Abstract
We developed an algorithm, HMZDelFinder, that uses whole exome sequencing (WES) data to identify rare and intragenic homozygous and hemizygous (HMZ) deletions that may represent complete loss-of-function of the indicated gene. HMZDelFinder was applied to 4866 samples in the Baylor–Hopkins Center for Mendelian Genomics (BHCMG) cohort and detected 773 HMZ deletion calls (567 homozygous or 206 hemizygous) with an estimated sensitivity of 86.5% (82% for single-exonic and 88% for multi-exonic calls) and precision of 78% (53% single-exonic and 96% for multi-exonic calls). Out of 773 HMZDelFinder-detected deletion calls, 82 were subjected to array comparative genomic hybridization (aCGH) and/or breakpoint PCR and 64 were confirmed. These include 18 single-exon deletions out of which 8 were exclusively detected by HMZDelFinder and not by any of seven other CNV detection tools examined. Further investigation of the 64 validated deletion calls revealed at least 15 pathogenic HMZ deletions. Of those, 7 accounted for 17–50% of pathogenic CNVs in different disease cohorts where 7.1–11% of the molecular diagnosis solved rate was attributed to CNVs. In summary, we present an algorithm to detect rare, intragenic, single-exon deletion CNVs using WES data; this tool can be useful for disease gene discovery efforts and clinical WES analyses.
INTRODUCTION
Copy number variants (CNVs) contribute to a substantial fraction of human genetic variation and are increasingly implicated in disease associations and human gene and genome evolution (1). CNVs have been found to be causal for many human disease phenotypes, including dozens of genomic disorders and hundreds of known Mendelian disease traits (2,3). Homozygous and hemizygous (HMZ) whole- and partial-gene deletions often result in null alleles and a complete loss of gene function (4). Although HMZ deletions constitute only a subset of all clinically relevant CNVs, they can play a major role in the discovery of novel Mendelian genes (5–9). In addition, heterozygous deletions involving recessive disease genes are an important part of an individual's recessive carrier status (10) and also directly contribute to disease by introducing compound heterozygous states where a deletion on one chromosome homologue coincides in genomic position with a loss of function or hypomorphic single nucleotide variant (SNV) allele on the other homologue (11–15).
Whole exome sequencing (WES) targets approximately 1% of the human genome (exons) coding for protein and it is enriched for disease-associated variants. The WES approach directly detects SNVs and very short (<50 bp) insertions or deletions (InDels), and also provides an opportunity for the detection of larger CNVs (16). The read depth information from WES data is a potential indicator of copy number information. However, unavoidable biases in exome capture technology and variability in sequencing efficiency in WES data of individual genomes present a challenge for inferring undistorted copy number information from simple summaries of sequencing data.
Current available tools for the detection of CNVs from WES data (17,18) are capable of identifying CNVs encompassing three or more exons, but can have high false positive rates (19). Distortions in read depth that vary by capture region and hybridization make detection of deletions and duplications as small as a single exon a difficult challenge; the former ‘single-exon HMZ CNV detection’ being the focus of the work presented here.
CNV calling methods from WES data try to remove the systematic experimental variations in capture and sequencing by normalization approaches. CNV-calling algorithms apply different normalization methods that include: (i) principal component analysis in XHMM (17), (ii) singular value decomposition in CoNIFER (18), (iii) a generalized additive model in CoNVex (ftp://ftp.sanger.ac.uk/pub/users/pv1/CoNVex/Docs/CoNVex.pdf), (iv) log-linear decomposition in CODEX (20), (v) selection of a highly correlated reference sample set for each sample in CANOES (21) and CLAMMS (22) and (vi) comparison of each exon's depth to its gene's median depth in ExonDel (23). These normalization methods enable a more linear correlation between read depth and inferred copy number. The drawbacks include a requirement for large sample collections as input, which can present computational challenges, and an increased risk of removing true signal from the data, which affects detection of small and rare CNVs.
Inherent depth-of-coverage fluctuations can be overcome by using excessive depth of coverage (for instance >850x) (24). However, this costly approach cannot be implemented retrospectively in the analyses of large-scale WES studies, which typically vary in average depth of coverage between 40x and 100x in both research and clinical diagnostic laboratories (25).
Here, we developed a new algorithm, HMZDelFinder, to identify intragenic rare variant HMZ deletion CNVs potentially contributing to Mendelian disease. This algorithm extracts different data sources from WES. These data include: (i) read count information from BAM files and (ii) zygosity information from VCF files. The read count information from BAM files is jointly called from all the samples in the cohort, which enables potential exonic rare HMZ deletions to be identified, whereas it allows exclusion of exons with a low depth-of-coverage. The VCF files are used to cull B-allele frequency information per exome, which enables the identification of regions of absence of heterozygosity (AOH) consistent with inherited copy number neutral genomic segments in which rare homozygous deletions may be embedded; i.e. identity by descent. Joint sample calling per exon aims to reduce false-negative calls for small (e.g. single exon) CNVs whereas information about AOH genomic intervals is anticipated to potentially further reduce false-positive calls.
We applied HMZDelFinder to the analysis of WES data from 4866 subjects (including 2580 males and 2286 females) enrolled in the Baylor–Hopkins Center for Mendelian Genomics (BHCMG) cohort. We identified 773 deletion calls including 567 homozygous and 206 hemizygous (i.e. X-chromosome in males) deletion CNVs with an estimated sensitivity of 86.5% (82% for single-exonic calls and 88% for multi-exonic calls) and precision of 78% (53% single-exonic calls and 96% for multi-exonic calls) as informed by orthogonal experimental validation of selected genomic deletion calls. Additional evaluation, performed on 50 samples from the 1000 Genomes Project (1000GP) data and analyses of inheritance using trio data confirmed the high sensitivity and precision of HMZDelFinder. Finally, the comparison of HMZDelFinder to other CNV calling algorithms (CoNIFER (18), CoNVex (ftp://ftp.sanger.ac.uk/pub/users/pv1/CoNVex/Docs/CoNVex.pdf), XHMM (17), ExonDel (23), CANOES (21), CLAMMS (22) and CODEX (20)) revealed that HMZDelFinder performed quantitatively better with respect to the detection of rare and small intragenic HMZ deletions; particularly those spanning only a single exon. The HMZDelFinder-detected rare intragenic CNV can have utility in research gene discovery efforts (14,15,26), and may be relevant to clinical genomic diagnostics.
MATERIALS AND METHODS
Input data
DNA samples were processed according to protocols previously described (27). Sequencing was performed in the Human Genome Sequencing Center (HGSC) using Illumina Hi-Seq (San Diego, CA, USA) instruments after exome capture with HGSC VCRome (1901 samples) or the HGSC CORE (2965 samples) designs. To minimize the influence of differences between the two designs on the results of the CNV detection method, we identified the intersection of the capture designs and excluded exons/targets located outside the regions of overlap. Personal genome sequence was achieved at an average depth-of-coverage of 95X, with >92% of the targeted bases having >20 reads. Raw sequence data were post-processed using the Mercury pipeline (28). The Mercury pipeline performs conversion of raw sequencing data (bcl files) to the fastq format using Casava, mapping of the short reads against a human genome reference sequence (GRCh37) by the Burrows–Wheeler alignment, recalibration using GATK (29) and variant calling using the Atlas2 suite (30). The Mercury pipeline is available in the cloud via DNAnexus (http://blog.dnanexus.com/2013-10-22-run-mercury-variant-calling-pipeline/).
Extraction of read depth from BAM files and preprocessing of VCFs to identify AOH segments
For input, the algorithm used BAM and corresponding VCF files generated on 4866 samples (2580 males and 2286 females) sequenced at the BHCMG, part of the Centers for Mendelian Genomics (31,32). Each individual genome BAM file was transformed into per-exon read depth (reads per thousand base pairs per million reads; RPKM) using a custom R script and the featureCount function implemented in the Bioconductor R package Rsubread (33). VCF files, from each individual personal genome, were used to identify regions of AOH using the following algorithm: first, from all SNVs that passed quality filters in the single VCF, we extracted a B-allele frequency (i.e. variant reads/total reads ratio); next, we transformed this ratio by subtracting 0.5 and taking the absolute value for each data point. After such a transformation, values > 0.45 were considered indicative of homozygous variants (expected value is 0.5) corresponding to either alternative or reference alleles, whereas lower values likely indicate heterozygous alleles. Transformed B-allele frequency data were then processed using circular binary segmentation (CBS) implemented in the DNAcopy R Bioconductor package (34). In summary, segments with the mean signal > 0.45 and size >1 kb were classified as AOH regions and submitted to further CNV analysis. Since the output of CBS may contain gaps between segments (i.e. regions with no SNVs that for example may represent HMZ deletions), the identified AOH regions were extended to include adjacent gaps.
