Abstract
N6-Threonylcarbamoyladenosine (t6A) and its derivatives are universally conserved modified nucleosides found at position 37, 3΄ adjacent to the anticodon in tRNAs responsible for ANN codons. These modifications have pleiotropic functions of tRNAs in decoding and protein synthesis. In certain species of bacteria, fungi, plants and protists, t6A is further modified to the cyclic t6A (ct6A) via dehydration catalyzed by TcdA. This additional modification is involved in efficient decoding of tRNALys. Previous work indicated that the chemical structure of ct6A is a cyclic active ester with an oxazolone ring. In this study, we solved the crystal structure of chemically synthesized ct6A nucleoside. Unexpectedly, we found that the ct6A adopted a hydantoin isoform rather than an oxazolone isoform, and further showed that the hydantoin isoform of ct6A was actually present in Escherichia coli tRNAs. In addition, we observed that hydantoin ct6A is susceptible to epimerization under mild alkaline conditions, warning us to avoid conventional deacylation of tRNAs. A hallmark structural feature of this isoform is the twisted arrangement of the hydantoin and adenine rings. Functional roles of ct6A37 in tRNAs should be reconsidered.
INTRODUCTION
Transfer (t) RNA is an adaptor molecule that links the codon in messenger (m) RNA to its corresponding amino acid during protein synthesis. tRNAs contain a number of modified nucleosides that are introduced enzymatically after transcription. To date, more than 100 species of modified nucleosides have been identified in tRNAs from all domains of life (1). These modifications help to ensure proper tRNA function by stabilizing tertiary structures and modulating decoding properties (2–6). A wide variety of chemical modifications are found in the tRNA anticodon loops, especially at the first letter of the anticodon (position 34), the so-called ‘wobble’ position. Modifications at this position play critical roles in modulating decoding capabilities (4,6,7). On the other hand, hypermodified purine bases are also found at position 37, 3΄ adjacent to the anticodon; these modifications contribute to decoding processes by ensuring reading-frame maintenance via stabilization of the codon–anticodon interaction on the ribosome (8,9).
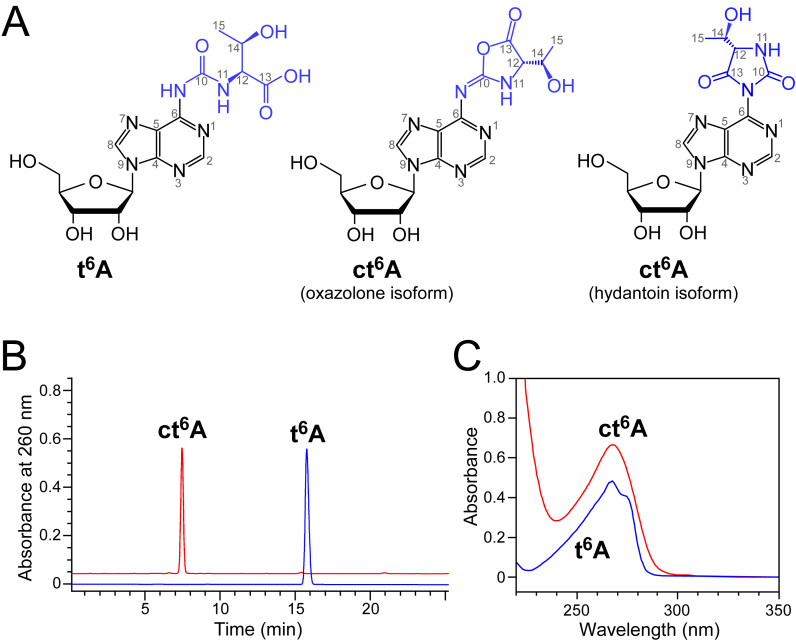
N6-Threonylcarbamoyladenosine (t6A) and its derivatives are modifications found at position 37 of tRNAs responsible for decoding A-starting codons (ANN) in all domains of life (Figure 1A) (8,10). Extensive in vitro and in vivo studies of the biological functions of t6A revealed that this modification has pleiotropic functions in translation, including aminoacylation of tRNAs (11), tRNA binding to the A-site codon (12), efficient translocation (13), reading-frame maintenance (14) and prevention of leaky scanning of initiation codons and read-through of stop codons (15). Structural studies revealed that t6A enhances anticodon–codon base pairing by cross-strand base stacking of the t6A base with the first adenine base of the codon, and stabilizes the anticodon stem–loop structure by stacking with A38 and preventing base-pairing with U33 (16–19).
Figure 1.
Chemical synthesis of ct6A. (A) Chemical structures of t6A, oxazolone isoform of cyclic t6A, and hydantoin isoform of cyclic t6A. (B) HPLC elution profiles of chemically synthesized t6A and ct6A. (C) UV spectra of chemically synthesized t6A and ct6A in H2O; for ct6A λmin = 233, λmax = 269; ε = 18 900 dm3·mol−1·cm−1.
