Abstract
The oncogenic transcription factor MYC and its binding partner MAX regulate gene expression by binding to DNA at enhancer-box (E-box) elements 5΄-CACGTG-3΄. In mammalian genomes, the central E-box CpG has the potential to be methylated at the 5-position of cytosine (5mC), or to undergo further oxidation to the 5-hydroxymethyl (5hmC), 5-formyl (5fC), or 5-carboxyl (5caC) forms. We find that MAX exhibits the greatest affinity for a 5caC or unmodified C-containing E-box, and much reduced affinities for the corresponding 5mC, 5hmC or 5fC forms. Crystallization of MAX with a 5caC modified E-box oligonucleotide revealed that MAX Arg36 recognizes 5caC using a 5caC–Arg–Guanine triad, with the next nearest residue to the carboxylate group being Arg60. In an analysis of >800 primary multiple myelomas, MAX alterations occurred at a frequency of ∼3%, more than half of which were single nucleotide substitutions affecting a basic clamp-like interface important for DNA interaction. Among these, arginines 35, 36 and 60 were the most frequently altered. In vitro binding studies showed that whereas mutation of Arg36 (R36W) or Arg35 (R35H/L) completely abolished DNA binding, mutation of Arg60 (R60Q) significantly reduced DNA binding, but retained a preference for the 5caC modified E-box. Interestingly, MAX alterations define a subset of myeloma patients with lower MYC expression and a better overall prognosis. Together these data indicate that MAX can act as a direct epigenetic sensor of E-box cytosine modification states and that local CpG modification and MAX variants converge to modulate the MAX-MYC transcriptional network.
INTRODUCTION
Postsynthetic modifications to DNA and histone proteins play a key role in defining the three dimensional organization of chromatin in the nucleus and DNA accessibility, shaping gene expression programs by enabling the expression of some genes while restricting that of others. Such epigenetic regulation controls cell fate decisions during development and the maintenance of cellular identity in differentiated cells, a program that is frequently altered in the progression to cancer. Among these, the methylation of cytosine residues (5mC) in the sequence context CpG is perhaps the best studied and affects gene regulation through both direct and indirect (secondary to changes in chromatin structure) effects on the binding of proteins to DNA. Most CpGs in the genome are methylated and in the context of intact chromatin, the absence of 5mC demarks accessible regions of the genome, including CpG island-containing gene promoters and active enhancers (1,2). While the methylation status of most CpG sites are largely invariant across cell types, the fraction that do vary (∼20%) overlap distal regulatory regions, and in particular enhancers and transcription factor binding sites, suggesting that the gain or loss of 5mC at such sites is a key defining feature of lineage-specific gene regulation (3,4).
It is now evident that 5mC patterns can be further shaped during development and even in post-mitotic cells by the ten-eleven translocation (Tet) family of Fe(II)- and α-ketoglutarate-dependent dioxygenases which convert 5mC to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) in successive oxidation steps (5–8). One fate of oxidized 5mC residues—particularly 5fC and 5caC—involves excision by thymine DNA glycosylase (TDG) followed by incorporation of an unmodified C, leading to a net DNA ‘demethylation’ (8). Accumulated evidence now support the view that 5hmC exists as a relatively stable modification unaffected by TDG depletion, constituting a distinct epigenetic signal that primarily marks lowly expressed genes, gene bodies and intragenic regions (9,10) (reviewed in (11,12)). In contrast, 5fC and 5caC are found at much lower levels at steady state (1–10% that of 5hmC), and preferentially accumulate at enhancers and other distal regulatory regions in the absence of TDG (13–18), implying that the dynamic turnover of 5mC may be particularly important in enhancer function. Recent work by our group and others showing that the binding of certain transcription factors to DNA is influenced by the presence of oxidized 5mC bases in vitro (19–23) and that 5fC/5caC pose a barrier to transcriptional elongation in cells (24) raise the question of whether 5fC/5caC may have functions in gene regulation beyond that of intermediates in the DNA demethylation cycle.
The oncogenic transcription factor MYC and its binding partner MAX are basic-helix–loop–helix (bHLH) transcription factors that preferentially recognize a subset of enhancer-box (E-box) sequences containing a central CpG dinucleotide (5΄-CACGTG-3΄) (25–28). MYC-MAX binding to E-box sequences is thought ‘amplify’ transcription by promoting elongation at a wide number of genes (29,30). MAX also dimerizes with a number of other bHLH proteins, including Mad1-4 (Mxd), Mnt, Mga and others (31,32). The MAX-Mad/Mnt/Mga heterodimers directly antagonize the function of MYC by instead recruiting corepressor complexes to mediate transcriptional repression from the same E-box sequences. Ultimately, it is the relative abundance of MYC and the other binding partners and their competition for MAX that mediates distinct biologic outputs (32,33). Dysregulation of the MYC-MAX transcriptional network is a common mechanism driving oncogenic progression in many human cancers. Most often this is achieved through translocations or amplification of the MYC locus, resulting in its overexpression. Alterations in MAX have also recently been reported (34–36), but the functional significance of these is not well understood. In this study, we sought to determine the influence of 5mC oxidation and the impact of cancer-associated MAX mutations on the biochemical and structural features of MAX binding with its cognate E-box.
Here, we provide evidence that MAX, and potentially other bHLH transcription factors, can ‘sense’ the oxidation status of 5mCpGs, and further that cancer-associated mutations in MAX differentially affect binding to these features. In a large collection of over 800 primary multiple myelomas, we establish that somatic MAX mutations occur at a frequency of ∼3% and are likely to represent loss of function alterations. Among myeloma patients, MAX mutations are associated with low expression of MYC and a better overall prognosis. Taken together the data suggest that 5mCpG oxidation may play a critical role in fine-tuning E-box recognition by the MYC family of transcription factors, and ultimately transcriptional output.
MATERIALS AND METHODS
Protein expression and purification
Human full length MAX (NP_002373.3) and the bHLH-leucine zipper domain (residues 22–107) were cloned into hexahistidine-SUMO (small ubiquitin-like modifier) tagged vector (pXC1385 and pXC1407, respectively) and expressed in Escherichia coli BL21 (DE3) Codon-plus RIL. Cells were cultured in LB medium at 37°C until OD600 reached 0.5 before induction by adding 0.4 mM isopropyl β-d-1-thiogalactopyranoside for 16 h at 16°C. Cells were harvested and lysed in 20 mM sodium phosphate, pH 7.4, 150 mM NaCl, 1 mM dithiothreitol (DTT), 50 mM imidazol and 0.3 mM phenylmethyl-sulphonyl fluoride by sonication. The lysate was clarified by centrifugation at 35 000 × g for 60 min at 4°C. The hexahistidine fusion protein was isolated on nickel-charged chelating column (GE Healthcare). The hexahistidine-SUMO tag was removed by digestion with Ulp1 protease (purified in-house) for 16 h at 4°C (37), resulting in the retention of two extraneous N-terminal residues (His-Met). The protein was further purified by tandem HiTrap Q and SP column (i.e. passing through Q and eluted from SP column). The protein domain used for crystallization was then loaded on a HiLoad Superdex 200 (16/60) column (GE Healthcare) and concentrated to approximately 87 mg/ml in 20 mM Tris–HCl, pH 8.0, 500 mM NaCl and 1 mM DTT.
Site-directed mutagenesis
Point mutations were introduced into the wild-type 6xHis-SUMO-tagged human full length MAX (pXC1385) using the QuickChange site-directed mutagenesis kit (Stratagene) to generate MAX H28R (pXC1506), R35H (pXC1580), R35L (pXC1581), R36W (pXC1500), R36K (pXC1507) and R60Q (pXC1533). All mutants were verified by sequencing. The mutant proteins were purified as described above for the wild type (WT) protein. With the exception of H28R which yielded a lower protein yield (0.3 mg/l), the remaining mutant proteins were all recovered a similar or higher protein yields as the WT protein (2 mg from 1 l culture for WT, 3.3 mg for R35H, 6.5 mg for R35L, 1.4 mg for R36W, 2.7 mg for R36K and 3.2 mg for R60Q). Size exclusion chromatography and dynamic light scattering measurements indicated a similar size and integrity for all (Supplementary Figure S1).
