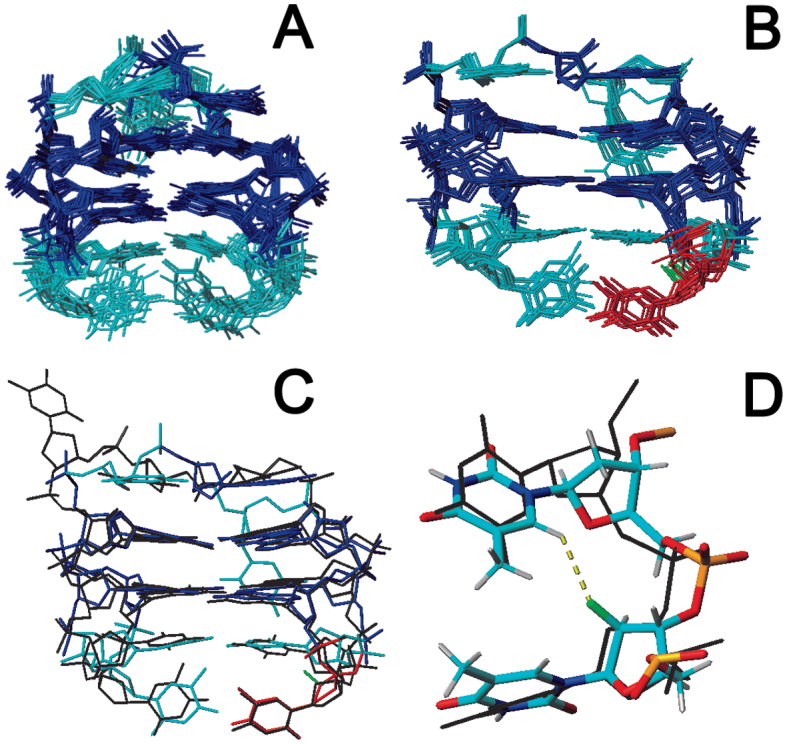
Solution structure ensembles of unmodified (A) TBA1, and (B) aptamer 5. Modified 2΄F-araT3 is shown in red. (C) Superimposition of a representative structure of aptamer 5, and the crystallographic structure of TBA1 in complex with alpha-thrombin (black colour, PDB code: 4DII). (D) Detail of the 2΄F-araT3-T4 region. Crystallographic structure is superimposed in black. The potential C-H6…F2΄ favourable interaction is shown in yellow. PDB code 5MJX.
DISCUSSION
The overwhelming majority of oligonucleotides arrays produced by photolithography are DNA, and despite the robustness of the phosphoramidite chemistry, attempts at expanding the technology to RNA have been infrequent (59–62), and reports on arrays containing nucleoside analogues have been rare (63). Considering the essential role of chemically-modified nucleosides in modern aptamer discovery (10,64), the prospect of extending the chemical repertoire of nucleic acids microarrays is highly appealing. We present here the fabrication of the first 2΄F-ANA microarrays and their use in exploring the binding landscape of TBA1 to its target thrombin and in identifying novel 2΄F-ANA aptamers. TBA1, being a 15mer sequence, requires a library of 32 768 oligonucleotides in order to explore all possible DNA-to-2΄F-ANA permutations. This challenge was partially undertaken via solid-phase synthesis (19) but requires a much higher-throughput method in order to be completed. Large libraries assembled with 2΄F-ANA are amenable to in vitro selection (65,66), but the fundamental bias of selection towards strong binders would offer a truncated view of the chemical space occupied by 2΄F-ANA in TBA1 and, in particular, the positions that are most sensitive to modification. However, in situ synthesized arrays provide the means to examine each permutation separately as a unique member of a library whose entire binding landscape can be probed simultaneously in a single experiment. Therefore, the binding affinities of all permutations are directly comparable as they derive from the fluorescence intensities recorded in the same assay and from oligonucleotides in close physical proximity on the same substrate. It has been shown that the fluorescent signals in this context can be used as an absolute measure of binding affinity (29,53). We also observe a close correlation between fluorescence intensity and Kd, but more careful calibration may be necessary in order to extrapolate a dissociation constant directly comparable to those obtained by other methods. Indeed, we seem to measure slightly stronger binding constants for the same sequences on microarrays than on nitrocellulose. Discrepancies are not unheard of (52) and may well be the consequence of variations in experimental setup and conditions or in the very nature of the assay. Nevertheless, the measured binding constant for the unmodified TBA1 oligonucleotide matches well with reported values (6,8,67) and, in our assay, serves as a reference point. The strong binders identified in this study have a common denominator (2΄F-araT3) and its effect on binding can be, at least partially, understood using UV-melting and NMR. However, 2΄F-ANA-modified G-quadruplexes may adopt unusual conformations that are better adapted to thrombin binding when covalently bound to a microarray surface. Whether structural differences exist and whether they also participate in the improved binding affinity is unclear at this point.
