Abstract
Transitions of B-DNA to alternative DNA structures (ADS) can be triggered by negative torsional strain, which occurs during replication and transcription, and may lead to genomic instability. However, how ADS are recognized in cells is unclear. We found that the binding of candidate anticancer drug, curaxin, to cellular DNA results in uncoiling of nucleosomal DNA, accumulation of negative supercoiling and conversion of multiple regions of genomic DNA into left-handed Z-form. Histone chaperone FACT binds rapidly to the same regions via the SSRP1 subunit in curaxin-treated cells. In vitro binding of purified SSRP1 or its isolated CID domain to a methylated DNA fragment containing alternating purine/pyrimidines, which is prone to Z-DNA transition, is much stronger than to other types of DNA. We propose that FACT can recognize and bind Z-DNA or DNA in transition from a B to Z form. Binding of FACT to these genomic regions triggers a p53 response. Furthermore, FACT has been shown to bind to other types of ADS through a different structural domain, which also leads to p53 activation. Thus, we propose that FACT acts as a sensor of ADS formation in cells. Recognition of ADS by FACT followed by a p53 response may explain the role of FACT in DNA damage prevention.
INTRODUCTION
The prevailing DNA conformation in living cells is the right-handed double helix known as B-DNA. However, DNA may be folded in several different ways forming so-called alternative DNA structures (ADS) or variants of non-B DNA, such as triple and quadruple helices, cruciform and hairpin structures, or a left-handed double helix known as Z-DNA. While B- to non-B DNA transitions are energy consuming and rarely happen spontaneously, DNA torsional stress, such as negative supercoiling generated during RNA synthesis may induce these ADS transitions. Wrapping of DNA into nucleosomes creates an additional risk of ADS formation. Eukaryotic DNA is wound 1.65 times around an octamer of histone proteins (core) approximately every 200bp. This process leads to over-twisting of the double-helix; however, topoisomerases in cells relax linker DNA between nucleosomes. Conversely, the uncoiling of nucleosomal DNA results in the accumulation of negative supercoiling. Although negative supercoils or under-twisting of DNA facilitate transcription by promoting easier strand separation, they also present a potential risk for DNA transition into alternative forms. Indeed, ADS have been detected at sites of active transcription (1–4). Moreover, some ADS are involved in regulation of transcription (e.g. FUSE element in MYC promoter (5,6)).
At the same time, ADS are known triggers of genomic instability. Sites with nucleotide composition permissive for non-B DNA transitions are often involved in deletions, expansions or translocations, and are associated with cancer and neurodegenerative diseases (for review, see (7)). Thus, it would be beneficial for cells to recognize ADS before DNA damaging events occur. However, although several ADS binding proteins have been identified, a specialized signaling response to ADS formation in cells is not known.
The most frequent reason for nucleosome loss in cells is their destabilization caused by transcribing RNA polymerase. There is a special class of proteins, known as histone chaperones, which control nucleosome stability in cells. Histone chaperones ensure proper formation of histone oligomers before their deposition on DNA, and also protects the histone core from falling apart when its contact with DNA is weakened, e.g. during transcription. However, there has been no known link between DNA topology and activity of histone chaperones except for one case. It has been shown that histone chaperone FACT (FAcilitates Chromatin Transcription) can bind DNA containing platinum adducts, UV-induced thymine dimers or cruciform DNA, which all represent cases of non-B DNA or ADS, through HMG domain of SSRP1 subunit (8–10). HMG domain proteins are known to bind bent or kinked DNA (for review, see (11)). Treatment of cells with cisplatin or UV results in FACT-dependent activation of p53. Therefore, FACT binding to non-B DNA was interpreted as a DNA damage response by cells (10,12). However, we previously discovered small molecules with prominent anti-cancer activity, curaxins, that activated p53 through FACT without causing any detectable DNA damage (13). Lead curaxin, CBL0137, is currently being tested in clinical trials as an anti-cancer agent (NCT01905228). A search for the mechanism of action of curaxins revealed that their anti-cancer activity depends on their ability to bind DNA and to induce tight binding of FACT to chromatin, which paralyzes the transcription elongation activity of FACT (13). However, the mechanism(s) of FACT trapping in chromatin were unclear.
FACT is a dimer of SSRP1 and SPT16 subunits, both of which contain several domains that can interact with different components of nucleosomes (14–17). SPT16 subunit can bind the surface of H3/H4 tetramer, via its middle domain (MD), and H2A/H2B dimer, via C-terminl acidic domain (18,19,14,20). Some data from yeast suggest that SSRP1 subunit can also bind H2A/H2B dimer (14), but no such data exist for metazoan SSRP1. SSRP1 can bind nucleosomal DNA via HMG domain but this binding is inhibited in metazoan cells by phosphorylation (21,22). At the same time, it was recently shown in cell-free system that human FACT does not bind intact nucleosome, but needs some accessory factor(s) to provide access to its inner parts (18). It looks like that in a situation when DNA contact with histone core is weakened, FACT can hold nucleosome with partially unwrapped DNA, prevent disassembly of a core, and facilitate access of RNA polymerase to nucleosomal DNA (23).
In this study, we elucidated the mechanism of FACT trapping in chromatin in curaxin treated cells, which we named c-trapping and propose a new functional role of FACT as a sensor of non-B DNA transition in cells. We found that curaxin binding to DNA destabilizes nucleosome. Via SPT16 subunit FACT binds the surface of the H3/H4 tetramer, which is normally bound by the H2A/H2B dimer (18). However, this is not the only mechanism of c-trapping. Disassembly of nucleosomes in cells in the absence of DNA breaks results in negative supercoiling and extensive transition of B-DNA into left-handed Z-DNA form. SSRP1 binds genomic regions prone to this transition in curaxin treated cells. Importantly, the isolated C-terminal intrinsically disordered domain (CID) of SSRP1, but not HMG domain, underwent c-trapping in curaxin-treated cells. Thus, FACT possesses at least two different domains that recognize ADS: HMG for bent and cruciform DNA (9,12,24), and CID for DNA prone to form Z-DNA. Taking into account the ability of FACT to activate p53, c-trapping of FACT may be considered a novel cellular response that detects an increased risk of genomic instability prior to the occurrence of DNA damage. Moreover, the mechanism by which FACT selects areas of transcription is not known. Z-DNA or DNA in transition from B to Z form, which is formed at regions of high transcription due to nucleosome loss (25), may serve as a signal to recruit FACT. Finally, destabilization of chromatin may present a novel mechanism of activity of anti-cancer agents that hit the same universal target, DNA, but without the mutagenic effect of conventional chemotherapy.
MATERIALS AND METHODS
Cells, reagents and constructs
HT1080, HeLa, DLD1 and MM1.S cells were obtained from ATCC and maintained according to ATCC recommendations. Cells expressing tagged SSRP1 and SPT16 were obtained through lentiviral transduction (26), and tagged histones through transfection using Lipofectamine 2000 (Invitrogen). Stable cell lines were obtained through sorting of cells positive for tag-specific fluorescent signal.
The curaxin CBL0137 was provided by Incuron, LLC. Cisplatin, ethidium bromide, propodium iodide, bovine serum albumin (BSA), and agarose were purchased from Sigma-Aldrich. Hoechst 33342 was purchased from Invitrogen. Unmodified oligonucleotides were ordered from IDT DNA Technology. Methylated oligonucleotides was custom synthesized by Sigma-Aldrich.
The following plasmids were used in the study: pH2B_mCherry_IRES_neo3 (27) was a gift from Daniel Gerlich (Addgene plasmid # 21044), mKate-H3-23 (Addgene plasmid # 56060) and mOrange2-H4-23 (Addgene plasmid # 57964) were gifts from Michael Davidson. N- and C-terminal GFP-tagged human SSRP1 constructs were previously described (13,28). Flag, GFP or mCherry tagged human codon-optimized SPT16 lentiviral constructs was ordered from Genecopoeia. Cloning of SSRP1 deletion mutant tagged with 3XFlag or GFP, provided with nuclear localization signal, is described in Supplementary Materials. Flag-tagged human SSRP1 was cloned into pLA-CMV-N-Flag vector. Recombinant proteins were overexpressed in HeLa cells using lentiviral transduction, and purified from cell lysates using ANTI-FLAG® M2 Affinity Gel purification system (Sigma-Aldrich, A2220). Human FACT complex was purified from Flag-SPT16 and His-tagged SSRP1expressing S9 cells as previously described (29) using constructs provided by D. Reinberg (New York University School of Medicine, New York, NY, USA).
Circular dichroism
Circular dichroism (CD) was done using cholesteric liquid-crystalline dispersion (CLD). Сalf thymus DNA (Sigma) was sonicated using ultrasonic dispergator UZDN-2T (SPE UKRROSPRIBOR Ltd, Ukraine) to obtain fragments of (0.5–0.8) × 106 Da. The CD spectra were registered by portative dichrometer SKD-2 (Institute of Spectroscopy, RAS, Troitsk, Russia). Quartz cuvettes with an optical path length of 1 cm were used. DNA liquid dispersion was formed by mixing equal volumes of aqueous solutions (with 0.3М NaCl; 2 mM Na–phosphate buffer, рН 6.8) of DNA and polyethylene glycol (PEG, M 4000Da, Fluka) with concentration of 340 mg/ml. The CD spectrum was registered 1 h after solution mixing. The formation of DNA liquid dispersion was confirmed by the appearance of an intensive band (λmax = 270 nm, Δε = 130–140 M–1 cm–1). 1–4 μl of CBL0137 stock solution were then added to the formed DNA liquid dispersion solution. After intensive stirring, the CD spectrum was registered again in the DNA and CBL0137 chromophores spectral absorption region (250–400 nm).
Effect of CBL0137 on DNA topology
DNA topoisomer formation induced by CBL0137 was assessed using pUC19 vector from New England BioLabs (NEB). DNA was nicked with Nb.BsrDI endonuclease (NEB). Nicked DNA was treated for 15 min with CBL0137 in T4 Ligase buffer at RT, and then T4 Ligase was added for 15 min. The ligation reaction was stopped by adding EDTA to a final concentration of 25 mM. DNA was isolated by phenol/chloroform extraction, followed by ethanol-based precipitation, loaded on 1% agarose gel and run in TAE buffer at 5 V/cm for 5 h. Gels were stained with 0.1 μl/ml Gel Red (Biotium) and imaged using UVP GelDoc-It TS imaging system equipped with a Series 6100 Camera (UVP).
DNA-ase I footprinting
DNA-ase I footprinting was performed as previously described (30). Details of nucleotide composition can be found in Supplementary Materials.
In vitro nucleosome reconstitution assays
Mononucleosome based assays: histones were prepared as previously described (31). DNA fragment based on the ‘601’ nucleosome positioning sequence (32) was used. Labeling mutations to H2B (T112C) and H4 (E63C) allowed for the addition of fluorescent tags to histone complexes of interest (33). Alexa488 C5-maleimide (Invitrogen) and Atto-647N maleimide (Sigma-Aldrich) was used to label H4 and H2B, respectively. Nucleosome particles were constructed using salt dialysis techniques (34). Reactions were run in buffer containing of 20 mM Tris pH 7.5-8, 150 mM NaCl, 1 mM TCEP, 0.01% Octyl-G and 0.01% CHAPS. For the nucleosome disassembly assay, 0.5 μM premade nucleosomes were incubated with increasing amounts of curaxin at RT for 20 min. For FACT binding assay, 0.1–0.2 uM of nucleosome was incubated with increasing concentrations of FACT (0, 25, 50, 100, 200 nM), in the presence or absence of CBL0137. For nucleosome assembly, stoichiometric ratios of fluorescently labeled H2A–H2B and H3–H4 complexes were mixed with 2-fold excess of DNA (0.5 μM) as described in (35). The mixtures were incubated at RT for 20 min. Flag-SPT16 and His-SSRP1 were titrated into a histone/DNA mixture (at constant concentration) and incubated at RT for an additional 20 min as previously described (36) in the absence or presence of 10μM CBL0137. Samples were run on a 5% native PAGE gel at 150 V for 1 h at 4°C. After electrophoresis, gels were scanned for fluorescent visualization and/or soaked in ethidium bromide and imaged with GelDoc-It TS imaging system equipped with a Series 6100 Camera (UVP).
Polynucleosome based assay: polynucleosomes were assembled using pUC19 circular plasmid DNA and Chromatin Assembly kit from Active Motiff (cat# 53500) according to manufacture instructions.
SSRP1 binding to dsDNA oligonucleotides
The following DNA oligonucleotides were ordered from IDT DNA Technology (unmodified) or Sigma-Aldrich (methylated) and annealed with corresponding complementary oligonucleotides: random nucleotide probe - AGCAGACCACGTGGTCTG; AT probe – T10(AT)10T10; AC probe – (AC)18; GC probe – (GC)18; AmeC probe – (A-5meC)15A; GmeC probe – (G-5meC)15G, labeled with 32P using PNK4 (NEB), gamma-32P-dATP (Perkin Elmer). The 20 μl binding reaction consisted of 250 pM DNA probe mixed with either SSRP1, SPT16 or CID domain of SSRP1 (0.6–2.4 μM), or Z-DNA antibody (Abcam, cat# ab2079) or control sheep IgG (Abcam, cat# ab37385) (4 μg, ∼1 μM). For the supershift, the following SSRP1 antibodies were used: mouse monoclonal 10D1 (BioLegend, cat# 609702), goat polyclonal D15 (Santa Cruz Biotechnology, cat# sc5909), mouse polyclonal D-7 (Santa Cruz Biotechnology, cat# sc74536), or mouse IgG (Abcam, cat# ab37355). Mixed reactions were incubated at RT for 15–30 min in binding buffer composed of 20 mM HEPES, 50 mM KCl, 10% glycerol, 0.1 mM EDTA, 1 mM DTT and 200 μg/ml BSA. Reactions were run in 8% PAGE at 150 V for 2 h.
