Abstract
The exosome complex is a major eukaryotic exoribonuclease that requires the SKI complex for its activity in the cytoplasm. In yeast, the Ski7 protein links both complexes, whereas a functional equivalent of the Ski7 has remained unknown in the human genome.
Proteomic analysis revealed that a previously uncharacterized short splicing isoform of HBS1L (HBS1LV3) is the long-sought factor linking the exosome and SKI complexes in humans. In contrast, the canonical HBS1L variant, HBS1LV1, which acts as a ribosome dissociation factor, does not associate with the exosome and instead interacts with the mRNA surveillance factor PELOTA. Interestingly, both HBS1LV1 and HBS1LV3 interact with the SKI complex and HBS1LV1 seems to antagonize SKI/exosome supercomplex formation. HBS1LV3 contains a unique C-terminal region of unknown structure, with a conserved RxxxFxxxL motif responsible for exosome binding and may interact with the exosome core subunit RRP43 in a way that resembles the association between Rrp6 RNase and Rrp43 in yeast. HBS1LV3 or the SKI complex helicase (SKI2W) depletion similarly affected the transcriptome, deregulating multiple genes. Furthermore, half-lives of representative upregulated mRNAs were increased, supporting the involvement of HBS1LV3 and SKI2W in the same mRNA degradation pathway, essential for transcriptome homeostasis in the cytoplasm.
INTRODUCTION
Messenger RNA (mRNA) molecules exist largely to produce proteins. Since steady-state mRNA levels are determined by the rates of synthesis and decay, cytoplasmic degradation of mRNA plays a fundamental role in the regulation of gene expression.
Multiple enzymes, particularly nucleases, participate in eukaryotic mRNA decay. Nucleases involved in 3΄-5΄ cytoplasmic mRNA degradation pathways can act individually (e.g. the DIS3L2 protein) (1,2), or be constituents of multisubunit complexes. The exosome is a major multisubunit nuclease that catalyzes the 3΄-5΄ degradation of RNA in both the nucleus and the cytoplasm (3,4).
The exosome comprises a nine-subunit, ring-shaped, catalytically inactive core that associates with one (EXOSC10 [RRP6] or DIS3 or DIS3L) or two (EXOSC10 and DIS3) 3΄-5΄ exoribonucleases (5,6). Several functional isoforms of the exosome are found in human cells (7) – each has distinct catalytic subunits and occupies a different intracellular location. The 10-subunit isoform encompassing the ring and only one nuclease – the EXOSC10 protein – is localized in the nucleoli. Meanwhile, the 11-subunit exosome complex has a nucleoplasmic localization and consists of the ring, EXOSC10 and an additional nuclease, DIS3. However, a small fraction of complexes associated with DIS3 and EXOSC10 proteins can also be found in the cytoplasm. The main cytoplasmic isoform containing a DIS3 paralogue, the DIS3-like protein (DIS3L), participates in 3΄-5΄ mRNA degradation in the cytoplasmic compartment (1,5).
The substrates of exosome complexes containing DIS3L or DIS3 pass through the central channel of the exosome ring to reach the exoribonuclease active site (8–12). Although the exosome complex efficiently degrades both single-stranded and partially double-stranded RNA substrates in vitro, studies in yeast showed that cooperation between the exosome and the SKI complex is indispensable for efficient cytoplasmic mRNA decay in vivo (13–15). The yeast SKI complex was initially identified as being essential for degradation of virus-like particles (16) and was since characterized as a hetero-tetramer composed of Ski2, Ski3 and two copies of the Ski8 protein. Through its helicase activity, Ski2 is the catalytic subunit of the SKI complex and forms part of the structure that contains the central channel. Ski3 and Ski8 are RNA-binding proteins that have regulatory functions (17). RNA substrates are delivered through the SKI complex structure directly to the central channel of the exosome ring (17).
Our current knowledge about the SKI complex in humans is very limited. The human orthologues of yeast SKI2, SKI3 and SKI8 genes are SKIV2L, TTC37 and WDR61, respectively (18). Mutations in TTC37 and SKIV2L cause the extremely rare congenital bowel disorder known as trichohepatoenteric syndrome (19,20). Moreover, the product of the SKIV2L gene (the SKI2W protein) is an important contributor to the regulation of the innate immune response that detects foreign nucleic acids (21). Despite the association between the SKI complex and genetic diseases, the exosome/SKI-dependent RNA decay pathways and RNA substrates that are targeted by this supercomplex have not been extensively studied in human cells. In particular, little is known about the mechanism that ensures the physical interaction between the human SKI and exosome complexes.
The yeast SKI complex interacts with the exosome via the auxiliary Ski7 protein, which is an integral component of the cytoplasmic exosome isoform (14,22,23). The phenotype of SKI7 deletion mutants is indistinguishable from that resulting from deletion of genes encoding SKI complex subunits, and leads to inactivation of cytoplasmic exosome-dependent mRNA turnover and quality control pathways as well as to elevated copy numbers of yeast virus-like particles (13,16,24,25). Yeast Ski7 contains domains that share homology with the Hbs1, a GTPase that interacts with Dom34 (PELOTA in humans). This two-subunit complex, structurally similar to eRF3-eRF1, is responsible for dissociation of stalled ribosomes (26). Hbs1 is also a ribosomal surveillance factor, involved in no-go and non-stop mRNA decay (26,27). Due to structural differences between the N-terminal regions of yeast Ski7 and Hbs1 proteins, only Ski7 interacts with the exosome core (14), whereas Hbs1 participates in translation-dependent RNA quality control pathways in Saccharomyces cerevisiae (28).
Since it was thought that no apparent orthologue of the yeast Ski7 protein is encoded in the human genome, the mechanism of cooperation between the SKI and exosome complexes in human cells remained unclear. Here, we show that a protein encoded by a short splicing isoform of HBS1L that contains a specific new region links the SKI complex with the exosome, thus functioning as a Ski7 analogue in humans.
MATERIALS AND METHODS
Cell culture
HEK293 Flp-In T-REx and derived stable cell lines were grown in cell culture dishes in Dulbecco's modified Eagle's medium (Gibco) supplemented with 10% fetal bovine serum (Gibco) at 37°C in a humidified 5% CO2 atmosphere.
Generation of stable cell lines
Stable inducible cell lines producing proteins of interest were obtained using the Flp-In T-REx system (Invitrogen) according to the manufacturer's instructions.
siRNA transfection
siRNA-mediated knockdowns were performed using siRNA (Eurogentec) and Lipofectamine RNAiMAX Reagent (Invitrogen) according to the manufacturer's instructions. Test and negative control siRNA molecules were used at a final concentration of 25 nM. To induce production of exogenous protein variants, cells were grown in the presence of doxycycline for an additional 48 h before harvesting.
The siRNA sequences that specifically target endogenous HBS1L, HBS1LV3 and SKIV2L mRNA are listed below. siRNA oligonucleotide sequences against HBS1LV1, SKIV2L and scrambled siRNA were described previously (29,30). siRNA oligonucleotides against HBS1LV3 were designed by Eurogentec.
siRNAs:
| HBS1LV1 siRNA | 5΄-r(CCAGUAGAUUCCCAGACAU)d(TT)-3΄ |
| #1 SKIV2L siRNA | 5΄-r(GGAGAUAGACUUUGAGAAA)d(TT)-3΄ |
| #2 SKIV2L siRNA | 5΄-r(GCCUUAGCUGUAUGUUGGA)d(TT)-3΄ |
| #1 HBS1LV3 siRNA | 5΄-r(CCUGUCACAAUUAGCAAAU)d(TT)-3΄ |
| #2 HBS1LV3 siRNA | 5΄-r(GUCAUUGGCAUUUCAUAAA)d(TT)-3΄ |
| #3 HBS1LV3 siRNA | 5΄-r(GGCAUUUCAUAAAGCUUCU)d(TT)-3΄ |
| scrambled siRNA | 5΄-r(UUCUUCGAACGUGUCACGU)d(TT)-3΄ |
We established HBS1LV1 siRNA, #2 SKIV2L siRNA and #3 HBS1LV3 siRNA as the most effective, and we used them in all consecutive experiments shown in this study.
Transient transfection with plasmid vectors, cross-linking and co-immunoprecipitation (Co-IP) assay
For transient transfection, HEK293 Flp-In T-REx cell lines were cultured as previously described and transfected using TransIT-2020 reagent (Mirus) according to the manufacturer's instructions. Cross-linking was performed using dithiobis[succinimidyl propionate] (DSP; final conc. 2 mM) (Pierce) according to the manufacturer's protocol. Immunoprecipitation was based on a previously described method (31) with modifications:
Expression of exogenous genes was induced simultaneously with transient transfection by the addition of tetracycline at a final concentration of 0.1 μg/ml. Following a 48 h incubation, cells were scraped off the plates and pelleted at 400 × g for 3 min. Pellets were resuspended in 0.7 ml lysis buffer (100 mM NaCl; 20 mM HEPES-KOH pH = 7.1; 3 mM MgCl2; 1 mM PMSF; 20 nM pepstatin; 6 nM leupeptin; 2 ng/ml chymostatin; 10% glycerol; 0.5% NP-40) and incubated for 30 min at 4°C in the presence of 0.1 mg/ml RNase A (Sigma-Aldrich). Lysates were then sonicated at 4°C in a BioRuptor sonicator (Diagenode) using 25 pulses of 25 s at setting H (interrupted by 25 s pauses) and centrifuged at 15 000 × g for 15 min at 4°C. Cleared extracts were mixed with 50–100 μl aliquots of magnetic beads (CNBr-activated SepFast Mag4F; Biotoolomics) coated with GFP-trap nanobodies (home-made) (32) freshly washed with lysis buffer and incubated for 1.5 h at 4°C with rotation. Immunoprecipitation was followed by five washes with washing buffer (0.5 M or 0.1 M NaCl; 20 mM HEPES-KOH pH = 7.1; 3 mM MgCl2; 1 mM PMSF; 20 nM pepstatin; 6 nM leupeptin; 2 ng/ml chymostatin; 10% glycerol; 0.5% NP-40) and one wash with lysis buffer (see above). Two methods of protein complex elution and analysis were used: (i) Protein complexes were eluted for 5 min at 95°C in sodium dodecylsulphate (SDS) sample buffer (64 mM Tris-HCl pH = 6.8; 15% 2-mercaptoethanol; 10% glycerol; 2% SDS; 0.1% bromophenol blue) and then analyzed by SDS-PAGE followed by Western blot analysis, or (ii) Protein complexes were eluted for 10 min at 95°C in stripping buffer (50 mM Tris-HCl pH 8.0; 50 mM DTT; 1 mM PMSF; 20 nM pepstatin; 6 nM leupeptin; 2 ng/ml chymostatin; 10% glycerol, 3% SDS), precipitated using a standard chloroform–methanol protocol (33) and analyzed by High Resolution Mass Spectrometry.
