Abstract
Histone demethylase PHF8 is upregulated and plays oncogenic roles in various cancers; however, the mechanisms underlying its dysregulation and functions in carcinogenesis remain obscure. Here, we report the novel functions of PHF8 in EMT (epithelial to mesenchymal transition) and breast cancer development. Genome-wide gene expression analysis revealed that PHF8 overexpression induces an EMT-like process, including the upregulation of SNAI1 and ZEB1. PHF8 demethylates H3K9me1, H3K9me2 and sustains H3K4me3 to prime the transcriptional activation of SNAI1 by TGF-β signaling. We show that PHF8 is upregulated and positively correlated with MYC at protein levels in breast cancer. MYC post-transcriptionally regulates the expression of PHF8 via the repression of microRNAs. Specifically, miR-22 directly targets and inhibits PHF8 expression, and mediates the regulation of PHF8 by MYC and TGF-β signaling. This novel MYC/microRNAs/PHF8 regulatory axis thus places PHF8 as an important downstream effector of MYC. Indeed, PHF8 contributes to MYC-induced cell proliferation and the expression of EMT-related genes. We also report that PHF8 plays important roles in breast cancer cell migration and tumor growth. These oncogenic functions of PHF8 in breast cancer confer its candidacy as a promising therapeutic target for this disease.
INTRODUCTION
Breast cancer is the most common cancer among American women. According to the American Cancer Society, in 2016 about 246 660 new cases of invasive breast cancer will be diagnosed and about 40 450 women will die from this disease (1). Although substantial efforts have been made to understand the mechanisms underlying both the metastasis of breast cancer and the emergence of drug resistance, these issues remain challenges to successful therapy. Thus new strategies are needed, and these will depend on the identification of more effective drug targets. Epigenetic mechanisms have proven to be important in cancer development (2). Therefore chromatin regulators and non-coding RNAs have been increasingly targeted in developing cancer therapies. For example, targeting of the bromodomain and extraterminal domain (BET) protein by the inhibitor JQ1 has been shown to antagonize the proliferation of multiple myeloma cells, and to do so by repressing c-Myc (hereafter termed MYC) and its downstream effectors (3). Similarly, targeting the KDM4 family member, NCDM-32B has been effective in reducing cell proliferation and transformation in breast cancer (4).
Histone methylation, a common form of epigenetic regulation, is controlled by both methyltransferases and demethylases and plays fundamental roles in many cellular processes (2). Recently, several histone demethylases were found to play roles in breast cancer development. For example, the H3K27me3 (trimethylated histone 3 lysine 27) histone demethylase KDM6B/JMJD3 is upregulated in invasive breast carcinoma and promotes TGF-β-induced EMT and tumor cell invasion by regulating the expression of SNAI1 (5). Similarly, the H3K4me3 demethylase family member KDM5A/RBP2 promotes breast cancer metastasis by regulating the pro-metastasis gene TNC (6).
Histone demethylase PHF8 (PHD finger protein 8) acts on monomethylated histone H4 lysine 20 (H4K20me1), monomethylated and dimethylated H3 lysine 9 (H3K9me1/2), and dimethylated H3 lysine 27 (H3K27me2), serving as a transcription coactivator (7–9). Truncations and a point mutation (F279S) affecting the JmjC domain, as well as total deletion of PHF8 are associated with intellectual disability (ID), autism and cleft lip/palate (CLP) (7). PHF8 can bind over one third of human genes (7,9,10); however, physiologically PHF8 regulates only 2–5% of these direct target genes, and such regulation seems to depend on cell type and cellular context (7,10). Notably, the mechanisms underlying such transcriptional selectivity are largely unknown. Bioinformatics analysis has shown that PHF8 binding sites overlap significantly with the consensus sequences of several transcription factors: E2F1, ETS-1 SP1, FOXO1, TCF and MYC-MAX (11). Moreover, PHF8 interacts with E2F1 to regulate the G0/G1-to-S transition (9), and with MYC to regulate cytoskeletal dynamics in HeLa cells (11). Collectively, these findings suggest that PHF8 functions as a transcriptional co-activator and engages in diverse cellular processes.
Emerging evidence has suggested that overexpression of PHF8 was associated with several types of cancers, including prostate cancer (12), esophageal squamous cell carcinoma (13), lung cancer (14) and breast cancer (15). Although high expression of PHF8 contributes to cell proliferation by regulating cell-cycle related genes, it is not clear how PHF8 regulates cell migration. A recent study showed that the deubiquitinase USP7 stabilizing PHF8 is linked to the upregulation of cyclin A2 in breast cancer, how PHF8 sustains its high expression level remains to be delineated (15). Moreover, PHF8 interacts with RARα to sensitize cells to ATRA (all-trans retinoic acid), an agent used in treating acute promyelocytic leukemia (APL) (16). In T-ALL (T cell Acute Lymphoblastic Leukemia), PHF8 is recruited to activated NOTCH1 to regulate its target genes (17). These data indicate that PHF8 exerts distinct functions in different types of cancer. However, the mechanisms underlying its specific functions in each case remain obscure.
In this study, we report a novel function of PHF8 in promoting EMT, the mechanism whereby PHF8 regulates SNAI1 expression in the context of TGF-β, and the oncogenic functions of PHF8 in breast cancer development in vitro and in vivo. We elucidate a novel regulatory axis of MYC/microRNAs/PHF8 involved in the regulation of PHF8 by TGF-β signaling and MYC with PHF8 in turn contributing to the oncogenic functions of MYC. These findings pave the way for further studies of the therapeutic significance by targeting PHF8 in breast cancer.
MATERIALS AND METHODS
Cell culture and treatment
MCF-7, SKBR3, BT474, MDA-MB-231, HaCaT, HEK293T, Phoenix A, and MCF10A cells were obtained from ATCC. MCF-7, MDA-MB-231, HaCaT, HEK293T and Phoenix A cells were grown in Dulbecco's Modified Eagle Medium (DMEM) containing 10% FBS (Gibco). A549, SKBR3 and BT474 cells were cultured in RPMI1640 medium containing 10% FBS. MCF10A cells were cultured in DMEM-F12 supplemented with 20 ng/ml Epidermal Growth Factor (EGF) (Sigma), 100 ng/ml cholera toxin (Sigma), 10 μg/ml insulin (Sigma), 500 ng/ml hydrocortisone (Sigma), and 5% horse serum. All of the cell lines used in this study were maintained in the specified medium supplemented with 1× Penicillin–Streptomycin (Gibco) and incubated in 5% CO2 at 37 °C. TGF-β1 (Calbiochem) was added into the medium directly at 5 ng/ml. For induction of shRNA expression, doxycycline (Sigma) was added to the culture medium at 1 μg/ml for at least 3 days.
Chromatin immunoprecipitation assays
MCF10A cells stably expressing a construct of interest (control or PHF8 shRNA; mock or PHF8 overexpressing) grown in 10-cm plates were cross-linked with 1% formaldehyde and quenched with 0.125M glycine. Cells were washed with cold PBS and then lysed and washed using the truChlP Chromatin Shearing Kit (Covaris). DNA was fragmented using the Covaris S2 Focused-ultrasonicator. Protein–DNA complexes were immunoprecipitated (IP) with the following antibodies: control IgG (A01008, GenScript), anti-PHF8 (ab36068, abcam), anti-H3K4me3 (07-473, EMD Millipore), anti-H3K9me1 (ab8896, Abcam), anti-H3K9me2 (ab1220, Abcam) and anti-H4K20me1 (ab9051, Abcam). The IP material was washed and de-crosslinked at 65°C overnight. The ChIP DNA was purified and qPCR was performed using primers designed to flank the consensus PHF8 binding sites at the promoters of target genes.
Xenograft mouse experiments
6-week-old FVB/N female mice, Athymic nude female mice and NOD SCID mice were obtained from the Jackson Laboratory and Charles River Laboratories, respectively. The protocols for the animal studies reported herein were reviewed and approved by the Institutional Review Board (IRB) and the Institutional Animal Care and Use Committee (IACUC) at The University of Iowa. For the xenograft model, MT2 murine breast cancer cells (1 × 106) overexpressing empty vector or human wide type PHF8 were resuspended in 100 μl Matrigel and PBS (1:1) and orthotopically injected into the fat pad the mammary gland of 6-week-old FVB/N mice. The MDA-MB-231 cells stably expressing luciferase with control or PHF8 (shPHF8-2) shRNAs were established. The clones (2 × 106) with best knockdown efficiency obtained by clonal selection were injected in 12-week-old athymic nude mice and 18-week-old NOD SCID mice, respectively. After the mice were sacrificed, tumor weight was measured and tumor volume was calculated according to the formula: 0.5 × length × width2. Tumors from MT2 cells were fixed, embedded in paraffin, and serially sectioned at a thickness of 4 μm, and hematoxylin and eosin (H&E) staining was performed. Tumor growth in NOD SCID mice implanted with MDA-MB-231 cells was monitored for 3 weeks by bioluminescent imaging.
Statistical analysis
We employed the GraphPad Prism software (v6.02) to conduct statistical analysis. Results are expressed as the mean ± SD. Unless otherwise indicated, differences between experimental groups were compared using an unpaired two-tailed Student's t-test (for two conditions) or one way of ANOVA (for more than three conditions). A P value <0.05 was considered statistically significant.
