Abstract
The FADS1 and FADS2 genes in the FADS cluster encode the rate-limiting enzymes in the synthesis of long-chain polyunsaturated fatty acids (LC-PUFAs). Genetic variation in this region has been associated with a large number of diseases and traits many of them correlated to differences in metabolism of PUFAs. However, the causative variants leading to these associations have not been identified. Here we find that the multiallelic rs174557 located in an AluYe5 element in intron 1 of FADS1 is functional and lies within a PATZ1 binding site. The derived allele of rs174557, which is the common variant in most populations, diminishes binding of PATZ1, a transcription factor conferring allele-specific downregulation of FADS1. The PATZ1 binding site overlaps with a SP1 site. The competitive binding between the suppressive PATZ1 and the activating complex of SP1 and SREBP1c determines the enhancer activity of this region, which regulates expression of FADS1.
INTRODUCTION
LC-PUFAs are essential fatty acids to humans that cannot be synthesized de novo. They include ω-6 arachidonic acid (AA), ω-3 eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA) that need to be obtained through diet either directly or as their precursors linoleic acid (LA) and alpha-linolenic acid (ALA). Perturbation of the metabolism of LC-PUFAs may affect the risk of many diseases, including cardiovascular disease, cancer, and inflammatory and autoimmune diseases (1). The rate-limiting enzymes in the endogenous biosynthesis of LC-PUFAs are δ-5 desaturase (D5D) and δ-6 desaturase (D6D), that are encoded by fatty acid desaturase 1 (FADS1) and fatty acid desaturase 2 (FADS2), respectively (2). The FADS1 and FADS2 genes are oriented head to head on chromosome 11q12-13.1 in a cluster together with FADS3, which has an unknown function.
Both candidate gene based and genome-wide association studies (GWAS) on complex blood lipid traits have yielded significant hits in the FADS cluster, e.g. a strong correlation with levels of PUFAs in blood (3–11). The association between FADS variants and PUFAs profiles was recently replicated in the liver, which is the major tissue for the synthesis and metabolism of LC-PUFAs (12). Out of the genes in the FADS cluster, only FADS1 is differently expressed between genotypes in liver tissue (12–14). Moreover, GWAS have shown association of the FADS cluster to 35 different traits (Supplementary Tables S1 and S2) including levels of numerous lipids and increased risk of many diseases such as rheumatoid arthritis (15), colorectal cancer (16), Crohn's disease (17), inflammatory bowel disease (18), laryngeal squamous cell carcinoma (19) and several heart rate traits (20,21). These findings suggest the hypothesis that genetic variants in the FADS cluster regulate FADS1 activity, which in turn affects the synthesis of LC-PUFAs and thereby disease risk.
The identification of genetic variants regulating FADS expression is hindered by the high linkage disequilibrium (LD). We previously fine mapped the FADS region by genome-wide genotyping and targeted re-sequencing and identified two major haplotypes based on a LD block defined by 28 closely linked SNPs (13). The two haplotypes show dramatic differences in allele frequencies among human populations. The derived (D) haplotype is almost fixed in Africa, whereas indigenous populations in America instead have very high frequencies of the ancestral (A) haplotype. In Europe and Asia, both haplotypes exist and the D haplotype mostly have higher allele frequencies. Intriguingly, it has been proposed that the FADS cluster has been under selective pressure, probably as a result of adaptation to available nutritional sources of LC-PUFAs during human evolution. A recent publication from the 1000 Genomes Project (22) showed a signal of selection at the FADS locus in East Asian populations, a finding that was also supported by data from Kothapalli et al. (23). In a separate study where 230 ancient Europeans were analyzed, the FADS region was one of the top hits for selection in Neolitic Europe (24). Taken together with previous signatures for selection in Africa (13) as well as in the indigenous people of Greenland (25), a population that has lived on a specialized diet rich in LC-PUFAs for a long time, it now seems apparent that the FADS cluster has played an important role during our evolutionary history.
The D haplotype shows evidence of positive selection and confers a more efficient biosynthesis of LC-PUFAs from the precursors. In the present study, we attempted to identify the functional regulatory variants, by systematically testing SNPs in putative regulatory regions using luciferase reporter assay. Our results show that genetic variation in an Alu element, at the rs174557 locus, is a major contributor to the difference in FADS activity between the two haplotypes.
MATERIALS AND METHODS
Samples
DNA samples from the NSHPS cohort (26) were used for SMRT sequencing on the Pacific Biosciences RSII (PacBio) instrument. Two samples, homozygous for the A or the D haplotypes respectively in the FADS cluster were used for hybridization based target capture of the FADS region. The same two samples were employed as templates for PCR amplification of putative regulatory regions for luciferase assay. To study which allele of rs174557 is present on the A and D haplotypes respectively, a pooled sample containing DNA from 500 individuals (27) was used for PCR amplification and PacBio sequencing. The same approach was performed on DNA samples from two chimpanzees (Pan troglodytes) and one bonobo (Pan paniscus).
Identification of alleles present at rs174557 in a pooled DNA sample and in chimpanzee
PCR amplification of a region containing rs174557 as well as the two flanking SNPs rs174556 and rs174560 that tag the A/D haplotypes was performed in a pooled sample containing DNA from 500 individuals from the NSHPS cohort (27). The amplification was performed using the specific primers rs57-Pac-seqF and rs57-Pac-seqR (Supplementary Table S3) and the high fidelity PrimeSTAR GXL DNA polymerase generating a 1232-bp amplicon. Following amplification, a sequencing library was produced using the Pacific Biosciences 1.0 template preparation kit according to the manufacturer's instructions. The library was loaded on one SMRT cell and sequenced on the PacBio RS II instrument using C4-P6 chemistry and a 150 min movie time. The same method for PCR amplification and sequencing was applied to two chimpanzee samples as well as a sample from bonobo. Each of these three samples was run on a separate SMRT cell.
Targeted long-read sequencing of the FADS region in homozygous individuals
To re-sequence the region harboring rs174557 using long reads, custom RNA probes (SureSelect, Agilent technologies) were designed to tile across the FADS1 gene. The RNA probes were then used for hybridization based sequence capture and long-read PacBio sequencing of the two samples homozygous for the A and D haplotypes. PacBio libraries for Sure Select target enrichment were prepared according to the manufacturer's instructions, except Agencourt AMPure XP magnetic bead system protocols and PCR protocols that were modified to suit 2 kb fragments. Sequencing was performed on the PacBio RSII system using the C4-P6 chemistry and polymerase and a 240 min movie time.
Cell culture
HepG2 cells were originally purchased from the American Type Culture Collection (ATCC) and maintained in RPMI1640 basal medium supplemented with 10% fetal bovine serum (FBS) and 2 mM l-glutamine. MCF7 cells were maintained in DMEM basal medium supplemented with 10% FBS and 2 mM l-glutamine. 293T cells were grown in DMEM supplemented with glutamax, 10% FBS, 1 mM sodium pyruvate and 500 μg/ml Geneticin. All the cells were also supplemented with 100 units of penicillin and 100 μg of streptomycin per 1 ml of culture medium.
Luciferase constructs and reporter assay
The putative regulatory regions in the FADS cluster were first amplified by PCR from two samples homozygous for either haplotype A or haplotype D, respectively. To test putative enhancer activities, the amplified fragments were inserted upstream of the minimal promoter sequence of plasmid pGL4.23 (Promega). The promoterless plasmid pGL4.10 (Promega) was employed to test the putative promoter activities of FADS1 and FADS2. Variations in the 3΄UTR region of FADS1 were validated by inserting the testing regions between the coding sequence of luciferase and polyA signal in luciferase constructs driven either by the SV40 promoter or the FADS1 promoter. All the fragments were cloned into luciferase plasmids by making the primers either containing suitable overhanging restriction enzyme sites or containing the 15-bp homologous recombination arms for SLiCE cloning (28). To verify that the rs174557 locus is working as enhancer, the minimal enhancer region encompassing rs174557 that named rs57-70bp was also inserted between BamHI and SalI sites downstream of the SV40 polyA signal by SLiCE cloing in pGL4.23. All the resulting plasmids were verified by Sanger sequencing. Mutations were introduced either by site-directed mutagen with Phusion Hot Start Flex DNA Polymerase (NEB) following the manufacturer's instructions or by primer annealing and extension. Detailed information on primer sequences and cloning methods are available in Supplementary Tables S4 and S5.
HepG2 cells were plated one day before transfection in 96-well plates. The confluency was 50–70% on transfection. For normal luciferase assay, each well was transfected with 0.3 μl X-tremeGENE HP DNA transfection reagent (Roche) and 100 ng of experimental firefly luciferase reporter plasmid, and 1 ng of pGL4.74 renilla luciferase reporter vector as internal control for monitoring transfection and lysis efficiency. If not otherwise specified in the figure, the results are demonstrated as fold changes of the experimental plasmid over the empty control plasmid pGL4.23. For cotransfection studies, the other renilla luciferase reporter vector pRL-MP was created by replacing the SV40 promoter of pGL4.73 with the minimal promoter sequence of pGL4.23 in BglII and HindIII sites. This vector was used as internal control to minimize the chance of the promoter sequence of the internal control being regulated by the same cotransfected TFs. Each well was transfected with 0.3 μl X-tremeGENE HP DNA transfection reagent (Roche), 40 ng of appropriate firefly luciferase reporter plasmid, 50 ng expression plasmids and 10 ng of pRL-MP. For experiments cotransfecting PATZ1 and SREBP1c in Figure 7B and Supplementary Figure S13, the total amount of transfected expression plasmid was equalized to 50 ng with empty expression vector pCDNA3.1 if needed. The results are expressed directly as the ratio of luciferase activity from experimental plasmids to renilla luciferase activity from pRL-MP.