Topoisomerase activity assays
150ng of supercoiled DNA (pHOT-1, TopoGEN, Port Orange, FL, USA) was mixed with 1.25 units of human Topoisomerase I or four units of human Topoisomerase IIα (both from TopoGEN, Port Orange, FL) and test compounds in a 20 μl final volume in assay buffers for either Topoisomerase I (10 mM Tris–HCl, 1 mM EDTA, 150 mM NaCl, 0.1% bovine serum albumin, 0.1 mM spermidine, 5% glycerol) or Topoisomerase II (50 mM Tris–HCl, 5 mM ATP, 150 mM NaCl, 30 μg/ml bovine serum albumin, 0.5 mM dithiotreitol, 10 mM MgCl2). Reactions were incubated at 37°С for 30 min and terminated with SDS (final concentration of 1%) and proteinase K (50 μg/ml) for 2 h at 50°С. Samples were mixed with DNA loading buffer, and loaded onto 1% agarose gel. The gel was run in 1× TAE buffer at 1 V/cm for 3–4 h. Following electrophoresis, the gel was stained with 0.5 μg/ml ethidium bromide and visualized using the GelDoc-It TS imaging system equipped with a Series 6100 Camera (UVP).
Micrococcal nuclease digestion assay
Micrococcal nuclease digestion assay was performed as previously described (37) with some modifications. HeLa cells were trypsinized, harvested, and washed once with 1× RSB buffer (10 mM Tris, pH7.6, 15 mM NaCl and 1.5 mM MgCl2). After centrifugation, the cell pellet was resuspended in 1× RSB buffer with 1% Triton X-100 and homogenized by five strokes with a loose-fitting pestle to release nuclei. Nuclei were collected by centrifugation and washed twice with 1 ml of buffer A (15 mM Tris, pH 7.5, 15 mM NaCl, 60 mM KCl, 0.34 M sucrose, 0.5 mM spermidine, 0.15 mM spermine, 0.25 mM PMSF and 0.1% β-mercaptoethanol). Finally, nuclei (from 20 × 106 cells) were resuspended in 1.5 ml of buffer A, and 15 μl of 0.1 M CaCl2 was added. Nuclei were treated for 15 min with CBL0137 at 37°C. For digestion, 1 μl of 200 U/ml micrococcal nuclease (NEB) was added to 0.5 ml nuclei suspension at 37°C. Aliquots (60 μl) were taken at each time point, and 1.5 μl 0.5 M gbv ouyuioo;' EDTA was added to stop the reaction (final concentration: 12.5 mM). Subsequently, 18 μl H2O, 12 μl of 10% SDS and 24 μl of 5 M NaCl were added to each tube. The mixtures were extracted with phenol–chloroform followed by ethanol-based precipitation. DNA was analyzed on a 1.5% agarose gel, stained with 0.5 μg/ml ethidium bromide in 1× TAE for 30 min, destained for 2 × 15 min in ddH2O, and imaged as described above.
S1 nuclease digestion
Sensitivity of cellular DNA to S1 nuclease digestion followed by flow cytometry assessment of DNA content was done as described by Prosperi et al. (38) with some modifications. Nuclei were prepared from HeLa cells as described above. Isolated nuclei were resuspended in S1 digestion buffer. Nuclei were treated for 15 min with 50 μM CBL0137 at 37°C. For digestion, 1500 U of S1 was added to suspension of 106 nuclei at 37°C for 30 min. Staining was done with 1 μg/ml propidium iodide for 30 min. Assessment of DNA content was done using LSRII flow cytometer (Becton Dickinson). Fluorescent intensity of unstained cells was taken as 0. Fluorescent intensity corresponding to G1 peak in untreated control cells was taken as 1.
Protein extraction and western blotting
Soluble protein extracts were prepared by incubation of cells in 1× Cell Culture Lysis Reagent (Promega, Madison, WI, USA) with Complete Protease Inhibitor Cocktail (Roche) on ice for 20 minutes. Samples were centrifuged at 4°C at 13 000 rpm for 20 min, and the supernatant was collected. Insoluble proteins were obtained by resuspension of the remaining pellets in 1× Cell Culture Lysis Reagent (Promega, Madison, WI) and sonicated three times for 30 s each using Bioruptor UCD-200 (Diagenode).
Nucleoplasm and intact chromatin were isolated from cells as described in (39) for the detection of c-trapping of C-terminal half of SSRP1, HMG or CID domains. Chromatin was resuspended in RIPA buffer – 50 mM Tris–HCl, pH 7.4, 1% NP-40, 0.5% Na-deoxycholate, 450 mM NaCl, 1 mM EDTA, 1 mM EGTA on ice for 30 min followed by soniction.
For histone detection, cells were lysed in RIPA buffer with 150 mM NaCl, 1 mM EDTA, 1 mM EGTA on ice for 30 min followed by centrifugation. The pellet was resuspended in the same buffer with the addition of 450 mM NaCl and processed using the same regimen followed by sonication. Extraction of chromatin proteins with different concentrations of NaCl was done as described in (39).
Protein concentrations were determined with the Bio-Rad DC protein assay kit (Hercules, CA, USA). Samples were mixed with Laemmli Gel Loading buffer and boiled for 15 min at 100°C. For total cell extracts, normalized numbers of cells were directly lysed in Laemmli Gel Loading buffer and boiled for 15 min before electrophoresis. Western blotting was run as previously described (13). Primary antibodies and their concentrations were used as follows: SSRP1, mouse monoclonal 10D1 (BioLegend, cat# 609702; 1:4000) or rabbit polyclonal (Abcam, cat# ab21584; 1:1000); SPT16, mouse monoclonal 8D2 (BioLegend, cat# 607002; 1:1000); GFP, rabbit polyclonal (Santa Cruz Biotechnology, cat# SC8334; 1:500); Flag, mouse monoclonal M2 (Sigma-Aldrich, cat# F3165; 1:2000); β-actin, (Sigma, cat# a1978; 1:20 000). Horseradish peroxidase-conjugated secondary antibodies were purchased from Santa Cruz Biotechnology. Antibody for histones—H2A mouse (cat# 3636S), H2B rabbit (cat# 12364S), H3 rabbit (cat# 4499S) and H4 rabbit (cat# 2592P) were purchased from Cell Signaling.
Immunofluorescence staining and live cell microscopy
Cells were plated in 35 mm glass bottom plates from MatTek Corporation (Ashland, MA, USA). For live cell fluorescent image acquisition, cells were treated with drugs and images were obtained at different time points with a Zeiss Axio Observer A1 inverted microscope with N-Achroplan 100×/1.25 oil lens, Zeiss MRC5 camera, and AxioVision Rel.4.8 software. Alternatively, cells were fixed in 4% paraformaldehyde for 10 min before imaging.
For immunofluorescence staining, cells were washed with PBS and fixed in 4% paraformaldehyde at room temperature for 15 min. For Z-DNA staining, a 4% paraformaldehyde solution containing 0.1% Triton-X100 in PBS was added to cells for 15 min immediately after removal of media. Cells were then washed three times with PBS. Blocking was done in 3% BSA, 0.1% Triton-X100 in PBS. Primary antibodies, SSRP1 from BioLegend (the same as for immunoblotting), for Z-DNA from Abcam (cat# ab2079) were used at 1:200 dilution. AlexaFluor 488 donkey anti-mouse (Invitrogen, cat# A21206; 1:1000) and AlexaFluor 594 donkey anti-sheep (Jackson ImmunoResearch, cat# 713-585-147; 1:500) were used as a secondary antibodies. Antibodies were diluted in 0.5% BSA + 0.05% Triton X100 in PBS. After each antibody incubation, cells were washed three times with 0.05% Triton X100 in PBS. For DNA counterstaining, 1 μg/ml solution of Hoechst 33342 in PBS was used.
Images of nuclei were slightly rescaled (no >30% of orginial size) in order to be of approximately the same size. Figure panels with re-scaled images are shown without scale bars. Contrast and brightness were adjusted for images that demonstrated c-trapping. No adjustment was done for Z-DNA staining. Image analyses and quantitification was done using ImageJ.
ChIP-sequencing and analysis
ChIP was performed using HT1080 as previously described (40). Experiments with treated (3 μM of CBL0137, 1 h) and untreated HT1080 cells were repeated three times. The ChIP libraries were single-end sequenced on an Illumina HiSeq2000 with 50 bp reads. Each sample was sequenced in a single flow cell lane and generated 89–190 million reads.
ChIP with MM1.S cells was performed using similar conditions in two replicates. The pooled ChIP libraries were sequenced on Illumina HiSeq2000 with 100 bp paired-end reads to generate ∼40 million reads per sample. The resulting raw sequencing reads were filtered for quality and aligned to the most recent build of the human genome (hg19) with BowTie.
The peak positions in relation to genome features were calculated using MACS (41). For this, the data from replicate samples were concatenated together. Sequences of peaks reproduced in HT1080 and MM1.S cells treated with curaxin were analyzed for motif enrichment using MEME motif-based sequence analysis tool.
Statistical analysis of co-localization between SSRP1 binding sites in curaxin-treated cells and non-B DNA
SSRP1 binding sites identified in curaxin-treated cells and seven non-B DNAs (a phased repeat, direct repeat, G quadruplex, inverted repeat, mirror repeat short tandem repeat and Z-DNA motif) downloaded from non-B DNA database were used to identify which non-B DNA significantly overlaps with the binding sites. Two different statistical methods, LOLA and ColoWeb, were employed. ColoWeb is a web based tool that calculates two different statistics for enrichment and lack of enrichment of genomic features using the histogram generated from feature density matrix. Also, it calculates a conservative empirical p-value by bootstrapping within the histogram and produces visualization of both feature density matrix and the histogram. LOLA is an R package that tests the enrichment between regions of interest against genomic feature databases using Fisher-exact test. LOLA requires the user to define the universe, which contains all possible genomic locations of region of interest; we used the union of all peaks generated using MACS with P-value <0.05 and all non-B DNA regions. The two methods generate consistent results.
Molecular modeling
A superimposition of 3D conformer of CBL0137 with DNA was analyzed by computer modelling. The molecule docking to DNA was performed with the MOLOC (Gerber Molecular Design) molecular mechanics tools and by GOLD software (Cambridge Crystallographic Data Center).
Accession numbers
Sequencing data in the form of bed files are available at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE45393.
RESULTS
FACT binds to unfolding chromatin in CBL0137-treated cells
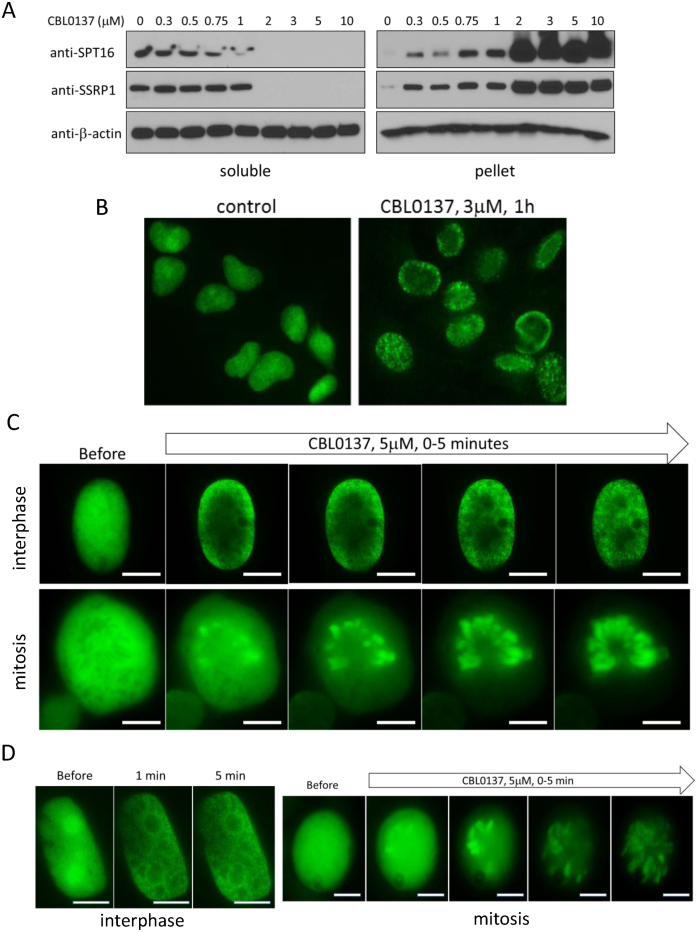
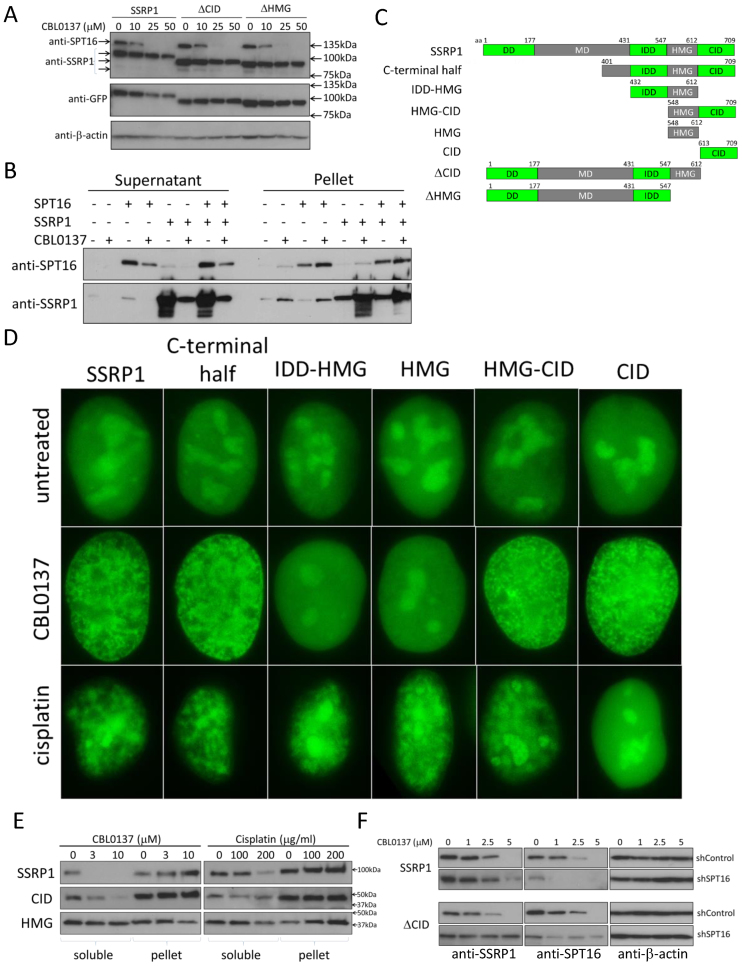
Using biochemical fractionation and fluorescent microscopy we previously demonstrated that curaxin treatment causes a rapid transition of FACT from the nucleoplasm to a state of tight association with chromatin (13). We named this phenomenon ‘chromatin trapping’ of FACT or c-trapping. To investigate the mechanism of c-trapping, we first examined the dose and time dependence as well as specificity of this phenomenon. To do this we used two methods, (i) immunoblotting of soluble, representing nucleoplasm, and pelleted, representing chromatin, fractions of total cell lysates ((Figure 1A) see details in Materials and Methods), and (ii) ectopic expression of GFP or mCherry tagged SSRP1 and SPT16 subunits of FACT, which undergo c-trapping in CBL0137-treated cells similarly to endogenous FACT subunits (Figure 1B–D and Supplemental Figure S1).
