Abstract
Biogenesis of messenger RNA is critically influenced by the phosphorylation state of the carboxy-terminal domain (CTD) in the largest RNA polymerase II (RNAPII) subunit. Several kinases and phosphatases are required to maintain proper CTD phosphorylation levels and, additionally, several other proteins modulate them, including Rpb4/7 and Sub1. The Rpb4/7 heterodimer, constituting the RNAPII stalk, promote phosphatase functions and Sub1 globally influences CTD phosphorylation, though its mechanism remains mostly unknown. Here, we show that Sub1 physically interacts with the RNAPII stalk domain, Rpb4/7, likely through its C-terminal region, and associates with Fcp1. While Rpb4 is not required for Sub1 interaction with RNAPII complex, a fully functional heterodimer is required for Sub1 association to promoters. We also demonstrate that a complete CTD is necessary for proper association of Sub1 to chromatin and to the RNAPII. Finally, genetic data show a functional relationship between Sub1 and the RNAPII clamp domain. Altogether, our results indicate that Sub1, Rpb4/7 and Fcp1 interaction modulates CTD phosphorylation. In addition, Sub1 interaction with Rpb4/7 can also modulate transcription start site selection and transcription elongation rate likely by influencing the clamp function.
INTRODUCTION
Eukaryotic RNA polymerase II (RNAPII) contains 12 subunits, Rpb1–Rpb12. Rpb1, the largest subunit, has a unique and highly conserved carboxy-terminal domain (CTD) with an essential role in transcription regulation in vivo (1–4). The RNAPII CTD is required for efficient capping, splicing and cleavage/polyadenylation of pre-mRNAs (5–7). It recruits RNA processing, export and histone modifying factors to the transcription complex, coupling mRNA biogenesis to other nuclear processes (3,8). Nearby the CTD and the RNA exit channel, two RNAPII subunits, Rpb4 and Rpb7, form a heterodimer that protrudes from the enzyme core like a stalk. From this location, the Rpb4/7 heterodimer regulates the interaction with factors important for RNA biogenesis, such as several components of the PIC and the Mediator (9–11), and for CTD modification. Indeed, we have shown that the Rpb4/7 heterodimer plays a key role in controlling phosphorylation of the CTD (12).
The CTD is composed of a repeated heptapeptide motif (26-52 depending upon the organism) with a consensus sequence of Tyr1-Ser2-Pro3-Thr4-Ser5-Pro6-Ser7 (13,14). Five of the seven residues can be phosphorylated, though Ser2 and Ser5 phosphorylation seem to be the predominant modifications (5,7,15). In mammals, the CTD can be also acetylated (16), glycosylated (17,18) and methylated (19). The multitude of possible CTD modifications, especially Ser phosphorylations, in combination with the numerous repetitions, generates a wide range of phosphorylation patterns that have been proposed to comprise a “CTD code” (20). The CTD modifications are dynamic and differentially orchestrate the recruitment of a number of factors required for transcriptional efficiency and RNA processing (21,22).
Several kinases and phosphatases regulate the levels of CTD phosphorylation (23). In Saccharomyces cerevisiae, four cyclin-dependent kinases (CDKs), Srb10, Kin28, Ctk1 and Bur1 (3,7) as well as four phosphatases, Rtr1, Ssu72, Glc7 and Fcp1 (23,24) determine CTD phosphorylation along the transcription cycle. Three additional yeast factors have been involved in the modulation of CTD phosphorylation: Ess1, Sub1 and Rpb4/7 (12,25,26). The peptidyl prolyl cis–trans isomerase Ess1 promotes the function of Ssu72 by isomerising CTD prolines in yeast, while human Pin1 isomerase assists Fcp1 activity (26–28). More recently, we have shown that Rpb4/7 heterodimer is important for recruiting Ssu72 and Fcp1 phosphatases (12). Sub1 globally modulates CTD phosphorylation all along the transcription cycle (25) although its mechanism remains essentially unknown. Sub1 was originally identified as a transcriptional stimulatory protein, homologous to the human positive coactivator PC4 (29–33) that physically interacts with TFIIB, arguing for a role as a coactivator in transcription initiation (34,35). Indeed, Sub1 has been identified as a component of the pre-initiation (PIC) complex (36), and having a role in the selection of transcription start site (TSS) (37). Additionally, Sub1 has also been implicated in other aspects of mRNA biogenesis, such as elongation (38), transcription termination, and 3΄end formation (39–41). Here, we present novel data demonstrating that Sub1 directly interacts with the Rpb4/7 heterodimer via Rpb7 and, in association with Fcp1, modulates the CTD phosphorylation levels. Moreover, our data suggest a role for Sub1 related to the Rpb1 clamp domain, in agreement with the role of Sub1 in the transcription start site (TSS) selection and in the regulation of the RNAPII transcription rate and its interaction with the elongation factor Spt5 (38).
MATERIALS AND METHODS
Yeast strains and media
The strains used are listed in Table 1 (Supplementary data). Strain construction and other genetic manipulations were performed following standard procedures (42). Oligonucleotide sequences are available upon request.
TAP purification and mass spectrometric analysis
Purification of Sub1–TAP was performed as described in (43). Sub1–TAP and associated proteins were recovered from cell extracts by affinity selection on an IgG matrix. After washing, TEV protease was added to release the bound material. The eluate was incubated with calmodulin-coated beads in the presence of calcium. After washing, the bound material was released by incubation with EGTA. The eluate was analyzed by Nano-ESI ion-trap mass spectrometry, or by trichloroacetic acid precipitation and analyzed by Western blotting.
Co-immunoprecipitation and western blot analysis
Cells containing TAP-tagged Sub1, and HA-tagged Rpb4/Rpb7 were grown in 200 ml of rich medium to an OD600 of 1.0, harvested, washed with water, followed by suspension in 2.5 ml of lysis buffer (20 mM HEPES pH 7.6, 200 mM potassium acetate, 1 mM EDTA pH 8.0, glycerol 10 %) containing protease and phosphatase inhibitors. The cell suspension was flash frozen in liquid nitrogen, and then ground in a chilled mortar to a fine powder. Afterwards, the cell lysate was thawed slowly on ice and transferred to pre-chilled tubes and centrifuged at 13 200 rpm for 20 min. The supernatant was collected and total protein concentration was estimated measuring absorbance at 280 nm in a nanodrop. In experiments where Sub1–TAP was precipitated, the volume of each cell extract containing 25 mg of protein was incubated with 50 μl of IgG Sepharose 6FF (GE Healthcare) slurry for 2 h at 4°C. In experiments where Rpb3 was immunoprecipitated, cell extracts were incubated with 5 μl of anti-Rpb3 antibody for 2 h at 4°C, followed by binding to 30 μl of Protein A Sepharose CL-4B (GE Healthcare) for another 2 h at 4°C. The IPs were extensively washed with lysis buffer and beads were suspended in SDS-PAGE sample buffer. Thereafter they were incubated at 65°C for 20 min and supernatants were loaded onto a SDS-PAGE gel. In the case of Rpb3 IP in whole cell extracts from wt and rpb4Δ, and wt and rpb1-CTD11 cells containing Sub1–TAP, we proceeded similarly, except that cell lysis was achieved at 4°C using a FastPrep System and 5 mg of total protein was immunoprecipitated. This was also the procedure used to IP Sub1–HA to study Fcp1-MYC/Sub1–HA association. In addition, RNase A and DNAse I treatments were done by incubating washed beads in lysis buffer with 10 mg/ml RNase A at room temperature for 15 minutes or with 10 units/μl DNAse I for 20 min at 37°C in its own buffer.
Western blot analysis was performed using the appropriate antibodies in each case. Anti-phosphoglycerate kinase (Pgk1, 459250; Invitrogen), anti-HA (12CA5, Roche); anti-TAP (CAB1001, Open Biosystems); anti-Rpb1: Y-80 (sc-25758, Santacruz) and 8WG16 (8WG16 (nonP-CTD, Covance); anti-Rpb4 (2Y14; Santa Cruz); anti-Rpb3 (1Y26; Santa Cruz); anti-MYC (9E10, Santa Cruz) were acquired from the indicated vendors. The ECL reagents were used for detection. The signal was acquired on film and/or with a ChemiDoc XRS (Bio-Rad) system and when necessary quantified with the Quantity One software (Bio-Rad).
Expression and purification of recombinant proteins
Recombinant Sub1
Transfection, virus amplification and protein expression
Sub1 recombinant bacmid was transfected into adherent High Five™ insect cells (Invitrogen). For virus amplification, recombinant baculovirus stocks were used to infect insect cells for four days. The amplified virus stock (25 ml) was obtained by cell pipetting and centrifugation at 2000 rpm for 5 min and then stored at 4°C. For protein expression, the virus amplification procedure was followed except that the flasks containing adherent insect cells were infected each with 70 μl of the amplified virus stock. Finally, the cells were harvested and frozen.
Purification
Frozen cell pellets were thawed and resuspended in lysis buffer (100 mM Tris pH 7.4, 300 mM NaCl, 6 mM MgCl2, 10% (v/v) glycerol, half tablet of EDTA-free Protease Inhibitor Cocktail (Complete™, Roche) and DNase I (Roche)). Cells were sonicated, centrifuged and the supernatant loaded into a HisTrap HP (GE Healthcare) affinity chromatography column equilibrated in binding buffer (100 mM Tris pH 7.4, 300 mM NaCl, 10% (v/v) glycerol). The column was extensively washed with HTA buffer (100 mM Tris pH 7.4, 300 mM NaCl, 10% (v/v) glycerol, 20 mM Imidazole) and washing buffer (100 mM Tris pH 7.4, 1.5 M NaCl, 10% (v/v) glycerol, 20 mM Imidazole), and then re-equilibrated in HTA buffer. Protein was eluted by running a gradient from 0% to 100% HTB buffer (100 mM Tris pH 7.4, 300 mM NaCl, 10% (v/v) glycerol, 250 mM imidazole) in 20 column volumes and collected fractions were analyzed by SDS-PAGE. Selected fractions were pooled, concentrated in a 10 kDa cut-off centrifugal filter (Millipore) and loaded onto a Superdex 200 10/300 GL (GE Healthcare) column equilibrated in GF buffer (20 mM Tris pH 7.4, 100 mM NaCl). The fractions containing the eluted protein were collected and analyzed by SDS-PAGE. The concentrated pool of the fractions containing the protein was frozen in liquid nitrogen and stored at −80°C.