The spectrum of binding signals recorded at a given thrombin concentration yields crucial information about the position-dependence of 2΄F-ANA in TBA1. Perhaps the strongest effect originates with replacing syn-oriented guanines G1, G5, G10 and/or G14 with 2΄F-ANA where the fluorine atom at the endo face of the arabinose sugar sterically clashes with guanine in a syn conformation. This observation may be useful as an indirect method to reveal the preferred orientation of a nucleobase in exotic nucleic acid structures. Anti-oriented guanines G2, G6, G11 and G15 seem to tolerate 2΄F-ANA chemistry but the substitution has no clear positive effect on the binding affinity. When modified with 2΄F-ANA, these positions do however participate in the increased stability of the aptamers 6–11, which can be ascribed to the conformational preorganization of 2΄F-araG nucleotides locking the nucleobase in an anti-orientation (51). The only guanine not involved in G-quartet formation is G8, located in the three-nucleotide loop facing away from the contact surface of TBA1 and thrombin. The base is found in an anti conformation and participate in stacking interactions with G10, however 2΄F-araG8 as a single modification has little impact on binding.
Positions T12 and particularly T3 clearly emerge as the ideal locations for the introduction of 2΄F-ANA nucleotides, since a single modification at either position significantly increases the binding affinity. Yet this effect seems not to be additive, as sequences modified at both positions do not bind thrombin twice as strongly as any single-modified sequence. T3 and T12 are parts of the two TT loops that recognize exosite I (fibrinogen binding site) of thrombin, and as such, constitute the binding motif of TBA1 (7). It is therefore surprising that the core region of thrombin interaction can accommodate the presence of nucleoside analogues and even strengthen the binding interaction. Our NMR studies show that 2΄-fluoroarabinose sugar substitution at position T3 (and also, presumably, at the equivalent position T12) provokes a preorganization of the TT loop residues into a conformation coinciding with that found in the TBA1-alpha thrombin complex, a plausible source for the increased binding properties of 2΄F-araT3-containing aptamers. The fact that both loops appear more rigid with the single modification of position T3 reasonably explains why further modification at position T12 has little additional effect on binding. As to whether the increased binding affinity of the 2΄F-ANA-modified aptamers can be attributed to the increased pre-organization of the aptamer into the binding conformation, or an increased affinity of that conformation, we cannot yet provide a definitive answer. Nevertheless, the increased Tm of 2΄F-ANA-rich quadruplexes together with the NMR results indicating structuring of the lateral loops into an appropriate conformation for thrombin recognition, suggests that pre-organization is likely to play an important role. Electrostatic interactions involving the substituents at the carbon 2΄ of the arabinose sugar may be an additional source of stabilization of the TBA1 structure. Indeed, it has been previously shown that, in 5΄-pyrimidine-purine-3΄ steps, the introduction of a 2΄-β-fluorine in the pyrimidine allows the formation of an internal pseudohydrogen bond between the fluorine atom and the exocyclic H8 proton of the 3΄ purine base (17,68,69). This pseudohydrogen bond was also observed in quadruplexes and was accompanied with an electrostatic interaction between the H2΄ of the same 2΄F-ANA residue and the O4΄ of the 3΄ neighbouring guanine, resulting in an overall considerable increase in the melting temperature of the structure (+12°C with a single 2΄F-ANA modification (18)). In the case of TBA1, positions T3 and T12 do not have a guanine residue as 3΄ neighbours, but the F2΄ in 2΄F-araT3 and 2΄F-araT12 may still participate in C-H6…F2΄ electrostatic interactions with the near thymine 4 or 13. Indeed, fluorine atoms at the 2΄ position of T3 and T12 are in close proximity with the H6 proton of T4 and T12, respectively (∼2.5 Å) and the angular geometry seems to allow for the formation of a C-H6…F2΄ pseudohydrogen bond (Figure 9D).
