Abstract
Owing to their great potentials in genetic code extension and the development of nucleic acid-based functional nanodevices, DNA duplexes containing HgII-mediated base pairs have been extensively studied during the past 60 years. However, structural basis underlying these base pairs remains poorly understood. Herein, we present five high-resolution crystal structures including one first-time reported C–HgII–T containing duplex, three T–HgII–T containing duplexes and one native duplex containing T–T pair without HgII. Our structures suggest that both C–T and T–T pairs are flexible in interacting with the HgII ion with various binding modes including N3–HgII–N3, N4–HgII–N3, O2–HgII–N3 and N3–HgII–O4. Our studies also reveal that the overall conformations of the C–HgII–T and T–HgII–T pairs are affected by their neighboring residues via the interactions with the solvent molecules or other metal ions, such as SrII. These results provide detailed insights into the interactions between HgII and nucleobases and the structural basis for the rational design of C–HgII–T or T–HgII–T containing DNA nanodevices in the future.
INTRODUCTION
The relative easy synthesis and modification of DNA oligonucleotides, in combination with their self-assembly property that is mainly attributed to the base pairing interactions, makes DNA ideal programmable scaffolds for the development of novel functional materials and nanodevices (1–6). DNAs are very flexible in pairing; besides the common base pairing modes (Watson–Crick, Wobble and Hoogsteen types of base pairs), they can form reverse Watson-Crick pair and reverse Hoogsteen pair, which also play important biological role in cell. In addition, various metal-mediated base pairs (metallo-base pairs) have also been discovered since early 1960s (7–9). These unusual base pairs, which are composed of either artificial and/or natural nucleobases, can diversify the overall nucleic acid structures and functions, therefore have great potentials to achieve genetic code expansion (10,11), to develop novel therapeutics (12–15) and to design new materials with metallic functions. To date, various metallo-base pairs have been successfully utilized in the design of molecular magnets (16–18) and electric transport nanowires (19–22). The unique binding specificities between the natural base pairs and certain metal ions have also been applied to develop metal ion sensors that have great significance in analytical chemistry (23–25).
Although the structural characteristics of several metal-mediated artificial base pairs in DNA duplexes have been achieved (16–18,26–31), surprisingly, there is only very limited structural information available for the detailed interactions between the metal ions and the natural base pairs. Particularly for the X-ray crystal structures, there is only one HIV-1 RNA dimerization initiation site that captured a G–AuIII–C pair (27), one RNA duplex containing C–AgI–C pair (32), and one DNA duplex (12TT) containing two consecutive T–HgII–T pairs (33) reported previously; interestingly, in the latter two structures, formation of the metallo-base pairs involves the homo-pyrimidine mispairs, C:C and T:T pairs. Similar to the homo-pyrimidine mispairs, C:T pair, the hetero-pyrimidne mispair, can also occur in vivo as a consequence of either replication errors or heteroduplex formation in the course of genetic recombination (34). The pyrimidine:pyrimidine mispairs can cause the spontaneous point mutations of the DNA genome (35) and they could not be efficiently repaired by the repair systems of certain species, such as Escherichia coli (34).
Owing to their biological importance, the pyrimidine:pyrimidine mispairs have been extensively studied (36,37) and it has been known for long time that both T:T and C:T pairs have very high binding affinities to the mercury ion (38–41); and, these interactions have been widely investigated and utilized to detect or decontaminate the HgII pollution in the environment (25,42,43). Besides the crystal structure (33), one solution NMR structure (44) of the 12TT DNA duplex containing two consecutive T–HgII–T pairs was also reported in 2014; both structures revealed a linear N3–HgII–N3 interaction between the T:T pair and the HgII ion (Figure 1A). However, it is not clear if the separated T–HgII–T pair takes the same interaction mode as the consecutive ones. Residues C and T are different in their chemical compositions, but C* (the imino tautomeric form of C) has very similar structure as the T in the canonical tautomeric form (36); the N3–HgII–N3 interaction has also been proposed for the C–HgII–T pair (Figure 1B) (45). However, the detailed interactions between the C–HgII–T pairs have not been structurally characterized. Herein, we present five high-resolution crystal structures of DNA duplexes including one containing the C–HgII–T pairs, three containing separated T–HgII–T pairs and one containing only T–T pair without HgII. Our structures suggest that the HgII ion is very flexible and dynamic in pairing with pyrimidine-pyrimidine pairs in DNA duplexes. More importantly, these structures also reveal that the C–HgII–T and T–HgII–T pairing can be stabilized by the neighboring residues via the interactions with the solvent molecules or other metal ions. Our studies provide the detailed structural information that is useful for the rational design of DNA nanodevices containing the C–HgII–T or T–HgII–T pairs.
Figure 1.
Schematic view showing (A) the N3–HgII–N3 interaction reported in the 12TT DNA structure and (B) the N3–HgII–N3 interaction proposed for the C–HgII–T pair.
MATERIALS AND METHODS
Crystallization and data collection
All DNAs utilized in the crystallization studies were purchased from the Shanghai GENERAY Biotech Co., Ltd, and dissolved in ddH2O without further purification. The crystallization samples were prepared at room temperature by mixing the DNA and HgCl2, which was also dissolved in ddH2O; the final concentrations of the DNA and HgCl2 were summarized in Supplementary Table S1. All the crystals were grown using the hanging-drop vapor diffusion method. The droplet contains equal volume of DNA sample and different reagents of the Nucleic Acid Mini (NAM) Screen™ kit ordered from the Hampton Research company, whereas, the reservoir solution is composed of 30% (v/v) 2-methyl-2,4-pentanediol (MPD) for all crystals. The crystals of DNA1–HgII, DNA3, DNA3–HgII, DNA3–HgII–BaII and DNA3–HgII–SrII were grown under the conditions of #18, #24, #17, #21 and #23, respectively; the detailed compositions of the reagents were also summarized in Supplementary Table S1. The crystallizations of DNA1–HgII, DNA3–HgII–SrII and DNA3–HgII–BaII were performed at room temperature; all the crystals appeared immediately after the samples were mixed with the crystallization reagents and reached their final sizes within 1 or 2 h. The DNA3–HgII crystals were grew at 18°C, and they appeared two weeks after the crystallization and reached the full sizes after another two weeks. Growth of the DNA3 alone sample required much lower temperature (4°C) and longer crystallization time (more than three months).
