Abstract
Double-stranded DNA minicircles of less than 1000 bp in length have great interest in both fundamental research and therapeutic applications. Although minicircles have shown promising activity in gene therapy thanks to their good biostability and better intracellular trafficking, minicircles down to 250 bp in size have not yet been investigated from the test tube to the cell for lack of an efficient production method. Herein, we report a novel versatile plasmid-free method for the production of DNA minicircles comprising fewer than 250 bp. We designed a linear nicked DNA double-stranded oligonucleotide blunt-ended substrate for efficient minicircle production in a ligase-mediated and bending protein-assisted circularization reaction at high DNA concentration of 2 μM. This one pot multi-step reaction based-method yields hundreds of micrograms of minicircle with sequences of any base composition and position and containing or not a variety of site-specifically chemical modifications or physiological supercoiling. Biochemical and cellular studies were then conducted to design a 95 bp minicircle capable of binding in vitro two NF-κB transcription factors per minicircle and to efficiently inhibiting NF-κB-dependent transcriptional activity in human cells. Therefore, our production method could pave the way for the design of minicircles as new decoy nucleic acids.
INTRODUCTION
Synthetic double-stranded DNA minicircles (dsMCs) are attractive nano-objects that received attention for their use in therapeutic applications. dsMCs from 250 to 1000 bp in length recently entered the field of gene therapy as minivectors with a better delivery efficiency than conventional exogenous plasmid-based vector used as nucleic acid therapeutics to express small interfering RNA (1,2). Reducing the size of conventional plasmids to small dsMCs offers several advantages such as increased resistance to the shearing forces associated with nucleic acids delivery system (3), significantly better biostability in human serum and in the living cell (1) and an improvement in DNA intracellular delivery and trafficking (4,5). To our knowledge, the potential of small dsMCs of sizes down to 250 bp as nucleic acid therapeutics has not yet been investigated because their quantitative production is a limiting step for biological assays. In more fundamental studies, dsMCs less than 200 bp in length have been used for several purposes. In the field of DNA nanotechnology, small dsMCs were employed as building objects to assemble nanoarchitectures after the incorporation of G-quadruplexes, RNA hairpins or chemically functionalized oligodeoxynucleotides leading to the possibility of creating molecular devices with a broad range of functions (6–8). dsMCs in the range of 100 bp in length also offer the unique opportunity to mimic short DNA loops containing permanent bending and topological constrains formed during essential DNA-dependent transactions such as transcription and recombination (9–12). Producing minicircles with customized DNA sequences, chemical functionalization and supercoiling remains a hurdle for further applications including the study of interactions between minicircle and other molecules such as proteins and drugs. Therefore, despite a growing interest for dsMCs less than 250 bp in length, no versatile method currently exists for the rationale, design and production of small minicircles to carry out more straightforward experiments by exploiting the unique structure of this DNA nanomaterial in studies from the test tube to the cell.
The available production methods for small dsMCs present several qualitative and quantitative limitations. A minicircle-producer plasmid through integrase-dependent DNA recombination in bacteria was unable to generate minicircles of less than 250 bp due to unpredictable sequence- and length-dependent negative effects on the recombination efficiency (2,13). In vitro, a recombination-based methodology involves time-consuming preparation and purification of both plasmids and recombinase protein. Furthermore, the minicircle recovery step is labor-intensive and yields low amounts of material, in the microgram range (14,15). Another approach consists in using a linear DNA fragment the ends of which need to be in close proximity to each other while leaving them accessible for covalent sealing by enzymatic ligation. The main hurdle to such an approach comes from the intrinsic stiffness of the DNA double helix that makes the cyclization reaction very inefficient for DNA lengths less than about 300 bp, as the DNA rigidity dominates in the order of its persistent length (∼150 bp) (16). In reported ligase-mediated DNA circularization assays which have been used extensively to study DNA bending through minicircle formation in the presence or absence of protein-induced bending (9), the DNA concentration was kept very low in the nanomolar range or less. Such a weak DNA concentration is indeed necessary in order to favor the formation of minicircles arising from the intramolecular circularization of flexible or bent linear DNA molecules and to minimize as much as possible linear DNA multimers formed competitively through DNA concentration dependent intermolecular reaction. Because a very low DNA concentration renders the circularization reaction unfit for the quantitative production of dsMC, a key requirement for minicircle production is to design a more efficient circularization reaction by increasing DNA bendability. Higher efficiency of the circularization reaction has been achieved using specific sequences endowed with intrinsic bendability (so called adenine tracts) but this approach precludes the possibility of incorporating freely any DNA sequence of interest (17). A smart strategy employed DNA substrates with long cohesive ends, the flexibility of which was exploited to produce a few tens of micrograms of ligaturable minicircles (18). This process requires labor-intensive preparation of plasmid-based linear DNA substrates. When using oligodeoxynucleotides, a low minicircle yield is observed due to the formation of byproducts, i.e. truncated DNA strands and circular multimers (19). Therefore, a versatile method for the production of minicircles having fewer than 250 bp in length with customized DNA sequences with or without chemical functionalities is not yet available. The production of sufficient quantities of material is another important criterion to be fulfilled with a view to design dsMCs performing functions inside the cell.
Here, we report a cell-free method for the production of dsMCs less than 250 bp in size. We designed a nicked double-stranded DNA oligonucleotide blunt-ended substrate for efficient minicircle production in a ligase-mediated and bending protein-assisted circularization reaction at high DNA concentration. Various minicircles can be produced containing a variety of site-specific chemical modifications or with controlled extent of physiological supercoiling. As a proof of principle of our method, we designed a minicircle endowed with the capacity to bind efficiently the transcription factor NF-κB and to inhibit its transcriptional activity in cellulo. This novel production method might be exploited to help in the design of minicircles as therapeutic nucleic acids.
MATERIALS AND METHODS
Single-stranded oligodeoxynucleotides from Eurogentec were synthesized with or without chemical modifications as indicated in the text. For the oligodeoxynucleotide sequence composition incorporated into the minicircles designed for NF-κB-dependent biochemical and biological studies, we used the Jaspar data base (http://jaspar.binf.ku.dk) to select minicircle sequence with the highest and lowest score for specific NF-κB binding and non-specific sequences including the design of scrambled minicircles, respectively. Proteinase K, T4 DNA ligase, T4 polynucleotide kinase and Exonuclease III were purchased from Thermo Fisher, T5 exonuclease and low molecular weight DNA ladder were from New England Biolabs and CircLigase from Epicentre. Full length rat NF-кB p50 homodimer and bacterial formamidopyrimidine (Fpg) repair protein were purchased from Promega and New England Biolabs, respectively. Bal31 nuclease was from New England Biolabs. Streptavidin and SYBR Green I nucleic acid gel stain were purchased from Sigma-Aldrich. Tumor Necrosis Factor α (TNFα) was from Miltenyi Biotec.