Electrophoretic mobility shift assay
The gel shift assays were performed by incubating 5 nM 6-carboxy-fluorescein (FAM) labeled double-stranded oligonucleotides [FAM-5΄-TAGGCCAXGTGACCGG-3΄/5΄-CCGGTCAYGTGGCCTA-3΄ where X or Y is C, 5-methyl-C (M), 5-hydroxymethyl-C (H), 5-formyl-C (F) or 5-carboxyl-C (5caC)] with an increasing amount (up to 0.5 μM) of full length MAX protein at room temperature (∼22°C) for 30 min under the conditions of 20 mM Tris–HCl (pH 8.0), 150 mM NaCl, 5% (v/v) glycerol, 1 mM DTT and 0.1 mg/ml bovine serum albumin (BSA). Protein/DNA complexes were loaded onto a 6% native 1× TBE polyacrylamide gel, and electrophoresed for 40 min at 80 V in 1× TBE buffer. Gels were scanned using Typhoon Trio+ imager (GE Healthcare).
Fluorescent polarization
Fluorescence polarization measurements were carried out at 25°C on a Synergy 4 Microplate Reader (BioTek) as described (38). A 5 nM of FAM-labeled double strand DNA was incubated for 10 min with increasing amounts of proteins (up to 1 μM) in 20 mM Tris–HCl, pH 7.5, 150 mM NaCl, 1 mM DTT, 5% (v/v) glycerol and 0.1 mg/ml BSA. No change in fluorescence intensity was observed with the addition of protein. Curves were fit individually using GraphPad PRISM 5.0d software. Binding constants (KD) were calculated as [mP] = [maximum mP] x [C] / (KD +[C]) + [baseline mP], where [mP] is milli-Polarization and [C] is protein concentration (39). Saturated [mP] was calculated as saturation = ([mP] − [baseline mP])/([maximum mP] − [baseline mP]). Curves were normalized as percentage of bound and reported is the mean ± SEM of the interpolated KD from two independent experiments performed in duplicate. For those binding curves that did not reach saturation, the lower limit of the binding affinity was estimated (39).
Crystallography
For co-crystallization, 1 mM MAX purified protein was mixed with 0.5 mM double-stranded oligonucleotide with nine different DNAs (Supplementary Figure S2). The best diffracting crystal used for data collection was obtained from 5΄-AGTAGCAXGTGCTACT-3΄ (where X = 5caC) (synthesized by New England Biolabs). Crystals appeared within 1-day under the conditions of 0.1 M sodium citrate-citric acid at pH 5.5, 20% (w/v) polyethylene glycol 3000. Crystals were cryoprotected by soaking in mother liquor supplemented with 30% ethylene glycol. X-ray diffraction datasets were collected at the SER-CAT beamline (22ID-D) at the Advanced Photon Source, Argonne National Laboratory and processed using XDS (40). The structure was solved by molecular replacement by PHENIX (41,42) using the Max–DNA complex structure (PDB 1AN2 (43)) as a search model. PHENIX refinement scripts were used for refinement, and the statistics shown in Supplementary Table S1 were calculated for the entire resolution range.
MAX mutations in multiple myeloma
The CoMMpass (Clinical outcomes in Multiple Myeloma to personal assessment) database interim analysis IA9 was interrogated for single nucleotide variants (SNVs) and structural variants in the MAX locus (https://research.themmrf.org). RNA-seq data and overall survival data were extracted for those patients harboring MAX variants and compared to those for whom MAX status could be definitively called but whom lacked MAX SNVs (wild-type). Overall survival data between the two groups were compared using a Log-rank (Mantel-Cox) test and Hazard Ratio determined using the Mantel–Haenszel test in GraphPad Prism 6.0. MYC expression levels were determined from RNAseq data that was processed with Cufflinks by the CoMMpass consortium and expressed as FPKM—fragments per kilobase of exon per million reads mapped. The distributions between the two groups were compared using the Mann–Whitney U test implemented in GraphPad Prism 6.0.
RESULTS
The bHLH DNA-binding domain together with leucine zipper dimerization domains (bHLH-LZ) of the Max–Max, Max–Myc and Max–Mad dimers in complex with E-box DNA have been structurally examined (43,44). However, in these in vitro studies it was necessary to use a chemical ligation approach to achieve homogeneous Max–Myc or Max–Mad heterodimers (44). Whereas the various Max partners exhibit marked differences in the leucine zipper regions, the mode of interaction of the bHLH domains with DNA is essentially identical between the Max homodimer and the heterodimers (44). Therefore, for simplicity, we used the full-length human MAX–MAX homodimer to biochemically characterize interactions with E-box DNA, and the bHLH-LZ region (residues 22–107) for structural analysis (Supplementary Figure S3A and B).
Binding of MAX to CpG-modified E-boxes
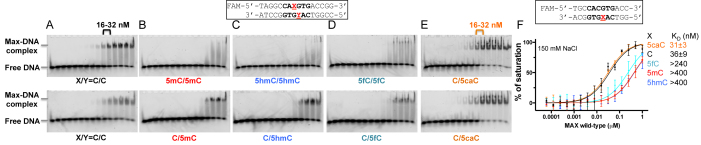
We first compared the binding of human full-length MAX protein to double-stranded oligonucleotides containing the E-box sequence in which the status of the central CpG was either unmodified, 5mC-modified, or oxidized to 5hmC, 5fC or 5caC by electrophoretic mobility shift assay. As expected, MAX bound to the unmodified E-box and methylation of the CpG greatly inhibited DNA binding, as observed previously (28) (Figure 1A and B). 5hmC further decreased binding significantly (Figure 1C). Surprisingly, the 5fC containing oligo showed a small amount of discrete complex, whereas 5caC restored binding to the level of unmodified C (Figure 1D and E). Due to synthetic limitations, the 5caC modification is not compatible with a FAM label on the same oligonucleotide, thus the 5caC-containing double-stranded oligonucleotide was only hemi-modified, with the 5caC base on the bottom strand only. In all other cases we compared both the fully modified setting in which the modified base was on both strands (Figure 1A–D, top) as well as the hemi-modified setting (Figure 1A–E, bottom). From the intensities of shifted bands, we estimated the apparent binding affinity to be between 16 and 32 nM for both completely unmodified C and hemi-modified 5caC-containing oligonucleotides. We expect that a fully modified 5caC oligo might have an even stronger binding affinity to the MAX homodimer.
Figure 1.
MAX binds 5caC DNA. (A–E) Electrophoretic mobility shift assay of full length MAX protein binding to oligonucleotides containing a single E-box where the central CpG are unmodified (C/C), fully modified (M/M, H/H, F/F), or hemi-modified (C/M, C/H, C/F or C/5caC). The protein concentrations used were a maximum of 0.5 μM (the right most lane of each panel) followed by serial 2-fold dilutions (from right to left). (F) DNA binding was quantified by fluorescence polarization. Data represent the mean ± SEM of two independent determinations performed in duplicate.
We repeated the binding assays using fluorescence polarization to quantitatively measure the dissociation constants (KD) between the MAX protein and the five E-box forms, comparing directly double-stranded oligonucleotides that were modified only on the bottom strand (Figure 1F). MAX bound the unmodified C and the 5caC-containing oligonucleotides equally well with a KD of ∼30 nM. Methylation (5mC) and 5-hydroxymethylation (5hmC) significantly reduced binding by a factor of >13, whereas 5-formycytosine (5fC) reduced affinity by a factor of 8. The affinity of MAX for the E-box is thus on the order 5caC ∼ C > 5fC > 5mC ∼ 5hmC. We also tested the effect of independent modification of the two DNA strands (i.e. strand-biased DNA modification) on binding affinity. As expected, MAX had a slightly better affinity for the hemi-modified C/5caC than it did for unmodified C/C, whereas addition of a single methyl group onto one strand (5mC/5caC or C/5mC) reduced binding, regardless of which it was opposite, and was nearly as detrimental as that observed with the fully methylated (5mC/5mC) site (Supplementary Figure S4).
