Abstract
ATP-dependent chromatin remodeling complexes are important regulators of gene expression in Eukaryotes. In plants, SWI/SNF-type complexes have been shown critical for transcriptional control of key developmental processes, growth and stress responses. To gain insight into mechanisms underlying these roles, we performed whole genome mapping of the SWI/SNF catalytic subunit BRM in Arabidopsis thaliana, combined with transcript profiling experiments. Our data show that BRM occupies thousands of sites in Arabidopsis genome, most of which located within or close to genes. Among identified direct BRM transcriptional targets almost equal numbers were up- and downregulated upon BRM depletion, suggesting that BRM can act as both activator and repressor of gene expression. Interestingly, in addition to genes showing canonical pattern of BRM enrichment near transcription start site, many other genes showed a transcription termination site-centred BRM occupancy profile. We found that BRM-bound 3΄ gene regions have promoter-like features, including presence of TATA boxes and high H3K4me3 levels, and possess high antisense transcriptional activity which is subjected to both activation and repression by SWI/SNF complex. Our data suggest that binding to gene terminators and controlling transcription of non-coding RNAs is another way through which SWI/SNF complex regulates expression of its targets.
INTRODUCTION
Genes are packed into chromatin, which explains the need for chromatin remodelling to ensure access to regulatory sequences. This activity is mediated by multi-subunit complexes that use the energy derived from ATP hydrolysis to alter the interactions between histone octamers and DNA (1,2). Initially discovered in S. cerevisiae, chromatin remodelling complexes belonging to the SWI/SNF class are probably the most comprehensively studied ones, with a well-established role in gene expression regulation. The SWI/SNF complexes are assembled around a central SNF2-type ATPase containing a C-terminal bromodomain. Purified ATPase proteins have been shown to remodel nucleosomes in vitro, reflecting the central function of this subunit (3). Binding sites of SNF2 ATPases have been repeatedly mapped genome-wide in yeast and animals, revealing a strong preference for promoters in yeast (4) and targeting to both promoters and other regulatory elements like enhancers and super enhancers in animals (5–9). SNF2 ATPases may act as activators or repressors of gene expression (1,10). Recently, SWI/SNF complexes have also been implicated in regulation of non-coding transcription originating from promoters, enhancers, intergenic regions, and transcription termination sites (TTS) of protein-coding genes (11,12,6).
In Arabidopsis, the SNF2 family is represented by four ATPases: BRAHMA (BRM), SPLAYED (SYD), CHR12 and CHR23 (13,14). BRM is the only subunit with the C-terminal bromodomain and is therefore considered to be the closest homolog of the yeast and animal ATPases. Arabidopsis brm null mutants display pleiotropic phenotypes (15), reflecting diverse functions of this protein in plant development and growth. Indeed, BRM was found to be essential for transcriptional control of key genes involved in: cotyledon separation, repression of seed maturation and embryogenesis, flower patterning, cell division during leaf development, root stem cell niche maintenance, inflorescence architecture, flowering (16–23), as well as abiotic stress responses and hormonal responses to abscisic acid, gibberellins, cytokinins and auxins (24–28). Other core subunits of Arabidopsis SWI/SNF complex like SWI3 and SWP73 proteins were also shown to participate in these processes (18,23,29–34), suggesting that BRM performs most of its functions as core ATPase subunit of SWI/SNF complex. BRM functions are partially redundant with those of SYD ATPase, as brm syd double mutant is lethal (35). Although many processes controlled by BRM at different developmental stages have been recently uncovered, the molecular network through which it regulates plant growth, as well as general mode of its action, have not been extensively analyzed and remain largely unknown. A recent work has demonstrated that BRM binding at some of its targets identified using BRM-GFP tagged line, requires an H3K27 demethylase REF6 (36), suggesting that those BRM targets (∼12%) might be specified by a REF6 DNA-binding properties (36).
In this study we have used a complementary approach of genome wide mapping of BRM binding, using highly specific anti-BRM antibodies. By combining this analysis with gene expression profiling of brm-1 knockout plants, we have identified genes that are likely direct targets of BRM transcriptional regulation. We have chosen to use Agronomics tiling arrays (37) that enable both detection of protein-bound genomic regions and sensitive measure of changes in the levels of sense and antisense transcripts using the same platform. Our analyses demonstrate that BRM is a direct regulator of key genes functioning in known and new signaling pathways, the mis-regulation of which is likely responsible for many of the phenotypes observed in brm mutants. Importantly, our data reveal that BRM can act as both positive and negative regulator of its targets. In addition, we find that while BRM predominantly regulates its target genes by binding to their promoters, many genes are bound by BRM at their 3΄ ends. We show that these 3΄ regions serve as promoters of non-coding transcription extensively regulated by BRM. Given that terminator-bound genes are mis-regulated in brm-1 mutant as often as the promoter-bound targets, our data are consistent with a view that Arabidopsis SWI/SNF complex, in addition to the promoter-centred gene expression regulation pathway, controls expression of many of its targets by binding to their 3΄ ends and directly controlling transcription of non-coding RNAs originating therefrom.
MATERIALS AND METHODS
Plant lines
Arabidopsis thaliana wild type and all mutant lines were of the Columbia-0 (Col-0) ecotype. The brm-1, brm-3, brm-5, swi3c-2 and swi3d-1 mutant alleles were characterized previously (15,17,38,34). Arabidopsis transgenic plants were generated using A. tumefaciens-mediated transformation following the vacuum infiltration method (39).
Cloning of genetic constructs
For the construction of the following constructs, pASAt1g18700::LUC, pASAt1g70300::LUC, pASAt2g23760::LUC, pASAt2g29090::LUC, pASAt3g16785::LUC, pASAt1g18690::LUC, pASAt2g29080::LUC, pASAt5g21326::LUC the respective genomic regions, we have selected and amplified: At1g18700 (+440; −1941), At1g70300 (+1747; −1273), At2g23760 (+3019; −413), At2g29090 (+3122; −364), At3g16785 (+2334; −391), At1g18690 (+793; −1245), At2g29080 (+506; −821), At5g21326 (+493; −1236) from Arabidopsis Col-0 genomic DNA and cloned into pGWB635 (40). As ncRNA genes do not have open reading frame we could not use conventional reporter systems to monitor ncRNA activity. To overcome this problem, the IRES sequence (Internal Ribosomal Entry Site) was fused between AS promoter sequence and reporter gene. This allows the translation of non-coding transcripts containing premature terminations in front of the open reading frame of the reporter gene. Promoters were amplified from genomic Col-0 DNA, cloned into modified pENTR-D/TOPO vector and sequenced. Corresponding entry clones, with sense and antisense promoters were used in LR recombination reactions to deliver them into a gateway compatible expression vector of pGWB series (40), pGWB635, containing luciferase marker gene (LUC). BRM cDNA in pENTR-D/Topo was kindly provided by Dr Xiuren Zhang (Texas A&M University) and used in LR reaction for delivering into pGWB602 expression vector. BRM cDNA was also PCR-amplified and cloned into pCambia1300 containing 35S promoter. To create BRM containing a mutation in the ATP binding site reproducing the human BRG1 ATPase-dead point mutation (Supplementary Figure S14D), gene specific BRM cDNA fragment was PCR amplified using primers with introduced mutation corresponding to lysine 1012 to arginine substitution in BRM protein. The fragment (BRMmut) was cloned into pJet 1.2 vector and sequenced to verify the presence of the mutation. Next, BRMmut fragment was directly integrated into 35S::BRM::pGWB602 by restriction cloning via NdeI-AgeI restriction sites to create 35S::BRMmut::pGWB602. All genetic constructs were transformed into Agrobacterium tumefaciens GV3101 strain and subsequently used for transient assays or generation of stable transgenic lines.
Transactivation experiments and transient expression in Arabidopsis seedlings
Data shown in Figure 5E and Supplementary Figure S13, S14 were generated using transient Arabidopsis transformation protocol based on cocultivation of young Arabidopsis seedlings with Agrobacterium tumefaciens. We used a modified Fast Agro-mediated Seedling Transformation method published by (41). This approach was successfully used to express constructs driven by different sense/antisense promoters in Arabidopsis seedling cotyledons. Transformed into Agrobacterium tumefaciens GV3101 strain promoter-luciferase fusion constructs were used as ‘reporter’ and 35S::BRM as ‘effectors’ constructs in our assays. For transient expression, about 35–40 5-days-old Arabidopsis seedlings were cocultivated with selected ‘reporter’ and ‘effectors’ constructs for 2 days in cocultivation media [0.25 MS, pH6; 100 μM acetosyringone; 0.005% Silwet]. After cocultivation, seedlings were surface sterilized with 0.1% bleach for 10 min, then washed twice in sterile ddH20 and transferred to MS solid plates for imaging. Transformed Arabidopsis seedlings were kept in Percival growth chamber for 2–3 h and then subjected to LUC imagining using NightSHADE LB 985 in vivo Plant Imaging System (Berthold). Taking into account variability in transfection, we counted only experiments with transfection rate 75% and higher. Similar results were obtained using both constructs expressing BRM.
Figure 5.
