Abstract
Homologous recombination (HR) is a DNA double-strand break (DSB) repair pathway that protects the genome from chromosomal instability. RAD51 mediator proteins (i.e. paralogs) are critical for efficient HR in mammalian cells. However, how HR-deficient cells process DSBs is not clear. Here, we utilized a loss-of-function HR-reporter substrate to simultaneously monitor HR-mediated gene conversion and non-conservative mutation events. The assay is designed around a heteroallelic duplication of the Aprt gene at its endogenous locus in isogenic Chinese hamster ovary cell lines. We found that RAD51D-deficient cells had a reduced capacity for HR-mediated gene conversion both spontaneously and in response to I-SceI-induced DSBs. Further, RAD51D-deficiency shifted DSB repair toward highly deleterious single-strand annealing (SSA) and end-joining processes that led to the loss of large chromosomal segments surrounding site-specific DSBs at an exceptionally high frequency. Deletions in the proximity of the break were due to a non-homologous end-joining pathway, while larger deletions were processed via a SSA pathway. Overall, our data revealed that, in addition to leading to chromosomal abnormalities, RAD51D-deficiency resulted in a high frequency of deletions advancing our understanding of how a RAD51 paralog is involved in maintaining genomic stability and how its deficiency may predispose cells to tumorigenesis.
INTRODUCTION
DNA double-strand breaks (DSBs) are highly toxic lesions that can arise during cellular DNA metabolic processes, such as DNA replication and programmed antigen receptor rearrangements, or upon exposure to endogenous and/or exogenous DNA damaging agents. In cancer therapy, ionizing radiation and chemotherapeutic agents are commonly utilized due to their toxicity via the formation of DSBs in highly proliferative cells (1,2). DSBs represent severe lesions for the cell to repair, and unrepaired DSBs stimulate cell-cycle checkpoints leading to cell death or genetic and chromosomal instability. Mammalian cells have evolved several DSB repair pathways to process these toxic lesions that are generally grouped into two categories: homologous recombination (HR) and non-homologous end-joining (NHEJ). NHEJ mechanisms essentially join two ends of broken DNA with minimal regard to sequence and can result in error-prone processing, while HR is considered a higher fidelity pathway, and uses stretches of homologous sequence to ensure accurate repair. Defects in either of these pathways can lead to a failure to properly repair damaged DNA and can manifest as spontaneous genomic instability such as chromosome breaks, chromosome translocations and rearrangements, gene amplifications, genetic deletions, aneuploidy, micronuclei, and loss of heterozygosity. This genomic instability plays a pivotal role in carcinogenesis and is a fundamental hallmark of cancer cells (3).
HR is a high-fidelity repair mechanism that utilizes one or both ends of a DSB to ‘invade’ an intact DNA molecule of identical or very similar sequence, typically the sister chromatid, followed by resynthesis of the damaged region using the undamaged molecule as a template (4,5). This allows the replacement of damaged DNA regions without loss or alteration of base sequence. How the cell decides to forgo NHEJ in favor of HR when repairing a two-ended DSB is not completely understood but appears to involve recognition of the break and giving license for processive nucleolytic resection to generate free 3΄ single-stranded DNA overhangs coated with single-stranded DNA binding protein, RPA (6). The next key step is to displace RPA in favor of the recombinase protein, RAD51, which polymerizes to form helical nucleoprotein filaments on the single-stranded DNA ends. The RAD51-coated single-stranded DNA then engages in a homology search by probing the homologous duplex DNA for the corresponding identical or highly homologous sequence (4,5,7).
Nucleation and stabilization of the presynaptic filament is tightly regulated and requires the concerted action of several mediator proteins (8). The primary mediator in mammalian cells is BRCA2, which appears to work with five RAD51 paralogs (RAD51B, RAD51C, RAD51D, XRCC2 and XRCC3) to facilitate RAD51 filament formation (9–12). Genetic deletion of any one of the RAD51 paralogs in genetically engineered mouse models produces severe developmental defects and early embryonic lethality with accumulation of unrepaired DNA damage (13–16). RAD51 paralogs are not essential, as mutant and knockout cells can grow with impaired viability. This characteristic has led to several RAD51 paralog-deficient model cell systems in mouse embryonic fibroblast, Chinese hamster ovary (CHO), and chicken DT40 B-lymphocyte cells (17–23). Cells deficient in any of the RAD51 paralogs are quite similar and have a general phenotype of a spontaneous increase in random mutagenesis and chromosomal aberrations, including substantial aneuploidy and chromosomal instability, which can lead to growth arrest and cell death (19,20,22–26). These cells are hypersensitive to various DNA-damaging agents, particularly DNA cross-linking agents and agents that cause replication stress (19,20,22–26). They also exhibit a modest sensitivity to ionizing radiation. Upon DNA damage, RAD51 paralog-deficient cells display elevated frequencies of gene mutation, chromosomal damage and chromosomal missegregation. In addition, DNA damage-induced DSB repair, sister chromatid exchange, and RAD51 nuclear foci formation at damaged sites are all severely reduced (19,22,23,25–28). RAD51-paralog deficient cells also have impaired HR-mediated gene targeting (20). Collectively, these traits suggest that RAD51 paralogs play a critical role in preserving genomic integrity through activities in the HR-associated DSB repair pathway. In humans, the function of the paralogs as mediators of HR demonstrate important tumor suppressor activity, as mutations in RAD51C lead to Fanconi anemia, mutations in RAD51C and RAD51D are associated with ovarian cancer, and a common RAD51B variant predispose individuals to breast cancer (29–32).
HR output is severely diminished in RAD51 paralog-deficient cells as evinced by decreases in damage-induced sister chromatid exchange. Studies have also measured gene conversion frequencies in these cells using HR-reporter systems consisting of homologous direct repeat substrates that measure intramolecular HR after I-SceI-induced DSBs (12,33–36). These studies demonstrated up to 25-fold reduction in HR repair in RAD51 paralog-deficient mammalian cells. Additionally, XRCC3-, RAD51C- and XRCC2-deficient cells showed altered HR product spectra with increased gene conversion tract lengths (37–39). XRCC3 deficiency also led to increased frequencies of discontinuous conversion tracts and local rearrangements near the site of the DSB (37). These assays have been integral to implicating the RAD51 paralogs to efficient homology-directed DSB repair. Most of these studies are limited however, as the output is a measure of successful HR leading to a gain-of-function event for selection. While effective for measuring the capacity of a cell to perform HR, these assays give little insight into how cells deficient for HR process DSBs and do not easily lend themselves to a quantitative analysis of alternative repair pathways in the processing of DSBs. In this study, we used an alternative HR-reporter system based on the loss-of-function phenotype of the adenine phosphoribosyl transferase (Aprt) gene in CHO cells. This assay allows simultaneous detection of accurate gene conversion events with estimated conversion tract length, single-strand annealing (SSA), large deletions, as well as other small events, such as point mutations, frameshifts, and small insertions or deletions (indels), which lead to loss of APRT function (40). With this comprehensive reporter system we investigated the outcome of DSB repair processing in isogenic CHO cell lines that were proficient or deficient for the RAD51 paralog, RAD51D, to determine the effect of RAD51D in DSB repair. We found that RAD51D-deficient CHO cells had a significantly higher frequency of spontaneous and I-SceI DSB-induced APRT marker loss. The repair spectra indicated that these cells had severely reduced capacity for conservative HR (gene conversion), and as a result, repair manifested as detrimental loss of large segments of the reporter locus through large NHEJ-derived deletions and SSA. Importantly, the frequency of these detrimental events was at or above that of gene conversion events in the RAD51D-proficient cells. These results indicate that, in addition to chromosomal instability resulting in aneuploidy, chromosome breakage, translocations, and fusion, HR deficiency (by loss of RAD51D function) leads to loss of large segments of chromosomes localized around DSBs. Thus, results from this study provide new insight into how RAD51-mediator proteins, specifically RAD51D, are involved in protecting the integrity of endogenous sequences surrounding DSBs.
MATERIALS AND METHODS
Plasmids and cell line constructions
The pAPRT-X plasmid was constructed from plasmid pGS100 [described in (40,41)], by integrating an 18-bp I-SceI site and a 30-bp triplex-forming oligonucleotide (TFO) target site into intron 2 of the Aprt sequence (Supplementary Table S1) using site-directed mutagenesis. The I-SceI site is 609-bp from exon 2 and 491-bp from exon 3. Other plasmids used in this study were the I-SceI expression vector, pCMV-I-SceI, and the control pCMV-Empty (pCMV-E) (42).
