Abstract
Expression of bacterial type II toxin-antitoxin (TA) systems is regulated at the transcriptional level through direct binding of the antitoxin to pseudo-palindromic sequences on operator DNA. In this context, the toxin functions as a co-repressor by stimulating DNA binding through direct interaction with the antitoxin. Here, we determine crystal structures of the complete 90 kDa heterooctameric VapBC1 complex from Caulobacter crescentus CB15 both in isolation and bound to its cognate DNA operator sequence at 1.6 and 2.7 Å resolution, respectively. DNA binding is associated with a dramatic architectural rearrangement of conserved TA interactions in which C-terminal extended structures of the antitoxin VapB1 swap positions to interlock the complex in the DNA-bound state. We further show that a pseudo-palindromic protein sequence in the antitoxin is responsible for this interaction and required for binding and inactivation of the VapC1 toxin dimer. Sequence analysis of 4127 orthologous VapB sequences reveals that such palindromic protein sequences are widespread and unique to bacterial and archaeal VapB antitoxins suggesting a general principle governing regulation of VapBC TA systems. Finally, a structure of C-terminally truncated VapB1 bound to VapC1 reveals discrete states of the TA interaction that suggest a structural basis for toxin activation in vivo.
INTRODUCTION
Toxin-antitoxin (TA) systems play important roles for many prokaryotic organisms in survival during stress, such as starvation and antibiotic pressure (1). In most cases, toxin activation causes a halt in growth leading to a dormant or persistent state, which appears to be highly relevant for understanding bacterial pathogenesis and behaviour during chronic and recurring infections (2,3). TA systems are found ubiquitously among bacteria and archaea and are present in surprisingly large numbers in some organisms (4,5). In the most well-characterized and abundant class, the type II TA systems, the toxin and antitoxin form a tight protein–protein complex in which the activity of the toxin is inhibited during normal growth. When the cell experiences stress, increased levels of the alarmone molecule (p)ppGpp causes activation of specific proteases that destroy the antitoxin by proteolytic cleavage, subsequently releasing and activating the toxin (6). At the genomic level, type II TA genes are arranged in bicistronic operons with the antitoxin preceding the toxin, ensuring rapid inhibition of the toxin through translational coupling. Expression from TA loci is further regulated at the transcriptional level through direct binding of the antitoxin to pseudo-palindromic sequences in the promoter region (Figure 1A) (7). During this process, the toxin acts as a co-repressor by structurally stabilizing and increasing the affinity of the antitoxin for DNA (7,8). In an additional layer of regulatory complexity, some TA systems display what is known as conditional cooperativity, whereby the toxin causes de-repression at high concentrations by binding to additional sites on the antitoxin and destabilizing the operator-DNA interaction (9–11).
Figure 1.
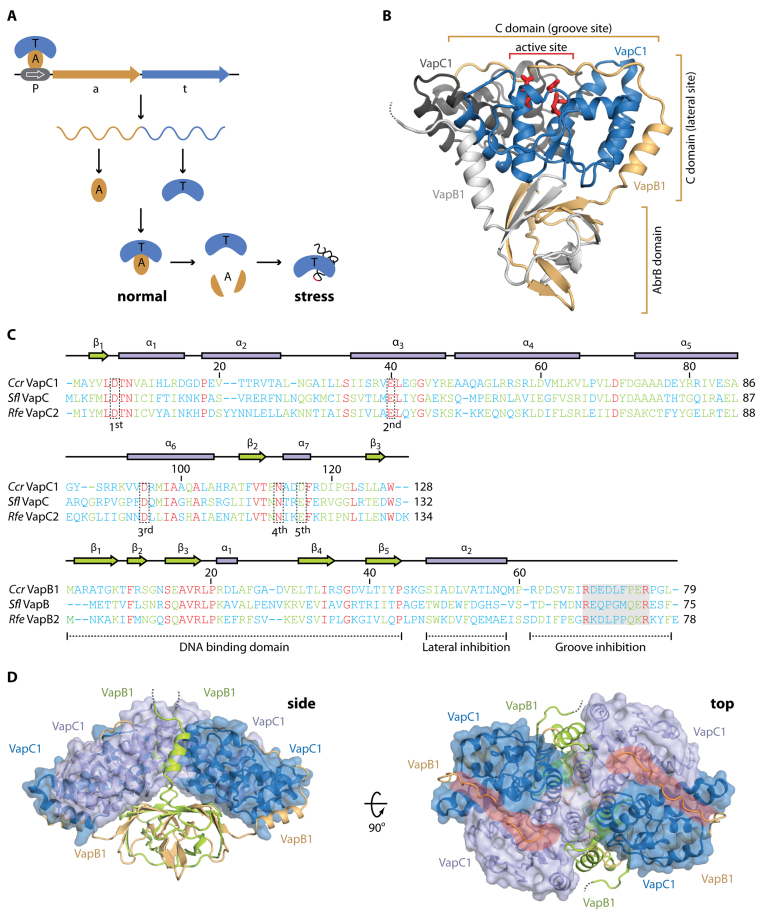
Overall structure of the C. crescentus CB15 VapBC1 complex. (A) Overview of the genomic organization and regulation of type II TA systems including VapBC. The TA locus consists of a promoter (P), antitoxin (a) and toxin (t) genes that give rise to a single mRNA expressing two separate proteins, A (light brown) and T (blue) that associate into a stable complex under normal growth conditions. During stress, the antitoxin is degraded by cellular proteases activating the toxin, which is a functional RNase targeting specific stable RNA species, including tRNA (lower right). Transcription from the TA locus is auto-regulated by binding of the TA complex to the promotor (P, top left). (B) Overview of the apo VapBC1 heterotetrameric structure with the location of the active site indicated by red sticks. One VapC1 toxin is shown in blue and the associated VapB1 antitoxin in light brown with the N-terminal AbrB domain and two interaction areas with the toxin indicated. The other VapC1 and VapB1 molecules of the tetramer are shown in dark and light grey, respectively, to highlight the TA interaction. The disordered VapB C terminus is shown with a dashed line. (C) Structure-based sequence alignment and secondary structure of the Caulobacter crescentus (Ccr) VapC1 and VapB1 proteins with the Shigella flexneri VapBC (Sfl, PDB ID 3TND) and Rickettsia felis VapBC2 (Rfe, PDB ID 3ZVK) structures (12,13). Sequence numbers correspond to the Caulobacter sequence. Conserved active site residues in VapC are indicated by dashed boxes (1st, 2nd, 3rd, 4th and 5th) and functional regions of VapB are indicated below the sequence (dashed lines). The region containing the pseudo-palindromic protein sequence in the VapB C-terminus is likewise indicated (shaded box). The sequences have been aligned using PROMALS3D (42) and are coloured by the level of similarity (blue = dissimilar, green = some similarity, red = identical). (D) Two orthogonal views of the heterooctameric apo VapBC1 complex formed by crystallographic symmetry. The VapC1 toxins are shown in light/dark blue with a semi-transparent surface and the VapB1 antitoxins in light brown/lime. The side view shows the protruding AbrB domains while in the top view, the positively charged grooves are shown with red. The disordered VapB C termini are shown with dashed lines.
In the most abundant type II TA family, Virulence associated proteins B and C (VapBC), the VapC toxin and the VapB antitoxin form a large, heterooctameric VapB4C4 complex in which the antitoxin inhibits the toxin by wrapping its C-terminus around it (12–14). Available crystal structures of VapBC complexes have revealed some of the molecular diversity of the interaction and shown that one VapB antitoxin can inhibit either one or two toxin molecules simultaneously using a 1:1 proximal, 1:1 distal or 1:2 mode of interaction (12–18). The N-terminus of VapB dimerizes to form a DNA binding domain belonging to either the AbrB, helix-turn-helix, ribbon-helix-helix or PhD/YefM type, which is required for transcriptional regulation (7). When activated, VapC toxins are functional endo-ribonucleases belonging to the PIN domain family (19,20) and appear to harbour RNase activity against specific cellular RNAs, including specific tRNA and rRNA species (21–24). PIN domain proteins consist of a parallel β-sheet with α-helices distributed on both sides that organize several conserved, acidic residues into an active site that binds two Mg2+ ions and is mechanistically similar to T4 phage RNase H (19,20). Upon interaction with VapB, positively charged residues from the antitoxin are inserted into the toxin active site, displacing the divalent metal ions thus rendering the PIN domain inactive (12,25).