In 2013, a cyclic form of t6A (ct6A) with a molecular mass of 394 Da (Figure 1A) was discovered in bacteria, fungi, protists, and plants (20). Because ct6A is formed via dehydration of t6A, it is easily hydrolyzed to t6A during handling of tRNA and preparation of nucleosides under mild alkaline conditions (21). Consequently, t6A was discovered >40 years (10) before the detection of ct6A. When total nucleosides are prepared under neutral conditions in the shortest possible period of time, ct6A can be clearly detected, and very little t6A is observed in Escherichia coli tRNAs (20), demonstrating that ct6A is a bona fide modification in tRNAs from species including E. coli and yeast. According to MS and NMR analyses, the chemical structure of ct6A was determined to be a cyclic active ester with an oxazolone ring (Figure 1A).
ct6A is synthesized by tRNA threonylcarbamoyladenosine dehydratase A (TcdA), which catalyzes ATP-dependent dehydration of t6A to form ct6A (20). CsdA and CsdE are additional factors required for efficient ct6A formation (20). The yeast homologs of tcdA, TCD1 and TCD2, are required for respiratory cell growth, indicating the physiological importance of ct6A (20). Reporter assays using a ΔtcdA strain revealed that ct6A is involved in efficient decoding of tRNALys, and possibly for other tRNAs containing ct6A. The crystal structures of TcdA in complex with AMP or ATP have been solved (22,23), providing insight into the molecular basis of ct6A formation.
The discovery of ct6A encouraged us to characterize the structural and biochemical contributions of this modification to tRNA functions. For this purpose, we chemically synthesized the 5΄,3΄,2΄-O-acetylated derivative of ct6A, which was stable under acidic and mild basic conditions (24). This advance made it feasible to synthesize model RNA oligonucleotides containing ct6A for use in further structural and biochemical studies.
In this study, we elaborated simple synthesis of the non-sugar-protected ct6A nucleoside using carbodiimide chemistry, and synthesized ct6A nucleoside at multi-milligram scale. The resultant ct6A was successfully crystallized, and the tertiary structure of the ct6A nucleoside was determined by X-ray crystallography. Contrary to our expectation, the crystal structure of ct6A contained a hydantoin isoform rather than the previously determined oxazolone isoform (Figure 1A). These two isoforms are not differentiated by mass spectrometric analysis and simple NMR studies. The hydantoin structure of synthetic ct6A in solution was further confirmed by detailed analysis of 15N NMR data and the presence of characteristic absorption bands in IR spectrum. Very surprising result of these structural studies prompted us to investigate whether the hydantoin isoform of ct6A is actually present in natural tRNAs. Careful LC/MS co-injection analysis revealed the identity of the synthetic hydantoin ct6A with the nucleoside in E. coli tRNAs. In addition, we found that hydantoin ct6A is susceptible to epimerization under mild alkaline conditions, warning us to avoid conventional deacylation procedure of tRNAs. Based on the structural features of this isoform, the structure–function relationship of ct6A37 in tRNAs should be reconsidered.
MATERIALS AND METHODS
General information
A Waters HPLC (515) system using XTerra® Waters column (MS C8, 5 μm, 4.6 × 150 mm, 100 Å) was used for analytical high-performance liquid column chromatography (HPLC). Chromatography was performed at room temperature, at a flow rate of 1 ml/min, in a gradient of acetonitrile (Solvent B) in 0.1% AcOH/water (Solvent A), as follows: 0% B to 30% B in 0–30 min, and 30% B to 50% B in 30–35 min. The elution profile was monitored by UV absorption at 254 nm using a Waters 996 photodiode array detector. High-resolution mass spectrometry with electrospray ionization was conducted on a Maldi SYNAPT G2-S HDMS (Waters). UV–vis spectra were obtained on an O-2800 Hitachi UV Digilab1 spectrophotometer. IR data were recorded on an FT-IR ALPHA (Bruker) equipped with a Platinum ATR QuickSnap™ module. NMR spectra were recorded in DMSO-d6 on Bruker Avance II Plus 700 and Bruker DPX-250 spectrometers. Chemical shifts are given as ppm values relative to DMSO, used as an internal reference (2.50 ppm for 1H NMR and 39.51 ppm for 13C NMR). Signals were assigned based on 1H–1H COSY, 1H–13C HSQC, 1H–13C HMBC and 13C DEPT 135 spectra. Signals shapes and multiplicities are indicated as follows: br s = broad singlet, s = singlet, d = doublet, dd = double doublets, ddd = double double doublets, t = triplet, q = quartet, m = multiplet. Scalar coupling constants J are given in Hertz (Hz). 15N chemical shifts were obtained from analysis of 1H–15N HSQC and 1H–15N HMBC spectra, and are given relative to liquid NH3. 1D NOE and 2D NOESY spectra were recorded with a mixing time of 300 ms.
Chemical synthesis of t6A
t6A nucleoside was obtained in good yield according to the procedure reported by Chheda et al. (25–27). In this method, the exo-amine group of sugar-acetylated adenosine was functionalized with an ethoxycarbonyl group using ethylchloroformate as the electrophile, affording a reactive N6-carbamate derivative. The ethoxy group of this compound was subsequently substituted with l-threonine to give sugar-protected t6A, which was de-protected by treatment with ammonium-saturated methanol (10 M), yielding the t6A nucleoside as the ammonium salt. The t6A ammonium salt was then purified by column chromatography on silica gel 60 (230–400 mesh, Sigma-Aldrich). Before cyclization, the t6A ammonium salt was converted to the nucleoside with a free carboxylate by cation ion exchange chromatography on Amberlite™ IR120 (H+ form). The purity and structure of the t6A nucleoside were unambiguously confirmed by analytical HPLC (Figure 1B), UV spectrum (Figure 1C), IR (Supplementary Figure S1A), 1H NMR (Supplementary Figure S2A) and 13C NMR (Supplementary Figure S3A), and the results were in full agreement with earlier publications (25–28).
t6A nucleoside bearing d-allo-threonine residue was synthesized according the same procedure as described above for l-t6A isomer. The spectral data of d-allo-t6A fully correlated with those reported previously (29), and they are shown in Supplementary Figure S4A (IR), Supplementary Figure S5A (1H NMR) and Supplementary Figure S6A (13C NMR).