Mass spectrometry analysis
Precipitated proteins were dissolved in 100 μl of 100 mM ammonium bicarbonate buffer, reduced in 100 mM DTT for 30 min at 57°C, alkylated in 55 mM iodoacetamide for 40 min at RT in the dark and digested overnight with 10 ng/ml trypsin (Promega) at 37°C. Finally, trifluoroacetic acid was added at a final concentration of 0.1%. MS analysis was performed by LC-MS in the Laboratory of Mass Spectrometry (IBB PAS, Warsaw) using a nanoAcquity UPLC system (Waters) coupled to a LTQ-Orbitrap Velos mass spectrometer (Thermo Scientific). Peptides were separated by a 180 min linear gradient of 95% solution A (0.1% formic acid in water) to 35% solution B (acetonitrile and 0.1% formic acid). The measurement of each sample was preceded by three washing runs to avoid cross-contamination. The final MS washing run was searched for the presence of cross-contamination between samples. If the protein of interest was identified in the washing run and in the next measured sample at the same or smaller intensity, then the sample was treated as cross-contamination. These samples were excluded from final graphs.
The mass spectrometer was operated in the data-dependent MS-MS2 mode, and data were acquired in the m/z range of 300–2000. Data were analyzed with the Max-Quant (Version 1.5.3.12) platform. The reference human proteome database from UniProt was used, and HBS1L protein sequences were removed and replaced with unique sequence fragments of HBS1LV1 and HBS1LV3. Label-Free-Quantification (LFQ) intensity values were calculated using the MaxLFQ algorithm. Identified proteins were analyzed as follows. Protein abundance was defined as the signal intensity calculated by MaxQuant software for a protein (sum of intensities of identified peptides of given protein) divided by its molecular weight. Specificity was defined as the ratio of the protein LFQ intensity measured in the bait purification to background level (i.e. protein LFQ intensity in the negative control purification with the background level arbitrarily set to 1 for proteins not detected in the negative control). High values of both protein abundance and specificity indicated proteins that were enriched in the purification, and thus suggested interactions. The exosome, the SKI complex components and HBS1L proteins are included in the Figure 1A, B, E, F and Supplementary Figure S1. Extended data are presented in the Supplementary Figure S2. Since the HBS1LV1 protein was identified in a washing run before the HBS1LV3 co-immunoprecipitation (Co-IP) measurement, the HBS1LV1 hit was deemed an artefact and as such was removed from Figure 1E.
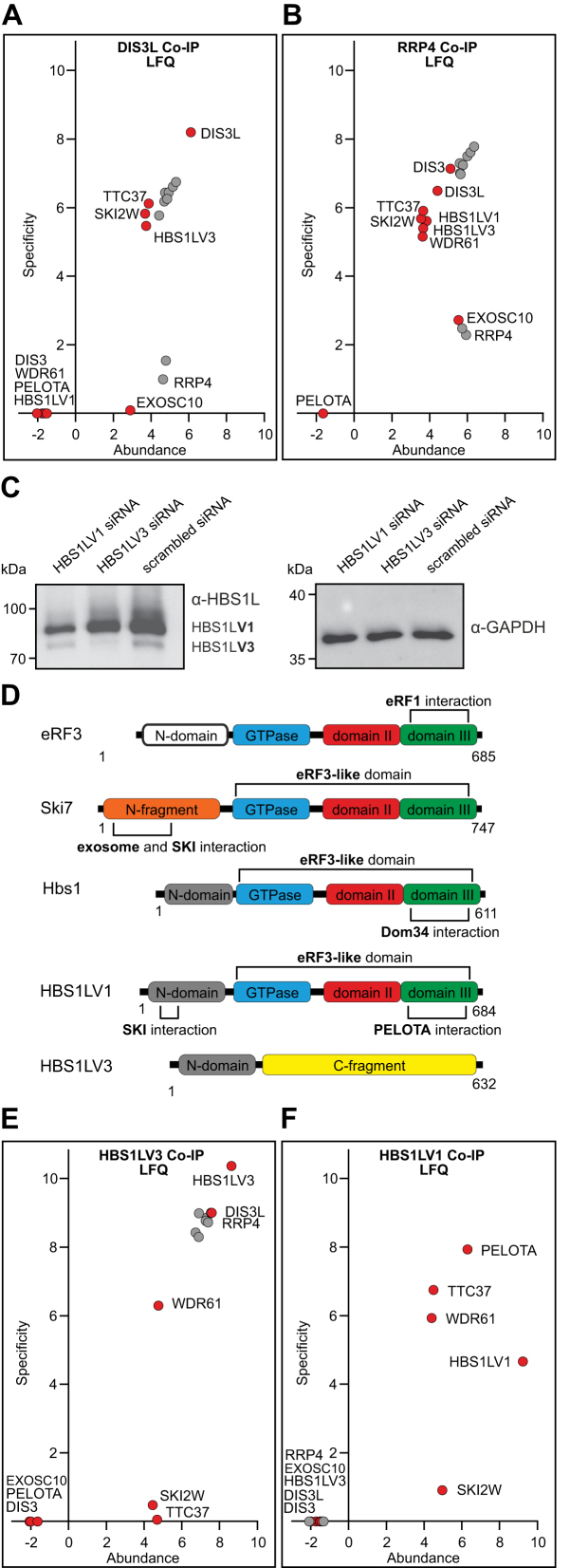
Figure 1.
The cytoplasmic exosome complex interacts with HBS1LV3 but not HBS1LV1 in human cells. Semi-quantitative analysis of co-immunoprecipitation-mass spectrometry (Co-IP-MS) experiments using EGFP-tagged (A) DIS3L, (B) RRP4, (E) HBS1LV3 or (F) HBS1LV1 proteins as baits under high salt washing buffer conditions. Plots present the exosome and SKI complex subunits identified in Co-IP-MS experiments while other interactors are listed in Data set S1 and visualized in Supplementary Figure S2. DSP cross-linker was used in experiments (A) and (B) at the final concentration of 2 mM. The exosome core components are colored grey. Estimated quantities of identified proteins were calculated using the Label Free Quantification (LFQ) algorithm and are represented graphically. Protein abundance was calculated as LFQ intensity of the protein signal divided by its molecular weight and is shown on the x-axis in a logarithmic scale (log10). Specificity was defined as the ratio of protein LFQ intensity measured in the bait Co-IP to the level of the background (protein LFQ intensity in HEK293 T-REx sample), and shown on the y-axis in a logarithmic scale. (C) Western blot showing HBS1LV1 and HBS1LV3 proteins in HEK293 cells. siRNA-mediated protein depletion confirms specific HBS1L isoforms. siRNAs specific to HBS1LV1 or HBS1LV3 were used. Proteins were analyzed using antibodies against the N-terminus of the HBS1L and α-GAPDH antibodies (loading control). Positions of the HBS1LV1 and HBS1LV3 proteins are indicated. (D) Schematic representation of the domain architecture of eRF3, Ski7 and Hbs1 from S. cerevisiae and human HBS1L isoforms. Protein regions sharing similar functions are shown in the same color. Two independent regions within the N-terminal fragment of Ski7, shown in orange, are known to interact with the exosome and the SKI complex. The N-domains of eRF3 and Hbs1 are colored white and grey, respectively. The GTPase domain is shown in blue. Domains II and III of the eRF3-like domain are colored red and green, respectively. The C-terminal fragment of HBS1LV3 is colored yellow.
Real-Time quantitative PCR (qPCR) validation
RNA for quantitative PCR (qPCR) was isolated from HEK293 Flp-In T-REx cell lines induced with doxycycline (0.1 μg/ml), using TRI Reagent (Sigma-Aldrich) according to the manufacturer's instructions. To eliminate genomic DNA, 10 μg of total RNA was treated with 4U TURBO DNase (Ambion) and incubated at 37°C for 2 h. RNA was then extracted using phenol:chloroform and precipitated with isopropanol. cDNA synthesis was performed using 2 μg of DNase-treated RNA, 250 ng random hexamers, 50 pmol oligo(dT) primer and 200 U SuperScript III Reverse Transcriptase (Life Technologies). qPCR reactions were performed according to the manufacturer's guidelines using 20 ng of the cDNA and a Roche LightCycler 480 system. Analyses were performed in triplicate. Reactions without reverse transcriptase (negative controls) were also carried out and showed an insignificant background. The specificity of each reaction was confirmed by a melting curve analysis. GAPDH mRNA was used for normalization. XIST and MALAT1 long non-coding RNAs were used for RNAi normalization because RNA-seq data showed that their levels were unaffected by HBS1LV3 depletion.
