Abstract
The DNA replication or S-phase checkpoint monitors the integrity of DNA synthesis. Replication stress or DNA damage triggers fork stalling and checkpoint signaling to activate repair pathways. Recovery from checkpoint activation is critical for cell survival following DNA damage. Recovery from the S-phase checkpoint includes inactivation of checkpoint signaling and restart of stalled replication forks. Previous studies demonstrated that degradation of Mrc1, the Saccharomyces cerevisiae ortholog of human Claspin, is facilitated by the SCFDia2 ubiquitin ligase and is important for cell cycle re-entry after DNA damage-induced S-phase checkpoint activation. Here, we show that degradation of Mrc1 facilitated by the SCFDia2 complex is critical to restart stalled replication forks during checkpoint recovery. Using DNA fiber analysis, we showed that Dia2 functions with the Sgs1 and Mph1 helicases (orthologs of human BLM and FANCM, respectively) in the recombination-mediated fork restart pathway. In addition, Dia2 physically interacts with Sgs1 upon checkpoint activation. Importantly, failure to target Mrc1 for degradation during recovery inhibits Sgs1 chromatin association, but this can be alleviated by induced proteolysis of Mrc1 after checkpoint activation. Together, these studies provide new mechanistic insights into how cells recover from activation of the S-phase checkpoint.
INTRODUCTION
Checkpoint recovery is central to accurate cell division. Checkpoint pathways monitor progress during cell division so that in the event of an error, the checkpoint is activated to block the cell cycle and activate repair pathways. Checkpoint recovery mechanisms contribute to the successful completion of a checkpoint. Checkpoint recovery encompasses the pathways responsible for inactivation of checkpoint signaling and cell cycle re-entry after the initial stress has been alleviated.
The DNA replication or S-phase checkpoint monitors the integrity of DNA synthesis. Replication stress such as limited free nucleotides or DNA damage in the form of DNA breaks or adducts leads to replication fork stalling and the initiation of the checkpoint-signaling pathway (1–4). In the budding yeast Saccharomyces cerevisiae, initiation of the S-phase checkpoint leads to activation and recruitment of the Mec1/ATR kinase (5–7). Mec1 activates the kinase Rad53 via mediator proteins that include Mrc1, Rad9, Tof1 and Csm3 (7–15). Rad53 activation results in replication fork stabilization and inhibition of further origin firing (2,4,16–18).
Recovery from the S-phase checkpoint requires inactivation of checkpoint signaling and restart of stalled replication forks, although it is not clear whether there is coordination between the pathways that inhibit checkpoint signaling and those that facilitate fork restart. In yeast, inactivation of the signaling kinase Rad53 by phosphatases Pph3 and Ptc2 contributes to fork restart during recovery from a methyl methane sulfonate (MMS)-induced checkpoint (19,20). In addition, fork restart during recovery from a hydroxyurea (HU)-induced checkpoint is dependent on the chromatin remodeling proteins Ino80 and Isw2 (21–23).
Protein degradation of checkpoint adaptor proteins is also important for recovery from the S-phase checkpoint. Our previous work showed that the degradation of the checkpoint adaptor and replication fork protein Mrc1, controlled by the SCFDia2 ubiquitin ligase complex, is important for cell cycle re-entry after cells are exposed to the alkylating agent MMS. Rad53 phophorylation levels persist longer in dia2Δ cells during recovery from an MMS-induced checkpoint and dia2Δ cells exhibit greater recovery delays when the Rad53 phosphatase Pph3 is also absent, suggesting that Mrc1 degradation contributes to checkpoint signaling inactivation (24). Further, degradation of Claspin, the human homolog of Mrc1, is linked to inhibition of Chk1 kinase signaling, after an HU-induced S-phase checkpoint (25–27).
Multiple mechanisms can lead to restart of stalled replication forks. In some cases, direct restart of a stable, stalled fork is possible, although little is known about direct restart mechanisms (28). Alternatively, there is evidence that compromised forks can be restarted after remodeling by fork reversal or nucleolytic processing of nascent DNA strands or recombination-mediated mechanisms (28–30). The Sgs1 helicase, a member of the RecQ helicase family and yeast ortholog of the Bloom Syndrome protein BLM, is important for recombination-mediated fork restart (31,32). Mph1, a member of the SF2 helicase family and the yeast homolog of FANCM, plays a role in fork maintenance and fork reversal (33,34), which is important for recombination-mediated fork restart.
In these studies, we reveal a mechanistic link between degradation of a checkpoint protein and recombination-mediated restart of stalled replication forks. We show that Dia2-dependent degradation of Mrc1 is required for replication fork restart in a pathway that includes Sgs1 and Mph1. Moreover, we find that Dia2 interacts with Sgs1 and that the recruitment of Sgs1 to chromatin during checkpoint recovery is dependent on Mrc1 degradation.
MATERIALS AND METHODS
Plasmids and strains
Tables 1 and 2 contain a list of strains and plasmids used in this study.
Table 1. Strains used in this study.
| Strain | Description | Reference |
|---|---|---|
| Y80 | ade2-1 ura3-1 leu2-3,112 his3-11,15 trp1-1 can1-100 MATa | (57) |
| DKY1032 | As Y80 but HIS3::BrdU-Inc | This study |
| DKY194 | As Y80 but dia2Δ::kanMX | (58) |
| DKY1033 | As DKY194 but HIS3::BrdU-Inc | This study |
| DKY1042 | As Y80 but sgs1Δ::kanMX HIS3::BrdU-Inc | This study |
| DKY1043 | As Y80 but mph1Δ::kanMX HIS3::BrdU-Inc | This study |
| DKY1044 | As Y80 but dia2Δ::kanMX sgs1Δ::kanMX HIS3::BrdU-Inc | This study |
| DKY1045 | As Y80 but dia2Δ::kanMX mph1Δ::kanMX HIS3::BrdU-Inc | This study |
| AKY149 | As DKY194 but dia2Δ::kanMX::9MYC-DIA2 URA3 | (36) |
| AKY188 | As AKY149 but DIA2-ΔF | (36) |
| AKY189 | As AKY149 but DIA2-ΔN200 | (36) |
| AKY190 | As AKY149 but DIA2-LRR | (36) |
| AKY192 | As AKY149 but DIA2-TPR | (36) |
| AKY193 | As AKY149 but DIA2-ΔN175 | (36) |
| AKY199 | As AKY149 but DIA2-ΔN134 | (36) |
| DKY1046 | As AKY149 but HIS3::BrdU-Inc | This study |
| DKY1047 | As AKY188 but HIS3::BrdU-Inc | This study |
| DKY702 | mrc1::HIS3::3HA-MRC1 LEU2 dia2::KanMX::9MYC-DIA2::URA3 his3 trp1 ade2 MATa | (24) |
| DKY909 | dia2::KanMX mrc1::HIS3::3HA-AID-MRC1::LEU2 ura3 leu2 his3 trp1 ade2 MATa | (24) |
| DKY1048 | As DKY702 but TRP1::BrdU-Inc | This study |
| DKY1049 | As DKY909 but URA3::BrdU-Inc | This study |
Table 2. Plasmids used in this study.
Yeast cell culture and treatments
Yeast strains were cultured by standard methods (35). For α-factor arrest, yeast cultures were grown in either liquid minimal medium or yeast extract-peptone-dextrose (YPD) and pulsed with 2 μg/ml α-factor peptide (Alpha Aesar) every hour for 2 h. HU or MMS treated cells were grown in minimal or YPD medium. For strains harboring galactose-inducible plasmids, cells were first treated with 2% raffinose for overnight followed by treatment with 2% galactose at 30°C.
Immunoblotting
Protein samples were separated by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred to nitrocellulose membranes (Pall Corporation, USA). After blocking in 5% milk, membranes were incubated with the following primary antibodies: 9E11 (Covance), anti-HA (Covance), anti-Sgs1 (Santa Cruz Biotech) and anti-Pgk1 (Invitrogen). Horseradish peroxidase conjugated secondary antibodies were used (anti-mouse, Jackson Immuno-Research or anti-goat, Santa Cruz Biotech). Protein bands were detected using a Pierce ECL kit (Thermo Fisher).