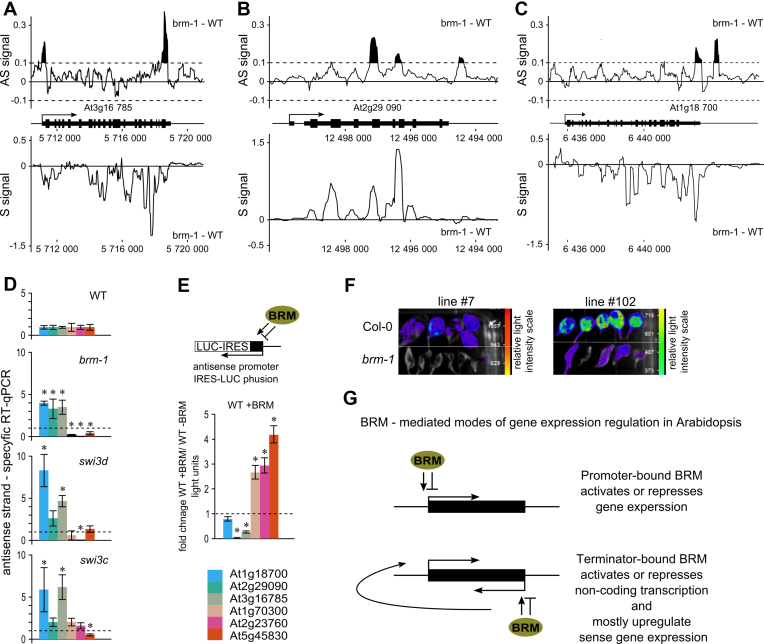
SWI/SNF complex regulates antisense transcription. (A, B and C) Difference (brm-1 – WT) in intensity for antisense (AS signal) and sense (S signal) expression values from whole genome tilling arrays was plotted along the selected target genes. Negative values indicate down and positive up regulation. For the antisense signal a cut-off value of 0.1 (black filled picks) was used to indicate changes in antisense expression above background. (D) Mutants in SWI/SNF subunits show misregulation of antisense transcription. Strand specific RT-qPCR quantification of antisense transcripts at selected 3΄BRM-bound genes. (E) BRM controls antisense transcription from TTS regions of 3΄BRM-occupied genes. TTS regions of selected genes were cloned into luciferase marker gene (LUC) reporter construct and used for transient co-transformation of Arabidopsis Col-0 (WT) seedlings with or without BRM expression construct. Data represent fold change in LUC activity upon BRM overexpression relative to seedlings transformed with empty expression vector; data are mean ±SE for at least 20 individually transformed plants. (F) pAS-At5g45830::LUC transgene was transformed into BRM/brm-1 plant line. brm-1 heterozygotes segregating 3:1 for the transgene were selected and their offspring were genotyped to select WT and brm-1 homozygous plants. Data for two independent transformants (line #7 and #102) are shown. (G) Model of 5΄BRM driven activation or repression of gene expression (Top), and 3΄BRM driven regulation of antisense transcription leading to concomitant sense gene regulation (Bottom). * significant change (t-test, P < 0.05).
BRM Western blot and immunoprecipitation
Nuclear extracts were obtained using nuclei isolated from 3 g of Arabidopsis seedlings according to the protocol of (42) except that the nuclei were re-suspended in IP1 buffer (20 mM Hepes-KOH pH 8.0, 0.15 M KCl, 5 mM MgCl2, 0.2% Triton X-100, 10% glycerol, 1 mM dithiothreitol, 0.1 mM ethylenediaminetetraacetic acid, 0.1 mM PMSF, protease inhibitors (Complete ethylenediaminetetraacetic acid-free, Roche), digested with DNase and RNase for 1 h at 4°C and lysed by sonication in a sonicator bath (Diagenode). Nuclear extracts were centrifuged for 5 min at 12 000 x g at 4°C and the supernatant was used for BRM immunodetection or immunoprecipitation experiment.
For BRM immunoprecipitation, 4 μg of anti-BRM antibody or pre-immune serum was bound to Dynabeads Protein A resin as recommended by the manufacturer (Invitrogen) and incubated with ∼100 μg of nuclear extract or total protein extract at 4°C. After 6 h incubation with gentle mixing, the beads were washed 4 times with IP1 buffer and bound protein was eluted with elution buffer (1% sodium dodecyl sulphate, 0.1 M NaHCO3) at room temperature. A portion of the recovered proteins was resolved by polyacrylamide gel electrophoresis and the presence of BRM was determined by Western blotting using anti-BRM antibodies or visualized on silver-stained gel. Bands corresponding to full length or truncated BRM protein present in brm-3 mutant were excised from the gel and proteins were identified using mass spectrometry.
For BRM detection after luciferase activity analysis, roughly 20 transformed seedlings were placed in a 1.5 ml eppendorf tube and homogenized by grinding with a plastic pestle in the presence of 50 μl of 2xNuPAGE sample buffer containing 2-Mercaptoethanol. Mixture was boiled for 10 min at 95°C. The supernatant after a centrifugation at 4000 x g for 5 min was loaded for sodium dodecyl sulphate-polyacrylamide gel electrophoresis analysis. Western analyses were performed as described (29).
ChIP-qPCR and ChIP-chip experiments
ChIP experiments were performed as described previously (26) with some modifications. Chromatin was isolated from 3-week-old seedlings of wild-type (Col-0) and brm-1 mutant grown under long-days conditions. For BRM ChIP, anti-BRM antibody was used (29). For histone modification ChIP experiments (Supplementary Figure S9), anti-H3 (ab1791, Abcam), anti-H3K4me3 (C15410003, Diagenode), anti-H3K27me3 (C15410195, Diagenode) or anti-H3K9me2 (ab1220, Abcam) were used. Antibodies were bound to Dynabeads Protein A (Invitrogen) and incubated overnight with isolated chromatin. The extracted DNA was resuspended in 100 μl of water. ChIP enrichment was determined by qPCR using LightCycler 480 SYBR Green I Master mix (Roche). Reactions were performed with 2 μl of immunoprecipitated DNA as template. The amount of ChIP DNA was calculated based on the standard curve and relative to input samples for each pair of primers. The TA3 retrotransposon and PP2A coding sequence served as controls. Primers used are listed in Supplementary Table S7.
For ChIP-chip experiment, DNA from BRM ChIP and input samples (two biological replicates) were subjected to two rounds of amplification using a WGA2 Kit (Sigma-Aldrich), according to the manufacturer's protocol. Four micrograms of DNA was used for fragmentation and labeling according to Affymetrix chromatin immunoprecipitation assay protocol (P/N 702238 Rev.4) using a GeneChip WT Terminal Labeling Kit (Affymetrix). Labeled DNA was hybridized to the Agronomics microarray (37) using a GeneChip Hybridization Wash and Stain Kit according to the manufacturer's recommendations (Affymetrix).
Microarray gene expression profiling
Material was collected from 3 weeks-old WT and brm-1 seedlings grown in soil under long-day conditions. Total RNA was extracted using RNeasy plant mini kit (Qiagen) according to the manufacturer's protocol, followed by treatment with TURBO DNase (Ambion) and RiboMinus™ Plant Kit (Invitrogen) to reduce the rRNA fraction. The quantity and quality of the isolated RNA was determined using a NanoDrop ND1000 spectrophotometer (Nanodrop technologies) and RNA integrity was assessed with a Bioanalyzer 2100 (Agilent Technologies). RNA (100 ng) was used for cDNA synthesis with an Ambion WT Expression Kit. After fragmentation and labeling with a GeneChip WT Terminal Labeling Kit (Affymetrix), 5.5 μg of cDNA was hybridized with an Agronomics array (37) using a GeneChip Hybridization Wash and Stain Kit, according to the manufacturer's recommendations (Affymetrix). Three biological replicates were examined for each genotype.
Data analysis
ChIP-chip microarray probe signals were extracted using Affymetrix apt-cel software. As AGRONOMICS1 tiling arrays contain probes from both genome strands, signals for both strands were merged for each replicate and then transformed to TAIR10 coordinates. The signal was then normalized separately for each of the two biological replicates. To determine the correlation between biological repeats, Pearson correlation coefficient was computed using python scipy.stats module on normalized signals from ChIP and Input samples. The Pearson r coefficient was 0.967 between ChIP replicates and r = 0.943 for Input (Supplementary Figure S1), indicating that the ChIP-chip experiments were reproducible. The average of the replicates for each probe was used to compute log2 (IP/Input) for every probe and the significant regions were then called by finding maximal regions of length > 20 probes with enrichment > 2 (log2 enrichment > 1), for all except at most 3 faulty probes. The resulting 8048 BRM binding sites that corresponded to 4832 nearest neighbor genes were used for calculating BRM profiles (Figure 1) and BRM direct targets selection (Figure 2).
Figure 1.
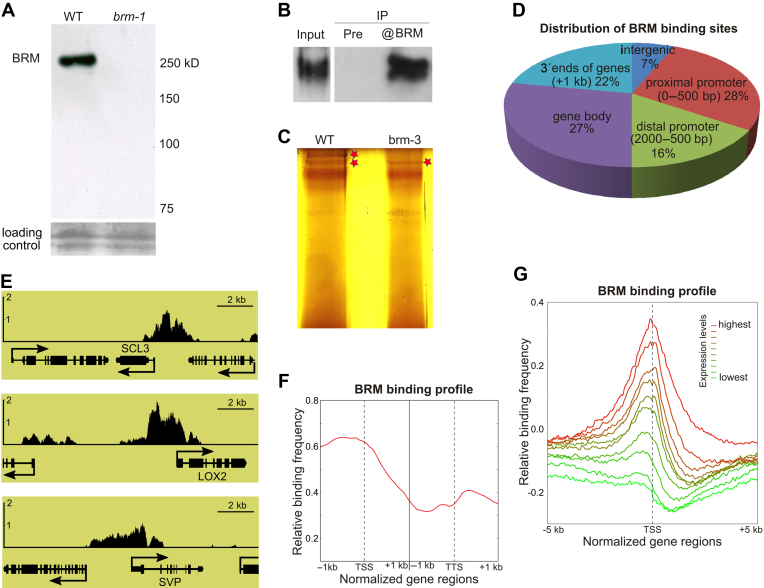
Genome-wide identification of BRAHMA (BRM) occupancy using ChIP-chip. (A–C) Specificity of anti-BRM antibody used for ChIP-chip. (A) Western blot of nuclear extracts from WT and brm-1 null mutant. (B) Western blot showing precipitation of BRM from WT nuclear extracts; pre-immunization serum (Pre) was used as a negative control. (C) Silver-stained gel showing immunoprecipitated BRM (asterisks) from WT and brm-3 whole-cell extracts. (D) Distribution of BRM-bound regions throughout the Arabidopsis genome. (E) BRM occupancy at selected regions around known BRM target genes SCL3, LOX2 and SVP. Y axis represents BRM enrichment. (F) Frequency of BRM-binding sites across a virtually normalized gene unit. TSS, transcription start site; TTS, transcription termination site. (G) Analysis of average BRM binding site frequency surrounding the TSS. Genes were classified into 10 groups based on expression levels.