Structure of MAX bound with 5caC DNA
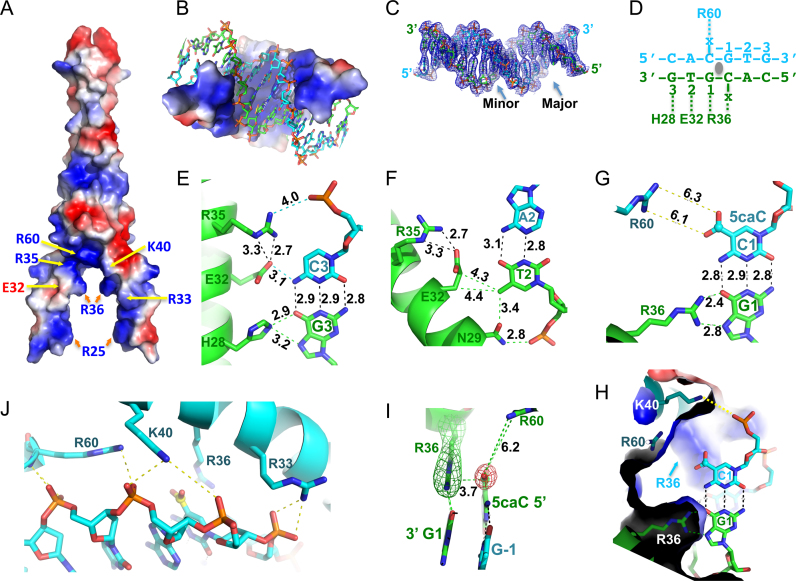
To understand how MAX binds 5caC DNA, we next co-crystallized the MAX bHLH-LZ (residues 22–107) with a 16-base pair palindromic sequence containing a fully modified 5caCpG within the E-box element (Supplementary Figure S3B). The MAX-DNA complex structure was determined to a resolution of 2.39 Å (Supplementary Table S1). The structure is composed of two long helices, α1 (residues 22–49) and α2 (residues 60–106) connected by a loop (residues 50–59) (Supplementary Figure S3C). The basic region of the N-terminal portion of helix α1 binds in the major groove of the DNA (Figure 2A–C), recognizing half of the palindromic E-box sequence (Figure 2D). His28 donates one hydrogen bond (H-bond) to either Guanine O6 or Guanine N7 of the outer C3:G3 base pair (Figure 2E). Glu32 accepts a H-bond from the N4 amino group (NH2) of the opposite cytosine (Figure 2E), while its side chain aliphatic carbons (Cβ and Cγ) are in the vicinity of the Thymine methyl group of the inner A2:T2 base pair (Figure 2F). The Thymine methyl group also forms a carbon-oxygen (C–H…O type) hydrogen bond (45) with the side chain carbonyl oxygen of Asn29 (Figure 2F).
Figure 2.
Structure of MAX-5caC complex. (A) The surface charge of MAX homodimer at neutral pH is displayed as blue for positive, red for negative, and white for neutral. (B) The inner surface of the clamp-like DNA binding domain is predominantly basic. (C) Electron density 2Fo – Fc, contoured at 1σ above the mean, is shown for the entire 16-bp DNA with major and minor grooves indicated. (D) Each MAX monomer recognizes one half of the palindromic E-box sequence (x = 5caC). (E) Interactions with the outer C3:G3 base pair. The numerical numbers indicate the inter-atom distance in angstrom. (F) Interactions with the inner A2:T2 base pair. (G) The bidentate hydrogen bonds formed between Arg36 and the central G1—a pattern specific to Guanine. (H) Close-up view of the central C1:G1 base pair, recognized by the pair of Arg36 residues. Additional neighboring residues near the carboxylate group of 5caC are Lys40 and Lys60. Two 5caC:G base pairs interact with two side chains of Arg-36 residues. Lys-40 interacts with the phosphate group of 5caC. (I) Arg36 forms a 5caC–Arg–Gua triad with the central CpG dinucleotide. Simulated annealing omit electron densities, contoured at 3σ and 5σ above the mean, are shown for Arg36 (green mesh) and the carboxylate moiety of 5caC (red mesh), respectively. (J) Phosphate interactions with Arg33, Lys40 and Arg60.
The central 5caCpG dinucleotide is involved in interactions with three charged residues, Arg36, Lys40 and Arg60 (Figure 2G and H). The guanidino group of Arg36 interacts with the central 5΄-CpG-3΄ dinucleotide by donating two hydrogen atoms to the O6 and N7 atoms of the 3΄ Guanine (G1), as well as stacks with the carboxylate group of the 5΄ 5caC (Figure 2D and I), forming a 5caC–Arg–Guanine triad. The next closest residues to the carboxylate group are Arg60 (∼6 Å) and Lys40 (∼7 Å) (Figure 2G and H)—too far to make direct contact (but potentially involved via water-mediated interactions). This creates a clamp-like basic environment ideal for electrostatic attraction with the two negatively charged carboxylate groups of central 5caC:G base pairs and the phosphate backbone (Figure 2A and J). The negatively charged Glu32 and the polar residue Asn29 are further away and separated by base pairs in between.
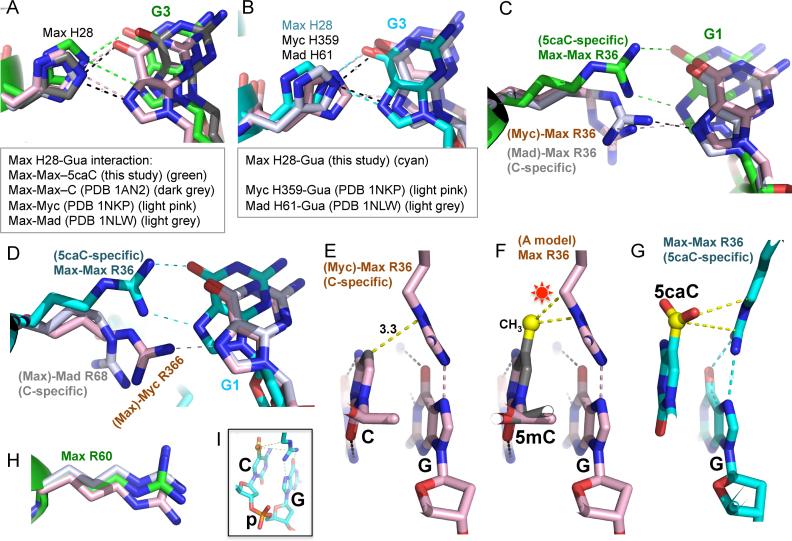
Arg36 adopts alternative conformations: C-specific and 5caC-specific
We compared our structure with previously characterized structures of the Max–Max homodimer, and the Max–Myc and Max–Mad heterodimers in complex with unmodified E-box DNA (43,44). Regardless of the partner, the corresponding His28-G3 and Glu32/Asn29-T2 interactions are conserved between Max, Myc and Mad (Figure 3A and B). Of the side chains involved in DNA base contact, Arg36 of Max has the largest conformational difference comparing 5caC and unmodified E-box DNA binding (Figure 3C and D). In the structures of the Max–Myc and Max–Mad bound with unmodified DNA, the guanidino group of Arg36 forms a singular H-bond with the Guanine N7 atom of central CpG (Figure 3C). The corresponding arginine in Myc (Arg366) likewise forms a single H-bond with Guanine N7, whereas the corresponding residue in Mad (Arg68) points away from the N7 atom of the Guanine (Figure 3D). In this conformation, which we term ‘C-specific’, the guanidino group of Arg36 (and the corresponding Arg336 of Myc or Arg68 of Mad) is quite close to the C5 atom of the CpG Cytosine (3.3–3.7 Å) (Figure 3E). Methylation or hydroxylmethylation at the C5 atom is likely to cause steric obstruction with the Arg36 in this conformation (Figure 3f), perhaps explaining the diminished binding to E-box DNA containing these modifications. In contrast, the negatively charged carboxylate group of 5caC is accommodated by inducing the positively charged Arg36 to adopt the rotated ‘5caC-specific’ conformation (Figure 3G), via a series of the side chain torsion angle rotations. In this conformation, Arg36 makes two H-bond interactions with Guanine O6 and N7. The conformation of Max Arg60 on the other hand, also involved in the distant binding of 5caC (Figure 2G), appears to be relatively constant regardless of whether Max is in the homodimer, or heterodimer with Myc or Mad, or whether the DNA is unmodified or 5-carboxylated (Figure 3H). The strong binding for 5caC/C (Figure 1) suggests that if the two DNA strands were modified independently, Arg36 from each monomer of the Max homodimer (or by extension, each monomer of the Max–Myc or Max–Mad heterodimers) could adopt strand-specific conformation: one for the 5caC-specific mode and the other for the C-specific mode.
