Abstract
JMJD6, a jumonji C (Jmj C) domain-containing protein demethylase and hydroxylase, has been implicated in an array of biological processes. It has been shown that JMJD6 interacts with and hydroxylates multiple serine/arginine-rich (SR) proteins and SR related proteins, including U2AF65, all of which are known to function in alternative splicing regulation. However, whether JMJD6 is widely involved in alternative splicing and the molecular mechanism underlying JMJD6-regulated alternative splicing have remained incompletely understood. Here, by using RASL-Seq, we investigated the functional impact of RNA-dependent interaction between JMJD6 and U2AF65, revealing that JMJD6 and U2AF65 co-regulated a large number of alternative splicing events. We further demonstrated the JMJD6 function in alternative splicing in jmjd6 knockout mice. Mechanistically, we showed that the enzymatic activity of JMJD6 was required for a subset of JMJD6-regulated splicing, and JMJD6-mediated lysine hydroxylation of U2AF65 could account for, at least partially, their co-regulated alternative splicing events, suggesting both JMJD6 enzymatic activity-dependent and independent control of alternative splicing. These findings reveal an intimate link between JMJD6 and U2AF65 in alternative splicing regulation, which has important implications in development and disease processes.
INTRODUCTION
Precursor messenger RNAs (Pre-mRNAs) are subjected to multiple steps of processing to produce mature messenger RNAs (mRNAs), which include but are not limited to capping at the 5΄ end, cleavage and polyadenylation at the 3΄ end, and intron removal by splicing (1). RNA splicing is the process by which introns are removed from the pre-mRNA and the remaining exons connected to form mature mRNA. A typical eukaryotic intron has GU dinucleotide at its 5΄ end and AG at its 3΄ end. In front of the AG sequence, there is a branch-point sequence (BPS) followed by a series of pyrimidines, known as the polypyrimidine tract (Py tract) (2). The splicing reaction is catalyzed by the spliceosome, a large RNA-protein complex composed of five small nuclear ribonucleoproteins (snRNPs) (U1, U2, U4, U5 and U6) (3,4). U1 snRNP recognizes the GU dinucleotide at the 5΄ splice sites (5΄ SSs) while the U2 snRNP binds to the 3΄ splice sites (3΄ SSs) by base-pairing with the branch-point sequence (BPS) in the intron and the remaining three snRNPs (U4/6 and U5) contribute to the formation of the catalytic core of spliceosome (3,4). Due to the fact that BPS is highly degenerative in higher eukaryotic cells, the addition of U2 snRNP requires a number of accessory proteins, including U2 small nuclear RNA auxiliary factor 1 (U2AF1/U2AF35), U2AF2 (U2AF65) and splicing factor 1 (SF1) (2,5). After U1 snRNP binding at the 5΄ SS and SF1 to the BPS, the U2AF65/35 heterodimer interacts with the Py tract to assist the recruitment of U2 snRNP (6–9).
Many pre-mRNAs are alternatively spliced to produce different mRNA isoforms that either encode different proteins or confer differential RNA stability. This process allows the human genome to produce many more proteins than would be expected from its limited number of protein-coding genes (5). In humans, ∼95% of multi-exonic genes are alternatively spliced (10). There are many modes of alternative splicing, of which five basic ones are generally recognized, including exon skipping or cassette exon, mutually exclusive exons, alternative donors, alternative acceptors, and differential intron retention (2,5,11). The most common mode is cassette exon, in which a particular exon may be included or skipped in a context specific manner (11). Alternative splicing is regulated by numerous trans-acting proteins (repressors and activators) via specific cis-acting elements (silencers and enhancers) they recognize in pre-mRNAs. Molecular mechanisms underlying alternative splicing regulation are very diversified and complicated, and new regulatory paradigms are continuously emerging with the use of high-throughput sequencing techniques (12). Abnormal splicing variants are thought to contribute to development of cancer as well as other diseases in humans (13–16).
U2AF65 has been implicated as a key regulator of alternative splicing (17–21). Recently, it was estimated that U2AF65 has the capacity to directly bind ∼88% of functional 3΄ splice sites in the human genome based on U2AF65 CLIP-Seq (cross-linking immunoprecipitation coupled with high-throughput sequencing), and in addition, numerous U2AF65 binding events also occur in exonic and intronic locations, thus providing additional mechanisms for the regulation of alternative splicing (21). Indeed, both RNA-Seq (RNA high-throughput sequencing) and RASL-Seq (RNA-mediated oligonucleotide annealing, selection and ligation coupled with high-throughput sequencing) experiments indicate that U2AF65 is extensively involved in the regulation of alternative splicing (21). However, how U2AF65 activity and thus its controlled alternative splicing events might be regulated still remains incompletely understood.
Jumonji C (Jmj C) domain-containing protein 6 (JMJD6), also known as PTDSR or PSR, was originally identified as a phosphatidylserine receptor on the surface of phagocytes (22–24). Subsequent sequence analysis of JMJD6 reveals that it has multiple nuclear localization signals as well as a JmjC domain, suggesting that it may possess novel nuclear functions (25–29). Indeed, JMJD6 has been shown to be an iron (Fe2+)- and 2-oxoglutarate (2-OG)-dependent dioxygenase that demethylates histone H3 at arginine 2 (H3R2) and histone H4 at arginine 3 (H4R3) in both biochemical and cell-based assays (30,31). JMJD6 arginine demethylase activity has also been demonstrated on a couple of non-histone proteins, including estrogen receptor (ERα) (32), RNA helicase A (RHA) (33), heat shock protein 70 (HSP70) (34), Paired Box 3 (PAX3) (35) and TNF receptor associated factor 6 (TRAF6) (36). JMJD6΄s lysyl-5-hydroxylation function has been demonstrated on both histone and non-histone proteins, such as U2AF65 and tumor protein 53 (TP53) (37–40). We recently reported a new transcriptional paradigm in which JMJD6 and its functional partner, bromo-domain-containing protein 4 (BRD4), regulate promoter-proximal Pol II pausing release (31). Besides its function in transcriptional control, we and others have shown that JMJD6 also interacts with multiple arginine–serine-rich (RS) domains of SR proteins and SR-like splicing factors, including U2AF65, suggesting JMJD6 may be involved in splicing regulation (31,39,41,42). Indeed, a recent study demonstrated that JMJD6 regulates splicing of vascular endothelial growth factor receptor 1 (VEGFR1/Flt1) in mouse endothelial cells, which is required for angiogenic sprouting (43). The effect of iron on splicing of a ferrochelatase has been linked to JMJD6 activity (44). Despite these rapid progresses, however, it has remained unclear how extensively JMJD6 is involved in regulated splicing and whether such regulatory function is directly linked to its interaction with and/or its activity on U2AF65. In addition, JMJD6 has been implicated in a multitude of biological processes, including embryonic development (24,45,46), cell cycle control (40), cellular proliferation and motility (47,48), adipocyte differentiation (49) and development of various types of cancers, such as breast (47,50), lung (51) and colon cancer (40). Whether the JMJD6΄s function in any of these biological processes is linked to its regulatory role in splicing is still an open question.
In the present study, we investigated the functional significance of the interaction between JMJD6 and U2AF65 by using a high-throughput sequencing technique, RASL-Seq (52,53). We found that JMJD6 regulated a large set of alternative splicing events, and importantly, its regulated splicing events were significantly overlapped those controlled by U2AF65. We further confirmed the function of JMJD6 in alternative splicing in jmjd6 knockout mice. Mechanistically, we demonstrated that JMJD6 and U2AF65 co-regulated a large subset of alternative splicing events, which were dependent on JMJD6 enzymatic activity and JMJD6-modified lysine residues (K15, 38 and 276) in U2AF65. These findings establish an U2AF65-mediated, JMJD6-regulated splicing pathway in mammalian cells.
MATERIALS AND METHODS
Plasmids and cloning procedures
JMJD6 and U2AF65 were PCR-amplified from cDNAs by using KOD Hot Start DNA Polymerase (Novagen) and then cloned into p3XFLAG-CMV-10 (Sigma) or pEGFP-C3 (Clontech) for expression. All point mutations were generated by using QuikChange Lightning Site-Directed Mutagenesis Kit (Stratagene).
siRNAs and antibodies
Two independent siRNAs against JMJD6 or U2AF65 were used in this study: siJMJD6(1) (Dharmacon siGENOME SMARTpool M-010363-03), siJMJD6(2) (QIAGEN, CAGCTATGGTGAACACCCTAA), siU2AF65(1) (Dharmacon siGENOME SMARTpool M-012380-01-0020), siU2AF65(2) (QIAGEN, ACCCAACTACCTGAACGATGA). Anti-U2AF65 (C-8) (SC-166695), anti-BRD4 (SC-48772) and anti-GFP (SC-8334) antibodies were purchased from Santa Cruz Biotechnology; anti-JMJD6 (ab10526) antibody was purchased from Abcam.
Plasmids, siRNA transfection, RNA isolation and RT-qPCR
Plasmid and siRNA transfection were performed using Lipofectamine 2000 (Invitrogen) according to manufacturer's protocols. Total RNA was isolated using RNeasy Mini Kit (Qiagen). First-strand cDNA synthesis from total RNA was carried out using iScript™ cDNA Synthesis Kit (Bio-Rad). All RT-qPCRs were repeated at least three times and representative results were shown. Sequence information for primers used to check the expression of JMJD6 or U2AF65 was as follows: JMJD6 mRNA (F) 5΄-CTGAATTCAAACCCCTGGAA-3΄; JMJD6 mRNA (R) 5΄- TACCGTCTTGTGCCATACCA-3΄; U2AF65 mRNA (F) 5΄-GATTCCAGCCACTGCTCTTC-3΄; U2AF65 mRNA (R) 5΄-CTGAGCGGAACTCCAAAAAG-3΄. Sequence information for other primers used in this study is available upon request.
Protein immunoblotting and immunoprecipitation
Protein immunoblotting and immunoprecipitation were performed following the protocol described previously (31).
Splicing profiling by RASL-seq and validation by RT-PCR
The RASL reaction (Supplementary Figure S1) was carried out as previously described (52–54). Briefly, a pool of oligonucleotides was designed to detect 3710 alternative splicing events (5610 in the expanded version). These oligonucleotides were mixed with 1 μg of total RNA followed by selection and ligation. Ligated oligos were amplified with barcoded primers, and PCR products were then subjected to high-throughput sequencing. Data analysis was performed as previously described (53,54). A minimal of five counts for each isoform (both long and short isoform) in a given splicing event was required to calculate an expressed isoform and significantly changed splicing events were selected based on triplicated experiments with P-value <0.05 and/or fold change in isoform ratio ≥2. Gene ontology (GO) analysis for genes with changed alternative spliced exons was performed with DAVID (55). To validate altered splicing events detected by RASL-Seq, RT (reverse transcriptase) reactions were carried out followed by standard PCR (RT-PCR) using primer sets specifically targeting the flanking exons of the alternatively spliced exon in individual genes.