The pET-28a-derived vector containing the ARS binding factor 2 protein (Abf2p) gene kindly provided by F. Culard (CNRS-CBM, Orléans, France), was used for the transformation of BL21 (DE3) pLysS (Novagen). The transformed strain was grown at 37°C to an OD600 of 0.5 and induced with isopropyl-1-thio-β-D-galactopyranoside (Calbiochem) at a final concentration of 100 μM for 6 h. After centrifugation, the cells were resuspended in a sonication buffer (50 mM Tris-HCl, 500 mM NaCl, 20 mM imidazole, 5 mM β-mercaptoethanol pH 8) supplemented with a protease inhibitors mixture (Sigma-Aldrich) and 100 mM phenylmethylsulfonylfluoride (Pierce) and then sonicated. All purification steps were performed at 4°C. The lysate obtained after extract centrifugation was applied to a nickel agarose column (Qiagen) and N-terminal histidine-tagged Abf2p was eluted with buffer containing 100 mM imidazole. Abf2p was concentrated to 3 mg/ml on an Amicon Ultra-4 10K (Millipore) and stored at −80°C in a buffer containing 25 mM Tris-HCl, 100 mM NaCl, 1 mM dithiothreitol, 5% glycerol pH 8 (protein purity > 95%). In addition to its high production yield, His-tagged-Abf2p presents the advantage of supporting several cycles of thawing and freezing and long storage time at −80°C for at least 2 years that make this protein a robust biochemical DNA bender.
Production of double-stranded closed relaxed and supercoiled minicircles
Preparation of nicked DNA templates was performed by mixing equimolar quantities of complementary single-stranded oligonucleotides in buffer containing 10 mM Tris-HCl and 25 mM NaCl, pH 7.5 at a concentration of 40 μM; hybridization was carried out by slowly cooling the oligonucleotides mixture from 80°C to 15°C. A standard circularization reaction was carried out by adding one of the overlapping nicked duplexes shown in Supplementary Data at a final concentration of 2 μM in a buffer containing 40 mM Tris-HCl, 10 mM MgCl2, 10 mM DTT, 0.5 mM ATP and 5% glycerol, pH 7.8. Abf2p was next added to a final concentration of 3 μM followed by T4 DNA ligase addition (10 units); the reaction mixture was then incubated at 20°C for 1 h yielding nicked minicircle intermediate. Next, the reaction mixture was sequentially incubated with proteinase K (0.3 units, 30 min at 55°C), T4 polynucleotide kinase with 1 mM ATP (100 units, 30 min at 37°C), T4 DNA ligase (25 units, 1 h at 20°C) and exonuclease T5 (200 units, 30 min at 37°C) to obtain covalently closed minicircle after carrying out a final incubation with proteinase K. The same protocol was followed to generate supercoiled minicircle in the presence of increasing concentrations of Ethidium Bromide (Sigma-Aldrich) as indicated in Figure 2A except that a final drug extraction step was carried out in the presence of butanol. The minicircle was then precipitated by addition of AcONH4 to a final concentration of 2.5 M followed by addition of 2 volumes of cold ethanol. After centrifugation at 14 000 rpm for 15 min, the pellet was washed by addition of 70% ethanol and then resuspended in buffer containing 10 mM Tris-HCl, 1 mM EDTA pH 7.5; the optical density of the dsMC sample was measured to determine its concentration and to control that OD260/280 was larger than 1.8 corresponding to high quality DNA. The overall yield for a one pot production of pure dsMC from in phase nicked linear duplex was 30% (50 μg of 95 bp minicircle produced per ml of circularization reaction with an initial amount of input DNA of 150 μg). As shown in pathway 3 on Figure 2A, the single nicked circular DNA was also treated with exonuclease III (2 h at 37°C) to yield single-stranded minicircle which was then purified using a silica bead DNA gel extraction kit (Thermo Scientific). Next, the single-stranded minicircle was allowed to hybridize with the complementary strand of 95 nucleotides (nt) or with both complementary strands of 40 and 55 nt yielding respectively single or double-nicked minicircle. A control single-stranded minicircle of 95 nt in length was produced by circularization of a single-stranded oligonucleotide by CircLigase enzyme according to the manual instructions.
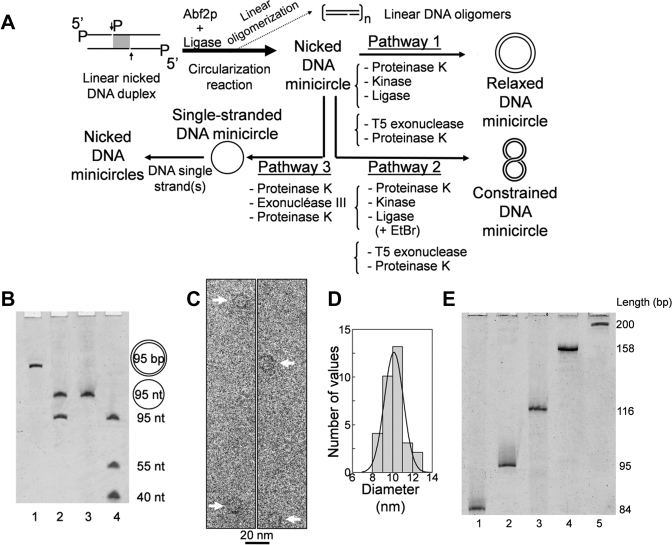
Figure 2.
CNDCA-based production of minicircles. (A) Scheme illustrating the method of DNA circularization through CNDCA and the derived pathways used to prepare relaxed, supercoiled dsMC and nicked minicircles. The filled space in grey within the central segment of the starting linear duplex refers to the overlapped region with nick indicated by an arrow. P indicates the presence of a phosphate group at DNA 5΄ end. The length of the starting linear DNA has an integral number of helical turns to prepare the relaxed covalently closed DNA minicircle. (B) Image of stained denaturing polyacrylamide gel showing the 95 bp double-stranded minicircle (lane 1) and the nicked DNA minicircle (lane 2) respectively generated according to pathways 1 and 3. The denaturing conditions enabled strand separation of the single nicked duplex showing that the covalently closed minicircle is free of strand nick as confirmed by the position of the migration of control DNA samples indicated on the right side. (C) Cryo-electron microscopy micrograph showing 95 bp closed minicircles produced according to pathway 1; scale bar indicates 20 nm. (D) Histogram of the longest diameters measured from cryo-electron micrograph of 95 bp minicircles (n = 32). The Gaussian fit provides a mean diameter value of 10.1 ± 1.2 nm. (E) Image of stained denaturing polyacrylamide gel showing DNA minicircles of various sizes as generated according to pathway 1; the sequence of input linear nicked DNA duplexes with integer multiples of the helical repeat is shown in Supplementary Data.
Gel electrophoresis analysis
The products formed in the circularization reaction were analyzed under native polyacrylamide gel electrophoresis (PAGE). A total of 130 ng of input DNA was loaded on a 5% native gel (19:1, acrylamide:bisacrylamide (w/w) in TBE buffer, i.e. 90 mM Tris borate, 1 mM EDTA, pH 8.3) followed by a migration at 12.5 V/cm for 90 min. Electrophoresis of dsMCs was also performed in the same conditions but in the presence of 10 mM MgCl2 to separate minicircle topoisomers. Each topoisomer is characterized by a decrease in linking number (Lk) (underwound molecules with the possibility of negative supercoiling formation). The first 95 bp minicircle topoisomer shown in Figure 3 with a value of ΔLk = −1 is characterized by a superhelical density (σ) equal to 0.11 as deduced from the formula ΔLk/Lk0 where Lk0 is the number of helical turns in the original linear molecule. Minicircles of various sizes generated in the circularization reaction were analyzed on 8% denaturing gel (19:1, acrylamide:bisacrylamide; 8 M urea; TBE); DNA samples in formamide were heat denatured and electrophoresed at 18 W for 90 min.
