Abstract
Yield of protein per translated mRNA may vary by four orders of magnitude. Many studies analyzed the influence of mRNA features on the translation yield. However, a detailed understanding of how mRNA sequence determines its propensity to be translated is still missing. Here, we constructed a set of reporter plasmid libraries encoding CER fluorescent protein preceded by randomized 5΄ untranslated regions (5΄-UTR) and Red fluorescent protein (RFP) used as an internal control. Each library was transformed into Escherchia coli cells, separated by efficiency of CER mRNA translation by a cell sorter and subjected to next generation sequencing. We tested efficiency of translation of the CER gene preceded by each of 48 natural 5΄-UTR sequences and introduced random and designed mutations into natural and artificially selected 5΄-UTRs. Several distinct properties could be ascribed to a group of 5΄-UTRs most efficient in translation. In addition to known ones, several previously unrecognized features that contribute to the translation enhancement were found, such as low proportion of cytidine residues, multiple SD sequences and AG repeats. The latter could be identified as translation enhancer, albeit less efficient than SD sequence in several natural 5΄-UTRs.
INTRODUCTION
Factors that influence mRNA translation efficiency are of primary value for translational control of gene expression (1,2). Here, we refer to relative translation efficiency as relative number of protein molecules synthesized per equally transcribed mRNAs. Understanding the rules that determine translation efficiency is indispensable for biotechnology (3). To a large extent, although not exclusively, the efficiency of mRNA translation is determined by the efficiency of translation initiation (4,5) that in turn depends primarily on the sequence of mRNA 5΄-UTR (5΄ untranslated region) (6–9). Although elongation may became limiting at very high translation initiation rates and for mRNAs carrying specific features, e.g. excess of suboptimal codons (10,11), we would not address the efficiency of elongation here suggesting the reader to follow the discussion of elongation efficiency published in the literature (5,12–14).
Our understanding of mRNA features that affect the translation efficiency in bacteria went a long way starting from the identification of the SD sequence in 1970's (15). The optimal position and length of this feature appeared to be essential for efficient initiation of translation (16–18). Later, AU-rich sequences were suggested to act as translation enhancer sites via binding by the S1 ribosomal protein (16,17,19,20). Sequestration of ribosome binding site by RNA secondary structure negatively affects initiation of translation (5,17,21). Further studies identified a 4 nt region 5΄ of the SD sequence, named the standby site, as one more determinant of the translation efficiency (6).
In our previous study (17) we used a dual fluorescent protein reporter plasmid to systematically study the influence of several known mRNA features on the translation efficiency. The results were in good agreement with previously published data, e.g. (16). However, after cloning of several natural 5΄-UTR sequences upstream of the CER reporter gene we noted that some of them were more efficient than expected based on mRNA elements (7). This could indicate the influence of additional motifs. Such assumptions are not unprecedented, e.g. in a study of translation efficiency in vitro using a large randomized mRNA library, a new C-rich enhancer element was discovered (22), although it appeared not to be active in vivo and was not observed in a follow-up study (23).
Multiple previous efforts were aimed at systematic analysis of factors that influence the translation efficiency (16,18). Genome-wide ribosome profiling allows one to monitor the translation efficiency of all transcribed mRNAs of an organism simultaneously (24). However, this approach is restricted to natural mRNAs whose 5΄-UTR composition is far from being random.
One option is to analyze artificially designed sets of reporter plasmids with varied length and location of the SD sequence, start codons, the length and location of RNA hairpins in the ribosome binding site and adjacent regions, the presence of AU-rich enhancers and existence an upstream cistron (17). However, the variety of manually designed and cloned artificial reporter plasmids is limited. Large libraries of reporter plasmids proved to be more useful for systematical studies of the translation efficiency (13,25,26). A highly efficient Flowseq analysis of translation efficiency was applied to combinatorial libraries composed by a set of natural and artificial promoters, ribosome binding sites and starting parts of coding areas (13,26). While the overall variety of reporter constructs in these studies were 14 234 (13) and 12 653 (26), the numbers of individual 5΄-UTR sequence variants were rather small, 140 and 111, respectively.
Here, we performed complete randomization of the entire 5΄-UTR in order to reveal previously overlooked elements that may contribute to the translation efficiency. We then analyzed in detail several 5΄-UTR sequences whose efficiency was badly predicted by known features. Further, we designed a set of reporter constructs where the CER gene was preceded by a set of 48 natural 5΄-UTRs from Escherichia coli mRNAs. This experiment led us to understanding that natural 5΄-UTRs possessing SD elements of suboptimal length and location could drive an unexpectedly efficient translation. Finally, we performed partial randomization of the 5΄-UTR of eno mRNA that was shown to be highly efficient in translation.
MATERIALS AND METHODS
Strains and media
E. coli strain BW25113 was grown at 37°C in LB media, supplied with 100 μg/ml ampicillin if required. The JM109 E. coli strain was used for cloning procedures.
Plasmids
pRFPCER (17,27), the dual fluorescent protein reporter made in our laboratory was used as the host vector for construction of reporter plasmids. pRFPCER was digested with BsmBI restriction enzyme, and the obtained linearized vector was directly ligated with pair of pre-annealed complementary oligonucleotides containing BamHI site used for insertion of randomized fragments. For cloning of individual 5΄-UTR variants a pairs of complementary oligonucleotides were synthesized (Supplementary Table S1). pRFPCER plasmid was digested with NdeI and SacII restriction enzymes, and the obtained linearized vector was directly ligated with pre-annealed complementary oligonucleotides containing necessary variant of 5΄-UTR.
Library construction and sorting
Randomized libraries construction
The library of plasmids with 20 or 30 nucleotides long randomized region in 5΄-UTR directly upstream the start codon was designed on the basis of the existing cloning methodology for small random sequence oligodeoxynucleotides (28,29). For library generation, two types of single-stranded oligonucleotides were synthesized and annealed by heating to 95°C for 1 min followed by cooling to room temperature. The first type of oligonucleotides contains SacII site at the 5΄-end flanked by a short extension for efficient recognition by restriction endonuclease, 20 or 30 nucleotides long randomized region or partially randomized eno 5΄-UTR, the start codon and first 19 nucleotides of the CER coding region:
5΄ – ACTG CCGCGGNNN…NATGAAAGAGACGGACGAGAGCG – 3΄
The second type of oligonucleotide is complementary to the start codon and first 19 nucleotides of the CER coding region and contains additional extension at the 5΄-end to generate a sticky end compatible with BamHI site:
5΄ – GATC CGCTCTCGTCCGTCTCTTTCAT – 3΄.
pRFPCER plasmid with additional BamHI restriction site at a distance of 18 nt after the start codon in the CER coding region was digested with BamHI restriction enzyme and ligated with adaptor duplexes described earlier. After ligation the recessive 3΄-ends were extended by the Klenow fragment of E. coli DNA polymerase I to generate blunt ends. The linear products were treated by SacII restriction enzyme and circularized by ligation. As a result, randomized fragment was introduced directly upstream of the start codon, while the 5΄-end of the transcript contained GG sequence immediately followed by the randomized sequence region. The libraries were electroporated into JM109 E. coli strain. Libraries were amplified by growing of the transformed cells without plating for 16 h in 100 ml of LB medium with ampicillin. The obtained cells were centrifuged, dissolved in a small volume of the medium with glycerol (20%) and antibiotic, and then frozen in liquid nitrogen and stored at −80°C. An aliquot of cells after electroporation was used to estimate the number of independent transformants that appeared to approach 106–107 different variants.
Sorting
Libraries grown in LB medium with ampicillin were washed in phosphate buffered saline, diluted in PBS to ca 0.004 A600 and used for cell sorting. Sorting was done by Becton Dickinson FACSAria III with monitoring of RFP fluorescence at 561/582 nm and CER fluorescence at 405/530 nm in logarithmic scale. The cells were collected to eight fractions on the basis of CER/RFP fluorescence ratio in logarithmic scale. For each fraction we collected the number of cells proportional to the number of cells in that fraction among entire library. In total across all fractions we collected 3 million cells that correspond to ca triplicate of total complexity of the library.
Amplicon preparation for sequencing
Sorted cells were incubated for at least 16 h at 37°C with vigorous shaking. After that the cells were prepared for the fluorescence measurement as follows: 200 μl overnight culture of cells after sorting was transferred to 96-well plate and centrifuged at 4000 rpm for 7–10 min and then, the supernatant was decanted and 200 μl of 0.9% NaCl was added and after resuspension was centrifuged again under the same conditions. This washing step was repeated 2 times. After the second centrifugation NaCl was decanted and cells were resuspended in fresh 200 μl of 0,9% NaCl. The samples were transferred to a black 96-well plate measured by Perkin-Elmer Victor X5 reader using wavelengths 531/595 nm for RFP, and 430/486 nm for CER. After subtraction of background, CER/RFP fluorescence ratio was calculated and normalized to that for reference plasmid where RFP and CER were preceded by identical 5΄-UTRs.
Cells from each fraction were collected and their plasmids were isolated. Then PCR with primers flanking the both sides of randomization region was conducted and its results were checked by electrophoresis in 1,5-2% agarose gel. The obtained amplicons were used for sequencing.
