Abstract
The CRISPR (clustered regularly interspaced short palindromic repeats) system protects archaea and bacteria by eliminating nucleic acid invaders in a crRNA-guided manner. The Sulfolobus islandicus type III-B Cmr–α system targets invading nucleic acid at both RNA and DNA levels and DNA targeting relies on the directional transcription of the protospacer in vivo. To gain further insight into the involved mechanism, we purified a native effector complex of III-B Cmr–α from S. islandicus and characterized it in vitro. Cmr–α cleaved RNAs complementary to crRNA present in the complex and its ssDNA destruction activity was activated by target RNA. The ssDNA cleavage required mismatches between the 5΄-tag of crRNA and the 3΄-flanking region of target RNA. An invader plasmid assay showed that mutation either in the histidine-aspartate acid (HD) domain (a quadruple mutation) or in the GGDD motif of the Cmr–2α protein resulted in attenuation of the DNA interference in vivo. However, double mutation of the HD motif only abolished the DNase activity in vitro. Furthermore, the activated Cmr–α binary complex functioned as a highly active DNase to destroy a large excess DNA substrate, which could provide a powerful means to rapidly degrade replicating viral DNA.
INTRODUCTION
The CRISPR-Cas (clustered regularly interspaced short palindromic repeats, CRISPR-associated) system provides an inheritable adaptive immunity against invasion of viruses or plasmids in archaea and bacteria (1–3). The system is comprised of two parts: CRISPR loci and cas gene cassettes. The former are composed of arrays of short repetitive DNA sequences (repeats) that are interspaced by unique DNA segments derived from invading genetic elements (spacers), whereas the latter encode structural proteins or enzymes associated with nucleic acids interference (4–8). All known CRISPR-Cas systems function in three distinct stages: (i) Adaptation—in which new spacers are acquired from invading nucleic acids, (ii) crRNA biogenesis—where CRISPR loci are transcribed and processed into mature CRISPR RNAs (crRNAs) and (iii) Interference—in which crRNAs form ribonucleoprotein complexes with Cas proteins, which identify invading nucleic acids for destruction.
At least six different types of CRISPR-Cas systems are known among which type I, II and III are widely investigated (3). Characterization of these systems shows that there is a general conservation in spacer acquisition (9,10), but the mechanisms of crRNA biogenesis and invader silencing are type-specific (11,12). Type I and II CRISPR-Cas target dsDNA and their effector complexes distinguish invading DNAs from self DNAs by recognition of a short motif immediately adjacent to the target sequence (protospacer adjacent motif, PAM) (13–19). Initial studies on type III-A and III-B systems showed that the Staphylococcus epidermidis Csm system mediates DNA interference in vivo, and the activity is independent of a PAM motif (20,21) whereas in vitro characterization of type III-B effector complexes purified from Pyrococcus furiosus and Sulfolobus solfataricus revealed their RNA interference activity (22–24).
A breakthrough in studying the function of type III CRISPR systems was made in our genetic analysis of a type III-B CRISPR RAMP module (Cmr) system in Sulfolobus islandicus. First, we found that the III-B system (Cmr–α) mediates transcription-dependent DNA interference (25) and subsequently, the same system was shown to confer RNA interference. Thus the Cmr–α system represents the first dual nucleic acids-targeting CRISPR-Cas system to be identified (26). Investigation of other type III CRISPR-Cas effector complexes has also revealed dual DNA/RNA interference, including III-A systems present in S. epidermidis, Streptococcus thermophilus and Thermus thermophilus (27–31), the type III-B systems of T. thermophilus and P. furiosus (32–36) and the type III-D system from S. solfataricus (37,38). To date, only two III-B systems are known to solely mediate RNA interference, including the Cmr7-containing S. solfataricus Cmr and S. islandicus Cmr-β systems (23,26,38–40).
Here we further characterized the S. islandicus Cmr–α system by purification of its wild-type and mutated effector complexes from the native host and testing for its DNA and RNA cleavage activity. We found that, upon activation by target RNA, the native Cmr–α complex exhibits very fast turnover on ssDNA substrate and is capable of degrading large amounts of DNA substrate.
MATERIALS AND METHODS
Strains, growth conditions and transformation of Sulfolobus strains
All Sulfolobus strains were derived from the original isolate S. islandicus REY15A (41) (Supplementary Table S1). Genetic host E233S1 and the Δcmr-β mutant were reported previously (42,43). S. islandicus MF1 was constructed with the E233S1 strain in two steps using a CRISPR-assisted gene deletion/mutagenesis procedure recently developed in our laboratory (Supplementary Figure S2) (44); (i) the genetic region encompassing the two cassettes of type I-A cas genes and the two CRISPR arrays was deleted and (ii) the promoter of csa5 and the coding sequence of cas6 were fused together, yielding an active cas6 gene. Four cmr-2α mutants were used in this work, two of which, cmr-2α-HD-M1 and -M2 were reported previously (44) whereas cmr-2αPalm-M1 and -M2 strains were constructed as for the two cmr-2αHD mutants (Supplementary Table S1). Sulfolobus strains were grown in SCV medium (basic salts and 0.2% sucrose, 0.2% casa amino acids, 1% vitamin solution) at 78°C. If appropriate, uracil was supplemented to 20 μg/ml. Transformation was performed by electroporation as previously described (42).
Construction of plasmids
Protospacer SS1 of the lacS gene was employed for gene silencing previously using a plasmid carrying an artificial CRISPR array containing SS1 spacer (26). DNA fragments of CRISPR array with multiple identical spacers were generated by polymerase chain reaction (PCR) with SS1-fwd and SS1-rev (Supplementary Table S3), and the resulting DNA fragments were digested by SalI and inserted into pSeSD1 between StuI and SalI sites. The ligation was used to transform Escherichia coli and the transformants were screened for the size of CRISPR array by PCR. A plasmid containing 10 copies of SS1 spacers (pAC10-SS1) was identified and used as the vector to clone cmr6α.
A C-terminal His-tagged version of cmr6α was obtained by inserting its coding sequence into the Sulfolobus expression vector pSeSD1 (45) at NdeI and StuI sites. Then, the tagged gene was amplified by PCR using the primer pair of MCS-up and MCS-dw (Supplementary Table S3). The PCR product was digested with SmaI and XhoI, and inserted into pAC10-SS1 at SalI and SmaI sites, giving pAC-cmr6α.
Plasmids for mutagenesis in the Palm domain of Cmr-2α were constructed as described previously (44) using the oligonucleotides listed in Supplementary Table S3.
All the primers to be used for DNA cloning were synthesized from TAG Copenhagen A/S (Copenhagen, Denmark). Sequences of all plasmid constructs were verified by DNA sequencing at the MacroGen Europe (Amsterdam, Netherlands).
Purification of Cmr–α crRNA ribonucleoprotein complex
The expression and purification procedure reported for the purification of a tagged Cmr-β from S. solfataricus (23) was followed with modification. S. islandicus strains carrying a cmr6α expression plasmid were grown in SCV at 78°C up to A600 = 0.7, and cells were collected from at least 10 liters of culture by centrifugation at 8000 rpm for 10 min. Cell pellet was re-suspended in Buffer A (20 mM HEPES pH 7.5, 30 mM Imidazole, 500 mM NaCl) and disrupted by French press. The cell extract was loaded onto a 1 ml HisTrap HP (GE Healthcare) and His-tagged protein was eluted by Buffer B (20 mM HEPES pH 7.5, 500 mM Imidazole, 500 mM NaCl). Five milliliters of Buffer B fractions were concentrated and further purified by size exclusion chromatography in Buffer C (20 mM Tris-HCl pH 7.5, 250 mM NaCl) with a Superdex 200 Hiload column (GE Healthcare). Sample fractions were analyzed by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and those containing the complete set of Cmr–α components were pooled together and used for further analysis.