To eliminate the His6-tag at the C-terminus, the protein was incubated with TEV protease (ratio Sub1:TEV 1:50) overnight at 4°C in a mixture containing 1 mM DTT. Digestion was confirmed by SDS-PAGE given the different electrophoretic mobilities of the His-tagged and untagged proteins. The digested protein was purified by recovering it from the supernatant after incubation of the reaction mix with a Ni resin (Ni Sepharose High Performance, Amersham Biosciences), where both undigested protein and TEV protease were captured through their polyhistidine tails. Fractions containing the digested protein were pooled and concentrated using Millipore centrifugal devices.
rRpb4/Rpb7-6His
We co-expressed and purified Rpb4/7 complex as described (44) from a plasmid containing Rpb4 and Rpb7, where a six-histidine tail was fused to the C-terminus of Rpb7.
Pull down-assays
Recombinant Rpb4/Rpb7-6His proteins (3 μg) were incubated in a 20 μl slurry of HisPur Cobalt Resine (Thermo Scientific) in binding buffer (20 mM Tris pH 7.5, 50 mM NaCl) for 2 h at 4°C. Thereafter, the resin with attached Rpb4/Rpb7-6His was washed three times with binding buffer and then Sub1 (3 μg) was added and incubated with the resin for 3 more hours at 4°C in binding buffer. As a negative control, the same amount of Sub1 was incubated in parallel under the same conditions with 20 μl slurry of Cobalt resin without Rpb4/7 proteins. After incubation, the resin was washed four times with washing buffer (20 mM Tris pH 7.5, 100 mM NaCl) and then treated with 15 μl of elution buffer (20 mM Tris pH 7.5, 100 mM NaCl, 150 mM imidazole) for 5 minutes at room temperature. The eluate was collected and analysed by SDS-PAGE. The gel was stained by immersion into Blue Safe staining solution (NZY tech) for several hours.
Chromatin immunoprecipitation (ChIP)
Chromatin purification, immunoprecipitation, quantitative real-time PCR (qPCR) amplification and data analysis were performed as described (12,25,45). Briefly, PCR of purified chromatin, following immunoprecipitation, was performed by quantitative real-time PCR with the CFX96 Detection System (Bio-Rad Laboratories, Inc.), using SsoAdvanced™ Universal SYBR® Green Supermix (Bio-Rad) following the manufacturer's instructions. Four serial 10-fold dilutions of genomic DNA were amplified using the same reaction mixture as the samples to construct the standard curves. Real-time PCR reactions were performed in triplicate using at least three independent ChIPs. Quantitative analysis was carried out using the CFX96 Manager Software (version 3.1, Bio-Rad). The values obtained for the IP'd PCR products were compared to those of the total input, and the ratio of the values from each PCR product from transcribed genes to a non-transcribed region of chromosome VII was calculated. Numbers on the y-axis of graphs are detailed in the corresponding figure legend.
RNA isolation and RT-PCR
Total RNA was extracted as described (46) and RT-PCR was performed using the iScript RT reagent Kit (Bio-Rad), following the manufacturer's instructions. PCR reactions were performed in triplicate with at least three independent cDNA samples.
Sub1 mutagenesis
The sub1-FRN54-56AGG triple mutation was generated by site-directed mutagenesis of a wild-type copy of SUB1 cloned into a centromeric plasmid under the control of its own promoter and with a 6x-HA epitope at the C-terminus of the protein. Plasmid expressing the ssDNA binding domain mutation or a wt copy of SUB1 was used to transform the sub1Δ mutant and to generate the sub1-FRN54-56AGG strain. As a control, the sub1Δ strain was transformed with an empty plasmid.
RESULTS
Sub1 interacts with RNAPII through the Rpb4/7 heterodimer
Sub1 influences RNAPII CTD phosphorylation via all four CTD kinases: Kin28, Srb10, Bur1 and Ctk1 (25). This was observed both genetically and biochemically, including Sub1 effects on kinase activity and/or recruitment to chromatin. Thus, Sub1 can act throughout the transcription cycle as a general regulator of CTD phosphorylation, though we do not understand the biochemical basis of the influence of Sub1 on the activity of all four CTD kinases. We considered two possible explanations: (i) Sub1 enhances the association (or dissociation) of an unidentified, common regulator with the kinases; or (ii) Sub1 influences kinase accessibility to the CTD.
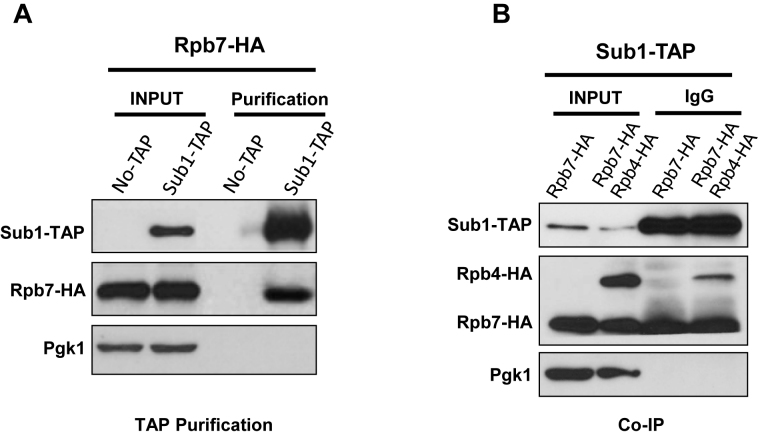
To further explore the first possibility, we decided to analyse the repertoire of Sub1 physical interactions using the TAP strategy (43). Accordingly, Sub1 was TAP-tagged and associated proteins were MS-analysed. We found among other proteins Spt5, consistent with previous work (38,47). More interestingly, we identified the Rpb7 subunit of RNAPII as a co-purifying protein. This discovery is extremely interesting since Rpb7, together with Rpb4, is near the CTD, and the heterodimer Rpb4/7 has been functionally related with CTD phosphorylation (12). To corroborate the specificity of Sub1-Rpb7 association, we purified Sub1–TAP complexes in a double tagged strain, Sub1–TAP Rpb7–HA, and analyzed the TAP purified complex by western blot using the corresponding antibodies to visualize Sub1 and Rpb7 (Figure 1A). We confirmed Sub1–TAP/Rpb7–HA association by co-immunoprecipitation (Co-IP), and demonstrated that Sub1 also associates with Rpb4, using a tripled tagged strain Sub1–TAP Rpb7–HA Rpb4–HA (Figure 1B). These data indicate that Sub1 associates with the Rpb4/7 heterodimer.
Figure 1.
Sub1 interacts with the RNAPII through the Rpb4/7 heterodimer. (A) Sub1 tandem affinity purification from Sub1–TAP and Sub1–TAP Rpb7–HA whole cell extracts (WCE). Input and the purified proteins were precipitated with trichloroacetic acid and analyzed by western blotting using anti-TAP and anti-HA. Anti-Pgk1 was used as a loading control. (B) Co-IP performed using WCEs from Sub1–TAP (Rpb7–HA and Rpb7–HA/Rpb4–HA) with IgG Sepharose. Input and IPs were analyzed by western blotting with antibodies to the indicated proteins.
Allelic specific interaction between SUB1 and RPB4/7
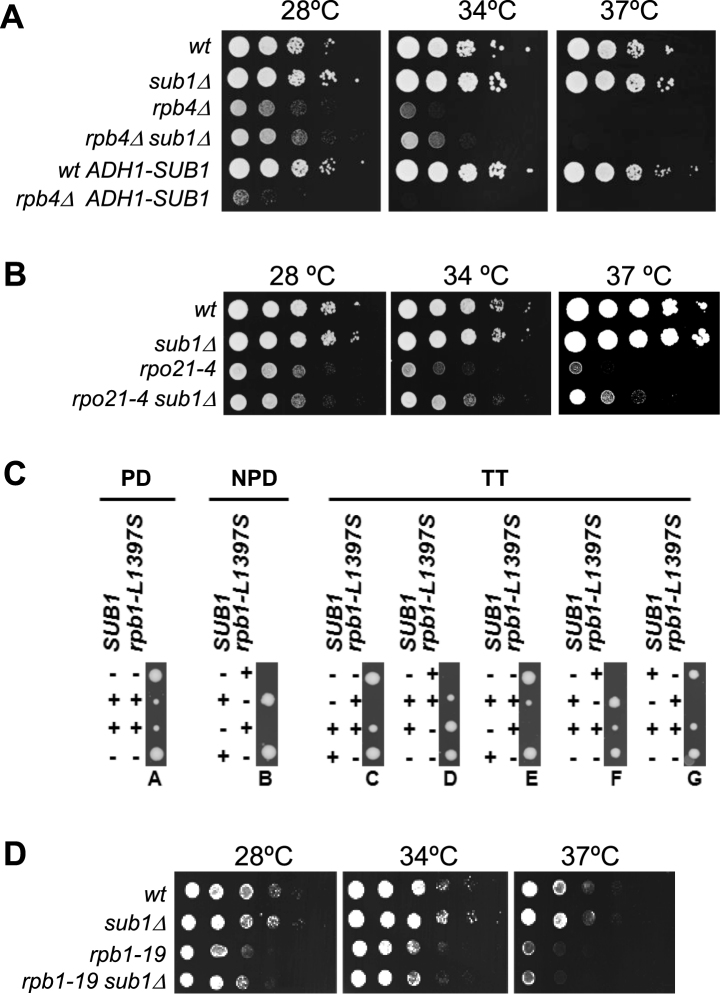
To gain more insight into the functional connection between Sub1 and the RNAPII stalk heterodimer, we analyzed genetic interaction between SUB1 and RPB4/7. As Rpb7, but not Rpb4, is essential for cell viability we worked with the rpb4Δ mutant that displays slow growth at 28°C and thermosensitivity at 37°C (48,49). Deletion of SUB1 partially suppresses the rpb4Δ growth defects at 28 and 34°C, but does not suppress rpb4Δ thermosensitivity at 37°C (Figure 2A). On the other hand, overexpression of SUB1 using a strong constitutive or inducible promoter (ADH1 and GAL1, respectively; Figure 2A and Supplementary Figure S1A), dramatically reduces rpb4Δ cell growth. In addition, we used an Rpb7-TAP strain where the C-terminal Rpb7-TAP tag causes growth defects (Supplementary Figure S1B). In this case, the growth defects at 28 and 34°C are also partially rescued by SUB1 deletion (Supplementary Figure 1B). Therefore, SUB1 genetically interacts with RPB4 and RPB7, in agreement with the association of Sub1 with the Rpb4/7 heterodimer (Figure 1). However, this genetic result was unexpected, because deletion of SUB1 mostly displays negative genetic interactions with mutations compromising transcription initiation and elongation, for instance mutations in TFIIB, TFIIE, TFIIH, Kin28, Ctk1, Bur1, Fcp1, Spt5/Spt4 (25,36,38,41,50). The positive genetic interaction between SUB1 and RPB4 and RPB7 suggests that Sub1 may be gaining a function when the heterodimer function is altered.