Overview of the deletion CNV detection analysis pipeline
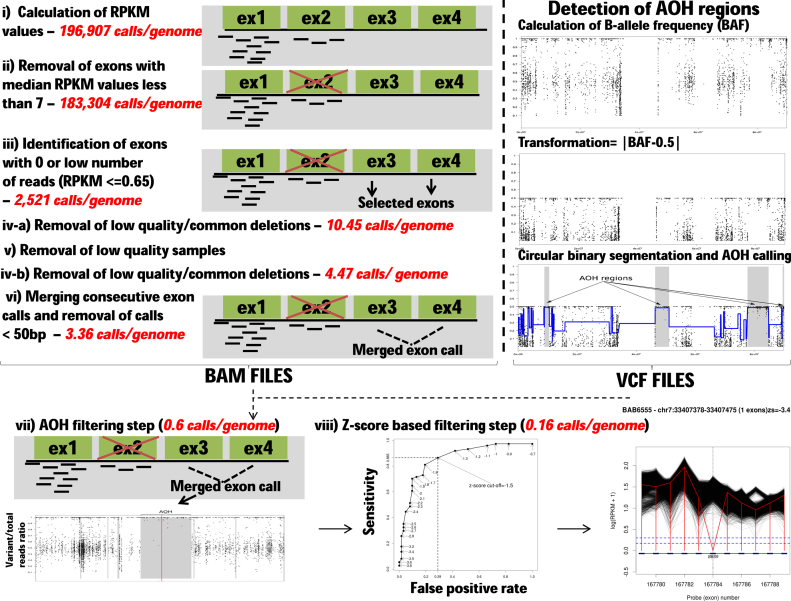
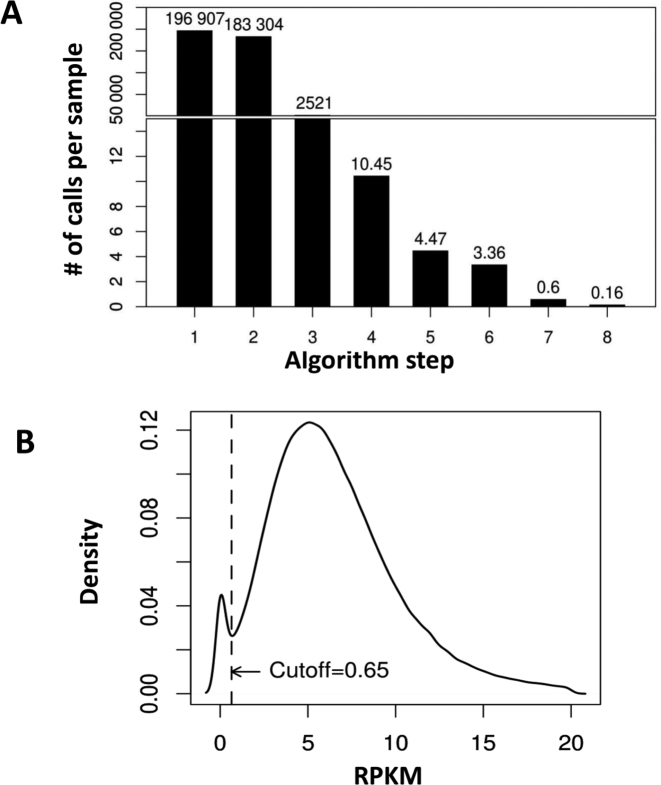
To identify potential HMZ deletions from WES data, we developed an algorithm to call such variants jointly across the entire sample data set. Joint calling allowed for rigorous control data at each captured exon and minimized the number of false positive calls that could emerge from low coverage regions. HMZ deletion CNVs were called in all 4866 WES samples (2580 males and 2286 females) using a procedure consisting of 8 steps (Figures 1 and 2A). First, the data from WES were transformed into per-exon read depth values, i.e. each sample was processed to calculate the RPKM values for each one of its 196,907 exons that were captured and sequenced. Second, all exons with median RPKM < 7 were removed from the analysis to avoid exons that presented with a low average depth-of-coverage value, ∼7% of exons (13,603 out of 196,907) were excluded. Third, in this next step the algorithm annotated a single exon as potentially deleted if it presented 0 or a low level of read depth (RPKM < 0.65) (please refer to the next section for details of how we performed selection of the RPKM threshold value). This filtering step identified 2521 potentially rare deleted exons on average per sample. In the fourth step, low quality and common deletion CNVs were parsed from further processing if the frequency found for a particular HMZ deletion was ≥0.5% in the BHCMG study cohort. This step decreased the average number of putative calls to 10.45 calls per personal genome sample. Fifth, to minimize the influence of low quality samples on algorithm output, we excluded outlier samples with the highest number of calls (i.e. the top 2% of the highest number of calls) and then repeated step 4 without these samples, which reduced the average number of calls to 4.47. In the sixth step, calls from consecutive exons were merged and then calls <50 bp were excluded. After this step the average number of putative HMZ deletions was 3.36. In the penultimate seventh step, potential CNV calls identified in a given sample were then intersected with AOH regions (determined by CBS) larger than 1 kb as defined in the previous section. Calls that do not overlap with any AOH region were parsed from further analysis as potential false positives (FPs). This is due to the expectation that rare, pathogenic homozygous deletions are likely to be located within larger AOH regions, because of the inheritance of common haplotype block from both parents (see also Supplementary Text for further justification of AOH filtering step). This step resulted in 2903 potential deletions (0.6 calls per sample on average). In the final eighth step, a z-RPKM value was derived for each individually identified deleted exon. The z-RPKM derivation occurred as follows: for a given deleted exon, from its original RPKM value the average RPKM in this exon across all samples was subtracted and divided by the standard deviation. The final score for a deletion CNV was computed as an average z-RPKM across all of its exons. This filtering step resulted in 773 best quality calls (0.16 calls per sample on average) having z-RPKM lower or equal to −1.5. The z-score threshold was determined based on the analysis of validation results described below. The final call set of 773 was used for evaluation of algorithm performance and comparison to other CNV calling methods.
Figure 1.
HMZDelFinder algorithm workflow. Different filtering steps were used for data processing of BAM files shown on the left. The number of calls after each filtering step are displayed in red and italicized. The specific BAM and VCF processing steps in the algorithm are: (i) Normalized read depth (RPKM) values are calculated for each exon captured with HGSC VCRome or the HGSC CORE designs. (ii) Low quality exons with their median RPKM values <7 are further removed from the analysis. (iii) Exons with low read numbers are identified (RPKM ≤ 0.65). The threshold was set to 0.65 based on the density distribution of 0.5% quantile of RPKM values for each exon. (iv) Common deletions are subtracted from the list of potential calls if the frequency of a particular homozygous and hemizygous (HMZ) deletion ≥0.5% in the whole cohort. (v) Samples with the highest number of deletions are removed from the analysis and step (iv) is repeated without these low quality samples. (vi) The consecutive exon deletion calls are merged if they are at most 10 exons apart from each other. (vii) In AOH filtering step, absence of heterozygosity (AOH) is calculated from VCF files and a representative AOH plot is displayed in the lower track (above right). In that plot, the y-axis shows the B-allele frequency (i.e. variant/total reads ratio) extracted from exome data VCF files. This B-allele frequency information is then processed using circular binary segmentation (CBS) implemented in the DNAcopy R Bioconductor package. The resulting segments (gray in color) in the AOH plot denote AOH regions identified by the above algorithm. As expected, the AOH regions consist of the variants (points) that have variant/total reads ratio around 1. After the identification of AOH regions from exome sequencing data, the deletion calls are removed if they do not reside in any AOH region larger than 1 kb. (viii) The final HMZ copy number variant (CNV) deletion calls are prioritized based on their average z-RPKM values. In the deletion plots, the loci that contain the deleted exons and its neighboring exons are shown. Y-axis displays the RPKM values on a log scale. The dashed vertical black line indicates the deleted exon. The red vertical line connects RPKM values at the deleted exon and neighboring exons in the sample. Each black line demonstrates the RPKM information for all of the other samples in the Baylor–Hopkins Center for Mendelian Genomics (BHCMG) cohort. The lower blue dashed line exhibits the threshold RPKM value used in the study. The details of the call (i.e. sample name, position, number of exons deleted and z-score) are provided at the top of each plot. The generated deletion plots are manually inspected further to eliminate potential false positive calls.
Figure 2.
HMZDelFinder algorithm yield and RPKM threshold value selection. (A) Bar graph documenting number of HMZ deletion calls after each filtering step. (B) Distribution of 0.5% quantiles of RPKM values across all the BHCMG samples is calculated for each exon from the capture target. The first mode of the distribution likely includes poorly covered and commonly deleted exons in our cohort. We selected an RPKM threshold between these two modes (at RPKM = 0.65) to initially annotate all of these exons as potentially deleted (step 3). In step 4, common deletions are subtracted from the list of deletion calls if the frequency of a particular HMZ deletion ≥0.5% in the whole cohort.
Selection of RPKM threshold value
To determine the optimal RPKM threshold (used in step 3 of the analysis pipeline described above), we analyzed the global distribution of RPKMs for all exons in all samples. For every exon (design target), we calculated the 0.5% quantile of all RPKM values for this exon (i.e. the maximum RPKM of the 0.5% of the lowest RPKM values in the exon). We found that the density distribution function derived for these values across all exons (Figure 2B) is bi-modal. The first mode corresponds to the population of exons with poor coverage in a significant fraction of study samples (i.e. in ≥0.5% of individuals). This could be due to technical artefact, repetitive sequences or because of the existence of common variant, and therefore likely non-pathogenic, heterozygous or homozygous deletions. We set the RPKM threshold at the local minimum between two modes (RPKM = 0.65) of the aforementioned distribution. Such a selection of the threshold forces our algorithm to include all of the poorly covered or commonly deleted exons (i.e. corresponding to the first mode of the distribution) into the first set of potential deletion calls generated in step 3. However, since these calls are present in >0.5% of the cohort, they can be jointly removed in step 4 of the algorithm, because they exceed the frequency cut-off value. Note that if the RPKM threshold would be lower, then only a part of these low quality calls originating from non-informative exons would be identified in the step 3 and as a potential consequence they could pass the frequency filter and increase the false positive rate of the algorithm. Similarly, the selection of higher threshold would result in additional false negative calls.
In order to estimate the influence of selected threshold (RPKM ≤ 0.65) on the sensitivity of HMZDelFinder, we computationally characterized 5 large experimentally identified homozygous deletions in 5 samples from 4 families (one deletion was shared between two individuals from the same family) spanning in total 86 exons. This control CNV ‘call set’ was validated as homozygous by polymerase chain reaction (PCR) or array comparative genomic hybridization (aCGH) (Supplementary Table S1). The presence of HMZ deletions in a given exon is determined independently from the copy number information retrieved from the adjacent exons. Therefore, for multi-exonic deletions, it is expected that the algorithm makes a call in every exon within the deletion. The threshold RPKM ≤ 0.65 resulted in 84 out of 86 exons being called (Supplementary Figure S2). This experiment enabled us to estimate the false negative rate as ∼2.3% and supported the notion that this selected threshold presents a minimum impact on algorithm sensitivity.
Sensitivity of the algorithm to the minimal size of AOH threshold
We empirically examined different thresholds for the minimal size of AOH region that can be used for filtering and we found virtually no difference in the number of calls overlapping with AOH regions when the minimal AOH size varies between 1 kb and 100 kb (Supplementary Figure S3). This may suggest that homozygous deletions are usually surrounded by AOH genomic intervals that are larger than 100 kb. On the other hand, this may also stem from the limited resolution of AOH that depends on the availability and the number of SNV variants in the VCF files from that particular genomic interval interrogated.
Tests on 1000GP data
We selected 50 sample data sets from the 1000GP for which both WES data and genome-wide CNV calls based on the low-coverage and trio-phased data sets were available. The consensus BED file containing the chromosomal positions of targets used for exome sequencing was downloaded from the following URL: (ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/phase1/analysis_results/supporting/exome_pull_down/20120518.analysis_exome_targets.consensus.annotation.bed) and BAM files were obtained from (ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/phase1/data/). The integrated genotype data including the information about deletion CNVs were retrieved from the UCSC genome browser (ftp://hgdownload.cse.ucsc.edu/gbdb/hg19/1000Genomes/). From the set of CNV calls obtained from whole genome sequencing (WGS) data, we extracted all deletions (i.e. with ‘SVTYPE = DEL’ in INFO column) for which at least one sample was homozygous or hemizygous. Next, we selected the subset of deletions that overlap with the targets included in the consensus BED file, so they could be potentially detected using WES data. We removed the deletion calls that were observed more than 3 times in the 1000GP data set (i.e. maximum frequency = 6%) and the calls within low-copy repeats (LCRs). On the same data set of 50 samples, we also implemented seven CNV calling algorithms including: CoNIFER (18), CoNVex (ftp://ftp.sanger.ac.uk/pub/users/pv1/CoNVex/Docs/CoNVex.pdf), XHMM (17), ExonDel (23), CANOES (21), CLAMMS (22) and CODEX (20).