Extraction and analysis of crRNA from the Cmr–α ribonucleoprotein complex
One hundred microliters of purified Cmr–α complex were mixed with 200 μl DEPC-H2O, 600 μl Trizol agent (Sigma) and 300 μl chloroform in the indicated order. The mixture was incubated at room temperature for 5 min and centrifuged at 12 000 rpm for 10 min. The upper phase was transferred into a new tube and re-extracted with 300 μl chloroform. RNA in the upper phase was precipitated with one volume of isopropanol and washed with 1 ml of 75% ethanol. The pellet was air-dried for 30 min at the room temperature and dissolved in 15 μl DEPC-H2O, giving crRNA preparations for further analysis. An aliquot of the purified crRNA was 5΄ labeled with 32P-γ-ATP (PerkinElmer) using T4 polynucleotide kinase (New England Biolabs) and separated on a 12% denaturing polyacrylamide gel. The labeled RNAs were identified by exposing the gel to a phosphor screen (GE Healthcare) and scanned with a Typhoon FLA 7000 (GE Healthcare).
About 1 μg of crRNA extracted from the Cmr–α complex purified from the wild-type S. islandicus strain was sent to Beijing Genomics Institute, China for RNA sequencing using a paired-end sequencing protocol with the maximum read length of 90 bases. The RNA sample was phosphorylated before the analysis since crRNAs carry a 5΄-OH group that are not a direct substrate for the RNA sequencing reaction. The reads were mapped to the genome of S. islandicus Rey15A (Genbank ID: CP002424) using CLC Genomics Workbench 7.5 (CLC Genomics, Denmark).
Labeling of DNA and RNA substrates
DNA and RNA substrates used in cleavage assays were 5΄ labeled with 32P using T4 polynucleotide kinase (New England Biolabs) or 3΄ labeled with [32P]pCp (PerkinElmer) using T4 RNA ligase (Invitrogen). Double strand DNA was generated by annealing of labeled SS1 ssDNA with unlabeled SS1T ssDNA; bubble DNA was made by annealing of labeled SS1 ssDNA and unlabeled S32T ssDNA; R-loop DNA was made by annealing of labeled SS1 ssDNA, unlabeled S32* RNA and unlabeled S32T ssDNA (Supplementary Table S4). For the annealing assay, 50 pmol of labeled substrate was mixed with an equivalent amount of unlabeled nucleic acid as indicated in the experiments and incubated at 90°C for 0.5 min. The mixture was allowed to slowly cool down to the room temperature.
All nucleic acids were purified by recovering the corresponding bands from either a native polyacrylamide gel (dsDNA) or denaturing polyacrylamide gel (ssDNA/RNA) PAGE. DNA and RNA oligonucleotides to be used as substrate for cleavage assays were purchased from IDT, USA.
RNA cleavage and binding assay and DNA cleavage assay
For both RNA and DNA cleavage assay, the reaction mixture (10 μl in total) contains 50 mM Tris-Cl (pH 7,6), 10 mM MgCl2, 5 mM DTT and indicated amount of complex and substrates. In the DNA cleavage assay, 200 nM (unless otherwise indicated) unlabeled RNA was supplemented into the reaction mixture to activate DNA cleavage activity. The reaction was performed at 70°C and stopped at the indicated time point by the addition of 2 × RNA loading dye (New England Biolabs) and cooling on ice. Finally, the samples were heated for 5 min at 95°C and then run on 18% polyacrylamide denaturing gels and visualized by phosphor imaging.
To test the association of RNA substrates/cleavage products with Cmr–α, cleavage reactions were set up and mixed with 2 × RNA loading dye solution at indicated time points and the samples were then loaded on a 10% non-denaturing polyacrylamide gel to detect the formation of Cmr–α-crRNA:target RNA tertiary complexes. RNA ladders were generated by Decade™ Marker RNA (Ambion) following the instructions, while 10/60 DNA ladders were purchased from IDT and labeled by 32P with T4 polynucleotide kinase.
RESULTS
Purification of tagged native Cmr–α complexes from S. islandicus
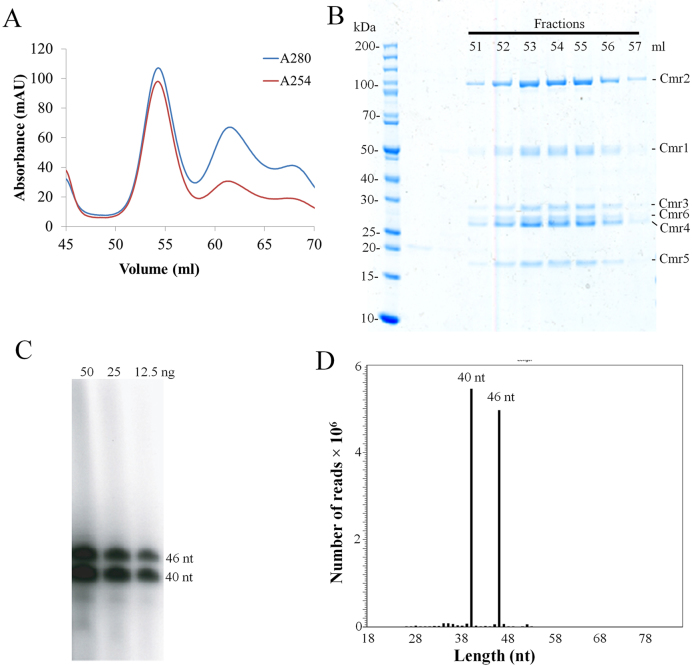
To purify a native crRNA ribonucleoprotein complex of the S. islandicus Cmr–α system, a His-tagged cmr-6α gene was inserted into the Sulfolobus expression vector pSeSD1 and the fusion protein was expressed in strain E233S1. Then, a native Cmr–α effector complex was purified in two-step purification following the procedure described previously (23). Nickel affinity chromatography showed only one peak of UV absorbance (Supplementary Figure S1A), and analyzing fractionated samples collected around the peak by SDS-PAGE revealed that several of them contained proteins of apparent sizes matching the six different subunits encoded by the Cmr–α system (Supplementary Figure S1B), suggesting that the Cmr–α complex was co-purified in the experiment. These fractions were pooled and subjected to size exclusion chromatography in which the largest protein complex eluted at 54 ml (Figure 1A). The UV254 profile of the gel filtration indicated that only this protein complex contained nucleic acids (Figure 1A) and SDS-PAGE analysis showed that fractions 52–57 contained all six Cmr–α proteins (Cmr–1α to 6α) (Figure 1B). To verify crRNA was indeed present in the protein complex, these fractions were pooled together and used to analyze the crRNA component by Trizol extraction, radiolabeling and denaturing polyacrylamide gel electrophoresis. RNAs of two different sizes were detected, differing by 6 nt (Figure 1C). The extracted RNA sample was sequenced by paired-end RNA sequencing, and this revealed that >99% sequencing reads matched the spacer sequences in the two CRISPR arrays of S. islandicus and that there was a very biased distribution of the abundance of crRNAs on the chromosomal spacers (Supplementary Figure S2). Furthermore, almost all crRNAs started with the 8 nt repeat tag at the 5΄-end and they are almost exclusively 40 or 46 nt in size regardless of the lengths of original spacers (Figure 1D), indicating that Cmr crRNAs in S. islandicus were also processed following the ruler mechanism as previously reported for P. furiosus and T. thermophilus Cmr crRNAs (22,32).
Figure 1.
Purification of a tagged native effector complex of Cmr–α from Sulfolobus islandicus. (A) UV spectrum of gel filtration chromatography of the His-tagged Cmr–α complex. Blue: UV absorbance at 280 nm; red: UV absorbance at 254 nm. (B) SDS-PAGE of the selected peak fractions after the size exclusion chromatography. (C) Denaturing gel electrophoresis analysis of 5΄-labeled RNA copurified with Cmr–α. (D) Histogram illustrating the size distribution of the mapped crRNA reads.