Dual-labeling DNA fiber assay
Yeast strains capable of incorporating thymidine analogs were generated by transforming with linearized HSV-TK plasmids (gift of O. Aparicio). Yeast strains were synchronized in G1 with α-factor. Cells were released from G1 arrest in fresh media containing 400 μg/ml IdU for 15 min. For replication fork restart and fork stability assays, after IdU treatment, cells were either untreated or treated with 0.05% MMS for 60 min followed by release in fresh media containing 400 μg/ml CldU for 15 min. For measurement of replication fork velocity, no MMS treatment was done and cells were immediately transferred to CldU containing media for 15 min after 15 min IdU treatment. DNA fibers were stretched on silanized glass slides and IdU (red fibers) and CldU (green fibers) incorporation was detected by using mouse anti-BrdU clone B44 antibody (Becton Dickinson, cat # 347580) and Rat anti-BrdU clone BU1/75 antibody (AbD Serotec, cat # OBT0030G), respectively. Images were captured on a Deltavision microscope (Applied Precision) and analyzed using Deltavision softWoRx 5.5 software. All DNA fiber results shown are the means of three independent experiments (100 DNA fibers/experiment). Error bars show standard error of the mean. P-values for restart experiments were derived from the Student's t-test. P-values for fork velocities and nascent strand degradation were determined using the Mann–Whitney test.
Viability and spotting assays
For viability assays, overnight cultures were first grown in YPD to log phase. Equal numbers of cells were plated on media containing the indicated amount of MMS or HU. Colony forming units (CFU) were counted after 4 days at 30°C. The percentage viability was calculated by dividing CFU on YPD with MMS or HU by CFU on YPD without MMS or HU. For spotting assays, 10-fold serial dilutions of 2 × 107 to 2 × 103 cells of the indicated strains were spotted on YPD, YPD + 0.007% MMS and YPD + 200 mM HU plates using replica plater and incubated at 30°C for 2–3 days.
Immunoprecipitation experiments
Immunoprecipitation experiments were performed using protocols as previously described (36). Briefly, cell pellets were resuspended in NETN lysis buffer (20 mM Tris, pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.5% Igepal, 10 mM NaF, 25 mM β–glycerophosphate, 1 mM phenylmethylsulfonyl fluoride and 1 mM pepstatin, as well as complete protease inhibitor cocktail (Roche Applied Science). After vortexing the cells for 10 min at 4°C with glass beads, lysates were microcentrifuged at 10 000 rpm at 4°C followed by transfer of clear supernatant to new tubes. One milligram of lysate was incubated with 1:200 anti-myc antibody. The samples were then incubated with protein A/G-agarose beads (Santa Cruz Biotechnology) for 2 h at 4°C. Beads were washed three times with same NETN lysis buffer, boiled in 1X Laemmli loading dye and loaded to SDS-PAGE for Western blotting.
Chromatin fractionation experiment
Chromatin fractionation was performed as previously described (36). Briefly, spheroplasts were isolated and lysed with 0.2% Triton-X100 and 150 mM NaCl. Whole cell extracts (WCE) were fractionated to supernatant (S) and pellet (P) using centrifugation at 15 000 ×g for 15 min at 4°C. Prior to centrifugation, WCEs were either untreated or treated with 150 kU DNaseI for 30 min. One volume of 2X Laemmli buffer was added in WCE/Input and supernatant (S) and for pellet (P) containing chromatin fraction, 2X Laemmli buffer to equal volume of S was added. Samples were boiled and then resolved by SDS-PAGE prior to immunoblotting.
Auxin-induced degradation of Mrc1
Auxin induced Mrc1 degradation was performed as previously described (24). For chromatin fractionation and DNA fiber assays, either vehicle (100% ethanol) or 1.5 mM indole-3-acetic acid (IAA, Alfa Aesar) was added to cell cultures in YPD at 30°C.
Flow cytometry
Flow cytometry was performed as previously described (24). Briefly, cells were harvested and fixed with 70% ethanol. Fixed cells were resuspended in 1x PBS, sonicated to break clumps and treated with 100 μg/ml RNase in Tris-EDTA overnight. Samples were stained with 50 μg/ml propidium iodide for 1 h and analyzed by flow cytometry using FACSCaliber (BD Biosciences) and FlowJo software (Tree Star). Cell cycle distribution graphs were prepared using Deltagraph software (Red Rock). 2C DNA content was quantitated using flow cytometry data gated for a standard 2C distribution developed from an asynchronous population and percentages were calculated by dividing the number of cells with 2C DNA content by the total number counted.
RESULTS
We have previously demonstrated that Dia2-mediated degradation of Mrc1 is important for cell cycle re-entry from an MMS-induced S-phase checkpoint (24). The absence of Dia2 leads to increased persistence of Rad53 phosphorylation during checkpoint recovery, suggesting that Mrc1 degradation could contribute to checkpoint signaling inactivation. However, a key transition during checkpoint recovery is the restart of stalled replication forks. As dia2Δ exhibits enhanced phenotypes in combination with known replication fork restart mutants, including sgs1Δ and pph3Δ (37,38), we investigated the role of Dia2 and Mrc1 degradation in the restart of stalled replication forks.
Dia2 is required for restart of stalled replication forks
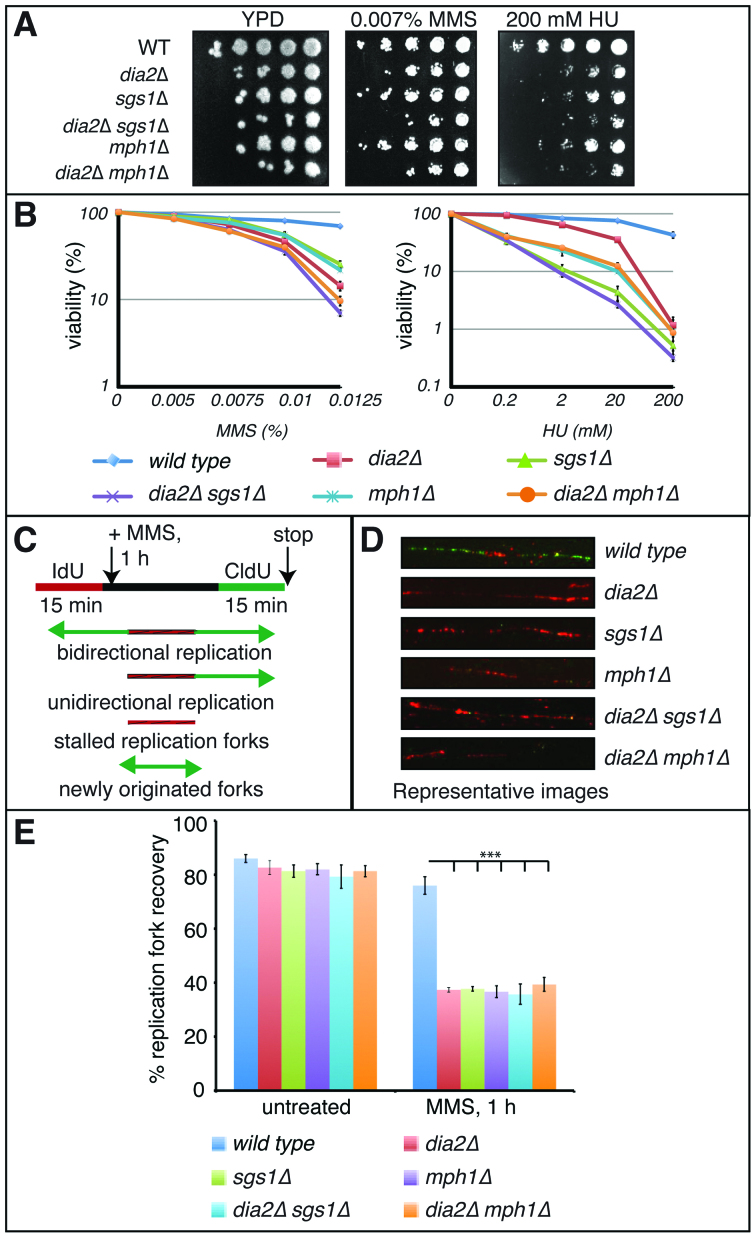
Given the known genetic links between Sgs1 and Dia2 (37,38), we chose to compare the restart of stalled forks in dia2Δ cells to the sgs1Δ mutant and the functionally similar mph1Δ mutant. Sgs1 and Mph1 are yeast orthologs of human Bloom Syndrome protein BLM helicase and Fanconi anemia protein FANCM (32,39), respectively. Both Sgs1 and Mph1 protect stalled replication forks during replication stress and strains lacking SGS1 or MPH1 are hypersensitive to the replication inhibitors HU and MMS (40–43). To determine if Dia2 is essential for stalled replication fork restart and if it works in the same pathway with Sgs1 and Mph1, we established wild type, dia2Δ, sgs1Δ, mph1Δ, dia2Δsgs1Δ and dia2Δmph1Δ strains bearing an integrated copy of HSV–TK (Herpes simplex virus–thymidine kinase) expression vector that allows efficient incorporation of thymidine analogs such as BrdU, IdU or CldU. We observed that dia2Δ cells exhibited reduced viability in combination with sgs1Δ, as expected (37,38), as well as with mph1Δ mutants when challenged with replication stress (Figure 1A and B).