Chemical synthesis of ct6A
t6A nucleoside in the free carboxylic form (300 mg, 0.72 mmol) was dissolved in 10 ml of water and mixed with 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide immobilized on a polymer support (EDC-P) (10 equiv, 5.14 g, 7.2 mmol, loading 1.4 mmol/g), followed by vigorous stirring at room temperature for 1 h. After consumption of t6A nucleoside was checked by TLC analysis (n-BuOH/H2O = 85/15, v/v), the reaction was quenched by filtering off the polymer-bounded EDC, followed by three careful washes of the polymer bed with H2O (10 ml each time). The filtrate was evaporated to dryness under reduced pressure, and the crude product was subjected to column chromatography on silica gel with a linear gradient of H2O (0–3%) in n-BuOH to purify ct6A nucleoside (140 mg, yield 49%). ct6A nucleoside was obtained with similar yield (153 mg, yield 51%) using dimethylformamide (DMF) as a solvent. The purity of ct6A and its different lipophilicity relative to t6A was confirmed by analytical HPLC (Figure 1B), UV spectroscopy (Figure 1C), IR (Supplementary Figure S1B), 1H NMR (Supplementary Figure S2B), 13C NMR (Supplementary Figure S3B), and high-resolution MS (Supplementary Figure S7).
Cyclization of d-allo-t6A was performed with the same procedure as described above to generate d-allo-ct6A with slightly lower yield (40%). The structure of d-allo-ct6A was confirmed by IR (Supplementary Figure S4B), 1H NMR (Supplementary Figure S5B) and 13C NMR (Supplementary Figure S6B). HPLC co-injection analysis of d-allo-ct6A and l-ct6A mixture showed that both isomers can be clearly separated (Supplementary Figure S8).
Single crystal X-ray diffraction analysis
Colorless crystals of ct6A (0.30 × 0.10 × 0.05 mm) were obtained from aqueous ethanol (1:1) solution. Single crystal X-ray diffraction data were collected on a Bruker Smart APEX2 diffractometer at 100 K, using an Incoatec Microfocus Source IμS Cu-Kα (λ = 1.54178 Å) as the source of radiation. Integration of the data yielded 14 087 reflections to a 2θmax angle of 142°, of which 3384 were independent (Rint = 0.040), and 3152 reflections with I ≥ 2σ (I). The crystal structure was solved by the intrinsic phasing method. Formula: C15H18N6O7·H2O, Mw = 412.37, Z = 4, crystal system: orthorhombic, space group: P212121, a = 6.355(1), b = 8.173(1), c = 33.492(3) Å, α = β = γ = 90°, V = 1739.4(2) Å3, ρcalcd = 1.57 g cm−3, μ = 1.11 mm−1. Semi-empirical absorption correction based on multiple scanned equivalent reflections (0.636 < T < 0.754) was applied. The final anisotropic full-matrix least-squares refinement on F2 with 342 variables converged at Robs = 0.041, Rall = 0.045, wR (F2) = 0.099 with residual electron density Δρmax = 0.23 eÅ−3 (Δρmin = -0.25 eÅ−3). Positions of all hydrogen atoms were located on a difference Fourier map, and their coordinates and isotropic displacement parameters were refined without restriction, with the exception of hydrogen atoms in water molecules, whose parameters were restrained to the oxygen atom. The correct absolute configuration was provided using a starting material with known configuration. Data collection, reduction, and absorption correction were performed using theAPEX2 (Version 2014.9, Bruker AXS), SAINT-PLUS (Version 8.34A, Bruker AXS), and Sadabs (Version 2014/4, Bruker AXS) programs, respectively. Structure solution and refinement were performed using the Shelxtl suite (Version 2014/6, Bruker AXS). The CDCC 2015 software package (CSD system 2015, Version 5.36, Conquest, Mercury, Mogul) (30) was utilized for subsequent molecular geometry analysis and visualizations.
Quantum chemistry computations
Ab initio energy scans around N1-C6-N6-C13 torsion angle for ct6A were performed using the Gaussian09 program with the DFT methodology (Gaussian 09, Revision C.01, Gaussian, Inc., Wallingford CT, 2010). The Truhlar exchange-correlation energy functional M062x and aug-cc-pvdz Dunning's basis set were applied to calculate the wave functions. Initial atomic coordinates were those determined for the crystal. For rigid energy scans, a 10° step was set, and then for every conformer an EM062x was computed without molecular geometry optimization. Differences in energies (ΔE) compared to EM062x for the initial atomic coordinates were calculated, and their highest values are defined as energies of the rotation barrier. For relaxed energy scans, the geometric parameter under investigation was set to a starting value and then iteratively increased by 20°. For each scan step, the optimised molecular geometry EM062x was calculated. Conformational energy (ΔE) was calculated as the difference between the particular conformer EM062x and the lowest value of EM062x determined during the energy scan.
Nucleoside analysis by mass spectrometry
Total RNA was extracted from E. coli strain BW25113 cultured in LB broth for 18 h, using the acidic phenol method as reported previously (20). Total nucleosides were prepared by digesting E. coli total RNA with nuclease P1 (Wako Pure Chemical Industries) and bacterial alkaline phosphatase (BAP) (E. coli strain C75, TAKARA BIO INC.) under acidic conditions (31). For these reactions, a solution (typically 40 μl) containing 1 μg/μl total RNA, 20 mM trimethylamine-acetate (pH 5.3), nuclease P1 (0.1 units for 40 μg of RNA), and BAP (0.16 units for 40 μg of RNA) was incubated at 37°C for 1 h.