Figure 2.
Allelic specific interaction between SUB1 and RPB4/7. (A) Genetic interaction between SUB1 and RPB4. SUB1 deletion partially suppresses the slow growth phenotype of the rpb4Δ strain at 28 and 34°C, while overexpression of SUB1 exacerbates it. SUB1 was overexpressed from a strong constitutive ADH1 promoter. Serial dilutions (1:10) of wt and mutant strains were spotted on selective SC media and grown for 2–3 days at the indicated temperatures. (B) Genetic interaction between SUB1 and the rpo21-4 mutation localized in the Rpb1 foot domain. Deletion of SUB1 partially suppresses the slow growth phenotype of rpo21-4 strain at 28, 34 and 37°C. Cells were assayed as in (A) in rich media. (C) sub1Δ and rpb1-L1397S are synthetically lethal. A diploid yeast strain heterozygous for both SUB1 and rpb1-L1397S was sporulated and the meiotic progeny were separated by tetrad dissection, and allowed to grow for 3 days. Forty-six tetrads were dissected, with thirty showing tetratype segregation pattern (TT, five of which are shown), nine showing paternal ditype (PD) and 7 a nonpaternal ditype (NPD). The genotype of the resulting colonies was inferred by growth or no growth on selective medium and 37°C. The sub1Δ deletion is indicated as (−), and wt (+); and in the case of RPB1, (+) means rpb1-L1397S and (−) wt. Cells deleted for rpb1-L1397S alone (− −) show a slow growth phenotype, as reported previously. Double mutants cells (− +) were unviable. (D) There is not genetic interaction between sub1Δ and the rpb1–19 mutation on the jaw domain of Rpb1. Serial dilution assay as in (B) showing that single and double mutants, rpb1–19 and rpb1-19sub1Δ respectively, display similar growth.
To investigate the specificity of this interaction, we analyzed the genetic interaction between SUB1 and RPB1. For that purpose we choose three rpb1 alleles. Two of them are functionally and structurally related to Rpb4/7, rpo21-4 and rpb1-L1397S, which are localized in the foot and clamp domains, respectively (51,52); and a third one, rpb1-19, is localized in the jaw domain (53), far from Rpb4/7 within the context of RNAPII complex (54).
The foot domain is a conserved region of RNAPII located at the surface of the complex, with poor or no conservation in their paralogs, Rpa190 (RNAPI) and Rpc160 (RNAPIII), or in their homologs in archaea and bacteria (55,56). The foot domain is crucial for the assembly and stability of RNAPII, by ensuring the correct association of Rpb1 with Rpb6 and Rpb4/Rpb7. In fact, assembly defects alter not only transcriptional activity but also RNAPII-DNA association and CTD phosphorylation (51). Thus, we used the rpo21-4 (rpb1-W954-LELE-955) foot mutant (57,58) where the association of the Rpb4/7 heterodimer with RNAPII is reduced (51). This mutant displays reduced growth at 28°C and exhibits slow growth at 34°C and 37°C (Figure 2B). Interestingly, we observed that deletion of SUB1 suppressed rpo21-4 growth defects at 34 and 37°C, whereas sub1Δ does not affect growth at these temperatures in a wt RPB1 background. Hence, rpo21-4 suppression recapitulates the growth defects observed in the double mutant rpb4Δsub1Δ. These data indicate that sub1Δ suppresses mutations compromising the function of the Rpb4/7 heterodimer, either by deletion of RPB4 or by a mutation on the Rpb1 foot domain.
The RNAPII clamp is a flexible structural element postulated to intervene in the regulation of DNA access to the cleft (59). An open clamp is observed in the absence of the Rpb4/7 heterodimer (56), while the complete enzyme presents a closed clamp conformation (54). Rpb4/7 plays a role in the mobility of this domain during transcription initiation (60,61). Therefore, we studied whether a sub1Δ deletion might genetically interact with the rpb1-L1397S clamp mutation. Specifically, this amino acid substitution occurs in the Rpb1 α-47b helix of the Switch 1 loop of the catalytic site within the structure of the RNAPII. In fact, the Switch 1 loop holds the DNA template strand at position +2/+3 downstream of the catalytic Mg2+ (62). The rpb1-L1397S mutant displays slow growth at 28°C, cold and thermosensitivity and reduced RNAPII association with genes (52). Both, rpb1-L1397S and sub1Δ show upregulation of IMD2 transcription (38,52), which is a hallmark of defects on TSS selection (63). Indeed, it has been shown that Sub1 participates in TSS selection (37). We then crossed the rpb1-L1397S mutant with the sub1Δ deletion mutant. The diploid cells were sporulated and the tetrads dissected. Interestingly, sub1Δ conferred synthetic lethality in combination with rpb1-L1397S (Figure 2C). This could be explained as a result of the additive effect of both mutations, sub1Δ and rpb1-L1397S, reducing RNAPII gene occupancy during transcription to levels that may be incompatible with cell viability (41,45,52). Also, synthetic lethality may be the result of the combined defect on elongation (38,64,65), and/or on the TSS selection (37,52). In any case and importantly, this result shows the essentiality of Sub1 in a context where the clamp function is impaired.
We also tested the effect of SUB1 deletion on the rpb1-19 jaw mutant. In this case no genetic interaction was observed (Figure 2D). As noted above, this domain is situated far from Rpb4/7 within the RNAPII structure (54). All of these data indicate an allele-specific interaction between sub1Δ and mutations localized within or proximal to the Rpb4/7 heterodimer in the context of the RNAPII 10 subunit complex. In addition, they corroborate the specificity of a functional relationship between Sub1 and the Rpb4/7 heterodimer consistent with their physical association (Figure 1).
A functional Rpb4/7 heterodimer is a requisite for Sub1 recruitment to gene promoters
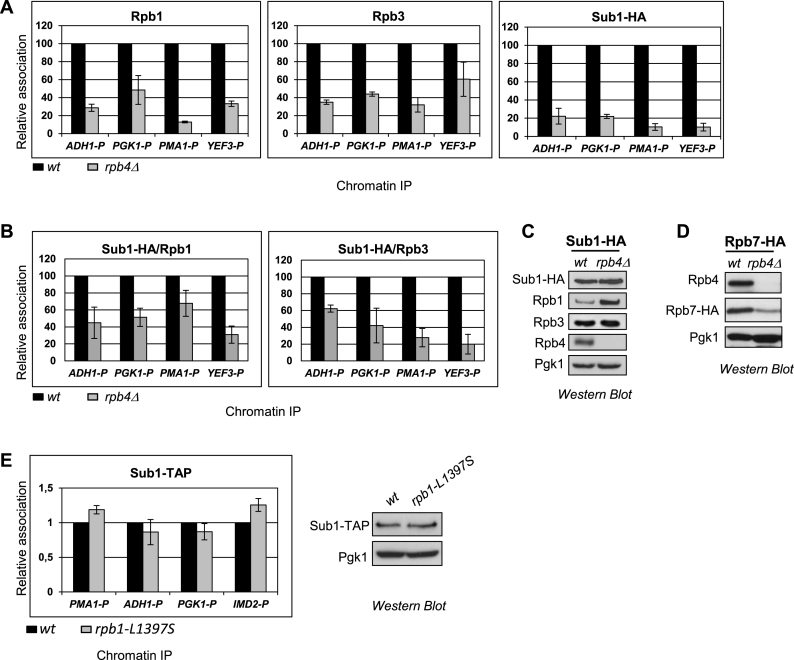
We next asked if Sub1 association with promoter DNA was affected in rpb4Δ cells, because in these cells Rpb7 levels are significantly reduced, along with diminished association of RNAPII with DNA (66–68). Thus, we performed chromatin immunoprecipitation (ChIP) using the wt and rpb4Δ Sub1–HA tagged strains, where in addition to Sub1, we also analyzed Rpb1 and Rpb3 occupancy at the promoters of several constitutively transcribed genes. In agreement with published data (12), Rpb4 is required for proper RNAPII occupancy to gene promoters as inferred from reduced Rpb1 and Rpb3 association levels (Figure 3A, left and middle panel, respectively). Interestingly, Sub1 occupancy was also dramatically decreased in rpb4Δ cells. Thus, <20% of Sub1 was associated with chromatin in cells lacking Rpb4 (Figure 3A, right panel). To determine if this was simply a consequence of reduced RNAPII occupancy, we calculated Sub1/Rpb1 and Sub1/Rpb3 ratios, and observed that this was not the case, because the levels of Sub1 association with chromatin were even lower than those of Rpb1 and Rpb3 (Figure 3B). We observed that these effects on chromatin association are not due to diminished Sub1–HA protein levels, as they are unaltered in rpb4Δ cells (Figure 3C). Although we did not know whether Sub1 directly interacts with Rpb7 and/or Rpb4, it is possible that reduced Rpb7 levels in rpb4Δ cells (Figure 3D, and (66–68)) could account for the Sub1 defect on promoter association. We also tested Sub1–TAP association with chromatin in the clamp mutant, rpb1-L1397S. No effect was observed: Sub1 was properly detected at gene promoters in the mutant cells (Figure 3E, left panel) with no change in Sub1–TAP levels (Figure 3E, right panel).
Figure 3.
A functional Rpb4/7 heterodimer is a requisite for Sub1 recruitment to gene promoters. (A) Chromatin immunoprecipitation (ChIP) analyses were performed using wt and rpb4Δ strains. Left panel: Rpb1 binding to the promoter (P) of four constitutively expressed genes, ADH1, PGK1, PMA1 and YEF3 was examined by qPCR. Results were quantified (see Materials and Methods), and relative Rpb1 binding in rpb4Δ cells is plotted relative to that in wt cells (set equal to 100). The data plotted correspond to mean values from at least three independent experiments, and the error bars represent standard deviations. Middle and Right panels show relative Rpb3 and Sub1–HA binding, respectively, plotted as for Rpb1. (B) Plots of Sub1–HA/Rpb1, Sub1–HA /Rpb3 ratios in wild-type (wt) and rpb4Δ using data from Figure 1A. (C) Whole cell extracts (WCE) were prepared from wild-type (wt) and rpb4Δ strains expressing Sub1–HA and analyzed by western blotting using the following antibodies: anti-HA, anti-Rpb1 (8W16G), anti-Rpb3, anti-Rpb4 (2Y14) and anti-Pgk1 as a control for total protein. (D) Whole cell extracts (WCE) were prepared from wild-type (wt) and rpb4Δ strains expressing Rpb7–HA and analyzed by Western blotting using anti-HA, anti-Rpb4 and anti-Pgk1. (E) Left panel: ChIP analysis in wt and rpb1-L1397S cells as in (A) to analyze Sub1–TAP occupancy at the promoter (P) of three constitutively expressed genes (PMA1, ADH1 and PGK1) and to the promoter of the IMD2 inducible gene, whose expression is regulated by Sub1 (38,82), and is upregulated in the rpb1-L1397S mutant (52). Right panel: WCE were prepared from wild-type (wt) and rpb1-L1397S strains expressing Sub1–TAP and analyzed by Western blotting using anti-TAP and anti-Pgk1 as loading control.