Sequencing
Amplicon libraries were sequenced with the genomic analyzer Ion Torrent PGM (Life Technologies) using Ion PGM™ Template OT2 200 Kit (Life Technologies) for emulsion PCR and Ion Chips 314 or 318 and the reagent kit Ion PGM™ Sequencing 200 Kit v2 (Life Technologies) for sequencing according to the manufacturer's instructions.
Data analysis
Raw sequencing data filtering and UTR's sequence extraction
Raw reads were processed from bulk FASTQ data. They were filtered by length from 50 nt to 150 nt for the libraries containing 20 and 30 randomized nucleotides or 140 nt for randomized eno 5΄-UTR. 5΄-UTR sequence was determined as the reverse complement to the region between constant sequences CTCGTCCGTCTCTTTCAT and CCGCGGCT. Up to two mismatches including Indels were allowed in detection of constant parts. All reads without those sequences were removed. After extraction of 5΄-UTR sequences, reads were filtered by length with only a single nucleotide deviation from the target length of 20, 30 or 27 nucleotides for the libraries containing 20 and 30 randomized nucleotides and randomized eno 5΄-UTR. For the control mix of plasmids 8/7, 6/10, 4/7AU, 4/7, 2/7 and 0 this filtering was not applied. After 5΄-UTR variant calling, variants were counted in every fraction and the counting data were saved in a simple tabulated file. For the control mix, counting of correct reads was done by keeping variants with the exact match for known sequences.
Error correction
For the correction of errors, we created a custom error correction workflow based on finding similar variants across a sparse set of dissimilar sequences. The Levenshtein distance (LD) with nucleotide substitutions and indels allowed was calculated for all variants. Then, the variants differing by LD at most 3 were merged. In the merging procedure, variants found more than 10 times were preferred to be considered as correct at the expense of those found more rarely; similarly, variants of the expected length were considered to be correct at the expense of those with a single indel. Error correction was applied for the libraries containing 20 and 30 randomized nucleotides only, for control mixture the correction was not needed, and for randomized eno 5΄-UTR the correction was not applicable.
Fraction assigning
Each variant distribution among fractions was fitted to the Gaussian distribution, the resulting effective (virtual) fraction was computed as the mean of this normal distribution. In most cases the virtual fraction is an integer because the distributions are narrow.
Analysis of 5΄-UTRs secondary structures
For sequence analysis, we used FASTA sequences of 5΄-UTR variants with constant GG before the randomized block and with the first 50 nt coding sequence. For calculation of the pairing scores we used Vienna RNA package version 2.2.4 (30), program RNAplfold, parameter ‘-W’ 30 nt. For each 5΄-UTR nucleotide, pairing scores to the 5΄ and 3΄ directions were calculated for the window of 41 nt (for the library containing 20 randomized nucleotides) and 51 nt (for the libraries containing 30 randomized nucleotides and randomized eno 5΄-UTR). For the calculation of ΔG, RNAfold was applied to fold the first 20 nt for the library containing 20 randomized nucleotides and 50 nt, for the library containing 30 randomized nucleotides. For the calculation of the folding energy within a sliding window, each (i, …, i+29) subsequence was folded by RNAfold for every start position i in the range (1, …, 20) for the library containing 20 randomized nucleotides and (1, …, 30) for the library containing 30 randomized nucleotides.
Analysis of SD like sequences in 5΄-UTRs
The SD were scored in sliding windows (i, …, i+8), with position specific score matrix (PSSM) weights taken from the Transterm Database (31), ID T0030, Score value scaled from 0 to 1, for i in the range (1, …, 22) the library containing 20 randomized nucleotides and (1, …, 32), for the library containing 30 randomized nucleotides and randomized eno 5΄-UTR. The position of the SD sequence in randomized 5΄-UTRs was detected as the position of a subsequence with the maximal SD score.
To analyze the influence of more than one SD sequence on the translation efficiency relative to that of a single SD sequence, we calculated the SD score for real 5΄-UTR sequences and for matching virtual 5΄-UTR sequences composed of an oligoC string with a single SD sequence detected as one with the maximal score in the real 5΄-UTR. After subtraction of the latter from the former, we obtained the SD scoring for 5΄-UTRs after virtual exclusion of the SD sequence with the best score.
Machine learning
To assess the influence of known factors on the translation efficiency, we applied two machine learning techniques, Random Forest regression with multidimensional output (32) assigning each 5΄-UTR to the class with the maximal predicted regression value, and logistic regression (32) with the one-versus-the-rest strategy (33) to handle multiclass data. Both techniques were used to predict one of eight classes defined by the CER/RFP ratio for each 5΄-UTR. Left and right pairing scores (RNAfold scores) for all 5΄-UTR nucleotides and first 18 nucleotides of the coding sequence, SD scores for all 5΄-UTR nucleotides and first 8 nucleotides of the coding sequence, and ΔG for all 5΄-UTR nucleotides and first 18 nucleotides of the coding sequence were used as the parameters for the library containing 20 randomized nucleotides. The prediction accuracy is estimated using standard classification accuracy measures, such as precision (proportion of true positives among predicted to be positive), recall (proportion of predicted to be positive among actually true positive) and f1-score (harmonic mean of precision and recall).
In order to estimate the predictive ability of the models, we used 10-fold cross-validation. The original sample was randomly partitioned into 10 equally-sized subsamples. Then one subsample was retained as the validation data, and the remaining nine subsamples were used as the training data. The process was repeated 10 times, with each of the 10 subsamples used exactly once as the validation data. We then averaged the obtained results to produce a single estimate of the accuracy measure.
General data processing and plotting
Raw sequence data processing, error correction, fraction assigning, statistical analysis and plotting were implemented in R (34), for profiling custom scripts in Python (35) were used.
RESULTS AND DISCUSSION
Validation of the flowseq method for large-scale analysis of the translation efficiency
Two fluorescent proteins, CER and RFP, possess readily distinguishable spectral properties that allow for their simultaneous detection in the same bacterial culture either in an ensemble measurement by a fluorimeter or at the level of individual cells by flow cytometry (17,27). Application of fluorescent cell sorting and next generation sequencing (Flowseq) for the analysis of combinatorial reporter libraries (Figure 1) demonstrated superior performance in several recently published studies (13,25,26,36,37). In order to compare the CER/RFP ratio measured by the fluorimeter and the cell sorter and to assess the efficiency of sorting, we performed a control experiment (Supplementary Figure S1A and B). We mixed equal numbers of cells transformed by reporter constructs carrying a 8 nt SD sequence located 7 nucleotides upstream of the start codon (8/7), a 6 nt SD located 10 nucleotides upstream of the start codon (6/10), a 4 nt SD located 7 nucleotides upstream of the start codon (4/7), the same construct additionally containing an AU-rich translation enhancer (4/7AU), a 2 nt SD located 7 nucleotides upstream of the start codon (2/7) and a construct that has no SD sequences (0). Their relative CER/RFP expression levels were previously determined to be 17, 13, 0.8, 3.7, 0.03 and 0.001, respectively (17). The mixture of the cells transformed by these plasmids was sorted to eight fractions differing by the CER/RFP ratio (Supplementary Figure S1A). After sorting, we measured the bulk CER to RFP fluorescence ratio in all fractions by the spectrofluorimeter (Supplementary Figure S2A) and determined the proportion of cells containing each of the reporter constructs in all fractions (Supplementary Figure S1B). Cells containing plasmids that encode more efficiently translated CER gene according to the bulk spectrofluorimeter measurements were found in fractions possessing higher CER/RFP ratio according to cell sorting. A majority of cells containing each of the reporter constructs were concentrated in just one or two fractions. We conclude that Flowseq may be used for reliable and precise evaluation of the CER/RFP ratio on the single-cell level with the measurements matching the bulk spectrofluorimeter measurements in a cell culture.
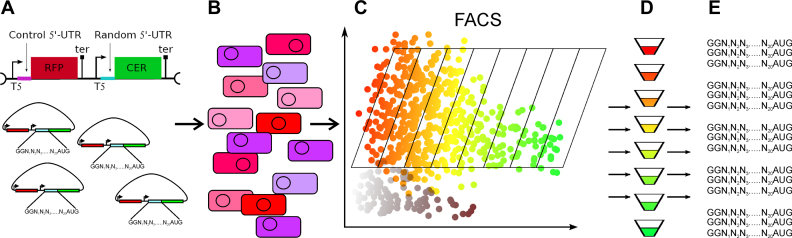
Figure 1.
The principal scheme of the Flowseq experiment. Presented are the steps of library construction, transformation, sorting and sequencing. (A) Cloning of the randomized DNA fragment into pRFPCER reporter vector in front of CER gene. RFP gene retains its constant 5΄-UTR. (B) Electroporation of entire plasmid library into E. coli cells. (C) Separation of cells on the basis of CER/RFP fluorescence by cell sorter. (D) Collection of cell pools (F1–F8) according to CER/RFP fluorescence ratio. (E) DNA extraction and amplification of randomized region followed by next generation sequencing.