Total nucleosides were analyzed using a Q Exactive hybrid Quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific) equipped with an ESI source and Ultimate 3000 liquid chromatography system (Dionex). For HILIC/ESI-MS (31) was performed using a ZIC-cHILIC column (3 μm particle size, 2.1 × 150 mm, Merck Millipore). The mobile phase consisted of 5 mM ammonium acetate (pH 5.3) (solvent A) and acetonitrile (solvent B). Total nucleosides (2 μg) or synthetic ct6A (5 pmol) dissolved in 90% acetonitrile were injected and chromatographed with a flow rate of 100 μl/min in a multistep linear gradient: 90–40% B from 0 to 30 min, 40% B for 10 min, followed by initialization to 90% B. Proton adducts of nucleosides were scanned in positive polarity mode over an m/z range of 110–700. ODS/ESI-MS (32) was performed on a Sunshell C18 column (2.6 μm particle size, 2.1 × 150 mm, ChromaNik Technologies) with the same solvent system described above. The gradient program was as follows: 0–40% B from 0 to 30 min, 40% B for 5 min, followed by initialization to 0% B, at a flow rate of 75 μl/min. Total nucleosides (4 μg) or synthetic ct6A (10 pmol) dissolved in LC/MS grade ultrapure water (Wako) were injected. Positively charged nucleosides were scanned over an m/z range of 200–500. For higher-energy collision-induced dissociation (CID) in ODS/ESI-MS, the following gradient program was used: 0–15% B from 0 to 30 min with a curved gradient, 15–60% B from 30 to 35 min, 60% B for 10 min, followed by initialization to 0% B, at a flow rate of 75 μl/min. Bases liberated from nucleosides by in-source fragmentation (20 eV) were scanned over an m/z range of 240–400, and further scanned by data-dependent HCD with a normalized collision energy of 30. Total nucleosides (12 μg) or synthetic ct6A (400 pmol) dissolved in LC/MS grade ultrapure water were subjected to this analysis.
Epimerization of ct6A
For synthetic nucleosides, 4 nmol of l-ct6A or d-allo-ct6A was dissolved in 400 μl of 100 mM sodium borate buffer (pH 9.0) and incubated at 37°C for 5 min, followed by adding 10 μl of 3 M NaOAc (pH 5.2) to stop the reaction. The nucleosides were desalted by using Oasis HLB cartridge (3 ml, 30 mg, Waters), dried in vacuo and dissolved in water. 10 pmol of each sample was subjected to ODS/ESI-MS as described above.
For natural ct6A, 160 μg of E. coli total RNA in 400 μl of 100 mM sodium borate buffer (pH 9.0) was incubated at 37°C for 15 min, followed by adding 40 μl of 3 M NaOAc (pH 5.2) to stop the reaction and subjected to ethanol precipitation to recover total RNA. Nucleoside preparation was carried out by one-step acidic digestion (31) as described above. Four microgram of total nucleoside was subjected to ODS/ESI-MS. For co-injection analysis, 10 pmol of d-allo-ct6A was mixed.
For amine adduct formation, 4 nmol of l-ct6A or d-allo-ct6A, or 160 μg of E. coli total RNA was dissolved in 400 μl of 100 mM Tris–HCl (pH 8.5) and incubated at 37°C for 3 h. Further preparation was performed as the same procedure described above.
RESULTS
Chemical synthesis of ct6A
As reported previously (24), 5΄,3΄,2΄-O-acetylated derivative of l-ct6A was synthesized by dehydration of sugar protected l-t6A with acetic anhydride, while for l-ct6A preparation carbodiimide chemistry was applied (20). Here, for multimilligram scale synthesis, we used polymer-bound EDC as the activating agent for the carboxyl group of t6A. After the reaction, the polymer-bound EDC and its urea form product were removed by filtration, and the product was separated by silica gel column chromatography. The purity of the products was confirmed by HPLC analysis (Figure 1B). Yields of l-ct6A nucleoside synthesized in water and DMF as solvents were 49% and 51%, respectively. The resultant l-ct6A nucleoside was characterized by UV (Figure 1C), IR (Supplementary Figure S1B), 1H NMR (Supplementary Figure S2B), 13C NMR (Supplementary Figure S3B) and MS (Supplementary Figure S7); the results were identical to those reported previously (20).
We also synthesized diastereoisomer of ct6A with d-allo-threonine (Cα epimer) to examine possible epimerization of l-ct6A under mild alkaline conditions. First, we obtained d-allo-t6A nucleoside that was subjected to the cyclization with the same procedure as for l-ct6A nucleoside. The resultant d-allo-ct6A nucleoside was characterized by IR (Supplementary Figure S4B), 1H NMR (Supplementary Figure S5B) and 13C NMR (Supplementary Figure S6B). l-ct6A and d-allo-ct6A nucleosides were clearly separated by HPLC analysis (Supplementary Figure S8).