RNA sequencing of the S. islandicus Cmr–α crRNAs showed that the crRNAs generated from spacer 32 in Cluster 1 (L1S32) is a dominant species. For this reason, we designed the RNA substrate corresponding to this spacer (S32 RNA) and used it to test nucleic acids cleavage by Cmr–α. Two RNA substrates, S10 and SS1, were designed: the former of which was designed based on the spacer L2S10 (array 2, spacer 10), and the latter was for the artificial spacer SS1 of the lacS gene, both of which were used in our genetic analyses of the CRISPR-Cas system in S. islandicus. RNA cleavage of the Cmr–α complex was tested with each of the three RNA substrates. Cmr–α only cleaved the S10 substrate but was apparently inactive with either SS1 or S32 RNA substrate (Figure 2). In addition, initial attempts failed to detect any DNA cleavage activity of the native effector complex on any designed DNA substrates.
Figure 2.
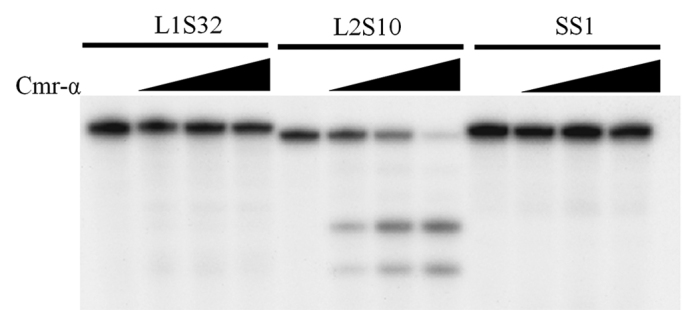
Native Cmr–α purified from the wild-type strain only cleaves S10 target RNA. The 5΄-end labelled substrates were incubated with Cmr–α at different concentration (0/25/50/100 ng/μl) for 1 h and then separated on 18% denaturing-PAGE gel.
Since the RNA seq data of the crRNAs present in the Cmr–α complex was not particularly useful in selecting a proper spacer for in vitro analysis, we decided to construct a S. islandicus host-plasmid system to produce a Cmr–α effector complex containing only a single defined crRNA. For this purpose, the mutant S. islandicus MF1 was constructed, lacking the genomic region containing two CRISPR loci and the interference and adaptation modules of type I-A CRISPR-Cas system except for the csa5 promoter and the cas6 coding sequence, which were fused together to produce a functional Cas6 for crRNA biogenesis (Supplementary Figure S3A). An expression plasmid was constructed, containing a His-tagged version of cmr-6α gene and an artificial mini-CRISPR locus of 10 copies of SS1 repeat-spacer units (Supplementary Figure S3B). After introducing the plasmid into the MF1 strain by electroporation, transformants were obtained and used for producing cell mass for purifying the native tagged Cmr–α complex. Since the purified effector complex only carried SS1 crRNA, the effector complex was designated as Cmr–α-SS1 and characterized.
RNA cleavage of the S. islandicus Cmr–α complex
First, RNA cleavage activity of Cmr–α-SS1 was tested using radiolabeled S10, S32 and SS1. The effector complex was mixed with each target RNA in 1:1 molar ratio and incubated at 70°C for 20 min. Analyzing cleavage products by denaturing polyacrylamide gel electrophoresis showed that only SS1 RNA substrate was cleaved whereas L2S10 and L1S32 RNAs were not substrates of the effector complex (Supplementary Figure S4). This indicated that complementarity between crRNA and target RNA is required for the RNase activity of Cmr–α.
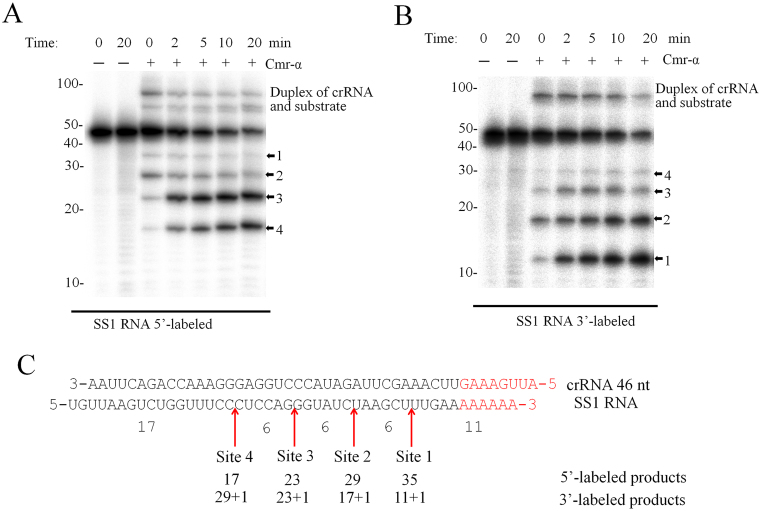
To characterize the RNA cleavage of Cmr–α-SS1 in more detail, an SS1 target RNA was radiolabeled either at the 5΄-end or at 3΄-end and used as substrate to test RNA cleavage in time-course experiments. As shown in Figure 3A and B, four cleavage products were observed already at time 0, for which all components were mixed together at the room temperature (RT) and the loading buffer was added ca. 1 min later to quench the reaction. These results indicated that the thermophilic Cmr–α complex is capable of cleaving the target RNA immediately after substrate binding at RT. In addition, the cleavage started predominantly at site 2, and after 2 min the 23- and 17-nt products of 5΄-labeled substrate and the 12-nt product of 3΄-labeled substrate were dominant. After 20 min's incubation, about 80% of target RNA was destructed. All cleavage products were positioned to the RNA substrate, and they differed in 6 nt from each other (Figure 3C).
Figure 3.
Cmr–α-SS1 cleaves SS1 substrate at four sites. (A and B) SS1 RNA was labeled at 5΄-end or 3΄-end respectively. 25 nM of the labelled substrates were incubated with (+) or without (−) 25 nM of Cmr–α-SS1 for the indicated time points. Then, the samples were analyzed by denaturing PAGE. The four cleavage sites are indicated with arrowheads. (C) Schematic depicting the cleavage sites on SS1 RNA. Note that 3΄ labeling introduced one more nucleotide at the 3΄-end of SS1 RNA.
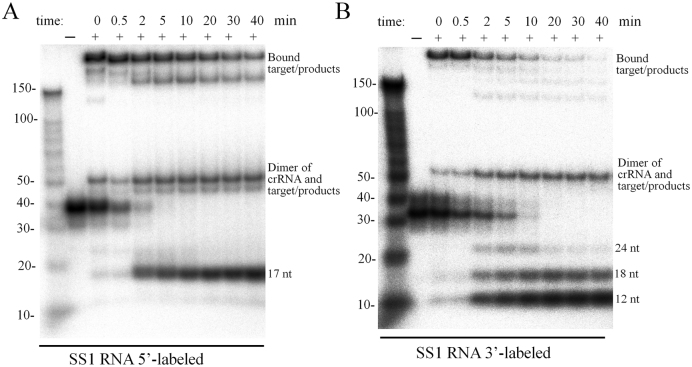
The same samples were also analyzed by non-denaturing gel electrophoresis to investigate the disassociation of the products from Cmr–α complex. As shown in Figure 4, after 2 min, the 17-nt product from the 5΄-end and the 12-nt, 18-nt, 24-nt products from the 3΄-end were observed at their corresponding position, suggesting that they could be released from the complex, while the 23-nt and 29-nt from the 5΄-end were still associated with the complex.
Figure 4.
Disassociation of target RNA and cleavage products from the Cmr–α complex. RNA cleavage was conducted as described in Figure 3 and samples withdrawn during incubation were analyzed by non-denaturing polyacrylamide gel electrophoresis.