Complete randomization of 5΄-UTR
The composition of natural 5΄-UTRs is a subject for biases as a consequence of evolution. To obtain an unbiased library of 5΄-UTRs, we cloned 20 or 30 completely randomized nucleotides upstream the CER reporter gene in the pRFPCER reporter plasmid. To minimize an influence of 5΄-UTR sequence on transcription most sensitive to the identity of the first nucleotides of the transcript we used constant two G residues as the 5΄-most transcript nucleotides. Thus, the libraries we created encoded 22 and 32 nt 5΄-UTRs composed by 5΄-GG and followed by the 20 and 30 completely randomized nucleotides followed by AUG start codon. E. coli cells transformed by this library were divided in two pools and sorted to eight fractions differing by the CER/RFP ratio (Figure 2, Supplementary Figure S3). As expected, a majority of randomized 5΄-UTR sequences performed poorly as translation initiation sites. Only 0.03% of cells transformed with the library containing 20 randomized nucleotides fall into fraction 8, which corresponds to the maximal translation efficiency of CER (Supplementary Figure S2C). Although the cells transformed with the library containing 30 randomized nucleotides also have very low number of efficiently translated variants, their abundance is substantially higher than that for 20 randomized nucleotides (Supplementary Figure S2D). As much as 0.45% of cells fall into the fraction F8 with maximal translation efficiency.
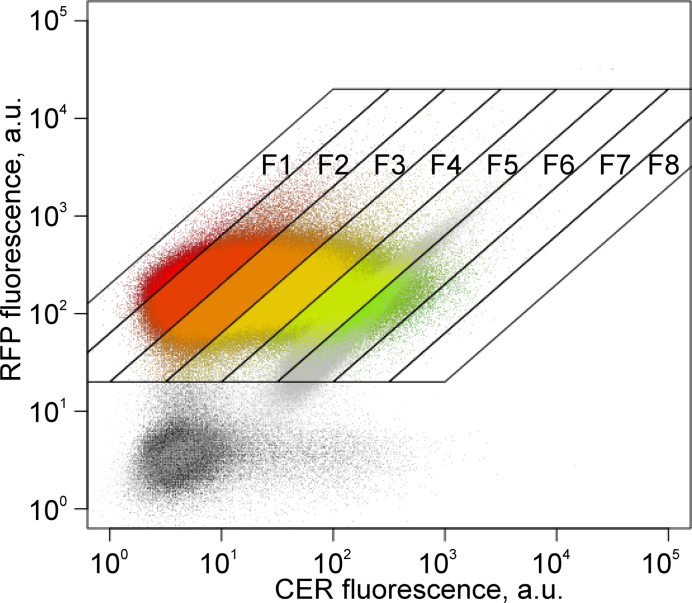
Figure 2.
Distribution of the CER and RFP fluorescence intensity for cells transformed by reporter construct libraries with 20 nt randomized fragments in the 5΄-UTRs of CER. Each dot corresponds to a single event. The CER fluorescence intensity increases along the X-axis; the RFP intensity used as a control increases along the Y-axis. An overlay of sorting results for cells transformed by a control 4/7 construct is shown in grey. Positions of all collected fractions differing by the CER/RFP ratio are shown. A related Supplementary Figure S3 contains the data on creation and sorting of a library with randomized 30 nt fragments in the 5΄-UTRs.
The small proportion of highly efficient 5΄-UTR variants in the randomized libraries demonstrates that ribosome movement along CER coding region, i.e. elongation does not limit translation in this system. Otherwise, one would observe an artificial increase of cell counts at the fraction corresponding to the maximal translation efficiency limited by elongation, followed by an abrupt decrease of cell counts, corresponding to more efficient translation. Since we have not observed this effect experimentally (see, Supplementary Figure S2C and D), we can conclude that translation elongation at CER coding region proceed sufficiently fast not to limit overall yield of CER.
All eight fractions for each library and each of the two pools were subjected to next generation sequencing. Sequences observed in both collection replicas were used to evaluate the reproducibility of sorting (Supplementary Figure S4). For libraries of reporter plasmids containing 20 or 30 randomized nucleotides in the 5΄-UTRs we observed 0.95 and 0.98 correlation coefficients, respectively. Since the reproducibility was sufficient, for further analysis we merged both replicas, yielding 11 692 unique 5΄-UTRs containing the 20 nt randomized fragment and 11 889 unique 5΄-UTRs containing the 30 nt randomized fragment. Most variants were represented by 10 to 1500 reads usually found in one or two fractions after sorting (see Supplementary Tables S2 and S3 for raw 5΄-UTR sequences and their distribution among the fractions). Thus, we obtained a uniquely representative collection of 5΄-UTRs that is roughly five times more diverse than a collection of 5΄-UTRs of natural E. coli genes (38).
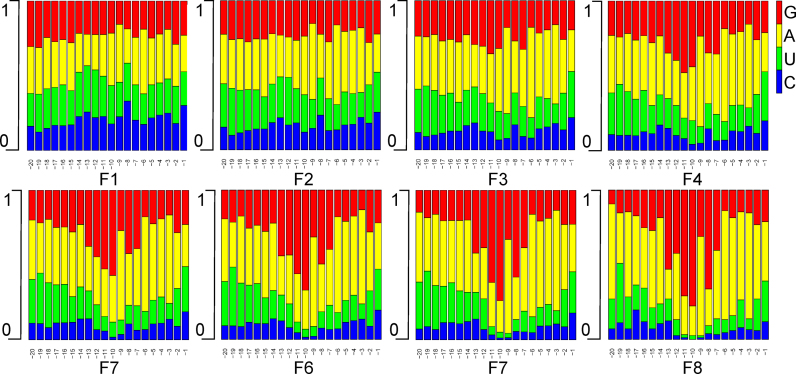
The analysis of the nucleotide composition of 5΄-UTRs differing in translation efficiency (Figure 3, Supplementary Figure S5) revealed nearly uniform nucleotide distribution in poorly active 5΄-UTRs (Figure 3, Supplementary Figure S5, panels F1). Higher efficiency of translation yielded more biased nucleotide composition of the 5΄-UTR (Figure 3, Supplementary Figure S5). A significant enrichment of A and G at positions −7 to −13 relative to the AUG start codon in highly efficient 5΄-UTRs (Figure 3, Supplementary Figure S5, panels F8) most likely reflected selection for SD-like sequences (see below). Other regions of 5΄-UTRs of efficiently translated mRNA variants also demonstrated biases in the nucleotide composition. A significant prevalence of A and to lesser extent U residues was characteristic for regions both up- and downstream the putative SD motifs of 5΄-UTR regions from the fraction of the most efficient translation (Figure 3, Supplementary Figure S5, panels F8). Overrepresentation of A and U residues in the 5΄-UTR sequences of mRNAs with the highest translation efficiency most likely reflects a presence of AU-rich translation enhancers (20) that were previously suggested to bind ribosomal protein S1 (16,17,19,20). Notably, all regions of highly efficient 5΄-UTRs have markedly reduced proportion of C residues (Figure 3, Supplementary Figure S5, panels F8).
Figure 3.
Influence of the nucleotide sequence of 20 nt fragments in 5΄-UTRs on the translation efficiency. Panels F1–F8 correspond to fractions sorted by the translation efficiency from the least efficient F1 to the most efficient F8. Each bar represents a proportion of nucleotides at the corresponding position in the 5΄-UTR relative to the AUG start codon, as shown below the graphs. Nucleotides are color-coded as shown in the legend at the upper right side of the figure G – red, A – yellow, U – green, C – blue. A related Supplementary Figure S5 contains the data on the positional nucleotide composition of 5΄-UTRs with for randomized 30 nt fragments.
Comparison of the composition of efficiently translated 5΄-UTR sequences from libraries containing 20 and 30 randomized nucleotides revealed that the composition of shorter 5΄-UTRs have much stronger sequence biases reflecting frequent presence of strong SD sequences (Figure 3, Supplementary Figure S5, panels F8). At the same time, the 5΄-part of longer 5΄-UTR sequence of efficiently translated mRNA variants, which are not present in the short 5΄-UTRs, have compositional bias toward AU over GC nucleotides reflecting an importance of this region for translation efficiency. In our previous work (16) we demonstrated that extension of short 5΄-UTR by AU-rich enhancer with formation of 29 nt long UTR resulted in 5-fold increase in translation efficiency. Natural transcripts of E. coli have on average 5΄-UTR of 25–35 nucleotides (39), which is quite in line with relatively better performance of 30 nt long 5΄-UTRs. Summarizing, majority of short 5΄-UTRs need to have large SD sequence to be efficient in translation, while longer 5΄-UTRs could achieve the same translation efficiency with smaller SD and various, e.g. AU-rich upstream translation enhancers.
