Abstract
Ribosome biogenesis occurs successively in the nucleolus, nucleoplasm, and cytoplasm. Maturation of the ribosomal small subunit is completed in the cytoplasm by incorporation of a particular class of ribosomal proteins and final cleavage of 18S-E pre-rRNA (18S-E). Here, we show that poly(A)-specific ribonuclease (PARN) participates in steps leading to 18S-E maturation in human cells. We found PARN as a novel component of the pre-40S particle pulled down with the pre-ribosome factor LTV1 or Bystin. Reverse pull-down analysis revealed that PARN is a constitutive component of the Bystin-associated pre-40S particle. Knockdown of PARN or exogenous expression of an enzyme-dead PARN mutant (D28A) accumulated 18S-E in both the cytoplasm and nucleus. Moreover, expression of D28A accumulated 18S-E in Bystin-associated pre-40S particles, suggesting that the enzymatic activity of PARN is necessary for the release of 18S-E from Bystin-associated pre-40S particles. Finally, RNase H–based fragmentation analysis and 3΄-sequence analysis of 18S-E species present in cells expressing wild-type PARN or D28A suggested that PARN degrades the extended regions encompassing nucleotides 5–44 at the 3΄ end of mature 18S rRNA. Our results reveal a novel role for PARN in ribosome biogenesis in human cells.
INTRODUCTION
The production of 60S and 40S ribosomal subunits in eukaryotic cells takes place mainly in the nucleolus and nucleoplasm (1). Along this production pathway, multiple pre-ribosomal particles (90S, pre-40S, pre-60S) are formed that transiently associate with hundreds of non-ribosomal factors in the nucleus prior to the formation of the mature 60S and 40S subunits. Eventually, the 60S and 40S subunits are exported to the cytoplasm independently of each other. In yeast cells, the nuclear export of pre-40S and pre-60S particles requires the GTPase Ran, a central component in the nuclear–cytoplasmic exchange apparatus, as well as the exportin Crm1p (2–4). During 60S biogenesis, Nmd3p, which associates with late pre-60S particles, recruits Crm1p (5,6). The nuclear export of pre-40S particles, on the other hand, depends on the ribosomal protein Rps15p (7) in addition to Crm1p, and indeed export is tightly coordinated with pre-rRNA maturation. Crm1p interacts with Ltv1p, which is a component of late pre-40S particles, and shuttles between the nucleus and cytoplasm. Ltv1p is required for efficient nuclear export of pre-40S subunits (8). In human cells, pre-40S particles are exported from the nucleus as an incomplete form containing premature 18S rRNA (18S-E), which has a 3΄-end extension that is cleaved in the cytoplasm; this process is believed to prevent premature translation initiation on pre-mRNAs within the nucleus (1). One of the well-known factors involved in the maturation of 18S-E is the cytoplasmic kinase RIO2, which is a component of late pre-40S particles that are indistinguishable from the late pre-40S particles associated with LTV1 in human cells (9). RIO2 is necessary for the release of ENP1 (Bystin; human homolog of yeast Enp1p) from cytoplasmic pre-40S particles and is essential for the recycling of LTV1 and of the small-subunit factors DIM2 and NOB1 during late 18S-E maturation in human cells (10). In yeast, another kinase Hrr25p (yeast homolog of human CKIε and CKIδ) is also a component of the late pre-40S particles associated with Ltv1p and the protein kinase Rio2p (11), and Hrr25p regulates the maturation of pre-40S particles through phosphorylation of Ltv1p (12). Enp1p is an additional component of late pre-40S particles, and it forms a stable complex with Ltv1p and Rps3p. Phosphorylation of Ltv1p causes their dissociation from late pre-40S particles (12), whereas a lack of phosphorylation induces formation of the characteristic ‘beak’ structure of mature 40S subunits and results in salt-resistant integration of Rps3p into the 40S subunit (9). Like Rio2p, Hrr25p is believed to function at the last step of 40S subunit synthesis; however, Hrr25p is involved in a distinctly different step from that of Rio2p. Minimally, Rio2p does not phosphorylate Ltv1p. Most of those proteins are involved in the nuclear exit of pre-40S subunits and are bound to the subunit region that is involved in the formation of the beak structure (9,11). Thus, 40S subunit maturation seems to occur via multiple steps that are coupled with nuclear export (9,11,12).
In eukaryotic cells, it has long been believed that a premature pre-40S particle is not functional for translation initiation, as mentioned above. Recently, however, several lines of evidence suggest that the translation initiation machinery can recruit pre-40S particles at least under particular conditions—probably as part of quality control during ribosome biogenesis (13–17). Therefore, the final steps of 40S maturation seem to be critical regulatory steps for ribosome production (18).
In this study, we demonstrate that poly(A)-specific ribonuclease (PARN) is a novel trans-acting factor that participates in the processing of pre-18S rRNA at late stages of small-subunit biogenesis in human cells. PARN is a 3΄-5΄ exoribonuclease that has multiple cellular functions: it participates in the degradation of poly(A) tails of mRNAs (19), the maturation of H/ACA box small nucleolar RNAs (20), the 3΄-end maturation of human telomerase RNA (21), and the trimming and metabolism of microRNAs (22,23). In those functions, PARN shows substrate specificity to RNA sequence other than Poly(A) sequence (20–23). Thus, this study describes a novel role for PARN in the maturation of 40S ribosomal subunits.
MATERIALS AND METHODS
Generation of doxycycline-inducible cells expressing an epitope-tagged protein
Flp-In T-Rex 293 cells (Thermo Fisher Scientific Inc.), seeded in 24-well plates, were transfected with 2 μl Lipofectamine 2000 (Thermo Fisher Scientific Inc.) mixed with 0.25 μg of each pcDNA5-FRT/TO construct together with 0.25 μg of vector pOG44 (Thermo Fisher Scientific Inc.). After a 24- to 48-h transfection (90–100% confluency), 100 μg/ml hygromycin B (Invitrogen) was added to DMEM supplemented with fetal bovine serum (Biowest). Thereafter, cells were selected for ∼2 weeks, during which the medium was replaced with fresh medium (with 100 μg/ml hygromycin B) every 2 days. Afterwards, growing cell colonies were expanded in large-scale dishes and used for subsequent analysis.
HA (hemagglutinin) and FLAG (HF)-tag two-step purification of LTV1-HF associated complex
At 48 h after 1 μg/ml doxycycline addition, doxycycline-inducible LTV1-HF-expressing cells and control parental Flp-In T-Rex 293 cells were harvested from ten 90-mm dishes and then washed once with phosphate-buffered saline and lysed by vigorous agitation for 30 s in 4 ml lysis buffer (50 mM Tris–HCl pH 8.0 containing 150 mM NaCl, 0.5% (w/v) IGEPAL-CA630 and 1 mM phenylmethylsulfonyl fluoride) and then incubated on ice for 30 min. The soluble whole-cell lysate was obtained by centrifugation at 20 000 × g for 30 min at 4°C. Whole-cell lysate was incubated with 40 μl anti-FLAG M2 agarose beads for 3 h at 4°C. After washing the beads five times with 1 ml lysis buffer and once with 50 mM Tris–HCl pH 8.0 containing 150 mM NaCl, the complexes bound to the beads were eluted with 500 μg/ml FLAG peptide. Eluates were then subjected to hemagglutinin (HA)-based immunoprecipitation. In brief, FLAG eluates were diluted in 400 μl lysis buffer and allowed to bind to anti-HA-conjugated agarose beads for 3 h at 4°C. Beads were washed five times with 1 ml lysis buffer, and then the complexes bound to the beads were eluted with 2× RNA extraction solution (10 mM Tris–HCl pH 8.0, 350 mM NaCl, 1% SDS, 7M Urea and 10 mM EDTA). Eluted complexes were used for experimentation.