RNA immunoprecipitation (RNA-IP)
RNA-IP without crosslinking was performed as previously described with minor modifications (56). Briefly, cells grown in a 10-cm dish were lysed in polysome lysis buffer (100 mM KCl, 5 mM MgCl2, 10 mM HEPES, pH 7.0, 0.5% NP-40, 1 mM DTT, 100 U/ml RNasin RNase inhibitor (Promega, N2511), 2 mM vanadyl ribonucleoside complexes solution (Sigma, 94742), 25 μl/ml protease inhibitor cocktail for mammalian tissues (Sigma, P8340)) and then subjected to IP followed by washing with polysome lysis buffer four times, and then polysome lysis buffer plus 1 M urea four times. RNAs was released by adding 150 μl of polysome lysis buffer with 0.1% SDS and 45 μg proteinase K (Ambion, AM2548) and incubated at 50°C for 30 min. RNA extracted with phenol–chloroform–isoamyl alcohol mixture (Sigma, 77618) was recovered by adding 2 μl GlycoBlue (15 mg/ml, Ambion, AM9516), 36 μl 3 M sodium acetate and 750 μl ethanol followed by incubation at −20°C for overnight. Precipitated RNAs were washed with 70% ethanol, air dried, and re-suspended in RNase-free water followed by DNase I (Promega, M6101) treatment to remove genomic DNA. The resultant RNAs were subjected to RT-qPCR analysis using primers specifically designed to detect specific pre-mRNAs.
UV-crosslinked RNA-IP was performed as previously described with minor modifications (57,58). Briefly, cells from a 10-cm dish were UV-irradiated at 254 nm, 400 mJ/cm2 (Stratagene Stratalinker) and then lysed in cell lysis buffer (10 mM Tris–HCl, 100 mM NaCl, 2.5 mM MgCl2, 35 mg/ml digitonin, 0.5% Triton X-100). Supernatant was subjected to IP with wash, once in low-stringency buffer (1× PBS (150 mM NaCl), 0.1% SDS, 0.5% sodium deoxycholate, 0.5% NP-40) and twice in high-stringency buffer (5× PBS (750 mM NaCl), 0.1% SDS, 0.5% sodium deoxycholate, 0.5% NP-40). RNA release, extraction, treatment with DNase I and RT-qPCR were performed as described above.
RESULTS
JMJD6 and U2AF65 co-regulate alternative splicing
Through affinity purification followed by mass spectrometry analysis, we previously reported that JMJD6 was associated with an array of proteins involved in RNA biology metabolism (31). Gene ontology (GO) analysis of potential JMJD6 interactors revealed that RNA processing, RNA metabolic process, and RNA splicing were among the most enriched GO terms, suggesting JMJD6 might play a regulatory role in RNA metabolism (Supplementary Table S1). One of the JMJD6 interactors was U2AF65, which has been reported earlier to be lysine hydroxylated by JMJD6 (21,39). In the present study, we focused on investigating JMJD6 function in the regulation of alternative splicing and the underlying molecular mechanism with respect to its interaction with U2AF65.
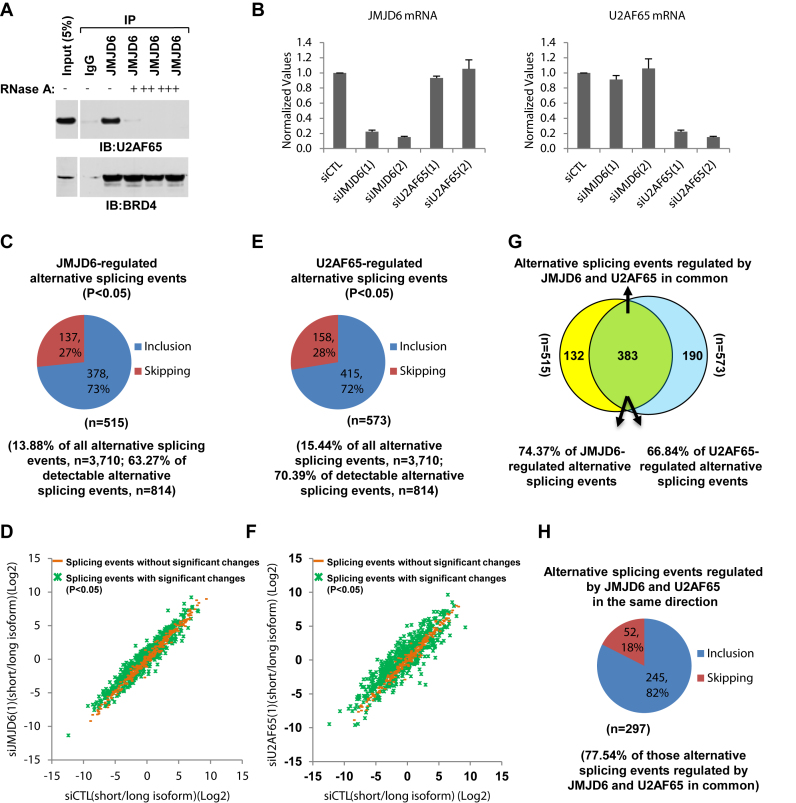
Firstly, JMJD6 interaction with U2AF65 was confirmed by immunoprecipitation (IP) with anti-JMJD6 antibody followed by immunoblotting (IB) with anti-U2AF65 antibody (Figure 1A, upper panel). Interestingly, the inclusion of RNase A during IP completely abolished the interaction, suggesting that JMJD6 associated with U2AF65 in an RNA-dependent manner, consistent with an early report (39). As a control, RNase A showed no significant impact on the interaction between JMJD6 and BRD4, a protein we reported previously to co-regulate transcriptional pause release (31) (Figure 1A, bottom panel). The RNA-dependent connection between JMJD6 and U2AF65 prompted us to investigate their functional relationship in regulated splicing. For this purpose, we performed RASL-Seq (RNA-mediated oligonucleotide annealing, selection and ligation coupled with high-throughput sequencing) (Supplementary Figure S1) with total RNA extracted from HEK293T cells transfected with control siRNA (small interfering RNA) or siRNAs specifically targeting JMJD6 or U2AF65. The knockdown efficiency of two independent siRNAs targeting JMJD6 or U2AF65 was confirmed by RT-qPCR (Figure 1B) and immunoblotting (see later in Figures 6A and 7A). The data also showed that knockdown of JMJD6 had no effect on the expression of U2AF65, and vice versa.
Figure 1.
JMJD6 and U2AF65 co-regulate alternative splicing. (A) JMJD6 interacted with U2AF65 in a RNA-dependent manner. Cell lysates from HEK293T cells were subjected to immunoprecipitation (IP) with control IgG or anti-JMJD6 antibody in the presence or absence of RNase A (+: 50 ng/μl; ++: 250 ng/μl; +++: 500 ng/μl) followed by immunoblotting (IB) with anti-U2AF65 (upper panel) or anti-BRD4 (bottom panel) antibodies. (B) HEK293T cells were transfected with control siRNA (siCTL) or two independent siRNAs specifically targeting JMJD6 (siJMJD6(1), siJMJD6(2)) or U2AF65 (siU2AF65(1), siU2AF65(2)) for 72 h followed by RNA extraction and RT-qPCR analysis using primers specifically targeting JMJD6 (left panel) or U2AF65 (right panel). Data shown was the relative fold change compared to control samples after normalization to actin. (C, E) Pie chart showing JMJD6- (C) or U2AF65- (E) regulated alternative splicing events, both exon inclusion and skipping, examined through RASL-Seq analysis using RNA samples described in (B) (P < 0.05). Data shown was from siJMJD6(1) (C) and siU2AF65(1) (E). (D, F) Scatter plot showing the isoform ratio (short versus long, log2) of all detectable alternative splicing events (read counts >5) in RASL-Seq when knocking down JMJD6 (D) or U2AF65 (F). (G) Venn diagram showing overlapping between JMJD6 and U2AF65-regulated alternative splicing events (P = 0.000785, hypergeometric test, expected background value: 52). (H) Pie chart showing alternative splicing events regulated by JMJD6 and U2AF65 in the same direction including exon inclusion and skipping (P < 0.05).
Figure 6.
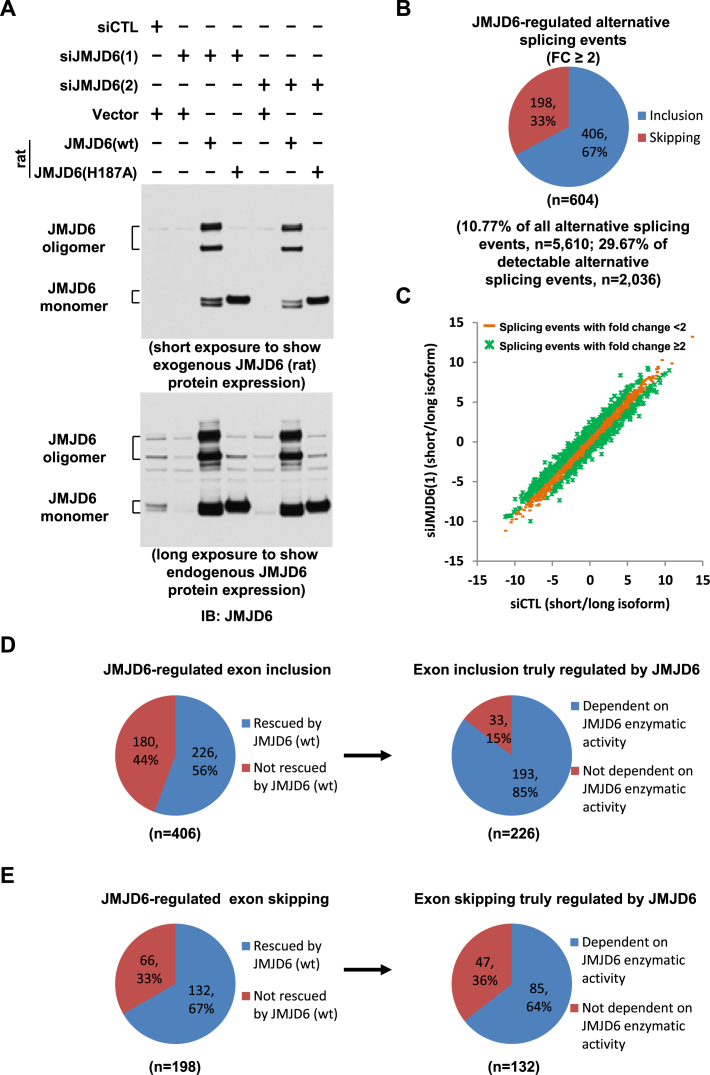
JMJD6 enzymatic activity is involved in its regulated alternative splicing. (A) HEK293T cells were transfected with control siRNA (siCTL) or two independent siRNAs specifically targeting JMJD6 (siJMJD6(1), siJMJD6(2)) in the presence or absence of a control vector or vectors expressing rat wild-type JMJD6 (wt) or its enzymatically deficient mutant with substitution of histidine 187 to alanine (H187A), followed by immunoblotting (IB) with anti-JMJD6 antibody. JMJD6 monomer and oligomer were shown as indicated. (B) Pie chart showing alternative splicing events regulated by JMJD6, both exon inclusion and skipping (FC≥2), examined through RASL-Seq analysis (expanded version) using RNA samples described in (A). Data shown was from siJMJD6(1). (C) Scatter plot showing the isoform ratio (short versus long, log2) of all detectable alternative splicing events in RASL-Seq (expanded version) when knocking down JMJD6. (D, E) Left panels: Pie chart showing exon inclusion (D) or skipping (E) regulated by JMJD6, including the ones could be rescued by JMJD6 protein (referred as exon inclusion (D) or skipping (E) truly regulated by JMJD6) and the ones could not; Right panels: Pie chart showing exon inclusion (D) or skipping (E) truly regulated by JMJD6 including the ones were dependent on JMJD6 enzymatic activity and the ones were not.