All the crystals were cryoprotected using 30% (v/v) MPD and flash-frozen by quickly dipping into liquid nitrogen. The X-ray diffraction data were collected on beamline BL17U1 and BL19U1 at Shanghai Synchrotron Radiation Facility (SSRF, Shanghai, China) at cryogenic temperature, maintained with cryogenic system. One single crystal was used for each structure; data processing was carried out using the HKL2000 or HKL3000 programs (46). The data collection and processing statistics were summarized in Table 1.
Table 1. Data collection and structural refinement statistics.
| DNA1–HgII | DNA3 | DNA3–HgII | DNA3–HgII–BaII | DNA3–HgII–SrII | |
|---|---|---|---|---|---|
| Data collection | |||||
| Wavelength (Å) | 0.97928 | 0.97928 | 0.97928 | 0.97928 | 0.97928 |
| Space group | P21212 | P43212 | P43212 | P43212 | P43212 |
| a, b, c (Å) | 37.8, 51.1, 21.6 | 42.5, 42.5, 25.1 | 42.5, 42.5, 25.0 | 41.0, 41.0, 24.9 | 40.3, 40.3, 25.0 |
| Resolution range (Å) | 30.0–1.05 | 30.0–1.50 | 30.0–1.5 | 30.0–1.45 | 30.0–1.05 |
| Outer shell (Å) | 1.09–1.05 | 1.55–1.50 | 1.58–1.50 | 1.50–1.45 | 1.09–1.05 |
| Completeness (%)α | 98.1 (96.4) | 99.7 (100.0) | 95.0 (93.8) | 99.8 (100.0) | 97.1 (96.4) |
| Rsym (%)α | 11.9 (45.3) | 6.9 (49.5) | 10.1 (44.0) | 10.4 (49.1) | 8.6 (41.2) |
| CC1/2 valueb | 0.653 | 0.891 | 0.959 | 0.828 | 0.637 |
| I/σ(I)α | 24.0 (2.0) | 37.2 (2.6) | 17.6 (2.3) | 14.5 (2.5) | 16.8 (2.6) |
| Multiplicity | 2.0 (1.9) | 22.5 (19.5) | 20.6 (10.2) | 7.6 (5.9) | 6.9 (4.5) |
| Refinement | |||||
| Resolution | 30.4–1.05 | 21.6–1.50 | 21.6–1.5 | 29.0–1.45 | 18.8–1.05 |
| No. of reflections | 19098 | 3932 | 3784 | 3792 | 8921 |
| Rwork (%) | 15.5 | 18.5 | 16.8 | 14.8 | 14.0 |
| Rfree (%) | 17.5 | 19.7 | 18.8 | 19.3 | 15.6 |
| r.m.s.d. bonds (Å) | 0.009 | 0.011 | 0.006 | 0.006 | 0.004 |
| r.m.s.d. angles (°) | 1.097 | 1.190 | 0.586 | 1.136 | 0.839 |
aValues in parentheses are for the outer shell.
bCC1/2 is the half-data-set correlation coefficient. The value listed in the table is for the outer shell of each data set.
Structure determination and refinement
The structures of DNA1–HgII, DNA3–HgII and DNA3–HgII–SrII were all solved by the SAD (single anomalous diffraction) method (47) using the hkl2map program (48); the Figure of Merit (FOM) values are 0.800, 0.658 and 0.755 for the DNA1–HgII, DNA3–HgII and DNA3–HgII–SrII structures, respectively. Based on the original electron density maps, the DNA models were manually built using the graphic program Coot (49). Then, the DNA models were refined against the diffraction data using Refmac5 program (50) embedded in the CCP4i suite (51). 5% randomly selected data was set aside for free R-factor cross validation calculations during the refinement. The 2Fo − Fc and Fo − Fc electron density maps were regularly calculated and used as guide for the building of the missing ions and solvent molecules using COOT. The DNA3 and DNA3–HgII–BaII structure were solved by the different Fourier method using the DNA3–HgII and DNA3–HgII–SrII structures as the starting model. The final refinement of the DNA3 and DNA3–HgII structures were done using the phenix.refine program (52) of Phenix (53). The anisotropic displacement factors were applied to all the complex structures during the refinement. The detailed refinement statistics are summarized in the Table 1.
RESULTS
C–HgII–T adopts an unexpected interacting mode
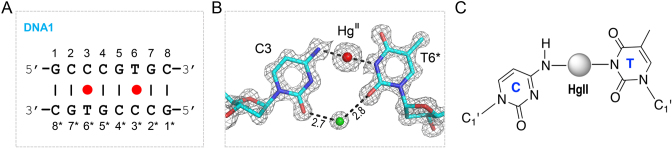
The C–HgII–T containing duplex is formed by DNA1 (Figure 2A); DNA1 is partially self-complementary, and it was designed to form two separated C–T pairs, one between the C3 and the T6* nucleobases and the other one between the C3* and the T6 nucleobases. The DNA1 (referred to as DNA1–HgII hereafter) crystal diffracted to 1.05 Å resolution, and the refinement resulted in the well-defined electron density for all the residues and the HgII ions, which forms two coordination (Figure 2B). As predicted, one coordination formed between the HgII ion and the endocyclic nitrogen atom N3 of the T6* residue; however, instead of the predicted N3 atom of the C3 residue, the second coordination formed between the exocyclic nitrogen atom N4 of the C3 residue and the HgII ion. The average distance is 2.13 Å for the N4–HgII bond and the HgII–N3 bond; the N4–HgII–N3 pair has a linear like geometry, the angle of N4–HgII–N3 is about 178°.
Figure 2.
Unexpected C–HgII–T pairing mode. (A) Sequences and secondary structure of DNA1. (B) The C–HgII–T pair observed in the DNA1–HgII structure. The 2Fo – Fc maps are contoured at 1.5σ level. Both C3 and T6* residues are shown as sticks in atomic colors (C, cyan; N, blue; O, red). The HgII ion and the C–T pair interacting water molecule are shown as spheres in red and green, respectively. (C) Schematic view showing the detailed C–HgII–T interaction.