Protein identification by liquid chromatography–coupled tandem mass spectrometry (LC-MS/MS)
Proteins were dissolved in Laemmli buffer and subjected to SDS-PAGE. Each gel band was cut into five pieces, each of which was subjected to in-gel digestion with trypsin (24). The resulting peptides were analyzed using a nanoscale LC-MS/MS system with a Q-TOF2 hybrid mass spectrometer (Q-Tof 2, Micromass Wythenshawe, UK) as described (25,26).
Construction of enzymatically inactive PARN
To generate an enzymatically inactive (enzyme-dead) PARN mutant bearing the mutation D28A (27,28), mutagenesis was performed with a PCR-based method in combination with a methylated DNA–specific endonuclease. In brief, PCR was executed using mutagenesis primer sets 5΄-CTTCTTCGCCATCGCTGGGGAGTTTTCAG-3΄ and 5΄-CTGAAAACTCCCCAGCGATGGCGAAGAAG-3΄ with the template pcDNA5-FRT/TO-PARN-Myc or the template pcDNA5-FRT/TO-HEF (hemagglutinin/TEV protease recognition sequence/FLAG)-PARN. The PCR product was digested with the methylated DNA-specific endonuclease, DpnI, to eliminate template DNA that contained methylated nucleotides. The resulting plasmid was amplified in DH5α Escherichia coli competent cells. Mutation was verified by DNA sequencing.
RNA interference
HeLa cells were cultured in 35-mm dishes until they reached 80% confluency and then transfected with 50 nM of each stealth small interfering RNA (siRNA) using Lipofectamine 2000, according to manufacturer's protocol (Thermo Fisher Scientific). The following stealth siRNA sequences were used: 5΄-GAGCUCUGUCCUAUGUAUCUCCUAA-3΄ and 5΄-UUAGGAGAUACAUAGGACAGAGCUC-3΄ for PARN knockdown, and 5΄-GAGUGUCAUCCAUGUCUCUCUCUAA-3΄ and 5΄-UUAGAGAGAGACAUGGAUGACACUC-3΄ for the negative control of PARN. The cells were transferred to 35- or 90-mm dishes 24 h after transfection. The transfected cells were then cultured until the desired time point in DMEM supplemented with 10% fetal bovine serum.
RNase H targeting
RNA precipitated from complexes was digested with RNase H to remove DNA/RNA hybrids. Briefly, RNA was dissolved in 20 mM Tris–HCl containing 10 mM MgCl2, 100 mM KCl, and 0.1 mM DTT with or without targeting probe 18S (1800–1823 of 18S rRNA: 5΄-ACTTCCTCTAGATAGTCAAGTTCG-3΄). For hybridization, each reaction was incubated at 65°C for 10 min, then incubated at 37°C for 5 min for annealing. Then, 1 U RNase H (Invitrogen) was added and incubated at 37°C for 30 min. RNA was precipitated with isopropanol, subjected to electrophoresis through a denaturing gel containing 9% acrylamide and 7.5 M urea, and the analyzed with northern blotting using probes 5΄ITS1 and 18S-3΄.
RESULTS
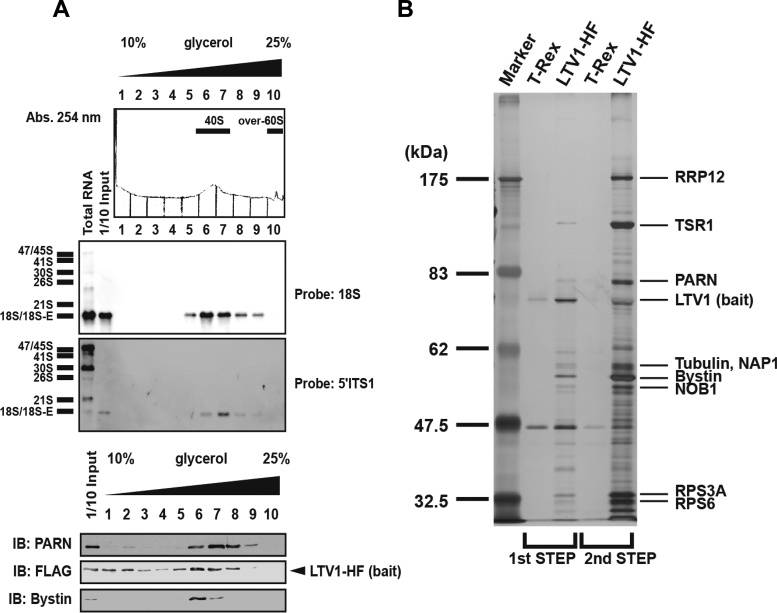
To examine the late steps of pre-18S rRNA maturation in human cells, we isolated pre-40S particles by tandem affinity purification with LTV1 that was expressed from the HF-tagged protein-coding transgene integrated into a common locus in the Flp-In T-Rex 293 genome. The isolated LTV1-HF–associated pre-40S particles predominantly contained 18S rRNA and/or 18S-E pre-rRNA as detected by ethidium bromide staining and northern blotting with DNA probes corresponding to nucleotide residues 358–382 (18S) and 1864–1891 (5΄ITS1; ITS = internal transcribed spacer) in 47S pre-rRNA (Supplementary Figure S1A–C). Upon fractionation by glycerol-gradient centrifugation (Figure 1A), the particles were found mainly in fractions 6 and 7 with an apparent sedimentation coefficient of 40S (40S fraction). These fractions contained 18S/18S-E and, as expected, Bystin (Figure 1A).
Figure 1.
Purification of human LTV1-HF-associated pre-40S particles using two-step affinity purification. (A) The LTV1-associated complex prepared by two-step purification was separated into 10 fractions by glycerol gradient centrifugation (10–25% glycerol, top). The eluate was monitored at 254 nm. RNAs in each fraction were detected by northern blotting with a probe for 18S or 5΄ITS1. RNA sizes are indicated to the left. Proteins eluted in each fraction were detected by immunoblotting (IB) with an antibody against PARN, FLAG, or Bystin. (B) LTV1-associated proteins isolated from T-Rex or T-Rex cells expressing LTV1 (LTV1-HF) were separated by SDS-PAGE and visualized by silver staining. Proteins were identified by LC–MS/MS after in-gel trypsinization. Molecular mass markers (kDa) are indicated to the left, and the identified protein names are shown on the right. Lane 1, marker proteins; lane 2, control for the first step of purification; lane 3, proteins isolated by the first step of purification (10%); lane 4, control for the second step of purification and lane 5, proteins isolated by the second step of purification (100%).