Figure 1.
C-trapping of FACT. (A) Immunoblotting of soluble protein extracts and chromatin pellets of HT1080 cells treated with CBL0137 for 1 h, probed with the indicated antibodies. (B) Immunofluorescence staining of HT1080 cells with antibodies to SSRP1. (C) Fluorescent imaging of two live cells expressing GFP-tagged SSRP1 before and after treatment with CBL0137. Top row—HT1080 cell in interphase, 3μM CBL0137, bottom row—DLD1 cell in mitosis, 5 μM CBL0137. (D) Fluorescent imaging of two live HT1080 cells, interphase and mitotic, expressing GFP-tagged SPT16 before and after treatment with CBL0137. Bars are 10μm.
C-trapping was observed in all tested cell lines (>20 cell lines, data not shown). Importantly, c-trapping and toxicity of CBL0137 occur at the same concentrations (∼0.3–0.5 μM depending on cell line, Figures 1A, 2 and (13)), however, c-trapping is observed within minutes after start of treatment (Figure 1C and D), while cell death after 48 h (13). Thus, these two effects of CBL0137 are probably connected, but c-trapping cannot be viewed as a sign of cell death.
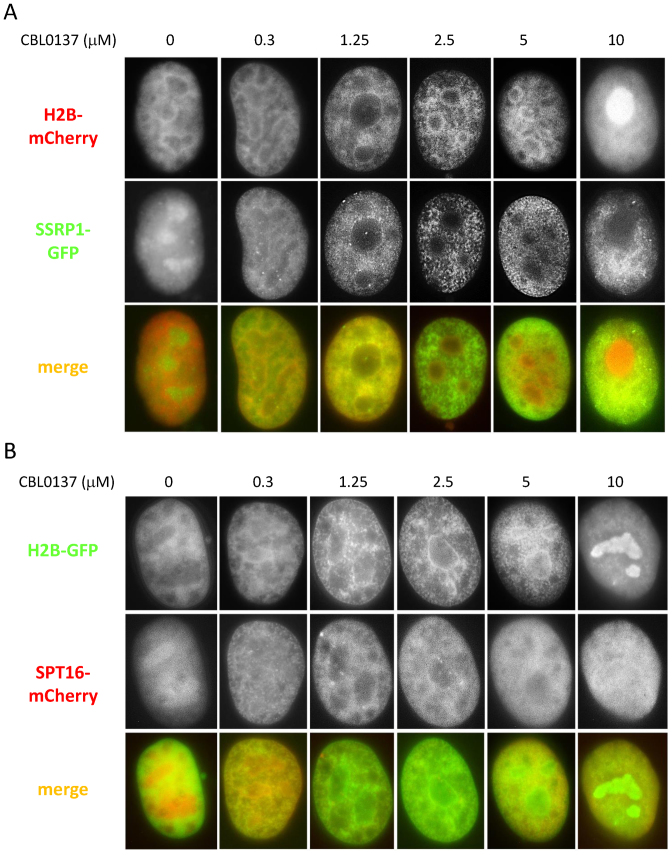
Figure 2.
Dose-dependent changes in the distribution of FACT subunits and histone H2B in CBL0137-treated cells. Fluorescent imaging of HT1080 cells expressing either mCherry tagged H2B and GFP-tagged SSRP1 (A) or GFP-tagged H2B and mCherry tagged SPT16. Cells were treated for 1 h and then fixed with 4% paraformaldehyde.
In untreated cells, FACT is diffusely spread throughout the nucleoplasm with some accumulation in nucleoli ((13,42) and Figure 2A and B). Changes in the distribution of FACT subunits begin 30–60 s after the addition of CBL0137 to cell culture medium (Figure 1C and D). In the presence of 0.3 μM CBL0137, both subunits become associated with thin fibers of chromatin, which is illustrated by their overlap with fluorescently labeled histone H2B (Figure 2A and B). At higher concentrations of CBL0137 (1–3 μM), FACT subunits are still associated with chromatin, however, the chromatin itself changes such that it appears as thicker fibers with bead like structures contouring nucleoli (Figure 2A and B). At concentrations of ≥5 μM CBL0137, the cellular distribution of SSRP1, SPT16, and H2B is dramatically changed. SSRP1 appears as thin fibers situated along the nuclear periphery and absent in and near nucleoli (Figure 2A). SPT16 becomes more diffuse (Figure 2B). Histone H2B loses its fiber-like appearance and accumulates in nucleoli (Figure 2A and B). Full transition takes 20–30 min (Figure 3A). Behavior similar to H2B was observed with fluorescently tagged histones H3 and H4 (Supplemental Figure S2).
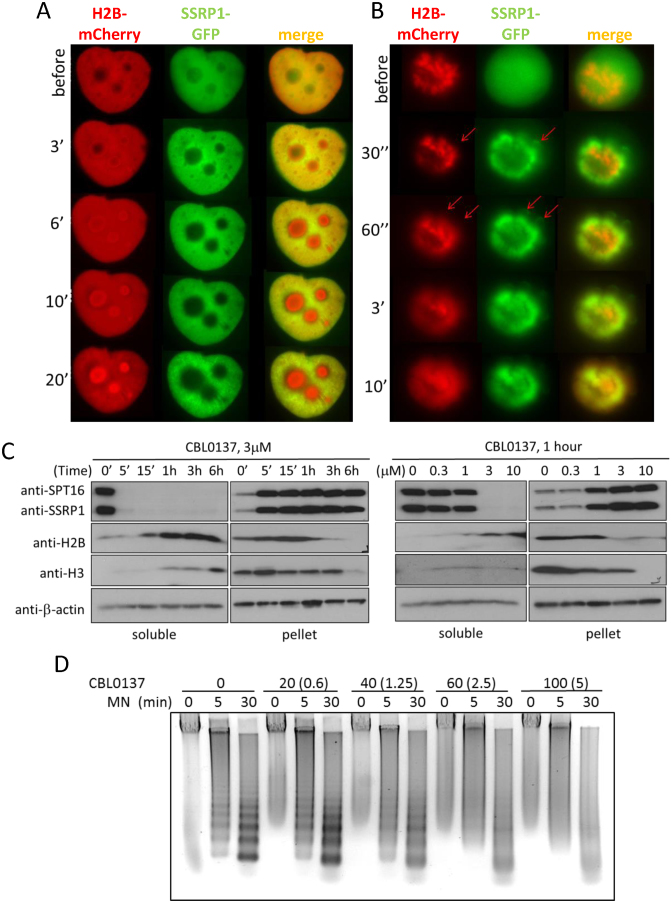
Figure 3.
Loss of histone from chromatin in CBL0137-treated cells. (A, B) Live cell images of one Hela-H2B-mCherry/GFP-SSRP1 cell in interphase (A) or undergoing mitosis (B) before and after CBL0137 treatment (5 μM). Red arrows indicate loops (possibly DNA) seen in GFP, but not mCherry channels. (C) Immunoblotting of soluble extracts and chromatin pellet of HeLa cells treated with CBL0137, probed with the indicated antibodies. (D) Gel electrophoresis of DNA isolated from nuclei of HeLa cells incubated with CBL0137 followed by digestion with micrococcal nuclease (MN). The actual concentration of CBL0137 in the incubation buffer and its concentration in the corresponding cell culture medium recalculated per nuclei (in the parentheses) are shown.
Similar to our results presented here, Musinova et al previously demonstrated that histone H2B, which is not incorporated into chromatin, accumulates in nucleoli due to the presence of a nucleoli localization signal within its nuclear localization signal (43). Therefore, we hypothesized that CBL0137 treatment leads to accumulation of histones in nucleoli due to their liberation from chromatin, which is consistent with the loss of visible chromatin fibers (Figure 2A and B, 10 μM).
C-trapping not only occurs in interphase cells but also in mitotic cells. As shown in Figure 1C and D, diffuse GFP-SSRP1 or GFP-SPT16 signals in mitotic cells became overlapped with mitotic chromosomes within a few minutes following the addition of CBL0137, which is indicative of a rapid CBL0137-induced binding of FACT to chromatin (Figure 1C and D). However, later on and in the presence of ≥3 μM of CBL0137, histone H2B was lost from chromosomes and appeared more like a diffuse cloud (Figure 3B). FACT subunits followed histones into the cloud-like shape, however, some SSRP1 was still associated with loops in the periphery, which were most likely leftover of decondensed and disassembled chromatin (Figure 3B).
If CBL0137 treatment causes chromatin disassembly with histone loss from chromatin, then free histones should be detectable in the soluble fraction of cell extracts upon CBL0137 treatment. Indeed, accumulation of core histones in the soluble fraction and loss from the chromatin pellet was detected via western blotting in cells treated with ≥3 μM CBL0137 10–20 minutes after start of treatment (Figure 3C).
Histone loss from chromatin should make genomic DNA more sensitive to digestion with nucleases. Therefore, we tested this hypothesis using micrococcal nuclease (MN) digestion assay, which demonstrated a dose-dependent loss of ordered nucleosome-protected ladder of DNA fragments in CBL0137-treated cells (Figure 3D). Interestingly, instead of complete digestion of nucleosome-free DNA into nucleotides, which was expected in the case of complete nucleosome disassembly at high dose of CBL0137 (>5 uM in cell-based experiments), we observed a DNA smear, which may indicate either loss of ordered nucleosome arrays in cells (different degree of nucleosome opening and loss) or inhibition of MN activity by CBL0137. To distinguish between these possibilities, we compared MN digestion of protein-free genomic DNA in the presence and absence of CBL0137. As shown on Supplemental Figure S3, the presence of CBL0137 significantly inhibited MN digestion of pure DNA, suggesting that CBL0137 inhibited nuclease activity as has been shown for other DNA binding compounds (44).
In summary, we found that CBL0137 treatment induces changes in chromatin structure, which may result in exposure of sites for FACT binding within inner part of nucleosome. With increasing dose of CBL0137 these changes lead to chromatin destabilization and appearance of free histones. At the same concentrations (≥5 μM of CBL0137), overlap of SSRP1 and SPT16 signals is lost, what can be interpreted as either separation of FACT subunits or/and their binding to different structures.
Curaxin binding to DNA destabilizes nucleosomes
It was recently shown that human FACT does not bind the intact nucleosome, but rather it can invade into nucleosomes with partially stripped DNA (18). SPT16 subunit binds to a hexasome, which is a nucleosome with one H2A/H2B dimer detached, via an interaction between its middle domain and the surface of H3/H4 tetramer that is normally bound by H2A/H2B dimer (18). Thus, we proposed that uncoiling of DNA from the histone core induced by CBL0137 treatment exposes SPT16 binding sites. To test effect of CBL0137 on nucleosome stability in cell-free system, we assembled mononucleosomes from recombinant histones and a 207 bp linear DNA fragment (‘601’ nucleosome positioning sequence (32)) or polynucleosomes using circular plasmid DNA. Incubation of mononucleosomes with increasing concentrations of CBL0137 led to the appearance of hexasomes, emergence of free DNA, and complete loss of nucleosome band at higher concentrations (Figure 4A). The effect of CBL0137 on artificial chromatin assembled in vitro using plasmid DNA and recombinant histones was very similar to its effect on chromatin in cells, resulting in loss of nucleosome protected bands upon MNase digestion of chromatin and appearance of a DNA smear (Figure 4B).
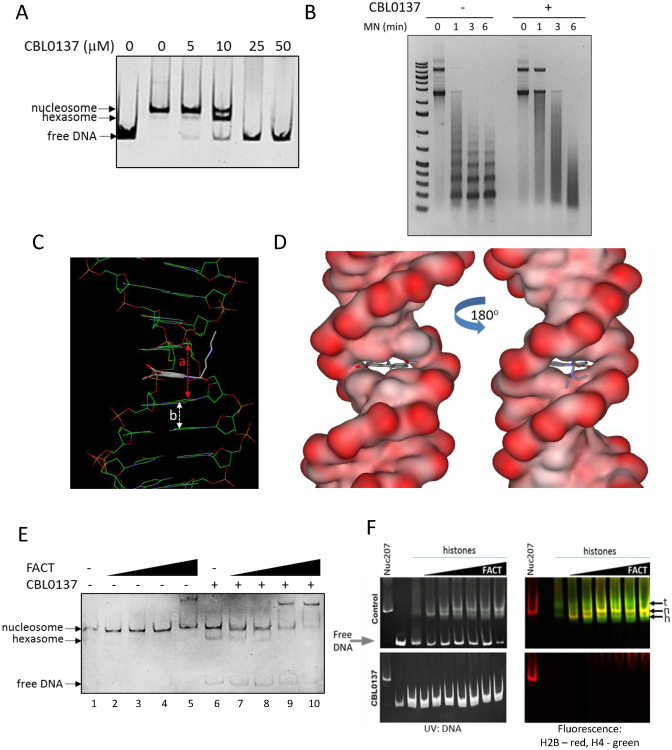
Figure 4.
