Abstract
Mutations in SPARTAN are associated with early onset hepatocellular carcinoma and progeroid features. A regulatory function of Spartan has been implicated in DNA damage tolerance pathways such as translesion synthesis, but the exact function of the protein remained unclear. Here, we reveal the role of human Spartan in facilitating replication of DNA–protein crosslink-containing DNA. We found that purified Spartan has a DNA-dependent protease activity degrading certain proteins bound to DNA. In concert, Spartan is required for direct DPC removal in vivo; we also show that the protease Spartan facilitates repair of formaldehyde-induced DNA–protein crosslinks in later phases of replication using the bromodeoxyuridin (BrdU) comet assay. Moreover, DNA fibre assay indicates that formaldehyde-induced replication stress dramatically decreases the speed of replication fork movement in Spartan-deficient cells, which accumulate in the G2/M cell cycle phase. Finally, epistasis analysis mapped these Spartan functions to the RAD6-RAD18 DNA damage tolerance pathway. Our results reveal that Spartan facilitates replication of DNA–protein crosslink-containing DNA enzymatically, as a protease, which may explain its role in preventing carcinogenesis and aging.
INTRODUCTION
DNA is constantly exposed to different exogenous and endogenous factors that cause DNA damage which, if left unrepaired, challenges the movement of the replication machinery. Stalling of the replication fork can lead to strand breaks and chromosomal rearrangements causing genome instability, early onset of aging and eventually cancer (1–3). To rescue the stalled replication fork, so-called DNA damage tolerance (DDT) pathways have evolved; the name reflects the belief that these pathways do not necessarily repair the actual lesion causing fork stalling but rather facilitate mechanisms that achieve replication across them such as translesion synthesis and template switching (4–6). Indeed, several types of DNA lesions do not require repair processing for their bypass such as the UV-crosslinked cys-syn T-T dimers, which can be efficiently bypassed by translesion synthesis polymerase η (7). However, there are lesions, such as interstrand-crosslinks or protein–DNA crosslinks (DPC), whose processing cannot be omitted before replication proceeds across them (8). DPCs are particularly challenging lesions due to their bulky size and sometimes hard-to-displace DNA-binding property and because they can inhibit the movement of not only polymerases but of the replicative helicase as well (9,10). However, until recently, replication-coupled DPC repair has not received particular attention.
Events at the stalled replication machinery are regulated by the damage-induced ubiquitylation of proliferating cell nuclear antigen (PCNA) (the sliding clamp of the replicative polymerase) performed predominantly by the Rad18 ubiquitin ligase (11,12). The so-called RAD6-RAD18 DDT pathway includes regulators such as other ubiquitin ligases and effectors like translesion polymerases for direct damage bypass as well as double-stranded DNA translocases for template switching (13–16). Monoubiquitylated PCNA can provide a binding platform for many DDT players to exchange the replicative polymerase at the 3΄-prime end and thus facilitate replication through the lesion. For example, the binding of translesion synthesis polymerases to ubiquitylated PCNA enables their access to the lesion. Monoubiquitin–PCNA can be further ubiquitylated; the generated polyubiquitin–PCNA is required for template switching—mediated by specialized dsDNA translocases such as HLTF—during which the newly replicated nascent strand of the sister duplex can serve as a template for DNA synthesis (17,18). However, immediate replication through the damage is not always possible, and gaps may remain opposite the lesions, which might be filled in only after the majority of the DNA has been replicated in the late S or G2 phases; thus, this process is frequently referred to as post-replication repair (19,20).
One of the puzzling questions is the decision making between various DDT pathways when the replication fork stalls at lesions. At least some elements of the question might be answered by studying Spartan (known also as DVC1) identified by our and other laboratories as a previously unrecognized member of the DDT pathway (21–26). Upon UV-induced DNA damage, Spartan is recruited to the site of the stalled replication fork, facilitated by its PCNA-interacting (PIP) and ubiquitin-binding (UBZ) motifs, which ensure direct interaction with ubiquitylated PCNA. Spartan increases the cellular level of ubiquitylated PCNA by either inhibiting USP1-dependent PCNA-deubiquitylation or by stimulating the Rad18 ubiquitin ligase and can facilitate the recruitment of translesion synthesis polymerase η to the lesion (21,22,24). Other studies did not find connection between Rad18-mediated PCNA ubiquitylation and Spartan recruitment but observed that upon binding to PCNA Spartan recruits the ubiquitin-selective chaperone p97 to blocked forks, which may facilitate p97-dependent removal of polymerase η from monoubiquitylated PCNA. Moreover, Spartan was reported to directly interact with POLD3, an accessory subunit of the replicative polymerase δ, and contribute to the suppression of damage-induced mutagenesis (24,27). Although the detailed function of Spartan in the regulation of PCNA ubiquitylation and polymerase switch is not clear yet, all previous studies point to a central role for Spartan in DDT (21,28,29). Its function in protecting genome stability is also supported by recent findings revealing that mutations in human SPARTAN cause early onset hepatocellular carcinoma, genomic instability and a progeroid feature known as Ruijs-Aalfs syndrome (29,30). Furthermore, Spartan insufficiency in mice causes chromosomal instability, cellular senescence and early onset of age-related phenotypes, whereas complete loss of SPARTAN results in early embryonic lethality (28).
Yeast Wss1 was suggested to be a Spartan homologue because the two proteins exhibit similar domain organization containing a metalloprotease domain in the form of an SprT domain in Spartan and a WLM domain in Wss1, an SHP motif and Spartan harbours a ubiquitin-binding-zinc-finger (UBZ) motif for ubiquitin binding while Wss1 has two SUMO-interacting motifs (31). Recent discovery revealed that Wss1 exhibits a DNA-dependent protease activity in degrading DPCs, and yeast genetic evidence provided strong support for its major role in eliminating DPCs in connection with replication (32). Jentsch et al. also suggested that Wss1 may be the first identified member of a so far hidden repair pathway for direct proteolytic removal of DNA-bound proteins and raised the possibility of the existence of this mechanism in higher eukaryotes as well (31–33).
DPCs can be generated by exogenous reactive agents such as reactive aldehydes, ionizing irradiation and UV light but are also quite commonly formed during cellular metabolism due to either the trapping of a normally transient covalent protein-DNA intermediate such as the topoisomerase-DNA reaction or chemical reactions caused by reactive aldehydes such as formaldehyde (FA), which can be generated in the course of histone demethylation or during the metabolism of ethanol (34–36). Exposure to FA has been reported to cause nasopharyngeal cancer and myeloid leukaemia; consequently, FA was classified as a Class I human carcinogen by the IARC in 2006 (37).
DPCs present a high threat to cellular life since they strongly inhibit transcription as well as replication, but, until recently, no specific mechanism has been proposed for their removal, and nucleotide excision repair and homologous recombination were believed to repair them (38). The discovery of yeast Wss1 as a protease directly targeting the protein component of DPCs has changed our thinking about DPC repair. However, until now, no clear orthologue of Wss1 has been described in higher eukaryotes, and the most timely question of whether Spartan has a protease activity and participates in DPC repair has not been answered.
Here, we provide biochemical evidence that human Spartan is a DNA-dependent protease for the specific removal of certain DNA-bound proteins. We also present in vivo evidence for the role of Spartan in direct DPC removal and replication of FA-induced DPCs by monitoring DPC-content, comet-assay and DNA-fibre techniques. Finally, we carried out epistasis analysis and mapped Spartan's function in the replication of DPC-containing DNA to the RAD6-RAD18 DDT pathway. Our study indicates the existence of a novel Spartan protease-based DPC-repair pathway in human cells.
MATERIAL AND METHODS
Spartan cleavage assay
TAP purification of Spartan and Mgs1 and purifications of the Fan1, HLTF, yRad5, Ub-PCNA, PCNA, RFC and RPA proteins, used as potential substrates for the Spartan protease, were carried out according to published protocols (18,39). Wild-type and mutant Spartan proteins were expressed in yeast, purified using Glutathione beads and eluted either with glutathione or by cleavage between the GST- and FLAG-Tags by the PreScission protease, respectively, as described previously (21). Protease assays were typically carried out in 10 μl buffer containing 20 mM Tris/HCl, pH: 7.5, 50 mM NaCl and 0.5 μg purified Spartan in the presence or absence of 1 μg ΦX174 single-stranded DNA. Reactions were incubated at 37°C for 2 h or as indicated otherwise in the figures, followed by adding Laemmli buffer before SDS-PAGE and subsequent Coomassie blue staining or Western blotting with anti-FLAG antibody against FLAG-Spartan. To test the DNA dependence of Spartan's protease activity, 1 μg of several types of DNA was added to the reaction: single-stranded ΦX174 virion, double-stranded nicked Bluescript plasmid or its enzymatically nicked version, single-stranded 75-mer oligonucleotide and partial heteroduplex 75/45-mer oligonucleotides.
Cell cultures and Western blots
Human embryonic kidney HEK 293 cells were cultured in Dulbecco's Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (Sigma) and antibiotics (100 μg/ml streptomycin sulphate and 100 U/ml penicillin) at 37°C. Transfections were carried out using the Lipofectamin 2000 (ThermoFischer Scientific) transfection reagent, according to the manufacturer's protocol. The level of ectopic expression was detected at 48 h after plasmid transfection by harvesting the cells and performing Western blot analysis. For immunoblots, mouse HRP-conjugated anti-FLAG antibody (Sigma, M2 A8592,1:3000) and for the detection of ectopic PCNA level mouse HRP-conjugated anti-PCNA antibody (Santa Cruz Biotechnology, sc-56 HRP, 1:3000) was used. For the detection of β-actin expression level in Western blots, rabbit anti β-actin antibody (Abcam, 1:300) was used as primary and HRP-conjugated goat anti-rabbit IgG antibody (DAKO, P 0448, 1:3000) was used as secondary antibody. To visualize HRP-conjugated antibodies, the Western Bright ECL (Advansta, DV-K12045-D50) detection kit was used.
Generation of stably silenced cell lines
To generate cell lines with stable knockdown of SPARTAN and SPARTAN/RAD18, HEK 293 WT cells were transfected with plasmids containing specific shDNA sequences: pIL2322 (containing SPARTAN shDNA): O2689: 5΄ AGCTTCTAC TTTCCTAGA GTATCATTC AAGAGATGA TACTCTAGG AAAGTAGTT TTTTG 3΄ and pIL2321 (carrying RAD18 shDNA): O2702: 5΄-AGCTTGTTC AGACATCAT AAGAGATTC AAGAGATCT CTTATGATG TCTGAACTG TTTTTG 3΄. Stably expressing cell lines were selected for G418 resistance.
Assay for monitoring in vivo DPC removal
Detection of DPCs was performed by the SDS/KCl precipitation assay (32,40) according to the patent of Costa et al. (US patent number 5,545,529), which is an adaptation of the SDS/KCl precipitation assay for human cells.