Influence of secondary structure in the 5΄-UTR on the translation efficiency
The folding energy of the secondary structure for the sliding window of 30 nucleotides (see Data analysis), the total 5΄-UTR folding energy and the RNAplfold score of each nucleotide forming a base pair in the 5΄ and 3΄ directions was calculated for all 5΄-UTR sequences in the data set (Supplementary Tables S4 and S5). We plotted the distribution of the folding energy for all 5΄-UTRs grouped by expression efficiencies (Figure 4A, Supplementary Figure S6A) and visualized the distribution of the pairing scores (Figure 4B, Supplementary Figure S6B–D) and the folding energy in 30 nt sliding windows (Supplementary Figure S6E and F). The analysis of secondary structure for all 5΄-UTR variants detected by Flowseq revealed that the folding energy positively correlates with the translation efficiency, which means that weak secondary structure is beneficial for translation. The proportion of structured 5΄-UTRs is maximal for 5΄-UTRs yielding low translation efficiency (Supplementary Figure S6E and F). The proportion of nucleotides with high RNAplfold scores steadily decreases with increase of translation efficiency (Figure 4B, Supplementary Figure S6B–D). Moreover, location of paired 5΄-UTR regions is non-random in mRNAs differing in translation efficiency. UTRs of inefficiently translated mRNAs are enriched in secondary structure elements in the ribosome binding region, while secondary structure elements of efficiently translated 5΄-UTRs, if present, tend not to overlap with the ribosome binding site (seen most clearly in Figure 4B, panel F8). We did not observe secondary structure patterns consistently amplifying the translational efficiency.
Figure 4.
Influence of the secondary structure on the translation efficiency. (A) Distribution of the folding energy for 5΄-UTRs with 20 nt randomized fragments of varying translation efficiencies. Each graph corresponds to a fraction from the highest (F8) to the lowest (F1) translation efficiency, marked below the graph. The 25–75 percentile is boxed, the median value is shown as a horizontal line, outliers less than Q1 – 1.5 Interquartile range (IQR) and more than Q3 + 1.5 IQR are shown as circles. (B) The plfold-score of forming a base pair in the 3΄ direction of a given nucleotide for 5΄-UTRs with 20 nt randomized fragments of varying translation efficiencies. Each graph corresponds to a fraction from the highest (F8) to the lowest (F1) translation efficiency. Position relative to mRNA sequence is shown above graphs; sketch of 5΄-UTR sequence is shown below graphs. Lines correspond to individual mRNAs grouped by the position of maximal plfold-score. The color represents the plfold-score of forming a base pair in the 3΄ direction from black (pairing is unlikely) to red (pairing is highly probable). A related Supplementary Figure S6 contains the data for 5΄-UTRs with randomized 30 nt fragments and the plfold-scores of forming a base pair in the 5΄ direction of a given nucleotide for both libraries.
Frequency and length of the SD sequences in 5΄-UTRs differing in translation efficiencies
The best known element of 5΄-UTR that enhances the translation efficiency is the SD sequence that is complementary to the 3΄-terminal region of the 16S rRNA. We calculated the score of similarity (PSSM score) to the canonical SD sequence (exact PSSM used is visualized on Supplementary Figure S7B) for each 9 nt sliding window in all 5΄-UTR variants present in our data set (Supplementary Tables S6 and S7; see Data Processing). For each 5΄-UTR, a window with the maximal similarity to the SD sequence was found and the distribution of maximal scores was plotted for mRNAs differing in translation efficiency (Figure 5A, Supplementary Figure S7A). As expected, mRNA pools with higher translation efficiencies contain sequences closer on average to the SD sequence.
Figure 5.
Frequency and distribution of SD-like subsequences in 5΄-UTRs with randomized 20 nt fragments differing in mRNA translation efficiency. (A) Distribution of maximal SD position specific score matrix (PSSM) scores (X-axis) in 5΄-UTR pools with varying translation efficiency. The colors of plots reflect the translation efficiency from the most efficient (green) to the least efficient (red). Height of bars corresponds to the frequency of motifs with indicated similarity to SD PSSM. (B) Positional distribution of SD sequence PSSM score for 5΄-UTR pools differing in translation efficiency. Position relative to mRNA sequence is shown above graphs; sketch of 5΄-UTR sequence is shown below graphs. Lines correspond to individual mRNAs grouped by the position of maximal PSSM score. The color indicates a similarity of subsequence starting at particular nucleotide to SD sequence (Supplementary Figure S7B). The translation efficiency increases from the least efficient (F1) to the most efficient (F8). A related Supplementary Figure S7 contains the same data for 5΄-UTRs with randomized 30 nt fragments.
Plotting similarity to the SD sequence for each position of the sliding window for all 5΄-UTR sequences in the data set (Figure 5B, Supplementary Figure S7B) revealed a steady increase in frequency of SD-like sequences from poorly translated mRNAs to efficiently translated ones. Moreover, in poorly translated mRNAs SD-like sequences could be on average more frequently found outside its preferred location in natural mRNAs, while efficiently translated mRNAs are enriched in favorably located SD sequences (compare Figure 5B, Supplementary Figure S7B, panels F1, showing even distribution of few SD-like sequences, with panels F8 indicating that the majority of SD-like sequences are located at optimal positions).
Unexpectedly, 5΄-UTRs that possesses several SD like sequences are enriched among efficiently translated mRNA. It can be seen from overall increase in motifs partially matching SD sequence along efficiently translated 5΄-UTRs in addition to single strong SD sequence. However, an accurate quantification of this phenomenon seems difficult. This effect may be explained in part by the fact that the SD sequence contains a repeat of the AGG motif, so it can form duplexes with the 3΄-end region of 16S rRNA in three relative orientations. This property could explain an apparent presence of two additional secondary maxima of similarity to SD sequence that surround the main maximum clearly visible in Figure 5B and Supplementary Figure S7B. However, even if we subtract this effect from the observed experimental data (Supplementary Figure S8), we still see that multiple SD sequences on average yield stronger increase of the translation efficiency than single SD sequence.
Automatic selection of features influencing translation enhancement
We then applied automatic machine learning methods to construct predictive models for translation efficiency and extract features of 5΄-UTR sequence that influence the translation efficiency most significantly. This analysis was performed for the library containing 30 randomized nucleotides. The following input features were considered: left and right pairing scores (RNAfold scores), SD scores and ΔG folding in a 30 nt window. In total, the number of 5΄-UTR sequence features was 230. The data set contained 6477 5΄-UTR sequences. Each 5΄-UTR sequence was supplied with an 8-dimensional output vector with components equal to numbers of occurrences of the sequence in each of eight fractions differed by their CER/RFP ratio. In some cases all occurrences belonged to the same fraction, while in other cases a minor number of occurrences belonged to adjacent fractions.
The predictive models were constructed in two different ways. First, the problem was considered as a regression one. We used the Random Forest regression method (32) with a multidimensional output. We modeled not the initial output, but its normalized version, transforming each component of the output into [0,1] by dividing it by the sum of all output components. The prediction of the fraction for a given 5΄-UTR sequence was the index of the output component for which the trained regression model provided the highest value.
Second, the problem was considered as a multiclass classification problem. For each 5΄-UTR sequence, the class label was the index of the output component with the maximal value, hence, we had eight classes corresponding to eight fractions. We used logistic regression (32) as the base classifier and the one-versus-the-rest strategy (33) to handle the multiclass data.
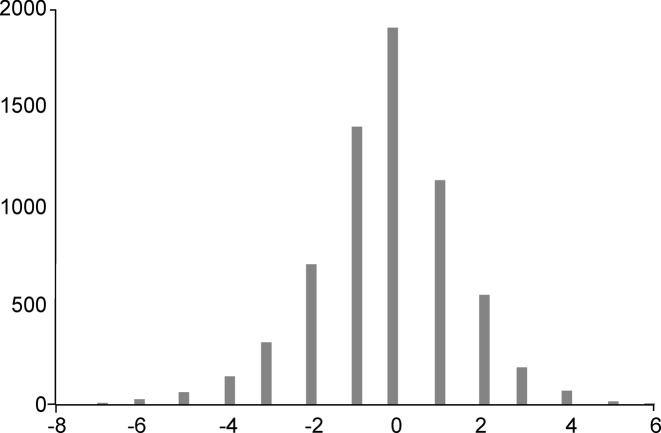
The prediction accuracies of the two methods were comparable (Supplementary Table S8). The number of large classification errors, when the predicted class differed sharply from the predicted one, is low, and most errors involve immediately adjacent classes (Figure 6). Hence, we further restricted the analysis to the multiclass classification by logistic regression.
Figure 6.
Histogram of differences between true and predicted integer class labels (fraction numbers) for the multiclass classification by logistic regression approach.
We then used the predictive model for automatic selection of the most important features that influence prediction accuracy. Again, we considered two approaches.
First, we used classification based on the logistic regression with l1-regularization (32,33) (the results below are for the case of regularization parameter equal to 0.05). We constructed a predictive model in two steps: (i) apply logistic regression with l1-regularization and select the most important features as those corresponding to non-zero coefficients of the regression equation, (ii) re-estimate coefficients of the logistic regression using only l2-regularization and the selected features. Second, we considered the integer class label (1, …, 8) as an ordinal variable and used the ordinal regression (40) that incorporates the ordinal nature of the dependent variable. The results for both approaches are shown in Supplementary Table S9. Again, there were no strong differences in the accuracy, although the ordinal regression performed slightly better (that had been expected, as it captures additional structure present in the data).