Figure 2.
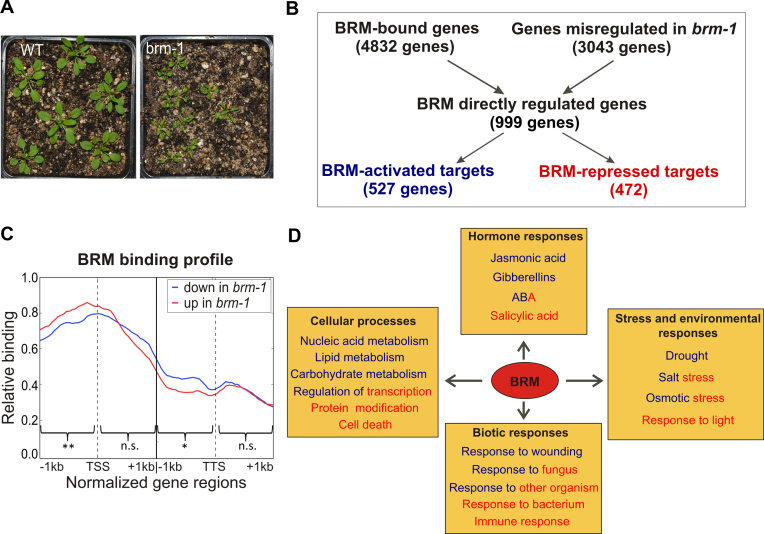
Analysis of direct transcriptional BRM targets. (A) Three-week old WT and brm-1 mutant plants used for ChIP-chip and transcriptome analyses. (B) BRM ChIP-chip and transcript profiling data of WT and brm-1 mutant were used to identify directly regulated genes. (C) Frequency of BRM-binding sites across a virtually normalized gene unit. Genes were classified into two groups based on expression changes in brm-1 mutant. TSS, transcription start site; TTS, transcription termination site. Asterisks indicate significant differences between the groups (Wilcoxon test, * P < 0.05; ** P < 0.01; n.s. not significant). (D) Functional classification of directly regulated genes based on gene ontology (GO) using AgriGo tool. GO categories enriched within BRM-activated and BRM-repressed targets are shown in blue and red color, respectively.
To identify different BRM-bound gene clusters (Supplementary Figure S4), BRM ChIP signal around features of interest (TSS or TTS) was clustered using the k-means algorithm Biopython (43), with k = 2, 3 or 4 and Pearson correlation-based distance function. For analyses of 5΄ and 3΄ BRM bound genes, genes were selected based on the average signal 2000 bp upstream to TSS (−) and 1000 bp downstream of TTS region. Gene clusters were defined by at least log2 enrichment of entire region >0.3 and >1.5 level of BRM enrichment for 5΄ over 3΄ and 3΄ over 5΄ for 5΄BRM and 3΄BRM-occupied genes, respectively. Heatmap images (Figure 3B and Supplementary Figure S6) were performed in python matplotlib with binning of the signal to bins of size 50 bp. The same data were used for boxplots (Supplementary Figure S12A) but only using the gene body part of the signal profile. Histone modification and DNA methylation profiles were computed as average signals for 50-bp windows, beginning at the 5΄ and 3΄ ends of genes.
Figure 3.
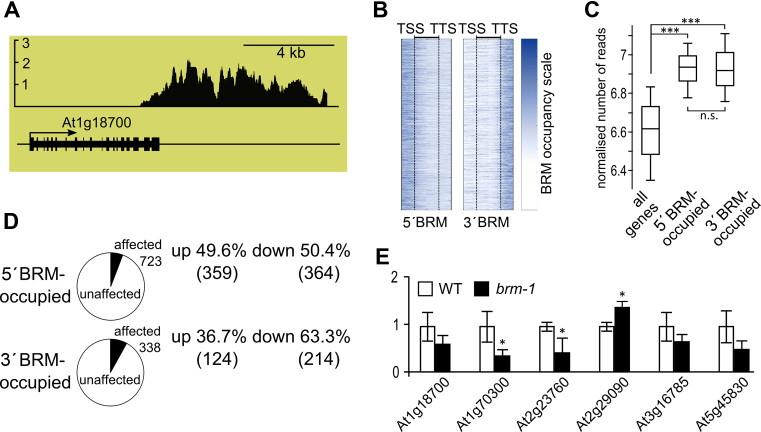
Characterization of 3΄ BRM bound genes. (A) Example of 3΄ BRM-occupied gene (At1g18700) from whole-genome ChIP-chip using anti-BRM antibodies, y-axis represents BRM enrichment. (B) BRM occupancy along genes; genes with BRM bound at 5΄ end or near 3΄ end are shown. TSS and TTS represent aligned transcription start and transcription termination sites, respectively, of genes in each class. (C) Expression levels of all, 5΄ and 3΄ BRM-occupied genes using RNAseq data from (47). Asterisks indicate significant differences between the 5΄BRM and 3΄BRM groups (Wilcoxon signed-rank p-test; *** P < 0.001; n.s. not significant). (D) Comparison of expression of 5΄ and 3΄ BRM-occupied genes in brm-1 null mutant and WT plants. (E) RT-qPCR analysis of mRNA expression for selected 3΄ BRM-bound genes in brm-1 mutant and WT. * significant change between brm-1 and WT (t-test, P < 0.05).
For transcript profiling, probe intensities for strand-specific signals were extracted using Affymetrix apt-cel software. The AGRONOMICS1 tiling array contains probes from both genome strands, and can be used to simultaneously estimate levels of sense and antisense transcripts. Probe positions were transformed to the TAIR10 genome assembly. The signal was normalized separately for each of the three biological replicates and the average of the replicates for each probe was used. The average of 20 probes with highest signal was counted for each gene in WT and brm-1 mutant and used to detect differentially expressed genes.
ChIP-chip and microarray expression data have been submitted to GEO database under accession number GSE71657.
GEO term analyses
Gene lists generated were analyzed using AgriGO (44) and GORILLA (45). Resulting gene ontology (GO) term lists are shown in Figure 2D, Supplementary Figure S7 and Supplementary Tables S4 and S5.
DNA motif analysis
Differential motif analysis was performed for 500 bp located behind TTS of 3΄BRM-occupied and 5΄BRM-occupied genes as positive and negative sets respectively, using Amadeus tool (46) with default parameters. Sense promoters showed similar number of TATA elements per sequence with 10,7 for BRM5΄ and 10,5 for BRM3΄-occupied genes. BRM-enriched terminator regions showed on average 8,8 TATA box elements compared to 7,9 for genes with 5΄ centered BRM occupancy (P-value< 0.0001, CHI square test).
Antisense quantification
We generated a deep RNA database by merging previously generated RNA seq (47) data for WT Arabidopsis accessions (SRR748577, SRR748578, SRR748581, SRR748582, SRR748583, SRR748584, SRR748585, SRR748616, SRR748632). Log2 of gene length-normalized number of reads was used for quantification after filtering outliers for 40–60% of distribution. Only genes located at least 750 bp from nearest annotated gene were used for Supplementary Figure S11.
RNA extraction and RT-qPCR analyses
Total RNA was extracted from seedlings using phenol–chloroform procedure (48). RNA samples were treated with TURBO DNase (Ambion), according to standard manufacturer protocol and efficiency of DNA removal was analyzed using PCR with PP2A primers. Quality and amount of RNA samples were tested on 1.2% agarose gel and Nanodrop 2000. A total of 3.5 µg RNA was used for cDNA synthesis from seedlings (sense and antisense); cDNA for sense transcript was synthesized using oligodT primers. cDNAs for antisense transcripts were synthetized using gene specific primers. Expression levels were assayed by qPCR in LightCycler ® 480 Instrument II device, using SYBR Green mix (Roche). Data were normalized using PP2A and UBC (49). Primers used are listed in Supplementary Table S7.
RESULTS
Genome-wide identification of BRM binding sites
We performed genome-wide localization study of BRM in 3-weeks-old Arabidopsis plants by ChIP-chip using Agronomics whole genome tiling arrays (37) and anti-BRM antibody (29) that allowed us to analyze endogenous protein. This antibody specifically detected ∼250 kD band in WT plants that could no longer be detected in brm-1 knockout line (15) (Figure 1A). Characterization of the antibody showed that it can also detect native protein in cell extract as we could efficiently immunoprecipitate BRM protein (Figure 1B and C). In agreement with recent finding that BRM can be phosphorylated by SnRK2.2/2.3 kinases at C-terminal region (50), two bands corresponding to wild-type BRM protein were often detected after BRM immunoprecipitation (Figure 1B and C), while in brm-3 mutant (38) only one band was detected corresponding to a truncated BRM protein lacking C-terminal fragment of 454 amino acids (Figure 1C).
