Abstract
The nonhomologous end-joining (NHEJ) pathway is the primary repair pathway for DNA double strand breaks (DSBs) in humans. Repair is mediated by a core complex of NHEJ factors that includes a ligase (DNA Ligase IV; L4) that relies on juxtaposition of 3΄ hydroxyl and 5΄ phosphate termini of the strand breaks for catalysis. However, chromosome breaks arising from biological sources often have different end chemistries, and how these different end chemistries impact the way in which the core complex directs the necessary transitions from end pairing to ligation is not known. Here, using single-molecule FRET (smFRET), we show that prior to ligation, differences in end chemistry strongly modulate the bridging of broken ends by the NHEJ core complex. In particular, the 5΄ phosphate group is a recognition element for L4 and is critical for the ability of NHEJ factors to promote stable pairing of ends. Moreover, other chemical incompatibilities, including products of aborted ligation, are sufficient to disrupt end pairing. Based on these observations, we propose a mechanism for iterative repair of DSBs by NHEJ.
INTRODUCTION
Chromosomal DNA double-strand breaks (DSBs) are associated with chemical adducts and damaged bases which prove challenging to repair (1). The free ends of the broken chromosome must be joined to preserve genomic integrity and suppress cellular senescence and apoptosis (2). However, break repair can occur incorrectly, resulting in translocation and rearrangements in chromosomes, potentially giving rise to neoplastic transformation (3,4). In humans, DSBs are predominantly repaired via the nonhomologous end-joining (NHEJ) pathway, a process involving the recruitment of various proteins that direct the synapsis and ligation of broken chromosomes. NHEJ is linked to numerous areas of human health including nervous system development, adaptive immunity, cancer, and aging (1,5). It relies on a ‘core’ complex, including the Ku70/86 (Ku) heterodimer, XRCC4, XLF and DNA Ligase IV (L4) (1). The core complex is sufficient for end recognition, end pairing (synapsis), and ligation of a large fraction of DSBs, including both ‘simple’ or ‘sticky’ DSBs, as well DSBs with subtle flanking helical distortion (e.g. flanked by some mispairs and oxidized bases) (6). Complex DSBs containing abasic sites, chemically non-ligatable ends and bulky adducts (such as carbohydrate groups) require the recruitment of additional repair factors by the core NHEJ machinery to effect repair; however, it presently remains unclear if such modified DNA ends can directly affect the bridging of ends (1). When the termini of either of the two strand breaks are correctly positioned, DNA ligases catalyze the formation of a phosphodiester bond to restore strand continuity (7).
Previously, we used super-resolution microscopy to show that the core NHEJ proteins form filaments adjacent to DSBs in cells, and that these filaments mediate dynamic pairing between DNAs that can be monitored using single-molecule FRET (smFRET) assays (8). Our smFRET analysis revealed that Ku, L4, XRCC4 and XLF promote the most efficient pairing of DNAs. The omission of any of these components leads to lower overall synapsis. Furthermore, we found that the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) did not appear to mediate efficient bridging of DNAs even in the presence of Ku. Filament-mediated bridging was further demonstrated in a recent study using optical trapping (9). Here, using a similar smFRET assay that relies on the bridging of two dsDNA molecules, we manipulated the end chemistry at the break via removal of the 5΄ phosphate and introduction of dideoxynucleotides. We found that the 5΄ phosphate acts as a recognition element for the retention of joined molecules, and that this relates to the activity L4, though not the final ligation step. Using this assay, we also noted two different modes of association, i.e. either transient or persistent interactions, for the NHEJ complex and DNAs. We propose that disruptions in the compatibility between ends could lead to dissociation of paired DNAs and subsequent iterative pairing attempts, potentially introducing errors into the repair process.
MATERIALS AND METHODS
Protein purification
Ku70/86
Purification of Ku was as previously described (10). Briefly, Hi-5 insect cells were co-infected with His-tagged Ku70 and Ku86 baculovirus constructs to express the proteins. Cells were lysed and the recombinant proteins purified using Ni-NTA chromatography (Qiagen), DNA affinity chromatography, and anion exchange chromatography (MonoQ; GE Healthcare) (Supplementary Figure S1).
LX (L4/XRCC4 of K273R L4/XRCC4) and XLF
Purification of LX and XLF were done as previously described (10). Briefly, protein extracts were prepared from Hi-5 insect cells infected with baculovirus constructs to over-express His-tagged L4 and untagged XRCC4, or His-tagged XLF, and L4/XRCC4 complexes or XLF the recombinant proteins purified via His-Trap and MonoQ anion exchange columns (GE Healthcare) chromatography (Supplementary Figure S1).
