Abstract
One typical mechanism to promote genomic instability, a hallmark of cancer, is to inactivate tumor suppressors, such as PALB2. It has recently been reported that mutations in PALB2 increase the risk of breast cancer by 8–9-fold by age 40 and the life time risk is ∼3–4-fold. To date, predicting the functional consequences of PALB2 mutations has been challenging as they lead to different cancer risks. Here, we performed a structure–function analysis of PALB2, using PALB2 truncated mutants (R170fs, L531fs, Q775X and W1038X), and uncovered a new mechanism by which cancer cells could drive genomic instability. Remarkably, the PALB2 W1038X mutant, harboring a mutation in its C-terminal domain, is still proficient in stimulating RAD51-mediated recombination in vitro, although it is unusually localized to the cytoplasm. After further investigation, we identified a hidden NES within the WD40 domain of PALB2 and found that the W1038X truncation leads to the exposure of this NES to CRM1, an export protein. This concept was also confirmed with another WD40-containing protein, RBBP4. Consequently, our studies reveal an unreported mechanism linking the nucleocytoplasmic translocation of PALB2 mutants to cancer formation.
INTRODUCTION
Breast and ovarian cancers are among the most common and lethal malignancies in women worldwide. Mutations of the BRCA1 (BReast CAncer 1, early onset) or BRCA2 (BReast CAncer 2, early onset) genes have been associated with an increased risk of breast and ovarian cancer—ranging from 55 to 85% and 10 to 45%, respectively. Although these genes play a major role in breast and ovarian cancer susceptibility, all inherited cases cannot only be explained by mutations in these genes. BRCA1 and BRCA2 genes are involved in a common pathway of DNA repair: homologous recombination, an error-free mechanism that relies on the recombinase activity of the RAD51 protein and leads to the repair of double-strand breaks during the S and G2 phases of the cell cycle (1). Recent studies have linked several other genes involved in this pathway, such as APRIN (2) or PALB2 (Partner and Localizer of BRCA2), to breast or ovarian cancer development (3,4).
PALB2 codes for a protein essential for BRCA2 intranuclear localization and function in DNA repair, which also acts as the link between BRCA2 and BRCA1 (5–7). The cell cycle controls the interaction of BRCA1 with PALB2–BRCA2 to constrain BRCA2 function to the S/G2 phases in human cells (8). Given its capacity to interact directly with RAD51 and help it to overcome RPA inhibition, PALB2 has been known as a RAD51 mediator (9). PALB2 is a 1186-amino acid nuclear protein with a molecular weight of 130 kDa, whose export/import properties remain unknown. Several functional domains of PALB2 have been mapped and characterized (Figure 1A). For instance, the N-terminus harbors a coiled-coil domain that allows: (i) physical interaction with BRCA1 (5); (ii) dimerization of wild-type PALB2 (10); (iii) interaction with RAD51 (9). Two DNA binding domains (DBD) have also been identified: one close to the coiled-coil domain and the other in the middle region of PALB2 (9). Furthermore, a small domain involved in chromatin anchorage has been located upstream of the second DNA binding domain (11). The PALB2 amino-acid sequence ends with a WD40 repeat-containing domain, a β-propeller known to be a second RAD51-interacting domain (9) and a BRCA2-interacting domain (12). The WD40 domain is one of the most abundant and interacting domains in the human genome (13). It can interact with proteins, peptides or nucleic acids using multiple surfaces or modes of interaction. The WD40 domain is thought to possess a scaffolding role, as no WD40 domain with intrinsic enzymatic activity has been found (14).
Figure 1.
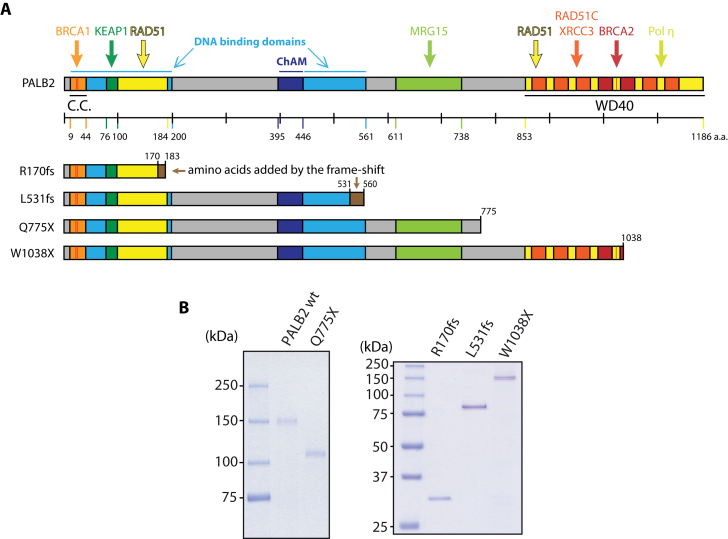
PALB2 wild-type and mutant proteins. (A) Schematic representation of PALB2 domains and truncated variant proteins analyzed in this study. C.C.: coiled-coil domain, this domain also allows dimerization of PALB2; WD40: WD40-repeat domain. (B) SDS-PAGE of purified wild-type PALB2 (wt) and truncated variants of PALB2 (250 ng). The proteins were stained with Coomassie blue.
Several cancer-causing mutations that predispose to breast and pancreatic cancer, as well as being implicated in the development of ovarian cancer, have been identified in the PALB2 gene. Some of these mutations have been predicted to lead to the expression of truncated proteins (6). In this study, we performed a structure–function analysis of PALB2 by using truncating PALB2 mutations found in breast or ovarian cancer patients. We selected four mutations according to the position of the truncation (Figure 1A), in order to study the effects of truncations all along the protein: c.509_510delGA (p.Arg170Ilefs*14, R170fs) (15), c.1592delT (p.Leu531Cysfs*30, L531fs) (16), c.2323C>T (p.Gln775*, Q775X) (17) and c.3113G>A (p.Trp1038*, W1038X) (18). We purified these PALB2 mutants and compared their biochemical properties to the full-length protein. We show that the PALB2 R170fs mutant fails to localize to DNA damage sites. Unexpectedly, PALB2 W1038X, the closest to the wild-type protein in length, presents a nuclear localization defect. This finding allowed us to uncover a Nuclear Export Sequence (NES), hidden in the wild-type PALB2 WD40 domain but exposed when a truncation into the WD40 domain occurs. Our results explain why a truncation in PALB2 could have a more severe effect than a complete inactivation of the gene.
MATERIALS AND METHODS
Chemicals
Neocarzinostatin (N9162) was purchased from Sigma. Selinexor (KPT-330) (S7252) was obtained from Selleckchem.
Western blotting
The antibodies used for immunodetection by western blot were: PALB2 antibody rabbit pAb (A301-246A, Bethyl), monoclonal ANTI-FLAG® M2 antibody produced in mouse (F3165, Sigma), GAPDH antibody mouse mAb (10R-G109a, Fitzgerald), anti-Histone H3 antibody—Nuclear Loading Control and ChIP Grade (ab1791, Abcam), purified mouse anti-human Exportin-1/CRM1 (611832, BD Biosciences), mouse anti-GFP (11814460001, Roche).
Protein purification, DNA binding and strand invasion assays
RAD51 was purified as described previously (19). PALB2 protein purifications, DNA binding and D-loop assays were performed as described previously (9).
Immunofluorescence studies
Localization of PALB2 mutants was visualized by immunofluorescence against the Flag-tag. HEK293T cells were fixed with 4% (w/v) paraformaldehyde for 15 min at room temperature, washed with TBS, and fixed again with ice-cold methanol for 5 min at −20°C. Cells were then permeabilized with 0.2% Triton X-100 in PBS for 5 min at room temperature. After three 5 min-washes in TBS, cells were quenched with 0.1% (w/v) sodium borohydride in PBS for 5 min. Cells were washed once with TBS and incubated for 45 min in blocking buffer (PBS, 10% (v/v) goat serum, 1% (w/v) BSA). Next, cells were incubated for 2 h at room temperature with the primary antibody (Monoclonal ANTI-FLAG® M2 antibody produced in mouse, F3165, Sigma) diluted 1:500 in blocking buffer. After three 5 min-washes with TBS, cells were incubated for 45 min at room temperature in the dark with the secondary antibody (Alexa Fluor® 568 Goat Anti-Mouse IgG (H+L) Antibody, A11004, Life Technologies) diluted 1:800 in TBS 1% (w/v) BSA. Coverslips were washed three times for 5 min with TBS and mounted onto slides using ProLong® Gold Antifade Mountant (P36931, Life Technologies). The result was visualized using an epifluorescence microscope. RAD51 immunofluoresence was performed using 14B4 anti-RAD51 monoclonal antibody (Novus).
