Abstract
Efficient non-viral gene delivery is highly desirable but often unattainable with some cell-types. We report here that non-viral DNA polyplexes can efficiently transfect differentiated neuronal and stem cells. Polyplex transfection centrifugation protocols was enhanced by including a simultaneous treatment with a DOPE/CHEMS lipid suspension and a microtubule inhibitor, Tubastatin A. Lipoplex transfection protocols were not improved by this treatment. This mechanism of action was unravelled by systematically identifying and rationally mitigating barriers limiting high transfection efficiency, allowing unexpected improvements in the transfection of mesenchymal stem cells (MSC), primary neuron and several hard-to-transfect cell types beyond what are currently achievable using cationic polymers. The optimized formulation and method achieved high transfection efficiency with no adverse effects on cell viability, cell proliferation or differentiation. High efficiency modification of MSC for cytokine overexpression, efficient generation of dopaminergic neuron using neural stem cells and enhanced genome editing with CRISPR-Cas9 were demonstrated. In summary, this study described a cost-effective method for efficient, rapid and scalable workflow for ex vivo gene delivery using a myriad of nucleic acids including plasmid DNA, mRNA, siRNA and shRNA.
INTRODUCTION
Gene delivery is extremely useful for investigating and manipulating cellular processes. Numerous preclinical and clinical studies have now shown that ex vivo genetic modification of human cells significantly improved their therapeutic potentials (1,2). To date, the vast majority of academic and clinical labs has exploited viral vectors as efficient nucleic acid delivery vehicles both in vitro and in vivo (3). While virus mediated gene delivery is highly efficient, the major drawback is the random integration of virus vector into the host genome, which may interrupt essential gene expression and cellular processes (3). The preparation procedure is both labor intensive and technically demanding, thus pose a challenge to scale up with increasing number of transgenes. For these reasons, much efforts have been made to develop non-viral transfection methods. Many cell lines can be transfected at relatively high efficiency with cationic polymers, but stem cells (1,4–6) and post-mitotic cells (7,8) are known to be recalcitrant (0–35% transfection efficiency).
Recent efforts to improve transfection of hard-to-transfect cell types by optimizing protocols using cationic polymer have met with limited success (9). Attempts have been made to identify the underlying mechanisms limiting efficient transfection in post-mitotic cells. A prevailing idea of why post-mitotic, differentiated cells including neuronal cells are difficult to transfect using non-viral polymer complexed with nucleic acids (polyplex) is presumed to be due to the inability of the nucleic acids to be internalized (10). It is believed that the lack of nuclear membrane breakdown in non-dividing cells is another important reason for poor transfection efficiency (11). However, even at high rate of cell division, the efficiencies of polymer based transfection of stem cells are typically poor (5,12). The low efficiency of polymer based method has led to the adoption of electroporation as a gene delivery method (4,6). While high transfection can be achieved with electroporation, a major drawback is the low cell viability post-transfection and the issue of scalability (6,12). Other physical methods including, microinjection, gene gun, electroporation, sonoporation, laser (2) and cell deformation (13,14) are attractive alternatives but require specialized setups.
To date, the goal of attaining high transfection efficiency in hard-to-transfect cell types using non-viral carriers remain elusive and efforts to produce even more novel polymers to enhance transfection continues (5,8,15). Here, we describe the development of a formulation and protocol using cationic polymers to efficiently transfect a variety of hard-to-transfect cell types. We speculated that by temporally re-configuring the intracellular trafficking of the genetic cargo from early endosomal compartment and stabilizing the microtubule network simultaneously may result in significant enhancement of transfection with DNA polyplexes. This study also provides useful insights into the rational design of scalable approaches for high efficiency of non-viral gene transfection using off-the-shelf cationic polymers.
MATERIALS AND METHODS
Cell culture
Neuro2A (ATCC: CCL-131TM) stably expressed GFRα2a, A375 (ATCC, CRL-1619) and MDA-MB-231 cell line (ATCC, HTB-26) were cultured and maintained following manufacturer's instructions. To generate differentiated cell lines, Neuro2A cells were differentiated with 50 ng/ml glial cell-line derived neurotrophic factor, GDNF (Biosource, Camarillo, CA, USA), or 10 μM all trans retinoic acid, RA (Sigma, St. Louis, MO, USA) in DMEM supplemented with 1% FBS for 48 h prior transfection. Rat primary cortical neurons were isolated and maintained in Neurobasal media supplemented with B-27 (Invitrogen).Non-neuronal cells comprise <0.5% of the cell population of neurons. On DIV 3 (3 days in vitro), primary neurons were transfected. Human adult bone marrow mesenchymal stem cells (RoosterBio) and NSF-60 cell line (400301-SF, Cell Line Service) were cultured and maintained according to manufacturer's instruction. Neural stem cells were derived from embryonic stem cells as described by Zhang et al. (16). Human dermal fibroblast was purchased from Cell Applications and cultured as instructed.
Transfection procedure
Plasmid DNA expressing EGFP (5600 bp pIRES-EGFP-EV71, pDNA) was purified according to manufacturer's instruction (Geneaid Biotech, Taiwan). For each well (24-well plate format), 1 mg/ml of LPEI (25-kDa; Polyscience, USA) was added to pDNA in 25 mM HEPES buffer at different N/P ratios and incubated at room temperature for 15 min. The final volume of the DNA complex is 50 μl. The N/P ratio (molar ratio of PEI nitrogen to pDNA phosphate) was calculated based on the PEI content and size of plasmid. The positive charge of PEI is due the nitrogen groups present in NHCH2CH2 (43 g/mol). The negative charge of pDNA is due to the presence of phosphate group in the deoxyribose nucleotide. The average molecular weight of the nucleotides is assumed to be 330 g/mol (17). LPEI/pDNA complex was then added to complete medium (1:10) to prepare the transfection mixture. The culture media was removed and replaced with the transfection mixture, with or without centrifugation. Cells were incubated for 48 h before analysis. For transfection of differentiated cells, transfection mixture was replaced with DMEM (1% FBS) containing corresponding differentiation reagent. To improve transfection in differentiated neurons, DOPE/CHEMS and Tubastatin A were used. Lipid film comprising DOPE/CHEMS (9:2 molar ratio) (Polar Avanti Lipid, Alabaster), was formed at the bottom of glass tube after evaporation of the solvent, chloroform. The lipid film was reconstituted in 25 mM HEPES buffer and sonicated for 2 min in a bath-type sonicator (Brandson 2200). The lipid solution was added to the cell culture at various periods of time post-transfection. Tubastatin A was purchased from Bio Vision, SF, USA. One hour post-transfection, Tubastatin A (5 or 16 μM) was added to the culture media. The inhibitor containing media were replaced by fresh media 24 h later. Transfection efficiency (percentage of EGFP positive cells) was quantified either through cell counting with ImageJ (http://rsb.info.nih.gov/ij/) or FACS analysis after incubation for indicated periods.
Flow cytometry analysis
After transfection, the cells were trypsinized, centrifuged and re-suspended in PBS. Cell clumps were removed by filtering through 40 μm mesh. The percentage of cells expressing EGFP was quantified by FACS (BD FACSCanto, BD Biosciences) and the raw data was analyzed using WinMDI (V2.9). At least 10 000 cells were analyzed per sample.