Figure 7.
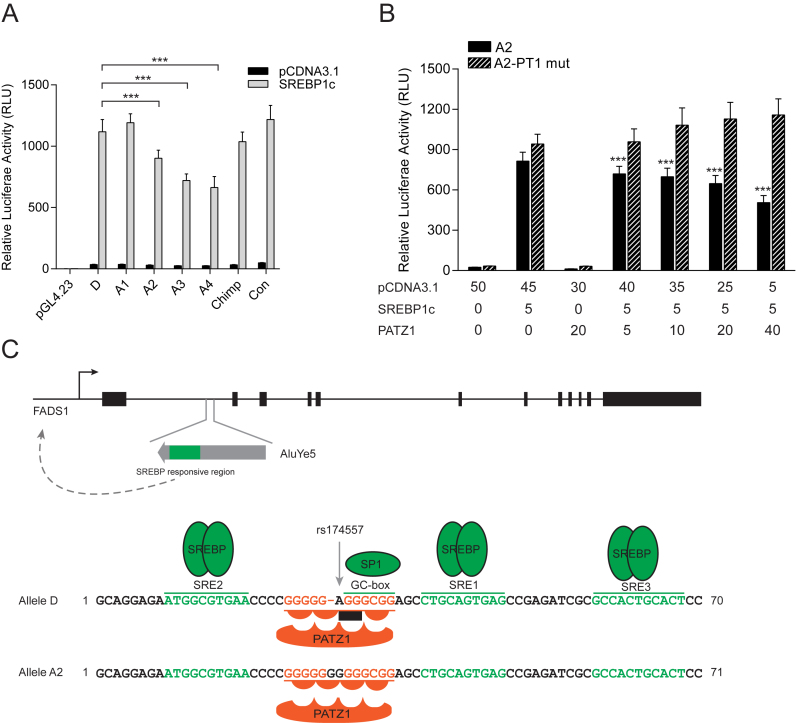
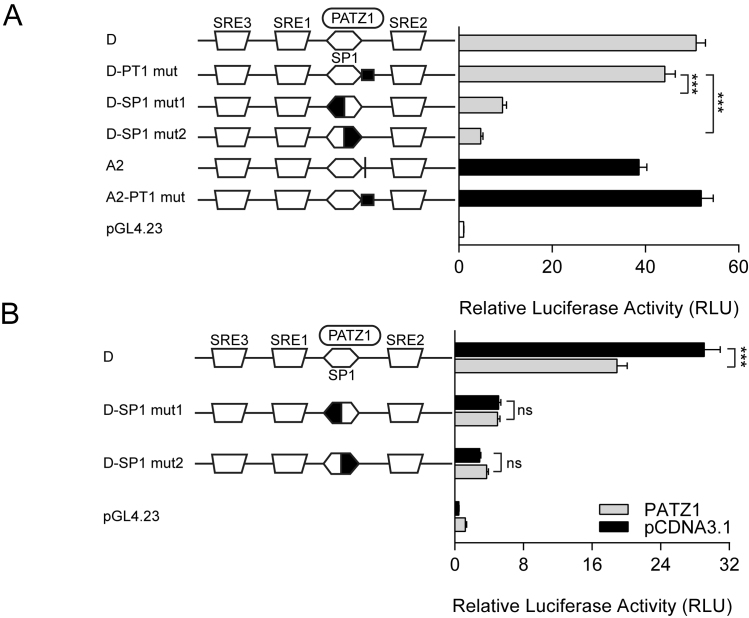
Competition between PATZ1 and SP1 for the overlapping GC-box element determines the extent of the rs174557 locus to be induced by SREBP1c. (A) The alleles of rs174557 respond differently to SREBP1 induction in luciferase assay. Abbreviations are as follows: Chimp, the chimpanzee allele; and con, the consensus sequence of AluYe5 element. ***P < 0.0001 compared with allele D of rs174557. (B) PATZ1 determines the degree of SREBP1c induction on allele A2 of rs174557 in a dosage dependent manner. A gradual increase of PATZ1 expression leads to a gradual decrease in enhancer activity. Disrupting the PATZ1 binding site (A2-PT1 mut) completely abolished the suppression by PATZ1. ***P < 0.0001 by one-way ANOVA when compared with allele A2 of rs174557 with only SREBP1c overexpressed. All the data in A and B came from three to four independent transfections each with six technical replicates. The bars represent mean±SD from all technical replicates. (C) Model of how FADS1 is regulated by PATZ1 binding to rs174557 with different affinities between alleles.
Cells were harvested 24 h after transfection and assayed with the Dual-Luciferase Reporter Assay System (Promega) either on an Infinite M200 pro reader (Tecan) or on a LB 9507 luminometer (Berthold Technologies). Luciferase results in Supplementary Figures S2–S4 and S7 came from one transfection experiment with each transfection has six technical replicates. All the other luciferase experiments came from at least three independent transfections i.e., unique plasmid preparations and transfections each with six technical replicates. Luciferase values are expressed as averages with error bars representing standard deviations (SDs) from all technical replicates. Statistical analyses were carried out by t tests except data in Figure 7B and S13 which were calculated by one-way ANOVA.
Plasmids construction and lentivirus production
The coding regions of human PATZ1 (long A isoform) was amplified from HepG2 cDNA and cloned into pCDNA3.1 by SLiCE cloning (28). pCDNA3.1-2xFLAG-SREBP-1c was a gift from Timothy Osborne (Addgene plasmid #26802) (29). Lentiviral plasmids overexpression PATZ1 and SREBP1c were constructed by fusing a 3xFLAG tag at the N-terminal of the coding sequence of PATZ1 and SREBP1c, respectively and inserted into pGreenPuro shRNA expression lentivector (SI505A-1, System Biosciences). The detailed primer information is listed in Supplementary Table S6. The identities of the plasmids were verified by Sanger sequencing. Lentivirus was produced in 293T cells by transfecting the constructed plasmid together with packaging plasmids pLP1, pLP2 and envelope plasmid pLP/VSVG (Life Technologies) using polyethylenimine (Polysciences) following the manufacturer's instructions. Cells were plated in 12-well plates and transduced with virus supernatant together with sequabrene (Sigma) at final concentration of 8 μg/ml. The cells stably overexpressing PATZ1 and SREBP1c were selected by Puromycin (Life Technologies) at concentration of 1 μg/ml. The selected cells were maintained in 0.5 μg/ml of Puromycin and regularly passaged for further analysis.
Quantitative real time PCR
At least three replicates were studied by quantitative real time PCR (qPCR) on cDNA samples and on genomic DNA samples from ChIP experiments each replicate with triplicated values. For each target, multiple primers were designed and tested to select the ones with the best specificity and the ideal efficiency. List of primers used in this paper is shown in Supplementary Table S3. The qPCR reactions were performed with SYBR Green JumpStart Taq ReadyMix (Sigma) or JumpStart Taq ReadyMix (Sigma) coupled with EvaGreen dye (Biotium).
For gene expression analysis, RNA was extracted from cells with TRIzol (Life Technologies) according to the manufacturer's instructions. The extracted RNA was dissolved in nuclease-free water and quantified using Nanodrop 2000 (Life Technologies). A total of 1 μg RNA was reverse transcribed into cDNA with Maxima First Strand cDNA synthesis kit (Life Technologies). Expression was normalized to three control transcripts including RPS18, ACTB and GAPDH. Statistical analyses were carried out by two-tailed t tests.
For ChIP experiments, a standard curve was generated for each primer pair with serial dilutions of sonicated input DNA each time when a ChIP was done. The amounts of immunoprecipitated DNA for different loci were first normalized to the input based on the standard curves. Then the relative enrichment of target DNA was calculated as fold increase over negative control regions. As rs174557 is homozygous for the D allele in HepG2 cells, specific primers targeting rs174557 were designed and optimized to validate its enhancer features. One gene desert region on chromosome 12 (GDCHR12) and one intronic region of CD36 gene were employed as negative controls and one region nearby the transcription start site of UROD gene was used as positive control. All the primer sequences used for ChIP-qPCR are listed in Supplementary Table S3.
Western blotting
Protein was extracted simultaneously with total RNA from samples lysed with TRIzol (Life Technologies) following the manufacturer's protocol with slight modification. The concentration of extracted protein was determined by Qubit 2.0 Fluorometer (Life Technologies). Protein samples were denatured in 1× NuPAGE LDS Sample Buffer, separated by NuPAGE Novex 4–12% Bis-Tris Protein gel (Life Technologies) and blot transferred onto PVDF transfer membranes by iBlot Dry Blotting System (Life Technologies). Membranes were blocked with 5% BSA in PBS, incubated with the appropriate antibody targeting PATZ1 (SC-292109, Santa Cruz Biotechnology), FADS1 (ab126706, Abcam), FADS2 (SC-98480, Santa Cruz Biotechnology), SREBP1 (SC-8984, Santa Cruz Biotechnology) and β-actin (SC-81178, Santa Cruz Biotechnology) (Supplementary Table S7). HRP-conjugated secondary antibody (SC-2004 or SC-2005, Santa Cruz Biotechnology) was employed to visualize the target protein under a CCD camera from ChemiDoc XRS System (Bio-Rad).