Establishment of PALB2-depleted and control cell lines
Lentiviruses carrying shRNA constructs were used in order to create cell lines stably expressing a shRNA scrambled control or targeting PALB2 mRNA. To produce the lentiviruses, 30–40%-confluent 10 cm culture dishes of HEK293T were transfected with 5 μg of pMD2G, 15 μg of psPAX2 and 20 μg of pLKO.1 carrying a shRNA targeting PALB2 mRNA (targeted sequence: 5΄-GATGCACATTGATGATTCTTA-3΄) or control (Sigma), using calcium-phosphate method overnight. The media were changed the next day to be harvested and filtered onto a 0.22 μM filter 24 h later. To establish the stable cell lines, lentivirus (1.5 ml) were added onto 30–40%-confluent 10 cm culture dishes of HEK293T for a total media volume of 3 ml, polybrene was added to the media at 10 ng/ml and cells were incubated overnight at 37°C, 5% CO2. Media was then replaced by 10 ml of fresh media containing 1 μg/ml puromycin. Once the selection completed, cells were harvested and the knockdown efficiency was estimated through western-blot analysis.
Fractionation
For each conditions, 3 millions of HEK293T cells stably expressing a shRNA targeting PALB2 were resuspended in 200 μl of Buffer A (10 mM HEPES pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 0.1% Triton X-100, 1 mM DTT, 1 mM PMSF, 0.125 μg/ml leupeptin, 5 mM NaF, 1 mM Na3VO4, 0.025 μg/ml aprotinin, complete protease inhibitor cocktail (Roche)) and incubated on ice for 5 min. Centrifugation (1300g at 4°C for 4 min) was then carried out to separate the nuclear (pellet) from the cytoplasmic (supernatants) fractions. The latter were centrifuged one more time at maximum speed for 15 min at 4°C and 160 μl of the supernatants, corresponding to cytosolic extracts, were added to 40 μl of Laemmli 5X buffer. Nuclear fractions were washed twice with 900 μl buffer A, three times with 500 μl of buffer A and resuspended in 200 μl of buffer B (3 mM EDTA, 0.2 mM EGTA, 1 mM DTT, 1 mM PMSF, 0.125 μg/ml leupeptin, 5 mM NaF, 1 mM Na3VO4, 0.025 μg/ml aprotinin). After a 30-min incubation on ice, centrifugation (1700g, 4 min at 4°C) was used to separate the chromatin (pellet) from the nuclear soluble fraction (supernatant); 160 μl of nuclear soluble fractions were added to 40 μl of Laemmli 5X buffer. Chromatin fractions were washed four times with 500 μl of Buffer B and resuspended in 160 μl of the same buffer prior to addition of 40 μl of Laemmli 5X buffer. Samples were then sonicated on ice for 30 s (20–40% amplitude) and boiled at 95°C for 10 min.
NES analysis
The predicted NES sequence was cloned in frame into the carboxyl terminal part of eGFP using the pEGFP-C1 vector and complementary oligos JYM3212/JYM3213 annealed together. The resulting plasmid was then transfected into HEK293T cells plated onto glass coverslips using the calcium phosphate method. Twenty four hours following transfection, cells were treated for 3 h with 20 ng/ml of Leptomycin B (L2913, Sigma) or vehicle (70% methanol). Cells were then fixed with 4% (w/v) paraformaldehyde in PBS for 15 min at room temperature and washed once with PBS. Coverslips were then mounted onto slides using ProLong® Gold Antifade Mountant (P36931, Life Technologies). Results were visualized using an epifluorescence microscope.
Immunoprecipitations
Anti-GFP immunoprecipitations were performed as follows: HEK293T were seeded into 10 cm cell culture dishes and transfected with 15 μg of peGFPC1-PALB2 828–945 or empty vectors (two dishes by condition) using the CaPO4 method. Forty eight hours later, cells were harvested and lysed using immunoprecipitation lysis buffer (IP buffer) as previously described (20). Total amount of protein in supernatants was quantified by Bradford assay. Two milligram of total lysate was precleared by a 20 min incubation with 30 μl of Protein A/G Ultralink® Resin (53133, Thermo Scientific) at 4°C. Supernatants were further incubated with 2 μg of anti-GFP mouse antibody (11814460001, Roche) and 30 μl of fresh resin, at 4°C overnight. Beads were then washed four times with 1 ml of IP buffer, once with 1 ml of PBS, and resuspended in 50 μl of SDS sample buffer (125 mM Tris–HCl pH 6.8, 2% SDS, 10% glycerol, 1 mg/ml Bromophenol blue, 100 mM DTT, 2% β-mercaptoethanol) prior to boiling at 95°C for 5 min.
Flag-tag immunoprecipitations were performed as follows. HEK293T were seeded into 10 cm cell culture dishes and transfected with 15 μg of pcDNA3-Flag PALB2 wt and mutants vectors, using the CaPO4 method. Twenty four hours later, cells were treated with 3 mM hydroxyurea for 16 h. Cells were then harvested and resuspended in 500 μl of IP150 buffer (50 mM Tris–HCl pH 7.4, 150 mM NaCl, 0.5% NP40, 1 mM PMSF, 5 mM NaF, 3.4 μg/ml Aprotinin, 0.4 mM Na3VO4, 1 ng/ml Leupeptin, Complete Protease Inhibitor Cocktail (Roche)). After a 25 min incubation on ice, cells were sonicated three times for 10 s (40–50% output). Lysates were supplemented with 1 mM MgCl2 and treated with 15 units of Benzonase for 1 h at 4°C. After centrifugation, protein concentration in soluble extracts were quantified by Bradford assay. 75 μl of ANTI-FLAG® M2 Affinity Gel beads (A2220, Sigma) were washed and then incubated with 2 mg of protein extracts in 500 μl of IP150 buffer, for 2 h at 4°C, under agitation. Beads were then washed four times with IP150 buffer, resuspended in 50 μl of SDS sample buffer (125 mM Tris–HCl pH 6.8, 2% SDS, 10% glycerol, 1 mg/ml bromophenol blue, 100 mM DTT, 2% β-mercaptoethanol) and boiled at 95°C for 2 min.
Immunoprecipitations with purified proteins were performed as follows. piBRCA2 (250 ng) with or without PALB2 WT or W1038X (150 ng) were incubated in a 100 μl final volume of IP buffer (20 mM Tris–acetate pH 8.0, 125 mM KOAc, 0.5 mM EDTA, 0.5% NP40 (Sigma), 0.1 mg/ml BSA, 10% glycerol, 1 mM DTT and protease inhibitors) during 10 min at 27°C. Complexes were incubated with PALB2 antibody (bethyl, 1:200) for 20 min at 4°C, followed by pulldown using 30 µl of Protein A/G Ultralink beads (ThermoFisher Scientific) during 20 min at 4°C. After four washes with IP buffer, beads were resuspended in SDS sample buffer, boiled at 95°C for 2 min, and loaded on SDS-PAGE for western blot analysis.
Cas9/mClover-LMNA1 homologous recombination assay
The pCR2.1-CloverLMNAdonor, pX330-LMNAgRNA1 and piRFP670-N1 plasmids were previously described (21). The pX330-LMNAgRNA1 plasmid used in this study was modified from the original plasmid by Q5 site-directed mutagenesis (NEB) in order to remove the 3XFlag tag from the Cas9 endonuclease, using the following primers: ATGGCCCCAAAGAAGAAGCG and GGTGGCACCGGTCCAACC.