The presence of 2΄F-ANA in the other two T nucleotides of the TT loops, namely T4 and T13, is much less beneficial to stability and binding; hence they are rarely found modified in the selected subset of good binders. In addition, it should be noted that T4 and T13 are the only T positions where 2΄F-ANA modification does not lead to increased thermal stability, which suggests that the introduction of a fluorine at 2΄F-araT4 and 2΄F-araT13 has local, yet adverse effects on the structure of TBA1. T4 and T13 participate in thrombin recognition through polar interactions with neighbouring arginine residues, and feature a coplanar arrangement of the guanidine side chains of Arg75 and Arg77 and the thymine base of T4 (7), which may be disrupted due to the presence of a 2΄F. Evidence from negative structural disruptions may come from the formation of repulsive electrostatic interactions between the 2΄-fluorine in T4 and the neighbouring 5΄-oxygen, as suggested by their close proximity in the solution structure (Figure S11, Supplementary Data). Nevertheless, additional NMR experiments will be necessary for a full understanding of the effect of a fluorine substitution at those positions.
CONCLUSION
The application of 2΄F-ANA chemistry in non-canonical nucleic acids topologies such as quadruplexes or i-motifs (70), beyond the historical realm of antisense and siRNA strategies, illustrates its appealing electronic and structural properties and by introducing 2΄F-ANA to in situ synthesized microarrays, we provide a method to investigate the chemical space occupied by 2΄F-ANA in a systematic way. We presented here the fabrication of a microarray populated with all possible deoxyribose-to-fluoroarabinose permutations of a 15mer quadruplex aptamer (TBA1) and its use in thrombin binding experiments using a ‘sandwich’ approach. In so doing, we extracted essential information on the positional effect of 2΄F-ANA in TBA1 and found that syn-oriented guanines do not tolerate the presence of a 2΄-β-fluorine atom while an increased binding and stability are observed upon modification of thymidines T3 and T12, residues directly participating in thrombin binding. Several combinations of 2΄F-ANA in TBA1, accommodating up to 10 2΄F-ANA modifications, were also identified as promising aptamer candidates, reaching dissociation constants up to three times lower than that of the DNA-only TBA1. In these cases, a high 2΄F-ANA content translated into a considerable increase in quadruplex stability (+20°C) and into stable folding even in the absence of potassium ions. Structural studies on the effect of 2΄F-ANA substitution at position 3 indicate that 2΄-fluoroarabinose provokes a preorganization of this key residue in the appropriate conformation for thrombin binding. Taken together, our results encourage further development of 2΄F-ANA-based quadruplexes in general, and TBA1 aptamers in particular, for example in thrombin sensing platforms (57,71,72), or in therapeutics (73), while enriching the collection of nucleoside analogues available for in situ microarray synthesis.