Chromatin immunoprecipitation
ChIP experiments were carried out as previously described with modifications (30,31). Cells were cultured in T175 flask with regular medium changed every other day until reaching confluency of >80%. Cells were crosslinked with 0.37% formaldehyde on a shaking platform for 10 min at room temperature and quenched with 125 mM glycine for 5 min. Cells were collected, washed twice with ice-cold PBS and resuspended in cell lysis buffer (10 mM Tris–HCl, pH 8.0, 10 mM NaCl and 0.2% NP-40) supplemented with protease inhibitor (Roche) to isolate nuclei. RIPA buffer (1 × PBS, 1%NP-40, 0.1% SDS, 0.5% sodium deoxycholate and 0.004% sodium azide) supplemented with protease inhibitor was used to isolate the cross-linked chromatin from nuclei. The isolated chromatin was sonicated to an average size of 300 bp using the Bioruptor UCD-300 sonication system (Diagenode). An aliquot was saved each time to be subjected to DNA extraction and used as input. The leftover was used for ChIP assay.
For ChIP experiments with antibody against histone modifications, the following antibodies were used which including H3K4me1 (ab8895, Abcam), H3K4me2 (ab7766, Abcam), H3K4me3 (ab8580, Abcam), H3K27ac (ab4729, Abcam) and H3K27me3 (ab6002, Abcam) (Supplementary Table S7). Normal rabit IgG (12-370, Millipore) was also included to check the background of antibody nonspecific binding. Essentially, 4 μg of antibody together with 40 μl Dynabeads protein G (Life Technologies) was added simultaneously into sonicated chromatin coming from 1 × 107 cells and incubated on a rotating platform overnight at 4°C. For ChIP experiments against transcription factors, chromatin from 1.5–4 × 107 cells was first precleared by incubating with Dynabeads for 2 h at 4°C. The supernatant was separated with a magnetic separation stand and incubated overnight at 4°C with 8 μg of antibody against SREBP1 (SC-8984, Santa Cruz Biotechnology) or 8 μg of antibody against SP1 (SC-59, Santa Cruz Biotechnology) or 4 μg of antibody against FLAG epitope (F7425, Sigma). The immune complexes were captured by incubation with 75 μl of Dynabeads protein G at room temperature for 1 h. For all ChIP assays, a series of washing steps was applied to Dynabeads protein G after immunoprecipitation including four times of washing with RIPA buffer, two times of washing with ChIP wash buffer (10 mM Tris–HCl, pH 8.0, 0.25 M LiCl, 10 mM EDTA, 1% NP-40 and 1% sodium deoxycholate) and once with TE buffer. The DNA–protein complexes were eluted in direct elution buffer (10 mM Tris–HCl, pH 8.0, 5 mM EDTA, 0.3 M NaCl and 0.5% SDS) and together with the previous saved sonicated lysates to be used as input were treated with RNase A and proteinase K respectively and incubated at 65°C overnight to reverse crosslink. DNA was purified with Agencourt AMPure XP beads (Beckman Coulter) following the manufacturer's instructions, eluted in 10 mM Tris–HCl (pH 8.0) and ready to be analyzed by quantitative real time PCR. Input DNA was quantified by Nanodrop 2000 (Life Technologies) and serial diluted to be used as templates for setting up stand curves of different primers targeting candidate regions.
Electrophoretic mobility shift assays
HepG2 cell nuclear extract with ectopically expressed 3xFLAG-PATZ1 was extracted using NucBuster Protein Extraction kit (Novagen) and quantified with Qubit 2.0 Fluorometer (Life Technologies). The oligonucleotides were minimized to be around 20 bp in length with several nucleotides flanking both sides of the motif sequence for PATZ1. A list of probes used in this assay can be found in Supplementary Table S3. Both biotinylated and non-biotinylated probes were annealed with reverse complementary nucleotide sequences by NEB2 buffer (NEB). A total of 6 μg nuclear extracts were incubated with 200 fmol of biotinylated double-stranded probes for 40 min on ice in 10 mM Tris–HCl, 80 mM KCl, 1 mM DTT, 1 μg of Poly (dA-dT), 7.5% glycerol, 0.063% NP-40 and 2 mM MgCl2. For competition assay, extra amounts of unlabeled probes were added to the binding reaction and further incubated at room temperature for 20 min. Samples were separated in Criterion 5% TBE Precast Gels (Bio-rad), and electro-transferred into a Genescreen plus hybridization transfer membrane (Perkin Elmer). DNA–protein complexes were cross-linked using UV-light and detected by LightShift Chemiluminescent EMSA Kit (Life Technologies).
RESULTS
rs174557 is the functional variant
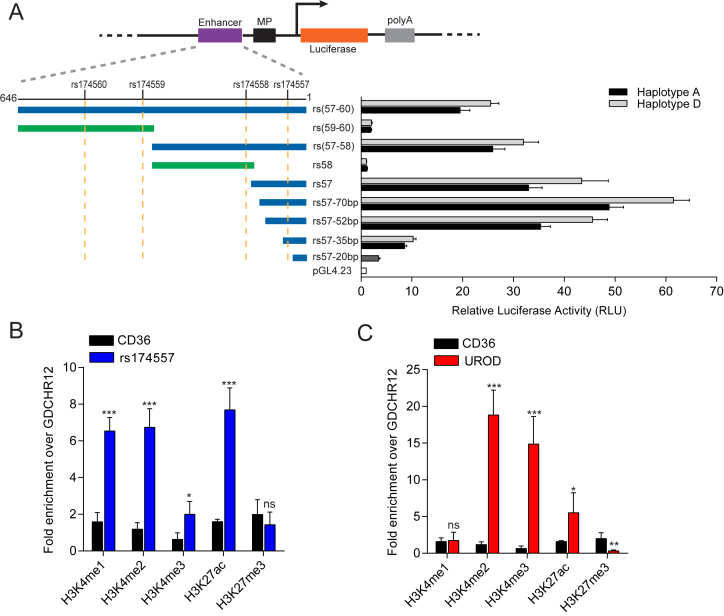
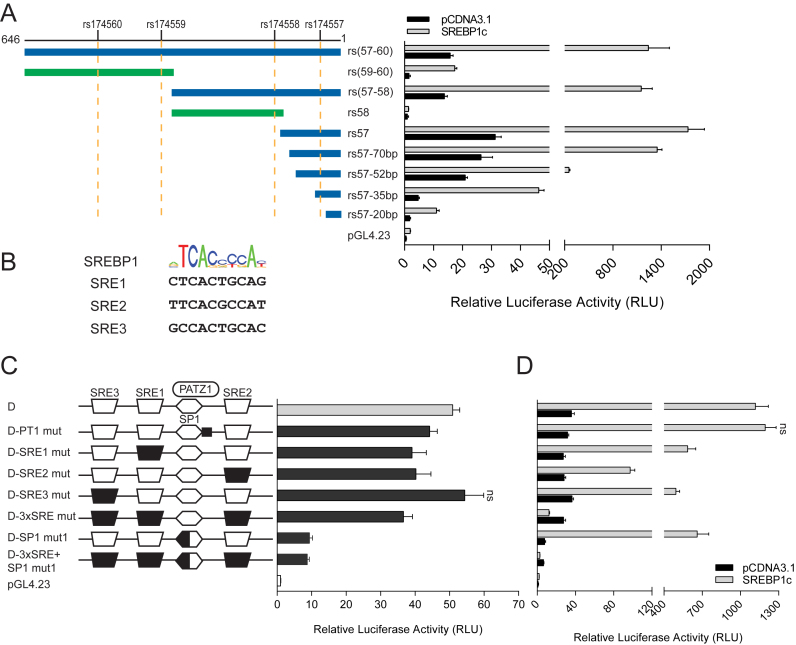
To identify functional variants that explain the difference in FADS activity between the two main haplotypes, putative regulatory regions were selected based on Chromatin Immunoprecipitation (ChIP) coupled to high-throughput sequencing (ChIP-seq) signals of histone modifications in liver tissue (32) and in HepG2 cells from the ENCyclopedia Of DNA Elements (ENCODE) project (33). The selected regions included promoters, introns, intergenic and 3΄UTR regions of FADS1 and FADS2 genes, which were evaluated by dual luciferase assay (Supplementary Figure S1). DNA was amplified from two samples homozygous for either haplotype D or A in the FADS cluster. Sanger sequencing showed a total of 19 SNPs in candidate regulatory regions (Supplementary Table S8). Out of the 19 SNPs tested by luciferase assay (Supplementary Figures S2–S4), a 646-bp fragment located in intron 1 of FADS1 consistently showed significant differences in luciferase assay between the two haplotypes and was selected for further study (Figure 1A and Supplementary Figure S3A). Strong enhancer activity was observed when the fragment was inserted upstream of the minimal promoter of pGL4.23 as compared to the control plasmid. Since there are four SNPs (rs174557, rs174558, rs174559 and rs174560) in this fragment, we generated a series of truncation constructs on both haplotypes to identify the functional variant. Only constructs containing rs174557 showed an enhancer activity comparable to the original 646-bp fragment. More importantly, significant differences between the alleles were still observed with constructs containing only rs174557 indicating that it is the functional SNP (Figure 1A). Verification that rs174557 is indeed functional was further obtained by site-directed mutagenesis in which a point mutation of rs174557 from allele A (later identified as allele A2 in Figure 2A) to allele D significantly increased the enhancer activity. In comparison, site-directed mutagenesis of rs174558 did not show any difference in luciferase assay (Supplementary Figures S5A).
Figure 1.