Orthogonal validation of CNV calls from BHCMG cohort
To evaluate results predicted by HMZDelFinder in the BHCMG cohort, we selected 196 HMZ deletion calls in 90 samples (see Results for details). The genomic coordinates for each predicted deletion were used to select probes included in the design of three custom high-density Agilent arrays (all in the 8 × 60K format, with ∼200 bp per probe coverage, AMADID 077567, 077569 and 080031). Hybridization controls were gender matched (HapMap individual NA10851 as male control and HapMap individual NA15510 as female control). Scanned array images were processed using Agilent Feature Extraction software (version 10) and extracted files were analyzed using Agilent Genomic Workbench (version 7.0.4.0). Array designs were based on the February 2009 genome build (GRCh37/hg19 assembly).
We performed long-range PCR using primers spanning deletion breakpoints validated by aCGH to confirm and more accurately map the experimentally identified CNVs. PCR reagents and concentrations have been described previously (35). The thermal cycler was programmed as follows: 94°C × 1 min; 30 cycles of 94°C × 30 s followed by 68°C × 7 min; 72°C × 10 min. PCR primers are listed in the Supplementary Methods. Breakpoint PCR products were treated with ExoSAP-IT (Affymetrix) according to the manufacturer's instructions, then sequenced by Sanger di-deoxynucleotide sequencing (Baylor College of Medicine Sequencing Core, Houston, TX, USA).
Testing genetic model for inheritance of homozygous deletion CNVs
To experimentally test the inheritance model for presumed homozygous deletions identified by HMZDelFinder, we selected the following families from the BHCMG cohort: ten trios (proband and unaffected parents) and three quartets (two unaffected parents and two affected siblings). In order to estimate the actual copy number state for the selected deletion calls and test the carrier status of parental samples, we performed digital droplet PCR (ddPCR) experiments for all individuals that had DNA available in these 13 families.
ddPCR was performed using the QX200™ AutoDG™ Droplet Digital™ PCR System from Bio-Rad following the manufacture's protocols. Briefly, a 20 µl mixture was constructed for each PCR reaction, containing 10 µl of 2x Q200 ddPCR EvaGreen Supermix, 0.25 µl of each primer (10 µM) and 20 ng of genomic DNA. The reaction mixture was subjected to automatic droplet generation, followed by PCR reaction and droplet reading. Cycling conditions for PCR are as the following: 5 min at 95°C, 40 cycles of 30 s at 95°C/1 min at 64°C/1 min at 72°C, 5 min at 4°C, 5 min at 90°C and finally infinite hold at 4°C. Ramp rate was set for 2°C per second for all steps. These data were analyzed using QuantaSoftTM Software from Bio-Rad, and concentrations of positive droplets (number of positive droplets per µl of reaction) were obtained for each PCR reaction. Primers to a control gene, RPPH1, were also included for each sample.
Determining performance of other CNV calling algorithms to detect HMZ deletions
To compare our results with other CNV calling tools, we implemented seven algorithms, including: CoNIFER (18), CoNVex (ftp://ftp.sanger.ac.uk/pub/users/pv1/CoNVex/Docs/CoNVex.pdf), XHMM (17), ExonDel (23), CANOES (21), CLAMMS (22) and CODEX (20) on a selected BHCMG data set including all samples with validated HMZ deletions and the 1000GP data set using default parameters. In addition, for algorithms that require control data (all except ExonDel), the control samples were selected from the BHCMG cohort. All deletion calls from CoNIFER, CoNVex, XHMM and ExonDel were compared to HMZDelFinder output. For CANOES, CLAMMS and CODEX that are able to distinguish HMZ from heterozygous deletions, only HMZ deletion calls were compared to HMZDelFinder output.
Alternative splicing analysis
Using the Bioconductor R transcript annotation package TxDb.Hsapiens.UCSC.hg19. knownGene, we downloaded all the information regarding the gene ids, gene names, transcript ids, transcript names and exon ids from the UCSC gene annotation table. For each potential deletion call and for each exon within such deletion, we determined the number of alternatively spliced transcripts, in which this exon is included. Next, we calculated the ratio of this number to the number of all transcripts of the associated gene. To generate a control data set, we computed the same ratios for all of the exons from the capture target regions.
RESULTS
Evaluation of the algorithm performance using 1000GP data
For evaluation of the algorithm performance, HMZDelFinder was applied to 50 samples extracted from 1000GP data using its default parameters as described in Materials and Methods. The 1000GP Consortium reported 7674 HMZ deletion calls in the set of 50 samples that map to 1180 different genomic intervals. In the 50 samples there were no HMZ deletions with frequency < 0.5%. Therefore, we relaxed the maximum frequency cut-off from 0.5% to 6% for the low quality/common deletion calls that were parsed from the analysis (step 4 of the algorithm as described in the Materials and Methods section). Since we focused on the detection of non-pathogenic and relatively common deletions (with frequency up to 6%), the AOH filtering step was not used. In total, 6 homozygous deletions in 6 individuals were reported by the 1000GP Consortium from which HMZDelFinder was able to detect all of them (Supplementary Table S4 and Figure S5). In addition, HMZDelFinder identified one deletion on chromosome X, i.e. a hemizygous deletion CNV, in a male subject spanning ZNF630 (Supplementary Figure S5G) that was not reported in the set of calls reported by the 1000GP. Analysis of this segment in the database of genomic variants (36) indicates that this particular deletion (nssv470162) was previously detected in the same individual using a SNP array further supporting the HMZDelFinder result.
On the same 50 sample data set we tested seven other CNV calling algorithms (CoNIFER, CoNVeX, XHMM, ExonDel, CANOES, CLAMMS and CODEX; Supplementary Table S4). None of the algorithms detected all of the six homozygous deletions detected by HMZDelFinder. The next most sensitive algorithm was CLAMMS, which detected five out of the six homozygous deletions – it missed a deletion that included six exons.
Evaluation of the algorithm performance using BHCMG data
The HMZDelFinder for calling rare and intragenic deletion CNV was implemented on WES data from 4866 samples (2580 males and 2286 females) in the BHCMG cohort (Figures 1 and 2). To measure the performance of the algorithm, we selected 196 deletion calls from 90 samples tested by aCGH/PCR. Out of 196 deletion calls, 74 were confirmed as HMZ deletions in 62 different sample genomes (see Supplementary Text and Supplementary Figure S6 for details on how we used empirical evidence to fine tune HMZDelFinder and the selection criteria for deletion calls and samples to be used for experimental validation). The fine-tuned HMZDelFinder was implemented for the BHCMG cohort and gave as output 2903 calls. Out of those, there were 134 deletion calls in 68 samples that underwent experimental validation, from which 72 had been confirmed (Supplementary Table S7). This allows an estimate of the algorithm precision as 53.7%.
To further optimize the analysis of the deletion calls and toward efforts to increase algorithm precision, we calculated z-score values as described in the Materials and Methods section. The z-score can be used to prioritize the list of deletion calls per sample. We evaluated this approach on the above-mentioned set of 72 true positive, 2 false negatives and 62 false positive calls validated by aCGH and/or PCR. We computed the sensitivity and false discovery rate (FDR = FP/(TP+FP), where TP stands for true positives) on the data filtered with different z-score cut-off values ranging from −0.7 to −3.8. These data show that by applying filtering (z-score < −1.5), the total number of calls can be reduced to 773. There were 82 deletion calls in 55 samples that underwent independent experimental validation. Out of those, 64 calls were confirmed. Using this filtering, the precision (1-FDR) of the algorithm increased from 53.7% to 78% (64 confirmed HMZDelFinder detected deletions out of 82 deletion calls), while still keeping the sensitivity at 86.5% (64 confirmed HMZDelFinder detected deletions out of 74 confirmed deletions) (Supplementary Figure S8A). The precision for the single and multi-exonic calls was 53% and 96%, respectively (see Supplementary Figure S8B and C). This set of 773 best-quality calls optimized for maximum sensitivity and precision were used for further analysis.
Comparison with other CNV detection methods
To compare HMZDelFinder output with other CNV detection tools, we processed WES data from the 62 BHCMG samples in which 74 HMZ deletions were confirmed by aCGH/PCR. The set of calls generated by HMZDelFinder was compared with the output from CoNIFER (18), CoNVex (ftp://ftp.sanger.ac.uk/pub/users/pv1/CoNVex/Docs/CoNVex.pdf), XHMM (17), ExonDel (23), CANOES (21), CLAMMS (22) and CODEX (20). Detailed comparisons of HMZ CNVs detected by these seven algorithms are presented in Supplementary Table S7 and the subset of those deletions, which were found to contribute to patients’ phenotypes is shown in Table 1.