MS-based analyses of the isolated LTV1-associated pre-40S particles identified seven trans-acting factors known to be involved in 40S biogenesis [TSR1, Bystin, RRP12, NOB1, DIM2, RIOK2 and C21orf70 (FAM207A)] (Figure 1B, Supplementary Tables S1 and S2) (10,29) as well as four rRNA processing factors [NCL, UTP14A, NOL6 (UTP22) and DDX21 (Guα)] and four RNA helicases [DDX9/DHX9, DDX5, DDX41 and DDX57 (DHX57)] (Supplementary Tables S1 and S2). Interestingly, the analyses also identified PARN as an additional component of the LTV1-associated pre-40S particles (Supplementary Tables S1 and S2, Figure 1B). PARN co-eluted with 18S-E in the centrifugation fractions, with a sedimentation coefficient slightly greater than the 40S fraction (Figure 1A).
To determine the 3΄-terminal sequence of 18S-E present in the LTV1-associated pre-40S particles, we prepared 3΄-terminal RNA fragments by RNase H degradation of RNAs extracted from the LTV1-associated pre-40S particles using a DNA oligonucleotide complementary to the sequence 1800–1823 of 18S rRNA as a guide sequence and carried out northern blotting with probes 18S-3΄ and 5΄ITS1 (Supplementary Figure S1D). We assumed that the longest fragment generated from 18S-E by this RNase H degradation had about 120 bases based on the report by Preti et al., in which site E cleavages were found to occur at residues 78 and 81 downstream of 3΄ end of mature 18S (31). However, we detected three RNA fragments of size ∼50, ∼60 and ∼90 ribonucleotide residues, which we designated pre-18S rRNAs containing 18S’, 18S'E1 and 18S'E2, respectively (Supplementary Figure S1E). SYBR Gold staining indicated that LTV1 was predominantly associated with 18S'E1 (Supplementary Figure S1E). The ∼50-base RNase H fragment (18S’) was detected unambiguously in total RNA, suggesting that this fragment was generated from mature 18S rRNA. An RNase H–based analysis of Bystin-associated pre-40S particles yielded similar results (Supplementary Figure S1E), suggesting that 18S'E1 is also the predominant pre-RNA in Bystin-associated pre-40S particles.
PARN is a component of both LTV1- and Bystin-associated pre-40S particles
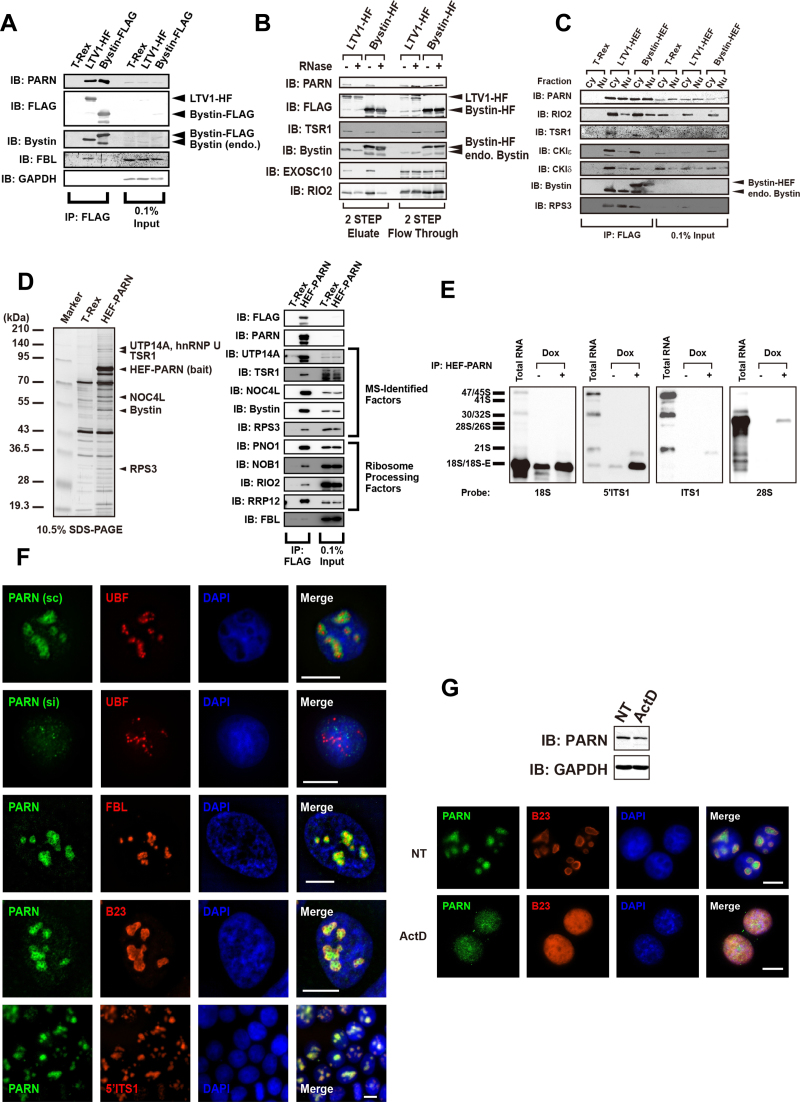
Immunoblotting using an antibody against PARN confirmed the MS result that PARN is a component of LTV1-associated pre-40S particles (Figure 2A). RNase treatment of the isolated pre-40S particles indicated that the association of LTV1 or Bystin with PARN was RNA dependent, whereas the binding of LTV1 or Bystin to RIO2 was RNA independent (Figure 2B). In addition, PARN was found associated with Bystin and LTV1 almost equally in the cytoplasmic and the nuclear extracts (Figure 2C), whereas RIO2, CKIε and CKIδ were predominantly associated with Bystin and LTV1 in the cytoplasm (9–11,30) (Figure 2C). The 18S-E was also associated with Bystin or LTV1 in both the cytoplasm and nucleus (Supplementary Figure S2A). Collectively, these results suggested that PARN is a component of both the LTV1- and Bystin-associated pre-40S particles present in both the cytoplasm and nucleus.
Figure 2.
Identification of PARN as a component of pre-40S particles present in the cytoplasm and nucleus. (A) Proteins immunoprecipitated (IP) with LTV1-HF or Bystin-FLAG as affinity bait were visualized by immunoblotting (IB) with antibodies against the proteins indicated to the left. Proteins in input (0.1%) are also indicated. (B) LTV1- or Bystin-associated pre-40S particles were isolated with (+) or without (–) RNase A treatment before the second-step elution from anti-HA-conjugated beads. Proteins associated with LTV1 or Bystin were detected by immunoblotting with antibodies against the proteins indicated to the left. (C) LTV1- or Bystin-associated pre-40S particles were isolated from cytoplasmic extract (Cy) or nuclear extract (Nu). Antibodies used for immunoblotting are indicated to the left. (D) Proteins pulled down with HEF-PARN as affinity bait were separated by SDS-PAGE and visualized with silver staining. The names of the proteins identified by MS are indicated to the right. Molecular mass markers are indicated to the left. HEF-PARN–associated proteins were also detected by immunoblotting with the antibodies against the proteins indicated. (E) At 48 h after induction with doxycycline (Dox), ribonucleoproteins (RNPs) were pulled down from extracts of doxycycline-inducible Flp-In T-Rex 293 cells expressing HEF-PARN that were treated with (+) or without (–) doxycycline. RNAs extracted from the HEF-PARN complex were separated by 0.8% agarose gel electrophoresis and detected by northern blotting with the DNA probes indicated under each blot. RNA sizes are indicated to the left. (F) PARN (green), UBF (red), fibrillarin (FBL; red) and B23 (red) were detected by immunocytochemistry with antibodies against the corresponding proteins. Staining of PARN with the antibody was reduced in siRNA (si)-treated 293EBNA cells but not in scRNA (sc)-treated 293EBNA control cells. DAPI was used to stain DNA in the nucleus; the nucleolus was stained weakly with DAPI. Cy3-labeled 5΄ITS1 probe was used for FISH. (G) HeLa cells were treated with 1 μg/ml actinomycin D (ActD) for 2 h. PARN (green) and B23 (red) were delocalized from the nucleolus upon treatment with ActD and dispersed into the nucleoplasm. PARN was detected by immunoblotting before and after ActD treatment of HeLa cells.