CBL0137 destabilizes nucleosomes and causes FACT binding to chromatin in vitro. (A) Gel electrophoresis of preassembled mononucleosome Nuc207 incubated with different concentrations of CBL0137. (B) Gel electrophoresis of the products of micrococcal nuclease (MN) digestion of polynucleosome, assembled from circular plasmid DNA and recombinant histones, in the presence and absence of 10μM CBL0137. (C and D) Computer modeling of CBL0137 binding to DNA. a and b – interbase pairs distance in the presence (a) and absence (b) of curaxin. (E) Gel electrophoresis of preassembled mononucleosome Nuc207 incubated with different concentrations of FACT (10, 50, 100, 200 nM) and 10 μM CBL0137. (F) In vitro FACT assisted nucleosome assembly is inhibited by CBL0137. Gel electrophoresis of 207 bp DNA fragment incubated with histones and increasing concentration of FACT, with or without CBL0137 (25 μM). The same gels were visualized using UV for ethidium bromide stained (in gel) DNA (left) and fluorescently labeled histones (right). Control panels show reduction of free DNA and increase in bands corresponding to fully assembled nucleosome (Nuc207). Nuc207 control was not incubated with CBL0137. Arrows indicate positions of nucleosome (n), hexasome (h) and tetrasome (t).
Pang et al showed that DNA intercalators from the antracycline family destabilize nucleosomes and this effect was not related to their DNA damaging activity (45). We already observed that curaxins bind DNA (13). In this current study, we aimed to establish the mode of their binding to DNA to better understand their effect on nucleosomes. Computer modeling suggested that the carbazole moiety of curaxins (e.g. CBL0137) intercalates between DNA base pairs. Symmetrical side chains of the molecule at positions 3 and 7 of carbazole protrude into the major groove of DNA, while carbazole N-side chain fills the minor grove (Figure 4C and D; Supplemental Figure S4A). Circular dichroism and DNAse I footprinting confirmed intercalation and change in DNA topology (Supplemental Figure S4B, C) in the presence of CBL0137. Although CBL0137 does not cause chemical modifications in DNA, intercalation of the carbazole moiety substantially increases the distance between base pairs (Figure 4C), leading to untwisting of DNA, a phenomenon observed with other intercalators (46). We demonstrated this effect in vitro as a change of mobility of circular nicked DNA incubated first with CBL0137, then ligated to lock it's twisting followed by removal of the small molecule (Supplemental Figure S4D). Using different concentrations of CBL0137 in this assay, we detected a dissociation constant (Kd) of CBL0137–DNA binding as 40 ± 20μM (see details in Supplementary Information ‘Calculations of Kd of CBL0137’). Based on this, there are between 0.015 and 0.04 molecules of CBL0137 bound to DNA per base pair at equilibrium or one molecule of ligand per 25–66 bp of DNA. In cells, c-trapping is developed in a range of ∼0.2–10 μM of CBL0137 (Figures 1B and 2, see details of calculations in Material and Methods), which is around ∼1 molecule per 5–100 bp of DNA. We used this range of DNA/ligand ratio in all further cell-free experiments. Thus, high affinity binding of CBL0137 to DNA changes the shape of the double helix, destabilizes nucleosomes and leads to DNA uncoiling from the nucleosome core.
Based on the recent data of Tsunaka et al., DNA uncoiling from the nucleosome core, induced by CBL0137, should expose sites for FACT binding normally shielded by DNA in intact nucleosome (18). To test this, we incubated mononucleosomes with recombinant FACT in the presence or absence of CBL0137. In the absence of CBL0137, no binding of FACT to the intact nucleosome was detected until the highest concentration of FACT used, where a shift of a fraction of the nucleosome band appeared (Figure 4E, lane 5). The addition of CBL0137 at the concentration that caused partial nucleosome disassembly (appearance of hexasome and some free DNA, while most of nucleosome band is still present (Figure 4E, lane 6)), resulted in FACT binding to both hexasomes and nucleosomes. Importantly, in line with the findings of Tsunaka et al. (18), FACT preferentially binds to the hexasome, since the hexasome band completely disappeared whereas the nucleosome band was only reduced (lanes 7–10).
Finally, we tested what happened with the nucleosome assembly function of FACT in the presence of CBL0137. We compared the in-gel mobility of the product of the reaction, which consisted of DNA, fluorescently labeled histones and increasing amounts of FACT. As shown on Figure 4F, addition of FACT leads to the appearance of the nucleosome band whereas in the presence of CBL0137 DNA stays unbound and free histones did not emerge in a native gel (Figure 4F). Thus, FACT was unable to assemble nucleosome with CBL0137 bound DNA.
Our data demonstrated that binding of CBL0137 to DNA alters the shape of the double helix and destabilizes nucleosomes, leading to separation of H2A/H2B dimer and formation of hexasome at lower and complete disassembly at higher concentrations of CBL0137. FACT binds nucleosome components in the process of disassembly.
SSRP1 and SPT16 subunits of FACT binds different components of disassembling nucleosome in cells
It was shown in cell free conditions that FACT binds different components of the nucleosome via separate domains (16,18,21,23,35,47,48). To better understand which part of the nucleosome FACT binds in cells during c-trapping, we aimed to establish which domain of FACT is involved in c-trapping.
First, we tested whether SSRP1 or SPT16 can bind chromatin in cells treated with CBL0137 in the absence of the another subunit, which was depleted using shRNA. Because SPT16 was unstable and undetectable in the absence of SSRP1, which is in line with the mechanism of regulation of FACT complex stability (28), we were able to do this experiment only with SSRP1. Depletion of SPT16 did not abrogate the c-trapping of SSRP1 (Figure 5A), suggesting that SSRP1 can bind to some component of chromatin in human cells. This finding was unexpected since, in contrast to yeast SSRP1 homolog, Pob3, for which direct binding to histones was demonstrated (14,16,49,50), no such data exist for mammalian SSRP1. SSRP1 can bind free DNA or DNA within the nucleosome, but only if SSRP1 is unphosphorylated (21,35) or DNA is bent, such as by the addition of platinum adducts in cis-configuration (24). In higher eukaryotes, most SSRP1 in cells is phosphorylated (21,22). The rate at which c-trapping occurs excludes the possibility of that CBL0137 induced the dephosphorylation of SSRP1 in cells. Moreover, in our previous study, we already tested the binding of SSRP1 to a DNA fragment as well as to reconstituted mononucleosomes in the presence of CBL0137. Independently of CBL0137, SSRP1 that was isolated from insect cells did not bind free DNA or mononucleosomes (13). Thus, we focused on confirming the observation that SSRP1 could undergo c-trapping in CBL0137-treated cells in the absence of SPT16 using two independent approaches.
Figure 5.
Different SSRP1 domains are responsible for c-trapping in CBL0137- and cisplatin-treated cells. (A) Immunoblotting of soluble extracts of HeLa- GFP-SSRP1 cells transduced with control shRNA, SPT16 shRNA or untransduced (mock), treated with CBL0137 for 1 h. (B) Immunoblotting of supernatant and chromatin pellet of reactions consisting of chromatin purified from HeLa cells, recombinant SSRP1 and SPT16 incubated for 20 min in the presence or absence of 30μM CBL0137, which is the equivalent of the cell-based concentration of 3μM at RT. (C) Scheme of domain organization of full length SSRP1 and designations of truncated mutants used in the study. (D) GFP-fluorescence in nuclei of CBL0137- or cisplatin-treated HeLa cells transduced with the indicated constructs. Cells were fixed with 4% paraformaldehyde either 30 min after start of treatment with 3 μM of CBL0137 or 12 h after start of treatment with 200 μg/ml of cisplatin. (E) Immunoblotting of extracts of HT1080 cells transduced with full length SSRP1 and either CID or HMG domain constructs fused with GFP and treated with different concentrations of CBL0137 for 1 h or cisplatin for 8 h. Antibodies to GFP were used for the detection of SSRP1 variants. (F) Immunoblotting of extracts of HT1080 cells expressing full length SSRP1 or ΔCID and transduced with control shRNA or shRNA to SPT16. Cells were treated with different concentrations of CBL0137 for 1 h.
First, we isolated chromatin from HeLa or HT1080 cells and washed it with a buffer containing 500mM NaCl, which removed most non-histone proteins, including FACT subunits. This isolated chromatin was incubated with recombinant SSRP1, recombinant SPT16, or both in the presence or absence of CBL0137. Similar to cell based experiments, incubation of chromatin with CBL0137 resulted in SSRP1 and SPT16 redistribution from the supernatant to the chromatin pellet independently of the presence of the other subunit (Figure 5B), which confirmed our observation made in cells that SSRP1 can undergo c-trapping in the absence of SPT16.
For the second approach, we used a SSRP1 variant that lacked the N-terminal dimerization domain (Figure 5C, C-terminal half construct) and, therefore, was unable to bind SPT16. This truncated SSRP1 also underwent c-trapping upon CBL0137 treatment (Figure 5D), however, the concentration of CBL0137 required for c-trapping was higher than for full length SSRP1 (Supplemental Figure S5). Moreover, the concentrations of NaCl needed to extract the C-terminal half of SSRP1 from chromatin was lower than for the full length protein (Supplemental Figure S6). These observations demonstrated that SSRP1 can bind chromatin in CBL0137-treated cells independently of SPT16, but this binding required higher concentrations of CBL0137 and had lower affinity.
Within the C-terminal half of human SSRP1, the HMG domain is the only domain known to interact with chromatin via binding to nucleosomal DNA under some specific conditions (9,21,24). Thus, we tested whether the HMG domain is responsible for the chromatin binding of SSRP1 during c-trapping. For this, we generated a set of SSRP1 deletion mutants harboring different combinations of C-terminal domains tagged with GFP and containing a nuclear localization signal (Figure 5C). As a positive control, we used platinated DNA since SSRP1 binding via the HMG domain has been shown in cell-free conditions (24). Because binding of SSRP1 to platinated DNA was never tested in cells, we first checked whether SSRP1 would bind to chromatin in cisplatin treated cells. We observed a phenomenon very similar to c-trapping in cells treated with cisplatin where both subunits of FACT were redistributed from nucleoplasm to pellet fraction via western blotting and cell imaging (Figure 5D and Supplemental Figure S7).
Further testing of SSRP1 truncated variants surprisingly demonstrated that only variants containing the C-terminal intrinsically disordered domain (CID, aa 613–709), located next to the HMG domain (Figure 5C), including a variant consisting of only the CID domain, underwent c-trapping in CBL0137-treated cells whereas no changes were observed with variants containing the HMG domain in the absence of CID. Fluorescent imaging and western blotting both demonstrated binding of CID to chromatin in CBL0137-treated cells and HMG in cisplatin-treated cells (Figure 5D, E). These experiments demonstrated that in CBL0137-treated cells, SSRP1 binds to chromatin via the CID domain.
No interactions was described so far the CID domain of SSRP1. To verify the role of the CID domain in c-trapping, we generated two additional SSRP1 constructs that lacked either only the CID domain (ΔCID) or both the CID and HMG domains (ΔHMG) (Figure 5C). Since these constructs contain the dimerization domain located on the N-terminus of SSRP1 (aa1-177) (Figure 5C), they retained the ability to bind SPT16 and therefore had the potential to undergo c-trapping via SPT16. To exclude this possibility, we tested c-trapping of ΔCID and ΔHMG variants in cells that were depleted of SPT16 by shRNA. These experiments clearly showed that SSRP1 variants that lacked CID are unable to undergo c-trapping in the absence of SPT16 whereas full length SSRP1 can do this on its own (Figure 5F and Supplemental Figure S8). Thus, using SSRP1 truncated mutants we established that c-trapping of SSRP1 may occur either through its binding to SPT16, which according to Tsunaka et al binds the H3/H4 surface of hexasome (18), or via the CID domain, which binds a yet unidentified target.
We observed both in vitro and in cells that chromatin disassembly induced by CBL0137 is a dose-dependent process. Hence, we investigated whether SPT16 or SSRP1-mediated c-trapping occurred with the same concentrations of CBL0137. Because SPT16 cannot be expressed in cells in the absence of SSRP1 (28), we used SSRP1 variants that undergo c-trapping either due to binding to SPT16 (variant expressed from ΔCID construct) or independently of SPT16 (the one expressed from CID) to address this question. This experiment demonstrated that SPT16-mediated c-trapping occurs at a lower concentration range of CBL0137 (0.3–5 μM) than c-trapping mediated by the CID- (2.5–25 μM) (Supplemental Figure S5).
These data suggest that both subunits of FACT bind chromatin disassembled due to CBL0137 treatment. It is likely that the two subunits recognize independently different targets. SPT16 via its middle domain binds the H3/H4 surface of hexasomes that become exposed upon H2A/H2B detachment induced by low concentrations of CBL0137, when DNA is only partially unwrapped from the nucleosome core. The SSRP1 target appeared at higher concentrations of CBL0137, when we observe complete nucleosome disassembly.