Although structures of VapBC complexes in a range of states are available from several organisms, the conformational changes occurring upon DNA binding as well as the molecular basis for toxin activation are still not understood. To address this, we decided to determine crystal structures of the intact VapBC1 complex from the Gram-negative, oligotrophic bacterium Caulobacter crescentus CB15, both in isolation and bound to cognate operator DNA. Both on and off DNA, VapBC1 forms a compact heterooctameric assembly with two protruding AbrB-type DNA-binding domains. C-terminal extensions of two of the four VapB1 antitoxins in the complex interact tightly with all four VapC1 active sites in a 1:2 ratio. Upon binding DNA, the complex expands to dock the two AbrB domains into adjacent major grooves, causing a distortion of the regular B-form DNA double helix. Remarkably, the VapB1 C-termini switch from cis to trans conformation in the DNA-bound form, effectively interlocking the complex on DNA through non-covalent cross-linking that may have important consequences for cooperativity. Moreover, we show that the structural switch and recognition of the 2-fold symmetric VapC1 dimers is achieved by a pseudo-palindromic protein sequence in VapB1, which is a widespread feature of both bacterial and archaeal VapB antitoxins. Finally, we present a structure of VapC1 bound to a truncated version of VapB1 that only contains one half of the palindromic motif, which suggests a possible mechanism for toxin activation in vivo.
MATERIALS AND METHODS
Protein purification and oligomer analysis
The Caulobacter crescentus CB15 vapB1 and vapC1 genes (locus tags CC0032 and CC0031) were amplified by PCR, digested with EcoRI/BamHI or BamHI/HindIII, and ligated together into EcoRI/HindIII-digested expression vector pMG25 generating pKW912HB, which was used for expressing seleno-methionine (Se-Met) substituted VapBC1 complex in E. coli B834 (DE3). Cells were initially grown in minimal media supplemented 25 μg/ml methionine at 25°C until OD600 = 1, then pelleted and grown in minimal media without methionine for an additional 6 h at 37°C. Upon methionine depletion, 50 μg/ml Se-Met was added to the cells, which were further incubated at 37°C for 30 min before protein expression was induced with 1 mM isopropyl β-D-1-thiogalactopyranoside and the cells grown overnight at 25°C under vigorous shaking. For purification, the cells were lysed by sonication and the raw extract cleared by centrifugation and purification on a 1 ml HisTrapTM column (GE Healthcare) eluting in 50 mM Tris-HCl pH 8.0, 500 mM KCl, 150 mM imidazole, 5 mM MgCl2, 10% glycerol and 5 mM β-mercaptoethanol (BME). The eluted protein was precipitated overnight in 66% saturated (NH4)2SO4, followed by ion exchange chromatography on both 1 ml Source 15Q and 1 ml MonoQ columns (GE Healthcare) using 50 mM Tris-Cl pH 8.8, 5 mM MgCl2, 5 mM BME and a gradient into 1 M KCl in both cases. Final separation was achieved on a 24 ml Superdex 75 10/300 GL column (GE Healthcare) equilibrated in 25 mM Tris-HCl pH 8, 500 mM NaCl, 5 mM MgCl2 and 5 mM BME. Peak fractions were concentrated to ∼9 mg/ml prior to crystallization. Se-Met substituted complex was used both for the apo and DNA-bound structures. For expression of the truncated VapB1ΔC:VapC1 complex, pKW912HB was modified by PCR by insertion of a stop codon at position 73 of VapB1. Native protein was expressed in Escherichia coli BL21 (DE3) growing at 37°C in LB media until OD600 = 0.6, followed by overnight expression at 25°C after induction with 1 mM isopropyl β-D-1-thiogalactopyranoside. The cells were lysed and protein purified as described above. Oligomer analysis of protein complexes was performed using a 24 ml Superdex 200 10/300 HR (GE Healthcare) column equilibrated in 25 mM Tris-HCl pH 8, 500 mM NaCl, 5 mM MgCl2 and 5 mM BME.
DNA oligos and binding assays
Two complementary 27-bp DNA oligos derived from the promoter region of the C. crescentus vapbc1 operon were synthesized (Eurofins Genomics) as follows: Fw, 5΄-GGAACGTATATACGCATATTGACGGAG-3΄ and Rv, 5΄-CTCCGTCAATATGCGTATATACGTTCC-3΄. The Fw oligo was synthesized both with (Fw-label) and without a 5΄ fluorescein label for visualization. For gel filtration and crystallography, the oligos were dissolved in standard TE Buffer to 80 μM each, mixed 1:1 and heated to 96°C for 2 min then cooled to 19°C to a final concentration of 40 μM duplex. To generate the labeled duplex DNA, equal volumes of Fw-label and Rv (both at 200 μM) were mixed, heated to 96°C for 2 min and cooled to 19°C, resulting in 100 μM final DNA duplex. Binding analysis by gel filtration was performed by mixing equal volumes of protein and DNA (both at 130 μM), or protein and DNA separately, and diluting 40x in binding buffer (25 mM Tris-HCl, pH 8, 150 mM NaCl and 5 mM MgCl2, 5 mM BME) before incubating at 37°C for 1 h. The samples were then loaded onto 24 ml Superdex200 10/300 GL column (GE Healthcare) pre-equilibrated in binding buffer and monitored by A280 and A260 on an ÄKTA system. For electrophoretic mobility shift assay (EMSA), samples containing a fixed concentration of 5΄-fluorescein labelled DNA duplex (10 nM) and protein complex concentrations varying from 6.25 nM to 1.6 μM were mixed in binding buffer (25 mM Tris-HCl, pH 8, 150 mM NaCl, 5 mM MgCl2, 5 mM BME) and incubated for 1 h at 37°C, before separation on a 8% native polyacrylamide gel electrophoresis gels and visualization by fluorescence scanning at 495/517 nm (absorption/emission, Typhoon Trio, GE Healthcare).
Crystallization and structure determination
Trigonal crystals of the isolated Se-Met VapBC1 complex appeared in 3 M NaCl and 0.1 M Bis-Tris pH 5.5, whereas plate-shaped crystals of VapB1ΔC:VapC1 appeared in 0.1 M Na malonate pH 4.0 and 12% (w/v) PEG 3350. For the DNA-bound structure, the Fw and Rv oligos were resuspended in TE buffer, mixed to 40 μM duplex, heated at 96°C for 2 min and slowly cooled to 19°C before co-crystallization with the protein. DNA and protein were mixed in a 4:1 molar ratio at 6 mg/ml protein, after which plate-shaped crystals appeared in 20% (w/v) PEG 3350 and 0.2 M succinic acid pH 7.0. All crystals were cryo-protected in reservoir solution supplemented with 25% glycerol and flash-frozen in liquid nitrogen prior to data collection. Complete and highly redundant Se-Met single-wavelength anomalous dispersion (SAD) data were collected at 0.972 Å for VapBC1 and VapBC1:DNA at the I911-3 beamline at Max-lab (Lund, Sweden), while VapB1ΔC:VapC1 native data were collected at the ID30B beamline at ESRF (Grenoble, France). All data sets were processed with XDS (26). For VapBC1, 12 Se sites were located by Phenix.autosol (27) and used for SAD phasing in PHASER. Phasing was followed by automatic model building in Phenix.autobuild (28), manual model building in Coot (29) and iterative refinement with Phenix.refine (28). The final coordinates were validated using MolProbity (28). The VapBC1:DNA and VapB1ΔC:VapC1 structures were determined by MR using isolated VapBC1 as a search model. Both structures were manually rebuilt in Coot, iteratively refined with Phenix.refine and validated with MolProbity.