To confirm the association of PARN with pre-40S particles, we performed a reverse pull-down assay with PARN as affinity bait and identified Bystin in the PARN-associated particles by MS-based and immunoblot analyses (Figure 2D and Supplementary Table S2). Although these analyses failed to detect LTV1, we confirmed the association of PARN with LTV1 by transient overexpression of LTV1-Myc in HEF-PARN-expressing cells (Supplementary Figure S2B), suggesting that PARN preferentially associates with Bystin rather than LTV1 in the cells. In addition, the MS-based analysis identified TSR1, UTP14A, and NOC4L (Figure 2D and Supplementary Table S3). PARN was also associated with 18S-E, and weakly with 21S pre-rRNA (21S), as determined by northern blotting (Supplementary Figure S1A and Figure 2E), though this 21S was smaller than that present in total RNA. Collectively, those data suggest that the PARN may be involved in both the early and late steps of 40S biogenesis.
We performed immunocytochemical analysis of PARN using anti-PARN; its specificity was confirmed using stealth siRNA–transfected cells and scrambled RNA (scRNA)-transfected control cells (Figure 2F). PARN co-localized partly with fibrillarin and B23, which are components of the dense fibrillar and granular compartments, respectively, of the nucleolus, whereas upstream binding factor (UBF), a component of the fibrillar center, was surrounded by PARN in the nucleolus (Figure 2F). We also found that PARN co-localized with pre-rRNAs hybridized with a 5΄ITS1 probe as assessed with fluorescence in situ hybridization (FISH) (Figure 2F). The staining with the 5΄ITS1 probe was no longer seen after RNase treatment of cells, confirming the specificity of the FISH analysis (Supplementary Figure S2B). In addition, upon treatment with actinomycin D, PARN was excluded from the nucleolus and was dispersed throughout the nucleoplasm (Figure 2G), which was similar to the behavior of B23, a nucleolar marker that re-localizes to the nucleoplasm in response to actinomycin D (25); notably, the total amount of PARN in the cells was not significantly affected (Figure 2G). Thus, PARN behaves in vivo as a component of pre-ribosomal particles in all aspects examined.
PARN deficiency results in the accumulation of 18S-E in both the cytoplasm and nucleus
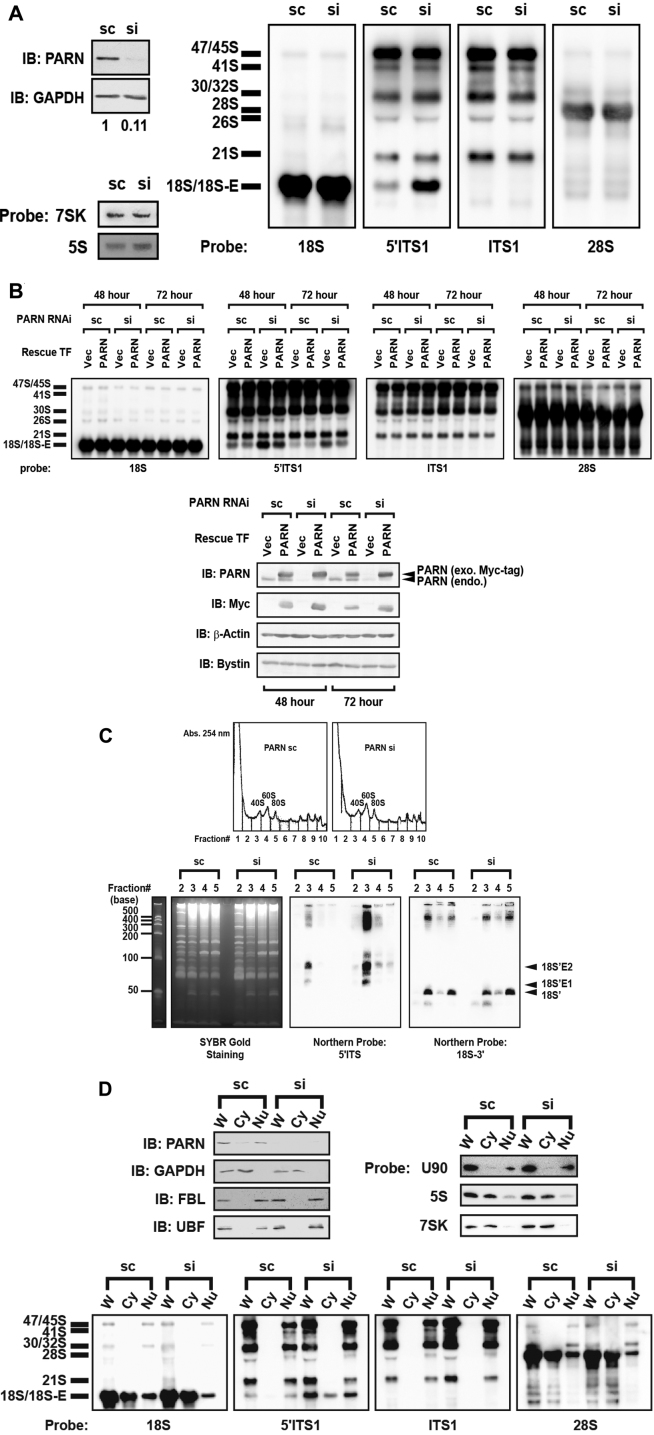
To assess the possible involvement of PARN in 40S ribosome biogenesis, we examined the effects of siRNA-mediated knockdown of PARN on cell proliferation and processing of pre-rRNAs in 293EBNA and HeLa cells. The cell proliferation assay with carboxyfluorescein diacetate ester showed that the knockdown slowed cell division (Supplementary Figure S3A), suggesting that PARN is required for cell proliferation. We next examined pre-rRNA processing by northern blotting with DNA probes for 28S, 18S, 5.8S, ITS1 and 5΄ITS1 (Supplementary Figure S1B). The highest molecular size precursor detected corresponded to a mixture of 45S and 47S pre-rRNAs that were detected strongly with probes 5΄ITS1 and ITS1 (Figure 3A and Supplementary Figure S3B). The 30S and 32S pre-rRNAs (indicated as 30S/32S in Figure 3A and Supplementary Figure S3B) were produced by cleavage of the 45S/47S RNAs in the 3΄ part of the ITS1 (site 2 in Supplementary Figure S1B); the 30S pre-rRNA was detected with probes 18S, 5΄ITS1, and ITS1 and the 32S pre-rRNA with probe 28S (Supplementary Figure S1B, Figure 3A, and Supplementary Figure S3B). Two other major 18S rRNA precursors (18S-E and 21S) were detected with probe 5΄ITS1; 18S-E was produced by cleavage at both sites 1 and E whereas 21S by cleavage at both 1 and C or 1 and 2 (Supplementary Figure S1B). The 21S was detected with probes 5΄ITS1 and ITS1 and seen above the 18S rRNA that was detected with probe 5΄ITS1 but not with ITS1 (Figure 3ASupplementary Figures S1B and S3B). Knockdown of PARN accumulated 18S-E in both 293EBNA and HeLa cells but appeared not to significantly affect the levels of 21S compared with cells treated with control scRNA, whereas PARN knockdown did not affect the production of the RNA polymerase III–transcribed RNAs (5S rRNA or 7SK RNA) in either cell line (Figure 3A and Supplementary Figure S3B). This 18S-E accumulation was partially rescued by transfection of cells with a siRNA-resistant plasmid encoding PARN (Figure 3B). In addition, the RNase H targeting method combined with northern blotting with probes 5΄ITS1 and 18S-3΄ revealed that PARN knockdown accumulated 18S'E1, 18S'E2, and other intermediate fragments mostly in the 40S fraction that had been separated by sucrose gradient centrifugation of whole-cell extracts, whereas the method using the 18S-3΄ probe detected the 18S’ fragment predominantly in the 40S and 80S fractions (Figure 3C). Finally, we prepared cytoplasmic and nuclear extracts from the scRNA- or siRNA-treated cells and detected pre-rRNAs by northern blotting. This analysis indicated that PARN knockdown accumulated 18S-E in both the cytoplasmic and nuclear extracts with preferential accumulation in the nucleus (Figure 3D).