Briefly, exponentially growing cells were treated with 500 μM formaldehyde in serum-free medium for 2 h, washed with PBS and harvested (0 h time point) or left to recover in complete DMEM for 3 h or 6 h. The collected cells were counted and 1×106 cells/aliquots were assayed in triplicates. Cells were lysed with a 2% SDS solution buffered with 20 mM Tris-HCl (pH 7.5) containing 1 mM PMSF proteinase inhibitor. Lysed samples were frozen in liquid nitrogen overnight. Thawed samples were subjected to vigorous vortexing for about 10 s followed by warming for 10 min at 65°C. Next, 0.5 ml of 200 mM KCl in 20 mM Tris-HCl, pH 7.5, was added, and the mixture was passed through a 1 ml pipette tip 5 times. SDS-KCl precipitate formation was induced by cooling the samples on ice for 5 min, and the precipitate was then collected by centrifugation at 3000 ×g for 5 min at 4°C, and the supernatant was saved to measure free DNA. The DPC-containing pellet was resuspended in 1 ml 100 mM KCl, 20 mM Tris-HCl, pH 7.5, heated for 10 min at 65°C followed by chilling on ice and centrifugation for 5 min at 3000xg. The washing step was repeated twice, and the 1 ml supernatant of the first centrifugation and the 2 ml of washing fractions were combined resulting in the free DNA fraction. The final SDS-KCl precipitate was resuspended in 1 ml 100 mM KCl, 10 mM EDTA, 20 mM Tris-HCl, pH 7.5 before Prot K was added to a final concentration of 0.2 mg/ml followed by incubation for 3 h at 50°C. SDS was precipitated by chilling samples on ice, and after addition of 10 μl BSA (50 mg/ml) the SDS precipitate was centrifuged, and the supernatant was used to determine the amount of DPC-containing DNA by electrophoresis on 0.7% agarose gel stained with ethidium-bromide for visualization. The gel was scanned and quantitated using a Typhoon Trio Imager (GE Healthcare). The amount of DNA-protein crosslinking was determined as the percentage of SDS/KCl-precipitable DNA in the total DNA (SDS/KCl precipitated plus soluble free DNA).
BrdU comet assay with Prot K treatment
The bromodeoxyuridin (BrdU) comet assay modified with Prot K treatment is a DPC-specific version of our formerly published method (41) with modifications based on a previously used comet method which was improved to specifically measure FA-induced DPCs (42,43). Our specific modifications are detailed below. The comet assay is a sensitive single-cell electrophoresis assay that can be used to detect various DNA lesions. The alkaline version of the assay measures single- and double-strand DNA breaks and alkali labile sites, while double-strand breaks can be detected by the neutral version of the assay (44,45). Pulse labelling the cells with BrdU, a thymidine analogue mainly used to distinguish newly synthesized DNA, renders this assay highly S phase specific (41).
Under standard alkaline conditions, crosslinks can be detected via the decrease in DNA migration, but DNA-DNA intra- or inter-strand crosslinks and DPCs cannot be distinguished. Since our aim was to detect DPCs in the S phase, we modified the standard BrdU alkaline protocol in a way that the assay became highly specific for detecting repair processes during DNA replication and DPCs.
Exponentially growing cells were plated in DMEM supplemented with 10% fetal bovine serum (FBS) (Sigma), 100 μg/ml streptomycin sulphate and 100 U/ml penicillin and grown for 24 h. Cells were transfected with Lipofectamine using different plasmid constructs (silencing-resistant N-terminally FLAG-tagged constructs: SPARTAN(4A) pIL 2854, SPARTAN(HEAA) pIL 2337, SPARTAN(PIP) pIL2338, SPARTAN(UBZ) pIL2336 and SPARTAN(PIP/UBZ) pIL2339 as specified in (21,48). The following day, cells were collected, replated in duplicate to be left untreated or for FA treatment. At 48 h post-transfection, the growth medium was changed to fresh DMEM containing 20 μM BrdU at 37°C for 20 min, and the cells were washed twice with PBS. After 2 h of FA treatment (250 μM, 500 μM, 750 μM), cells were washed, left for 3 h recovery where indicated, or harvested immediately and resuspended in ice-cold PBS. Cells were pelleted by centrifugation, resuspended in 150 μM ice-cold H2O2 diluted in PBS, and kept on ice for 5 min. The H2O2 treatment was employed to induce strand breaks, as an alternative to gamma irradiation which is also used to detect cisplatin, mitomycin C and FA-induced DNA interstrand crosslinks (42,46). For the rescue experiment illustrated in Figure 3A, the BrdU comet analysis was performed after 500 μM FA treatment for 2 h and 3 h of recovery and the release of non-crosslinked DNA was achieved using 100 μM ice-cold H2O2 diluted in PBS.
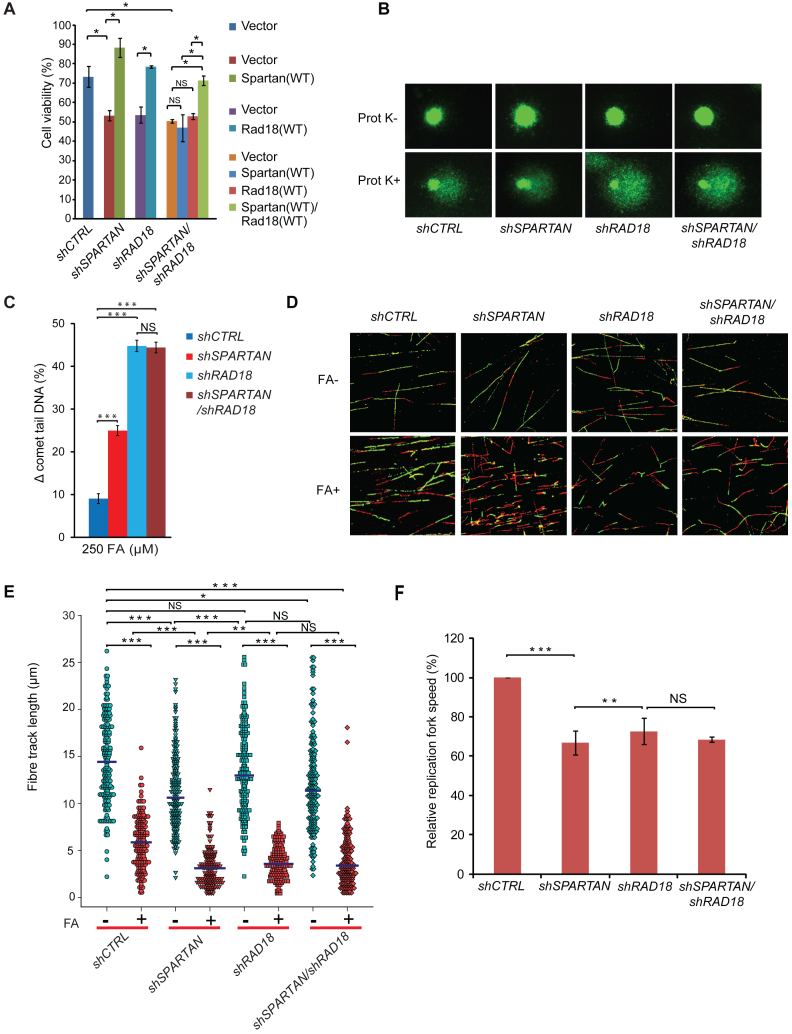
Figure 3.
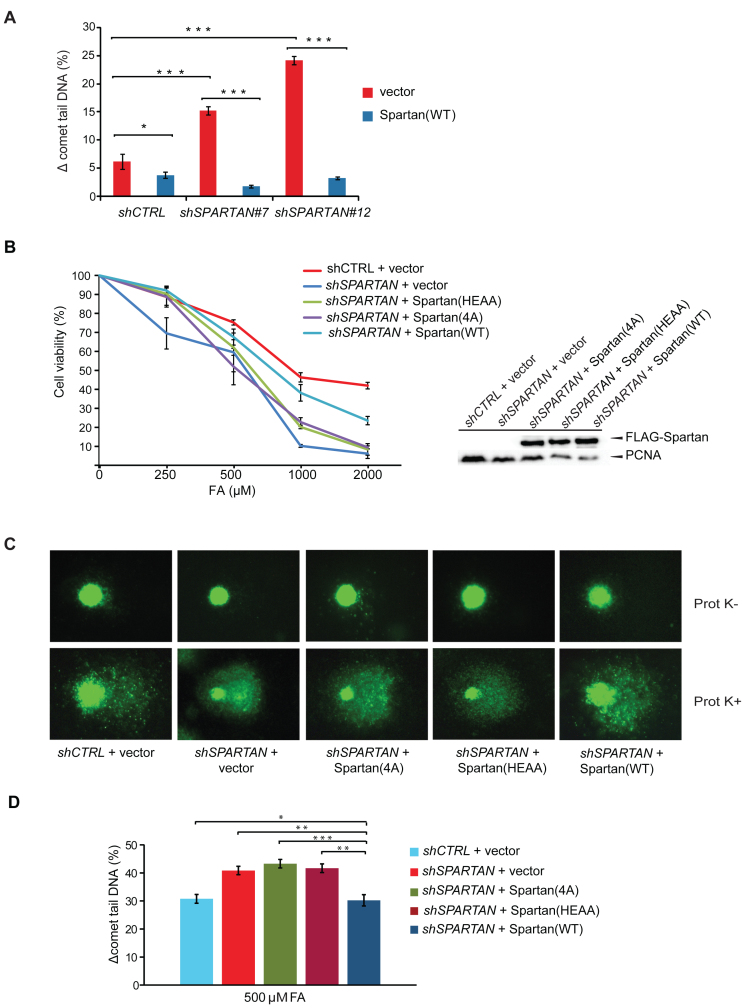
The DNA-binding and the protease domains of Spartan contribute to cell resistance against DPC. (A) Complementation of Spartan-silenced cells. Ectopic expression of silencing-resistant Spartan complemented the DPC-repair deficiency of Spartan-silenced cells as revealed by the Prot K-modified BrdU comet assay. Cells were treated with 500 μM FA for 2 h, harvested after 3 h recovery followed by 100 μM H2O2 treatment. Two independent SPARTAN stable knockdown cell lines (Spartan #7 and #12) were assayed, and ± standard error of the mean was calculated from three independent experiments. Statistical analysis was made by Student's t-test. Statistical significance was labelled by *P < 0.05, **P < 0.01, ***P < 0.001. (B) Cell viability assay of HEK 293 cells depleted of endogenous Spartan expressing siRNA-resistant FLAG-tagged Spartan or Spartan(4A) mutant or Spartan(HEAA) mutant proteins. Resazurin Fluorometric Cell Viability Assay was performed after 2 days of mock or FA treatment. Percentage of viability was calculated by defining the viability of untreated HEK 293 cells stably expressing the corresponding shRNA as 100%. Error bars indicate ± standard error of the mean from three independent experiments. The expression of the siRNA-resistant wild-type and mutant Spartan proteins are verified with α-FLAG antibody in Western blot analysis. (C) The DNA-binding and the SprT domains of Spartan are required for DPC-removal. Representative images of FA-treated cells analysed by the DPC-specific BrdU comet assay. HEK 293 cells stably expressing control shRNA were transfected with FLAG-empty vector expressing FLAG and cells stably expressing SPARTAN shRNA were transfected with FLAG-empty vector, siRNA-resistant FLAG-tagged SPARTAN(4A) mutant, siRNA-resistant SPARTAN(HEAA) mutant or siRNA-resistant SPARTAN(WT). (D) Quantitation of post-replication repair of DPC-containing DNA in cells expressing DNA-binding- and SprT-mutant Spartan proteins. Three independent experiments as shown in Figure 3C were quantitated, and ± standard error of the mean was calculated. Statistical analysis was made by Student's t-test. Statistical significance was labelled by *: P < 0.05, **: P < 0.01, ***: P < 0.001.
H2O2 was removed by washing twice with ice-cold PBS and, after the last centrifugation, cells were resuspended in 0.75% low melting agarose diluted in PBS kept at 37°C.
The lysis solution (2.5 M NaCl, 0.1 M EDTA, 10 mM Tris [pH 10], 1% Triton X-100 and 0.5% sarcosyl) was supplemented with Prot K (Sigma-Aldrich) to a final concentration of 0.8% w/v. Samples were kept in Prot K-containing lysis solution at 4°C overnight. The lysis was performed overnight at 4°C in Prot K-containing lysis solution to eliminate possible artificial break induction at AP sites, which can be generated during Prot K treatment at the regularly used temperature of 37°C (47).