We then selected a set of the most important features, which corresponded to the highest coefficients both in the logistic regression model and in the ordinal regression model, restricting the initial set of 230 features to 57 ones. We then again applied ordinal regression, and observed no drop in performance (Supplementary Table S10). Finally, we checked whether we could improve the accuracy of prediction by adding specially generated features to the initial set of 230 features. We generated new features by summing adjacent 3–4 features of type L and R. For example, a new feature 9RRRR is the sum of four values of features 9R, 10R, 11R, 12R. The extended feature set contained 473 features, and after performing feature selection procedure as above, we obtained a set of the most important 53 features without loss of accuracy (Supplementary Table S11).
From this analysis we conclude that there is a statistically significant dependence between considered features and translation, although it is not sufficient to predict the fraction with a high accuracy. A set of the most important features can be selected, reducing the input dimension from 230 to only 53, without the loss in prediction accuracy. The selected features are in agreement with the subject domain knowledge. Comparison of observed and predicted translation efficiency for randomly selected 80 sequences from the data set is presented on a Supplementary Figure S9. The accuracy of prediction is on the same range with that of the previously published algorithm (7). The difference with published algorithm (7), however, is that we used machine learning and not construction of the knowledge based model.
Mutational analysis of 5΄-UTRs with unexpectedly high translation efficiencies
While all known determinants were found to influence translation efficiency, their presence could not explain all variability of translation efficiency across the entire data set. In order to identify new translation enhancers beyond SD sequences we selected three 5΄-UTRs with surprisingly high translation efficiency and minimal SD-like sequences for detailed mutational analysis.
Among efficiently translated mRNAs, belonging to top fraction 8, sorted by translation efficiency we observed mRNAs with an A-rich 5΄-UTR (Table 1, A-rich 5΄-UTR) that, on the other hand, had at most 4 nt SD sequence. To study it in detail, we created a reporter plasmid containing this 5΄-UTR sequence upstream the CER fluorescent protein gene. The measured translation efficiency of the CER mRNAs encoded by this reporter construct appeared to be 6-fold higher than that for the control RFP mRNA encoded in the same reporter construct (Table 1, Control), matching what had been expected based on the Flowseq experiment. An A-rich 5΄-UTR contained two short SD sequences (AAGG and GGA) and an additional AUG codon located in-frame with CER gene. Mutation of the additional AUG codon decreased the translation efficiency about 2-fold (Table 1, A-rich 5΄-UTR noAUG), indicating that upstream AUG codon might be used for translation initiation in addition to one starting the CER gene. Substitution of the shorter SD sequence, GGA, by AAA (Table 1, A-rich 5΄-UTR -2G) resulted in a 1.3-fold decrease in the translation efficiency indicating that this additional SD sequence contributed moderately to the efficiency of translation. Mutation of both SD sequences and the additional AUG codon resulted in 16-fold decrease in the translation efficiency (Table 1, A-rich 5΄-UTR -5G). It should be noted that the latter mutant possesses at most two nucleotides complementary to the 16S rRNA 3΄-terminal region, but its translation efficiency is still an order of magnitude higher than that for other reporter mRNA with 2 nt SD sequences (11). Thus, the efficient translation of this ‘A-rich 5΄-UTR’ mRNA could be explained by the additive effect of two SD sequences, one contributing more than the other, additional AUG codon, and yet unknown sequence elements that increase the translation efficiency in addition to those listed above. Of note, the observed translation efficiency of ‘A-rich 5΄-UTR’ mRNA and its mutants may not be explained based of features known to be important for initiation of translation (Table 1, last column). The ‘A-rich 5΄-UTR’ has a notably reduced number of cytosine residues, as do many 5΄-UTRs with high efficiency of translation (Figure 3).
Table 1. Translation efficiency of ‘A-rich 5΄-UTR’ mRNA and its mutant forms.
| Name | 5΄-UTR sequence | Relative translation efficiency* | Predicted translation efficiency# |
|---|---|---|---|
| Control | GGAGAAGGAGAUAUCAU | 1 | 1 |
| A-rich 5΄-UTR | GGGAUUUAAAAAAAGGCGGAAAAAUAAUGCAU | 6.02 | 0.68 |
| A-rich 5΄-UTR noAUG | GGGAUUUAAAAAAAGGCGGAAAAAUAAUACAU | 2.59 | 0.82 |
| A-rich 5΄-UTR -2G | GGGAUUUAAAAAAAGGCAAAAAAAUAAUGCAU | 4.50 | 0.68 |
| A-rich 5΄-UTR -5G | GGGAUUUAAAAAAAAACAAAAAAAUAAUACAU | 0.38 | 1.57 |
| 2/7 | CACACAACACCUGAUCAACU | 0.03 | 0.14 |
*Relative values of fluorescence of CER protein, whose mRNA contained 5΄-UTR shown and RFP protein whose mRNA contained ‘Control’ 5΄-UTR.
#Relative translation efficiencies predicted by RBS calculator on the basis of known mRNA features affection translation.
Another unusually efficient mRNA possessed an SD sequence of 4 nt (Table 2, Short SD 5΄-UTR) located at a distance 13 nt from AUG. A similar artificial mRNA (Table 2, 4/13) had almost two orders of magnitude weaker translation. Several mutations were introduced to this 5΄-UTR (Table 2) to identify regions that affect translation. To check for possibility that unusually efficient translation of Short SD 5΄-UTR mRNA is due to additional in-frame GUG that may act as potential start codon we converted it to GUC and GCG (Table 2, ShortSD-GUC and ShortSD-GCG). No or insignificant reduction of translation efficiency resulted from these substitutions speaks in favor of insignificance of this GUG codon for translation efficiency.
Table 2. Translation efficiency of ‘Short SD 5΄-UTR’ mRNA and its mutant forms.
| Name | 5΄-UTR sequence | Relative translation efficiency* | Predicted translation efficiency# |
|---|---|---|---|
| Control | GGAGAAGGAGAUAUCAU | 1 | 1 |
| Short SD 5΄-UTR | GGUUUCUUAUUUGGUUCGGAGUGAGAUGCGAU | 4.31 | 0.68 |
| Short SD 5΄-UTR -SD | GGUUUCUUAUUUGGUUCCCUCUGAGAUGCGAU | 0.076 | 0.13 |
| Short SD 5΄-UTR -9UA | GGCCCCCCGCCCGGUUCGGAGUGAGAUGCGAU | 0.073 | 1.18 |
| Short SD 5΄-UTR -3U | GGUUUCUUACCCGGUUCGGAGUGAGAUGCGAU | 0.51 | 0.65 |
| Short SD 5΄-UTR -3΄ | GGUUUCUUAUUUGGUUCGGAGUCACAUCCCAU | 2.35 | 0.18 |
| Short SD 5΄-UTR +SD | GGUUUCUUAUUUGGUUCCACAUGGAGUCCCAU | 0.41 | 0.92 |
| Short SD 5΄-UTR -GUC | GGUUUCUUAUUUGGUUCGGAGUCAGAUGCGAU | 3.21 | 0.11 |
| Short SD 5΄-UTR -GCG | GGUUUCUUAUUUGGUUCGGAGCGAGAUGCGAU | 4.32 | 0.56 |
| 4/13 | CACCCGGAGCAACAACAACU | 0.06 | 0.04 |
| 4/7 | CACACAACACCGGAGCAACU | 0.8 | 0.22 |
*Relative values of fluorescence of CER protein, whose mRNA contained 5΄-UTR shown and RFP protein whose mRNA contained ‘Control’ 5΄-UTR.
#Relative translation efficiencies predicted by RBS calculator on the basis of known mRNA features affection translation.
Mutations of the SD sequence (Table 2, Short SD 5΄-UTR -SD) practically abolished translation. However, mutations of the 5΄-proximal region, leaving the SD sequence intact, also decreased translation to the same extent (Table 2, Short SD 5΄-UTR -9UA). Even substitution of three uridine residues upstream to the SD sequence significantly decreased translation (Table 2, Short SD 5΄-UTR -3U). Mutations in the region between the SD sequence and the start codon affected translation only moderately (Table 2, Short SD 5΄-UTR -3΄). Moving the SD sequence closer to the start codon (Table 2, Short SD 5΄-UTR +SD), which has been highly beneficial for the artificial mRNA with a 4 nt SD sequence (Table 2, 4/7), decreased the translation efficiency more than 5-fold. The unexpected translation efficiency of this 5΄-UTR may be explained by a combination of previously uncharacterized translation enhancers. Notably, the translation efficiency is highly sensitive to mutations within this 5΄-UTR. Not only the presence, but also the location of the SD sequence proved to be important for the translation efficiency; and the most puzzling finding is that the SD location distant to the start codon, unfavorable for other 5΄-UTRs, is beneficial in this case. The U-rich sequence located upstream the distant SD was shown to be an enhancer of translation, according to the translation efficiency of Short SD 5΄-UTR -3U and Short SD 5΄-UTR -9UA mutants.