Bioinformatic analysis
Ribo-depleted total RNA was used to prepare strand-specific libraries (dUTP RNA) that were subsequently processed using an Illumina HiSeq sequencing platform in the 75-nt pair-end mode. Experiments were performed in triplicate and reads were mapped to the reference human genome (hg38) using the STAR short read aligner (version STAR_2.4.0b) (34). Quality control, read processing and filtering, visualization of the results and counting of reads for the Genecode v22 comprehensive annotation were performed using custom scripts and elements of the RSeQC, BEDtools and SAMtools packages. Differential expression analyses were performed using the DESeq2 Bioconductor R package (35).
Localization studies
Stable cell lines expressing EGFP-tagged HBS1LV1, HBS1LV3 or SKI2W were cultured on Lab-Tek II Chamber Slides (Thermo Scientific) coated with poly-L-lysine (Sigma-Aldrich), for 24 h in the presence of tetracycline (0.1 μg/ml). Cells were incubated with nucleic acid stain Hoechst 33342 (Invitrogen) at a final concentration of 1 μg/ml for 30 min, then washed two times with phosphate buffered saline (PBS) and fixed with 3.7% formaldehyde for 30 min at room temperature. Cells were gently washed three times with PBS and then mounted in ProLong Gold antifade reagent (Invitrogen). Laser scanning confocal microscopy imaging was performed on a FV1000 confocal system (Olympus) with a 60x/1.4 oil immersion objective.
Co-sedimentation assay
Cell pellets were resuspended in 0.5 ml lysis buffer (50 mM NaCl; 20 mM HEPES-KOH pH = 7.1; 3 mM MgCl2; 1 mM PMSF; 20 nM pepstatin; 6 nM leupeptin; 2 ng/ml chymostatin; 5% glycerol; 0.5% NP-40) and incubated for 30 min at 4°C in the presence of 0.1 mg/ml RNase A (Sigma-Aldrich). Lysates were then sonicated at 4°C in a BioRuptor sonicator (Diagenode) using 25 pulses of 25 s at setting H (with 25 s intervals), and centrifuged at 15 000 × g for 15 min at 4°C. Cleared extracts were loaded onto 13 ml continuous 10–30% glycerol gradients containing 50 mM NaCl, 20 mM HEPES-KOH pH = 7.1, 3 mM MgCl2, 1 mM PMSF, 20 nM pepstatin, 6 nM leupeptin, 2 ng/ml chymostatin and centrifuged at 274 000 × g for 18 h at 4°C; fractions were collected using an ÄKTA FPLC machine. Collected fractions were precipitated with trichloroacetic acid (final concentration 10%) overnight. Samples were then centrifuged at 15 000 × g for 15 min at 4°C, washed with 1 ml of cold acetone and centrifuged again under the same conditions. Proteins were resuspended in SDS sample buffer (64 mM Tris-HCl pH = 6.8, 15% 2-mercaptoethanol, 10% glycerol, 2% SDS, 0.1% bromophenol blue) and used for Western blotting analysis.
Structural analysis
HBS1LV3 helix models were created and visualized using Coot (36) and PyMOL (The PyMOL Molecular Graphics System, Schrödinger, LLC). Initial rigid-body docking was performed using the ZDOCK server (37) in an unbiased way (i.e. without selection of blocking and contacting residues). For homology modeling, available structures of S. cerevisiae Rrp43 in complex with Rrp6 were used (PDB codes: 4IFD, 4OO1, 5C0W, 5C0X) (9,38). In this study we also used the human exosome structure (PDB code: 2NN6) (39).
Western blot analysis
Samples were processed using α-HBS1L (1:2000; HPA029729; Sigma-Aldrich), α-EXOSC3 (1:3000; 15062-1-AP; Proteintech Europe), α-SKI2W (1:600; 11462-1-AP; Proteintech Europe), α-DIS3L (1:2000; HPA041805 Sigma-Aldrich), α-GFP (1:2000; sc-9996; Santa Cruz) and α-GAPDH (1:5000, NB300-327; Novus Biologicals) antibodies. Primary antibodies were detected with goat α-rabbit or α-mouse secondary antibodies (Calbiochem) conjugated with horseradish peroxidase and visualized using an Immun-Star WesternC Chemiluminescence Kit (Bio-Rad) according to the manufacturer's protocol. Signals from ECL substrates were detected and documented using X-ray film (developed with AGFA CP1000) or by the charge-coupled device camera in the FluorChem SP Superior Performance Imaging System (Alpha Innotech).
Oligonucleotides
Oligonucleotides used in cloning, site-directed mutagenesis, qPCR and Northern blotting are listed in Supplementary Tables S1, S2, S3 and S4, respectively.
Northern blot analysis
Total RNA was extracted using the standard TRI Reagent protocol (Sigma-Aldrich), according to manufacturer's instructions. RNA (13 μg) was separated by electrophoresis on a 1% denaturing formaldehyde-agarose gel. RNA was then transferred onto Hybond N+ (GE Healthcare) membranes by overnight capillary transfer in 20x SSC solution (3 M NaCl, 0.3 M sodium citrate) and fixed on the membrane by UV cross-linking at 254 nm. Hybridization procedures were carried out in PerfectHyb Plus hybridization buffer (Sigma-Aldrich). The blots were hybridized overnight at 63°C with respective PCR-based probes (see Supplementary Table S4 for primers used for amplifications with human cDNA as a template), labeled by random-priming with [α-32P]dATP and a DecaLabel DNA Labeling Kit (Thermo Scientific) according to the manufacturer's instructions. After hybridization, membranes were washed twice with 2x SSC, 0.1% SDS at 63°C for 45 min and exposed to a Phosphorimager screen (FujiFilm), which was then scanned using a FLA7000 scanner (FujiFilm). Between hybridizations, probes were stripped off the membranes at 65°C using boiling 0.1% SDS (two washes, 45 min each).
RESULTS
A short isoform of HBS1L interacts with the exosome complex
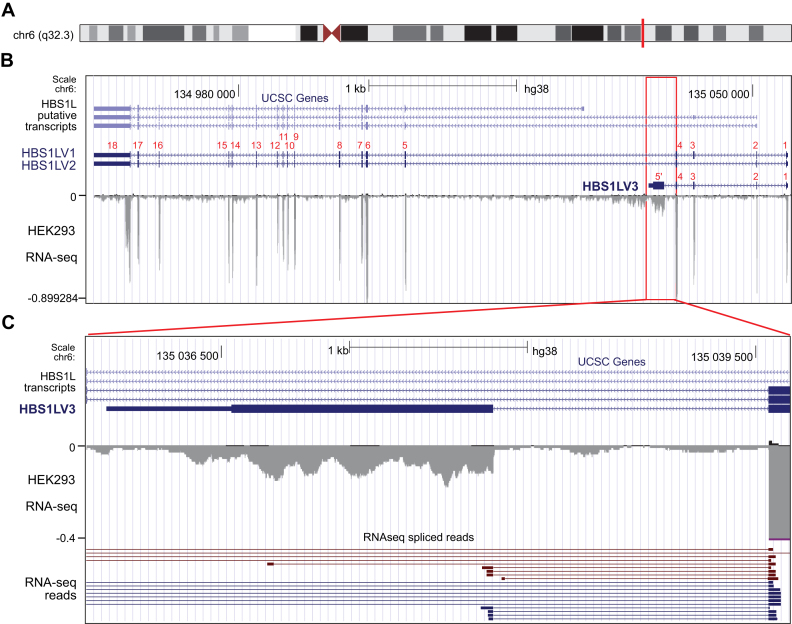
To identify proteins interacting with the cytoplasmic form of the exosome, Co-IP experiments were performed using stable HEK293 cell lines producing an EGFP-tagged DIS3L protein or RRP4 exosome core subunit (Figure 1A and B, Supplementary Figure S2). In addition to high-salt conditions that enabled reductions in background signals, we conducted Co-IPs from cells treated with the cell-permeable cross-linker dithiobis[succinimidyl propionate] (DSP), to increase the likelihood of detecting weak, transient interactions. The enriched proteins were then identified by high-resolution MS and analyzed by LFQ using MaxQuant software (40,41) (Figure 1A and B; Supplementary Figures S1; S2; Data set S1). As expected, using DIS3L-EGFP as bait we recovered high amounts of all exosome core components, whereas the amount of the nucleus-specific subunits DIS3 and EXOSC10 was not significant. For RRP4-EGFP Co-IP assays carried out in parallel, we observed 2–3 orders of magnitude higher levels of nucleus-specific components. Importantly, all subunits of the SKI complex were detected in DIS3L-EGFP cross-linked samples, although the amount of the smallest one, WDR61, was relatively low, because of which it was discarded by the LFQ algorithm. This indicates that the human exosome indeed physically interacts with the SKI complex. Interestingly, we also identified the HBS1L protein in the DIS3L-EGFP and RRP4-EGFP Co-IP experiments. Closer inspection of the peptides revealed that for the DIS3L-EGFP Co-IP, only the N-terminal fragment of the canonical HBS1L (HBS1LV1) was represented in the MS spectrum. A previous study indicated that HBS1L pre-mRNA may be alternatively spliced (42). Indeed, we observed several peptides that mapped to the putative protein encoded by the short splicing isoform of the HBS1L, which we will hereafter refer to as HBS1LV3. Analysis of cDNA clones and RNA-seq data revealed that, apart from the canonical variant, additional transcripts that potentially coded for HBS1LV3 indeed existed in human cells (Figure 2). In contrast, a putative HBS1LV2 mRNA, which is annotated in databases, was undetectable in HEK293 cells using RT-PCR. To further confirm the existence of HBS1LV3, we attempted to raise specific antibodies that recognized HBS1LV3, but were unable to obtain antibodies with sufficient specificity for this isoform. However, commercial antibodies raised against a fragment located within the N-terminal region that is common to both HBS1LV1 and HBS1LV3 produced two bands on a Western blot. siRNA-mediated silencing in HEK293 cell lines confirmed that the main, upper band corresponded to the canonical HBS1LV1 variant while the less intense, lower band was consistent with the HBS1LV3 variant (Figure 1C).