Figure 7.
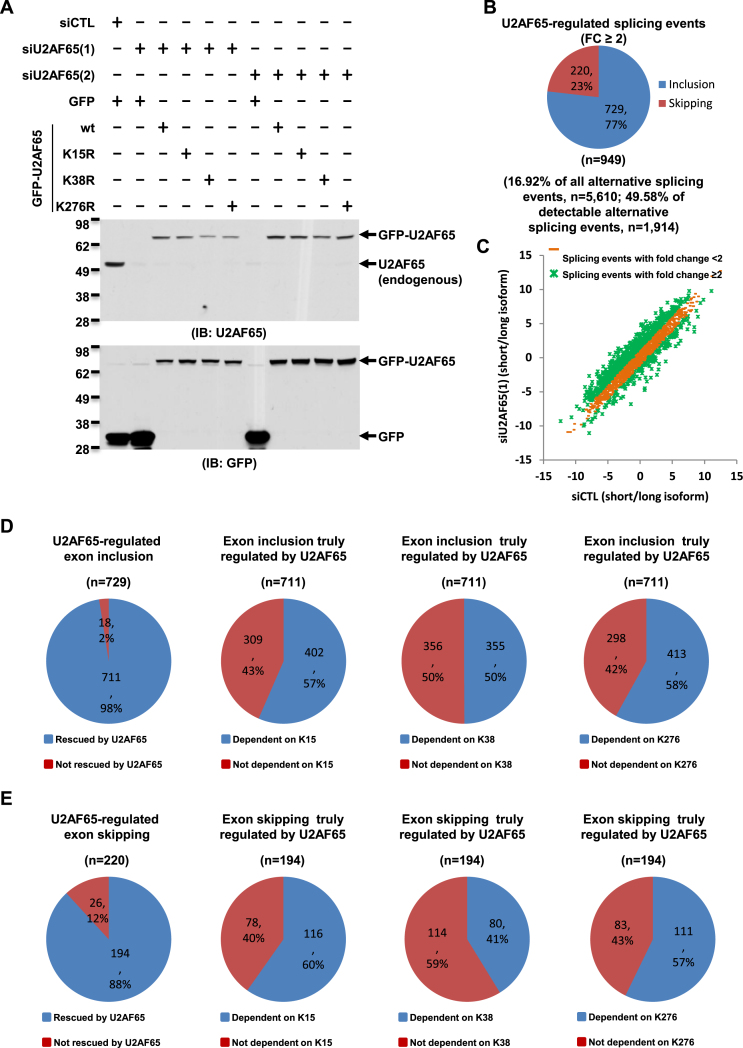
JMJD6-mediated lysine hydroxylation of U2AF65 is involved in alternative splicing regulation. (A) HEK293T cells were transfected with control siRNA (siCTL) or two independent siRNAs specifically targeting U2AF65 (siU2AF65(1), siU2AF65(2)) in the presence or absence of a control vector or vectors expressing wild-type U2AF65 (wt) or its mutants with substitution of lysine 15, 38 or 276 to arginine (K15R, K38R or K276R), followed by immunoblotting (IB) with anti-U2AF65 (upper panel) or anti-GFP antibody (bottom panel).(B) Pie chart showing alternative splicing events regulated by U2AF65, both exon inclusion and skipping (FC≥2), examined through RASL-Seq analysis (expanded version) using RNA samples described in (A). Data shown was from siU2AF65(1). (C) Scatter plot showing the isoform ratio (short versus long, log2) of all detectable alternative splicing events in RASL-Seq (expanded version) when knocking down U2AF65. (D, E) Far left panels: Pie chart showing exon inclusion (D) or skipping (E) regulated by U2AF65, including the ones could be rescued by U2AF65 protein (referred to as exon inclusion (D) or skipping (E) truly regulated by U2AF65) and the ones could not; Right three panels: Pie chart showing exon inclusion (D) or skipping (E) truly regulated by U2AF65, including the ones were dependent on K15, K38 or K276 as indicated, and the ones were not.
The human RASL-Seq platform was initially designed to target 3710 alternative splicing events. Although this technology focuses on selected and annotated targets compared to completely unbiased profiling of alternative splicing by RNA-Seq, RASL-Seq has been proven to be robust in quantitative measurement of alternative splicing, which enables large-scale comparison of regulated splicing events (21,53,54,59,60). By filtering events with counts <5, we identified 814 alternative splicing events in HEK293T cells in the current experiment. Changes in isoform ratio (counts for long isoform versus short isoform) between control and knockdown cells were then calculated and statistical significance was determined based on triplicated experiments. We found that 515 alternative splicing events showed significant changes in isoform ratio upon knockdown of JMJD6 (siJMJD6(1), P < 0.05), which represented 14% and 63% of total (n = 3710) and detectable (n = 814) alternative splicing events, respectively, on the platform (Figure 1C and D and Supplementary Table S2). Interestingly, among these 515 altered alternative splicing events induced by JMJD6 knockdown, 378 and 137 events showed increased exon skipping and inclusion, respectively, suggesting JMJD6 preferentially promoted exon inclusion (Figure 1C and D and Supplementary Table S2). The impact of the second independent siRNA targeting JMJD6 (siJMJD6(2)) on alternative splicing was similarly assessed, again revealing splicing ratio changes in 12% and 54% of total and detectable alternative splicing events, respectively, with prevalent induction of exon skipping (Supplementary Figure S2A and S2B). Altered splicing events were extensively overlapped between the experiments using these two independent siRNAs against JMJD6 (Supplementary Figure S2C and S2D).
Similarly, knockdown of U2AF65 (siU2AF65(1)) led to significant changes in the isoform ratio of 573 alternative splicing events, corresponding to 15% and 70% of total (n = 3710) and detectable (n = 814) events, respectively (Figure 1E and F and Supplementary Table S2). U2AF65 also appeared to preferentially promote exon inclusion as nearly three-quarters of altered events showed increased exon skipping upon U2AF65 knockdown, fully consistent with a previous report (Figure 1E and F and Supplementary Table S2) (21). The second independent siRNA targeting U2AF65 (siU2AF65(2)) gave similar results (Supplementary Figure S2E and S2F) and the induced splicing events by both siRNAs targeting U2AF65 were significantly overlapped and correlated (Supplementary Figure S2G and S2H). For simplicity, we focused on the alternative splicing events regulated by JMJD6 and U2AF65 based on the use of siJMJD6(1) and siU2AF65(1). We also assessed the impact of knocking down JMJD6 and U2AF65 on alternative splicing by using percentage spliced in (PSI, long isoform divided by the sum of both short and long isoforms), and found that 517 events were significantly changed upon JMJD6 knockdown, among which 381 and 136 events exhibited increased exon skipping and inclusion, respectively (Supplementary Figure S3A). The data are highly comparable to those obtained based on changes in splicing ratio (Supplementary FigureS3B and S3C). Similarly, U2AF65-regulated splicing events calculated based on changes in PSI or isoform ratio were also highly concordant (Supplementary Figure S3D-F).
Comparing the alternative splicing events regulated by JMJD6 and U2AF65, we found that 383 events were regulated by both proteins, which were 74% and 67% of JMJD6 and U2AF65-regulated events, respectively, suggesting a functional interaction between these two proteins (Figure 1G). Given the large number of splicing events regulated by both proteins, we determined that the overlap was statistically significant (P = 0.000785, hypergeometric test). More importantly, 77% of the alternative splicing events regulated by both JMJD6 and U2AF65 were altered in the same directions (Figure 1H).
If a more stringent cutoff was taken (P < 0.05 and fold change (FC) of isoform ratio between control and knockdown samples >2), knockdown of JMJD6 altered the isoform ratio of 6.6% and 30% of total and detectable alternative splicing events, respectively (Supplementary Figure S4A and S4B). The extent of JMJD6΄s impact on alternative splicing was comparable to other classic RNA-binding proteins that have been assessed on the RASL-Seq platform, such as SRSF1, SRSF2 and RBFox2 (54,59). When the same cutoff was applied to determine the alternative splicing events regulated by U2AF65 (Supplementary Figure S4C-D) and then compared to that of JMJD6, a significant overlap could still be observed (Supplementary Figure S4E and S4F). The high degree of overlap between JMJD6 and U2AF65-regulated alternative splicing events could also be confirmed by comparing data from any of the two siRNAs targeting JMJD6 or U2AF65. Taken together, these data suggested that JMJD6 and U2AF65 co-regulate a large set of alternative splicing events.