Curaxin causes SSRP1 binding to microsatellite regions of cellular DNA
Based on the previous findings, we proposed three possible targets of SSRP1 binding in the process of c-trapping: (i) SSRP1 binds a non-histone protein associated with chromatin, whose availability for binding has been changed by CBL0137 treatment. We tested this hypothesis by comparison of the protein complexes immunoprecipitated with SSRP1 antibody from control and CBL0137-treated cells using mass spectrometry. We did not find significant changes in their protein composition except for a reduction in the amount of core histones (data not shown). (ii) SSRP1 binds histone-free DNA regions that are exposed due to nucleosome disassembly. However, the absence of SSRP1 binding to DNA in cell-free experiments contradicted this hypothesis. (iii) SSRP1 binds non-B DNA. Uncoiling of nucleosomal DNA from histones in cells could result in very strong negative supercoiling with σ up to –0.1 (51). DNA supercoiling is relaxed through rotation of the double helix. In cells, this process is hampered by the length of genomic DNA and the presence of multiple points of ‘fixation’ of DNA on nuclear structures. In principle, DNA breaks and topoisomerases could release superhelical tension, however, curaxin, unlike many other known intercalators, does not induce DNA breaks (13) but does block the activity of topoisomerases (Supplemental Figure S9). This situation creates strong superhelical stress, the reduction of which may require base unpairing and formation of alternative DNA structures (ADS) (51). Indeed, nuclear DNA in cells treated with CBL0137 become more sensitive to digestion with the single strand-specific nuclease S1, which is consistent with an increase in base unpairing (Supplemental Figure S10). Knowing that the HMG domain of SSRP1 binds ADS in the form of bent and cruciform DNA (8,9,52), we proposed that the CID domain might bind another form of non-B DNA.
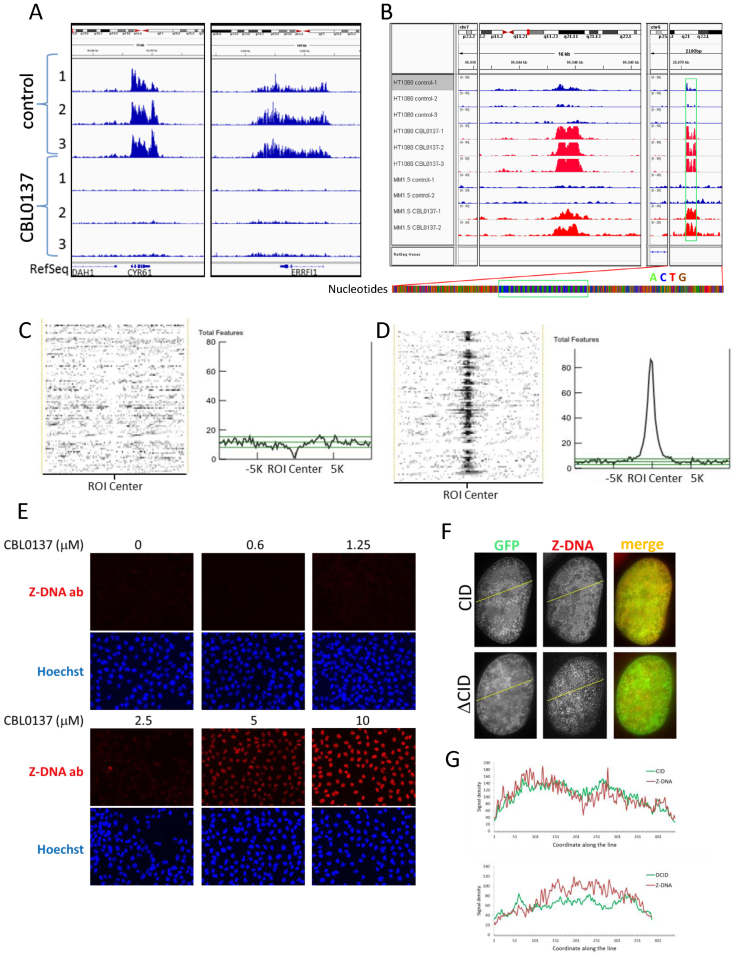
To predict what type of ADS may be bound by SSRP1, we sought to define sequences of genomic regions bound by SSRP1 in CBL0137-treated cells using chromatin immunoprecipitation (ChIP) with SSRP1 antibodies followed by next generation sequencing (ChIP-seq). First, we observed an almost complete change in the genomic location of SSRP1 between control and treated cells (3 μM CBL0137, 1 h) (Figure 6A, B; Supplemental Figure S11A and B) with the reproducible appearance of significantly enriched (>5-fold) CBL0137-specific peaks that were absent in control cells in several independent experiments using two different cell lines—HT1080 and MM1.S (Figure 6B), which supports the idea that SSRP1 binds certain types of genomic DNA regions.
Figure 6.
CBL0137 treatment causes binding of SSRP1 to genomic region predicted to form Z-DNA and induces conversion of DNA into Z form in cells. (A, B) Genome browser views of SSRP1 binding in control and CBL0137-treated cells detected using ChIP-sequencing approach. Examples of alignment of NGS reads from three independent experiments (1–3) with HT1080 and two (1–2) with MM1.S cells to (A) a region of chromosome 1 showing loss of peaks from gene coding region in HT1080 cells; (B) appearance of a peaks in treated HT1080 and MM1.S cells at a region of chromosomes 7 and 6 with no known genetic features (RefSeq line). Magnified region of chromosome 6 shown below with each nucleotide represented by colored bar, what reveals uniform di-nucleotide sequence under the peak (green box) versus more variable nucleotide composition at the adjacent region. (C, D) Heat plots and histograms of distribution of sequences predicted to form G-quadruplexes (C) or Z-DNA (D) in relation to the center of SSRP1 bound regions in CBL0137-treated cells. Statistical evaluation and correlation with other forms of predicted non-B DNA regions are shown in Supplemental Tables S3 and S4. (E) Staining of HeLa cells with antibody to Z-DNA. Immunofluorescence imaging. (F) Immunofluorescence staining with Z-DNA antibody (red) of HT1080 cells expressing either nuclear GFP tagged CID or ΔCID treated with CBL0137 (3 μM, 1 h). (G) ImageJ generated fluorescence intensity profiles along yellow lines shown on panel F (from right to left) for GFP and Z-DNA antibody signals in HT1080 cells expression GFP-tagged CID domain of SSRP1 (upper panel) or SSRP1 lacking CID (ΔCID, lower panel).
Model-based Analysis of ChIP-Seq (MACS (41)) revealed significant (P < 10−5, fold enrichment > 5) CBL0137-induced redistribution of SSRP1 from coding to non-coding areas of the genome (Supplemental Figure S11B, Table S1). Multiple EM for Motif Elicitation (MEME, (53)) analysis did not identify a single consensus element in SSRP1 bound regions in either control or in CBL0137-treated cells. However, MEME revealed that in CBL0137-treated cells, sites of SSRP1 binding precisely coincided with the regions of more homogenous nucleotide arrangement than in regions flanking SSRP1 peaks or in SSRP1-enriched regions in control cells (Figure 6B; Supplemental Figure S11C and D). For a more accurate assessment of nucleotide composition of SSRP1-bound regions in CBL0137-treated cells, we selected all 30 bp genomic intervals that have a sum of read coverage in three replicates of CBL0137-treated HT1080 cells at least 300 times more than the sum of coverage in three control replicates (∼10 times enrichment per each control/treatment pair) to run ‘bi-clustering’ analysis (see details in Supplemental Material and Methods). This analysis demonstrated significant enrichment of purine/pyrimidine pairs of AC and their complementary GT nucleotides, which in many cases were embedded in more complex repetitive patterns, enriched also for short tandem AT repeats (Supplemental Table S2). Thus, CBL0137 treatment causes binding of FACT to AC and AT rich genomic regions known as mini- or microsatellites (54), depending on their length.
It is known from literature that (AC/GT)n tandem dinucleotide repeats of various length are frequent in the mammalian genome (55–57) and that they are prone to transition from the canonical B-DNA conformation to non-B-DNA forms in response to negative superhelical stress (58) and increased concentration of ions (59,60). The presence of (AT)n regions additionally destabilize (AC/GT)n repeats due to the lower stability of this base pairing (61–63). To understand which type of non-B DNA can be expected at genomic regions bound by SSRP1 in CBL0137-treated cells, we used NON-B DB database, which provides the most complete list of ADS predictions available for the human genome (64). Two different statistical methods, LOLA and ColoWeb (65,66), generated consistent results and identified a significant correlation with several types of elements predisposed to form non-B DNA, such as Z-DNA, direct, mirrored and inverted repeats (Supplemental Tables S3 and S4). No correlation was found with G-quadruplex motif, A-phased or short tandem repeats (Figure 6C; Supplemental Tables S3 and S4). The highest correlation was found with sequences prone to form a left-handed helix or Z-DNA (Figure 6D; Supplemental Tables S3 and S4). Thus, in cells treated with CBL0137, SSRP1 is bound to genomic regions that have a high probability to transition to Z-DNA under conditions of negative superhelical stress. Since CBL0137 induces uncoiling of nucleosomal DNA, which leads to the accumulation of negative supercoiling and inhibits the topoisomerases able to resolve this condition, we proposed that SSRP1 binds Z-DNA.
CBL0137 treatment induces transition of cellular B-DNA into left-handed Z-form
Z-DNA is highly immunogenic (67–69) and Z-DNA recognizing antibodies, generated upon immunization of animals with chemically stabilized dsDNA oligonucleotides of (GC)n content are available from multiple vendors (68,70,71). Immunofluorescence staining of untreated cells has minimal background, which is similar to the control lacking primary antibody. Treatment of cells with CBL0137 caused the appearance of nuclear Z-DNA staining in a dose and time dependent manner that coincided with the c-trapping observed with the CID domain of SSRP1 and reached a peak at 15–60 min after start of treatment with ≥2.5 μM CBL0137 (Figure 6E and Supplemental Figure S12). Importantly, closer overlap was observed between the Z-DNA signal and the location of the GFP-tagged CID domain of SSRP1, but not ΔCID, which is consistent with the proposed independent mechanisms driving the c-trapping of these two SSRP1 variants (Figure 6F and G). Although, positive staining with Z-DNA antibody cannot be used as a final proof of Z-DNA formation in curaxin treated cells, selective binding of SSRP1 to the genomic regions with the very high probability of Z-DNA formation together with appearance of Z-DNA antibody staining in CBL0137 treated cells, made Z-DNA the primary candidate substrate for SSRP1 binding during c-trapping.
SSRP1 binds DNA with high propensity to Z-DNA transition
We proposed that SSRP1 in cells can bind Z-DNA via CID domain based on (i) the occurrence of CID-dependent c-trapping at the same concentrations of CBL0137 at which Z-DNA formation is observed via immunofluorescence staining; and (ii) selective binding of SSRP1 to genomic regions with high probability of Z-DNA formation detected via ChIP-seq. The next logical step would be testing the binding of purified recombinant SSRP1 to Z-DNA in cell-free system. The problem with this approach is the difficulty in getting a stable Z-DNA probe under physiological conditions. Z-DNA may be stabilized by negative supercoiling, binding of Z-DNA recognizing proteins or some chemical modifications, such as methylation of cytidines (72). Theoretically, it can be expected that alternating purine/pyrimidine repeats, which occasionally may form Z-DNA spontaneously (73), would be bound by SSRP1 if this transition occurred and this binding could be detected using a gel shift assay.
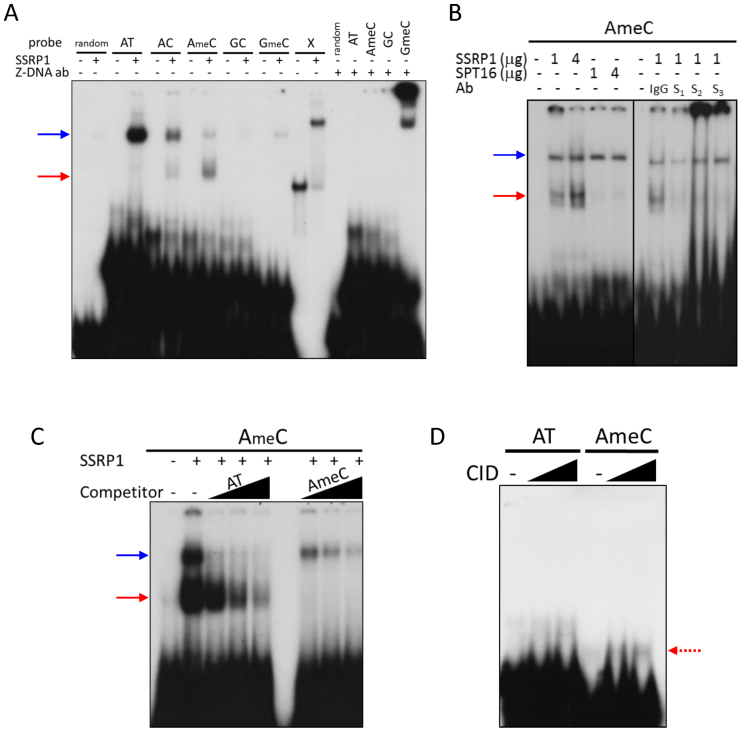
Based on cell-free experiments, methylated dinucleotide repeat of d(G–C)n*d(G–C)n (further mentioned as GC or GmeC, if methylated) has the highest probability of Z-DNA transition among simple naturally occurring oligonucleotide repetitive sequences (59,74,75). However, ChIP-seq data did not show enrichment of GC repeats within SSRP1 bound regions in CBL0137-treated cells. At the same time, the major constituent of these regions was tandem d(A–C)n*d(G–T)n repeats (further—AC or AmeC if methylated), which also have very high tendency to spontaneous transition into Z-DNA (59,74,75). Some minor proportion of these regions was also presented by d(A–T)n*d(A–T)n (further—AT), which is more resistant to Z-DNA transition than GC or AC (76), but provide the higherst probability of base unpairing (among all other combinations of nucleotides), necessary for this transition. Thus, we tested the binding of recombinant SSRP1 purified from HeLa cells to several methylated and unmethylated oligonucleotides with different propensities to form Z-DNA in cell-free conditions. We also used a stable cruciform probe prepared as described in (9) as a positive control for SSRP1 binding. As a control of Z-DNA formation, we used Z-DNA antibody and as a negative control, SPT16, which does not bind DNA (35). In these experiments, the Z-DNA antibody formed a complex with GmeC probe, most of which remained stuck in the gel well. This may be explained by the tendency of Z-DNA to self-aggregate (77,78). A weak, but reproducible, band shifted by Z-DNA antibody was also observed with the AmeC probe, while no band was observed when non-specific antibody was used (see longer exposure image on Supplementary Figure S13A, and different experiment on Supplementary Figure S13B).