RESULTS
PHF8 gain-of-function induces an EMT-like process and facilitates EMT induction by TGF-β
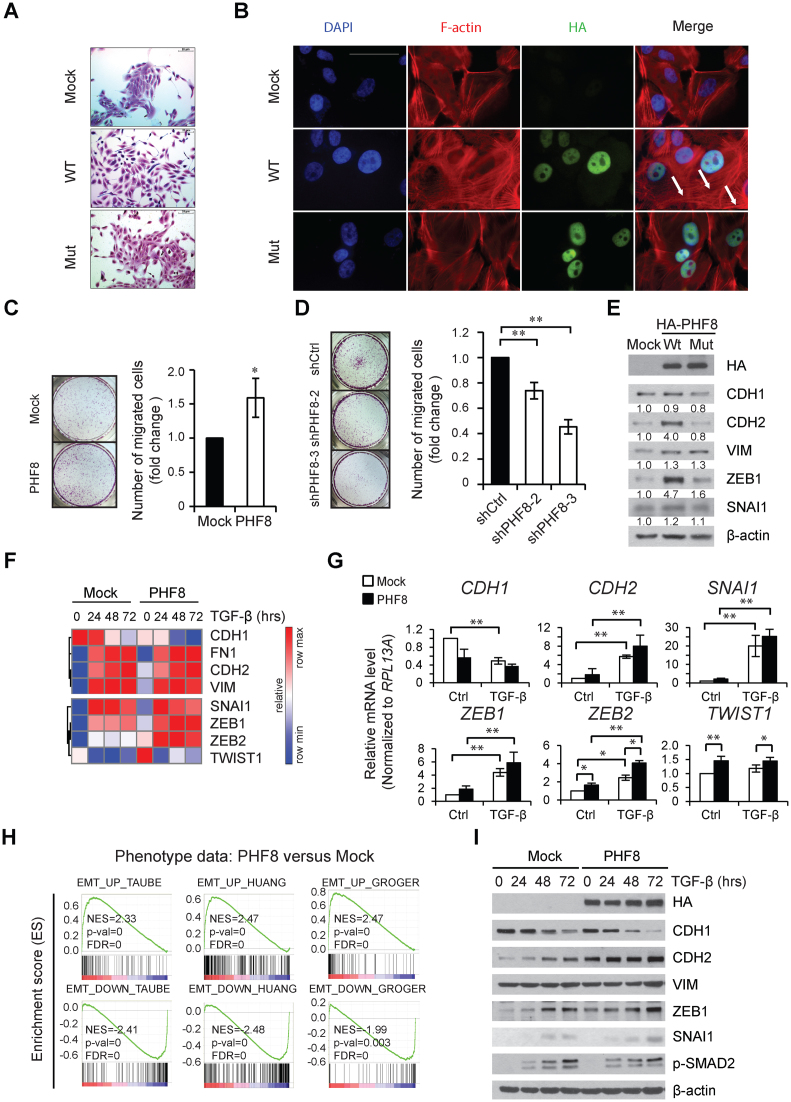
The oncogenic functions of PHF8 in several types of cancer prompted us to investigate the mechanism whereby PHF8 contributes to cancer development. We first examined whether PHF8 possesses malignant transformation ability using 3D acinar formation assay, which was validated for this purpose (18). Overexpression of wild-type PHF8 in MCF10A cells (MCF10A-wtPHF8) significantly increased the acini sizes without inducing invasive phenotype; whereas knockdown of PHF8 by shRNAs decreased the acini sizes (Supplementary Figure S1A and S1B). Interestingly, MCF10A-wtPHF8 cells displayed a spindle-like morphology with a scattered distribution indicative of an EMT phenomenon; whereas control cells expressing empty vector (MCF10A-Mock) or the X-linked mental retardation-associated and demethylase activity-dead PHF8 F279S mutant (mutPHF8) retained a cobblestone-like appearance with tight cell-cell contact (Figure 1A). This morphological change induced by wild type PHF8 indicates that the demethylation activity of PHF8 plays a major role in the induction of the EMT-like process. As transforming growth factor beta (TGF-β) is one of the most potent EMT inducers (19), we treated these stable cell lines with TGF-β1 for 48 h and found that the rearrangement of cytoplasmic F-actin stress fibers was enhanced by overexpressing wild type but not mutant PHF8 (Figure 1B). Since EMT has been recognized as a mechanism promoting cell migration and cancer metastasis, we performed transwell migration assays to address whether PHF8 promotes cell migration. In this context, MCF10A-wtPHF8 cells have an approximately 1.5-fold enhancement of migration compared with MCF10A-Mock cells (Figure 1C). In contrast, knockdown of PHF8 resulted in 60% decrease in cell migration (Figure 1D). Examination of EMT markers by immunoblotting in the context of wild type PHF8 overexpression in MCF10A cells revealed upregulation of the mesenchymal markers N-Cadherin (CDH2) and VIMENTIN (VIM), as well as key transcription factors SNAI1 and ZEB1 in the EMT (Figure 1E). To confirm whether this EMT-promoting function is conserved in cancer cells, we used the human lung adenocarcinoma A549 cells, as they undergo TGF-β induced EMT (20). We observed the similar upregulation of CDH2, VIM and SNAI1, and the downregulation of the epithelial marker E-Cadherin (CDH1) in A549 cells overexpressing wild type PHF8 (Supplementary Figure S2A). Knockdown of PHF8 in A549 cells by shRNAs antagonized the induction of spindle-like morphology changes by TGF-β1 for 24, 48 and 72 h (Supplementary Figures S2B and S2C). Overexpression of PHF8 downregulated CDH1 in A549 cells, but not in MCF10A cells, implicating the fine-tuning roles of PHF8 can be cell type specific. We also found that the overexpression of mutPHF8 elevated VIM in both cell lines, indicative of the demethylase-independent function of PHF8 in regulating the expression of specific genes. However, the demethylase dependent function is still dominant as only the wild type PHF8 causes morphological changes in both MCF10A and A549 cells.
Figure 1.
PHF8 overexpression promotes an EMT-like process. (A) Morphological change of MCF10A cells overexpressing vector (Mock), wild type PHF8 (WT) and the catalytically lethal PHF8 F279S mutant (Mut). The cells were stained with 0.1% of crystal violet. (B) The same MCF10A cells as in (A) were treated with TGF-β1 for 48 hours followed by immunofluorescence staining of F-actin. White scale bar in the upper left micrograph represents 50 μm. The white arrows indicate F-actin staining. (C and D) Transwell assays in MCF10A cells with PHF8-overexpression (C) and PHF8-knockdown (D). The number of migrated cells was quantified and shown at the right panels. (E) Western blotting for proteins encoded by select EMT genes in MCF10A cells overexpressing empty vector (Mock), wtPHF8 (WT) and mutPHF8 (Mut). The relative signal intensities for proteins of interest are normalized to β-actin and shown below each lane. (F) A heat map of expression of select EMT-related genes, extracted from the normalized RNA-seq data from MCF10A-Mock and MCF10A-wtPHF8 cells treated with TGF-β1. (G) qPCR analysis of expression of the selected EMT-related genes in MCF10-Mock and MCF10-wtPHF8 cells with TGF-β1 treatment for 0 and 72 h. (H) GSEA of the genes from MCF10A-Mock and MCF10A-wtPHF8 cells at steady state, using the EMT gene signatures of Taube et al. (22), Huang et al. (24), Groger et al. (23) as references, respectively. (I) Western blotting of selected proteins in MCF10A-Mock and MCF10A-wtPHF8 cells at different time points after TGF-β1 treatment. SD was obtained from three independent experiments.*P < 0.05, **P < 0.01; (C) unpaired two-tailed Student's t-test; (D and G) one-way ANOVA.
To gain a global view on whether overexpression of PHF8 regulates the TGF-β induced EMT, we treated MCF10A-Mock and MCF10A-wtPHF8 cells with TGF-β1 for 0, 24, 48 and 72 h and performed RNA-seq in biological duplicates. The raw reads of each RNA-seq data are shown in Supplementary Table S1. In MCF10A-Mock cells, we observed an upregulation of CDH2, FN1, VIM, SNAI1, ZEB1, ZEB2 and a downregulation of CDH1, confirming that the TGF-β1 treatment was effective (Figure 1F). At basal level (in the absence of TGF-β1), overexpression of wtPHF8 enhanced the expression of CDH2, VIM, TWIST1, ZEB1 and ZEB2. The real-time qPCR results further supported that ectopic expression of PHF8 did not change the expression of RPLP0 (as control, Supplementary Figure S2D), but significantly enhanced ZEB2 expression and reduced CDH1 expression both at steady state and upon TGF-β1 treatment, and that it slightly enhanced TGF-β1 induced upregulation of CDH2 and TWIST1 (Figure 1G). A potential reason for the discrepancy in regulation of the CDH1 protein and mRNA is that PHF8 may indirectly regulate other factors that affect its processing and/or post-translational regulation (21).
The EMT gene signatures have been demonstrated in different cell lines and cancer samples (22–24). We analyzed our gene expression data using the EMT gene signature from the human mammary epithelial (HMLE) cell line (22) as reference and found that more than 75% of TGF-β1 regulated genes in our MCF10A-Mock cells exhibit a pattern similar to those in HMLE cells (Supplementary Figure S2E). These results not only validated that TGF-β induces EMT in our approach, but also supported the notion that TGF-β induced gene regulation is dependent on cell type. We next performed gene set enrichment analysis (GSEA) (25,26) for those genes whose expression differed between the MCF10-Mock and MCF10A-PHF8 cells. This analysis indicated that the three EMT signature genes (22–24) were significantly enriched in MCF10A-PHF8 cells, strongly indicating that PHF8 overexpression induces a sustained EMT signaling program (Figure 1H). To determine whether PHF8 has positive effects on TGF-β regulated genes, we also carried out GSEA at different times following TGF-β treatment. PHF8 overexpression significantly enhanced the regulations of EMT genes at all time points. The lower enrichment scores at 24, 48 and 72 h of TGF-β treatment than at 0 h were due to the basal induction of an EMT-like process by PHF8 overexpression (Supplementary Figure S2F). We next examined the protein levels of selected EMT-related genes and found that PHF8 overexpression enhanced the downregulation of CDH1 and general elevation of CDH2, ZEB1 and SNAI1 (Figure 1I), supporting the gene set enrichment analysis (GSEA) of RNA-seq data. Taken together, these data indicate that overexpression of PHF8 induces an EMT-like process, and it fine-tunes the regulation of select EMT genes.