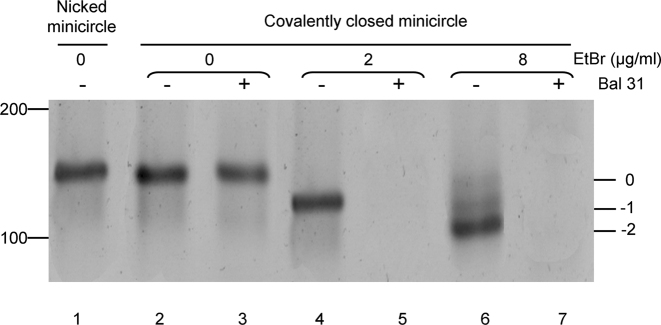
Figure 3.
CNDCA-based production of supercoiled minicircle. Image of stained native polyacrylamide gel showing 95 bp minicircle topoisomers of decreased linking number (ΔLk values of −1 and −2) (lanes 4, 6) as indicated on the right and obtained by religation of the nicked minicircle in the presence of EtBr. The slowest migrating band corresponds to the relaxed minicircle that migrates expectedly at the same rate as the nicked minicircle (lanes 1 and 2). Digestion of minicircle samples by the nuclease Bal31 is indicated on the right. The position of the molecular mass markers in bp is indicated on the left.
Different binding assays were used for interaction studies of the linear DNA substrate and dsMCs according to the DNA interacting proteins. NF-κB p50/p50 protein was incubated with the 95 bp minicircle (10 ng) as a function of the protein concentration in 10 mM Tris-HCl, 50 mM NaCl, 3 mM DTT, 0.2 mM EDTA, 2% glycerol, 50 μg/ml acetylated BSA, pH 7.5 for 30 min at 4°C. The protein concentration used was 5, 20, 40, 100, 120 nM and 2, 5, 20, 40, 60, 180 nM in the presence of minicircle containing 1 and 2 κB binding sites, respectively. The DNA binding activity of Abf2p and NF-κB was analyzed by electrophoretic mobility shift assay (EMSA). The reaction mixture was loaded on a native 5% polyacrylamide gel (19:1, acrylamide:bisacrylamide (w/w); TBE) and then electrophoresed for 1 h. After staining and quantification of band intensity, the fraction of DNA bound to protein for each retarded band was calculated according to the following equation: fraction minicircle bound = [protein-minicircle complex/total minicircle]. NF-κB binding data obtained with minicircle containing a single κB binding site were fitted to a square hyperbola using non-linear least squares analysis with the origin 9.0 software (Microcal). When the minicircle contained two κB binding sites, the binding curves corresponding to unbound, singly bound and doubly bound forms were fitted as previously described (20) where the dissociation constant for each binding site was assumed to be identical since these two binding sites are identical. The cooperativity parameter was determined by measuring the maximum value of the singly bound form using equation numbered 16 (20).
For the gel-based methodology, gel staining was carried out by using SYBR Green before exposition to a Typhoon phosphorimager to determine bands intensity using ImageQuant 5.1 software. The DNA was visualized by fluorescence and thus the molar ratio of the starting linear duplex over oligomers was calculated by taking into account the fact that band intensity is increased as a function of the oligomer length.
Cryo-electron microscopy
A 3 μl drop of 95 bp minicircle solution at 248 μg/ml concentration in buffer containing 10 mM Tris-HCl, 1 mM EDTA, pH 8 was deposited onto a glow-discharged quantifoil R2/2 grid (Jena) and vitrified in liquid ethane using a cryo-fixation device (UMR 8502 CNRS-LPS, Orsay, France). The cryo-specimens were transferred to a Gatan 626 cryo-holder (Gatan Inc.) and examined at −180°C with a JEM 2011 cryo-transmission electron microscope (Jeol) using an acceleration voltage of 200 kV and a 50 000-fold magnification. Images were recorded with 1 to 2.5 μm of defocus under low-electron-dose micrograph conditions on a Kodak SO163. The calculated diameter of minicircle was derived from the formula P/ where P is the perimeter as determined by multiplying the bp number of the linear DNA with the bp width equal to 0.34 nm. The longest diameters measurement was performed from analysis of 32 imaged minicircles using Image J software. The histogram shown in Figure 2D was obtained by splitting the range of diameter values into 5 equal-sized classes as derived from Scott's normal reference rule. The distribution was fitted with a Gaussian function using the origin 9.0 software (Microcal) providing a mean value of the diameter as indicated further. It was also verified whether the diameter values measured in the sample were normally distributed by using the Shapiro–Wilks normality test. As we failed to reject the null hypothesis with a P-value of 0.35, the diameter values were normally distributed. We then performed a parametric test to determine whether the mean value of diameters was statistically different from the diameter value calculated for a perfect round-shaped minicircle. The one-sample t-test with null hypothesis that the difference between measured diameters and calculated diameter values is zero, failed to reject the null hypothesis with a large P-value of 0.39 indicating no statistical difference between the mean diameter value and the calculated diameter value. Statistical analysis was derived from NIST/SEMATECH e-Handbook of statistical methods at (http://www.itl.nist.gov/div898/handbook/index.htm).
where P is the perimeter as determined by multiplying the bp number of the linear DNA with the bp width equal to 0.34 nm. The longest diameters measurement was performed from analysis of 32 imaged minicircles using Image J software. The histogram shown in Figure 2D was obtained by splitting the range of diameter values into 5 equal-sized classes as derived from Scott's normal reference rule. The distribution was fitted with a Gaussian function using the origin 9.0 software (Microcal) providing a mean value of the diameter as indicated further. It was also verified whether the diameter values measured in the sample were normally distributed by using the Shapiro–Wilks normality test. As we failed to reject the null hypothesis with a P-value of 0.35, the diameter values were normally distributed. We then performed a parametric test to determine whether the mean value of diameters was statistically different from the diameter value calculated for a perfect round-shaped minicircle. The one-sample t-test with null hypothesis that the difference between measured diameters and calculated diameter values is zero, failed to reject the null hypothesis with a large P-value of 0.39 indicating no statistical difference between the mean diameter value and the calculated diameter value. Statistical analysis was derived from NIST/SEMATECH e-Handbook of statistical methods at (http://www.itl.nist.gov/div898/handbook/index.htm).