Table 1. Algorithm performance of WES-CNV tools compared to HMZDelFinder for validated pathogenic calls.
| Sample ID | Gene | # of Exons Deleted/ Total | CNV Size (kb) | CoNIFER | CoNVex | XHMM | ExonDel | CANOES | CLAMMS | CODEX |
|---|---|---|---|---|---|---|---|---|---|---|
| BAB4090 | BBS9 | 1/23 | 0.14 | N [0] | N [117] | N [6] | N [413] | N [0] | N [5] | N [4] |
| BAB4091 | BBS9 | 1/23 | 0.14 | N [0] | N [324] | N [9] | N [417] | N [0] | N [6] | N [6] |
| BAB6555 | BBS9 | 1/23 | 0.1 | N [0] | Y,0.1 [146] | N [184] | N [146] | N [2] | N [2] | N [9] |
| BAB6557 | BBS9 | 1/23 | 0.1 | N [2] | N [158] | N [66] | N [157] | N [0] | N [2] | N [3] |
| BAB4984 | DOCK8* | 18/63 | 214 | Y,344 [5] | Y,210 [249] | Y,359 [21] | Y,41 [424] | N [0] | N [5] | Y,212 [4] |
| BAB4985 | DOCK8* | 18/63 | 214 | Y,350 [6] | Y,214 [197] | Y,359 [26] | Y,39 [496] | N [0] | Y,90 [5] | Y,212 [4] |
| BAB4212 | WWOX | 3/9 | 46 | N [3] | Y,46 [554] | Y,46 [26] | N [747] | N [1] | Y,38 [6] | Y,46 [6] |
| BAB3498 | SNX14 | 25/29 | 64 | Y,80 [3] | Y,64 [304] | Y,64 [26] | N [426] | N [4] | Y,42 [7] | Y,64 [9] |
| BAB5029 | AP4E1** | 23/24 | 193 | Y,467 [2] | Y,193 [372] | Y,193 [18] | N [627] | N [1] | Y,146 [5] | Y,196 [8] |
| BAB5866 | DMD | 2/79 | 2.6 | N [1] | N [451] | Y,93 [12] | N [143] | N [2] | N [6] | Y,2.6 [5] |
| BAB5867 | DMD | 2/79 | 2.6 | N [2] | N [156] | Y,2.6 [9] | N [137] | N [2] | N [7] | Y,2.6 [7] |
| LAT0248 | RIPPLY1 | 2/4 | 0.8 | N [6] | N [264] | N [32] | N [220] | N [0] | N [3] | Y,0.8 [2] |
| BAB3747 | CNTNAP2 | 1/24 | 0.17 | N [5] | N [28] | N [5] | N [525] | N [3] | Y,0.17 [7] | N [9] |
| BAB3748 | CNTNAP2 | 1/24 | 0.17 | N [4] | N [448] | N [26] | N [672] | N [4] | Y,0.17 [7] | N [8] |
| BAB6883 | GRID2 | 2/16 | 26 | N [1] | N [18] | Y,26 [125] | Y,25 [282] | N [1] | Y,26 [6] | Y,26 [9] |
Y: pathogenic call detected; N: pathogenic call not detected; followed by corresponding CNV size; [ ]: the number of HMZ+heterozygous deletions (CoNIFER, CoNVex, XHMM, ExonDel) or the number of HMZ deletions (CANOES, CLAMMS, CODEX) detected in total for a given sample; * deletion includes CBWD1; ** deletion includes TNFAIP8L3.
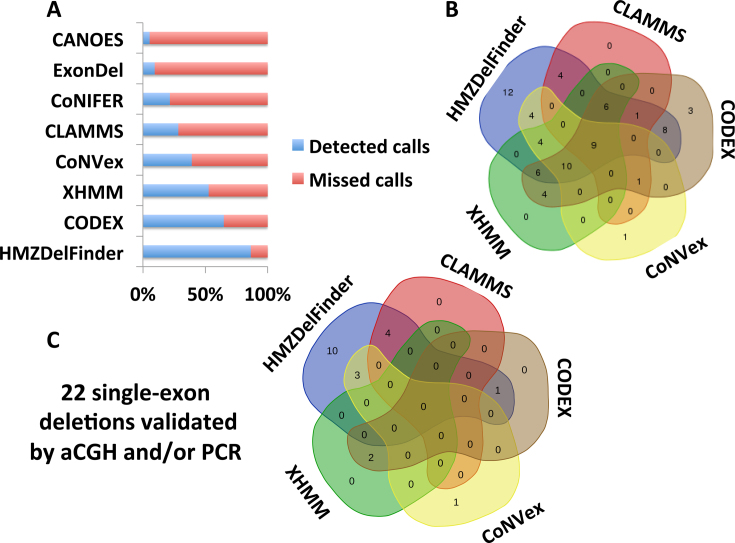
WES analyses using CoNIFER, CoNVex, XHMM, ExonDel, CANOES, CLAMMS, CODEX and HMZDelFinder detected 16 (22%), 29 (39%), 39 (53%), 7 (9%), 4 (5%), 21 (28%), 48 (65%) and 64 (86.5%) out of those 74 validated deletions, respectively (Figure 3A and B). Single-exon deletions were particularly underrepresented as CoNVex detected 4 (18%), CANOES detected 3 (14%), CLAMMS detected 4 (18%), CODEX detected 3 (14%), XHMM detected 2 (9%) out of 22 validated in our cohort (Figure 3C); whereas HMZDelFinder detected 18 (82%) single-exon deletions (Figure 3C). None of the validated single-exon deletion calls were identified by CoNIFER or ExonDel, despite the fact that the latter has been specifically developed to detect homozygous deletions at the single exon level. The ExonDel algorithm detects CNVs based on the distribution of the median read depth information across all the exons in a given sample, in contrast, HMZDelFinder performs joint-calling of read depth information per exon retrieved from multiple samples. ExonDel generated an average number of 394.7 calls per exome on which 7 out of 74 validated deletions (0 of 22 single-exon deletions) were partially detected.
Figure 3.
Comparative analysis of HMZDelFinder and seven other CNV calling algorithms for empirically verified deletion CNVs. (A) Horizontal barplot shows the fractions of calls detected by HMZDelFinder, CODEX, XHMM, CoNVex, CLAMMS, CoNIFER, ExonDel and CANOES out of 74 confirmed HMZ deletions by array comparative genomic hybridization (aCGH) and/or polymerase chain reaction (PCR). (B) The Venn diagram depicts the number of calls detected by the top five performing algorithms (HMZDelFinder, CODEX, XHMM, CoNVex and CLAMMS) out of the 74 validated deletions by aCGH and/or PCR. (C) Out of the 22 experimentally validated single-exon deletions, the Venn diagram shows the number of calls detected by top five performing algorithms (HMZDelFinder, CODEX, XHMM, CoNVex and CLAMMS). Of note, HMZDelFinder detected 18/22 single-exon deletions.
Sensitivity of the algorithm to the cohort size
HMZDelFinder is tailored to find rare and intragenic variant events. To analyze the impact of the cohort size on the deletion detection rate, we implemented HMZDelFinder using subsets of the BHCMG cohort (4866 samples in total including 2580 males (53%) and 2286 (47%) females) consisting of different sample sizes, i.e. using the WES data from 100, 200, 500, 1000, 2000, 3000, 4000 and 4866 individuals. WES data from the 62 samples with the 74 HMZ validated deletions were included in all of these data subsets. The remaining samples in each subset were selected randomly from the entire BHCMG cohort. The gender ratio was kept equivalent in each subset, 53% males and 47% females, to that of the total BHCMG cohort. This analytical approach enabled estimation of the sensitivity and precision as samples were processed together with cohorts of different sizes (Supplementary Figure S9). While the precision is mostly stable across the set of experiments, we observed an increase in the sensitivity, from 46% to 72%, when the number of samples is enlarged from 200 to 300. This reflects the main limitation of using small cohorts while searching for rare and pathogenic CNVs using HMZDelFinder. This could be exemplified by the lack of ability to detect HMZ deletions that occur more than once in the set of validated 62 samples and therefore are relatively too common to pass the maximum frequency threshold. Furthermore, sensitivity increases to 74% for the cohort size of 500 samples and finally achieves 86.5% for 1000 samples.
Consanguinity rate in the BHCMG cohort
Rare and pathogenic homozygous deletions are likely to be located within larger AOH regions due to the inheritance of a shared haplotype block from both parents (inherited by descent - IBD). To identify consanguineous families in the BHCMG cohort, the fraction of the genome with genomic intervals presenting large regions of AOH (>5 Mb) was calculated for each sample. To differentiate the individuals with a relatively high coefficient of consanguinity and consanguineous families from non-consanguineous families, we determined a threshold for the fraction of the genome covered by AOH regions as >2% of the genome based on the training data of samples in BHCMG cohort with parents who are first-degree and second-degree relatives. Based on these calculations, ∼13% of samples from the BHCMG cohort (654 out of 4866) could be considered as consanguineous; given that the cumulative size of AOH regions larger than 5 Mb exceeds 2% of their genomes.
Inheritance of predicted CNV calls
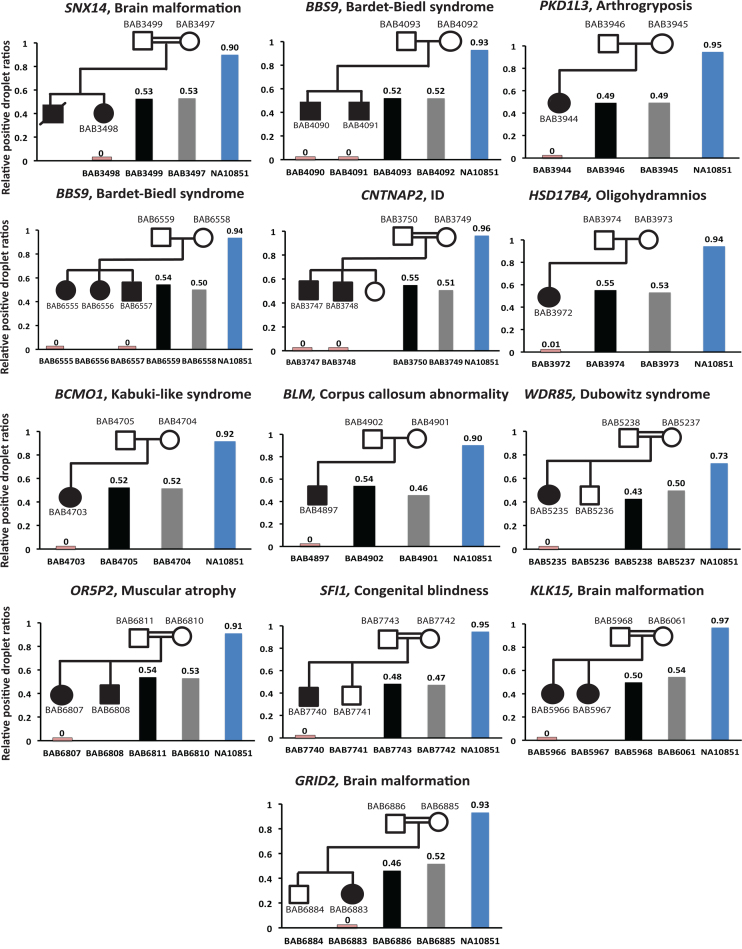
For algorithmically identified homozygous deletion CNVs, we investigated a selected sample set in which DNA was available for both parents for empirical ‘wet-bench’ experimental verification of Mendelian expectations. We confirmed the presence of homozygous deletions in 16 out of 16 affected individuals in 13 families by ddPCR (Figure 4). Importantly, ddPCR experiments also experimentally demonstrated that, consistent with Mendelian recessive expectations, the parents of these 16 individuals are heterozygous carriers of the experimentally analyzed exonic deletion calls. In particular, we determined that among all parental samples the relative positive droplet ratios (target genes compared to control genes) varied between 43–55%, i.e. the ratios were close to the expected value (50%) for heterozygous deletion carriers. In summary, these experimental validations confirmed the homozygous deletions of 16 homozygous deletion calls originally identified by HMZDelFinder as well as fulfilment of Mendelian expectations – the heterozygous deletions in parents of these 13 families (Figure 4).