Identification of rs174557 as the functional variant. (A) Only luciferase constructs harboring rs174557 showed strong enhancer activity with the minimal enhancer region mapped to fragment rs57-70bp. Fragments containing each haplotype or allele were inserted upstream of the minimal promoter sequence of pGL4.23. The luciferase activities were normalized to the empty control vector pGL4.23. All the constructs containing rs174557 showed significant differences (P < 0.0001) between haplotype D (light gray bars) and haplotype A (black bars) in luciferase assay. Values are expressed as the mean from three independent transfections each with six technical replicates. Error bars represent SD from all technical replicates. Abbreviations are as follows: MP, minimal promoter; and polyA, the SV40 polyA signal. (B) The rs174557 locus showed classical enhancer features in HepG2 cells identified by ChIP-qPCR on different histone modifications. (C) The transcription start site of UROD gene was employed as the positive control for ChIP. For B and C, two genomic regions CD36 and GDCHR12 were used as negative controls. All the values are expressed as fold changes over GDCHR12. The error bars represent SD from five replicates with P value calculated by two-tailed t tests. *P < 0.05; **P < 0.01; ***P < 0.001 and ns, not significant compared with GDCHR12.
Figure 2.
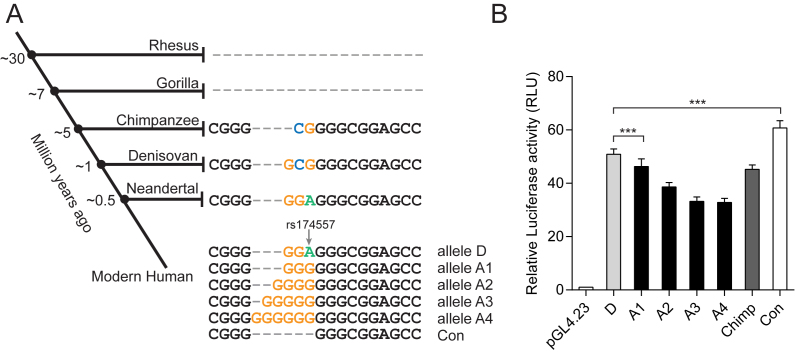
Evolution of the rs174557 locus. (A) Illustration of how the AluYe5 element and SNP rs174557 has evolved with multiple alleles detected in modern humans determined by PacBio sequencing in pooled genomic DNA from 500 samples. (B) Enhancer activity of all the alleles of rs174557 in human, the chimpanzee allele (Chimp) and the consensus sequence of AluYe5 element (Con). All the data came from three independent transfections each with six technical replicates. Error bars represent SD with ***P < 0.0001 calculated by two-tailed t tests when compared with the D allele of rs174557.
Enhancers are presumed to work in a position-independent fashion (34). The minimal enhancer region named rs57-70bp identified through truncation constructs (Figure 1A) was therefore inserted downstream of the SV40 polyA signal in pGL4.23. This fragment showed enhancer activity with a significant difference between the two alleles although the extent of activation was less when compared to insertions upstream of the minimal promoter (Supplementary Figures S5B).
The SNP rs174557 is located in a transposable element (TE) that belongs to a subfamily of Alu elements named AluYe5 (35). There is growing evidence that TEs could work as enhancers (36,37). Four other AluYe5 elements homologous to the rs174557 locus were cloned and inserted upstream of the minimal promoter sequence of pGL4.23 to test their activity. Three out of the four inserts showed enhancer activity in HepG2 cells supporting the idea that rs174557 can be a functional variant located in a TE (Supplementary Figure S5C). The rs174557 locus is devoid of ChIP-seq signals in ENCODE likely due to difficulties in unique alignment of reads to this region. To validate that the locus harboring rs174557 has an enhancer activity in vivo, we sought to characterize its enhancer features in HepG2 cells by ChIP followed by quantitative real time PCR (ChIP-qPCR) of different histone modifications. A fragment named GDCHR12 from one gene desert region of chromosome 12 and one intronic region from CD36 were employed as negative controls not enriched with any active histone modification markers. Compared with the two negative control regions, the rs174557 locus showed classical enhancer features characterized by enrichment of H3K4me1, H3K4me2 and H3K27ac, but not of H3K4me3 (Figure 1B). One region near the transcription start site of UROD was used as positive control and showed active promoter features with high signals for H3K4me3, H3K4me2 and H3K27ac, but not for H3K4me1 (Figure 1C).
Alleles and haplotypes at rs174557
Haplotype D has been associated with higher FADS1 expression at both the mRNA and the protein level (12,13). Because of its location in an AluYe5 element, rs174557 was not included in the previously defined haplotypes (13). To determine the linkage between alleles of rs174557 and the two haplotypes, a region of 1.2 kb in length, including the whole AluYe5 element with rs174557 and the flanking SNPs rs174556 and rs174560 from the 28-SNP haplotype, was PCR amplified in pooled genomic DNA from 500 European individuals and sequenced on the PacBio instrument. The pooled sequencing of these 1000 chromosomes confirmed that rs174557 was linked to the A/D haplotypes in 95.8% of cases (Supplementary Table S9). In a separate experiment, long-read targeted re-sequencing of the FADS region was performed using a hybridization-based capture method and PacBio sequencing in two individuals homozygous for the D or A haplotype, respectively. These experiments further verified that the D allele of rs174557 conferring higher enhancer activity was located on the D haplotype. In the pooled DNA, we detected multiple alleles on the A haplotype, and named them based on the length of the consecutive guanosine stretches at rs174557 (Figure 2A). The presence of multiple alleles for rs174557 was supported both by the recently annotated variant rs375569409 that corresponds to allele D and A3 and by direct Sanger sequencing of PCR products with genomic DNA from different sources as templates which verified alleles D, A1 and A2 (Supplementary Figure S6). At present, we cannot determine if allele A1–A4 are tagged by other SNPs.
The AluYe5 element containing rs174557 in intron 1 of FADS1 has only been detected in human and chimpanzee, while it is absent from rhesus and gorilla, indicating that this insertion has occurred relatively recently in primate evolution (Figure 2A). PacBio sequencing of the 1.2-kb PCR amplicon from DNA of two chimpanzees and one bonobo confirmed the chimpanzee reference sequence (panTro4), which is different from all the alleles observed in rs174557 in human. Neanderthals may have the D allele and Denisovans a sequence similar to chimpanzee, although the observations from these hominids are uncertain since they are based on relatively low numbers of sequence reads. The AluYe5 insertion event thus is likely to have occurred 5–7 million years (MY) before present, while the D allele of rs174557 in humans has previously been dated to about 300 000 years ago (13).
To test if all the identified alleles at rs174557 could function as enhancers, we amplified and inserted the minimal enhancer fragment rs57-70bp of each allele into pGL4.23 upstream of the minimal promoter sequence. All the constructs showed strong enhancer activity compared to the control plasmid. Significant differences were observed between allele D and all the A alleles (alleles A1–A4). Intriguingly, a gradual decrease in enhancer activity accompanied a gradual increase in the number of consecutive guanosine nucleotides in allele A1 through A4 (Figure 2). This is in accordance with a significant increase in enhancer activity when the consensus sequence of AluYe5, which has the least number of repeated guanosines, was inserted suggesting possible involvement of a suppressor binding differently to rs174557.
PATZ1 binds directly to rs174557 and regulates FADS1 expression
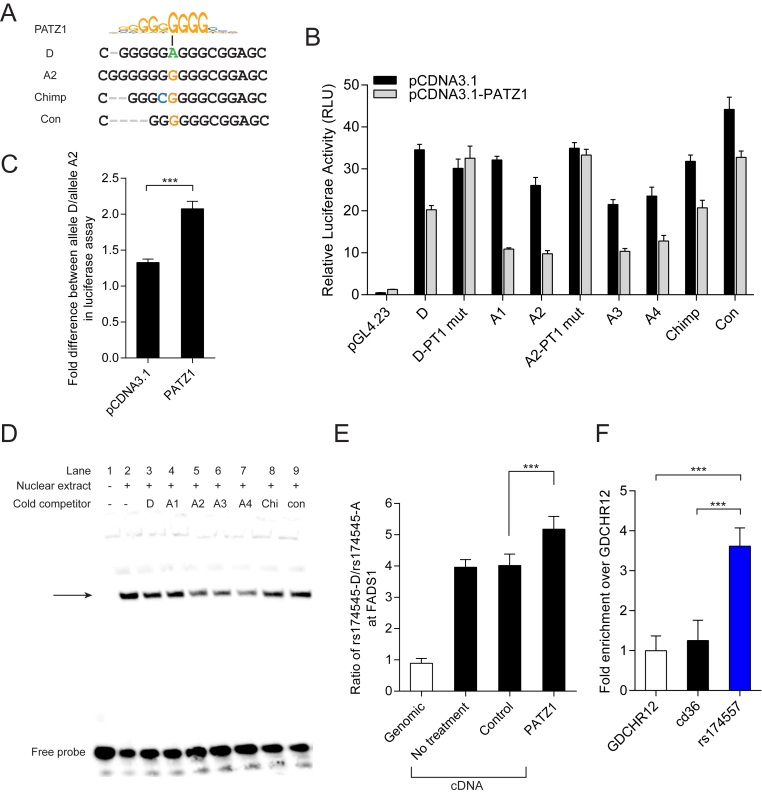
We searched for transcription factors (TFs) predicted to bind differently to rs174557 using the TRAP tool (38). Among the three most promising candidates SP1, MAZ and PATZ1, only PATZ1 induced allele specific responses in luciferase assay and was selected for further study. PATZ1 was predicted to preferentially bind to all the A alleles (Figure 3A). In accordance with the motif prediction, overexpression of PATZ1 significantly suppressed the enhancer activity (Supplementary Figure S7A). Strikingly, allele A2 was significantly more suppressed by PATZ1 compared to allele D in different luciferase constructs containing rs174557 (Figure 3C and Supplementary Figure S7B). To test the specificity of PATZ1 binding to different alleles, all the alleles of rs174557 were subjected to PATZ1 overexpression in luciferase assay and all the A alleles were more potently suppressed compared to allele D (Figure 3B). The construct containing the consensus AluYe5 sequence, which has a partially disrupted PATZ1 binding site, showed a strongly attenuated response to PATZ1 suppression as compared to all the other alleles. Introduction of point mutations in the PATZ1 binding site with allele D and allele A2 as templates (luciferase constructs D-PT1 mut and A2-PT1 mut, respectively in Figure 3B with detailed mutations listed in Supplementary Figure S8), completely abolished binding of PATZ1 to rs174557 and the suppression caused by PATZ1 overexpression.