Since 46 nt crRNA is more abundant in the Cmr–α complex of the early fractions whereas 40 nt crRNA predominates those from the later fractions in the gel filtration (Supplementary Figure S5A), we investigated the correlation between the length of crRNA and the RNA cleavage patterns using F51 and F56. Whereas more cleavage products were produced at site 4 for F51 sample, more site 3 products were obtained with F56 (Supplementary Figure S5C), and this is consistent with the relative contents of 46 and 40 nt crRNA in the two fractions. Therefore, the Cmr–α complex carrying 46-nt crRNAs could contain one more pair of Cmr4–Cmr5 compared to those containing 40 nt crRNA, consistent with the current model of type III-B systems in which a larger and a smaller effector complexes co-exist, differing in one pair of Cmr4–Cmr5 proteins (46) and the active sites reside on Cmr4 subunits in the complex (36,47).
We quantified the uncleaved target RNA in Figure 3 and this revealed that ca. 20% of target RNA remained uncleaved after 20 min incubation. To study the interaction between target RNA and Cmr–α in more details, they were mixed in different molar ratios (RNA:Cmr = 1:1, 1:0.5, 1:2 and 1:4) and incubated for 60 min during which samples were taken and analyzed for RNA cleavage. We found that: (i) doubling the amount of target RNA in the reaction reduced the percentage of uncleaved RNA from 18 to 11% (Supplementary Figure S6A and B), indicating that the incomplete cleavage did not result from a contamination of any non-cleavable RNAs in the substrate, and (ii) adding excess amounts of Cmr–α-SS1 accelerated RNA cleavage at time 0, but the presence of 2 or 4-fold excess Cmr–α complex did not yield any impact on target RNA cleavage (Supplementary Figure S6C), revealing an interesting feature between the interaction between Cmr–α and its target RNA.
Target RNA-activated DNA cleavage
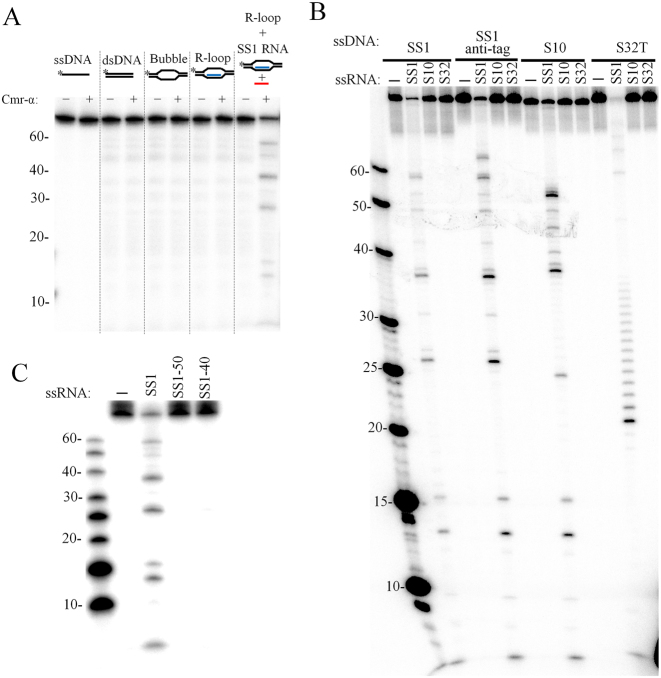
To investigate DNA cleavage of Cmr–α, we designed several different DNA substrates, including ssDNA, dsDNA, bubble DNA and R-loop DNA, the last of which was used to mimic the transcription structure (Supplementary Table S4 and Figure S7). Furthermore, the SS1 RNA target was also included in another R-loop DNA sample to mimic the transcript of protospacer. Since all these DNA substrates contain the DNA strand complementary to SS1 crRNA (SS1 DNA), this DNA was radio-labeled in order to follow the DNA cleavage. As shown in Figure 5A, Cmr–α-SS1 only cleaved the R-loop DNA in the presence of the cognate target RNA. Cleavage of ssDNA, dsDNA and bubble DNA was then tested in the presence of the cognate target RNA. This showed that ssDNA and the ssDNA region of bubble DNA were cleaved whereas dsDNA was not a substrate for the Cmr–α complex (Supplementary Figure S8).
Figure 5.
Target RNA activates the ssDNA cleavage activity of Cmr–α. (A) ssDNA, dsDNA, bubble DNA, R-loop and R-loop together with 200 nM of SS1 RNA were incubated with (+) or without (−) 50 nM of Cmr–α-SS1 for 1 h. Then, the samples were analyzed by denaturing PAGE. (B) Different labeled ssDNA substrates (25 nM) were incubated with 50 nM Cmr–α in the presence of 200 nM of different ssRNA for 1 h and then analyzed by denaturing PAGE. (C) Requirement of the non-complementary 3΄-flanking region of the target RNA for DNA cleavage activity. A total of 25 nM of labeled SS1 ssDNA substrate was incubated with 50 nM Cmr–α in the presence of 200 nM of indicated ssRNA for 1 h and then analyzed by denaturing PAGE.
The Cmr–α DNase activity was further characterized with 4 different ssDNA substrates in combination with three different RNAs. The results showed that DNA shredding occurred for all four ssDNA substrates tested but the DNase activity showed a strict dependence on the presence of the SS1 target RNA (Figure 5B), meaning that the activation of DNA cleavage by Cmr–α requires the complementarity between its target RNA and the crRNA residing in Cmr–α. Once activated, the Cmr–α DNase was active in shredding all four tested DNA substrates, and the cleavage sites were mapped to the positions after all thymidine bases in all tested ssDNA substrates (Supplementary Figure S9). This indicates that the DNA cleavage might not involve the recognition of DNA substrate via base paring between the crRNA and its ssDNA target.
To investigate how target DNA was to be discriminated by Cmr–α, three variants of SS1 target RNA, including SS1 (mismatch at the anti-tag region), SS1-50 (with the anti-tag sequence) and SS1-40 (lacking the anti-tag region), were tested for their capability to support the Cmr–α DNase activity. All the three RNAs were equally cleaved by Cmr–α–SS1 (Supplementary Figure S10). However, only SS1 activated the Cmr–α DNase (Figure 5C), indicating that Cmr–α not only depends on the binding a cognate RNA to the crRNA, but also on the mismatches between the crRNA and the target RNA at the anti-tag region to activate the DNase activity, suggesting that DNA target discrimination occurs at the RNA level.
Cmr–2α contained the active sites for the DNA interference
Cas10 is the signature protein for type III CRISPR-Cas systems and this protein is named as Cmr-2 in III-B systems. The protein contains two conserved motifs, i.e. the histidine-aspartate acid (HD) nuclease domain and the GGDD motif in the Palm domain (33,48–51), which were implicated in DNA cleavage. To investigate the function of Cmr–2α in the Cmr–α DNA interference, two mutants carrying substitutions in the HD domain reported previously (44) and two mutants of PALM domains constructed in this work (Supplementary Table S1) were employed for invader plasmid assay to test their DNA interference activity. As shown in Figure 6A, substitution of H14N and D15N in the HD nuclease domain of Cmr–2α did not affect the DNA interference by the mutant effector complex, but a quadruple mutation (H14N, D15N, K19A and I23A) in the same domain abolished the in vivo DNA targeting. For the Palm domain, either a double mutation (G666K and D667K) or octal mutation (662—IYlGGDDiLA—671 to AAlAAAAiAS) in the GGDD motif region inactivated the DNA interference, indicating that both domains are essential for the in vivo Cmr–α DNA interference.
Figure 6.