SSRP1 shifted the cruciform probe as expected based on Gariglio et al. (9) (Figure 7A). With linear probes, SSRP1 formed either one or two bands, migrating with different velocities, and a slower migrating band (blue arrow on Figure 7A and Supplemental Figure S13D) was observed with all linear probes used. This pattern was not observed previously when we used SSRP1 purified from insect cells (13). A similar band was also observed when we used SPT16 rather than SSRP1 in the binding reaction. SPT16 and SSRP1 were both purified from Hela cells using the same method (Supplemental Figure S13A and B). Thus, we tested the specificity of this band using several antibodies to SSRP1. Neither of the tested SSRP1 antibodies affected this slower migrating band (Figure 7B, blue arrow), however, they all shifted the faster migrating band (Figure 7B, red arrow), with two out of three antibodies forming complexes stuck in the gel wells similarly to Z-DNA antibodies. Thus, we concluded that the slower migrating band was due to non-specific DNA binding activity that contaminated SSRP1 and SPT16 purified from HeLa cells whereas the faster migrating band (red arrow) was formed by SSRP1. The strongest binding of SSRP1 was observed with AmeC probe followed by AC (Figure 7A, red arrow). The specificity of SSRP1 binding to AmeC was confirmed in competition experiment with cold oligonucleotides (Figure 7C). Finally we tried to detect binding of recombinant Flag-tagged CID domain to DNA under the same conditions. A band shift in the presence of the CID domain was more prominent with the AmeC than with the AT probe, which suggests weak, but selective binding (Figure 7D).
Figure 7.
Binding of SSRP1 to different types of DNA oligonucleotides under cell-free conditions. (A–D) Gel shift assays of (A) SSRP1 or Z-DNA antibody incubated for 20 min at RT with different types of 32P-labeled double stranded linear oligonucleotides: random—17 bp non-repetitive DNA fragment, indicated repetitive fragments (composition provided in Materials and Methods) or cruciform DNA probe (X); (B) SSRP1 or SPT16 with AmeC probe in the presence or absence of antibodies to SSRP1 (S1 – 10D1 from Biolegend, S2 – D-15 and S3 – D-7 from Santa Cruz Biotechnology); (C) SSRP1 with 32P-labeled AmeC probe and 10, 30 or 100 times excess of unlabeled AT or AmeC oligonucleotides; (D) CID domain of SSRP1 with 32P-labeled AT or AmeC probes. Blue arrows – non-specific band, red arrows – SSRP1 shifted band.
Thus, we found that SSRP1 can bind DNA consisting mostly of tandem dinucleotide d(A-C)n*d(G-T)n repeats (AC) in both cells and in cell-free conditions. Our cumulative data suggest that SSRP1 binds AC in Z-form or in its transition between B- and Z-DNA, most probably via the CID domain. First, binding of AC probe by SSRP1 or CID was far from complete, with only a minor proportion of the probe being bound, which is consistent with the dynamic nature of the B- to Z transition. In contrast, the binding of SSRP1 to the stable cruciform probe was almost complete. Second, binding was increased when the probability of a probe transitioning into Z-DNA was also increased by the methylation of cytidines. Third, this probe was also bound, albeit weakly, by the antibody to Z-DNA. The weak binding of the antibody may be explained by the fact that the antibody was raised against a chemically stabilized GC, not AC, probe (http://www.abcam.com/z-dna-antibody-ab2079.html). Finally, we saw a similar tendency for self-aggregation of the AmeC probe in the presence of SSRP1 antibodies as was observed for the GmeC probe in the presence of Z-DNA antibody, which is in line with the nature of Z-, but not B-DNA.
DISCUSSION
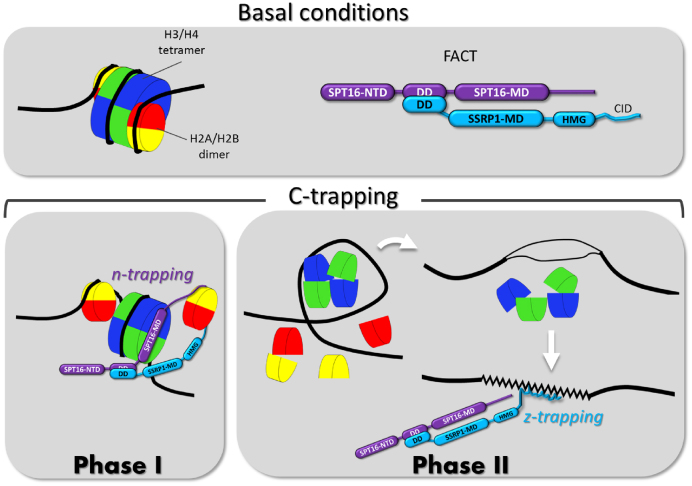
In the present study, we described the ability of the small molecule to cause unfolding of chromatin in cells, resulting in the phenomenon of chromatin trapping of the histone chaperone FACT, which we named c-trapping. Below, we propose a model to explain the mechanism of chromatin unfolding and c-trapping in response to CBL0137 (Figure 8). We built this model based on three sets of data: (i) current knowledge of the structural organization of a unit of chromatin, the nucleosome, which consists of an octamer of histones, known as the histone or nucleosome core that is bound by 147 base pairs of DNA. The stability of the nucleosome depends on several points of contact between nucleic and amino acids of DNA and histones, respectively (79); (ii) recently described binding preferences of several domains of mammalian FACT subunits (14,15,18,48,50); (iii) and findings described in this manuscript and discussed below.
Figure 8.
Model of the mechanism of c-trapping. The upper panel shows the scheme of the nucleosome with a standard color code for core histones (H2A-yellow, H2B – red, H3 – blue, H4 – green) and the domain structure of FACT subunits (NTD – N-terminal domain, DD – dimerization domain, MD – middle domain). The lower panels show two phases of c-trapping: i) n(nucleosome)-trapping that occurs via the SPT16 subunit binding to the hexasome of the partially uncoiled nucleosome when one molecule of CBL0137 is bound per ∼10–100 bp of DNA; ii) z(Z-DNA)-trapping that occurs via SSRP1 subunit binding to DNA when the nucleosome is disassembled upon binding of one or more molecules of CBL0137 to every 10 bp of DNA.
CBL0137 binds to DNA via intercalation, i.e. insertion of the planar carbazole body between base pairs and protruding of carbazole side chains into the major and minor grooves of DNA (Figure 4A, B). This high affinity, non-covalent binding does not modify DNA chemically, but changes the topology of the DNA helix and most probably its flexibility as has been shown for other intercalators (80). The modified DNA helix starts unwrapping from the histone core. There may be several reasons for the unwrapping: loss of precise position of amino acids in histones and base pairs in DNA; increased rigidity of the DNA helix that makes its curving around the nucleosome core difficult, or direct interference of the small molecule with the amino acid – DNA interaction. The exact cause of DNA unwrapping requires further investigation.
At relatively low doses (∼1 molecule per >10<100 base pairs of DNA), CBL0137 causes partial unwrapping of DNA from the nucleosome core, which leads to the dissociation of the H2A/H2B dimer and exposure of the H3/H4 tetramer docking surface for SPT16 binding (Figure 8, Phase 1). Alternatively, SPT16 may provoke dimer detachment as soon as dimer's contact with DNA and the core is weakened by CBL0137 binding. In either case, at this stage FACT binds to the hexasome or tetrasome via SPT16 middle domain and possibly, as suggested by Kemble et al., holds the dimer via C-terminal domains (14) (Figure 8, Phase 1). With increasing amounts of CBL0137 molecules bound to DNA (>1 per each 10 bp), complete nucleosome disassembly occurs. As a results of this, supercoils of DNA that lose their histone cores appear as under-twisted helix that becomes prone to base unpairing and transition from B- to non-B DNA to consume the excessive energy of negative supercoiling (Figure 8, Phase 2). Our data suggest that the prevailing form of non-B DNA present in CBL0137 treated cells is Z-DNA. SSRP1 subunit of FACT binds DNA prone to Z-DNA transition most probably when it already adapted a left handed helical conformation. The CID domain of SSRP1 plays a role in either Z-DNA recognition or/and Z-DNA binding (Figure 8).
Thus, based on this model, we propose to differentiate two phases in both processes: chromatin unpacking and c-trapping, although in reality there may be more phases and they may not be fully discrete especially in the cells. Phase one is partial nucleosome unfolding accompanied by FACT binding to the open nucleosome or hexasome via SPT16 (‘n (nucleosome)-trapping’). Phase two is complete nucleosome disassembly with SSRP1 binding to alternative DNA structures (‘z(Z-DNA)-trapping’, Figure 8).
Although not every detail in this model is fully proven, we believe that it is reliably justified due to the following considerations. CBL0137 binding to DNA and destabilization of nucleosomes is observed in the cells and cell-free conditions using different methods ((13) and Supplemental Figure S3). Although we did not establish the exact mechanism of CBL0137-bound DNA unwrapping from the nucleosome, the change in the shape and flexibility of DNA following its binding by small molecules is well supported by existing literature (reviewed in (81–83)). Moreover, the destabilizing effect of other DNA intercalators, e.g. anthracyclines, on nucleosome was also demonstrated (45). In principle, we cannot exclude that CBL0137 binds histones in addition to DNA, as it was shown for ethidium bromide and propidium iodide (84), what may change their ability to form nucleosomes. To what extend (if at all) this mechanism contributes to the observed effects remains to be determined.
It was difficult to dissect the individual role of the two FACT subunits in c-trapping since SSRP1 and SPT16 are always in a complex in cells and, therefore, replicate the behavior of each other. Nevertheless, the use of SSRP1 mutants lacking SPT16-binding domain and enforced overexpression of individual subunits in cells as well as use of recombinant proteins and FACT depleted chromatin made it possible to understand the roles of the subunits. We found that each of the subunits is capable of c-trapping independently of the other one. Our observations indicate that during gradual increase in the concentrations of CBL0137, SPT16 starts binding chromatin first, before nucleosome disassembly, while c-trapping of SSRP1 occurs at higher CBL0137 concentrations, upon appearance of free outer and inner histones in the nucleoplasm of cells and disappearance of nucleosome protected DNA fragments upon MN digestion.
Binding of SPT16 to uncoiled nucleosomes was expected based on the recent report of Tsunaka et al., who convincingly demonstrated that weakening of the contact between DNA and the histone core makes it possible for SPT16 to access the H3/H4 dimer, either via invasion of FACT into yet complete nucleosome with partially unwrapped DNA or via binding of the exposed hexamer that already lost the H2A/H2B dimer (18).
The ability of the SSRP1 subunit to undergo c-trapping in the absence of SPT16 was less expected. C-trapping of SSRP1 demonstrated in the cells with depleted SPT16 (due to gene expression knockdown) or cell-free system with recombinant SSRP1 did not fully exclude the role of SPT16, due to potentially incomplete depletion of SPT16 from the cells or chromatin. However, c-trapping of the C-terminal half of SSRP1 that is incapable of binding SPT16 convincingly excluded the need for SPT16. Our initial hypothesis was that the HMG domain is responsible for c-trapping since this was the only domain of mammalian SSRP1 that had an established role in binding a component of chromatin, DNA (17,21). However, as much as we tried, we were unable to detect SSRP1 binding to linear DNA with CBL0137 in a cell-free system or to see c-trapping of the SSRP1's HMG domain in cells treated with CBL0137 (13). Unexpectedly, the CID domain underwent c-trapping in CBL0137-treated cells.
Microscopically, c-trapping of CID was firmly established, however western blotting, as well as the results of gel-shift experiments, showed that CID binding to ‘curaxinized’ chromatin is substantially weaker than that of the whole FACT complex. This can be explained by a cooperative nature of binding of the whole complex or by the stabilizing effect of SPT16 or N-terminal domains of SSRP1 on the CID/chromatin binding.
The most unexpected finding of these current studies is the emergence of Z-DNA in CBL0137-treated cells. Even though Z-DNA was described shortly after B-DNA (reviewed in (70), its biological relevance and existence in vivo remain questionable and debatable. The major challenge is the absence of tools for accurate assessment of DNA shape in cells. Alternative DNA structures (ADS) are transient in nature, unstable and cannot be isolated from cells in their native form by biochemical methods. Z-DNA has been undetectable even with the most powerful microscopic methods available. Antibodies raised against artificial oligonucleotide chemically stabilized in Z form is practically the only tool to visualize Z-DNA in vivo. Indirect ways to detect Z-DNA utilized reporter plasmids with Z-DNA-prone inserts or boosting rate of transcription at certain genomic regions. In both cases, however, Z-DNA emerges locally and its detection is either indirect (mutations in reporter (85) or poorly detectable (1,86). Therefore, finding that CBL0137 induces massive stable and long-lasting transition of genomic DNA into the Z-form at a scale that enables easy detection by specific antibodies as whole nucleus staining in CBL0137-treated cells was surprising. Although, we understand the limitations of staining with Z-DNA antibody, taking into account other data, we hope that CBL0137 will serve as a valuable tool that will facilitate studies of Z-DNA in vivo. Similarly, Z-DNA detection in curaxin-treated cells can be considered as a biomarker of activity of this class of anticancer drugs.
Even though Z-DNA occurrence in CBL0137-treated cells was an unprecedented finding, it is not that unexpected based on earlier predictions of a theoretical link between nucleosome disassembly, negative supercoiling and Z-DNA transition (70). Possibility of Z-DNA transition was also demonstrated for some genomic regions such as promoter zones with nucleosome removed by RNA polymerase or chromatin remodeling factors (1,87–89). Treatment of cells with CBL0137 provides the necessary conditions for Z-DNA transition, i.e. CBL0137 binds to DNA and causes nucleosome disassembly. At concentrations when Z-DNA staining is observed, nucleosome disassembly is quite massive since we see histone redistribution between chromatin and nucleoli microscopically (Figures 2 and 3A and B) and loss of nucleosome protected bands via nuclease digestion (Figure 3D). We also observed increased sensitivity of CBL0137 treated cells to ssDNA nuclease S1 (Supplemental Figure S10). In the absence of single strand breaks (data not shown), this result is interpreted as an increase in base unpairing, a sign of accumulated negative supercoiling, and B- to non-B DNA transition. Next, CBL0137 inhibits topoisomerase (I and II) cleavage activity, therefore enzymes that are responsible for releasing superhelical stress cannot release this stress either through normal cleavage and religation or through generation of DNA breaks. Based on these data, formation of Z-DNA is highly probable and may be expected.