Figure 2.
Transcript variant coding for putative HBS1LV3 protein is present in human HEK293 cells. Illustrative screen shots from the University of California Santa Cruz (UCSC) Genome Browser. Exons are shown as blue rectangles and numbered in red. HBS1LV1 mRNA consists of 18 exons, HBS1LV2 mRNA lacks the third exon and HBS1LV3 mRNA contains an alternative fifth exon (designated as 5΄). Introns are shown as lines connecting exons. Arrowheads indicate the direction of transcription. Mapped reads are shown as grey peaks. (A) Chromosomal localization of the HBS1L gene. (B) The UCSC Genes track of gene predictions. (C) Zoomed-in view of the junction between exon 4 and alternative exon 5. The junction was confirmed by overlapping RNA-seq reads.
The DIS3L Co-IP-MS experiments recovered not only the SKI complex and the exosome but also many other putative interactors (Supplementary Figure S2; Data set S1). However, the majority of these other proteins was highly sub-stoichiometric or had relatively low specificity. Although we cannot exclude the possibility that the cytoplasmic exosome and/or the SKI complex have other functionally relevant interacting partners, their analysis is outside the scope of this study.
Together, our analyses revealed that the human cytoplasmic exosome interacts with the HBS1LV3, a protein encoded by the short splicing variant of HBS1L. Thus, like Ski7 in yeast, in humans HBS1LV3 likely links the human exosome with the SKI complex.
HBS1LV3 links the exosome with the SKI complex
The yeast Ski7 is a multi-domain protein. The N-terminal fragment of Ski7 participates in interactions with both the exosome and the SKI complex (23). The C-terminal region of Ski7 shares similarities with the eRF3 (eukaryotic release factor 3) protein, and consists of 3 domains: a GTP-binding domain that recognizes an empty A site in ribosomes during non-stop decay, followed by two beta barrels (43,44). Interestingly, the C-terminal domain Ski7 shares similarity with the yeast Hbs1 protein (Figure 1D). HBS1LV3 in humans has an N-terminal region that is identical to the canonical HBS1LV1 variant and shares sequence similarity with the yeast Ski7 (Figure 1D). In contrast, the HBS1LV3 C-terminal part displays no similarity to any known protein, apart from its orthologues in other vertebrate species (Supplementary Figure S4). Although in silico predictions revealed the presence of some secondary structure features in the HBS1LV3 C-terminal part (Supplementary Figure S5), making reliable predictions of the structure in this part of the protein was difficult.
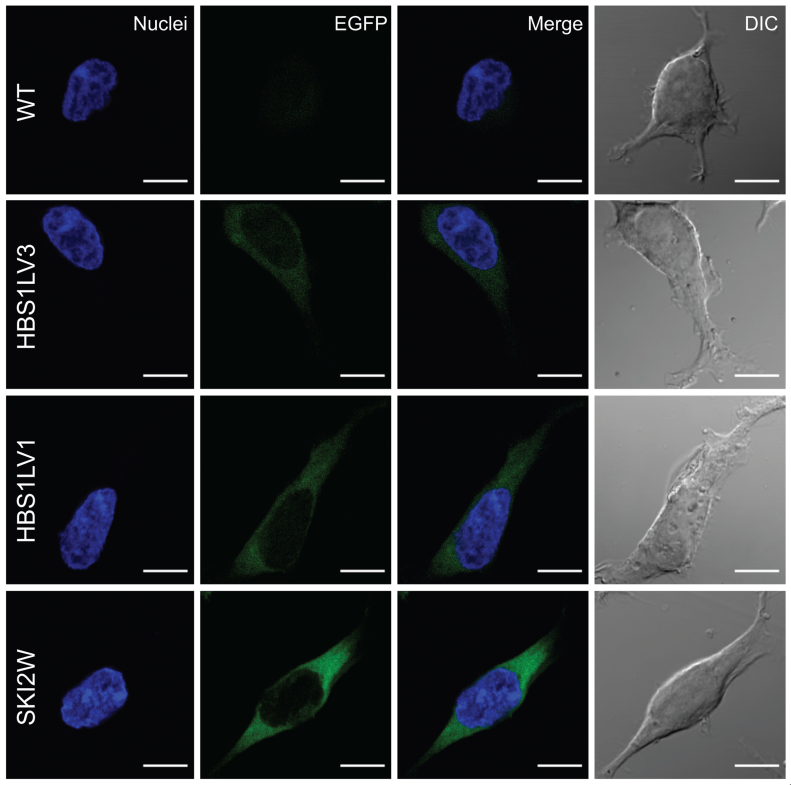
In order to further characterize HBS1LV3 and HBS1LV1, the respective cDNAs were cloned and stable cell lines producing EGFP-tagged proteins were constructed. In parallel with HBS1LV1 and HBS1LV3, a cell line producing EGFP-tagged SKI2W was similarly generated to serve as a control for subsequent experiments. Importantly, confocal imaging revealed that EGFP-tagged HBS1LV3 localized to the cytoplasm in human HEK293 cells, which is in agreement with its co-purification with DIS3L (Figure 3).
Figure 3.
EGFP-tagged HBS1LV3, HBS1LV1 and SKI2W proteins localized to the cytoplasm. Fluorescence imaging of stable HEK293 cell lines expressing EGFP-tagged HBS1LV3, HBS1LV1 or SKI2W proteins. Nuclei were stained with Hoechst. Scale bars represent 10 μm.
Subsequent Co-IP-MS experiments revealed that the newly discovered HBS1LV3 protein specifically interacts with the cytoplasmic exosome complex, since, in addition to the exosome core subunits, we identified the exclusively cytoplasmic DIS3L exoribonuclease but not its nuclear paralogue, the DIS3 protein, which localizes mainly in the nucleoplasm of human cells (Figure 1E and F, Supplementary Figure S2). Finally, SKI complex components also co-purified with HBS1LV3. Meanwhile, canonical HBS1LV1 did not seem to interact with the exosome, but did co-purify with the SKI complex (Figure 1E and F, Supplementary Figure S2). This result indicated that the N-terminal fragment shared by both analyzed HBS1L isoforms is responsible for interactions with the SKI complex but not with the exosome. Importantly, the well-known HBS1LV1 binding partner and Dom34 homologue, the PELOTA protein, did not co-purify with HBS1LV3 (Figure 1E and F; Supplementary Figure S2), suggesting that the two HBS1L variants play different roles. Finally, MS analyses showed that SKI2W co-purified with all predicted SKI complex subunits as well as HBS1LV3 and the exosome (Data set S1).
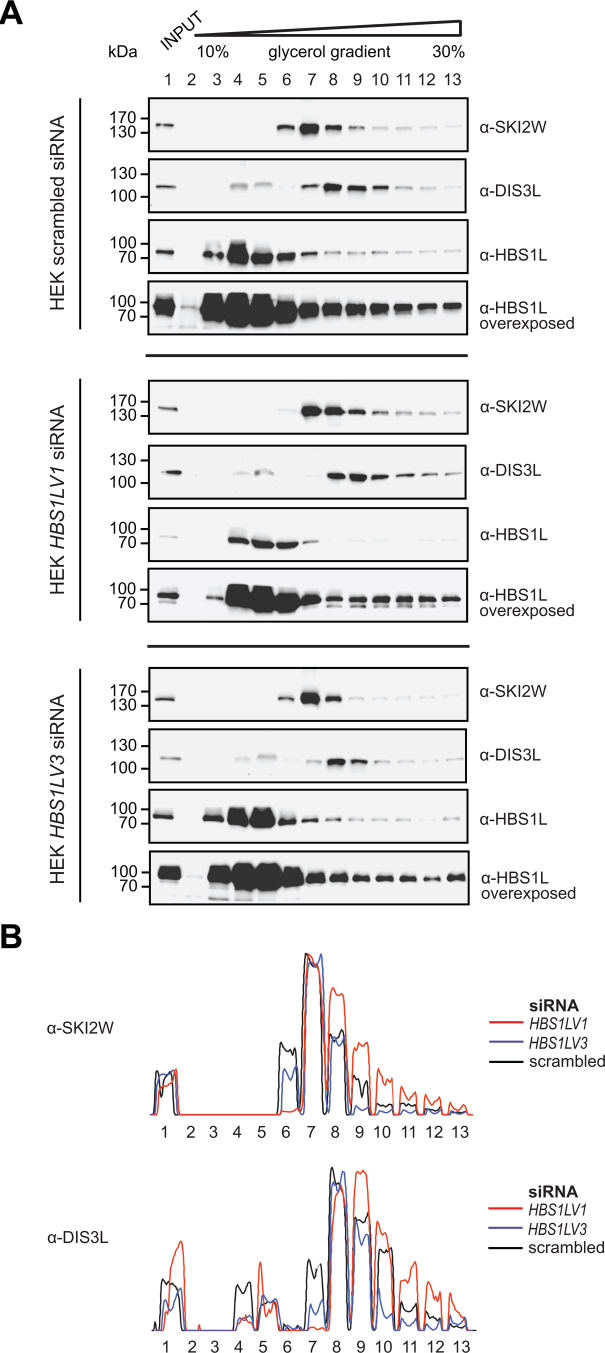
In order to verify if HBS1LV3, like Ski7 in yeast, bridges the exosome with the SKI complex in humans, we performed co-sedimentation experiments using extracts from control cells and cells depleted of HBS1LV3 or HBS1LV1. The extracts were separated in linear glycerol gradient, followed by Western blot detection of SKI2W, DIS3L and HBS1L (Figure 4 and Supplementary Figure S3, where the second biological replica is shown). First of all, it is clearly visible that gross amounts of SKI2W and DIS3L migrate as separate assemblies with peaks at fractions 6–8 for SKI2W and 8–10 for DIS3L, which is in agreement with the transient nature of the SKI/exosome supercomplex formation. However, they are also partially migrating as higher molecular weight species (lanes 9–13). Importantly, depletion of HBS1LV3 strongly reduces that fraction (Figure 4; compare upper and bottom panels). Interestingly, upon HBS1LV1 depletion, there is an opposite effect, i.e. more DIS3L and SKI2W migrate as larger assemblies (Figure 4; compare upper and middle panels). This is probably because HBS1LV1 competes with HBS1LV3 for interactions with the SKI complex, thus inhibiting exosome complex association with the SKI. Finally, HBS1LV3 (which unfortunately is well-visible only in HBS1LV1 depleted samples) co-migrates with DIS3L, enriching toward higher molecular weight species. Altogether, these data strongly suggest that HBS1LV3 indeed bridges SKI complex with the exosome, since HBS1LV3 levels determine the efficiency of the SKI/exosome supercomplex formation.