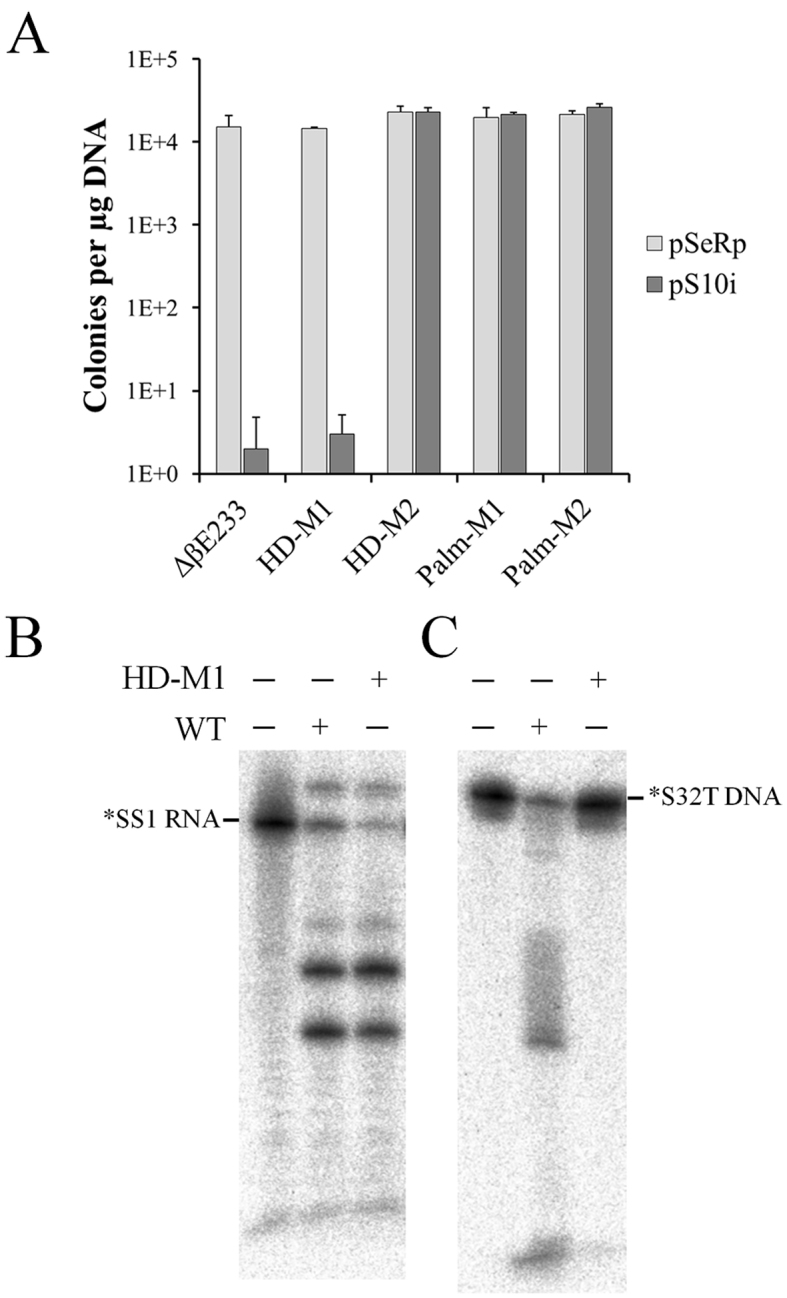
Functional analyses of Cmr-2α. (A) In vivo DNA interference activity of cmr-2α mutants carrying substitutions in the HD or Palm domain. pSe-Rp—a repeat cloning vector for Sulfolobus, pS10i—an invader plasmid carrying a target sequence of spacer 10 in CRISPR locus 2 in S. islandicus REY15A. Following Cmr2α mutants were used: HD-M1—a double mutation (H14N, D15N) in the HD domain; HD-M2—a quadruple HD mutation (H14N, D15N, K19A, I23A), Palm-M1—a double mutation (G666K, D667K) in the GGDD motif and Palm-M2—an octal PALM mutation (662—IYlGGDDiLA—671 to AAlAAAAiAS). (B) In vitro RNA/DNA cleavage by the Cmr-2HDM1 effector complex containing Cmr-2 HDM1 mutant protein. RNA cleavage assay was conducted with 25 nM labeled SS1 ssRNA, 50 nM Cmr–α complex of either the wild-type (WT) or the mutant complex (HD-M1) harboring Cmr2αH14N, D15N and incubated for 20 min. DNA cleavage was conducted with 25 nM labeled S32T ssDNA substrate, 200 nM SS1 RNA and 50 nM WT or HD-M1 Cmr–α and incubated for 1 h.
All four mutants were then subjected to purification of Cmr–α effector complex by the affinity purification and size exclusion chromatography as described for the wild-type strain. Effector complex was not obtainable from them except the cmr–2α HDM1 mutant from which the effector complex Cmr–HDM1 was isolated containing Cmr–2αH14N,D15N. RNA cleavage and DNA cleavage by Cmr–HD–M1 was then analyzed and compared with the activities of the wild-type Cmr–α complex. We found that, while both effector complexes showed a similar pattern of RNA cleavage, DNA cleavage by the mutated Cmr–α complex was attenuated (Figure 6B), reinforcing the conclusion that both HD and GGDD motifs of Cmr–2α are important for DNA interference in vivo by the CRISPR-Cas system.
Large excess amounts of DNA substrate greatly accelerate DNA cleavage by Cmr–α
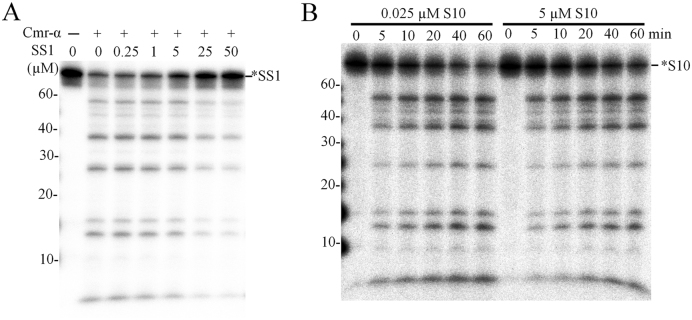
To further characterize DNA cleavage by Cmr–α, 25 nM labeled SS1 or S10 ssDNA was mixed with a 10-fold excess of unlabeled SS1, S10 or S32 ssDNA (250 nM) and tested for the DNase activity. As shown in Supplementary Figure S11, Cmr–α DNase cleaved similar amounts of labeled SS1 and S10 ssDNA regardless whether there was 10-fold extra unlabeled ssDNA in the reaction or not, suggesting that increasing DNA substrate concentration could have accelerated the DNA shredding. This was then investigated systematically by testing DNA destruction in a wide range of SS1 ssDNA concentrations (25 nM to 50 μM), which were prepared by mixing different amounts of unlabeled SS1-ssDNA with a constant amount of labeled one (25 nM). These substrate mixtures were assayed for DNA cleavage by Cmr-SS1 in the presence of a 4-fold excess of target RNA (200 nM). As shown in Figure 7A, very similar patterns of DNA cleavage were observed for substrate concentrations of 2000-fold difference. Uncleaved ssDNA was quantitated for each sample and a positive correlation was seen for the concentrations of the input substrate and cleaved products (Table 1). More specifically, at the lowest substrate concentration (25 nM), 17 nM of DNA substrate was cleaved, while 13.6 μM DNA was destroyed when the substrate was amounted to 50 μM, indicating that the DNase activity of the same amount of Cmr–α complex was increased for >700-fold (Table 1). To yield an insight into the superactive DNA cleavage activity, we did a time-course experiment to compared the dynamics of the cleavage of 25 nM ssDNA substrate with that of 200-fold excess substrate (25 nM labeled + 5 μM unlabeled ssDNA substrate) and very similar patterns of DNA cleavage were obtained (Figure 7B). Therefore, we conclude that Cmr–α could degrade a large amount of viral DNA at a low expression level.
Figure 7.
ssDNA substrate accelerated DNA cleavage of Cmr–α–SS1. (A) 25 nM of labeled SS1 ssDNA substrate, as well as unlabeled SS1 ssDNA at the indicated concentrations, were incubated with 50 nM Cmr–α–SS1 and 200 nM SS1 RNA at 70°C for 1 h. Then, the samples were analyzed by denaturing PAGE. (B) 25 nM of labeled S10 ssDNA substrate was incubated with 50 nM Cmr–α–SS1 and 200 nM SS1 RNA, in the presence or absence of 5 μM unlabeled S10 ssDNA for 1 h during which aliquots of samples were taken at indicated time points, cleavage reactions were stopped by adding 2× loading buffer and analyzed by denaturing PAGE.