HEK293T cells were seeded and transfected with control or PALB2 siRNA using Lipofectamine RNAiMAX (Invitrogen). Twenty-four hours post-transfection, 3 millions cells were pelleted and resuspended in complete nucleofector solution (SF Cell Line 4D-Nucleofector™ X Kit, Lonza) to which 1 μg of pCR2.1-CloverLMNAdonor, 1 μg pX330-LMNAgRNA1, 1 μg of the indicated PALB2 construct, 0.1 μg of piRFP670-N1 (used as transfection control) and 200 pmol of siRNA was added. Once transferred to a Lonza certified cuvette, cells were transfected using the 4D-Nucleofector X-unit (program DG-130), immediately resuspended in culture media and transferred to a 10 cm dish. After 48 h, cells were trypsinized and plated into glass coverslips. Clover expression was assayed by fluorescence microscopy the next day, that is 72 h post-nucleofection. Data represent the mean percentages (±SEM) of Clover-positive cells over the iRFP670-positive population from independent experiments performed in triplicates (n > 800 cells per condition).
Homology modelling structure
FASTA sequences were entered in the website http://swissmodel.expasy.org/ and the model building option was chosen. Built models were downloaded, and the one with the higher score of identity was chosen to work with PyMOLE software. For each model, cartoon representation of the structure where preferred. The NES sequence was selected with the option: ‘select resi xxx-xxx’ and colored in red for each model.
RESULTS
PALB2 DNA binding is required to stimulate RAD51 activity
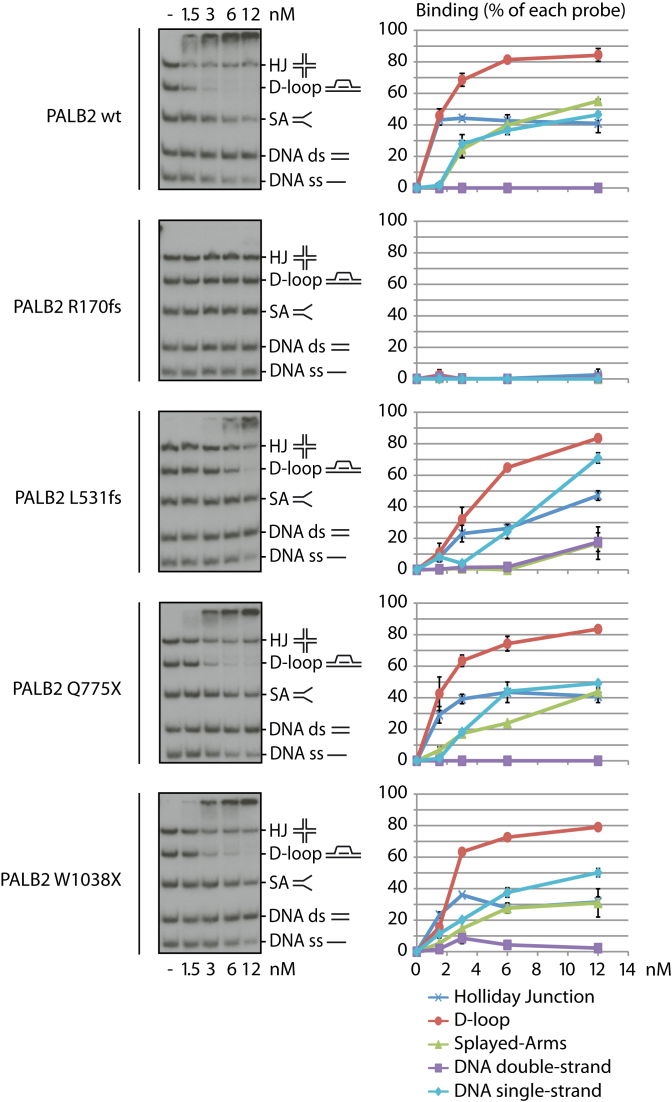
Cancer-associated mutations in human PALB2 occur throughout its sequence, with some mutations being associated to high risk of breast cancer. Here, we propose a systematic approach to characterize PALB2 biochemical functions, using PALB2 mutants designed based on truncating mutations found in breast or ovarian cancer patients. We selected four mutations allowing us to study the effects of truncations all along the protein (Figure 1A): c.509_510delGA (p.Arg170Ilefs*14) (15), c.1592delT (p.Leu531Cysfs*30) (16), c.2323C>T (p.Gln775*) (17) and c.3113G>A (p.Trp1038*) (18), named R170fs, L531fs, Q775X and W1038X, respectively, to simplify the reading (6). We previously reported that PALB2 contains two distinct DNA binding domains (DBD), allowing the protein to bind several DNA substrates such as single-stranded (ss), double-stranded (ds), Holliday junction (HJ), replication fork (splayed-arms, SA) and preferentially to displacement loop (D-loop) DNAs. Moreover, PALB2 stimulates RAD51-mediated D-loop formation (9,22). To investigate the biochemical properties of PALB2 R170fs, L531fs, Q775X and W1038X, we first purified the proteins to homogeneity (Figure 1B). Our hypothesis was that Q775X and W1038X, which both contain intact DBD, and L531fs, which bears a frame-shift close to the end of the second DBD, should all bind DNA substrates in a similar way to PALB2. In contrast, R170fs, a truncated form of PALB2, harboring a frame-shift altering the end of the first DBD should be impaired in DNA binding. Consistent with our predictions, Q775X, W1038X and L531fs were able to bind the same DNA substrates as wild-type PALB2 (wt) with some differences in affinity, however. L531fs and W1038X bound SA with lower affinity than PALB2 wt or Q775X (15–20% of binding versus 40–50% for 6 nM of protein, Supplementary Figure S1); these three mutants also have reduced affinity for the D-loop (50–85% versus 90% for 12 nM of protein). Finally, L531fs bound less HJ at low concentration of protein (20% versus 50–60% for 6 nM of protein). In these experimental conditions, R170fs weakly bound DNA substrates at the highest concentration used, suggesting that the loss of the second DBD affects the affinity for DNA (Supplementary Figure S1). Our previous work showed that PALB2 has a preference for D-loop substrates when gel mobility shifts assays are performed using a mixture of DNA substrates. In competition experiments, all proteins bound preferentially the D-loop substrate, except PALB2 R170fs, which did not bind DNA in these conditions (Figure 2). Together, these results indicate that the second DBD is critical for PALB2 DNA binding properties and suggest that any truncations occurring in the second DBD might have an effect on PALB2 DNA binding properties.
Figure 2.
Competition EMSAs with five different DNA substrates and PALB2 mutant proteins. Left: Analysis of the preference of wild-type PALB2 (wt) and mutant proteins to bind specific DNA structures in competition. EMSAs were performed using single-strand DNA, double-strand DNA, splayed arms, Holliday junctions, and D-loops, altogether. Right: quantification of the data from three distinct experiments.
PALB2 stimulates RAD51-mediated strand invasion of a single-stranded DNA into a supercoiled plasmid to form a D-loop structure (9,22). We used this assay to assess whether PALB2 mutants were able to stimulate RAD51. Based on the PALB2 amino acid sequence (Figure 1A), all mutants still contain at least one RAD51 binding domain. In the conditions tested, 12 nM PALB2 stimulated RAD51 activity by approximately 7-fold (Supplementary Figure S2). L531fs, Q775X and W1038X were still able to stimulate D-loop formation but with less efficiency. Surprisingly, R170fs failed to stimulate RAD51 although it still contains the first RAD51 interaction domain.