Figure 3.
Arg36 adopts alternative conformations: C-specific and 5caC-specific. (A) Comparison of Max His28-Gua interaction among Max–Max homodimer, Max–Myc and Max–Mad heterodimers. (B) Comparison of Max His28 and its corresponding residue in Myc (His359) and Mad (His61). (C) Superimposition of Max Arg36-Gua interaction in Max–Max homodimer (in complex with 5caC DNA), Max–Myc (unmodified DNA) and Max–Mad (unmodified DNA) heterodimers. (D) Superimposition of Max Arg36 (5caC DNA) and its corresponding residue in Myc (Arg366) and Max (Arg68) in complex with unmodified DNA. (E) With unmodified DNA, Max Arg36 forms a van der Waals contact with the ring C5 atom of unmodified Cyt and a hydrogen bond with Gua of the central CpG dinculeotide, adopted from PDB 1NKP (44). (F) Modeling a methyl group (in yellow ball) onto unmodified CpG site potentially results in repulsion (indicated by a star) with the Arg36 of Max in the C-specific conformation. (G) The 5caC–Arg–Gua triad in the 5caC-specific interactions observed by Max Arg36. (H) Max Arg60 conformation is conserved among Max–Max, Max–Myc and Max–Mad dimers. (I) A close-up view of 5caCpG dinucleotide connected by the sugar-backbone (p: phosphate group).
Cancer-associated MAX mutations have altered DNA binding properties
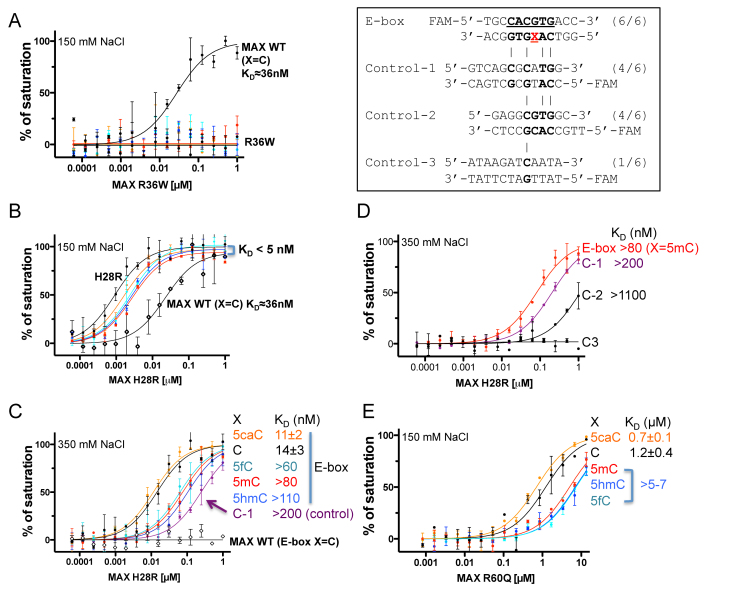
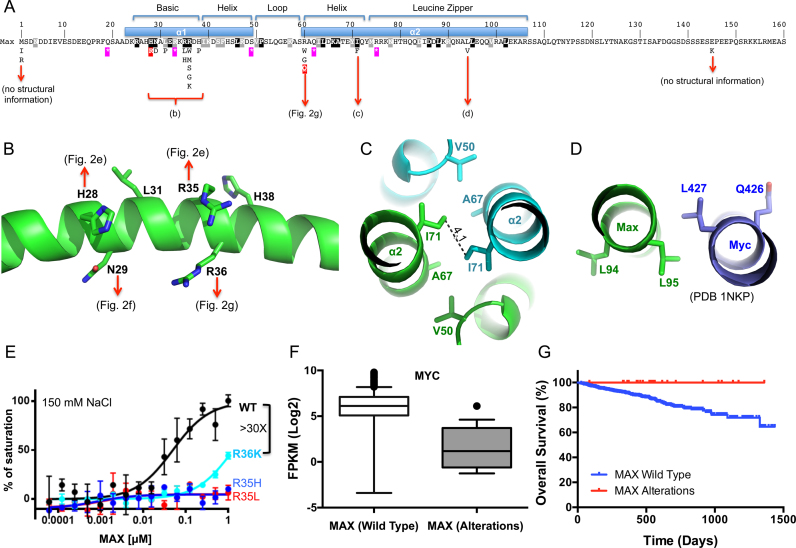
While the oncogenic consequences of MYC overexpression/amplification are well documented, recent work indicates that MAX is also a target of mutation in human cancers. Germline mutations in MAX confer a hereditary predisposition to pheochromocytoma and paragangliomas (34,35) and somatic mutations have been described in a number of other sporadic tumor types (cBioportal, http://www.cbioportal.org/) (36,46,47). We characterized three such cancer related MAX mutations: R36W has been found in a myeloma, H28R found primarily in endometrial cancers, and R60Q found in a wide variety of cancers including endometrial, glioma, acute myeloid leukemia, colorectal and stomach cancers. Among these, H28R and R60Q were recently identified as recurrent ‘hotspot’ mutations predicted to have functional consequences in cancer development (48).
Consistent with the central role for Arg36-mediated hydrogen bonding to the central CpG base pair noted above, the change from a positively charged Arg36 to an aromatic tryptophan (R36W) resulted in complete loss of DNA binding (Figure 4A). In contrast, substitution of His28, which recognizes the outer guanine (G3) (Figure 2E), with arginine (H28R), resulted in significantly higher affinity for all five forms of E-box DNA (Figure 4B). This observation is in agreement with previous studies showing that both Arg and His can be used in Guanine recognition (49) (e.g. see (50)). Indeed, H28R bound DNA so tightly that the estimated KD values were below the probe concentration of assay conditions regardless of CpG modification status (5 nM probe, 150 mM NaCl) (Figure 4B). We thus increased the NaCl concentration to 350 mM. Under these more stringent conditions, H28R retained the same relative order of binding preference as wild-type MAX (5caC∼C > 5fC > 5mC∼5hmC), with the KD values ranging from 11 to 110 nM (Figure 4C). Moreover, H28R was able to bind two control sequences (C-1 and C-2) that retain one or both of the outer G3:C3 base pairs, but lack the cognate E-box sequence—albeit with a reduced affinity relative to the E-box containing sequence. Wild-type MAX showed no detectable binding to the canonical E-box sequence or the control sequences under the same high salt condition (Figure 4C). It appears that the retention of both of the outer G3:C3 base pairs renders the control-1 sequence a reasonable target for the H28R mutant, with an affinity only about 2-fold lower than that of the hemi-methylated E-box sequence and 14-fold less than the unmodified sequence (Figure 4D). These data suggest that the H28R mutation may confer very tight, potentially indiscriminate DNA binding of MAX to degenerate E-box sequences.
Figure 4.