Cell culture, intracellular delivery of DNA minicircle and NF-κB transcriptional activity
The NF-κB luciferase reporter cells were stable NF-κB-RE-luc2P Human Embryonic Kidney (HEK) 293 cell line purchased from Promega. Cells were grown at 37°C with 5% CO2 in Dulbecco's modified Eagle's medium (DMEM) supplemented with fetal bovine serum (FBS) (10%), hygromycin B (50 μg/ml), 100 IU penicillin and streptomycin (100 μg/ml). One day before transfection, stable HEK293-NF-κB-luciferase reporter cells were seeded onto 24 well plates. When the cells reached 60% confluence, they were transfected in triplicate with either an unspecific or specific minicircle containing two κB binding sites using the cationic polymer histidinylated linear polyethyleneimine (His-lPEI) (Polytheragène) as transfection agent (21). Briefly, His-lPEI (1 mg/ml) was diluted two times in 10 mM Hepes pH 7.4 and 25 μl of the diluted transfection agent was added to a tube containing dsMC at a concentration of 50 μg/ml in 100 μl of 10 mM Hepes pH 7.4. After gentle mixing, the solution was kept at room temperature for 30 min to form polyplex-containing minicircle. The size and ζ potential of the polyplex-containing minicircle was respectively 129 nm and 34 mV as measured with nano Partica SZ-100 (Horiba). The final dsMC concentration for each well plate is as indicated in the text. After 4 h at 37°C in 5% CO2 incubator, 400 μl of DMEM with FBS (18%) with or without human TNFα (50 ng/ml) was added as previously described (22); cell viability was quantitated 12 h later, using Alamar Blue assay (Invitrogen) and luciferase expression was monitored after cell lysis. Firefly luciferase gene expression was measured using luciferin as described elsewhere (23). Luciferase activity was normalized to total protein using BCA protein assay kit (Uptima). All measurements were performed in triplicate and are presented as mean ± S.D. based on six independent experiments. A paired Student's t-test was used to evaluate statistically significant differences in the luciferase activity of cells treated with control versus κB minicircle (Figure 5B). P-values < 0.05 were considered statistically significant.
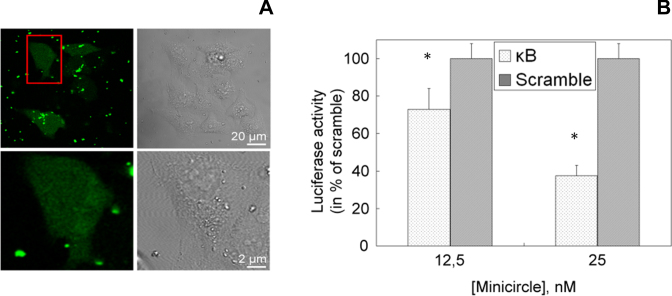
Figure 5.
95 bp κB minicircle delivered into cells inhibits NF-κB transcriptional activity. (A) Confocal microscopy images show that the minicircle is internalized in HEK293 cells; the red rectangular frame indicates the selected cell whose size is magnified 10 times as represented in the lower part (right: bright-field image of HEK293 cells; left: the fluorescence channel of fluorescein). (B) Responsiveness of the luciferase reporter gene driven by a NF-κB responsive promoter in the HEK293 cell line delivered with a minicircle containing two κB sequences (κB) as compared to the control non-specific minicircle (scramble). A specific decrease of luciferase activity is observed as a function of κB minicircle delivering concentration indicating κB minicircle-dependent NF-κB transcriptional inhibition. Data are means ± S.D. obtained from triplicate wells and are representative of six independent experiments (*P < 0.05).
For the intracellular localization of dsMCs, HEK293 cells were seeded in a 4-well Lab-Teck chambered coverglass (Nunc). The following day, cells were transfected using His-lPEI and fluorescein-labeled minicircles prepared from fluorescent linear duplexes (Supplementary Data) by our circularization-based method. Subsequently, imaging of living cells was carried out using a Zeiss Axiovert 200M inverted microscope (equipped with a Plan-Apochromat 63x objective, NA = 1.4) coupled with LSM 510 scanning confocal head (Carl Zeiss).
RESULTS AND DISCUSSION
DNA circularization reaction at high DNA concentration
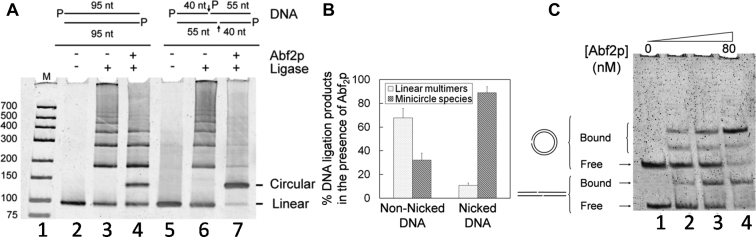
We were first interested in revisiting the ligase-mediated multimerization–circularization reaction at high DNA concentration (2 μM) in the absence or presence of a bending protein. The electrophoresis experiment shown in Figure 1A indicates that a blunt-ended 95 bp duplex with phosphorylated 5΄-ends that co-migrates expectedly at similar distance to the molecular weight marker of 100 bp (lanes 1 and 2), formed mainly linear dimers and also higher molecular weight multimers in ligase-mediated reaction in the absence of bending protein (lane 3); no monomer circular species was formed as deduced from the absence of any ligation product migrating at the expected position as indicated (Figure 1A, right side) (24). This result shows that circularization of linear DNA smaller than the persistence length is inefficient as expected (25) and indicates that T4 DNA ligase efficiently joined blunt-ended DNA duplexes (26). Next, we studied the high DNA concentration circularization reaction in the presence of a bending protein to tentatively enable an intramolecular circularization reaction. For that purpose, we used the well-known bending protein Abf2p that has two conserved boxes named box A and box B like the high-mobility-group (HMG) box domain bending protein HMGB1 (27). Abf2p was found to induce DNA circularization as the major ligation reaction in the low DNA nanomolar concentration range (Supplementary Data), in agreement with the circularization efficiency of other bending proteins including HMGB1 (A+B) (28). In contrast, when the circularization reaction is carried out at high DNA concentration of 2 μM in the presence of Abf2p (Figure 1A, lane 4), 95 bp monomer minicircle species represents only 30 ± 6% of total ligation products (Figure 1B). Therefore, the high DNA concentration promotes the contaminating intermolecular reaction over the cyclization intramolecular reaction the efficiency of which is thus not sufficient to generate a high amount of minicircle species.
Figure 1.
Concentrated Nicked DNA Circularization Assay (CNDCA) allows high yield formation of minicircle species. (A) Image of stained native polyacrylamide gel analyzing the differential formation of DNA minicircles and DNA multimers in a ligase-assisted circularization reaction with non-nicked (lanes 2–4) versus nicked (lanes 5–7) linear DNA substrates of 95 bp in size with an identical sequence. The circularization reactions were carried out at elevated DNA concentration (2 μM) in the presence and absence of the bending protein Abf2p as indicated. M denotes molecular weight DNA ladder and the size in bp is indicated on the left. Arrow and P indicate strand nick and phosphorylation at the 5΄ terminus, respectively. (B) Quantification of overall DNA multimers and minicircle species formed in the cyclization reaction in the presence of Abf2p with input nicked versus non-nicked DNA. (C) Electrophoretic mobility shift assay showing comparative binding activity of increasing concentration of the bending protein Abf2p with 95 bp linear nicked DNA used in CNDCA and nicked 95 bp minicircle of identical sequence.