Figure 3.
PATZ1 suppresses FADS1 expression by directly binding to rs174557. (A) The D allele of rs174557 creates a less conserved PATZ1 motif compared with allele A2 (43). (B) The alleles of rs174557 were differently suppressed by PATZ1 overexpression in luciferase assay. Disrupting the predicted PATZ1 motif with both allele D (D-PT1 mut) and allele A2 (A2-PT1 mut) as templates completely abolished the suppression. Values are expressed as mean (±SD) from three independent transfections each with six technical replicates. (C) Allele A2 of rs174557 is significantly more suppressed by PATZ1 overexpression than allele D in luciferase assay. Values are expressed as fold changes of allele D versus allele A2 in luciferase activity in both control condition and under PATZ1 overexpression from B. ***P < 0.0001 with error bars represent SD. (D) The binding of biotinylated allele A2 to nuclear protein extract from HepG2 cells overexpressed with PATZ1 was differently competed by alleles of rs174557 in EMSA. The arrow indicates the shifted DNA–protein complex which was most potently competed by allele A2 (lane 5), allele A3 (lane 6) and allele A4 (lane 7). Abbreviations are as follows: Chi, chimpanzee allele; and con, the consensus sequence of AluYe5 element. (E) PATZ1 suppresses FADS1 expression in an allele specific manner with a preference for the A allele in MCF7 cells. The bars indicate allelic expression imbalance of FADS1 (mean±SD) from six replicates. ***P < 0.001 when comparing cells overexpressing PATZ1 with cells transduced with control virus. (F) ChIP-qPCR of PATZ1 chromatin binding at the rs174557 locus in HepG2 cells overexpressed with PATZ1 with antibody against FLAG epitope. The bars indicate fold changes over the negative control region GDCHR12. ***P < 0.001 calculated by two-tailed t tests. The experiments were repeated six times with error bars representing SD.
To characterize the binding preference of PATZ1 to different alleles of rs174557, we further employed electrophoretic mobility shift assay (EMSA). The biotinylated probe allele A2 was incubated with nuclear extract from HepG2 cells overexpressing PATZ1 and the specificity of the band where PATZ1 bound was verified by adding an extra amount of non-biotinylated probes to the protein-DNA complex (Supplementary Figure S9). In accordance with the luciferase assay, the binding of PATZ1 to biotinylated A2 allele was more efficiently competed by non-biotinylated A2, A3 and A4 alleles compared to the other alleles, which indicates that they bind PATZ1 more strongly (Figure 3D).
To determine if PATZ1 can directly regulate FADS1 expression, both HepG2 and MCF7 cells were transduced with lentivirus overexpressing PATZ1 with a 3xFLAG tag fused to its N-terminus. The expression level of PATZ1 was detected by western blot, and a dramatic increase was observed compared to the cells transduced with the control virus (Supplementary Figure S10). As the FADS1 locus in MCF7 cells is heterozygous D/A2 at rs174557, we examined whether the PATZ1 allele-specific binding on rs174557 caused allelic imbalance in FADS1 transcription using allele specific qPCR of one coding SNP, rs174545, in the 3΄UTR of FADS1 and in LD with rs174557. The D allele of rs174545 is preferentially transcribed in normal MCF7 cells. After PATZ1 overexpression the allelic imbalance in FADS1 expression was significantly increased further in favor of the D allele which is in accordance with the A alleles of rs174557 showing stronger PATZ1 binding (Figure 3E). To validate that PATZ1 indeed binds directly to rs174557 in vivo, we performed ChIP-qPCR with an antibody against the FLAG epitope in HepG2 cells overexpressing PATZ1. Compared to the two negative control regions, the rs174557 locus was significantly enriched with PATZ1 binding, which supports the hypothesis that PATZ1 regulates FADS1 expression by directly binding to the rs174557 locus (Figure 3F).
SREBP1c and SP1 mediate the enhancer activity at rs174557
Even though PATZ1 was identified to act as a suppressor binding directly to rs174557, this region showed strong enhancer activity. SREBP1c is a key TF targeting genes involved in fatty acid synthesis including both FADS1 and FADS2 (2). The enhancer activity of the 646-bp fragment encompassing rs174557, rs174558, rs174559 and rs174560 was highly induced by SREBP1c overexpression in luciferase assay (Figure 4A). To identify the minimal region responsible for the observed SREBP1c induction, we investigated the same set of truncation constructs on the D haplotype used to identify the minimal enhancer region of rs174557 (Figure 1A). The region responsible for SREBP1c induction was mapped to the minimal enhancer fragment rs57-70bp, which showed similar enhancer activity after SREBP1c overexpression as the original 646-bp fragment (Figure 4A). Three canonical sterol regulatory element (SRE) sites flanking rs174557 (Figure 4B) and one GC-box element bound by SP1 which overlaps with the PATZ1 binding site (Figure 7C) were predicted with the position weight matrices from the JASPAR database (39). Under normal conditions, the enhancer activity of the rs57-70bp fragment was mostly attenuated by point mutations in the GC-box element predicted to be bound by SP1. Significantly decreased enhancer activity was also observed when either the SRE1 or SRE2 site or all the three SRE sites were mutated (Figure 4C). There was no significant decrease in SREBP1c induction when the PATZ1 binding site was mutated. However, significantly attenuated responses to SREBP1c overexpression were observed in luciferase assay when mutations were introduced to any of the three SRE sites or the GC-box element (Figure 4D). Moreover, the response to SREBP1c induction was completely lost when all the three SRE sites were mutated. This suggests that all the three SRE sites and the GC-box element are functional and are essential for the full SREBP1c response. This conclusion is further supported by the results from the serial truncation constructs subjected to SREBP1c overexpression. A consecutive increase in luciferase activity was observed after SREBP1c overexpression among fragments rs57-20bp containing only one SRE site, rs57-35bp containing one SRE site and the GC-box element, rs57-52bp containing two SRE sites and the GC-box element and rs57-70bp containing all the three SRE sites and the GC-box element (Figure 4A).
Figure 4.
The enhancer activity observed in the rs174557 locus is mediated by SREBP1c and SP1. (A) The enhancer activity of the rs174557 locus is highly induced by SREBP1c. The minimal region responsive to SREBP1c induction was mapped to fragment rs57-70bp. The same luciferase constructs as in Figure 1A were subjected to SREBP1c overexpression in luciferase assay. (B) Three conserved SREBP1c binding sites were predicted in fragment rs57-70bp. (C and D) Disrupting the binding sites for SREBP1c and SP1 in fragment rs57-70bp attenuated its enhancer activity in normal condition (C) and under overexpression of SREBP1c (D) in luciferase assay. ns, not significant compared with the wild type D allele. All the other point mutations significantly decreased the enhancer activity (P < 0.0001 by two-tailed t tests) compared with the wild type D allele both in normal condition and under overexpression of SREBP1c. The mutations introduced to different motifs are shadowed in black in demonstration in C with the names of the mutation constructs listed. Details of point mutations made to each construct are shown in Supplementary Figure S8. All the data in A, C and D came from three independent transfections each with six technical replicates. Error bars, SD.
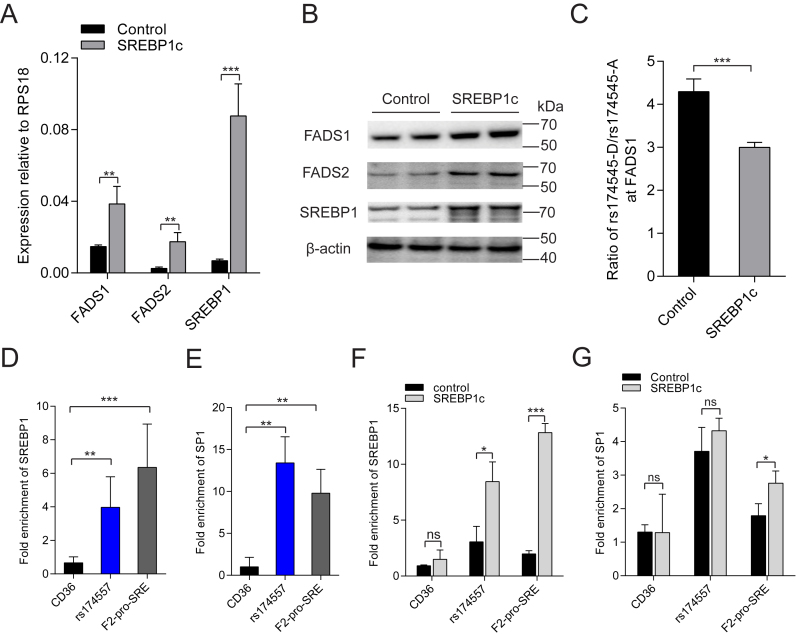
HepG2 cells were transduced with lentivirus overexpressing the mature form of SREBP1c. This resulted in significantly increased SREBP1c expression compared to cells transduced with the control virus. In accordance with previous reports (2,40), both FADS1 and FADS2 were significantly induced by SREBP1c overexpression at mRNA level validated by RT-qPCR with transcripts RPS18 (Figure 5A), ACTB and GAPDH (Supplementary Figure S11) as controls. Western blot analysis also showed that overexpression of SREBP1c significantly increased the translation of both FADS1 and FADS2 in HepG2 cells (Figure 5B). In order to evaluate the effect of SREBP1c on the allelic expression imbalance of FADS1 observed in MCF7 cells, MCF7 cells overexpressing SREBP1c were established by lentiviral transduction. Overexpression of SREBP1c in MCF7 cells led to a significant decrease in the allelic expression imbalance of FADS1 (Figure 5C) which is contrary to a significant increase in the allelic expression imbalance of FADS1 caused by PATZ1 overexpression (Figure 3E).