ACCESSION NUMBER
PDB code 5MJX.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank ChemGenes Corporation for providing us with 5΄-NPPOC 2΄-F-araGiPac phosphoramidite.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Swiss National Science Foundation [PBBEP2-146174 to J.L.]; Natural Sciences and Engineering Research Council of Canada [to M.J.D.]; Austrian Science Fund [FWF P23797 and FWF P27275 to J.L. and M.M.S.]; Spanish MINECO [BFU2014-52864-R to C.G.]. Funding for open access charge: Austrian Science Fund [FWF P23797 and FWF P27275 to J.L. and M.M.S.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Rhodes D., Lipps H.J.. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015; 43:8627–8637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Murat P., Balasubramanian S.. Existence and consequences of G-quadruplex structures in DNA. Curr. Opin. Genet. Dev. 2014; 25:22–29. [DOI] [PubMed] [Google Scholar]
- 3. De Cian A., Lacroix L., Douarre C., Temime-Smaali N., Trentesaux C., Riou J.F., Mergny J.L.. Targeting telomeres and telomerase. Biochimie. 2008; 90:131–155. [DOI] [PubMed] [Google Scholar]
- 4. Balasubramanian S., Hurley L.H., Neidle S.. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy. Nat. Rev. Drug Discov. 2011; 10:261–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bock L.C., Griffin L.C., Latham J.A., Vermaas E.H., Toole J.J.. Selection of single-stranded-DNA molecules that bind and inhibit human thrombin. Nature. 1992; 355:564–566. [DOI] [PubMed] [Google Scholar]
- 6. Macaya R.F., Waldron J.A., Beutel B.A., Gao H.T., Joesten M.E., Yang M.H., Patel R., Bertelsen A.H., Cook A.F.. Structural and functional-characterization of potent antithrombotic oligonucleotides possessing both quadruplex and duplex motifs. Biochemistry. 1995; 34:4478–4492. [DOI] [PubMed] [Google Scholar]
- 7. Russo Krauss I., Merlino A., Randazzo A., Novellino E., Mazzarella L., Sica F.. High-resolution structures of two complexes between thrombin and thrombin-binding aptamer shed light on the role of cations in the aptamer inhibitory activity. Nucleic Acids Res. 2012; 40:8119–8128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tasset D.M., Kubik M.F., Steiner W.. Oligonucleotide inhibitors of human thrombin that bind distinct epitopes. J. Mol. Biol. 1997; 272:688–698. [DOI] [PubMed] [Google Scholar]
- 9. Russo Krauss I., Pica A., Merlino A., Mazzarella L., Sica F.. Duplex-quadruplex motifs in a peculiar structural organization cooperatively contribute to thrombin binding of a DNA aptamer. Acta. Crystallogr. D Biol. Crystallogr. 2013; 69:2403–2411. [DOI] [PubMed] [Google Scholar]
- 10. Wang R.E., Wu H., Niu Y., Cai J.. Improving the stability of aptamers by chemical modification. Curr. Med. Chem. 2011; 18:4126–4138. [DOI] [PubMed] [Google Scholar]
- 11. Avino A., Fabrega C., Tintore M., Eritja R.. Thrombin binding aptamer, more than a simple aptamer: chemically modified derivatives and biomedical applications. Curr. Pharm. Design. 2012; 18:2036–2047. [DOI] [PubMed] [Google Scholar]
- 12. Wilds C.J., Damha M.J.. 2΄-Deoxy-2΄-fluoro-beta-D-arabinonucleosides and oligonucleotides (2΄F-ANA): synthesis and physicochemical studies. Nucleic Acids Res. 2000; 28:3625–3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Watts J.K., Damha M.J.. 2΄F-arabinonucleic acids (2΄F-ANA) - History, properties, and new frontiers. Can. J. Chem. 2008; 86:641–656. [Google Scholar]