ChIP-chip showed that BRM occupies thousands of sites in Arabidopsis genome, as we determined 8048 BRM binding sites that were at least 2-fold enriched compared to input. Most of them (93%) were localized close to or inside annotated genes. In summary 4832 genes were found to be bound by BRM (Supplementary Table S1). ChIP-chip replicates were highly reproducible as IP signal from individual probes and individual inputs showed Pearson correlation coefficient higher than 0.96 and 0.94, respectively (Supplementary Figure S1). Majority of identified BRM binding sites were located in the promoters (Figure 1D), of which 28% localized in proximal (defined as 0–500 bp upstream of transcription start site (TSS)) and 16% in distal (500–2000 bp upstream of TSS) promoter regions. The rest of BRM binding sites were divided nearly equally between gene bodies and 3΄ ends of genes (0–1000 bp downstream of TTS) with each of those accounting for one quarter of genome-wide BRM binding sites (Figure 1D).
Inspection of BRM binding pattern at known BRM targets confirmed published results. In particular, we clearly observed BRM binding at SCL3, LOX2, SVP (26,27,23) and number of other known direct targets (Figure 1E and Supplementary Figure S2A). These results were confirmed for selected genes by ChIP-qPCR using WT and brm-1 plants (BRM knock-out mutant), as negative control (Supplementary Figure S2B). Importantly, BRM binding pattern on these genes revealed by ChIP-chip closely matched the published BRM patterns including high BRM occupancy at promoter and inside first exon, and low occupancy inside exon 2 of SCL3 and SVP genes (Figure 1E) (26,23).
In a recent work Li et al. (36) mapped BRM binding sites in Arabidopsis genome using a BRM-GFP line. We have therefore compared the BRM bound genes defined by us with the targets defined by Li et al. This analysis revealed 3026 genes bound by BRM in both ChIP experiments, representing respectively 62.5% and 57.5% of all gene targets reported in our study and in Li et al. (Supplementary Figure S3). The overlap is statistically significant (P-value < 1 × 10−5), despite different developmental stage analyzed, and differences in ChIP approach (GFP tagged versus anti-BRM antibody) and analysis platform (next generation sequencing versus microarrays). This confirms the robustness of our analysis and suggests that BRM can stably bind to the same group of genes in different plant tissues and growth conditions. Analysis of average BRM binding distribution across gene units revealed that majority of BRM binding is centered near the TSS (Figure 1F). Clustering of Arabidopsis genes based on their expression levels showed that the highly expressed genes are more likely to be bound by BRM than the low expressed genes (Figure 1G). We also noted a smaller second peak of BRM binding signal at TTS (Figure 1F). Indeed, unsupervised clustering of BRM localization produced three different classes of BRM-bound genes, with BRM localized at the 5΄ end, 3΄ end or both ends (Supplementary Figure S4). Clustering profiles for 4 clusters did not reveal additional distinct BRM binding patterns (Supplementary Figure S4).
Analysis of direct transcriptional BRM targets
To further characterize the role of BRM in regulation of gene expression, we performed transcript profiling of 3 weeks-old WT and brm-1 mutant plants (Figure 2A) using strand-specific whole genome Agronomics tilling arrays (the same as used for ChIP-chip). This type of arrays allows measuring expression of more than 90% of the annotated Arabidopsis genes (37). We have used this data to find direct transcriptional BRM targets, defined as genes occupied by BRM and displaying altered expression in brm-1 mutant (Figure 2B). Approximately 20% of genes bound by BRM showed significantly changed expression in the mutant relative to WT with applied criteria (Fc > 1.25, FDR < 0.01) (Supplementary Table S2). This relatively small number of affected genes is in line with published reports showing that the vast majority of genes occupied by animal SWI/SNF do not show altered expression in SWI/SNF knock-outs (51). Roughly half of so defined direct transcriptional BRM targets (472 out of 999 genes) were up- and another half (527 genes) downregulated in brm-1 mutant (Supplementary Table S3), suggesting that in plants BRM works as both activator and repressor of gene expression. This is in agreement with the data published so far. For example, BRM direct targets SCL3 and SVP were down-regulated, while ABI5 was up-regulated in brm mutants (23,24,26). The ability to both activate and repress gene expression has also been demonstrated for BRM homologs in animals (10). Interestingly, although meta-gene profiles for target genes up- and down-regulated in brm-1 are overall similar, genes up-regulated in brm-1 are more likely to be bound by BRM around TSS when compared to down-regulated targets. In contrast, target genes down-regulated in brm-1 show a relative increase in BRM binding at TTS compared to up-regulated genes. These differences, although small, were statistically significant for regions 1kb upstream of TSS and 1 kb upstream of TTS (Figure 2C).
Analysis of BRM-activated and BRM-repressed direct targets showed an enrichment in GO terms related to hormone responses – 42 and 30 genes (P-value 2.4 × 10−10 and 8.6 × 10−6), stress and environmental responses – 42 and 42 genes (P-value 9.8 × 10−6 and 7.4 × 10−7), biotic stresses – 33 and 42 genes (P-value 2.5 × 10−10 and 9.5 × 10−18) and diverse cellular processes – 78 and 73 genes (P-value 4.6 × 10−6 and 2.3 × 10−6) for down- and up-regulated genes, respectively (Figure 2D, Supplementary Figure S5 and Supplementary Tables S4 and S5). Interestingly, some of the GO categories were enriched exclusively in BRM-activated or BRM-repressed groups of targets. The BRM-activated genes were primarily enriched in categories of jasmonic acid and gibberellic acid responses, as well as response to drought, while BRM-repressed genes were primarily enriched in categories of salicylic acid and light responses (Figure 2D).
In summary, our whole genome profiling of BRM showed that BRM binds almost 5000 of gene regions and is as likely to repress, as it is to up-regulate its direct targets. The majority of BRM binding sites are located at promoter regions. However, almost 1/4 of BRM binding sites are located at 3΄ ends of genes (TTS regions). Interestingly, genes down regulated in brm-1 mutant showed a more pronounced BRM binding at TTS compared to up regulated genes.
Genes occupied by BRM at 3΄ ends
It is known that genes are often structured as gene loops (52–54). In agreement, multiple factors bound to promoter regions are also found at transcription terminators (55–57). Our analysis revealed that BRM is often localized simultaneously at 5΄ and 3΄ ends of genes (Supplementary Figure S4). Despite this, we found a relatively large number of genes with predominant BRM binding at either 5΄ or 3΄ ends (Supplementary Figure S4). To investigate the role of BRM binding at 3΄ versus 5΄ ends of genes, we selected a set of genes with BRM present preferentially at the 3΄ or the 5΄ gene regions (Figure 3A). To this end, BRM ChIP-chip data were re-analyzed, so that genes were divided into three parts (2 kb upstream of TSS, gene body, and 1 kb downstream of TTS), and the average BRM signal for each entire region was calculated (Supplementary Figure S6). To focus exclusively on 5΄ and 3΄-bound targets, genes bound by BRM at both 5΄ and 3΄ ends were excluded. Such re-analysis gave 3835 genes with the BRM signal enriched around their 5΄ end and 1759 genes with BRM detected predominantly at the 3΄ end (Figure 3B and Supplementary Table S6).
Comparison of GO terms enrichment among 5΄ and 3΄BRM-bound targets showed similar GO categories in both groups, mostly associated with responses to different types of signals (Supplementary Figure S7), in agreement with our previous analysis (Figure 2C). Also comparison of mRNA levels showed that the 3΄ and 5΄BRM-bound genes represent similarly expressed, mostly active genes (Figure 3C). We concluded that BRM target genes bound at 5΄ and 3΄ end are similar in regard to their expression levels and functions.
BRM controls mRNA expression of 3΄-bound genes
To test if 3΄BRM-bound genes are regulated by BRM in a similar or different way than 5΄BRM targets, we compared expression changes within 3΄ and 5΄BRM-bound genes using our brm-1 mutant microarray data. Out of the 3835 5΄BRM-occupied genes 723 (18.9%) showed significantly changed expression relative to WT plants (Figure 3D). Notably, 338 (19.2%) of the 1759 3΄BRM-occupied genes were affected as well (Figure 3D). This result has been validated by RT-qPCR for a set of randomly selected 3΄BRM targets. RT-qPCR showed that 3 out of 6 tested 3΄BRM-bound genes were significantly mis-regulated in brm-1 mutant (Figure 3E). Together, these data show that 3΄BRM-occupied genes are as likely to be misregulated in brm mutant as are the 5΄BRM bound genes, suggesting that they represent legitimate, direct targets of the BRM-containing SWI/SNF complex.
BRM binding profiles indicated that BRM occupancy for target genes down-regulated in brm-1 was relatively higher at TTS region compared to genes up-regulated in brm-1 (Figure 2C). In agreement, whereas close to half of 5΄BRM occupied genes were down- and another half up-regulated in brm-1 mutant, as much as 63% of 3΄BRM-bound genes that were mis-regulated showed down regulation in brm-1 mutant (Figure 3D). This suggests that BRM is more likely to up-regulate gene expression when it is bound at 3΄ end of a gene.