Figure 5.
SREBP1c and SP1 bind to the rs174557 locus in vivo to regulate FADS expression. (A and B) The expression of FADS1 and FADS2 in HepG2 cells were significantly increased by overexpression of SREBP1c detected by both RT-qPCR (A) and western blot (B). The qPCR results are expressed as mean±SD from three replicates with **P < 0.01 and ***P < 0.001 calculated by two-tailed t tests. (C) The allelic expression imbalance of FADS1 in MCF7 cells was significantly decreased by SREBP1c overexpression. The results came from four replicates with the error bars represent SD. (D–G) The rs174557 locus is bound by both SREBBP1 and SP1 in vivo detected by ChIP-qPCR with antibody against SREBP1 (D) and SP1 (E) in HepG2 cells. The binding of SREBP1 (F), but not SP1 (G) to the rs174557 locus was significantly increased by SREBP1c overexpression. The region F2-pro-SRE targeting the SREBP1 responsive region in the FADS2 promoter was used as the positive control for both SREBP1 and SP1 binding. All the results are expressed as fold enrichments over the negative control region GDCHR12 with three to six replicates. Error bars represent SD. ns, not significant; *P < 0.05; **P < 0.01 and ***P < 0.001 compared with GDCHR12.
To test if the identified SREBP1c responsive region in rs174557 locus was bound by SREBP1c and SP1 in vivo, ChIP-qPCR with antibodies against SREBP1 or SP1 was carried out. In normal HepG2 cells, the rs174557 locus was enriched with both SREBP1c (Figure 5D) and SP1 (Figure 5E) signals compared to negative control regions. One region in the FADS2 promoter where the reported SRE sites were located was employed as a positive control (40), and showed a similar degree of enrichment for both SREBP1 and SP1. Overexpression of the mature nuclear form of SREBP1c in HepG2 cells led to a significant increase in the binding of SREBP1c to both the rs174557 locus and the FADS2 promoter region compared to cells transduced with control virus (Figure 5F). The binding of SP1 to the rs174557 locus was slightly but non-significantly increased by overexpression of SREBP1c, while the binding to the FADS2 promoter was significantly increased (Figure 5G). There are three regions in the FADS cluster that are enriched with SREBP1 signal in ChIP-seq experiments in ENCODE (Supplementary Figure S12A). The FADS2 promoter has previously been reported to be induced by SREBP1 (40), the other two regions including the promoter of FADS1 and the intronic region of FADS1 where rs174561 resides did not show any increased response to SREBP1c overexpression in luciferase assay (Supplementary Figure S12B–D). This suggests that the newly identified SREBP1c responsive region at the rs174557 locus might be actively involved in the regulation of FADS1 by SREBP1c.
Competition between PATZ1 and SREBP1c-SP1 determines the enhancer activity at rs174557
SREBP1c is virtually unable to bind DNA with high affinity unless in complex with other partners, most commonly NFY and SP1 (41). All three SRE sites and the GC-box element identified in the rs174557 locus were proved to be functional suggesting that the enhancer activity in this region is mainly mediated by the SREBP1c-SP1 complex. In contrast to the activating SREBP1c-SP1 complex, PATZ1 acts as a suppressor by directly binding to rs174557 which overlaps with the binding site of SP1. In addition, overexpression of PATZ1 or SREBP1c had directly opposite effect on the allelic imbalance in FADS1 expression in MCF7 cells (Figures 3E and 5C). We proposed that PATZ1 and SP1 might compete for the same binding site with the differences observed among the rs174557 alleles being determined by the different affinities of PATZ1 binding (Figure 7C). Indeed, the sequence essential for SP1 binding was finely mapped to a canonical GC-box element (GGGCGG) as shown by the sharp decreases in enhancer activities when either half of the motif sequence was mutated compared to mutations to the three SRE sites (Figure 4C) and to the adjacent sequence predicted to be bound by PATZ1 but not overlapping with the GC-box element (Figure 6A).
Figure 6.
The GC-box element is essential for both SP1 and PATZ1 Binding. (A) Disrupting either half of the GC-box element bound by SP1 sharply decreased the enhancer activity of the rs174557 locus in comparison with disrupting the nearby non-overlapping PATZ1 site. The grey bars indicate the luciferase constructs with the D allele as template and the black bars indicate the constructs with the A2 allele as template. ***P < 0.0001 calculated by two-tailed t tests. (B) The whole GC-box element is also essential for PATZ1 binding. Mutations introduced to either half of the GC-box element completely abolished the suppression of PATZ1 in luciferase assay. ns, not significant and ***P < 0.0001 calculated by one-tailed t tests. All the data in A and B came from three independent transfections each with six technical replicates. The bars represent mean ± SD from all technical replicates.
PATZ1 preferentially binds to simply repeated G/C sequences (42,43). The only difference among all the A alleles of rs174557 and the consensus AluYe5 sequence is the length of the consecutive repeat of guanine nucleotides (Figure 2A). In accordance with the motif sequence of PATZ1, a gradual increase in the number of consecutive guanine nucleotides was associated with a gradual decrease in enhancer activity (Figure 2B). Moreover, when PATZ1 was overexpressed, the consensus AluYe5 sequence which has the least number of repeated guanine nucleotides had dramatically attenuated response to PATZ1 suppression compared to all the A alleles (Figure 3B). The binding of PATZ1 to this site was further proven by introducing mutations to the repeated guanine nucleotides adjacent to the GC-box element, which completely abolished the PATZ1 suppression in both allele D and allele A2 (Luciferase constructs D-PT1 mut and A2-PT1 mut, respectively in Figure 3B). In addition, the whole GC-box element bound by SP1 is also essential for the PATZ1 binding verified by a complete loss of PATZ1 suppression on the enhancer activity of allele D when either half of the GC-box element is mutated (Figure 6B). Thus the whole repeated guanine nucleotides sequence is essential for PATZ1 binding and it is overlapping with the GC-box element (Figure 7C).
The competition between PATZ1 and SP1 binding to the overlapping GC-box element seems to determine the enhancer activity of the rs174557 locus even when SREBP1c is overexpressed. Although all the alleles of rs174557 are highly induced by SREBP1c overexpression, significant differences were observed among different alleles (Figure 7A). Alleles A2, A3 and A4, which were previously shown to be more strongly bound by PATZ1 in EMSA (Figure 3D), showed significantly decreased response to SREBP1c induction compared to other alleles. We wanted to evaluate if the differences in binding affinity of PATZ1 to different alleles of rs174557 determines the extent to which each allele can be activated by the SREBP1c-SP1 complex. A set of luciferase experiments were conducted with a stable amount of SREBP1c and increasing amounts of PATZ1 cotransfected with different alleles of rs174557. PATZ1 decreased the responses of allele A1 and A2 to SREBP1c overexpression in a dose-dependent manner, whereas no decrease was observed when the binding site for PATZ1 of allele A2 was mutated (Figure 7B and Supplementary Figure S13A). For allele A3 and allele A4, even though their response to SREBP1c overexpression was dramatically lower before PATZ1 overexpression, a further significant suppression by PATZ1 was observed when the maximum amount of plasmid overexpressing PATZ1 was added (Supplementary Figure S13C and D). Under the same SREBP1c overexpression conditions, the D allele that has a less conserved binding site for PATZ1 failed to be suppressed by increasing amount of PATZ1 (Supplementary Figure S13B). A model for how the activating complex SREBP1c-SP1 interacts with the suppressive PATZ1 in the rs174557 locus is depicted in Figure 7C.
DISCUSSION
Only 10–15% of the genetic variants associated with traits in GWAS are located in coding regions (33). Most of the remaining top hits are predicted to be due to LD with regulatory variants, but only a limited number of these causative variants have been identified to date. In this study, based on the LD block from our previous work, we identified rs174557 as an important functional variant in the FADS cluster by systematic luciferase assay on all putative regulatory variants that separate the two main haplotypes. It should also be noted that our previous analysis on the LD blocks were originally conducted in Europeans based on array data and subsequent investigations using sequencing approaches have found additional SNPs present in populations with other genetic backgrounds that are also in strong LD with the A/D haplotypes such as rs174537. This SNP has previously been associated with fatty acid metabolism (44) and DNA methylation status of an enhancer region upstream of FADS1 (45). Although not part of the original 28 SNPs, the rs174537 is in complete linkage based on sequencing data (D’ = 1.0) with rs1535, that is part of the 28 SNPs, in both European and Asian populations (46). There is thus strong evidence that many of the effects seen both on molecular characteristics and in higher order traits linked to SNPs in the FADS cluster can be linked to the A/D-haplotypes and therefore likely can be attributed to rs174557 investigated here.