- 14. Deleavey G.F., Watts J.K., Alain T., Robert F., Kalota A., Aishwarya V., Pelletier J., Gewirtz A.M., Sonenberg N., Damha M.J.. Synergistic effects between analogs of DNA and RNA improve the potency of siRNA-mediated gene silencing. Nucleic Acids Res. 2010; 38:4547–4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Anzahaee M.Y., Deleavey G.F., Le P.U., Fakhoury J., Petrecca K., Damha M.J.. Arabinonucleic Acids: 2΄-stereoisomeric modulators of siRNA activity. Nucleic Acid Ther. 2014; 24:336–343. [DOI] [PubMed] [Google Scholar]
- 16. Souleimanian N., Deleavey G.F., Soifer H., Wang S.J., Tiemann K., Damha M.J., Stein C.A.. Antisense 2΄-Deoxy, 2΄-Fluoroarabino nucleic acid (2΄F-ANA) oligonucleotides: in vitro gymnotic silencers of gene expression whose potency is enhanced by fatty acids. Mol. Ther. NucleicAcids. 2012; 1:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Anzahaee M.Y., Watts J.K., Alla N.R., Nicholson A.W., Damha M.J.. Energetically important C-H center dot center dot center dot F-C Pseudohydrogen bonding in water: evidence and application to rational design of oligonucleotides with high binding affinity. J. Am. Chem. Soc. 2011; 133:728–731. [DOI] [PubMed] [Google Scholar]
- 18. Martin-Pintado N., Yahyaee-Anzahaee M., Deleavey G.F., Portella G., Orozco M., Damha M.J., Gonzalez C.. Dramatic effect of Furanose C2΄ substitution on structure and stability: directing the folding of the human telomeric quadruplex with a single fluorine atom. J. Am. Chem. Soc. 2013; 135:5344–5347. [DOI] [PubMed] [Google Scholar]
- 19. Peng C.G., Damha M.J.. G-quadruplex induced stabilization by 2΄-deoxy-2΄-fluoro-D-arabinonucleic acids (2΄F-ANA). Nucleic Acids Res. 2007; 35:4977–4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Singh-Gasson S., Green R.D., Yue Y., Nelson C., Blattner F., Sussman M.R., Cerrina F.. Maskless fabrication of light-directed oligonucleotide microarrays using a digital micromirror array. Nat. Biotech. 1999; 17:974–978. [DOI] [PubMed] [Google Scholar]
- 21. Fodor S.P.A., Read J.L., Pirrung M.C., Stryer L., Lu A.T., Solas D.. Light-directed, spatially addressable parallel chemical synthesis. Science. 1991; 251:767–773. [DOI] [PubMed] [Google Scholar]
- 22. Trevino V., Falciani F., Barrera-Saldaña H.A.. DNA microarrays: a powerful genomic tool for biomedical and clinical research. Mol. Med. 2007; 13:527–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Witt M., Walter J.-G., Stahl F.. Aptamer microarrays—current status and future prospects. Microarrays. 2015; 4:115–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Berger M.F., Bulyk M.L.. Universal protein-binding microarrays for the comprehensive characterization of the DNA-binding specificities of transcription factors. Nat. Prot. 2009; 4:393–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chang Y.C., Kao W.C., Wang W.Y., Wang W.Y., Yang R.B., Peck K.. Identification and characterization of oligonucleotides that inhibit Toll-like receptor 2-associated immune responses. FASEB J. 2009; 23:3078–3088. [DOI] [PubMed] [Google Scholar]
- 26. Li Y., Lee H.J., Corn R.M.. Fabrication and characterization of RNA aptamer microarrays for the study of protein-aptamer interactions with SPR imaging. Nucleic Acids Res. 2006; 34:6416–6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Platt M., Rowe W., Knowles J., Day P.J., Kell D.B.. Analysis of aptamer sequence activity relationships. Integr. Biol. 2009; 1:116–122. [DOI] [PubMed] [Google Scholar]