To implement DNA circularization efficiency in a sequence-independent manner, we introduced a DNA structural distortion to stimulate the protein-assisted circularization reaction with a catalyst-like activity. A DNA nick which is known to induce flexible bending (17,29,30), is specifically recognized by HMGB1 in a structure-specific manner (31) indicating that a DNA nick could increase the bending protein assisted-circularization reaction efficiency. In addition, DNA nick is efficiently sealed by T4 DNA ligase (32) that fulfills our requirement to generate a final minicircle product devoid of nicks. We therefore designed a nicked double-stranded 95 bp blunt-ended duplex containing one internal nick per strand, the base sequence being identical to the non-nicked duplex used above. This nicked duplex possesses a centrally placed overlapping region of 15 bp as schematically drawn in Figure 1A, indicating the nucleotide length of DNA strands used to build it. In the absence of bending protein, both nicked and non-nicked duplexes, which have identical electrophoretic mobility, yielded only linear multimers arising from the high DNA concentrated circularization reaction (Figure 1A, lanes 3,6). In contrast to the non-nicked duplex (lane 4), the circularization reaction of the nicked duplex in the presence of Abf2p generated a huge amount of circular species with a faint amount of linear multimers and almost complete consumption of the starting duplex (lane 7). As shown in Figure 1B, minicircle species represent near 90% of all ligation products when circularization reaction is carried out with the nicked DNA substrate. Thus, our assay named Concentrated Nicked DNA Circularization Assay (CNDCA) gave a high yield of DNA circular species that has never been reported previously. This circularization efficiency can be explained by several key physico-chemical parameters specific to our CNDCA. Abf2p exhibits the same order of binding activity with the starting linear nicked DNA substrate and the minicircle end product (Figure 1C); this could be due to the fact that Abf2p has no C-terminal acidic tail (27) that was previously shown to be responsible for the well-known preferential recognition of minicircles by an HMG-box domain protein (33). This Abf2p binding activity is different from previous data that reported that bending protein exhibited a large binding preference for dsMC over linear DNA by at least one order of magnitude, resulting in a weak circularization efficiency (34). Thus, Abf2p is likely not significantly displaced from the nicked DNA substrate in the course of minicircle formation, contributing to CNDCA efficiency. We also determined whether the yield of circularization reaction was dependent on nicks position. For that purpose, the centrally positioned overlapping region containing nicks was displaced close to one or other 95 bp duplex end. Circularization reaction was found to be decreased near 5-fold with nicks situated close to DNA ends as compared to nicks in the central position of DNA (Supplementary Data). CNDCA is thus effective when nicks are in central position of the linear DNA substrate that strongly suggests that DNA bending induced by Abf2p occurs in the center of DNA bringing its two ends into close proximity.
Methodology for one pot CNDCA-based DNA production of DNA minicircles
We then designed a CNDCA-based strategy allowing one pot preparation of DNA minicircles. The production of minicircles of increasing size was achieved by using starting blunt-ended nicked duplexes containing one or several overlapping regions to avoid the use of long oligodeoxynucleotides, since full length oligodeoxynucleotide synthesis by the solid-phase phosphoramidite method enables high yield and purity in the range of less than 65 nucleotides in length. Therefore, the presence of nicks within the DNA substrates is relevant not only for high circularization efficiency but also for producing minicircles of increasing size without the need to use long oligodeoxynucleotides. As shown in Figure 2A, the starting linear nicked duplex having phosphorylated 5΄-ends except at one internal nick position yielded an intermediate nicked dsMC used in turn for the production of either relaxed (pathway 1), constrained (pathway 2) dsMCs or controlled nicked minicircles from purified single-stranded minicircles (pathway 3). Final minicircle products arising from pathways 1 and 3 using an overlapping 95 bp duplex with an integer number of helical repeats as starting linear DNA substrate were analyzed by denaturing PAGE (Figure 2B). dsMC obtained from pathway 1 migrated as one band indicating that it is entirely covalently closed (lane 1) while the single nicked minicircle assembled according to pathway 3 expectedly yielded two bands arising from strand dissociation under the denaturing conditions. Migration of the 95 nt single-stranded minicircle control and starting linear oligodeoxynucleotides (lanes 3 and 4) confirmed the double- and single-stranded nature of the minicircles generated from pathways 1 and 3, respectively. Next, the 95 bp dsMC generated from pathway 1 was directly visualized by cryo-electron microscopy (cryo-EM) in similar conditions to those previously used (35). Our purpose is to know whether this minicircle is close to perfect circle and thus without constrain which should be the case as DNA used here has an integer number of helical repeats. As can be seen on the micrograph shown in Figure 2C, 95 bp dsMCs have an apparently regular circular shape. Next the longest diameters were measured from minicircles in the sample (n = 32) and histogram of measured diameters is shown in Figure 2D. The measured diameters distribution was then fitted with a Gaussian function providing a mean diameter value of 10.1 ± 1.2 nm. Conducting a one-sample-t test (see Material and Methods), we next verified that there was no statistical difference between this mean diameter value and the calculated diameter for perfect round-shaped 95 bp minicircle (10.3 nm). These cryo-EM data are in agreement with the formation of near perfect round-shaped 95 bp dsMC through pathway 1 as indicated by the mean diameter value close to that for perfect minicircle and by the normal distribution of diameter values which is likely resulting from error distribution and not from conformational variability of minicircles. Consistent with our cryo-EM observation of near perfect round circles that are expected to be torsionally unconstrained, the 95 bp dsMC exhibited the same electrophoretic mobility as control relaxed nicked 95 bp minicircle and was insensitive to Bal31 endonuclease activity indicating that the 95 bp dsMC is relaxed (Figure 3, lanes 1–3). We conclude that the 95 bp dsMC generated from pathway 1 is close to perfect regular circle and consequently unconstrained, in agreement with previous cryo-EM-based studies indicating that DNA minicircles with a different length but with an integer number of helical repeats, are also relaxed (19,36).
As analyzed by denaturing PAGE (Figure 2D), this strategy can generate relaxed minicircles of various sizes (84–200 bp) from correspondingly linear nicked DNA duplexes with an integer number of helical repeats (Supplementary Data, duplexes D2 to D6). The overall yield to generate double-stranded minicircles according to pathways 1 was nearly 30% allowing dsMC production in the range of several hundreds of micrograms with the possibility to easily scale up the production to milligram quantity by simply increasing the reaction volume to 20 ml. Decreased efficiency for minicircles production was observed down to 84 bp indicating the lower limit to the size of minicircles generated by this method. We also determined whether CNDCA-based strategy could generate minicircles from linear nicked duplexes whose length is no longer an integer multiple of the helical repeat to produce intermediate-size DNA minicircles. Linear nicked duplexes of 90 and 100 bp with semi-integer number of helix turns generate minicircle with an overall production yield of 18%. This lower production efficiency was due to a decrease in the circularization yield as indicated by a 2-fold decrease of circular species amount formed from out of phase nicked DNA duplexes of 90 and 100 bp in size. When the circularization reaction was carried out with non-nicked linear DNA of 90 and 100 bp, circular species formation was nearly completely inhibited (Supplementary Data). This differential circularization efficiency indicates that linear nicked DNA substrate makes circularization reaction still effective for minicircles production from out of phase DNA duplexes.