Cell lines in this study were all derived from the 51D1Lox CHO AA8 cells described by Hinz et al. (23). These cells contain gene-targeted modifications that placed LoxP sites flanking exon 4 of the Rad51D gene and a neomycin resistance gene at one allele, and exon 4, and a puromycin resistance gene at the second allele. From these cells we isolated and characterized a hemizygous APRT-deficient and spontaneous HPRT-deficient clone by sequentially selecting with 8-azaadenine and 6-thioguanine. This clone was then modified to create the RAD51D-WT-APRT-X (RAD51D-WT) cell line using the pAPRT-X plasmid in a two-step gene modification procedure described in detail by Merrihew et al. (41). Briefly, gene targeting using pGS100 followed by selection for ‘pop-out’ recombinants was used to insert a yeast FLP recombination target (FRT) site in intron 2 of the Aprt gene. An Aprt gene duplication was created by utilizing the site-specific FLP recombination system to target the vector pAPRT-X into the FRT site. This resulted in the direct repeat heteroallelic Aprt intrachromosomal recombination substrate at the endogenous Aprt genomic locus (Figure 1A). The reporter locus structure was confirmed by Southern blot analysis and the I-SceI/TFO target sites were confirmed intact by PCR followed by direct DNA sequencing. The RAD51D-WT cells were then treated with the Cre recombinase plasmid pBS185 as described by Hinz et al. (23) to create two separately isolated RAD51D-deficient cell lines: RAD51D-KO-APRT-X [RAD51D(−)]; clones 1 and 7. The knockouts were identified by Mitomycin-C, puromycin, and G418 sensitivity, and exon 4 removal was confirmed by PCR analysis with primers flanking the LoxP sequences (Supplementary Figure S1). The end result was a set of isogenic cell lines, differing only in RAD51D status, otherwise each containing the same recombination substrate.
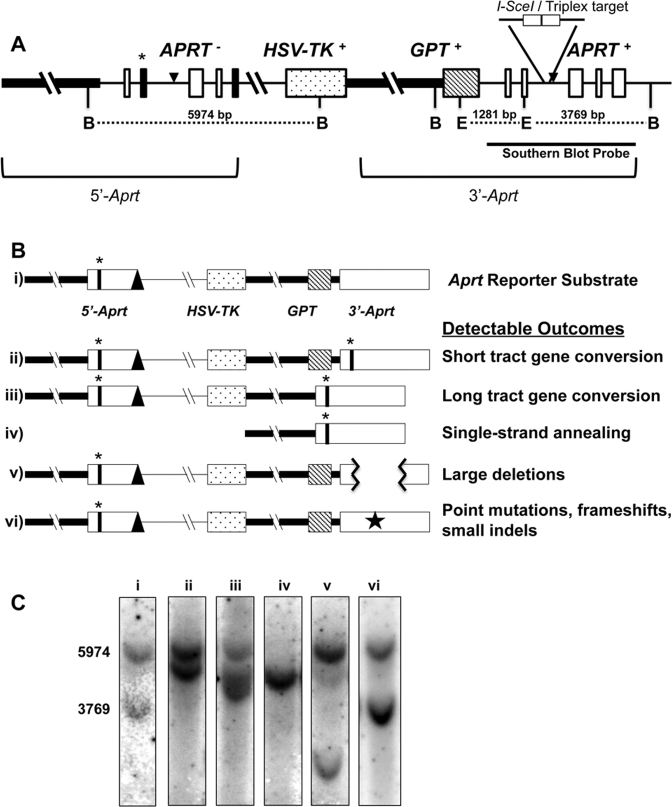
Figure 1.
Aprt intrachromosomal homologous recombination reporter substrate. (A) Schematic structure of the tandem Aprt gene-repeat reporter locus. The thick horizontal line represents the Aprt promoter region. Thin horizontal lines represent non-coding introns. Open white boxes represent non-mutated Aprt exons. Black filled boxes represent mutated exons (exon 2-point mutation and exon 5-truncation) that inactive APRT function. The dot-filled box represents the HSV-TK gene, and the diagonal-line filled box, the gpt gene. The asterisk (*) highlights the mutated EcoRV restriction endonuclease site (in exon 2). The TFO target/I-SceI sites are indicated in intron 2 of 3΄-Aprt repeat. The black triangles represent FRT sites used for locus construction. B: BglII, E: EcoRV restriction endonuclease sites. Representative repair endpoint structures are shown schematically in (B), and expected Southern blot outcomes are shown in (C) for the original substrate (i), Short tract gene conversion (STGC) (ii), Long tract gene conversion (LTGC) (iii), Single-strand annealing (SSA) (iv), an example of a Large Deletion (v), and Point mutations, frameshifts, small indels (Other) (vi) [represented by a star in panel (B)] categories when selecting against APRT function. A Southern blot representation of non-treated/non-selected cells (C-i) was included for comparison and size standards with band sizes (bp) to the left.
Cell culture and selection conditions
All cell lines were cultured in alpha-modified Minimal Essential Medium (α-MEM) (Sigma-Aldrich, St. Louis, MO, USA) containing 10% dialyzed fetal bovine serum (DFBS) (Atlanta Biologicals, Lawrenceville, GA, USA) in a humidified atmosphere of 5% CO2/95% air at 37°C. APRT(−), GPT(−) and HSV-TK(−) clones were selected for using 0.2 mM 8-azaadenine (MP Biomedicals, Solon, OH, USA), 10 μg/ml 6-thioguanine (Sigma-Aldrich, St. Louis, MO, USA), and 0.4 mM ganciclovir (InvivoGen, San Diego, CA, USA) in α-MEM containing 10% DFCS, respectively. Co-selection with both 8-azaadenine and ganciclovir was used to isolate APRT(−)/HSV-TK(−) clones.
Determination of spontaneous and DSB-induced frequencies of selectable marker loss
Independent populations of each cell line [RAD51D-WT, RAD51D(−) clone 1 and RAD51D(−) clone 7] were initiated at 50 cells per well in 12-well cell culture dishes. After 5–6 days, each independent population was trypsinized, counted, and divided into two subpopulations by replating into 12-well dishes. The first subpopulation was transfected with pCMV-E, an empty vector containing the CMV promoter. The second subpopulation was treated with the I-SceI expression vector pCMV-I-SceI to induce a site-specific DSB in intron 2 of the downstream Aprt heteroallele. Transfections were initiated 24 h after division of the cell cultures into subpopulations using 0.3 μl Xfect transfection reagent (Clontech, Mountain View, CA, USA) and 1.0 μg plasmid DNA per well complexed per manufacturers instructions in 25 μl Opti-MEM (ThermoFisher Scientific Waltham, MA, USA). Subpopulations were expanded to ∼90% confluency in 10-cm plates then split into four selections using drug concentrations outlined above; 8-azaadenine for APRT(−), 6-thioguanine for GPT(−), ganciclovir for HSV-TK(−) and 8-azaadenine+ganciclovir for APRT(−)/HSV-TK(−). All selections consisted of 3 plates/subpopulation with each plate containing ∼5 × 105 cells. An additional 3 plates were plated with ∼500 cells per plate in non-selective media to determine the number of viable cells plated. For each independent population, a single clone from each selection condition, for both pCMV-E and pCMV-I-SceI treated subpopulations, was selected at random and expanded for analysis by Southern blotting and PCR analysis. Corrected I-SceI-induced frequency was determined by subtracting the spontaneous frequency from the I-SceI-induced frequency for each independent population. Frequencies for each selection condition were determined by dividing the total number of recombinant clones by the total number of viable cells plated. Frequencies are represented as the mean ± standard error of the mean (S.E.M.) with significance (P) determined by an analysis of variance (ANOVA) test followed by Tukey's post hoc analysis.
Analysis of recombinant structures
Individual, randomly selected clones from each selection of each subpopulation were expanded to ∼90% confluency, and harvested for DNA isolation. A subset of cells from each clone was also interrogated for function of each selectable marker. DNA was prepared by proteinase K digestion followed by phenol/chloroform extraction and ethanol precipitation. The DNA was then subjected to Southern blot and PCR analyses to determine DNA repair/mutation end-products. For Southern analysis, the genomic DNA was digested with BglII and EcoRV restriction enzymes, resolved on an 0.8% agarose gel, and DNA fragments were detected using a Southern probe that recognizes the entire Aprt gene (Figure 1A). From non-treated cells, there are three diagnostic DNA fragments recognized by the probe: the upstream Aprt (5974-bp); the 3΄ portion of the downstream Aprt (3769-bp); and the 5΄ portion of the downstream Aprt gene (1281-bp) (Figure 1B–i and C–i). The smaller 1281-bp band, however, was usually run off the gel to obtain better resolution of the larger bands. For a description of Southern blot output for different repair categories, please refer to the Results section. PCR analysis of the gpt and Aprt regions (Supplementary Figure S2) was used to confirm results obtained from drug resistance and Southern blotting using primers (listed in Supplementary Table S1) flanking those regions. Gene conversion and other small mutation events in the Aprt gene were distinguished by PCR using primers flanking Aprt exon 2 followed by EcoRV digestion. Statistical analysis of the proportional distribution of recombinants from RAD51D-WT and RAD51D(−) cells was performed using Fisher's exact test.