Cancer associated MAX mutants with altered DNA binding property. (A) R36W diminished DNA binding as measured by fluorescence polarization assays. Inset within the box: Sequences of the various modified E-box and control (C-1, C-2 and C-3) DNA substrates used. (B and C) H28R enhanced DNA binding with oligos containing varied forms of cytosine. The estimated dissociation constant KD values are uniformly low in 150 mM NaCl (panel B) but considerably increased in 350 mM NaCl (panel C). Note that the smaller the calculated KD value, the higher the binding affinity. (D) The enhanced DNA binding affinity of H28R might allow the mutant to bind near-cognate sequence. (E) The effect of the R60Q mutation on DNA binding. Despite significant reduction of affinity, 5caC DNA is still preferred over unmodified DNA by the R60Q mutant. Data represent the mean ± SEM of two independent determinations performed in duplicate.
Substitution of the positively charged Arg60 with a polar glutamine residue (R60Q) reduced DNA binding significantly, by a factor of ∼20. Nevertheless, even at the lower affinity, the mutant retained its preference for the 5caC modification over 5mC or the other oxidized forms (5hmC, 5fC) (Figure 4E). The weakened DNA binding by the mutant might provide an opportunity for other enzymes (such as the DNA glycosylase TDG (8)) to compete and excise 5caC base, resulting in demethylation.
MAX mutations in Multiple Myeloma
Multiple myeloma (MM) is a malignancy of the long-lived antibody-producing plasma cells of the bone marrow. While myeloma cells maintain many of the features of their normal counterparts (e.g. antibody production, dependence on interactions with the bone marrow stroma for survival), they acquire proliferative capacity and the dysregulation of MYC is thought to play a central role in driving their clonal expansion (51). Rearrangements that juxtapose MYC to the immunoglobulin loci or other super-enhancers drive high levels of MYC expression in ∼20–50% of cases (52,53). In addition to dysregulated MYC, MAX mutations are also observed in myeloma. The R36W mutation described above was originally uncovered in a study of ∼200 multiple myelomas, three of whom harbored MAX mutations (1.5%, cBioportal (54)). To determine the frequency and scope of MAX mutations, we queried the CoMMpass project (https://research.themmrf.org/), a prospective collection of purified tumor samples from over 1000 newly diagnosed myeloma patients, the majority of which have been characterized by exome sequencing and have corresponding RNA-seq and clinical outcomes data. Among the 805 patients for whom exome sequence was available, we identified 24 primary patient samples with 25 unique mutations affecting the MAX locus (16 missense, 5 nonsense, 2 splice site and 2 deletion/rearrangements; Supplementary Table S2) suggesting an overall frequency of ∼3%. The finding that many tumors harbor nonsense mutations, start codon mutations, or deletion/rearrangements predicted to result in a loss of all or most of the MAX gene product (n = 10), as well as several patients with more than one mutation (n = 5), strongly suggests that it is loss of function alterations in MAX that are important in myeloma. Indeed, of the remaining missense mutations, more than half affected amino acids 29–36 (Figure 5A), and in particular conserved residues R35 (n = 4) or R36 (n = 8). The finding that R36 can be mutated to a number of other residues (G, S, M, W or K), and R35 to L or H, suggested that the loss of the positively charge side chain(s) in the basic clamp region is particularly important in these cases. These mutations are likely to directly interfere with the Arg side chain interaction with the central guanine G1 (Figure 2G) resulting in a complete loss of binding affinity, as shown for the R36W mutant above (Figure 4A). Although the R36K mutation retains the positive charge, we found it reduced the mutant MAX binding affinity by >30-fold (Figure 5E). Likewise, mutation at R35 to a hydrophobic residue (L) or a shorter side chain (H), which could affect interactions with the negatively charged DNA phosphate group and E32 side chain (Figure 2E), also resulted in a total loss of binding (Figure 5E). An additional 25% (n = 4) affected R60, which was again changed to a hydrophobic residue (W) or the smallest residue (G), which might introduce flexibility and affect protein stability. We predict that this might weaken overall DNA binding affinity by abolishing electrostatic interaction with the phosphate backbone (Figure 2J) as exemplified by the reduced binding affinity of the R60Q mutation (Figure 4E) described above. Two additional missense mutations were observed in the helical leucine zipper region (I71W and L94V) (Figure 5C and D), which might affect dimerization or other protein–protein interactions.
Figure 5.
MAX alterations in multiple myeloma. (A) Relative position of MAX variants detected in multiple myeloma and their relationship to the structural features of the MAX protein. Features of the DNA binding (bHLH) and dimerization domains (leucine zipper) are shown. Missense mutations are shown below. Purple stars show the position of nonsense mutations. Black/gray shaded residues are those conserved across the MAX bHLH family (see Supplementary Figure S3D). Those ‘hotspot’ mutations that were biochemically analyzed (H28R, R60Q) are marked in red. (B–D) Structural impacts of MAX variants in different regions of the MAX protein. (B) The detailed impact of missense mutations in the basic region—detailed interactions can be observed in panels of Figure 2E, F and G. (C) An I71 to F variant is likely to interfere with the dimer interaction via the leucine zipper. (D) The L94V, located on the outer surface of helix α2, has the potential to alter interactions with regions outside of structured region of the dimer or other proteins. (E) R36K reduced DNA binding significantly, whereas the R35H and R35L completely abolished binding under the same conditions. Data represent two independent determinations performed in duplicate. (F) Comparison of MYC expression in multiple myeloma samples from patients with wild type MAX (n = 625) or MAX alterations (n = 18). Shown are box plots of the MYC expression levels between samples in each class. Median is indicated by a line, box is the 1st and 3rd quartiles, whiskers are the most extreme data point that is 1.5 times the interquartile range. Significance (P < 0.0001) was assessed by the Mann–Whitney U test. (G) Survival outcomes among Multiple Myeloma patients with wild-type MAX (n = 740) or MAX alterations (n = 24). Patients with MAX alterations exhibit a significantly better outcomes [Mantel–Cox log rank test, P = 0.041 (95% CI, 0.13–0.96)].
The above data suggested that MAX inactivation plays an important role in the pathogenesis of at least some myelomas. As noted above, MYC translocation/overexpression is also common in multiple myeloma, and has been associated with a poorer outcome (52,53). We therefore next examined the relationship between MAX mutations and MYC expression and outcomes among MM patients. Interestingly, among the 643 patients for whom both mutation status and RNA-seq data were available, there was an inverse relationship between MYC expression and MAX status, in that tissue from patients harboring MAX alterations expressed significantly lower levels of MYC transcript than those with wild-type MAX (Figure 5F). Such patients are also trending towards a more favorable outcome; among the 764 patients for whom mutation status and outcomes data were available, the 24 patients with MAX alterations had a significantly better prognosis, with a hazard ratio for overall survival of 0.35 (Figure 5G), which may be a reflection of the tendency toward low MYC expression among this group of patients.
DISCUSSION
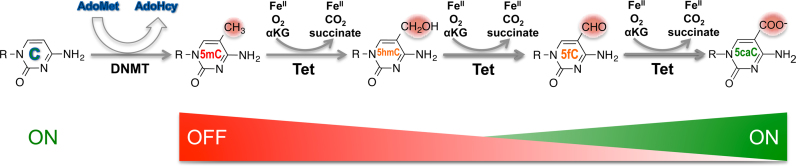
DNA 5mC is a major epigenetic signal that acts to regulate chromatin structure and ultimately gene expression. The Tet-mediated 5mC oxidation products, 5hmC, 5fC and 5caC are now recognized as an additional component of the ‘methylome’. While accumulating evidence supports a role for 5hmC as a stable epigenetic mark, 5fC and 5caC are the preferred substrates among the five forms of cytosine for excision by TDG and thus have been largely considered an intermediate in the demethylation cycle. Our results show that the DNA binding bHLH domain of MAX is responsive to all forms of modified CpG within its E-box recognition sequence, displaying the highest affinity for cognate sequences containing a central 5caC and unmodified C, with reduced affinity for 5fC and much lower affinity for 5mC and 5hmC. Taken at face value, this result could be interpreted to mean that the progressive Tet-mediated oxidation of 5mC may be a way to titrate in transcriptional activity in a graded and cyclical fashion, with the 5mC form of such sites being the ‘off’ position and Tet-mediated oxidation steps a way of progressively increasing affinity for the binding site while moving towards 5caC (Figure 6). Given the similarities between the bHLH domains of MAX and its binding partners, and the structural conservation of the critical amino acids whether partnered with MYC or Mad (Mxd), it is likely that the ability to ‘sense’ the 5mC oxidation state translates to most (if not all) MAX heterodimers and the extended MYC family of bHLH transcription factors. The balance between Tet-mediated generation of 5caC and TDG-mediated removal and replacement by unmodified C could define an equilibrium state that ensures the maintenance of a favorable (5caC or C) platform for MAX binding and regulatory activity. Dynamic competition between MAX binding and the local action of de novo DNA methyltransferase(s) might then represent the switch back to the ‘off’ position. In this context, one hypothesis is that the reduced binding affinity to the C/5caC forms exhibited by certain MAX variants (R36, R60) provides an increased opportunity for TDG-mediated turnover and subsequent methylation, whereas the H28 variant (H28R) with increased binding affinity may have the opposite effect. In all, these data raise the possibility that the role of DNA modification (methylation and/or oxidation) in regulating transcription factor activity may be more refined than a simple on-off switch.