BRM-bound terminators resemble promoters and show high antisense transcription
In order to characterize the mechanism by which BRM controls expression of terminator-bound genes, we performed de novo motif discovery using the 3΄ regions of those genes, and as a negative set, the corresponding regions of genes occupied by BRM at the 5΄ end (Figure 4A). Several overrepresented motifs were found (Supplementary Figure S8), including a motif highly reminiscent of a TATA box (Figure 4B). Re-analysis of the putative TATA box elements in promoter region of 5΄ and terminator region of 3΄BRM-occupied genes showed no statistical differences, suggesting that the TATA boxes enriched around TTS of 3΄BRM-occupied genes may represent genuine TSS.
Figure 4.
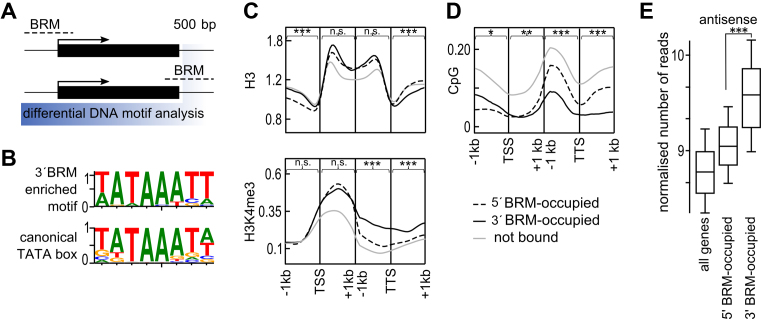
TTS regions occupied by BRM act as promoters of antisense transcription. (A) Schematic of de novo motif discovery procedure used to define motifs enriched in 500 bp region behind TTS (highlighted in blue) of 3΄BRM-occupied genes. Black rectangles represent genes and dashed lines indicate BRM binding sites. (B) Weblogo of discovered motif, with canonical TATA box motif shown below, y-axis – information context. (C) Genes were grouped based on BRM occupancy into 5΄, 3΄ and BRM – not bound, and for each class occupancy profiles along a 1 kb region around TSS and TTS were plotted. Top to bottom occupancy profile of H3 and H3K4me3, using published ChIP-seq data (58–59). (D) CG DNA methylation profiles for 5΄BRM, 3΄BRM and BRM – not bound genes for a 1 kb region around TSS and TTS were plotted based on (61). (E) 3΄BRM-occupied genes show high antisense levels. RNAseq data (47) were combined, normalized for gene length and average for each gene category was plotted. Asterisks in C, D and E indicate significant differences between the 5΄BRM and 3΄BRM groups (Wilcoxon test, *P < 0.05; **P <0.01; ***P < 0.001; n.s. not significant).
To learn more about 3΄BRM-bound genes, we used previously generated whole genome data for several histone marks (58–60) and DNA methylation (61), and profiled their occupancy across 5΄ and 3΄BRM-bound genes as well as genes not bound by BRM (Figure 4C and D). We noticed that TTS regions of 3΄BRM-bound genes show many features of promoters, as exemplified by TSS of 5΄BRM-occupied genes. In particular, the H3 level at TTS of 3΄BRM-bound genes showed a localized and statistically significant decrease compared with that of TTS of either 5΄BRM-bound or BRM not bound genes (Figure 4C). Transcription initiation sites of actively transcribed genes are often marked by specific chromatin modifications, of which histone H3 lysine 4 tri-methylation (H3K4me3) is the most studied one. The 5΄BRM-occupied genes showed a canonical H3K4me3 profile with a strong enrichment around TSS. In contrast, 3΄BRM-occupied genes showed a much broader pattern of H3K4me3 distribution, suggesting the presence of a second peak around the TTS (Figure 4C). Statistical analysis showed that this difference is significant (Figure 4C). In addition we performed independent ChIP qPCR experiment and tested the levels of H3K4me3 at selected 5΄BRM and 3΄BRM-bound targets. This analysis showed that selected 3΄BRM but not the 5΄BRM bound genes show high H3K4me3 signal at their TTS (Supplementary Figure S9), confirming our whole genome analysis. Consistent with the observation that both 5΄ and 3΄BRM targets are active genes with relatively high expression levels (Figure 3C), they showed low levels of H3K27me3 and H3K9me2 repressive marks (Supplementary Figures S9 and S10).
We have also analyzed DNA methylation profile of 5΄ and 3΄BRM bound genes. The most pronounced changes could be observed in case of CpG methylation. Both 5΄ and 3΄BRM occupied genes showed lower methylation levels than all other genes (Figure 4D and Supplementary Figure S10). Interestingly, 5΄ and 3΄BRM-bound genes showed the most pronounced reduction of methylation in all contexts along the promoter and terminator regions, respectively. In addition, 3΄ BRM-bound genes are characterized by low level of CG methylation in gene bodies (Figure 4D). The significance of this observation is unclear at present, as the role of gene body methylation is poorly understood (62).
To conclude, the TTS regions of 3΄BRM-occupied genes show promoter-like features, including low H3 and high H3K4me3 levels that are absent from the TTS of 5΄BRM-bound genes. Together with low levels of repressive histone marks and DNA methylation, this suggests that BRM-occupied gene terminators may represent active promoters of antisense transcription. Consistent with this hypothesis, quantification of sense and antisense reads across 5΄BRM-occupied and 3΄BRM-occupied genes revealed a significantly higher level of antisense reads for the latter (Wilcoxon signed-rank p-test < 2.2e−16) and no difference in the sense transcripts levels (Wilcoxon signed-rank p-test 0.4845) (compare Figures 4E and 3C), when analyzed using RNA sequencing data (47). Since that observation could in principle be explained by the proximity of the 3΄BRM-occupied genes to other genes, we repeated the analysis on a subset of genes selected to be positioned clearly away from nearby genes, reaching the same conclusions (Supplementary Figure S11).
SWI/SNF complex regulates antisense transcription from gene terminators
The design of our whole genome study allowed us to interrogate not only sense transcription but also antisense signals along the genes affected in brm-1 mutant. We did not detect any global changes in antisense signal in 3΄ or 5΄BRM bound genes when we compared WT and brm-1 mutant (Supplementary Figure S12A). However, inspection of 3΄BRM-occupied targets showed that at interrogated genes brm-1 strongly affects antisense transcripts at 3΄ end of genes (Figure 5A, B and C). The examination of the sense strand indicated a concomitant change of sense signal that could either occur in the same or opposite direction compared to antisense transcript change (compare Figure 5A and B). The concomitant change of antisense and sense transcription at 3΄BRM bound genes indicates a possible role of BRM-dependent antisense transcripts in regulation of sense expression.
To confirm those findings, we analyzed levels of six antisense transcripts originating from terminators using strand specific RT-qPCR in WT and brm-1 knock-out plants. In agreement with the lack of global change in antisense signal from microarrays, brm-1 mutant showed both up- and down-regulation of antisense transcripts. In three cases antisense transcripts were significantly upregulated in the mutant and in three other cases the antisense levels were reduced (Figure 5D). Similar results were obtained for an extended set of genes (Supplementary Figure S12B). These results confirm that 3΄BRM-bound genes indeed tend to produce antisense transcripts and suggest that BRM is able to regulate TTS-derived antisense transcription. Changes observed at individual genes by RT-qPCR (Figure 5D) were in good agreement with our tilling array data (Figure 5A, B and C). Next we analyzed expression of those antisense transcripts in swi3c and swi3d mutants, defective in homologs of the conserved SWI3 subunit of SWI/SNF complex (Figure 5D) (34). We detected mis-regulation of antisense transcripts similar to that observed in brm-1, suggesting that the BRM-dependent control of the antisense transcription originating at TTS requires the activity of the whole SWI/SNF complex.
Antisense transcription is often linked to sense transcription in a complicated loop where they affect each other expression (63). We therefore detached the TTS-localized antisense promoters from the effect of sense transcription by cloning selected transcription terminator regions in front of an IRES-LUC reporter (Internal Ribosome Entry Site). Putative antisense promoter regions were selected to include 2 kb downstream from TTS or to beginning of next gene and 3 kb inside gene body unless there was a clear polyA signal detected (64). For this analysis we selected six target genes that showed 3΄BRM binding pattern (Figure 3B) and presence of antisense signal (Figure 4E). The TTS of three out of six 3΄BRM-occupied genes tested showed clear antisense transcriptional activation upon transient BRM overexpression (Figure 5E and Supplementary Figure S13A). Transcriptional activity of two other TTS regions was suppressed by BRM. Data from mutant analysis and transactivation studies are mostly consistent, as the antisense transcripts up-regulated in the absence of BRM (brm-1 mutant) were down-regulated by addition of BRM (2 out of 3 tested), whereas the opposite could be seen for antisense transcripts down-regulated in brm-1 mutant (3 out of 3) (compare Figure 5D and E). Notably, putative antisense transcripts driven by two randomly selected terminators did not show BRM-dependent response (Supplementary Figure S13B), indicating specificity of our assay. Stable lines generated for the 3΄ region of one of the targets showed strongly reduced LUC reporter activity in brm-1 and brm-3 backgrounds (Figure 5F and Supplementary Figure S14A), in good agreement with the observed transactivation of this construct by BRM in the transient assay (Figure 5E).
Finally, we sought to test if the transactivation by BRM is dependent on its ATPase activity, as it was shown before to be required for chromatin remodeling in human (65–67). Transient transformation with constructs encoding either BRM-targeted sense or antisense promoter fused with LUC, showed reduced activity in hypomorphic brm-3 (38) as well as in brm-5 mutant (17) containing a point mutation in the ATPase domain (Supplementary Figure S14B). This was further corroborated by independently generated point mutant in the BRM catalytic center that was unable to activate both sense and antisense expression in the transient transactivation system (Supplementary Figure S14C and D). These results suggest that both promoter-bound and terminator-bound BRM targets require BRM ATPase activity for sense and antisense transcription regulation.