- 28. Chen Y., Nakamoto K., Niwa O., Corn R.M.. On-chip synthesis of RNA aptamer microarrays for multiplexed protein biosensing with SPR imaging measurements. Langmuir. 2012; 28:8281–8285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Warren C.L., Kratochvil N.C.S., Hauschild K.E., Foister S., Brezinski M.L., Dervan P.B., Phillips G.N., Ansari A.Z.. Defining the sequence-recognition profile of DNA-binding molecules. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:867–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lao Y.-H., Peck K., Chen L.-C.. Enhancement of aptamer microarray sensitivity through spacer optimization and avidity effect. Anal. Chem. 2009; 81:1747–1754. [DOI] [PubMed] [Google Scholar]
- 31. Edwards K.A., Wang Y., Baeumner A.J.. Aptamer sandwich assays: human alpha-thrombin detection using liposome enhancement. Anal. Bioanal. Chem. 2010; 398:2645–2654. [DOI] [PubMed] [Google Scholar]
- 32. Tennico Y.H., Hutanu D., Koesdjojo M.T., Bartel C.M., Remcho V.T.. On-chip aptamer-based sandwich assay for thrombin detection employing magnetic beads and quantum dots. Anal. Chem. 2010; 82:5591–5597. [DOI] [PubMed] [Google Scholar]
- 33. Hecht A., Kumar A.A., Kopelman R.. Label-acquired magnetorotation as a signal transduction method for protein detection: aptamer-based detection of thrombin. Anal. Chem. 2011; 83:7123–7128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sosic A., Meneghello A., Cretaio E., Gatto B.. Human thrombin detection through a sandwich aptamer microarray: interaction analysis in solution and in solid phase. Sensors (Basel). 2011; 11:9426–9441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lee S.J., Tatavarty R., Gu M.B.. Electrospun polystyrene-poly(styrene-co-maleic anhydride) nanofiber as a new aptasensor platform. Biosens. Bioelectron. 2012; 38:302–307. [DOI] [PubMed] [Google Scholar]
- 36. Romhildt L., Pahlke C., Zorgiebel F., Braun H.G., Opitz J., Baraban L., Cuniberti G.. Patterned biochemical functionalization improves aptamer-based detection of unlabeled thrombin in a sandwich assay. ACS Appl. Mater. Interfaces. 2013; 5:12029–12035. [DOI] [PubMed] [Google Scholar]
- 37. Agbavwe C., Kim C., Hong D., Heinrich K., Wang T., Somoza M.M.. Efficiency, error and yield in light-directed maskless synthesis of DNA microarrays. J. Nanobiotechnology. 2011; 9:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sack M., Holz K., Holik A.K., Kretschy N., Somoza V., Stengele K.P., Somoza M.M.. Express photolithographic DNA microarray synthesis with optimized chemistry and high-efficiency photolabile groups. J. Nanobiotechnology. 2016; 14:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sack M., Kretschy N., Rohm B., Somoza V., Somoza M.M.. Simultaneous light-directed synthesis of mirror-image microarrays in a photochemical reaction cell with flare suppression. Anal. Chem. 2013; 85:8513–8517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hölz K., Lietard J., Somoza M.M.. A high-power 365 nm UV LED mercury arc lamp replacement for photochemistry and chemical photolithography. ACS Sustainable Chem. Eng. 2016; 5:828–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schneider T.D., Stephens R.M.. Sequence Logos - a New Way to Display Consensus Sequences. Nucleic Acids Res. 1990; 18:6097–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Goddard T.D., Kneller D.G.. SPARKY 3. San Francisco: University of California. [Google Scholar]