Liver is the most important organ for cholesterol and fatty acid synthesis. SREBP1c is recognized as a key regulator of fatty acid synthesis in the liver (2,47) and is known to regulate FADS2 expression by directly binding to SRE elements in its promoter (40). However, to our knowledge it is not known how SREBP1c regulates FADS1. We have shown that the rs174557 locus in intron 1 of FADS1 is the major region responding to SREBP1c induction (Figure 4A and Supplementary Figure S12). We propose a model in which the activating complex SREBP1c-SP1 competes with PATZ1 for an overlapping binding site to influence FADS1 activity (Figure 7C). The D allele of rs174557 attenuates PATZ1 chromatin binding, thereby enabling SP1 to bind more potently to the GC-box element overlapping with the PATZ1 motif. Insulin and other stimuli (41) lead to increased expression of SREBP1c which can then bind the three SRE sites and cooperate with SP1 at the GC-box element to compete with PATZ1, which increases the enhancer activity and upregulates FADS1.
Alu, as a subfamily of TEs, comprising > 1 million copies, makes up 10% of the human genome (48). The functional SNP rs174557 resides in an AluYe5 element that belongs to a subfamily of Alu. Recent studies have demonstrated that a portion of TEs were hypomethylated and enriched with histone modifications signals resembling active enhancers in a tissue specific manner (36,37). In accordance with these observations, our work provides a detailed example of an Alu element directly involved in gene regulation by functioning as an enhancer. The AluYe5 element makes FADS1 respond to the SREBP1c signal more powerfully, which enables LC-PUFAs to be more efficiently synthesized.
As LC-PUFAs play essential roles in neuron growth and brain development (49,50), the insertion of this AluYe5 element into intron 1 of the FADS1 gene may have contributed to an evolutionary advantage for both humans and chimpanzee. The insertion can be dated to 5–7 MY ago. Our data suggest that several A alleles have appeared since then, possibly due to slippage in the poly-G stretch during replication (51). A major event was the appearance of the D allele through a G>A mutation, which may render the sequence less prone to new mutations in the oligo-Gs. We have previously estimated the age of the D haplotype to about 300 000 years ago. The D allele of rs174557, which enables LC-PUFAs to be more efficiently synthesized from its precursors, might have enabled its carriers to survive in environments with limited access to essential LC-PUFAs.
Nowadays, the human diet has dramatically changed with an excess of food resources and a striking shift in the ratio of dietary LA to ALA (1). This high dietary intake of LA increases the amount of AA synthesized. This may further increase the synthesis of AA-derived proinflammatory eicosanoids implicated in chronic inflammation and cancer (52). For example, persistent chronic inflammation has been suggested to contribute to numerous human diseases such as cardiovascular disease, arthritis, asthma, colorectal cancer and the metabolic syndrome (52–54). This hypothesis is supported by several recent GWAS that have associated variations in the FADS cluster with increased risk of diseases related to inflammation including rheumatoid arthritis (15), colorectal cancer (16), Crohn's disease (17), inflammatory bowel disease (18) and laryngeal squamous cell carcinoma (19). Considering our current diet, the evolutionary advantage of a more efficient biosynthesis of LC-PUFAs in carriers of the D allele of rs174557 might make them more susceptible to diseases associated with inflammation. For example, the D haplotype were associated with increased risk of colorectal cancer (both the D allele of rs174550 and rs1535) (16), and laryngeal squamous cell carcinoma (the D allele of rs174549) (19). Our findings open the possibility to investigate the disease risk associated with each allele of rs174557. Misregulation of PATZ1 has also been implicated in several cancers but the involvement of FADS regulation by PATZ1 in these diseases warrants further study (55). In conclusion, we have identified a novel functional regulatory variant for the FADS cluster for which the derived allele was advantageous during evolution, but modern diet may have turned it into a disadvantage by increasing the risk of metabolic and inflammatory diseases.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to acknowledge support of the National Genomics Infrastructure (NGI)/Uppsala Genome Center and UPPMAX for providing assistance in massive parallel sequencing and computational infrastructure. We thank Berith Nilsson for technical assistance during lentivirus production.
Footnotes
Present address: Madhusudhan Bysani, Department of Clinical Sciences, Lund University Diabetes Center, Malmö, Sweden., Helena Nord, Galderma, Uppsala, Sweden.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Work performed at NGI/Uppsala Genome Center has been funded by RFI/VR and Science for Life Laboratory, Sweden; Swedish Research Council [CW 521-2010-3505, 621-2011-6052, UG 521-2012-2884]; Swedish Diabetes Foundation (CW), Diabetes Wellness Network Sweden (CW), the Family Ernfors Fund (CW), Uppsala University (CW), EXODIAB (CW) and the Swedish Cancer Foundation [CW 15 0878]. Funding for open access charge: EXODIAB.
Conflict of interest statement. None declared.
REFERENCES
- 1.Simopoulos A.P. The importance of the omega-6/omega-3 fatty acid ratio in cardiovascular disease and other chronic diseases. Exp. Biol. Med. 2008; 233:674–688. [DOI] [PubMed] [Google Scholar]
- 2.Nakamura M.T., Nara T.Y.. Structure, function, and dietary regulation of Delta 6, Delta 5, and Delta 9 desaturases. Annu. Rev. Nutr. 2004; 24:345–376. [DOI] [PubMed] [Google Scholar]
- 3.Gieger C., Geistlinger L., Altmaier E., Hrabé de Angelis M., Kronenberg F., Meitinger T., Mewes H.-W., Wichmann H.E., Weinberger K.M., Adamski J. et al. Genetics meets metabolomics: a genome-wide association study of metabolite profiles in human serum. PLoS Genet. 2008; 4:e1000282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kettunen J., Tukiainen T., Sarin A.-P., Ortega-Alonso A., Tikkanen E., Lyytikainen L.-P., Kangas A.J., Soininen P., Wurtz P., Silander K. et al. Genome-wide association study identifies multiple loci influencing human serum metabolite levels. Nat. Genet. 2012; 44:269–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Illig T., Gieger C., Zhai G., Romisch-Margl W., Wang-Sattler R., Prehn C., Altmaier E., Kastenmuller G., Kato B.S., Mewes H.-W. et al. A genome-wide perspective of genetic variation in human metabolism. Nat. Genet. 2010; 42:137–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tanaka T., Shen J., Abecasis G.R., Kisialiou A., Ordovas J.M., Guralnik J.M., Singleton A., Bandinelli S., Cherubini A., Arnett D. et al. Genome-wide association study of plasma polyunsaturated fatty acids in the InCHIANTI study. PLoS Genet. 2009; 5:e1000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lemaitre R.N., Tanaka T., Tang W., Manichaikul A., Foy M., Kabagambe E.K., Nettleton J.A., King I.B., Weng L.-C., Bhattacharya S. et al. Genetic loci associated with plasma phospholipid n-3 fatty acids: a meta-analysis of genome-wide association studies from the CHARGE Consortium. PLoS Genet. 2011; 7:e1002193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shin S.-Y., Fauman E.B., Petersen A.-K., Krumsiek J., Santos R., Huang J., Arnold M., Erte I., Forgetta V., Yang T.-P. et al. An atlas of genetic influences on human blood metabolites. Nat. Genet. 2014; 46:543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schaeffer L., Gohlke H., Muller M., Heid I.M., Palmer L.J., Kompauer I., Demmelmair H., Illig T., Koletzko B., Heinrich J.. Common genetic variants of the FADS1 FADS2 gene cluster and their reconstructed haplotypes are associated with the fatty acid composition in phospholipids. Hum. Mol. Genet. 2006; 15:1745–1756. [DOI] [PubMed] [Google Scholar]
- 10.Suhre K., Shin S.Y., Petersen A.K., Mohney R.P., Meredith D., Wagele B., Altmaier E., Deloukas P., Erdmann J., Grundberg E. et al. Human metabolic individuality in biomedical and pharmaceutical research. Nature. 2011; 477:54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raffler J., Romisch-Margl W., Petersen A.K., Pagel P., Blochl F., Hengstenberg C., Illig T., Meisinger C., Stark K., Wichmann H.E. et al. Identification and MS-assisted interpretation of genetically influenced NMR signals in human plasma. Genome Med. 2013; 5:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang L., Athinarayanan S., Jiang G., Chalasani N., Zhang M., Liu W.. Fatty acid desaturase 1 gene polymorphisms control human hepatic lipid composition. Hepatology. 2015; 61:119–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ameur A., Enroth S., Johansson Å., Zaboli G., Igl W., Johansson Anna C., Rivas Manuel A., Daly Mark J., Schmitz G., Hicks Andrew A. et al. Genetic adaptation of fatty-acid metabolism: a human-specific haplotype increasing the biosynthesis of long-chain omega-3 and omega-6 fatty acids. Am. J. Hum. Genet. 2012; 90:809–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schadt E.E., Molony C., Chudin E., Hao K., Yang X., Lum P.Y., Kasarskis A., Zhang B., Wang S., Suver C. et al. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008; 6:e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okada Y., Wu D., Trynka G., Raj T., Terao C., Ikari K., Kochi Y., Ohmura K., Suzuki A., Yoshida S. et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014; 506:376–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang B., Jia W.-H., Matsuda K., Kweon S.-S., Matsuo K., Xiang Y.-B., Shin A., Jee S.H., Kim D.-H., Cai Q. et al. Large-scale genetic study in East Asians identifies six new loci associated with colorectal cancer risk. Nat. Genet. 2014; 46:533–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franke A., McGovern D.P., Barrett J.C., Wang K., Radford-Smith G.L., Ahmad T., Lees C.W., Balschun T., Lee J., Roberts R. et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat. Genet. 2010; 42:1118–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jostins L., Ripke S., Weersma R.K., Duerr R.H., McGovern D.P., Hui K.Y., Lee J.C., Philip Schumm L., Sharma Y., Anderson C.A. et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012; 491:119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei Q.Y., Yu D.K., Liu M.B., Wang M.Y., Zhao M.Q., Liu M., Jia W.H., Ma H.X., Fang J.G., Xu W. et al. Genome-wide association study identifies three susceptibility loci for laryngeal squamous cell carcinoma in the Chinese population. Nat. Genet. 2014; 46:1110–1114. [DOI] [PubMed] [Google Scholar]