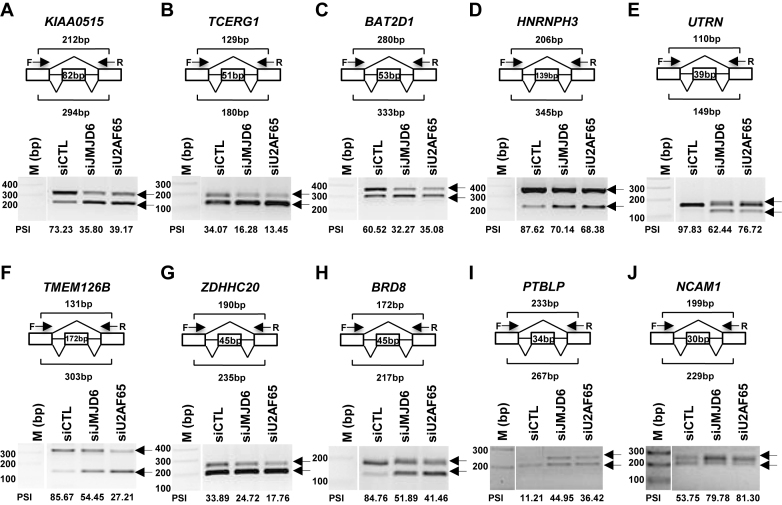
Next, we sought to validate the altered alternative splicing events identified through RASL-Seq by RT-PCR (reverse transcription polymerase chain reaction). Specifically, total RNA extracted from HEK293T cells transfected with control siRNA or siRNAs targeting JMJD6 or U2AF65 were subjected to RT followed by PCR using specific primer sets, which were designed to target the two exons flanking the alternatively spliced exon to detect the expression of both isoforms (depicted in top panels of Figure 2A–J). In total, we validated 20 events co-regulated by both JMJD6 and U2AF65 with a successful rate of 90% (two failed), and showed a representative set of validated events (Figure 2A–J). Of note, some alternative splicing events appear to express an isoform at the levels too low to be detectable with RASL-Seq (i.e. too few counts after sequencing), but still subjected to the regulation by JMJD6 and U2AF65, as exemplified by exon 6 in RBM5 and exon 2 in MADD, indicating the impact of both proteins on alternative splicing could be larger than what had been detected by RASL-Seq (Supplementary Figure S5A and S5B).
Figure 2.
Validation of alternative splicing events co-regulated by JMJD6 and U2AF65 through RT-PCR. (A–J) First-strand cDNA synthesis was done from RNA samples described in Figure 1B followed by standard PCR analysis (RT-PCR) using primer sets specifically targeting to the upstream and downstream exon relative to the cassette exon (alternatively spliced exon) to detect the expression of both short and long isoforms of representative genes as indicated. The length of the alternatively spliced exon as well as the expected length of the PCR products was shown as indicated. DNA marker (M) was included on the left. Percentage spliced in (PSI) was calculated as the ratio of the density of the long isoform versus that of the sum of the long and short isoforms. The position of the alternatively spliced exon in each gene was as follows: KIAA0515 (NM_013318, exon 30) (A); TCERG1 (BI091338, exon 3) (B); BAT2D1 (AV650960, exon 6) (C); HNRNPH3 (NM_012207, exon 3) (D); UTRN (NM_007124, exon 66) (E); TMEM126B (NM_018480, exon 2) (F); ZDHHC20 (uc001uod.1, exon 13) (G); BRD8 (NM_006696, exon 21) (H); PTBLP (NM_021190, exon 10) (I); NCAM1 (NM_181351, exon 9) (J). F: forward primer; R: reverse primer; bp: base pair.
JMJD6 regulates alternative splicing in the mouse
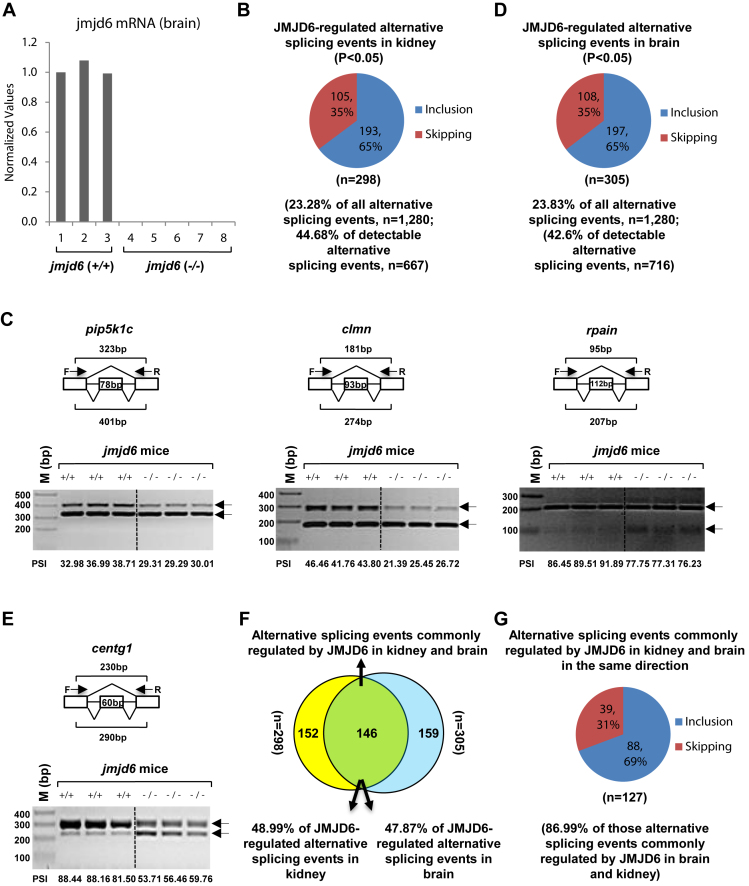
JMJD6 is highly conserved throughout the animal kingdom and functionally essential for embryonic development (24,45,46). Jmjd6 knockout mice died prenatally and jmjd6 deficient embryos manifested severe developmental defects in multiple organs, including lung, kidney, liver, intestine, eye and brain. For example, a delay in differentiation of kidney was observed in jmjd6-deficient embryos, exhibiting primitive glomeruli and poorly-developed collecting tubules at E16.5(24). Jmjd6-deficient embryos also showed severe brain malformation characterized by exencephaly, disrupted forebrain proliferative zones, expanded midbrain, and a disrupted cortical plate as well as altered morphology of the olfactory bulb, brainstem-spinal cord junction, and cerebellum (45). We took advantage of this existing mouse model to test JMJD6-regulated alternative splicing in vivo. We isolated total RNA from kidney and brain from wild type (+/+) (n = 3) and knockout (−/−) (n = 5) mice (E17.5) for RASL-Seq analysis (Figure 3A). The mouse RASL-Seq platform was designed to target 1280 alternative splicing events conserved between mouse and human for the experiments presented in this report. We found that jmjd6 knockout led to significant changes in around 23% and 45% of total and detectable alternative splicing events, respectively in kidney compared to wild-type mice, confirming an extensive impact of JMJD6 in alternative splicing in vivo (Figure 3B and Supplementary Table S3). We found that JMJD6-regulated the expression of isoforms from many genes known to be critical for kidney development, such as clmn, pkd1, ptpro, fip1l1, bat3, fgfr2 and lef1, suggesting their contributions to developmental defects in kidney observed in jmjd6 knockout mice. Similar to what observed in cultured HEK293T cells, jmjd6 knockout also preferentially induced exon skipping (Figure 3B). Representative JMJD6-regulated alternative splicing events were validated by RT-PCR, including pip5k1c, clmn, rpain, phf20l1 and unc84a (Figure 3C and Supplementary Figure S6A). Of note, PSI between individual wild type or knockout groups was highly consistent (Figure 3C and Supplementary Figure S6A). To further demonstrate JMJD6΄s impact on alternative splicing, we also performed RASL-Seq with RNAs extracted from the brain of wild type or jmjd6 knockout mice, finding that around 24% and 43% of total and detectable alternative splicing events, respectively, were significantly altered by jmjd6 knockout and 65% of the altered events displayed induced exon skipping (Figure 3D and Supplementary Table S3). JMJD6-regulated splicing was found on many genes involved in brain development, such as centg1, kras, lhx6, pax6, grin1, grik1 and grik2. Representative JMJD6-regulated alternative splicing events in the brain were validated by RT-PCR (Figure 3E and Supplementary Figure S6B).
Figure 3.
JMJD6 regulates alternative splicing in vivo. (A) RNAs were extracted from kidney and brain samples of wild type (jmjd6 (+/+)) (n = 3) or jmjd6 knockout (jmjd6 (−/−)) (n = 5) mice, and the expression of jmjd6 was examined through RT-qPCR. Data shown was jmjd6 expression in brain samples. (B,D) Pie chart showing alternative splicing events, both exon inclusion and skipping, regulated by jmjd6 in kidney (B) or brain (D) samples examined through RASL-Seq analysis using RNA samples described in (A) (P < 0.05). (C, E) RT-PCR was done using RNA samples from kidney (C) or brain (E), both wild type (+/+, n = 3) and jmjd6 knockout (−/−, n = 3), as described in Figure 3A to validate alternative splicing events (exon inclusion) regulated by JMJD6 detected by RASL-Seq. Representative examples were shown as follows: pip5k1c (NM_008844, exon 17) (Figure 3C, left panel); clmn (NM_053155, exon 13) (Figure 3C, middle panel); rpain (NM_027186, exon 4) (Figure 3C, right panel); centg1 (NM_001033263, exon 14) (E). F: forward primer; R: reverse primer; bp: base pair. (F) Venn diagram showing overlapping between JMJD6-regulated alternative splicing events in kidney and brain (P = 0.002229, hypergeometric test, expected background value: 24). (G) Pie chart showing alternative splicing events commonly regulated by JMJD6 in kidney and brain in the same direction, including exon inclusion and skipping (P < 0.05).
To determine whether JMJD6-regulated alternative splicing has tissue specificity, the altered alternative splicing events in kidney were compared to those in brain. We noted that 146 events were commonly regulated, which was 49% and 48% of JMJD6-regulated alternative splicing events in kidney and brain, respectively, suggesting that JMJD6 regulates both tissue specific alternative splicing events as well as a core set of alternative splicing events in different tissues (Figure 3F). Importantly, the vast majority of this common set of events (∼87%) was altered in the same direction in both kidney and brain, suggesting the direct impact of JMJD6 ablation on these alternative splicing events (Figure 3G and Supplementary Table S3). As the alternative splicing events included in the mouse RASL-Seq platform were conserved between mouse and human, JMJD6-regulated alternative splicing events in mouse kidney and brain were compared to those in HEK293T cells. JMJD6-regulated alternative splicing events in both kidney and brain in mice were first combined and duplicates removed, resulting in a list of 457 unique events, which were then compared to the set of 515 JMJD6-regulated events detected in HEK293T cells. We noted 141 events in common (Supplementary Figure S6C). Importantly, around 78% of these 141 events were co-regulated by JMJD6 and U2AF65 in HEK293T cells, suggesting overlaps between the two species were enriched for co-regulation by JMJD6 and U2AF65 in human (Supplementary Figure S6D). If a more stringent cutoff was taken (P < 0.05, FC of isoform ratio > 1.5), 22% and 26% of detectable alternative splicing events displayed significant changes in kidney and brain from jmjd6 knockout mice, respectively (Supplementary Figure S6E and S6F). Similarly, under this more stringent cutoff, JMJD6΄s function in alternative splicing appeared to have tissue specificity as well as commonality (Supplementary Figure S6G and S6H). Taken together, our data described above suggested a widespread role of JMJD6 in alternative splicing in both cultured cells and animals.