The third 5΄-UTR with unexpectedly high translation efficiency contained six AG repeats in a row (Table 3, AG-rich 5΄-UTR). Additionally it contained a candidate GGAG SD sequence located at a long distance, 20 nt to AUG start codon, making it unlikely to be functional. Cells carrying this reporter construct were found in fraction 7, the second best in the translation efficiency. In agreement with it, when this 5΄-UTR sequence was cloned upstream the CER gene in an individual reporter construct (Table 3, AG-rich 5΄-UTR), the latter was translated 5-fold more efficiently than the control RFP gene (Table 3, Control). Mutations that disrupt the first (Table 3, AG-rich 5΄-UTR –AG1-3) and the last (Table 3, AG-rich 5΄-UTR –AG4-6) three AG repeats resulted in 34 and 3.6-fold decrease in the translation efficiency, respectively. One possibility was that the GAG sequence found in the AG repeats could serve as an SD sequence. We mutated the proximal (Table 3, AG-rich 5΄-UTR +SD1) and the distal (Table 3, AG-rich 5΄-UTR +SD2) parts of the AG repeat sequence to create stronger SD sequences AGGAG and GGAGG. Surprisingly, introduction of a SD sequence at any of these two positions decreased the translation efficiency about 3.3-fold (Table 3). Moreover, even elimination of a single G in the middle of the AG repeat region (Table 3, AG-rich 5΄-UTR -G) decreased translation 4.5-fold. Thus, the AG-repeated region serves as a unique translation enhancer element. Even after elimination of the most functionally important part of it (Table 3, AG-rich 5΄-UTR –AG1-3) the residual translation efficiency was considerably higher than that of an artificial reporter mRNA (Table 3, 4/16) containing a comparable SD sequence.
Table 3. Translation efficiency of ‘AG-rich 5΄-UTR’ mRNA and its mutant forms.
| Name | 5΄-UTR sequence | Relative translation efficiency* | Predicted translation efficiency# |
|---|---|---|---|
| Control | GGAGAAGGAGAUAUCAU | 1 | 1 |
| AG-rich 5΄-UTR | GGAGUCUAAAGAGAGAGAGAGU | 5.09 | 2.05 |
| AG-rich 5΄-UTR –AG1-3 | GGAGUCUAAACACACAGAGAGU | 0.15 | 0.13 |
| AG-rich 5΄-UTR –AG4-6 | GGAGUCUAAAGAGAGACACACU | 1.40 | 0.58 |
| AG-rich 5΄-UTR +SD1 | GGAGUCUAAAGAGAGGAGAAGU | 1.52 | 4.18 |
| AG-rich 5΄-UTR +SD2 | GGAGUCUAAAGGAGGAGAGAGU | 1.56 | 12.37 |
| AG-rich 5΄-UTR -G | GGAGUCUAAAGAGACAGAGAGU | 1.14 | 0.31 |
| 4/16 | CCGGAGCACACACAACAACU | 0.02 | 0.04 |
*Relative values of fluorescence of CER protein, whose mRNA contained 5΄-UTR shown and RFP protein whose mRNA contained ‘Control’ 5΄-UTR.
#Relative translation efficiencies predicted by RBS calculator on the basis of known mRNA features affection translation.
To rule out a possibility that unusual translation efficiency is an artificial result of an interaction between 5΄-UTR and CER coding region, e.g. via long-range secondary structure formation, we switched CER and RFP coding regions. Thus, created pCERRFP reporter plasmid was used for insertion of unusually efficient A-rich, Short SD and AG-rich 5΄-UTR sequences and several of their mutant variants in front of RFP reporter gene, this time using CER gene as a control (Supplementary Tables S14–S16). We found that translation efficiencies follow the same tendencies for either of CER and RFP reporter genes. Switching CER and RFP coding regions even exaggerate observed differences between the control, highly efficient 5΄-UTRs and their mutant forms.
We searched through the 5΄-UTR sequences of natural E. coli genes to see if we could identify similar AG-rich translation enhancers that are present at a place of SD sequence. Among the genes that possesses AG-rich 5΄-UTR sequences instead of SD we found ybaB (conserved DNA-binding protein, ribosome binding site GAGAGAGAAACCUAUG), lon (protease, ribosome binding site GAGAGAGCUCUAUG), rplY (gene encoding ribosomal protein L25, ribosome binding site AGAGAGAAAGAAAUG) and lrp (transcriptional regulator, ribosome binding site AGAGAGACAAUAAUAAUG), while other genes possessed shorter AG tracks. We inserted complete 5΄-UTR sequences of ybaB, lon, rplY and lrp genes upstream of CER reporter gene (Table 4). If several 5΄-UTR forms were annotated for a gene, we used the shortest annotated form. In addition, to evaluate the significance of AG-tracks for translation efficiency we substituted AG- with AC-repeats and, separately, mutated AG-tracks to strong SD sequences (Table 4). Although for the tested natural AG-rich 5΄-UTRs we found SD sequence to perform better than naturally occurring AG-repeats (Table 4, compare lon and lon+SD, lrp and lrp+SD, ybaB and ybaB+SD, rplY and rplY+SD), mutations eliminating AG-repeats in all tested cases resulted in an order of magnitude lower translation (Table 4, compare lon and lon-AG, lrp and lrp-AG, ybaB and ybaB-AG, rplY and rplY-AG). As one can see an efficiency of AG repeat containing 5΄UTR related to that of SD sequence containing depends on a context. Invariably, AG-repeats performed better to stimulate translation than similar 5΄-UTR sequence without this element. Thus, we can suggest regarding them as context-dependent translation enhancers.
Table 4. Translation efficiency of natural AG-rich 5΄-UTR mRNAs and their mutant forms.
| Name | 5΄-UTR sequence | Relative translation efficiency* | Predicted translation efficiency# |
|---|---|---|---|
| Control | GGAGAAGGAGAUAUCAU | 1 | 1 |
| lon | TTAAACTAAGAGAGAGCTCT | 0.25 | 0.36 |
| lon-AG | TTAAACTAACACACAGCTCT | 0.03 | 0.06 |
| lon+SD | TTAAACTAAGGAGGAGCTCT | 1.84 | 3.14 |
| lrp | AATACAGAGAGACAATAATT | 1.04 | 0.67 |
| lrp-AG | AATACACACACACAATAATT | 0.13 | 0.12 |
| lrp+SD | AATACAGGAGGACAATAATT | 5.66 | 1.91 |
| ybaB | CGTGATTGAGAGAGAAACCT | 0.8 | 2.86 |
| ybaB-AG | CGTGATTCACACAGAAACCT | 0.04 | 0.32 |
| ybaB+SD | CGTGATTGGAGGAGAAACCT | 4.84 | 9.31 |
| rplY | TTAAACTAAGAGAGAGCTCT | 0.19 | 0.36 |
| rplY-AG | TTAAACTAACACACAGCTCT | 0.03 | 0.06 |
| rplY+SD | TTAAACTAAGGAGGAGCTCT | 1.81 | 3.14 |
*Relative values of fluorescence of CER protein, whose mRNA contained 5΄-UTR shown and RFP protein whose mRNA contained ‘Control’ 5΄-UTR.
#Relative translation efficiencies predicted by RBS calculator on the basis of known mRNA features affection translation.
Unusually high efficiency of translation if the reporter gene is preceded by some natural E. coli 5΄-UTRs
We used the dual fluorescent protein reporter to study the influence of known features of mRNA, such as variable start codons, SD sequences of variable length and location, secondary structure elements and AU-rich sequences on the translation efficiency. Previously we have noticed that the majority of natural mRNA of E. coli possesses relatively short SD sequences, most common being four nucleotides long. Artificial reporter mRNAs with SD sequences of similar length and position were translated at a moderate level in our system (17). Identification of AG-rich translation enhancer in an artificial and a set of natural 5΄-UTRs prompted us to analyze larger set of natural 5΄-UTR sequences for their translation effciency.