RESULTS
The endogenous chromosomal HR reporter substrate
To investigate how a deficiency in HR affects the overall repair of DSBs, we constructed an intrachromosomal recombination reporter in isogenic CHO cell lines either proficient (RAD51D-WT) or deficient (RAD51D(−)) for the RAD51 paralog, RAD51D. The reporter assay is designed around a tandem repeat of the Aprt gene, at its endogenous locus, where the upstream Aprt (5΄-Aprt) gene results in a non-functional protein due to a mutation of an endogenous EcoRV restriction enzyme site in exon 2, as well as a 3΄ truncation of exon 5. The downstream Aprt (3΄-Aprt) is intact and fully functional. The reporter construct contains two additional selectable markers: the Escherichia coli guanine-hypoxanthine phosphoribosyltransferase (gpt) gene imbedded within homologous sequence of the 3΄-Aprt promoter region; and the Herpes simplex virus-thymidine kinase (HSV-TK) gene residing in non-homologous sequence between the Aprt repeats. Within intron 2 of the 3΄-Aprt gene, we integrated the 18-bp I-SceI recognition site directly adjacent to a 30-bp TFO target site, which can be used to target site-specific DNA damage to the endogenous Aprt locus (43). The structure of the genomic heteroallelic Aprt construct is shown in Figure 1A.
This is a loss-of-function assay and recombination and/or mutation events are detected by selecting against the function of the APRT, GPT and HSV-TK selectable markers with 8-azaadenine, 6-thioguanine and ganciclovir, respectively. Selection can be against individual markers, or a combination of markers, and results of selection are reported as the frequency of surviving colonies per number of viable cells selected. Individual clones are interrogated for function of each selectable marker and genomic DNA subjected to Southern blot and PCR analyses to determine the spectra of recombination and/or mutation end-products. Southern analysis was designed to determine the overall structure of the Aprt reporter locus as well as to interrogate gene conversion of the EcoRV site within exon 2. Southern results for possible repair end-products are discussed briefly below. As a loss-of-function assay, this recombination-reporter system allows simultaneous detection of a wide range of outcomes and can distinguish conservative gene conversions from non-conservative events such as SSA, large deletions, and smaller mutational events within coding regions. The smaller mutation events may comprise point mutations, frameshifts and small indels. We categorized the mutants/recombinants into five categories:
Short-tract gene conversion (STGC): events where the overall structure of the reporter locus remains intact, while the 3΄-Aprt gene has gained the deactivating EcoRV-site mutation from exon 2 of the upstream repeat (Figure 1B-ii). These events were detected as a 5΄-Aprt repeat (5974-bp) and 3΄-Aprt repeat (5050-bp) on Southern blots (Figure 1C-ii).
Long-tract gene conversion (LTGC): represented by co-conversion events where the 3΄-Aprt gene has acquired the deactivating polymorphism in exon 2 and lost the gpt gene due to longer repair synthesis into the promoter region of the upstream repeat (Figure 1B-iii). These events were detected as a full-length 5΄-Aprt (5974-bp) and, due to loss of the gpt region and its EcoRV site, a shorter 3΄-Aprt (4783-bp) than STGCs on a Southern blot (Figure 1C-iii).
Single-strand annealing (SSA): events that result in the loss of all three selectable markers and have a structure reflecting the precise deletion of the sequences between the Aprt repeats, while retaining a combination of the 5΄ region of the upstream repeat and the 3΄ region of the downstream repeat, resulting in only one copy of the Aprt gene (Figure 1B-iv). These events were detected as a single (4783-bp) band on Southern blots (Figure 1C-iv).
Large deletions: are losses of substantial sequence from the reporter locus that deactivate one or more of the selectable marker genes. These were detected as unpredictable size changes in or loss of diagnostic bands by Southern blot or PCR analysis (Figure 1B–v and 1C-v). These events are presumably derived from NHEJ or other end-joining processes.
Other: all small mutations that lead to deactivation of selected markers that are not detectable by Southern or PCR analysis. These may include point mutations, frameshifts, and small indels (Figure 1B-vi), which appeared similar to the untreated cells on Southern blots (Figure 1C-vi).
Spontaneous HR and/or mutation events in RAD51D-proficient vs. RAD51D-deficient mammalian cells
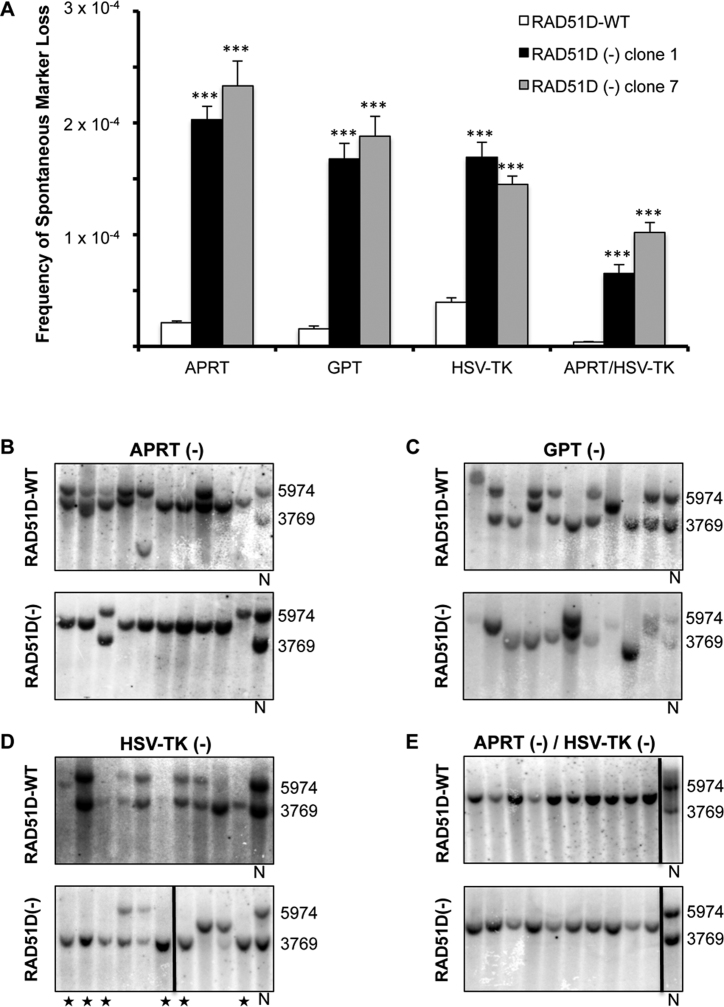
Spontaneous or non-induced HR events were measured by resistance to drug selection in cells transfected with the control, pCMV-E empty vector. These events are the result of endogenous processes resulting in mutation and/or HR and can be initiated throughout the reporter locus. As seen in Figure 2A, the frequency of spontaneous loss of any of the three markers was ∼4–11-fold higher (P < 0.001) in both RAD51D(−) cell lines when compared to the RAD51-WT cells. Similarly, simultaneous selection against APRT and HSV-TK revealed ∼18–28-fold higher frequencies (P < 0.001) in the RAD51D(−) cells compared to the RAD51-WT cells. As Aprt and HSV-TK are ∼5500-bp apart, double selection is intended to detect large events that affect a major portion of the reporter locus. Strikingly, the frequency of these large APRT(−)/HSV-TK(−) events in RAD51D(−) cells was higher than any of the frequencies measured in RAD51D-WT cells for the smaller localized events associated with individual marker loss (Figure 2A). When observing the repair spectrum of resistant clones, STGC, LTGC, SSA and large deletions accounted for ∼63–100% of the spontaneous events resulting in marker deficiency, regardless of the RAD51D status of the cells and selected marker (Figure 2B–E and Table 1). This suggests that the majority of marker loss in this system was mediated by DSB repair mechanisms. Collectively, these results demonstrate the role of RAD51D as an important factor in maintaining genome stability in response to spontaneous DNA damage.
Figure 2.
Spontaneous loss of selectable markers indicating HR/mutation events in CHO cells. (A) Frequency of spontaneous marker loss (mean ± S.E.M). Both RAD51D-deficient cell lines (clones 1 and 7) showed significantly higher marker loss than RAD51D-proficient cells for all selections based on ANOVA and Tukey's post hoc analysis (***P < 0.001). Representative Southern blots from clones selected for loss of APRT (B), GPT (C), HSV-TK (D), and APRT/HSV-TK (E) from RAD51D-proficient (top panels) and RAD51D-deficient cell lines (bottom panels). Each lane on a blot represents an individually isolated clone, and N represents non-treated cells used as a control and size standard. Numbers to the right of the Southern blots represent sizes (bp) of the non-treated control DNA bands. ★ indicate examples of events where deletion and SSA were indistinguishable by Southern blot. Vertical black lines within the Southern blots indicate samples that were separated on the same blot or were combined from different blots for ease of presentation.