JMJD6 and U2AF65 bind pre-mRNA directly to regulate alternative splicing
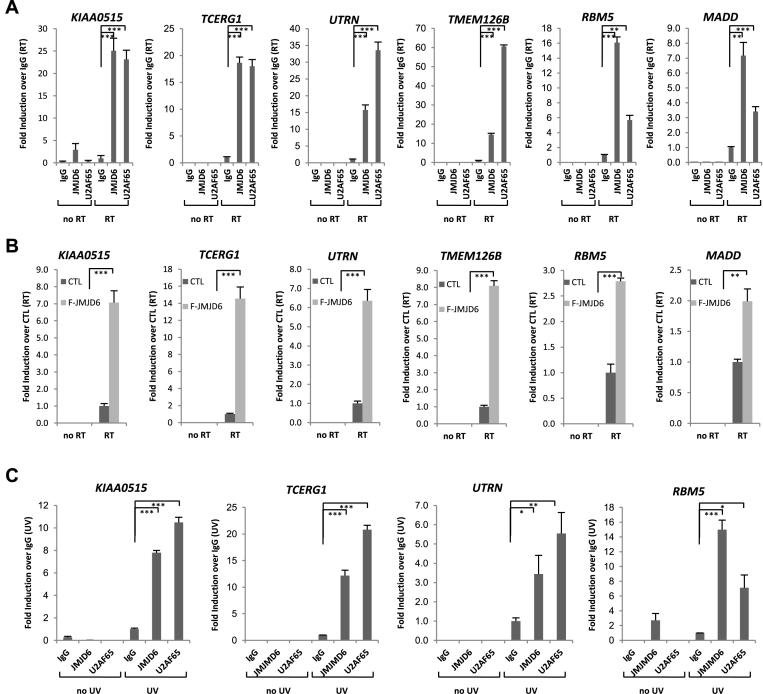
We next sought to understand the molecular mechanism underlying JMJD6-regulated alternative splicing. Recently, a structural study revealed a novel substrate binding groove and two positively charged surfaces in JMJD6, suggesting its capability to directly bind RNA (61). To test this, we first performed RIP (RNA immunoprecipitation) on HEK293T cells using control IgG or specific antibodies against JMJD6 or U2AF65, and examined resultant RNAs by RT-PCR with primers that specifically target JMJD6 and U2AF65 regulated exons (Supplementary Figure S7A). For all altered alternative splicing events shown in Figure 2 and Supplementary Figure S5, we detected robust JMJD6 and U2AF65 association with RNA compared to IgG (Figure 4A and Supplementary Figure S7B). We performed PCR amplification without reverse transcriptase (no RT) to rule out contamination of genomic DNA. We also tested several alternatively spliced genes (DNAJC7, ATP5C1 and AASDHPPT) included in our RASL-Seq, but their splicing unaffected by knockdown of JMJD6 or U2AF65 based on our RNA-Seq results, finding no binding of their pre-mRNAs with either JMJD6 or U2AF65 (Supplementary Figure S7C). For further comparison, we examined two other genes (MAPKAP1 and BCL11A), which were included in our RASL-Seq pool and shown to be sensitive to knockdown of JMJD6 but not U2AF65, for interaction with JMJD6 and U2AF65. We detected binding of JMJD6, but not U2AF65, with both of these pre-mRNAs (Supplementary Figure S7D). Conversely, analysis of the two other genes (N-PAC and OPA1), which were included in our RASL-Seq pool and detected to be sensitive to knockdown of U2AF65, but not JMJD6, showed the association of their pre-mRNAs with U2AF65, but not JMJD6 (Supplementary Figure S7E). Together, these data demonstrated a degree of selectivity for JMJD6 and U2AF65 to interact with RNA.
Figure 4.
JMJD6 and U2AF65 co-bind with the pre-mRNAs they regulate. (A) HEK293T cells were subjected to RIP using control IgG, anti-JMJD6 or U2AF65 antibodies, and the resultant RNAs were examined by RT-qPCR with primer sets specifically targeting to the upstream and downstream intron region relative to the alternatively spliced exon to determine JMJD6 and U2AF65 binding with pre-mRNA. The position of the alternatively spliced exon in each gene was described in Figure 2. RT: with reverse transcriptase; no RT: without adding reverse transcriptase. RIP signals were presented as fold change over IgG (RT) (± S.E.M., **P < 0.01, ***P < 0.001). (B) HEK293T cells stably expressing a control vector (CTL) or Flag-tagged JMJD6 (F-JMJD6) were subjected to RIP using anti-Flag antibody. Flag-tagged JMJD6 binding with pre-mRNAs of representative genes was shown. RIP signals were presented as fold change over control sample (RT) (± S.E.M., **P < 0.01, ***P < 0.001). (C) HEK293T cells were treated with or without UV (ultraviolet) before RIP followed by RT-qPCR as described in (A) to determine JMJD6 and U2AF65 binding with pre-mRNA. RIP signals were presented as fold change over IgG (UV) (± S.E.M., *P < 0.05, **P < 0.01, ***P < 0.001).
To further confirm JMJD6 binding with RNA, we subjected HEK293T cells stably expressing a control vector or Flag-tagged JMJD6 to RIP using anti-Flag specific antibody. In accordance with RIP using anti-JMJD6 antibody, we detected robust binding of Flag-tagged JMJD6 with specific pre-mRNAs using the same set of primers described above (Figure 4B and Supplementary Figure S7F). The observed JMJD6 binding with RNA could be direct or indirect. To distinguish between these possibilities, we performed UV (ultraviolet)-crosslink RIP in HEK293T cells with control IgG or anti-JMJD6 antibody and separated resultant RNAs on a gel (57). We detected a large amount of RNA associated with JMJD6 in UV-treated cells, but not in untreated cells, suggesting JMJD6 binding with RNA was direct. No RNA was detected in IgG pulldown in either untreated or UV-treated cells (Supplementary Figure S8A). RT-PCR analysis of resultant RNAs revealed that, compared to IgG controls, both JMJD6 and U2AF65 were significantly enriched on a selective set of pre-mRNAs examined, including KIAA0515, TCERG1, UTRN and RBM5 (Figure 4C), but not DNAJC7 expressed at comparable levels in either UV-treated or untreated cells, which served as a negative control (Supplementary Figure S8B). We similarly confirmed direct binding of JMJD6 with these pre-mRNAs with anti-Flag antibody in HEK293T cells that stably express Flag-tagged JMJD6 (Supplementary Figure S8C).
U2AF65 regulates JMJD6 interaction with pre-mRNA
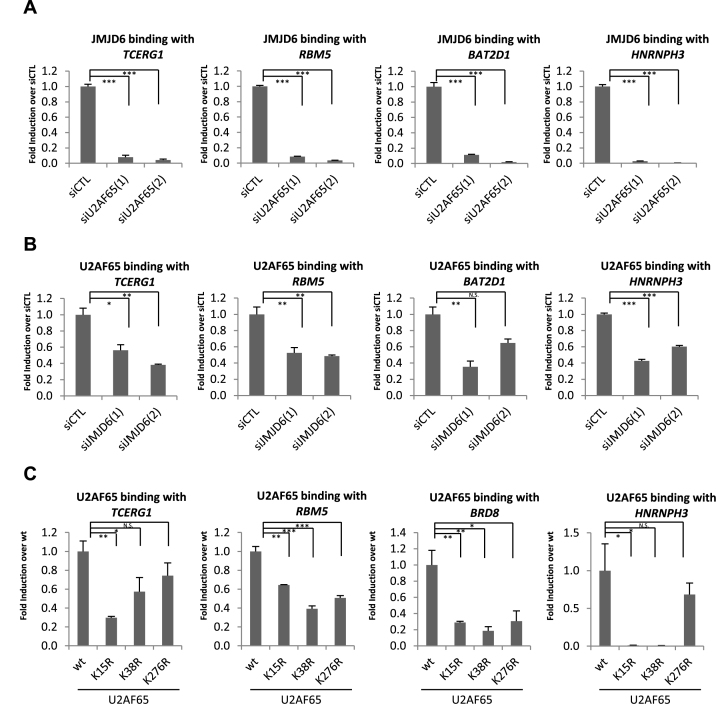
The RNA-dependent interaction between JMJD6 and U2AF65 (Figure 1A), and the high degree of overlap between JMJD6 and U2AF65-regulated alternative splicing programs prompted us to examine whether JMJD6 binding with RNA was regulated by U2AF65. HEK293T cells were transfected with control siRNA or two independent siRNAs specifically targeting JMJD6 or U2AF65 followed by RIP with control IgG or antibody specifically against JMJD6 or U2AF65. We found that knockdown of U2AF65 nearly abolished JMJD6 binding with pre-mRNAs co-regulated by JMJD6 and U2AF65, as illustrated on representative examples, such as TCERG1, HNRNPH3, BAT2D1 and RBM5 (Figure 5A). Of note, in accordance with the fact that knockdown of splicing factors may trigger changes in expression levels of some pre-mRNAs, knockdown of U2AF65 led to decreased levels of the pre-mRNA from two specific genes RBM5 and BAT2D1 based on input normalized data in Figure 5A (Supplementary Figure S9A). As a control, JMJD6 interaction with pre-mRNA of TPP2, which was occupied by JMJD6 but not U2AF65, was unaffected upon U2AF65 knockdown (Supplementary Figure S9B). The amount of immunoprecipitated JMJD6 in each condition was similar (Supplementary Figure S9C). To our surprise, knockdown of JMJD6 also resulted in a significant decrease in U2AF65 binding with pre-mRNAs they co-regulated (Figure 5B). The amount of U2AF65 obtained in each immunoprecipitation performed was similar (Supplementary Figure S9D). Importantly, despite that JMJD6 was efficiently knocked down, which largely abolished JMJD6 binding, the impact of JMJD6 on U2AF65 binding was not as dramatic as that of U2AF65 on JMJD6 (Figure 5A and B and Supplementary Figure S9E). Potential reasons might account for these observations: (i) Enzymes redundant to JMJD6 may also regulate U2AF65; (ii) JMJD6 enzymatic activity was not proportionally affected to its protein level, which has been demonstrated on other epigenetic regulators, such as CARM1 (62) and SET7/9 (63). As a control, JMJD6 interaction with N-PAC and OPA1 pre-mRNA, which was occupied by U2AF65 but not JMJD6, was unaffected upon knockdown of JMJD6 (Supplementary Figure S9F).