In conclusion, our data indicate that a functional Rpb4/7 heterodimer is required for Sub1 association with gene promoters. It has been suggested that Sub1, together with TFIIB, functions in the recruitment of RNAPII to constitutively transcribed genes. Hence, the reduced level of Sub1 bound to promoters in rpb4Δ cells is likely not due to reduced RNAPII recruitment. Rather, it is more likely that Sub1 cannot be maintained stably associated to the DNA, and/or disassociates faster from the promoter, in the absence of the Rpb4/7 heterodimer.
Sub1 associates with the Rpb4/7 heterodimer via direct interaction with Rpb7
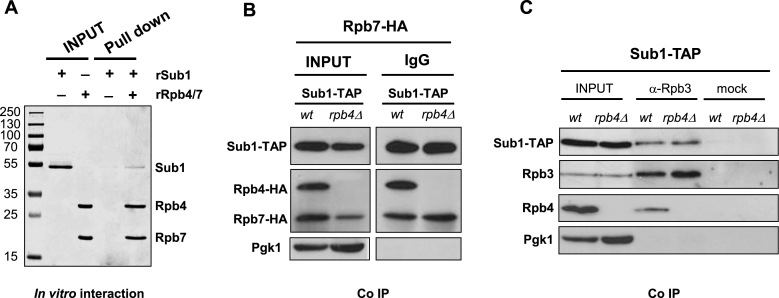
We next proceeded with in vitro assays to investigate whether the Rpb4/7-Sub1 interaction may be direct. For that purpose we carried out pull down assays using recombinant Sub1 and Rpb4/7 proteins. As shown in Figure 4A, Sub1 remains associated with Rpb4/7 after extensive washing, indicating that it directly interacts with the heterodimer. This result demonstrates for the first time that Sub1 interacts directly with the RNAPII complex. However, we did not know the direct target of Sub1: Rpb7, Rpb4 or both.
Figure 4.
Sub1 associates with the Rpb4/7 heterodimer via direct interaction with Rpb7. (A) Pull down assay. 15% SDS-PAGE gel showing bands corresponding to recombinant proteins rSub1 and rRpb4/Rpb7-6His. (lanes 2 and 3, respectively) and pull down assays where rSub1 and a Co2+ resin have been incubated in the absence (lane 4) and in the presence of rRpb4/Rpb7-6His (lane 5). (B) Co-IP performed with IgG Sepharose using WCEs from wt strains expressing Sub1–TAP and Rpb7–HA/Rpb4–HA and from rpb4Δ cells expressing Sub1–TAP and Rpb7–HA. Input and IPs were analyzed by Western blotting with antibodies to the indicated proteins. (C) Co-IP performed on WCEs from Sub1–TAP cells (wt and rpb4Δ) using anti-Rpb3 antibody. As a control, the same amount of cell extracts was incubated only with Protein A sepharose. Inputs and IPs were analyzed by Western blotting with antibodies to the indicated proteins.
To identify the direct target of Sub1, we co-immunoprecipitated Sub1–TAP and Rpb7–HA using whole cell extracts from a wt strain expressing Rpb4–HA or from the rpb4Δ strain, and analyzed its association by western blot to determine the contribution of Rpb4 to Sub1 interaction with the heterodimer. Surprisingly, we observed that Sub1 still interacts with Rpb7 when lacking Rpb4 (Figure 4B). In fact, the interaction between Sub1 and Rpb7 is similar in isogenic RPB4 wt and rpb4Δ cells, though Rpb7–HA levels are reduced in the absence of Rpb4 as previously described (66–68). Therefore, from all these data we conclude that the target of Sub1 is Rpb7, in agreement with identifying only Rpb7 in the Sub1–TAP purifications (Figure 1A). This result also supports our conclusion that reduced association of Sub1 to chromatin in rpb4Δ cells is due to decreased Rpb7 levels (Figure 3A, D). In summary, our data clearly demonstrate that Sub1 interacts with Rpb4/7 and indicate that this interaction could explain how Sub1 is able to modulate CTD phosphorylation.
Additionally, we tested if Sub1 association with the core polymerase is influenced by Rpb4. For that purpose we immunoprecipitated Rpb3 from wt and rpb4Δ whole cell extracts expressing Sub1–TAP, and analyzed Sub1 association by western blot. As shown in Figure 4C, Sub1–TAP associates with RNAPII in the absence of Rpb4. Therefore, our results indicate that Sub1 targets Rpb7 to be associated with the RNAPII, though we cannot exclude that it can also interact with other RNAPII subunits. On the other hand, and consistent with sub1Δ suppression of rpb4Δ (Figure 2A and Supplementary Figure S1A), Sub1 may have a negative effect during transcription when interacting with Rpb7 in the absence of Rpb4.
Sub1 carboxy-terminal region is important for the functional interaction with Rpb4/7
Sub1 is a 292-residue polypeptide showing strong similarity to its human homolog PC4 (127 amino acids) over a 65-residue region (amino acids 41–105) that includes a single-stranded DNA binding domain (DBD) and sequences essential for co-activator function (33,34,69). Although Sub1 is highly related to PC4, yeast Sub1 is much larger than human PC4 (33 KDa versus 15 KDa (34,35,39). Specifically, Sub1 has an extra carboxy-terminal (CT) region of ∼190 amino acids with unknown function(s), suggesting that Sub1 might have functional differences from PC4 due to this additional domain. Like PC4, Sub1 has the capacity to tightly bind melted DNA and single-stranded DNA in vitro (34) and both have been implicated in DNA-dependent processes other than transcription, such as DNA repair and replication (69–72).
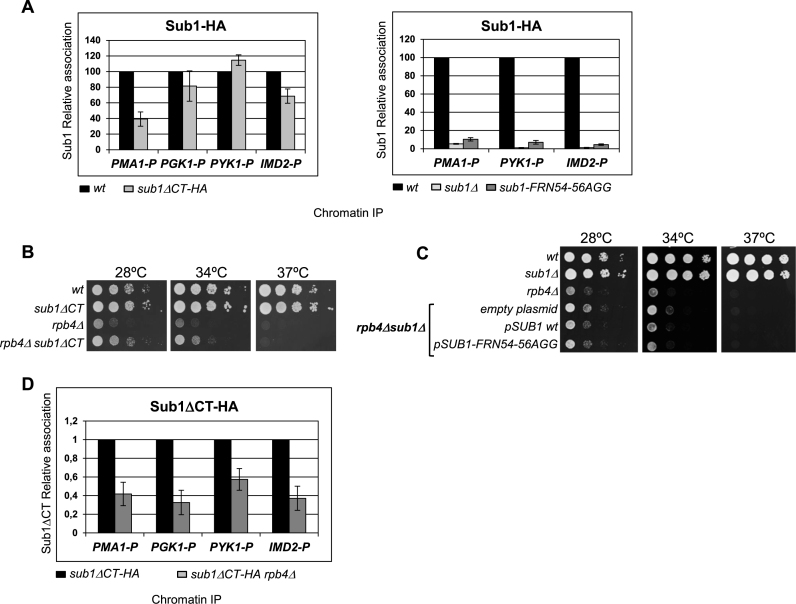
To explore which Sub1 region is involved in Rpb4/7 interaction, we investigated the importance of the Sub1 CT region and DNA-binding domain (DBD) relative to its interaction with Rpb4/7. For that purpose, two mutants were generated: (i) a strain where the chromosomal CT region of Sub1 (from amino acid 105 to stop) was substituted by a 6xHA epitope; and (ii) a triple-mutant, sub1-FRN54-56AGG, which encodes Phe54 → Ala, Arg55 → Gly and Asn56 → Gly replacements. Mutations in the corresponding residues in PC4 (Phe77, Lys78 and Lys79) severely affected the binding of PC4 to ssDNA in vitro (33). Protein expression of all the mutants was confirmed by western blot (Supplementary Figure S2A, upper panel). As shown, the levels of the Sub1ΔCT-HA protein are very low compared to that of Sub1 levels in the isogenic wt and sub1-FRN54-56AGG cells, and this effect is not due to defective transcription (Supplementary Figure S2A, lower panel). In sub1ΔCT cells, where Sub1 contains only the DNA binding domain, the protein becomes very unstable.
We next analyzed the association of Sub1–HA with the promoters of several constitutively transcribed genes (PMA1, PGK1, PYK1), and with the promoter of the Sub1 target gene IMD2, in normal growth conditions (Figure 5A). For that purpose, we performed ChIP assays first in wt, and sub1ΔCT expressing Sub1–HA and Sub1ΔCT-HA, respectively (Figure 5A, left panel). As expected, Sub1–HA was efficiently recruited to the promoter of all of genes tested in wt cells. However, unexpectedly and very interestingly, Sub1 association was either unaffected or only slightly affected in sub1ΔCT cells (Figure 5A, left panel), although Sub1ΔCT-HA protein levels were extremely low (Supplementary Figure S2A). This result indicates that the C-terminal domain of Sub1 is not required for Sub1 occupancy of gene promoters, but can contribute to it. Thus, the CT domain, which is not present in PC4, might be involved in other functions, such as the regulation of Sub1 binding to the DNA and/or in the interaction with other factors. In contrast, and as expected, Sub1 association was almost abolished in sub1-FRN54-56AGG cells (Figure 5A, right panel), which is consistent with the importance of these residues for PC4 DNA binding capacity and with the clear homology between Sub1 and PC4 in the DBD (33,69,73).
Figure 5.