Imaging studies
FITC- and Rho-pDNA were prepared according to manufacturer recommendation (Mirus Bio, USA). Expression of EGFP was observed in cells transfected with fluorescently labeled pDNA. FITC- or Rho-pDNA was used for visualization of internalized polyplex. Four hour post-transfection, the extracellular fluorescence of FITC or Rho-pDNA was quenched with EtBr (20 μg/ml) or 0.4% trypan blue respectively. Cell images were taken before and after quenching with an inverted Zeiss microscrope equipped with fluorescence detection (Zeiss cell observer Z1). The images were processed with Axio Vision Rel. 4.7. Quenching efficiency of trypan blue was examined. To study co-localization of Rho-pDNA and the acidic compartment, cells were incubated with lysotracker green DND-26 (50 nM) for 5 min. Images were captured with a Zeiss confocal microscopy (LSM710, Oberkochen, Germany) or EVOS FL cell imaging system (ThermoScientific). Colocalized pixel was analyzed with Zeiss ZEN software (v2010).
Human GCSF modification of mesenchymal stem cell
One day prior to transfection, 60 000 human BM MSC were seeded in the tissue culture plate (24-well format). Cells were modified with complexes of Turbofect (ThermoScientific) with 200 ng Human Granulocyte Colony Stimulating Factor (GCSF) expression vector (SC306744, Origene) or 500 ng mRNA (Trilink). Following centrifugation, the transfection mixtures were replaced with fresh media in the presence or absence of DOPE/CHEMS and 10 μM Tubastatin A. The non-modified and modified MSC was then subjected to the following analysis:
Western blot: One day post-transfection, cells were harvested and lysed with lysis buffer (150 mM sodium chloride, 1.0% NP-40, 50 mM Tris pH 8.0) supplemented with protease inhibitor cocktail (Roche). Twenty microgram of the whole cell lysate were resolved on 8% polyacrylamide sodium dodecyl sulfate gels and analyzed by means of immunoblotting with goat anti-human GCSF (AF-214-NA, R&D systems) and mouse anti α-tubulin, respectively.
Elisa: GCSF levels in the conditioned media were determined with the use of the Human GCSF Quantikine ELISA Kit (SCS50, R&D Systems). To ensure that the modification of MSC did not affect the expression of other cytokine, VEGF levels of the conditioned media of non-modified and modified MSC were compared. The VEGF levels were measured by the Human VEGF PicoKine ELISA Kit (MyBiosource).
Proliferation assay: The bioactivity of GCSF secreted by MSC was assayed based on the proliferative activity of GCSF-dependent NFS60 cells. NSF60 cells were washed twice with sterile PBS and then resuspended in serum free RPMI 1640 medium. The conditioned media of non-modified and modified MSC was added to the NSF60 culture (100× dilution of the conditioned media). After 3 days incubation, the cell number of NSF-60 was determined with NC-3000 cell viability analyzer following manufacturer's instruction (Chemometec).
RESULTS
Preferential localization of pDNA in acidic compartments in differentiated cells
We used neuronal development as a model to initially identify and mitigate critical barriers to polymer-based transfection. Neuronal cells are exceptionally sensitive to changes in environment (7) and hence, serve to define experimental parameters that would be useful for other cell types. As expected, the prolonged exposure of neuronal cells to polyplex at high N/P ratios reduced cell viability (Supplementary Data). However, attempts to shorten the period of exposure so as to reduce toxicity, substantially reduced transfection efficiency (Supplementary Data). Low speed centrifugation (18) resulted in transfection efficiency comparable to prolonged periods of incubation with polyplex (Supplementary Data) with no significant cell death (Supplementary Data). This workflow was then used for all subsequent studies.
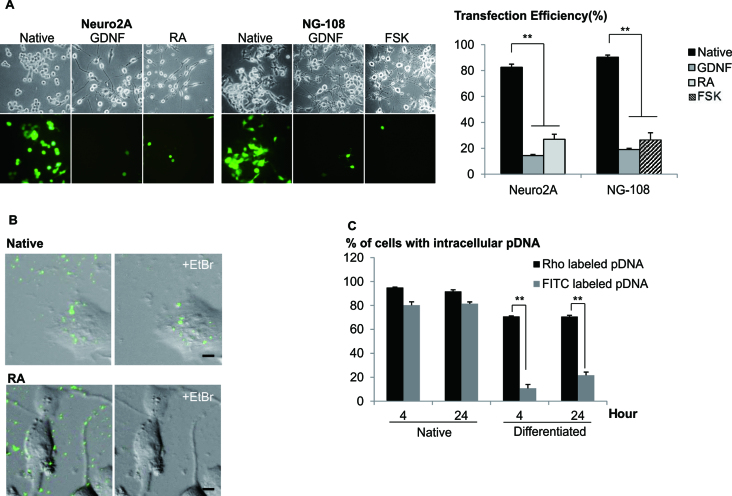
Differentiated neuronal cells are known to be recalcitrant to transfection using cationic polymers (7) and this was obvious when compared to the native undifferentiated cells (Figure 1A). This contrast in transfection efficiencies raised the interesting possibility that the mechanism of uptake of DNA may be different with distinct states of the cells. In order to test the hypothesis that inefficient transfection in differentiated cells may simply be due to the lack of DNA uptake, qualitative image analyses was used to examine the presence of intracellular fluorescence of Rhodamine labeled plasmid DNA (Rho-pDNA) in both native and differentiated cells (Supplementary Data). The amounts of intracellular pDNA in native and differentiated cells were comparable, as measured by quantitative PCR after dissociation of pDNA from polyplex (Supplementary Data), indicative that the internalization processes may not pose a significant barrier in transfection.
Figure 1.
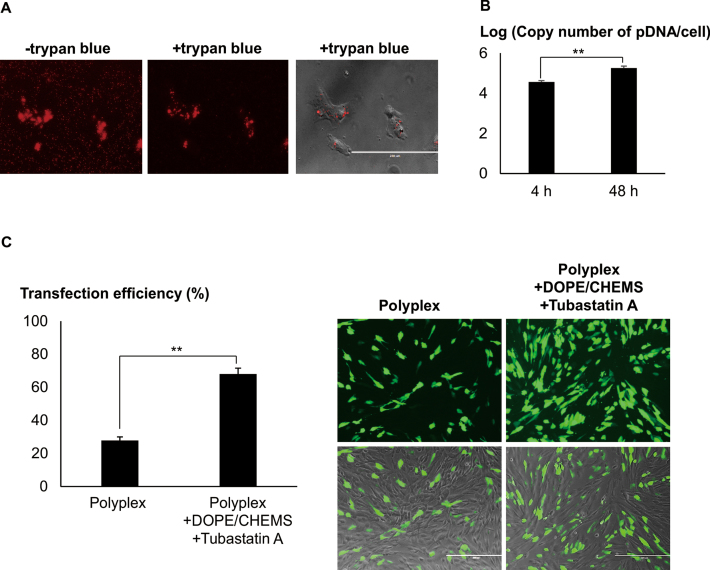
Native but not differentiated neuronal cells were transfected with high efficiencies. (A) Cells were differentiated with either GDNF, RA or FSK, 48 h prior to transfection. Cells were transfected with LPEI/2 μg pDNA at N/P = 10. Following centrifugation, transfection mixture was replaced with fresh media. Representative images were acquired 48 h later. Cells bearing neurites twice the cell body length were considered as differentiated. Transfection efficiency was measured using FACS. Results were presented as mean ± SD (n = 4). (B) Native and differentiated (RA) Neuro2A cells were transfected with LPEI/FITC-pDNA. EtBr/1xPBS (20 μg/ml, quenching reagent) was added 4 h post-transfection. Images were taken before and after (+EtBr) quenching. Bar represents 5 μm. (C) Cells were transfected with LPEI/FITC- or Rho-pDNA. EtBr and 0.4% Trypan blue were added (4 or 24 h post-transfection) to quench the extracellular FITC and Rho fluorescence, respectively. Percentages of cells with internalized labeled-pDNA (normalized to the total number of cells per image) were obtained through cell counting. Results were presented as mean ± SEM. Significant differences between the points were calculated using two tailed Student's t-test. **P < 0.005.