Figure 5.
U2AF65 regulates JMJD6 binding with pre-mRNAs they co-regulate. (A, B) HEK293T cells were transfected with control siRNA (siCTL) or two independent siRNAs specifically targeting JMJD6 (siJMJD6(1), siJMJD6(2)) or U2AF65 (siU2AF65(1), siU2AF65(2)) followed by RIP with control IgG or antibody specifically against JMJD6 or U2AF65. JMJD6 (A) or U2AF65 (B) binding with pre-mRNAs of representative genes was shown as indicated. RIP signals were first normalized to input and then presented as fold change over control (siCTL) (± S.E.M., *P < 0.05, **P < 0.01, ***P < 0.001). (C) HEK293T cells were transfected with vectors expressing GFP-tagged wild-type U2AF65 (wt) or U2AF65 mutants with lysine 15, 38 or 276 substituted with arginine (K15R, K38R or K276R), followed by RIP with anti-GFP antibody. U2AF65 binding with pre-mRNAs of representative genes was shown as indicated. RIP signals were first normalized to input and then presented as fold change over control (wt) (± S.E.M., *P < 0.05, **P < 0.01, ***P < 0.001).
It has been previously reported that JMJD6 catalyzes hydroxylation on lysine residues 15, 38 and 276 in U2AF65 (39). To see whether the detected impact of JMJD6 on U2AF65 binding can be linked to its modification on these lysine residues, we transfected HEK293T cells with vectors expressing GFP-tagged wild-type U2AF65 (wt) or U2AF65 mutants in which lysine 15, 38 or 276 was individually substituted with arginine (K15R, K38R or K276R), and then performed RIP with anti-GFP specific antibody. We found that substitution of lysine 15 or 38 with arginine largely diminished U2AF65 binding with pre-mRNAs from TCERG1, HNRNPH3, BRD8 and RBM5, whereas substitution of lysine 276 with arginine significantly altered U2AF65 binding with pre-mRNAs from RBM5 and BRD8, but not TECRG1 or HNRNPH3 (Figure 5C). Serving as controls, binding of U2AF65 with pre-mRNAs of two other genes, N-PAC and OPA1, which were found to be associated with U2AF65 but not JMJD6 (Supplementary Figure S7E), was unaffected by any of the mutants (Supplementary Figure S9G). However, we do not exclude the possibility that these mutants will affect U2AF65 binding to pre-mRNAs without JMJD6 binding as they also seemed to be able to affect U2AF65 binding independent of JMJD6 enzymatic activity (vide infra). U2AF65 (wt), K15R, K38R and K276R were found to be expressed at similar levels (Supplementary Figure S9H). Taken together, these data suggested that U2AF65 was required for JMJD6 binding with RNA, and conversely, JMJD6 also modulated U2AF65 binding with RNA, which could potentially link to JMJD6 hydroxylase activity on several lysine residues (K15, 38 and 276) in U2AF65.
JMJD6 regulation of alternative splicing is linked to its enzymatic activity
We next sought to address whether the enzymatic activity of JMJD6 was important for its regulatory function in alternative splicing. Several published studies suggest that JMJD6 function in alternative splicing depends on its enzymatic activity (39,43), but others indicated that the enzymatic activity of JMJD6 was not essential (41,49). Because only a reporter gene or a very limited number of genes were examined in each of these studies, we revisited this critical issue in a much larger scale by using our RASL-Seq platform. We transfected HEK293T cells with control siRNA or two independent siRNAs specifically targeting JMJD6 in the presence or absence of a control vector or vectors expressing rat wild-type JMJD6 (wt) or its enzymatically deficient mutant containing a substitution of histidine 187 to alanine (H187A), followed by RNA extraction and RASL-Seq analysis. The RASL-Seq platform was expanded in this set of experiments to target 5610 alternative splicing events. The knockdown efficiency of two independent siRNAs targeting JMJD6 (Figure 6A, top panel, compared lanes 2 and 5 to lane 1) and the expression of exogenous rat JMJD6 (wt) and JMJD6 (H187A) (Figure 6A, bottom panel, columns 3, 4, 6 and 7) were determined by using specific anti-JMJD6 antibody. We observed oligomerization of JMJD6 (wt), but not JMJD6 (m) protein, as reported previously (31). RASL-Seq analysis revealed that around 11% and 30% of total and detectable alternative splicing events, respectively, displayed fold changes ≥2 (FC ≥ 2) in JMJD6 knockdown cells compared to control samples, which was strikingly consistent with our initial observation (Figure 6B and C and Supplementary Figure S4A, S4B). Of note, due to the increased number of alternative splicing events included in this extended RASL-Seq pool and increased sequencing depth for this experiment, both the absolute number of detectable (i.e. events with a minimal of 5 counts for each isoform) and JMJD6-regulated alternative splicing events increased. Among those JMJD6-dependent exon inclusion events (n = 406), 56% of them could be rescued by exogenous JMJD6 (P = 6.48E−28) (i.e. siJMJD6 versus siCTL, FC > 2 and siJMJD6 + JMJD6 (wt) versus siJMJD6, FC > 1), which were thus referred to as exon inclusion truly regulated by JMJD6 (n = 226) (Figure 6D, left panel). Interestingly, the vast majority of those exon inclusion truly regulated by JMJD6 (n = 226) was dependent on its enzymatic activity, as JMJD6 (H187A) failed to rescue them (P = 4.88E−24) (i.e. siJMJD6 versus siCTL, FC>2 and siJMJD6 + JMJD6 (wt) versus siJMJD6, FC > 1 and siJMJD6+JMJD6 (H187A) versus siJMJD6+JMJD6 (wt), FC < 1) (Figure 6D, right panel). For JMJD6-affected exon skipping events (n = 198), 67% of them seemed to be truly regulated by JMJD6 (P = 1.68E−16) (Figure 6E, left panel). Furthermore, 64% of those JMJD6 truly regulated exon skipping events were also dependent on the JMJD6 enzymatic activity (Figure 6E, right panel). The percentage of JMJD6 truly regulated alternative splicing events and those dependent on the JMJD6 enzymatic activity were also confirmed with the second independent siRNA targeting JMJD6 (data not shown).
A subset of JMJD6 and U2AF65 co-regulated alternative splicing events were linked to JMJD6-mediated lysine hydroxylation on U2AF65
As the JMJD6 enzymatic activity appeared to be involved, at least partially, in its regulated alternative splicing, and JMJD6 functionally interacted with U2AF65, we further tested whether JMJD6-mediated U2AF65 lysine hydroxylation was linked to JMJD6 and U2AF65 co-regulated alternative splicing events. We transfected HEK293T cells with control siRNA or two independent siRNAs targeting U2AF65 in the presence or absence of control GFP vector or vectors expressing GFP-tagged wild-type U2AF65 (wt) or its mutants with substitution of lysine 15, 38 or 276 to arginine (K15R, K38R or K276R) followed by RNA extraction and RASL-Seq analysis. The knockdown efficiency of two independent siRNAs targeting U2AF65 was determined by immunoblotting using anti-U2AF65 antibody (Figure 7A, top panel, compared lanes 2 and 7 to lane 1), and the expression of GFP control vector and exogenous GFP-tagged U2AF65 (wt), U2AF65 (K15R), U2AF65 (K38R) and U2AF65 (K276R) (Figure 7A, bottom panel) were determined with antibody specifically against GFP. We found that around 17% and 50% of total and detectable alternative splicing events, respectively, displayed fold changes larger than 2 (FC > 2) in U2AF65 knocking down cells compared to control samples, consistent with our initial observation (Figure 7B and C and Supplementary Figure S4C, S4D). Among those U2AF65-dependent exon inclusion events (n = 729), nearly all of them (98%) could be rescued by exogenous U2AF65 (P = 5.77E−42), which we referred to as exon inclusion truly regulated by U2AF65 (Figure 7D, far left panel). We overlapped JMJD6 enzymatic activity-dependent exon inclusion events (n = 193) with those truly regulated by U2AF65 and found that there were 75 events shared by both, suggesting that JMJD6 might target U2AF65 to regulate a subset of alternative splicing events, meanwhile, there are other substrates in addition to U2AF65 through which JMJD6 exerts its function in exon inclusion regulation (Supplementary Figure S10A). Interestingly, mutation of each lysine residue (K15, 38 or 276) significantly impaired U2AF65 function in alternative splicing (Figure 7D, right three panels). For example, U2AF65 (K15R), U2AF65 (K38R) and U2AF65 (K276R) failed to rescue 57%, 50% and 58% of U2AF65 truly regulated exon inclusion events, respectively, compared to the rescue effect of U2AF65 (wt) (P = 4.63E−170, 5.25E−207 and 6.05E−167, respectively). Removing the duplicated events regulated by these three lysines, 584 out of 711 exon inclusion events truly regulated by U2AF65 were linked to either lysine 15, 38 or 276 (Supplementary Figure S10B). Importantly, for those 75 exon inclusion events dependent on JMJD6 enzymatic activity and truly regulated by U2AF65, 60 of them were dependent on K15, 38 or 276 in U2AF65, suggesting that, as long as an exon inclusion event was dependent on JMJD6 enzymatic activity and U2AF65, it was often linked to K15, 38 or 276 in U2AF65 (Supplementary Figure S10C). However, for those exon inclusion events affected by either lysine 15, 38 or 276, a large number of them were not mediated by JMJD6 enzymatic activity, and the mechanisms through which they regulate exon inclusion remain unknown.
Similarly, the vast majority (∼88%) of those exon skipping events regulated by U2AF65 could be rescued by exogenous U2AF65 (P = 2.02E−12) (Figure 7E, far left panel). When we overlapped JMJD6 enzymatic activity-dependent exon skipping events (n = 85) with those truly regulated by U2AF65 (n = 194), it was found that there were 30 events shared by both, suggesting that, similar as exon inclusion, JMJD6 might target U2AF65 to regulate a subset of alternative splicing events, meanwhile, there are other substrates in addition to U2AF65 through which JMJD6 exerts its function in exon skipping regulation (Supplementary Figure S10D). Each of the three U2AF65 mutants significantly impaired U2AF65 function in exon skipping (K15R, P = 0.00231; K38R, P = 0.00271; K276R, P = 0.00066) (Figure 7E, right three panels). Removing duplicated events regulated by these three lysines, 155 out of 194 exon skipping events truly regulated by U2AF65 were linked to either lysine 15, 38 or 276 in U2AF65 (Supplementary Figure S10E). Importantly, for those 30 exon skipping events dependent on JMJD6 enzymatic activity and truly regulated by U2AF65, 26 of them were dependent on K15, 38 or 276 in U2AF65, suggesting that, similar as exon inclusion, as long as an exon skipping event was dependent on JMJD6 enzymatic activity and U2AF65, it was often linked to K15, 38 or 276 in U2AF65 (Supplementary Figure S10F). Taken together, the impact of JMJD6 and lysine 15, 38 or 276 in U2AF65 having on a subset of alternative splicing events was consistent with JMJD6΄s function as a U2AF65 lysine hydroxylase.