Table 1. Distribution of recombination/mutation events obtained after spontaneous marker loss.
| APRT (−)*** | GPT (−)*** | HSV-TK (−) | APRT (−)/HSV-TK (−) | |||||
|---|---|---|---|---|---|---|---|---|
| RAD51D-WT n (%) | RAD51D (−) n (%) | RAD51D-WT n (%) | RAD51D (−) n (%) | RAD51D-WT n (%) | RAD51D (−) n (%) | RAD51D-WT n (%) | RAD51D (−) n (%) | |
| STGC | 20 (30%) | 1 (2%) | 1 (3%) | 0 (0%) | NA | NA | NA | NA |
| LTGC | 5 (8%) | 0 (0%) | 3 (10%) | 0 (0%) | NA | NA | NA | NA |
| SSA | 19 (29%) | 21 (42%) | 13 (43%) | 12 (40%) | 11 (29%) | 14 (38%) | 41 (98%) | 38 (97%) |
| Deletion | 4 (6%) | 11 (22%) | 3 (10%) | 18 (60%) | 13 (34%) | 14 (38%) | 1 (2%) | 1 (3%) |
| Othera | 18 (27%) | 17 (34%) | 10 (33%) | 0 (0%) | 14 (37%) | 9 (24%) | 0 (0%) | 0 (0%) |
| N | 66 | 50 | 30 | 30 | 38 | 37 | 42 | 39 |
aOther indicates small mutations to coding sequence, such as point mutations, frameshifts, and small indels that do not change the overall structure of the region and are not readily discernable by Southern blot or PCR analysis.
***Indicates significant difference (P < 0.001) in endpoint spectra between RAD51D-WT and RAD51D(−) cells for that particular selection pressure as determined by Fisher's exact test.
NA—not applicable.
The 3΄-Aprt and gpt regions of the reporter locus allow detection of conservative as well as non-conservative repair events that result in the loss of marker function. Gene conversions (STGC + LTGC) comprised the largest category (∼38%) of events that resulted in APRT deficiency in the RAD51D-WT cells; hence, spontaneous DNA lesions were predominantly repaired through HR. The gpt gene was only lost in ∼13% of the RAD51-WT cells due to gene conversion. The lower incidence of gene conversion events in the gpt region compared to the Aprt region was presumably due to the lack in homology, as the gpt region has no homology to the 5΄-Aprt region but is flanked on both sides by Aprt sequence. Large deletions made up only a small portion of events leading to APRT and GPT loss (∼6% and 10%, respectively) in the RAD51D-WT cells, suggesting that NHEJ was not a major contributor to spontaneous mutation events. SSA events made up a substantial percentage of repair events, however, and contributed to ∼29% of APRT deficiency and ∼43% of GPT deficiency. Additionally, the inefficiency of gene conversion at the gpt region corresponded to a marked increase in SSA when compared to events at the 3΄-Aprt region. SSA in this system represents large events leading to a loss of ∼9-kb of sequence. The prevalence of HR and SSA in our assay implies that the majority of spontaneous DNA damage underwent resection prior to repair.
The RAD51D(−) cells had a significantly different repair spectra (P < 0.001) than the RAD51D-WT cells for both the Aprt and gpt regions (Figure 2B and C, and Table 1). As expected, they showed a severely reduced capacity for HR with only one detectable spontaneous gene conversion event. This corresponded with a much higher incidence of SSA (∼42%) in APRT-deficient clones and large deletions in APRT-deficient (∼22%) and GPT-deficient (∼60%) clones. This is a >10-fold increase in both SSA and NHEJ-derived deletion frequencies at these loci (Supplementary Table S2). By simultaneously measuring HR along with other repair pathways, the results revealed that RAD51D plays a substantial role in conservative repair mechanisms, thus preventing repair events leading to deletions (SSA + large deletions). The fact that SSA was the predominant repair endpoint resulting in APRT deficiency in the absence of RAD51D further suggests that the majority of spontaneous repair events underwent DNA end resection. This implies that resection is upstream of the role of RAD51D in DNA repair. RAD51D-deficiency also resulted in a substantial decrease in ‘Other’ events in favor of deletions at the gpt locus. We are uncertain of the cause of this result, although it may be related to the type of damage or repair pathway choice occurring at this locus.
The HSV-TK gene is part of the non-homologous region located between the Aprt repeats, and no gene conversion events can be detected using this marker. As mentioned above, both RAD51D(−) clones demonstrated a much higher spontaneous HSV-TK deficiency than the RAD51D-WT cells. Their repair spectra, however, were not significantly different (P = 0.481) undergoing predominantly SSA and large deletion events (Figure 2D and Table 1). The inability to perform HR corresponded to an increase in large deletion events compared to the APRT and GPT markers, especially in the RAD51D-WT cells. This may be due to the type of detectable events in this region, as both WT and knockout profiles shifted from smaller and internal deletions to loss of the entire 5΄ region of the reporter locus [See Figure 2D RAD51D(−) samples marked with ★ for examples]. These events could also presumably be the result of SSA events between the promoter regions of the Aprt repeats, but we cannot determine this by our Southern analysis, and so we categorized these events as NHEJ deletion events (Table 1). Simultaneous selection against APRT and HSV-TK enriches for large events that affect markers ∼5500-bp apart. The most common repair product detected by this selection was SSA; however, in some previous studies, large deletions and rearrangements were also detectable (40,44–46). In our experiments, the repair endpoints were almost exclusively SSA for both RAD51D-WT and RAD51D(−) lines (Figure 2E and Table 1). Selecting against HSV-TK or APRT/HSV-TK can only detect non-conservative repair events, and the high mutation frequencies in RAD51D-deficienct cells indicate that HR is important for preventing these events. The protective effects of HR, however, did not reveal differences in the repair spectra in these contexts, suggesting that the resultant repair processes were similar regardless of the HR status of the cells.
Overall, RAD51D-deficient cells suffered a significantly higher frequency of spontaneous marker loss. Repair events in these HR-deficient cells predominantly manifested as SSA and large NHEJ-derived deletions. These results indicate that the inability to efficiently repair spontaneous/endogenous DNA damage by HR forces repair into non-conservative pathways (SSA + large deletions), with the vast majority of repair events resulting in sizable deletions (>100-bp based on PCR analysis) within the reporter locus. When assessing loss of a selectable marker in a non-homologous region (HSV-TK) or for expected large events (APRT/HSV-TK), RAD51D-proficient and RAD51D-deficient cells showed similar repair spectra, primarily consisting of large deletions and SSA. However, this occurred in RAD51D-deficient cells at substantially higher frequencies. Taken together, these data suggest that RAD51D is an important factor for protecting against large deletion events.
DSB-induced HR and/or mutation events in RAD51D-proficient vs. RAD51D-deficient mammalian cells
The cause of spontaneous DNA damage is uncertain and can occur at any location within the reporter locus. To determine the effects of RAD51D on DSB processing and repair, we induced site-specific DSBs using the yeast homing endonuclease I-SceI. I-SceI targets a site located in intron 2 of the 3΄-Aprt and is 609 bp from exon 2 and 491 bp from exon 3. The location of the target site between exons requires a sizable deletion to affect coding sequence and, consequently, the APRT marker. Smaller deletions localized around the break would be undetectable unless they interfered with mRNA splicing. This enriches for HR (gene conversions) and larger deletion (SSA and large deletions) events, but precludes us from a direct measure of small NHEJ mutational events. Because damage is driven from a specified site within the homologous Aprt sequence, this assay allows for more specific information on HR-associated DSB repair and how markers that are proximal versus distal to the break are affected by the presence or absence of RAD51D.