PHF8 facilitates the transactivation of SNAI1 by TGF-β through regulations of H3K4me3 and H3K9me2/1
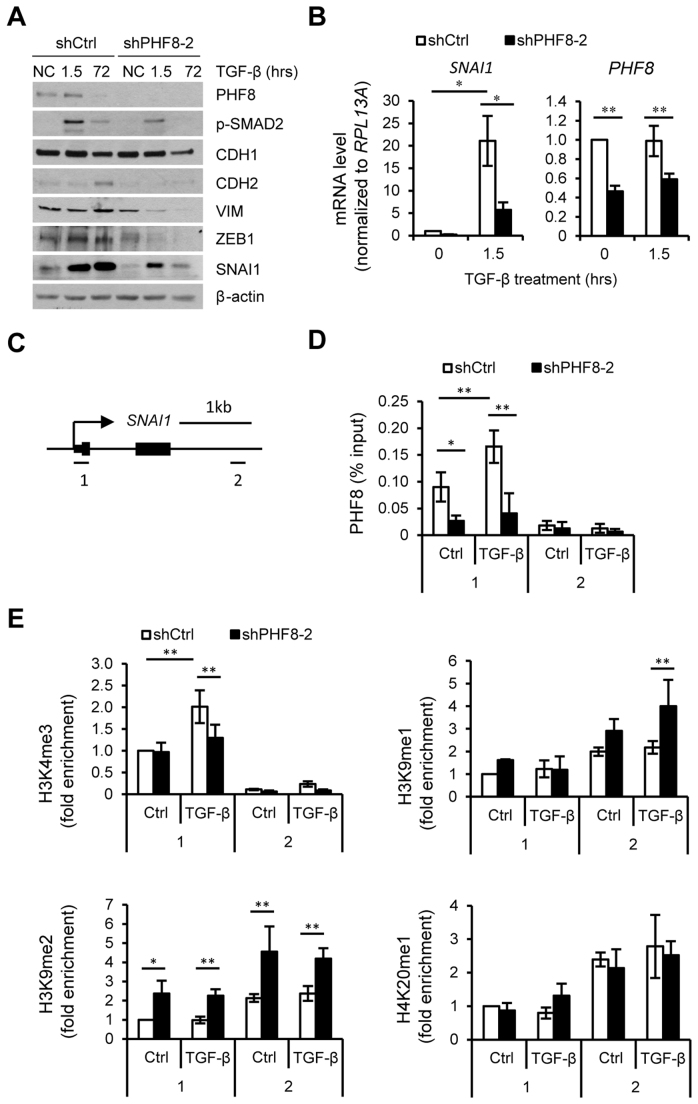
To further investigate the mechanisms by which PHF8 regulates TGF-β signaling and TGF-β induced EMT, we treated MCF10A cells expressing control or PHF8 shRNAs with TGF-β1 for 1.5 and 72 h to evaluate early and late TGF-β1 responsive genes. TGF-β1 induced a biphasic regulation of PHF8, e.g. a slight upregulation at 1.5 h and downregulation at 72 h (Figure 2A). The early upregulation of PHF8 suggests that PHF8 may have a function in regulating TGF-β1 responsive genes. Indeed, knockdown of PHF8 significantly decreased the phosphorylation of SMAD2, and antagonized the elevations of CDH2, VIM, SNAI1and ZEB1 proteins (Figure 2A) and SNAI1 mRNA induced by TGF-β (Figure 2B). In contrast, knockdown of PHF8 and TGF-β1 treatment did not significantly change the expression levels of GAPDH and CHGA (Supplementary Figure S2G). Knockdown of PHF8 did not significantly affect the TGF-β induced downregulation of CDH1 protein, consistent with our western blotting results indicating that the overexpression had no obvious effect on the CDH1 protein. Similar gene regulation affected by PHF8 knockdown was observed in A549 cells with TGF-β1 treatment at different time points (Supplementary Figure S2H). To further verify if PHF8 demethylase activity is important for the EMT induction, we generated double stable MCF10A cells expressing control or PHF8 shRNAs combined with empty vector (Mock), PHF8 shRNA resistant wtPHF8 or mutPHF8. wtPHF8 strongly induced a spindle-like morphological change (Supplementary Figure S2I) and EMT-related gene expression (Supplementary Figure S2J). Notably, the mutPHF8 slightly upregulated the expression of ZEB1 and CDH2, supporting the demethylase independent function of PHF8.
Figure 2.
PHF8 is required for the regulation of TGF-β responsive genes and regulates H3K4me3 and H3K9me2/1 on SNAI1. (A) Western blotting of TGF-β regulated genes in MCF10A cells stably expressing a doxycycline-inducible shRNAs (control or PHF8). (B) qPCR analysis of SNAI1 and PHF8 expression. (C) The SNAI1 amplicons for ChIP-qPCR were illustrated. (D) ChIP assays with PHF8 antibody were performed in the same cells as in (A). qPCR assay on TSS (amplicon 1) and gene body (amplicon 2) for SNAI1 was shown. (E) Similar ChIP-qPCR assays as (D) with indicated antibodies against different histone modifications were performed for SNAI1 gene. The cells were incubated without (Ctrl) or with TGF-β1 for 1.5 h. Numbers on the x-axes represent the two amplicons. Error bars, ± SD obtained from at least three independent experiments. *P < 0.05, **P < 0.01; one-way ANOVA.
We next performed PHF8 ChIP (chromatin immunoprecipitation) in the control cells. The ChIP-qPCR showed that PHF8 binds to the TSSs (transcription starting site) of SNAI1 (amplicon 1) at steady state (Figure 2C and D), and this occupancy was increased by 70% by TGF-β1 treatment for 1.5 h. The facts that knockdown of PHF8 abolished the binding and low occupancy of PHF8 at the gene body regions (Amplicon 2) support the specificity of PHF8 binding to the TSS of SNAI1. Knockdown of PHF8 also abolished its occupancy at the TSS of GAPDH and ZEB1, but not for CHGA which is not targeted by PHF8 (as control, Supplementary Figure S3).
We next performed ChIP assay of the active chromatin marker H3K4me3 and the demethylation substrates of PHF8: H3K9me1, H3K9me2 and H4K20me1 (Figure 2E). Without TGF-β treatment, PHF8 knockdown upregulated the levels of H3K9me1 and H3K9me2 at the TSS and gene body region of SNAI1, but it did not change the levels of H3K4me3 and H3K20me1 at the TSS of SNAI1. Upon TGF- β treatment, only H3K4me3 was significantly increased at the TSS of SNAI1, supporting the recruitment of PHF8 as it binds to this methylated histone via its PhD domain (7). We also found that PHF8 knockdown decreased the H3K4me3 level but increased the levels of H3K9me2 and H4K20me1 at the TSS of SNAI1. Notably, the H3K9me2 level was also significantly increased at the gene body of SNAI1 in PHF8 depleted cells at steady state and upon TGF-β treatment, which is consistent with the notion that H3K9me2 is associated with transcriptional repression. However, it is very likely that the increase of H3K9me2 at the gene body region is a secondary event as PHF8 does not bind there. Taken together, our results show that TGF-β1 treatment recruits PHF8 to the TSS of SNAI1, and PHF8 is critical to maintain the level of H3K4me3 at the TSS of SNAI1 and to suppress the levels of H3K9me1 and H3K9me2 at the TSS and gene body regions of SNAI1, and thus facilitating its trans-activation by TGF-β signaling.
TGF-β signaling regulates PHF8 partially through MYC
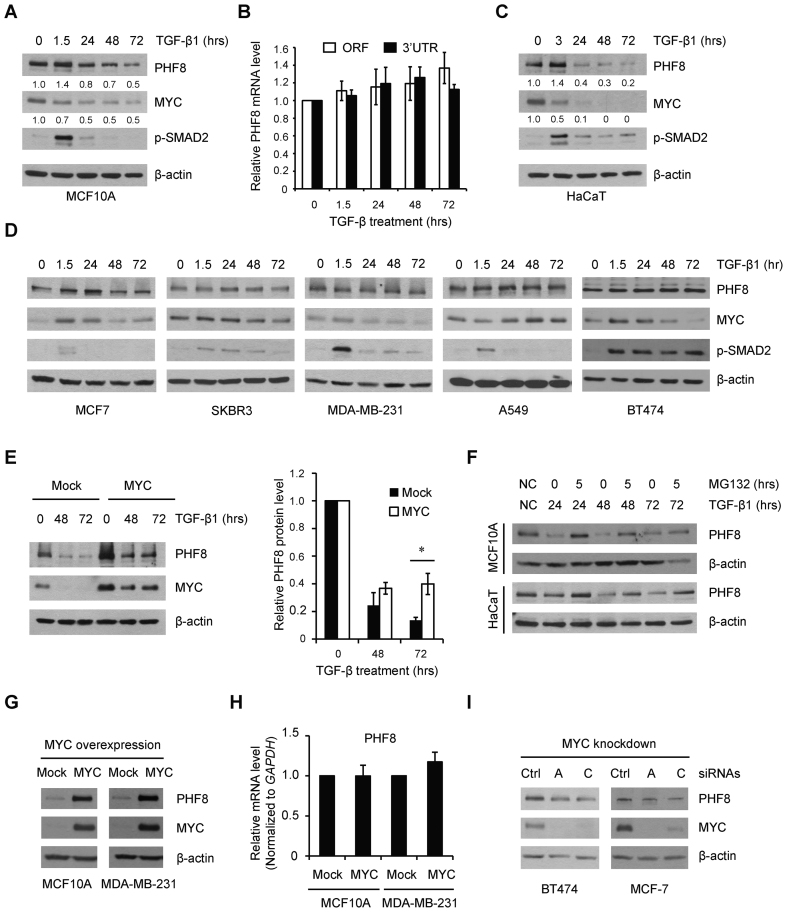
TGF-β exerts growth inhibitory function in normal and early cancer cells, however, such function is often lost along cancer development (27). We next sought to determine whether PHF8 is regulated by TGF-β signaling. Again, TGF-β1 induced biphasic regulation of PHF8 protein in MCF10A cells, starting with a 1.4-fold upregulation at 1.5 h and ending with a gradual downregulation from 24 h (Figure 3A); whereas PHF8 mRNA remained relatively stable (Figure 3B). Such regulation was also observed in another non-malignant, TGF-β responsive HaCaT cells (Figure 3C). Notably, TGF-β1 failed to significantly downregulate PHF8 in several cancer cell lines (Figure 3D). A hallmark mechanism in the growth inhibitory function of TGF-β signaling is the repression of MYC in non-transformed cells, but not in cancer cells (28). We found that the downregulation of MYC by TGF-β1 is similar to that of PHF8 in MCF10A and HaCaT cells at later time points. In cancer cells, MYC and PHF8 share co-regulation pattern in MCF7 and SKBR3 cells. Similar regulation pattern was observed, except that MYC was downregulated at 72 h in MDA-MB-231, and at 1.5 h in A549 cells. In BT474 cells, TGF-β treatment did not change PHF8, but upregulated MYC at 24 h and gradually downregulated it at 72 h (Figure 3D). Notably, PHF8 expression is low in MCF10A, but high in breast cancer cell lines including MCF7, SKBR3, BT474, MDA-MB-468, MDA-MB-231, SUM149 and SUM159 cells (Supplementary Figure S4). This phenomenon prompted us to investigate whether there is an essential link between MYC and PHF8. Indeed, constitutive overexpression of MYC in MCF10A cells partially restored the expression of PHF8 (increased from 50% to 70%) upon TGF-β1 treatment (Figure 3E), suggesting that the repression of MYC contributes to TGF-β1 induced downregulation of PHF8. Moreover, addition of the proteasome inhibitor MG132 partially restored PHF8 protein levels in the context of TGF-β1 treatment in both MCF10A and HaCaT cells (Figure 3F), demonstrating that PHF8 may also be regulated through protein degradation. Furthermore, constitutive overexpression of MYC resulted in increased PHF8 expression in MCF10A and MDA-MB-231 cells (Figure 3G); whereas levels of the PHF8 mRNA remained unchanged (Figure 3H). In BT474 and MCF-7 cells, which express higher levels of PHF8, knockdown of MYC by small interfering RNAs (siRNAs) moderately downregulated PHF8 (Figure 3I). These data demonstrate that MYC is a positive regulator of PHF8.