Why Z-DNA, and not other forms of ADS, which also can discharge negative supercoiling, such as cruciform DNA? Theoretically, any other ADS consuming excess of free energy of negative supercoiling can be formed. Indeed, cruciform DNA was our first suspect and we expected to find significant enrichment of SSRP1 in CBL0137-treated cells in genomic regions with inverted repeats. Although there was enrichment of inverted repeats within SSRP1 bound regions, the enrichment of tandem purine/pyrimidine dinucleotide repeats by far exceeded inverted repeats (Supplemental Tables S3 and S4). Moreover, AC/GT tandem dinucleotide repeats occupy a significant proportion of the mammalian genome, much higher than that in lower eukaryotes (e.g. yeast (90)). Since yeasts also lack CID domain in their bipartite SSRP1 homologs, Pob3 and Nph6, it would be interesting to correlate an expansion of AC/GT repeats with the appearance of CID domain in eukaryotes.
Theoretically, we cannot exclude SSRP1 binding to DNA in a sequence, but not structure, dependent manner. In untreated cells, these regions may be buried deep in heterochromatin (most of them are in non-coding regions of the genome), while nucleosome disassembly makes them accessible to SSRP1 binding. Since these regions are prone to Z-DNA transition, FACT binding may be a mechanism to prevent the transition. Complex structural studies are needed to firmly establish the form of DNA to which SSRP1 binds via the CID domain. Nevertheless, the binding pattern of SSRP1 to DNA probes seen using gel shift is more in line with binding to unstable transient structures (only part of the probe is bound, binding is increased with time of incubation), than to stable sequence. The same pattern is seen with Z-DNA antibody which also binds only a fraction of the probe in line with the expected dynamic transitions of the probes between B- and Z-DNA states.
In the literature, Z-DNA is described as a good immunogen and antibodies to Z-DNA are frequently detected in animals or patients with auto-immune diseases (67,69). Therefore, this antibody is considered a reliable tool to detect Z-DNA. Z-DNA staining appears at concentrations and times when CID domain undergoes c-trapping. The overlap of CID and Z-DNA staining is also higher than for CID-less SSRP1 (ΔCID) or SPT16. Still the overlap is not full (Figure 6F). Moreover, in gel shift assays, we saw a differential preference in binding between SSRP1 (AC) and Z-DNA antibody (GC). In line with this, there is no GC tandem dinucleotide repeats in SSRP1 enriched genomic regions in cells. Although in vitro short nucleotides composed of repetitive GCs demonstrated the highest propensity to Z-DNA transition (78), in cells the situation may be different. This transition requires base unpairing, therefore the stronger bonding of GC/CG sequences than that of AC/GT may make the latter the first candidate for this transition in cells. Thus in cells we may have most of antibody bound to GC repetitive regions and SSRP1 to AC, or, as we proposed earlier, SSRP1 may bind DNA before transition to Z-DNA or even force Z-DNA transit back to B-DNA. Genome wide mapping of Z-DNA antibody, SSRP1 and nucleosomes at single nucleotide resolution in control and treated cells may help to explain this discrepancy in the future.
What is the biological significance of c-trapping? We propose several implications: (i) c-trapping may be a mechanism to recruit FACT to certain areas of the genome with increased risk of nucleosome loss to perform its basic nucleosome stabilizing function. This hypothesis provides the missing mechanism for selective FACT enrichment at certain genomic regions in cells. (ii) C-trapping followed by casein kinase 2 mediated phosphorylation and activation of p53 may present a novel type of cell stress response to the problem with chromatin stability and/or DNA topology, since the former is a serious threat for epigenetic stability and the latter is a well-known trigger of genomic instability. (iii) Induction of c-trapping with anti-cancer small molecule depletes cells of functional FACT, which is essential for tumor, but not normal cell viability (40) and this may be proposed as a novel approach to cancer treatment. Finally, CBL0137 may become a valuable tool to study chromatin dynamics in vivo, to understand the mechanism of why chromatin opening is more toxic for tumor than for normal cells, and potentially for therapeutic erasure of epigenetic signatures in cells, e.g. during reprogramming.
ACCESSION NUMBERS
Sequencing data in the form of bed files are available at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE45393.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Drs. Andrei Gudkov, Daria Fleyshman, Vasiliy Studitsky, Alexander Vologodskii, and Dominic Smiraglia for the critical discussion of the work and the manuscript; Bruce Specht for the administrative help, Catherine Burkhart from Burkhart Document Solutions for critical review and editing of the manuscript.
Footnotes
Present address: Mahadeb Pal, Division of Molecular Medicine, Bose Institute, 93/1, Acharya Prafulla Chandra Road, Kolkata 700009, West Bengal, India.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Incuron LLC (to K.G.); National Cancer Institute [R01CA197967 to K.G. and P30CA016056 to Roswell Park Cancer Center]; Komen Foundation award [CCR13264604 to K.G.]. Funding for open access charge: Incuron LLC (to K.G.); National Cancer Institute [R01CA197967 to K.G. and P30CA016056 to Roswell Park Cancer Center]; Komen Foundation award [CCR13264604 to K.G.].
Conflict of interest statement. Katerina Gurova is the co-author on the patent WO 2010/042445A1 describing composition of matter and use of curaxin molecules. She also received grants and consulting payments from Incuron, LLC.
REFERENCES
- 1. Maruyama A., Mimura J., Harada N., Itoh K.. Nrf2 activation is associated with Z-DNA formation in the human HO-1 promoter. Nucleic Acids Res. 2013; 41:5223–5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Du X., Wojtowicz D., Bowers A.A., Levens D., Benham C.J., Przytycka T.M.. The genome-wide distribution of non-B DNA motifs is shaped by operon structure and suggests the transcriptional importance of non-B DNA structures in Escherichia coli. Nucleic Acids Res. 2013; 41:5965–5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Naughton C., Avlonitis N., Corless S., Prendergast J.G., Mati I.K., Eijk P.P., Cockroft S.L., Bradley M., Ylstra B., Gilbert N.. Transcription forms and remodels supercoiling domains unfolding large-scale chromatin structures. Nat Struct Mol Biol. 2013; 20:387–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Krasilnikov A.S., Podtelezhnikov A., Vologodskii A., Mirkin S.M.. Large-scale effects of transcriptional DNA supercoiling in vivo. J Mol Biol. 1999; 292:1149–1160. [DOI] [PubMed] [Google Scholar]
- 5. Liu J., Kouzine F., Nie Z., Chung H.J., Elisha-Feil Z., Weber A., Zhao K., Levens D.. The FUSE/FBP/FIR/TFIIH system is a molecular machine programming a pulse of c-myc expression. EMBO J. 2006; 25:2119–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kouzine F., Liu J., Sanford S., Chung H.J., Levens D.. The dynamic response of upstream DNA to transcription-generated torsional stress. Nat Struct Mol Biol. 2004; 11:1092–1100. [DOI] [PubMed] [Google Scholar]
- 7. Wang G., Vasquez K.M.. Impact of alternative DNA structures on DNA damage, DNA repair, and genetic instability. DNA Repair (Amst). 2014; 19:143–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bruhn S.L., Pil P.M., Essigmann J.M., Housman D.E., Lippard S.J.. Isolation and characterization of human cDNA clones encoding a high mobility group box protein that recognizes structural distortions to DNA caused by binding of the anticancer agent cisplatin. Proc Natl Acad Sci U S A. 1992; 89:2307–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gariglio M., Ying G.G., Hertel L., Gaboli M., Clerc R.G., Landolfo S.. The high-mobility group protein T160 binds to both linear and cruciform DNA and mediates DNA bending as determined by ring closure. Exp Cell Res. 1997; 236:472–481. [DOI] [PubMed] [Google Scholar]
- 10. Keller D.M., Zeng X., Wang Y., Zhang Q.H., Kapoor M., Shu H., Goodman R., Lozano G., Zhao Y., Lu H.. A DNA damage-induced p53 serine 392 kinase complex contains CK2, hSpt16, and SSRP1. Mol Cell. 2001; 7:283–292. [DOI] [PubMed] [Google Scholar]
- 11. Stros M., Launholt D., Grasser K.D.. The HMG-box: a versatile protein domain occurring in a wide variety of DNA-binding proteins. Cell Mol Life Sci. 2007; 64:2590–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Keller D.M., Lu H.. p53 serine 392 phosphorylation increases after UV through induction of the assembly of the CK2.hSPT16.SSRP1 complex. J Biol Chem. 2002; 277:50206–50213. [DOI] [PubMed] [Google Scholar]
- 13. Gasparian A.V., Burkhart C.A., Purmal A.A., Brodsky L., Pal M., Saranadasa M., Bosykh D.A., Commane M., Guryanova O.A., Pal S. et al. . Curaxins: Anticancer Compounds That Simultaneously Suppress NF-{kappa}B and Activate p53 by Targeting FACT. Sci Transl Med. 2011; 3:95ra74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kemble D.J., McCullough L.L., Whitby F.G., Formosa T., Hill C.P.. FACT Disrupts Nucleosome Structure by Binding H2A-H2B with Conserved Peptide Motifs. Mol Cell. 2015; 60:294–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang W., Zeng F., Liu Y., Shao C., Li S., Lv H., Shi Y., Niu L., Teng M., Li X.. Crystal Structure of Human SSRP1 Middle Domain Reveals a Role in DNA Binding. Sci Rep. 2015; 5:18688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. VanDemark A.P., Xin H., McCullough L., Rawlins R., Bentley S., Heroux A., Stillman D.J., Hill C.P., Formosa T.. Structural and functional analysis of the Spt16p N-terminal domain reveals overlapping roles of yFACT subunits. J Biol Chem. 2008; 283:5058–5068. [DOI] [PubMed] [Google Scholar]
- 17. Winkler D.D., Luger K.. The histone chaperone FACT: structural insights and mechanisms for nucleosome reorganization. J Biol Chem. 2011; 286:18369–18374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tsunaka Y., Fujiwara Y., Oyama T., Hirose S., Morikawa K.. Integrated molecular mechanism directing nucleosome reorganization by human FACT. Genes Dev. 2016; 30:673–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Are V.N., Ghosh B., Kumar A., Gadre R., Makde R.D.. Crystal structure and dynamics of Spt16N-domain of FACT complex from Cicer arietinum. Int. J. Biol Macromol. 2016; 88:36–43. [DOI] [PubMed] [Google Scholar]
- 20. Zheng S., Crickard J.B., Srikanth A., Reese J.C.. A highly conserved region within H2B is important for FACT to act on nucleosomes. Mol Cell Biol. 2014; 34:303–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tsunaka Y., Toga J., Yamaguchi H., Tate S., Hirose S., Morikawa K.. Phosphorylated intrinsically disordered region of FACT masks its nucleosomal DNA binding elements. J Biol Chem. 2009; 284:24610–24621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li Y., Keller D.M., Scott J.D., Lu H.. CK2 phosphorylates SSRP1 and inhibits its DNA-binding activity. J Biol Chem. 2005; 280:11869–11875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hsieh F.K., Kulaeva O.I., Patel S.S., Dyer P.N., Luger K., Reinberg D., Studitsky V.M.. Histone chaperone FACT action during transcription through chromatin by RNA polymerase II. Proc Natl Acad Sci U S A. 2013; 110:7654–7659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yarnell A.T., Oh S., Reinberg D., Lippard S.J.. Interaction of FACT, SSRP1, and the high mobility group (HMG) domain of SSRP1 with DNA damaged by the anticancer drug cisplatin. J Biol Chem. 2001; 276:25736–25741. [DOI] [PubMed] [Google Scholar]
- 25. Cerna A., Cuadrado A., Jouve N., Diaz de la Espina S.M., De la Torre C.. Z-DNA, a new in situ marker for transcription. Eur J Histochem. 2004; 48:49–56. [PubMed] [Google Scholar]
- 26. Gurova K.V., Hill J.E., Razorenova O.V., Chumakov P.M., Gudkov A.V.. p53 pathway in renal cell carcinoma is repressed by a dominant mechanism. Cancer Res. 2004; 64:1951–1958. [DOI] [PubMed] [Google Scholar]
- 27. Steigemann P., Wurzenberger C., Schmitz M.H., Held M., Guizetti J., Maar S., Gerlich D.W.. Aurora B-mediated abscission checkpoint protects against tetraploidization. Cell. 2009; 136:473–484. [DOI] [PubMed] [Google Scholar]
- 28. Safina A., Garcia H., Commane M., Guryanova O., Degan S., Kolesnikova K., Gurova K.V.. Complex mutual regulation of facilitates chromatin transcription (FACT) subunits on both mRNA and protein levels in human cells. Cell Cycle. 2013; 12:2423–2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Belotserkovskaya R., Oh S., Bondarenko V.A., Orphanides G., Studitsky V.M., Reinberg D.. FACT facilitates transcription-dependent nucleosome alteration. Science. 2003; 301:1090–1093. [DOI] [PubMed] [Google Scholar]