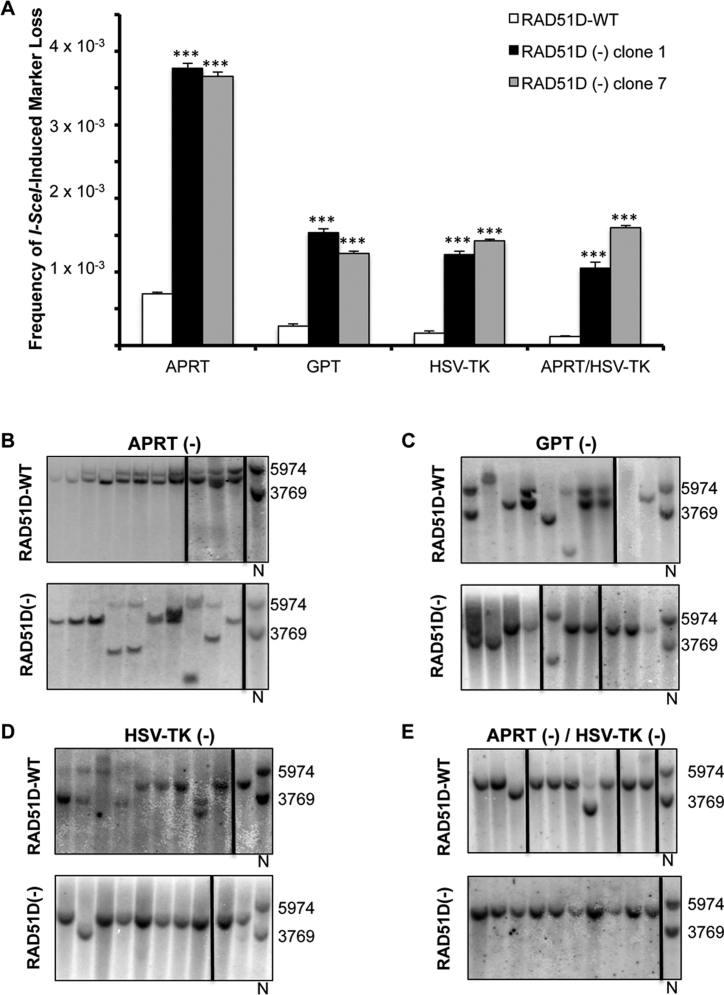
Upon I-SceI DSB induction, there was a significant increase (P <0.001) of marker loss in both RAD51D-WT and RAD51D-deficient cells (compare Figures 2A and 3A), demonstrating the ability of I-SceI to induce DSBs in this system. The RAD51D(−) cells demonstrated an ∼5- to 13-fold higher (P <0.001) frequency of resistant clones for all four selections than the RAD51D-WT cells. I-SceI DSB induction specifically increased the loss of APRT function in RAD51D-WT cells by ∼33-fold over spontaneous APRT loss (Figure 3A). Most of these events (∼45%) were gene conversions (STGC + LTGC) indicating a preference for HR repair (Figure 3B and Table 2). Induced DSBs also drove a high percentage of non-conservative DSB repair events with SSA (29%) and large deletions (24%) accounting for ∼53% of marker loss. The higher incidence of large deletions, with a concomitant lower incidence of ‘Other’ events compared to non-induced cells highlights the difference in repair of frank two-ended DSBs verses spontaneous DNA damage. The repair spectra were significantly different (P <0.001) between the RAD51D(−) and the RAD51D-WT cells (Figure 3B and Table 2) with gene conversions (STGC + LTGC) accounting for only ∼10% of repair, while SSA (39%) and large deletions (49%) represented the vast majority of repair endpoints in the RAD51D(−) cells. To further investigate deletion events, we sequenced the Aprt PCR amplicons that demonstrated internal deletions of the 3΄-Aprt region. In all deletion events (10 for RAD51D-WT and 17 for RAD51D(−)), we found either blunt-end ligation or single-base homology at the junction, suggesting these repair events were due to canonical-NHEJ mechanisms. This further demonstrates the importance of RAD51D in guiding high-fidelity repair processes to prevent deletions in response to frank DSBs in mammalian cells. Further, none of these deletion events contained a gene conversion event in exon 2 and are thus not a product of mixed HR/NHEJ events as reported previously in XRCC3-deficient cells and in interchromosomal recombination (37,47).
Figure 3.
I-SceI DSB-induced loss of selectable markers indicating HR/mutation events in CHO cells. (A) Frequency of corrected (spontaneous frequency subtracted from DSB-induced frequency)I−SceI-induced marker loss (mean ± S.E.M). Both RAD51D-deficient cell lines (clones 1 and 7) showed significantly higher marker loss compared to the RAD51D-proficient cells for all selections based on ANOVA and Tukey's post hoc analysis (***P < 0.001). Representative Southern blots from clones selected for loss of APRT (B), GPT (C), HSV-TK (D) and APRT/HSV-TK (E) from both RAD51D-proficient (top panels) and RAD51D-deficient cell lines (bottom panels). Each lane on a blot represents an individually isolated clone, and N represents non-treated cells used as a control and size standard. Numbers to the right of Southern blots represent sizes (bp) of non-treated control DNA bands. Vertical black lines within Southern blots indicate samples that were separated on the same blot or were from different blots for ease of presentation.
Table 2. Distribution of recombination/mutation events obtained after I-SceI DSB-induced marker loss.
| APRT (−)*** | GPT (−)*** | HSV-TK (−) | APRT (−)/HSV-TK (−) | |||||
|---|---|---|---|---|---|---|---|---|
| RAD51D-WT n (%) | RAD51D (−) n (%) | RAD51D-WT n (%) | RAD51D (−) n (%) | RAD51D-WT n (%) | RAD51D (−) n (%) | RAD51D-WT n (%) | RAD51D (−) n (%) | |
| STGC | 24 (39%) | 6 (9%) | 0 (0%) | 0 (0%) | NA | NA | NA | NA |
| LTGC | 4 (6%) | 1 (1%) | 20 (44%) | 0 (0%) | NA | NA | NA | NA |
| SSA | 18 (29%) | 26 (39%) | 14 (31%) | 30 (77%) | 29 (78%) | 21 (91%) | 61 (85%) | 34 (94%) |
| Deletion | 15 (24%) | 33 (49%) | 5 (11%) | 9 (23%) | 4 (11%) | 2 (9%) | 11 (15%) | 2 (6%) |
| Othera | 1 (2%) | 1 (1%) | 6 (13%) | 0 (0%) | 4 (11%) | 0 (0%) | 0 (0%) | 0 (0%) |
| N | 62 | 67 | 45 | 39 | 37 | 23 | 72 | 36 |
aOther indicates small mutations to coding sequence, such as point mutations, frameshifts, and small indels that do not change the overall structure of the region and are not readily discernable by Southern blot or PCR analysis.
***Indicates significant difference (P < 0.001) in endpoint spectra between RAD51D-WT and RAD51D(−) cells for that particular selection pressure as determined by Fisher's exact test.
NA—not applicable.
Previous reports have implicated RAD51 paralogs in regulating gene conversion tract length (37–39). Thus, we measured the incidence of LTGC in our assay by monitoring GPT loss in response to I-SceI DSB induction. The gpt gene is located 1320-bp from the I-SceI recognition site, while theEcoRV site in exon 2 of Aprt that is changed upon gene conversion, is only 709-bp from the I-SceI site. Thus, the length of the tract required for LTGC is almost double that required for Aprt conversion. The overall frequency of GPT loss after I-SceI DSB induction was lower than that of APRT loss for both RAD51D-proficient and RAD51D-deficient cells indicating that sites further from the I-SceI recognition site are less susceptible to DSB-induced loss of sequence (Figure 3A). Most of the repair in the RAD51D-proficient cells was via gene conversion (∼44%), all of which were classified as LTGC. In contrast, RAD51D-deficient cells were completely deficient in LTGC events within the gpt gene and 100% of GPT loss was due to detectable deletions (SSA + large deletions) of the reporter substrate (Figure 3C and Table 2). The considerable decrease of LTGC in our assay suggests a strict requirement for RAD51D in this process. The majority of GPT loss was due to SSA (∼77%) in these cells. These shifts in repair endpoints result in a significant difference in overall repair spectra between RAD51D-proficient and deficient cells (P < 0.001) at the gpt locus (Figure 3C and Table 2). Interestingly, both RAD51D-proficent and RAD51D-deficient cells suffered fewer large deletions and more SSA events for GPT loss compared to APRT loss. This suggests that for events that resulted in marker loss near the I-SceI-induced DSB, end-joining mechanisms were important for maintaining locus integrity by minimizing the size of deletion, while marker loss further from the DSB appeared to have undergone SSA annealing events, at the expense of end-joining pathways, resulting in much larger loss of sequence. This was exacerbated in the RAD51D-deficient cells where HR was unavailable.
The HSV-TK marker is located 6911-bp from the I-SceI DSB-target site and, as mentioned previously, is located within a non-homologous region of the reporter substrate that is unable to undergo intrachromosomal HR. The repair spectra did not differ significantly between the RAD51D-proficient and RAD51D-deficient cell types in this region (Figure 3D and Table 2). It was readily apparent that the I-SceI-induced DSBs drove SSA repair in both RAD51D-proficient and RAD51D-deficient cells, further indicating that repair leading to loss of distal markers was not due to end-joining mechanisms. Similar to selection against HSV-TK, the spectra of repair endpoints of clones obtained from simultaneous selection against APRT and HSV-TK after I-SceI-induced DSBs were not different between the cell lines (Figure 3E and Table 2), with SSA representing the vast majority of repair endpoints for both RAD51D-proficient and RAD51D-deficient cells.