Figure 4.
Density-based separation of protein complexes by glycerol gradient centrifugation. (A) Protein–protein complexes were separated by glycerol gradient centrifugation. Precipitated proteins were then resolved in SDS-PAGE and analyzed by Western blotting. α-SKI2W, α-DIS3L and α-HBS1L antibodies were used. (B) SKI2W and DIS3L band intensities are shown as a line graph.
The unique C-terminal region of HBS1LV3 binds to the exosome core
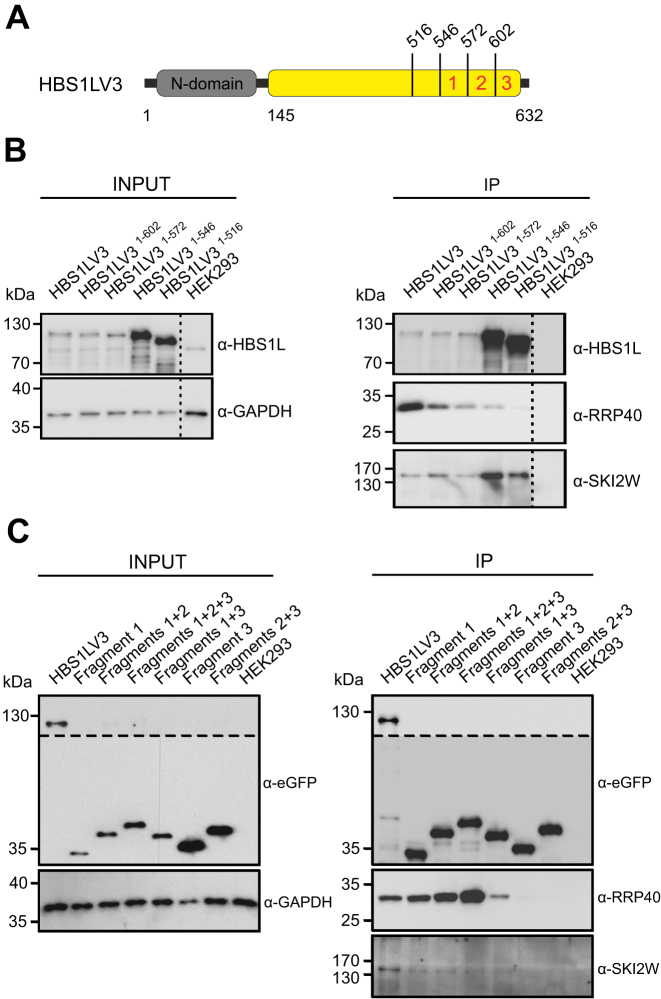
The exosome-interacting fragments in the HBS1LV3 variant likely reside within its unique C-terminal region. In order to determine the approximate location of exosome-binding motifs within HBS1LV3, we carried out Co-IP experiments with EGFP-tagged, C-terminally truncated proteins. We observed that full-length and minimally truncated proteins (HBS1LV3, HBS1LV31-602 and HBS1LV31-572) co-purified with both RRP40 (exosome core protein) and the SKI2W helicase (Figure 5A and B). Meanwhile, longer truncations (HBS1LV31-546 and HBS1LV31-516) substantially weakened interaction with the exosome RRP40 protein (Figure 5A and B), indicating that HBS1LV3 residues that participate in interactions with the exosome complex are located downstream of position 546. Interestingly, this region is also the most conserved one (Supplementary Figure S4), and contains a previously identified motif PFDFxxxSPDDIVKxNQ (aa 609–625) (42).
Figure 5.
The HBS1LV3 C-terminus interacts with the cytoplasmic exosome complex. (A) Schematic representation of the HBS1LV3 protein. The N-terminal domain and the C-terminus of HBS1LV3 are colored grey and yellow, respectively. Positions of protein truncations are indicated. Locations of Fragments 1, 2 and 3 used for Co-IP experiments are depicted by red numbers. Total cell extracts were prepared using HEK293 cells transiently transfected with vectors bearing EGFP-tagged full-length HBS1LV3 and (B) truncated HBS1LV3 proteins or (C) fragments of the HBS1LV3 C-terminus. Co-IPs were performed using GFP trap resin. Both the complete cell extracts (INPUT) and precipitated proteins (IP) were separated by SDS-PAGE and analyzed by Western blotting. α-EGFP, α-RRP40 and α-SKI2W antibodies were used to visualize precipitated proteins with α-GAPDH antibodies serving as a loading control. The dashed lines in (B) indicate that the gel was cropped. The upper part of blots in (C) was overexposed due to the weak chemiluminescence signal.
To further characterize the exosome-interacting motifs, we divided the extreme C-terminal amino acids (aa 546–632) of the HBS1LV3 protein into three fragments referred to as: Fragment 1 (aa 546–572), 2 (aa 572–602) and 3 (aa 602–632). Co-IP experiments using EGFP-tagged single (1 and 3) and fused (1+2, 1+2+3, 1+3 and 2+3) fragments were then conducted (Figure 5A and C). We demonstrated that the core exosome interacts with an increasing efficiency, as evidenced by anti-RRP40 Western blot detection, with constructs comprising Fragments 1, 1+2 and 1+2+3, respectively. The interaction is weaker for Fragments 1+3 and there were no interactions seen with Fragments 3 and 2+3 (Figure 5A and C). In addition, these fragments do not contribute to the HBS1LV3/SKI interactions, which is consistent with previous observations. Thus, we conclude that Fragment 1 (aa 546–572) is crucial and sufficient to mediate interactions with the exosome, while other regions of the HBS1LV3 C-terminus, including the PFDFxxxSPDDIVKxNQ motif, play a minor role in the interaction.
The HBS1LV3 peptide docks into the RRP43 exosome ring subunit
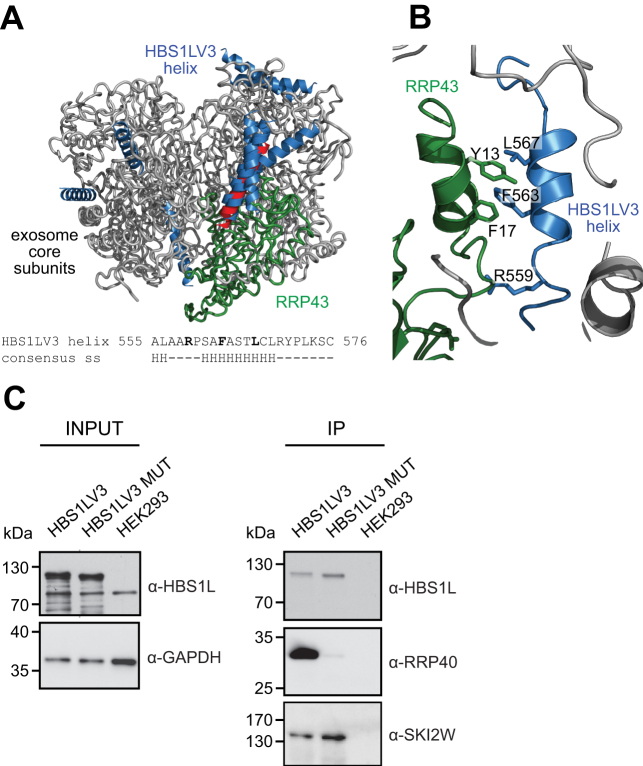
We next analyzed the amino acid sequence of the HBS1LV3 exosome-interacting fragment (aa 546–572), and found that these residues may adopt an alpha-helical conformation (Figure 6A, Supplementary Figures S4 and S5), which allowed us to model its structure based on an ideal helix conformation.
Figure 6.
Model of HBS1LV3-exosome interactions. HBS1LV3 helix (aa 555–576) docks into the surface of RRP43. (A) Modeled human HBS1LV3 helices docked into the exosome core (PDB code: 2NN6, (39)) are shown in blue. The RRP43 protein is shown in green. The remainder of the human exosome complex is colored grey. The 10 best predictions are shown, with 5 predictions docking into the same position on the surface of the RRP43 exosome subunit. The highest ranking prediction is colored red. Bottom: secondary structure prediction of HBS1LV3 fragment. (B) A magnified view of interactions. (C) R559, F563 and L567 HBS1LV3 amino acids are crucial for interactions with the exosome complex. Total cell extracts were prepared from HEK293 cells transiently transfected with vectors expressing either EGFP-tagged HBS1LV3 (WT) or HBS1LV3 MUT (R559A, F563A, L567A) proteins. Co-IPs were performed using GFP trap resin. The complete cell extracts (INPUT) and precipitated proteins (IP) were separated by SDS-PAGE and analyzed by Western blotting. α-HBS1L, α-RRP40 and α-SKI2W antibodies were used to visualize precipitated proteins. α-GAPDH antibodies were utilized as a loading control.