Sub1 carboxy-terminal region is important for the functional interaction with Rpb4/7. (A) Left panel: Chromatin immunoprecipitation (ChIP) analyses were performed using wt and sub1ΔCT-HA strains expressing Sub1–HA. Sub1 binding to the promoter (P) of three constitutively expressed genes, PMA1, PGK1, PYK1 and the inducible gene IMD2 was examined by qPCR. Results were quantitated (as in figure 3), and relative Sub1–HA binding in sub1ΔCT-HA cells is plotted relative to that in wt cells (set equal to 100). Right panel, Chromatin immunoprecipitation (ChIP) analyses were performed using wt, SUB1 deletion mutant (sub1Δ), and the DNA binding mutant sub1-FRN54-56AGG. (B) Genetic interaction between sub1ΔCT and rpb4Δ. SUB1 deletion partially suppresses the slow growth phenotype of rpb4Δ strain at 28 and 34°C in rich medium. Serial dilutions (1:10) of wt and mutant strains were spotted on rich medium and grown for 2–3 days at the indicated temperatures. (C) There is no genetic interaction between the ssDNA binding domain of Sub1 and RPB4. Serial dilutions (1:10) of the indicated strains were spotted on selective SC medium and grown for 2–3 days at the indicated temperatures. (D) ChIP analyses were performed in wt and rpb4Δ cells expressing Sub1ΔCT-HA protein.
We further tested the genetic interaction between RPB4 and the Sub1 CT region and Sub1 DBD. Interestingly, deletion of the Sub1 CT region in rpb4Δ cells rescues their growth defects at 28 and 34°C (Figure 5B), recapitulating the effects of the SUB1 complete deletion (see Figure 2A). In contrast, no effect was observed when the SUB1 DBD mutation was expressed in the rpb4Δ sub1Δ cells (Figure 5C, Supplementary Figure S2B). Thus, it is likely that Sub1 may need the CT region to interact with Rpb4/7 upon its association to promoters through the DBD. This agrees with some decrease of Sub1 crosslinking at the promoters in the absence of the CT region (Figure 5A, left panel). If our hypothesis is correct, we should observe that association of Sub1ΔCT-HA with gene promoters decreases when lacking RPB4. Indeed, this is what we observed when we performed ChIP assays in sub1ΔCT-HA and sub1ΔCT-HA rpb4Δ cells (Figure 5D). Sub1ΔCT-HA association with the gene promoters is significantly reduced in rpb4Δ cells. Though this reduction (∼60–50%) is lower than the one observed for Sub1–HA wt in the absence of Rpb4 (∼80%, Figure 3A), it supports our idea that Sub1-CT region may interact with Rpb4/7 to be stably associated with the chromatin. Moreover, our data corroborate the genetic interaction between SUB1 and RPB4 and offer insight into the function(s) of Sub1 CT region, which has not been identified previously.
Sub1 and Fcp1 are components of the same complex
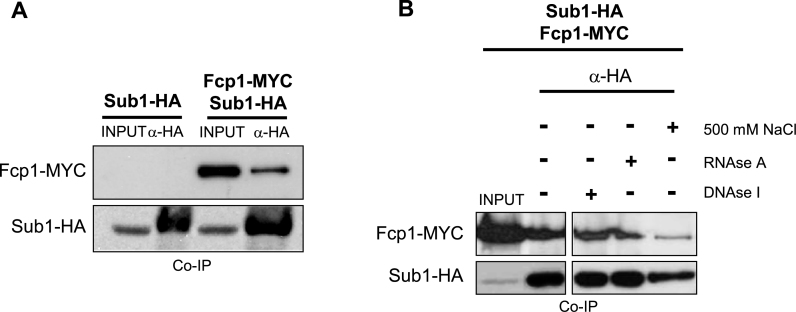
Several facts suggested that Sub1 and Fcp1 interact, at least as part of the same complex where the heterodimer Rpb4/7 is present. First, we reported that Sub1 is a regulator of Fcp1 and our data indicated that a function of Sub1 is to facilitate accumulation of Fcp1, likely by directly or indirectly increasing its stability. Additionally we showed that Fcp1 crosslinking to DNA was significantly reduced in the sub1Δ strain, with comparable reductions at both 5΄ coding and terminator regions (41). Second, a number of findings suggested that Rpb4/7 might recruit Fcp1 to regulate CTD modifications, among them our more recent work showing that Fcp1 association to chromatin is impaired during the entire transcription cycle in the absence of Rpb4 (12). Furthermore, in vitro binding and yeast two-hybrid assays in S. pombe and D. melanogaster, respectively, showed that Rpb4 interacts with Fcp1 (74,75). Structural and biochemical studies also suggest that in S. cerevisiae Fcp1 might also interact with RNAPII through Rpb4/7 (76). Third, here we have clearly shown that Rpb4/7 interact with Sub1.
To investigate if Sub1 and Fcp1 interact, we performed CoIPs assays. We detected Fcp1-MYC by IP of Sub1–HA (Figure 6A). Furthermore, CoIPs assays showed that Sub1-Fcp1 interaction is resistant to treatment with either DNAse I or RNAse A, and only strong washing conditions, such as high salt (500 mM NaCl), weakened their association (Figure 6B). These data strengthen the conclusion that Sub1 is functionally related to RNAPII CTD phosphorylation and indicates that this function is likely to be executed via its interaction with Rpb4/7 and Fcp1. Moreover, our data uncover the existence of a physical and functional connection between Sub1, Rpb4/7 and Fcp1 which is important to maintain proper RNAPII phosphorylation levels, key in the biogenesis of mRNAs.
Figure 6.
Sub1 and Fcp1 are components of the same complex. (A) Co-IP performed on WCEs from Sub1–HA and Sub1–HA Fcp1-MYC cells using an anti-HA antibody. Inputs and IPs were analyzed by Western blotting with antibodies to the indicated proteins (anti-MYC and anti-HA for Fcp1 and Sub1, respectively). (B) Same as in (A), except that in the fourth IP the beads were washed with lysis buffer containing 500 mM NaCl, and second and third IPs were treated with DNAse I and RNAse A, respectively.
A full length Rpb1-CTD is required for efficient Sub1 association to gene promoters
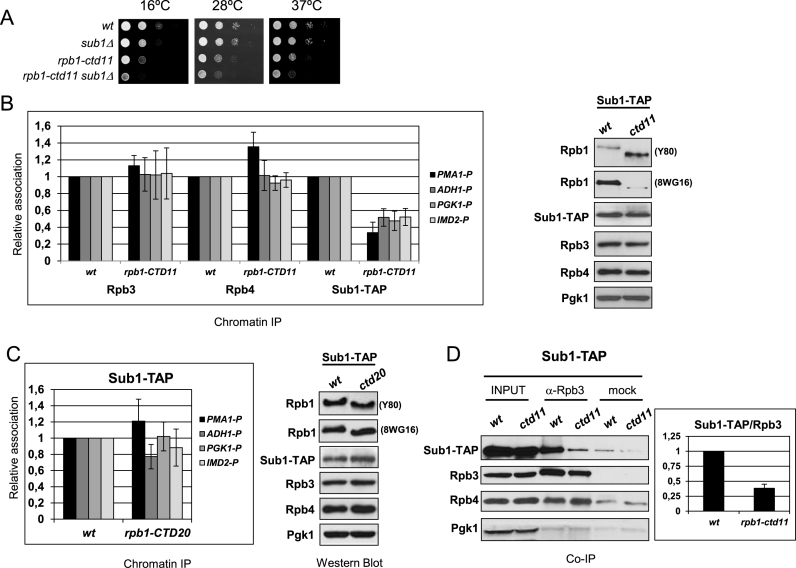
Our data suggest that Sub1 may regulate CTD phosphorylation through interaction with Rpb4/7 and Fcp1, based on demonstration that Rpb4/7 is required for CTD dephosphorylation by CTD phosphatases (12). In addition, in an extensive genetic interaction network, SUB1 and the CTD interact (77). This study revealed a number of significant genetic interactions as the CTD was progressively shortened. Indeed a negative genetic interaction between rpb1-CTD11 (with a CTD containing only eleven heptad repeats) and the SUB1 deletion was observed. Although we did not find a direct interaction between Sub1 and the CTD, we wondered if the CTD, as well as Rpb4/7, may contribute to Sub1 occupancy at promoters and to its association with RNAPII. To answer the question we took advantage of several CTD truncation mutants containing 11 and 20 repeats (rpb1-CTD11 and rpb1-CTD20), respectively (77). First, we deleted SUB1 in the rpb1-CTD11 mutant to confirm the genetic interaction. As shown in Figure 7A, deletion of SUB1 exacerbates the growth phenotype of the rpb1-CTD11 cells. We next decided to investigate if Sub1 association to gene promoters was dependent on the length of the CTD. For that purpose, we TAP tagged Sub1 in the RPB1-CTD wt, rpb1-CTD11 and rpb1-CTD20 strains and we analyzed Sub1–TAP occupancy by ChIP in several gene promoters (Figure 7B and C). Interestingly, the occupancy of Sub1 in these regions gets worse as the number of CTD repeats decreases. Thus, in rpb1-CTD11 cells, Sub1 occupancy of gene promoters is significantly diminished when compared to wt cells (Figure 7B, left panel) and this is not due to reduced Sub1 levels (Figure 7B, right panel). On the other hand, in rpb1-CTD20 cells, Sub1 association with chromatin diminishes only slightly (Figure 7C) compared to wt cells. Moreover, the effect on Sub1 occupancy dependent on CTD length is specific for Sub1, as no effect is observed for Rpb4 and Rpb3 crosslinking in rpb1-CTD11 cells (Figure 7B).
Figure 7.
A full length Rpb1-CTD is required for efficient Sub1 association to gene promoters. (A) Genetic interaction between sub1Δ and rpb1-CTD11. The sub1Δ deletion increases the slow growth phenotype of rpb1-CTD11 strain at 16, 28 and 37°C. Serial dilutions (1:10) of wt and mutant strains were spotted on rich medium and grown for 2–3 days at the indicated temperatures. (B) Chromatin immunoprecipitation (ChIP) analyses were performed using wt and rpb1-CTD11 strains. Left panel: Rpb3, Rpb4 and Sub1–TAP binding to the promoter (P) of three constitutively expressed genes, PMA1, ADH1 and PGK1, and to the promoter of the IMD2 inducible gene, was examined by qPCR. Results were quantitated (see Material and Methods), and relative Rpb3, Rpb4 and Sub1–TAP binding in rpb1-CTD11cells is plotted relative to that in wt cells (set equal to 1). Right panel: WCE were prepared from wild-type (wt) and rpb1-CTD11 strains expressing Sub1–TAP and analyzed by Western blotting using the indicated antibodies. (C) Same as in (B) for wt and rpb1-CTD20 strains. (D) Left panel: Co-IP performed on WCEs from Sub1–TAP cells (wt and rpb1-CTD11) using an anti-Rpb3 antibody. As a control, the same amount of cell extract was incubated with Protein A Sepharose alone. Inputs and IPs were analyzed by Western blotting with antibodies to the indicated proteins. Right panel: Values from the quantification of Sub1-TAP and Rpb3 immunoreactive signals from three experiments were calcuated and Sub1-TAP/Rpb3 mean ratios were plotted (arbitrary units), where error bars represent standard deviations.