Overall, I-SceI-induced DSBs resulted in a high level of selectable marker loss in both cell types, especially in the proximal Aprt region. In homologous Aprt regions, the RAD51D-proficient cells lost the APRT and GPT markers predominantly via gene conversion. The repair spectra also suggest that RAD51D-deficient cells were unable to efficiently perform gene conversion; however, the frequency of STGC at the Aprt locus was not different from the RAD51D-proficient cells (Supplementary Table S3). There was a much higher incidence of SSA and large deletion events with frequencies increasing by ∼7.5- and 11-fold, respectively (Supplementary Table S3). This suggests that in response to I-SceI-induced DSBs RAD51D may not be as important in facilitating HR as it is to protecting the genome from deleterious repair events. In these cells, marker loss was predominantly due to large deletions in proximity of the break; however, marker loss further from the break resulted from SSA. Intriguingly, in the gpt region, the RAD51D-proficient cells lost GPT largely due to LTGC, while these events were not detected in the RAD51D-deficient cells. The vast majority of GPT marker loss was instead due to SSA (∼77%). This suggests that in HR-deficient cells, SSA and not NHEJ (or other end-joining mechanisms), was the predominant mechanism for loss of sequence at a distance no farther than 1320-bp from the I-SceI-induced break. As with the spontaneous events, it is important to note the high frequency of marker loss due to deleterious deletion events (SSA + large deletions) in the RAD51D-deficient cells compared to the RAD51D-proficient cells, as this demonstrates the importance of RAD51D in maintaining genomic structure localized around the DSBs.
DISCUSSION
Previous studies have established that RAD51-mediator proteins, including BRCA2 and the five RAD51 paralogs, are vital for efficient HR. However, these studies provide little information on how the cells actually manage to process DSBs in the absence of these mediator proteins. Because our assay is a loss-of-function assay, we are able to quantitatively measure a spectrum of repair events ranging from conservative HR-mediated gene conversion to large detrimental deletions. We found that RAD51D-deficient cells had a significantly higher frequency of spontaneous and I-SceI DSB-induced selectable marker loss. These cells also had a severely reduced capacity for HR-mediated gene conversion events. Further, higher marker loss and inefficiency to complete HR manifested as detrimental loss of large segments of the reporter locus via large deletions and SSA events.
Spontaneous breaks are thought to result primarily from stalled and/or collapsed replication forks and are often repaired via HR (48,49). In this study and previous studies using this assay, we observed that the majority of spontaneous DSB repair in wild-type cells was indeed due to HR-mediated gene conversion (40,45,46). Here, we found that RAD51D deficiency resulted in considerably higher spontaneous loss of all three selectable markers at the reporter locus in our assays. The majority of this loss was due to deleterious DSB repair, leading to SSA/deletion events. This is consistent with previous studies where HR-deficient cells were more mutation prone due to an increase in spontaneous deletions. Hinz et al. (23,50), using the same RAD51D CHO cells used in our study, reported an ∼12-fold increase in spontaneous mutation rates with deletions increasing from 36% of the mutations in wild-type cells to 86% in the knockout cells at the endogenous Hprt locus (23,50). Similarly BRCA2-deficient CHO cells showed a ∼4-fold increase in mutation rate of the Hprt gene with deletions increasing from 13% of the mutations in wild-type cells to 52% in the knockout cells (51). The authors suggested that these events were due to end-joining at persisting breaks when broken replication forks were not restarted. We did not detect a high number of NHEJ-derived deletions, indicating that NHEJ was not the major repair pathway for spontaneous events in our assay. This is consistent with most spontaneous breaks resulting from collapsed forks, thus initially being one-ended, and inappropriate substrates for end-joining. In areas of high homology (3΄-Aprt region), the wild-type cells underwent a much higher incidence of spontaneous SSA events compared to large deletions. SSA was even more frequent in regions of lower homology (e.g. the gpt region) and much more frequent in the RAD51D-deficient cells. The shift from gene conversion events to SSA events was also seen previously in RAD51-paralog deficient yeast and in BRCA2-deficient mammalian cells (52–55). The prevalence of HR and SSA in our assay suggests that the majority of DSBs are being resected to provide the necessary 3΄-ends for these events. The fact that this occurs at such high frequencies in the RAD51-deficient cells may indicate that RAD51D has a protective roll against hyper-resection at spontaneous DSBs. This supports a recent report that the RAD51 paralogs are important for protecting stalled replication forks from collapse as well as reinitiating replication at stalled sites (56). Our results also support the finding that the RAD51 paralogs were protecting the nascent DNA from degradation at these stalled/collapsed forks (56). We conclude that the majority of spontaneous breaks are being resected in preparation for HR repair. When these breaks persist or when HR is unavailable, these ends containing 3΄ single-stranded overhangs are inefficient NHEJ substrates and are thus captured as SSA end-points in our assay.
Overall, I-SceI-induced DSBs resulted in a high level of selectable marker loss in both cell types, especially in the proximal Aprt region. As seen in previous studies, the majority of these events were gene conversions in the wild-type cells (40,45,46). I-SceI-induced DSBs also resulted in substantial SSA events and large deletions in proximity of the target site. Due to the location of the I-SceI site, the reporter system was only able to detect larger deletions (≥100 bp), thus we were unable to detect small NHEJ (or other end-joining) events that may have occurred near the break. This precluded us from completely monitoring fidelity of end-joining. Nevertheless, I-SceI-DSB induction drove a high level of large deletions in the proximal 3΄-Aprt region, displaying the importance of end-joining in mutagenesis of frank two-ended DSBs. This was especially evident in the RAD51D(−) cells where HR deficiency led to almost half of APRT loss resulting from large deletion events (Figure 3B). The high number of large deletions in the RAD51D-deficient cells suggests that a large number of DSB ends are being processed prior to end-joining, either passively due to delayed repair or through programmed resection in the initial steps of HR. The latter possibility is supported by the fact that SSA, which requires substantial end resection in this assay, accounts for the remaining APRT loss in RAD51D(−) cells.
In the gpt region, ∼1300-bp from the I-SceI site, the majority of repair events in the wild-type cells were the result of gene conversion, and there were considerably fewer large deletions in these cells following DSB induction. The RAD51D(−) cells showed no detectable gene conversion events in this region, yet large deletions were also substantially decreased. The loss of the more distal HSV-TK region also included fewer deletions in both RAD51D-proficient and RAD51D–deficient cells. SSA was increased in each of these cases and a deficiency for RAD51D, and consequently HR, led to almost exclusive use of SSA in the loss of distal sequences. Thus, end-joining (e.g. NHEJ) was not the main repair pathway involved in the mutation events that led to deletions of sequences distant from the DSB, rather repair was the result of SSA in our assay.
Taken together our data suggest that in our assay, wild-type and RAD51D-deficient cells repair frank two-ended DSBs primarily through end-joining (e.g. NHEJ) resulting in conservative repair or in small undetectable mutations near the DSB. The remaining breaks either persist leading to end degradation and larger detectable deletions, or are signaled for HR repair. In wild-type cells, HR resulted in gene conversions via intra- or interchromosomal HR, point mutations from HR repair synthesis, or error-free repair via sister chromatid HR. We speculate that RAD51D acts downstream of resection, therefore RAD51D(−) cells likely initiate HR at a rate similar to wild-type cells leading to processive resection of DSB ends. In the absence of RAD51D, the cells are unable to efficiently form RAD51 nucleofilaments on the 3΄ single-stranded DNA ends (23) to engage in strand invasion leading to the abortion of HR. The majority of DSBs then appear to be repaired by NHEJ or other end-joining mechanisms, but result in substantial deletions due to end processing and delayed repair. The remaining breaks undergo hyper-resection, possibly due to the lack of RAD51 filament formation to protect the ends. These hyper-resected breaks result in SSA in our assay, however, may have larger implications for DNA regions without tandem repeat sequences.
Previous reports indicate that the RAD51 paralogs XRCC3, RAD51C and, XRCC2 as well as BRCA2 are important for regulating gene conversion tract length, with their deficiency leading to a bias toward LTGC (37–39,57). RAD51C and XRCC3 interact in a complex (CX3) believed to be involved at later stages of HR. As XRCC2, RAD51C, and RAD51D are all members of the RAD51B/RAD51C/RAD51D/XRCC2 (BCDX2) complex, we investigated how RAD51D might be involved in this process. We monitored LTGC in our assay by observing co-conversion of Aprt and gpt as driven from I-SceI-induced DSBs. We found that RAD51D-deficent cells were severely inefficient at LTGC and only identified one clone in the entire study. This suggests that RAD51D has a different role in determining conversion tract length; XRCC3, RAD51C, XRCC2 and BRCA2 have apparent protective roles against LTGC, while RAD51D is required for LTGC. RAD51D may be working upstream of this process to initiate or stabilize strand invasion or repair synthesis, although further experimentation is required to confirm its exact role. Our assay also measures LTGC in a different manner than some of the previous studies, as we measure a loss of sequence instead of a gain. This may be an inherently different process and may explain the differences between our and previous LTGC results concerning the RAD51 paralogs. It will be interesting to determine the epistatic relationship between RAD51D and these other mediator proteins in determining gene conversion tract length.