Single-molecule FRET assay
DNA preparations
All oligonucleotides (Supplementary Methods) were purchased from Integrated DNA Technologies. To anneal oligonucleotides, appropriate mixtures were heated or 10 min at 95°C followed by slow cooling to room temperature. A complete list of substrates is available in the Supplementary Information.
NHEJ reactions and analysis
Our end-joining assay was performed as described previously (8). Briefly, 50 nM Ku, LX, XLF, gloxy (0.5 mg/ml glucose oxidase and 0.40 μg/ml catylase), and 1 nM dsDNA were added stepwise to NEB4 (20 mM pH 7.5 TrisAc, 50 mM KAc, 10 mM MgAc) with 0.8% glucose, ∼5 mM Trolox (11), 1 mg/mL BSA and 2 mM DTT. The reaction was immediately flowed into an imaging chamber that had been prepared with the surface DNA (∼250 pM). Movies consisting of 1000 frames (33 Hz) were acquired for analysis in custom Matlab software. Analysis of smFRET trajectories and histograms was performed as previously described (8). Autocorrelation of smFRET trajectories and energies calculations are described in the Supplementary Methods.
5΄ Adenylation Reaction
A 5΄ adenylation kit from NEB (E2610L) was used to adenylate 4nt complementary oligos attached to a PEG coverslip. Neutravadin was added to the slides, followed by annealed oligonucleotides. The slide was washed and then a 6× adenylation reaction was carried out at 37°C for 2 h. Following the reaction, the chamber was washed with 1 M NaCl for 1 h. The chamber was then washed with T50 buffer (10 mM Tris–HCl and 50 mM NaCl) and normal sample preparation was resumed.
L4 deadenylation reaction
L4 was treated with 15 mM pyrophosphate for 20 min on ice prior to incorporation in NHEJ reactions (12).
RESULTS AND DISCUSSION
Prior studies of NHEJ fidelity have used ensemble assays to measure the ligation efficiency of various DNA ends (13–15). These assays have relied on the ligation of various DNA substrates using recombinant proteins, cellular extracts, or cells, with a subsequent quantification of ligated products via gel shift or polymerase chain reaction. We recently developed a smFRET assay for NHEJ which uses the purified components Ku, LX (XRCC4/L4 heteromultimer) and XLF to join together two ∼80 bp dsDNA molecules (Figure 1A, Supplementary Figure S2A) (8). The NHEJ proteins together with the DNAs make up a paired end complex (PEC), a highly dynamic nucleoprotein complex that is responsible for maintaining synapsis of the ends and subsequently ligating together the DNA ends. Our smFRET assay allowed us to monitor the formation of PECs (via detection of acceptor fluorescence), quantify the yield in terms of the number of FRET pairs formed, and directly observe the dynamics of the synapsed ends for each of a large number of individual end-pairs (8). Furthermore, we could monitor the stability of association (via dwell time analysis of FRET pairs) and the transition between FRET states (11).
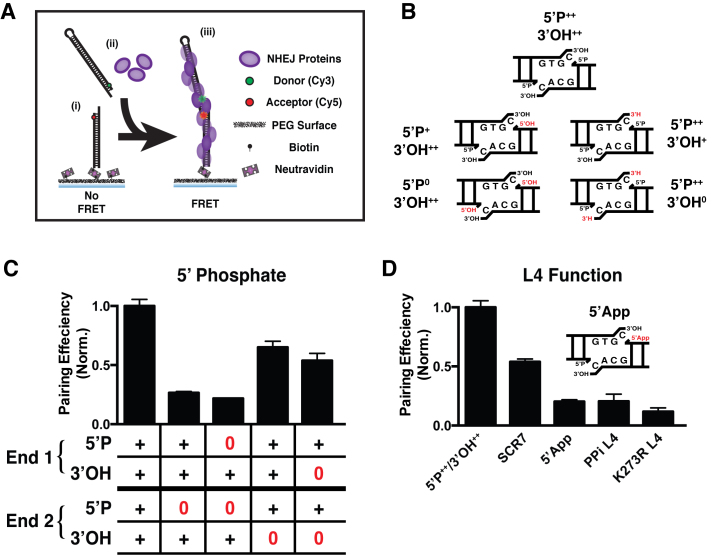
Figure 1.