In order to test the hypothesis that the trafficking mechanism/s may have altered when cells were differentiated, we exploited the pH sensitive property of fluorescein (19) to examine the intracellular localization of pDNA. Distinct fluorescence was observed in majority of native cells but significantly reduced in differentiated cells transfected with polyplex containing FITC-pDNA (Figure 1B). The detection of intracellular Rho-pDNA (pH insensitive) in both the native and differentiated cells suggested that pDNA were uptaken similarly (Supplementary Data). Interestingly, the percentage of cells containing FITC- or Rho-pDNA was comparable in native cells but the FITC/Rho ratio was significantly lower in differentiated cells (Figure 1C), suggesting that polyplex was sequestered efficiently in acidic compartments in the differentiated cells. Similar observations were found in NG108 neuronal model and primary cortical neurons (Supplementary Data).
To further validate the observation of pDNA residing in acidic compartment in differentiated cells, co-localization of pDNA with lysotracker green labeled acidic compartment was visualized using confocal imaging. Rho-pDNA was found to be localized to the acidic compartment as early as 2 h post-transfection and increased over time (Supplementary Data).
Re-routing from acidic compartment resulting in enhanced transfection
Next, we speculated that the transfection efficiency should be enhanced by facilitating the escape of polyplex from the acidic compartment. As predicted, the addition of pH sensitive DOPE/CHEMS (20) resulted in significant augmentation of transfection efficiency in differentiated neurons (Figure 2). Confocal microscopy of labelled DNA (Supplementary Data) revealed the effect of DOPE/CHEM in facilitating the escape from these acidic compartments. Notably, the effect of DOPE/CHEMS was temporal, where the optimal effect was achieved when the reagent was added soon after the cells were exposed to the polyplex (Supplementary Data). It is also worthy to note that chloroquine, a lysosomotropic compound known to inhibit fusion of the endosome and reduce enzymatic degradation of DNA by buffering the vesicular interior (21), was highly cytotoxic and less effective than DOPE/CHEMS. Similarly, PLUS™ Reagent and INF7 fusogenic peptide were not as effective in enhancing transfection either used immediately after the cells were exposed to the polyplex (Supplementary Data) or when used together with the polyplex (Supplementary Data).
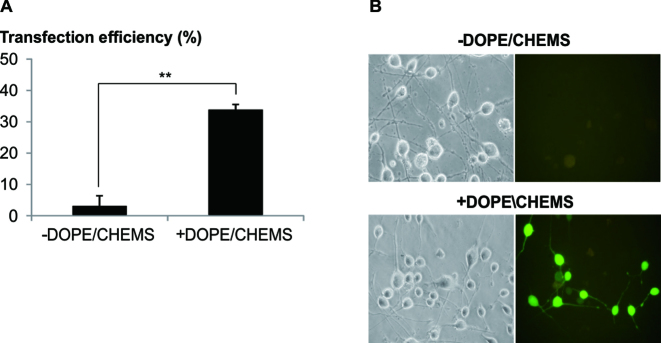
Figure 2.
Addition of DOPE/CHEMS significantly enhanced transfection. (A) Polyplex of LPEI/pDNA (N/P = 10, 2 μg of pDNA) was added to differentiated Neuro2A cells (10 μM RA) and centrifuged at 280 g (5 min). After which, transfection mixture was replaced with fresh media. One hour post-transfection, 50 μl of 25mM HEPES (–) or 12 μg of DOPE/CHEMS prepared at 9:2 molar ratio in 25 mM HEPES (+) was added to the culture medium. Twenty-four hours later, transfection efficiency was analyzed by counting the fluorescent and bright field images. To determine the transfection efficiency of the differentiated population, cells bearing neurites twice the cell body length were counted. Transfection efficiency was calculated as the percentage of EGFP positive cells normalized to the total number of differentiated cells per image. Data are shown as mean ± SEM of biological triplicates. Significant differences between data points were calculated using two tailed Student's t-test. **P < 0.005. (B) Representative images captured (20x magnification) at the end of incubation were presented.
Improved intracellular trafficking of polyplex resulting in efficient transfection
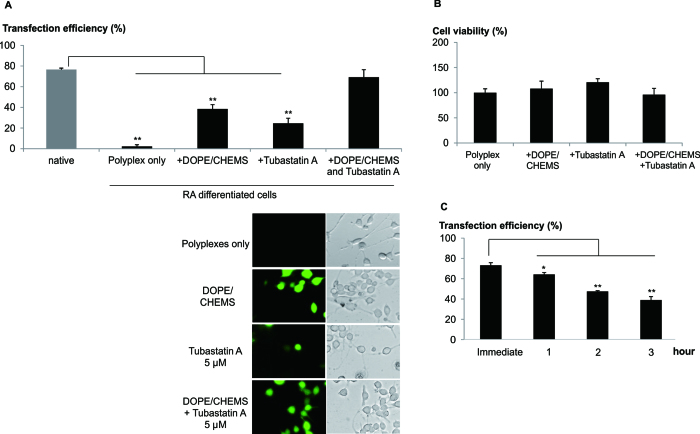
Microtubule mediated transportation is known to be involved in the trafficking of polyplex to the nucleus (22). We next explored the influence of chemotherapeutic mediators of intracellular trafficking as a strategy to further enhance transfection efficiency. In particular, Tubastatin A, a histone deacetylase-6 (HDAC6) inhibitor which enhances microtubule mediated intracellular transport of cargo was evaluated (22). The co-administration of DOPE/CHEMS and Tubastatin A resulted in the remarkable increase in the number of EGFP positive differentiated neuronal cells (∼ 70%, Figure 3A).
Figure 3.