A mutation leading to a truncation in the WD40 domain induces a cellular mislocalization
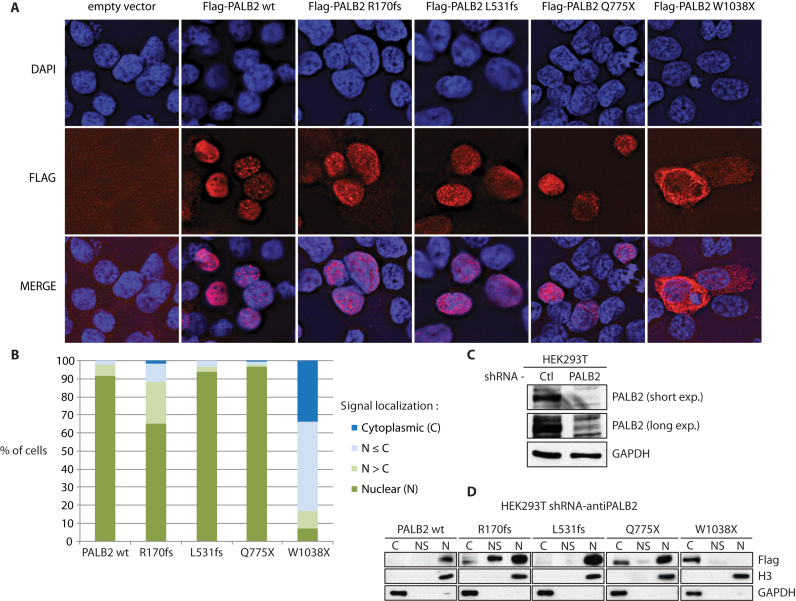
The fact that the PALB2 W1038X mutation creates a high risk of breast cancer (18), but that our mutant protein still had activity in D-loop formation, led us to investigate other mechanisms explaining a possible defect by the PALB2 W1038X mutation. PALB2 has been shown to interact with BRCA1 to help relocate BRCA2 to DNA damage sites (5). To assess if the PALB2 mutants were able to localize at DNA damage sites, we performed immunofluorescence using HEK293T cells treated or untreated with neocarzinostatin, a radiomimetic DNA damaging agent. Spontaneous or induced DNA damage was visualized using the phosphorylation on Ser139 of the histone variant H2AX (γ-H2AX), a widely used DNA double-strand breaks (DSB) marker. By using a Flag-tagged version of each protein, PALB2 as well as L531fs, Q775X, and W1038X mutants were able to form foci in the nucleus (Supplementary Figure S3A). These foci co-localized with γ-H2AX, suggesting the localization of PALB2 proteins at DSBs. On the contrary, R170fs presented a diffuse signal in the nucleus and failed to form foci even in the presence of neocarzinostatin. This is consistent with our observed loss of DNA binding ability but also the fact that PALB2 R170fs does not bear a Chromatin-Associated Motif (ChAM) (11). Interestingly, we observed that W1038X was able to form nuclear foci, but this was in a minority of cells (<10%). In fact, later analysis revealed that PALB2 W1038X displayed predominantly a cytoplasmic distribution, even following treatment with neocarzinostatin (Supplementary Figure S3B and S4A).
PALB2 is known to anchor BRCA2 to nuclear structures, however, its nucleocytoplasmic transport has been unexplored. We investigated the cellular localization of the PALB2 mutants by over-expressing a Flag-tagged version of each of them in HEK293T cells followed by immunofluorescence analysis. R170fs, L531fs and Q775X were found mainly localized in the nucleus as the wild-type protein. However, W1038X was detected in the cytoplasm (Figure 3). More precisely, four patterns of nuclear/cytoplasmic localizations were observed: (i) a nuclear PALB2-Flag signal, (ii) a PALB2-Flag signal stronger in the nucleus but still detected in the cytoplasm, (iii) a PALB2-Flag signal stronger in the cytoplasm but still detected in nucleus and (iv) a cytoplasmic PALB2-Flag signal (Supplementary Figure S4B). The proportion of cells showing a strong cytoplasmic Flag signal was greater for the W1038X protein (Figure 3B). In a further attempt to confirm the cytoplasmic localization of W1038X, fractionation analysis was performed using HEK293T cells stably expressing a shRNA targeting PALB2, to remove any interference with endogenous PALB2 (Figure 3C). Less than 10% PALB2 protein remained after PALB2 shRNA knockdown. All mutant proteins were detected in the chromatin fraction but W1038X was found predominantly in the cytosolic fraction (Figure 3D), thus confirming our immunofluorescence results.
Figure 3.
Cellular localization of PALB2 mutant proteins. (A) Immunofluorescence of mutant forms of Flag-tagged PALB2. DAPI (blue), anti-Flag (red), and the merge picture are shown. All mutants except W1038X were mainly nuclear. (B) Quantification of the cytoplasmic and nuclear accumulation of PALB2 mutants. Experiments were performed in quadruplicate. (C) Knockdown of endogenous PALB2 in HEK293T cells by expressing constitutively a shRNA against PALB2. (D) Analysis of the cellular localization of PALB2 mutant forms by cellular fractionation. The cytoplasmic (C), nuclear soluble (NS) and chromatin (N) fractions are shown. Blots against the GAPDH and histone H3 proteins are shown as controls.
The cytoplasmic localization of PALB2 W1038X should affect its interaction with BRCA2 and RAD51. PALB2 is a chromatin-bound protein that recruits BRCA2 which in turn recruits RAD51 for homologous recombination (23). Since PALB2 W1038X is mainly cytoplasmic, we expected that the interaction with RAD51 and BRCA2 would be weak. Consistent with the abnormal distribution of the protein, BRCA2 and RAD51 interacted weakly with PALB2 W1038X (Supplementary Figure S4C). We observed that purified piBRCA2 interacted to a much lesser extent to PALB2 W1038X compared to the wild-type protein (Supplementary Figure S4D). This suggests that the BRCA2 binding site, previously identified as A1025 (12), might not be optimal within PALB2 W1038X conformation.
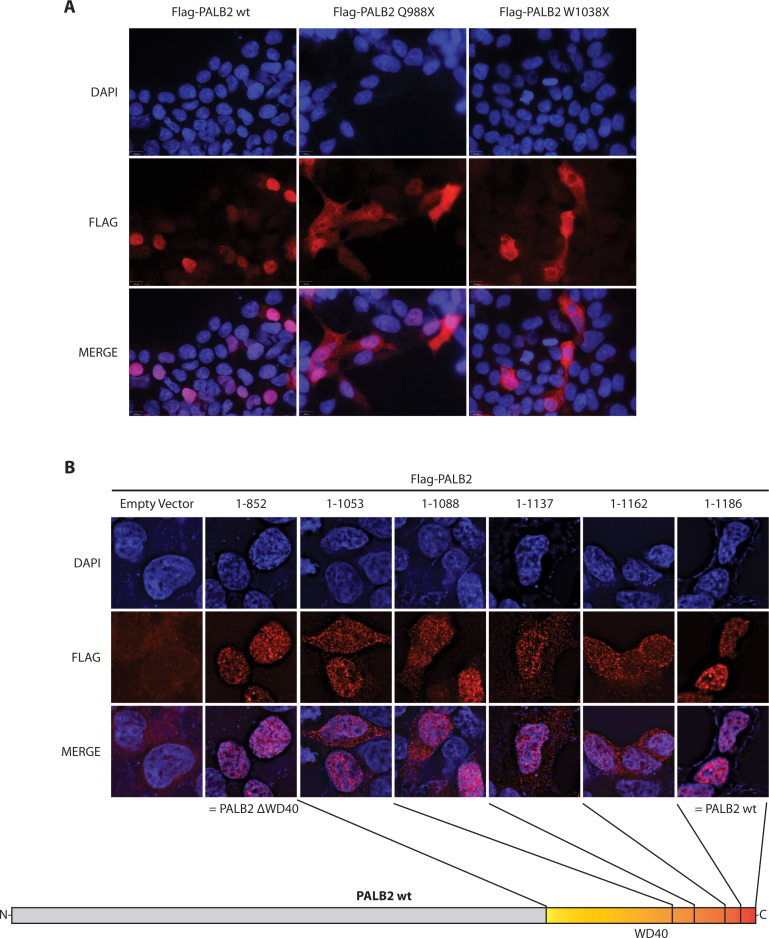
The Q988X cancer-causing mutation also causes cytoplasmic accumulation
We hypothesized that the mislocalization encountered for W1038X could apply also for any truncations occurring in the WD40-repeat domain located at amino acids 853–1186. PALB2 Q988X (c.2962C>T, p.Gln988*) is a mutation found in breast cancer and Fanconi Anemia patients (24,25). Remarkably, PALB2 Q988X displayed the same mislocalization pattern as W1038X (Figure 4A). This defect was further confirmed by performing immunofluorescence on several arbitrary truncations in the WD40 of PALB2 (Figure 4B). Interestingly, when the WD40 domain was completely deleted (truncation at amino acid 852), the protein was localized in major proportion back to the nucleus, as for the wild-type protein. This last observation suggested that a sequence in the WD40, located between amino acids 852 and 987, caused the mislocalization of WD40-truncated forms of PALB2.