Figure 3.
Involvement of PARN in processing of 18S-E. (A) RNAs in HeLa cells were detected by northern blotting with probes for 18S, 5΄ITS1, 28S and ITS1 after treatment with control scRNA (sc) or siRNA (si). rRNAs were separated via 0.8% agarose gel electrophoresis. Northern blotting was also done using probes for 7SK and 5S. Cellular PARN and GAPDH were detected by immunoblotting (IB). (B) RNAs were extracted from HeLa cells that had been transfected (TF) with scRNA or siRNA for PARN knockdown. The cells were co-transfected with empty vector (Vec) or expression vector containing an siRNA-resistant sequence encoding PARN for the rescue experiment for 48 and 72 h. RNAs were detected by northern blotting with the probes indicated under each blot. Proteins were detected by immunoblotting with the antibodies indicated. (C) RNase H targeting analysis was carried out in combination with sucrose gradient centrifugation. Extracts prepared from scRNA-treated or siRNA-treated cells were subjected to sucrose gradient centrifugation. RNAs that were collected from the 40S, 60S or 80S fraction were subjected to RNase H targeting analysis followed by SYBR Gold staining or northern blotting with probes for 5΄ITS1 and 18S-3΄. The RNA fragments generated by RNase H (18S'E1, 18S'E2, 18S’) are indicated to the right. RNA sizes (bases) are indicated to the left. (D) Northern blotting was carried out for RNAs extracted from the cytoplasmic (Cy) and nuclear (Nu) fractions prepared from scRNA- or siRNA-treated cells. The analyses were also carried out for RNAs prepared from whole-cell extract (W). The DNA probes used are indicated below the corresponding blots. RNAs were separated by 0.8% agarose gel electrophoresis for subsequent northern blotting analyses. To assign cell fractions, immunoblotting was carried out with antibodies against PARN, GAPDH, fibrillarin (FBL) and UBF as well as northern blotting with probes for U90, 7SK and 5S RNAs. For these purposes, proteins were subjected to SDS-PAGE and RNAs to electrophoresis through a denaturing urea-PAGE gel.
Expression of an enzyme-dead mutant of PARN accumulates 18S-E on Bystin-associated pre-40S particles
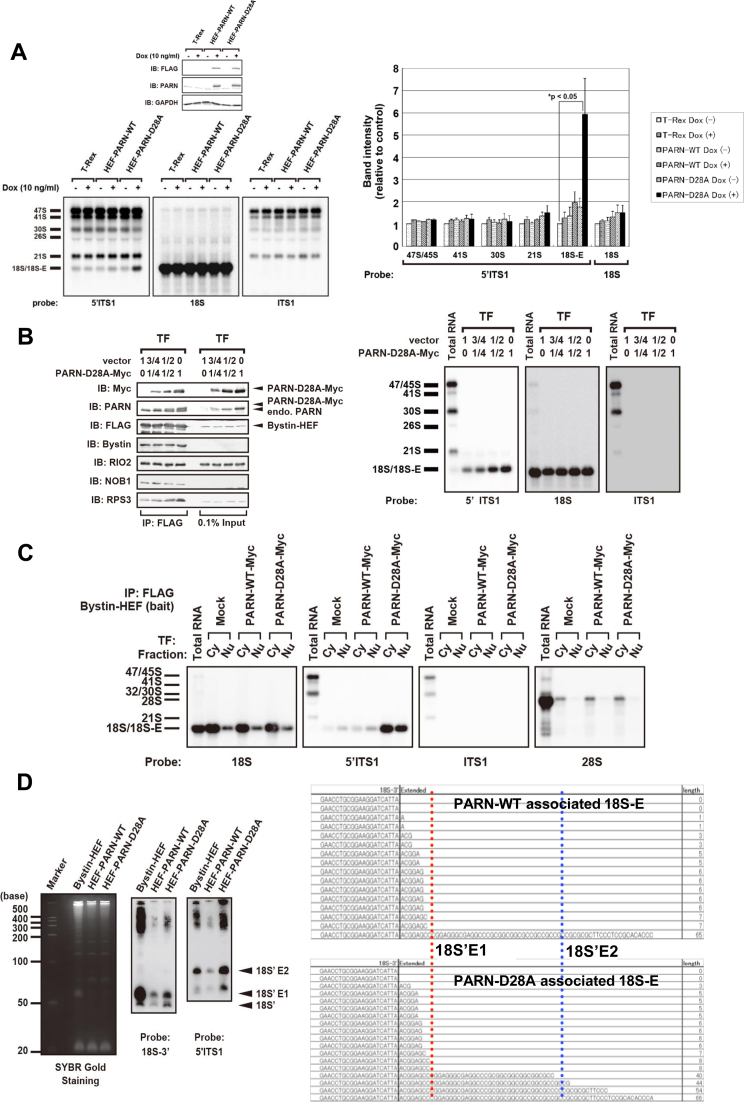
To examine the possible requirement for PARN enzymatic activity in the processing of 18S-E, we constructed a doxycycline-inducible cell line expressing an enzyme-dead mutant of HEF-PARN, named PARN-D28A, in which PARN aspartic acid residue 28 was replaced with alanine (27,28). We assumed that, if PARN enzymatic activity is required for 18S-E processing, the expression of PARN-D28A should cause accumulation of 18S-E in the cell via a dominant-negative effect. As expected, the expression of PARN-D28A resulted in the accumulation of cellular 18S-E compared with cells that expressed wild-type PARN (PARN-WT) or cells that were not induced with doxycycline (Figure 4A). We also noted that the expression of PARN-D28A did not affect the level of 21S compared with cells expressing PARN-WT (Figure 4A). Also, as expected, PARN-D28A did not rescue the effect of siRNA-mediated knockdown of PARN on 18S-E but rather resulted in additive accumulation of 18S-E (Supplementary Figure S4A), supporting strongly that PARN enzymatic activity is required for 18S-E processing.