Figure 4.
Examining inheritance of homozygous deletions experimentally by ddPCR. The segregation of HMZDelFinder-detected deletion calls is confirmed by digital droplet PCR (ddPCR) in 16 individuals in 13 families. Each family is presented with its pedigree structure using standardized symbols (squares = males; circles = females; filled symbols show affected individuals). The gene and the proband's phenotype are depicted above each pedigree. Each bar graph shows the relative positive droplet ratios (target gene compared to control gene) in each available family member (blue vertical bar = ddPCR counts in control DNA; grey = counts in mother; black = father; pink = counts observed in affected child with homozygous deletion). The affected individuals with the deletion calls detected by HMZDelFinder are experimentally verified to have homozygous deletion CNV (the relative positive droplet ratios ≈ 0) and the parents are confirmed to be heterozygous carriers (relative positive droplet ratios ≈ 0.5).
Investigation of small, rare exonic deletions in 4866 BHCMG samples
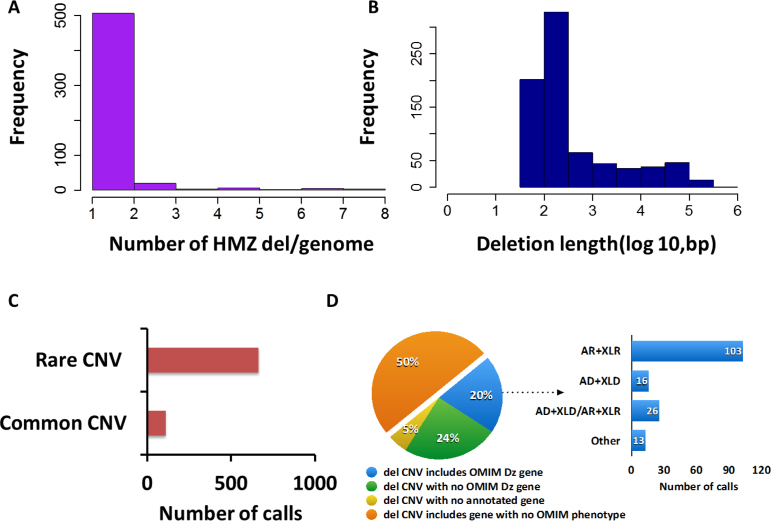
After algorithm optimization, HMZDelFinder detected 773 deletions, comprising either homozygous (567) or hemizygous (206) calls, thus yielding an average of 0.16 deletions per genome (Figure 5A). The length of these deletions ranges from 51 bp (1 exon) to 371.1 kb (39 exons) with a median length of 179 bp (Figure 5B). Importantly, 572 out of 773 calls (∼74%) correspond to single-exon deletions. To evaluate the frequency of those potential HMZ deletions in our cohort we compared the genomic coordinates of those 773 calls with high resolution CNV data extracted from an array containing 42 million oligos performed in 450 individuals comprised of samples with European, African and East Asian ancestry (37), 1000GP pilot phase data (38,39) and Deciphering Developmental Disorders data (40). Our analysis revealed that 85.9% were rare variant CNVs (MAF ≤ 1%) and 14.1% of 773 identified CNVs reside in a genomic interval containing a common CNV (MAF > 1%) (Figure 5C).
Figure 5.
Summary statistics of 773 deletion calls detected by HMZDelFinder. (A) The distribution of HMZ deletion calls per genome. The average number of deletion calls per genome is calculated as 0.16. X axis displays the number of deletion calls per genome. Y-axis exhibits the number of samples that have the corresponding number of deletion calls. (B) The length distribution of HMZ deletion calls per genome. The median length of deletion calls is calculated as 179 bp. X-axis displays the length of deletion calls on a log scale. Y-axis exhibits the number of deletion calls that have the corresponding size. (C) Common CNVs are retrieved from an array data containing 42 million oligos, 1000 Genomes Project (1000GP) pilot phase data, Deciphering Developmental Disorders (DDD) data (MAF ≥ 1%). Then, they are intersected with 773 deletion calls. The percentage of the deletion calls involved in common CNVs is displayed as a column plot. The plot shows that 85.9% of the detected HMZ deletion calls do not reside in common CNVs (MAF ≥ 1%). (D) (Left) The 773 deletion calls are examined based on their involvement of any disease-associated gene in OMIM. The pie chart conveys the information that 20% of these calls include at least one-disease associated gene in OMIM. (Right) Out of those deletions that involve at least one-disease associated gene, 65.1% of them encompass a gene with a recessive inheritance pattern as shown in the barplot (AR: Autosomal recessive, XLR: X-linked recessive, AD: Autosomal dominant, XLD: X-linked dominant).
Discovery of potential pathogenic gene deletions in patients with Mendelian disorders
To examine for a potential disease contribution for these computationally predicted HMZ intragenic deletions, we investigated whether genes disrupted or encompassed by these deletions are associated with a disease phenotype in OMIM (http://www.omim.org). Our analysis revealed that 20% (158) of 773 predicted deletions encompass at least one gene, which is associated with a disease trait in the OMIM database (http://www.omim.org) (Figure 5D). Of these, 65.1% (103 out of 158 HMZ deletions) encompass a gene, in which the phenotype is associated with a recessive inheritance pattern in OMIM. Of the 206 hemizygous deletions, 42 (20.3%) deletions occurred in a gene associated with a known X-linked recessive disease.
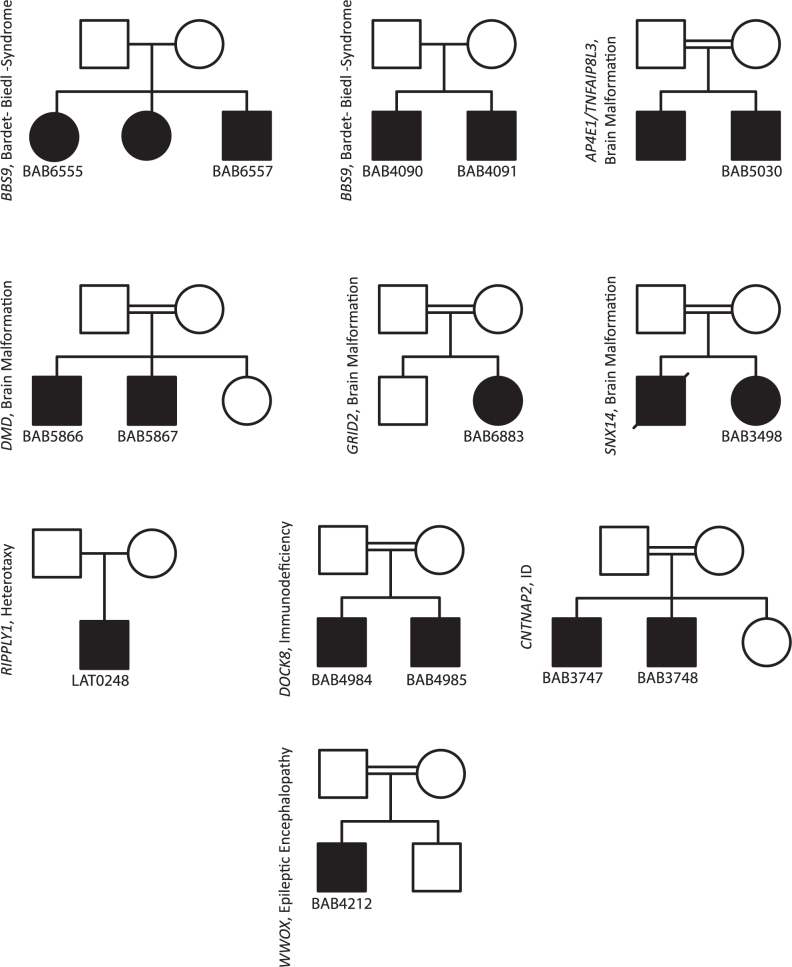
From the list of 64 validated calls predicted by HMZDelFinder, a subset of homozygous deletions (N = 12) likely explain or contribute to the subjects assessed phenotypes in 8 families (Figure 6, Table 1). These potentially disease associated variants/genes include: (i) DOCK8 (2 patients (14), Hyper-IgE recurrent infection syndrome, autosomal recessive, MIM #243700, (41)), (ii) SNX14 (1 patient (26), autosomal recessive cerebellar ataxia and intellectual disability, MIM #616354, (8)), (iii) WWOX (1 patient, epileptic encephalopathy, early infantile with microcephaly, MIM #616211, (42)), (iv) AP4E1 (1 patient (26), spastic quadriplegic cerebral palsy 4, MIM #613744), (v) CNTNAP2 (2 patients (26), cortical dysplasia-focal epilepsy syndrome, MIM #610042), (vi) BBS9 (4 patients, Bardet-Biedl syndrome 9, MIM #615986), and (vii) GRID2 (1 patient (15), spinocerebellar ataxia, autosomal recessive 18, MIM #616204). For hemizygous deletions (N = 3), HMZDelFinder detected DMD (2 patients (26), Becker muscular dystrophy, MIM #300376, which was an incidental finding and explained a part of patients’ phenotypes) and a novel candidate disease gene RIPPLY1 (Figure 7).
Figure 6.
Pedigree structures of 10 families with 15 confirmed HMZ deletions initially identified by HMZDelFinder. All of the 15 known disease gene deletions are subjected to aCGH or deletion CNV breakpoint junction PCR for orthogonal confirmations of the bioinformatically identified deletion calls. The presence and zygosity of 15/15 known disease gene deletions are confirmed by at least one orthogonal experimental validation platform. The gene and cohort names are indicated next to the pedigrees. In a subset of the families that carry BBS9, DOCK8, DMD and CNTNAP2, the gene deletions are confirmed in an another affected family member in addition to the probands.
Figure 7.