Figure 1.
Dia2 is essential for restart of stalled replication forks. (A) Ten fold serial dilutions of the indicated strains were spotted on yeast extract-peptone-dextrose (YPD), YPD + 0.007% methyl methane sulfonate (MMS) or 200 mM hydroxyurea (HU) and incubated at 30°C. (B) Equal number of cells were plated on media containing the indicated amounts of MMS or HU and colony-forming units were counted after 4 days at 30°C. Error bars represent standard deviations from three independent experiments. (C) Schematic of DNA fiber assay depicting sites of replication. Red tracts, IdU; green tracts, CldU. (D) Representative figure showing inability to restart replication after 0.05% MMS treatment in absence of Dia2, Sgs1 or Mph1. (E) Dia2, Sgs1 and Mph1 work in concert to mediate replication fork restart after MMS-induced fork stalling. The replication restart efficiencies of wild type, dia2Δ, sgs1Δ, mph1Δ, dia2Δsgs1Δ and dia2Δmph1Δ strains were measured as the number of restarted replication forks (IdU–CldU tracts) compared with the total number of IdU-labeled tracts (IdU only tracts plus IdU–CldU tracts). ***P < 0.001.
We next monitored replication events in individual chromosomes during recovery from an MMS-induced checkpoint using a dual-labeling DNA fiber assay. Cells synchronized in G1 were allowed to enter S-phase and then were pulse-labeled with IdU (red label) for 15 min. Following IdU labeling, cells were either untreated or treated with 0.05% MMS for 1 h, followed by a second pulse-labeling with CldU (green label) for 15 min in the absence of MMS (Figure 1C). DNA fibers were isolated and incubated with anti-IdU and anti-CldU antibodies to visualize DNA replication. Wild type cells were able to restart the majority of MMS-induced stalled forks (76%), as shown by the green-labeled DNA tracts (Figure 1D and E). However, dia2Δ cells exhibited severe deficiency in stalled replication fork restart (37%), suggesting that Dia2 is important for efficient recovery of the MMS-stalled replication forks. Interestingly, depletion of Dia2 in sgs1Δ or mph1Δ strains did not exaggerate the phenotype observed in sgs1Δ or mph1Δ single-deficient strains (36% and 39% in dia2Δsgs1Δ and dia2Δmph1Δ strains versus 38% and 37% in sgs1Δ and mph1Δ strains) suggesting that Dia2, Sgs1 and Mph1 are epistatic for restart of MMS-stalled replication forks.
Dia2 and Sgs1 are important for maintenance of normal replication fork velocity
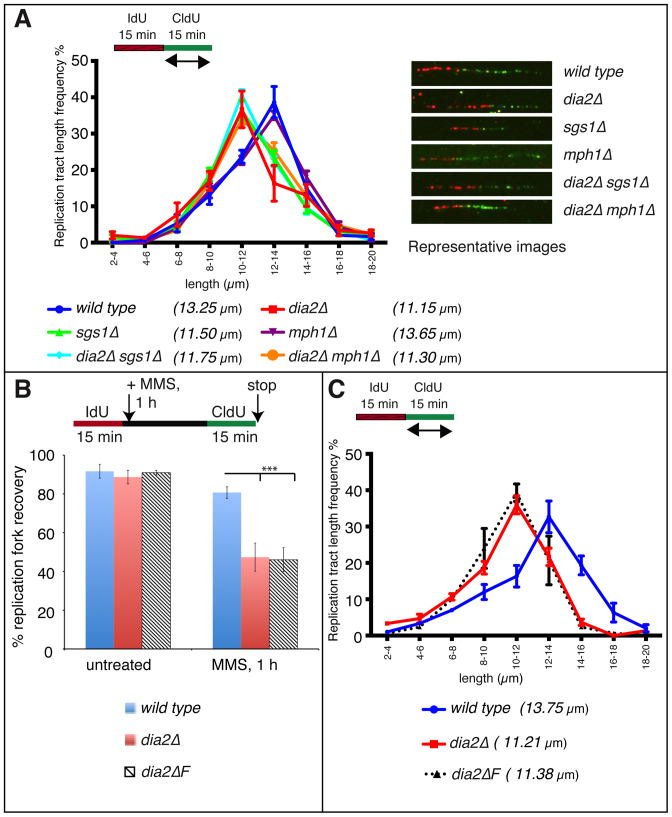
In addition to its role in recombination-mediated replication fork restart, human BLM helicase is also essential for maintaining normal replication fork velocity. Thus, we investigated whether Sgs1, Dia2 and Mph1 also affect normal replication fork velocity. We pulse labeled G1-synchronized wild type, dia2Δ, sgs1Δ, mph1Δ, dia2Δsgs1Δ and dia2Δmph1Δ strains with IdU for 15 min after release into S phase, followed immediately by another pulse labeling with CldU for 15 min. To assess ongoing replication fork velocity, we measured CldU (green label) tract lengths in IdU-CldU double-labeled (red-green labels) tracts. The distribution pattern of tract lengths was similar between wild type and mph1Δ strains with median tract lengths being 13.25 μm and 13.65 μm, respectively. By contrast, the dia2Δ and sgs1Δ strains had shorter tract length distribution with median tract lengths of 11.15 μm and 11.50 μm, respectively, suggesting that Dia2 and Sgs1, but not Mph1, maintain normal replication fork velocity. The dia2Δsgs1Δ strain also had a similar tract length distribution (median tract length 11.75 μm) to dia2Δ and sgs1Δ strains, further suggesting that Dia2 and Sgs1 are epistatic for maintaining normal fork velocity (Figure 2A).
Figure 2.
Dia2 and Sgs1 maintains normal replication velocity and F-Box domain of Dia2 is important for both fork restart as well as fork speed. (A) Distributions of CldU tract lengths were determined on IdU–CldU double-labeled DNA fibers isolated from untreated wild type, dia2Δ, sgs1Δ, mph1Δ, dia2Δsgs1Δ and dia2Δmph1Δ strains. P < 0.05 for wild type versus dia2Δ, sgs1Δ, dia2Δsgs1Δ or dia2Δmph1Δ strains and P-value is not significant for wild type versus mph1Δ strain. (B) Replication fork restart efiiciencies of wild type, dia2Δ and the dia2 F-Box domain deletion strains (dia2ΔF) were measured as the number of restarted replication forks (IdU–CldU tracts) compared with the total number of IdU-labeled tracts (IdU only and IdU–CldU tracts). ***P < 0.001. (C) CldU tract length distributions were determined in wild type, dia2Δ and dia2 F-Box domain deletion strains (dia2ΔF). P < 0.05 for wild type versus dia2Δ or dia2ΔF.
The Dia2 F-box domain is essential for stalled replication fork restart and maintenance of replication velocity
To determine whether the function of Dia2 in replication fork restart and fork velocity was due to its role as a part of a ubiquitin ligase, we examined a Dia2 mutant lacking the F-box domain using the same methods as described above. The absence of the F-box domain resulted in a severe deficiency of restart of MMS-stalled replication forks (Figure 2B) and reduced fork velocity (Figure 2C). The defects were comparable to the phenotypes in strains lacking Dia2, suggesting that Dia2's role as a ubiquitin ligase is essential to promote stalled fork restart as well as maintenance of unperturbed replication fork velocity.