Enhanced trafficking and endosomal escape of polyplex greatly improved transfection. Cells were transfected with LPEI/pDNA using low speed centrifugation procedure. Post-transfection, DOPE/CHEMS and/or Tubastatin A were added to the culture medium. (A) One day post-transfection, bright field and fluorescent images of the native and differentiated (RA) Neuro2A cells were taken for cell counting. For differentiated cells, the analysis excluded non differentiated cells. Transfection efficiency was calculated as the percentage of EGFP+ cells normalized to the total number of differentiated cells per image. Data presented as mean ± SEM (n = 4). Representative images of differentiated cells were presented. (B) One day post-transfection, cell viability of the primary cortical neurons was obtained by counting total number of cells per bright field image and normalized to control (polyplex only). Data presented group mean ± SEM (n = 4). (C) In the presence of DOPE/CHEMS, Tubastatin A was added to the primary cortical neurons culture immediately, 1, 2 or 3 h post-transfection. Transfection efficiency was quantified by FACS analysis 24 h later and presented as mean ± SD, n = 3. Significant differences in transfection efficiencies were calculated using two tailed Student's t-test. *P < 0.05; **P < 0.005.
Additionally, Trichostatin A, an alternative HDAC6 inhibitor (HDAC6i), similarly enhanced transfection efficiency in the presence of DOPE/CHEMS (Supplementary Data). A time course study revealed that exposure of differentiated neuronal cells to DOPE/CHEMS and Tubastatin A for at least 12 h was required for high transfection efficiency (Supplementary Data). Next, the effects of DOPE/CHEM and Tubastatin A were examined using primary cortical neurons in vitro. Remarkably, close to 80% of these post-mitotic cortical neurons were transfected (Supplementary Data) without significant cytotoxicity (Figure 3B). It should be noted that high transfection efficiency occurred when these reagents were added immediately after transfection (Figure 3C).
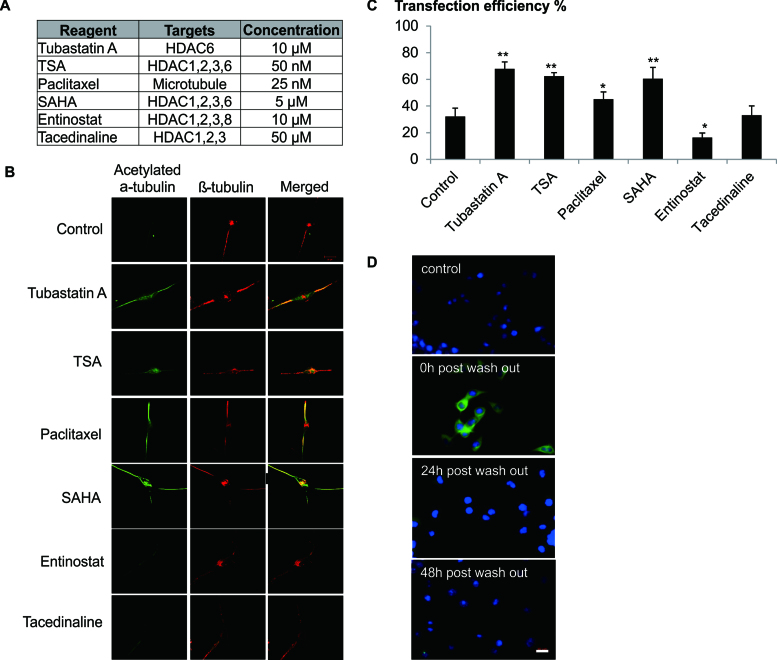
In order to provide further evidence of the contribution of microtubular stability to the increase in transfection efficiency, the effects of various HDACi on tubulin acetylation and transfection were examined. In line with previous reports (23), HDAC6 targeted small molecules (Tubastatin A, TSA, Varinostat/SAHA) and paclitaxel but not Entinostat or Tacedenaline markedly enhanced tubulin acetylation (Figure 4A). As expected, the efficiency of transfection was enhanced when cells were treated with DOPE/CHEMS and small molecules that induced tubulin acetylation (Figure 4B).
Figure 4.
Effect of HDAC6 inhibition on the microtubule stabilization is specific and temporal. (A) Cells were treated with HDACi and Palitaxel at the indicated concentration. (B) Differentiated Neuro2A cells (RA) were exposed to various HDAC inhibitors and Paclitaxel for 2 h. Cells were then fixed with 4% formaldehyde and co-stained for acetylated α-tubulin (Green) and β-tubulin (Red). Confocal images were shown (100× magnification). (C) Differentiated Neuro2A cells were transfected in the presence of DOPE/CHEMS and various microtubule stabilizers for 12 h. Cell treated with only DOPE/CHEMS served as the control. Transfection efficiency was quantified by FACS analysis 48 h later and presented as mean ± SD (n = 3). Significant differences in transfection efficiencies between control and HDACi/Palitaxel treated cultures were calculated using the two tailed Student's t-test. *P < 0.05; **P < 0.005. (D) Neuro2A cells were treated with Tubastatin A for 12 h (0 h washout) and the media was then replaced with complete media. At various time points, cells were fixed with 4% formaldehyde and stained for acetylated α-tubulin (green) and nucleus (Hoechst stain, blue). Cells not treated with Tubastatin A were used as a control. Representative images were shown. Bar represents 20 μm.
HDAC6 inhibition is known to affect microtubule dynamics in non-neuronal cells and neurite outgrowth in neuronal cells (24). Thus, the possibility of long term adverse effects of Tubastatin A on cellular processes of neuronal cells including microtubule dynamics, neurogenesis and global cellular metabolism were evaluated. Evidently, tubulin acetylation was transient (<24 h) on removal of Tubastatin A (Figure 4C). With the transfection protocol used herein no significant long term changes were observed on the ability of the cells to differentiate (Supplementary Data) or on global metabolism (Supplementary Data).
Enhanced transfection efficiencies in a broad spectrum of cell types
We next extended the study to include other cell types known to be recalcitrant to polymer based transfection. Similar to the findings with differentiated neuron, majority of the MSC in the culture showed efficient uptake of pDNA (Figure 5A and B), indicating that the internalization process is unlikely to pose a significant barrier in transfection. Similarly, the intracellular pDNA was localized to acidic compartments in MSC and re-routing from acidic compartment significantly enhanced transfection using DOPE/CHEMS. Furthermore, the co-administration of DOPE/CHEMS and Tubastatin A resulted in a highly significant increase in the number of EGFP positive MSC (∼70%, Figure 5C), which was not due to low speed deposition of polyplexes. Hence, using the method and the formulation of DOPE/CHEMS/HDAC6i, MSC was successfully transfected with high efficiency. In order to address the possibility that MSCs phenotype may have been altered by transfection with polyplex and the treatment with DOPE/CHEMS/HDAC6i, cells were transfected with a plasmid encoding HSV1-Thymidine kinase and subsequently analyzed for changes in biochemical marker expressions. No significant changes were observed with the expressions of the haematopoietic markers (CD14, CD20, CD34 and CD45) and other MSC markers (Supplementary Data). These genetically modified cells were also able to differentiate into different lineages, similar to that of the native, non-transfected MSC cells (Supplementary Data).
Figure 5.