Table 1. ssDNA cleavage with increasing amounts of unlabeled ssDNA substrate.
| Original SS1 (μM) | 0.025 | 0.275 | 1 | 5 | 25 | 50 |
| Cleaved SS1 (μM) | 0.017 | 0.19 | 0.63 | 2.76 | 9.41 | 13.6 |
The values of ‘Original SS1’ are the sum of total input substrate including labeled and unlabeled SS1 ssDNA. To obtain the values of ‘Cleaved SS1’, the residual amount of SS1 substrate was estimated by image quantification for each sample in Figure 7A and deducing the residual SS1 from the total input value yielded the amount of the cleaved products (Cleaved SS1).
DISCUSSION
In this study, a native Cmr–α effector complex was purified from S. islandicus and characterized. The purified complex contains 6 different subunits, Cmr–1α to 6α that form two different stoichiometries to accommodate crRNAs of two different sizes (40 and 46 nt, respectively), which is consistent with the results obtained for III-B Cmr complexes of T. thermophilus and P. furiosus (32–34,46,47). The natural species of crRNAs carried by Cmr–α complex were generated from nearly all the spacers from the two CRISPR arrays. Unexpectedly, the crRNA of L1S32 comprised up to 69% of the total amounts of crRNAs. Since the RNA substrate based on L1S32 cannot be cleaved by the Cmr–α complex, it is more likely that the extremely biased over-presentation of L1S32 crRNA is due to an artefact occurred in the RNA sequencing procedure. Apart from the unusual L1S32 dominance, crRNAs incorporated into the Cmr–α complex still show an uneven distribution among the spacers of the two CRISPR arrays as their levels can be mounted up to a difference of 10 000-fold, consistent with the results obtained from T. thermophilus (32). Recently, the transcriptome of S. islandicus has been determined by RNA sequencing (52), showing that CRISPR locus 2 yield higher levels of crRNA than locus 1, consistent with the distribution of the crRNA from Cmr–α complex in general (Supplementary Figure S2). However, the transcriptomic analysis also shows that spacers 1–13, 26–44, 55–63 and 66–72 from locus 2 and spacers 21–22 from locus 1 are expressed in higher abundance, which is different from our RNA sequencing data, suggesting that the maturation and incorporation of crRNAs are also biased.
Among all known CRISPR-Cas systems, those of type III exhibit a huge diversity in sequence similarity and subunit composition (1,53) such that the original discoveries of a III-A Csm system conferring DNA interference (20,21) and III-B Cmr systems possessing RNA cleavage activity (22–24) were considered as a general distinction between the two subclasses. This view was first challenged by the discovery of the transcription-dependent DNA interference and dual DNA/RNA targeting by the S. islandicus Cmr–α (type III-B, SisCmr–α) (25,26). Subsequently, dual nucleic acid interference has been demonstrated for the S. epidermidis Csm (type III-A, SeCsm) (27,30) and the P. furiosus III-B Cmr system (PfuCmr) (54). Moreover, in vitro studies show that several III CRISPR-Cas systems show both RNA and DNA cleavage activities including the reconstituted T. maritima III-B Cmr (TmCmr), PfuCmr, SeCsm, StCsm and SsoCsm (30,31,38,54,55), the last of which is also classified as a III-D system (53). Here we show that these general results also apply for the S. islandicus Cmr–α system, suggesting that most type III CRISPR-Cas systems possess the RNA-activated DNA targeting activity although the activity remains to be tested for SeCsm and SsoCsm. The only known exceptions are the closely related S. solfataricus Cmr (23,39) and S. islandicus Cmr-β (26), which only exhibit RNA interference.
Characterization of a few III-A, III-B and III-D systems in vitro (30,31,38,54,55) clearly indicates that Cas10 bears the active sites for DNA cleavage for type III CRISPR-Cas systems, i.e. the HD nuclease domain and GGDD motif in the Palm domain. So far, P. furiosus Cmr2 is the only Cas10 protein for which both domains have been functionally characterized in vivo and in vitro (54). The authors found that alanine substitutions of HD motif of Cmr2 abolishes the DNA cleavage by the mutant Cmr complex in vitro but the same substitutions for DD in the GGDD motif of the protein does not affect the DNA cleavage, however, mutations in both motifs are required to silence the DNA interference assayed by invader plasmid (54). We found that while H14N and D15N substitutions (HDM1) in Cmr–2α abolished the DNA cleavage of the mutant Cmr–α complex in vitro, the system still functions in DNA interference in vivo as plasmid transformation rates were very similar for the wild-type strain and the mutant. This either suggests that the mutated H14N D15N motif could still function in vivo or there could be another active site of DNA cleavage in Cmr–2α, such as the GGDD motif in the Palm domain. The latter scenario is likely to be true since the Palm domain of PfuCmr and SeCsm functions as an active site of DNA interference (30,54). Interestingly, we found that substitution of additional two conserved amino acids (K19A and I23A) in the HD domain, i.e. a quadruple mutation in the Cmr–2α HD domain (HD-M2) attenuated the SisCmr–α in vivo DNA targeting (Figure 6A). The mutant complex is very likely to be inactive in vitro since it carries the HD mutation that abolishes the DNA cleavage activity in the Cmr–HDM1 complex. This was not tested thus far because our attempts failed to purify a Cmr–α complex from the HDM2 mutant by the Cmr-6 co-purification. Furthermore, Cmr–α complex was not obtainable from two Palm mutants of Cmr–2α, including a double and an octal mutation in the Palm domain (PalmM1 or PalmM2). This could be due to that the resulting Cmr–α complexes were either less stable than the wild-type one such that the components disassociated during purification, alternatively Cmr–α subunits could also lost the capability of forming the effector complex in vivo. In each scenario, this suggests both HD and Palm domains of Cmr2–α play a structural role in maintaining the integrity of the effector complex.
The importance of the mismatch between the target and crRNA for type III DNA interference was first demonstrated for SeCsm (21). The same principle also functions in SisCmr–α (25) as revealed from its in vivo analysis and this was further confirmed by the in vitro study of SeCsm (30). However, analysis of PfuCmr indicates that the self versus non-self DNA distinction occurs at RNA level (54). We also investigated how the Cmr–α system distinguishes target and non-target nucleic acids in vitro and found that it is the mismatch between the 5΄-handle of crRNA and 3΄-sequence of the target RNA that triggers the RNA-activated Cmr–α DNase and removing the sequence corresponding to the 5΄-handle inactivates the DNase, which is consistent with the results obtained with StCsm (31), but different from the results obtained with TmCmr systems (55), suggesting a diversification in molecular mechanisms of DNA interference by type III systems. Indeed, SisCmr–α cleaves ssDNA after thymidine nucleotides, in a similar fashion as observed with the reconstituted TmCmr (55); PfuCmr also cleaves dsDNA (54), further arguing for functional diversification of type III CRISPR-Cas systems as predicted from bioinformatics analyses (1,53).