Mph1 but not Dia2 or Sgs1 protects nascent DNA strands from degradation after fork stalling
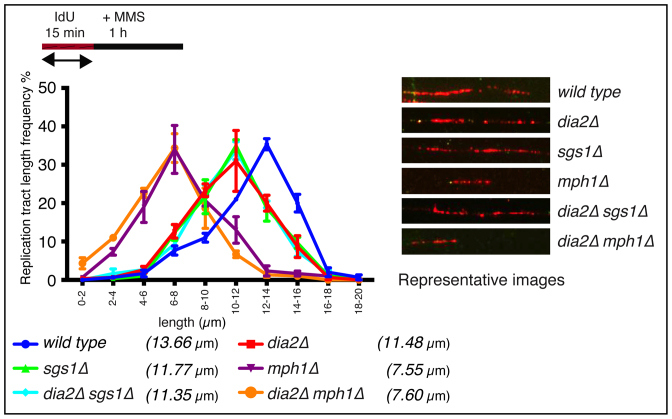
In human cells, Fanconi Anemia proteins have dual roles in replication fork restart and protection of DNA strands from degradation after fork stalling whereas Human BLM is dispensable for nascent DNA strand stability at stalled forks (44,45). As we observed replication fork restart defects in dia2Δ, sgs1Δ and mph1Δ cells, we asked if Dia2, Sgs1 or Mph1 have any role in nascent DNA strand protection after MMS-induced fork stalling. For this, cells were labeled with IdU (red label) for 15 min followed by 1 hour 0.05% MMS treatment. DNA tract length distributions were analyzed in wild type, dia2Δ, sgs1Δ, mph1Δ, dia2Δsgs1Δ and dia2Δmph1Δ strains. As Dia2 and Sgs1 maintain normal replication fork velocity (Figure 2A), dia2Δ and sgs1Δ strains exhibited slightly shorter tract length distribution patterns (median of 11.48 μm and 11.77 μm, respectively) compared to the wild type (median of 13.66 μm; Figure 3). Strikingly, mph1Δ strains showed significantly shorter tract length frequency (median of 7.55 μm) suggesting that Mph1, but not Dia2 or Sgs1, is important for protection of nascent DNA strands from degradation after MMS-induced fork stalling. The dia2Δmph1Δ strain showed tract length patterns similar to mph1Δ strains (median of 7.60 μm compared to 7.55 μm) further suggesting that the phenotype observed in the double-deficient strain is due to the absence of Mph1 alone (Figure 3).
Figure 3.
Mph1 protects nascent DNA strands from nucleolytic degradation after fork stalling. Nascent replication fork tract lengths (labeled with IdU only) were measured after 1 h of 0.05% MMS treatment in dia2Δ, sgs1Δ, mph1Δ, dia2Δsgs1Δ and dia2Δmph1Δ strains. P < 0.05 for wild type versus dia2Δ, sgs1Δ or dia2Δsgs1Δ and P < 0.001 for wild type versus mph1Δ or dia2Δmph1Δ strains.
Dia2 physically interacts with Sgs1 during replication stress
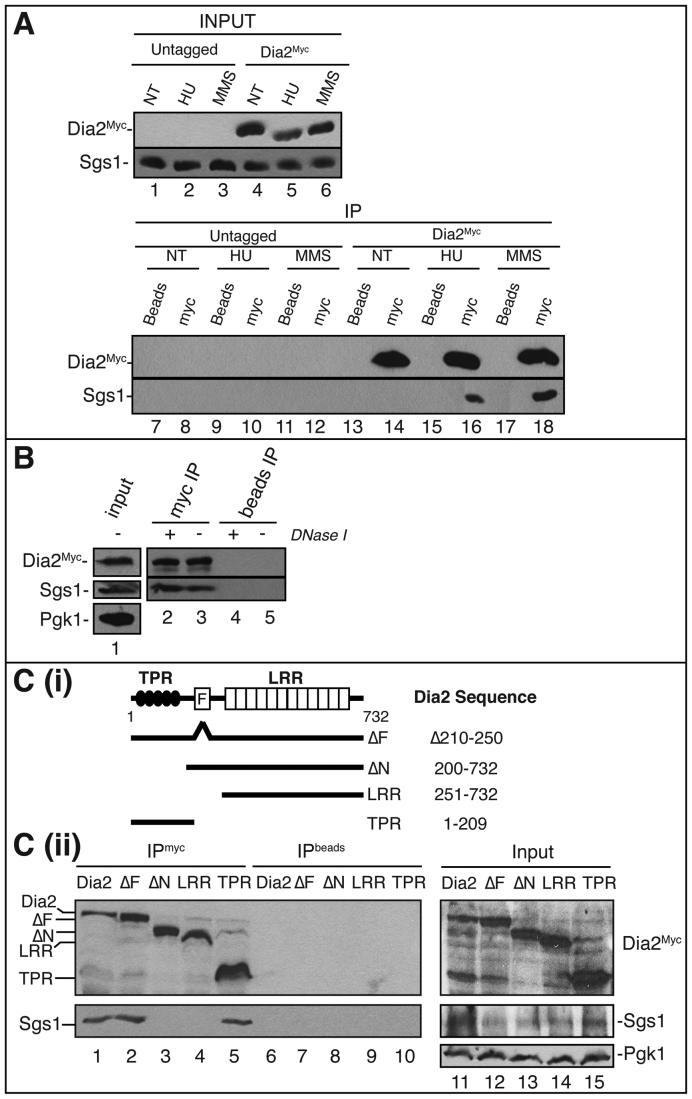
We next asked if Dia2 physically interacts with Sgs1. To determine this, we used a strain expressing full-length Myc-tagged Dia2 (Dia2Myc) at the endogenous locus. Cells were either untreated or treated with MMS or HU to induce replication fork stalling. Immunoprecipitation of Dia2Myc protein was carried out by incubating whole cell lysates with anti-Myc antibody. As shown in Figure 4A, Dia2Myc co-precipitated with Sgs1 only during MMS or HU treatment. Moreover, Sgs1 did not co-precipitate with the anti-Myc antibody in wild-type cells, indicating that the Dia2–Sgs1 interaction is specific. To determine if this interaction was direct or via DNA-bridges, we repeated the immunoprecipitation with and without DNase I. Interaction between Dia2 and Sgs1 was similar in both untreated or DNase I-treated cell lysates suggesting that the Dia2-Sgs1 interaction is not mediated by DNA-bridges (Figure 4B).
Figure 4.
Dia2-Sgs1 interaction is induced by stalled replication forks. (A) Dia2 physically interacts with Sgs1. Untreated (NT), 0.05% MMS treated or 200 mM HU treated cell lysates prepared from wild type as well as 9myc-tagged Dia2 strains were used in anti-myc immunoprecipitations (lanes 8, 10, 12, 14, 16 and 18) or incubated with protein A/G beads without antibody as a negative control (lanes 7, 9, 11, 13, 15 and 17). Immunoblots were probed with anti-myc or anti-Sgs1 antibodies. (B) Dia2 interaction with Sgs1 is not via DNA bridges. Cell lysates from wild-type cells (lane 1) were either untreated or treated with DNaseI prior to immunoprecipitation with anti-Myc antibody. (C) Sgs1 interacts with the TPR domain in Dia2. (i) Schematics showing Dia2 protein domains and mutants used in the immunoprecipitation study. TPR, tetratricopeptide repeats; F, F-Box domain; LRR, leucine-rich repeats. (ii) Strains expressing Myc-tagged Dia2 protein segments were treated with 0.05% MMS. Cell lysates (lanes 11–15) were prepared for immunoprecipitations with an anti-myc antibody (lanes 1–5) or protein A/G beads without antibody (lanes 6–10). Immunoblots were probed with anti-myc and anti-Sgs1 antibodies.
Dia2 interacts with Sgs1 via an N-terminal domain
Next, we mapped the interaction site of Sgs1 on Dia2. For this we used strains containing full-length as well as truncated versions of Myc-tagged Dia2, expressed from the endogenous locus, using co-immunoprecipation conditions as previously described ((36) and Figure 4C(i)). As shown, full-length Dia2 coimmunoprecipitated Sgs1 (Figure 4C(ii)). The Dia2 mutant lacking the F-box domain (ΔF) and an N-terminal fragment (amino acids 1–209), which contains the TPR domain and NLS, also co-precipitated with Sgs1. By contrast, the ΔN mutant, which lacks the first 200 amino acids of the protein, including the TPR domain, did not co-precipitate with Sgs1. Likewise, a C-terminal Dia2 fragment containing the LRR domain but lacking other known domains did not co-precipitate with Sgs1. Taken together, these results indicate that the N-terminal portion of Dia2 containing the TPR domain and NLS interacts with Sgs1 (Figure 4C(ii)).