Alkaline electrophoresis, immunostaining and imaging were performed as published previously. For immunostaining, rat anti-BrdU primary antibody (BioRad, OBT0030G, 1:300) and goat anti-rat AlexaFluor488 conjugated secondary antibody (Molecular Probes Inc. A11006, 1:400) were applied. Imaging was performed using Zeiss Axioscope fluorescent microscopy, and quantitation of images was done with the Komet 5.0 software (Kinetic Imaging Ltd., Liverpool, UK).
DNA fibre assay
Exponentially growing HEK 293 cells were pulse labelled with 20 μM iododeoxyuridine (IdU) at 37°C for 20/30 min, washed twice with prewarmed PBS and labelled with 200 μM BrdU for 40/60 min (for untreated control) or exposed to 500 μM FA supplemented with 200 μM BrdU for 40/60 min. In the experiment illustrated in Figure 4E, cells were plated at 80–90% confluency in 6-well plates and were transiently transfected with Turbofect (Thermo Scientific) transfection reagent for 5 h using 4 μg of human expression vector pRK2F expressing FLAG-Spartan(WT) (pIL2335), HEAA-mutant FLAG-Spartan (pIL2337) in siRNA resistant form or empty vector (pIL 1440), as published earlier (21,48). Fiber assay was performed 48 h after transfection.
Figure 4.
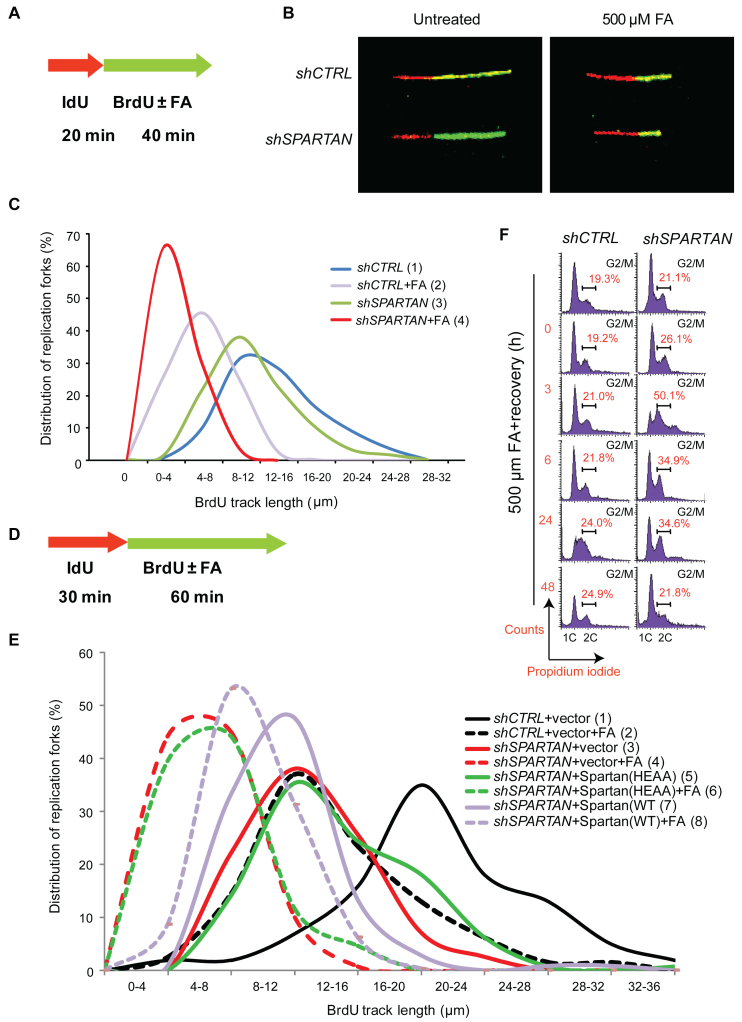
SPARTAN knockdown leads to deficiency during replication of FA-induced DNA damage. (A) Schematic illustration of the strategy of replication fork analysis via the DNA fibre assay. Exponentially growing HEK 293 cells were pulse labelled with 20 μM IdU (red label) at 37°C for 20 min then mock treated or exposed to 500 μM FA supplemented with 200 μM BrdU (green label) for 40 min. DNA fibres were imaged using a confocal laser scanning microscope. The lengths of DNA tracts were measured using Olympus Fluoroview FV1000 2.0 software. Minimum 100 fibres/experiment were measured, and at least three independent experiments were carried out. (B) Representative images of fibres taken from an undisturbed and an FA-treated sample of HEK 293 cells stably expressing control or SPARTAN shRNA as indicated. (C) Replication fork movement on FA-damaged DNA is inhibited in Spartan-depleted cells. DNA fibre experiment was carried out as detailed in (A). Data from the measurement of second track lengths are illustrated by distribution curves. Statistical analysis was made by Student's t-test p(1-2)< 0.001, p(1-3) < 0.001, p(2-4) < 0.001. (D) Schematic illustration of the strategy of replication fork analysis via the DNA fibre assay to detect the effect of the ectopic expression of the Spartan(HEAA) mutant. (E) Inhibited replication fork movement on FA-damaged DNA in Spartan-depleted cells was not rescued by Spartan(HEAA). Exponentially growing HEK 293 cells were pulse labelled with 20 μM IdU (red label) at 37°C for 30 min then mock treated or exposed to 500 μM FA supplemented with 200 μM BrdU (green label) for 60 min. Data from the measurement of second track lengths are illustrated by distribution curves. Statistical analysis was made by Student's t-test p(1-2) < 0.001, p(2-4) < 0.001, p(3-4) < 0.001, p(4-6) > 0.05 (non-significant), p(3-5) < 0.05, p(5-6) < 0.001, p(6-8) < 0.001, p(4-8) < 0.001. (F) Spartan silencing affects cell cycle progression and leads to a G2/M accumulation after FA exposure. Cell cycle analysis was carried out employing the propidium iodide staining method to measure the DNA content by flow cytometry after 2 h of FA treatment followed by 0–24 h recovery. Cells were harvested at the indicated time points, fixed with ice-cold 70% ethanol overnight, and DNA was stained with propidium-iodide.
Isolation of DNA fibres and immunolabelling were carried out as described previously (49). Briefly, 2 μl of cells resuspended in PBS (106 cells/ml) were diluted 1:2 with unlabelled cells and spotted onto clean glass slides. Cells were lysed with lysis solution (0.5% sodium dodecyl sulphate (SDS) in 200 mM Tris-HCl (pH 5.5), 50 mM EDTA). Slides were tilted at 15° to the horizontal, allowing a stream of DNA to run slowly down the slide. Next, slides were air-dried and fixed in methanol-acetic acid (3:1). Fixed fibres were rehydrated in water and denatured (2.5 M HCl for 1 h). BrdU incorporation was detected using rat anti-BrdU antibody (1:300; BioRad, OBT0030G), and IdU incorporation was followed using mouse anti-IdU antibody (BD Biosciences, 347580, 1:400). Slides were washed and incubated with fluorescently labelled secondary antibodies (Alexa Fluor488-conjugated goat anti-rat IgG antibody (Molecular Probes Inc. A11006, 1:400,) and Cy3-labeled sheep anti-mouse IgG antibody (Sigma Aldrich, C2181, 1:400) to detect BrdU and IdU, respectively. After extensive washing with PBS, slides were stained in mounting medium containing glycerol and DAPI. DNA fibres were imaged using an Olympus confocal laser scanning microscope. The lengths of DNA tracks corresponding to IdU and BrdU labelling were measured using the Olympus Fluoroview 2.0 software. In each experiment, minimum 100 independent fibres were analysed. Results are illustrated as ±SEM of three independent experiments.
Cell viability assay
Exponentially growing HEK 293 cells were plated at 70% confluency in 6-well plates. After 24 h, cell transfections were carried out according to the instructions of the manufacturer, using the Lipofectamine 2000 (Invitrogen) transfection reagent. Cell competition-based cell viability assay was performed as described earlier (21). The cells were transfected with the following siRNA-resistant N-terminally FLAG-tagged plasmids: pIL2335 containing WT SPARTAN, pIL2615 containing RAD18 and pIL1440 containing FLAG-empty vector. After 24 h, the cells were treated with FA (Sigma) for 2 h and washed briefly three times with PBS. After 7 days of culturing, the ratio of GFP-positive and GFP-negative cells was determined by flow cytometry (FACSCalibur, BD Biosciences). The percentage of viability was calculated by defining the viability of untreated HEK 293 cells stably expressing control shRNA as 100%.
Resazurin fluorometric cell viability assay
Exponentially growing HEK 293 cells were plated at 60% confluency in 6-well plates. After 24 h, cell transient transfections were carried out according to the instructions of the manufacturer, using the Lipofectamine 2000 (Invitrogen) transfection reagent. The cells were transfected with the following siRNA-resistant N-terminally FLAG-tagged plasmids: pIL2335 containing WT SPARTAN, pIL2854 containing SPARTAN(4A), pIL2331 containing SPARTAN(HEAA) and pIL1440 containing FLAG-empty vector. One day after transfection, siRNA-transfected cells were treated for 2 h with different concentrations of FA (0 μM, 250 μM, 500 μM, 1000 μM, 2000 μM) in serum-free DMEM. After 2 h treatment, cells were washed three times with prewarmed PBS, harvested and seeded in triplicate in 96-well plates at a density of 10 000 cells/well in a volume of 100 ml of DMEM containing fetal calf serum and penicillin/ streptomycin. After 2 days of culturing, 0.15 mg/ml of resazurin (Sigma, R7017-1G) solution in PBS was mixed with DMEM (100 ml) in a ratio of 20: 100 and was added to each well. After 5 h of incubation at 37°C, cell viability was monitored by measuring fluorescence with excitation wavelength at 565 nm and emission wavelength at 580 nm in a Fluoroskan Ascent FL (Thermo Scientific) fluorimeter. The fluorescent signal generated from the assay is directly proportional to the number of living cells in the sample. Percentages of living cells were calculated according to the calibration curves of the appropriate cell lines (50).
Cell cycle analysis by flow cytometry
Cell cycle analysis was carried out using flow cytometry. Cells were plated in 100 mm plates in complete medium. After 24 h, treatment with 500 μM FA was applied for 2 h, and the cells were washed three times with PBS. Cells were trypsinized, harvested and fixed with 70% ethanol overnight at -20°C. The following day, ethanol was removed by washing with 1xPBS, and the cells were resuspended in phosphate buffer containing 10 μg/ml RNaseA and 20 μg/ml propidium iodide (Sigma) and incubated at room temperature for 30 min. For flow cytometry measurements, the BD FACSCalibur (BD Biosciences) flow cytometer and for quantitative analysis CellQuestTM (BD Biosciences) was used.
RESULTS
Spartan has a DNA-dependent protease activity
Spartan has four well-defined distinct motifs including an N-terminal metalloprotease-like domain (SprT), a DNA-binding domain labelled as KKGK (48), a PCNA-interacting (PIP) and a C-terminal ubiquitin-interacting (UBZ) domains (Figure 1A). Our previous study (21) using various point-mutant SPARTAN constructs in the complementation study of Spartan-silenced cells revealed the importance of the SprT motif in providing UV-resistance; however, no particular function has been assigned to this domain.
Figure 1.