Figure 4.
Cellular localization of WD40-mutant PALB2 and WD40-truncated variant proteins. (A) Immunofluorescence of Flag-tagged PALB2 wt, Q988X (c.2962C>T) and W1038X (c.3113G>A). Both mutant forms present a similar mislocalization. DAPI (blue), anti-Flag (red), and the merge picture are shown. (B) Immunofluorescence of Flag-tagged PALB2 proteins truncated in the WD40-domain as indicated on the schematic representation. PALB2 1–852 corresponds to a complete deletion of the WD40 domain, while PALB2 1–1186 corresponds to the wild-type protein. All proteins truncated between position 1038 and 1162 present a similar mislocalization. DAPI (blue), anti-Flag (red) and the merge picture are shown.
PALB2 WD40 masks a nuclear export sequence
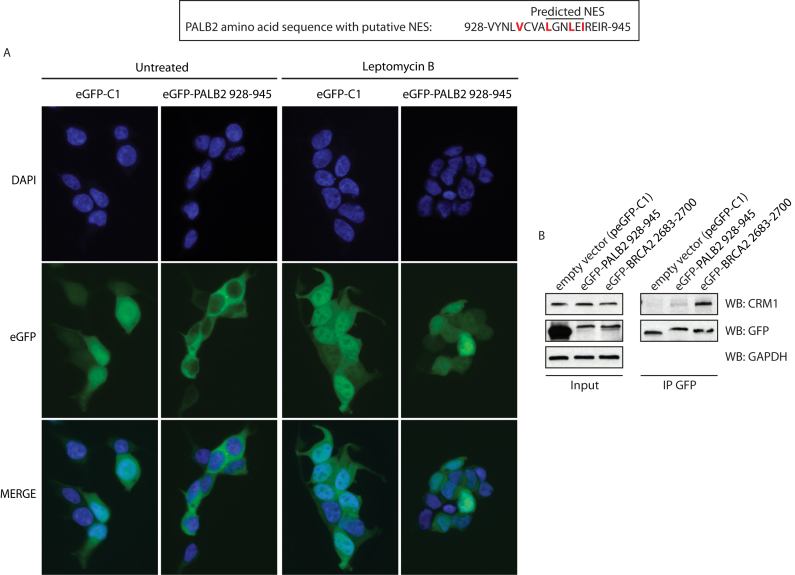
Cytoplasmic accumulation can be governed by nuclear export signals (NES) (26). Because the WD40-deleted protein was nuclear and that truncations from amino acid 1038 failed to recover the localization of truncated proteins to the nucleus (Figure 4B), the hypothesis of a nuclear localization signal (NLS) located in C-terminal part of PALB2 was excluded. Hence, we searched for the existence of a NES within the WD40 domain using NetNES 1.1 Server and ValidNESs (Supplementary Figure S5). Analysis of common predictions led us to identify a putative NES located in the WD40 domain, between the amino acids 928 and 945 with a relatively high score. This NES is conserved in pluricellular organisms (Supplementary Figure S6A). In order to test whether the sequence 928-VYNLVCVALGNLEIREIR-945 can act as a NES, we fused it to enhanced Green Fluorescent Protein (eGFP) and monitored its cellular localization by immunofluorescence assays. While eGFP alone harbored often a stronger signal in the nucleus, eGFP-putative PALB2 NES fusion was found in the cytoplasm, validating that this sequence is indeed a NES (Figure 5A, left panel). The main mediator of nuclear export in many cell types is the Chromosome Region Maintenance 1 (CRM1) protein, also referred to as Exportin 1 or Xpo1. Hence, we examined if the W1038X export could be a CRM1-mediated transport. We analyzed the eGFP-NES localization in presence or absence of leptomycin B, a CRM1 inhibitor. We observed that, following CRM1 inhibition, the eGFP-PALB2 NES fusion protein became nuclear. Moreover, treatment with Selinexor (KPT-330), a CRM1-specific inhibitor, led to nuclear accumulation of PALB2 W1038X (Supplementary Figure S7), inferring that nuclear exclusion of PALB2 Q988X and W1038X is a CRM1-dependent pathway (Figure 5A, right panel). This was further confirmed by co-immunoprecipitation, where CRM1 interacts with eGFP-PALB2 NES and eGFP-BRCA2 NES (20), while the control experiment with eGFP failed to do so (Figure 5B).
Figure 5.
A sequence contained in PALB2 WD40-domain acts as a CRM1-dependent nuclear export signal. (A, Top) The sequence predicted as a NES by ValidNESs is underlined. The hydrophobic amino acids that could constitute a NES are highlighted in red. Bottom: Immunofluorescence analysis of eGFP alone or fused to the NES sequence (amino acids 928–945). The analysis was performed with or without the CRM1-inhibitor leptomycin B. DAPI (blue), anti-Flag (red) and the merge picture are shown. (B) Co-immunoprecipitation of endogenous CRM1 with eGFP alone or fused to the putative NES of PALB2, or to BRCA2 NES (as a positive control).
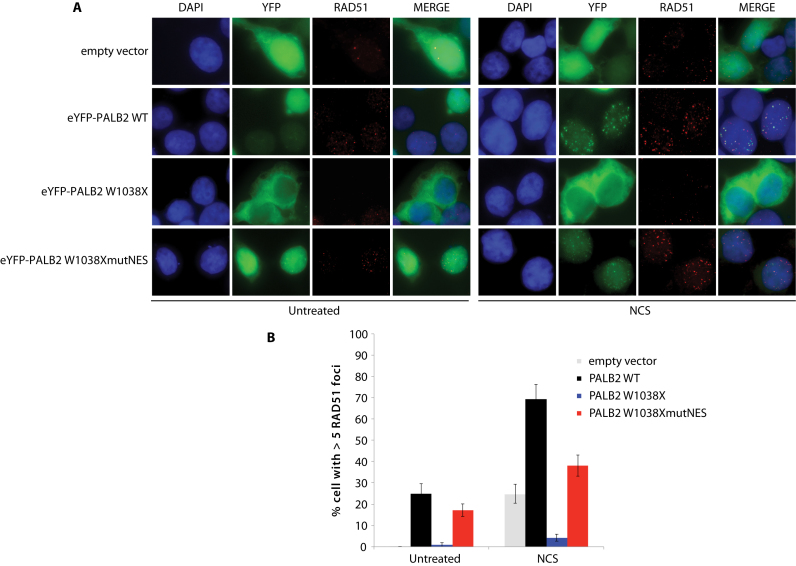
We performed some additional characterization of PALB2 W1038X protein function in cells, using immunofluorescence staining against RAD51 to ensure that HR cannot be activated in the presence of cytoplasmic W1038X PALB2 in PALB2-knockdown cells (Figure 6). HEK293 shPALB2 knockdown cells transfected with a vector alone showed poor RAD51 foci induction after neocarzinostatin treatment, consistent with limited amounts of PALB2 left in the knockdown. In contrast, ∼70% of HEK293 shPALB2 cells complemented with the PALB2 wild-type protein harbored more than five RAD51 foci. Consistent with the cytoplasmic accumulation of PALB2 W1038X, we observed that RAD51 foci formation was strongly impaired in the presence of the PALB2 W1038X mutation. Very interestingly, when the cryptic NES in PALB2 W1038X was mutated (Supplementary Figure S8A; 928-VYNLVCVALGNLEIREIR-945 to 928-VYNLACVAAGNAEAREIR-945), it significantly restored PALB2 W1038X nuclear localization (Supplementary Figure S8B) and RAD51 foci formation (Figure 6B). Moreover, deletion of the NES sequence in PALB2 W1038X led to nuclear accumulation (Supplementary Figure S8B).