Hydantoin isoform of ct6A revealed by crystal structure
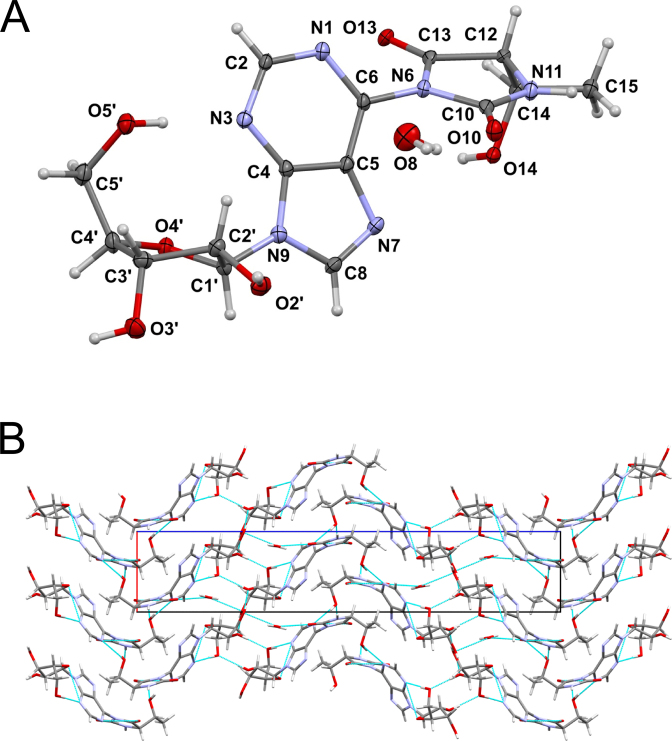
The ct6A nucleoside crystallized as the monohydrate in an orthorhombic crystal system of space group P212121. Additional X-ray crystallography data are summarized in Supplementary Tables S1–S5. Strikingly, the crystal structure showed that the N6-substituent of ct6A adopts the hydantoin form (Figure 2A), rather than the previously assigned oxazolone form (Figure 1A). This observation prompted us to check whether the isomerization occurs during the crystallization. However, careful analysis of ct6A nucleoside by TLC and HPLC before and after crystallization (data not shown) revealed that no isomerization occurred during this process.
Figure 2.
Crystal structure of chemically synthesized ct6A. (A) Crystal structure of the hydantoin isoform of ct6A. Displacement ellipsoids were drawn at the 50% probability level. Hydrogen atoms are represented by circles with an arbitrary radius. (B) Crystal packing of ct6A. Hydrogen bonds are indicated by dashed blue lines.
In this structure, the hydrogen bonds between molecules play a vital role in crystal packing (Supplementary Table S6). In particular, the N3 nitrogen forms a strong intermolecular bond with 5΄-OH in the ribose, stabilizing the syn conformation of the ct6A base (Figure 2A). The water molecule localized in the crystal links O3΄ of the ribose and O10 in the hydantoin ring from another molecule. These interactions enable formation of unique crystal packing with an infinite layer of sugar rings perpendicular to the [001] direction (Figure 2B).
Detailed structure of hydantoin ct6A
The bond lengths and valency angles in the adenosine and hydantoin moieties of ct6A (Supplementary Tables S3 and S4) are consistent with the respective values observed in crystal structures of similar chemical compounds deposited in the Cambridge Structural Database (CSD) (Supplementary Table S5) (30). The only significant difference in the ct6A nucleoside is the C6–N6 bond, which is longer (1.407(4) Å) than the corresponding bond in the structure of t6A nucleoside (1.3784(3) Å). This difference can be explained by repulsive interactions between the carbonyl oxygens of the ct6A hydantoin ring and the N1 and N7 nitrogens of the adenine moiety (Figure 1A). Similarly, elongated C6–N6 bond (1.413 Å) was found in the crystal structure of adenyl-N6-tetramethylsuccinimide (CSD-PULQIX), a modified adenosine with a cyclic imide containing the N6 nitrogen (33).
The ureido functionality in the ct6A hydantoin ring exhibits a significant electron density delocalization (Supplementary Figure S9). The elongated C10–N6 bond (1.428(4) Å) accompanied by the short C10–N11 bond (1.337(5) Å) (Supplementary Table S3) indicate a possibility of tautomeric conversion to the enol form with protonation at O10, which is additionally stabilized by hydrogen bonding with a nearby water molecule. Similarly, in the t6A structure, the ureido group is also influenced by electron density delocalization as indicated by relatively long C10–N6 (1.4089(3) Å) and short C10–N11 ((1.3216(3) Å)) bonds (34,35).
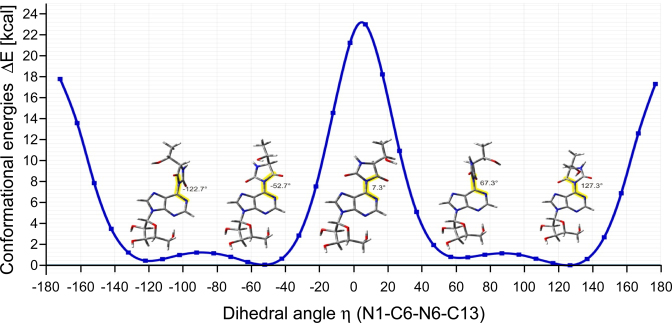
The relative orientation of the hydantoin and adenine rings in the ct6A crystal structure is described by the η(N1–C6–N6–C13) dihedral angle. It adopts value of -52.7(5)°, clearly indicating twisting arrangement of those rings (Figure 2A and Table 1). It is stabilized by the repulsive interactions between carbonyl oxygens of the hydantoin ring and the N1 and N7 nitrogens of the adenine moiety (Supplementary Figure S9). This repulsion restricts free rotation around C6–N6 bond. Indeed, an ab initio energy rigid scan over the η angle revealed two rotation barriers around η = 0° (ΔE = 23.0 kcal) and η = 180° (ΔE = 17.7 kcal) arising from the plane arrangement of these two rings (Figure 3). The dihedral angle η = −52.7(5)° observed in the crystal is close to the most stable conformation determined by the rigid energy scans (Figure 3). Similar rotation barriers were also observed by the relaxed energy scan (Supplementary Figure S10). The corresponding twisted arrangement of two rings was found in the crystal structure of adenyl-N6-tetramethylsuccinimide (CSD-PULQIX) with a corresponding dihedral angle η = −43.3° (33).