Figure 3.
MYC regulates PHF8 and mediates the regulation of PHF8 by TGF-β. (A) Western blotting analysis of PHF8 and MYC protein levels in MCF10A cells treated with TGF-β for different lengths of time. The relative signal intensity for PHF8 and MYC, normalized to β-actin, is shown beneath each lane. (B) RT-qPCR analysis of PHF8 mRNA levels over time following treatment with TGF-β1. RT-qPCR primers complementary to PHF8 ORF (open reading frame) and 3΄UTR (untranslated region) sequences were used. (C) Western blotting analysis of PHF8 and MYC protein levels in HaCaT cells treated with TGF-β. (D) Protein levels of PHF8, MYC and phosphorylated SMAD2 in the indicated cell lines treated with TGF-β1 for the indicated time, as assessed by western blotting. (E) PHF8 and MYC protein levels in MCF10A-Mock and MCF10A-MYC cells over time following treatment with TGF-β1, as determined by western blotting (left panel). PHF8 protein levels were quantified by densitometry from three independent experiments (right panel). *P < 0.05; unpaired two-tailed Student's t-test. (F) PHF8 protein levels in MCF10A and HaCaT cells treated with TGF-β for different time points with or without MG132, as assessed by western blotting. (G) Western blotting analysis of PHF8 protein levels in MCF10A and MDA-MB-231 cells that stably overexpress MYC. (H) qPCR analysis of PHF8 expression in MCF10A and MDA-MB-231 cells overexpressing empty vector or MYC. (I) Western blotting analysis of PHF8 and MYC expression in the context of siRNA-mediated MYC knockdown in BT474 and MCF-7 cells. Error bars, ± SD obtained from at least three independent experiments.
We next asked whether the downregulation of PHF8 contributes to cell cycle regulation by TGF-β, as the downregulation of MYC is critical in this context. In MCF10A-Mock cells, TGF-β treatment dramatically increased G0/G1 phase and shortened S phase at 48 and 72 h, and PHF8 overexpression significantly blocked these effects with extended G2/M phase (Supplementary Figure S5). These findings support the hypothesis that the deregulation of PHF8, as a potential MYC downstream effector, may contribute to the functional switch of TGF-β signaling from growth inhibition to cancer promotion.
MYC upregulates PHF8 by repressing microRNAs
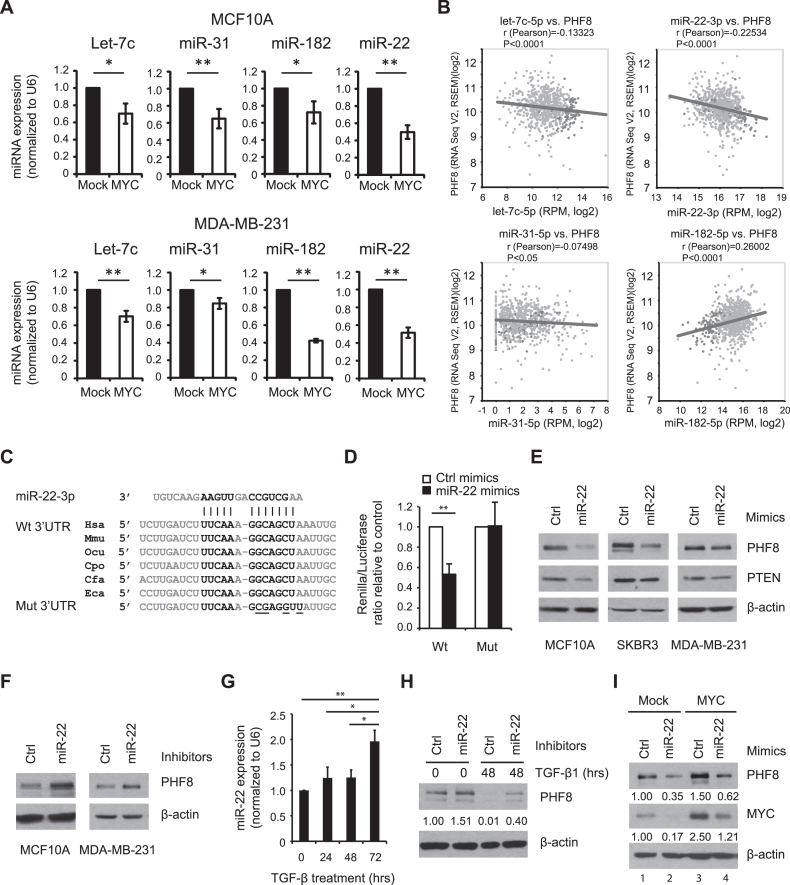
The fact that MYC overexpression did not change PHF8 mRNA levels suggests that MYC regulates PHF8 via post-transcriptional and/or post-translational regulatory mechanisms. MYC has been reported to repress microRNAs (miRNAs), which mediate MYC functions in tumorigenesis (29). We sought to identify MYC-regulated miRNAs that target PHF8. MiRNA target prediction using TargetScan (30), PITA (31) and CLIP-seq data using starBase (32) showed that several miRNAs, such as miR-22, miR-31, miR-182 and the let-7 family members, have highly conserved binding sites in the PHF8 3΄UTR (Supplementary Figure S6). Notably, miR-22 is one of the MYC-repressed miRNAs (29), and our qPCR results showed that it was repressed more than the other miRNAs when MYC was constitutively overexpressed in MCF10A and MDA-MB-231 cells (Figure 4A). Among these miRNAs, our bioinformatics analysis of the data from The Cancer Genome Atlas (TCGA) showed that miR-22 is more inversely correlated with PHF8 expression in breast cancer samples than let-7c or miR-31 (Figure 4B). Interestingly, miR-182 showed a positive correlation with PHF8. It is possible that miR-182 is more subject to activation by other factors than that to repression by MYC. Taken together, miR-22 appears to be a promising candidate miRNA to repress PHF8 in the context of regulation by MYC. We then focused on miR-22 and carried out a dual luciferase reporter assay for miR-22. This approach revealed that miR-22 mimics significantly decreased luciferase activies of wild type PHF8 3΄UTR by about 50%, but not that of the mutant 3΄UTR (Figure 4C and D). Thus, miR-22 appears to directly downregulate PHF8 by targeting its 3΄UTR. To verify that miR-22 regulates PHF8 in vivo, we transiently transfected MCF10A, SKBR3 and MDA-MB-231 cells with miR-22 mimics or inhibitors, using the known miR-22 target PTEN (33) as a positive control. The mimics substantially decreased PHF8 expression in all three cell lines (Figure 4E). Moreover, the inhibitor increased PHF8 levels in MCF10A and MDA-MB-231 cells (Figure 4F). These results supported the hypothesis that PHF8 is a miR-22 target.