We concluded that BRM acts as a regulator of antisense transcription at genes where it shows a 3΄-centered occupancy profile. Our data show that in Arabidopsis SWI/SNF not only suppresses but can also act as a positive regulator of antisense transcription similarly to what has been established for SWI/SNF activity toward sense promoters of protein coding genes (Figure 5G).
DISCUSSION
Functions of SWI/SNF complexes have been extensively characterized in yeast and animals, including their global binding patterns (10,68,69). Recently, understanding of SWI/SNF roles in plant growth and development has also greatly improved, including a proposed role of histone demethylase REF6 in target specification (13,36). In this work we undertook global approach to gain more extensive view into the biological roles and general mode of action of SWI/SNF catalytic subunit BRM.
We have mapped over 8000 binding sites of endogenous BRM protein using specific anti-BRM antibody. Analysis of BRM binding pattern indicates that BRM is almost exclusively associated with genes, as nearly 93% of detected binding sites were found to locate within or close to gene units (from 2 kb upstream of TSS to 1 kb downstream of TTS). This binding pattern has a number of interesting similarities with published whole genome patterns of animal BRM homologs. Firstly, both human ATPase BRG1 and Arabidopsis BRM display preferential binding at highly expressed, active genes (7). Moreover, unlike yeast SNF2 ATPases that bind almost exclusively to promoters near the TSS site (4), Arabidopsis BRM and mammalian BRG1 bind not only proximal regions of promoters, but also distal promoter regions, gene bodies and gene terminators (5,7,51). In addition, like mammalian homologs, BRM is able to both activate and repress its target genes. On the other hand, only 7% of BRM binding sites could be classified as intergenic regions (Figure 1D), while animal SWI/SNF-like complexes have been shown to frequently bind enhancers and other regulatory elements (10). This could reflect the relatively small size of Arabidopsis genome and its dense gene arrangement. Our data are in good agreement with recently generated data using GFP tagged BRM (36) that showed BRM binding at 5278 genes, very similar number to 4832 genes determined by us using a highly specific BRM antibody.
Previous genetic studies describing brm mutant phenotypes and expression profiles using ATH1 microarrays suggested that BRM directly or indirectly regulates hundreds of genes. By comparing BRM-bound genes with genes displaying altered expression in brm-1 null mutant we obtained a list of genes that are likely direct transcriptional targets of BRM. Analysis of so defined BRM transcriptional direct target genes revealed both known and new pathways in which BRM play significant roles (Supplementary Figure S5). Hormone and stress responses were the most enriched GEO categories in our analysis (Figure 2D). In agreement with previous findings, BRM appears to regulate gibberellin and ABA responses, as well as response to drought (26,24). Other abiotic stress responses found to be regulated by BRM are osmotic and salt-stress responses (Figure 2D). Our data reveal that BRM is also involved in positive regulation of jasmonic acid (JA) responses, a function previously reported for BRM ortholog, SYD (70). In agreement, among BRM-activated targets we found a number of JA biosynthetic and signaling genes, including six JAZ regulators (JAZ3, JAZ5, JAZ6, JAZ7, JAZ9, JAZ10) (71). Moreover, BRM appears to negatively regulate salicylic acid responses, and responses against microbial pathogens, known to be regulated by this hormone (72); light responses, and cell death (Figure 2D), by regulating expression of key transcription factors. In summary, we found 65 and 50 TFs that were BRM-activated and BRM-repressed, respectively, that are likely to be major components of transcriptional network regulated by BRM. This conclusion is consistent with animal studies, showing that BRM homologs often regulate TFs critical for important biological processes including cell pluripotency, differentiation and signaling (73,74). We expect that the actual number of BRM-regulated TFs involved in developmental processes is much higher than we detected, as we used in this study 3-weeks-old seedlings composed mainly of vegetative, fully differentiated cells.
In addition to promoter – bound genes, many other targets have BRM occupancy profiles centred around terminators rather than promoters (Figure 3A and B). BRM-bound terminators show high H3K4me3, low H3 and DNA methylation levels, accompanied by the presence of TATA boxes, resembling classical promoters. Moreover, 3΄BRM-bound genes show extensive non-coding transcription originating from their 3΄ end regions. Transient BRM overexpression can either activate or suppress antisense transcription from selected 3΄ bound targets. Expression changes of those non-coding antisense transcripts observed in brm-1, as well as mutants in other subunits of the SWI/SNF complex were mostly complementary to changes observed after BRM overexpression (Figure 5D). As 5΄BRM-bound genes tend to have low levels of antisense transcription (Figure 4E and Supplementary Figure S11), we did not test the ability of BRM to regulate antisense transcripts derived from their terminators. Instead control terminators from BRM - not bound genes were analyzed, and did not show responsiveness to BRM. Perhaps the ability to both activate and repress antisense transcription should not be unexpected, as SWI/SNF complexes have been implicated in both activation and repression of protein coding gene expression through sense promoters.
Given that BRM binds to gene terminators, the observed effects of mutations in SWI/SNF subunits and BRM transient expression suggest that BRM regulates antisense transcription directly. However, we note here that we were unable to directly test BRM binding to the used transgenes in transient experiments. Although we recognize the need of performing the above described experiments, the insufficient amounts of plant material from transiently transformed 5-days-old Arabidopsis seedlings have currently hindered this approach. Nonetheless, we provided multiple additional evidence supporting our conclusions. Firstly, stable transgenic lines in brm-1 and brm-3 mutant backgrounds validate results from the transient assays (Figure 5F and Supplementary Figure S14A). In addition, the ability of BRM to regulate its 5΄ and 3΄ target in transient system seems to be dependent on the ATPase activity of BRM, as the ability of BRM to activate these promoters was strongly impaired when generated by us BRM point mutant in the ATP-binding site was used (Supplementary Figure S14C and D). This result is consistent with reduced activity of antisense promoter in brm-5 mutant containing a point mutation in the ATPase domain (Supplementary Figure S14B).
Involvement of chromatin remodelers in regulation of non-coding transcription has not been reported in plants, although it has been shown that SWI3B, a SWI/SNF complex subunit interacts directly with IDN2 protein helping to reinforce long-ncRNA mediated gene silencing (75). The recruitment/modulation of SWI/SNF complex activity has also been recently supported in mammals by the discovery of the 7SK ncRNA that interacts with human SWI/SNF complex, to supress non-coding transcription at enhancers (76). Other recent reports also implicate non-plant SWI/SNF complexes in regulation of ncRNA expression. Yeast SWI/SNF-type RSC complex have been reported to suppress non-coding transcription from TTS and other genomic sites (11). Similarly, human SWI/SNF-type complex specific for embryonic stem cells suppresses noncoding transcription in ESC (6). Importantly, SWI/SNF can also activate antisense transcription from divergent promoters in yeast cells (12). Our data suggest that Arabidopsis BRM and SWI/SNF complex both activate and repress ncRNA transcription from TTS regions to regulate expression of protein-coding genes.
We show that in addition to a promoter centred gene regulation, in plants SWI/SNF complex controls expression of a large number of its direct targets through their 3΄ ends. The significance of 3΄ centred BRM-dependent gene expression control is supported by our observation that 3΄BRM-occupied genes are as likely to be mis-regulated in brm-1 mutant, as are the 5΄BRM-occupied genes. Interestingly, the BRM mediated regulation of ncRNA originating from TTS is independent of the presence of linked sense promoter (Figure 5E and F). This suggests the possibility that 3΄-bound BRM uses antisense promoters to control sense expression of those genes (Figure 5G). Further studies will be required to elucidate the precise molecular mechanism involved in this regulation.