Figure 4.
Effects of PARN-D28A on binding of 18S-E to Bystin-associated pre-40S particles. (A) RNAs extracted from Flp-In T-Rex 293 cells expressing PARN-WT or PARN-D28A that were induced with (+) or without (–) doxycycline (Dox) were analyzed by northern blotting with the probes indicated below of the corresponding blots. The bar graph shows the proportion of staining intensity for each pre-rRNA relative to the corresponding pre-rRNA of control Flp-In T-Rex 293 cells. Data represent the mean ± SEM of three independent experiments. The P value was calculated with the unpaired t-test. Immunoblotting (IB) was carried out with the antibodies indicated. (B) The plasmid encoding PARN-D28A was mixed with vector pcDNA3.1 (proportion: 0:1, 1:4, 1:2 or 1:0) and transfected into Flp-In T-Rex 293 cells expressing Bystin-HEF. Pre-40S particles were immunoprecipitated (IP) with Bystin-HEF as affinity bait, and the protein components were detected by immunoblotting with the antibodies indicated or RNA components were detected by northern blotting with probes for 18S ITS1 and 5΄ITS1. (C) Bystin-HEF–expressing cells were transiently transfected with empty vector (Mock) or vector expressing PARN-WT-Myc or PARN-D28A-Myc. Cells were fractionated, and the Bystin-HEF–associated pre-40S complex was immunoprecipitated with anti-FLAG–conjugated beads from cytoplasmic (Cy) or nuclear (Nu) extract. RNA associated with Bystin-HEF in each cellular fraction was separated by 0.8% agarose gel electrophoresis and detected by northern blotting with the probes indicated. (D) RNA fragments prepared from PARN-WT– or PARN-D28A–associated pre-40S particles were detected by SYBR Gold staining (left) or northern blotting with a probe for 18S-3΄ or 5΄ITS1 after denaturing urea-PAGE (7.5 M urea, 9% acrylamide gel) after targeted RNase H digestion as described in Supplementary Figure S1D. Arrows indicate RNA fragments originating from 18S’, 18S'E1, and 18S'E2 produced by RNase H digestion. Right: 3΄-RACE analysis of PARN-WT– or PARN-D28A–associated pre-18S rRNA. The 3΄-end sequences of the pre-18S rRNAs associated with PARN-WT or PARN-D28A were determined by 3΄-RACE.
Given that PARN was found to associate preferentially with Bystin-associated pre-40S particles (Figure 2D), we explored a possible role for PARN in the function of Bystin-associated pre-40S particles. We isolated Bystin-associated pre-40S particles from extracts of T-Rex 293 cells stably expressing HEF-tagged Bystin (Bystin-HEF), with transient expression of PARN-D28A or PARN-WT. This analysis showed that PARN-D28A, via increased binding to pre-40S particles, caused accumulation of 18S-E on Bystin-associated pre-40S particles (Figure 4B), whereas PARN-WT did not cause such accumulation (Supplementary Figure S4B). In addition, this 18S-E accumulation occurred in both the cytoplasm and nucleus upon expression of PARN-D28A but not PARN-WT (Figure 4C and Supplementary Figure S4C). Collectively, these data suggested that PARN enzymatic activity is required for dissociation of 18S-E from Bystin-associated pre-40S particles in both the cytoplasm and nucleus.
Finally, based on our result that the association between PARN and Bystin is RNA dependent (Figure 2B), we characterized the 18S-E molecules associated with PARN-D28A or PARN-WT via targeted RNase H cleavage. We found that 18S'E2 associated with PARN-D28A preferentially over PARN-WT (Figure 4D). Nucleotide sequence analysis revealed that the PARN-D28A–associated 18S-E contained sequence extensions of 3–8, 40–54 and 66 nucleotides in the ITS1 at the 3΄ end of 18S rRNA, whereas the PARN-WT–associated 18S-E had sequence extensions of 3–8 and 65 nucleotides but lacked the molecular species with the extension of 40–54 nucleotides (Figure 4D). Thus, we concluded that PARN cleaves 18S'E2 to yield 18S'E1 on Bystin-associated pre-40S.
DISCUSSION
Our results demonstrate that PARN has a previously unknown role in regulating pre-rRNA processing at late stages of 40S subunit maturation in human cells. This conclusion was drawn based on our four principal findings: (i) PARN is a component of LTV1- and Bystin-associated pre-40S particles that contain 18S-E; (ii) PARN associates reciprocally with Bystin and 18S-E; (iii) PARN localizes in nucleoli along with 5΄ITS1 probe-reactive pre-rRNAs, and PARN is excluded from nucleoli upon inhibition of transcription with actinomycin D and (iv) PARN deficiency results in the accumulation of 18S-E.
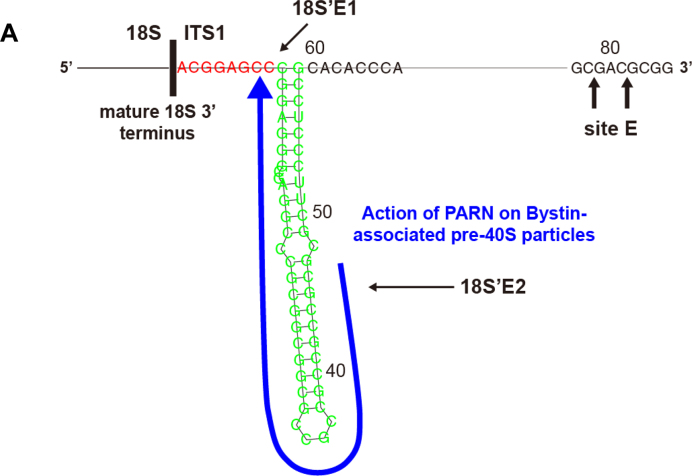
PARN deficiency induced accumulation of 18S-E primarily in the nucleus rather than the cytoplasm (Figure 3D). This is in contrast to the effect of RIO2 deficiency, which results in 18S-E accumulation mainly in the cytoplasm (10), suggesting that PARN initiates 18S-E processing in the nucleus and thus functions at an earlier stage than RIO2. This is consistent with our result that Bystin localized in the nucleus, including the nucleolus. In addition, the analysis using the PARN knockdown cells (Figure 3A–D) or cells expressing PARN-D28A (Figure 4A–D) suggests that PARN is involved in the processing of 18S'E2 (∼+40–44) to yield 18S'E1 (∼+5–7) on Bystin-associated pre-40S particles (Figure 5).
Figure 5.
Schematic representation of PARN action during 18S-E processing. (A) The secondary structure was predicted by an RNA folding program (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi). The 3'-terminal sequence of the mature 18S rRNA and the oligonucleotide sequences corresponding to the 3΄ region of 18S'E1 and 18S'E2 are indicated. The arrow denotes the proposed region processed by PARN.