Minicircles functionalized with chemical modifications or containing physiological supercoiling
Commercially available synthetic DNA oligonucleotides functionalized with various base analogs and chemical modifications were used to generate functionalized minicircle for various applications as biochemical tools and potential therapeutics. Firstly, we were interested in the production of 95 bp minicircle containing a base modification such as 8-oxoguanine residues that is formed naturally when DNA is subjected to oxidative conditions or ionizing radiations. Such lesion is substrate for the well known bacterial-derived formamidopyrimidine (Fpg) repair protein that catalyzes the excision of 8-oxoguanine yielding a strand nick (37), the formation of which can be easily exploited to directly reveal the presence and the location of such base residue within functionalized minicircle. The production yield of single and double 8-oxoguanine-modified minicircles was insensitive to the presence of base modification within starting overlapping modified duplex (Supplementary Data). When the minicircle containing a single or double base modification was incubated in the presence of Fpg, we observed a pattern of DNA fragments on denaturating PAGE that corresponds to cleavage of DNA strand at expected position of 8-oxoguanine present within minicircles allowing to validate our strategy for production of minicircles with site specifically base modification (Supplementary Data). Secondly, dsMCs with site-specifically placed labels were produced. After completion of our protocol using input nicked duplex containing one biotin residue linked by a spacer to the C5-atom of pyrimidin ring, we obtained a pure closed dsMC with similar yield to that obtained for unmodified minicircle production. The biotine-labeled minicircle was able to bind to streptavidin as shown by the slower migration of complex (Supplementary Data). Biotinylated minicircles could be used for further biochemical applications based on the use of streptavidin coated surfaces.
As schematically shown in pathway 2 on Figure 2A, our method enabled also production of constrained dsMC based on religation of nicked dsMC in the presence of Ethidium Bromide (EtBr), an intercalating drug inducing a double helix linking number decrease by lowering the base pairs twist. Because biophysical characterization of small constrained 95 bp minicircle was not previously reported, we used native PAGE together with Bal31 nuclease assay that are well known technical approaches previously used to determine the formation of topoisomers and the presence of DNA distortions, respectively (13,15,18,36). A total of 95 bp dsMCs produced in the presence of EtBr exhibit an increase in electrophoretic mobility as a function of drug concentration, yielding two major bands corresponding to negatively supercoiled topoisomers of decreased linking number (ΔLk values of −1 and −2) (Figure 3, lanes 4, 6). This indicates that the overall compaction of minicircles is increased as a consequence to DNA writhing (13,15) as confirmed by magnesium dependent effect on the electrophoretic migration of topoisomers (Figure 3 and Supplementary Data). The existence of writhe structure was previously observed with a constrained 178 bp minicircle (15) but this is the first time to our knowledge that it is also observed with small 95 bp minicircles. We next used Bal31 nuclease to detect the presence of DNA distortions within our constrained minicircles. We found that both minicircle topoisomers are nuclease sensitive on the contrary to relaxed closed minicircle formed by religation in absence of EtBr (Figure 3 lanes 3,5,7) indicating helix destabilization in constrained minicircle with near physiological superhelical densities (σ values in the range of −0.1 to −0.2). Our data indicate that pre-bent DNA within small 95 bp minicircle is still capable to writhe likely through kink out of plane bending that corresponds to a new upper limit of DNA bending strain and also confirm the capacity of our production method to generate supercoiled minicircles.
DNA minicircles containing one to several recognition sites for transcription factor binding activity
To exemplify the versatility of our method regarding the production of dsMCs with various sequence compositions and in order to design minicircles for potential therapeutic applications, we next produced relaxed minicircles containing one or several consensus sequences 5΄-GGGACTTTCC/5΄-GGAAAGTCCC of the immunoglobulin К-light-chain gene enhancer (κB). This sequence is part of numerous important nuclear factor К-light-chain enhancers of activated B cells (NF-κB) transcription factor-responsive genes (38). As anticipated from the molecular basis of our CNDCA-based minicircle production, the yield for minicircles containing one or several κB motifs was unchanged compared to minicircles with a random sequence composition and in particular was independent of the κB sequence position as described below. Our method for dsMC production thus provides the opportunity to study extensively the recognition mechanism of various DNA-binding proteins with pre-bent DNA as exemplified below with the transcription factor NF-κB using 95 bp dsMCs.
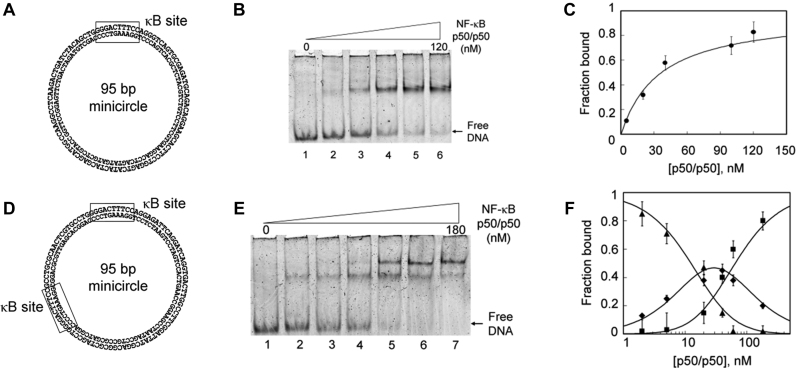
A relaxed closed 95 bp minicircle containing a single κB binding site (Figure 4A) was incubated with increasing concentrations of NF-κB p50/p50 protein and the reaction mixture analyzed by EMSA. As shown in Figure 4B, a retarded band appeared upon addition of the NF-κB protein indicating the formation of a NF-κB/minicircle complex. Under these conditions, no gel-shifted complexes were detectable when the minicircle has no κB binding site revealing the specific nature of NF-κB interactions (Supplementary Data). As deduced from the half-maximal binding equilibrium (Figure 4C), NF-κB exhibits a significant affinity for κB minicircle with a Kd value of 30 ± 3.6 nM that is similar to that reported with specific linear DNA substrates (39). Our data indicate that the pre-bent κB sequence did not substantially modulate NF-κB DNA binding activity that suggests that DNA bending is involved in indirect readout of κB sites by NF-κB. Our observation is in line with the fact that several linear κB sequences adopt a pre-bent conformation (40).
Figure 4.
Design of minicircles containing one and two κB binding sites efficiently bound by NF-κB p50/p50. (A and D) Sequence of 95 bp minicircles containing one and two κB binding sequences as indicated by the rectangular frame. (B and E) EMSA showing titration of minicircle by increased concentrations of NF-κB p50/p50 as indicated under Materials and Methods. (C and F) Plot of data from panels B and E, respectively; data were fitted to a square hyperbola using non-linear least squares analysis and according to previously reported equations (20) related to unbound (▴), singly bound (♦) and doubly bound (▪) DNA.
In order to test the capacity for a minicircle to attract specifically more than one NF-κB p50/p50 protein, we therefore prepared a 95 bp minicircle containing two κB sites (Figure 4D). The binding activity of NF-κB with this minicircle was carried out as described above. As shown in Figure 4E, two retarded bands are now present on the EMSA gel upon addition of the NF-κB protein, indicating the binding of two proteins per minicircle. To investigate the possible influence of the first bound protein on the specific binding of the second one, we used the previously described calculation that defines a protein binding cooperativity parameter (see Materials and Methods). This parameter was experimentally determined from the representation shown in Figure 4F where the fraction of each DNA species detected from EMSA was plotted as a function of the protein concentration. We found that the cooperativity parameter is 1.2, indicating that NF-κB binding to the first site did not notably distort the second κB binding site structure to modulate the protein affinity. When the second specific site was placed in a nearly opposite position to the first one, we still found that NF-κB bound with identical affinity to each binding site. However, when a third binding site was present, there was a significant decrease activity in binding to the minicircle that could partly be explained by protein hindrance and/or conformational change of the third binding site. Overall, this protein–DNA minicircle interaction study illustrates the potential of our methodology for the design of minicircles with various sequence compositions and shows for the first time that NF-κB efficiently binds pre-bent recognition sequences in a topologically closed DNA domain.