Figure 4.
miR-22 regulates PHF8 and mediates the regulation of PHF8 by TGF-β and MYC. (A) Constitutive overexpression of MYC downregulates miRNAs. RT-qPCR analysis of the expression levels of let-7 family (let-7c), miR-31, miR-182 and miR-22 in MCF10A and MDA-MB-231 cells overexpressing empty vector (Mock) or MYC. (B) Correlation expression of miRNAs and PHF8 in breast cancer samples of TCGA. (C) Comparison of miR-22 binding sites in PHF8 3΄UTR sequence of human (Hsa), mouse (Mmu), rabbit (Ocu), guinea pig (Cpo), dog (Cfa) and horse (Eca). The nucleotides mutated in the PHF8 3΄UTR are underlined. (D) miR-22 targets the PHF8 3΄UTR. Dual luciferase reporter assay on HEK293T cells cotransfected with Luc-PHF8 3΄-UTR-WT or Luc-PHF8 3΄-UTR-MUT in combination with the miR-22 or control mimics. (E and F) Western blotting analysis of PHF8 expression of the cells transfected with miR-22 mimics (E) or inhibitors (F) as indicated. (G) RT-qPCR analysis of miR-22 expression in MCF10A cells with TGF-β1 treatment. (H) Western blotting analysis of PHF8 expression in MCF10A cells transiently transfected with control or miR-22 inhibitors and treated with TGF-β1 for 48 h. (I) Western blotting analysis of PHF8 expression in MCF10A-Mock and MCF10A-MYC cells transfected with control and miR-22 mimics. The relative signal intensity for PHF8, normalized to β-actin, is shown beneath each lane (H and I). Error bars, ± SD obtained from at least three independent experiments. *P < 0.05, **P < 0.01; (A and D) unpaired two-tailed Student's t-test; (G) one-way ANOVA.
We next examined the regulation of miR-22 by TGF-β1. In MCF10A cells, miR-22 was upregulated by TGF-β1 (Figure 4G), and a gradual increase in miR-22 levels was inversely correlated with downregulation of PHF8. When MCF10A cells were transiently transfected with miR-22 inhibitors, PHF8 protein was partially restored under 48-h exposure to TGF-β1 (Figure 4H). To determine whether miR-22 mediates the regulation of PHF8 by MYC, we transfected miR-22 mimics in MCF10A cells overexpressing MYC. As expected, miR-22 mimics attenuated MYC-mediated upregulation of PHF8 (Figure 4I). Interestingly, miR-22 mimics also downregulated endogenous MYC in MCF10A cells. As the 3΄UTR of MYC doesn't have the potential binding sites for miR-22 (34), the downregulation of MYC seems as an indirect effect of miR-22 mimics. Nevertheless, the total level of MYC (lane 4) was slightly higher than the endogenous MYC (lane 1). PHF8 was upregulated by 50% by MYC overexpression (lane 3) and was reduced by 38% in the presence of miR-22 mimics (lane 4) than that in lane 1. However, the PHF8 level (lane 4) was still nearly 2-fold higher than that treated with miR-22 mimics along (lane 2), indicating that miR-22 mimics partially rescued the upregulation of PHF8 by the overexpressed MYC. Collectively, these results support that PHF8 is a direct target of miR-22 and that MYC regulates PHF8 by repressing miR-22, thereby partially contributing to TGF-β induced regulation of PHF8.
PHF8 contributes to MYC-induced cell proliferation and EMT
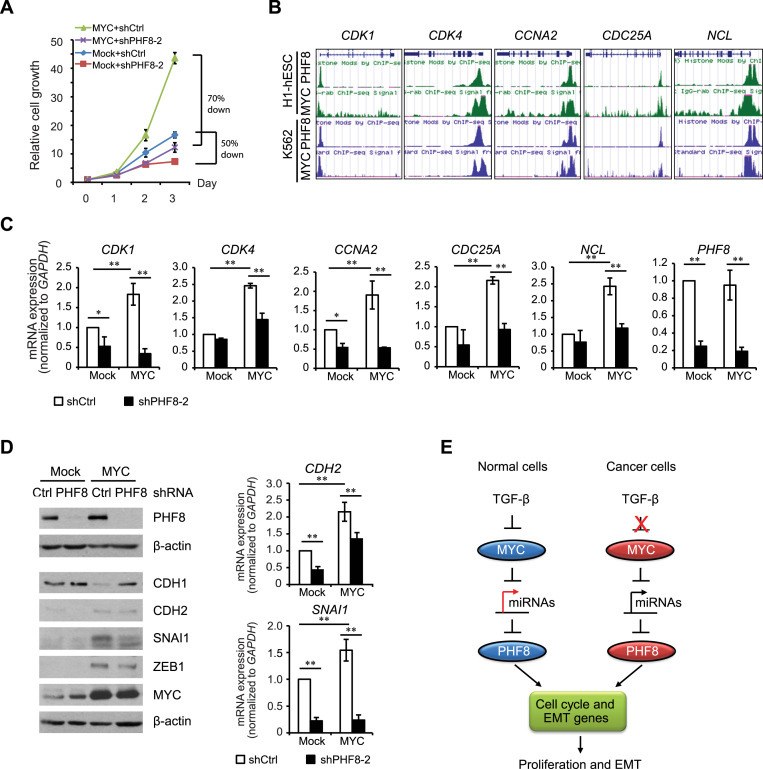
Having demonstrated that MYC regulates PHF8 post-transcriptionally by repressing miR-22, we next investigated whether PHF8 contributes to the cellular functions of MYC. As both PHF8 and MYC play important roles in cell-cycle regulation (9,35), we first asked whether PHF8 facilitates MYC-driven cell proliferation. In MCF10A cells stably expressing both MYC and an inducible PHF8 shRNA, PHF8 knockdown reduced MYC-driven cell proliferation by 70%. In the absence of MYC overexpression, PHF8 knockdown reduced cell proliferation by 50% (Figure 5A). These results indicated that PHF8 contributes for MYC-driven cell proliferation.
Figure 5.
PHF8 plays important roles in MYC-induced cell proliferation and EMT. (A) MTT assay of the relative cell proliferation in MCF10A-Mock and MCF10A-MYC cell lines with doxycycline-induced PHF8 knockdown. (B) Analysis of ChIP-seq data of PHF8 and MYC in H1 and K562 cells using the UCSC Genome Browser. (C) qPCR analysis of the expression of select genes. In MCF10A-ER (Mock) and MCF10A-MYC-ER cell lines with doxycycline-induced control or PHF8 shRNAs, PHF8 knockdown and MYC overexpression were induced by doxycycline and tamoxifen for 96 and 12 h, respectively. (D) Western blotting and qPCR analysis of expression of select EMT genes in the cell lines described as in (A). (E) Schematic illustration of the regulations and functions of PHF8 in the context of TGF-β and MYC signaling. Error bars, ± SD obtained from at least three independent experiments. *P < 0.05, **P < 0.01; one-way ANOVA.
To dissect the underlying mechanisms, we focused on select cell cycle genes CDK1, CDK4, CCNA2, CDC25A and NCL. Analysis of PHF8 and MYC ChIP-seq data from H1 and K562 cells using ENCODE data (36) showed the co-occupancy of PHF8 and MYC at the TSSs of these genes (Figure 5B). Such co-occupancy on cytoskeleton genes was also observed in HeLa cells (11). We next established the double-stable MCF10A cell lines overexpressing tamoxifen-inducible MYC and doxycycline-inducible shRNA against PHF8 to evaluate the effect of PHF8 on these genes. The qPCR results showed that MYC activation by 12-h treatment with 4-hydroxytamoxifen (4-OHT) significantly upregulated the expression of CDK1, CDK4, CCNA2, CDC25A and NCL (Figure 5C). However, such upregulation was abolished in the cells with PHF8 knockdown. Moreover, PHF8 knockdown selectively downregulated the basal expression of CDK1, CDK4 and CCNA2 in MCF10A cells (Figure 5C). These results indicate that PHF8 has a fundamental function in cell proliferation and such function also applies to pro-proliferative role of MYC.
As both PHF8 (our data) and MYC promote the EMT (37,38), we next asked whether PHF8 is required for the MYC-induced EMT. Constitutive overexpression of MYC in MCF10A cells led to a decrease in basal level of the CDH1 protein and increases in those of CDH2, SNAI1 and ZEB1 (Figure 5D). Importantly, PHF8 knockdown antagonized MYC-induced upregulation of the ZEB1, SNAI1 and CDH2 proteins in MCF10A-MYC cells (Figure 5D). At transcriptional level, we confirmed that levels of both SNAI1 and CDH2 mRNAs were significantly reduced by the loss of PHF8 with and without MYC overexpression. Taken together, our results suggest that PHF8 and MYC synergistically contribute to cell proliferation and the EMT. The discovery of the MYC/microRNAs/PHF8 regulatory axis and its connection to TGF-β signaling in cancer development are illustrated in Figure 5E.