Bioinformatics analysis
A manually curated database of VapBC pairs from a previous study (30) was used to determine typical size distributions for VapB and VapC proteins, respectively. To generate a pool of potential vapBC loci for the analysis of VapB C-terminal motifs, pairs of adjacently annotated open reading frames matching the expected size distributions in the correct order were then extracted from all publicly available bacterial and archaeal genomes as of May 2016. For gene pairs in which the second gene encoded a protein matching to any of the PIN domain pfam models (PF01850, PF08745, PF10130, PF13470, PF13638 or PF17146) (31), the first gene was considered a valid vapB and included in the analysis. To identify pseudo-palindromic protein sequence motifs in this pool, for each protein sequence s, we identified all pairs of occurrences of arginine residues at positions i < j, where i > = 55 and j-i+1 was between 3 and 12, i.e. all sub-sequences s[i..j] = R…R of the protein sequence of lengths between 5 and 14, where the two arginine residues were separated by between 3 and 12 residues. Each such substring s[i..j] was then considered a ‘seed’ of a pseudo-palindromic motif (u, uRev), where u = s[i-lbest]…s[i]…s[i+rbest] and uRev= s[j-rbest]s[j]…s[j+lbest], such that distance Hamming-Blossum62 (u,uRev) is maximum, i+rbest < j-rbest and lbest ≤ 4. The Hamming-Blossum62 (x[1..n], y[1..n]) distance between two strings x and y (of equal length) is the sum of Blossum62 (x[i], y[i]) for i = 1,..,n (32).
RESULTS
C. crescentus VapB1 inhibits two VapC1 toxins simultaneously
The Gram-negative, oligotrophic bacterium Caulobacter crescentus CB15 contains a total of 14 type II TA loci, including 4 relBE, 2 vapBC, 2 parDE, 2 hipBA, 1 hicBA, 1 higBA, in addition to two uncharacterized loci (4,5). To understand the structural basis for toxin and DNA binding among distantly related VapBC orthologues, we decided to perform an in-depth analysis of one of the two VapBC systems found in this organism identified by the genes CC_0031 (vapC1) and CC_0032 (vapB1). Based on both sequence conservation and the ability of VapB1 to inhibit the toxicity of VapC1 (Supplementary Figure S1A), this locus represents a bona fide VapBC TA system (4,5). Se-Met substituted C. crescentus CB15 VapBC1 apo complex was generated by overexpression of the full-length operon in the Met-auxotroph E. coli strain B834 (DE3) in minimal media, purified to homogeneity and crystallised, and the structure determined to 1.6 Å using Se-Met SAD followed by automatic structure building and iterative refinement to Rwork/Rfree = 15.8%/16.8% (Supplementary Table S1, apo-VapBC1). The final model contains a single VapB2C2 heterotetramer in the crystallographic asymmetric unit (ASU) and is complete for both VapC1 toxins (residues 2–128) and one VapB1 antitoxin (2-79), while the other antitoxin molecule only could be traced up to residue 63 (Figure 1B, dashed line, Figure 1C and Supplementary Figure S1B). The tetramer interacts closely with another tetramer related by a crystallographic 2-fold axis and analysis of the interaction energy using PISA (33) strongly suggests that the relevant, biological entity is a VapB4C4 heterooctamer, as observed previously for other VapBC complexes (Figure 1D) (12–13,15–16,18). The complex has a compact structure and is organized with the toxins tightly interacting and forming a large, curved α-helical array with the antitoxin DNA binding domains protruding below. The complex is stabilised by tight toxin–antitoxin, toxin–toxin and antitoxin–antitoxin interactions.
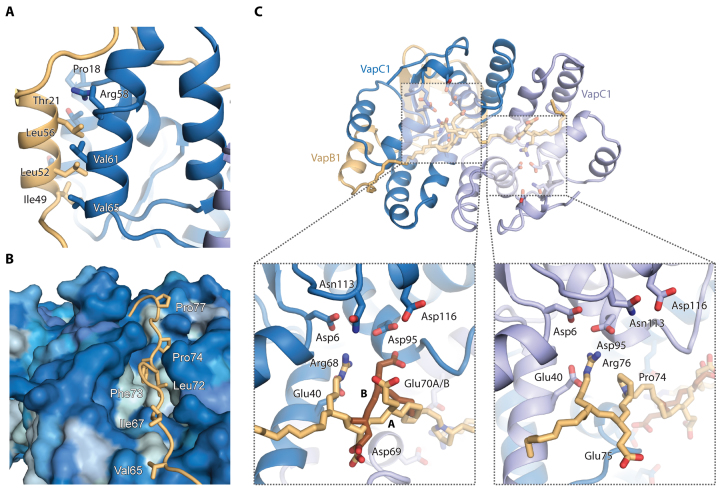
C. crescentus VapC1 displays a classical PIN domain RNase fold with the conserved and mostly acidic residues (Asp6, Glu40, Asp95, Asn113 and Asp116) in close proximity in the active site (Figure 1B, red sticks and C, dashed boxes). Like other VapC PIN domain proteins, four VapC1 toxins form two symmetrical homodimers held together by hydrophobic interactions generating a continuous and positively charged groove across the top surface near the active site cavities (Figure 1D, top). VapB1 consists of an N-terminal DNA-binding domain that dimerizes to form an AbrB-type motif (34) and a C-terminal extended region that interacts with the toxin active site in the complex (Figure 1B). Toxin–antitoxin interactions are primarily hydrophobic and distributed at two sites, which we term the ‘lateral’ and ‘groove’ sites (Figure 1B). At the lateral site, an α-helix from VapB1 packs against several α-helices of VapC1 using a leucine zipper-like motif (Figure 2A). At the groove site, the extended C-terminal extension of a single VapB1 antitoxin molecule locates into the continuous groove running across an entire toxin dimer where hydrophobic interactions anchor it to the surface and conserved Arg and Glu residues make specific interactions with residues in both VapC1 active sites (Figure 2B, C and Supplementary Figure S1C). One VapB1 molecule thus inhibits two VapC1 active sites simultaneously (the light brown molecule in Figure 1D) and the interaction can be described as 1:2. Consequently, the other VapB1 antitoxin is less tightly associated with the toxin dimer and only interacts at the lateral site (the green molecule in Figure 1D), explaining why this molecule only is visible to residue 63 in the electron density (Supplementary Figure S1B).
Figure 2.
Details of toxin–antitoxin interactions. (A) The lateral interaction site is dominated by hydrophobic interactions between VapB1 (light brown) and VapC1 (blue) in a leucine zipper-like motif. (B) At the groove site, the extended C-terminus of VapB1 (light brown) lies in a hydrophobic groove spanning the surface of the VapC1 dimer (blue). The VapC1 surface is coloured by hydrophobicity (light blue = hydrophobic, dark blue = hydrophilic) and relevant, interacting residues in VapB1 are labeled. (C) Overview of the 1:2 antitoxin:toxin interaction of the extended C-terminus of VapB1 (top) and close-up views of interactions at the two active sites of the VapC1 homodimer (bottom). Conserved acidic residues in the VapC1 active sites are shown in blue sticks and interacting residues in VapB1 with light brown sticks. VapB1 displays two discrete main chain conformations (A and B) at one site, which is illustrated with light/dark brown backbone colours.
VapC1 inhibition is mediated by a pseudo-palindromic motif in VapB1
Intriguingly, the C-terminus of VapB1 contains two motifs of equivalent arginine residues (Arg68 and Arg76) flanked by two acidic residues on opposite sides (Asp69 and Glu75) that interact in very similar ways in the two active sites of a VapC1 dimer (Figure 2C). In this motif, which is part of a region previously described as conserved (12), the arginine residues are separated by seven residues of extended protein structure, which matches the distance between the two active sites (22 Å). Moreover, the flanking acidic residues on either side result in an overall pseudo-palindromic motif that complements the internal, structural 2-fold symmetry of the VapC1 dimer (…R68D69EDLFPE75R76…). The VapB1 C-terminus has an extended, β-strand like conformation inside the VapC active site with side chains on alternate sides of the peptide backbone (Figure 2C). In the first part of the motif (R68D69), Arg68 thus points into the VapC1 active site whilst Asp69 points away and apparently serves to neutralize a number of positively charged surface residues. This structural motif is then repeated in the other VapC1 monomer only in reverse orientation (E75R76), with Glu75 pointing away and Arg76 pointing into the VapC active site. To understand if this type of interaction is unique to C. crescentus VapBC1 or a more widespread phenomenon, we first compared our structure to those of R. felis VapBC2 (13), which exhibits a similar 1:2 binding of the antitoxin to the toxin and S. flexneri VapBC, which displays 1:1 binding (12) (Supplementary Figure S2A). With sequence identities of 21.3%/25.0% (S. flexneri VapB / VapC) and 25.6%/30.5% (R. felis VapB/VapC), these proteins are homologous to C. crescentus VapBC1 (Figure 1C) and their crystal structures reveal very similar heterooctameric assemblies. Intriguingly, in both cases, the VapB C-termini contain similar pseudo-palindromic motifs based around two arginine residues exactly eight residues apart (R. felis VapB2, …R66KDLPPQKR74…; S. flexneri VapB, …R64EQPGMQER72…). In the R. felis VapBC2 structure, the two arginine residues from a single antitoxin molecule are located into the active sites of two VapC2 toxins similarly to what we observe in C. crescentus. In the S. flexneri VapBC structure, however, only the first arginine motif of both antitoxins is used in a 1:1 interaction, despite the presence of a perfect three-letter palindromic sequence (REQ…QER). Both VapB C-termini display a β-strand like fold as observed in C. crescentus VapBC1, with the arginine residue pointing into the active site and the following charged residue (K/E) pointing away (Supplementary Figure S2B). Furthermore, the third residue in the pseudo-palindromic motif appears to have a conserved acidic or amide functional group (D/E/Q) that points into the active site alongside the arginine (Supplementary Figure S2B).