DNA End Chemistry Modulates PEC Formation. (A) SmFRET assay: (i) dsDNA with an Cy5 near the end of the DNA is tethered to the surface via a biotin/neutravadin linkage, (ii) dsDNA with a Cy3 and NHEJ proteins (purple) are added to the sample chamber, and (iii) NHEJ proteins will pair together the surface and solution dsDNAs such that FRET is observed. (B) Diagram of the dsDNA ends we use to explore how changes in chemical compatibility for ligation affect pairing efficiency. Changes are shown in red (removal of 5΄ phosphate to yield 5΄OH or removal of 3΄OH to yield 3΄H). (C) Quantification of pairing efficiency in ends lacking 5΄P or 3΄OH, where ‘+’ denotes the presence of a chemical group, and ‘0’ denotes an absence. (D) Quantification of pairing efficiency in reactions where L4 activity has been disrupted. SCR7 indicates reactions treated with 10 μM of the ligase inhibitor, 5΄App is an adenylated substrate (see cartoon), PPi L4 is deadenylated L4, and K273R L4 is a catalytic mutant not capable of being adenylated. Error bars are the SEM of two pairing experiments.
We first examined the effect that DNA end chemistry has on the overall pairing efficiency of different DNA substrates. We used DNA ends with a 4-nucleotide 3΄ overhang that contained 5΄ phosphate (5΄P) and 3΄ hydroxyl (3΄OH) groups, and compared this control with ends in which the 5΄P and 3΄OH groups had been systematically removed (Figure 1B, Supplementary Figure S2B). We observed a substantial reduction in the pairing efficiency for substrates lacking even a single 5΄P, while substrates where a 3΄OH was missing showed only a small reduction in pairing (Figure 1C). The 3΄ overhang improves the overall pairing efficiency when compared with blunt end substrates and ends with 5΄ overhangs; however, 5΄ overhangs lacking a 5΄P do show diminished pairing efficiency (Supplementary Figure S2B and C). DNA substrates lacking both 5΄P and 3΄OH showed pairing efficiencies similar to substrates lacking only 5΄P (Supplementary Figure S2D-E), indicating that the presence of a 5΄P dominates the pairing process.
We used EMSA analysis to determine the contribution that the omission of a 5΄P has on steps prior to end pairing/PEC formation. This measurement determined that 5΄P had a small impact on the ability of Ku, XLF and LX to form a complex at a single DNA end (Supplementary Figure 2F; compare lowest mobility species with and without 5΄P). Similarly, the presence of a 5΄P had only a slight impact on the ability of LX alone to bind a DNA strand break (Supplementary Figure S3B). Together, these results are consistent with our observations that the severe effects of 5΄P omission on PEC formation are largely due to the disrupted end pairing, and not to defects in earlier protein assembly steps.
To directly assess L4 function on synapsis, we used the ligase inhibitor SCR7 to disrupt pairing (16,17). The presence of this weak inhibitor resulted in a modestly reduced pairing efficiency, similar to the pan-ligase inhibitor L189 (Supplementary Figure S3A). In sum, this supports a critical role for DNA binding of L4, but especially the ability of L4 to engage 5΄P in the context of the entire NHEJ core machinery (Ku, XRCC4, L4 and XLF) so as to mediate the most efficient synapsis of an end pair (Figure 1D). Ligation involves the (i) transfer of an adenyl group from the ligase active site to the 5΄ phosphate strand break terminus, followed by (ii) the nucleophilic attack of the 3΄OH strand break terminus and formation of a phosphodiester bond. Completion of step ii) is normally fast and efficient, but can be disrupted by altered end chemistry or flanking nucleotide damage, resulting in the accumulation of a 5΄ adenylated intermediate (7). Such products of aborted ligation can be toxic. To assess the effect on aborted ligation on end pairing we used both a 5΄ adenylated DNA substrate (5΄App) and two examples of LX preparations missing the adenyl group (PPi L4, L4 de-adenylated by pyrophosphate treatment), and K273R L4 (L4 mutated at the catalytic site so as to block adenylation). Pairing was severely reduced in all three cases (Figure 1D). We confirmed by EMSA analysis that the K273R mutation does not significantly impact either the ability of L4 to form a complex with other NHEJ factors at a single end (Supplementary Figure S2F), or its intrinsic ability to bind DNA strand breaks (Supplementary Figure S3B). Furthermore, given that the filament forming proteins XRCC4 and XLF are largely thought to mediate pairing, we tested these proteins on ends lacking 5΄P or 3΄OH and observed no measureable change in their pairing efficiency based on end chemistry (Supplementary Figure S3C). Either the substrate or enzyme product of aborted ligation is thus sufficient to block end pairing, even though de-adenylation is not sufficient to disrupt binding of L4 to DNA (as shown using the K273R ligase in Supplementary Figure S3B) (12).