Recently, Preti et al. (31) proposed that 18S-E is processed gradually to produce mature 18S by the action of 3΄-5΄ exonucleases. Their results from a 3΄-RACE (rapid amplification of cDNA ends) analysis revealed that 18S-E ends at the approximate position +35–40 and, at higher frequency, at position +24 in total RNA of HeLa cells; interestingly, however, the same analysis also revealed that 18S-E ends at the approximate position +42–45 of 18S-E present in the nucleolus and the nucleus (where its level is higher) whereas it ends at approximately +24 in the cytoplasm. Our present data are consistent with their observations and extends their proposal; namely, PARN precedes the processing of 18S'E2 that pauses at ∼+40–44 during the presumptive exo- and/or endonucleolytic processing to produce 18S'E1 in the nucleolus or nucleus (Figure 5). In turn, this suggests that the action of PARN halts at ∼+5–7 (corresponding to 18S'E1) (Figure 5). Our present data also suggest that this processing occurs on Bystin-pre-40S particles. The further maturation of 18S'E1 may take place by NOB1-mediated cleavage at the 3΄ end of 18S in the cytoplasm (32) or by processing with other 3΄-5΄ exonucleases. Because RIO2 is required for the processing of pre-18S-E, which contains 20- to 30-nucleotide extensions at the 3΄ end of ITS1 (1), it will be interesting to address whether RIO2 and PARN act in the same or different pathways during 18S-E maturation.
Given the report by Preti et al (31) that 18S-E cleaved at site E (+78 or +81) is present in the nucleolus, it is possible that PARN can participate in the processing of this 18S-E molecular species. This is consistent with our result that PARN is present in the nucleolus (Figure 2F). In this case, it is intriguing to speculate that PARN associates with Bystin after processing of 18S-E to 18S'E2. In addition, PARN is associated with the 21S with smaller size (Figure 2E), despite that the deficiency of PARN did not affect the level of 21S (Figure 3A). PARN may be recruited to pre-40S particles formed at early stages of 40S biogenesis in the nucleolus. Since involvement of exosome in the processing of 21S has been reported (31–33), it is likely that PARN acts after the processing of 21S by the exosome. However, we do not exclude a possibility that PARN is involved in processing of the smaller 21S in the nucleolus. Those possibilities may be clarified by further analyses of pre-40S particles present in the nucleolus.
In yeast, under certain conditions, the pre-40S small subunit is incorporated into the translation initiation machinery to initiate translation (13–17). This incorporation has been proposed to be a critical step in determining the cytoplasmic fate of 20S; i.e. the pre-rRNA is processed further to produce mature 18S rRNA or is degraded by the so-called ‘no-go’ RNA decay machinery (18). Recently, Strunk et al. proposed a detailed model in which pre-40S particles are checked by translation-like cycle via formation of at least eight intermediates including ones joining 60S and pre-40S in the cytoplasm before the maturation of 40S ribosome subunit in yeast cells (34). In this model, conversion of intermediate I containing minimally six late factors including Enp1p and Ltv1p to intermediate II lacking Ltv1p, is a rate-limiting step in cytoplasmic 40S ribosome maturation. During this conversion step, Hrr25 causes dissociation of Ltv1p via its phosphorylation and coincidences incorporation of Rps3 into Enp1-containing pre-40S particles (12). Given our results that the enzyme-dead mutant of PARN accumulates 18S'E2 and Rps3 on Bystin-associated pre-40S particle, it is intriguing to speculate that PARN is involved in the rate-limiting step in the conversion of the human counterparts of those yeast intermediates during 40S ribosome maturation in human cells. In addition, extended 3΄end region of pre-18S rRNA is expected to locate around the mRNA entry side on yeast pre-40S particle before Nob1p cleavage and/or formation of translation-like pre-40S–60S complex (35), it is also tempting to speculate that extended 3΄end region of human 18S'E2 is involved in formation or inhibition of translation-like pre-40S–60S complex in human cells and that action of PARN is required for steps dissociating pre-40S from translation-like pre-40S–60S complex or producing mature 18S by the action of NOB1.
Given that 18S-E is the human counterpart of yeast 20S present in the cytoplasm, our study suggests that human cells utilize a similar but more complicated mechanism than yeast cells to determine the fate of pre-40S small subunits. Our present results also suggest that PARN maintains a steady-state cellular level of 18S-E via its catalytic activity toward 18S-E, thereby implying a role for 18S-E in determining the fate of pre-40S small subunits.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Sally-Fujiyama-Nakamura for her technical assistance and discussion.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Core Research for Evolutional Science and Technology (CREST) from the Japan Science and Technology Agency (JST) [13415564]; Grant-in-Aid for Scientific Research, Ministry of Education, Culture, Sports, Science & Technology of Japan (MEXT) [24241075]; Global Innovation Research Organization of Tokyo University of Agriculture & Technology. Funding for open access charge: CREST/JST.
Conflict of interest statement. None declared.
REFERENCES
- 1. Rouquette J., Choesmel V., Gleizes P.E.. Nuclear export and cytoplasmic processing of precursors to the 40S ribosomal subunits in mammalian cells. EMBO J. 2005; 24:2862–2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hurt E., Hannus S., Schmelzl B., Lau D., Tollervey D., Simos G.. A novel in vivo assay reveals inhibition of ribosomal nuclear export in ran-cycle and nucleoporin mutants. J. Cell. Biol. 1999; 144:389–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Moy T.I., Silver P.A.. Nuclear export of the small ribosomal subunit requires the ran-GTPase cycle and certain nucleoporins. Genes Dev. 1999; 13:2118–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gleizes P.E., Noaillac-Depeyre J., Leger-Silvestre I., Teulieres F., Dauxois J.Y., Pommet D., Azum-Gelade M.C., Gas N.. Ultrastructural localization of rRNA shows defective nuclear export of preribosomes in mutants of the Nup82p complex. J. Cell Biol. 2001; 155:923–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ho J.H., Kallstrom G., Johnson A.W.. Nmd3p is a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit. J. Cell Biol. 2000; 151:1057–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gadal O., Strauss D., Petfalski E., Gleizes P.E., Gas N., Tollervey D., Hurt E.. Rlp7p is associated with 60S preribosomes, restricted to the granular component of the nucleolus, and required for pre-rRNA processing. J. Cell Biol. 2002; 157:941–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Leger-Silvestre I., Milkereit P., Ferreira-Cerca S., Saveanu C., Rousselle J.C., Choesmel V., Guinefoleau C., Gas N., Gleizes P.E.. The ribosomal protein Rps15p is required for nuclear exit of the 40S subunit precursors in yeast. EMBO J. 2004; 23:2336–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Seiser R.M., Sundberg A.E., Wollam B.J., Zobel-Thropp P., Baldwin K., Spector M.D., Lycan D.E.. Ltv1 is required for efficient nuclear export of the ribosomal small subunit in Saccharomyces cerevisiae. Genetics. 2006; 174:679–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schafer T., Maco B., Petfalski E., Tollervey D., Bottcher B., Aebi U., Hurt E.. Hrr25-dependent phosphorylation state regulates organization of the pre-40S subunit. Nature. 2006; 441:651–655. [DOI] [PubMed] [Google Scholar]