Pseudo-palindromic motifs are widespread among VapB antitoxins
To investigate if such pseudo-palindromic motifs are widely present among VapB antitoxins, we generated a large set of 4127 putative VapB sequences extracted from all known bacterial and archaeal genomes and searched for C-terminal sequence patterns containing two arginine residues separated by 3 to 12 residues. The sequences were then sorted by the strength of their pseudo-palindromic character using a standard BLOSUM substitution matrix to up-weigh entries with similar functional groups at equivalent positions with respect to the palindrome centre (Table 1 and Supplementary Table S2) (32). The analysis reveals that unique palindromic as well as pseudo-palindromic motifs are surprisingly common and found among VapB antitoxins from a wide range of microbial species including Acidobacteria, Cyanobacteria, Proteobacteria, Spirochaetes, thermophilic bacteria as well as both branches of archaea, Crenarchaeota and Euryarchaeota. Roughly 45% of all sequences contain a motif in their C-terminus that matches the R…R search pattern and half of these (21.9% of all sequences) contain a more complex pattern involving one or more glutamate or aspartate residues on either side of the arginine residues (Table 1). The most prominent subclass of these consists of the sequences that contain ‘RE…ER’ as part of the pattern (7.3%) and this group contains some remarkable examples of perfect palindromes of the type ‘REQ…QER’ as also observed in S. flexneri VapBC (12) and even in some cases ‘EREQ…QERE’. The most complex patterns are found in Proteobacteria (e.g. Pseudomonas, Acetobacter, Haemophilus, Nitrosococcus) while the ‘REQ…QER’ pattern occurs more widely in both Cyanobacteria, Spirochaetes and Actinobacteria. Taken together, our sequence analysis demonstrates that subtle (pseudo-) palindromic sequence patterns appear to be widespread in the C-terminal extensions of VapB antitoxins across both bacteria and archaea. This discovery supports that the antitoxin C terminus plays an important, functional role in toxin–antitoxin biology and our structural data suggest this most likely involves modulation of either toxin inhibition or regulation of operator binding, or both.
Table 1. Pseudo-palindromic protein sequence motifs in VapB C-termini. The table shows a hierarchical view of (pseudo-) palindromic motifs and their subclasses found in the C-termini of 4127 putative VapB sequences extracted from all known bacterial and archaeal genome sequences. For each motif, the number of observations in the set is listed in the right column along with the percentage of the total.
| Motif or subclass | No. of observations (% of all) |
|---|---|
| R…R | 909 (22.03%) |
| RR…RR | 68 (1.65%) |
| RE…ER | 267 (6.47%) |
| REQ…QER | 23 (0.56%) |
| EREQ…QERE | 9 (0.22%) |
| ER…RE | 144 (3.49%) |
| R.R…R.R | 117 (2.83%) |
| R.E…E.R | 197 (4.77%) |
| E.R…R.E | 79 (1.91%) |
VapBC1 distorts operator DNA upon binding
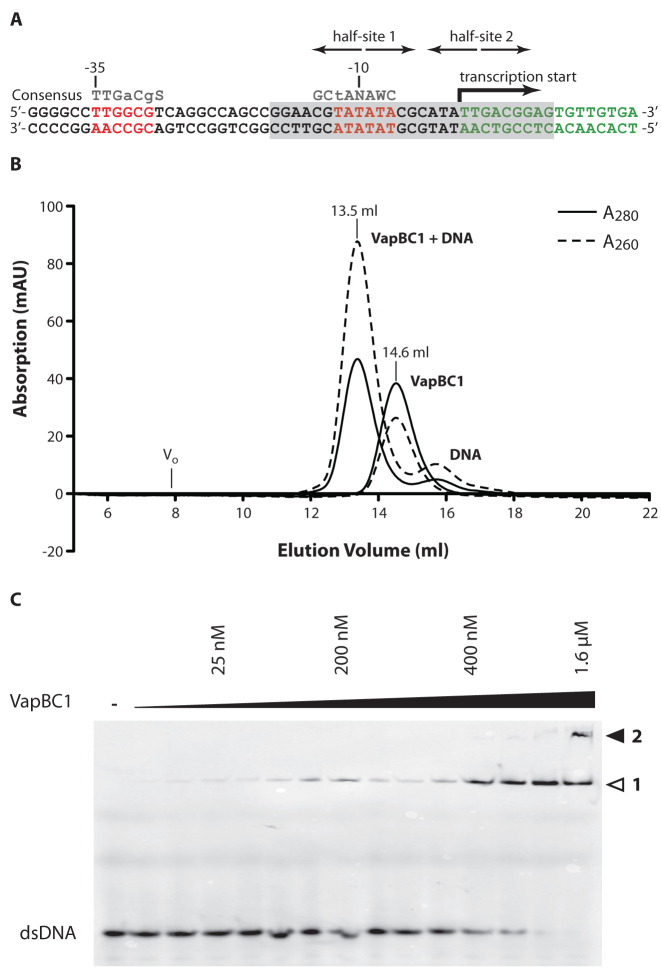
The overall structure of heterooctameric C. crescentus VapBC1 with two exposed DNA-binding domains suggests the complex is able to bind directly to the TA promoter, like other type II TA systems (7). Promoter elements in C. crescentus vary somewhat from the canonical sequences found in E. coli, however, both the −35 and −10 regions required for σ factor binding can be located (35,36). The vapBC1 operator region has been mapped by 5΄ RACE and found to be of the RpoD (σ73) type (37) (Figure 3A). This region contains a perfectly palindromic sequence (half-site 1) overlapping with the −10 element (TATA box), while the putative half-site 2 is not clearly palindromic. To test if VapBC1 can bind directly to DNA in this region, we generated a 27-bp DNA duplex covering both the −10 element and transcription start site and analysed the affinity of the intact VapBC1 complex in vitro using gel filtration chromatography and EMSA. In gel filtration, separation of VapBC1 pre-incubated with DNA results in a new, monodisperse peak with reversed A260/A280 ratio eluting earlier than the isolated heterooctamer suggesting formation of a protein-DNA complex (Figure 3B). In EMSA, titration of the protein complex into a fixed, low amount of fluorescently labelled DNA duplex (10 nM), caused formation of a higher band already in the low-nanomolar range, which was completely shifted around 500 nM VapBC1 (Figure 3C). At higher protein concentrations (>1 μM) we additionally observed a super-shifted complex suggesting formation of a higher-order complex. Together, these experiments conclusively demonstrate that C. crescentus VapBC1 forms a tight and stable complex with its operator region with an affinity in the nanomolar range.
Figure 3.
Operator structure and analysis of DNA binding. (A) Sequence and organization of the Caulobacter crescentus CB15 vapBC1 operator region. The −35 and −10 elements are shown with red letters with the consensus from analysis of a range of Caulobacter promoters shown above (S = C or G, W = A or T, N = any nucleotide, lowercase letters indicate partially conserved positions) (36). The location of the transcription start site, the palindromic half-site 1 overlapping the −10 region as well as the putative downstream half-site 2 are indicated with arrows. The region included in the 27-bp DNA duplex used for crystallography and binding studies is shown on a grey background. (B) Analysis of binding of a 27-bp DNA duplex covering half-sites 1+2 to VapBC1 using gel filtration chromatography. For both the isolated VapBC and VapBC-DNA, the absorption was monitored at A280 (solid lines) and A260 (dashed lines). The void volume (Vo) is approximately 8 ml. (C) DNA binding analysed by EMSA. A 27-bp fluorescein-labelled DNA duplex was present in a constant concentration of 10 nM while the VapBC complex was titrated from 6.25 nM to 1.6 μM. The gel shows formation of a stable VapBC-DNA complex with an affinity in the nanomolar range (band marked 1) as well as a higher-order complex at high protein concentrations (band marked 2).