Another common feature for type III systems is that they often coexist with other systems (type I or II) (1), suggesting that type III and I/II CRISPR-Cas systems have their preferable nucleic acid targets in antivirus immunity. Indeed, different from type I and II systems, a number of type III CRISPR-Cas systems show RNA-activated generic DNA degradation that is not based on base-pairing between crRNA and DNA substrate. Here we have unraveled a unique feature for the DNA cleavage; the turnover of ssDNA cleavage by Cmr–α is very fast and an increase of up to 700-fold of Cmr–α DNase has been observed for 1 h incubation. Interestingly, transcriptome analyses of genome expression in S. islandicus LAL14/1 and REY15A have revealed that whereas type I genes are significantly up-regulated after virus infection, type III genes are not induced or even repressed (52,56,57). This raises an important question as to what role type III CRISPR-Cas systems play in the arms race between microbial hosts and their viruses during evolution. Since it takes time from infection of a new virus to the acquisition of new spacers and eventually the production of functional antiviral effector complexes, the lag between the virus invasion and the onset of CRISPR immunity against the new virus should allow virus to replicate, producing massive replication intermediates. Indeed, recently it has been shown that the Sulfolobus SIRV2 virus produces an exceptionally large amount of ssDNA intermediates (58), a phenomenon that have not been observed for other Sulfolobus viruses such as fuselloviruses and their satellites (59,60). Therefore, the capability of massive ssDNA destruction by type III immunity would greatly limit the replication of SIRV2-like viruses whereas the DNA interference by the type I-A system would not be effective. In this regard, it is advantageous to have different CRISPR-Cas systems in an organism to lead the arms race toward the microbe side during evolution.
Supplementary Material
ACKNOWLEDGEMENTS
We thank members in the German CRISPR consortium and colleagues in the Archaea Centre for stimulating discussions.
Footnotes
Present addresses:
Ling Deng, Department of Food Science, University of Copenhagen, Frederiksberg, Denmark.
Wenfang Peng, Hubei Collaborative Innovation Center for Green Transformation of Bioresources, Hubei Key Laboratory of Industrial Biotechnology, College of Life Science, Hubei University, Wuhan 430062, P.R. China.
Jing Zhang, J. Michael Bishop Institute of Cancer Research, 9-1 Keyuan Road South, Chengdu, 610041 Sichuan, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Danish Council for Independent Research [DFF-4181-00274, DFF-1323-00330 to Q.S.]; Scientific and Technological Self-Innovation Foundation of Huazhong Agricultural University [2014RC011 to Q.S.]; Carlsberg Foundation; Biotechnology and Biological Sciences Research Council [BB/M000400/1 to M.F.W.]; China Scholarship Council PhD studentship (to W.H.). Funding for open access charge: University of Copenhagen.
Conflict of interest statement. None declared.
REFERENCES
- 1.Makarova K.S., Wolf Y.I., Alkhnbashi O.S., Costa F., Shah S.A., Saunders S.J., Barrangou R., Brouns S.J., Charpentier E., Haft D.H. et al. An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol. 2015; 13:722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marraffini L.A. CRISPR-Cas immunity in prokaryotes. Nature. 2015; 526:55–61. [DOI] [PubMed] [Google Scholar]
- 3.Mohanraju P., Makarova K.S., Zetsche B., Zhang F., Koonin E.V., van der Oost J.. Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems. Science. 2016; 353:aad5147. [DOI] [PubMed] [Google Scholar]
- 4.Bolotin A., Quinquis B., Sorokin A., Ehrlich S.D.. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005; 151:2551–2561. [DOI] [PubMed] [Google Scholar]
- 5.Mojica F.J., Diez-Villasenor C., Garcia-Martinez J., Soria E.. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 2005; 60:174–182. [DOI] [PubMed] [Google Scholar]
- 6.Lillestol R.K., Redder P., Garrett R.A., Brugger K.. A putative viral defence mechanism in archaeal cells. Archaea. 2006; 2:59–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jansen R., Embden J.D., Gaastra W., Schouls L.M.. Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol. 2002; 43:1565–1575. [DOI] [PubMed] [Google Scholar]
- 8.Haft D.H., Selengut J., Mongodin E.F., Nelson K.E.. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput. Biol. 2005; 1:e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heler R., Marraffini L.A., Bikard D.. Adapting to new threats: the generation of memory by CRISPR-Cas immune systems. Mol. Microbiol. 2014; 93:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sternberg S.H., Richter H., Charpentier E., Qimron U.. Adaptation in CRISPR-Cas Systems. Mol. Cell. 2016; 61:797–808. [DOI] [PubMed] [Google Scholar]
- 11.Charpentier E., Richter H., van der Oost J., White M.F.. Biogenesis pathways of RNA guides in archaeal and bacterial CRISPR-Cas adaptive immunity. FEMS Microbiol. Rev. 2015; 39:428–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Plagens A., Richter H., Charpentier E., Randau L.. DNA and RNA interference mechanisms by CRISPR-Cas surveillance complexes. FEMS Microbiol. Rev. 2015; 39:442–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mojica F.J., Diez-Villasenor C., Garcia-Martinez J., Almendros C.. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009; 155:733–740. [DOI] [PubMed] [Google Scholar]
- 14.Gudbergsdottir S., Deng L., Chen Z., Jensen J.V., Jensen L.R., She Q., Garrett R.A.. Dynamic properties of the Sulfolobus CRISPR/Cas and CRISPR/Cmr systems when challenged with vector-borne viral and plasmid genes and protospacers. Mol. Microbiol. 2011; 79:35–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sashital D.G., Wiedenheft B., Doudna J.A.. Mechanism of foreign DNA selection in a bacterial adaptive immune system. Mol. Cell. 2012; 46:606–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sinkunas T., Gasiunas G., Waghmare S.P., Dickman M.J., Barrangou R., Horvath P., Siksnys V.. In vitro reconstitution of Cascade-mediated CRISPR immunity in Streptococcus thermophilus. EMBO J. 2013; 32:385–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayes R.P., Xiao Y., Ding F., van Erp P.B., Rajashankar K., Bailey S., Wiedenheft B., Ke A.. Structural basis for promiscuous PAM recognition in type I-E Cascade from E. coli. Nature. 2016; 530:499–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gasiunas G., Barrangou R., Horvath P., Siksnys V.. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:E2579–E2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E.. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012; 337:816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marraffini L.A., Sontheimer E.J.. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008; 322:1843–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marraffini L.A., Sontheimer E.J.. Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature. 2010; 463:568–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hale C.R., Zhao P., Olson S., Duff M.O., Graveley B.R., Wells L., Terns R.M., Terns M.P.. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009; 139:945–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang J., Rouillon C., Kerou M., Reeks J., Brugger K., Graham S., Reimann J., Cannone G., Liu H., Albers S.V. et al. Structure and mechanism of the CMR complex for CRISPR-mediated antiviral immunity. Mol. Cell. 2012; 45:303–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hale C.R., Majumdar S., Elmore J., Pfister N., Compton M., Olson S., Resch A.M., Glover C.V. 3rd, Graveley B.R., Terns R.M. et al. Essential features and rational design of CRISPR RNAs that function with the Cas RAMP module complex to cleave RNAs. Mol. Cell. 2012; 45:292–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deng L., Garrett R.A., Shah S.A., Peng X., She Q.. A novel interference mechanism by a type IIIB CRISPR-Cmr module in Sulfolobus. Mol. Microbiol. 2013; 87:1088–1099. [DOI] [PubMed] [Google Scholar]