In the DNA1–HgII structure, each asymmetric unit contains one DNA1 duplex, which captured two HgII ions. Interestingly, none of the two HgII ions are fully occupied; the occupancy is around 0.8 for both HgII ions. Increasing the HgCl2 concentration in the crystallization sample has no clear improvement on the HgII occupancy, suggesting that the low HgII occupancy may be caused by other factors, such as the dynamic interaction between the HgII ion and the C and T residues. The dynamic interaction can be further supported by the even lower HgII ion occupancy observed in the DNA3–HgII and DNA3–HgII–BaII structures, which will be described in the later section.
Based on the 12TT DNA structure, it was predicted that the N3 atom of the T residue (in the canonical tautomeric form) will release one imino proton and directly bind with the HgII ion (Figure 1A). The bond distances and the linear-like geometries of the C–HgII–T pairs in the DNA1–HgII structure are similar to those of the T–HgII–T pairs observed in the 12TT DNA structure. Based on these similarities, we hypothesized that both the C and T residues exist in their canonical forms in forming the C(N4)–HgII–T(N3) interaction (Figure 2C), and one proton was released from their N4 and N3 atoms, respectively. Besides the canonical forms, the C and T residues can also exist in the imino form (C*) or enol form (T*), respectively; these tautomers have also been extensively studied and characterized, owing to their involvement in the biological important transition mispairs (A:C and G:T) and transversion mispairs (C:C, T:T and C:T) (34,35). The slow equilibration of the C and C* tautomers may play certain role during the deprotonation of the C residue and its dynamic interaction with the HgII ion.
C–HgII–T pairing is stabilized by the solvent and the flanking residues
In the reported 12TT DNA structure (33) and the C–AgI–C pair containing RNA structure (32), the metallo-base pairs are mainly stabilized by the N3–HgII–N3 and N3–AgI–N3 interactions, respectively. Interestingly, besides the HgII-mediated interactions, we observed that the conformation of the C3–T6* pair is also stabilized by one water molecule located at the minor grove of the DNA1–HgII structure (Figure 2B). The water molecule forms two strong hydrogen bonds (H-bond), one (2.7 Å) with the O2 atom of C3 residue and the other (2.8 Å) with the O2 atom of the T6* residue.
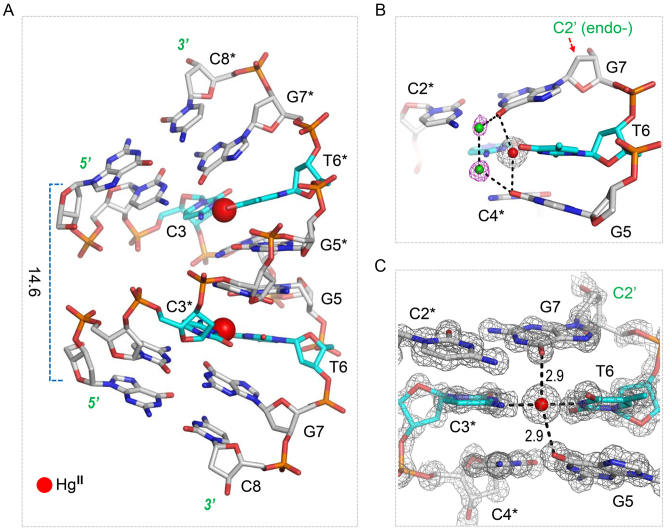
The local base pair parameters, pseudorotation phase angles of the sugar rings, and the sugar ring conformations (Supplementary Tables S2 and S3) of the DNA1 duplex all indicated that the DNA1–HgII structure adopts an A-form conformation, which has also been observed in several other DNA duplexes with similar sequences, including a 8-bp DNA duplex (5΄-GCCCGGGC-3΄, PDB_ID: 9DNA) (54). There is only one difference (G6 versus T6) between this native d(GCCCGGGC) and our DNA1; however, structural comparison revealed that the overall structures are significantly different from each other (Supplementary Figure S1A). Owing to the nearly perpendicular orientation between base pairs and the helix axis, and the relatively smaller average helix twist angle, the d(GCCCGGGC) duplex is more extended than the ideal A-form DNA along the helix axis. However, in contrast to the d(GCCCGGGC) structure, the DNA1–HgII structure is much compressed than ideal A-form DNA along the helix axis, indicated by the very short distance (14.6 Å) between the C1΄ atom of G1 and the C1΄ atom of G1* (Figure 3A).
Figure 3.
H-bond interactions and the C–HgII–T pairing. (A) The stick and sphere view showing the overall fold of the DNA1–HgII structure. (B) The water-mediated H-bond interactions observed in the DNA1–HgII structure. (C) The detailed conformations of the C–HgII–T and the two neighboring C–G pairs. The DNAs are shown as sticks, the C atoms of the C–HgII–T pairs and other residues are colored in cyan and gray, respectively. The water molecules and the HgII ion are shown as green and red spheres, respectively. The 2Fo – Fc maps are contoured at 1.5σ level in C).
Indicated by the low root mean square deviations (RMSD) value (0.3 Å), the DNA1–HgII and d(GCCCGGGC) structures are very similar at the first four residues (G1, C2, C3 and C4, Supplementary Figure S1B). However, the structural comparison revealed that the conformations of the last four residues are different in the two structures (Supplementary Figure S1C). In the d(GCCCGGGC) structure, the sugar puckers of both G6 and G7 residues adopt C3΄-endo conformations, which are common for the A-form DNA duplex. However, in the DNA1–HgII structure, the sugar pucker of G7 adopts C2΄-endo conformations (Figure 3B), which is common for B-form DNA duplexes. The presence of the B-form sugar pucker leads to the significant tilting of G7 nucleobase toward the G5:C4* base pair in the middle of the duplex.
Previous studies suggested that the stability of HgII containing metallo-base pairs, including both C–HgII–T and T–HgII–T pairs, is affected by the flanking residues (45). In the DNA1–HgII structure, the T6–HgII–C3* pair is flanked by the G5:C4* and G7:C2* pairs. The nucleobases of G5 and G7 interact with each other via the H-bonds mediated by two water molecules in the major groove. Supported by the clear electron density (Figure 3B), these water molecules are well ordered; the distances between them and between the water and the O6 atoms of G5 and G7 are all within the range of 2.8–3.0 Å. These H-bond interactions are right on top of the T6–HgII–C3* pair, suggesting that they may involve in the T6–HgII–C3* pairing stabilization. In the DNA1–HgII structure, the distances (2.9 Å) between the HgII ions and the O6 atoms of the two flanking G residues are also very short; the orientations of the O6–HgII–O6 and N4–HgII–N3 are approximately perpendicular to each other (Figure 3C). The close contacts between the HgII and the O6 atoms of neighboring G residues may also contribute to the T6–HgII–C3* pairing and structural stabilization.