The TTS regions of genes or antisense transcripts originating therefrom have been repeatedly implicated in sensing environmental signals in many systems, including sulfur sensing by 3΄ UTR of SULTR2;1 (77) and cold sensing by FLC 3΄ region (48) in plants, or the requirement for the 3΄ region of KCS1 for phosphate sensing in yeast (78). Our finding that a large fraction of the 3΄ SWI/SNF targets are stress-related genes (Supplementary Figure S7) is in good agreement with those observations.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank K. Sosnowska for help with experiments and X. Zhang for kindly providing BRM cDNA clone.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Centre [UMO-2011/01/D/NZ1/01614 to R.A.] and [2011/01/D/NZ8/03690 to S.S.]; Polish National Center for Research and Development [ERA-NET-NEURON/10/2013 to B.W]. Funding for open access charge: National Science Centre [UMO-2011/01/D/NZ1/01614 to R.A.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Clapier C.R., Cairns B.R.. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 2009; 78:273–304. [DOI] [PubMed] [Google Scholar]
- 2. Narlikar G.J., Sundaramoorthy R., Owen-Hughes T.. Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes. Cell. 2013; 154:490–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Phelan M.L., Sif S., Narlikar G.J., Kingston R.E.. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol. Cell. 1999; 3:247–253. [DOI] [PubMed] [Google Scholar]
- 4. Yen K., Vinayachandran V., Batta K., Koerber R.T., Pugh B.F.. Genome-wide nucleosome specificity and directionality of chromatin remodelers. Cell. 2012; 149:1461–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. De S., Wurster A.L., Precht P., Wood W.H., Becker K.G., Pazin M.J.. Dynamic BRG1 recruitment during T helper differentiation and activation reveals distal regulatory elements. Mol. Cell. Biol. 2011; 31:1512–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hainer S.J., Gu W., Carone B.R., Landry B.D., Rando O.J., Mello C.C., Fazzio T.G.. Suppression of pervasive noncoding transcription in embryonic stem cells by esBAF. Genes Dev. 2015; 29:362–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ho L., Jothi R., Ronan J.L., Cui K., Zhao K., Crabtree G.R.. An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:5187–5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Riedel C.G., Dowen R.H., Lourenco G.F., Kirienko N.V., Heimbucher T., West J.A., Bowman S.K., Kingston R.E., Dillin A., Asara J.M. et al. DAF-16 employs the chromatin remodeller SWI/SNF to promote stress resistance and longevity. Nat. Cell Biol. 2013; 15:491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yu Y., Chen Y., Kim B., Wang H., Zhao C., He X., Liu L., Liu W., Wu L.M.N., Mao M. et al. Olig2 targets chromatin remodelers to enhancers to initiate oligodendrocyte differentiation. Cell. 2013; 152:248–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hargreaves D.C., Crabtree G.R.. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 2011; 21:396–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Alcid E.A., Tsukiyama T.. ATP-dependent chromatin remodeling shapes the long noncoding RNA landscape. Genes Dev. 2014; 28:2348–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Marquardt S., Escalante-Chong R., Pho N., Wang J., Churchman L.S., Springer M., Buratowski S.. A chromatin-based mechanism for limiting divergent noncoding transcription. Cell. 2014; 157:1712–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Han S.-K., Wu M.-F., Cui S., Wagner D.. Roles and activities of chromatin remodeling ATPases in plants. Plant J. 2015; 83:62–77. [DOI] [PubMed] [Google Scholar]
- 14. Knizewski L., Ginalski K., Jerzmanowski A.. Snf2 proteins in plants: gene silencing and beyond. Trends Plant Sci. 2008; 13:557–565. [DOI] [PubMed] [Google Scholar]
- 15. Hurtado L., Farrona S., Reyes J.C.. The putative SWI/SNF complex subunit BRAHMA activates flower homeotic genes in Arabidopsis thaliana. Plant Mol. Biol. 2006; 62:291–304. [DOI] [PubMed] [Google Scholar]
- 16. Farrona S., Hurtado L., March-Díaz R., Schmitz R.J., Florencio F.J., Turck F., Amasino R.M., Reyes J.C.. Brahma is required for proper expression of the floral repressor FLC in Arabidopsis. PloS One. 2011; 6:e17997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tang X., Hou A., Babu M., Nguyen V., Hurtado L., Lu Q., Reyes J.C., Wang A., Keller W.A., Harada J.J. et al. The Arabidopsis BRAHMA chromatin-remodeling ATPase is involved in repression of seed maturation genes in leaves. Plant Physiol. 2008; 147:1143–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kwon C.S., Hibara K., Pfluger J., Bezhani S., Metha H., Aida M., Tasaka M., Wagner D.. A role for chromatin remodeling in regulation of CUC gene expression in the Arabidopsis cotyledon boundary. Development. 2006; 133:3223–3230. [DOI] [PubMed] [Google Scholar]
- 19. Vercruyssen L., Verkest A., Gonzalez N., Heyndrickx K.S., Eeckhout D., Han S.-K., Jégu T., Archacki R., Van Leene J., Andriankaja M. et al. ANGUSTIFOLIA3 binds to SWI/SNF chromatin remodeling complexes to regulate transcription during Arabidopsis leaf development. Plant Cell. 2014; 26:210–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wu M.-F., Sang Y., Bezhani S., Yamaguchi N., Han S.-K., Li Z., Su Y., Slewinski T.L., Wagner D.. SWI2/SNF2 chromatin remodeling ATPases overcome polycomb repression and control floral organ identity with the LEAFY and SEPALLATA3 transcription factors. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:3576–3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yang S., Li C., Zhao L., Gao S., Lu J., Zhao M., Chen C.-Y., Liu X., Luo M., Cui Y. et al. The arabidopsis SWI2/SNF2 chromatin remodeling ATPase BRAHMA targets directly to PINs and is required for root stem cell niche maintenance. Plant Cell. 2015; 27:1670–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhao M., Yang S., Chen C.-Y., Li C., Shan W., Lu W., Cui Y., Liu X., Wu K.. Arabidopsis BREVIPEDICELLUS interacts with the SWI2/SNF2 chromatin remodeling ATPase BRAHMA to regulate KNAT2 and KNAT6 expression in control of inflorescence architecture. PLoS Genet. 2015; 11:e1005125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li C., Chen C., Gao L., Yang S., Nguyen V., Shi X., Siminovitch K., Kohalmi S.E., Huang S., Wu K. et al. The arabidopsis SWI2/SNF2 chromatin remodeler BRAHMA regulates polycomb function during vegetative development and directly activates the flowering repressor gene SVP. PLOS Genet. 2015; 11:e1004944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Han S.-K., Sang Y., Rodrigues A., Wu M.-F., Rodriguez P.L., Wagner D. BIOL425 F2010 . The SWI2/SNF2 chromatin remodeling ATPase BRAHMA represses abscisic acid responses in the absence of the stress stimulus in arabidopsis. Plant Cell. 2012; 24:4892–4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Buszewicz D., Archacki R., Palusiński A., Kotliński M., Fogtman A., Iwanicka-Nowicka R., Sosnowska K., Kuciński J., Pupel P., Olędzki J. et al. HD2C histone deacetylase and a SWI/SNF chromatin remodelling complex interact and both are involved in mediating the heat stress response in Arabidopsis. Plant Cell Environ. 2016; 39:2108–2122. [DOI] [PubMed] [Google Scholar]
- 26. Archacki R., Buszewicz D., Sarnowski T.J., Sarnowska E., Rolicka A.T., Tohge T., Fernie A.R., Jikumaru Y., Kotlinski M., Iwanicka-Nowicka R. et al. BRAHMA ATPase of the SWI/SNF chromatin remodeling complex acts as a positive regulator of gibberellin-mediated responses in arabidopsis. PloS One. 2013; 8:e58588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Efroni I., Han S.-K., Kim H.J., Wu M.-F., Steiner E., Birnbaum K.D., Hong J.C., Eshed Y., Wagner D.. Regulation of leaf maturation by chromatin-mediated modulation of cytokinin responses. Dev. Cell. 2013; 24:438–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wu M.-F., Yamaguchi N., Xiao J., Bargmann B., Estelle M., Sang Y., Wagner D.. Auxin-regulated chromatin switch directs acquisition of flower primordium founder fate. eLife. 2015; 4:e09269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Archacki R., Sarnowski T.J., Halibart-Puzio J., Brzeska K., Buszewicz D., Prymakowska-Bosak M., Koncz C., Jerzmanowski A.. Genetic analysis of functional redundancy of BRM ATPase and ATSWI3C subunits of Arabidopsis SWI/SNF chromatin remodelling complexes. Planta. 2009; 229:1281–1292. [DOI] [PubMed] [Google Scholar]
- 30. Jégu T., Latrasse D., Delarue M., Hirt H., Domenichini S., Ariel F., Crespi M., Bergounioux C., Raynaud C., Benhamed M.. The BAF60 subunit of the SWI/SNF chromatin-remodeling complex directly controls the formation of a gene loop at FLOWERING LOCUS C in Arabidopsis. Plant Cell. 2014; 26:538–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sacharowski S.P., Gratkowska D.M., Sarnowska E.A., Kondrak P., Jancewicz I., Porri A., Bucior E., Rolicka A.T., Franzen R., Kowalczyk J. et al. SWP73 subunits of arabidopsis SWI/SNF chromatin remodeling complexes play distinct roles in leaf and flower development. Plant Cell. 2015; 27:1889–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Saez A., Rodrigues A., Santiago J., Rubio S., Rodriguez P.L.. HAB1-SWI3B interaction reveals a link between abscisic acid signaling and putative SWI/SNF chromatin-remodeling complexes in Arabidopsis. Plant Cell. 2008; 20:2972–2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sarnowska E.A., Rolicka A.T., Bucior E., Cwiek P., Tohge T., Fernie A.R., Jikumaru Y., Kamiya Y., Franzen R., Schmelzer E. et al. DELLA-interacting SWI3C core subunit of switch/sucrose nonfermenting chromatin remodeling complex modulates gibberellin responses and hormonal cross talk in Arabidopsis. Plant Physiol. 2013; 163:305–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sarnowski T.J., Ríos G., Jásik J., Swiezewski S., Kaczanowski S., Li Y., Kwiatkowska A., Pawlikowska K., Koźbiał M., Koźbiał P. et al. SWI3 subunits of putative SWI/SNF chromatin-remodeling complexes play distinct roles during Arabidopsis development. Plant Cell. 2005; 17:2454–2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bezhani S., Winter C., Hershman S., Wagner J.D., Kennedy J.F., Kwon C.S., Pfluger J., Su Y., Wagner D.. Unique, shared, and redundant roles for the Arabidopsis SWI/SNF chromatin remodeling ATPases BRAHMA and SPLAYED. Plant Cell. 2007; 19:403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li C., Gu L., Gao L., Chen C., Wei C.-Q., Qiu Q., Chien C.-W., Wang S., Jiang L., Ai L.-F. et al. Concerted genomic targeting of H3K27 demethylase REF6 and chromatin-remodeling ATPase BRM in Arabidopsis. Nat. Genet. 2016; 48:687–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rehrauer H., Aquino C., Gruissem W., Henz S., Hilson P., Laubinger S., Naouar N., Patrignani A., Rombauts S., Shu H. et al. AGRONOMICS1 - A new resource for Arabidopsis transcriptome profiling. Plant Physiol. 2009; 152:487–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Farrona S., Hurtado L., Reyes J.C.. A nucleosome interaction module is required for normal function of arabidopsis thaliana BRAHMA. J. Mol. Biol. 2007; 373:240–250. [DOI] [PubMed] [Google Scholar]