Mesenchymal stem cells were efficiently transfected in the presence of DOPE/CHEMS and HDACi. (A) MSCs were transfected with Rho-pDNA polyplexes (complexation of 350 ng Rho-pDNA at the ratio of 1 μg pDNA to 2 μl Turbofect) using low speed centrifugation. Four hour post-transfection, the media was replaced with 0.4% trypan blue/1× PBS. Cell images were taken before (–) and after (+) quenching with 0.4% trypan blue. Bar represents 200 μm. (B) Polyplex of Turbofect/pDNA (350 ng of pDNA) was added to MSC and centrifuged at 280 g (5 min). At various time points, post-transfection, cells were treated with pAA/DNAse to effectively remove extracellular pDNA. Following trypsinization, cells were collected for pAA/urea lysis buffer treatment. The absolute copy numbers of EGFP DNA was quantified using the standards by real-time qPCR. Bar graph shows Log10 mean and ± SEM (n = 4). (C) MSC was transfected with Turbofect/pDNA in the presence or absence of DOPE/CHEMS plus 10 μM Tubastatin A. After twenty four hours incubation, transfection efficiency (percentage of EGFP+ cells normalized to the total number of cells) was acquired using FACS. Results were presented as mean ± SD (n = 3). Significant differences between data points were calculated using two tailed Student's t-test. **P < 0.005.
Another cell type, mouse embryonic fibroblast (MEF) cells, are also known to be poorly transfected by polyplex (25). The transfection efficiencies using various polymers in MEF were evaluated in the presence or absence of DOPE/CHEMS and HDAC6i, (Supplementary Data). The combinatorial effect of DOPE/CHEMS and Tubastatin A was also found to enhance the transfection efficiencies of a number of commercially available polymers beside LPEI. Interestingly, certain polymers (eg. Jetprime) were superior in comparison to the others, suggesting that the structure or chemical property of polymers may further contribute to the transfection outcome. It is known that the DNA polyplexes and lipoplexes (lipid/DNA complexes) transfection proceed through mechanisms that are fundamentally different during endosomal escape (26). As the method was designed to facilitate the escape of DNA polyplexes from the endosome, it was not surprising that DOPE/CHEMS did not enhance lipid (X-tremeGene 9 and Fugene) based transfection effectively. Hence, the addition of DOPE/CHEMS and HDAC6i only resulted in the moderate improvement of transfection with lipid based carriers (Supplementary Data).
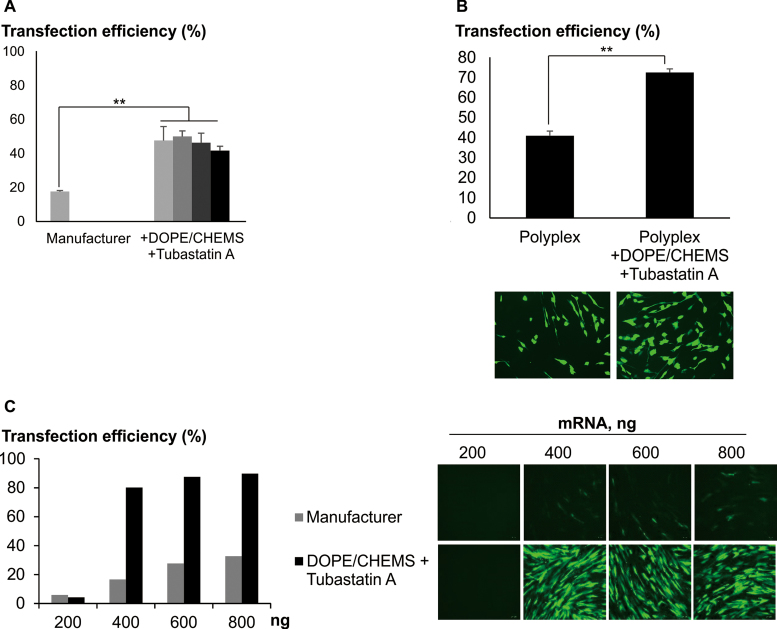
Extending the principle findings further, significant enhancements of transfection were also observed with other cell types including cancer cell lines (Supplementary Data), neural stem cells (NSC) and human dermal fibroblast (Figure 6A and B). The extent of enhancements in transfection efficiencies varied between cell types and polymers used. Delivery of mRNA has several advantages over pDNA which include adjustable and rapid expression of transgenes, avoiding integration into host genome and cell cycle dependent transfection effects (27,28). A pronounced enhancement in transfection was observed with the addition of DOPE/CHEMS and Tubastatin, resulting in ∼90% of MSC expressing EGFP with low amounts of mRNA used (Figure 6C). This effect was likely to be due solely to the effect of DOPE/CHEMS as the addition of another HDAC6i alone was modest in enhancing mRNA transfection (Supplementary Data).
Figure 6.
Coordinating endosomal escape and intracellular trafficking for efficient ex vivo modification. (A) Neural stem cells (16) were transfected with Transficient/pmaxGFP (MBL International Corp./Lonza) at various amount of DNA. Cells were transfected following manufacturer's instruction or centrifugation procedure. Following centrifugation, cells were treated with DOPE/CHEMS and 10 μM Tubastatin A. Data presented as mean ± SD (n = 3). For cells transfected at >250 ng of pDNA (manufacturer's instruction), transfection efficiencies were not analyzed due to the low remaining cell number. (B) Human dermal fibroblast (Cell Applications) was transfected with Transficient/500 ng pmaxGFP using centrifugation procedure, with or without DOPE/CHEMS plus 10 μM Tubastatin A. Data presented as mean ± SD (n = 3). Representative images are presented. Significant differences in transfection efficiencies were calculated using two tailed Student's t-test. **P < 0.005. (C) Complexes of Transficient/EGFP_mRNA (Trilink), at various amount, were prepared. MSC were transfected using manufacturer protocol or centrifugation method. After centrifugation, cells were treated with DOPE/CHEMS plus Tubastatin A (10 μM). Data presented as mean of biological duplicates. Representative images are presented. In all cases, transfection efficiency was acquired with FACS, 24 h post-transfection. Transfection efficiency was calculated as the percentage of EGFP+ cells normalized to the total number of cells as quantified by FACS.
Efficient genetic modification of mammalian cells
Next, we examined the functional consequences of increased transfection efficiency in the modification of MSC for cytokine production (29,30), in differentiation of neural stem cells (NSC) into dopaminergic neuron (31) and in genome targeted editing using CRISPR/Cas9 (13,32).