- 30. Bailly C., Tardy C., Wang L., Armitage B., Hopkins K., Kumar A., Schuster G.B., Boykin D.W., Wilson W.D.. Recognition of ATGA sequences by the unfused aromatic dication DB293 forming stacked dimers in the DNA minor groove. Biochemistry. 2001; 40:9770–9779. [DOI] [PubMed] [Google Scholar]
- 31. Luger K., Rechsteiner T.J., Richmond T.J.. Expression and purification of recombinant histones and nucleosome reconstitution. Methods Mol Biol. 1999; 119:1–16. [DOI] [PubMed] [Google Scholar]
- 32. Lowary P.T., Widom J.. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol. 1998; 276:19–42. [DOI] [PubMed] [Google Scholar]
- 33. Park Y.J., Dyer P.N., Tremethick D.J., Luger K.. A new fluorescence resonance energy transfer approach demonstrates that the histone variant H2AZ stabilizes the histone octamer within the nucleosome. J Biol Chem. 2004; 279:24274–24282. [DOI] [PubMed] [Google Scholar]
- 34. Luger K., Rechsteiner T.J., Richmond T.J.. Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 1999; 304:3–19. [DOI] [PubMed] [Google Scholar]
- 35. Winkler D.D., Muthurajan U.M., Hieb A.R., Luger K.. Histone chaperone FACT coordinates nucleosome interaction through multiple synergistic binding events. J Biol Chem. 2011; 286:41883–41892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Winkler D.D., Luger K., Hieb A.R.. Quantifying chromatin-associated interactions: the HI-FI system. Methods Enzymol. 2012; 512:243–274. [DOI] [PubMed] [Google Scholar]
- 37. Telford D.J., Stewart B.W.. Micrococcal nuclease: its specificity and use for chromatin analysis. Int. J. Biochem. 1989; 21:127–137. [DOI] [PubMed] [Google Scholar]
- 38. Prosperi E., Supino R., Bottiroli G.. Increased sensitivity of damaged DNA to digestion with nuclease S1 as assessed in single cells by flow cytometry. Cytometry. 1993; 14:53–58. [DOI] [PubMed] [Google Scholar]
- 39. Shechter D., Dormann H.L., Allis C.D., Hake S.B.. Extraction, purification and analysis of histones. Nature protocols. 2007; 2:1445–1457. [DOI] [PubMed] [Google Scholar]
- 40. Garcia H., Miecznikowski J.C., Safina A., Commane M., Ruusulehto A., Kilpinen S., Leach R.W., Attwood K., Li Y., Degan S. et al. . Facilitates chromatin transcription complex is an ‘accelerator’ of tumor transformation and potential marker and target of aggressive cancers. Cell reports. 2013; 4:159–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W. et al. . Model-based analysis of ChIP-Seq (MACS). Genome biology. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dejmek J., Iglehart J.D., Lazaro J.B.. DNA-dependent protein kinase (DNA-PK)-dependent cisplatin-induced loss of nucleolar facilitator of chromatin transcription (FACT) and regulation of cisplatin sensitivity by DNA-PK and FACT. Mol Cancer Res. 2009; 7:581–591. [DOI] [PubMed] [Google Scholar]
- 43. Musinova Y.R., Lisitsyna O.M., Golyshev S.A., Tuzhikov A.I., Polyakov V.Y., Sheval E.V.. Nucleolar localization/retention signal is responsible for transient accumulation of histone H2B in the nucleolus through electrostatic interactions. Biochim Biophys Acta. 2011; 1813:27–38. [DOI] [PubMed] [Google Scholar]
- 44. Biebricher A.S., Heller I., Roijmans R.F., Hoekstra T.P., Peterman E.J., Wuite G.J.. The impact of DNA intercalators on DNA and DNA-processing enzymes elucidated through force-dependent binding kinetics. Nat Commun. 2015; 6:7304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pang B., Qiao X., Janssen L., Velds A., Groothuis T., Kerkhoven R., Nieuwland M., Ovaa H., Rottenberg S., van Tellingen O. et al. . Drug-induced histone eviction from open chromatin contributes to the chemotherapeutic effects of doxorubicin. Nat Commun. 2013; 4:1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hendry L.B., Mahesh V.B., Bransome E.D. Jr, Ewing D.E.. Small molecule intercalation with double stranded DNA: implications for normal gene regulation and for predicting the biological efficacy and genotoxicity of drugs and other chemicals. Mutation research. 2007; 623:53–71. [DOI] [PubMed] [Google Scholar]
- 47. Hondele M., Stuwe T., Hassler M., Halbach F., Bowman A., Zhang E.T., Nijmeijer B., Kotthoff C., Rybin V., Amlacher S. et al. . Structural basis of histone H2A-H2B recognition by the essential chaperone FACT. Nature. 2013; 499:111–114. [DOI] [PubMed] [Google Scholar]
- 48. Hsieh F.K., Kulaeva O.I., Studitsky V.M.. Experimental analysis of hFACT action during Pol II transcription in vitro. Methods Mol Biol. 2015; 1276:315–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nair D.M., Ge Z., Mersfelder E.L., Parthun M.R.. Genetic interactions between POB3 and the acetylation of newly synthesized histones. Curr Genet. 2011; 57:271–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hoffmann C., Neumann H.. In Vivo Mapping of FACT-Histone Interactions Identifies a Role of Pob3 C-terminus in H2A-H2B Binding. ACS Chem Biol. 2015; 10:2753–2763. [DOI] [PubMed] [Google Scholar]
- 51. Vologodskii A. Biophysics of DNA. 2015; Cambridge, UK: Cambridge University Press. [Google Scholar]
- 52. Bruhn S.L., Housman D.E., Lippard S.J.. Isolation and characterization of cDNA clones encoding the Drosophila homolog of the HMG-box SSRP family that recognizes specific DNA structures. Nucleic Acids Res. 1993; 21:1643–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bailey T.L., Elkan C.. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994; 2:28–36. [PubMed] [Google Scholar]
- 54. Ellegren H. Microsatellites: simple sequences with complex evolution. Nat Rev Genet. 2004; 5:435–445. [DOI] [PubMed] [Google Scholar]
- 55. Hamada H., Kakunaga T.. Potential Z-DNA forming sequences are highly dispersed in the human genome. Nature. 1982; 298:396–398. [DOI] [PubMed] [Google Scholar]
- 56. Schroth G.P., Chou P.J., Ho P.S.. Mapping Z-DNA in the human genome. Computer-aided mapping reveals a nonrandom distribution of potential Z-DNA-forming sequences in human genes. J Biol Chem. 1992; 267:11846–11855. [PubMed] [Google Scholar]
- 57. Tripathi J., Brahmachari S.K.. Distribution of simple repetitive (TG/CA)n and (CT/AG)n sequences in human and rodent genomes. Journal of biomolecular structure & dynamics. 1991; 9:387–397. [DOI] [PubMed] [Google Scholar]
- 58. Dobi A., Mahan M.A., v Agoston D.. Detection of conformation bias between the complementary strands of deoxynucleotide repeat (TG/AC)n. Electrophoresis. 1997; 18:12–16. [DOI] [PubMed] [Google Scholar]
- 59. Haniford D.B., Pulleyblank D.E.. Facile transition of poly[d(TG) x d(CA)] into a left-handed helix in physiological conditions. Nature. 1983; 302:632–634. [DOI] [PubMed] [Google Scholar]
- 60. Herbert A., Rich A.. The biology of left-handed Z-DNA. J Biol Chem. 1996; 271:11595–11598. [DOI] [PubMed] [Google Scholar]
- 61. Kim D., Reddy S., Kim D.Y., Rich A., Lee S., Kim K.K., Kim Y.G.. Base extrusion is found at helical junctions between right- and left-handed forms of DNA and RNA. Nucleic Acids Res. 2009; 37:4353–4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ha S.C., Lowenhaupt K., Rich A., Kim Y.G., Kim K.K.. Crystal structure of a junction between B-DNA and Z-DNA reveals two extruded bases. Nature. 2005; 437:1183–1186. [DOI] [PubMed] [Google Scholar]
- 63. Bothe J.R., Lowenhaupt K., Al-Hashimi H.M.. Sequence-specific B-DNA flexibility modulates Z-DNA formation. J Am Chem Soc. 2011; 133:2016–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Cer R.Z., Donohue D.E., Mudunuri U.S., Temiz N.A., Loss M.A., Starner N.J., Halusa G.N., Volfovsky N., Yi M., Luke B.T. et al. . Non-B DB v2.0: a database of predicted non-B DNA-forming motifs and its associated tools. Nucleic Acids Res. 2013; 41:D94–D100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kim R., Smith O.K., Wong W.C., Ryan A.M., Ryan M.C., Aladjem M.I.. ColoWeb: a resource for analysis of colocalization of genomic features. BMC Genomics. 2015; 16:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sheffield N.C., Bock C.. LOLA: enrichment analysis for genomic region sets and regulatory elements in R and Bioconductor. Bioinformatics. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Bergen H.R. 3rd, Losman M.J., O'Connor T., Zacharias W., Larson J.E., Accavitti M.A., Wells R.D., Koopman W.J.. Specificity of monoclonal anti-Z-DNA antibodies from unimmunized MRL/Mp-lpr/lpr mice. J Immunol. 1987; 139:743–748. [PubMed] [Google Scholar]
- 68. Lafer E.M., Moller A., Nordheim A., Stollar B.D., Rich A.. Antibodies specific for left-handed Z-DNA. Proc Natl Acad Sci U S A. 1981; 78:3546–3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Edgington S.M., Stollar B.D.. Immunogenicity of Z-DNA depends on the size of polynucleotide presented in complexes with methylated BSA. Mol Immunol. 1992; 29:609–617. [DOI] [PubMed] [Google Scholar]
- 70. Rich A., Zhang S.. Timeline: Z-DNA: the long road to biological function. Nat Rev Genet. 2003; 4:566–572. [DOI] [PubMed] [Google Scholar]
- 71. Moller A., Gabriels J.E., Lafer E.M., Nordheim A., Rich A., Stollar B.D.. Monoclonal antibodies recognize different parts of Z-DNA. J Biol Chem. 1982; 257:12081–12085. [PubMed] [Google Scholar]
- 72. Rich A., Nordheim A., Wang A.H.. The chemistry and biology of left-handed Z-DNA. Annu Rev Biochem. 1984; 53:791–846. [DOI] [PubMed] [Google Scholar]
- 73. Behe M., Felsenfeld G.. Effects of methylation on a synthetic polynucleotide: the B–Z transition in poly(dG-m5dC).poly(dG-m5dC). Proc Natl Acad Sci U S A. 1981; 78:1619–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Nordheim A., Rich A.. The sequence (dC-dA)n X (dG-dT)n forms left-handed Z-DNA in negatively supercoiled plasmids. Proc Natl Acad Sci U S A. 1983; 80:1821–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wang A.H., Quigley G.J., Kolpak F.J., Crawford J.L., van Boom J.H., van der Marel G., Rich A.. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979; 282:680–686. [DOI] [PubMed] [Google Scholar]
- 76. Ellison M.J., Feigon J., Kelleher R.J. 3rd, Wang A.H., Habener J.F., Rich A.. An assessment of the Z-DNA forming potential of alternating dA-dT stretches in supercoiled plasmids. Biochemistry. 1986; 25:3648–3655. [DOI] [PubMed] [Google Scholar]
- 77. van de Sande J.H., Jovin T.M.. Z* DNA, the left-handed helical form of poly[d(G-C)] in MgCl2-ethanol, is biologically active. EMBO J. 1982; 1:115–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Jovin T.M., McIntosh L.P., Arndt-Jovin D.J., Zarling D.A., Robert-Nicoud M., van de Sande J.H., Jorgenson K.F., Eckstein F.. Left-handed DNA: from synthetic polymers to chromosomes. J Biomol Struct Dyn. 1983; 1:21–57. [DOI] [PubMed] [Google Scholar]
- 79. Luger K., Mader A.W., Richmond R.K., Sargent D.F., Richmond T.J.. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997; 389:251–260. [DOI] [PubMed] [Google Scholar]
- 80. Ha Duong T., Zakrzewska K.. Influence of drug binding on DNA flexibility: a normal mode analysis. J Biomol Struct Dyn. 1997; 14:691–701. [DOI] [PubMed] [Google Scholar]
- 81. Rescifina A., Zagni C., Varrica M.G., Pistara V., Corsaro A.. Recent advances in small organic molecules as DNA intercalating agents: synthesis, activity, and modeling. Eur J Med Chem. 2014; 74:95–115. [DOI] [PubMed] [Google Scholar]
- 82. Strekowski L., Wilson B.. Noncovalent interactions with DNA: an overview. Mutation research. 2007; 623:3–13. [DOI] [PubMed] [Google Scholar]
- 83. Pindur U., Jansen M., Lemster T.. Advances in DNA-ligands with groove binding, intercalating and/or alkylating activity: chemistry, DNA-binding and biology. Curr Med Chem. 2005; 12:2805–2847. [DOI] [PubMed] [Google Scholar]
- 84. Banerjee A., Majumder P., Sanyal S., Singh J., Jana K., Das C., Dasgupta D.. The DNA intercalators ethidium bromide and propidium iodide also bind to core histones. FEBS Open Bio. 2014; 4:251–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Wang G., Gaddis S., Vasquez K.M.. Methods to detect replication-dependent and replication-independent DNA structure-induced genetic instability. Methods. 2013; 64:67–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Gulis G., Silva I.C., Sousa H.R., Sousa I.G., Bezerra M.A., Quilici L.S., Maranhao A.Q., Brigido M.M.. Characterization of an In Vivo Z-DNA Detection Probe Based on a Cell Nucleus Accumulating Intrabody. Mol Biotechnol. 2016; 58:585–594. [DOI] [PubMed] [Google Scholar]
- 87. Shin S.I., Ham S., Park J., Seo S.H., Lim C.H., Jeon H., Huh J., Roh T.Y.. Z-DNA-forming sites identified by ChIP-Seq are associated with actively transcribed regions in the human genome. DNA Res. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Liu H., Mulholland N., Fu H., Zhao K.. Cooperative activity of BRG1 and Z-DNA formation in chromatin remodeling. Mol Cell Biol. 2006; 26:2550–2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Liu R., Liu H., Chen X., Kirby M., Brown P.O., Zhao K.. Regulation of CSF1 promoter by the SWI/SNF-like BAF complex. Cell. 2001; 106:309–318. [DOI] [PubMed] [Google Scholar]
- 90. Katti M.V., Ranjekar P.K., Gupta V.S.. Differential distribution of simple sequence repeats in eukaryotic genome sequences. Molecular biology and evolution. 2001; 18:1161–1167. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.