- 39. Logemann E., Birkenbihl R.P., Ulker B., Somssich I.E.. An improved method for preparing Agrobacterium cells that simplifies the Arabidopsis transformation protocol. Plant Methods. 2:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nakamura S, Mano S., Tanaka Y., Ohnishi M., Nakamori C., Araki M., Niwa T., Nishimura M., Kaminaka H., Nakagawa T. et al. Gateway binaryvectors with the bialaphos resistance gene, bar, as a selection marker forplant transformation. Biosci Biotechnol Biochem. 2010; 74:1315–1324. [DOI] [PubMed] [Google Scholar]
- 41. Li J.-F., Park E., von Arnim A.G., Nebenführ A.. The FAST technique: a simplified Agrobacterium-based transformation method for transient gene expression analysis in seedlings of Arabidopsis and other plant species. Plant Methods. 2009; 5:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gendrel A.-V., Lippman Z., Martienssen R., Colot V.. Profiling histone modification patterns in plants using genomic tiling microarrays. Nat. Methods. 2005; 2:213–218. [DOI] [PubMed] [Google Scholar]
- 43. Cock P.J.A., Antao T., Chang J.T., Chapman B.A., Cox C.J., Dalke A., Friedberg I., Hamelryck T., Kauff F., Wilczynski B. et al. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009; 25:1422–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Du Z., Zhou X., Ling Y., Zhang Z., Su Z.. agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 2010; 38:W64–W70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Eden E., Navon R., Steinfeld I., Lipson D., Yakhini Z.. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009; 10:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Linhart C., Halperin Y., Shamir R.. Transcription factor and microRNA motif discovery: The Amadeus platform and a compendium of metazoan target sets. Genome Res. 2008; 18:1180–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schmitz R.J., Schultz M.D., Urich M.A., Nery J.R., Pelizzola M., Libiger O., Alix A., McCosh R.B., Chen H., Schork N.J. et al. Patterns of population epigenomic diversity. Nature. 2013; 495:193–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Swiezewski S., Liu F., Magusin A., Dean C.. Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature. 2009; 462:799–802. [DOI] [PubMed] [Google Scholar]
- 49. Czechowski T., Stitt M., Altmann T., Udvardi M.K., Scheible W.-R.. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005; 139:5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Peirats-Llobet M., Han S.-K., Gonzalez-Guzman M., Jeong C.W., Rodriguez L., Belda-Palazon B., Wagner D., Rodriguez P.L.. A direct link between abscisic acid sensing and the chromatin-remodeling ATPase BRAHMA via core ABA signaling pathway components. Mol. Plant. 2016; 9:136–147. [DOI] [PubMed] [Google Scholar]
- 51. Euskirchen G.M., Auerbach R.K., Davidov E., Gianoulis T.A., Zhong G., Rozowsky J., Bhardwaj N., Gerstein M.B., Snyder M.. Diverse roles and interactions of the swi/snf chromatin remodeling complex revealed using global approaches. PLoS Genet. 2011; 7:e1002008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. O'Sullivan J.M., Tan-Wong S.M., Morillon A., Lee B., Coles J., Mellor J., Proudfoot N.J.. Gene loops juxtapose promoters and terminators in yeast. Nat. Genet. 2004; 36:1014–1018. [DOI] [PubMed] [Google Scholar]
- 53. Crevillén P., Sonmez C., Wu Z., Dean C.. A gene loop containing the floral repressor FLC is disrupted in the early phase of vernalization. EMBO J. 2013; 9:140–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. O'Reilly D., Greaves D.R.. Cell-type-specific expression of the human CD68 gene is associated with changes in Pol II phosphorylation and short-range intrachromosomal gene looping. Genomics. 2007; 90:407–415. [DOI] [PubMed] [Google Scholar]
- 55. Németh A., Guibert S., Tiwari V.K., Ohlsson R., Längst G.. Epigenetic regulation of TTF-I-mediated promoter-terminator interactions of rRNA genes. EMBO J. 2008; 23:1255–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gu M., Naiyachit Y., Wood T.J., Millar C.B.. H2A.Z marks antisense promoters and has positive effects on antisense transcript levels in budding yeast. BMC Genomics. 2015; 16:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Murray S.C., Serra Barros A., Brown D.A., Dudek P., Ayling J., Mellor J.. A pre-initiation complex at the 3΄-end of genes drives antisense transcription independent of divergent sense transcription. Nucleic Acids Res. 2012; 40:2432–2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kim S.Y., Lee J., Eshed-Williams L., Zilberman D., Sung Z.R.. EMF1 and PRC2 cooperate to repress key regulators of arabidopsis development. PLoS Genet. 2012; 8:e1002512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Moissiard G., Cokus S.J., Cary J., Feng S., Billi A.C., Stroud H., Husmann D., Zhan Y., Lajoie B.R., McCord R.P. et al. MORC family ATPases required for heterochromatin condensation and gene silencing. Science. 2012; 336:1448–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Luo C., Sidote D.J., Zhang Y., Kerstetter R.A., Michael T.P., Lam E.. Integrative analysis of chromatin states in Arabidopsis identified potential regulatory mechanisms for natural antisense transcript production. Plant J. 2013; 73:77–90. [DOI] [PubMed] [Google Scholar]
- 61. Rutowicz K., Puzio M., Halibart-Puzio J., Lirski M., Kotliński M., Kroteń M.A., Knizewski L., Lange B., Muszewska A., Śniegowska-Świerk K. et al. A specialized histone H1 variant is required for adaptive responses to complex abiotic stress and related DNA methylation in arabidopsis. Plant Physiol. 2015; 169:2080–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Jones P.A. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012; 13:484–492. [DOI] [PubMed] [Google Scholar]
- 63. Wang Z.-W., Wu Z., Raitskin O., Sun Q., Dean C.. Antisense-mediated FLC transcriptional repression requires the P-TEFb transcription elongation factor. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:7468–7473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Duc C., Sherstnev A., Cole C., Barton G.J., Simpson G.G.. Transcription termination and chimeric RNA formation controlled by arabidopsis thaliana FPA. PLoS Genet. 2013; 9:e1003867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Khavari P.A., Peterson C.L., Tamkun J.W., Mendel D.B., Crabtree G.R.. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature. 1993; 366:170–174. [DOI] [PubMed] [Google Scholar]
- 66. de la Serna I.L., Carlson K.A., Hill D.A., Guidi C.J., Stephenson R.O., Sif S., Kingston R.E., Imbalzano A.N.. Mammalian SWI-SNF complexes contribute to activation of the hsp70 gene. Mol. Cell. Biol. 2000; 20:2839–2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Johnson T.A., Elbi C., Parekh B.S., Hager G.L., John S.. Chromatin remodeling complexes interact dynamically with a glucocorticoid receptor-regulated promoter. Mol. Biol. Cell. 2008; 19:3308–3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Euskirchen G., Auerbach R.K., Snyder M.. SWI/SNF chromatin-remodeling factors: multiscale analyses and diverse functions. J. Biol. Chem. 2012; 287:30897–30905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kadoch C., Crabtree G.R.. Mammalian SWI/SNF chromatin remodeling complexes and cancer: Mechanistic insights gained from human genomics. Sci. Adv. 2015; 1:e1500447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Walley J.W., Rowe H.C., Xiao Y., Chehab E.W., Kliebenstein D.J., Wagner D., Dehesh K.. The chromatin remodeler SPLAYED regulates specific stress signaling pathways. PLoS Pathog. 2008; 4:e1000237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wasternack C., Hause B.. Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 2013; 111:1021–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Vlot A.C., Dempsey D.A., Klessig D.F.. Salicylic Acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 2009; 47:177–206. [DOI] [PubMed] [Google Scholar]
- 73. Kidder B.L., Palmer S., Knott J.G.. SWI/SNF-Brg1 Regulates Self-Renewal and Occupies Core Pluripotency-Related Genes in Embryonic Stem Cells. Stem Cells. 2009; 27:317–328. [DOI] [PubMed] [Google Scholar]
- 74. Seo S. Geminin regulates neuronal differentiation by antagonizing Brg1 activity. Genes Dev. 2005; 19:1723–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zhu Y., Rowley M.J., Böhmdorfer G., Wierzbicki A.T.. A SWI/SNF chromatin-remodeling complex acts in noncoding RNA-mediated transcriptional silencing. Mol. Cell. 2013; 49:298–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Flynn R.A., Do B.T., Rubin A.J., Calo E., Lee B., Kuchelmeister H., Rale M., Chu C., Kool E.T., Wysocka J. et al. 7SK-BAF axis controls pervasive transcription at enhancers. Nat. Struct. Mol. Biol. 2016; 23:231–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Maruyama-Nakashita A., Watanabe-Takahashi A., Inoue E., Yamaya T., Saito K., Takahashi H.. Sulfur-responsive elements in the 3΄-nontranscribed intergenic region are essential for the induction of SULFATE TRANSPORTER 2;1 gene expression in arabidopsis roots under sulfur deficiency. Plant Cell. 2015; 27:1279–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Nishizawa M., Komai T., Katou Y., Shirahige K., Ito T., Toh-e A.. Nutrient-regulated antisense and intragenic RNAs modulate a signal transduction pathway in yeast. PLoS Biol. 2008; 6:e326. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.