T–HgII–T pair has multiple interaction modes
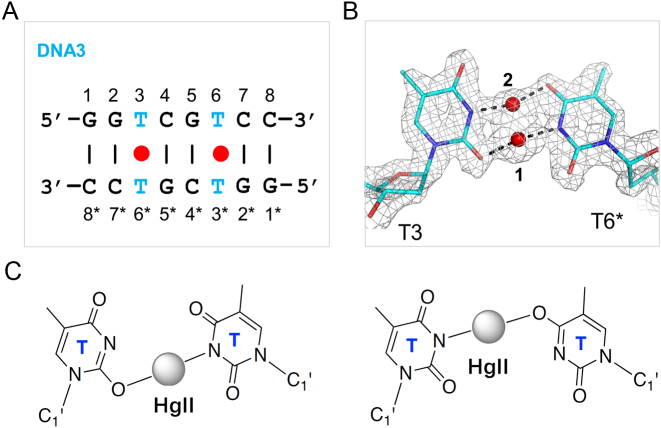
C and T residues are different in their chemical composition, but they could form similar structure via the tautomerization. In the previous studies (45), it was predicted that the HgII-mediated pyrimidine-pyrimidine pairs (including T–T, T–C and C–T) are all formed through the N3–HgII–N3 interaction. However, the unexpected N4–HgII–N3 pairing mode observed in our DNA1–HgII structure suggested that the C residue is flexible in interacting with the HgII ion. Probably due to the relative low resolution (2.7 Å), the conformation of the T–HgII–T pair is not well defined in the previous 12TT DNA structure (33). To investigate whether the T residue is also flexible in interacting with the HgII ion and to better characterize the T–HgII–T pairing, we carried out the crystallographic studies using DNA3 (5΄-GGTCGTCC-3΄), which can form 8-bp duplex containing two separated T–T pairs (Figure 4A).
Figure 4.
The flexible T–HgII–T pairing mode. (A) Sequence and secondary structure of DNA3. (B) The detailed conformation of the T–HgII–T pair observed in the DNA3–HgII structure. The 2Fo – Fc maps are contoured at 1.5σ level. DNA residues are shown as sticks in atomic color (C, cyan; N, blue; O, red). The HgII ions are shown as red spheres. The HgII ion is bound in two alternative geometries, HgII-1 and HgII-2, in the DNA3–HgII structure. (C) Schematic view showing the detailed interaction between the T residues and the HgII-1 (left panel) or HgII-2 ions (right panel).
Totally, three DNA3–HgII complex structures have been solved with high resolutions (1.05–1.50 Å, the structure is referred as DNA3–HgII). In addition to the HgII ion, one of the structures also captured a SrII ion and another structure captured a BaII ion, both of which are existed in the crystallization buffers. These two structures are referred to as DNA3–HgII–SrII and DNA3–HgII–BaII, respectively. Unlike the B-form conformation of the 12TT DNA structure, our DNA3–HgII, DNA3–HgII–SrII and DNA3–HgII–BaII structures all have an A-form like conformation (Supplementary Figure S2 and Supplementary Tables S4–S7). Though the sequence of DNA3 is very different from the d(GCCCGGGC) DNA, the overall structures of DNA3–HgII and d(GCCCGGGC) DNA duplex are very similar (Supplementary Figure S3), with the RMSD value between the two structures only 0.5 Å.
Interestingly, in the DNA3–HgII structure, two HgII ions (HgII-1 and HgII-2) were captured in-between the T:T mispair (Figure 4B) with relatively very short distance (1.8 Å) between them, suggesting that the two HgII ions might be alternative to each other. The HgII-1 coordinates with the exocyclic oxygen atom O2 of the T3 residue and the endocyclic nitrogen atom N3 of the T6* residue; the average distance is 2.03 Å for the O2–HgII and HgII–N3 bonds. The HgII-2 coordinates with the N3 atom of the T3 residue and the exocyclic oxygen O4 of the T6* residue; the average distance between the N3–HgII and HgII–O4 bonds is also 2.03 Å. Similar T(N3)-HgII–T(O4) interaction has been predicted for the free thymine bases in previous theoretic study (55). In the DNA3–HgII structure, both the O2–HgII–N3 and the N3–HgII–O4 interactions have a linear like geometry, formation of such interactions may require the deprotonation and the tautomerization (into T*) of the T residue (Figure 4C).
SrII ion can stabilize the T–HgII–T pairing
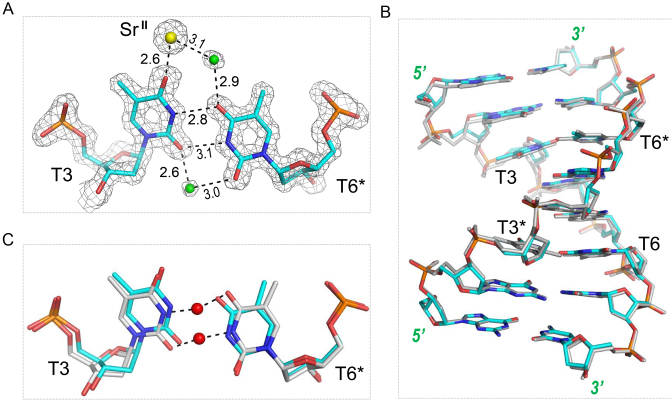
In the DNA3–HgII–SrII structure, the HgII ion coordinates with the N3 atoms of T3 and T6* (Figure 5A), the average distance between the HgII ion and the N3 atoms is 2.05 Å; similar N3–HgII–N3 interactions are also observed in the DNA3–HgII–BaII structure (Figure 5B). The conformations of the T6* residues are identical in the two structures. However, the T3 residue in the DNA3–HgII–BaII structure has two alternative conformations: one (colored in magenta) is similar to the one in the DNA3–HgII structure, and the other (colored in cyan) is similar to the T3 of the DNA3–HgII–SrII structure. These observations also suggest that the HgII coordinating T residues are highly dynamic and the interactions between the HgII ion and the T–T pairs can be very flexible.
Figure 5.