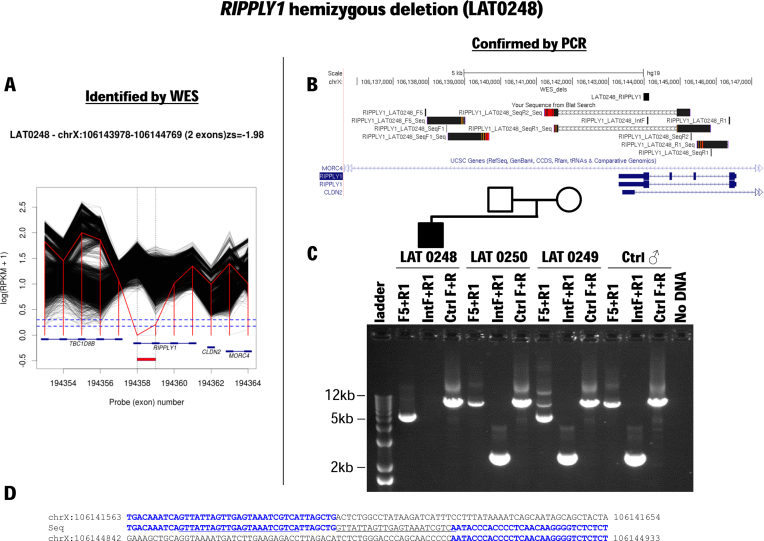
Hemizygous partial deletion RIPPLY1, a novel candidate heterotaxy gene, in a male patient with heterotaxy. (A) Whole exome sequencing (WES) read count data (RPKM) are plotted for subject LAT0248 (red line) and all other BHCMG subjects (black lines) in the region of chromosome X containing RIPPLY1. Near-zero RPKM values suggest a hemizygous deletion of the final exon of RIPPLY1 and possibly also the penultimate exon. (B) The minimum CNV size estimated from RPKM data (‘WES_dels’) is shown along with breakpoint sequence data (‘Your Sequence from Blat Search’) and the RIPPLY1 gene structure. (C) PCR with primers spanning the deletion breakpoint confirms the deletion and demonstrates that it was inherited from the proband's mother. (D) Breakpoint sequencing demonstrates that the final two exons of RIPPLY1 are deleted and offers clues concerning the mutational mechanism generating this CNV. Note the 22 bp insertion that matches near-upstream sequence (underline).
Novel candidate Mendelian genes
RIPPLY1 is a novel candidate disease gene for a heterotaxy syndrome. We recently identified RIPPLY2 SNV mutations in association with a novel syndrome consisting of heterotaxy and segmentation defects of the cervical vertebrae clinically diagnosed as Klippel–Feil syndrome (MIM #613702) (43). Recent studies reveal that 8–11% of congenital scoliosis, a common/complex trait, in the Chinese population is explained by a simple Mendelian recessive model at the TBX6 locus. This recessive model consists of one rare variant null allele (due to either a 16p11.2 deletion CNV or a loss of function allele caused by a nonsense/frameshift SNV of TBX6) in combination with a hypomorphic allele consisting of a noncoding upstream SNV haplotype that is a common variant in the Chinese population. These congenital scoliosis patients have vertebral segmentation defects (11). RIPPLY1 and RIPPLY2 act to regulate TBX6 during somitogenesis and development of the embryo's body plan.
To facilitate novel disease gene discovery, we integrated BHCMG SNV and CNV data. In this way, we found a number of candidate genes in which we observe a HMZ point mutation or HMZ deletion in subjects with similar phenotypes. This integrative approach led to the identification of a hemizygous splicing mutation in RGN (OMIM #300212) and a hemizygous deletion in exon 4 of RGN in two unrelated male subjects presenting osteoporosis. The protein encoded by this gene is a highly conserved, calcium-binding protein and anticipated to have an important role in calcium homeostasis. The aggregate data suggest RGN as a potential candidate causative gene for osteoporosis.
HMZ deletions are overrepresented in alternatively spliced exons
We performed alternative splicing analysis on the exons included in the predicted 773 deletion calls. This analysis revealed that 18% of deleted exons are observed in less than one-fourth of their associated genes alternatively spliced transcripts; whereas when we considered all of the targeted exons in the genome this number was 12% (Binomial test, P-value = 1.043 × 10−8) (Supplementary Figure S10). This analysis suggests that a larger than expected fraction of HMZ deletions identified in this BHCMG disease cohort affect exclusively an alternative transcript.
DISCUSSION
CNVs have been causally associated with a significant number of inherited genetic and genomic disorders (3,35,44). These pathogenic CNVs include a substantial number of exonic deletions that have been shown to contribute to various diseases including both genomic disorders and common complex traits (2,44–46). Studies by Boone et al. (10,35), Retterer et al. (48) and Feng et al. (24) have reported that pathogenic exonic deletions may occur more commonly in the human genome than recognized by currently utilized molecular diagnostic techniques. Likewise, the analysis of tiling oligonucleotide microarray data of genomes from individuals not selected for rare phenotypes reveals that each person carries >1000 CNVs over 500 bp in size and approximately 2% of these deletions encompass exon-level deletions (37). Multiplex ligation-dependent probe amplification and targeted aCGH (35,49–58) data indicate that exon-level deletions can be the molecular cause of diverse genetic diseases, nonetheless detection of exon-deletion CNV genome-wide, in a cost effective and unbiased manner, remains a challenge. Additionally, the sheer volume of WES data generated in the past few years provide an invaluable resource for rare variant CNV detection, including smaller exon-level and even single-exon intragenic deletions. The expanded use of WES in clinical diagnostics (25,59,60) and research (31,32,61,62) for the detection of exon-level SNVs and small InDels renders it a powerful tool for genome-wide rare variant assessment (14,15,26). In addition, the versatility of WES for large and rare CNV detection suggests its future usage as a potential comprehensive mutation detection assay in clinical and research labs. However, it requires investment in data analysis tools in order to diminish its limitations regarding CNV detection (63).
Despite the availability of several CNV calling tools to identify large CNVs, detection of rare and intragenic CNVs from WES data is a challenge. Other CNV detection algorithms including CoNIFER (18), CoNVex (ftp://ftp.sanger.ac.uk/pub/users/pv1/CoNVex/Docs/CoNVex.pdf), XHMM (17), CANOES (21), CLAMMS (22) and CODEX (20) are optimized to identify mainly heterozygous CNVs encompassing at least three consecutive exons. There remain limitations for the detection of single exon-level deletion CNV from WES data in currently utilized analysis tools because there is only one data point for each exon. We constructed an analytical tool, HMZDelFinder to enable small sized CNV detection from WES data by developing an algorithm to identify rare, potentially encompassing only a single exon, HMZ deletions. First, utilization of RPKM values for each exon of all samples contributes extensively to the normalization of experimental raw data by exon length and total read number. In addition, the joint calling implemented in HMZDelFinder described here enables evaluation of the distribution of 4866 sample data points for each single exon simultaneously. Examination of this distribution for each exon allows detection of outlier exons (RPKM ≤ 0.65) and development of further filtering steps eliminates low-coverage exons from the analysis, for instance exons with high GC content that may have evaded capture or those encompassing LCRs. In this manner, we reduce the batch effect inherent to WES data analysis (47). Furthermore, the large sample size of the BHCMG cohort (4866 samples) has elevated the power of the tool, diminishing the limitations to retrieving single exon-level CNV information from WES data. As a result HMZDelFinder could identify 18/22 experimentally validated single-exon deletions while the other programs identified at most 4 (CoNVex and CLAMMS; see Figure 3 and Supplementary Table S7).
In addition to the importance of the joint calling approach for individual exons across multiple samples, the AOH information was a key to filter the false positive calls. The Clan Genomics hypothesis posits that pathogenic variants tend to arise in the recent history of a family or the more extended clan. In the case of recessive disorders, an intragenic heterozygous deletion that arose in an autosomal recessive disease gene may occur at a locus a few generations later in the form of a homozygous CNV due to consanguinity. In this scenario, it is likely that the deletion is inherited as a part of a larger haplotype block common to both parents that is visible in the proband as an AOH region surrounding the homozygous deletion. Similarly, if the affected child inherits two heterozygous deletions of different sizes that arose independently in each parental ancestor, then the overlapping part of these deletions will form a homozygous CNV and the non-overlapping deletion intervals will constitute the AOH region on one or both sides of this CNV (64). Thus, large AOH regions may surround many rare pathogenic homozygous deletions (see e.g. Supplementary Figure S11). This assumption was used to calculate B-allele frequency information from the VCF files and extracted additional information content from the WES data, which added further strength to the exonic read depth information provided by BAM files. Both parameters were shown here to efficiently refine (i.e. an observed 5-fold reduction) the list of potentially pathogenic CNVs and decrease false-positive calls.
Application of HMZDelFinder to the analysis of the BHCMG cohort enabled identification of pathogenic HMZ deletions in 15 individuals (Figure 6). Out of these, 7 pathogenic HMZ deletions in 7 individuals were shown to be causative of the subject's clinical phenotype in three distinct disease cohort studies, i.e. primary immunodeficiency disorders, Mendelian neurological disorders and neurogenetic disorders (14,15,26), where CNVs contributed to 11%, 7.1% and 9.1%, of the molecular diagnoses, respectively. Importantly, HMZDelFinder detected alleles accounted for 17–50% of pathogenic CNVs reported in those studies. In addition, we identified two potential novel disease genes: RIPPLY1, a novel candidate gene for heterotaxy, and RGN, a candidate gene for osteoporosis. We also identified a number of HMZ deletion calls that are not clearly associated with the patient's clinical phenotype. Those deletions presumably encompass genes that do not cause the expressed disease phenotype or they affect disease genes that are tolerant to loss of function variants. For the non-validated calls, one possibility is that they affect non-pathogenic or non-functional alternative isoforms of their associated gene. In fact, our analysis revealed that 18% of 773 deletion calls map to minor isoform exons (Supplementary Figure S10).
The evaluation of HMZDelFinder performance was limited to BHCMG and 1000GP cohorts due to the lack of other data sets that could be used as a reference for rare and small CNV detection from WES data. In particular, the validated CNV data set from the BHCMG cohort is a subset of HMZDelFinder deletion calls, which may lead to a bias in comparison of the algorithm performance versus that of other tools. Therefore, we performed additional evaluation on 50 samples from 1000GP data using deletion calls generated from WGS data as a potential ‘gold standard’ to test novel approaches to detect CNVs from exome. HMZDelFinder was able to detect 6 out of 6 rare homozygous deletions, including a single-exon deletion in 3 patients, reported by the 1000GP Consortium. Moreover, HMZDelFinder identified a hemizygous deletion spanning ZNF630 in the sample NA18856 that was not reported by the 1000GP Consortium (Supplementary Figure S5G). Such deletion was previously reported in this HapMap sample as inferred by SNP array and reported in database of genomic variants. Interestingly, the SNP array data indicate that one out of two deletion breakpoints maps to a non-unique sequence, whereas the second one maps to a LCR. Based on this observation we suggest that deletions with one or two breakpoints within LCRs are likely more difficult to be detected by the analysis of low coverage WGS, which usually rely on breakpoint detection using read pair and split-read data. Because of the poor mappability of reads within LCRs, this information may be unavailable resulting in false negative calls.