We used the previously determined structure of the human exosome ring (PDB ID: 2NN6) (39) and performed automatic docking of HBS1LV3 exosome-interacting fragment using the ZDOCK server (37) (Figure 6A, Supplementary Figure S6). Strikingly, 5 out of the 10 best predictions docked into the surface of the RRP43 exosome core subunit in exactly the same position as where the DIRIRAGNFKSALANLE peptide of yeast Rrp6 maps into yeast Rrp43 (Figure 6A and B) (9,38). In contrast, Ski7 residues located in proximity of the abovementioned Rrp43 surface are mainly disordered and do not make contacts with Rrp43 (23).
The critical residues of the yeast Rrp6 peptide DIRIRAGNFKSALANLE involved in the interaction with Rrp43 are R602, F606 and L610 (Figure 6B, Supplementary Figure S6D) (9,38). Remarkably, the HBS1LV3 fragment (aa 546–572) contains a conserved RxxxFxxxL motif in which the R559, F563 and L567 residues are spaced similarly to yeast Rrp6 (Supplementary Figure S6F). However, the predicted helical region of HBS1LV3 could be smaller than that of yeast Rrp6 due to the presence of two prolines at positions 560 and 572. The first proline is after a conserved arginine (R559), so this arginine is likely to be located just outside the helix. In the crystal structure of the yeast Rrp6/exosome complex, this conserved arginine (R559) makes contacts with main chain atoms as well as with the side chain of glutamic acid (E48), which is conserved in the human exosome (E31) (Supplementary Figure S6). In yeast Rrp6, phenylalanine 606 (F606, equivalent to F563 in HBS1LV3) is part of a stacking interaction with an Rrp43 histidine (H34). In human RRP43 this histidine is substituted with a phenylalanine (F17) that can also engage in a stacking interaction (Figure 6A and B; Supplementary Figure S6). Strikingly, a model of the HBS1LV3 C-terminal peptide further optimized by homology modeling, docked into the human exosome structure, fits well with the corresponding binding site in human RRP43. In particular, the R559 and F563 residues of HBS1LV3 that are crucial for HBS1LV3/exosome interactions docked well into the RRP43 binding site (Figure 6A and B). Furthermore, there is an additional aromatic residue tyrosine (Y13) in the human RRP43 exosome subunit that is located near the conserved F17 in such a way that the helix's phenylalanine (F563) is located between the human RRP43 F17 and Y13 residues, thus this tyrosine can also take part in a stacking interaction (Figure 6A and B). In the case of the yeast, conserved leucine (L610) of Rrp6 makes contact with the side-chain of a Rrp43 threonine (T16) (Supplementary Figure S6C). This amino acid is also present in the HBS1LV3 RxxxFxxxL motif (L567). Although the precise role of this amino acid in the exosome/HBS1LV3 linkage is difficult to predict, because T16 is not conserved, the interaction could involve hydrophobic contacts.
To provide experimental support for our model, we performed site directed mutagenesis to substitute alanines at crucial residues in the HBS1LV3 C-terminal peptide (R559A, F563A, L567A; HBS1LV3 MUT). Mutation of these residues largely abolished interactions of HBS1LV3 with the exosome (Figure 6C). Thus, the identified peptide and its crucial amino acids are both essential and sufficient for HBS1LV3/exosome interactions.
The HBS1LV3 protein participates in the cytoplasmic exosome-dependent mRNA decay pathway
To identify transcripts that are degraded by cytoplasmic SKI/HBS1LV3/exosome supercomplexes, we used specific siRNAs to knock down expression of genes coding for HBS1L proteins and SKI2W helicase in HEK293 cells. In order to reduce the risk of possible siRNA off-target effects, parallel rescue experiments were performed in which cell lines exogenously producing siRNA-insensitive protein variants were treated with respective siRNAs. Slight overexpression of exogenous proteins compared to the endogenous proteins’ levels was observed (Supplementary Figure S7). qPCR-based mRNA quantification in siRNA-treated cells showed a 50–91% decrease in siRNA-targeted transcript levels (Supplementary Figure S8A–C). Moreover, these results were validated at the protein level by Western blot analysis (Supplementary Figure S8D), which demonstrated that even relatively modest decreases in transcript levels resulted in a strong decrease in the protein amounts. Total ribo-depleted RNA-seq libraries were prepared from all cell lines treated with siRNAs in triplicate. On average, 13.9 million reads were uniquely mapped to the genome for each library.
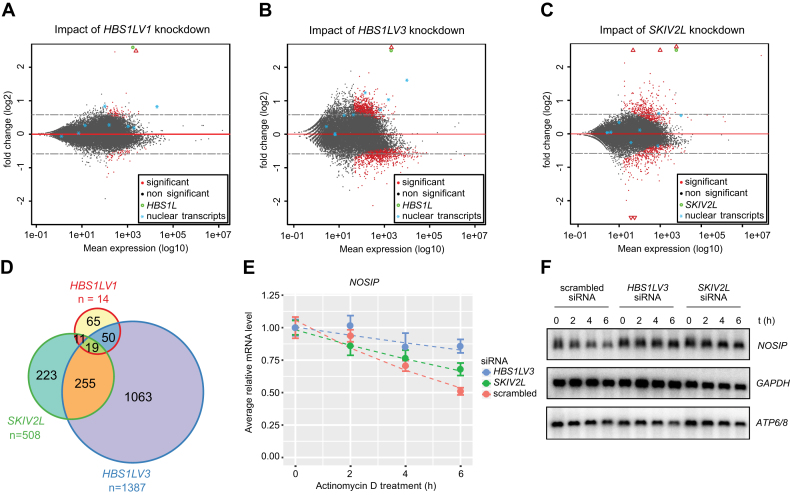
To identify genes with altered expression, we compared the effect of siRNA silencing of HBS1LV1, HBS1LV3 or SKIV2L with respective rescue cell lines treated with the same siRNAs. Differential expression analysis revealed varying degrees of influence of siRNA-mediated protein depletion on steady-state levels of known transcripts. Importantly, depletion of HBS1LV3 had the strongest effect on the transcriptome with 2142 genes (11 301 transcripts) deregulated by more than 1.5-fold, whereas depletion of SKI2W altered expression of 608 genes (4135 transcripts). Depletion of canonical HBS1LV1 had the weakest effect on gene expression, giving only 199 statistically significant hits (1483 transcripts) (Figure 7A–C), despite the fact that depletion of both mRNA and protein appeared to be equal or even more efficient for HBS1LV1 than for HBS1LV3 and SKIV2L (Supplementary Figure S8).
Figure 7.
HBS1LV3 depletion leads to global changes in mRNA levels. MA-plots showing log2 fold change as a function of mean expression levels on a logarithmic scale. Differential expression analysis of (A) HBS1LV1, (B) HBS1LV3, (C) SKIV2L knockdown in HEK293 cell lines rescued with overexpression of the protein of interest expressed from a siRNA-insensitive construct versus endogenous protein knockdown were analyzed. Points located below the red line represent genes with more pronounced expression in the knockdown cells relative to the rescue cell lines. Points located above the line reflect transcripts with relatively higher steady-state levels in the rescue cell lines relative to the knockdown cell lines. Using a FDR threshold of 5%, events were colored red and black for significant and not significant, respectively. Genes falling outside of the set range were plotted as empty triangles on the edge of the plot area. Nuclear transcripts shown on the plot: KCNQ1OT1, NEAT1, MALAT, XIST, TSIX, MIAT, U6 and 7SK. (D) Venn diagram showing the overlap between genes that significantly accumulated in knock down cell lines (FDR threshold of 10%). RNA-seq libraries were prepared in triplicate. (E) Relative mRNA level (y-axis) of NOSIP mRNA was measured by Real-Time qPCR following 0, 2, 4 and 6 h actinomycin D treatment. GAPDH mRNA was used for normalization. Error bar represents SEM (five biological replicates). (F) Dashed lines represent exponential decay fits. Northern blot analysis was performed using RNA from actinomycin D-treated cells, with DNA probes complementary to NOSIP, GAPDH and mitochondrial ATP6/8 transcripts (negative control).
Direct mRNA substrates of the cytoplasmic exosome would be expected to be stabilized when the components of this complex are depleted, and indeed more genes were upregulated than downregulated in the presence of specific siRNAs (1387 for HBS1LV3, 508 for SKIV2L and 145 for HBS1LV1). We then looked at the overlap of these hits to assess the functional intersection of the analyzed proteins. A more pronounced intersection between HBS1LV3 and SKIV2L (54% of the SKIV2L set and 20% of the HBS1LV3 set were common) occurred relative to canonical HBS1LV1 with the other sets (8–10% SKIV2L-HBS1LV1 common hits and 10–15% HBS1LV3-HBS1LV1 common hits) (Figure 7D). Because the overlap of considerably different-sized sets (over 8-fold different) may be confusing to interpret, we also compared sets of fixed sizes that contained the most significantly deregulated genes rather than choosing an absolute significance threshold. For the top 200 most significantly upregulated genes, there were twice as many common targets for HBS1LV3 compared to SKI2W than for HBS1LV1 compared to SKI2W or to HBS1LV3.
Together, our data demonstrated that the lack of the HBS1LV3 or the SKI2W protein resulted in similar global effects, which strongly suggests their involvement in the same mRNA degradation pathway. However, since the effect of HBS1LV3 depletion was overall stronger than that of SKI2W, we cannot exclude that HBS1LV3 has a SKI complex-independent role in RNA decay. Finally, the function of HBS1LV1 in transcriptome homeostasis appears to be less important.
The HBS1LV3 protein enables proper cytoplasmic mRNA decay
Data obtained from high-throughput experiments were further validated by qPCR (Supplementary Figure S9). The transcripts most significantly affected by HBS1LV3 downregulation (NOSIP, TRAPPC2L splicing variant and TNFRSF12A) were selected and tested. We demonstrated that levels of the selected transcripts increased on average 2- to 2.5-fold in the HBS1LV3 isoform knockdown compared to control samples. Rescue experiments conducted simultaneously restored the wild-type phenotype, thereby confirming the previously obtained results.