The detailed conformations of the T–HgII–T pairs observed in (A) the DNA3–HgII–SrII structure, and (B) the DNA3–HgII–BaII structure, respectively. (C) The SrII ion-capping cage observed in the DNA3–HgII–SrII structure. (D) the H-bonds formed between W1–W4 and the G2 and G5* residues. All the 2Fo – Fc maps are contoured at 1.5σ level. DNA residues are shown as sticks. The HgII ions are shown as red spheres in all structures. The SrII ion and water molecule are shown as yellow and green spheres in the DNA3–HgII–SrII structure. The BaII ion is shown as yellow spheres in the DNA3–HgII–BaII structure.
The N3–HgII–N3 interaction modes are similar in our DNA3–HgII–SrII structure and the 12TT DNA structure (33), which was depicted in Figure 1A; however, due to the sequence difference, no mercury-mercury metallophilic attraction, which helps stabilize the 12TT structure, is observed in our structures. Instead, as indicated by the short distance (2.27 Å), the HgII ion in the DNA3:HgII:SrII structure binds tightly to one water molecule, which forms one H-bond (2.45 Å) with the O2 atom of T3 and one H-bond (2.52 Å) with the O2 atom of T6* (Figure 5A). These H-bonds all form at the minor groove side of the T–HgII–T pair and are well defined in the DNA3:HgII:SrII structure. The SrII ion is located at the major groove side of the T–HgII–T pair and form eight-coordination: in addition to the O4 atoms of T3 and T6*, it also coordinates with four water molecules (W1–W4) and the O6 atoms of G2 and G5* (Figure 5C). The nucleobase O atoms and the water molecules arrange like a cage, with the former sitting on the bottom and the latter forming the cap; the SrII ion is trapped in the central, which is about 2.5, 2.7 and 2.7 Å away from the O4 atoms (of T3 andT6*), O6 atoms (of G2 and G5*), and the water molecules, respectively. Interestingly, in addition to the interactions with the SrII ion, the water molecules also form four H-bonds with the G2 and G5* residues (Figure 5D), including one between the W1 and the N7 atom of G2, one between the W2 and the O6 atom of G2, one between the W3 and the N7 atom of G5*, and one between the W4 and the O6 atom of G5*.
In all the DNA3–HgII structures, the T3–HgII–T6* pairs are flanked by two C:G pairs, G2:C7* and C4:G5*; however, the detailed conformations and geometries of the flanking C:G pairs are different (Supplementary Figure S4, Supplementary Tables S4–S7). When the SrII (or BaII) ion is absence, the G2:C7* and C4:G5* pairs have regular Watson–Crick shape, the distance between the O6 atoms of G2 and G5* residues is 6.1 Å; whereas, when the SrII (or BaII) ion is presence, the nucleobases of both G2 and G5* are tilted towards each other, resulting in shortened distance (5.2 Å) between the O6 atoms of G2 and G5*. Similar conformational changes also occur to the G5:C4* and C7:G2*, flanking the T6–HgII–T3* pairs. Besides the local conformational changes, the presence of SrII (or BaII) ion also leads to more compacted folding of the structure, indicated by the distances (18.3 versus 21.1 Å) between the C1΄ atoms of G1 and G1* residues of the DNA3–HgII–SrII and DNA3–HgII structures (Supplementary Figure S5).
Characterization of the T:T mispair
In addition to the complex structures, we also solved one DNA3 structure in the absence of HgII ion at 1.5 Å resolution. The T3 and T6* residues form a wobble base pair in the DNA3 structure; as depicted in Figure 6A, there are two direct H-bonds formed between the T3 and T6* residues, one (2.8 Å) is between the N3 atom of T3 and the O4 atom of T6*, and the other (3.1 Å) is between the O2 atom of T3 and the N3 atom of T6*. Besides, the T3 and T6* residues can also interact with each other through water-mediated H-bonds; the water is located at the minor groove of the T3:T6* pair, and the distances between the water and the O2 atoms of T3 and T6* are 2.6 Å and 3.0 Å, respectively. One SrII ion was also captured in the DNA3 structure; it coordinates with the T3 residue and interacts with T6* through water-mediated H-bonds, but it does not form any direct interaction with the flanking G2:C7* and C4:G5* pairs.
Figure 6.
The T:T mispair. (A) The detailed conformation of the T:T mispair observed in the apo-DNA3 structure. (B) Superposition of the apo- DNA3 and the DNA3–HgII complex structures. (C) Superposition of the T:T mispair and the T–HgII–T pair observed in the apo- and complex structures, respectively. The 2Fo – Fc maps are contoured at 1.5σ level. The DNA residues are shown as sticks with the C atoms colored in cyan and gray for the apo- and complex structures, respectively. The SrII ion and the water molecule of the apo- structure are shown as yellow and green spheres, respectively. The HgII ions are shown as red spheres in the complex structure.
In the DNA3 structure, the DNA3 duplex adopts A-form like conformation (Supplementary Tables S8 and S9) and its overall conformation is very similar to that of the DNA3–HgII complex (Figure 6B), the RMSD between the two structures is 0.2 Å. Compared to the DNA3–HgII structure, the presence of the T:T mispair only caused very mild conformational change at the local region (Figure 6C). The nucleobases of the T6* residues have a slight tilting in the two structures, but the backbone and the sugar conformations are almost identical in the two residues. Introducing of the HgII ion slightly pushed the T3 residue toward the outside in the DNA3–HgII structure, but this subtle perturbation (∼0.5 Å) can be well accommodated by the duplex, via the adjustment of the flanking residues.
Compared to the HgII ions captured in the DNA1–HgII structure, the occupancies of the HgII ions in the DNA3–HgII structure are even lower; the total occupancy of the two HgII ions is only 0.45 (0.25 for the HgII-1 and 0.2 for the HgII-2). During the refinement of the DNA3–HgII structure, some positive electron densities were observed around the two T residues; alignment of the DNA3 structure and the DNA3–HgII structure suggested that the extra densities was caused by the T:T mispair, which coexists with the T–HgII–T pair but not modeled in the structure (Supplementary Figure S6). The co-existence of the T:T mispair and the T–HgII–T pair in the duplex further indicated that the T–HgII–T pairing is highly dynamic.