Inhibition of NF-κB cellular transcriptional activity by the κB minicircle
The two κB sites minicircle produced by our CNDCA-based strategy was evaluated for its capacity to modulate in cellulo NF-κB-dependent transcription by its specific NF-κB binding activity. For that purpose, we next designed a new experiment based on the so-called decoy approach. The classical decoy strategy consists in the intracellular delivery of a short and linear DNA double-stranded oligonucleotide that contains one specific binding site able to trap a transcription factor of interest that in turn inhibits its activity (41). Here, our strategy consists in using a minicircle instead of linear DNA. We delivered either the κB minicircle or a control minicircle intracellularly using HEK293 cells that stably expressed a luciferase-reporter gene driven by a NF-κB responsive promoter.
In order to determine whether the DNA minicircle was properly internalized in HEK293 cells, a fluorescein-labeled 95 bp κB minicircle was delivered in cells thanks to His-lPEI, a transfection agent known to transfer DNA inside cells and to efficiently trigger its endosome escape into cytosol (42). The transfected cells exhibited a few fluorescent spots and diffuse fluorescence throughout the cytoplasm that likely correspond respectively to κB minicircles complexed with His-lPEI inside endosomes and to released minicircles that are located in both the cytosol and nucleus compartments (Figure 5A). Indeed, molecules comprising fewer than 100 bp are known to diffuse easily inside the cytosol and can enter the nucleus (4). A similar observation was made with the non-specific minicircle (data not shown). Interestingly, the κB minicircle delivered intracellularly induced a specific and significant decrease in luciferase activity in a dose-dependent manner as shown in Figure 5B. The results show a 60% decrease in luminescence in cells transfected with a 25 nM concentration of κB minicircle. For comparison, a linear anti-NF-κB decoy DNA oligonucleotide was previously shown to induce a similar decrease in luciferase activity at a 2-fold higher concentration than that used here with the κB minicircle in the same experimental conditions (22). Our data indicate that the κB minicircle was able to efficiently inhibit NF-κB-dependent transcriptional activity and hence acted as a ligand for cellular NF-κB. The CNDCA-based methodology opens the possibility of designing DNA minicircles as new decoy nucleic acids directed against NF-κB as shown here and also against other relevant transcription factors known as targets for the treatment of several human diseases (41).
CONCLUSION
Our novel method enables the quantitative production of a variety of DNA minicircles containing customized sequence and including the possibility of chemical functionalizations. The minicircles produced herein support the binding and the enzymatic activity of proteins such as transcription factor and DNA repair protein. The capacity of DNA minicircle bearing κB motifs to inhibit NF-κB transcriptional activity in human cells paves the way for the design of minicircles as new decoy nucleic acids. In this regard, minicircles containing various transcription factor binding sites could be developed for multitargeting as many promoters are known to be regulated by several transcription factors in a compensatory manner.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank F. Culard for providing Abf2p expression vector and for helpful discussions; we are indebted to C. Gonçalves for her technical assistance in the design of cell-based experiments and to David Gosset for confocal microscopy experiments. The authors would like also to thank S. Bourg and F. Piazza for helpful discussions.
Footnotes
Present address: Thomas Thibault, Universidade Estadual de Campinas, Hemocentro, Campinas-São Paulo, Brazil.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Ministère de l'Enseignement Supérieur et de la Recherche and the University of Orléans, Ph.D funding [to T.T.]; The Ligue contre le cancer Région Centre et Loiret [to P.M., P.B., J.-M.M.]; CNRS-CEA ‘METSA’ French network (FR CNRS 3507) [to J.-M.M., J.D.]. Funding for open access charge: The Ligue contre le cancer Région Centre et Loiret.
Conflict of interest statement. None declared.
REFERENCES
- 1. Zhao N., Fogg J.M., Zechiedrich L., Zu Y.. Transfection of shRNA-encoding Minivector DNA of a few hundred base pairs to regulate gene expression in lymphoma cells. Gene Ther. 2011; 18:220–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stenler S., Wiklander O., Badal-Tejedor M., Turunen J., Nordin J.Z., Hallengärd D., Wahren B., Andaloussi S.E., Rutland M.W., Smith C.I.E. et al. . Micro-minicircle gene therapy: implications of size on fermentation, complexation, shearing resistance, and expression. Mol. Ther. Nucleic Acids. 2014; 2:e140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Catanese D.J. Jr, Fogg J.M., Schrock D.E. 2nd, Gilbert B.E., Zechiedrich L.. Supercoiled minivector DNA resists shear forces associated with gene therapy delivery. Gene Ther. 2012; 19:94–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lukacs G., Haggie P., Seksek O., Lechardeur D., Freedman N., Verkman A.S.. Size-dependent DNA mobility in cytoplasm and nucleus. J. Biol. Chem. 2000; 275:1625–1629. [DOI] [PubMed] [Google Scholar]
- 5. Kreiss P., Cameron B., Rangara R., Mailhe P., Aguerre-Charriol O., Airiau M., Scherman D., Crouzet J., Pitard B.. Plasmid DNA size does not affect the physicochemical properties of lipoplexes but modulates gene transfer efficiency. Nucleic Acids Res. 1999; 27:3792–3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gonçalves D.P., Schmidt T.L., Koeppel M.B., Heckel A.. DNA minicircles connected via G-quadruplex interaction modules. Small. 2010; 6:1347–1352. [DOI] [PubMed] [Google Scholar]
- 7. Mayer G., Ackermann D., Kuhn N., Famulok M.. Construction of DNA architectures with RNA hairpins. Angew. Chem. Int. Ed Engl. 2008; 47:971–973. [DOI] [PubMed] [Google Scholar]
- 8. Rasched G., Ackermann D., Schmidt T., Broekmann P., Heckel A., Famulok M.. DNA minicircles with gaps for versatile functionalization. Angew. Chem. Int. Ed Engl. 2008; 47:967–970. [DOI] [PubMed] [Google Scholar]
- 9. Bates A.D., Noy A., Piperakis M.M., Harris S.A., Maxwell A.. Small DNA circles as probes of DNA topology. Biochem. Soc. Trans. 2013; 41:565–570. [DOI] [PubMed] [Google Scholar]
- 10. Witz G., Stasiak A.. DNA supercoiling and its role in DNA decatenation and unknotting. Nucleic Acids Res. 2010; 38:2119–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Peters J.P., Maher L.J.. DNA curvature and flexibility in vitro and in vivo. Q. Rev. Biophys. 2010; 43:23–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kahn J.D. Topological effects of the TATA Box binding protein on minicircle DNA and a possible thermodynamics linkage to chromatin remodeling. Biochemistry. 2000; 39:3520–3524. [DOI] [PubMed] [Google Scholar]