Utilization of WES data as a single comprehensive assay for both the detection of point mutations, InDels and CNVs is urgently needed (65). First, using a single method for detection of a more comprehensive set of variants facilitates the integration of the results. Integration of SNVs and CNVs may lead to a higher diagnostic yield such as 58% diagnostic rate of intellectual disability (66–68). It also may stimulate an elevated rate of novel disease gene discovery as exemplified in the BHCMG sample set by allowing detection of different classes of variant types. Among these classes, HMZ deletions play an important role in pathogenesis of recessive and X-linked disorders. In summary, we demonstrated that our tool facilitates identification of rare variant HMZ deletions, even those encompassing just a single exon, which may contribute to a patients’ clinical phenotype and are likely to be missed by other CNV calling approaches.
AVAILABILITY
The source code and usage example of HMZDelFinder are freely available at https://github.com/BCM-Lupskilab/HMZDelFinder.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank the family members and the collaborators that participated in this study.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Supported in part by the National Human Genome Research Institute/National Heart Lung and Blood Institute [U54HG006542 to BHCMG]; National Human Genome Research Institute grant to Baylor College of Medicine Human Genome Sequencing Center [U54HG003273]; National Institute of Neurological Disorders and Stroke [R01NS05829 to J.R.L.]; NHGRI, NHBLI, and NINDS, all Institutes of the United States National Institutes of Health. Also funded in part by the Polish National Science Centre [2014/13/B/NZ2/01248], and the Cancer Prevention & Research Institute of Texas training Program [RP140102 to W.-L. C.].
Conflict of interest statement. J.R.L. has stock ownership in 23andMe and Lasergen, is a paid consultant for Regeneron Pharmaceuticals, and is a coinventor on multiple United States and European patents related to molecular diagnostics for inherited neuropathies, eye diseases and bacterial genomic fingerprinting. The Department of Molecular and Human Genetics at Baylor College of Medicine derives revenue from the chromosomal microarray analysis (CMA) and clinical exome sequencing offered in the Baylor Genetics Laboratory (http://bmgl.com/). Other authors have no disclosures relevant to the manuscript.
REFERENCES
- 1.Lupski J.R. Structural variation mutagenesis of the human genome: impact on disease and evolution. Environ. Mol. Mutagen. 2015; 56:419–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang F., Gu W., Hurles M.E., Lupski J.R.. Copy number variation in human health, disease and evolution. Annu. Rev. Genomics Hum. Genet. 2009; 10:451–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stankiewicz P., Lupski J.R.. Structural variation in the human genome and its role in disease. Annu. Rev. Med. 2010; 61:437–455. [DOI] [PubMed] [Google Scholar]
- 4.Alkuraya F.S. Natural human knockouts and the era of genotype to phenotype. Genome Med. 2015; 7:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lupski J.R., Belmont J.W., Boerwinkle E., Gibbs R.A.. Clan genomics and the complex architecture of human disease. Cell. 2011; 147:32–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hjeij R., Lindstrand A., Francis R., Zariwala M.A., Liu X., Li Y., Damerla R., Dougherty G.W., Abouhamed M., Olbrich H. et al. . ARMC4 mutations cause primary ciliary dyskinesia with randomization of left/right body asymmetry. Am. J. Hum. Genet. 2013; 93:357–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Day-Williams A.G., Sun C., Jelcic I., McLaughlin H., Harris T., Martin R., Carulli J.P.. Whole genome sequencing reveals a chromosome 9p deletion causing DOCK8 deficiency in an adult diagnosed with hyper IgE syndrome who developed progressive multifocal leukoencephalopathy. J. Clin. Immunol. 2015; 35:92–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thomas A.C., Williams H., Setó-Salvia N., Bacchelli C., Jenkins D., O'Sullivan M., Mengrelis K., Ishida M., Ocaka L., Chanudet E. et al. . Mutations in SNX14 cause a distinctive autosomal-recessive cerebellar ataxia and intellectual disability syndrome. Am. J. Hum. Genet. 2014; 95:611–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valduga M., Philippe C., Lambert L., Bach-Segura P., Schmitt E., Masutti J.P., François B., Pinaud P., Vibert M., Jonveaux P.. WWOX and severe autosomal recessive epileptic encephalopathy: first case in the prenatal period. J. Hum. Genet. 2015; 60:267–271. [DOI] [PubMed] [Google Scholar]
- 10.Boone P.M., Campbell I.M., Baggett B.C., Soens Z.T., Rao M.M., Hixson P.M., Patel A., Bi W., Cheung S.W., Lalani S.R. et al. . Deletions of recessive disease genes: CNV contribution to carrier states and disease-causing alleles. Genome Res. 2013; 23:1383–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu N., Ming X., Xiao J., Wu Z., Chen X., Shinawi M., Shen Y., Yu G., Liu J., Xie H. et al. . TBX6 null variants and a common hypomorphic allele in congenital scoliosis. N. Engl. J. Med. 2015; 372:341–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lalani S.R., Liu P., Rosenfeld J.A., Watkin L.B., Chiang T., Leduc M.S., Zhu W., Ding Y., Pan S., Vetrini F. et al. . Recurrent muscle weakness with rhabdomyolysis, metabolic crises, and cardiac arrhythmia due to bi-allelic TANGO2 mutations. Am. J. Hum. Genet. 2016; 98:347–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kremer L.S., Distelmaier F., Alhaddad B., Hempel M., Iuso A., Küpper C., Mühlhausen C., Kovacs-Nagy R., Satanovskij R., Graf E. et al. . Bi-allelic truncating mutations in TANGO2 Cause infancy-onset recurrent metabolic crises with encephalocardiomyopathy. Am. J. Hum. Genet. 2016; 98:358–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stray-Pedersen A., Sorte H.S., Samarakoon P., Gambin T., Chinn I.K., Coban Akdemir Z.H., Erichsen H.C., Forbes L.R., Gu S., Yuan B. et al. . Primary immunodeficiency diseases: Genomic approaches delineate heterogeneous Mendelian disorders. J. Allergy Clin. Immunol. 2016; doi:10.1016/j.jaci.2016.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Charng W.-L., Karaca E., Coban Akdemir Z., Gambin T., Atik M.M., Gu S., Posey J.E., Jhangiani S.N., Muzny D.M., Doddapaneni H. et al. . Exome sequencing in mostly consanguineous Arab families with neurologic disease provides a high potential molecular diagnosis rate. BMC Med. Genomics. 2016; 9:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carvalho C.M.B., Lupski J.R.. Mechanisms underlying structural variant formation in genomic disorders. Nat. Rev. Genet. 2016; 17:224–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fromer M., Moran J.L., Chambert K., Banks E., Bergen S.E., Ruderfer D.M., Handsaker R.E., McCarroll S.A., O'Donovan M.C., Owen M.J. et al. . Discovery and statistical genotyping of copy-number variation from whole-exome sequencing depth. Am. J. Hum. Genet. 2012; 91:597–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krumm N., Sudmant P.H., Ko A., O'Roak B.J., Malig M., Coe B.P., Quinlan A.R., Nickerson D.A., Eichler E.E., NHLBI Exome Sequencing Project . Copy number variation detection and genotyping from exome sequence data. Genome Res. 2012; 22:1525–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Ligt J., Boone P.M., Pfundt R., Vissers L.E.L.M., Richmond T., Geoghegan J., O'Moore K., de Leeuw N., Shaw C., Brunner H.G. et al. . Detection of clinically relevant copy number variants with whole-exome sequencing. Hum. Mutat. 2013; 34:1439–1448. [DOI] [PubMed] [Google Scholar]
- 20.Jiang Y., Oldridge D.A., Diskin S.J., Zhang N.R.. CODEX: a normalization and copy number variation detection method for whole exome sequencing. Nucleic Acids Res. 2015; 43:e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Backenroth D., Homsy J., Murillo L.R., Glessner J., Lin E., Brueckner M., Lifton R., Goldmuntz E., Chung W.K., Shen Y.. CANOES: detecting rare copy number variants from whole exome sequencing data. Nucleic Acids Res. 2014; 42:e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Packer J.S., Maxwell E.K., O'Dushlaine C., Lopez A.E., Dewey F.E., Chernomorsky R., Baras A., Overton J.D., Habegger L., Reid J.G.. CLAMMS: a scalable algorithm for calling common and rare copy number variants from exome sequencing data. Bioinformatics. 2016; 32:133–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo Y., Zhao S., Lehmann B.D., Sheng Q., Shaver T.M., Stricker T.P., Pietenpol J.A., Shyr Y.. Detection of internal exon deletion with exon Del. BMC Bioinformatics. 2014; 15:332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng Y., Chen D., Wang G.-L., Zhang V.W., Wong L.-J.C.. Improved molecular diagnosis by the detection of exonic deletions with target gene capture and deep sequencing. Genet. Med. 2015; 17:99–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Y., Muzny D.M., Reid J.G., Bainbridge M.N., Willis A., Ward P.A., Braxton A., Beuten J., Xia F., Niu Z. et al. . Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N. Engl. J. Med. 2013; 369:1502–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karaca E., Harel T., Pehlivan D., Jhangiani S.N., Gambin T., Coban Akdemir Z., Gonzaga-Jauregui C., Erdin S., Bayram Y., Campbell I.M. et al. . Genes that affect brain structure and function identified by rare variant analyses of mendelian neurologic disease. Neuron. 2015; 88:499–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lupski J.R., Gonzaga-Jauregui C., Yang Y., Bainbridge M.N., Jhangiani S., Buhay C.J., Kovar C.L., Wang M., Hawes A.C., Reid J.G. et al. . Exome sequencing resolves apparent incidental findings and reveals further complexity of SH3TC2 variant alleles causing Charcot-Marie-Tooth neuropathy. Genome Med. 2013; 5:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reid J.G., Carroll A., Veeraraghavan N., Dahdouli M., Sundquist A., English A., Bainbridge M., White S., Salerno W., Buhay C. et al. . Launching genomics into the cloud: deployment of Mercury, a next generation sequence analysis pipeline. BMC Bioinformatics. 2014; 15:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M. et al. . The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010; 20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Challis D., Yu J., Evani U.S., Jackson A.R., Paithankar S., Coarfa C., Milosavljevic A., Gibbs R.A., Yu F.. An integrative variant analysis suite for whole exome next-generation sequencing data. BMC Bioinformatics. 2012; 13:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bamshad M.J., Shendure J.A., Valle D., Hamosh A., Lupski J.R., Gibbs R.A., Boerwinkle E., Lifton R.P., Gerstein M., Gunel M. et al. . The centers for mendelian genomics: a new large-scale initiative to identify the genes underlying rare Mendelian conditions. Am. J. Med. Genet. A. 2012; 158A:1523–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chong J.X., Buckingham K.J., Jhangiani S.N., Boehm C., Sobreira N., Smith J.D., Harrell T.M., McMillin M.J., Wiszniewski W., Gambin T. et al. . The genetic basis of mendelian phenotypes: Discoveries, challenges and opportunities. Am. J. Hum. Genet. 2015; 97:199–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liao Y., Smyth G.K., Shi W.. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014; 30:923–930. [DOI] [PubMed] [Google Scholar]