We next asked if Sub1–TAP association to the RNAPII was also affected when the CTD has only eleven repeats. In this case, we performed CoIP assays using wt and rpb1-CTD11 whole cell extracts (Figure 7D). First, we analyzed levels of Rpb1 by western blot using whole cell extracts from wt, rpb1-CTD11 and rpb1-CTD20 cells. We used two different antibodies: 8WG16 directed against the CTD and Y-80 that recognizes the N-terminus of Rpb1 (Figure 7B, right panel and Figure 7C, right panel). We observed that levels of Rpb1 were significantly decreased in rpb1-CTD11 when analyzed with the 8WG16 antibody, as previously shown (77) and in agreement with the number of CTD repetitions (11 versus 25–26 in an otherwise wt cell). However, Y-80 antibody showed increased Rpb1 levels. These effects were not observed in the rpb1-CTD20 whole cell extracts, where Rpb1 levels are similar to the wt cells independently of the Rpb1 antibody used. We then performed Co-IP experiments using the anti-Rpb3 antibody, as no differences in Rpb3 levels were observed in the isogenic wt and rpb1-CTD11 cells. Very interestingly, and in agreement with the ChIP data, there is less Sub1–TAP associated to the RNAPII in the rpb1-CTD11 mutant than in the wt cells (Figure 7D). These results underscore the conclusion that Sub1 and the Rpb1-CTD are functionally related, which support the role of Sub1 in CTD phosphorylation (25). We conclude that Rpb4/7 and the Rpb1-CTD are important for Sub1 association with DNA. Furthermore, at least during transcription initiation, Sub1 appears to be localized to a region near the CTD and Rpb4/7 of RNAPII. These results support the conclusion that Sub1 affects clamp function and modulates CTD phosphorylation throughout the transcription cycle.
DISCUSSION
The data presented here provide novel insight into how the transcriptional coactivator Sub1 modulates RNAPII transcription through interaction with Rpb4/7. Our findings that Sub1 directly interacts with the Rpb4/7 heterodimer, associates with Fcp1, and is genetically and functionally related to the CTD, explain how Sub1 modulates RNAPII phosphorylation, which crucially regulates the biogenesis of mRNAs. We also provide evidence indicating that Sub1 contributes to RNAPII clamp function elucidating Sub1 role during the transition from the open to the closed complex formation, thus facilitating transcription elongation.
Sub1, as a PIC component, directly interacts with Rpb4/7
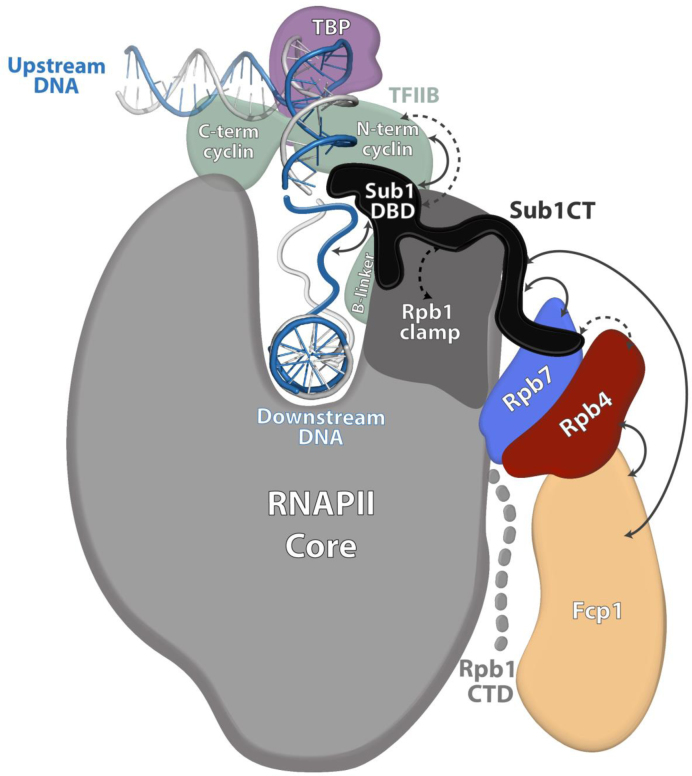
We show that Sub1 interacts with Rpb4/7, most likely via Rpb7 because Sub1 binds RNAPII in the absence of Rpb4 (Figure 4). However, a fully functional heterodimer seems to be important for Sub1 association with chromatin, as Sub1 crosslinking to gene promoters is significantly reduced in the rpb4 null mutant (Figure 3). It is likely that the reduced levels of Rpb7 in the rpb4Δ cells account for Sub1 chromatin association defects. The apparent discrepancy between the CoIP and ChIP experiments suggests that, while Rpb4 is not required for Sub1 interaction with RNAPII, a functional Rpb4/7 is necessary for Sub1 to stably associate with chromatin after RNAPII recruitment to the PIC. We propose a model where Sub1 is recruited to RNAPII through interaction with Rpb4/7, TFIIB (36,50,78) and DNA. Once at the PIC, Sub1 interacts with Rpb4/7 via its C-terminal region (Figures 5 and 8) to keep associated to RNAPII and chomatin.
Figure 8.
Schematic model showing the hypothetical localization of Sub1 during transcription initiation. Sub1 is bound to the promoter by interacting with upstream DNA at the junction between single- and double-stranded DNA (36) through its DNA Binding Domain (Sub1 DBD) (Figure 5). The proposed localization of Sub1 in this model explains the reported physical (solid arrow) and genetic (dashed arrow) interaction of Sub1 with TFIIB (35,50). The intrinsically-disordered C-terminal domain of Sub1 (Sub1 CT) may extend to directly interact with the Rpb4/7 heterodimer, as suggested by the specific genetic interaction between this domain and RPB4 (Figure 5), and the physical direct interaction with Rpb7 (Figure 4). The genetic interaction between Sub1 and Rpb1 clamp (Figure 2C) is also illustrated. In addition, the model shows the connections of Fcp1 phosphatase with Rpb4 and Sub1 revealed by our genetic (Allepuz-Fuster et al., Calvo & Manley, 2005) and CoIP assays (Figure 6), and consistent with the structural data (76). Our proposed model suggests that Sub1 may influence Rpb1-CTD phosphorylation (25) and Rpb1 clamp functionality through its interaction with Rpb4/7, which could also explain the already suggested role of Sub1 both during transcription initiation (36,37) and elongation (38).
SUB1 was originally isolated as an allele-specific suppressor of two TFIIB mutations (E62G and R78H) (35), located within the B-reader region (helix and loop, respectively) (79). Deletion of SUB1 is synthetically lethal when combined with these two mutations and, in agreement, overexpression of SUB1 is required for suppression of their cold sensitive phenotypes (35). Moreover, a specific interaction between Sub1 and TFIIB has also been described (35); and TFIIB is required for Sub1 recruitment to the promoters of constitutively transcribed genes (78).
Sub1 was subsequently identified as a PIC component (36). In this elegant study, ChIP data suggest that Sub1 mainly localizes to the promoter region of active genes in a manner dependent upon TBP. Additionally, FeBABE cleavage experiments indicate that Sub1 is located near the leading edge of the HIS4 transcription bubble, which is also close to the TFIIB-linker helix (79,80). This is in agreement with a direct interaction between Sub1 and TFIIB (35). They proposed a model where Sub1 is first recruited to the PIC by protein interactions, likely with TFIIB, and at that point both factors would cooperate in promoter melting. Then, the interaction of Sub1 with DNA would stabilize the open complex, thus promoting transcription initiation and promoter clearance. Hence, in this model, Sub1, upon promoter melting, can interact with the non-template strand or perhaps both strands at the upstream junction between single- and double-stranded DNA. In fact, the residues mutated in TFIIB that specifically interact with SUB1 are in the B-reader, which has been proposed to act in the capture of the template strand within the RNAPII active site (79).
Genetic data also suggest that Sub1, once bound to gene promoters, could help TFIIE and TFIIH to maintain the PIC in a stable but inactive conformation in the open complex (36). At this point, we propose that Sub1 may interact with Rpb4/7. One hypothesis is that the interaction is mediated by the carboxy-terminal region of Sub1. The intrinsically-disordered nature of the Sub1 C-terminal domain could allow the protein to span the distance between the bubble upstream junction and the RNAPII stalk (Figure 8). This interaction would help to maintain Sub1 associated to gene promoters until the next step in transcription. Supporting this idea, we identified a specific genetic interaction between sub1ΔCT and rpb4Δ (Figure 5B), and showed that the association of Sub1ΔCT with gene promoters is significantly reduced in the absence of RPB4 (Figure 5D). In contrast, a mutation altering Sub1 DNA binding does not affect growth of rpb4Δ mutants (Figure 5D). This is the first evidence of a role for the CT region of Sub1, which is not conserved in human PC4, and that also provides insight into the mechanism used by Sub1 to function beyond the PIC, later on transcription. Furthermore, this is also the first demonstration that Sub1 directly interacts with RNAPII at least through the Rpb4/7 stalk.
Additionally, our data showing a genetic interaction between SUB1, RPB4 and the RPB1 foot domain (Figure 2A and B), suggest that the function of Sub1 is negative when the integrity of RNAPII is compromised, such as in the case of rpb4Δ and rpo21-4 mutants (51). One likely hypothesis is that Sub1 has a negative role during transcription initiation, repressing transcription when PIC complexes are not well assembled and/or the integrity of the polymerase is altered. Therefore, in the rpb4Δ sub1Δ and rpo21-4 sub1Δ double mutants this repression would be abolished, enabling RNAPII to circumvent initiation defects due to rpb4Δ (81) and rpo21-4 (51), allowing better cell growth (Figure 2A and B). On the other hand, and in agreement with this idea, overexpression of SUB1 significantly aggravates rpb4Δ growth (Figure 2A). Our genetic data are also consistent with the proposed models for Sub1 function helping to maintain a stable PIC conformation (25,36), and its putative localization within the RNAPII-DNA complex at the initiation step (Figure 8).