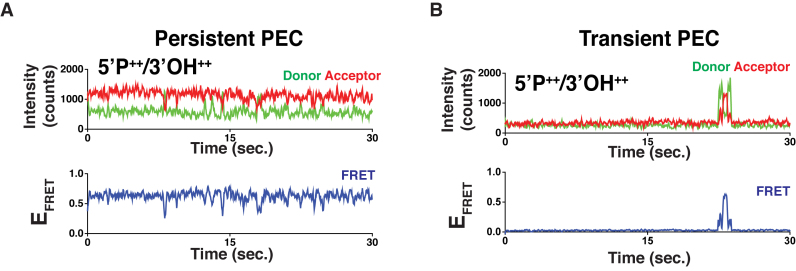
Examination of smFRET trajectories of these core NHEJ complex pairing reactions revealed two readily distinguishable modes of association; ‘Transient’ PECs that were only briefly associated (i.e. for <5 s; Figure 2B), and ‘Persistent’ PECs that remained stably associated for >30 s (Figure 2A) and are the basis for all prior and subsequent analysis (Figures 1 and 3). We therefore propose that productive filament-based pairing can occur following an end-to-end interaction governed by L4 generated Persistent PECs, or result in the rapid dissociation apparent in Transient PECs.
Figure 2.
Pairing modes of NHEJ complex. (A) Representative smFRET trajectory of a Persistent PEC. The PEC remains associated for the duration of our observation time. (B) Representative smFRET trajectory of a Transient PEC. This associates and dissociates in the course of our observation.
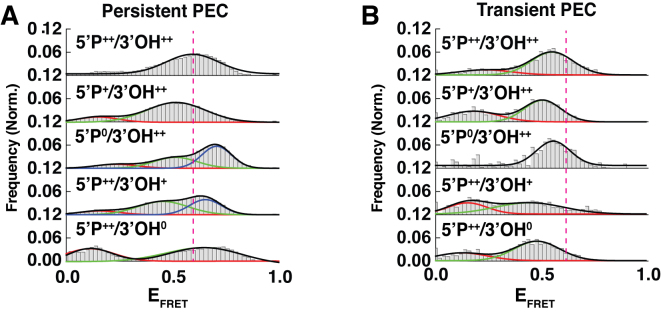
Figure 3.
Population Distributions of PECs. (A) FRET histograms for Persistent PECs of the various changes in end chemistry described in Figure 1. Populations shift based on small changes in the end chemistry. (B) FRET histograms for Transient PECs of the various changes in end chemistry described in Figure 1.
To determine the population dynamics for NHEJ reactions with different DNA end chemistries, we examined the different states observed in smFRET histograms for Persistent PECs (Figure 3A). For various reaction conditions the resulting FRET histograms showed very different distributions, suggesting that the DNA end chemistry modulates the possible conformational states that the PEC can assume (Figure 3A, Supplementary Figure S4A and B). Ends lacking a 5΄P showed a shift in the FRET population to lower FRET states, while substrates lacking both 5΄P resulted in an overall higher FRET state (Supplementary Figure S2A). Ends containing a single 3΄OH led to a narrower distribution than ends lacking both 3΄OH groups, which exhibited a wide distribution of FRET states (Figure 3A, panels 4 and 5). Ends lacking both 5΄P and 3΄OH resembled a convolution of histograms where the ends lack 5΄P and 3΄OH (Supplementary Figure S2A, panel 2). While reactions where both DNAs lack 5΄P and 3΄OH had FRET populations that resemble distribution where ends only lack 5΄P groups (Supplementary Figure S4A, panel 3). The FRET populations also shifted considerably when L4 activity is disrupted (Supplementary Figure S4B), further demonstrating that L4 drives the retention of bridged DNAs. The FRET histograms for the K273R L4 PECs showed a narrow distribution of states. In order to determine if different end chemistry might alter the pairing behavior, we compared pairing efficiency on substrates that lacked either 5΄P or 3΄OH and observed no difference in pairing (Supplementary Figure S4C). The difference between the Transient and Persistent PECs was further probed by examining the smFRET histograms of Transient PEC populations (Figure 3B). From this analysis, we found that Transient PECs predominately occupy different states compared with Persistent PECs. The ends lacking 5΄P are both slightly shifted to a lower primary FRET population peak (Figure 3B, panels 2 and 3), while those without a 3΄OH undergo larger shifts to lower FRET peaks (Figure 3B, panels 4 and 5).