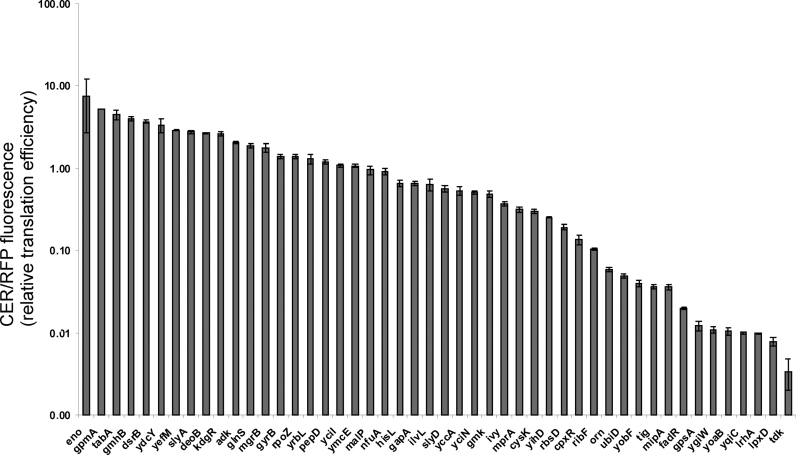
To this end we cloned shortest, according to RegulonDB, 5΄-UTR parts of tabA, ydcY, slyA, gmhB, yciN, ivy, yihD, pepD, yciI, cysK, cpxR, mipA, yrbL, gpmA, kdgR, adk, yefM, slyD, mprA, yobF, tig, glnS, mgrB, rpoZ, deoB, eno, rbsD, ubiD, gpsA, lrhA, lpxD, gyrA, malP, nfuA, ilvL, hisL, gapA, yccA, gmk, ribF, orn, fadR, ygiW, ymcE, yqiC, yoaB, tdk, dsrB genes (Supplementary Table S12) upstream of CER. This set covers a range of SD sequence lengths from 2 to 7 and a distance from the central guanosine in the aagGagg sequence to the start codon from 1 to 17; some extreme examples most likely being non-functional. All of cloned natural 5΄-UTR sequences lack known regulatory protein of RNA binding site, except for gpmA and yobF (41). The translation efficiencies were measured as the CER fluorescence normalized to that of the RFP, used as a control (Figure 7, Supplementary Table S12). While the overall range of translation efficiencies was comparable for natural and artificial 5΄-UTRs, ‘superefficient’ artificial 5΄-UTRs required long SD sequences of 6 to 8 nucleotides, while some natural 5΄-UTRs having SD sequences of 3 to 5 nucleotides demonstrated superior translation efficiencies.
Figure 7.
Dependence of the CER/RFP translation efficiency on preceding natural 5΄-UTR sequences normalized to that of control RFP mRNA.
Masking of a translation initiation site by a secondary structure may dramatically decrease translation efficiency. Artificial reporter mRNAs listed in Supplementary Table S12 were created to minimize the secondary structure of the ribosome binding sites, while natural mRNAs might possess secondary structure elements that inhibit translation. Hence, while lower translation rates of natural 5΄-UTRs could be explained by masking of translation initiation sites by secondary structures, this does not explain increased translation efficiencies of natural, relative to artificial, mRNAs. The most unexpected among tested natural 5΄-UTRs was highly efficient translation induced by the eno 5΄-UTR. This 5΄-UTR possesses a SD sequence of 4 nt located 9 nt upstream the start codon. Artificial reporter mRNAs possessing SD elements with similar features demonstrated an order of magnitude lower translation efficiencies (17).
The influence of each nucleotide in the eno 5΄-UTR on the translation efficiency
To determine the contribution of each nucleotide of the eno 5΄-UTR to the translation efficiency, we created a library of plasmids encoding randomly mutated eno 5΄-UTR. To this end we synthesized a DNA fragment corresponding to this 5΄-UTR using, for each position, a mixture of nucleotides containing 90% of the nucleotide naturally occurring at this position and ∼10% formed by equal amounts of the other three nucleotides. The created DNA library was cloned upstream the CER reporter gene to the pRFPCER reporter vector (17,27). E. coli cells transformed with this plasmid library and divided into two replicates were sorted to eight fractions differing by their CER/RFP ratio (Supplementary Figure S10). The CER and RFP fluorescence and proportion of cells in each fraction are shown in Supplementary Figure S2B. The pools differing by the translation efficiency were collected and used for next generation sequencing (Supplementary Table S13).
A good correlation between two replicas (Supplementary Figure S4A) allowed us to merge them and analyze the complete data set. In most cases an individual 5΄-UTR sequence was found in one or two adjacent fractions sorted by translation efficiencies. However, sometimes a given sequence was seen simultaneously in an efficiently translated pool and a pool of very inefficient mRNAs. The most probable explanation of this effect is accumulation of secondary mutations inactivating expression of the reporter CER gene elsewhere in a plasmid. In such cases we ignored the occurrence of a sequence in the poorly translated pool.
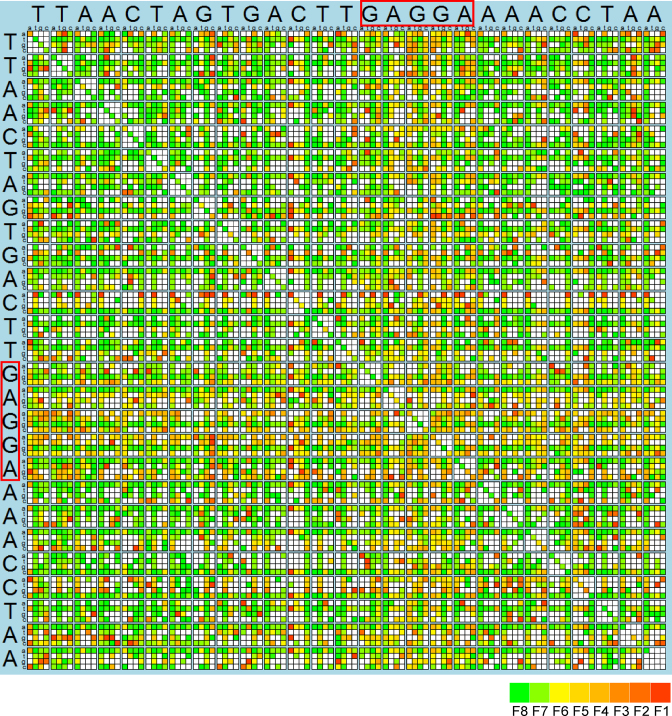
Totally, 4820 unique mutants of eno 5΄-UTR was found in the data set, and among them we found all 81 single nucleotide substitutions and 1292 out of 3402 possible double nucleotide substitutions. Their translation efficiencies are presented in Figure 8. As expected, mutations that disrupt the SD sequence either alone or in combination with other mutations diminish the translation efficiency (see the yellowish band in Figure 8 corresponding to substitutions in the SD sequence). However, we noted that the eno 5΄-UTR contains two overlapping SD sequences (GAGG, AGGA) each of four nucleotides long, forming a single GAGGA motif. Mutations that disrupted one, but not the other still moderately affected translation, but a strong effect was observed only for mutations that disrupted both SD sequences. These results might be interpreted to suggest that overlapping SD sequences are more beneficial for translation than a single one.
Figure 8.
Translation efficiency of CER mRNAs with 5΄-UTR corresponding to single and double mutants of eno 5΄-UTR. The axes correspond to nucleotides of eno 5΄-UTR shown by large letters. Coordinates of each square correspond to the first and second mutation of eno 5΄-UTR. Nucleotide variants are shown by small letters. Each square represents a 5΄-UTR variant with translation efficiency shown by color from green (the most efficient translation) to red (the least efficient one). White denotes lack of a 5΄-UTR variant in the data set. Two overlapping SD sequences are boxed. Color code is shown below the plot.
While studying the significance of individual nucleotides for the translation efficiency of the eno 5΄-UTR (Figure 8) along with the SD sequence, we identified a number of other residues crucial for the efficient translation. Only in a subset of mutants, such as C12A, the drop in the translation efficiency could be explained by formation of an unfavorable secondary structure. For other critical nucleotides, such as U2, G8 and A26, other explanations are needed. The negative effect of mutations sometimes is additive, e.g. with SD sequence mutations. Quite frequently drop in translation was caused by a combination of mutations that did not yield a significant reduction in translation efficiency alone (isolated red spots in the map in Figure 8). We also observed mutations that suppressed negative effects of other substitutions. Two of them are U14G, extending the SD sequence and C24U, destabilizing secondary structure formed by the C12A mutation.
Notably, there are no positions in the eno 5΄-UTR where all substitutions were inhibitory for translation. The likely explanation of this is that there are no strictly sequence-specific contacts of mRNA upstream from SD sequence with the ribosome, but there are positions where particular nucleotides are unfavorable. It seems that translation efficiency of eno 5΄-UTR could not be explained by a single enhancer-like region, but rather by a combination of multiple factors.
CONCLUSION
Despite decades of intensive study, we still lack a complete understanding of the rules that determine the translation efficiency. Here, we analyzed the translation efficiency of 48 natural E. coli 5΄-UTR, more than 4000 of mutant variants of natural eno 5΄-UTR, and over a 20 000 of 5΄-UTRs from randomized sequence libraries. All known factors that affect translation efficiency, such as SD sequence of the optimal length and location, lack of secondary structure and AU-rich enhancers were clearly revealed in our study. A remarkable feature of efficient 5΄-UTRs is reduced proportion of cytosine residues.
We found many examples of 5΄-UTRs, both artificial and natural, whose efficiency could not be easily explained by the known features, and in general, a precise prediction of the translation efficiency of an arbitrary 5΄-UTR seems problematic. We demonstrated the beneficial role of multiple SD sequences and start codons for translation and found an unusual ribosome binding site composed of AG repeats. Such repeats were found in several natural E. coli ribosome binding sites, indicating that it may represent a natural translation enhancer substituting for the SD sequence. The results of this study could be applied for creation of vectors for protein production in biotechnology and help to develop frameworks for fine tuning gene regulation systems.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by Russian Foundation for Basic Research [grant numbers 16-04-01100, 14-04-01061, 15-34-20139] (library preparation and sorting), Russian Science Foundation [grant 14-14-00072] (individual mutants cloning and analysis) and grant [14-50-00150] (data analysis) and Moscow University Development Program [grant PNR 5.13] (purchasing sorter).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Russian Foundation for Basic Research [16-04-01100, 14-04-01061, 15-34-20139]; Russian Science Foundation [14-14-00072 and 14-50-00150]; Moscow University Development Program [PNR 5.13]. Funding for open access charge: Russian Science Foundation [14-14-00072].