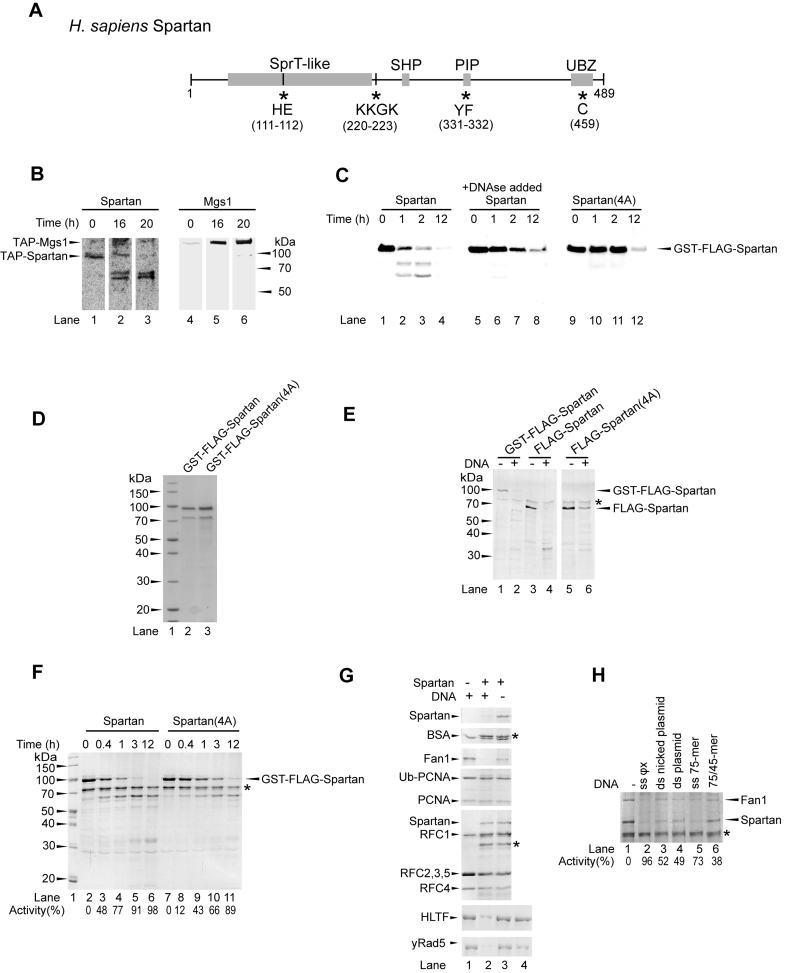
Spartan exhibits DNA-dependent protease activity. (A) Schematic representation of the domain structure of human Spartan. Spartan contains an SprT-like metalloprotease, an SHP, a PCNA-binding (PIP) and a ubiquitin-binding (UBZ) domain. Asterisks and the displayed amino acids indicate the sites mutated to generate the protease-deficient Spartan mutant Spartan(HEAA) (HE to AA), the DNA-binding Spartan(4A) (KKGK to AAAA), the PCNA-binding Spartan(PIP) (YF to AA), the UBZ-binding Spartan(UBZ) (C to S) and the PCNA-binding and UBZ-binding double mutant Spartan(PIP/UBZ) (YF/C to AA/S). (B) Spartan is cleaved rapidly in human cell extract. Total cell lysates (lanes 1 and 4) generated from human cells stably expressing tandem affinity-tagged Spartan or control Mgs1 were used for the purification on two subsequent affinity beads for 16 h (lanes 2 and 5) and an additional 4 h (lanes 3 and 6). The positions of the FLAG-tagged proteins were visualized by Western blot using anti-FLAG antibody. (C) Stability of human Spartan overexpressed in yeast. GST-FLAG-tagged wild-type Spartan (lanes 1–8) and DNA-binding mutant Spartan(4A) (lanes 9–12) were expressed in yeast, and the generated cell lysates were incubated for the indicated lengths of time at room temperature. In lanes 5–8, DNaseI was added during cell lysis. The stability of Spartan was followed by Western blot using anti-FLAG antibody. (D) Purity of purified Spartan, sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) of purified GST-FLAG-Spartan(WT) and GST-FLAG-Spartan(4A) stained with Coomassie blue (E) DNA-dependent self-cleavage of Spartan. Spartan and Spartan(4A) were purified as GST-FLAG- (lanes 1–2) or FLAG-fusion (lanes 3–6) proteins. Purified proteins were incubated in the absence or presence of ΦX174 single-stranded DNA (ssDNA) for 2 h at 37°C followed by SDS-PAGE and Coomassie blue staining. The asterisk indicates the position of a co-purifying contaminating protein which can serve as a loading control. (F) Comparison of the self-cleavage activity of the wild-type and the DNA-binding mutant Spartan proteins. Purified Spartan (lanes 2–6) and Spartan(4A) (lanes 7–11) were incubated for various lengths of time in the presence of ΦX174 ssDNA. (G) Spartan can cleave certain DNA-binding proteins in a strictly DNA-dependent manner. Purified BSA, Fan1, ubiquitin-PCNA, PCNA, RFC, HLTF and yRad5 were incubated without (control) or with Spartan for 2 h at 37°C in the absence or presence of ΦX174 ssDNA as indicated. Lane 4 of the lower two panels shows the no reaction control, in which all the reaction components were mixed and then boiled immediately. (H) Comparison of the activating potential of various DNA structures on Spartan protease. Purified Spartan was mixed with purified Fan1 and incubated either alone or with various types of DNA (ΦX174 ssDNA, nicked plasmid dsDNA, plasmid dsDNA, 75-mer oligonucleotide, 75/45-mer partial heteroduplex oligonucleotides, each at 100 ng/μl) for 2 h at 37°C. The percentage of cleavage activity was quantitated compared to the amount of Spartan and Fan1 in the sample containing no DNA.
During our effort to purify Spartan-interacting proteins from human cells stably expressing tandem-affinity tagged Spartan or various other proteins, we realized that the full-length Spartan becomes quite rapidly degraded in the cell extract, resulting in at least two distinct bands the molecular sizes of which indicate that the cleavage might occur after the N-terminal SprT domain (Figure 1B), but due to the unusual electrophoretic mobility of TAP-Spartan this conclusion is not definite. Moreover, when we overexpressed human Spartan in yeast cells for purification purposes, we also noticed its instability (Figure 1C, lanes 1–4). Importantly, adding DNase to the cell breaking buffer inhibited the cleavage of Spartan very significantly (Figure 1C, lanes 5–8). Moreover, when we purified the Spartan(4A) mutant, in which the DNA-binding KKGK amino-acids were mutated to AAAA, we realized that the stability of this mutant is much higher than that of the wild-type protein (Figure 1C, lanes 9–12). Finally, under cold conditions and in the presence of DNAse we managed to obtain Spartan wild-type and Spartan(4A) proteins with high degree of purity (Figure 1D). These experiments suggested that the cleavage of Spartan is DNA-dependent. As further support, we found that wild-type purified GST-FLAG-Spartan as well as FLAG-Spartan are stable when incubated in the absence of DNA (Figure 1E, lanes 1 and 3); however, when we added long single-stranded DNA (ssDNA) to the reaction all of the Spartan protein present became degraded (Figure 1E, lanes 2 and 4). Importantly, we noticed that in the parallel reaction using purified Spartan(4A), the degradation of this DNA-binding mutant was significantly impaired (Figure 1E, lanes 5 and 6). We kinetically compared the wild-type and the mutant protein for their self-degrading activity in the presence of DNA (Figure 1F, compare lanes 2–6 to 7–11) and noticed that the wild-type protein is several-fold more prone to degradation. We concluded that Spartan exhibits an intrinsic DNA-dependent protease activity and is able to cleave itself. Next, we asked whether the cleavage activity of Spartan is limited to itself or it can degrade other proteins as well. To this end, we assayed the protease Spartan on various highly purified potential protein substrates in the absence or presence of DNA (Figure 1G). Remarkably, Spartan was able to cleave in a DNA-dependent manner not only itself but also the DNA-binding proteins Fan1, HLTF and yRad5 while many proteins such as BSA, PCNA and RFC were inaccessible to its protease activity. We note that certain proteins that have high affinity to DNA, such as RFC and RPA, exhibited inhibitory activity against the protease Spartan, which also depended on their concentration (Figure 1G and data not shown) suggesting that the efficiency of competing with other proteins for DNA binding could be a rate-limiting step in the action of the protease Spartan. This could be particularly important at the stalled replication fork where the exposed ssDNA tracks rapidly become covered by RPA. To systematically check DNA structural requirements, we tested various DNAs for stimulation (Figure 1H). We found that various DNA structures could stimulate the protease activity at some level, but ΦX174 ssDNA stimulated the most, and the ss75-mer oligonucleotide was also quite effective, while double-stranded or nicked plasmid DNA or the 75/45-mer partial heteroduplex stimulated quite weakly. Taken together, our observations are consistent with a model depicting that Spartan is targeted to exposed ssDNA such as that found in case of fork stalling, where its protease activity can remove some DNA-bound proteins.
Spartan is required for DPC removal
To reveal whether Spartan is involved in DPC removal in vivo, we monitored the DPC content of genomic DNA by separating the total genomic DNA into two fractions, a free-DNA- and a DPC-containing one, using a previously established SDS-KCl protein precipitation technique. We induced DPCs in control and Spartan-silenced cell lines by FA treatment and followed DPC appearance after 2 h of treatment and subsequently DPC removal at 3 h as well as 6 h after washing out FA from the medium of the cells. As shown in Figure 2A, FA-treatment highly increased DPC formation (∼20-fold), and the repair by direct-removal of DPC from the genomic DNA in the recovery period was apparent. Importantly, we found that Spartan-silenced cells exhibited a somewhat higher amount of DPC even at the end of the 2 h FA-treatment and retained higher amounts of DPC than the WT control cells during the 3 h and 6 h recovery time following FA treatment (Figure 2A). From this experiment, we conclude that in vivo Spartan has a role in the removal of DPCs.
Figure 2.