Figure 6.
A hypothetical model of the cyclical impact of CpG modification on transcription factor binding. Schematic of chemical reactions of DNA cytosine methylation and 5mC oxidation. DNA methyltransferases convert a proportion of the cytosines (Cs) into 5mC in a S-adenosyl-l-methionine (AdoMet)-dependent reaction. The Tet dioxygenases then convert a fraction of 5mC to 5hmC, 5fC and 5caC in three consecutive Fe(II)- and α-ketoglutarate (αKG)-dependent oxidation reactions without releasing any formaldehyde. The potential consequence of modifications is indicated by color: green for activities associated with an increased level of gene expression (‘ON’) and red for those tending to decrease the level of expression (‘OFF’).
MAX and other transcription factors that can sense the oxidation status of 5mC and preferentially bind the Tet-mediated oxidation products may play a role in continually recruiting Tet enzymes to their genomic targets to maintain their open, active status and/or to initiate a shift in cell state. Indeed, in the absence of Tet function, DNA methylation accumulates predominantly at enhancers and other DNase hypersensitive regions (55,56). To this end, Wilms tumor protein WT1, which displays a preference for C, 5mC or 5caC over the 5hmC or 5fC forms (21), physically interacts with Tet2 and may recruit Tet2 to its target genes and/or bind to the products of Tet2 enzymatic activity (57,58). Moreover, recent genome-wide mapping of full-length Tet3 binding sites in mouse neural progenitor cells selectively enriched for sequences containing an E-box motif (5΄-tCACGTGa-3΄) that matches the recognition sequence of TFEB, another bHLH transcription factor (59) (Supplementary Figure S3D).
We provide evidence that for a subset of patients, somatic alterations in MAX play a role in the pathogenesis of multiple myeloma. An examination of the largest collection of multiple myeloma available to date showed MAX mutations to occur in ∼ 3% of cases, with missense mutations affecting R35/R36, and/or nonsense mutations being the most frequent. These alterations are unlikely to be therapy-related but rather appear to occur early in disease progression as the majority of CoMMpass samples (and all of those detected with MAX mutations) are derived from newly diagnosed patients. The finding that many tumors harbor nonsense mutations, start codon mutations, or deletion/rearrangements predicted to result in a loss of all or most of the MAX gene product (n = 10) and that several patients (n = 5) have more than one mutation strongly suggests that it is the inactivation of MAX that contributes to myelomagenesis, similar to what has been observed in other settings (34–36). Our in vitro binding experiments showing that alteration at R36 (R36W/K), R35 (R35H/L) or R60 (R60Q) (another one third of patients) significantly reduces or abolishes MAX DNA binding altogether is consistent with this suggestion.
Interestingly, MAX mutations marked a subset of patients with low levels of MYC expression and a better prognosis overall, as compared to those with wild-type MAX. One interpretation is that the consequences of MAX inactivation are functionally redundant with that of MYC overexpression in promoting myelomagenesis, but that the long-term disease course of a MYC-promoted versus a MAX-promoted myeloma ultimately differ. For example, when MAX is absent, MYC activity may go unrestrained. Analysis of the genome-wide occupancy of MYC and MAX in a myeloma cell line in which MYC is translocated and overexpressed (MM.1S (29)) indicates that whereas nearly all (97%) of MAX sites are co-occupied by MYC, only 3% of MYC binding sites are co-occupied by MAX (Supplementary Figure S5A and B). This is consistent with the recent suggestions that once MYC's high affinity sites are saturated, it can ‘invade’ other low affinity sites in areas of open chromatin, eventually including the majority of promoters (29,60,61) potentially facilitated through binding to other factors (e.g. WDR5) (61,62). Loss of MAX may shift the balance towards these lower affinity, non-physiologic sites. It is interesting to note that whereas MYC-MAX co-occupied sites are enriched in canonical E-box sequences, the MYC-only sites show reduced specificity for the central CpG (29) (Supplementary Figure S5C), which would potentially remove any impact of CpG methylation status. Alternatively, loss of MAX function could promote myelomagenesis by alleviating MAX-mediated repression of select genes normally mediated by its interaction with other partners. Ectopic expression of the repressive MAX partners directly antagonizes MYC-driven transformation in cell-based assays, and deletion of certain partners (e.g. MNT) is sufficient to promote tumor formation in mouse models (63), and is observed in human leukemias (64,65). It is possible that both consequences contribute.
Taken together, our data suggest that bHLH transcription factors join a growing group of transcriptional regulators that have adapted to respond to different cytosine modification states, potentially acting as direct epigenetic sensors to instruct downstream events. The TET enzymes are altered in human cancers, particularly hematologic malignancies, and are associated with altered genomic patterns of 5mC/5hmC (reviewed in (12,66)). Inactivating mutations in TET2 are observed in a wide variety of myeloid and lymphoid tumors, and although mutations in TET1/3 are relatively rare, mice deficient in Tet1 or the combination of Tet1/Tet2 develop late-onset B-cell lymphomas that exhibit gene expression patterns consistent with human B-cell malignancies (67,68). Thus, the convergence of altered patterns of oxidized 5mC and acquired mutations in the factors that read them hint at a new level of epigenetic complexity in cancer progression.
ACCESSION NUMBER
The X-ray structure (coordinates and structure factor files) of MAX residues 22–107 with bound 5caC DNA has been submitted to PDB under accession number 5EYO.
Supplementary Material
ACKNOWLEDGEMENTS
We thank B. Baker of New England Biolabs for synthesizing the oligonucleotides. The Department of Biochemistry of Emory University School of Medicine supported the use of SER-CAT beamlines. We also thank Daniel Auclair and Joan Levy from the Multiple Myeloma Research Foundation, the MMRF CoMMpass Network and Jonathan Keats (TGEN) for support and advice regarding the CoMMpass trial and associated data.