In this study, we detected a substantial difference in the frequencies of marker loss in the RAD51D(−) cells versus the RAD51D-WT cells. This may implicate RAD51D in processes that protect areas surrounding DSBs, such as involvement in early DSB repair pathway choice, controlling end resection, or reversing early steps in HR. These differences may also be a result of the limitations of the reporter system. A large percentage of HR was certainly directed to the sister chromatid and we expect a large majority of these events to be undetectable in HR-proficient cells, as repair would be directed to the 3΄-Aprt and completely conservative. Only HR events that result in gene conversion due to intra- or intermolecular interaction with the 5΄-Aprt region would be detectable. Repair of DSBs in the HR-deficient cells, regardless of which Aprt repeat the repair was directed to, resulted in aborted HR and deletions. Thus, the frequencies represent all possible deleterious events and only a portion of possible HR events. In this study, we were also unable to detect mixed HR/NHEJ repair events that have been previously reported (37,42,47,58). However, the reporter systems of these studies contained heterologous sequences or dealt with targeting vector integration, which may impact the recombination mechanisms involved. In previous studies, our system has been able to detect other complex repair endpoints such as aberrant recombinants containing deletions and rearrangements (44,59). Another consideration of our system is that SSA can be generated by spontaneous DSBs throughout the entire reporter locus, and thus may inflate spontaneous SSA numbers. Given these restrictions, the Aprt-reporter assay used in this study provides a comprehensive overview of repair at an endogenous genomic locus in response to DSBs.
In our study, we confirmed that RAD51D is important for efficient HR. By simultaneously monitoring multiple DSB repair outcomes, we were able to expand on this observation to describe how cells process DSBs in the absence of RAD51D. We observed that RAD51D/HR-deficiency promotes chromosome instability by shifting the repair of DSBs toward highly deleterious end-joining processes, leading to excessive loss of large segments of the chromosome localized around the DSBs. These large deletions occurred at an exceptionally high frequency in the absence of RAD51D. In addition to previously described chromosomal instability in RAD51D-deficient cells resulting in aneuploidy, chromosome breakage, translocations, and fusion, our new results emphasize the importance of RAD51D in protecting the fidelity of endogenous DNA sequences. These results also further our understanding of how the RAD51 paralogs are involved in maintaining genomic stability and how their deficiency may predispose cells to tumorigenesis.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank the Larry Thompson lab for kindly providing the initial CHO cells. We thank Darius Bordbar for technical assistance on Southern blotting and PCR analysis. We also thank members of the Nairn lab for assistance in cell line construction. We would also like to acknowledge members of the Vasquez lab for discussions and advice.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institute of Health/National Cancer Institute (NIH/NCI) [CA093729 to K.M.V., CA097175 to K.M.V. and R.S.N., T32 CA09480 to W.A.R.]. Funding for open access charge: NIH/NCI [CA093729 to K.M.V.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Helleday T., Petermann E., Lundin C., Hodgson B., Sharma R.A.. DNA repair pathways as targets for cancer therapy. Nat. Rev. Cancer. 2008; 8:193–204. [DOI] [PubMed] [Google Scholar]
- 2.Swift L.H., Golsteyn R.M.. Genotoxic anti-cancer agents and their relationship to DNA damage, mitosis, and checkpoint adaptation in proliferating cancer cells. Int. J. Mol. Sci. 2014; 15:3403–3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanahan D., Weinberg R.A.. Hallmarks of cancer: the next generation. Cell. 2011; 144:646–674. [DOI] [PubMed] [Google Scholar]
- 4.Heyer W.D., Ehmsen K.T., Liu J.. Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet. 2010; 44:113–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kowalczykowski S.C. An Overview of the Molecular Mechanisms of Recombinational DNA Repair. Cold Spring Harb. Perspect. Biol. 2015; 7:a016410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Symington L.S. End resection at double-strand breaks: mechanism and regulation. Cold Spring Harb. Perspect. Biol. 2014; 6:a016436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holthausen J.T., Wyman C., Kanaar R.. Regulation of DNA strand exchange in homologous recombination. DNA Repair (Amst.). 2010; 9:1264–1272. [DOI] [PubMed] [Google Scholar]
- 8.Heyer W.D. Regulation of recombination and genomic maintenance. Cold Spring Harb. Perspect. Biol. 2015; 7:a016501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jensen R.B., Carreira A., Kowalczykowski S.C.. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature. 2010; 467:678–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu J., Doty T., Gibson B., Heyer W.D.. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat. Struct. Mol. Biol. 2010; 17:1260–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jensen R.B., Ozes A., Kim T., Estep A., Kowalczykowski S.C.. BRCA2 is epistatic to the RAD51 paralogs in response to DNA damage. DNA Repair (Amst.). 2013; 12:306–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chun J., Buechelmaier E.S., Powell S.N.. Rad51 paralog complexes BCDX2 and CX3 act at different stages in the BRCA1-BRCA2-dependent homologous recombination pathway. Mol. Cell. Biol. 2013; 33:387–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shu Z., Smith S., Wang L., Rice M.C., Kmiec E.B.. Disruption of muREC2/RAD51L1 in mice results in early embryonic lethality which can Be partially rescued in a p53(-/-) background. Mol. Cell. Biol. 1999; 19:8686–8693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deans B., Griffin C.S., Maconochie M., Thacker J.. Xrcc2 is required for genetic stability, embryonic neurogenesis and viability in mice. EMBO J. 2000; 19:6675–6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pittman D.L., Schimenti J.C.. Midgestation lethality in mice deficient for the RecA-related gene, Rad51d/Rad51l3. Genesis. 2000; 26:167–173. [DOI] [PubMed] [Google Scholar]
- 16.Kuznetsov S.G., Haines D.C., Martin B.K., Sharan S.K.. Loss of Rad51c leads to embryonic lethality and modulation of Trp53-dependent tumorigenesis in mice. Cancer Res. 2009; 69:863–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones N.J., Cox R., Thacker J.. Isolation and cross-sensitivity of X-ray-sensitive mutants of V79-4 hamster cells. Mutat. Res. 1987; 183:279–286. [DOI] [PubMed] [Google Scholar]
- 18.Fuller L.F., Painter R.B.. A Chinese hamster ovary cell line hypersensitive to ionizing radiation and deficient in repair replication. Mutat. Res. 1988; 193:109–121. [DOI] [PubMed] [Google Scholar]
- 19.Takata M., Sasaki M.S., Sonoda E., Fukushima T., Morrison C., Albala J.S., Swagemakers S.M., Kanaar R., Thompson L.H., Takeda S.. The Rad51 paralog Rad51B promotes homologous recombinational repair. Mol. Cell. Biol. 2000; 20:6476–6482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takata M., Sasaki M.S., Tachiiri S., Fukushima T., Sonoda E., Schild D., Thompson L.H., Takeda S.. Chromosome instability and defective recombinational repair in knockout mutants of the five Rad51 paralogs. Mol. Cell. Biol. 2001; 21:2858–2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deans B., Griffin C.S., O'Regan P., Jasin M., Thacker J.. Homologous recombination deficiency leads to profound genetic instability in cells derived from Xrcc2-knockout mice. Cancer Res. 2003; 63:8181–8187. [PubMed] [Google Scholar]
- 22.Smiraldo P.G., Gruver A.M., Osborn J.C., Pittman D.L.. Extensive chromosomal instability in Rad51d-deficient mouse cells. Cancer Res. 2005; 65:2089–2096. [DOI] [PubMed] [Google Scholar]
- 23.Hinz J.M., Tebbs R.S., Wilson P.F., Nham P.B., Salazar E.P., Nagasawa H., Urbin S.S., Bedford J.S., Thompson L.H.. Repression of mutagenesis by Rad51D-mediated homologous recombination. Nucleic Acids Res. 2006; 34:1358–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu N., Lamerdin J.E., Tebbs R.S., Schild D., Tucker J.D., Shen M.R., Brookman K.W., Siciliano M.J., Walter C.A., Fan W. et al. XRCC2 and XRCC3, new human Rad51-family members, promote chromosome stability and protect against DNA cross-links and other damages. Mol. Cell. 1998; 1:783–793. [DOI] [PubMed] [Google Scholar]