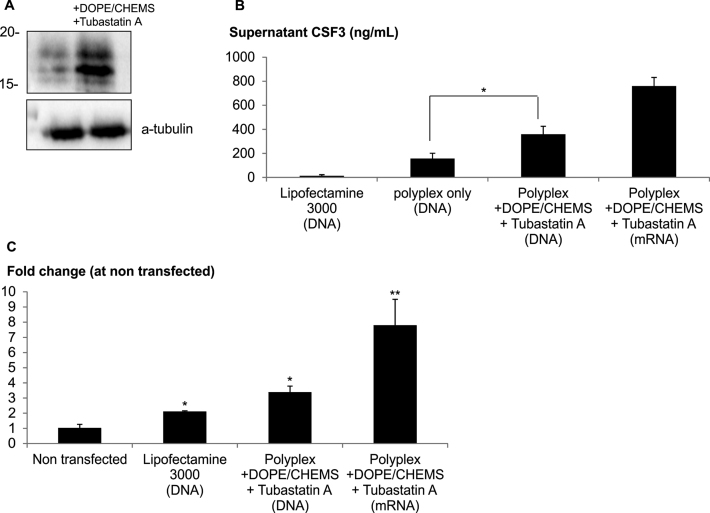
MSC was transfected with GCSF in the presence or absence of DOPE/CHEMS and HDACi as described above. The GCSF level in cellular lysates and conditioned media was determined with immunoblot and Elisa analysis, respectively. The MSC-GCSF lysate showed two bands that were smaller than the predicted size of 18.8 kDa, probably due to proteolytic cleavage (33). Interestingly, the presence of fusogenic lipid and HDACi significantly enhanced the expression of all three forms of GCSF (Figure 7A). The enhancing effect was further confirmed by Elisa measurements (Figure 7B). Furthermore, the method resulted in 26-fold higher GCSF expression in comparison to the transfection using Lipofectamine 3000. We have also shown that this method is highly efficient in mRNA delivery as indicated by the exceptionally high expression of GCSF. The bioactivity of GCSF produced by MSC was determined with the proliferative stimulation with GCSF dependent NSF60 cell line (34). The proliferative activity of NSF60 cells was highly correlated to the GCSF level in the conditioned media (Figure 7B and C). In a similar fashion, improved functional outcomes were observed in the differentiation of NSC into dopaminergic neuron (Supplementary Data) and genome editing (Supplementary Data) with transfections carried out in the presence of DOPE/CHEMS and HDACi.
Figure 7.
High productivity of GCSF-MSC generated with polymer based method. MSC were transfected with 200 ng GCSF DNA or 500 ng mRNA complexed with Turbofect or Lipofectamine using centrifugation method or manufacturer's instruction, respectively. Following centrifugation, the transfection mixture was replaced with media with or without DOPE/CHEMS plus 10 μM Tubastatin A. (A) One day later, the cells were lysed for immunoblotting analysis. (B) The GCSF level in the conditioned media was determined with Elisa analysis. Significant differences in GCSF level (ng/ml) were calculated using the two tailed Student's t-test. *P < 0.05. (C) To access the bioactivity of GCSF, the conditioned media was collected and diluted by 100 times in NSF60 culture (in serum free RPMI). After 3 day stimulation, the number of NSF60 was counted using automated cell counter (chemometec). Data presented as mean ± SD of biological triplicates. Significant differences in conditioned media from the transfected and non transfected cultures were calculated using the two tailed Student's t-test. *P < 0.05; **P < 0.005.
DISCUSSION
This study demonstrated the use of a scalable formulation and protocol that significantly enhanced the transfection efficiencies of a variety of polymers and in a variety of cells, including differentiated neuronal cell-lines, primary cortical neurons, NSC, fibroblast and MSC. The consequences of enhanced transfection were demonstrated in the significant functional improvements in cytotoxicity of modified MSC to cancer cells, the generation of dopaminergic neuron from NSC and in genome editing with CRISPR-Cas9. The rationale underlying the design of the formulation was to facilitate endosomal escape and enhance microtubule mediated trafficking in a temporally controlled manner using specific reagents.
It was thought that internalization of polyplex posed a significant barrier restricting high transfection efficiency in a variety of ‘hard-to-transfect cell including differentiated neuronal cells and attempts to increase the uptake of polyplex only resulted in marginal improvements in the number of neuron transfected (10). Unexpectedly, we found that the uptake of pDNA in the native and differentiated neuronal cells were not significantly different, an observation inconsistent with this hypothesis. Similarly, pDNA was found to be accumulated in MSC despite the low number of cells transfected, indicative that internalization of polyplex may not be a significant barrier to transfection.
It is well documented that endosomal escape of DNA complexes appeared to be a bottleneck for successful delivery and transfection (35–37). Efforts to improve endosomal escape of polyplexes through functionalization of polymers with fusogenic peptides (10), co-polymers (37) and liposomes such as DOPE/CHEMS (lipopolyplex) (38,39) have been described. Unlike the previous studies, we approached by direct influence of acidic compartment with DOPE/CHEMS. This is the first study, to our knowledge, to demonstrate effects of DOPE/CHEMS, a mixture of pH-sensitive lipids which is stable at extracellular pH but fusogenic at endosomal pH (40,41), in enhancing polyplexes but not lipoplexes mediated transfection. The localization of pDNA to acidic compartment in differentiated neurons is consistent with previous report (42) and re-routing with DOPE/CHEMS enhanced transfection. Our approach is coherent with prior studies that demonstrated fusion of liposomes (43) and intracellular acidic compartment (44) as a critical step in cytoplasmic delivery of their content. Similarly, by directly adding DOPE/CHEMS to many other cell types including stem cells and cell lines, significantly higher levels of transfection were observed.
Cytoplasmic trafficking of polyplex to the nucleus is dependent on microtubules and is mediated by dynein and kinesin motors (22,45–47). The coordinated efficient endosomal escape using fusogenic lipids with the stabilization of the microtubule network serves as an attractive synergistic strategy for enhancing polymer-mediated transfection. Evidently, high transfection efficiencies were observed in various cell types and with various polymers examined by such a rational approach. Consistent with other reports, stabilization of microtubule with HDACi had no effect on lipoplex transfection (48). In contrast, depolymerization of microtubule has been shown to enhance lipoplex transfection by inhibition of transport of lipoplex to lysosomes (48,49). These data suggest that the lipoplex and polyplex utilize different mechanisms of endosomal escape and distinct cytoskeleton trafficking pathway.
While the enhancement in transfection was obvious in a variety of cell types, one anomalous observation remained yet to be explained. It is puzzling that almost 100% of the cells internalized DNA but yet the number of cells expressing the transgenes was lower. This may be due to the existence of other barriers yet to be characterized, poor accessibility of DNA/mRNA by transcription/translation complex (50), heterogeneity in the cell population and may also be dependent on the cell cycle, an issue previously suggested (11,51), where dramatic differences in transfection were observed (11). It is now known that besides histones as the major substrates, many cellular proteins are acetylated by HDAC (52) and that changes in acetylation of these nonhistone proteins are known to have diverse cellular effects (53). An intriguing possibility arising from this study is that besides stabilizing the microtubule network by inhibiting β-tubulin deactylase, HDAC6i may also have other yet to be characterized mechanisms of action, resulting in the enhancement of transfection.
This study described a useful tool to enhance the delivery of nucleid acids to hard-to-transfect cells with off-the shelf reagents, avoiding the use of virus. Our method is useful for polymer based ex vivo gene modification where scalable and safe gene carrier is required for modification of a variety of cells including, MSC and human fibroblast (54,55). It is also worthy to note that this method significantly enhanced mRNA transfection, a promising, footprint-free strategy for ex vivo gene therapy (27,28). It has not escape our notice that the chemosensitizing effect of HDAC6 inhibition in transformed but not normal cells (56) may be exploited to achieve a synergistic therapeutic effect with the enhanced expression of a relevant transgene, an issue yet to be investigated.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Dr Azlinda Bte Anwar for providing lysotracker green DND-26 and Dr Zou RuiYang for LC/MS analysis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Exploit Technologies, Agency for Science, Technology and Research [ETPL/15-R15GAP-007]. Funding for open access charge: Exploit Technologies, Agency for Science, Technology and Research [ETPL/15-R15GAP-007].