DISCUSSION
We solved five high-resolution crystal structures of DNA duplexes in this study. The three DNA3–HgII complex structures revealed that the T–T mispair is very flexible in interacting with the HgII ion in the DNA duplex; besides the endocyclic N3 atom, the exocyclic O2 and O4 atoms can all participate in the coordination with the HgII ion, forming N3–HgII–N3, O2–HgII–N3 and N3–HgII–O4 interactions. The DNA1–HgII represents the first DNA structure carrying the C–HgII–T pairs to date; it reveals that the exocyclic N4 atom can also interact with the HgII ion. Though we failed to capture it in our study, the structure of the RNA duplex carrying the C–AgI–C pair suggested that the N3 atom of the C residue has the potential to coordinate with the HgII ion (32). Together, these observations advanced our understanding on the HgII-mediated metallo-base pairs.
More importantly, our results indicated that the T–HgII–T pairing is highly dynamic. In the DNA3–HgII structure, the total occupancy of the two HgII ions is 0.45; in the DNA3–HgII–BaII structure, the occupancy (0.3) of the HgII ion is even lower. Such properties will certainly decrease the efficiency of the molecular magnets, electric transport nanowires, metal ion sensors, and other DNA nonodevices, which depend on the formation of the T–HgII–T pair. Previous studies revealed that certain cations could affect the tautomerism of the T residue and change its binding mode with the ligands (56). Though the overall structures of DNA3–HgII–BaII and DNA3–HgII–SrII are very similar, the very low HgII ion occupancy indicated that BaII ion has lesser impact on the T–HgII–T stabilization. In contrast, the SrII ion can significantly enhance the binding between the HgII ion and the T:T mispair, indicated by the much higher occupancy (0.8) of the HgII ion. In addition to the direct interactions with the T–T mispair, the SrII ion also coordinates with both G2 and G5* (Figure 5C and D).
Previous 12TT DNA studies suggested that the formation of T–HgII–T pair can switch the 12TT DNA structure from an unusual nonhelical conformation back to the regular B-form duplex (33). Though all of our complex structures adopt A-form like conformation, structural comparison revealed that the formation of C–HgII–T or T–HgII–T pairs could significantly compress or stretch the duplex along the helix axis. All those conformational changes can drive the mechanical operations of DNA nanostructures, such as ‘tweezers’, ‘walkers’, ‘gears’ and more (57–59); our results, including the detailed C–HgII–T and T–HgII–T pairing, and the unique stabilization effect of the SrII ion on the T–HgII–T pairing, will guide the sequence design of the DNA nanodevices containing the C–HgII–T or T–HgII–T pair.
The C:C mispair has been structural characterized previously (32); with the help of two water molecules, the C:C mispair adopts a Watson–Crick like geometry. Though it did not form in the apo- 12TT DNA structure, the T–T mispair was captured in our apo- DNA3 structure at high resolution (Figure 6); in the structure, the T–T mispair adopts a wobble like geometry, and the presence of this mispair does not cause significant conformational perturbation comparing to the native DNA duplex. These observations provided a structural explanation for the spontaneous point mutations (35) caused by the homo-pyrimidine mispairs. Though we failed to crystallize any apo- DNA duplexes containing the hetero-pyrimidine mispair (the C:T mispair, which can also cause the spontaneous point mutations) in this work, we believe that the overall geometry of the C:T mispair should be similar to that of the C:C or T:T mispairs, and it will be very interesting to further verify the C:T mispair by structure (especially with high resolution crystal structures) in the future.
ACCESSION NUMBERS
Structure factors and coordinates have been deposited in the Protein Data Bank under accession codes 5WSQ, 5WSP, 5WSR, 5WSS, and 5GSK for DNA1–HgII, DNA3, DNA3–HgII, DNA3–HgII–BaII and DNA3–HgII–SrII, respectively.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the BL17U and BL19U beamline staff at the Shanghai Synchrotron Radiation Facility for help during data collection.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Key Research and Development Project of China [2016YFA0500600]; National Natural Science Foundation of China [31370728]; University at Albany, SUNY (to J. S). Funding for open access charge: National Natural Science Foundation of China [31370728].
Conflict of interest statement. None declared.
REFERENCES
- 1.Aldaye F.A., Palmer A.L., Sleiman H.F.. Assembling materials with DNA as the guide. Science. 2008; 321:1795–1799. [DOI] [PubMed] [Google Scholar]
- 2.Feldkamp U., Niemeyer C.M.. Rational design of DNA nanoarchitectures. Angew. Chem. Int. Ed. Engl. 2006; 45:1856–1876. [DOI] [PubMed] [Google Scholar]
- 3.Xiao L., Fu Z.R., Ding G.S., Fu H., Ni Z.J., Wang Z.X., Shi X.M., Guo W.Y., Ma J.. Prediction of survival after liver transplantation for chronic severe hepatitis B based on preoperative prognostic scores: a single center's experience in China. World J. Surg. 2009; 33:2420–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuzuya A., Ohya Y.. Nanomechanical molecular devices made of DNA origami. Acc. Chem. Res. 2014; 47:1742–1749. [DOI] [PubMed] [Google Scholar]
- 5.Linko V., Kostiainen M.A.. Automated design of DNA origami. Nat. Biotechnol. 2016; 34:826–827. [DOI] [PubMed] [Google Scholar]