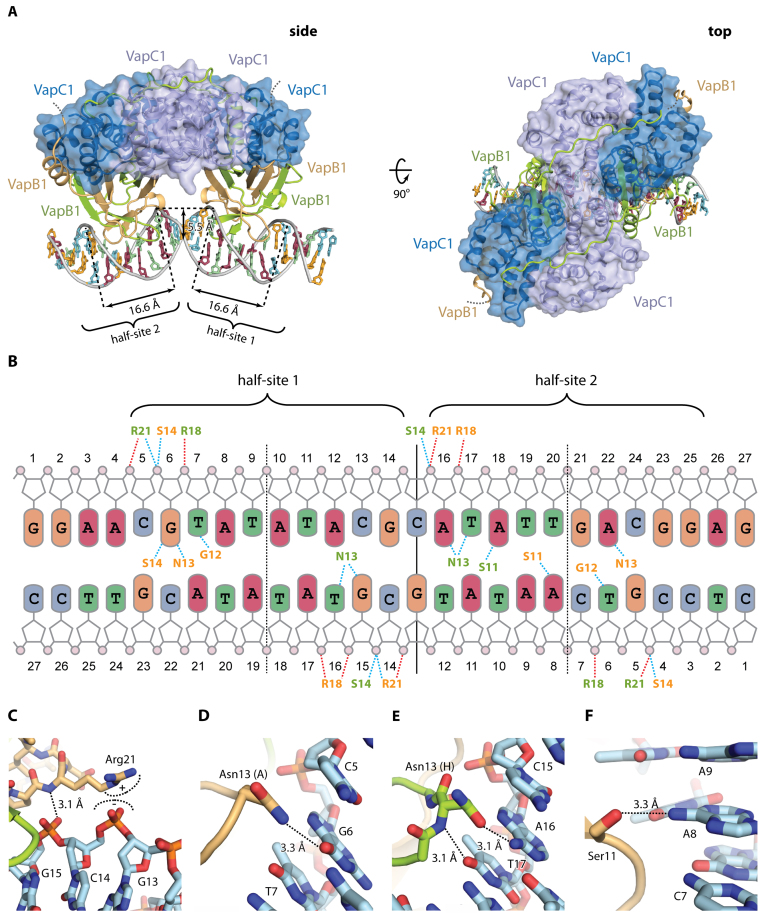
To understand the possible involvement of the antitoxin C-termini in VapBC operator binding, we next crystallised this C. crescentus VapBC1-DNA complex and determined its structure to 2.7 Å by molecular replacement using the apo structure as search model. The structure was iteratively refined to Rwork/Rfree = 17.8%/24.0% (Supplementary Table S1, VapBC1-DNA) and contains a complete VapB4C4 heterooctamer bound to the 27-bp DNA duplex in the crystallographic ASU with an overall architecture similar to the apo structure (Figure 4A). The model is complete for all VapC molecules (2-28), the DNA duplex (1-27) and two VapB antitoxins (residues 2–79) while the two remaining antitoxin molecules (similarly to the apo structure) only could be traced to around residue 63 (light brown molecules in Figure 4A). The two half-sites of the vapBC operator region are located ∼10 nucleotides apart (Figure 3A) and thus match the phase of the DNA duplex allowing the TA complex to interact with both half-sites in adjacent major grooves approaching DNA from the same direction (Supplementary Figure S3A). This is markedly different from the structure of R. felis VapBC2 bound to DNA, which has the same overall architecture but is much more open and approaches the DNA duplex from opposing sides (Supplementary Figure S3B) (13). In the only other known structure of a VapBC-type TA system bound to DNA, N. gonorrhoeae FitAB, the heterooctamer is even more open with no interactions between the two VapC homodimers (Supplementary Figure S3C) (14). This comparison reveals a surprising variety in DNA interaction modes among homologous TA complexes that structurally and architecturally appear very similar.
Figure 4.
Structure of VapBC1 bound to operator DNA. (A) Two orthogonal views of the VapBC1 heterooctamer (blue/green/light brown) bound to DNA with the VapC homotetramer in a semi-transparent surface representation, and DNA bases coloured as in B. In the side view (left), the positions of half-sites 1 and 2 are indicated with the widths of the major grooves and depth of the minor groove shown. The disordered VapB C termini are shown with dashed lines. (B) Schematic overview of the interactions between VapBC1 and DNA. Dashed lines indicate hydrogen bonds (blue) and electrostatic interactions (red) to phosphate groups (pink circles) and bases (coloured rounded squares). Residues in VapB1 are shown in colours matching panel A (light brown/green). Vertical lines indicate the operator pseudo 2-fold axis separating the two half sites (solid) as well as the half site 2-fold axes (dashed), and the extent of the two half-sites are indicated as well. (C) Interactions of VapB1 Arg21 with DNA. (D) Interactions of VapB1 Asn13 (chain A) with DNA. (E) Interactions of VapB1 Asn13 (chain H) with DNA, showing that it flips the side chain to reverse the hydrogen bond polarity in the other molecule. (F) Interactions of VapB1 Ser11 with DNA. Ser11 forms a base-specific hydrogen bond with A8 in half-site 2.
In the VapBC1-DNA structure, we observe an overall B-DNA like conformation of the operator region, however, with significantly expanded major groove widths (16.6 Å versus 11.6 Å in canonical B-DNA) and a diminished minor grooves depth (5.5 Å versus 8.2 Å in B-DNA) (Figure 4A). Despite half-site 2 not being perfectly palindromic, the structures of the two protein-bound half-sites overlay nearly perfectly suggesting that the DNA conformation is a direct result of VapB binding and not an intrinsic feature of the operator DNA sequence (Supplementary Figure S4A). Direct contacts between the VapB1 AbrB domains and DNA are sparse and symmetrically arranged in the two major grooves (Figure 4B). Arg21 is responsible for the most significant electrostatic interaction and this position contacts the phosphate backbone of DNA in all four antitoxin molecules of the complex (Figure 4C). Sequence-specific base interactions are found in a few places, most significantly Asn13, which contacts two guanine bases in half-site 1 (G6 and G15, Figure 4B and D) while the same side chains of the corresponding residues in half-site 2 have flipped 180° to be able to interact with adenine (A16 and A22) with reversed hydrogen bond polarity (Figure 4B and E). Likewise, Ser11 forms a sequence-specific hydrogen bond to DNA nucleotide A8 in half-site 2, an interaction which is not possible half-site 1 where this nucleotide is a pyrimidine (T18) (Figure 4F). Together, these examples illustrate some of the flexibility that allows VapB to adapt to an imperfect pseudo-palindromic DNA sequence using variable hydrogen bond patterns.