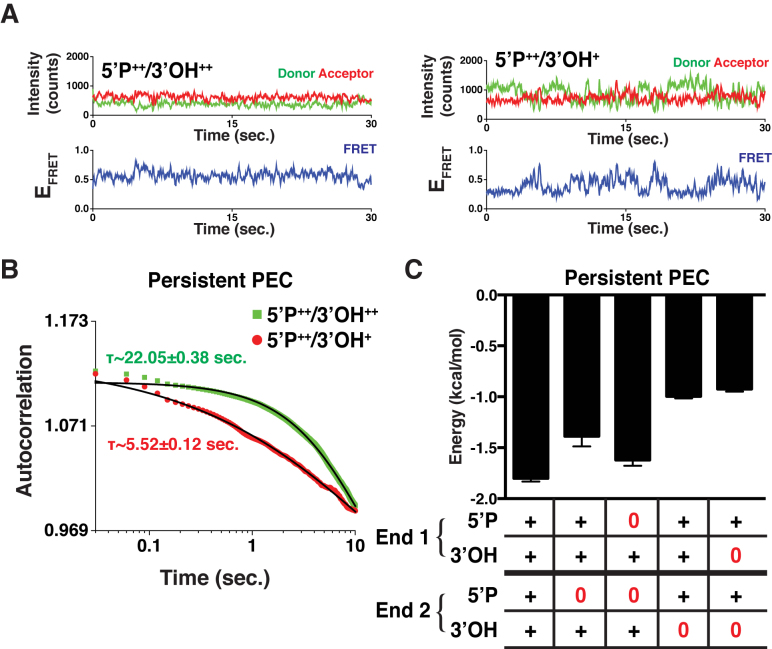
Next, we sought to determine how DNA end chemistry affects the stability of the DNA ends within Persistent PECs as observed in their smFRET trajectories (Figure 4A, Supplementary Figure S5A). We therefore quantified the time characteristics of the transitions observed in single trajectories as a function of the substrate's end chemistry using autocorrelation analysis (18), which yields the transition time between FRET states (Figure 4B). After determining autocorrelation lag times, we used Boltzmann inversion to derive the energetic stability of the ends within the PEC (19) (Supplementary Methods) (Figure 4C). This analysis showed that substrates whose ends result in PECs with faster transition rates have lower stability between FRET states than those with slower transition rates. We observed that DNA ends lacking 5΄P have greater stability than ends lacking a 3΄OH (Figure 4C, Supplementary Figure S3). The stability is likely derived from organization of the PEC, as well as the catalytic cycle of ligases. As opposed to other PECs where the high energy intermediate 5΄P-AMP has formed, ends lacking a 5΄P are unable to assume this intermediate state and do not need to form a phosphodiester bond and release AMP. Importantly, we found that disruption of the L4 catalytic cycle by addition of SCR7 resulted in less stable DNA ends within the PEC (Supplementary Figure S3B). These analyses revealed that the energetic barriers to transition between FRET states are not directly linked to the overall pairing efficiency of the substrates, suggesting that the dynamics of both Persistent and Transient PECs, as well as the conformations they are able to occupy, are more important to the overall synapsis process.
Figure 4.
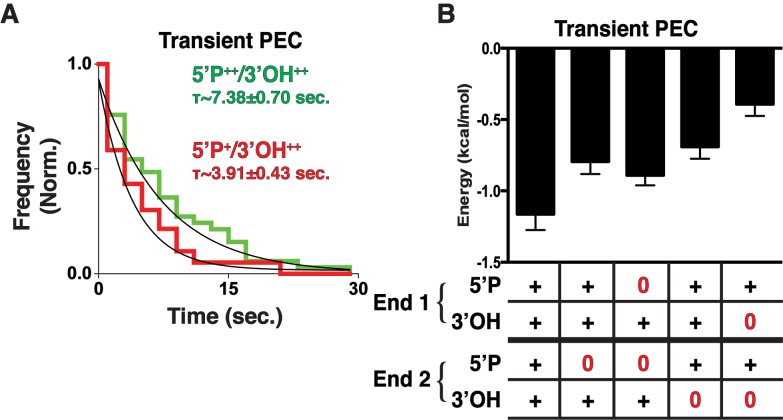
Analysis of Persistent PECs. (A) Two smFRET trajectories comparing the frequency of transition, or repositioning of the DNA ends within the PEC, where ‘+’ denotes only one DNA having the chemical feature and ‘++’ denotes both have the chemical group. 5΄P++/3΄OH++ undergoes fewer transitions than 5΄P++/3΄OH+. (B) Autocorrelation curves for the two populations 5΄P++/3΄OH++ (green) and 5΄P++/3΄OH+ (red) and the corresponding fits (black). (C) Using the time constants from autocorrelation, we calculated the average energetic barriers to transition. Error bars are the SEM of the autocorrelation fit.