- 6.Benson E., Mohammed A., Bosco A., Teixeira A.I., Orponen P., Hogberg B.. Computer-aided production of scaffolded DNA nanostructures from flat sheet meshes. Angew. Chem. Int. Ed. Engl. 2016; 55:8869–8872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simpson R.B. Association constants of methylmercuric and mercuric ions with nucleosides. J. Am. Chem. Soc. 1964; 86:2059–2065. [Google Scholar]
- 8.Yamane T., Davidson N.. On the complexing of deoxyribonucleic acid (DNA) by mercuric ion. J. Am. Chem. Soc. 1961; 3:2599–2607. [Google Scholar]
- 9.Katz S. The reversible reaction of Hg (II) and double-stranded polynucleotides. A step-function theory and its significance. Biochim. Biophys. Acta. 1963; 68:240–253. [DOI] [PubMed] [Google Scholar]
- 10.Funai T., Miyazaki Y., Aotani M., Yamaguchi E., Nakagawa O., Wada S., Torigoe H., Ono A., Urata H.. Ag(I) ion mediated formation of a C-A mispair by DNA polymerases. Angew. Chem. Int. Ed. Engl. 2012; 51:6464–6466. [DOI] [PubMed] [Google Scholar]
- 11.Funai T., Nakamura J., Miyazaki Y., Kiriu R., Nakagawa O., Wada S., Ono A., Urata H.. Regulated incorporation of two different metal ions into programmed sites in a duplex by DNA polymerase catalyzed primer extension. Angew. Chem. Int. Ed. Engl. 2014; 53:6624–6627. [DOI] [PubMed] [Google Scholar]
- 12.Xing H., Wong N.Y., Xiang Y., Lu Y.. DNA aptamer functionalized nanomaterials for intracellular analysis, cancer cell imaging and drug delivery. Curr. Opin. Chem. Biol. 2012; 16:429–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schubert S., Kurreck J.. Ribozyme- and deoxyribozyme-strategies for medical applications. Curr. Drug Targets. 2004; 5:667–681. [DOI] [PubMed] [Google Scholar]
- 14.Deleavey G.F., Damha M.J.. Designing chemically modified oligonucleotides for targeted gene silencing. Chem. Biol. 2012; 19:937–954. [DOI] [PubMed] [Google Scholar]
- 15.Alvarez-Salas L.M. Nucleic acids as therapeutic agents. Curr. Top. Med. Chem. 2008; 8:1379–1404. [DOI] [PubMed] [Google Scholar]
- 16.Clever G.H., Reitmeier S.J., Carell T., Schiemann O.. Antiferromagnetic coupling of stacked Cu(II)-salen complexes in DNA. Angew. Chem. Int. Ed. Engl. 2010; 49:4927–4929. [DOI] [PubMed] [Google Scholar]
- 17.Mallajosyula S.S., Pati S.K.. Conformational tuning of magnetic interactions in metal-DNA complexes. Angew. Chem. Int. Ed. Engl. 2009; 48:4977–4981. [DOI] [PubMed] [Google Scholar]
- 18.Tanaka K., Tengeiji A., Kato T., Toyama N., Shionoya M.. A discrete self-assembled metal array in artificial DNA. Science. 2003; 299:1212–1213. [DOI] [PubMed] [Google Scholar]
- 19.Liu S., Clever G.H., Takezawa Y., Kaneko M., Tanaka K., Guo X., Shionoya M.. Direct conductance measurement of individual metallo-DNA duplexes within single-molecule break junctions. Angew. Chem. Int. Ed. Engl. 2011; 50:8886–8890. [DOI] [PubMed] [Google Scholar]
- 20.Joseph J., Schuster G.B.. Long-distance radical cation hopping in DNA: the effect of thymine-Hg(II)-thymine base pairs. Org. Lett. 2007; 9:1843–1846. [DOI] [PubMed] [Google Scholar]
- 21.Ito T., Nikaido G., Nishimoto S.. Effects of metal binding to mismatched base pairs on DNA-mediated charge transfer. J. Inorg. Biochem. 2007; 101:1090–1093. [DOI] [PubMed] [Google Scholar]
- 22.Carell T., Behrens C., Gierlich J.. Electrontransfer through DNA and metal-containing DNA. Org. Biomol. Chem. 2003; 1:2221–2228. [DOI] [PubMed] [Google Scholar]
- 23.Song Y., Wei W., Qu X.. Colorimetric biosensing using smart materials. Adv. Mater. 2011; 23:4215–4236. [DOI] [PubMed] [Google Scholar]
- 24.Nolan E.M., Lippard S.J.. Tools and tactics for the optical detection of mercuric ion. Chem. Rev. 2008; 108:3443–3480. [DOI] [PubMed] [Google Scholar]
- 25.Ono A., Togashi H.. Highly selective oligonucleotide-based sensor for mercury(II) in aqueous solutions. Angew. Chem. Int. Ed. Engl. 2004; 43:4300–4302. [DOI] [PubMed] [Google Scholar]
- 26.Atwell S., Meggers E., Spraggon G., Schultz P.G.. Structure of a copper-mediated base pair in DNA. J. Am. Chem. Soc. 2001; 123:12364–12367. [DOI] [PubMed] [Google Scholar]
- 27.Ennifar E., Walter P., Dumas P.. A crystallographic study of the binding of 13 metal ions to two related RNA duplexes. Nucleic Acids Res. 2003; 31:2671–2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schlegel M.K., Essen L.O., Meggers E.. Duplex structure of a minimal nucleic acid. J. Am. Chem. Soc. 2008; 130:8158–8159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johannsen S., Megger N., Bohme D., Sigel R.K., Muller J.. Solution structure of a DNA double helix with consecutive metal-mediated base pairs. Nat. Chem. 2010; 2:229–234. [DOI] [PubMed] [Google Scholar]
- 30.Kumbhar S., Johannsen S., Sigel R.K., Waller M.P., Muller J.. A QM/MM refinement of an experimental DNA structure with metal-mediated base pairs. J. Inorg. Biochem. 2013; 127:203–210. [DOI] [PubMed] [Google Scholar]
- 31.Kaul C., Muller M., Wagner M., Schneider S., Carell T.. Reversible bond formation enables the replication and amplification of a crosslinking salen complex as an orthogonal base pair. Nat. Chem. 2011; 3:794–800. [DOI] [PubMed] [Google Scholar]
- 32.Kondo J., Tada Y., Dairaku T., Saneyoshi H., Okamoto I., Tanaka Y., Ono A.. High-resolution crystal structure of a silver(I)-RNA hybrid duplex containing Watson-Crick-like C-Silver(I)-C metallo-base pairs. Angew. Chem. Int. Ed. Engl. 2015; 54:13323–13326. [DOI] [PubMed] [Google Scholar]
- 33.Kondo J., Yamada T., Hirose C., Okamoto I., Tanaka Y., Ono A.. Crystal structure of metallo DNA duplex containing consecutive Watson-Crick-like T–Hg(II)–T base pairs. Angew. Chem. Int. Ed. Engl. 2014; 53:2385–2388. [DOI] [PubMed] [Google Scholar]