- 20.den Hoed M., Eijgelsheim M., Esko T., Brundel B.J., Peal D.S., Evans D.M., Nolte I.M., Segre A.V., Holm H., Handsaker R.E. et al. Identification of heart rate-associated loci and their effects on cardiac conduction and rhythm disorders. Nat Genet. 2013; 45:621–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eijgelsheim M., Newton-Cheh C., Sotoodehnia N., de Bakker P.I., Muller M., Morrison A.C., Smith A.V., Isaacs A., Sanna S., Dorr M. et al. Genome-wide association analysis identifies multiple loci related to resting heart rate. Hum. Mol. Genet. 2010; 19:3885–3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R. et al. A global reference for human genetic variation. Nature. 2015; 526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kothapalli K.S.D., Ye Kaixiong, Gadgil M.S., Carlson S.E., O'Brien K.O., Zhang J.Y., Park H.G., Ojukwu K., Zou J., Hyon S.S.. Positive selection on a regulatory insertion–deletion polymorphism in FADS2 influences apparent endogenous synthesis of arachidonic acid. Mol. Biol. Evol. 2016; 33:1726–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mathieson I., Lazaridis I., Rohland N., Mallick S., Patterson N., Roodenberg S.A., Harney E., Stewardson K., Fernandes D., Novak M. et al. Genome-wide patterns of selection in 230 ancient Eurasians. Nature. 2015; 528:499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fumagalli M., Moltke I., Grarup N., Racimo F., Bjerregaard P., Jorgensen M.E., Korneliussen T.S., Gerbault P., Skotte L., Linneberg A. et al. Greenlandic Inuit show genetic signatures of diet and climate adaptation. Science. 2015; 349:1343–1347. [DOI] [PubMed] [Google Scholar]
- 26.Igl W., Johansson A., Gyllensten U.. The Northern Swedish Population Health Study (NSPHS) – a paradigmatic study in a rural population combining community health and basic research. Rural Rem. Health. 2010; 10:1363. [PubMed] [Google Scholar]
- 27.Zaboli G., Ameur A., Igl W., Johansson A., Hayward C., Vitart V., Campbell S., Zgaga L., Polasek O., Schmitz G. et al. Sequencing of high-complexity DNA pools for identification of nucleotide and structural variants in regions associated with complex traits. Europ. J. Hum. Genet. 2012; 20:77–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y., Werling U., Edelmann W.. SLiCE: a novel bacterial cell extract-based DNA cloning method. Nucleic Acids Res. 2012; 40:e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Toth J.I., Datta S., Athanikar J.N., Freedman L.P., Osborne T.F.. Selective coactivator interactions in gene activation by SREBP-1a and -1c. Mol. Cell. Biol. 2004; 24:8288–8300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blecher-Gonen R., Barnett-Itzhaki Z., Jaitin D., Amann-Zalcenstein D., Lara-Astiaso D., Amit I.. High-throughput chromatin immunoprecipitation for genome-wide mapping of in vivo protein-DNA interactions and epigenomic states. Nat. Protocols. 2013; 8:539–554. [DOI] [PubMed] [Google Scholar]
- 31.Motallebipour M., Ameur A., Reddy Bysani M.S., Patra K., Wallerman O., Mangion J., Barker M., McKernan K., Komorowski J., Wadelius C.. Differential binding and co-binding pattern of FOXA1 and FOXA3 and their relation to H3K4me3 in HepG2 cells revealed by ChIP-seq. Genome Biol. 2009; 10:R129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bysani M., Wallerman O., Bornelov S., Zatloukal K., Komorowski J., Wadelius C.. ChIP-seq in steatohepatitis and normal liver tissue identifies candidate disease mechanisms related to progression to cancer. BMC Med. Genomics. 2013; 6:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dunham I., Kundaje A., Aldred S.F., Collins P.J., Davis C., Doyle F., Epstein C.B., Frietze S., Harrow J., Kaul R. et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bulger M., Groudine M.. Enhancers: The abundance and function of regulatory sequences beyond promoters. Dev. Biol. 2010; 339:250–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wheeler T.J., Clements J., Eddy S.R., Hubley R., Jones T.A., Jurka J., Smit A.F.A., Finn R.D.. Dfam: a database of repetitive DNA based on profile hidden Markov models. Nucleic Acids Res. 2013; 41:D70–D82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xie M., Hong C., Zhang B., Lowdon R.F., Xing X., Li D., Zhou X., Lee H.J., Maire C.L., Ligon K.L. et al. DNA hypomethylation within specific transposable element families associates with tissue-specific enhancer landscape. Nat. Genet. 2013; 45:836–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Su M., Han D.L., Boyd-Kirkup J., Yu X.M., Han J.D.J.. Evolution of Alu elements toward enhancers. Cell Rep. 2014; 7:376–385. [DOI] [PubMed] [Google Scholar]
- 38.Thomas-Chollier M., Hufton A., Heinig M., O'Keeffe S., Masri N.E., Roider H.G., Manke T., Vingron M.. Transcription factor binding predictions using TRAP for the analysis of ChIP-seq data and regulatory SNPs. Nat. Protoc. 2011; 6:1860–1869. [DOI] [PubMed] [Google Scholar]
- 39.Mathelier A., Zhao X., Zhang A.W., Parcy F., Worsley-Hunt R., Arenillas D.J., Buchman S., Chen C.-Y., Chou A., Ienasescu H. et al. JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res. 2014; 42:D142–D147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nara T.Y., He W.S., Tang C., Clarke S.D., Nakamura M.T.. The E-box like sterol regulatory element mediates the suppression of human Δ-6 desaturase gene by highly unsaturated fatty acids. Biochem. Biophys. Res. Commun. 2002; 296:111–117. [DOI] [PubMed] [Google Scholar]
- 41.Reed B.D., Charos A.E., Szekely A.M., Weissman S.M., Snyder M.. Genome-wide occupancy of SREBP1 and its partners NFY and SP1 reveals novel functional roles and combinatorial regulation of distinct classes of genes. PLoS Genet. 2008; 4:e1000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ow J.R., Ma H., Jean A., Goh Z., Lee Y.H., Chong Y.M., Soong R., Fu X.-Y., Yang H., Wu Q.. Patz1 regulates embryonic stem cell identity. Stem Cells Dev. 2013; 23:1062–1073. [DOI] [PubMed] [Google Scholar]
- 43.Kobayashi A., Yamagiwa H., Hoshino H., Muto A., Sato K., Morita M., Hayashi N., Yamamoto M., Igarashi K.. A combinatorial code for gene expression generated by transcription factor Bach2 and MAZR (MAZ-related factor) through the BTB/POZ domain. Mol. Cell. Biol. 2000; 20:1733–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hester A.G., Murphy R.C., Uhlson C.J., Ivester P., Lee T.C., Sergeant S., Miller L.R., Howard T.D., Mathias R.A., Chilton F.H.. Relationship between a common variant in the fatty acid desaturase (FADS) cluster and eicosanoid generation in humans. J. Biol. Chem. 2014; 289:22482–22489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Howard T.D., Mathias R.A., Seeds M.C., Herrington D.M., Hixson J.E., Shimmin L.C., Hawkins G.A., Sellers M., Ainsworth H.C., Sergeant S. et al. DNA methylation in an enhancer region of the FADS cluster Is associated with FADS activity in human liver. PLoS One. 2014; 9:e97510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johnson A.D., Handsaker R.E., Pulit S.L., Nizzari M.M., O'Donnell C.J., de Bakker P.I.W.. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008; 24:2938–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sampath H., Ntambi J.M.. Polyunsaturated fatty acid regulation of genes of lipid metabolism. Annu. Rev. Nutri. 2005; 25:317–340. [DOI] [PubMed] [Google Scholar]
- 48.Cordaux R., Batzer M.A.. The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 2009; 10:691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lattka E., Illig T., Heinrich J., Koletzko B.. Do FADS genotypes enhance our knowledge about fatty acid related phenotypes. Clin. Nutr. 2010; 29:277–287. [DOI] [PubMed] [Google Scholar]
- 50.Marszalek J.R., Lodish H.E.. Docosahexaenoic acid, fatty acid-interacting proteins, and neuronal function: breastmilk and fish are good for you. Annu. Rev. Cell. Dev. Biol. 2005; 21:633–657. [DOI] [PubMed] [Google Scholar]
- 51.Fan H., Chu J.Y.. A brief review of short tandem repeat mutation. Genomics Proteomics Bioinformatics. 2007; 5:7–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang D., DuBois R.N.. Eicosanoids and cancer. Nat. Rev. Cancer. 2010; 10:181–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chilton F.H., Murphy R.C., Wilson B.A., Sergeant S., Ainsworth H., Seeds M.C., Mathias R.A.. Diet-gene interactions and PUFA metabolism: a potential contributor to health disparities and human diseases. Nutrients. 2014; 6:1993–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hardwick J.P., Eckman K., Lee Y.K., Abdelmegeed M.A., Esterle A., Chilian W.M., Chiang J.Y., Song B.-J.. David RW. Eicosanoids in metabolic syndrome. Adv. Pharmacol. 2013; 66, Academic Press; 157–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Valentino T., Palmieri D., Vitiello M., Pierantoni G.M., Fusco A., Fedele M.. PATZ1 interacts with p53 and regulates expression of p53-target genes enhancing apoptosis or cell survival based on the cellular context. Cell Death Dis. 2013; 4:e963. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.