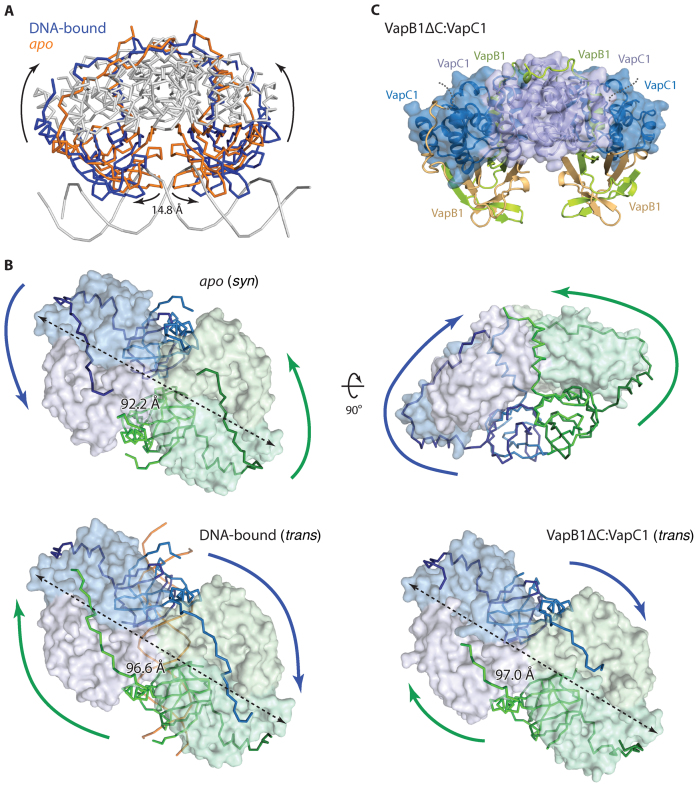
The VapB1 C-terminal extension is a flexible cross-linking module
In the apo state, the two AbrB domains of the VapBC1 complex are close together and physically interact (Supplementary Figure S4B) while upon binding DNA, the complex expands nearly 15 Å to accommodate the domains into adjacent major grooves (Figure 5A). Opening of the complex is effectively achieved by rotation of one VapC dimer by ∼25° with respect to the other and occurs without disturbing interactions between the toxin and the antitoxin AbrB domain (Supplementary Figure S4C and Supplementary Movie). In contrast, DNA binding causes a dramatic rearrangement of the VapB1 C-terminal extensions located in the groove sites, which swap places as the complex opens up (Figure 5B). More precisely, the ‘outermost’ VapB1 termini reach around and interact with both molecules of a toxin dimer in the apo state (Figure 5B, apo), whereas the ‘innermost’ VapB1 termini interact in the DNA-bound state (Figure 5B, DNA-bound). An important structural consequence of this is that the VapC1 toxin dimers are inhibited in syn by their cognate VapB1 antitoxins in the apo state (i.e. those antitoxins that belong to the same tetramer), while they interact in trans with antitoxins belonging to the other heterotetramer in the DNA-bound state. This rearrangement is apparently a direct result of the overall stretching of the TA complex by nearly 5 Å in length as required for DNA binding and made possible by relatively small differences in the number of interactions between the two states. Once the VapB1 C-termini have swapped positions, interactions between the pseudo-palindromic sequence in the antitoxin and the VapC1 active sites are nearly identical to the apo state, but occur in the reverse direction. Together, these observations suggest that the VapB1 C-terminal extension is a flexible module that is able to cross-link the VapC1 dimers in at least two different ways in the heterooctamer depending on whether DNA is bound or not and is therefore likely important for regulation of DNA binding.
Figure 5.
The VapB1 C-terminal extension is a flexible cross-linking module. (A) Superposition of the VapC homotetramers of the apo (orange) and DNA-bound (blue) crystal structures of VapBC1 showing that the AbrB domains move 14.8 Å apart upon DNA binding. For both structures, the antitoxin molecules are shown as coloured ribbons while the toxin molecules and DNA are in grey. (B) Comparison of the positions of the VapB1 C-terminal extensions in the apo, DNA-bound and VapB1ΔC:VapC1 structures annotated as syn or trans. In all cases the antitoxins are colour-coded corresponding to their cognate toxin dimer (blue or green) to highlight how they cross over in the DNA-bound and truncated structures (i.e. blue C-termini bind green toxin dimers). The DNA duplex is shown with orange ribbons, coloured arrows indicate the directions of the VapB1 C-termini while dashed arrows indicate the total length of the octamers. (C) Crystal structure of the VapB1ΔC:VapC1, which has a DNA-bound like conformation, with colouring like in Figure 1D. The disordered VapB C termini are shown with dashed lines.
C-terminal truncation of VapB1 does not induce a 1:1 toxin:antitoxin interaction
Since the observed 2:1 interaction of toxin to antitoxin only utilizes half of the toxin-binding potential of the available VapB1 C-termini, we finally asked what would happen if the C-terminus was truncated to only contain a single VapC binding site, which would correspond to a 1:1 molar ratio between binding sites on the antitoxins and VapC active sites in the complex. To answer this question, we introduced a stop codon at position 73 in the gene coding for VapB1 and purified the resulting truncated complex to homogeneity. In vivo, the truncated antitoxin displayed reduced activity (Supplementary Figure S1A), nevertheless, the oligomeric state of the VapBC complex remained stable as judged from gel filtration chromatography (Supplementary Figure S5). We next proceeded to crystallize the complex between this truncated version of VapB1 and VapC1 and determined the structure to 1.9 Å by molecular replacement using the apo structure as search model, built and iteratively refined the model to Rwork/Rfree = 19.6%/23.9% (Supplementary Table S1, VapB1ΔC:VapC1). The structure contains 12 protein chains per ASU, organized in a VapB4C4 heterooctamer (Figure 5C) and a VapB2C2 heterotetramer. The heterotetramer is related by 2-fold crystallographic symmetry to a neighbouring complex generating a second, complete heterooctamer, which is identical to the first within experimental error (Supplementary Figure S4D, r.m.s.d Cα atoms = 0.73 Å). All six toxin chains (residues 2–128) and three of the antitoxin chains (2-7) are essentially complete, while the remaining three antitoxin chains only could be traced to the end of the lateral binding site around residue 65 (light brown molecule in Figure 5C) as seen before. The overall conformation of the complex is somewhere in-between the apo and DNA-bound forms but closest to the DNA-bound form with the VapB1 AbrB domains well separated and with an overall stretched VapC1 complex (Figure 5B, VapB1ΔC:VapC1). Consistent with this, the VapB1 C-terminal extensions interact with the VapC1 active sites in trans like observed in the DNA-bound form, which suggests that the free TA complex exists in an equilibrium between the tight (apo) and more open (DNA-bound/truncated) conformations. Surprisingly, we find a 1:2-like interaction to be preserved in the truncated structure, i.e. only one VapB1 C-terminal extension interacts with each toxin dimer while the other C-terminal extension remains disordered. Since the C-terminus is truncated, this means that only half the VapC1 active sites are inhibited, while the other are not interacting with an antitoxin at all. Overall, the electron density for the antitoxin molecules is significantly poorer than for the full-length structure, particularly near the C-termini. Most significantly, the helix at one of the lateral sites appears to have become partly unstructured (left, light brown molecule in Figure 5C), suggesting that VapB in the cleaved state is less tightly bound than in the intact form.
DISCUSSION
In this paper, we show that C. crescentus VapBC1 forms a flexible macromolecular complex able to adapt to DNA binding via conformational changes, including a dramatic reorganization of stable toxin–antitoxin interactions. Flexibility in toxin and antitoxin interactions is not uncommon and switching of antitoxin C-termini have also been observed in other structurally unrelated TA systems such as ccdAB and MazEF suggesting it may be a general principle in TA biology (10,38–39). For the antitoxin VapB1, we identify a sequence pattern in the C-terminus that is directly involved in toxin inactivation and show that this is a widespread feature of both bacterial and archaeal VapB antitoxins. We also show that the pseudo-palindromic nature of this sequence is the structural basis for inhibition of the VapC toxin in a 1:2 ratio, with each half of the motif locating into individual VapC1 active sites (13,15). One might therefore speculate that the presence of such pseudo-palindromic sequences can be used to predict whether VapB antitoxins interact in a 1:1 or 1:2 ratio with their cognate toxins. However, analysis of available TA structures reveals that this does not appear to be the case: S. flexneri VapB binds in a 1:1 ratio to its cognate VapC toxin despite containing a perfect REQ…QER palindromic sequence, thus leaving the second part of the palindromic motif of both antitoxin molecules disordered in the structure (12). Nevertheless, we can conclude the sequence signature observed in S. flexneri VapBC is perfectly compatible with 1:2 binding and suggests that another conformation of the complex might exist that makes use of the full C-terminus, e.g. during operator DNA binding. Likewise, it was demonstrated that M. tuberculosis VapBC-15 can display both 1:1 and 1:2 interactions, which adds further support to the notion that the interaction is highly dynamic and depends on the functional state of the TA complex (15). The structure of C-terminally truncated VapB1 bound to VapC1 sheds further light on this surprisingly complex structural flexibility, showing that one of the two VapB1 antitoxin molecules does not bind to VapC despite the presence of a nearby unoccupied groove site. This strongly suggests that it is the overall conformation of the TA complex, not the availability of binding sites that dictates the location of the C-termini. The observation that the TA complex stretches upon binding DNA in order to locate the AbrB motifs into adjacent major grooves further highlights this.