Table 1. Selected dihedral angles (°) of ct6A and t6A.
| Dihedral angles | ct6A | t6A |
|---|---|---|
| N6 substituent arrangement | ||
| N1–C6–N6–C13 (η) | −52.7(5) | −1.54(5) |
| Nucleoside conformation | ||
| C4΄–O4΄–C1΄C2΄ (ν0) | −23.1(4) | −21.31(5) |
| O4΄–C1΄–C2΄–C3΄ (ν1) | 35.9(3) | 31.72(5) |
| C1΄–C2΄–C3΄–C4΄ (ν2) | −34.1(3) | −29.49(5) |
| C2΄–C3΄–C4΄–O4΄ (ν3) | 22.0(4) | 17.99(5) |
| C3΄–C4΄–O4΄–C1΄ (ν4) | 0.4(4) | 1.95(5) |
| O5΄–C5΄–C4΄–C3΄ (γ) | 48.4(4) | 57.23(5) |
| O5΄–C5΄–C4΄–O4΄ (γ-) | −71.0(4) | −63.76(5) |
| C4–N9–C1΄–O4΄ (χ) | 51.0(4) | −153.41(5) |
Figure 3.
Ab initio energy calculation for the rigid scan over the dihedral angle η. The rigid energy scan over the dihedral angle η(N1–C6–N6–C13) was performed at 10° steps using the hybrid functional M062x and aug-cc-pvdz Dunning's basis set. Initial atomic coordinates were obtained from the crystal structure. ΔE is calculated as the EM062x difference between the coordinates of each conformer and the initial coordinates. Structural models of ct6A are depicted at the indicated η angles.
The ribose of ct6A adopts the C2΄-endo pucker conformation (Figure 2A) with the values of endocyclic torsion angles ν0 − ν4 shown in Table 1. The pseudorotation phase angle and puckering amplitude (36,37) are P = 160.9(3)° and τm = 37.2(2)°, respectively. Because the torsion angle about the C4΄–O4΄ bond is only 0.4°, the conformation of the ribose moiety is almost exactly 2E. Similar sugar ring puckering was also observed in the t6A crystal structure, with P = 157.8° and τm = 31.8° (35).
Conformation of the 5΄-hydroxymethyl group of the ct6A nucleoside is described by the dihedral angle γ and γ(–) (37) and their values (Table 1) define a gauche(+) conformation around the exocyclic C4΄–C5΄ bond. Similar conformation was also found in the t6A crystal structure (35).
The glycosidic dihedral angle χ = 51.0(4)° (Table 1) clearly indicates the syn conformation of the ct6A base. It is stabilized by the intramolecular hydrogen bond between 5΄OH of sugar moiety and N3 nitrogen of heterobase. This type of interaction has not been found in crystal structure of the t6A nucleoside which adopts the anti conformation around the N-glycosidic bond (35).
Spectroscopic characterization of ct6A
The ct6A nucleoside was further characterized by NMR spectroscopy. All 1H and 13C resonances of ct6A were unambiguously assigned by a combination of 1H,1H-COSY, 1H,1H-NOESY, 1H,13C-HSQC and 1H,13C-HMBC experiments, all of which are identical to our published data (20). However, these NMR analyses did not provide direct evidence allowing differentiation of the two isoforms of ct6A. To obtain a signature of the hydantoin isoform of ct6A, we further recorded 1H,15N-HSQC and 1H,15N-HMBC spectra for the ct6A nucleoside to determine 15N chemical shifts (Table 2 and Supplementary Figure S11). The chemical shifts of N6 (163 ppm) and N11 (87 ppm) of ct6A were in the range characteristic to urea-type nitrogens, consistent with the analogous nitrogens in the N3-phenyl hydantoin structure (38), supporting the idea that the hydantoin isoform actually exists in solution. In the case of the oxazolone isoform, the C=N–Ar type nitrogen would appear at much lower magnetic field (39).
Table 2. 15N chemical shifts of ct6A and t6A in DMSO.
| ct6A | t6A | |
|---|---|---|
| N-1 | 275 | 233 |
| N-3 | 254 | 236 |
| N-7 | 241 | 239 |
| N-9 | 171 | 172 |
| N-6 | 163 | 116 |
| N-11 | 87 | 93 |
15N chemical shifts (ppm) relative to liquid NH3.
We next obtained IR spectra of t6A (Supplementary Figure S1A) and ct6A (Supplementary Figure S1B). The characteristic ureido carbonyl absorption at 1680 cm−1 in t6A disappeared in the spectrum of ct6A, and was replaced by two absorption bands at 1780 and 1730 cm−1. These absorptions are in the same range as the hydantoin ring of 3-purin-6-yl-hydantoins (40), further supporting the idea that the synthetic ct6A adopts the hydantoin isoform.
Solution structure of hydantoin ct6A
Conformation of hydantoin ct6A in solution was determined using NMR spectroscopy. The conformation of the ribose was inferred by vicinal spin–spin coupling constants determined by 1H NMR spectra in D2O (Supplementary Figure S12). At room temperature, the C2΄-endo and C3΄-endo conformers of ct6A were equally populated in solution, while in the crystal it was C2΄-endo sugar pucker (Figure 2A). Regarding exocyclic C4΄–C5΄ bond, we found that gauche (+) is the preferred conformation (∼62%) of ct6A in solution which is consistent with the result of the crystal structure.