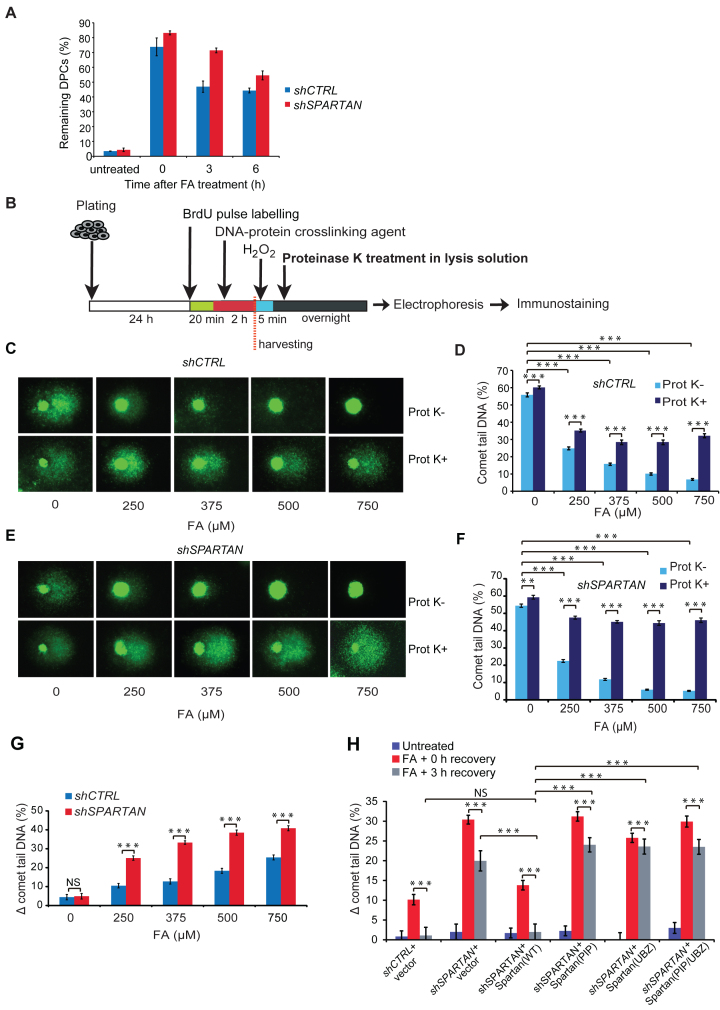
Spartan facilitates protein–DNA crosslink (DPC) repair. (A) Monitoring of in vivo DPC removal. Exponentially growing cells were treated with 500 μM FA for 2 h, washed and left to recover until 3 and 6 h. After an SDS/KCl precipitation assay, the quantification of the DPC-containing and the free DNA-containing fractions was carried out, and the percentage of DPCs in total DNA (DPC+free DNA) was graphed. Error bars indicate ± standard error of the mean of three independent experiments. (B) Schematic illustration of the experimental set-up of the Prot K-modified alkaline BrdU comet assay. Asynchronously growing HEK 293 cells were pulse labelled with BrdU for 20 min and treated with FA for 2 h. Cells were harvested, and DNA strand breaks were induced with H2O2 exposure to release DNA. For the detection of the amount of proteins crosslinked to DNA, the lysis solution was supplemented with Prot K (Prot K+) or lysed without Prot K (Prot K-) before electrophoresis and immunostaining using BrdU-recognizing antibody. (C) Effect of FA exposure on post-replication repair using the Prot K-modified BrdU comet assay. Representative images of control HEK 293 cells illustrate the dose-dependence of FA exposure and the difference caused by Prot K treatment. (D) Quantitation of the experiment shown in (C). Error bars indicate ± standard error of the mean of three independent experiments. Statistical analysis was made by Student's t-test. Statistical significance was labelled by ***: P < 0.001. (E) Dose dependence of FA exposure on HEK 293 cells stably expressing SPARTAN shRNA. Representative images illustrate the effect of FA and Prot K treatments. (F) Quantitation of the experiment shown in (E). Error bars indicate ± standard error of the mean of three independent experiments. Statistical analysis was made by Student's t-test. Statistical significance was labelled by **: P < 0.01, ***: P < 0.001. (G) Quantitative comparison of FA-induced post-replicative repair delay in cells stably expressing control or SPARTAN shRNA. The percentage of DPC-caused DNA maturation delay was calculated as a difference between Prot K+ and Prot K- samples (Δ comet tail DNA (%)). The data were obtained from minimum 300 cells measured in three independent experiments. Statistical analysis was made by Student's t-test. Statistical significance was labelled by ***: P < 0.001, NS: not significant. Corresponding data are shown in Figure 2D and F. (H) The PCNA-binding and the Ubiquitin-binding domains of Spartan are required for DPC-repair. HEK 293 cells stably expressing control shRNA were transfected with FLAG-empty vector expressing FLAG and cells stably expressing SPARTAN shRNA were transfected with FLAG-empty vector expressing FLAG, and siRNA-resistant FLAG-tagged SPARTAN(PIP) mutant, SPARTAN(UBZ) mutant, SPARTAN(PIP/UBZ) or SPARTAN(WT). Cells were treated for 2 h with 500 μM FA and analysed after 0 and 3 h of recovery by comet assay Statistical analysis was made by Student's t-test. Statistical significance was labelled by ***: P < 0.001, NS: not significant.
Spartan has been suggested to act when replication stalls at a DNA lesion. To reveal whether Spartan deficiency causes impairment in the replication of DPC-containing DNA, first we used the BrdU comet assay (41) combined with a Proteinase K (Prot K) treatment. Including Prot K treatment was necessary since no specific agent is known that causes only DPCs without any other types of damage such as DNA–DNA crosslinks or single-stranded breaks, and we wanted to make our assay highly specific for monitoring only DPC-caused events. For comet assay, which is a single-cell electrophoresis assay, we pulse labelled the cells with the thymidine analogue BrdU to distinguish replicating cells.During electrophoresis, migration of crosslinked DNA is reduced, and determination of DPCs is accomplished by the measurement of the reduction in induced DNA migration. To distinguish between crosslinked and intact DNA that is not damaged by FA, DNA migration can be induced by H2O2 following the FA treatment, and the reduction in induced DNA migration relative to the FA-untreated but H2O2-treated control can be detected as a measure of crosslinking.
The outlines of the DPC-specific comet assay method we employed are shown in Figure 2B. First, cells were pulse labelled with BrdU to visualize replicating cells only, and FA treatment was applied to induce DPCs (Figure 2B). Since during electrophoresis the migration of DPC-containing DNA is highly reduced, keeping the whole DNA molecule in the comet head, we applied a DNA fragmenting H2O2 treatment, which enabled the resulting DNA fragments to migrate to the tail of the comet. After H2O2 treatment, cells were embedded in agarose and immediately lysed, thus, no further repair process was allowed to remove H2O2-induced strand breaks before single-cell electrophoresis. To further sensitize the comet assay for DPCs and distinguish between FA-induced DNA–DNA and protein–DNA crosslinks, in a parallel sample, we added Prot K to the lysis buffer to remove crosslinked protein residues from the DNA. Due to the removal of DPCs, the protein-free high molecular weight DNA—which is not able to migrate in agarose—behaves as a single-stranded break having free ends at the stalled replication fork. Under denaturing alkaline conditions, it is able to relax, unwind, and unhook in the vicinity of free ends, and the forming free DNA loops migrate into the tail, resulting in a higher percentage of comet tail DNA. After the removal of the protein part of the DPC, the shorter, newly replicated still non-ligated DNA fragments like the Okazaki fragments are also capable of migrating into the tail, forming higher amounts of tail DNA.
Using the setup detailed above, the difference between the comet tail DNA percentage of Prot K-treated (Prot K+) and untreated (Prot K-) samples reflects the amount of DPCs left unrepaired. To test the sensitivity of the method, we treated cells with increasing concentrations of FA and monitored the changes in comet tail DNA percentage with and also without Prot K treatment (Figure 2C). We found that FA treatment causes a concentration-dependent decrease in DNA migration, and, as expected, the difference in the percentage of tail DNA of Prot K+ and Prot K- samples gradually increases, reflecting the presence of increasing amounts of DPCs correlating well with the applied dose of FA (Figure 2D). Thus, the Prot K-modified BrdU comet assay we employed is suitable for the precise detection of DPC removal in replicating cells.
Next, we used this technique to investigate whether Spartan has a role in DPC removal in replicating cells using Spartan-silenced HEK 293 cell lines generated by the stable expression of Spartan-specific shRNA in the DPC-specific BrdU comet assay (Figure 2E and Supplementary Figure S1A). As a result of increasing doses of FA, higher amounts of DNA remained in the comet head in the Spartan-silenced cell line than in the control cell line (Supplementary Figure S1B). However, the difference between the Spartan-knockdown and the control cell line can be observed more prominently after Prot K treatment (Figure 2E and F). We calculated the difference between the comet tails of Prot K-treated and untreated samples (Δ comet tail DNA (%)) for all FA concentrations we used (250–750 μM) and compared these data points between the control and the Spartan-silenced cell lines. This analysis revealed a high degree of deficiency in DPC-removal in Spartan-silenced cells at all examined FA concentrations (Figure 2G). We also monitored repair after 0 and 3 h of recovery following FA treatment and compared the effect of expression of shRNA-resistant PIP and UBZ mutant Spartan proteins in Spartan-silenced cells on DPC removal in replicating cells (Figure 2H, Supplementary Figure S1C and D). We found that wild-type Spartan was able to complement the deficiency of Spartan knockdown cells, but the PIP and UBZ mutant Spartans were unable to restore the deficiency of DPC-repair even after 3 h of recovery. Taken together, SPARTAN is required for DPC-repair in replicating cells, in which its PCNA- and ubiquitin-binding domains play an essential role.
The DNA-binding and the protease domains of Spartan contribute to cell resistance against DPCs
To test our hypothesis that the protease activity of Spartan is indeed required for DPC removal and to further confirm that the observed phenotype is Spartan-dependent, we employed a complementation assay (Figure 3A). First, to rescue the effect of Spartan silencing on FA-induced DPC accumulation, we ectopically expressed silencing-resistant FLAG-Spartan in two independently generated stably Spartan-depleted cell lines (#7 and #12) and carried out the DPC-specific comet assay applying 2 h of FA treatment and 3 h of recovery (Supplementary Figure S2A and B). We found that ectopic expression of silencing-resistant FLAG-Spartan resulted in the restoration of DPC-repair, indicating that the impairment in the silenced cell lines was indeed caused by the absence of Spartan (Figure 3A). Interestingly, we noticed that the ectopic expression of Spartan conferred some additional protection against crosslink accumulation as compared to the control cell line (indicated by the Δ comet tail DNA (%) value in Figure 3A and Supplementary Figure S2A), which can be explained by the dependence of DPC repair on the cellular concentration of Spartan.
Next, we tested the effect of the inactivation of the protease and DNA-binding activities of Spartan by a cell viability assay employing FA treatment. We ectopically expressed the siRNA-resistant forms of wild-type, DNA-binding mutant Spartan(4A), and SprT-mutant Spartan(HEAA) in Spartan-silenced cells and compared their FA sensitivity (Figure 3B). We found that while wild-type Spartan compensated the negative effect of Spartan depletion on cell survival quite well, neither Spartan(4A) nor Spartan(HEAA) was able to significantly rescue the deleterious effect of Spartan silencing, though their ectopic expression levels were similar (Figure 3B, right panel).
We also used the BrdU comet assay in complementation study. FA-induced crosslinks highly reduced tail DNA formation (Figure 3C upper row), and as a result of Prot K treatment free DNA could migrate into the comet tail (Figure 3C lower row). Compared to the control cells transfected with an empty vector, ectopic expression of siRNA-resistant Spartan decreased Prot K-induced tail formation significantly (see Figure 3CshSPARTAN+Spartan(WT), lower row last panel), as revealed by the lower amount of tail DNA and bigger head of comets, reflecting much lower amounts of unrepaired, FA-induced DPCs. In contrast, expression of siRNA-resistant Spartan(4A) and Spartan(HEAA) could not reduce tail DNA formation, and the phenotype observed was similar to that of the empty vector-transfected Spartan-silenced sample. Quantitative comparison of the difference in the tail DNA percentage of Prot K-treated and untreated samples revealed that in contrast to wild-type Spartan, expression of Spartan(4A) and Spartan(HEAA) was not able to rescue the DPC-repair deficiency caused by Spartan knockdown (Figure 3D). From these results, we conclude that the DNA-binding- and protease-domain mediated activities of Spartan are important determinants of the function Spartan has in the protection against the genotoxic effects of FA-induced DPCs.
Spartan facilitates immediate bypass of FA-induced DNA damage
One of the main hallmarks of replication stress is the abnormal slowing of replication fork movement, which can eventually lead to the stalling of the replication fork. Having established the requirement for Spartan in DPC-repair in replicating cells, our next question was whether Spartan has a role in the immediate bypass of DPCs, which can pose strong barriers to the movement of not only the replicative polymerases but the translesion synthesis polymerases as well. To monitor the speed of replication, we used the DNA fibre assay with which we can follow the elongation of the nucleotide analogue-labelled nascent DNA track at the single molecule level. As shown in Figure 4A, replicating DNA was visualized by first pulse labelling the cells with iododeoxyuridine (IdU, red label) followed by mock or FA treatment, performed simultaneously with BrdU pulse labelling before immunodetection of the nucleotide analogues via microscopy of the individual, newly replicated DNA tracks. We found that FA treatment significantly decreased replication fork speed as revealed by the shorter second tracks (green label) shown in the representative images of our DNA fibre experiments (Figure 4B). To unravel if Spartan can facilitate the bypass of FA-induced DNA damage, we compared Spartan-depleted and control cells and noticed that the lengths of the second tracks of Spartan-silenced cells are significantly shorter compared to those of control cells after FA treatment, reflecting a significantly stronger inhibitory effect of FA-induced damage on immediate bypass in the absence of Spartan (Figure 4B). Measuring the length distribution of the second DNA tracks of individual replication forks (Figure 4C) revealed that Spartan knockdown led to somewhat slower replication fork movement under unchallenged conditions, which might reflect the presence of some endogenous damage in the absence of exogenous FA treatment. However, more significantly reduced fork progression could be detected after FA treatment, and in Spartan-silenced cells more than a two-fold shortening of the average second track length was detected.