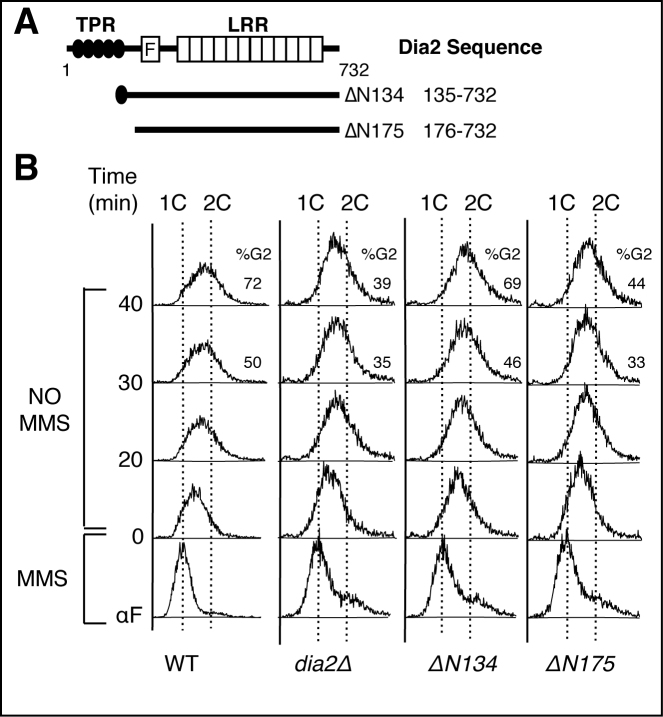
Given that the N-terminal domain of Dia2 interacts with both Mrc1 (46,47) and Sgs1, we asked whether this domain was important for both MMS sensitivity and recovery from an MMS-induced checkpoint. We have previously generated two N-terminal truncations of Dia2, ΔN134 and ΔN175 (36) (note that the numbering has been updated to reflect recognition of verified Dia2 start site (46)). The ΔN175 mutant lacks the entire TPR repeat region (amino acids 48–144) plus a small section of the protein prior to the F-box domain (residues 145–175) and is MMS-sensitive like dia2Δ cells, whereas the ΔN134 mutant is missing most but not all of the TPR domain and is not MMS sensitive (36) (Figure 5). We tested whether these mutants can re-enter the cell cycle efficiently during recovery from an MMS-induced checkpoint. Cells were arrested in G1, released into MMS-containing media for 40 min to induce fork stalling and then allowed to re-enter the cell cycle in the absence of MMS. Cell cycle progression was monitored by flow cytometry. We observed that the ΔN175 mutant exhibited delayed cell cycle re-entry kinetics with 44% of cells in G2/M at the 40-minute timepoint, which was similar to the dia2Δ strain with 39% of cells at G2/M at the same time. By contrast, the ΔN134 mutant finished S phase with kinetics similar to wildtype cells, with 69% and 72%, respectively, in G2/M phase at the 40 min timepoint (Figure 5). Together, these data suggest that the MMS-sensitivity of dia2Δ cells is linked, at least in part, to the function of Dia2 in checkpoint recovery. Further, the majority of the TPR domain is not required for checkpoint recovery, but a section of the protein between residues 134 and 175 is required.
Figure 5.
An N-terminal domain of Dia2 is required for checkpoint recovery. (A) Schematic showing N-terminal deletion mutants of Dia2. (B) Cells were arrested in G1 phase of cell cycle by α-factor (αF), released into YPD + 0.033% MMS for 40 min followed by release into YPD without MMS for indicated time points to allow checkpoint recovery. Samples were analyzed by flow cytometry. 1C and 2C indicate DNA content and percentage of cells with 2C DNA content is shown on the right side of the selected profiles.
Upon stalled fork recovery, Dia2 dissociates from Sgs1
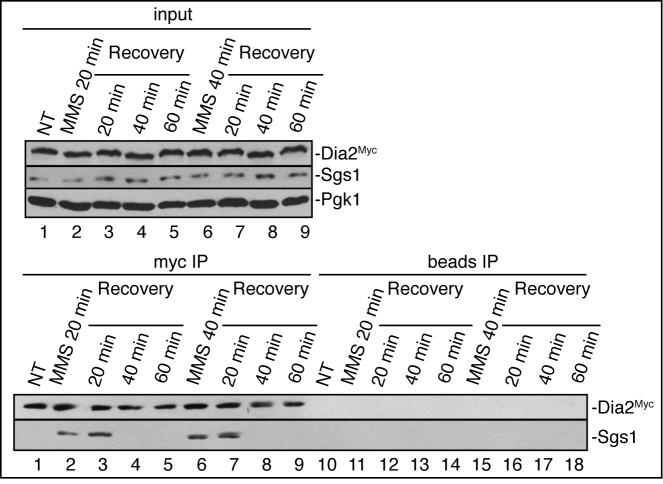
To determine the association dynamics between Dia2 and Sgs1 during checkpoint recovery from MMS-induced fork stalling, we used wild-type cells expressing Myc-tagged Dia2 at the endogenous locus. Cells were either untreated or treated with 0.05% MMS for 20 or 40 min and released to fresh media for 20, 40 or 60 min. Co-immunoprecipitation with Myc-Dia2 showed that Dia2 interacts with Sgs1 after MMS treatment through at least the first 20 min of recovery from MMS. At later time points during recovery, we were unable to detect any co-precipitation between Dia2 and Sgs1 (Figure 6). Together, these results suggest that the interaction of Dia2 with Sgs1 is important after checkpoint activation through the beginning of recovery from an MMS-induced checkpoint, consistent with their roles in replication fork restart.
Figure 6.
Upon stalled fork recovery, Dia2 dissociates from Sgs1. Wild-type cells were either untreated or treated with 0.05% MMS for 20 or 40 min and allowed to recover in the fresh media for 20, 40 and 60 min. Whole cell lysates were prepared and used in immunoprecipitations with an anti-myc antibody (lanes 1–9) or protein A/G beads without antibody (lanes 10–18). Immunoblots were probed with anti-myc or anti-Sgs1 antibodies.
Failure to degrade Mrc1 inhibits replication fork restart
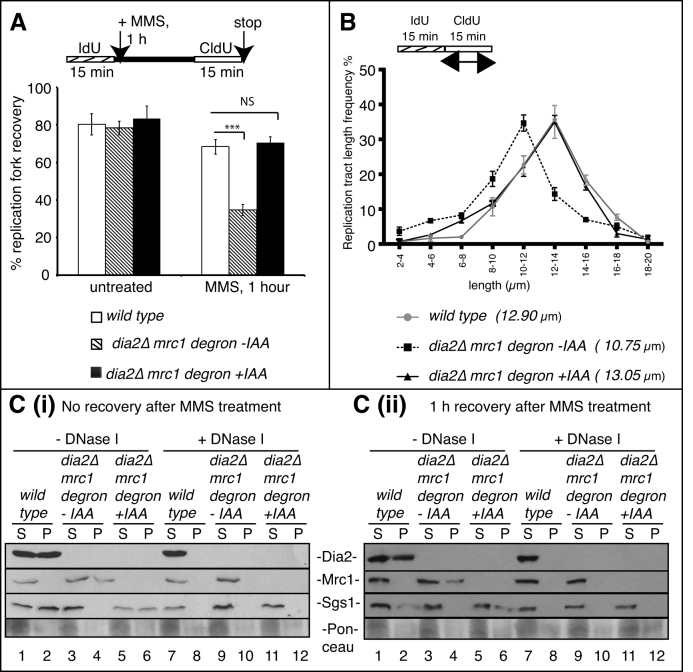
Our previous studies have shown that Dia2 mediated degradation of Mrc1 is important for cell cycle re-entry during recovery from an MMS-induced checkpoint (24). However, it is not clear whether Mrc1 contributes to restart of MMS-stalled replication forks. To address this question, we performed DNA fiber assays using wild type and a dia2Δ-Mrc1-IAA-inducible degron strain, which allows for temporal control of Mrc1 degradation. Cells synchronized in G1 were allowed to enter S phase and then were pulse-labeled with IdU (red label) for 15 min. Following IdU labeling, cells were either untreated or treated with 0.05% MMS for 1 h, followed by a second pulse-labeling with CldU (green label) for 15 min in the absence of MMS. Auxin (IAA) was added 15 min prior to CldU labeling to induce Mrc1 protein degradation. DNA fibers were isolated and incubated with anti-IdU and anti-CldU antibodies to visualize DNA replication. Strikingly, IAA-induced degradation of Mrc1 in dia2Δ cells completely rescued the deficiency of replication fork restart during recovery from an MMS-induced checkpoint (Figure 7A) and replication velocity (Figure 7B) phenotypes observed in the dia2Δ strain. Thus, the failure to appropriately target Mrc1 for degradation in the absence of Dia2 is the key constraint to restart of MMS-stalled replication forks in dia2Δ cells.
Figure 7.