- 34.Fazakerley G.V., Quignard E., Woisard A., Guschlbauer W., van der Marel G.A., van Boom J.H., Jones M., Radman M.. Structures of mismatched base pairs in DNA and their recognition by the Escherichia coli mismatch repair system. EMBO J. 1986; 5:3697–3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brown T.C., Jiricny J.. Different base/base mispairs are corrected with different efficiencies and specificities in monkey kidney cells. Cell. 1988; 54:705–711. [DOI] [PubMed] [Google Scholar]
- 36.Brovarets O.O., Hovorun D.M.. A novel conception for spontaneous transversions caused by homo-pyrimidine DNA mismatches: a QM/QTAIM highlight. Phys. Chem. Chem. Phys. 2015; 17:21381–21388. [DOI] [PubMed] [Google Scholar]
- 37.Brovarets O.O., Hovorun D.M.. Atomistic understanding of the C.T mismatched DNA base pair tautomerization via the DPT: QM and QTAIM computational approaches. J. Comput. Chem. 2013; 34:2577–2590. [DOI] [PubMed] [Google Scholar]
- 38.Uchiyama T., Miura T., Takeuchi H., Dairaku T., Komuro T., Kawamura T., Kondo Y., Benda L., Sychrovsky V., Bour P. et al. . Raman spectroscopic detection of the T–Hg II–T base pair and the ionic characteristics of mercury. Nucleic Acids Res. 2012; 40:5766–5774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Torigoe H., Ono A., Kozasa T.. Hg(II) ion specifically binds with T:T mismatched base pair in duplex DNA. Chemistry. 2010; 16:13218–13225. [DOI] [PubMed] [Google Scholar]
- 40.Tanaka Y., Oda S., Yamaguchi H., Kondo Y., Kojima C., Ono A.. 15N-15N J-coupling across Hg(II): direct observation of Hg(II)-mediated T–T base pairs in a DNA duplex. J. Am. Chem. Soc. 2007; 129:244–245. [DOI] [PubMed] [Google Scholar]
- 41.Miyake Y., Togashi H., Tashiro M., Yamaguchi H., Oda S., Kudo M., Tanaka Y., Kondo Y., Sawa R., Fujimoto T. et al. . MercuryII-mediated formation of thymine-HgII-thymine base pairs in DNA duplexes. J. Am. Chem. Soc. 2006; 128:2172–2173. [DOI] [PubMed] [Google Scholar]
- 42.Xue X., Wang F., Liu X.. One-step, room temperature, colorimetric detection of mercury (Hg2+) using DNA/nanoparticle conjugates. J. Am. Chem. Soc. 2008; 130:3244–3245. [DOI] [PubMed] [Google Scholar]
- 43.Chen G., Guo Z., Zeng G., Tang L.. Fluorescent and colorimetric sensors for environmental mercury detection. Analyst. 2015; 140:5400–5443. [DOI] [PubMed] [Google Scholar]
- 44.Yamaguchi H., Sebera J., Kondo J., Oda S., Komuro T., Kawamura T., Dairaku T., Kondo Y., Okamoto I., Ono A. et al. . The structure of metallo-DNA with consecutive thymine-HgII-thymine base pairs explains positive entropy for the metallo base pair formation. Nucleic Acids Res. 2014; 42:4094–4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ono A., Torigoe H., Tanaka Y., Okamoto I.. Binding of metal ions by pyrimidine base pairs in DNA duplexes. Chem. Soc. Rev. 2011; 40:5855–5866. [DOI] [PubMed] [Google Scholar]
- 46.Minor W., Cymborowski M., Otwinowski Z., Chruszcz M.. HKL-3000: the integration of data reduction and structure solution–from diffraction images to an initial model in minutes. Acta Crystallogr. D Biol. Crystallogr. 2006; 62:859–866. [DOI] [PubMed] [Google Scholar]
- 47.Giacovazzo C., Siliqi D.. Phasing via SAD/MAD data: the method of the joint probability distribution functions. Acta Crystallogr. D Biol. Crystallogr. 2004; 60:73–82. [DOI] [PubMed] [Google Scholar]
- 48.Pape T., Schneider T.R.. HKL2MAP: a graphical user interface for macromolecular phasing with SHELX programs. J. Appl. Cryst. 2004; 37:843–844. [Google Scholar]
- 49.Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 50.Murshudov G.N., Skubak P., Lebedev A.A., Pannu N.S., Steiner R.A., Nicholls R.A., Winn M.D., Long F., Vagin A.A.. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 2011; 67:355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Potterton E., Briggs P., Turkenburg M., Dodson E.. A graphical user interface to the CCP4 program suite. Acta Crystallogr. D Biol. Crystallogr. 2003; 59:1131–1137. [DOI] [PubMed] [Google Scholar]
- 52.Afonine P.V., Grosse-Kunstleve R.W., Echols N., Headd J.J., Moriarty N.W., Mustyakimov M., Terwilliger T.C., Urzhumtsev A., Zwart P.H., Adams P.D.. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 2012; 68:352–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Adams P.D., Grosse-Kunstleve R.W., Hung L.W., Ioerger T.R., McCoy A.J., Moriarty N.W., Read R.J., Sacchettini J.C., Sauter N.K., Terwilliger T.C.. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002; 58:1948–1954. [DOI] [PubMed] [Google Scholar]
- 54.Heinemann U., Lauble H., Frank R., Blocker H.. Crystal structure analysis of an A-DNA fragment at 1.8 A resolution: d(GCCCGGGC). Nucleic Acids Res. 1987; 15:9531–9550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sebera J., Burda J., Straka M., Ono A., Kojima C., Tanaka Y., Sychrovsky V.. Formation of a thymine-Hg(II)-thymine metal-mediated DNA base pair: proposal and theoretical calculation of the reaction pathway. Chemistry. 2013; 19:9884–9894. [DOI] [PubMed] [Google Scholar]
- 56.Samijlenko S.P., Yurenko Y.P., Stepanyugin A.V., Hovorun D.M.. Tautomeric equilibrium of uracil and thymine in model protein-nucleic acid contacts. Spectroscopic and quantum chemical approach. J. Phys. Chem. B. 2010; 114:1454–1461. [DOI] [PubMed] [Google Scholar]
- 57.Hwang M.T., Landon P.B., Lee J., Mo A., Meckes B., Glinsky G., Lal R.. DNA nano-carrier for repeatable capture and release of biomolecules. Nanoscale. 2015; 7:17397–17403. [DOI] [PubMed] [Google Scholar]
- 58.Omabegho T., Sha R., Seeman N.C.. A bipedal DNA Brownian motor with coordinated legs. Science. 2009; 324:67–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang Z.G., Elbaz J., Willner I.. DNA machines: bipedal walker and stepper. Nano Lett. 2011; 11:304–309. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.