Then, we conducted 1D NOE experiments to examine orientation of the ct6A base relative to the ribose (41,42). When H8 was irradiated, strong NOE effect was observed to H2΄ rather than to H1΄ (Supplementary Figure S13 and Supplementary Table S7), suggesting that ct6A predominantly takes anti conformation in solution. Supporting this finding, a strong cross peak of H8–H2΄, and a weak cross peak of H8–H1΄ were also observed in 2D NOESY spectrum in D2O (Supplementary Figure S14). All of these results confirmed the anti conformation of ct6A. This observation is not consistent with that found in the crystal structure where the ct6A base adopts the syn conformation. The relative orientation of the hydantoin and adenine rings could not be provided by NMR data analyses as there are no vicinal coupling constants available for the Karplus-like relationship.
Presence of hydantoin ct6A in E. coli tRNAs
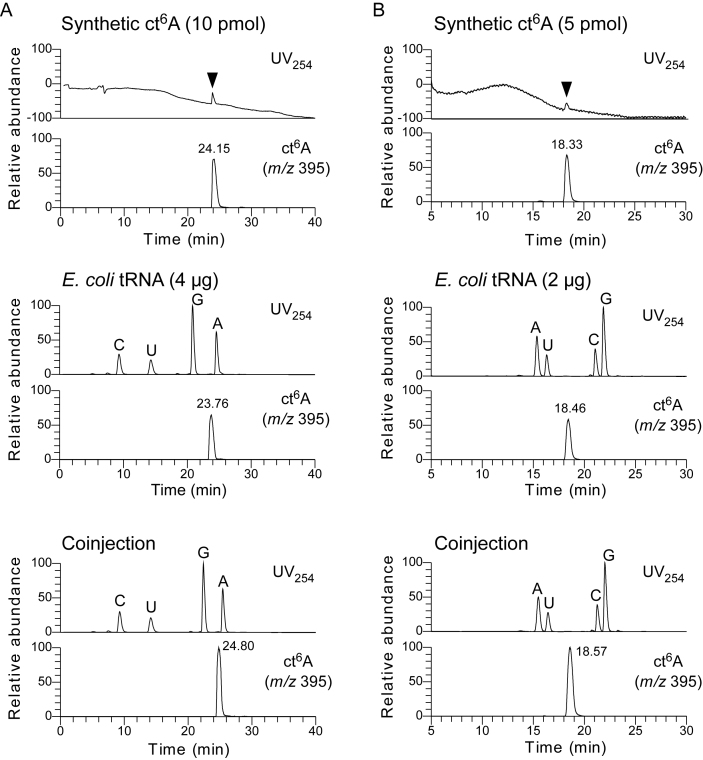
To determine whether the hydantoin isoform of ct6A is actually present in cellular tRNAs, we conducted LC/MS analyses to compare synthetic and natural ct6A. In reverse-phase column chromatography (ODS) coupled with mass spectrometry, the synthetic ct6A and natural ct6A in total nucleosides in E. coli tRNAs eluted at similar retention times, ∼24 min (Figure 4A). When both specimens were co-injected, we observed a single peak of mass chromatogram at m/z 394 (Figure 4A). In addition, we conducted the same MS analysis using hydrophilic interaction chromatography (HILIC). The synthetic and natural ct6A co-eluted at the same retention time as well (Figure 4B).
Figure 4.
LC/MS co-injection analyses of synthetic and natural ct6A nucleosides. LC/MS analyses of synthetic ct6A and E. coli total nucleosides by reverse-phase chromatography using octadecylsilyl resin (ODS, A), and by hydrophilic interaction chromatography (HILIC, B). UV traces (254 nm) and mass chromatograms (m/z 395) of synthetic ct6A (top), natural ct6A in E. coli total RNA (middle), and co-injected natural and synthetic ct6A (bottom). ct6A peaks in the UV trace are indicated by arrows.
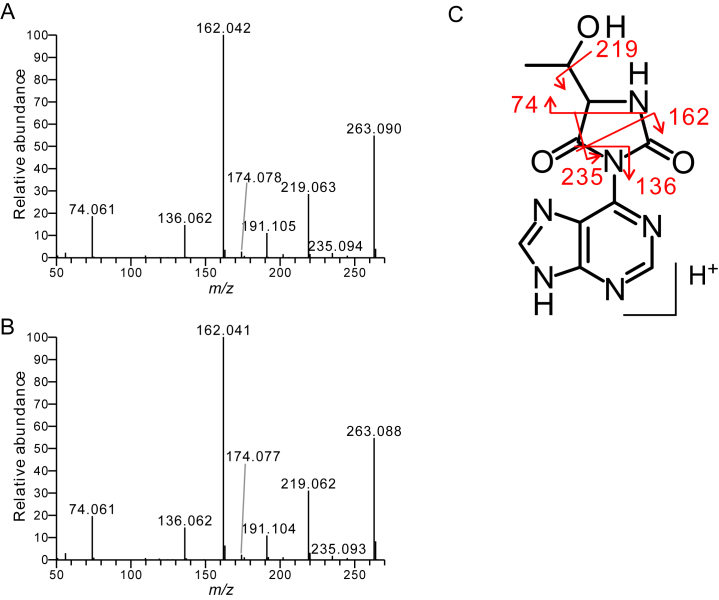
Next, we performed higher-energy collision-induced dissociation (CID) analysis of the base-related ion (BH2+) of both synthetic and natural ct6A. Product ions generated from the BH2+ of ct6A (m/z 263) were almost identical for both specimens (Figure 5A and B). These product ions could be assigned in the chemical structure of the hydantoin isoform of the ct6A base (Figure 5C).
Figure 5.
CID spectra of the ct6A base. CID spectra of the ct6A base (m/z 263) derived from synthetic ct6A nucleoside (A) and E. coli total nucleosides (B). (C) Product ions are assigned in the chemical structure of the hydantoin isoform of the ct6A base.