We also tested whether the predicted protease domain (SprT) of Spartan is required during replication of DPC-containing DNA (Figure 4D and E, and Supplementary Figure S2E) using 30 min IdU labelling followed by 60 min of FA treatment with a simultaneous BrdU labelling. We found that in contrast to the wild-type Spartan, expression of the Spartan(HEAA) protein was not able to rescue the Spartan knockdown cells from the replication inhibitory effect of FA treatment (Figure 4E) indicating the requirement for the Spartan protease-like domain. These findings are consistent with a model in which Spartan degrades the protein component of the replication fork-movement-blocking DPC, leaving only a short peptide bound to the DNA thus rendering the DNA accessible to immediate bypass by translesion synthesis polymerases.FA exposure-induced DNA damage results in G2/M accumulation of the cell cycle progression in cells with certain DNA repair deficiencies (51,52). Moreover, knockout of the potential Spartan homologue yeast WSS1 results in a strong G2 cell cycle arrest after FA exposure (32). Thus, impairment in DPC repair and replication of DPC-containing DNA caused by Spartan deficiency can also be expected to disturb normal cell cycle. To explore this hypothesis and to determine whether FA exposure leads to altered cell cycle progression, we compared control and Spartan-depleted cells by flow cytometry analysis using propidium iodide staining (Figure 4F). We detected some G2/M accumulation in Spartan-depleted cells compared to control cells even without any treatment. However, after FA treatment a more marked difference was observed. At 3 h post-treatment, a late S/G2 accumulation was found and, remarkably, cells with more than 2C DNA content also appeared in Spartan-depleted cells. These binucleated cells possibly reflect cells with micronuclei and aberrant unresolvable mitotic structures such as chromatin bridges, which we also noticed during the microscopic analysis of the BrdU comet assay (Supplementary Figure S2D). Moreover, after 24 h of FA exposure, control cells showed an increased S-phase blockage, which inhibits exit from the S phase with damaged DNA; in contrast, Spartan-silenced cells displayed a noticeable accumulation of G2/M-phase cells. These changes reflect that Spartan-depleted FA-treated cells leave the S phase faster than control cells, possibly with unrepaired DPCs and abnormal replication intermediates, which might explain Spartan deficiency-caused genomic instability.
Spartan and Rad18 act together in protecting the genome from DPCs
No particularly defined pathway has been assigned to DPC repair yet, and the discovery of the requirement for the protease Spartan in the replication of DPC-containing DNA raised the question whether it constitutes an independent DPC repair pathway or acts together with other fork rescue pathways. In a wide variety of species, Rad6-Rad18-dependent PCNA-monoubiquitylation constitutes a predominant pathway for the rescue of stalled replication forks, and, similarly, human Rad18 is required for the post-replication repair of UV-damaged DNA (41). Independent laboratories have also reported that RAD18 and SPARTAN cooperate in the protection against UV-irradiation though the role of Rad18 in regulating Spartan's function is still contradictory. To address whether Spartan cooperates with Rad18 in DPC repair, we carried out epistasis analysis comparing the phenotypes of single Spartan- and Rad18- and double Spartan/Rad18-silenced cell lines employing three different assays, namely, cell survival, DPC-specific BrdU comet assay and the DNA fibre analysis (Figure 5). First, we confirmed the efficiency of silencing and, for complementation studies, the expression of FLAG-Spartan and FLAG-Rad18 from vectors resistant to shRNA silencing by Western blot analysis (Supplementary Figure S3A and B). When comparing the FA sensitivity of the single- and double-silenced cell lines, we observed that the Spartan/Rad18-silenced cells did not exhibit higher FA sensitivity than the Spartan-silenced or the Rad18-silenced cells, indicating that Spartan and Rad18 act together in protecting cells from FA-induced damage. To further confirm the epistatic relationship between SPARTAN and RAD18, we tested whether ectopic expression of the siRNA-resistant Spartan or Rad18 proteins can rescue the observed phenotype. While the expression of Spartan in Spartan-silenced cells and the expression of Rad18 in Rad18-silenced cells could fully rescue FA sensitivity, the expression of either Spartan or Rad18 alone could not rescue the sensitivity of the Spartan/Rad18 double-silenced cells. However, expression of Spartan and Rad18 together readily rescued the sensitivity of Spartan/Rad18-silenced cells, supporting further that their cooperation is required to achieve DPC resistance (Figure 5A).
Figure 5.
Spartan and Rad18 act together in protecting genome integrity from FA-induced genotoxic stress. (A) Epistasis analysis of SPARTAN and RAD18 using cell viability assay for FA treatment. HEK 293 cells stably expressing shRNAs of SPARTAN, RAD18 and RAD18/SPARTAN or control were transfected with empty vector or N-terminally Flag-tagged SPARTAN, RAD18 or SPARTAN-RAD18 siRNA-resistant plasmid constructs. After 48 h, cells were treated with FA, and after 7 days of treatment cell viability was determined. (B) SPARTAN and RAD18 show epistatic relationship in DPC-removal. Representative images of the epistasis analysis of SPARTAN and RAD18 performed with the BrdU comet assay. The indicated cell lines were treated with 500 μM FA. (C) Quantitation of the experiment shown in (B). Statistical analysis was made by Student's t-test. Statistical significance was labelled by ***: P < 0.001, NS: not significant. (D) SPARTAN and RAD18 show epistatic relationship in the replication of DPC-containing DNA. Representative images of fibres from Spartan, Rad18 and Rad18/Spartan shRNA-silenced cells treated with FA or left untreated. (E) Quantitative measurements of second track shortening after FA treatment of different depleted cell lines, detected in the fibre assay detailed in (D), are illustrated in a dot plot diagram. Each dot represents a single measurement of the second label of a single ongoing replication fork. Three independent experiments were carried out, and at least 100 fibres per experiment per cell line were measured. Horizontal lines indicate mean values. (F) Relative replication fork speed was calculated from the quantitative measurements of the fibre assay detailed in (D) for Spartan-, Rad18- and Spartan/Rad18-depleted cells.
We also employed the comet assay to reveal the functional interplay of Spartan and Rad18. We found that the DPC-repair deficiency of the shSPARTAN/shRAD18 double-silenced cell line was not higher than that of the shRAD18 cell line, indicating again the requirement for their interaction (Figure 5B, C and Supplementary Figure S3C). Interestingly, the deficiency of shRAD18 was higher than that of shSPARTAN, suggesting that elimination of the RAD6-RAD18 pathway affects not only the Spartan-dependent DPC-repair pathway but other, yet to be identified DPC-repair pathways as well.
Finally, we tested for interaction between Spartan and Rad18 in the immediate bypass of FA-induced lesions via the DNA fibre assay (Figure 5D), dot-plotting the results (Figure 5E) where every dot represents a single fibre. We also calculated the mean of replication track lengths under unchallenged conditions as well as after FA exposure (shown by horizontal blue lines in Figure 5E). Comparison revealed a slower replication fork movement after FA treatment in the shSPARTAN, shRAD18 and shSPARTAN/shRAD18 cells than in the control shCTRL cells. Importantly, fork slowdown was not additive when Spartan and Rad18 silencing was combined in shSPARTAN/shRAD18 cells, indicating that Rad18 and Spartan function together in lesion bypass of FA-induced DNA damage (Figure 5F and Supplementary Figure S3D). Taken together, all three independent methods confirmed an epistatic relationship between SPARTAN and RAD18 in providing cellular resistance against DPC.
DISCUSSION
DPCs are toxic lesions that are heterogeneous in size and origin and interfere with most of the enzymatic processes that take place on the DNA such as replication and transcription. For a long time, the repair of DPCs was believed to be carried out only by canonical repair pathways such as nucleotide excision repair, the Fanconi anaemia pathway and homologous recombination (9,53). In contrast to this general view, a so far hidden DNA repair mechanism has been recognized recently in yeast with the finding that yeast Wss1 has a protease activity directly eliminating the protein components of DPCs (32), and a replication-dependent proteolysis mechanism acting on DPC, which was recapitulated using Xenopus egg extract in vitro (54), has also been described. In our study, we investigated whether human cells exhibit similar repair mechanisms. The presence of a metalloprotease-like domain in Spartan and its domain structure suggested some similarity to yeast Wss1; however, the questions whether Spartan has a protease activity, whether it facilitates DPC repair, and whether a yet unrecognized direct mechanism exists in human cells for DPC repair have not been addressed.
Here, we show that human Spartan is required for facilitating DPC-repair and replication of DPC-containing DNA and that it acts together with the RAD6-RAD18 DDT pathway. We are the first to report that purified Spartan has a DNA-dependent protease activity degrading certain proteins bound to DNA. In concert, our in vivo findings reveal that Spartan deficiency causes a slower DPC removal and that Spartan's protease- or DNA-binding deficiency impairs highly the DPC-repair of replicating cells as detected by a DPC-specific alkaline BrdU comet assay. Moreover, we demonstrate, employing the DNA fibre method, that Spartan deficiency dramatically decreases the speed of replication forks under FA-induced replication stress, indicating a role for Spartan in the immediate replication of DPC derivatives. Our results are fully consistent with the findings on the protease activity of Spartan and its role in DPC-repair published very recently at the final stage of the revision of our paper (55–57). In addition to these results, we provide genetic evidence for the interaction of SPARTAN with the RAD6-RAD18 DDT pathway in DPC repair.
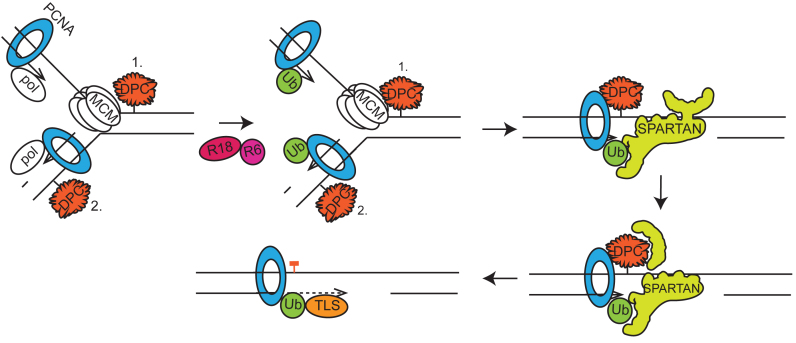
Based on our results, we propose a model shown in Figure 6 for the function of the protease Spartan in the completion of the replication of DPC-containing DNA. We suggest that when replication stalls upon encountering a DPC, Spartan can gain access to the fork, where its newly discovered DNA-dependent protease activity can remove the protein part of the DPC thereby facilitating the completion of replication. If the protein was bound covalently to the DNA, a short peptide derivative remains on the DNA the subsequent bypass of which may require translesion polymerase or template switching. Previous studies suggested that Spartan can facilitate the recruitment of the translesion synthesis polymerase η (21,22,24,48) to the stalled fork, thus, it is possible that Spartan can perform multiple tasks at DPCs and, in addition to the digestion of DPCs it can facilitate the exchange of the replicative polymerase to a damage bypass player. In doing so, the Spartan protease may remove some components of the replication machinery such as the catalytic subunit of the replicative polymerase thus providing access for a TLS polymerase to the remaining subunits of the replication machinery containing ubiquitin-PCNA. Previous studies also indicate that Spartan interacts with and recruits the ubiquitin-selective chaperone p97 to the blocked replication fork (25,26); thus, it would be interesting to study whether p97 is required for DPC-repair, and, if so, whether it acts together with Spartan in the proteolytic step by restructuring the substrates for proteolytic cleavage and removing the proteolytic products.