- 13. Fogg J.M., Kolmakova N., Rees I., Magonov S., Hansma H., Perona J.J., Zechiedrich E.L.. Exploring writhe in supercoiled minicircle DNA. J. Phys. Condens. Matter Inst. Phys. J. 2006; 18:S145–S159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hoess R., Wierzbicki A., Abremski K.. Formation of small circular DNA molecules via an in vitro site-specific recombination system. Gene. 1985; 40:325–329. [DOI] [PubMed] [Google Scholar]
- 15. Bednar J., Furrer P., Stasiak A., Dubochet J., Egelman E.H., Bates A.D.. The twist, writhe and overall shape of supercoiled DNA change during counterion-induced transition from a loosely to a tightly interwound superhelix. Possible implications for DNA structure in vivo. J. Mol. Biol. 1994; 235:825–847. [DOI] [PubMed] [Google Scholar]
- 16. Hagerman P.J. Flexibility of DNA. Annu. Rev. Biophys. Biophys. Chem. 1988; 17:265–286. [DOI] [PubMed] [Google Scholar]
- 17. Zhang Y., Crothers D.M.. High-throughput approach for detection of DNA bending and flexibility based on cyclization. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:3161–3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Du Q., Kotlyar A., Vologodskii A.. Kinking the double helix by bending deformation. Nucleic Acids Res. 2008; 36:1120–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Demurtas D., Amzallag A., Rawdon E.J., Maddocks J.H., Dubochet J., Stasiak A.. Bending modes of DNA directly addressed by cryo-electron microscopy of DNA minicircles. Nucleic Acids Res. 2009; 37:2882–2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cho S., Wensink P.C.. Linkage between oligomerization and DNA binding in Drosophila doublesex proteins. Biochemistry. 1998; 37:11301–11308. [DOI] [PubMed] [Google Scholar]
- 21. Billiet L., Gomez J.-P., Berchel M., Jaffrès P.-A., Le Gall T., Montier T., Bertrand E., Cheradame H., Guégan P., Mével M. et al. . Gene transfer by chemical vectors, and endocytosis routes of polyplexes, lipoplexes and lipopolyplexes in a myoblast cell line. Biomaterials. 2012; 33:2980–2990. [DOI] [PubMed] [Google Scholar]
- 22. Kasparkova J., Thibault T., Kostrhunova H., Stepankova J., Vojtiskova M., Muchova T., Midoux P., Malinge J.-M., Brabec V.. Different affinity of nuclear factor-kappa B proteins to DNA modified by antitumor cisplatin and its clinically ineffective trans isomer. FEBS J. 2014; 281:1393–1408. [DOI] [PubMed] [Google Scholar]
- 23. de Wet J.R., Wood K.V., DeLuca M., Helinski D.R., Subramani S.. Firefly luciferase gene: structure and expression in mammalian cells. Mol. Cell. Biol. 1987; 7:725–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huo Y.-X., Zhang Y.-T., Xiao Y., Zhang X., Buck M., Kolb A., Wang Y.-P.. IHF-binding sites inhibit DNA loop formation and transcription initiation. Nucleic Acids Res. 2009; 37:3878–3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Du Q., Smith C., Shiffeldrim N., Vologodskaia M., Vologodskii A.. Cyclization of short DNA fragments and bending fluctuations of the double helix. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:5397–5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Grasser K.D., Teo S.H., Lee K.B., Broadhurst R.W., Rees C., Hardman C.H., Thomas J.O.. DNA-binding properties of the tandem HMG boxes of high-mobility-group protein 1 (HMG1). Eur. J. Biochem. FEBS. 1998; 253:787–795. [DOI] [PubMed] [Google Scholar]
- 27. Diffley J.F., Stillman B.. A close relative of the nuclear, chromosomal high-mobility group protein HMG1 in yeast mitochondria. Proc. Natl. Acad. Sci. U.S.A. 1991; 88:7864–7868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Czapla L., Peters J.P., Rueter E.M., Olson W.K., Maher L.J.. Understanding apparent DNA flexibility enhancement by HU and HMGB architectural proteins. J. Mol. Biol. 2011; 409:278–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Furrer P., Bednar J., Stasiak A.Z., Katritch V., Michoud D., Stasiak A., Dubochet J.. Opposite effect of counterions on the persistence length of nicked and non-nicked DNA. J. Mol. Biol. 1997; 266:711–721. [DOI] [PubMed] [Google Scholar]
- 30. Protozanova E., Yakovchuk P., Frank-Kamenetskii M.D.. Stacked-unstacked equilibrium at the nick site of DNA. J. Mol. Biol. 2004; 342:775–785. [DOI] [PubMed] [Google Scholar]
- 31. Prasad R., Liu Y., Deterding L.J., Poltoratsky V.P., Kedar P.S., Horton J.K., Kanno S.-I., Asagoshi K., Hou E.W., Khodyreva S.N. et al. . HMGB1 is a cofactor in mammalian base excision repair. Mol. Cell. 2007; 27:829–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bullard D. R., Bowater R. P.. Direct comparison of nick-joining activity of the nucleic acid ligases from bacteriophage T4. Biochem. J. 2006; 398:135–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lee K.B., Thomas J.O.. The effect of the acidic tail on the DNA-binding properties of the HMG1,2 class of proteins: insights from tail switching and tail removal. J. Mol. Biol. 2000; 304:135–149. [DOI] [PubMed] [Google Scholar]
- 34. Kahn J.D., Crothers D.M.. Protein-induced bending and DNA cyclization. Proc. Natl. Acad. Sci. U.S.A. 1992; 89:6343–6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Amzallag A., Vaillant C., Jacob M., Unser M., Bednar J., Kahn J.D., Dubochet J., Stasiak A., Maddocks J.H.. 3D reconstruction and comparison of shapes of DNA minicircles observed by cryo-electron microscopy. Nucleic Acids Res. 2006; 34:e125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lionberger T.A., Demurtas D., Witz G., Dorier J., Lillian T., Meyhöfer E., Stasiak A.. Cooperative kinking at distant sites in mechanically stressed DNA. Nucleic Acids Res. 2011; 39:9820–9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Prakash A., Doublié S., Wallace S.S.. The Fpg/Nei family of DNA glycosylases: substrates, structures, and search for damage. Prog. Mol. Biol. Transl. Sci. 2012; 110:71–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chen F. E., Huang D.B., Chen Y. Q., Ghosh G.. Crystal structure of p50/p65 heterodimer of transcription factor NF-kappaB bound to DNA. Nature. 1998; 391:410–413. [DOI] [PubMed] [Google Scholar]
- 39. Phelps C.B., Sengchanthalangsy L.L., Malek S., Ghosh G.. Mechanism of kappa B DNA binding by Rel/NF-kappa B dimers. J. Biol. Chem. 2000; 275:24392–24399. [DOI] [PubMed] [Google Scholar]
- 40. Chen F.E., Ghosh G.. Regulation of DNA binding by Rel/NF-kappaB transcription factors: structural views. Oncogene. 1999; 18:6845–6852. [DOI] [PubMed] [Google Scholar]
- 41. Ahmad M.Z., Akhter S., Mallik N., Anwar M., Tabassum W., Ahmad F.J.. Application of decoy oligonucleotides as novel therapeutic strategy: a contemporary overview. Curr. Drug Discov. Technol. 2013; 10:71–84. [DOI] [PubMed] [Google Scholar]
- 42. Pichon C., Billiet L., Midoux P.. Chemical vectors for gene delivery: uptake and intracellular trafficking. Curr. Opin. Biotechnol. 2010; 21:640–645. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.