Conflict of interest statement. None declared.
REFERENCES
- 1. Baek K., Tu C., Zoldan J., Suggs L.J.. Gene transfection for stem cell therapy. Curr. Stem Cell Rep. 2016; 2:52–61. [Google Scholar]
- 2. Ramamoorth M., Narvekar A.. Non viral vectors in gene therapy—an overview. J. Clin. Diagn. Res. 2015; 9:GE01–GE06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Oliveira C., Ribeiro A.J., Veiga F., Silveira I.. Recent advances in nucleic acid-based delivery: from bench to clinical trials in genetic diseases. J. Biomed. Nanotechnol. 2016; 12:841–862. [DOI] [PubMed] [Google Scholar]
- 4. Haleem-Smith H., Derfoul A., Okafor C., Tuli R., Olsen D., Hall D.J., Tuan R.S.. Optimization of high-efficiency transfection of adult human mesenchymal stem cells in vitro. Mol. Biotechnol. 2005; 30:9–20. [DOI] [PubMed] [Google Scholar]
- 5. Neuhaus B., Tosun B., Rotan O., Frede A., Westendorf A.M., Epple M.. Nanoparticles as transfection agents: a comprehensive study with ten different cell lines. RSC Adv. 2016; 6:18102–18112. [Google Scholar]
- 6. Luo C., Lu D., Pan J., Long M.. Improving the gene transfection in human embryonic stem cells: balancing with cytotoxicity and pluripotent maintenance. ACS Appl. Mater. Interfaces. 2016; 8:8367–8375. [DOI] [PubMed] [Google Scholar]
- 7. Karra D., Dahm R.. Transfection techniques for neuronal cells. J. Neurosci. 2010; 30:6171–6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Morris V.B., Labhasetwar V.. Arginine-rich polyplexes for gene delivery to neuronal cells. Biomaterials. 2015; 60:151–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yu-Wai-Man P. Genetic manipulation for inherited neurodegenerative diseases: myth or reality?. Br. J. Ophthalmol. 2016; 100:1322–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Suk J.S., Suh J., Choy K., Lai S.K., Fu J., Hanes J.. Gene delivery to differentiated neurotypic cells with RGD and HIV Tat peptide functionalized polymeric nanoparticles. Biomaterials. 2006; 27:5143–5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brunner S., Sauer T., Carotta S., Cotten M., Saltik M., Wagner E.. Cell cycle dependence of gene transfer by lipoplex, polyplex and recombinant adenovirus. Gene Ther. 2000; 7:401–407. [DOI] [PubMed] [Google Scholar]
- 12. Abdul Halim N.S., Fakiruddin K.S., Ali S.A., Yahaya B.H.. A comparative study of non-viral gene delivery techniques to human adipose-derived mesenchymal stem cell. Int. J. Mol. Sci. 2014; 15:15044–15060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Han X., Liu Z., Jo M.C., Zhang K., Li Y., Zeng Z., Li N., Zu Y., Qin L.. CRISPR-Cas9 delivery to hard-to-transfect cells via membrane deformation. Sci. Adv. 2015; 1:e1500454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sharei A., Zoldan J., Adamo A., Sim W.Y., Cho N., Jackson E., Mao S., Schneider S., Han M.J., Lytton-Jean A. et al. A vector-free microfluidic platform for intracellular delivery. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:2082–2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhu K., Li J., Wang Y., Lai H., Wang C.. Nanoparticles-assisted stem cell therapy for ischemic heart disease. Stem Cells Int. 2016; 2016:1384658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang X.Q., Zhang S.C.. Differentiation of neural precursors and dopaminergic neurons from human embryonic stem cells. Methods Mol. Biol. 2010; 584:355–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ko Y.T., Bickel U., Huang J.. Polyethylenimine/oligonucleotide polyplexes investigated by fluorescence resonance energy transfer and fluorescence anisotropy. Oligonucleotides. 2011; 21:109–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Boussif O., Zanta M.A., Behr J.P.. Optimized galenics improve in vitro gene transfer with cationic molecules up to 1000-fold. Gene Ther. 1996; 3:1074–1080. [PubMed] [Google Scholar]
- 19. Ritter S.C., Milanick M.A., Meissner K.E.. Encapsulation of FITC to monitor extracellular pH: a step towards the development of red blood cells as circulating blood analyte biosensors. Biomed. Opt. Express. 2011; 2:2012–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Draffehn S., Kumke M.U.. Monitoring the collapse of pH-sensitive liposomal nanocarriers and environmental pH simultaneously: A fluorescence-based approach. Mol. Pharm. 2016; 13:1608–1617. [DOI] [PubMed] [Google Scholar]
- 21. Ciftci K., Levy R.J.. Enhanced plasmid DNA transfection with lysosomotropic agents in cultured fibroblasts. Int. J. Pharm. 2001; 218:81–92. [DOI] [PubMed] [Google Scholar]
- 22. Barua S., Rege K.. The influence of mediators of intracellular trafficking on transgene expression efficacy of polymer-plasmid DNA complexes. Biomaterials. 2010; 31:5894–5902. [DOI] [PubMed] [Google Scholar]
- 23. Dokmanovic M., Clarke C., Marks P.A.. Histone deacetylase inhibitors: overview and perspectives. Mol. Cancer Res.: MCR. 2007; 5:981–989. [DOI] [PubMed] [Google Scholar]
- 24. Tapia M., Wandosell F., Garrido J.J.. Impaired function of HDAC6 slows down axonal growth and interferes with axon initial segment development. PLoS One. 2010; 5:e12908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. El-Guendy N., Sinai A.P.. Potential problems inherent in cell-based stable NF-kappaB-GFP reporter systems. Mol. Cell Biochem. 2008; 312:147–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. ur Rehman Z., Hoekstra D., Zuhorn I.S.. Mechanism of polyplex- and lipoplex-mediated delivery of nucleic acids: real-time visualization of transient membrane destabilization without endosomal lysis. ACS Nano. 2013; 7:3767–3777. [DOI] [PubMed] [Google Scholar]
- 27. Levy O., Zhao W., Mortensen L.J., Leblanc S., Tsang K., Fu M., Phillips J.A., Sagar V., Anandakumaran P., Ngai J. et al. mRNA-engineered mesenchymal stem cells for targeted delivery of interleukin-10 to sites of inflammation. Blood. 2013; 122:e23–e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Borestrom C., Simonsson S., Enochson L., Bigdeli N., Brantsing C., Ellerstrom C., Hyllner J., Lindahl A.. Footprint-free human induced pluripotent stem cells from articular cartilage with redifferentiation capacity: a first step toward a clinical-grade cell source. Stem Cells Transl. Med. 2014; 3:433–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Choi J.J., Yoo S.A., Park S.J., Kang Y.J., Kim W.U., Oh I.H., Cho C.S.. Mesenchymal stem cells overexpressing interleukin-10 attenuate collagen-induced arthritis in mice. Clin. Exp. Immunol. 2008; 153:269–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jang Y.H., You D.H., Nam M.J.. Protective effects of HGF gene-expressing human mesenchymal stem cells in acetaminophen-treated hepatocytes. Growth Factors. 2015; 33:319–325. [DOI] [PubMed] [Google Scholar]