For those VapBC pairs for which VapB contains a pseudo-palindromic motif, we can conclude that regardless of whether 1:1 or 1:2 binding is observed, only 50% of the VapC binding capacity of VapB is used at any time. Either both antitoxin molecules interact with one VapC active site each (proximal/distal 1:1 interaction) and the other half of the palindromic motif is left unused, or one molecule uses both its interaction sites and the other none of them (1:2). One possible rationale for this apparent excess of VapC binding sites is that they are important for cooperativity in TA complex formation or toxin activation, or both. For example, the unused binding sites would allow additional toxin molecules to bind during activation and thus promote further destabilization of TA complex. For some TA systems, such as Phd/Doc and RelBE, a phenomenon known as ‘conditional cooperativity’ in DNA binding has been described, in which additional toxin molecules join the TA complex at high concentration, causing destabilization of DNA binding and consequently de-repression (9,10). A similar mechanism, by which excess VapC is able to break a VapBC DNA complex has been demonstrated for S. enterica LT2 VapBC, however, the structural basis for this mechanism, remains to be elucidated (40).
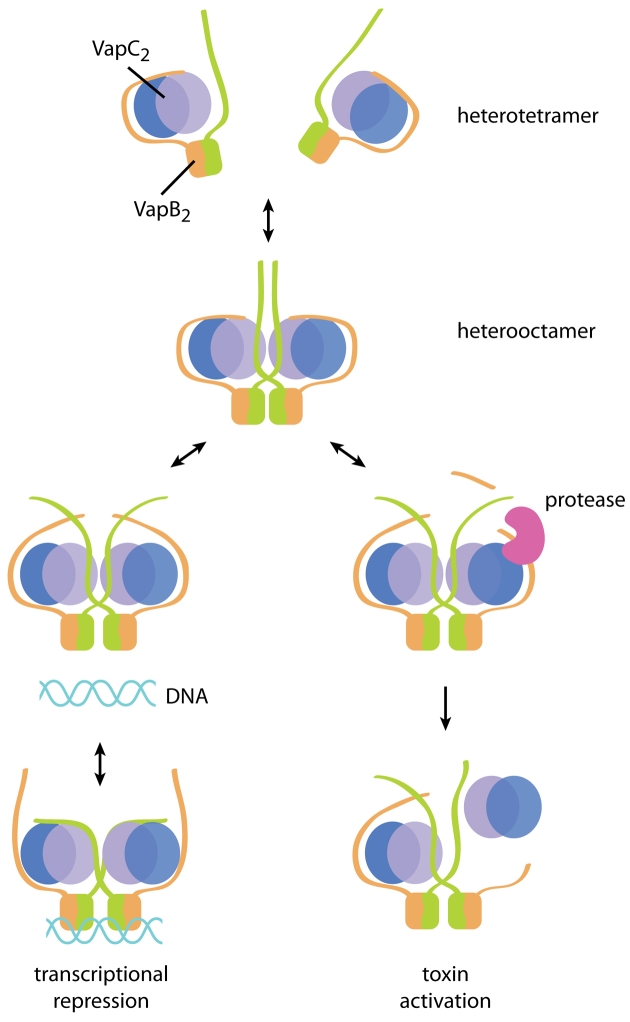
This paper presents for the first time structures of the same TA complex both on and off DNA and the structures surprisingly reveal that the C-terminal extensions of the antitoxin molecules switch place upon DNA binding. The result of this is that a C-terminal extension of one T2A2 heterotetramer stretches across and binds the toxin dimer in the other heterotetramer of the complex in the DNA bound state in trans, thus effectively cross-linking the complex while bound to DNA (Figure 5B, DNA-bound, blue ribbons). Conversely, in the apo state, the C-terminal extensions of VapB1 interact with their cognate VapC1 dimer in a syn interaction (Figure 5B, apo, green ribbons). In this state, the two heterotetramers are essentially free to dissociate, while they are cross-linked and interlocked in the DNA-bound state. The structure of C-terminally truncated VapB1 bound to VapC1 suggests that a destabilization of one of the VapC1-interacting helices of the antitoxin molecules occurs (Figure 5C, left, light-brown molecule). In vivo, the antitoxins are degraded by cellular proteases during toxin activation. It is therefore possible that the truncated structure represents a distinct step on the path to activation where the antitoxin is starting to dissociate. It is also possible that the observed flexibility of full-length VapB plays a role in toxin activation by allowing access of a stress-induced protease to the antitoxin C-terminus. In fact, it was shown for the ccdAB TA system that the protease cleaves throughout the C-terminus of the antitoxin, suggesting that the C-terminal region of VapB could be a target for the protease (41). From a structural perspective it also makes sense that the C-terminus, being extended and partly flexible, would be the prime target during activation.
Taken together, our observations can be summarised in a model for transcriptional repression and toxin activation in the VapBC TA class (Figure 6). Under normal growth conditions (when the toxin is inhibited), the VapB C-terminal extensions inhibit toxins in syn in a 1:2 ratio, which would allow the heterotetramers to dissociate and re-associate. DNA binding, on the other hand, stretches the complex and forces the C-termini to swap places, thus interlocking the TA complex while bound to the operator region and possibly at the same time preventing attack of the protease. During the structural rearrangements, the C-termini would be vulnerable to proteolytic cleavage and such an event would destabilise the TA complex as visualised in the VapB1ΔC:VapC1 complex structure, eventually leading to release of VapC toxin dimers. With these important new concepts in hand, functional studies should now be undertaken in vivo and in vitro produce a complete picture of the molecular mechanisms underlying DNA binding, cross-linking and toxin activation for VapBC-type TA systems. In particular, it would be worthwhile to demonstrate to which extent and by which mechanism conditional cooperativity is involved in regulating VapBC transcription.
Figure 6.
Model for DNA binding and activation of the VapBC-type toxins. Non-crosslinked VapBC tetramers can associate into heterooctamers that are able to bind DNA, which causes the extended C-termini of VapB to switch position and interlock the complex while bound to operator DNA. The flexibility of the C-termini also allows access of a stress-induced protease to the extended protein segments and this cleavage further destabilizes the complex, eventually leading to toxin activation.
ACCESSION NUMBERS
Atomic coordinates and crystallographic structure factors have been deposited in the Protein Data Bank with ID codes 5K8J (apo form), 5L6L (DNA-bound form) and 5L6M (VapB1ΔC:VapC1 form).
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank the beamline staff at MAX-Lab, Lund, Sweden and ESRF, Grenoble, France for help during data collection, Jeppe Achton Nielsen for help with data processing and refinement and Prof. Kenn Gerdes for insightful comments on the manuscript.
Footnotes
Present address: Kehan Xu, Anhui Medical University, Meishan Road 81, Hefei, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Lundbeck Foundation [R173-2014-1182 to D.E.B.]; Danish National Research Foundation's Centre for Bacterial Stress Response and Persistence, BASP [DNRF120 to D.E.B]; Chinese Scholarship Council postgraduate [to K.X.]. Funding for open access charge: Danish National Research Foundation (Grundforskningsfonden) [DNRF120].
Conflict of interest statement. None declared.