Figure 6.
Model for the role of the Spartan protease in the replication of DPC-containing DNA. Stalling of the helicase on the leading strand and the DNA polymerases (pol) on both the leading (1.) and the lagging (2.) strands due to DPC adducts activates the Rad6-Rad18 protein complex (R18-R6), which marks the stalled replication complex by PCNA monoubiquitylation (Ub). In both cases, the replication fork can be converted to a gapped form, either by the convergence of the opposing replication forks in the case of the leading strand or by leaving unreplicated regions opposite the DPC between Okazaki fragments. Often, DPCs are still impassable even for TLS DNA polymerases due to their size and strong, non-displaceable DNA binding. Next, the stalled monoubiquitylated PCNA and the exposed ssDNA attract Spartan, which simultaneously binds Ub-PCNA through its UBZ and PIP domains and ssDNA through its DNA-binding domain. The binding of ssDNA results in the conformational change of Spartan, which in turn activates Spartan's protease activity. The protease Spartan can cleave the DPC leaving free DNA or, if the protein was covalently bound, a short peptide bound to the DNA. Spartan's activity may also facilitate polymerase exchange somehow, for example, by removing some subunits of the replication complex and thus giving access to a TLS polymerase, which can then bypass the remaining short peptide-DNA crosslink adduct. These events might happen immediately when the fork stalls at a DPC or occur only after the majority of the DNA has replicated during the so-called post-replicative repair process. Since the replicative helicase might be inhibited only on the leading strand by a DPC, on the lagging strand the proteolytic cleavage of the DPC by Spartan may leave short peptides bound to the DNA that are accessible to immediate bypass by TLS polymerases and can be investigated by the DNA fibre assay.
Timely activation and targeting of the protease activity of Spartan must be particularly important since it should remove only DPCs or particular components of the replication machinery but keep the various protein components of the replication machinery intact so that they are able to resume replication after bypass. Our biochemical studies provided a few hints when we tested certain replication complex components as potential substrates and found that Spartan did not cleave PCNA, ubiquitin-PCNA, RFC and RPA, rather, RFC and RPA inhibited the protease activity of Spartan, most probably by outcompeting its binding to the DNA. Thus, successful competition for ssDNA binding might be a rate limiting step for Spartan activation, and one can envision that certain Spartan-interacting proteins such as ubiquitin–PCNA or the p97 chaperone may facilitate its successful competition with other ssDNA-binding proteins. Some of the current models suggest that the exposed ssDNA at stalled replication forks becomes rapidly covered by RPA (58), signalling for Rad18-dependent PCNA ubiquitylation, which then provides a binding surface for Spartan recruitment. Thus, it is possible that Spartan becomes activated upon binding to ubiquitin-PCNA by its PIP and UBZ domains, in the vicinity of which—at the stalled replication fork—protein free ssDNA becomes available, and Spartan cleaves only the DPCs neighbouring these ssDNA regions.
DPCs can be formed during normal enzymatic reactions involving transient covalent linkage between the enzyme and the DNA such as when topoisomerase 1 nicks one strand of the DNA to relieve DNA torsional stress, and for some of these special DPCs a dedicated repair pathway exist; e.g. DNA-trapped topoisomerase 1 is repaired by TDP1 cleaving the protein–DNA binding (59). In addition, reactive agents like aldehydes, UV, certain metal ions, and radiation can potentially crosslink any protein operating on the DNA (60); the repair of these requires a more general repair mechanism. Besides exogenous exposure, many agents are generated endogenously, in particular FA during histone demethylation (34). The importance of endogenously produced aldehyde was revealed by the finding that in mice the lack of aldehyde-detoxifying enzymes causes anaemia and leukaemia when the Fanconi anaemia pathway is not functional (61). In human blood, the normal endogenous FA concentration is as high as 100 μM (36). Furthermore, FA concentration in the metabolically active liver of rats was reported to be 2- to 4-fold higher than that found in the blood (62). Thus, DPC repair can be particularly important in liver, which might explain why the familial SPARTAN mutation specifically leads to early onset hepatocellular carcinoma. In addition, SPARTAN mutant patients were also affected by a progeroid syndrome characterized by genomic instability (29). Spartan insufficiency in transgenic mice also causes chromosomal instability and early onset of age-related phenotypes (28). It is of particular importance that mutations of the active site of the metalloprotease SprT domain of mouse Spartan impaired both lesion bypass and cell viability while the phenotypes of mutations in other domains of Spartan, such as in the SHP-domain mediating its interaction with the p97 chaperone, were not so prominent (28). In accordance with these findings, our complementation assay performed by monitoring FA-induced DPC-repair and using Spartan proteins mutant in the active site of the metalloprotease domain or in the DNA-binding site suggested a role for the protease activity of Spartan in the completion of replication. Also, similarly to the observations in mice, we found that Spartan-deficient human cells leave the S phase containing abnormal replication intermediates such as chromatin bridges, reflecting unresolved replication, which may explain Spartan deficiency-caused genomic instability. These findings are in good agreement with the clastogenic effect of FA reported formerly; SCE elevation with simultaneous micronucleus formation parallel to DPC induction was observed (43).
The recently described DNA-dependent metalloprotease Wss1 represents a DPC-specific pathway that is independent from NER and homologous recombination, as detected by epistasis analysis using rad4Δ NER-deficient and rad52Δ homologous recombination-deficient strains. Based on our results, it would be interesting to explore the epistatic relationship between WSS1 and RAD18 and Ubiquitin-PCNA as well. This might require a more specific phenotype than cell survival, with a more specialized assay that is able to visualize events at the stalled replication fork such as the DNA fibre analysis or the BrdU comet assay we employed.
We anticipate our results on the protease Spartan to be a starting point for more sophisticated assays on the existence of a direct DPC repair pathway in human cells. For example, the involvement and the functions of Spartan-interacting partners such as ubiquitylated-PCNA, Rad18 and chaperon p97 in DPC repair and the connection of this direct repair pathway with other repair pathways operating in the S phase, in particular homologous recombination, can now be tested. Also, it opens the way to in vitro assays aiming to reconstitute DPC repair with site-specific covalent DPC substrates and purified proteins and to examining specific elementary steps such as Spartan activation on RPA-covered DNA, the modifying effect of ubiquitin–PCNA binding, how Spartan is kept under control to avoid unwanted proteolysis, and Spartan-mediated exchange of replicative and translesion synthesis polymerases at DPCs and their derivatives. Finally, it will be interesting to explore the level to which the DPC repair function of Spartan contributes to protection against hepatocellular carcinoma and progeroid diseases.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Gabriella Tick for help in preparing the manuscript and Katalin Kovacs and Anita Németh for technical assistance.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Research, Development and Innovation Office [GINOP-2.3.2-15-2016-00020 and GINOP-2.3.2-15-2016-00001]; Scientific Excellence Cooperation Grant of the Hungarian Academy of Sciences [2016/2017]. Funding for open access charge: National Research, Development and Innovation Office [GINOP-2.3.2-15-2016-00020].
Conflict of interest statement. None declared.
REFERENCES
- 1. Negrini S., Gorgoulis V.G., Halazonetis T.D.. Genomic instability–an evolving hallmark of cancer. Nat. Rev. Mol. Cell Biol. 2010; 11:220–228. [DOI] [PubMed] [Google Scholar]
- 2. Vijg J., Suh Y.. Genome instability and aging. Annu. Rev. Physiol. 2013; 75:645–668. [DOI] [PubMed] [Google Scholar]
- 3. Lenart P., Krejci L.. DNA, the central molecule of aging. Mutat. Res. 2016; 786:1–7. [DOI] [PubMed] [Google Scholar]
- 4. Cohen I.S., Bar C., Paz-Elizur T., Ainbinder E., Leopold K., de Wind N., Geacintov N., Livneh Z.. DNA lesion identity drives choice of damage tolerance pathway in murine cell chromosomes. Nucleic Acids Res. 2015; 43:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Izhar L., Ziv O., Cohen I.S., Geacintov N.E., Livneh Z.. Genomic assay reveals tolerance of DNA damage by both translesion DNA synthesis and homology-dependent repair in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:E1462–E1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Torres-Ramos C.A., Prakash S., Prakash L.. Requirement of RAD5 and MMS2 for postreplication repair of UV-damaged DNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 2002; 22:2419–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sale J.E. Translesion DNA synthesis and mutagenesis Eukaryotes. Cold Spring Harb Perspect Biol. 2013; 5:a012708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang A.T., Sengerová B., Cattell E., Inagawa T., Hartley J.M., Kiakos K., Burgess-Brown N.A., Swift L.P., Enzlin J.H., Schofield C.J. et al. Human SNM1a and XPF-ERCC1 collaborate to initiate DNA interstrand cross-link repair. Genes Dev. 2011; 25:1859–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ide H., Nakano T., Shoulkamy M.I., Salem A.M.H.. Chen C. Formation, Repair, and Biological Effects of DNA–Protein Cross-Link Damage, Advances in DNA Repair. InTech; DOI: 10.5772/59683. Available from:http://www.intechopen.com/books/advances-in-dna-repair/formation-repair-and-biological-effects-of-dna-protein-cross-link-damage. [Google Scholar]
- 10. de Graaf B., Clore A., McCullough A.K.. Cellular pathways for DNA repair and damage tolerance of formaldehyde-induced DNA-protein crosslinks. DNA Repair (Amst). 2009; 8:1207–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bailly V., Lauder S., Prakash S., Prakash L.. Yeast DNA repair proteins Rad6 and Rad18 form a heterodimer that has ubiquitin conjugating, DNA binding, and ATP hydrolytic activities. J. Biol. Chem. 1997; 272:23360–23365. [DOI] [PubMed] [Google Scholar]
- 12. Yoon J.-H., Prakash S., Prakash L.. Requirement of Rad18 protein for replication through DNA lesions in mouse and human cells. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:7799–7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gangavarapu V., Haracska L., Unk I., Johnson R.E., Prakash S., Prakash L.. Mms2-Ubc13-dependent and -independent roles of Rad5 ubiquitin ligase in postreplication repair and translesion DNA synthesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 2006; 26:7783–7790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Blastyák A., Pintér L., Unk I., Prakash L., Prakash S., Haracska L.. Yeast Rad5 protein required for postreplication repair has a DNA helicase activity specific for replication fork regression. Mol. Cell. 2007; 28:167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xue X., Choi K., Bonner J., Chiba T., Kwon Y., Xu Y., Sanchez H., Wyman C., Niu H., Zhao X. et al. Restriction of replication fork regression activities by a conserved SMC complex. Mol. Cell. 2014; 56:436–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Burkovics P., Sebesta M., Balogh D., Haracska L., Krejci L.. Strand invasion by HLTF as a mechanism for template switch in fork rescue. Nucleic Acids Res. 2014; 42:1711–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Unk I., Hajdú I., Blastyák A., Haracska L.. Role of yeast Rad5 and its human orthologs, HLTF and SHPRH in DNA damage tolerance. DNA Repair (Amst). 2010; 9:257–267. [DOI] [PubMed] [Google Scholar]