To verify whether the observed upregulation of mRNA levels in HBS1LV3-depleted cells resulted from inhibition of mRNA decay, we measured the half-lives of selected transcripts by experiments using actinomycin D treatment to inhibit transcription. As in the previous experiments, we depleted proteins of interest using RNAi and then assessed NOSIP, TRAPPC2L splicing variant, TNFRSF12A and mitochondrial ATP6/8 (negative control) transcript levels following 0, 2, 4 and 6 h of transcription inhibition by 4 μg/ml of actinomycin D. All three tested mRNAs were stabilized in HBS1LV3-depleted cells with half-lives up to 5.5-fold longer than in control cells (Figure 7E; Supplementary Figure S10). SKI2W depletion resulted in stabilization of NOSIP and TNFRSF12A mRNAs, with up to 2-fold higher half-lives, but not of the alternative TRAPPCL2 mRNA. ATP6/8 remained largely unchanged in both cases. These results were further confirmed by Northern blot analysis (Figure 7F).
Together, these data strongly suggest that the inhibition of cytoplasmic mRNA decay occurs upon downregulation of HBS1LV3 or SKIV2L. Gene ontology analysis of transcripts upregulated following HBS1LV3 depletion did not identify any particular cellular process as being more significantly affected, suggesting that HBS1LV3 plays a general rather than specific role in cytoplasmic exosome-mediated mRNA decay.
DISCUSSION
The structure-function relationship of Hbs1 and Ski7 proteins deserves an evolutionary consideration. Saccharomyces species have two separate genes, probably as a consequence of a genome duplication event, one encoding Hbs1 and the other Ski7 (42). Both proteins have a C-terminal eRF3-like domain (Figure 1D), postulated to interact with the ribosome A site and important to translation-based quality control pathways (29). Due to sequence differences, the longer N-terminal region of Ski7 can bind the exosome and SKI while the C-terminal domain of Hbs1 interacts with Dom34, another factor involved in releasing stalled ribosomes. However, in other fungi alternative mRNA splicing of a single HBS1/SKI7 gene gives rise to two proteins that differ in their N-terminal region and only the longer one can bind the exosome and SKI complex; the short form is thought to perform Hbs1 functions and the long one – Ski7 functions (42,45). In these species the C-terminal eRF3-like domain is identical in both proteins (22,42) and thus presumably both can interact with Dom34. A similar phenomenon may occur in plants, where the HBS1 seems to be alternatively spliced and previous studies suggested that one of the gene's products may have Ski7-like properties (42). Here, we show an analogous situation in humans, where the HBS1L mRNA undergoes alternative splicing to yield two proteins. The canonical Hbs1-like protein HBS1LV1 can interacts with SKI and PELOTA (human homolog of Dom34) (Figure 1E and F; Supplementary Figure S2) (29). The newly discovered Ski7-like HBS1LV3 has the same N-terminal region, facilitating interaction with SKI, but the eRF3-like domain is replaced by a unique C-terminal region that binds to the exosome. Notably, this arrangement is specific to vertebrates (Supplementary Figure S4) as the HBS1LV3 protein seems to be absent from all other metazoans and the nature of the SKI-exosome interaction in those animals remains unknown.
In yeast, Ski7 is a stable component of the cytoplasmic exosome, while its interactions with the SKI complex are more transient (46). In this study, quantification of the Co-IP-MS experiments as well as the fact that cross-linking was essential for efficient SKI complex recovery in the cytoplasmic exosome pull-down assays strongly suggested that, like yeast Ski7, human HBS1LV3 is more strongly associated with the exosome core than the SKI. Although the exact molecular mechanism of cooperation between the exosome and the SKI complex in humans awaits further characterization, the molecular modeling experiments supported by mutagenesis that we presented in this study support an association between HBS1LV3 and the exosome core that is mediated by the RxxxFxxxL motif located between residues 559 and 567 of HBS1LV3. This interaction between HBS1LV3 and the RRP43 ring subunit resembles Rrp6 binding to the exosome core in S. cerevisiae. Another conserved motif with the sequence PFDFxxxSPDDIVKxNQ is located at the extreme C-terminus of human HBS1LV3 (aa 609–625) (42). Although the function of this motif is unclear, our data showed that these residues play a minor role in HBS1LV3/exosome interactions. Since the RxxxFxxxL motif identified in HBS1LV3 lies in a region that the yeast Ski7 protein lacks, the interaction of Ski7 with the yeast exosome core likely differs from that of the HBS1LV3/exosome interaction in humans. Indeed, a recently published structure of yeast Ski7/exosome complex revealed that Ski7 does not bind the Rrp43 region predicted by our homology modeling to be responsible for HBS1LV3 helix recognition (23) pointing to a similar but distinct mode of binding that is utilized by HBS1LV3. This difference would be another example of the rapid evolution of protein–protein interaction motifs between different species, with a preservation of a general interaction network that is quite common for proteins involved in RNA decay (47).
The eRF3-like domain of yeast Ski7 is not essential for exosome-mediated 3΄-5΄ decay, but is important for other RNA quality control pathways such as no-go decay, and is postulated to interact with the ribosomal A-site (29). The human Ski7-like protein HBS1LV3 lacks an eRF3-like domain, suggesting that HBS1LV3 may not directly participate in translation-dependent surveillance pathways. Indeed, we could not detect overrepresentation of mRNAs that are predicted to be subject to such quality control pathways among the genes that were upregulated upon HBS1LV3 depletion. Thus, our transcriptomic analysis strongly suggests that HBS1LV3 is mainly involved in bulk mRNA turnover, although it is highly probable that in HEK293 cells XRN1-mediated 5΄-3΄ degradation or DIS3L2-mediated 3΄-5΄ decay also play an important role in shaping the global transcriptomic landscape (1).
Although the canonical human HBS1LV1 does not bind to the exosome, it can still interact with the SKI complex, which is not the case for yeast Hbs1. Thus, HBS1LV1 can compete with HBS1LV3 for SKI complex and thus inhibit SKI/exosome supercomplex formation, although the exact role of HBS1LV1/SKI association remains to be determined. The relatively small effect of HBS1LV1 depletion on the steady-state mRNA levels suggests, however, that this protein does not play an important role in the decay of normal mRNA. Consistent with this possibility, canonical HBS1LV1 in humans was recently proposed to be involved in a quality control mechanism (e.g. non-stop mRNA decay) in mammalian cells (29).
In HEK293 cells, the HBS1LV3 protein is produced at much lower levels than the canonical variant HBS1LV1 (Figure 1C, Supplementary Figure S8D). In contrast to HEK293 cells, available data show that high amounts of HBS1LV3 expression occur in chronic myeloid leukemia cell lines and in bone marrow (www.proteinatlas.org), which suggests that the SKI/HBS1LV3/exosome supercomplex may play a primary role in mRNA decay in hematopoietic cells. These conjectures are consistent with the fact that exosome-dependent RNA decay involving the cytoplasmic exoribonuclease DIS3L controls primary erythroid cell maturation (48).
In conclusion, here we have identified HBS1LV3, an analogue of the yeast Ski7 protein, which is a long-sought linker between the human cytoplasmic exosome and the SKI complex, and in vertebrates is encoded by a specific splicing isoform of the HBS1L surveillance factor. The identification of HBS1LV3 provides new insights into exosome-mediated mRNA decay in humans.
ACCESSION NUMBERS
RNA-seq data supporting the conclusions of this article are available in the NCBI GEO repository http://www.ncbi.nlm.nih.gov/geo/, accession number GSE77926.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Ambro van Hoof for inspiration, all the members of the AD laboratory for their support and Szymon Świeżewski for critical reading of this manuscript. Author contributions: A.D. conceived and directed the studies. K. Kalisiak designed most of the experiments, constructed the majority of the vectors, established stable human cell lines, performed all biochemical assays and validation experiments, prepared RNA-seq libraries and participated in localization studies and statistical analysis. T.M.K. performed bioinformatics analyses. R.T. participated in designing the experiments, in the Northern blot and co-sedimentation assays. D.C. performed high resolution mass spectrometry analysis. Z.P. performed docking, homology modeling and structural analyses. A.C. participated in localization studies and statistical analysis. K. Kowalska participated in vector construction. A.D. and K. Kalisiak wrote the paper with contributions from R.T.,. T.K., Z.P. and A.C.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Centre [NCN Maestro: UMO-2011/02/A/NZ1/00001 and NCN Harmonia: UMO-2013/10/M/NZ4/00299]; Foundation for Polish Science fellowships [Master and Ideas for Poland to A.D.]; Experiments were carried out with the use of CePT infrastructure that was financed by the European Union via the European Regional Development Fund [Innovative economy 2007–13, Agreement POIG.02.02.00-14-024/08-00]. Funding for open access charge: National Science Centre [NCN Maestro: UMO-2011/02/A/NZ1/00001 and NCN Harmonia: UMO-2013/10/M/NZ4/00299].
Conflict of interest statement. None declared.