- 43. Macaya R.F., Schultze P., Smith F.W., Roe J.A., Feigon J.. Thrombin-binding DNA aptamer forms a unimolecular quadruplex structure in solution. Proc. Natl. Acad. Sci. U.S.A. 1993; 90:3745–3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Soliva R., Monaco V., Gomez-Pinto I., Meeuwenoord N.J., Marel G.A., Boom J.H., Gonzalez C., Orozco M.. Solution structure of a DNA duplex with a chiral alkyl phosphonate moiety. Nucleic Acids Res. 2001; 29:2973–2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Case D.A.V.B., Berryman J.T., Betz R.M., Cai Q., Cerutti D.S., Cheatham T.E. III, Darden T.A., Duke R.E.H.G., Goetz A.W., Gusarov S. et al. . AMBER 14. 2014; San Francisco: University of California. [Google Scholar]
- 46. Ivani I., Dans P.D., Noy A., Perez A., Faustino I., Hospital A., Walther J., Andrio P., Goni R., Balaceanu A. et al. . Parmbsc1: a refined force field for DNA simulations. Nat. Methods. 2016; 13:55–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Noy A., Luque F.J., Orozco M.. Theoretical analysis of antisense duplexes: determinants of the RNase H susceptibility. J. Am. Chem. Soc. 2008; 130:3486–3496. [DOI] [PubMed] [Google Scholar]
- 48. Koradi R., Billeter M., Wuthrich K.. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 1996; 14, 51–55, 29–32. [DOI] [PubMed] [Google Scholar]
- 49. Elzagheid M.I., Viazovkina E., Damha M.J.. Current Protocols in Nucleic Acid Chemistry. 2001; Hoboken NJ: John Wiley & Sons, Inc. [Google Scholar]
- 50. Franssen-van Hal N.L.W., van der Putte P., Hellmuth K., Matysiak S., Kretschy N., Somoza M.M.. Optimized light-directed synthesis of aptamer microarrays. Anal. Chem. 2013; 85:5950–5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sapse A.M., Snyder G.. Ab initio studies of the antiviral drug 1-(2-Fluoro-2-Deoxy-Beta-D-Arabinofuranosyl) thymine. Cancer Invest. 1985; 3:115–121. [DOI] [PubMed] [Google Scholar]
- 52. Katilius E., Flores C., Woodbury N.W.. Exploring the sequence space of a DNA aptamer using microarrays. Nucleic Acids Res. 2007; 35:7626–7635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Puckett J.W., Muzikar K.A., Tietjen J., Warren C.L., Ansari A.Z., Dervan P.B.. Quantitative microarray profiling of DNA-binding molecules. J. Am. Chem. Soc. 2007; 129:12310–12319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mergny J.L., Phan A.T., Lacroix L.. Following G-quartet formation by UV-spectroscopy. FEBS Lett. 1998; 435:74–78. [DOI] [PubMed] [Google Scholar]
- 55. Williamson J.R. G-quartet structures in telomeric DNA. Annu. Rev. Biophys. Biomol. Struct. 1994; 23:703–730. [DOI] [PubMed] [Google Scholar]
- 56. Nagatoishi S., Tanaka Y., Tsumoto K.. Circular dichroism spectra demonstrate formation of the thrombin-binding DNA aptamer G-quadruplex under stabilizing-cation-deficient conditions. Biochem. Biophys. Res. Commun. 2007; 354:837–838. [DOI] [PubMed] [Google Scholar]
- 57. Baldrich E., Restrepo A., O'Sullivan C.K.. Aptasensor development: elucidation of critical parameters for optimal aptamer performance. Anal. Chem. 2004; 76:7053–7063. [DOI] [PubMed] [Google Scholar]
- 58. Wang K.Y., McCurdy S., Shea R.G., Swaminathan S., Bolton P.H.. A DNA aptamer which binds to and inhibits thrombin exhibits a new structural motif for DNA. Biochemistry. 1993; 32:1899–1904. [DOI] [PubMed] [Google Scholar]
- 59. Lackey J.G., Mitra D., Somoza M.M., Cerrina F., Damha M.J.. Acetal levulinyl ester (ALE) groups for 2΄-hydroxyl protection of ribonucleosides in the synthesis of oligoribonucleotides on glass and microarrays. J. Am. Chem. Soc. 2009; 131:8496–8502. [DOI] [PubMed] [Google Scholar]