- 18. Unk I., Hajdú I., Fátyol K., Hurwitz J., Yoon J.-H., Prakash L., Prakash S., Haracska L.. Human HLTF functions as a ubiquitin ligase for proliferating cell nuclear antigen polyubiquitination. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:3768–3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Smirnova M., Klein H.L.. Role of the error-free damage bypass postreplication repair pathway in the maintenance of genomic stability. Mutat. Res. 2003; 532:117–135. [DOI] [PubMed] [Google Scholar]
- 20. Diamant N., Hendel A., Vered I., Carell T., Reißner T., De Wind N., Geacinov N., Livneh Z.. DNA damage bypass operates in the S and G2 phases of the cell cycle and exhibits differential mutagenicity. Nucleic Acids Res. 2012; 40:170–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Juhasz S., Balogh D., Hajdu I., Burkovics P., Villamil M.A., Zhuang Z., Haracska L.. Characterization of human Spartan/C1orf124, an ubiquitin-PCNA interacting regulator of DNA damage tolerance. Nucleic Acids Res. 2012; 40:10795–10808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Centore R.C., Yazinski S.A., Tse A., Zou L.. Spartan/C1orf124, a Reader of PCNA Ubiquitylation and a Regulator of UV-Induced DNA Damage Response. Mol. Cell. 2012; 46:625–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Machida Y., Kim M.S., Machida Y.J.. Spartan/C1orf124 is important to prevent UV-induced mutagenesis. Cell Cycle. 2012; 11:3395–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ghosal G., Leung J.W.C., Nair B.C., Fong K.W., Chen J.. Proliferating Cell Nuclear Antigen (PCNA)-binding protein C1orf124 is a regulator of translesion synthesis. J. Biol. Chem. 2012; 287:34225–34233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Davis E.J., Lachaud C., Appleton P., Macartney T.J., Näthke I., Rouse J.. DVC1 (C1orf124) recruits the p97 protein segregase to sites of DNA damage. Nat. Struct. Mol. Biol. 2012; 19:1093–1100. [DOI] [PubMed] [Google Scholar]
- 26. Mosbech A., Gibbs-Seymour I., Kagias K., Thorslund T., Beli P., Povlsen L., Nielsen S.V., Smedegaard S., Sedgwick G., Lukas C. et al. DVC1 (C1orf124) is a DNA damage–targeting p97 adaptor that promotes ubiquitin-dependent responses to replication blocks. Nat. Struct. Mol. Biol. 2012; 19:1084–1092. [DOI] [PubMed] [Google Scholar]
- 27. Kim M.S., Machida Y., Vashisht A.A., Wohlschlegel J.A., Pang Y.P., Machida Y.J.. Regulation of error-prone translesion synthesis by Spartan/C1orf124. Nucleic Acids Res. 2013; 41:1661–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Maskey R.S., Kim M.S., Baker D.J., Childs B., Malureanu L.A., Jeganathan K.B., Machida Y., van Deursen J.M., Machida Y.J.. Spartan deficiency causes genomicinstability and progeroid phenotypes. Nat. Commun. 2014; 5:5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lessel D., Vaz B., Halder S., Lockhart P.J., Marinovic-Terzic I., Lopez-Mosqueda J., Philipp M., Sim J.C.H., Smith K.R., Oehler J. et al. Mutations in SPRTN cause early onset hepatocellular carcinoma, genomic instability and progeroid features. Nat. Genet. 2014; 46:1239–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ruijs M.W.G., van Andel R.N.J., Oshima J., Madan K., Nieuwint A.W.M., Aalfs C.M.. Atypical progeroid syndrome: An unknown helicase gene defect?. Am. J. Med. Genet. 2003; 116A:295–299. [DOI] [PubMed] [Google Scholar]
- 31. Stingele J., Habermann B., Jentsch S.. DNA-protein crosslink repair: proteases as DNA repair enzymes. Trends Biochem. Sci. 2015; 40:67–71. [DOI] [PubMed] [Google Scholar]
- 32. Stingele J., Schwarz M.S., Bloemeke N., Wolf P.G., Jentsch S.. A DNA-dependent protease involved in DNA-protein crosslink repair. Cell. 2014; 158:327–338. [DOI] [PubMed] [Google Scholar]
- 33. Stingele J., Jentsch S.. DNA–protein crosslink repair. Nat. Rev. Mol. Cell Biol. 2015; 16:455–460. [DOI] [PubMed] [Google Scholar]
- 34. Shi Y., Lan F., Matson C., Mulligan P., Whetstine J.R., Cole P.A., Casero R.A., Shi Y.. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1 77 avenue louis pasteur. 2004; 119:941–953. [DOI] [PubMed] [Google Scholar]
- 35. Baker D.J., Wuenschell G., Xia L., Termini J., Bates S.E., Riggs A.D., O'Connor T.R.. Nucleotide excision repair eliminates unique DNA-protein cross-links from mammalian cells. J. Biol. Chem. 2007; 282:22592–22604. [DOI] [PubMed] [Google Scholar]
- 36. Swenberg J.A., Lu K., Moeller B.C., Gao L., Upton P.B., Nakamura J., Starr T.B.. Endogenous versus exogenous DNA adducts: Their role in carcinogenesis, epidemiology, and risk assessment. Toxicol. Sci. 2011; 120:130–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Beane Freeman L.E., Blair A., Lubin J.H., Stewart P.A., Hayes R.B., Hoover R.N., Hauptmann M.. Mortality from lymphohematopoietic malignancies among workers in formaldehyde industries: the national cancer institute cohort. J. Natl. Cancer Inst. 2009; 101:751–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nakano T., Morishita S., Katafuchi A., Matsubara M., Horikawa Y., Terato H., Salem A.M.H., Izumi S., Pack S.P., Makino K. et al. Nucleotide Excision Repair and Homologous Recombination Systems Commit Differentially to the Repair of DNA-Protein Crosslinks. Mol. Cell. 2007; 28:147–158. [DOI] [PubMed] [Google Scholar]
- 39. Han J., Liu T., Huen M.S.Y., Hu L., Chen Z., Huang J.. SIVA1 directs the E3 ubiquitin ligase RAD18 for PCNA monoubiquitination. J. Cell Biol. 2014; 205:811–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Costa M., Zhitkovich A.V.. Assay for detectingcovalent DNA-protein complexes. US patent US5545529.
- 41. Mórocz M., Gali H., Raskó I., Downes C.S., Haracska L.. Single cell analysis of human RAD18-dependent DNA post-replication repair by alkaline bromodeoxyuridine comet assay. PLoS One. 2013; 8:e70391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Merk O., Speit G.. Detection of crosslinks with the comet assay in relationship to genotoxicity and cytotoxicity. Environ. Mol. Mutagen. 1999; 33:167–172. [DOI] [PubMed] [Google Scholar]
- 43. Merk O., Speit G.. Significance of formaldehyde-induced DNA-protein crosslinks for mutagenesis. Environ. Mol. Mutagen. 1998; 32:260–268. [DOI] [PubMed] [Google Scholar]
- 44. Wojewódzka M., Buraczewska I., Kruszewski M.. A modified neutral comet assay: elimination of lysis at high temperature and validation of the assay with anti-single-stranded DNA antibody. Mutat. Res. 2002; 518:9–20. [DOI] [PubMed] [Google Scholar]
- 45. Collins A.R. The comet assay for DNA damage and repair: principles, applications, and limitations. Mol. Biotechnol. 2004; 26:249–261. [DOI] [PubMed] [Google Scholar]
- 46. Speit G., Schütz P., Högel J., Schmid O.. Characterization of the genotoxic potential of formaldehyde in V79 cells. Mutagenesis. 2007; 22:387–394. [DOI] [PubMed] [Google Scholar]
- 47. Lundin C., North M., Erixon K., Walters K., Jenssen D., Goldman A.S.H., Helleday T.. Methyl methanesulfonate (MMS) produces heat-labile DNA damage but no detectable in vivo DNA double-strand breaks. Nucleic Acids Res. 2005; 33:3799–3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Toth A., Hegedus L., Juhasz S., Haracska L., Burkovics P.. The DNA-binding box of human SPARTAN contributes to the targeting of Polη to DNA damage sites. DNA Repair (Amst). 2016; pii: S1568-7864(16)30171-9. doi:10.1016/j.dnarep.2016.10.007. [Epub ahead of print] PubMed PMID: 27838458. [DOI] [PubMed] [Google Scholar]
- 49. Jansen J.G., Tsaalbi-Shtylik A., Hendriks G., Gali H., Hendel A., Johansson F., Erixon K., Livneh Z., Mullenders L.H.F., Haracska L. et al. Separate domains of Rev1 mediate two modes of DNA damage bypass in mammalian cells. Mol. Cell. Biol. 2009; 29:3113–3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kanagaraj R., Huehn D., MacKellar A., Menigatti M., Zheng L., Urban V., Shevelev I., Greenleaf A.L., Janscak P.. RECQ5 helicase associates with the C-terminal repeat domain of RNA polymerase II during productive elongation phase of transcription. Nucleic Acids Res. 2010; 38:8131–8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kumari A., Owen N., Juarez E., McCullough A.K.. BLM protein mitigates formaldehyde-induced genomic instability. DNA Repair (Amst). 2015; 28:73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kumari A., Lim Y.X., Newell A.H., Olson S.B., McCullough A.K.. Formaldehyde-induced genome instability is suppressed by an XPF-dependent pathway. DNA Repair (Amst). 2012; 11:236–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rosado I.V., Langevin F., Crossan G.P., Takata M., Patel K.J.. Formaldehyde catabolism is essential in cells deficient for the Fanconi anemia DNA-repair pathway. Nat. Struct. Mol. Biol. 2011; 18:1432–1434. [DOI] [PubMed] [Google Scholar]
- 54. Duxin J.P., Dewar J.M., Yardimci H., Walter J.C.. Repair of a DNA-protein crosslink by replication-coupled proteolysis. Cell. 2014; 159:349–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lopez-Mosqueda J., Maddi K., Prgomet S., Kalayil S., Marinovic-Terzic I., Terzic J., Dikic I.. SPRTN is a mammalian DNA-binding metalloprotease that resolves DNA-protein crosslinks. Elife. 2016; 5:5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Stingele J., Bellelli R., Alte F., Hewitt G., Sarek G., Maslen S.L., Tsutakawa S.E., Borg A., Kjær S., Tainer J.A. et al. Mechanism and regulation of DNA-Protein crosslink repair by the DNA-dependent metalloprotease SPRTN. Mol. Cell. 2016; 64:688–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Vaz B., Popovic M., Newman J.A., Fielden J., Aitkenhead H., Halder S., Singh A.N., Vendrell I., Fischer R., Torrecilla I. et al. Metalloprotease SPRTN/DVC1 orchestrates replication-coupled DNA-Protein crosslink repair. Mol. Cell. 2016; 0:137–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Davies A.A., Huttner D., Daigaku Y., Chen S., Ulrich H.D.. Activation of Ubiquitin-Dependent DNA Damage Bypass Is Mediated by Replication Protein A. Mol. Cell. 2008; 29:625–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Desai S.D., Liu L.F., Vazquez-abad D., Arpa P.D.. Ubiquitin-dependent destruction of Topoisomerase I is stimulated by the antitumor drug camptothecin. J. Biol. Chem. 1997; 272:24159–24164. [DOI] [PubMed] [Google Scholar]
- 60. Barker S., Weinfeld M., Murray D.. DNA-protein crosslinks: Their induction, repair and biological consequences. Mutat. Res. 2005; 589:111–135. [DOI] [PubMed] [Google Scholar]
- 61. Garaycoechea J.I., Crossan G.P., Langevin F., Daly M., Arends M.J., Patel K.J.. Genotoxic consequences of endogenous aldehydes on mouse haematopoietic stem cell function. Nature. 2012; 489:571–575. [DOI] [PubMed] [Google Scholar]
- 62. Heck H.D.A., Casanova M.. The implausibility of leukemia induction by formaldehyde: a critical review of the biological evidence on distant-site toxicity. Regul. Toxicol. Pharmacol. 2004; 40:92–106. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.