In summary, our data and that of others (35,36,50) allow us to propose a model of Sub1 within the initiation complex (Figure 8). Sub1 binds DNA through its N-terminal DBD, while is stably maintained in the RNAPII-DNA complex by binding to Rpb4/7 via its CT region (Figure 8). Association of Sub1 with Fcp1 explains the role of Sub1 in the modulation of CTD phosphorylation during the entire transcription cycle (25). Our results here strongly suggest that Sub1 directly interacts with Rpb7, while Fcp1 interacts with Rpb4 to modulate RNAPII CTD phosphorylation (12). In addition, Sub1 interaction with Rpb4/7 and the Rpb1 clamp domain may help to explain how it may influence TSS selection (37) and transcription elongation rate together with Spt5 (38). This is supported by the genetic interactions between SUB1 deletion and the rpb1-L1397S mutation localized within the clamp domain (Figure 2C, and (52), and by the fact that Spt5 associates with Sub1, and interacts with Rpb4/7 and the Rpb1 clamp domain (64,65).
Our study provides significant information about RNAPII transcription regulation by Sub1 and Rpb4/7, and suggests a model that could be very helpful for future structural and functional studies on RNAPII transcription machinery.
Supplementary Material
ACKNOWLEDGEMENTS
We thank A. García for technical help with Sub1–TAP purification, and A. Collin for sharing information about sub1-ΔCT strain and Sub1 bacmid construction. Y. Takagi for plasmid to co-express recombinant Rpb4/7 proteins, and M. Kobor and F. Navarro for yeast strains. We also thank M. Hampsey for helpful discussions on the manuscript, and J.L. Manley for advice.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Spanish Ministry of Economy and Competitiveness (MINECO) [BFU2013-48374-P]; Predoctoral fellowships from MINECO (to J.A.L.); Technician Formation Program from the Spanish National Research Council (CSIC) (to N.G.-P.). Funding for open access charge: MINECO [BFU2013-48374-P].
Conflict of interest statement. None declared.
REFERENCES
- 1.Carlson M. Genetics of transcriptional regulation in yeast: connections to the RNA polymerase II CTD. Annu. Rev. Cell Dev. Biol. 1997; 13:1–23. [DOI] [PubMed] [Google Scholar]
- 2.Dahmus M.E. Reversible phosphorylation of the C-terminal domain of RNA polymerase II. J. Biol. Chem. 1996; 271:19009–19012. [DOI] [PubMed] [Google Scholar]
- 3.Phatnani H.P., Greenleaf A.L.. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006; 20:2922–2936. [DOI] [PubMed] [Google Scholar]
- 4.Nonet M., Sweetser D., Young R.A.. Functional redundancy and structural polymorphism in the large subunit of RNA polymerase II. Cell. 1987; 50:909–915. [DOI] [PubMed] [Google Scholar]
- 5.Cho E.J. RNA polymerase II carboxy-terminal domain with multiple connections. Exp. Mol. Med. 2007; 39:247–254. [DOI] [PubMed] [Google Scholar]
- 6.Hirose Y., Manley J.L.. RNA polymerase II and the integration of nuclear events. Genes Dev. 2000; 14:1415–1429. [PubMed] [Google Scholar]
- 7.Meinhart A., Kamenski T., Hoeppner S., Baumli S., Cramer P.. A structural perspective of CTD function. Genes Dev. 2005; 19:1401–1415. [DOI] [PubMed] [Google Scholar]
- 8.Buratowski S. Connections between mRNA 3΄ end processing and transcription termination. Curr. Opin. Cell Biol. 2005; 17:257–261. [DOI] [PubMed] [Google Scholar]
- 9.Murakami K., Tsai K.L., Kalisman N., Bushnell D.A., Asturias F.J., Kornberg R.D.. Structure of an RNA polymerase II preinitiation complex. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:13543–13548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Plaschka C., Lariviere L., Wenzeck L., Seizl M., Hemann M., Tegunov D., Petrotchenko E.V., Borchers C.H., Baumeister W., Herzog F. et al. . Architecture of the RNA polymerase II-Mediator core initiation complex. Nature. 2015; 518:376–380. [DOI] [PubMed] [Google Scholar]
- 11.Robinson P.J., Trnka M.J., Bushnell D.A., Davis R.E., Mattei P.J., Burlingame A.L., Kornberg R.D.. Structure of a complete mediator-RNA polymerase II pre-initiation complex. Cell. 2016; 166:1411–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allepuz-Fuster P., Martinez-Fernandez V., Garrido-Godino A.I., Alonso-Aguado S., Hanes S.D., Navarro F., Calvo O.. Rpb4/7 facilitates RNA polymerase II CTD dephosphorylation. Nucleic Acids Res. 2014; 42:13674–13688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eick D., Geyer M.. The RNA polymerase II carboxy-terminal domain (CTD) code. Chem. Rev. 2013; 113:8456–8490. [DOI] [PubMed] [Google Scholar]
- 14.Heidemann M., Hintermair C., Voss K., Eick D.. Dynamic phosphorylation patterns of RNA polymerase II CTD during transcription. Biochim. Biophys. Acta. 2013; 1829:55–62. [DOI] [PubMed] [Google Scholar]
- 15.Chapman R.D., Heidemann M., Hintermair C., Eick D.. Molecular evolution of the RNA polymerase II CTD. Trends Genet. 2008; 24:289–296. [DOI] [PubMed] [Google Scholar]
- 16.Schroder S., Herker E., Itzen F., He D., Thomas S., Gilchrist D.A., Kaehlcke K., Cho S., Pollard K.S., Capra J.A. et al. . Acetylation of RNA polymerase II regulates growth-factor-induced gene transcription in mammalian cells. Mol. Cell. 2013; 52:314–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kelly W.G., Dahmus M.E., Hart G.W.. RNA polymerase II is a glycoprotein. Modification of the COOH-terminal domain by O-GlcNAc. J. Biol. Chem. 1993; 268:10416–10424. [PubMed] [Google Scholar]
- 18.Ranuncolo S.M., Ghosh S., Hanover J.A., Hart G.W., Lewis B.A.. Evidence of the involvement of O-GlcNAc-modified human RNA polymerase II CTD in transcription in vitro and in vivo. J. Biol. Chem. 2012; 287:23549–23561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sims R.J., Rojas L.A., Beck D., Bonasio R., Schuller R., Drury W.J., Eick D., Reinberg D.. The C-terminal domain of RNA polymerase II is modified by site-specific methylation. Science. 2011; 332:99–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buratowski S. The CTD code. Nat. Struct. Biol. 2003; 10:679–680. [DOI] [PubMed] [Google Scholar]
- 21.Buratowski S. Progression through the RNA polymerase II CTD cycle. Mol. Cell. 2009; 36:541–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsin J.P., Manley J.L.. The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev. 2012; 26:2119–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jeronimo C., Bataille A.R., Robert F.. The writers, readers, and functions of the RNA polymerase II C-terminal domain code. Chem. Rev. 2013; 113:8491–8522. [DOI] [PubMed] [Google Scholar]
- 24.Schreieck A., Easter A.D., Etzold S., Wiederhold K., Lidschreiber M., Cramer P., Passmore L.A.. RNA polymerase II termination involves C-terminal-domain tyrosine dephosphorylation by CPF subunit Glc7. Nat. Struct. Mol. Biol. 2014; 21:175–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garcia A., Rosonina E., Manley J.L., Calvo O.. Sub1 globally regulates RNA polymerase II C-terminal domain phosphorylation. Mol. Cell. Biol. 2010; 30:5180–5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanes S.D. The Ess1 prolyl isomerase: Traffic cop of the RNA polymerase II transcription cycle. Biochim. Biophys. Acta. 2014; 1839:316–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verdecia M.A., Bowman M.E., Lu K.P., Hunter T., Noel J.P.. Structural basis for phosphoserine-proline recognition by group IV WW domains. Nat. Struct. Biol. 2000; 7:639–643. [DOI] [PubMed] [Google Scholar]
- 28.Wu X., Wilcox C.B., Devasahayam G., Hackett R.L., Arevalo-Rodriguez M., Cardenas M.E., Heitman J., Hanes S.D.. The Ess1 prolyl isomerase is linked to chromatin remodeling complexes and the general transcription machinery. EMBO J. 2000; 19:3727–3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ge H., Roeder R.G.. Purification, cloning, and characterization of a human coactivator, PC4, that mediates transcriptional activation of class II genes. Cell. 1994; 78:513–523. [DOI] [PubMed] [Google Scholar]
- 30.Kaiser K., Stelzer G., Meisterernst M.. The coactivator p15 (PC4) initiates transcriptional activation during TFIIA-TFIID-promoter complex formation. EMBO J. 1995; 14:3520–3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kretzschmar M., Kaiser K., Lottspeich F., Meisterernst M.. A novel mediator of class II gene transcription with homology to viral immediate-early transcriptional regulators. Cell. 1994; 78:525–534. [DOI] [PubMed] [Google Scholar]
- 32.Malik S., Guermah M., Roeder R.G.. A dynamic model for PC4 coactivator function in RNA polymerase II transcription. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:2192–2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Werten S., Stelzer G., Goppelt A., Langen F.M., Gros P., Timmers H.T., Van der Vliet P.C., Meisterernst M.. Interaction of PC4 with melted DNA inhibits transcription. EMBO J. 1998; 17:5103–5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Henry N.L., Bushnell D.A., Kornberg R.D.. A yeast transcriptional stimulatory protein similar to human PC4. J. Biol. Chem. 1996; 271:21842–21847. [DOI] [PubMed] [Google Scholar]
- 35.Knaus R., Pollock R., Guarente L.. Yeast SUB1 is a suppressor of TFIIB mutations and has homology to the human co-activator PC4. EMBO J. 1996; 15:1933–1940. [PMC free article] [PubMed] [Google Scholar]
- 36.Sikorski T.W., Ficarro S.B., Holik J., Kim T., Rando O.J., Marto J.A., Buratowski S.. Sub1 and RPA associate with RNA polymerase II at different stages of transcription. Mol. Cell. 2011; 44:397–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Braberg H., Jin H., Moehle E.A., Chan Y.A., Wang S., Shales M., Benschop J.J., Morris J.H., Qiu C., Hu F. et al. . From structure to systems: high-resolution, quantitative genetic analysis of RNA polymerase II. Cell. 2013; 154:775–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garcia A., Collin A., Calvo O.. Sub1 associates with Spt5 and influences RNA polymerase II transcription elongation rate. Mol. Biol. Cell. 2012; 23:4297–4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Calvo O., Manley J.L.. Evolutionarily conserved interaction between CstF-64 and PC4 links transcription, polyadenylation, and termination. Mol. Cell. 2001; 7:1013–1023. [DOI] [PubMed] [Google Scholar]