PHF8 promotes cancer cell proliferation, migration and tumor growth
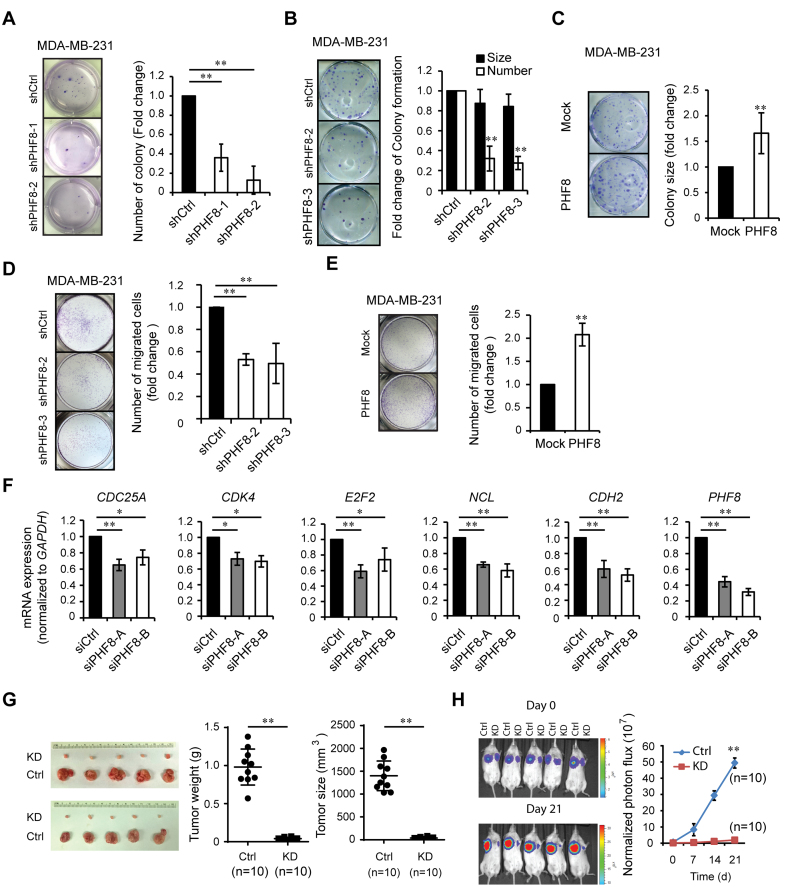
The roles of PHF8 in TGF-β induced EMT and in the oncogenic functions of MYC prompted us to investigate the functional impact of PHF8 on breast carcinogenesis. Doxycycline inducible scramble (control) and different PHF8 shRNAs were stably expressed in MDA-MB-231 and BT474 cells (Supplementary Figure S7A). The soft-agar assay showed that knockdown of PHF8 significantly reduced the formation of colonies (Figure 6A and Supplementary Figure S7B), suggesting that PHF8 is critical for the anchorage independent growth of MDA-MB-231 and BT474 cells. A colony formation assay in MDA-MB-231 cells showed that knockdown of PHF8 led to a slight decrease in average colony size by 15% but a drastic decrease in colony number by 70% (Figure 6B); whereas overexpression of PHF8 (Supplementary Figure S7C) showed a significant increase in the average size of colonies by 60% but not in the number of colonies of MDA-MB-231 cells (Figure 6C). It should be noted that the phenotype induced by PHF8 overexpression was not caused by modulating the MYC levels, because overexpression of PHF8 didn't change the MYC levels in MDA-MB-231, MCF10A and HaCaT cells (Supplementary Figure S7C). To confirm that PHF8 can promote the cancer cell migration, we performed Transwell migration assays using MDA-MB-231 cells. PHF8 inhibition resulted in 50% decrease in cell migration (Figure 6D); whereas overexpression of wild type PHF8 led to an ∼2-fold increase of migration compared with the cells expressing empty vector (Mock) (Figure 6E). Quantitative PCR (qPCR) assay showed that knockdown of PHF8 by shRNAs or two siRNAs downregulated the expression of cell cycle genes such as CDK4, E2F2 and NCL, and migration-related gene CDH2 in MDA-MB-231 cells (Supplementary Figure S7D and Figure 6F). The regulatory role of PHF8 on these gene expressions supports its cellular functions.
Figure 6.
PHF8 positively regulates proliferation, migration and tumor growth of breast cancer cells in vitro and in vivo. (A) Soft agar colony assays in MDA-MB-231 cells with doxycycline inducible PHF8 knockdown. Quantifications are shown at the right panel. (B and C) Colony formation assays in MDA-MB-231 cells with PHF8-knockdown (B) or with PHF8-overexpression (C). The number and size of colonies were quantified and shown at the right panel. (D and E) Transwell migration assays in MDA-MB-231 cells with PHF8-knockdown (D) or with PHF8-overexpression (E). Quantifications are shown at the right panels (A to E). (F) Knockdown of PHF8 by two siRNAs downregulated the expression of cell proliferation and migration related genes in MDA-MB-231, as detected by qPCR. Error bars denote SD obtained from three independent experiments (A to F). (G) Representative image of tumors from MDA-MB-231 cells stably expressing control and PHF8 shRNA implanted in NOD SCID mice. Tumors collected at week 6 after injection were weighed and assessed for volume (right panels). (H) Representative images of tumor growth in NOD SCID mice measured by bioluminescent imaging. The graph (right panel) showed the normalized photon flux at the indicated times. *P < 0.05, **P < 0.01; (A, D and F) one-way ANOVA; (B, C, E, G and H) unpaired two-tailed Student's t-test.
To further establish whether PHF8 facilitates tumorigenesis in vivo, we generated the MDA-MB-231 cells stably expressing luciferase with control or PHF8 (shPHF8-2) shRNAs and injected these cells in NOD SCID mice and athymic nude mice, respectively. Knockdown of PHF8 significantly reduced the tumor size and weight after injection at week 7 and week 6 in NOD SCID mice (Figure 6G) and nude mice (Supplementary Figure S7E), respectively. Meanwhile, the quantitative imaging of tumor growth in NOD SCID mice showed that knockdown of PHF8 effectively suppressed the tumor growth (Figure 6H).
We also used the MMTV-Erbb2 breast cancer derived cell line MT2 (39) to further address whether overexpression of PHF8 promotes tumor growth in vivo. The specificity of antibody to mouse PHF8 was confirmed with shRNA mediated knockdown and IHC staining (Supplementary Figure S7F and S7G). Notably, MT2 cells express relatively lower level of PHF8 protein than that in SKBR3 cells (Supplementary Figure S7F), so they are appropriate for PHF8 gain-of-function studies in mice. We then established MT2 stable cell lines expressing empty vector (Mock) or HA-tagged PHF8 (Supplementary Figure S7H) and implanted the two cell lines into the fat pad of mammary gland of FVB/N mice. The growth of the tumors overexpressing PHF8 was significantly increased at day 60 compared with that of control (Mock); this was reflected by an ∼4-fold increase in mean tumor weight and an ∼6-fold increase in mean tumor size (Supplementary Figure S7I). Therefore, PHF8 overexpression enhanced tumor growth in vivo in this MT2 model system.
Collectively, these in vitro and in vivo data from different mice models indicated the oncogenic functions of PHF8 in breast cancer by regulating proliferation- and migration-associated genes.
PHF8 is upregulated in breast cancer and associated with metastasis
To determine whether our findings are clinically relevant, we first analyzed RNA-seq data from The Cancer Genome Atlas (TCGA) and found that breast cancer is among the malignancies in which PHF8 expression is generally upregulated (Supplementary Figure S8A). Further analysis of 998 breast cancer samples in TCGA using starBase (32) revealed that levels of PHF8 expression are moderately but significantly higher (log2 fold change 1.19) in breast cancers than normal tissues (Supplementary Figure S8B). Mata-analysis in the ONCOMINE (40) revealed that PHF8 is more frequently upregulated in breast cancers than other cancer types, and the upregulation of PHF8 in breast cancers predominates over that of its subfamily members (JHDM1D and PHF2) (Supplementary Figure S8C). Among breast cancer, the significant upregulation of PHF8 was observed in invasive ductal and lobular breast carcinoma, invasive breast carcinoma stroma and rare types of breast cancer such as cribriform breast adenocarcinoma, male breast carcinoma and mucinous breast carcinoma (Supplementary Figure S8D). Further analysis showed that PHF8 expression is moderately elevated in breast cancers positive for human epidermal growth factor receptor 2 (ERBB2, also called HER2), estrogen receptor (ER), or progesterone receptor (PR) (Supplementary Figure S8E).
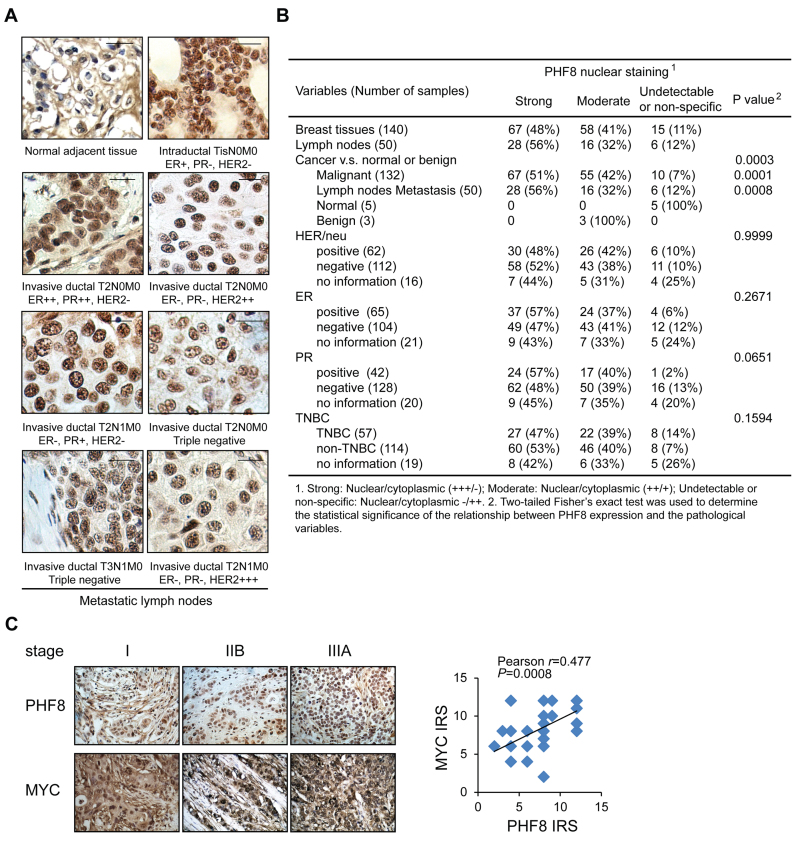
Since PHF8 expression is regulated at post-transcriptional and post-translational levels, we investigated whether the level of PHF8 protein is associated with any subtypes of breast cancer and the other pathological factors. We carried out immunohistochemical (IHC) staining of four breast cancer tissue arrays (Figure 7A), which comprised 190 samples representing in situ breast carcinomas, metastatic lymph nodes and cancer-adjacent tissues. As shown in Figure 7B, 95 samples showed strong and exclusively nuclear staining, 74 showed moderate nuclear staining accompanied by certain cytoplasmic staining, and 21 showed no staining. In all strong nuclear staining cases, PHF8 protein expression is significantly upregulated in the breast carcinoma samples (P < 0.001). High expression of PHF8 was detected in 48%, 57%, 57% and 47% of HER2-, ER-, PR-positive and triple negative breast cancers (TNBC), respectively; however, it was not significantly associated with all these subtypes of breast cancer. In contrast, staining was weak and non-specific in all three benign and five cancer-adjacent breast tissues. Notably, the elevated expression of PHF8 correlates positively with lymph-node metastasis (P < 0.001). Furthermore, we examined MYC protein expression in 46 cases of breast cancer samples and found positive correlation between MYC and PHF8 (Figure 7C), supporting our in vitro finding that MYC drives PHF8 expression.