Figure 6.
Effect of PALB2 W1038X mutant proteins on RAD51 foci formation. (A) Representative immunofluorescence images of RAD51 staining (red) in HEK293T-shPALB2 cells expressing the empty vector or the indicated peYFP-c1-PALB2 constructs (green). DAPI (blue) and merge pictures are also shown. (B) Quantification of RAD51 foci in the PALB2 mutant cells with or without NCS treatment. HEK293T-shPALB2 cells were treated with 100 ng/ml neocarzinostatin for 1 hour, followed by a 3-hour release. Data are represented as mean percentage ± S.E. of cells with more than five foci (n > 100).
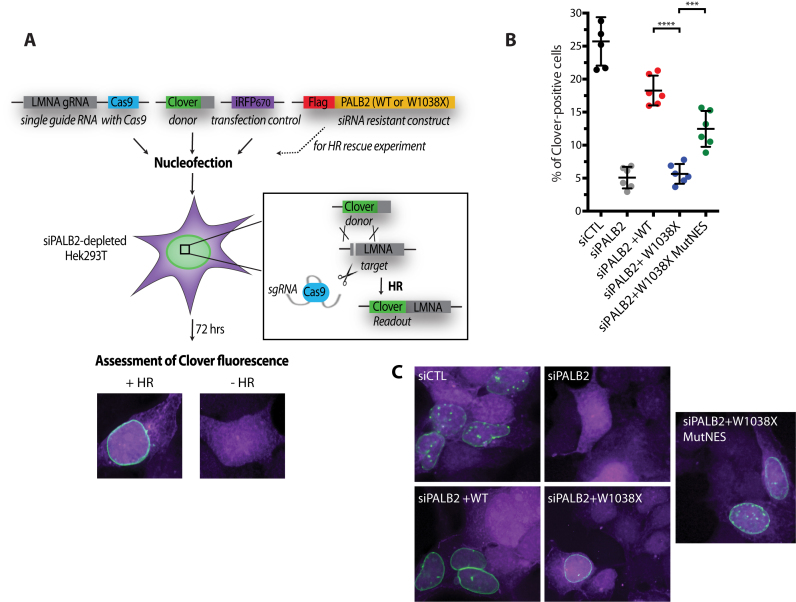
To corroborate these results, we performed Cas9/mClover-LMNA HR assays in HEK293T cells to measure gene targeting efficiency after depletion of endogenous PALB2 and complementation with siRNA resistant plasmid constructs (Figure 7A). siRNA depletion of PALB2 led to a 5-fold reduction of clover positive cells (Figure 7B). These cells were transfected with siRNA resistant wild-type PALB2, PALB2 W1038X, or PALB2 W1038X MutNES. PALB2 W1038X did not complement siRNA PALB2 cells. However, PALB2 W1038X MutNES increased significantly gene targeting. The reversion of RAD51 foci (Figure 6B) or gene targeting by PALB2 W1038X MutNES was not complete, and we believe that additional mutations are required for complete reversion. Mutational analysis of contributions made by residues in the NES motif is difficult because the exact nature of the NES sequences required for CRM1 binding are reported to be variable (27,28).
Figure 7.
Effect of PALB2 W1038X mutant proteins on homologous recombination using a Cas9/mClover-LMNA1 homologous recombination assay. (A) Schematic representation of the mClover-LMNA1 homologous recombination assay. Following nucleofection, Cas9 creates a double-strand break in the LMNA locus leading to integration of the mClover gene by homologous recombination. Clover-labeled Lamin A/C proteins exhibit green fluorescence enriched at the nuclear periphery, which is indicative of successful gene targeting by homologous recombination. (B) Gene-targeting efficiency of siRNA PALB2 cells complemented with wild-type, W1038X and W1038XMutNES siRNA resistant constructs. mClover positive cells were quantified. ***P < 0.01 and ****P < 0.001. (C) Representative images of the quantifications shown in (B). iRFP positive cells are in purple and mClover integrated at the LMNA locus leads to green fluorescence in and around the nucleus.
Unfolding the WD40 repeat of other proteins
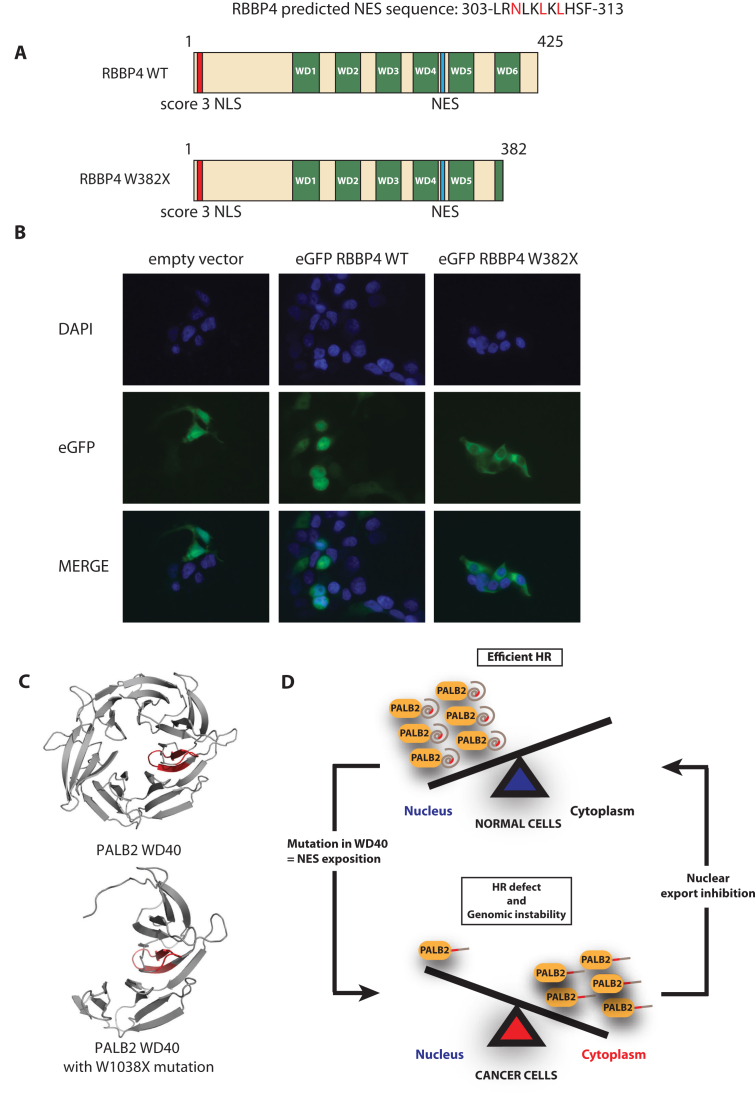
We wanted to test whether our model, implicating an exposed NES following a truncation in the WD40 domain is restricted to PALB2. Thus, using a bioinformatics search, we noticed that the nuclear protein RBBP4, shared similar structural properties as PALB2 (Figure 8A). Histone-binding protein RBBP4 (also known as RbAp48, or NURF55) contains a C-terminal WD40 repeat, which is truncated by the W382X mutation found in malignant melanoma (COSMIC database - http://cancer.sanger.ac.uk/cosmic). We found a NES predicted sequence (from amino acids 303–313) between the WD4 and WD5 repeats of RBBP4, that remains in the W382X protein (Figure 8A and Supplementary Figure S8C). Remarkably, upon introduction of the C-terminal truncation, the RBBP4 was found to be abnormally cytoplasmic (Figure 8B). Thus, these results suggest that our model can be applied to other proteins and highlights a novel mode of nucleocytoplasmic regulation in cancer.
Figure 8.