Conflict of interest statement. None declared.
REFERENCES
- 1. Li G.W. How do bacteria tune translation efficiency?. Curr. Opin. Microbiol. 2015; 24:66–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Li G.W., Burkhardt D., Gross C., Weissman J.S.. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell. 2014; 157:624–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Reeve B., Hargest T., Gilbert C., Ellis T.. Predicting translation initiation rates for designing synthetic biology. Front. Bioeng. Biotechnol. 2014; 2:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Laursen B.S., Sorensen H.P., Mortensen K.K., Sperling-Petersen H.U.. Initiation of protein synthesis in bacteria. Microbiol. Mol. Biol. Rev. 2005; 69:101–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kudla G., Murray A.W., Tollervey D., Plotkin J.B.. Coding-sequence determinants of gene expression in Escherichia coli. Science. 2009; 324:255–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Salis H.M., Mirsky E.A., Voigt C.A.. Automated design of synthetic ribosome binding sites to control protein expression. Nat. Biotechnol. 2009; 27:946–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Salis H.M. The ribosome binding site calculator. Methods Enzymol. 2011; 498:19–42. [DOI] [PubMed] [Google Scholar]
- 8. Seo S.W., Yang J.S., Kim I., Yang J., Min B.E., Kim S., Jung G.Y.. Predictive design of mRNA translation initiation region to control prokaryotic translation efficiency. Metab. Eng. 2013; 15:67–74. [DOI] [PubMed] [Google Scholar]
- 9. Na D., Lee D.. RBSDesigner: software for designing synthetic ribosome binding sites that yields a desired level of protein expression. Bioinformatics. 2010; 26:2633–2634. [DOI] [PubMed] [Google Scholar]
- 10. Mitarai N., Sneppen K., Pedersen S.. Ribosome collisions and translation efficiency: optimization by codon usage and mRNA destabilization. J. Mol. Biol. 2008; 382:236–245. [DOI] [PubMed] [Google Scholar]
- 11. Tuller T., Carmi A., Vestsigian K., Navon S., Dorfan Y., Zaborske J., Pan T., Dahan O., Furman I., Pilpel Y.. An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell. 2010; 141:344–354. [DOI] [PubMed] [Google Scholar]
- 12. Tuller T., Waldman Y.Y., Kupiec M., Ruppin E.. Translation efficiency is determined by both codon bias and folding energy. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:3645–3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Goodman D.B., Church G.M., Kosuri S.. Causes and effects of N-terminal codon bias in bacterial genes. Science. 2013; 342:475–479. [DOI] [PubMed] [Google Scholar]
- 14. Pop C., Rouskin S., Ingolia N.T., Han L., Phizicky E.M., Weissman J.S., Koller D.. Causal signals between codon bias, mRNA structure, and the efficiency of translation and elongation. Mol. Syst. Biol. 2014; 10:770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shine J., Dalgarno L.. Determinant of cistron specificity in bacterial ribosomes. Nature. 1975; 254:34–38. [DOI] [PubMed] [Google Scholar]
- 16. Vimberg V., Tats A., Remm M., Tenson T.. Translation initiation region sequence preferences in Escherichia coli. BMC Mol. Biol. 2007; 8:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Osterman I.A., Evfratov S.A., Sergiev P.V., Dontsova O.A.. Comparison of mRNA features affecting translation initiation and reinitiation. Nucleic Acids Res. 2013; 41:474–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen H., Bjerknes M., Kumar R., Jay E.. Determination of the optimal aligned spacing between the Shine-Dalgarno sequence and the translation initiation codon of Escherichia coli mRNAs. Nucleic Acids Res. 1994; 22:4953–4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Boni I.V., Isaeva D.M., Musychenko M.L., Tzareva N.V.. Ribosome-messenger recognition: mRNA target sites for ribosomal protein S1. Nucleic Acids Res. 1991; 19:155–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Komarova A.V., Tchufistova L.S., Dreyfus M., Boni I.V.. AU-rich sequences within 5΄ untranslated leaders enhance translation and stabilize mRNA in Escherichia coli. J. Bacteriol. 2005; 187:1344–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. de Smit M.H., van Duin J.. Secondary structure of the ribosome binding site determines translational efficiency: a quantitative analysis. Proc. Natl. Acad. Sci. U.S.A. 1990; 87:7668–7672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Barendt P.A., Shah N.A., Barendt G.A., Sarkar C.A.. Broad-specificity mRNA-rRNA complementarity in efficient protein translation. PLoS Genet. 2012; 8:e1002598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Barendt P.A., Shah N.A., Barendt G.A., Kothari P.A., Sarkar C.A.. Evidence for context-dependent complementarity of non-Shine-Dalgarno ribosome binding sites to Escherichia coli rRNA. ACS Chem. Biol. 2013; 8:958–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li G.W., Oh E., Weissman J.S.. The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature. 2012; 484:538–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Holmqvist E., Reimegard J., Wagner E.G.. Massive functional mapping of a 5΄-UTR by saturation mutagenesis, phenotypic sorting and deep sequencing. Nucleic Acids Res. 2013; 41:e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kosuri S., Goodman D.B., Cambray G., Mutalik V.K., Gao Y., Arkin A.P., Endy D., Church G.M.. Composability of regulatory sequences controlling transcription and translation in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:14024–14029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Osterman I.A., Prokhorova I.V., Sysoev V.O., Boykova Y.V., Efremenkova O.V., Svetlov M.S., Kolb V.A., Bogdanov A.A., Sergiev P.V., Dontsova O.A.. Attenuation-based dual-fluorescent-protein reporter for screening translation inhibitors. Antimicrob. Agents Chemother. 2012; 56:1774–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Oliphant A.R., Nussbaum A.L., Struhl K.. Cloning of random-sequence oligodeoxynucleotides. Gene. 1986; 44:177–183. [DOI] [PubMed] [Google Scholar]
- 29. Worthington M.T., Pelo J., Luo R.Q.. Cloning of random oligonucleotides to create single-insert plasmid libraries. Anal. Biochem. 2001; 294:169–175. [DOI] [PubMed] [Google Scholar]
- 30. Lorenz R., Bernhart S.H., Honer Zu Siederdissen C., Tafer H., Flamm C., Stadler P.F., Hofacker I.L.. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011; 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jacobs G.H., Chen A., Stevens S.G., Stockwell P.A., Black M.A., Tate W.P., Brown C.M.. Transterm: a database to aid the analysis of regulatory sequences in mRNAs. Nucleic Acids Res. 2009; 37:D72–D76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hastie T., Tibshirani R., Friedman J.. Elements of Statistical Learning. Springer Series in Statistics. 2009; http://www.springer.com/in/book/9780387848570. [Google Scholar]
- 33. Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V. et al. Scikit-learn: Machinelearning in Python. J Machine Learning Res. 2011; 12:2825–2830. [Google Scholar]
- 34. Dessau R.B., Pipper C.B.. [‘R’–project for statistical computing]. Ugeskr. Laeger. 2008; 170:328–330. [PubMed] [Google Scholar]
- 35. Sanner M.F. Python: a programming language for software integration and development. J. Mol. Graph. Model. 1999; 17:57–61. [PubMed] [Google Scholar]
- 36. Noderer W.L., Flockhart R.J., Bhaduri A., Diaz de Arce A.J., Zhang J., Khavari P.A., Wang C.L.. Quantitative analysis of mammalian translation initiation sites by FACS-seq. Mol. Syst. Biol. 2014; 10:748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dvir S., Velten L., Sharon E., Zeevi D., Carey L.B., Weinberger A., Segal E.. Deciphering the rules by which 5΄-UTR sequences affect protein expression in yeast. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:E2792–E2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Blattner F.R., Plunkett G. 3rd, Bloch C.A., Perna N.T., Burland V., Riley M., Collado-Vides J., Glasner J.D., Rode C.K., Mayhew G.F. et al. The complete genome sequence of Escherichia coli K-12. Science. 1997; 277:1453–1462. [DOI] [PubMed] [Google Scholar]
- 39. Kim D., Hong J.S., Qiu Y., Nagarajan H., Seo J.H., Cho B.K., Tsai S.F., Palsson B.O.. Comparative analysis of regulatory elements between Escherichia coli and Klebsiella pneumoniae by genome-wide transcription start site profiling. PLoS Genet. 2012; 8:e1002867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Agresti A. Analysis of ordinal categorical data. 2010; Hoboken: Wiley. [Google Scholar]
- 41. Keseler I.M., Mackie A., Peralta-Gil M., Santos-Zavaleta A., Gama-Castro S., Bonavides-Martinez C., Fulcher C., Huerta A.M., Kothari A., Krummenacker M. et al. EcoCyc: fusing model organism databases with systems biology. Nucleic Acids Res. 2013; 41:D605–D612. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.