Figure 7.
PHF8 is upregulated in breast cancers. (A) IHC analysis of PHF8 expression in human breast cancer samples. Status of markers associated with locations and stages are listed for each tumor. Scale bar: 10 μm. (B) Summary of PHF8 immunohistochemical staining of 190 breast cancer, metastatic lymph nodes and adjacent normal tissues. (C) IHC analysis of the expression levels of PHF8 and MYC in 46 breast cancer samples. Representative images (original magnification, ×400) from breast carcinoma samples at different stages are shown. The immunoreactive score (IRS) was determined by staining intensity and distribution. The correlation coefficient (r) and P values were analyzed as indicated (right panel).
We also examined PHF8 expression in spontaneous breast cancer and metastatic lung cancer specimens from MMTV-Erbb2 mouse breast cancer model. PHF8 was detectable at low levels in normal breast tissues; however, it was greatly elevated in small breast tumors of both early and late stages (Supplementary Figure S9). PHF8 expression was elevated in large tumors of early stage, and it maintained high levels in the peripheral region of large tumors of late stages. In metastasized tumors in lung, PHF8 was also detected at higher levels in small tumors and in the peripheral regions of large tumors. These findings showed that the elevated levels of PHF8 is associated with proliferating tumors as the tumor cells inside of large tumor mass may suffer from insufficient nutrition and oxygen supplies. Moreover, these data also suggested that PHF8 may exert its functions in early tumor development both in situ and de novo metastasized tumors in lung.
Collectively, these data on PHF8 expression show an association between elevations at both mRNA and protein levels and breast-cancer progression, supporting that PHF8 may play a role in breast carcinogenesis.
DISCUSSION
PHF8 has been implicated to play oncogenic roles in various types of malignancies. Although deregulated expression of PHF8 is usually linked to cell proliferation and migration, the detailed mechanism by which PHF8 contributes to migration and tumorigenesis is not well understood. In this study, we showed that PHF8 promotes an EMT-like process in non-malignant and cancer cell lines, and the role of PHF8 plays in promoting the expression of EMT genes such as CDH2 and key EMT transcription factors including SNAI1 and ZEB1 may contribute to cell migration and thus is associated with its oncogenic activity during cancer metastasis. PHF8 has demethylation activities on H3K9me2, H3K9me1, H3K27me2 and H4K20me1, depending on cell or gene context. Such activities have been demonstrated in transcription regulations: PHF8 regulates the gene transcription by demethylating H3K9me1 and H4K20me1 in HeLa cells (7,9); depletion of PHF8 increases the levels of H3K9me1 and H3K9me2 at rDNA (41) and PHF8 removes H3K9me2 in neurons (42). In MCF10 cells, we found that PHF8 knockdown did not significantly regulate H4K20me1, but consistently upregulated H3K9me1 and H3K9me2 around the TSS and at the 3΄ gene body region of SNAI1. We and others showed that PHF8 predominantly locates around the TSSs (7,9). Thus, the increases of H3K9me1 and H3K9me2 around the TSS of SNAI1 in PHF8 knockdown cells might result from PHF8 demethylase activity. However, how PHF8 knockdown upregulates H3K9me1 and H3K9me2 at the 3΄ gene body region of SNAI1 remains to be investigated. One of possible explanations is that PHF8 deficiency recruits other histone methyltransferases, for example EHMT2 (also known as G9a), to the promoters, facilitating the association with the other factors such as the Cockayne syndrome group B protein (CSB) and results in the increase of the H3K9me1/2 levels at the promoter and the coding region (43).
A recent study reported that PHF8 target genes are upregulated in more than 60% of breast cancers (44). Our results demonstrated that PHF8 expression is upregulated along breast cancer progression and other types of cancer. One of the major goals in this study is to understand how PHF8 expression is dysregulated in cancer. Recently, Wang et al. (15) showed that USP7 promotes deubiquitination and stabilization of PHF8 in breast cancer MCF7 cells, accounting for a mechanism underlying PHF8 deregulation. MYC plays a key regulatory role in cell proliferation and mediates key signaling events in many types of cancer (35). We demonstrated that MYC upregulates PHF8 protein level at least partially by repressing miR-22, and PHF8 contributes to MYC-driven cell proliferation and EMT. Thus, we identify a novel MYC/microRNAs/PHF8 regulatory axis, which represents an alternative explanation for the dysregulation of PHF8 in breast cancer and potentially in the other types of cancer. Moreover, PHF8 is subjected to proteasome-mediated protein degradation (15) (Figure 3F). Whether USP7 regulates PHF8 in the context of TGF-β signaling remains to be studied. Given that PHF8 protein level is regulated at post-transcriptional and post-translational levels, PHF8 IHC in large cohort of breast cancer samples with clinical follow-up data are still required to reveal the correlation of PHF8 expression with subtypes and prognosis of breast cancers.
It should be noted that Wang et al. (15) identified the USP7/PHF8/CCNA2 regulatory axis in proliferation and carcinogenesis of breast cancer, their xenograft experiments using MCF7 cells demonstrated the proliferative role of PHF8. In this study, we found that PHF8 regulates CCNA2 and the other cell cycle genes co-regulated by MYC. We performed the xenograft experiments to validate the oncogenic functions of PHF8 using human TNBC MDA-MB-231 cells and mouse breast cancer MT2 cells in different mouse models. Thus, our studies well complemented the work by Wang et al. Furthermore, our IHC results not only confirmed that PHF8 is upregulated in breast cancer, which is consistent with the conclusion from Wang et al. (15), but also provided evidence to support the positive correlation between PHF8 and MYC in breast cancer samples.
The MYC/microRNAs/PHF8 regulatory axis is supported by the similar expression patterns of MYC and PHF8 following TGF-β treatment in multiple cancer cell lines. In fact, PHF8 synergizing with MYC to regulate cell adhesion and cytoskeleton organization genes was reported in HeLa cells (11). Although whether such mechanisms play a role in breast cancer cells remains to be studied, identification of other factors including miRNAs involved in the regulation of PHF8 will improve our understanding of the MYC/miRNAs/PHF8 regulatory axis in TGF-β signaling and the other pathological pathways.
MYC and TGF-β have antagonistic actions in the control of cytostatic program, and the ability of TGF-β to repress MYC is often lost in cancers (45). In non-malignant cells, the regulation of PHF8 by MYC through miRNAs suggests the downregulation of PHF8 in late TGF-β treatment facilitates the cell growth inhibition by TGF-β. Indeed, our data demonstrated that PHF8 overexpression attenuated the TGF-β induced cell cycle arrest, supporting our hypothesis that the deregulation of PHF8 in cancer cells contributes to the switch from cytostatic function to cancer promoting of TGF-β signaling. However, the early upregulation of PHF8 by TGF-β accounts for the functional requirement of the induction of SNAI1 expression. Such functional involvement of MYC in early TGF-β regulated genes was also reported (37). Therefore, our studies provide an explanation for the TGF-β induced biphasic regulation of the PHF8 protein in non-malignant cells.
The oncogenic functions of PHF8 in different types of cancer suggests this enzyme could potentially be used as therapeutic agents for cancers in which PHF8 is overexpressed. Indeed, Compound 9 was developed and has a stronger inhibitory effect on PHF8 than KDM2A and JHDM1D, and it inhibits the proliferation of both HeLa and KYSE150 cells (46). Moreover, this inhibitor downregulates expression of the cell cycle-promoting transcription factor E2F1 and lengthens G0/G1 phase in both cell lines, consistent with a role for PHF8 in cell-cycle regulation (46). Although the effects of compound 9 on other cellular processes or other types of cancer and its cytotoxicity were not investigated, such initiative is already a major step towards developing more efficient and selective inhibitors of epigenetic factors that are relevant to cancer. Our discovery of the oncogenic functions of PHF8 in breast cancer paves the way to develop novel therapeutics for breast cancer patients.
ACCESSION NUMBERS
The RNA-seq data in this study have been deposited in NCBI's Gene Expression Omnibus (GEO) (www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO Series accession number GSE74377.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs Songhai Chen, Yuanchao Ye and Qing Xie for technical assistance and helpful discussions on the mouse work. We'd like to thank Dr Brad Amendt for his advice; the Flow Cytometry Facility at Carver College of Medicine, The University of Iowa for their technical assistance and Dr Sonia Sugg, Dr Amani Bashir for their assistance in analyzing the IHC data. We also thank Dr Christine Blaumueller for editorial consultation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Department of Anatomy and Cell Biology, the Carver College of Medicine, the University of Iowa [Lab startup to H.H.Q.]; Carver Trust Young Investigator Award from the Roy J. Carver Charitable Trust [01-224 to H.H.Q]; The National Institutes of Health and CTSA grant UL1RR024979 through ICTS (Institute for Clinical and Translational Science) at the University of Iowa [Pilot grant to H.H.Q]; ACS-IRG seed grant [IRG-77-004-34 to H.H.Q.] from the American Cancer Society, administrated through the Holden Comprehensive Cancer Center at the University of Iowa; Breast Cancer Research Award [to H.H.Q.] by the Holden Comprehensive Cancer Center at the University of Iowa; The National Institutes of Health grant [P30 CA86862] to the Genomics and Flow Cytometry core facilities at the University of Iowa; Libraries and Provost's Open Access Fund at the University of Iowa for the open access charge.
Conflict of interest statement. None declared.