Depletion of Mrc1 in dia2Δ strains rescues deficiencies in fork restart and maintenance of replication velocity by recruiting Sgs1 to chromatin. (A) Replication fork restart efficiencies of wild-type and a dia2Δmrc1 degron strain with or without IAA were measured as number of restarted forks (IdU-CldU tracts) compared with the total number of IdU-labeled tracts (IdU only plus IdU–CldU tracts). ***P < 0.001, NS = not significant. (B) CldU tract length distributions were determined in wild type, and a dia2Δmrc1 degron strain with or without IAA. P < 0.05 for wild type versus dia2Δmrc1 degron - IAA, and P-value is not significant for wild type versus dia2Δmrc1 degron + IAA. (C (i) and (ii)) Crude pellet containing chromatin fractions were separated from soluble cytoplasmic fractions from the untreated (lanes 1–6) or DNaseI-treated (lanes 7–12) whole cell lysates of wild type and a dia2Δmrc1 degron strain with or without IAA. Samples were resolved by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotted for Dia2Myc, Mrc1HA and Sgs1 using anti-Myc, anti-HA or anti-Sgs1 antibodies, respectively. S, soluble cytoplasmic fraction; P, pellet containing chromatin fraction.
Mrc1 degradation during checkpoint recovery promotes Sgs1 chromatin association
Given that both Mrc1 and Sgs1 interact with Dia2 (46,47), we sought to determine how Dia2-mediated Mrc1 degradation might affect Sgs1 function. As Mrc1 and Sgs1 interact with the N-terminal TPR-containing region in Dia2 and this region is required for Dia2 chromatin association, we initially asked if Mrc1 degradation affected the dynamics of Sgs1 or Dia2 recruitment to chromatin. For this, we used the same wild type and dia2Δ-Mrc1-IAA-inducible degron strains, synchronized to examine chromatin association during recovery from an MMS-induced checkpoint, in the presence or absence of auxin (IAA) to induce Mrc1 degradation. Cell lysates were either untreated or treated with DNase I and then pellets containing chromatin fractionations were separated from soluble cytoplasmic lysates. In wild-type cells, both Dia2 and Sgs1 were detected on chromatin during checkpoint activation (Figure 7C(i)) and after 1 h of checkpoint recovery (Figure 7C (ii)). By contrast, Mrc1 was not detected in the chromatin fraction at these times (Figure 7C (i) and (ii)). Strikingly, in the absence of Dia2, Mrc1 was detected in the chromatin fraction at both time points, suggesting that Dia2-dependent degradation of Mrc1 prevents large-scale chromatin association of Mrc1. Interestingly, Sgs1 chromatin association was severely inhibited in the absence of Dia2. However, IAA-induced degradation of Mrc1 in dia2Δ cells completely rescued Sgs1 chromatin association. Together, these results suggest that Dia2-mediated degradation of Mrc1 promotes Sgs1 chromatin association during recovery from an MMS-induced checkpoint (Figure 7C (i) and (ii)).
DISCUSSION
Our previous work demonstrated that Dia2-dependent degradation of Mrc1 was important for cell cycle re-entry during recovery from an MMS-induced S-phase checkpoint, but the molecular impact of this degradation remained unclear. In these studies, we find that degradation of Mrc1 facilitated by the SCFDia2 complex is critical to restart of MMS-stalled replication forks (Figure 8). Moreover, Dia2 functions in conjunction with the Sgs1 and Mph1 recombination-mediated fork restart pathway and physically interacts with Sgs1 specifically after checkpoint activation. Although the human homologs of Sgs1 and Mph1 have been shown to be required for fork restart (44,48,49), our DNA fiber studies are the first direct demonstration that these roles are conserved in budding yeast. Importantly, we find that failure to target Mrc1 for degradation during recovery from an MMS-induced checkpoint inhibits Sgs1 chromatin association but this can be alleviated by induced proteolysis of Mrc1 after checkpoint activation.
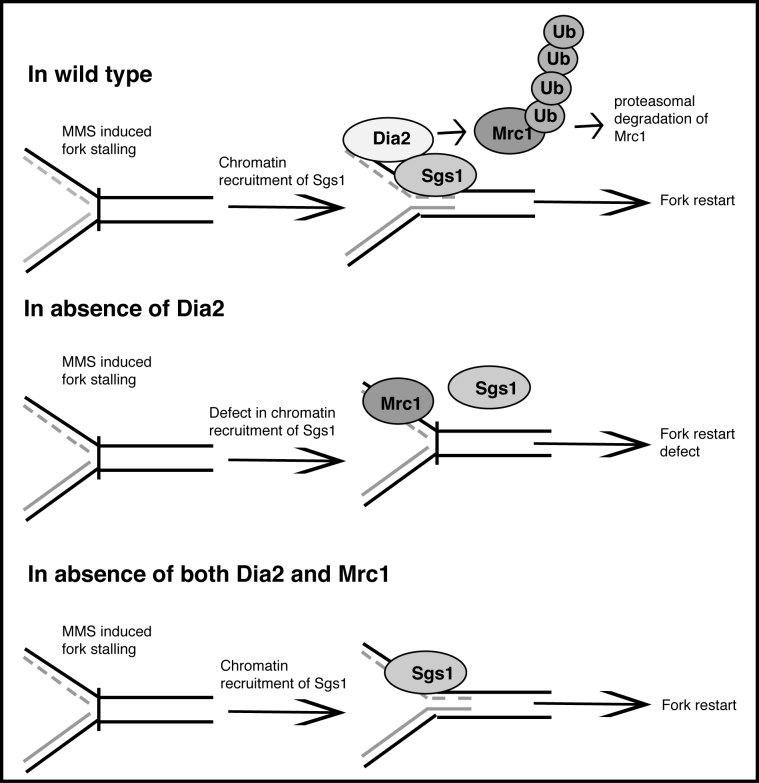
Figure 8.
Model depicting role of Dia2 in replication fork restart. Dia2 mediates stalled replication fork recovery by promoting recruitment of Sgs1 and degradation of Mrc1. In response to MMS induced replication fork stalling, Dia2 interacts with Sgs1. Dia2 and Sgs1 work in concert to restart stalled fork. Dia2 mediates ubiquitination of Mrc1 to facilitate Mrc1 degradation via the proteasome. In the absence of Dia2, Mrc1 degradation, Sgs1 chromatin association and replication fork recovery are inhibited. These defects can be reversed by induced degradation of Mrc1 in dia2Δ cells.
Successful recovery from the S-phase checkpoint requires both inactivation of the checkpoint signaling pathway and restart of stalled or collapsed forks to complete DNA replication. Degradation of Mrc1 likely contributes to signaling inactivation as phosphorylated Rad53 levels persist in dia2Δ cells and the checkpoint recovery defect in dia2Δ cells is exacerbated by the absence of the Rad53 phosphatase Pph3 (24). Here we demonstrate that degradation of Mrc1 is also required for a recombination-mediated fork restart pathway, suggesting that there may be coordination between signaling inactivation and fork restart pathways. One straightforward explanation is that signaling inactivation might lead to rearrangement of protein complexes at the fork that facilitate the transition from a stalled or damaged fork configuration to an active fork. This is consistent with previous studies that have demonstrated that inactivation of the Rad53 kinase is required for fork restart (19,20,50).
Dia2 associates specifically with the helicase Sgs1 upon S-phase checkpoint activation and both proteins function in the same recombination-mediated fork restart pathway after an MMS-induced checkpoint. Sgs1 also has a role in S-phase checkpoint signaling by promoting Rad53 activation (41,51–54). It is unlikely that Dia2 is required for this aspect of Sgs1 function, as Rad53 is constitutively activated in dia2Δ cells (37,38) and dia2Δ exhibits no enhanced phenotype with a sgs1 mutant that cannot activate Rad53, but does with a sgs1Δ strain (54). We anticipate that Dia2 interacts with Sgs1 on chromatin, as Dia2's N-terminal fragment containing the TPR domain is required for Dia2 chromatin association and this section of the protein is also required for interaction with Sgs1, as well as Mrc1. The coordination of Dia2 interaction with both Mrc1 and Sgs1 remains to be determined in future studies.
Our data suggest that MMS sensitivity can be linked to defects in recovery from the S-phase checkpoint. We find that dia2 mutants that fail to efficiently re-enter the cell cycle during recovery from an MMS-induced checkpoint also exhibit growth defects in the presence of MMS. A segment of the N-terminal section of Dia2 is required for both viability in the presence of MMS and recovery from an MMS-induced checkpoint. It seems likely that the TPR domain is not the main domain responsible for the MMS checkpoint response, as the ΔN134 mutant, which rescues MMS sensitivity and checkpoint recovery, lacks all but the last 10 amino acids of the TPR domain. Likewise, a ΔN144 mutant that lacks the entire TPR domain still exhibits growth in the presence of MMS (46), as does a ΔN159 mutant, albeit when overexpressed (47). We think that there may be an as yet undetermined domain following the TPR section but prior to the F-box domain that is specifically linked to the checkpoint recovery pathway, although future studies will be needed to verify this idea.