Given the observations that Persistent PECs exhibited different dynamics based on end chemistry, we wanted to define the contribution of end chemistry to the behavior of Transient PEC populations. We therefore carried out dwell time analysis on the smFRET trajectories of Transient PECs. This provided the time intervals of transient PEC association and their derived dwell time histograms, which were then fitted to provide the mean dwell time (Supplementary Methods). This analysis revealed that end chemistry strongly modulates the duration of synapsis even for Transient PECs (Figure 5A). Using the measured dwell times, we calculated the Transient PEC energetic stability (Figure 5B, Supplementary Figure S4). These metrics showed that substrates lacking a 5΄P or 3΄OH reduced the dwell times and stability of pairing compared with ligation compatible ends (Figure 5B). Interestingly, it appears that ends lacking a 5΄P exhibit longer dwell times, and consequently are more energetically stable than ends lacking a 3΄OH. This outcome is likely due to the inability of ends lacking a 5΄P to accept an AMP group transferred from L4, thus not forming a highly energetic intermediate. Ends lacking a 3΄OH are able to form these intermediates, but are unable to complete the reaction. Thus, the high-energy intermediate from the reaction persists and must be accommodated.
Figure 5.
Analysis of Transient PECs. (A) Dwell time histograms and fit (black) of transient PECs for 5΄P++/3΄OH++ (green) and 5΄P+/3΄OH++ (red) and fit (black). Ends containing both 5΄P have longer associations than those lacking a 5΄P. (B) Energy stability of Transient PECs for ends lacking 5΄P and 3΄OH. Error bars are the SEM of the dwell time exponential fit.
It has been well established that DSBs have damaged or missing nucleotides flanking the strand breaks, in addition to unique end chemistry that must be corrected prior to rejoining. Numerous accessory factors, such as APTX (aprataxin), PNKP (polynucleotide kinase phosphatase), TDP1 and TDP2 participate in NHEJ to clear ends and make them compatible for rejoining (6,20). End joining is also impaired by potential base mismatches and gaps, which are resolved via the action of nucleases (WRN, Artemis) and polymerases λ and μ (6). However, the role that damaged ends play in DNA end bridging is presently not well understood. We have determined that even simple changes in end chemistry (such as phosphate removal from the ends or the incorporation of bases incapable of forming phosphodiester bonds) trigger disruption of PECs and loss of end synapsis, such that overall bridging of DNAs is reduced. Furthermore, our results suggest that although filaments composed of XRCC4, XLF and L4 drive the bridging of DNAs, L4 plays a key role in determining whether ends remain paired. Our observations are consistent with an increasing body of in vitro and in vivo work in support of the specific functions of NHEJ ligation complex proteins in synapsis and the early stages of end-joining, which were previously attributed solely to DNAPKcs (8,9). We note that a recent study of NHEJ using Xenopus extracts favored a model in which DNA-PKcs mediates synapsis (21). Given the inherent uncertainty of the Xenopus extract system, these observations are likely to arise from the activity of other factors (such as DNA nucleases) which would then require the presence of Artemis/DNA-PKcs for efficient joining, as was recently shown using recombinant NHEJ proteins (22).
Our findings present a unique picture of the dynamics involved in the pairing process of the NHEJ complex. These observations, together with our prior observations of the spatiotemporal organization of NHEJ complexes, provide a basis for the selection of appropriately paired DNAs for DSB repair via the NHEJ pathway (Figure 6). Free DNA ends are recognized by Ku, which then recruits the core ligation complex components XRCC4, XLF, with L4 nucleating a filament on DNA adjacent to the break (8,23,24). These filaments allow pairing of the broken ends and alignment and proofreading of the ends within the PEC. The recognition of compatible ends is carried out by L4, which is able to sense chemical mismatches through disruption of its catalytic cycle. If errors are detected, the PEC will favor dissociation, providing access to the ends by processing enzymes recruited by the core complex (1). This allows for potential end pairing with a new dsDNA end to find an alternative match should there be more than a single DSB in close proximity. In cells, this search will be governed by the mobility of the DNA ends within the confines of the nucleus, chromosome territory and local chromatin environment (25,26). Given the large platform that NHEJ filaments provide for pairing, it is unlikely that forming new synapsis is a limiting factor. DNA ends that remain associated in a metastable state despite errors and with a lower probability of forming are likely subject to repair via synthesis through the actions of polymerases λ and μ or short resection via Artemis/DNA-PKcs or APLF (1). Ultimately, NHEJ is an iterative process where the ends are treated independently until they find a compatible match (1).