- 31. Sanchez-Danes A., Consiglio A., Richaud Y., Rodriguez-Piza I., Dehay B., Edel M., Bove J., Memo M., Vila M., Raya A. et al. Efficient generation of A9 midbrain dopaminergic neurons by lentiviral delivery of LMX1A in human embryonic stem cells and induced pluripotent stem cells. Hum. Gene Ther. 2012; 23:56–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. High K., Gregory P.D., Gersbach C.. CRISPR technology for gene therapy. Nat. Med. 2014; 20:476–477. [DOI] [PubMed] [Google Scholar]
- 33. Hunter M.G., Druhan L.J., Massullo P.R., Avalos B.R.. Proteolytic cleavage of granulocyte colony-stimulating factor and its receptor by neutrophil elastase induces growth inhibition and decreased cell surface expression of the granulocyte colony-stimulating factor receptor. Am. J. Hematol. 2003; 74:149–155. [DOI] [PubMed] [Google Scholar]
- 34. Hara K., Suda T., Suda J., Eguchi M., Ihle J.N., Nagata S., Miura Y., Saito M.. Bipotential murine hemopoietic cell line (NFS-60) that is responsive to IL-3, GM-CSF, G-CSF, and erythropoietin. Exp. Hematol. 1988; 16:256–261. [PubMed] [Google Scholar]
- 35. de Bruin K.G., Fella C., Ogris M., Wagner E., Ruthardt N., Brauchle C.. Dynamics of photoinduced endosomal release of polyplexes. J. Control Release. 2008; 130:175–182. [DOI] [PubMed] [Google Scholar]
- 36. Cho Y.W., Kim J.D., Park K.. Polycation gene delivery systems: escape from endosomes to cytosol. J. Pharm. Pharmacol. 2003; 55:721–734. [DOI] [PubMed] [Google Scholar]
- 37. Gillard M., Jia Z., Hou J., Song M., Gray P.P., Munro T.P., Monteiro M.J.. Intracellular trafficking pathways for plasmid DNA complexed with highly efficient endosome escape polymers. BMC Proc. 2015; 9:P69–P69. [DOI] [PubMed] [Google Scholar]
- 38. Chen W., Li H., Liu Z., Yuan W.. Lipopolyplex for therapeutic gene delivery and its application for the treatment of parkinson's disease. Front. Aging Neurosci. 2016; 8:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Guo W., Lee R.J.. Efficient gene delivery using anionic liposome-complexed polyplexes (LPDII). Biosci Rep. 2000; 20:419–432. [DOI] [PubMed] [Google Scholar]
- 40. Han M.R., Kwon M.C., Lee H.Y., Kim J.C., Kim J.D., Yoo S.K., Sin I.S., Kim S.M.. pH-dependent release property of alginate beads containing calcium carbonate particles. J. Microencapsul. 2007; 24:787–796. [DOI] [PubMed] [Google Scholar]
- 41. Chu C.J., Dijkstra J., Lai M.Z., Hong K., Szoka F.C.. Efficiency of cytoplasmic delivery by pH-sensitive liposomes to cells in culture. Pharm Res. 1990; 7:824–834. [DOI] [PubMed] [Google Scholar]
- 42. Suk J.S., Suh J., Lai S.K., Hanes J.. Quantifying the intracellular transport of viral and nonviral gene vectors in primary neurons. Exp. Biol. Med. (Maywood). 2007; 232:461–469. [PubMed] [Google Scholar]
- 43. Ellens H., Bentz J., Szoka F.C.. H+- and Ca2+-induced fusion and destabilization of liposomes. Biochemistry. 1985; 24:3099–3106. [DOI] [PubMed] [Google Scholar]
- 44. Simoes S., Moreira J.N., Fonseca C., Duzgunes N., de Lima M.C.. On the formulation of pH-sensitive liposomes with long circulation times. Adv. Drug Deliv. Rev. 2004; 56:947–965. [DOI] [PubMed] [Google Scholar]
- 45. Vaughan E.E., Dean D.A.. Intracellular trafficking of plasmids during transfection is mediated by microtubules. Mol. Ther. 2006; 13:422–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Suh J., Wirtz D., Hanes J.. Efficient active transport of gene nanocarriers to the cell nucleus. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:3878–3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kulkarni R.P., Castelino K., Majumdar A., Fraser S.E.. Intracellular transport dynamics of endosomes containing DNA polyplexes along the microtubule network. Biophys J. 2006; 90:L42–L44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang L., MacDonald R.C.. Effects of microtubule-depolymerizing agents on the transfection of cultured vascular smooth muscle cells: enhanced expression with free drug and especially with drug-gene lipoplexes. Mol. Ther. 2004; 9:729–737. [DOI] [PubMed] [Google Scholar]
- 49. Coppola S., Cardarelli F., Pozzi D., Estrada L.C., Digman M.A., Gratton E., Bifone A., Marianecci C., Caracciolo G.. The role of cytoskeleton networks on lipid-mediated delivery of DNA. Ther. Deliv. 2013; 4:191–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yoshihara C., Shew C.Y., Ito T., Koyama Y.. Loosening of DNA/polycation complexes by synthetic polyampholyte to improve the transcription efficiency: effect of charge balance in the polyampholyte. Biophys. J. 2010; 98:1257–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Vink T., Oudshoorn-Dickmann M., Roza M., Reitsma J.J., de Jong R.N.. A simple, robust and highly efficient transient expression system for producing antibodies. Methods. 2014; 65:5–10. [DOI] [PubMed] [Google Scholar]
- 52. Choudhary C., Kumar C., Gnad F., Nielsen M.L., Rehman M., Walther T.C., Olsen J.V., Mann M.. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009; 325:834–840. [DOI] [PubMed] [Google Scholar]
- 53. Falkenberg K.J., Johnstone R.W.. Histone deacetylases and their inhibitors in cancer, neurological diseases and immune disorders. Nat. Rev. Drug Discov. 2014; 13:673–691. [DOI] [PubMed] [Google Scholar]
- 54. Buckland K.F., Bobby Gaspar H.. Gene and cell therapy for children–new medicines, new challenges?. Adv. Drug Deliv. Rev. 2014; 73:162–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Haider H.K., Elmadbouh I., Jean-Baptiste M., Ashraf M.. Nonviral vector gene modification of stem cells for myocardial repair. Mol. Med. 2008; 14:79–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Namdar M., Perez G., Ngo L., Marks P.A.. Selective inhibition of histone deacetylase 6 (HDAC6) induces DNA damage and sensitizes transformed cells to anticancer agents. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:20003–20008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.