RBBP4, another WD40 domain containing protein, becomes cytoplasmic due to a cancer causing mutation. (A) Schematic representation of wild-type RBBP4 and RBBP4 W382. (B) Immunofluorescence analysis of eGFP-RBBP4 or eGFP-RBBP4W382. DAPI (blue), eGFP fluorescence (green), and the merge picture are shown. (C) Modelling of the wild-type PALB2 and PALB2 W1038X WD40 domains. (D) Model. In normal cells, PALB2 is a nuclear protein that promotes homologous recombination and DNA repair. In cancer cells, mutations arise in the WD40 repeat (such as the Q988X and W1038X mutations), which leads to NES unmasking and accumulation of PALB2 mutants in the cytoplasm. In the cytoplasm, PALB2 mutants, can no longer promote DNA repair, leading to genomic instability.
DISCUSSION
Aberrant cellular localization of proteins is a common feature of cancerous cells, leading to the inactivation of tumor suppressor proteins. High levels of the nuclear export protein CRM1 have been detected in various cancers and lead to cytoplasm relocalization of diverse nuclear proteins, namely p53, BRCA1 and ATM (29). Here, we describe a novel and general mechanism by which tumor suppressors, as PALB2, can be mislocalized by a mutation in the WD40 repeat sequence. We have found that two cancer-causing mutations, Q988X and W1038X, cause a similar defect. Furthermore, we have evidence that this concept can be generalized to other proteins, such as RBBP4, another WD40-containing protein.
Our initial goal was to investigate the impact of cancer-associated truncating mutations on the biochemical functions of PALB2. Surprisingly, we noticed that the most aggressive mutation analysed, PALB2 W1038X, displayed no significant defect in D-loop formation, suggesting that W1038X retained functionality. However, the mutant was unusually localized to the cytoplasm. It is important to note that PALB2 W1038X confers a risk of developing breast cancer of 49% by age 50 (18), while the cumulative risk for all mutations in PALB2 has been recently estimated to be 14% by the same age (4). The W1038X mutation occurs in the WD40 repeat, a highly structured barrel motif. Hence, we suggest that the PALB2 W1038X mutation could unfold or open a cavity in the WD40 repeat, therefore exposing a nuclear export signal. Using different predicting tools and truncations of the WD40 repeat, we identified a NES in PALB2 (VYNLVCVALGNLEIREIR) located between amino acids 852 and 987. We showed that this NES is functional, as it interacts with CRM1 and it relocalizes eGFP in the cytoplasm. To gain insight into the impact of the W1038X mutation on the WD40 repeat structure, we used the homology modelling website SWISS-MODEL, the PyMOLE software and the crystal structure of the PALB2 WD40 domain, and localized the NES inside the core of the PALB2 seven bladed propeller. Interestingly, structure predictions reveal that half of the WD40 domain is removed in the PALB2 W1038X mutant, leading to a cavity and an exposed NES (Figure 8C). Hence, the W1038X mutation undoubtedly has an impact on the WD40 structure, subsequently leading to aberrant cytoplasmic localization and function. Indeed, we have found that RAD51 foci formation and gene targeting is affected when PALB2 knockdown cells are transfected with PALB2 W1038X. In support of our model, introducing mutations in the exposed NES of PALB2 W1038X, restores significantly PALB2 W1038X nuclear accumulation, RAD51 foci formation, and gene targeting efficiency.
We envision that mutation in the WD40 domain can have several consequences. PALB2 interacts with several DNA repair proteins, including BRCA1 (30) (5) (31), BRCA2 (12), MRG15 (31), RAD51 and RAD51AP1 (9) (22), RAD51C (32), KEAP1 (33) (8), and polymerase eta (34). For instance, if mutations occur in the WD40 domain and PALB2 mutants become cytoplasmic, it might prevent the access of these associated proteins to DNA damage sites as these proteins could also become cytoplasmic. In this situation, a truncation in PALB2 could have a more severe effect than a complete inactivation of the gene as it will tether several factors outside of the nucleus. On the other hand, depending on the position of the PALB2 mutations, PALB2 could also be impaired for functional interactions with these repair factors. For instance, we observed that the PALB2 W1038X mutant interacts weakly to BRCA2 compared to the wild-type protein. Another possibility is that missense mutations in the PALB2 WD40 region can decrease the stability of the encoded protein. This was observed for the PALB2 T1030I mutation (32).
The human genome encodes 349 predicted WD40 repeat-containing proteins (35), including proteins associated with Sensenbrenner syndrome, Kallmann syndrome, Jeune syndrome, Crohn's disease, Van der Woude syndrome, Amelogenesis imperfecta and cancer. Therefore, mutations in the WD40 repeats of these proteins could potentially affect their localization and promote disease formation. We can envision several other mechanisms by which unfolding a WD40 could affect cellular function. First, unfolding of the WD40 through a mutation, amino acid substitution or deletion could expose a NLS, which could drive a cytoplasmic protein into the nucleus. Second, unfolding of the WD40 through mutations or deletions could abolish sites for protein-protein interactions leading to unmasked NES or NLS sequences. For instance, although BRCA2 does not contain any WD40 repeat, it has been reported that a breast-cancer-associated germline mutation, BRCA2 D2723H (c.Asp2723His), impairs the cellular localization of BRCA2 through disruption of its interaction with DSS1. Since DSS1 binding hides a NES sequence, located between position 2686 and 2696, loss of DSS1 interaction exposes the NES, resulting in BRCA2 export from the nucleus (20).
Of all the potential targets in nuclear-cytoplasmic transport, the nuclear export receptor CRM1 remains the most promising therapeutic target. Selinexor (KPT-330), a CRM1 inhibitor, is currently in Phase I/II clinical trials (for review see (36)) and is efficient in tumor xenografts including kidney, pancreas, prostate, breast, lung, melanoma, colon, gastric, ovarian, neuroblastoma, and sarcomas (36). Since PALB2 W1038X encodes a nearly functional DNA repair protein in biochemical assays, our work suggests that patients harboring such a mutation could be treated with Selinexor. Consequently, PALB2 W1038X would stay in the nucleus and accurately promote DNA repair and genome stability (Figure 8D). Remarkably, targeting PALB2 nuclear export might open the door to a new therapeutic strategy for personalized medicine.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Guy Poirier for advice and Jacques Côté for generous support. We are grateful to Dr Isabelle Brodeur for critical reading the manuscript. We thank Niraj Joshi, Mandy Ducy, and other members of the Masson lab for suggestions. We also thank Katherine O'Malley and Zhou Wang (The University of Pittsburgh) for the gift of the RBBP4 cDNA. All authors have contributed to the analysis and interpretation of results as well as to the revision of the manuscript. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Quebec Breast Cancer Foundation (joint grant with Jacques Côté) and CIHR [MOP-84260 to G.D. (in part) or MOP-111063 to J.Y.M.]. Anthony M. Couturier was a Luc Bélanger scholar. J.Y.M. was a FRQS Chercheur National Investigator and is a FRQS Chair in genome stability. Funding for open access charge: CIHR.
Conflict of interest statement. None declared.