Author Information: D.W. performed protein expression and purification, DNA binding assays and crystallization. H.H. performed X-ray data collection and structure determination. B.G.B. performed bioinformatics analysis; S.L., L.H.B. and P.M.V. participated in discussion and myeloma data analyses. X.Z., P.M.V. and X.C. organized and designed the scope of the study. All were involved in analyzing data and in preparing the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) [GM049245-23 to X.C. and CA077337-15 to P.M.V.]; T.J. Martell Foundation [to L.H.B.]; Georgia Research Alliance Eminent Scholar [to X.C.]. The open access publication charge for this paper has been waived by Oxford University Press – NAR Editorial Board members are entitled to one free paper per year in recognition of their work on behalf of the journal.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lister R., Pelizzola M., Dowen R.H., Hawkins R.D., Hon G., Tonti-Filippini J., Nery J.R., Lee L., Ye Z., Ngo Q.M. et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009; 462:315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roadmap Epigenomics C., Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J. et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015; 518:317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ziller M.J., Gu H., Muller F., Donaghey J., Tsai L.T., Kohlbacher O., De Jager P.L., Rosen E.D., Bennett D.A., Bernstein B.E. et al. Charting a dynamic DNA methylation landscape of the human genome. Nature. 2013; 500:477–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hon G.C., Rajagopal N., Shen Y., McCleary D.F., Yue F., Dang M.D., Ren B.. Epigenetic memory at embryonic enhancers identified in DNA methylation maps from adult mouse tissues. Nat. Genet. 2013; 45:1198–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tahiliani M., Koh K.P., Shen Y., Pastor W.A., Bandukwala H., Brudno Y., Agarwal S., Iyer L.M., Liu D.R., Aravind L. et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009; 324:930–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ito S., D'Alessio A.C., Taranova O.V., Hong K., Sowers L.C., Zhang Y.. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010; 466:1129–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ito S., Shen L., Dai Q., Wu S.C., Collins L.B., Swenberg J.A., He C., Zhang Y.. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011; 333:1300–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He Y.F., Li B.Z., Li Z., Liu P., Wang Y., Tang Q., Ding J., Jia Y., Chen Z., Li L. et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011; 333:1303–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bachman M., Uribe-Lewis S., Yang X., Williams M., Murrell A., Balasubramanian S.. 5-Hydroxymethylcytosine is a predominantly stable DNA modification. Nat. Chem. 2014; 6:1049–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bachman M., Uribe-Lewis S., Yang X., Burgess H.E., Iurlaro M., Reik W., Murrell A., Balasubramanian S.. 5-Formylcytosine can be a stable DNA modification in mammals. Nat. Chem. Biol. 2015; 11:555–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hashimoto H., Zhang X., Vertino P.M., Cheng X.. The mechanisms of generation, recognition, and erasure of DNA 5-methylcytosine and thymine oxidations. J. Biol. Chem. 2015; 290:20723–20733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rasmussen K.D., Helin K.. Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev. 2016; 30:733–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen L., Wu H., Diep D., Yamaguchi S., D'Alessio A.C., Fung H.L., Zhang K., Zhang Y.. Genome-wide analysis reveals TET- and TDG-dependent 5-methylcytosine oxidation dynamics. Cell. 2013; 153:692–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song C.X., Szulwach K.E., Dai Q., Fu Y., Mao S.Q., Lin L., Street C., Li Y., Poidevin M., Wu H. et al. Genome-wide profiling of 5-formylcytosine reveals its roles in epigenetic priming. Cell. 2013; 153:678–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu H., Wu X., Shen L., Zhang Y.. Single-base resolution analysis of active DNA demethylation using methylase-assisted bisulfite sequencing. Nat. Biotechnol. 2014; 32:1231–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu X., Han D., Zhao B.S., Song C.X., Zhang L.S., Dore L.C., He C.. Base-resolution maps of 5-formylcytosine and 5-carboxylcytosine reveal genome-wide DNA demethylation dynamics. Cell Res. 2015; 25:386–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun Z., Dai N., Borgaro J.G., Quimby A., Sun D., Correa I.R. Jr, Zheng Y., Zhu Z., Guan S.. A sensitive approach to map genome-wide 5-hydroxymethylcytosine and 5-formylcytosine at single-base resolution. Mol. Cell. 2015; 57:750–761. [DOI] [PubMed] [Google Scholar]
- 18.Neri F., Incarnato D., Krepelova A., Rapelli S., Anselmi F., Parlato C., Medana C., Dal Bello F., Oliviero S.. Single-base resolution analysis of 5-formyl and 5-carboxyl cytosine reveals promoter DNA methylation dynamics. Cell Rep. 2015; 10:674–683. [DOI] [PubMed] [Google Scholar]
- 19.Liu Y., Toh H., Sasaki H., Zhang X., Cheng X.. An atomic model of Zfp57 recognition of CpG methylation within a specific DNA sequence. Genes Dev. 2012; 26:2374–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y., Olanrewaju Y.O., Zheng Y., Hashimoto H., Blumenthal R.M., Zhang X., Cheng X.. Structural basis for Klf4 recognition of methylated DNA. Nucleic Acids Res. 2014; 42:4859–4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hashimoto H., Olanrewaju Y.O., Zheng Y., Wilson G.G., Zhang X., Cheng X.. Wilms tumor protein recognizes 5-carboxylcytosine within a specific DNA sequence. Genes Dev. 2014; 28:2304–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Golla J.P., Zhao J., Mann I.K., Sayeed S.K., Mandal A., Rose R.B., Vinson C.. Carboxylation of cytosine (5caC) in the CG dinucleotide in the E-box motif (CGCAG|GTG) increases binding of the Tcf3|Ascl1 helix-loop-helix heterodimer 10-fold. Biochem. Biophys. Res. Commun. 2014; 449:248–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khund Sayeed S., Zhao J., Sathyanarayana B.K., Golla J.P., Vinson C.. C/EBPbeta (CEBPB) protein binding to the C/EBP|CRE DNA 8-mer TTGC|GTCA is inhibited by 5hmC and enhanced by 5mC, 5fC, and 5caC in the CG dinucleotide. Biochim. Biophys. Acta. 2015; 1849:583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang L., Zhou Y., Xu L., Xiao R., Lu X., Chen L., Chong J., Li H., He C., Fu X.D. et al. Molecular basis for 5-carboxycytosine recognition by RNA polymerase II elongation complex. Nature. 2015; 523:621–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prendergast G.C., Ziff E.B.. A new bind for Myc. Trends Genet. 1992; 8:91–96. [DOI] [PubMed] [Google Scholar]
- 26.Torres R., Schreiber-Agus N., Morgenbesser S.D., DePinho R.A.. Myc and Max: a putative transcriptional complex in search of a cellular target. Curr. Opin. Cell Biol. 1992; 4:468–474. [DOI] [PubMed] [Google Scholar]
- 27.Blackwood E.M., Eisenman R.N.. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991; 251:1211–1217. [DOI] [PubMed] [Google Scholar]
- 28.Prendergast G.C., Lawe D., Ziff E.B.. Association of Myn, the murine homolog of max, with c-Myc stimulates methylation-sensitive DNA binding and ras cotransformation. Cell. 1991; 65:395–407. [DOI] [PubMed] [Google Scholar]
- 29.Lin C.Y., Loven J., Rahl P.B., Paranal R.M., Burge C.B., Bradner J.E., Lee T.I., Young R.A.. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012; 151:56–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nie Z., Hu G., Wei G., Cui K., Yamane A., Resch W., Wang R., Green D.R., Tessarollo L., Casellas R. et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 2012; 151:68–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ayer D.E., Kretzner L., Eisenman R.N.. Mad: a heterodimeric partner for Max that antagonizes Myc transcriptional activity. Cell. 1993; 72:211–222. [DOI] [PubMed] [Google Scholar]