DISCUSSION
JMJD6 has been described as an important regulator of gene transcription and pre-mRNA splicing, with implications in a large array of development and disease processes. However, the scope and the molecular mechanism of JMJD6 function in splicing regulation have remained incompletely understood. In the present study, we provide evidence that JMJD6 is widely involved in the regulation of alternative splicing in conjunction with one of its interacting partners/substrates, U2AF65. Our data suggest that JMJD6 may be a general regulator of the spliceosome. Mechanistically, JMJD6 and U2AF65 appear to co-bind with the pre-mRNAs they regulate, and a subset of JMJD6 and U2AF65 co-regulated alternative splicing events seem to depend on both JMJD6 enzymatic activity and lysine residues K15, K38 and/or K276 in U2AF65, which is consistent with the fact that JMJD6 functions as a lysine hydroxylase targeting U2AF65.
Initially, the RNA-dependent JMJD6 interaction with U2AF65 prompted us to examine a potential splicing program co-regulated by these two proteins. Instead of looking at specific genes as in previous reports (39,41,43,44), we applied a large-scale and robust method for detecting differential expression of mRNA isoforms to uncover such a program. Strikingly, we found that over 74% (P < 0.05) (53% if a more stringent cutoff was taken, P < 0.05 and FC > 2) of JMJD6-regulated alternative splicing events were co-regulated by U2AF65, suggesting a functional interaction between these two proteins. More interestingly, both JMJD6 and U2AF65 seem to preferentially promote exon inclusion, the mechanism for which has been described previously for U2AF65 through genome-wide mapping of U2AF65 binding sites with RNA using CLIP-Seq (21). Although we showed a few pre-mRNAs co-bound by JMJD6 and U2AF65, uncovering the genome-wide map of JMJD6 binding with RNA and therefore the underlying mechanistic insights for JMJD6 regulation of exon inclusion awaits similar approaches. To further demonstrate the functional role of JMJD6 in regulating alternative splicing, we also analyzed RNA samples from brain and kidney of wild type and jmjd6 knock out mice. In accordance with our observation on cultured cells, jmjd6 knockout mice displayed significant changes in a large cohort of alternative splicing events compared to wild type, suggesting the developmental abnormality of jmjd6 knock out mice may be linked to its function in regulated splicing. Future studies will investigate the contribution of specific isoform switching event(s) to a defined developmental defect.
To investigate the mechanism underlying JMJD6 regulated alternative splicing, we first focus on the RNA binding characteristic of JMJD6 revealed by the published structural study (61). Despite we have not yet mapped the exact binding sites, we have used RIP to show that JMJD6 and U2AF65 co-bind multiple pre-mRNAs they jointly regulate. JMJD6 is a member of the superfamily of iron- and 2-oxoglutarate-dependent dioxygenases. What is the ‘true’ substrate of JMJD6 and whether its enzymatic activity is linked to its biological functions has been a major focus for all studies related to this enzyme. We have determined whether the enzymatic activity of JMJD6 is involved in its regulated alternative splicing events by knocking down JMJD6 and then rescuing with JMJD6 (wt) and JMJD6 (H187A) (enzymatically-dead mutant) followed by RASL-Seq analysis. Our data demonstrated that the vast majority of JMJD6-regulated alternative splicing events is dependent on H187. Since H187A mutant also loses its ability for oligomerization, it is possible that the inefficient rescue by this mutant might result from, in addition to enzymatic deficiency, its inability to interact with other proteins, which are functional important in JMJD6-regulated alternative splicing. However, our previous work indicates that most proteins associated with wild-type and mutant (H187A) JMJD6 are nearly identical (31). Among all of the substrates reported for JMJD6 so far, we have focused on hydroxylation on multiple lysine residues in U2AF65 including lysine 15, 38 and 276. Our rescue experiments demonstrated that each of these lysines is involved in a large percentage of U2AF65-regulated alternative splicing events. However, despite these splicing events are co-regulated by JMJD6 and U2AF65, a significant portion of K15, K38 or K276 affected alternative splicing events appear to be independent of JMJD6 enzymatic activity, and the mechanism(s) through which these lysine residues affect alternative splicing remains as an interesting topic for future investigation.
We recently report a new paradigm in gene transcriptional pausing release, in which enhancers (anti-pause enhancers)-bound JMJD6 regulates p-TEFb activation and Pol II promoter-proximal pausing release of a substantial set of genes (31). In the present study, we show that JMJD6 plays a widespread role in alternative splicing. We propose that JMJD6 function in gene transcription and splicing control may account for, at least in part, various developmental abnormalities and pathological complications when it is dysregulated. Elucidation of JMJD6-regulated transcription and splicing programs and the underlying molecular mechanisms has important implications in JMJD6-assocaited human diseases, such as cancer.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Yoshinori Fukui at Kyushu University for kindly providing jmjd6 knockout mouse.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Thousand Young Talents Program Funds; Fundamental Research Funds for the Central University [2013121036, 20720160066 to W.L.]; National Natural Science Foundation of China [31371292, 31422030 and 91440112 to W.L.]; Natural Science Foundation of Fujian Province, China [2015J06007 to W.L.]; NIH [GM049369, HG004659 to X.D.F.]. Funding for open access charge: National Natural Science Foundation of China.
Conflict of interest statement. None declared.
REFERENCES
- 1. Moore M.J., Proudfoot N.J.. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009; 136:688–700. [DOI] [PubMed] [Google Scholar]
- 2. Matlin A.J., Clark F., Smith C.W.. Understanding alternative splicing: towards a cellular code. Nat. Rev. Mol. Cell Biol. 2005; 6:386–398. [DOI] [PubMed] [Google Scholar]
- 3. Jurica M.S., Moore M.J.. Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell. 2003; 12:5–14. [DOI] [PubMed] [Google Scholar]
- 4. Wahl M.C., Will C.L., Luhrmann R.. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009; 136:701–718. [DOI] [PubMed] [Google Scholar]
- 5. Black D.L. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003; 72:291–336. [DOI] [PubMed] [Google Scholar]
- 6. Guth S., Valcarcel J.. Kinetic role for mammalian SF1/BBP in spliceosome assembly and function after polypyrimidine tract recognition by U2AF. J. Biol. Chem. 2000; 275:38059–38066. [DOI] [PubMed] [Google Scholar]
- 7. Singh R., Valcarcel J., Green M.R.. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science. 1995; 268:1173–1176. [DOI] [PubMed] [Google Scholar]
- 8. Valcarcel J., Gaur R.K., Singh R., Green M.R.. Interaction of U2AF65 RS region with pre-mRNA branch point and promotion of base pairing with U2 snRNA [corrected]. Science. 1996; 273:1706–1709. [DOI] [PubMed] [Google Scholar]
- 9. Graveley B.R., Hertel K.J., Maniatis T.. The role of U2AF35 and U2AF65 in enhancer-dependent splicing. RNA. 2001; 7:806–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pan Q., Shai O., Lee L.J., Frey B.J., Blencowe B.J.. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008; 40:1413–1415. [DOI] [PubMed] [Google Scholar]
- 11. Sammeth M., Foissac S., Guigo R.. A general definition and nomenclature for alternative splicing events. PLoS Comput. Biol. 2008; 4:e1000147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. David C.J., Manley J.L.. The search for alternative splicing regulators: new approaches offer a path to a splicing code. Genes Dev. 2008; 22:279–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Skotheim R.I., Nees M.. Alternative splicing in cancer: noise, functional, or systematic?. Int. J. Biochem. Cell Biol. 2007; 39:1432–1449. [DOI] [PubMed] [Google Scholar]
- 14. He C., Zhou F., Zuo Z., Cheng H., Zhou R.. A global view of cancer-specific transcript variants by subtractive transcriptome-wide analysis. PLoS One. 2009; 4:e4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fackenthal J.D., Godley L.A.. Aberrant RNA splicing and its functional consequences in cancer cells. Dis. Models Mech. 2008; 1:37–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cieply B., Carstens R.P.. Functional roles of alternative splicing factors in human disease. Wiley Interdiscipl. Rev. RNA. 2015; 6:311–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hastings M.L., Allemand E., Duelli D.M., Myers M.P., Krainer A.R.. Control of pre-mRNA splicing by the general splicing factors PUF60 and U2AF(65). PLoS One. 2007; 2:e538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Park J.W., Parisky K., Celotto A.M., Reenan R.A., Graveley B.R.. Identification of alternative splicing regulators by RNA interference in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:15974–15979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Moore M.J., Wang Q., Kennedy C.J., Silver P.A.. An alternative splicing network links cell-cycle control to apoptosis. Cell. 2010; 142:625–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wu T., Fu X.D.. Genomic functions of U2AF in constitutive and regulated splicing. RNA Biol. 2015; 12:479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shao C., Yang B., Wu T., Huang J., Tang P., Zhou Y., Zhou J., Qiu J., Jiang L., Li H. et al. Mechanisms for U2AF to define 3΄ splice sites and regulate alternative splicing in the human genome. Nat. Struct. Mol. Biol. 2014; 21:997–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fadok V.A., Bratton D.L., Rose D.M., Pearson A., Ezekewitz R.A., Henson P.M.. A receptor for phosphatidylserine-specific clearance of apoptotic cells. Nature. 2000; 405:85–90. [DOI] [PubMed] [Google Scholar]
- 23. Wang X., Wu Y.C., Fadok V.A., Lee M.C., Gengyo-Ando K., Cheng L.C., Ledwich D., Hsu P.K., Chen J.Y., Chou B.K. et al. Cell corpse engulfment mediated by C. elegans phosphatidylserine receptor through CED-5 and CED-12. Science. 2003; 302:1563–1566. [DOI] [PubMed] [Google Scholar]