- 60. Lackey J.G., Somoza M.M., Mitra D., Cerrina F., Damha M.J.. In-situ chemical synthesis of rU-DNA chimeras on chips and enzymatic recognition. Chim. Oggi-Chem. Today. 2009; 27:30–33. [Google Scholar]
- 61. Wu C.-H., Holden M.T., Smith L.M.. Enzymatic fabrication of high-density RNA arrays. Angew. Chem. Int. Ed. 2014; 53:13514–13517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Holden M.T., Carter M.C.D., Wu C.H., Wolfer J., Codner E., Sussman M.R., Lynn D.M., Smith L.M.. Photolithographic synthesis of high-density DNA and RNA arrays on flexible, transparent, and easily subdivided plastic substrates. Anal. Chem. 2015; 87:11420–11428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yang F., Dong B., Nie K., Shi H., Wu Y., Wang H., Liu Z.. Light-Directed Synthesis of High-Density Peptide Nucleic Acid Microarrays. ACS Comb. Sci. 2015; 17:608–614. [DOI] [PubMed] [Google Scholar]
- 64. Keefe A.D., Cload S.T.. SELEX with modified nucleotides. Curr. Opin. Chem. Biol. 2008; 12:448–456. [DOI] [PubMed] [Google Scholar]
- 65. Peng C.G., Damha M.J.. Polymerase-directed synthesis of 2΄-deoxy-2΄-fluoro-beta-D-arabinonucleic acids. J. Am. Chem. Soc. 2007; 129:5310–5311. [DOI] [PubMed] [Google Scholar]
- 66. Pinheiro V.B., Taylor A.I., Cozens C., Abramov M., Renders M., Zhang S., Chaput J.C., Wengel J., Peak-Chew S.-Y., McLaughlin S.H. et al. . Synthetic Genetic Polymers Capable of Heredity and Evolution. Science. 2012; 336:341–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Pasternak A., Hernandez F.J., Rasmussen L.M., Vester B., Wengel J.. Improved thrombin binding aptamer by incorporation of a single unlocked nucleic acid monomer. Nucleic Acids Res. 2011; 39:1155–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Watts J.K., Martin-Pintado N., Gomez-Pinto I., Schwartzentruber J., Portella G., Orozco M., Gonzalez C., Damha M.J.. Differential stability of 2΄F-ANA.RNA and ANA.RNA hybrid duplexes: roles of structure, pseudohydrogen bonding, hydration, ion uptake and flexibility. Nucleic Acids Res. 2010; 38:2498–2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Martin-Pintado N., Yahyaee-Anzahaee M., Campos-Olivas R., Noronha A.M., Wilds C.J., Damha M.J., Gonzalez C.. The solution structure of double helical arabino nucleic acids (ANA and 2΄F-ANA): effect of arabinoses in duplex-hairpin interconversion. Nucleic Acids Res. 2012; 40:9329–9339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Abou Assi H., Harkness R.W., Martin-Pintado N., Wilds C.J., Campos-Olivas R., Mittermaier A.K., González C., Damha M.J.. Stabilization of i-motif structures by 2΄-β-fluorination of DNA. Nucleic Acids Res. 2016; 44:4998–5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Sosic A., Meneghello A., Antognoli A., Cretaio E., Gatto B.. Development of a multiplex sandwich aptamer microarray for the detection of VEGF(165) and thrombin. Sensors. 2013; 13:13425–13438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Jung Y.K., Lee T., Shin E., Kim B.-S.. Highly Tunable Aptasensing Microarrays with Graphene Oxide Multilayers. Sci. Rep. 2013; 3:3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Yu Y., Liang C., Lv Q., Li D., Xu X., Liu B., Lu A., Zhang G.. Molecular selection, modification and development of therapeutic oligonucleotide aptamers. Int. J. Mol. Sci. 2016; 17:358. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.