- 26.Peng W., Feng M., Feng X., Liang Y.X., She Q.. An archaeal CRISPR type III-B system exhibiting distinctive RNA targeting features and mediating dual RNA and DNA interference. Nucleic Acids Res. 2015; 43:406–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goldberg G.W., Jiang W., Bikard D., Marraffini L.A.. Conditional tolerance of temperate phages via transcription-dependent CRISPR-Cas targeting. Nature. 2014; 514:633–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tamulaitis G., Kazlauskiene M., Manakova E., Venclovas C., Nwokeoji A.O., Dickman M.J., Horvath P., Siksnys V.. Programmable RNA shredding by the type III-A CRISPR-Cas system of Streptococcus thermophilus. Mol. Cell. 2014; 56:506–517. [DOI] [PubMed] [Google Scholar]
- 29.Staals R.H., Zhu Y., Taylor D.W., Kornfeld J.E., Sharma K., Barendregt A., Koehorst J.J., Vlot M., Neupane N., Varossieau K. et al. RNA targeting by the type III-A CRISPR-Cas Csm complex of Thermus thermophilus. Mol. Cell. 2014; 56:518–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Samai P., Pyenson N., Jiang W., Goldberg G.W., Hatoum-Aslan A., Marraffini L.A.. Co-transcriptional DNA and RNA Cleavage during Type III CRISPR-Cas immunity. Cell. 2015; 161:1164–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kazlauskiene M., Tamulaitis G., Kostiuk G., Venclovas C., Siksnys V.. Spatiotemporal control of Type III-A CRISPR-Cas immunity: coupling DNA degradation with the target RNA recognition. Mol. Cell. 2016; 62:295–306. [DOI] [PubMed] [Google Scholar]
- 32.Staals R.H., Agari Y., Maki-Yonekura S., Zhu Y., Taylor D.W., van Duijn E., Barendregt A., Vlot M., Koehorst J.J., Sakamoto K. et al. Structure and activity of the RNA-targeting Type III-B CRISPR-Cas complex of Thermus thermophilus. Mol. Cell. 2013; 52:135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benda C., Ebert J., Scheltema R.A., Schiller H.B., Baumgartner M., Bonneau F., Mann M., Conti E.. Structural model of a CRISPR RNA-silencing complex reveals the RNA-target cleavage activity in Cmr4. Mol. Cell. 2014; 56:43–54. [DOI] [PubMed] [Google Scholar]
- 34.Hale C.R., Cocozaki A., Li H., Terns R.M., Terns M.P.. Target RNA capture and cleavage by the Cmr type III-B CRISPR-Cas effector complex. Genes Dev. 2014; 28:2432–2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramia N.F., Spilman M., Tang L., Shao Y., Elmore J., Hale C., Cocozaki A., Bhattacharya N., Terns R.M., Terns M.P. et al. Essential structural and functional roles of the Cmr4 subunit in RNA cleavage by the Cmr CRISPR-Cas complex. Cell Rep. 2014; 9:1610–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu X., Ye K.. Cmr4 is the slicer in the RNA-targeting Cmr CRISPR complex. Nucleic Acids Res. 2015; 43:1257–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rouillon C., Zhou M., Zhang J., Politis A., Beilsten-Edmands V., Cannone G., Graham S., Robinson C.V., Spagnolo L., White M.F.. Structure of the CRISPR interference complex CSM reveals key similarities with cascade. Mol. Cell. 2013; 52:124–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang J., Graham S., Tello A., Liu H., White M.F.. Multiple nucleic acid cleavage modes in divergent type III CRISPR systems. Nucleic Acids Res. 2016; 44:1789–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zebec Z., Manica A., Zhang J., White M.F., Schleper C.. CRISPR-mediated targeted mRNA degradation in the archaeon Sulfolobus solfataricus. Nucleic Acids Res. 2014; 42:5280–5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zebec Z., Zink I.A., Kerou M., Schleper C.. Efficient CRISPR-mediated post-transcriptional gene silencing in a hyperthermophilic archaeon using multiplexed crRNA expression. G3 (Bethesda). 2016; 6:3161–3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo L., Brugger K., Liu C., Shah S.A., Zheng H., Zhu Y., Wang S., Lillestol R.K., Chen L., Frank J. et al. Genome analyses of Icelandic strains of Sulfolobus islandicus, model organisms for genetic and virus-host interaction studies. J. Bacteriol. 2011; 193:1672–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.She Q., Zhang C., Deng L., Peng N., Chen Z., Liang Y.X.. Genetic analyses in the hyperthermophilic archaeon Sulfolobus islandicus. Biochem. Soc. Trans. 2009; 37:92–96. [DOI] [PubMed] [Google Scholar]
- 43.Peng W., Li H., Hallstrom S., Peng N., Liang Y.X., She Q.. Genetic determinants of PAM-dependent DNA targeting and pre-crRNA processing in Sulfolobus islandicus. RNA Biol. 2013; 10:738–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Y., Pan S., Zhang Y., Ren M., Feng M., Peng N., Chen L., Liang Y.X., She Q.. Harnessing Type I and Type III CRISPR-Cas systems for genome editing. Nucleic Acids Res. 2016; 44:e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peng N., Deng L., Mei Y., Jiang D., Hu Y., Awayez M., Liang Y., She Q.. A synthetic arabinose-inducible promoter confers high levels of recombinant protein expression in hyperthermophilic archaeon Sulfolobus islandicus. Appl. Environ. Microbiol. 2012; 78:5630–5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taylor D.W., Zhu Y., Staals R.H., Kornfeld J.E., Shinkai A., van der Oost J., Nogales E., Doudna J.A.. Structural biology. Structures of the CRISPR-Cmr complex reveal mode of RNA target positioning. Science. 2015; 348:581–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Osawa T., Inanaga H., Sato C., Numata T.. Crystal structure of the CRISPR-Cas RNA silencing Cmr complex bound to a target analog. Mol. Cell. 2015; 58:418–430. [DOI] [PubMed] [Google Scholar]
- 48.Zhu X., Ye K.. Crystal structure of Cmr2 suggests a nucleotide cyclase-related enzyme in type III CRISPR-Cas systems. FEBS Lett. 2012; 586:939–945. [DOI] [PubMed] [Google Scholar]
- 49.Cocozaki A.I., Ramia N.F., Shao Y., Hale C.R., Terns R.M., Terns M.P., Li H.. Structure of the Cmr2 subunit of the CRISPR-Cas RNA silencing complex. Structure. 2012; 20:545–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Osawa T., Inanaga H., Numata T.. Crystal structure of the Cmr2-Cmr3 subcomplex in the CRISPR-Cas RNA silencing effector complex. J. Mol. Biol. 2013; 425:3811–3823. [DOI] [PubMed] [Google Scholar]
- 51.Shao Y., Cocozaki A.I., Ramia N.F., Terns R.M., Terns M.P., Li H.. Structure of the Cmr2-Cmr3 subcomplex of the Cmr RNA silencing complex. Structure. 2013; 21:376–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leon-Sobrino C., Kot W.P., Garrett R.A.. Transcriptome changes in STSV2-infected Sulfolobus islandicusREY15A undergoing continuous CRISPR spacer acquisition. Mol. Microbiol. 2016; 99:719–728. [DOI] [PubMed] [Google Scholar]
- 53.Vestergaard G., Garrett R.A., Shah S.A.. CRISPR adaptive immune systems of Archaea. RNA Biol. 2014; 11:156–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Elmore J.R., Sheppard N.F., Ramia N., Deighan T., Li H., Terns R.M., Terns M.P.. Bipartite recognition of target RNAs activates DNA cleavage by the Type III-B CRISPR-Cas system. Genes Dev. 2016; 30:447–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Estrella M.A., Kuo F.T., Bailey S.. RNA-activated DNA cleavage by the Type III-B CRISPR-Cas effector complex. Genes Dev. 2016; 30:460–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Quax T.E., Voet M., Sismeiro O., Dillies M.A., Jagla B., Coppee J.Y., Sezonov G., Forterre P., van der Oost J., Lavigne R. et al. Massive activation of archaeal defense genes during viral infection. J. Virol. 2013; 87:8419–8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Okutan E., Deng L., Mirlashari S., Uldahl K., Halim M., Liu C., Garrett R.A., She Q.X., Peng X.. Novel insights into gene regulation of the rudivirus SIRV2 infecting Sulfolobus cells. RNA Biol. 2013; 10:875–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Martinez-Alvarez L., Bell S.D., Peng X.. Multiple consecutive initiation of replication producing novel brush-like intermediates at the termini of linear viral dsDNA genomes with hairpin ends. Nucleic Acids Res. 2016; 44:8799–8809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Contursi P., Fusco S., Cannio R., She Q.X.. Molecular biology of fuselloviruses and their satellites. Extremophiles. 2014; 18:473–489. [DOI] [PubMed] [Google Scholar]
- 60.Wang H., Peng N., Shah S.A., Huang L., She Q.. Archaeal extrachromosomal genetic elements. Microbiol. Mol. Biol. Rev. 2015; 79:117–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.