- 25.French C.A., Masson J.Y., Griffin C.S., O'Regan P., West S.C., Thacker J.. Role of mammalian RAD51L2 (RAD51C) in recombination and genetic stability. J. Biol. Chem. 2002; 277:19322–19330. [DOI] [PubMed] [Google Scholar]
- 26.Godthelp B.C., Wiegant W.W., van Duijn-Goedhart A., Scharer O.D., van Buul P.P., Kanaar R., Zdzienicka M.Z.. Mammalian Rad51C contributes to DNA cross-link resistance, sister chromatid cohesion and genomic stability. Nucleic Acids Res. 2002; 30:2172–2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O'Regan P., Wilson C., Townsend S., Thacker J.. XRCC2 is a nuclear RAD51-like protein required for damage-dependent RAD51 focus formation without the need for ATP binding. J. Biol. Chem. 2001; 276:22148–22153. [DOI] [PubMed] [Google Scholar]
- 28.Bishop D.K., Ear U., Bhattacharyya A., Calderone C., Beckett M., Weichselbaum R.R., Shinohara A.. Xrcc3 is required for assembly of Rad51 complexes in vivo. J. Biol. Chem. 1998; 273:21482–21488. [DOI] [PubMed] [Google Scholar]
- 29.Vaz F., Hanenberg H., Schuster B., Barker K., Wiek C., Erven V., Neveling K., Endt D., Kesterton I., Autore F. et al. Mutation of the RAD51C gene in a Fanconi anemia-like disorder. Nat. Genet. 2010; 42:406–409. [DOI] [PubMed] [Google Scholar]
- 30.Meindl A., Hellebrand H., Wiek C., Erven V., Wappenschmidt B., Niederacher D., Freund M., Lichtner P., Hartmann L., Schaal H. et al. Germline mutations in breast and ovarian cancer pedigrees establish RAD51C as a human cancer susceptibility gene. Nat. Genet. 2010; 42:410–414. [DOI] [PubMed] [Google Scholar]
- 31.Loveday C., Turnbull C., Ramsay E., Hughes D., Ruark E., Frankum J.R., Bowden G., Kalmyrzaev B., Warren-Perry M., Snape K. et al. Germline mutations in RAD51D confer susceptibility to ovarian cancer. Nat. Genet. 2011; 43:879–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Golmard L., Caux-Moncoutier V., Davy G., Al Ageeli E., Poirot B., Tirapo C., Michaux D., Barbaroux C., d'Enghien C.D., Nicolas A. et al. Germline mutation in the RAD51B gene confers predisposition to breast cancer. BMC Cancer. 2013; 13:484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson R.D., Liu N., Jasin M.. Mammalian XRCC2 promotes the repair of DNA double-strand breaks by homologous recombination. Nature. 1999; 401:397–399. [DOI] [PubMed] [Google Scholar]
- 34.Pierce A.J., Johnson R.D., Thompson L.H., Jasin M.. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999; 13:2633–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brenneman M.A., Weiss A.E., Nickoloff J.A., Chen D.J.. XRCC3 is required for efficient repair of chromosome breaks by homologous recombination. Mutat. Res. 2000; 459:89–97. [DOI] [PubMed] [Google Scholar]
- 36.French C.A., Tambini C.E., Thacker J.. Identification of functional domains in the RAD51L2 (RAD51C) protein and its requirement for gene conversion. J. Biol. Chem. 2003; 278:45445–45450. [DOI] [PubMed] [Google Scholar]
- 37.Brenneman M.A., Wagener B.M., Miller C.A., Allen C., Nickoloff J.A.. XRCC3 controls the fidelity of homologous recombination: roles for XRCC3 in late stages of recombination. Mol. Cell. 2002; 10:387–395. [DOI] [PubMed] [Google Scholar]
- 38.Nagaraju G., Odate S., Xie A., Scully R.. Differential regulation of short- and long-tract gene conversion between sister chromatids by Rad51C. Mol. Cell. Biol. 2006; 26:8075–8086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nagaraju G., Hartlerode A., Kwok A., Chandramouly G., Scully R.. XRCC2 and XRCC3 regulate the balance between short- and long-tract gene conversions between sister chromatids. Mol. Cell. Biol. 2009; 29:4283–4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sargent R.G., Brenneman M.A., Wilson J.H.. Repair of site-specific double-strand breaks in a mammalian chromosome by homologous and illegitimate recombination. Mol. Cell. Biol. 1997; 17:267–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Merrihew R.V., Sargent R.G., Wilson J.H.. Efficient modification of the APRT gene by FLP/FRT site-specific targeting. Somat. Cell Mol. Genet. 1995; 21:299–307. [DOI] [PubMed] [Google Scholar]
- 42.Rouet P., Smih F., Jasin M.. Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol. Cell. Biol. 1994; 14:8096–8106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vasquez K.M. Targeting and processing of site-specific DNA interstrand crosslinks. Environ. Mol. Mutagen. 2010; 51:527–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sargent R.G., Rolig R.L., Kilburn A.E., Adair G.M., Wilson J.H., Nairn R.S.. Recombination-dependent deletion formation in mammalian cells deficient in the nucleotide excision repair gene ERCC1. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:13122–13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Talbert L.L., Coletta L.D., Lowery M.G., Bolt A., Trono D., Adair G.M., Nairn R.S.. Characterization of CHO XPF mutant UV41: influence of XPF heterozygosity on double-strand break-induced intrachromosomal recombination. DNA Repair (Amst.). 2008; 7:1319–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sargent R.G., Meservy J.L., Perkins B.D., Kilburn A.E., Intody Z., Adair G.M., Nairn R.S., Wilson J.H.. Role of the nucleotide excision repair gene ERCC1 in formation of recombination-dependent rearrangements in mammalian cells. Nucleic Acids Res. 2000; 28:3771–3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Richardson C., Jasin M.. Coupled homologous and nonhomologous repair of a double-strand break preserves genomic integrity in mammalian cells. Mol. Cell. Biol. 2000; 20:9068–9075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saleh-Gohari N., Bryant H.E., Schultz N., Parker K.M., Cassel T.N., Helleday T.. Spontaneous homologous recombination is induced by collapsed replication forks that are caused by endogenous DNA single-strand breaks. Mol. Cell. Biol. 2005; 25:7158–7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arnaudeau C., Lundin C., Helleday T.. DNA double-strand breaks associated with replication forks are predominantly repaired by homologous recombination involving an exchange mechanism in mammalian cells. J. Mol. Biol. 2001; 307:1235–1245. [DOI] [PubMed] [Google Scholar]
- 50.Hinz J.M., Nham P.B., Urbin S.S., Jones I.M., Thompson L.H.. Disparate contributions of the Fanconi anemia pathway and homologous recombination in preventing spontaneous mutagenesis. Nucleic Acids Res. 2007; 35:3733–3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kraakman-van der Zwet M, Overkamp W.J., van Lange R.E., Essers J., van Duijn-Goedhart A., Wiggers I., Swaminathan S., van Buul P.P., Errami A., Tan R.T. et al. Brca2 (XRCC11) deficiency results in radioresistant DNA synthesis and a higher frequency of spontaneous deletions. Mol. Cell. Biol. 2002; 22:669–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Larminat F., Germanier M., Papouli E., Defais M.. Deficiency in BRCA2 leads to increase in non-conservative homologous recombination. Oncogene. 2002; 21:5188–5192. [DOI] [PubMed] [Google Scholar]
- 53.Tutt A., Bertwistle D., Valentine J., Gabriel A., Swift S., Ross G., Griffin C., Thacker J., Ashworth A.. Mutation in Brca2 stimulates error-prone homology-directed repair of DNA double-strand breaks occurring between repeated sequences. EMBO J. 2001; 20:4704–4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stark J.M., Pierce A.J., Oh J., Pastink A., Jasin M.. Genetic steps of mammalian homologous repair with distinct mutagenic consequences. Mol. Cell. Biol. 2004; 24:9305–9316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McDonald J.P., Rothstein R.. Unrepaired heteroduplex DNA in Saccharomyces cerevisiae is decreased in RAD1 RAD52-independent recombination. Genetics. 1994; 137:393–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Somyajit K., Saxena S., Babu S., Mishra A., Nagaraju G.. Mammalian RAD51 paralogs protect nascent DNA at stalled forks and mediate replication restart. Nucleic Acids Res. 2015; 43:9835–9855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saleh-Gohari N., Helleday T.. Strand invasion involving short tract gene conversion is specifically suppressed in BRCA2-deficient hamster cells. Oncogene. 2004; 23:9136–9141. [DOI] [PubMed] [Google Scholar]
- 58.Scheerer J.B., Adair G.M.. Homology dependence of targeted recombination at the Chinese hamster APRT locus. Mol. Cell. Biol. 1994; 14:6663–6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rahn J.J., Lowery M.P., Della-Coletta L., Adair G.M., Nairn R.S.. Depletion of Werner helicase results in mitotic hyperrecombination and pleiotropic homologous and nonhomologous recombination phenotypes. Mech. Ageing Dev. 2010; 131:562–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.