- 34.Olshen A.B., Venkatraman E.S., Lucito R., Wigler M.. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004; 5:557–572. [DOI] [PubMed] [Google Scholar]
- 35.Boone P.M., Bacino C.A., Shaw C.A., Eng P.A., Hixson P.M., Pursley A.N., Kang S.-H.L., Yang Y., Wiszniewska J., Nowakowska B.A. et al. . Detection of clinically relevant exonic copy-number changes by array CGH. Hum. Mutat. 2010; 31:1326–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.MacDonald J.R., Ziman R., Yuen R.K.C., Feuk L., Scherer S.W.. The database of genomic variants: a curated collection of structural variation in the human genome. Nucleic Acids Res. 2014; 42:D986–D992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Conrad D.F., Pinto D., Redon R., Feuk L., Gokcumen O., Zhang Y., Aerts J., Andrews T.D., Barnes C., Campbell P. et al. . Origins and functional impact of copy number variation in the human genome. Nature. 2010; 464:704–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mills R.E., Walter K., Stewart C., Handsaker R.E., Chen K., Alkan C., Abyzov A., Yoon S.C., Ye K., Cheetham R.K. et al. . Mapping copy number variation by population-scale genome sequencing. Nature. 2011; 470:59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu J., Grzeda K.R., Stewart C., Grubert F., Urban A.E., Snyder M.P., Marth G.T.. Copy number variation detection from 1000 genomes project exon capture sequencing data. BMC Bioinformatics. 2012; 13:305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Firth H.V., Wright C.F., DDD Study . The deciphering developmental disorders (DDD) study. Dev. Med. Child Neurol. 2011; 53:702–703. [DOI] [PubMed] [Google Scholar]
- 41.Engelhardt K.R., McGhee S., Winkler S., Sassi A., Woellner C., Lopez-Herrera G., Chen A., Kim H.S., Lloret M.G., Schulze I. et al. . Large deletions and point mutations involving the dedicator of cytokinesis 8 (DOCK8) in the autosomal-recessive form of hyper-IgE syndrome. J. Allergy Clin. Immunol. 2009; 124:1289–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abdel-Salam G., Thoenes M., Afifi H.H., Körber F., Swan D., Bolz H.J.. The supposed tumor suppressor gene WWOX is mutated in an early lethal microcephaly syndrome with epilepsy, growth retardation and retinal degeneration. Orphanet J. Rare Dis. 2014; 9:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karaca E., Yuregir O.O., Bozdogan S.T., Aslan H., Pehlivan D., Jhangiani S.N., Akdemir Z.C., Gambin T., Bayram Y., Atik M.M. et al. . Rare variants in the notch signaling pathway describe a novel type of autosomal recessive Klippel-Feil syndrome. Am. J. Med. Genet. A. 2015; 167:2795–2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lupski J.R. Genomic disorders ten years on. Genome Med. 2009; 1:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Altshuler D., Daly M.J., Lander E.S.. Genetic mapping in human disease. Science. 2008; 322:881–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee J.A., Lupski J.R.. Genomic rearrangements and gene copy-number alterations as a cause of nervous system disorders. Neuron. 2006; 52:103–121. [DOI] [PubMed] [Google Scholar]
- 47.Karakoc E., Alkan C., O'Roak B.J., Dennis M.Y., Vives L., Mark K., Rieder M.J., Nickerson D.A., Eichler E.E.. Detection of structural variants and indels within exome data. Nat. Methods. 2012; 9:176–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Retterer K., Scuffins J., Schmidt D., Lewis R., Pineda-Alvarez D., Stafford A., Schmidt L., Warren S., Gibellini F., Kondakova A. et al. . Assessing copy number from exome sequencing and exome array CGH based on CNV spectrum in a large clinical cohort. Genet. Med. 2015; 17:623–629. [DOI] [PubMed] [Google Scholar]
- 49.Lai K.K.S., Lo I.F.M., Tong T.M.F., Cheng L.Y.L., Lam S.T.S.. Detecting exon deletions and duplications of the DMD gene using Multiplex Ligation-dependent Probe Amplification (MLPA). Clin. Biochem. 2006; 39:367–372. [DOI] [PubMed] [Google Scholar]
- 50.De Luca A., Bottillo I., Dasdia M.C., Morella A., Lanari V., Bernardini L., Divona L., Giustini S., Sinibaldi L., Novelli A. et al. . Deletions of NF1 gene and exons detected by multiplex ligation-dependent probe amplification. J. Med. Genet. 2007; 44:800–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang F., Khajavi M., Connolly A.M., Towne C.F., Batish S.D., Lupski J.R.. The DNA replication FoSTeS/MMBIR mechanism can generate genomic, genic and exonic complex rearrangements in humans. Nat. Genet. 2009; 41:849–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang F., Seeman P., Liu P., Weterman M.A.J., Gonzaga-Jauregui C., Towne C.F., Batish S.D., De Vriendt E., De Jonghe P., Rautenstrauss B. et al. . Mechanisms for nonrecurrent genomic rearrangements associated with CMT1A or HNPP: rare CNVs as a cause for missing heritability. Am. J. Hum. Genet. 2010; 86:892–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boone P.M., Liu P., Zhang F., Carvalho C.M.B., Towne C.F., Batish S.D., Lupski J.R.. Alu-specific microhomology-mediated deletion of the final exon of SPAST in three unrelated subjects with hereditary spastic paraplegia. Genet. Med. 2011; 13:582–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Beunders G., Voorhoeve E., Golzio C., Pardo L.M., Rosenfeld J.A., Talkowski M.E., Simonic I., Lionel A.C., Vergult S., Pyatt R.E. et al. . Exonic deletions in AUTS2 cause a syndromic form of intellectual disability and suggest a critical role for the C terminus. Am. J. Hum. Genet. 2013; 92:210–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Amarillo I.E., Li W.L., Li X., Vilain E., Kantarci S.. De novo single exon deletion of AUTS2 in a patient with speech and language disorder: a review of disrupted AUTS2 and further evidence for its role in neurodevelopmental disorders. Am. J. Med. Genet. A. 2014; 164:958–965. [DOI] [PubMed] [Google Scholar]
- 56.Boone P.M., Yuan B., Campbell I.M., Scull J.C., Withers M.A., Baggett B.C., Beck C.R., Shaw C.J., Stankiewicz P., Moretti P. et al. . The Alu-rich genomic architecture of SPAST predisposes to diverse and functionally distinct disease-associated CNV alleles. Am. J. Hum. Genet. 2014; 95:143–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.DiVincenzo C., Elzinga C.D., Medeiros A.C., Karbassi I., Jones J.R., Evans M.C., Braastad C.D., Bishop C.M., Jaremko M., Wang Z. et al. . The allelic spectrum of Charcot-Marie-Tooth disease in over 17,000 individuals with neuropathy. Mol. Genet. Genomic Med. 2014; 2:522–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lindstrand A., Frangakis S., Carvalho C.M.B., Richardson E.B., McFadden K.A., Willer J.R., Pehlivan D., Liu P., Pediaditakis I.L., Sabo A. et al. . Copy-number variation contributes to the mutational load of Bardet-Biedl syndrome. Am. J. Hum. Genet. 2016; 99:318–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee H., Deignan J.L., Dorrani N., Strom S.P., Kantarci S., Quintero-Rivera F., Das K., Toy T., Harry B., Yourshaw M. et al. . Clinical exome sequencing for genetic identification of rare Mendelian disorders. JAMA. 2014; 312:1880–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang Y., Muzny D.M., Xia F., Niu Z., Person R., Ding Y., Ward P., Braxton A., Wang M., Buhay C. et al. . Molecular findings among patients referred for clinical whole-exome sequencing. JAMA. 2014; 312:1870–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bamshad M.J., Ng S.B., Bigham A.W., Tabor H.K., Emond M.J., Nickerson D.A., Shendure J.. Exome sequencing as a tool for Mendelian disease gene discovery. Nat. Rev. Genet. 2011; 12:745–755. [DOI] [PubMed] [Google Scholar]
- 62.Gilissen C., Hoischen A., Brunner H.G., Veltman J.A.. Disease gene identification strategies for exome sequencing. Eur. J. Hum. Genet. 2012; 20:490–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Teo S.M., Pawitan Y., Ku C.S., Chia K.S., Salim A.. Statistical challenges associated with detecting copy number variations with next-generation sequencing. Bioinformatics. 2012; 28:2711–2718. [DOI] [PubMed] [Google Scholar]
- 64.Harel T., Yoon W.H., Garone C., Gu S., Coban-Akdemir Z., Eldomery M.K., Posey J.E., Jhangiani S.N., Rosenfeld J.A., Cho M.T. et al. . Recurrent de novo and biallelic variation of ATAD3A, encoding a mitochondrial membrane protein, results in distinct neurological syndromes. Am. J. Hum. Genet. 2016; 99:831–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hehir-Kwa J.Y., Pfundt R., Veltman J.A.. Exome sequencing and whole genome sequencing for the detection of copy number variation. Expert Rev. Mol. Diagn. 2015; 15:1023–1032. [DOI] [PubMed] [Google Scholar]
- 66.de Ligt J., Willemsen M.H., van Bon B.W.M., Kleefstra T., Yntema H.G., Kroes T., Vulto-van Silfhout A.T., Koolen D.A., de Vries P., Gilissen C. et al. . Diagnostic exome sequencing in persons with severe intellectual disability. N. Engl. J. Med. 2012; 367:1921–1929. [DOI] [PubMed] [Google Scholar]
- 67.Rauch A., Wieczorek D., Graf E., Wieland T., Endele S., Schwarzmayr T., Albrecht B., Bartholdi D., Beygo J., Di Donato N. et al. . Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet. 2012; 380:1674–1682. [DOI] [PubMed] [Google Scholar]
- 68.Mefford H.C., Batshaw M.L., Hoffman E.P.. Genomics, intellectual disability and autism. N. Engl. J. Med. 2012; 366:733–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.