REFERENCES
- 1.Gerdes K., Maisonneuve E.. Bacterial persistence and toxin-antitoxin loci. Annu. Rev. Microbiol. 2012; 66:103–123. [DOI] [PubMed] [Google Scholar]
- 2.Maisonneuve E., Gerdes K.. Molecular mechanisms underlying bacterial persisters. Cell. 2014; 157:539–548. [DOI] [PubMed] [Google Scholar]
- 3.Page R., Peti W.. Toxin-antitoxin systems in bacterial growth arrest and persistence. Nat. Chem. Biol. 2016; 12:208–214. [DOI] [PubMed] [Google Scholar]
- 4.Makarova K.S., Wolf Y.I., Koonin E.V.. Comprehensive comparative-genomic analysis of type 2 toxin-antitoxin systems and related mobile stress response systems in prokaryotes. Biol. Direct. 2009; 4:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pandey D.P., Gerdes K.. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 2005; 33:966–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maisonneuve E., Castro-Camargo M., Gerdes K.. (p)ppGpp controls bacterial persistence by stochastic induction of toxin-antitoxin activity. Cell. 2013; 154:1140–1150. [DOI] [PubMed] [Google Scholar]
- 7.Gerdes K., Christensen S.K., Lobner-Olesen A.. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 2005; 3:371–382. [DOI] [PubMed] [Google Scholar]
- 8.Wilbur J.S., Chivers P.T., Mattison K., Potter L., Brennan R.G., So M.. Neisseria gonorrhoeae FitA interacts with FitB to bind DNA through its ribbon-helix-helix motif. Biochemistry. 2005; 44:12515–12524. [DOI] [PubMed] [Google Scholar]
- 9.Overgaard M., Borch J., Jorgensen M.G., Gerdes K.. Messenger RNA interferase RelE controls relBE transcription by conditional cooperativity. Mol. Microbiol. 2008; 69:841–857. [DOI] [PubMed] [Google Scholar]
- 10.Garcia-Pino A., Balasubramanian S., Wyns L., Gazit E., De Greve H., Magnuson R.D., Charlier D., van Nuland N.A., Loris R.. Allostery and intrinsic disorder mediate transcription regulation by conditional cooperativity. Cell. 2010; 142:101–111. [DOI] [PubMed] [Google Scholar]
- 11.Cataudella I., Trusina A., Sneppen K., Gerdes K., Mitarai N.. Conditional cooperativity in toxin-antitoxin regulation prevents random toxin activation and promotes fast translational recovery. Nucleic Acids Res. 2012; 40:6424–6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dienemann C., Boggild A., Winther K.S., Gerdes K., Brodersen D.E.. Crystal structure of the VapBC toxin-antitoxin complex from Shigella flexneri reveals a hetero-octameric DNA-binding assembly. J. Mol. Biol. 2011; 414:713–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mate M.J., Vincentelli R., Foos N., Raoult D., Cambillau C., Ortiz-Lombardia M.. Crystal structure of the DNA-bound VapBC2 antitoxin/toxin pair from Rickettsia Felis. Nucleic Acids Res. 2012; 40:3245–3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mattison K., Wilbur J.S., So M., Brennan R.G.. Structure of FitAB from Neisseria gonorrhoeae bound to DNA reveals a tetramer of toxin-antitoxin heterodimers containing pin domains and ribbon-helix-helix motifs. J. Biol. Chem. 2006; 281:37942–37951. [DOI] [PubMed] [Google Scholar]
- 15.Das U., Pogenberg V., Subhramanyam U.K., Wilmanns M., Gourinath S., Srinivasan A.. Crystal structure of the VapBC-15 complex from Mycobacterium tuberculosis reveals a two-metal ion dependent PIN-domain ribonuclease and a variable mode of toxin-antitoxin assembly. J. Struct. Biol. 2014; 188:249–258. [DOI] [PubMed] [Google Scholar]
- 16.Lee I.G., Lee S.J., Chae S., Lee K.Y., Kim J.H., Lee B.J.. Structural and functional studies of the Mycobacterium tuberculosis VapBC30 toxin-antitoxin system: implications for the design of novel antimicrobial peptides. Nucleic Acids Res. 2015; 43:7624–7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miallau L., Faller M., Chiang J., Arbing M., Guo F., Cascio D., Eisenberg D.. Structure and proposed activity of a member of the VapBC family of toxin-antitoxin systems. VapBC-5 from Mycobacterium tuberculosis. J. Biol. Chem. 2009; 284:276–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Min A.B., Miallau L., Sawaya M.R., Habel J., Cascio D., Eisenberg D.. The crystal structure of the Rv0301-Rv0300 VapBC-3 toxin-antitoxin complex from M. tuberculosis reveals a Mg (2) (+) ion in the active site and a putative RNA-binding site. Protein Sci. 2012; 21:1754–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arcus V.L., Backbro K., Roos A., Daniel E.L., Baker E.N.. Distant structural homology leads to the functional characterization of an archaeal PIN domain as an exonuclease. J. Biol. Chem. 2004; 279:16471–16478. [DOI] [PubMed] [Google Scholar]
- 20.Clissold P.M., Ponting C.P.. PIN domains in nonsense-mediated mRNA decay and RNAi. Curr. Biol. 2000; 10:R888–R890. [DOI] [PubMed] [Google Scholar]
- 21.Cruz J.W., Sharp J.D., Hoffer E.D., Maehigashi T., Vvedenskaya I.O., Konkimalla A., Husson R.N., Nickels B.E., Dunham C.M., Woychik N.A.. Growth-regulating Mycobacterium tuberculosis VapC-mt4 toxin is an isoacceptor-specific tRNase. Nat. Commun. 2015; 6:7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winther K., Tree J.J., Tollervey D., Gerdes K.. VapCs of Mycobacterium tuberculosis cleave RNAs essential for translation. Nucleic Acids Res. 2016; 44:9860–9871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Winther K.S., Brodersen D.E., Brown A.K., Gerdes K.. VapC20 of Mycobacterium tuberculosis cleaves the sarcin-ricin loop of 23S rRNA. Nat. Commun. 2013; 4:2796. [DOI] [PubMed] [Google Scholar]
- 24.Winther K.S., Gerdes K.. Enteric virulence associated protein VapC inhibits translation by cleavage of initiator tRNA. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:7403–7407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu K., Dedic E., Brodersen D.E.. Structural analysis of the active site architecture of the VapC toxin from Shigella flexneri. Proteins. 2016; 84:892–899. [DOI] [PubMed] [Google Scholar]
- 26.Kabsch W. Xds. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Terwilliger T.C., Adams P.D., Read R.J., McCoy A.J., Moriarty N.W., Grosse-Kunstleve R.W., Afonine P.V., Zwart P.H., Hung L.W.. Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr. D Biol. Crystallogr. 2009; 65:582–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. et al. . PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 30.Shah S.A., Vestergaard G., Garrett R.A.. Regulatory RNAs in Prokaryotes. 2011; Wien: Springer-Verlag. [Google Scholar]
- 31.Punta M., Coggill P.C., Eberhardt R.Y., Mistry J., Tate J., Boursnell C., Pang N., Forslund K., Ceric G., Clements J. et al. . The Pfam protein families database. Nucleic Acids Res. 2012; 40:D290–D301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Henikoff S., Henikoff J.G.. Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. U.S.A. 1992; 89:10915–10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krissinel E., Henrick K.. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007; 372:774–797. [DOI] [PubMed] [Google Scholar]
- 34.Coles M., Djuranovic S., Soding J., Frickey T., Koretke K., Truffault V., Martin J., Lupas A.N.. AbrB-like transcription factors assume a swapped hairpin fold that is evolutionarily related to double-psi beta barrels. Structure. 2005; 13:919–928. [DOI] [PubMed] [Google Scholar]
- 35.Malakooti J., Ely B.. Principal sigma subunit of the Caulobacter crescentus RNA polymerase. J. Bacteriol. 1995; 177:6854–6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Malakooti J., Wang S.P., Ely B.. A consensus promoter sequence for Caulobacter crescentus genes involved in biosynthetic and housekeeping functions. J. Bacteriol. 1995; 177:4372–4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou B., Schrader J.M., Kalogeraki V.S., Abeliuk E., Dinh C.B., Pham J.Q., Cui Z.Z., Dill D.L., McAdams H.H., Shapiro L.. The global regulatory architecture of transcription during the Caulobacter cell cycle. PLoS Genet. 2015; 11:e1004831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Jonge N., Garcia-Pino A., Buts L., Haesaerts S., Charlier D., Zangger K., Wyns L., De Greve H., Loris R.. Rejuvenation of CcdB-poisoned gyrase by an intrinsically disordered protein domain. Mol. Cell. 2009; 35:154–163. [DOI] [PubMed] [Google Scholar]
- 39.Zorzini V., Mernik A., Lah J., Sterckx Y.G., De Jonge N., Garcia-Pino A., De Greve H., Versees W., Loris R.. Substrate recognition and activity regulation of the Escherichia coli mRNA endonuclease MazF. J. Biol. Chem. 2016; 291:10950–10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Winther K.S., Gerdes K.. Regulation of enteric vapBC transcription: induction by VapC toxin dimer-breaking. Nucleic Acids Res. 2012; 40:4347–4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Melderen L., Thi M.H., Lecchi P., Gottesman S., Couturier M., Maurizi M.R.. ATP-dependent degradation of CcdA by Lon protease. Effects of secondary structure and heterologous subunit interactions. J. Biol. Chem. 1996; 271:27730–27738. [DOI] [PubMed] [Google Scholar]
- 42.Pei J., Grishin N.V.. PROMALS3D: multiple protein sequence alignment enhanced with evolutionary and three-dimensional structural information. Methods Mol. Biol. 2014; 1079:263–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.