- 32.Diolaiti D., McFerrin L., Carroll P.A., Eisenman R.N.. Functional interactions among members of the MAX and MLX transcriptional network during oncogenesis. Biochim. Biophys. Acta. 2015; 1849:484–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang L.H., Baker N.E.. E proteins and ID proteins: helix-loop-helix partners in development and disease. Dev. Cell. 2015; 35:269–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Comino-Mendez I., Gracia-Aznarez F.J., Schiavi F., Landa I., Leandro-Garcia L.J., Leton R., Honrado E., Ramos-Medina R., Caronia D., Pita G. et al. Exome sequencing identifies MAX mutations as a cause of hereditary pheochromocytoma. Nat. Genet. 2011; 43:663–667. [DOI] [PubMed] [Google Scholar]
- 35.Burnichon N., Cascon A., Schiavi F., Morales N.P., Comino-Mendez I., Abermil N., Inglada-Perez L., de Cubas A.A., Amar L., Barontini M. et al. MAX mutations cause hereditary and sporadic pheochromocytoma and paraganglioma. Clin. Cancer Res. 2012; 18:2828–2837. [DOI] [PubMed] [Google Scholar]
- 36.Romero O.A., Torres-Diz M., Pros E., Savola S., Gomez A., Moran S., Saez C., Iwakawa R., Villanueva A., Montuenga L.M. et al. MAX inactivation in small cell lung cancer disrupts MYC-SWI/SNF programs and is synthetic lethal with BRG1. Cancer Discov. 2014; 4:292–303. [DOI] [PubMed] [Google Scholar]
- 37.Lan F., Collins R.E., De Cegli R., Alpatov R., Horton J.R., Shi X., Gozani O., Cheng X., Shi Y.. Recognition of unmethylated histone H3 lysine 4 links BHC80 to LSD1-mediated gene repression. Nature. 2007; 448:718–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hashimoto H., Liu Y., Upadhyay A.K., Chang Y., Howerton S.B., Vertino P.M., Zhang X., Cheng X.. Recognition and potential mechanisms for replication and erasure of cytosine hydroxymethylation. Nucleic Acids Res. 2012; 40:4841–4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Motulsky H., Christopoulos A.. Fitting Models to Biological Data Using Linear and Nonlinear Regression: A Practical Guide to Curve Fitting. 2004; NY: Oxford University Press. [Google Scholar]
- 40.Kabsch W. Xds. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adams P.D., Grosse-Kunstleve R.W., Hung L.W., Ioerger T.R., McCoy A.J., Moriarty N.W., Read R.J., Sacchettini J.C., Sauter N.K., Terwilliger T.C.. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002; 58:1948–1954. [DOI] [PubMed] [Google Scholar]
- 43.Ferre-D'Amare A.R., Prendergast G.C., Ziff E.B., Burley S.K.. Recognition by Max of its cognate DNA through a dimeric b/HLH/Z domain. Nature. 1993; 363:38–45. [DOI] [PubMed] [Google Scholar]
- 44.Nair S.K., Burley S.K.. X-ray structures of Myc-Max and Mad-Max recognizing DNA. Molecular bases of regulation by proto-oncogenic transcription factors. Cell. 2003; 112:193–205. [DOI] [PubMed] [Google Scholar]
- 45.Horowitz S., Trievel R.C.. Carbon-oxygen hydrogen bonding in biological structure and function. J. Biol. Chem. 2012; 287:41576–41582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013; 6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012; 2:401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chang M.T., Asthana S., Gao S.P., Lee B.H., Chapman J.S., Kandoth C., Gao J., Socci N.D., Solit D.B., Olshen A.B. et al. Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat. Biotechnol. 2016; 34:155–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luscombe N.M., Laskowski R.A., Thornton J.M.. Amino acid-base interactions: a three-dimensional analysis of protein-DNA interactions at an atomic level. Nucleic Acids Res. 2001; 29:2860–2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Patel A., Horton J.R., Wilson G.G., Zhang X., Cheng X.. Structural basis for human PRDM9 action at recombination hot spots. Genes Dev. 2016; 30:257–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boise L.H., Kaufman J.L., Bahlis N.J., Lonial S., Lee K.P.. The Tao of myeloma. Blood. 2014; 124:1873–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Affer M., Chesi M., Chen W.D., Keats J.J., Demchenko Y.N., Tamizhmani K., Garbitt V.M., Riggs D.L., Brents L.A., Roschke A.V. et al. Promiscuous MYC locus rearrangements hijack enhancers but mostly super-enhancers to dysregulate MYC expression in multiple myeloma. Leukemia. 2014; 28:1725–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Walker B.A., Wardell C.P., Brioli A., Boyle E., Kaiser M.F., Begum D.B., Dahir N.B., Johnson D.C., Ross F.M., Davies F.E. et al. Translocations at 8q24 juxtapose MYC with genes that harbor superenhancers resulting in overexpression and poor prognosis in myeloma patients. Blood Cancer J. 2014; 4:e191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lohr J.G., Stojanov P., Carter S.L., Cruz-Gordillo P., Lawrence M.S., Auclair D., Sougnez C., Knoechel B., Gould J., Saksena G. et al. Widespread genetic heterogeneity in multiple myeloma: implications for targeted therapy. Cancer Cell. 2014; 25:91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hon G.C., Song C.X., Du T., Jin F., Selvaraj S., Lee A.Y., Yen C.A., Ye Z., Mao S.Q., Wang B.A. et al. 5mC oxidation by Tet2 modulates enhancer activity and timing of transcriptome reprogramming during differentiation. Mol. Cell. 2014; 56:286–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lu F., Liu Y., Jiang L., Yamaguchi S., Zhang Y.. Role of Tet proteins in enhancer activity and telomere elongation. Genes Dev. 2014; 28:2103–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rampal R., Alkalin A., Madzo J., Vasanthakumar A., Pronier E., Patel J., Li Y., Ahn J., Abdel-Wahab O., Shih A. et al. DNA hydroxymethylation profiling reveals that WT1 mutations result in loss of TET2 function in acute myeloid leukemia. Cell Rep. 2014; 9:1841–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Y., Xiao M., Chen X., Chen L., Xu Y., Lv L., Wang P., Yang H., Ma S., Lin H. et al. WT1 recruits TET2 to regulate its target gene expression and suppress leukemia cell proliferation. Mol. Cell. 2015; 57:662–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jin S.G., Zhang Z.M., Dunwell T.L., Harter M.R., Wu X., Johnson J., Li Z., Liu J., Szabo P.E., Lu Q. et al. Tet3 reads 5-carboxylcytosine through Its CXXC domain and is a potential guardian against neurodegeneration. Cell Rep. 2016; 14:493–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kress T.R., Sabo A., Amati B.. MYC: connecting selective transcriptional control to global RNA production. Nat. Rev. Cancer. 2015; 15:593–607. [DOI] [PubMed] [Google Scholar]
- 61.Lorenzin F., Benary U., Baluapuri A., Walz S., Jung L.A., von Eyss B., Kisker C., Wolf J., Eilers M., Wolf E.. Different promoter affinities account for specificity in MYC-dependent gene regulation. eLife. 2016; 5:e15161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thomas L.R., Wang Q., Grieb B.C., Phan J., Foshage A.M., Sun Q., Olejniczak E.T., Clark T., Dey S., Lorey S. et al. Interaction with WDR5 promotes target gene recognition and tumorigenesis by MYC. Mol Cell. 2015; 58:440–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Link J.M., Hurlin P.J.. The activities of MYC, MNT and the MAX-interactome in lymphocyte proliferation and oncogenesis. Biochim. Biophys. Acta. 2015; 1849:554–562. [DOI] [PubMed] [Google Scholar]
- 64.Vermeer M.H., van Doorn R., Dijkman R., Mao X., Whittaker S., van Voorst Vader P.C., Gerritsen M.J., Geerts M.L., Gellrich S., Soderberg O. et al. Novel and highly recurrent chromosomal alterations in Sezary syndrome. Cancer Res. 2008; 68:2689–2698. [DOI] [PubMed] [Google Scholar]
- 65.Edelmann J., Holzmann K., Miller F., Winkler D., Buhler A., Zenz T., Bullinger L., Kuhn M.W., Gerhardinger A., Bloehdorn J. et al. High-resolution genomic profiling of chronic lymphocytic leukemia reveals new recurrent genomic alterations. Blood. 2012; 120:4783–4794. [DOI] [PubMed] [Google Scholar]
- 66.Ko M., An J., Pastor W.A., Koralov S.B., Rajewsky K., Rao A.. TET proteins and 5-methylcytosine oxidation in hematological cancers. Immunol. Rev. 2015; 263:6–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cimmino L., Dawlaty M.M., Ndiaye-Lobry D., Yap Y.S., Bakogianni S., Yu Y., Bhattacharyya S., Shaknovich R., Geng H., Lobry C. et al. TET1 is a tumor suppressor of hematopoietic malignancy. Nat. Immunol. 2015; 16:653–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhao Z., Chen L., Dawlaty M.M., Pan F., Weeks O., Zhou Y., Cao Z., Shi H., Wang J., Lin L. et al. Combined loss of Tet1 and Tet2 promotes B cell, but not myeloid malignancies, in mice. Cell Rep. 2015; 13:1692–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.