REFERENCES
- 1.Velic D., Couturier A.M., Ferreira M.T., Rodrigue A., Poirier G.G., Fleury F., Masson J.Y.. DNA damage signalling and repair inhibitors: the long-sought-after achilles' heel of cancer. Biomolecules. 2015; 5:3204–3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Couturier A.M., Fleury H., Patenaude A.-M., Bentley V.L., Rodrigue A., Coulombe Y., Niraj J., Pauty J., Berman J.N., Dellaire G. et al. . Roles for APRIN (PDS5B) in homologous recombination and in ovarian cancer prediction. Nucleic Acids Res. 2016; 44:10879–10897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tischkowitz M., Xia B., Sabbaghian N., Reis-Filho J.S., Hamel N., Li G., van Beers E.H., Li L., Khalil T., Quenneville L.A. et al. . Analysis of PALB2/FANCN-associated breast cancer families. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:6788–6793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Antoniou A.C., Casadei S., Heikkinen T., Barrowdale D., Pylkas K., Roberts J., Lee A., Subramanian D., De Leeneer K., Fostira F. et al. . Breast-cancer risk in families with mutations in PALB2. N. Engl. J. Med. 2014; 371:497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang F., Ma J., Wu J., Ye L., Cai H., Xia B., Yu X.. PALB2 links BRCA1 and BRCA2 in the DNA-damage response. Curr. Biol.: CB. 2009; 19:524–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pauty J., Rodrigue A., Couturier A., Buisson R., Masson J.Y.. Exploring the roles of PALB2 at the crossroads of DNA repair and cancer. Biochem. J. 2014; 460:331–342. [DOI] [PubMed] [Google Scholar]
- 7.Buisson R., Masson J.Y.. PALB2 self-interaction controls homologous recombination. Nucleic Acids Res. 2012; 40:10312–10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Orthwein A., Noordermeer S.M., Wilson M.D., Landry S., Enchev R.I., Sherker A., Munro M., Pinder J., Salsman J., Dellaire G. et al. . A mechanism for the suppression of homologous recombination in G1 cells. Nature. 2015; 528:422–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buisson R., Dion-Cote A.M., Coulombe Y., Launay H., Cai H., Stasiak A.Z., Stasiak A., Xia B., Masson J.Y.. Cooperation of breast cancer proteins PALB2 and piccolo BRCA2 in stimulating homologous recombination. Nat. Struct. Mol. Biol. 2010; 17:1247–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sy S.M., Huen M.S., Zhu Y., Chen J.. PALB2 regulates recombinational repair through chromatin association and oligomerization. J. Biol. Chem. 2009; 284:18302–18310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bleuyard J.Y., Buisson R., Masson J.Y., Esashi F.. ChAM, a novel motif that mediates PALB2 intrinsic chromatin binding and facilitates DNA repair. EMBO Rep. 2012; 13:135–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oliver A.W., Swift S., Lord C.J., Ashworth A., Pearl L.H.. Structural basis for recruitment of BRCA2 by PALB2. EMBO Rep. 2009; 10:990–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu C., Min J.. Structure and function of WD40 domain proteins. Protein Cell. 2011; 2:202–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stirnimann C.U., Petsalaki E., Russell R.B., Muller C.W.. WD40 proteins propel cellular networks. Trends Biochem. Sci. 2010; 35:565–574. [DOI] [PubMed] [Google Scholar]
- 15.Dansonka-Mieszkowska A., Kluska A., Moes J., Dabrowska M., Nowakowska D., Niwinska A., Derlatka P., Cendrowski K., Kupryjanczyk J.. A novel germline PALB2 deletion in Polish breast and ovarian cancer patients. BMC Med. Genet. 2010; 11:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Erkko H., Xia B., Nikkila J., Schleutker J., Syrjakoski K., Mannermaa A., Kallioniemi A., Pylkas K., Karppinen S.M., Rapakko K. et al. . A recurrent mutation in PALB2 in Finnish cancer families. Nature. 2007; 446:316–319. [DOI] [PubMed] [Google Scholar]
- 17.Foulkes W.D., Ghadirian P., Akbari M.R., Hamel N., Giroux S., Sabbaghian N., Darnel A., Royer R., Poll A., Fafard E. et al. . Identification of a novel truncating PALB2 mutation and analysis of its contribution to early-onset breast cancer in French-Canadian women. Breast Cancer Res.: BCR. 2007; 9:R83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Southey M.C., Teo Z.L., Dowty J.G., Odefrey F.A., Park D.J., Tischkowitz M., Sabbaghian N., Apicella C., Byrnes G.B., Winship I. et al. . A PALB2 mutation associated with high risk of breast cancer. Breast Cancer Res.: BCR. 2010; 12:R109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baumann P., Benson F.E., Hajibagheri N., West S.C.. Purification of human Rad51 protein by selective spermidine precipitation. Mut. Res. DNA Repair. 1997; 384:65–72. [DOI] [PubMed] [Google Scholar]
- 20.Jeyasekharan A.D., Liu Y., Hattori H., Pisupati V., Jonsdottir A.B., Rajendra E., Lee M., Sundaramoorthy E., Schlachter S., Kaminski C.F. et al. . A cancer-associated BRCA2 mutation reveals masked nuclear export signals controlling localization. Nat. Struct. Mol. Biol. 2013; 20:1191–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pinder J., Salsman J., Dellaire G.. Nuclear domain ‘knock-in’ screen for the evaluation and identification of small molecule enhancers of CRISPR-based genome editing. Nucleic Acids Res. 2015; 43:9379–9392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dray E., Etchin J., Wiese C., Saro D., Williams G.J., Hammel M., Yu X., Galkin V.E., Liu D., Tsai M.S. et al. . Enhancement of RAD51 recombinase activity by the tumor suppressor PALB2. Nat. Struct. Mol. Biol. 2010; 17:1255–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xia B., Sheng Q., Nakanishi K., Ohashi A., Wu J., Christ N., Liu X., Jasin M., Couch F.J., Livingston D.M.. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol. Cell. 2006; 22:719–729. [DOI] [PubMed] [Google Scholar]
- 24.Hellebrand H., Sutter C., Honisch E., Gross E., Wappenschmidt B., Schem C., Deissler H., Ditsch N., Gress V., Kiechle M. et al. . Germline mutations in the PALB2 gene are population specific and occur with low frequencies in familial breast cancer. Hum. Mutat. 2011; 32:E2176–2188. [DOI] [PubMed] [Google Scholar]
- 25.Hofstatter E.W., Domchek S.M., Miron A., Garber J., Wang M., Componeschi K., Boghossian L., Miron P.L., Nathanson K.L., Tung N.. PALB2 mutations in familial breast and pancreatic cancer. Fam. Cancer. 2011; 10:225–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hung M.C., Link W.. Protein localization in disease and therapy. J. Cell Sci. 2011; 124:3381–3392. [DOI] [PubMed] [Google Scholar]
- 27.Guttler T., Madl T., Neumann P., Deichsel D., Corsini L., Monecke T., Ficner R., Sattler M., Gorlich D.. NES consensus redefined by structures of PKI-type and Rev-type nuclear export signals bound to CRM1. Nat. Struct. Mol. Biol. 2010; 17:1367–1376. [DOI] [PubMed] [Google Scholar]
- 28.Xu D., Farmer A., Collett G., Grishin N.V., Chook Y.M.. Sequence and structural analyses of nuclear export signals in the NESdb database. Mol. Biol. Cell. 2012; 23:3677–3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mendonca J., Sharma A., Kim H.S., Hammers H., Meeker A., De Marzo A., Carducci M., Kauffman M., Shacham S., Kachhap S.. Selective inhibitors of nuclear export (SINE) as novel therapeutics for prostate cancer. Oncotarget. 2014; 5:6102–6112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang F., Fan Q., Ren K., Andreassen P.R.. PALB2 functionally connects the breast cancer susceptibility proteins BRCA1 and BRCA2. Mol. Cancer Res. 2009; 7:1110–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sy S.M., Huen M.S., Chen J.. PALB2 is an integral component of the BRCA complex required for homologous recombination repair. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:7155–7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park J.Y., Singh T.R., Nassar N., Zhang F., Freund M., Hanenberg H., Meetei A.R., Andreassen P.R.. Breast cancer-associated missense mutants of the PALB2 WD40 domain, which directly binds RAD51C, RAD51 and BRCA2, disrupt DNA repair. Oncogene. 2014; 33:4803–4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma J., Cai H., Wu T., Sobhian B., Huo Y., Alcivar A., Mehta M., Cheung K.L., Ganesan S., Kong A.N. et al. . PALB2 interacts with KEAP1 to promote NRF2 nuclear accumulation and function. Mol. Cell. Biol. 2012; 32:1506–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buisson R., Niraj J., Pauty J., Maity R., Zhao W., Coulombe Y., Sung P., Masson J.Y.. Breast cancer proteins PALB2 and BRCA2 stimulate polymerase eta in recombination-associated DNA synthesis at blocked replication forks. Cell Rep. 2014; 6:553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Letunic I., Doerks T., Bork P.. SMART: recent updates, new developments and status in 2015. Nucleic Acids Res. 2015; 43:D257–D260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gravina G.L., Senapedis W., McCauley D., Baloglu E., Shacham S., Festuccia C.. Nucleo-cytoplasmic transport as a therapeutic target of cancer. J. Hematol. Oncol. 2014; 7:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.