REFERENCES
- 1.Siegel R.L., Miller K.D., Jemal A.. Cancer statistics, 2016. CA. 2016; 66:7–30. [DOI] [PubMed] [Google Scholar]
- 2.Greer E.L., Shi Y.. Histone methylation: a dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 2012; 13:343–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Delmore J.E., Issa G.C., Lemieux M.E., Rahl P.B., Shi J., Jacobs H.M., Kastritis E., Gilpatrick T., Paranal R.M., Qi J. et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011; 146:904–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ye Q., Holowatyj A., Wu J., Liu H., Zhang L., Suzuki T., Yang Z.Q.. Genetic alterations of KDM4 subfamily and therapeutic effect of novel demethylase inhibitor in breast cancer. Am. J. Cancer Res. 2015; 5:1519–1530. [PMC free article] [PubMed] [Google Scholar]
- 5.Ramadoss S., Chen X., Wang C.Y.. Histone demethylase KDM6B promotes epithelial-mesenchymal transition. J. Biol. Chem. 2012; 287:44508–44517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao J., Liu Z., Cheung W.K., Zhao M., Chen S.Y., Chan S.W., Booth C.J., Nguyen D.X., Yan Q.. Histone demethylase RBP2 is critical for breast cancer progression and metastasis. Cell Rep. 2014; 6:868–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qi H.H., Sarkissian M., Hu G.Q., Wang Z., Bhattacharjee A., Gordon D.B., Gonzales M., Lan F., Ongusaha P.P., Huarte M. et al. Histone H4K20/H3K9 demethylase PHF8 regulates zebrafish brain and craniofacial development. Nature. 2010; 466:503–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kleine-Kohlbrecher D., Christensen J., Vandamme J., Abarrategui I., Bak M., Tommerup N., Shi X., Gozani O., Rappsilber J., Salcini A.E. et al. A functional link between the histone demethylase PHF8 and the transcription factor ZNF711 in X-linked mental retardation. Mol. Cell. 2010; 38:165–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu W., Tanasa B., Tyurina O.V., Zhou T.Y., Gassmann R., Liu W.T., Ohgi K.A., Benner C., Garcia-Bassets I., Aggarwal A.K. et al. PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression. Nature. 2010; 466:508–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fortschegger K., de Graaf P., Outchkourov N.S., van Schaik F.M., Timmers H.T., Shiekhattar R.. PHF8 targets histone methylation and RNA polymerase II to activate transcription. Mol. Cell. Biol. 2010; 30:3286–3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Asensio-Juan E., Gallego C., Martinez-Balbas M.A.. The histone demethylase PHF8 is essential for cytoskeleton dynamics. Nucleic Acids Res. 2012; 40:9429–9440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bjorkman M., Ostling P., Harma V., Virtanen J., Mpindi J.P., Rantala J., Mirtti T., Vesterinen T., Lundin M., Sankila A. et al. Systematic knockdown of epigenetic enzymes identifies a novel histone demethylase PHF8 overexpressed in prostate cancer with an impact on cell proliferation, migration and invasion. Oncogene. 2012; 31:3444–3456. [DOI] [PubMed] [Google Scholar]
- 13.Sun X., Qiu J.J., Zhu S., Cao B., Sun L., Li S., Li P., Zhang S., Dong S.. Oncogenic features of PHF8 histone demethylase in esophageal squamous cell carcinoma. PLoS One. 2013; 8:e77353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen Y., Pan X., Zhao H.. The histone demethylase PHF8 is an oncogenic protein in human non-small cell lung cancer. Biochem. Biophys. Res. Commun. 2014; 451:119–125. [DOI] [PubMed] [Google Scholar]
- 15.Wang Q., Ma S., Song N., Li X., Liu L., Yang S., Ding X., Shan L., Zhou X., Su D. et al. Stabilization of histone demethylase PHF8 by USP7 promotes breast carcinogenesis. J. Clin. Invest. 2016; 126:2205–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arteaga M.F., Mikesch J.H., Qiu J., Christensen J., Helin K., Kogan S.C., Dong S., So C.W.. The histone demethylase PHF8 governs retinoic acid response in acute promyelocytic leukemia. Cancer Cell. 2013; 23:376–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yatim A., Benne C., Sobhian B., Laurent-Chabalier S., Deas O., Judde J.G., Lelievre J.D., Levy Y., Benkirane M.. NOTCH1 nuclear interactome reveals key regulators of its transcriptional activity and oncogenic function. Mol. Cell. 2012; 48:445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Debnath J., Muthuswamy S.K., Brugge J.S.. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003; 30:256–268. [DOI] [PubMed] [Google Scholar]
- 19.Lamouille S., Xu J., Derynck R.. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014; 15:178–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kasai H., Allen J.T., Mason R.M., Kamimura T., Zhang Z.. TGF-beta1 induces human alveolar epithelial to mesenchymal cell transition (EMT). Respir. Res. 2005; 6:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Roy F., Berx G.. The cell-cell adhesion molecule E-cadherin. Cell. Mol. Life Sci. 2008; 65:3756–3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taube J.H., Herschkowitz J.I., Komurov K., Zhou A.Y., Gupta S., Yang J., Hartwell K., Onder T.T., Gupta P.B., Evans K.W. et al. Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:15449–15454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Groger C.J., Grubinger M., Waldhor T., Vierlinger K., Mikulits W.. Meta-analysis of gene expression signatures defining the epithelial to mesenchymal transition during cancer progression. PLoS One. 2012; 7:e51136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang S., Holzel M., Knijnenburg T., Schlicker A., Roepman P., McDermott U., Garnett M., Grernrum W., Sun C., Prahallad A. et al. MED12 controls the response to multiple cancer drugs through regulation of TGF-beta receptor signaling. Cell. 2012; 151:937–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mootha V.K., Lindgren C.M., Eriksson K.F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstrale M., Laurila E. et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003; 34:267–273. [DOI] [PubMed] [Google Scholar]
- 27.Siegel P.M., Massague J.. Cytostatic and apoptotic actions of TGF-beta in homeostasis and cancer. Nat. Rev. Cancer. 2003; 3:807–821. [DOI] [PubMed] [Google Scholar]
- 28.Chen C.R., Kang Y., Massague J.. Defective repression of c-myc in breast cancer cells: A loss at the core of the transforming growth factor beta growth arrest program. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:992–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang T.C., Yu D., Lee Y.S., Wentzel E.A., Arking D.E., West K.M., Dang C.V., Thomas-Tikhonenko A., Mendell J.T.. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat. Genet. 2008; 40:43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Agarwal V., Bell G.W., Nam J.W., Bartel D.P.. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015; 4:e05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kertesz M., Iovino N., Unnerstall U., Gaul U., Segal E.. The role of site accessibility in microRNA target recognition. Nat. Genet. 2007; 39:1278–1284. [DOI] [PubMed] [Google Scholar]
- 32.Li J.H., Liu S., Zhou H., Qu L.H., Yang J.H.. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014; 42:D92–D97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poliseno L., Salmena L., Riccardi L., Fornari A., Song M.S., Hobbs R.M., Sportoletti P., Varmeh S., Egia A., Fedele G. et al. Identification of the miR-106b∼25 microRNA cluster as a proto-oncogenic PTEN-targeting intron that cooperates with its host gene MCM7 in transformation. Sci. Signal. 2010; 3:ra29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiong J., Du Q., Liang Z.. Tumor-suppressive microRNA-22 inhibits the transcription of E-box-containing c-Myc target genes by silencing c-Myc binding protein. Oncogene. 2010; 29:4980–4988. [DOI] [PubMed] [Google Scholar]
- 35.Dang C.V. MYC on the path to cancer. Cell. 2012; 149:22–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Consortium E.P. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith A.P., Verrecchia A., Faga G., Doni M., Perna D., Martinato F., Guccione E., Amati B.. A positive role for Myc in TGFbeta-induced Snail transcription and epithelial-to-mesenchymal transition. Oncogene. 2009; 28:422–430. [DOI] [PubMed] [Google Scholar]
- 38.Cho K.B., Cho M.K., Lee W.Y., Kang K.W.. Overexpression of c-myc induces epithelial mesenchymal transition in mammary epithelial cells. Cancer Lett. 2010; 293:230–239. [DOI] [PubMed] [Google Scholar]
- 39.Tan W., Zhang W., Strasner A., Grivennikov S., Cheng J.Q., Hoffman R.M., Karin M.. Tumour-infiltrating regulatory T cells stimulate mammary cancer metastasis through RANKL-RANK signalling. Nature. 2011; 470:548–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rhodes D.R., Yu J., Shanker K., Deshpande N., Varambally R., Ghosh D., Barrette T., Pandey A., Chinnaiyan A.M.. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004; 6:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feng W., Yonezawa M., Ye J., Jenuwein T., Grummt I.. PHF8 activates transcription of rRNA genes through H3K4me3 binding and H3K9me1/2 demethylation. Nat. Struct. Mol. Biol. 2010; 17:445–450. [DOI] [PubMed] [Google Scholar]
- 42.Oey N.E., Leung H.W., Ezhilarasan R., Zhou L., Beuerman R.W., VanDongen H.M., VanDongen A.M.. A neuronal activity-dependent dual function chromatin-modifying complex regulates arc expression. eNeuro. 2015; 2, doi:10.1523/ENEURO.0020-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yuan X., Feng W., Imhof A., Grummt I., Zhou Y.. Activation of RNA polymerase I transcription by cockayne syndrome group B protein and histone methyltransferase G9a. Mol. Cell. 2007; 27:585–595. [DOI] [PubMed] [Google Scholar]
- 44.Jiang P., Freedman M.L., Liu J.S., Liu X.S.. Inference of transcriptional regulation in cancers. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:7731–7736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Massague J. TGFbeta signalling in context. Nat. Rev. Mol. Cell Biol. 2012; 13:616–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suzuki T., Ozasa H., Itoh Y., Zhan P., Sawada H., Mino K., Walport L., Ohkubo R., Kawamura A., Yonezawa M. et al. Identification of the KDM2/7 histone lysine demethylase subfamily inhibitor and its antiproliferative activity. J. Med. Chem. 2013; 56:7222–7231. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.