REFERENCES
- 1.Lubas M., Damgaard C.K., Tomecki R., Cysewski D., Jensen T.H., Dziembowski A.. Exonuclease hDIS3L2 specifies an exosome-independent 3΄-5΄ degradation pathway of human cytoplasmic mRNA. EMBO J. 2013; 32:1855–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Malecki M., Viegas S.C., Carneiro T., Golik P., Dressaire C., Ferreira M.G., Arraiano C.M.. The exoribonuclease Dis3L2 defines a novel eukaryotic RNA degradation pathway. EMBO J. 2013; 32:1842–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mitchell P., Petfalski E., Shevchenko A., Mann M., Tollervey D.. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3΄–>5΄ exoribonucleases. Cell. 1997; 91:457–466. [DOI] [PubMed] [Google Scholar]
- 4.Chlebowski A., Lubas M., Jensen T.H., Dziembowski A.. RNA decay machines: the exosome. Biochim. Biophys. Acta. 2013; 1829:552–560. [DOI] [PubMed] [Google Scholar]
- 5.Tomecki R., Kristiansen M.S., Lykke-Andersen S., Chlebowski A., Larsen K.M., Szczesny R.J., Drazkowska K., Pastula A., Andersen J.S., Stepien P.P. et al. The human core exosome interacts with differentially localized processive RNases: hDIS3 and hDIS3L. EMBO J. 2010; 29:2342–2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Staals R.H.J., Bronkhorst A.W., Schilders G., Slomovic S., Schuster G., Heck A.J.R., Raijmakers R., Pruijn G.J.M.. Dis3-like 1: A novel exoribonuclease associated with the human exosome. EMBO J. 2010; 29:2358–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lykke-Andersen S., Tomecki R., Jensen T.H., Dziembowski A.. The eukaryotic RNA exosome: same scaffold but variable catalytic subunits. RNA Biol. 2011; 8:61–66. [DOI] [PubMed] [Google Scholar]
- 8.Malet H., Topf M., Clare D.K., Ebert J., Bonneau F., Basquin J., Drazkowska K., Tomecki R., Dziembowski A., Conti E. et al. RNA channelling by the eukaryotic exosome. EMBO Rep. 2010; 11:936–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Makino D.L., Baumgärtner M., Conti E.. Crystal structure of an RNA-bound 11-subunit eukaryotic exosome complex. Nature. 2013; 495:70–75. [DOI] [PubMed] [Google Scholar]
- 10.Wasmuth E.V., Lima C.D.. Exo- and endoribonucleolytic activities of yeast cytoplasmic and nuclear RNA exosomes are dependent on the noncatalytic core and central channel. Mol. Cell. 2012; 48:133–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drazkowska K., Tomecki R., Stodus K., Kowalska K., Czarnocki-Cieciura M., Dziembowski A.. The RNA exosome complex central channel controls both exonuclease and endonuclease Dis3 activities in vivo and in vitro. Nucleic Acids Res. 2013; 41:3845–3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Makino D.L., Schuch B., Stegmann E., Baumgärtner M., Basquin C., Conti E.. RNA degradation paths in a 12-subunit nuclear exosome complex. Nature. 2015; 524:54–58. [DOI] [PubMed] [Google Scholar]
- 13.Anderson J.S., Parker R.P.. The 3΄ to 5΄ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3΄ to 5΄ exonucleases of the exosome complex. EMBO J. 1998; 17:1497–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Araki Y., Takahashi S., Kobayashi T., Kajiho H., Hoshino S., Katada T.. Ski7p G protein interacts with the exosome and the Ski complex for 3΄-to-5΄ mRNA decay in yeast. EMBO J. 2001; 20:4684–4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Hoof A., Staples R.R., Baker R.E., Parker R.. Function of the ski4p (Csl4p) and Ski7p proteins in 3΄-to-5΄ degradation of mRNA. Mol. Cell. Biol. 2000; 20:8230–8243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ridley S.P., Sommer S.S., Wickner R.B.. Superkiller mutations in Saccharomyces cerevisiae suppress exclusion of M2 double-stranded RNA by L-A-HN and confer cold sensitivity in the presence of M and L-A-HN. Mol. Cell. Biol. 1984; 4:761–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Halbach F., Reichelt P., Rode M., Conti E.. The yeast ski complex: crystal structure and RNA channeling to the exosome complex. Cell. 2013; 154:814–826. [DOI] [PubMed] [Google Scholar]
- 18.Zhu B., Mandal S.S., Pham A.-D., Zheng Y., Erdjument-Bromage H., Batra S.K., Tempst P., Reinberg D.. The human PAF complex coordinates transcription with events downstream of RNA synthesis. Genes Dev. 2005; 19:1668–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fabre A., Charroux B., Martinez-Vinson C., Roquelaure B., Odul E., Sayar E., Smith H., Colomb V., Andre N., Hugot J.-P. et al. SKIV2L mutations cause syndromic diarrhea, or trichohepatoenteric syndrome. Am. J. Hum. Genet. 2012; 90:689–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hartley J.L., Zachos N.C., Dawood B., Donowitz M., Forman J., Pollitt R.J., Morgan N.V., Tee L., Gissen P., Kahr W.H.A. et al. Mutations in TTC37 cause trichohepatoenteric syndrome (phenotypic diarrhea of infancy). Gastroenterology. 2010; 138:2388–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eckard S.C., Rice G.I., Fabre A., Badens C., Gray E.E., Hartley J.L., Crow Y.J., Stetson D.B.. The SKIV2L RNA exosome limits activation of the RIG-I-like receptors. Nat. Immunol. 2014; 15:839–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kowalinski E., Schuller A., Green R., Conti E.. Saccharomyces cerevisiae Ski7 Is a GTP-binding protein adopting the characteristic conformation of active translational GTPases. Struct. Lond. Engl. 1993. 2015; 23:1336–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kowalinski E., Kögel A., Ebert J., Reichelt P., Stegmann E., Habermann B., Conti E.. Structure of a cytoplasmic 11-Subunit RNA exosome complex. Mol. Cell. 2016; 63:125–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benard L., Carroll K., Valle R.C.P., Masison D.C., Wickner R.B.. The Ski7 antiviral protein is an EF1-α Homolog that blocks expression of Non-Poly(A) mRNA in Saccharomyces cerevisiae. J. Virol. 1999; 73:2893–2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Hoof A., Lennertz P., Parker R.. Yeast exosome mutants accumulate 3΄-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol. Cell. Biol. 2000; 20:441–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsuboi T., Kuroha K., Kudo K., Makino S., Inoue E., Kashima I., Inada T.. Dom34:hbs1 plays a general role in quality-control systems by dissociation of a stalled ribosome at the 3΄ end of aberrant mRNA. Mol. Cell. 2012; 46:518–529. [DOI] [PubMed] [Google Scholar]
- 27.Doma M.K., Parker R.. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 2006; 440:561–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Atkinson G.C., Baldauf S.L., Hauryliuk V.. Evolution of nonstop, no-go and nonsense-mediated mRNA decay and their termination factor-derived components. BMC Evol. Biol. 2008; 8:290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saito S., Hosoda N., Hoshino S.. The Hbs1-Dom34 protein complex functions in non-stop mRNA decay in mammalian cells. J. Biol. Chem. 2013; 288:17832–17843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsuchiya F., Kano R., Sano J., Oguma K., Hasegawa A.. Apoptosis of canine mammary tumor cells induced by small interfering RNA (siRNA) against Bcl-xL gene. J. Vet. Med. Sci. 2006; 68:1199–1201. [DOI] [PubMed] [Google Scholar]
- 31.Girard C., Will C.L., Peng J., Makarov E.M., Kastner B., Lemm I., Urlaub H., Hartmuth K., Lührmann R.. Post-transcriptional spliceosomes are retained in nuclear speckles until splicing completion. Nat. Commun. 2012; 3:994. [DOI] [PubMed] [Google Scholar]
- 32.Płociński P., Laubitz D., Cysewski D., Stoduś K., Kowalska K., Dziembowski A.. Identification of protein partners in mycobacteria using a single-step affinity purification method. PloS One. 2014; 9:e91380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wessel D., Flügge U.I.. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 1984; 138:141–143. [DOI] [PubMed] [Google Scholar]
- 34.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pierce B.G., Wiehe K., Hwang H., Kim B.-H., Vreven T., Weng Z.. ZDOCK server: Interactive docking prediction of protein-protein complexes and symmetric multimers. Bioinformatics. 2014; 30:1771–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wasmuth E.V., Januszyk K., Lima C.D.. Structure of an Rrp6-RNA exosome complex bound to poly(A) RNA. Nature. 2014; 511:435–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Q., Greimann J.C., Lima C.D.. Reconstitution, activities and structure of the eukaryotic RNA exosome. Cell. 2006; 127:1223–1237. [DOI] [PubMed] [Google Scholar]
- 40.Cox J., Mann M.. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008; 26:1367–1372. [DOI] [PubMed] [Google Scholar]
- 41.Cox J., Hein M.Y., Luber C.A., Paron I., Nagaraj N., Mann M.. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics. 2014; 13:2513–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marshall A.N., Montealegre M.C., Jiménez-López C., Lorenz M.C., van Hoof A.. Alternative splicing and subfunctionalization generates functional diversity in fungal proteomes. PLoS Genet. 2013; 9:e1003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Hoof A., Frischmeyer P.A., Dietz H.C., Parker R.. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science. 2002; 295:2262–2264. [DOI] [PubMed] [Google Scholar]
- 44.Frischmeyer P.A., van Hoof A., O'Donnell K., Guerrerio A.L., Parker R., Dietz H.C.. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science. 2002; 295:2258–2261. [DOI] [PubMed] [Google Scholar]
- 45.Klauer A.A., van Hoof A.. Degradation of mRNAs that lack a stop codon: a decade of nonstop progress. Wiley Interdiscip. Rev. RNA. 2012; 3:649–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gavin A.-C., Aloy P., Grandi P., Krause R., Boesche M., Marzioch M., Rau C., Jensen L.J., Bastuck S., Dümpelfeld B. et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006; 440:631–636. [DOI] [PubMed] [Google Scholar]
- 47.Jonas S., Izaurralde E.. The role of disordered protein regions in the assembly of decapping complexes and RNP granules. Genes Dev. 2013; 27:2628–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McIver S.C., Kang Y.-A., DeVilbiss A.W., O'Driscoll C.A., Ouellette J.N., Pope N.J., Camprecios G., Chang C.-J., Yang D., Bouhassira E.E. et al. The exosome complex establishes a barricade to erythroid maturation. Blood. 2014; 124:2285–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.