- 24. Bose J., Gruber A.D., Helming L., Schiebe S., Wegener I., Hafner M., Beales M., Kontgen F., Lengeling A.. The phosphatidylserine receptor has essential functions during embryogenesis but not in apoptotic cell removal. J. Biol. 2004; 3:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hahn P., Bose J., Edler S., Lengeling A.. Genomic structure and expression of Jmjd6 and evolutionary analysis in the context of related JmjC domain containing proteins. BMC Genomics. 2008; 9:293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cikala M., Alexandrova O., David C.N., Proschel M., Stiening B., Cramer P., Bottger A.. The phosphatidylserine receptor from Hydra is a nuclear protein with potential Fe(II) dependent oxygenase activity. BMC Cell Biol. 2004; 5:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hahn P., Wegener I., Burrells A., Bose J., Wolf A., Erck C., Butler D., Schofield C.J., Bottger A., Lengeling A.. Analysis of Jmjd6 cellular localization and testing for its involvement in histone demethylation. PLoS One. 2010; 5:e13769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cui P., Qin B., Liu N., Pan G., Pei D.. Nuclear localization of the phosphatidylserine receptor protein via multiple nuclear localization signals. Exp. Cell Res. 2004; 293:154–163. [DOI] [PubMed] [Google Scholar]
- 29. Tibrewal N., Liu T., Li H., Birge R.B.. Characterization of the biochemical and biophysical properties of the phosphatidylserine receptor (PS-R) gene product. Mol. Cell. Biochem. 2007; 304:119–125. [DOI] [PubMed] [Google Scholar]
- 30. Chang B., Chen Y., Zhao Y., Bruick R.K.. JMJD6 is a histone arginine demethylase. Science. 2007; 318:444–447. [DOI] [PubMed] [Google Scholar]
- 31. Liu W., Ma Q., Wong K., Li W., Ohgi K., Zhang J., Aggarwal A.K., Rosenfeld M.G.. Brd4 and JMJD6-associated anti-pause enhancers in regulation of transcriptional pause release. Cell. 2013; 155:1581–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Poulard C., Rambaud J., Hussein N., Corbo L., Le Romancer M.. JMJD6 regulates ERalpha methylation on arginine. PLoS One. 2014; 9:e87982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lawrence P., Conderino J.S., Rieder E.. Redistribution of demethylated RNA helicase A during foot-and-mouth disease virus infection: role of Jumonji C-domain containing protein 6 in RHA demethylation. Virology. 2014; 452-453:1–11. [DOI] [PubMed] [Google Scholar]
- 34. Gao W.W., Xiao R.Q., Peng B.L., Xu H.T., Shen H.F., Huang M.F., Shi T.T., Yi J., Zhang W.J., Wu X.N. et al. Arginine methylation of HSP70 regulates retinoid acid-mediated RARbeta2 gene activation. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E3327–E3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wu T.F., Yao Y.L., Lai I.L., Lai C.C., Lin P.L., Yang W.M.. Loading of PAX3 to mitotic chromosomes is mediated by arginine methylation and associated with waardenburg syndrome. J. Biol. Chem. 2015; 290:20556–20564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tikhanovich I., Kuravi S., Artigues A., Villar M.T., Dorko K., Nawabi A., Roberts B., Weinman S.A.. Dynamic arginine methylation of TNF receptor associated factor 6 regulates Toll-like receptor signaling. J. Biol. Chem. 2015; 290:22236–22249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mantri M., Loik N.D., Hamed R.B., Claridge T.D., McCullagh J.S., Schofield C.J.. The 2-oxoglutarate-dependent oxygenase JMJD6 catalyses oxidation of lysine residues to give 5S-hydroxylysine residues. Chembiochem. 2011; 12:531–534. [DOI] [PubMed] [Google Scholar]
- 38. Unoki M., Masuda A., Dohmae N., Arita K., Yoshimatsu M., Iwai Y., Fukui Y., Ueda K., Hamamoto R., Shirakawa M. et al. Lysyl 5-hydroxylation, a novel histone modification, by Jumonji domain containing 6 (JMJD6). J. Biol. Chem. 2013; 288:6053–6062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Webby C.J., Wolf A., Gromak N., Dreger M., Kramer H., Kessler B., Nielsen M.L., Schmitz C., Butler D.S., Yates J.R. 3rd et al. Jmjd6 catalyses lysyl-hydroxylation of U2AF65, a protein associated with RNA splicing. Science. 2009; 325:90–93. [DOI] [PubMed] [Google Scholar]
- 40. Wang F., He L., Huangyang P., Liang J., Si W., Yan R., Han X., Liu S., Gui B., Li W. et al. JMJD6 promotes colon carcinogenesis through negative regulation of p53 by hydroxylation. PLoS Biol. 2014; 12:e1001819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Heim A., Grimm C., Muller U., Haussler S., Mackeen M.M., Merl J., Hauck S.M., Kessler B.M., Schofield C.J., Wolf A. et al. Jumonji domain containing protein 6 (Jmjd6) modulates splicing and specifically interacts with arginine-serine-rich (RS) domains of SR- and SR-like proteins. Nucleic Acids Res. 2014; 42:7833–7850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rahman S., Sowa M.E., Ottinger M., Smith J.A., Shi Y., Harper J.W., Howley P.M.. The Brd4 extraterminal domain confers transcription activation independent of pTEFb by recruiting multiple proteins, including NSD3. Mol. Cell. Biol. 2011; 31:2641–2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Boeckel J.N., Guarani V., Koyanagi M., Roexe T., Lengeling A., Schermuly R.T., Gellert P., Braun T., Zeiher A., Dimmeler S.. Jumonji domain-containing protein 6 (Jmjd6) is required for angiogenic sprouting and regulates splicing of VEGF-receptor 1. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:3276–3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Barman-Aksozen J., Beguin C., Dogar A.M., Schneider-Yin X., Minder E.I.. Iron availability modulates aberrant splicing of ferrochelatase through the iron- and 2-oxoglutarate dependent dioxygenase Jmjd6 and U2AF(65.). Blood Cells Mol. Dis. 2013; 51:151–161. [DOI] [PubMed] [Google Scholar]
- 45. Li M.O., Sarkisian M.R., Mehal W.Z., Rakic P., Flavell R.A.. Phosphatidylserine receptor is required for clearance of apoptotic cells. Science. 2003; 302:1560–1563. [DOI] [PubMed] [Google Scholar]
- 46. Kunisaki Y., Masuko S., Noda M., Inayoshi A., Sanui T., Harada M., Sasazuki T., Fukui Y.. Defective fetal liver erythropoiesis and T lymphopoiesis in mice lacking the phosphatidylserine receptor. Blood. 2004; 103:3362–3364. [DOI] [PubMed] [Google Scholar]
- 47. Lee Y.F., Miller L.D., Chan X.B., Black M.A., Pang B., Ong C.W., Salto-Tellez M., Liu E.T., Desai K.V.. JMJD6 is a driver of cellular proliferation and motility and a marker of poor prognosis in breast cancer. Breast Cancer Res: BCR. 2012; 14:R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chen C.F., Feng X., Liao H.Y., Jin W.J., Zhang J., Wang Y., Gong L.L., Liu J.J., Yuan X.H., Zhao B.B. et al. Regulation of T cell proliferation by JMJD6 and PDGF-BB during chronic hepatitis B infection. Sci. Rep. 2014; 4:6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hu Y.J., Belaghzal H., Hsiao W.Y., Qi J., Bradner J.E., Guertin D.A., Sif S., Imbalzano A.N.. Transcriptional and post-transcriptional control of adipocyte differentiation by Jumonji domain-containing protein 6. Nucleic Acids Res. 2015; 43:7790–7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Poulard C., Rambaud J., Lavergne E., Jacquemetton J., Renoir J.M., Tredan O., Chabaud S., Treilleux I., Corbo L., Le Romancer M.. Role of JMJD6 in breast tumourigenesis. PLoS One. 2015; 10:e0126181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang J., Ni S.S., Zhao W.L., Dong X.C., Wang J.L.. High expression of JMJD6 predicts unfavorable survival in lung adenocarcinoma. Tumour Biol. 2013; 34:2397–2401. [DOI] [PubMed] [Google Scholar]
- 52. Li H., Qiu J., Fu X.D.. RASL-seq for massively parallel and quantitative analysis of gene expression. Current Protocols in Molecular Biology. 2012; doi:10.1002/0471142727.mb0413s98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhou Z., Qiu J., Liu W., Zhou Y., Plocinik R.M., Li H., Hu Q., Ghosh G., Adams J.A., Rosenfeld M.G. et al. The Akt-SRPK-SR axis constitutes a major pathway in transducing EGF signaling to regulate alternative splicing in the nucleus. Mol. Cell. 2012; 47:422–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wei C., Qiu J., Zhou Y., Xue Y., Hu J., Ouyang K., Banerjee I., Zhang C., Chen B., Li H. et al. Repression of the central splicing regulator RBFox2 is functionally linked to pressure overload-induced heart failure. Cell Rep. 2015; doi:10.1016/j.celrep.2015.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Huang da W., Sherman B.T., Lempicki R.A.. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009; 4:44–57. [DOI] [PubMed] [Google Scholar]
- 56. Peritz T., Zeng F., Kannanayakal T.J., Kilk K., Eiriksdottir E., Langel U., Eberwine J.. Immunoprecipitation of mRNA-protein complexes. Nat. Protoc. 2006; 1:577–580. [DOI] [PubMed] [Google Scholar]
- 57. Ule J., Jensen K., Mele A., Darnell R.B.. CLIP: a method for identifying protein-RNA interaction sites in living cells. Methods. 2005; 37:376–386. [DOI] [PubMed] [Google Scholar]
- 58. Zhao J., Ohsumi T.K., Kung J.T., Ogawa Y., Grau D.J., Sarma K., Song J.J., Kingston R.E., Borowsky M., Lee J.T.. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol. Cell. 2010; 40:939–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Pandit S., Zhou Y., Shiue L., Coutinho-Mansfield G., Li H., Qiu J., Huang J., Yeo G.W., Ares M. Jr, Fu X.D.. Genome-wide analysis reveals SR protein cooperation and competition in regulated splicing. Mol. Cell. 2013; 50:223–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Komeno Y., Huang Y.J., Qiu J., Lin L., Xu Y., Zhou Y., Chen L., Monterroza D.D., Li H., DeKelver R.C. et al. SRSF2 is essential for hematopoiesis, and its myelodysplastic syndrome-related mutations dysregulate alternative Pre-mRNA splicing. Mol. Cell. Biol. 2015; 35:3071–3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hong X., Zang J., White J., Wang C., Pan C.H., Zhao R., Murphy R.C., Dai S., Henson P., Kappler J.W. et al. Interaction of JMJD6 with single-stranded RNA. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:14568–14572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wang L., Zhao Z., Meyer M.B., Saha S., Yu M., Guo A., Wisinski K.B., Huang W., Cai W., Pike J.W. et al. CARM1 methylates chromatin remodeling factor BAF155 to enhance tumor progression and metastasis. Cancer Cell. 2014; 25:21–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhang W.J., Wu X.N., Shi T.T., Xu H.T., Yi J., Shen H.F., Huang M.F., Shu X.Y., Wang F.F., Peng B.L. et al. Regulation of transcription factor Yin Yang 1 by SET7/9-mediated lysine methylation. Sci. Rep. 2016; 6:21718. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.