There are conflicting reports about whether Mrc1 associated with the replisome is targeted for degradation during DNA replication (46,47,55). Our studies do not directly address this question in a normal S phase. However, during recovery from an S-phase arrest induced by MMS, we do observe that Mrc1 accumulates on chromatin in dia2Δ cells when fork restart is defective and substantial depletion of Mrc1 is necessary to rescue fork restart in these cells. The depletion of Mrc1, including the chromatin-associated fraction, coincides with the accumulation of the helicase Sgs1 on chromatin. Thus, we favor a model in which degradation of chromatin-associated Mrc1 leads to rearrangements at the fork that are critical to recruitment of Sgs1 to chromatin to achieve restart of a substantial fraction of MMS-stalled replication forks. However, it is possible that a small fraction of Mrc1 remains associated with the replisome under these conditions, but that we are unable to detect it in our assay. Future studies will be necessary to comprehensively determine protein complex composition and changes at individual forks during recovery from an MMS-induced checkpoint.
We found that both Sgs1 and Dia2, but not Mph1, are also important for the maintenance of replication speed in an unperturbed replication fork during S-phase. This is consistent with work showing that the BLM helicase is important for maintaining replication speed (44,56). By contrast, Mph1, but neither Dia2 nor Sgs1, protects nascent DNA strands from nucleolytic degradation after MMS treatment. In human cells, Fanconi Anemia (FA) proteins FANCD2 and FANCD1 (also known as BRCA2) but not BLM maintain stability of the nascent DNA strands after replication stress (44,45,57). The eight FA proteins FANC(A/B/C/E/F/G/L/M) form a core complex that mediates monoubiquitination of the central FA effector proteins FANCD2 and FANCI, which then contribute to the repair of damaged DNA. Mph1 is the only known ortholog of the FA core complex protein FANCM, and there are no known homologs of other FA core complex proteins or effector proteins FANCD2 and FANCI in budding yeast (39). Thus, it appears that the functional separation of Sgs1 and Mph1 roles in fork restart and protection of nascent DNA strands is similar to what is observed in human cells between Fanconi Anemia and Bloom Syndrome pathways.
Together, these studies expand our understanding of how cells recover from genotoxin-induced checkpoints. As chemotherapy and radiotherapy induce genotoxic stress to trigger cell death in cancer cells, checkpoint recovery components may serve as alternative targets of treatment to sensitize cancer cells to antitumor therapies. With a link between checkpoint signaling inactivation and recombination-mediated replication fork restart, we have the tools to begin to delineate the molecular mechanisms required for recovery from agents that damage DNA or cause replication stress.
ACKNOWLEDGEMENTS
The authors thank the laboratory of O. Aparicio (University of Southern California) for the gift of the HSV-TK cassette plasmids and the D. Kirkpatrick laboratory (University of Minnesota) for the gift of strains and use of equipment.
FUNDING
Department of Genetics, Cell Biology and Development at the University of Minnesota. Funding for open access charge: Department of Genetics, Cell Biology and Development at the University of Minnesota.
Conflict of interest statement. None declared.
REFERENCES
- 1.Boddy M.N., Russell P.. DNA replication checkpoint. Curr. Biol. 2001; 11:R953–R956. [DOI] [PubMed] [Google Scholar]
- 2.Tercero J.A., Diffley J.F.. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature. 2001; 412:553–557. [DOI] [PubMed] [Google Scholar]
- 3.Nyberg K.A., Michelson R.J., Putnam C.W., Weinert T.A.. Toward maintaining the genome: DNA damage and replication checkpoints. Annu. Rev. Genet. 2002; 36:617–656. [DOI] [PubMed] [Google Scholar]
- 4.Branzei D., Foiani M.. The DNA damage response during DNA replication. Curr. Opin. Cell Biol. 2005; 17:568–575. [DOI] [PubMed] [Google Scholar]
- 5.Kondo T., Wakayama T., Naiki T., Matsumoto K., Sugimoto K.. Recruitment of Mec1 and Ddc1 checkpoint proteins to double-strand breaks through distinct mechanisms. Science. 2001; 294:867–870. [DOI] [PubMed] [Google Scholar]
- 6.Melo J.A., Cohen J., Toczyski D.P.. Two checkpoint complexes are independently recruited to sites of DNA damage in vivo. Genes Dev. 2001; 15:2809–2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Osborn A.J., Elledge S.J.. Mrc1 is a replication fork component whose phosphorylation in response to DNA replication stress activates Rad53. Genes Dev. 2003; 17:1755–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alcasabas A.A., Osborn A.J., Bachant J., Hu F., Werler P.J., Bousset K., Furuya K., Diffley J.F., Carr A.M., Elledge S.J.. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat. Cell Biol. 2001; 3:958–965. [DOI] [PubMed] [Google Scholar]
- 9.Foss E.J. Tof1p regulates DNA damage responses during S phase in Saccharomyces cerevisiae. Genetics. 2001; 157:567–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Navas T.A., Sanchez Y., Elledge S.J.. RAD9 and DNA polymerase epsilon form parallel sensory branches for transducing the DNA damage checkpoint signal in Saccharomyces cerevisiae. Genes Dev. 1996; 10:2632–2643. [DOI] [PubMed] [Google Scholar]
- 11.Tong A.H., Lesage G., Bader G.D., Ding H., Xu H., Xin X., Young J., Berriz G.F., Brost R.L., Chang M. et al. . Global mapping of the yeast genetic interaction network. Science. 2004; 303:808–813. [DOI] [PubMed] [Google Scholar]
- 12.Vialard J.E., Gilbert C.S., Green C.M., Lowndes N.F.. The budding yeast Rad9 checkpoint protein is subjected to Mec1/Tel1-dependent hyperphosphorylation and interacts with Rad53 after DNA damage. EMBO J. 1998; 17:5679–5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwartz M.F., Duong J.K., Sun Z., Morrow J.S., Pradhan D., Stern D.F.. Rad9 phosphorylation sites couple Rad53 to the Saccharomyces cerevisiae DNA damage checkpoint. Mol. Cell. 2002; 9:1055–1065. [DOI] [PubMed] [Google Scholar]
- 14.Sun Z., Hsiao J., Fay D.S., Stern D.F.. Rad53 FHA domain associated with phosphorylated Rad9 in the DNA damage checkpoint. Science. 1998; 281:272–274. [DOI] [PubMed] [Google Scholar]
- 15.Pellicioli A., Lucca C., Liberi G., Marini F., Lopes M., Plevani P., Romano A., Di Fiore P.P., Foiani M.. Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J. 1999; 18:6561–6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Santocanale C., Diffley J.F.. A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature. 1998; 395:615–618. [DOI] [PubMed] [Google Scholar]
- 17.Shirahige K., Hori Y., Shiraishi K., Yamashita M., Takahashi K., Obuse C., Tsurimoto T., Yoshikawa H.. Regulation of DNA-replication origins during cell-cycle progression. Nature. 1998; 395:618–621. [DOI] [PubMed] [Google Scholar]
- 18.Sogo J.M., Lopes M., Foiani M.. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science. 2002; 297:599–602. [DOI] [PubMed] [Google Scholar]
- 19.O'Neill B.M., Szyjka S.J., Lis E.T., Bailey A.O., Yates J.R., Aparicio O.M., Romesberg F.E.. Pph3-Psy2 is a phosphatase complex required for Rad53 dephosphorylation and replication fork restart during recovery from DNA damage. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:9290–9295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szyjka S.J., Aparicio J.G., Viggiani C.J., Knott S., Xu W., Tavare S., Aparicio O.M.. Rad53 regulates replication fork restart after DNA damage in Saccharomyces cerevisiae. Genes Dev. 2008; 22:1906–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Travesa A., Duch A., Quintana D.G.. Distinct phosphatases mediate the deactivation of the DNA damage checkpoint kinase Rad53. J. Biol. Chem. 2008; 283:17123–17130. [DOI] [PubMed] [Google Scholar]
- 22.Shimada K., Oma Y., Schleker T., Kugou K., Ohta K., Harata M., Gasser S.M.. Ino80 chromatin remodeling complex promotes recovery of stalled replication forks. Curr. Biol. 2008; 18:566–575. [DOI] [PubMed] [Google Scholar]