Figure 6.
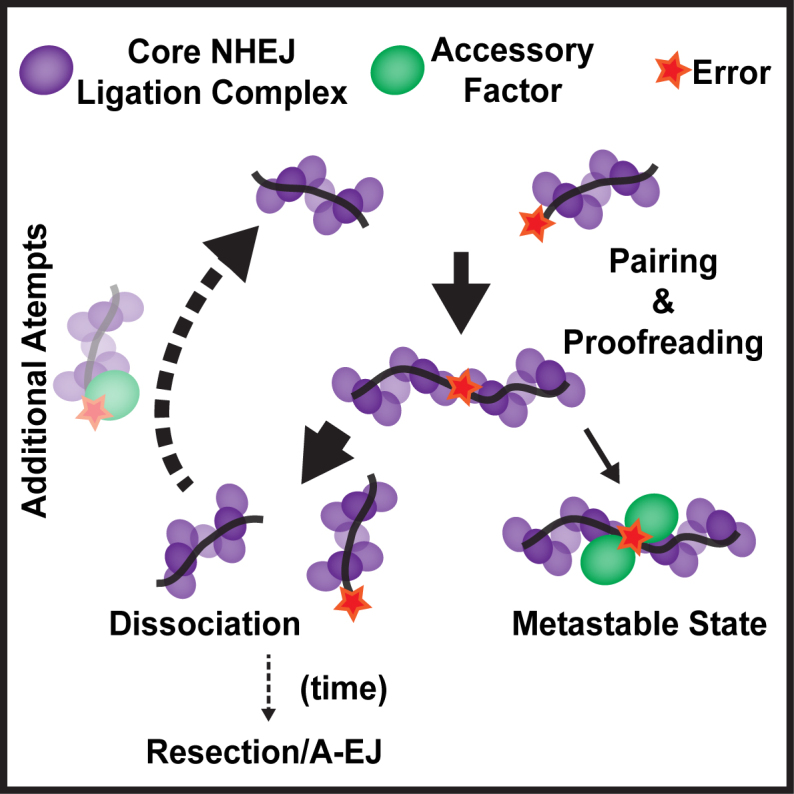
Model for uncoupled repair by NHEJ. Components of the NHEJ core ligation complex assemble on the DNA adjacent to DSBs to facilitate pairing of the free DNA ends and subsequent end-to-end alignment. When the ends are closely aligned, L4 is capable of proofreading the ends for chemical compatibility for ligation. If the ends are found to be insufficiently compatible due to their end chemistry, the ends have a large chance of dissociating. This dissociation step allows accessory factors controlled access to the ends, and reattempted repair. This iterative process can occur many times, although the chance that resection and alternative end-joining occurring increases with time. In some cases, ends can become stuck in a metastable state where they remain associated despite errors. This state likely attracts various accessory factors to assist in the repair and subsequent ligation.
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to acknowledge J.A. Borowiec, T.T. Huang and N.J. Cowan for careful reading of the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Cancer Institute [CA100504, CA084442]; National Institute of General Medical Sciences [GM108119, GM018818]. Funding for open access charge: NIH/NYU School of Medicine.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lieber M.R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 2010; 79:181–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hasty P. Is NHEJ a tumor suppressor or an aging suppressor?. Cell Cycle. 2008; 7:1139–1145. [DOI] [PubMed] [Google Scholar]
- 3.Roix J.J., McQueen P.G., Munson P.J., Parada L.A., Misteli T.. Spatial proximity of translocation-prone gene loci in human lymphomas. Nat. Genet. 2003; 34:287–291. [DOI] [PubMed] [Google Scholar]
- 4.Lieber M.R. Mechanisms of human lymphoid chromosomal translocations. Nat. Rev. Cancer. 2016; 16:387–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woodbine L., Gennery A.R., Jeggo P.A.. The clinical impact of deficiency in DNA non-homologous end-joining. DNA Repair. 2014; 16:84–96. [DOI] [PubMed] [Google Scholar]
- 6.Waters C.A., Strande N.T., Wyatt D.W., Pryor J.M., Ramsden D.A.. Nonhomologous end joining: a good solution for bad ends. DNA Repair. 2014; 17:39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ellenberger T., Tomkinson A.E.. Eukaryotic DNA ligases: structural and functional insights. Annu. Rev. Biochem. 2008; 77:313–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reid D.A., Keegan S., Leo-Macias A., Watanabe G., Strande N.T., Chang H.H., Oksuz B.A., Fenyo D., Lieber M.R., Ramsden D.A. et al. Organization and dynamics of the nonhomologous end-joining machinery during DNA double-strand break repair. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E2575–E2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brouwer I., Sitters G., Candelli A., Heerema S.J., Heller I., de Melo A.J., Zhang H., Normanno D., Modesti M., Peterman E.J. et al. Sliding sleeves of XRCC4-XLF bridge DNA and connect fragments of broken DNA. Nature. 2016; 535:566–569. [DOI] [PubMed] [Google Scholar]
- 10.Ma Y., Lu H., Tippin B., Goodman M.F., Shimazaki N., Koiwai O., Hsieh C.L., Schwarz K., Lieber M.R.. A biochemically defined system for mammalian nonhomologous DNA end joining. Mol. Cell. 2004; 16:701–713. [DOI] [PubMed] [Google Scholar]
- 11.Rothenberg E., Ha T.. Single-molecule FRET analysis of helicase functions. Methods Mol. Biol. 2010; 587:29–43. [DOI] [PubMed] [Google Scholar]