- 10. Zemp I., Wild T., O'Donohue M.F., Wandrey F., Widmann B., Gleizes P.E., Kutay U.. Distinct cytoplasmic maturation steps of 40S ribosomal subunit precursors require hRio2. J. Cell Biol. 2009; 185:1167–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schafer T., Strauss D., Petfalski E., Tollervey D., Hurt E.. The path from nucleolar 90S to cytoplasmic 40S pre-ribosomes. EMBO J. 2003; 22:1370–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ghalei H., Schaub F.X., Doherty J.R., Noguchi Y., Roush W.R., Cleveland J.L., Stroupe M.E., Karbstein K.. Hrr25/CK1δ-directed release of Ltv1 from pre-40S ribosomes is necessary for ribosome assembly and cell growth. J. Cell Biol. 2015; 208:745–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ford C.L., Randal-Whitis L., Ellis S.R.. Yeast proteins related to the p40/laminin receptor precursor are required for 20S ribosomal RNA processing and the maturation of 40S ribosomal subunits. Cancer Res. 1999; 59:704–710. [PubMed] [Google Scholar]
- 14. Jakovljevic J., de Mayolo P.A., Miles T.D., Nguyen T.M., Leger-Silvestre I., Gas N., Woolford J.L. Jr. The carboxy-terminal extension of yeast ribosomal protein S14 is necessary for maturation of 43S preribosomes. Mol. Cell. 2004; 14:331–342. [DOI] [PubMed] [Google Scholar]
- 15. Ferreira-Cerca S., Poll G., Gleizes P.E., Tschochner H., Milkereit P.. Roles of eukaryotic ribosomal proteins in maturation and transport of pre-18S rRNA and ribosome function. Mol. Cell. 2005; 20:263–275. [DOI] [PubMed] [Google Scholar]
- 16. Granneman S., Nandineni M.R., Baserga S.J.. The putative NTPase Fap7 mediates cytoplasmic 20S pre-rRNA processing through a direct interaction with Rps14. Mol. Cell. Biol. 2005; 25:10352–10364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lacombe T., Garcia-Gomez J.J., de la Cruz J., Roser D., Hurt E., Linder P., Kressler D.. Linear ubiquitin fusion to Rps31 and its subsequent cleavage are required for the efficient production and functional integrity of 40S ribosomal subunits. Mol. Microbiol. 2009; 72:69–84. [DOI] [PubMed] [Google Scholar]
- 18. Soudet J., Gelugne J.P., Belhabich-Baumas K., Caizergues-Ferrer M., Mougin A.. Immature small ribosomal subunits can engage in translation initiation in Saccharomyces cerevisiae. EMBO J. 2010; 29:80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gao M., Fritz D.T., Ford L.P., Wilusz J.. Interaction between a poly(A)-specific ribonuclease and the 5΄ cap influences mRNA deadenylation rates in vitro. Mol. Cell. 2000; 5:479–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Berndt H., Harnisch C., Rammelt C., Stohr N., Zirkel A., Dohm J.C., Himmelbauer H., Tavanez J.P., Huttelmaier S., Wahle E.. Maturation of mammalian H/ACA box snoRNAs: PAPD5-dependent adenylation and PARN-dependent trimming. RNA. 2012; 18:958–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nguyen D., Grenier St-Sauveur V., Bergeron D., Dupuis-Sandoval F., Scott M.S., Bachand F.. A Polyadenylation-dependent 3΄ end maturation pathway is required for the synthesis of the human telomerase RNA. Cell Rep. 2015; 13:2244–2257. [DOI] [PubMed] [Google Scholar]
- 22. Katoh T., Hojo H., Suzuki T.. Destabilization of microRNAs in human cells by 3΄ deadenylation mediated by PARN and CUGBP1. Nucleic Acids Res. 2015; 43:7521–7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yoda M., Cifuentes D., Izumi N., Sakaguchi Y., Suzuki T., Giraldez A.J., Tomari Y.. Poly(A)-specific ribonuclease mediates 3΄-end trimming of Argonaute2-cleaved precursor microRNAs. Cell Rep. 2013; 5:715–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yanagida M., Miura Y., Yagasaki K., Taoka M., Isobe T., Takahashi N.. Matrix assisted laser desorption/ionization-time of flight-mass spectrometry analysis of proteins detected by anti-phosphotyrosine antibody on two-dimensional-gels of fibrolast cell lysates after tumor necrosis factor-alpha stimulation. Electrophoresis. 2000; 21:1890–1898. [DOI] [PubMed] [Google Scholar]
- 25. Fujiyama-Nakamura S., Yoshikawa H., Homma K., Hayano T., Tsujimura-Takahashi T., Izumikawa K., Ishikawa H., Miyazawa N., Yanagida M., Miura Y. et al. . Parvulin (Par14), a peptidyl-prolyl cis-trans isomerase, is a novel rRNA processing factor that evolved in the metazoan lineage. Mol. Cell Proteomics. 2009; 8:1552–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kaji H., Kamiie J., Kawakami H., Kido K., Yamauchi Y., Shinkawa T., Taoka M., Takahashi N., Isobe T.. Proteomics reveals N-linked glycoprotein diversity in Caenorhabditis elegans and suggests an atypical translocation mechanism for integral membrane proteins. Mol. Cell Proteomics. 2007; 6:2100–2109. [DOI] [PubMed] [Google Scholar]
- 27. Kim J.H., Richter J.D.. Opposing polymerase-deadenylase activities regulate cytoplasmic polyadenylation. Mol. Cell. 2006; 24:173–183. [DOI] [PubMed] [Google Scholar]
- 28. Ren Y.G., Martinez J., Virtanen A.. Identification of the active site of poly(A)-specific ribonuclease by site-directed mutagenesis and Fe(2+)-mediated cleavage. J. Biol. Chem. 2002; 277:5982–5987. [DOI] [PubMed] [Google Scholar]
- 29. Wyler E., Zimmermann M., Widmann B., Gstaiger M., Pfannstiel J., Kutay U., Zemp I.. Tandem affinity purification combined with inducible shRNA expression as a tool to study the maturation of macromolecular assemblies. RNA. 2011; 17:189–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zemp I., Wandrey F., Rao S., Ashiono C., Wyler E., Montellese C., Kutay U.. CK1delta and CK1epsilon are components of human 40S subunit precursors required for cytoplasmic 40S maturation. J. Cell Sci. 2014; 127:1242–1253. [DOI] [PubMed] [Google Scholar]
- 31. Preti M., O'Donohue M.F., Montel-Lehry N., Bortolin-Cavaille M.L., Choesmel V., Gleizes P.E.. Gradual processing of the ITS1 from the nucleolus to the cytoplasm during synthesis of the human 18S rRNA. Nucleic Acids Res. 2013; 41:4709–4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sloan K.E., Mattijssen S., Lebaron S., Tollervey D., Pruijn G.J., Watkins N.J.. Both endonucleolytic and exonucleolytic cleavage mediate ITS1 removal during human ribosomal RNA processing. J. Cell Biol. 2013; 200:577–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tafforeau L., Zorbas C., Langhendries J.L., Mullineux S.T., Stamatopoulou V., Mullier R., Wacheul L., Lafontaine D.L.. The complexity of human ribosome biogenesis revealed by systematic nucleolar screening of Pre-rRNA processing factors. Mol. Cell. 2013; 51:539–551. [DOI] [PubMed] [Google Scholar]
- 34. Strunk B.S., Novak M.N., Young C.L., Karbstein K.. A translation-like cycle is a quality control checkpoint for maturing 40S ribosome subunits. Cell. 2012; 150:111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lebaron S., Schneider C., van Nues R.W., Swiatkowska A., Walsh D., Böttcher B., Granneman S., Watkins N.J., Tollervey D.. Proofreading of pre-40S ribosome maturation by a translation initiation factor and 60S subunits. Nat. Struct. Mol. Biol. 2012; 19:744–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.