- 23.Lee L., Rodriguez J., Tsukiyama T.. Chromatin remodeling factors Isw2 and Ino80 regulate checkpoint activity and chromatin structure in S phase. Genetics. 2015; 199:1077–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fong C.M., Arumugam A., Koepp D.M.. The Saccharomyces cerevisiae F-box protein Dia2 is a mediator of S-phase checkpoint recovery from DNA damage. Genetics. 2013; 193:483–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peschiaroli A., Dorrello N.V., Guardavaccaro D., Venere M., Halazonetis T., Sherman N.E., Pagano M.. SCF beta TrCP-mediated degradation of claspin regulates recovery from the DNA replication checkpoint response. Mol. Cell. 2006; 23:319–329. [DOI] [PubMed] [Google Scholar]
- 26.Mailand N., Bekker-Jensen S., Bartek J., Lukas J.. Destruction of claspin by SCF beta TrCP restrains Chk1 activation and facilitates recovery from genotoxic stress. Mol. Cell. 2006; 23:307–318. [DOI] [PubMed] [Google Scholar]
- 27.Bennett L.N., Clarke P.R.. Regulation of Claspin degradation by the ubiquitin-proteo some pathway during the cell cycle and in response to ATR-dependent checkpoint activation. FEBS Lett. 2006; 580:4176–4181. [DOI] [PubMed] [Google Scholar]
- 28.Petermann E., Helleday T.. Pathways of mammalian replication fork restart. Nat. Rev. Mol. Cell Biol. 2010; 11:683–687. [DOI] [PubMed] [Google Scholar]
- 29.Hanada K., Budzowska M., Davies S.L., van Drunen E., Onizawa H., Beverloo H.B., Maas A., Essers J., Hickson I.D., Kanaar R.. The structure-specific endonuclease Mus81 contributes to replication restart by generating double-strand DNA breaks. Nat. Struct. Mol. Biol. 2007; 14:1096–1104. [DOI] [PubMed] [Google Scholar]
- 30.Segurado M., Diffley J.F.. Separate roles for the DNA damage checkpoint protein kinases in stabilizing DNA replication forks. Genes Dev. 2008; 22:1816–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Larsen N.B., Hickson I.D.. RecQ helicases: conserved guardians of genomic integrity. Adv. Exp. Med. Biol. 2013; 767:161–184. [DOI] [PubMed] [Google Scholar]
- 32.Ashton T.M., Hickson I.D.. Yeast as a model system to study RecQ helicase function. DNA Repair (Amst). 2010; 9:303–314. [DOI] [PubMed] [Google Scholar]
- 33.Whitby M.C. The FANCM family of DNA helicases/translocases. DNA Repair (Amst). 2010; 9:224–236. [DOI] [PubMed] [Google Scholar]
- 34.Zheng X.F., Prakash R., Saro D., Longerich S., Niu H., Sung P.. Processing of DNA structures via DNA unwinding and branch migration by the S. cerevisiae Mph1 protein. DNA Repair (Amst). 2011; 10:1034–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rose M.D., Winston F., Hieter P.. Methods in Yeast Genetics: A Laboratory Course Manual. 1990; NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- 36.Kile A.C., Koepp D.M.. Activation of the S-phase checkpoint inhibits degradation of the F-box protein Dia 2. Mol. Cell Biol. 2010; 30:160–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pan X., Ye P., Yuan D.S., Wang X., Bader J.S., Boeke J.D.. A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell. 2006; 124:1069–1081. [DOI] [PubMed] [Google Scholar]
- 38.Blake D., Luke B., Kanellis P., Jorgensen P., Goh T., Penfold S., Breitkreutz B.J., Durocher D., Peter M., Tyers M.. The F-box protein Dia2 overcomes replication impedance to promote genome stability in Saccharomyces cerevisiae. Genetics. 2006; 174:1709–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Daee D.L., Ferrari E., Longerich S., Zheng X.F., Xue X., Branzei D., Sung P., Myung K.. Rad5-dependent DNA repair functions of the Saccharomyces cerevisiae FANCM protein homolog Mph1. J. Biol. Chem. 2012; 287:26563–26575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ui A., Satoh Y., Onoda F., Miyajima A., Seki M., Enomoto T.. The N-terminal region of Sgs1, which interacts with Top3, is required for complementation of MMS sensitivity and suppression of hyper-recombination in sgs1 disruptants. Mol. Genet. Genomics. 2001; 265:837–850. [DOI] [PubMed] [Google Scholar]
- 41.Cobb J.A., Bjergbaek L., Shimada K., Frei C., Gasser S.M.. DNA polymerase stabilization at stalled replication forks requires Mec1 and the RecQ helicase Sgs1. EMBO J. 2003; 22:4325–4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mankouri H.W., Ngo H.P., Hickson I.D.. Esc2 and Sgs1 act in functionally distinct branches of the homologous recombination repair pathway in Saccharomyces cerevisiae. Mol. Biol. Cell. 2009; 20:1683–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singh S., Shemesh K., Liefshitz B., Kupiec M.. Genetic and physical interactions between the yeast ELG1 gene and orthologs of the Fanconi anemia pathway. Cell Cycle. 2013; 12:1625–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chaudhury I., Sareen A., Raghunandan M., Sobeck A.. FANCD2 regulates BLM complex functions independently of FANCI to promote replication fork recovery. Nucleic Acids Res. 2013; 41:6444–6459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schlacher K., Wu H., Jasin M.. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 2012; 22:106–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morohashi H., Maculins T., Labib K.. The amino-terminal TPR domain of Dia2 tethers SCF(Dia2) to the replisome progression complex. Curr. Biol. 2009; 19:1943–1949. [DOI] [PubMed] [Google Scholar]
- 47.Mimura S., Komata M., Kishi T., Shirahige K., Kamura T.. SCF(Dia2) regulates DNA replication forks during S-phase in budding yeast. EMBO J. 2009; 28:3693–3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Davies S.L., North P.S., Hickson I.D.. Role for BLM in replication-fork restart and suppression of origin firing after replicative stress. Nat. Struct. Mol. Biol. 2007; 14:677–679. [DOI] [PubMed] [Google Scholar]
- 49.Schwab R.A., Blackford A.N., Niedzwiedz W.. ATR activation and replication fork restart are defective in FANCM-deficient cells. EMBO J. 2010; 29:806–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morafraile E.C., Diffley J.F., Tercero J.A., Segurado M.. Checkpoint-dependent RNR induction promotes fork restart after replicative stress. Sci. Rep. 2015; 5:7886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Frei C., Gasser S.M.. The yeast Sgs1p helicase acts upstream of Rad53p in the DNA replication checkpoint and colocalizes with Rad53p in S-phase-specific foci. Genes Dev. 2000; 14:81–96. [PMC free article] [PubMed] [Google Scholar]
- 52.Cobb J.A., Schleker T., Rojas V., Bjergbaek L., Tercero J.A., Gasser S.M.. Replisome instability, fork collapse, and gross chromosomal rearrangements arise synergistically from Mec1 kinase and RecQ helicase mutations. Genes Dev. 2005; 19:3055–3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bjergbaek L., Cobb J.A., Tsai-Pflugfelder M., Gasser S.M.. Mechanistically distinct roles for Sgs1p in checkpoint activation and replication fork maintenance. EMBO J. 2005; 24:405–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hegnauer A.M., Hustedt N., Shimada K., Pike B.L., Vogel M., Amsler P., Rubin S.M., van Leeuwen F., Guenole A., van Attikum H. et al. . An N-terminal acidic region of Sgs1 interacts with Rpa70 and recruits Rad53 kinase to stalled forks. EMBO J. 2012; 31:3768–3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maculins T., Nkosi P.J., Nishikawa H., Labib K.. Tethering of SCF(Dia2) to the replisome promotes efficient ubiquitylation and disassembly of the CMG helicase. Curr. Biol. 2015; 25:2254–2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rao V.A., Conti C., Guirouilh-Barbat J., Nakamura A., Miao Z.H., Davies S.L., Sacca B., Hickson I.D., Bensimon A., Pommier Y.. Endogenous gamma-H2AX-ATM-Chk2 checkpoint activation in Bloom's syndrome helicase deficient cells is related to DNA replication arrested forks. Mol. Cancer Res. 2007; 5:713–724. [DOI] [PubMed] [Google Scholar]
- 57.Schlacher K., Christ N., Siaud N., Egashira A., Wu H., Jasin M.. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011; 145:529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koepp D.M., Kile A.C., Swaminathan S., Rodriguez-Rivera V.. The F-box protein Dia2 regulates DNA replication. Mol. Biol. Cell. 2006; 17:1540–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Viggiani C.J., Aparicio O.M.. New vectors for simplified construction of BrdU-incorporating strains of Saccharomyces cerevisiae. Yeast. 2006; 23:1045–1051. [DOI] [PubMed] [Google Scholar]