- 12.Jayaram S., Ketner G., Adachi N., Hanakahi L.A.. Loss of DNA ligase IV prevents recognition of DNA by double-strand break repair proteins XRCC4 and XLF. Nucleic Acids Res. 2008; 36:5773–5786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Budman J., Chu G.. Processing of DNA for nonhomologous end-joining by cell-free extract. EMBO J. 2005; 24:849–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsai C.J., Kim S.A., Chu G.. Cernunnos/XLF promotes the ligation of mismatched and noncohesive DNA ends. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:7851–7856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waters C.A., Strande N.T., Pryor J.M., Strom C.N., Mieczkowski P., Burkhalter M.D., Oh S., Qaqish B.F., Moore D.T., Hendrickson E.A. et al. The fidelity of the ligation step determines how ends are resolved during nonhomologous end joining. Nat. Commun. 2014; 5:4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Srivastava M., Nambiar M., Sharma S., Karki S.S., Goldsmith G., Hegde M., Kumar S., Pandey M., Singh R.K., Ray P. et al. An inhibitor of nonhomologous end-joining abrogates double-strand break repair and impedes cancer progression. Cell. 2012; 151:1474–1487. [DOI] [PubMed] [Google Scholar]
- 17.Greco G.E., Matsumoto Y., Brooks R.C., Lu Z., Lieber M.R., Tomkinson A.E.. SCR7 is neither a selective nor a potent inhibitor of human DNA ligase IV. DNA Repair. 2016; 43:18–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rothenberg E., Trakselis M.A., Bell S.D., Ha T.. MCM forked substrate specificity involves dynamic interaction with the 5΄-tail. J. Biol. Chem. 2007; 282:34229–34234. [DOI] [PubMed] [Google Scholar]
- 19.Rothenberg E., Grimme J.M., Spies M., Ha T.. Human Rad52-mediated homology search and annealing occurs by continuous interactions between overlapping nucleoprotein complexes. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:20274–20279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xing M., Yang M., Huo W., Feng F., Wei L., Jiang W., Ning S., Yan Z., Li W., Wang Q. et al. Interactome analysis identifies a new paralogue of XRCC4 in non-homologous end joining DNA repair pathway. Nat. Commun. 2015; 6:6233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Graham T.G., Walter J.C., Loparo J.J.. Two-stage synapsis of DNA ends during non-homologous end joining. Mol. Cell. 2016; 61:850–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei P.C., Chang A.N., Kao J., Du Z., Meyers R.M., Alt F.W., Schwer B.. Long neural genes harbor recurrent DNA break clusters in neural stem/progenitor cells. Cell. 2016; 164:644–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lan L., Ui A., Nakajima S., Hatakeyama K., Hoshi M., Watanabe R., Janicki S.M., Ogiwara H., Kohno T., Kanno S. et al. The ACF1 complex is required for DNA double-strand break repair in human cells. Mol. Cell. 2010; 40:976–987. [DOI] [PubMed] [Google Scholar]
- 24.Ogiwara H., Ui A., Otsuka A., Satoh H., Yokomi I., Nakajima S., Yasui A., Yokota J., Kohno T.. Histone acetylation by CBP and p300 at double-strand break sites facilitates SWI/SNF chromatin remodeling and the recruitment of non-homologous end joining factors. Oncogene. 2011; 30:2135–2146. [DOI] [PubMed] [Google Scholar]
- 25.Roukos V., Misteli T.. The biogenesis of chromosome translocations. Nat. Cell Biol. 2014; 16:293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalousi A., Soutoglou E.. Nuclear compartmentalization of DNA repair. Curr. Opin. Genet. Dev. 2016; 37:148–157. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.