- 40.He X., Khan A.U., Cheng H., Pappas D.L. Jr, Hampsey M., Moore C.L.. Functional interactions between the transcription and mRNA 3΄ end processing machineries mediated by Ssu72 and Sub1. Genes Dev. 2003; 17:1030–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Calvo O., Manley J.L.. The transcriptional coactivator PC4/Sub1 has multiple functions in RNA polymerase II transcription. EMBO J. 2005; 24:1009–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burke D., Dawson D., Stearns T. Methods in Yeast Genetics. 2000; NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- 43.Pascual-Garcia P., Govind C.K., Queralt E., Cuenca-Bono B., Llopis A., Chavez S., Hinnebusch A.G., Rodriguez-Navarro S.. Sus1 is recruited to coding regions and functions during transcription elongation in association with SAGA and TREX2. Genes Dev. 2008; 22:2811–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sakurai H., Mitsuzawa H., Kimura M., Ishihama A.. The Rpb4 subunit of fission yeast Schizosaccharomyces pombe RNA polymerase II is essential for cell viability and similar in structure to the corresponding subunits of higher eukaryotes. Mol. Cell. Biol. 1999; 19:7511–7518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Calvo O.G., García A. RNA polymerase II phosphorylation and gene expression regulation. Protein Phosphorylation in Human Health. 2012; Rijeka: InTech; 151–194. [Google Scholar]
- 46.Schmitt M.E., Brown T.A., Trumpower B.L.. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990; 18:3091–3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lindstrom D.L., Squazzo S.L., Muster N., Burckin T.A., Wachter K.C., Emigh C.A., McCleery J.A., Yates J.R., Hartzog G.A.. Dual roles for Spt5 in pre-mRNA processing and transcription elongation revealed by identification of Spt5-associated proteins. Mol. Cell. Biol. 2003; 23:1368–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McKune K., Richards K.L., Edwards A.M., Young R.A., Woychik N.A.. RPB7, one of two dissociable subunits of yeast RNA polymerase II, is essential for cell viability. Yeast. 1993; 9:295–299. [DOI] [PubMed] [Google Scholar]
- 49.Woychik N.A., Young R.A.. RNA polymerase II subunit RPB4 is essential for high- and low-temperature yeast cell growth. Mol. Cell. Biol. 1989; 9:2854–2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu W.H., Pinto I., Chen B.S., Hampsey M.. Mutational analysis of yeast TFIIB. A functional relationship between Ssu72 and Sub1/Tsp1 defined by allele-specific interactions with TFIIB. Genetics. 1999; 153:643–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garrido-Godino A.I., Garcia-Lopez M.C., Navarro F.. Correct assembly of RNA polymerase II depends on the foot domain and is required for multiple steps of transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 2013; 33:3611–3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kwapisz M., Wery M., Despres D., Ghavi-Helm Y., Soutourina J., Thuriaux P., Lacroute F.. Mutations of RNA polymerase II activate key genes of the nucleoside triphosphate biosynthetic pathways. EMBO J. 2008; 27:2411–2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scafe C., Martin C., Nonet M., Podos S., Okamura S., Young R.A.. Conditional mutations occur predominantly in highly conserved residues of RNA polymerase II subunits. Mol. Cell. Biol. 1990; 10:1270–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Armache K.J., Mitterweger S., Meinhart A., Cramer P.. Structures of complete RNA polymerase II and its subcomplex, Rpb4/7. J. Biol. Chem. 2005; 280:7131–7134. [DOI] [PubMed] [Google Scholar]
- 55.Garcia-Lopez M.C., Pelechano V., Miron-Garcia M.C., Garrido-Godino A.I., Garcia A., Calvo O., Werner M., Perez-Ortin J.E., Navarro F.. The conserved foot domain of RNA pol II associates with proteins involved in transcriptional initiation and/or early elongation. Genetics. 2011; 189:1235–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cramer P., Bushnell D.A., Kornberg R.D.. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science. 2001; 292:1863–1876. [DOI] [PubMed] [Google Scholar]
- 57.Archambault J., Schappert K.T., Friesen J.D.. A suppressor of an RNA polymerase II mutation of Saccharomyces cerevisiae encodes a subunit common to RNA polymerases I, II, and III. Mol. Cell. Biol. 1990; 10:6123–6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Archambault J., Drebot M.A., Stone J.C., Friesen J.D.. Isolation and phenotypic analysis of conditional-lethal, linker-insertion mutations in the gene encoding the largest subunit of RNA polymerase II in Saccharomyces cerevisiae. Mol. Gen. Genet. 1992; 232:408–414. [DOI] [PubMed] [Google Scholar]
- 59.Armache K.J., Kettenberger H., Cramer P.. Architecture of initiation-competent 12-subunit RNA polymerase II. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:6964–6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Grohmann D., Werner F.. Hold on!: RNA polymerase interactions with the nascent RNA modulate transcription elongation and termination. RNA Biol. 2010; 7:310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Grohmann D., Werner F.. Cycling through transcription with the RNA polymerase F/E (RPB4/7) complex: structure, function and evolution of archaeal RNA polymerase. Res. Microbiol. 2011; 162:10–18. [DOI] [PubMed] [Google Scholar]
- 62.Gnatt A.L., Cramer P., Fu J., Bushnell D.A., Kornberg R.D.. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 A resolution. Science. 2001; 292:1876–1882. [DOI] [PubMed] [Google Scholar]
- 63.Jenks M.H., O'Rourke T.W., Reines D.. Properties of an intergenic terminator and start site switch that regulate IMD2 transcription in yeast. Mol. Cell. Biol. 2008; 28:3883–3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li W., Giles C., Li S.. Insights into how Spt5 functions in transcription elongation and repressing transcription coupled DNA repair. Nucleic Acids Res. 2014; 42:7069–7083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Martinez-Rucobo F.W., Sainsbury S., Cheung A.C., Cramer P.. Architecture of the RNA polymerase-Spt4/5 complex and basis of universal transcription processivity. EMBO J. 2011; 30:1302–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Edwards A.M., Kane C.M., Young R.A., Kornberg R.D.. Two dissociable subunits of yeast RNA polymerase II stimulate the initiation of transcription at a promoter in vitro. J. Biol. Chem. 1991; 266:71–75. [PubMed] [Google Scholar]
- 67.Sheffer A., Varon M., Choder M.. Rpb7 can interact with RNA polymerase II and support transcription during some stresses independently of Rpb4. Mol. Cell. Biol. 1999; 19:2672–2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Runner V.M., Podolny V., Buratowski S.. The Rpb4 subunit of RNA polymerase II contributes to cotranscriptional recruitment of 3΄ processing factors. Mol. Cell. Biol. 2008; 28:1883–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brandsen J., Werten S., van der Vliet P.C., Meisterernst M., Kroon J., Gros P.. C-terminal domain of transcription cofactor PC4 reveals dimeric ssDNA binding site. Nat. Struct. Biol. 1997; 4:900–903. [DOI] [PubMed] [Google Scholar]
- 70.Wang Z., Roeder R.G.. DNA topoisomerase I and PC4 can interact with human TFIIIC to promote both accurate termination and transcription reinitiation by RNA polymerase III. Mol. Cell. 1998; 1:749–757. [DOI] [PubMed] [Google Scholar]
- 71.Mortusewicz O., Roth W., Li N., Cardoso M.C., Meisterernst M., Leonhardt H.. Recruitment of RNA polymerase II cofactor PC4 to DNA damage sites. J. Cell Biol. 2008; 183:769–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pan Z.Q., Ge H., Amin A.A., Hurwitz J.. Transcription-positive cofactor 4 forms complexes with HSSB (RPA) on single-stranded DNA and influences HSSB-dependent enzymatic synthesis of simian virus 40 DNA. J. Biol. Chem. 1996; 271:22111–22116. [DOI] [PubMed] [Google Scholar]
- 73.Werten S., Wechselberger R., Boelens R., van der Vliet P.C., Kaptein R.. Identification of the single-stranded DNA binding surface of the transcriptional coactivator PC4 by NMR. J. Biol. Chem. 1999; 274:3693–3699. [DOI] [PubMed] [Google Scholar]
- 74.Kimura M., Suzuki H., Ishihama A.. Formation of a carboxy-terminal domain phosphatase (Fcp1)/TFIIF/RNA polymerase II (pol II) complex in Schizosaccharomyces pombe involves direct interaction between Fcp1 and the Rpb4 subunit of pol II. Mol. Cell. Biol. 2002; 22:1577–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tombacz I., Schauer T., Juhasz I., Komonyi O., Boros I.. The RNA Pol II CTD phosphatase Fcp1 is essential for normal development in Drosophila melanogaster. Gene. 2009; 446:58–67. [DOI] [PubMed] [Google Scholar]
- 76.Kamenski T., Heilmeier S., Meinhart A., Cramer P.. Structure and mechanism of RNA polymerase II CTD phosphatases. Mol. Cell. 2004; 15:399–407. [DOI] [PubMed] [Google Scholar]
- 77.Aristizabal M.J., Negri G.L., Benschop J.J., Holstege F.C., Krogan N.J., Kobor M.S.. High-throughput genetic and gene expression analysis of the RNAPII-CTD reveals unexpected connections to SRB10/CDK8. PLoS Genet. 2013; 9:e1003758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rosonina E., Willis I.M., Manley J.L.. Sub1 functions in osmoregulation and in transcription by both RNA polymerases II and III. Mol. Cell. Biol. 2009; 29:2308–2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kostrewa D., Zeller M.E., Armache K.J., Seizl M., Leike K., Thomm M., Cramer P.. RNA polymerase II-TFIIB structure and mechanism of transcription initiation. Nature. 2009; 462:323–330. [DOI] [PubMed] [Google Scholar]
- 80.Liu X., Bushnell D.A., Wang D., Calero G., Kornberg R.D.. Structure of an RNA polymerase II-TFIIB complex and the transcription initiation mechanism. Science. 2010; 327:206–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sharma N., Kumari R.. Rpb4 and Rpb7: multifunctional subunits of RNA polymerase II. Crit. Rev. Microbiol. 2013; 39:362–372. [DOI] [PubMed] [Google Scholar]
- 82.Koyama H., Sumiya E., Nagata M., Ito T., Sekimizu K.. Transcriptional repression of the IMD2 gene mediated by the transcriptional co-activator Sub1. Genes Cells. 2008; 13:1113–1126. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.