Epimerization of hydantoin ct6A under mild alkaline conditions
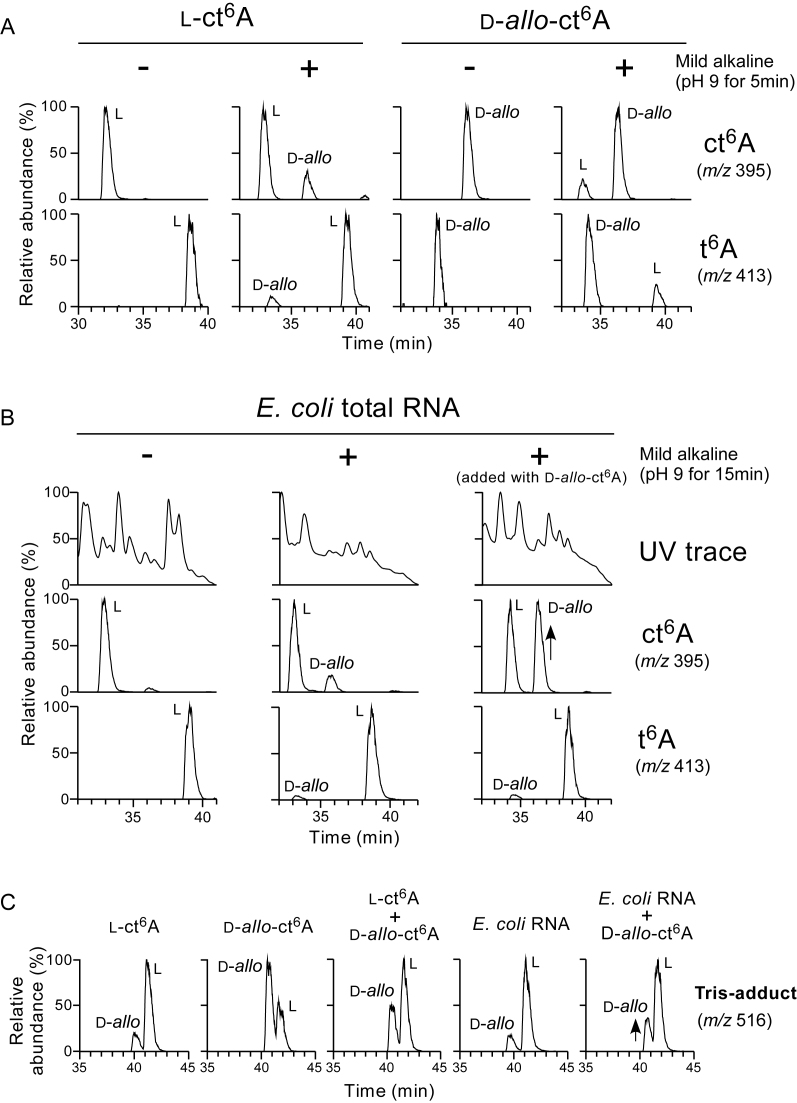
When total RNA was digested into nucleosides by conventional condition (pH 8.2), most of ct6A was hydrolyzed and converted to t6A as reported (20). In this condition, we noticed appearance of second peak of t6A that elutes faster than the original peak of t6A in LC/MS chromatogram (20). In addition, rest of ct6A also split and gave second peak that elutes slower than the original ct6A peak (20). When ct6A was incubated with Tris buffer, two peaks of Tris-adduct of t6A appeared (20). These findings prompted us to speculate that α-carbon of threonine residue in hydantoin ring is epimerized under mild alkaline treatment (Supplementary Figure S15). To confirm this speculation, ct6A epimer (d-allo-ct6A) was synthesized chemically, and compared to l-ct6A treated under mild alkaline condition and/or incubated with Tris buffer. As shown in Figure 6A, the synthetic l-ct6A gave a single peak in LC/MS. In addition, l-t6A, a hydrolyzed product of l-ct6A, was also detected. When l-ct6A was incubated with a buffer adjusted at pH 9 for 5 min, second peak of ct6A that eluted slower than the l-ct6A peak clearly appeared, while second peak of t6A that eluted faster than the l-t6A peak was also detected. Judging from the retention times of the synthetic d-allo-ct6A and its hydrolysate, d-allo-t6A, these second peaks were found to be d-allo-ct6A and d-allo-t6A, respectively (Figures 6A and Supplementary Figure S15). Furthermore, when the synthetic d-allo-ct6A was incubated under mild alkaline conditions (pH 9 for 5 min), l-ct6A and l-t6A clearly appeared. To confirm these findings, l-ct6A treated with mild alkaline was coinjected with d-allo-ct6A which was treated or untreated with mild alkaline (Supplementary Figure S16). Second peaks of l-ct6A appeared under mild alkaline conditions coeluted with d-allo-ct6A and d-allo-t6A, respectively.
Figure 6.
Epimerization of ct6A. (A) Epimerization of synthetic l-ct6A and D-allo-ct6A under mild alkaline condition. Each nucleoside was treated with (+) or without (−) mild alkaline (pH 9 for 5 min), and subjected to ODS/ESI-MS analysis to detect ct6A (m/z 395, upper panels) and t6A (m/z 413, lower panels). A peak corresponding to each epimer is indicated in the chromatographs. (B) Epimerization of natural ct6A in E. coli total RNA under mild alkaline condition. E. coli total RNA was treated with (+) or without (−) mild alkaline (pH 9 for 15 min), digested into nucleoside under acidic condition, and subjected to ODS/ESI-MS analysis to detect UV trace at 254 nm (top panels), ct6A (m/z 395, middle panels) and t6A (m/z 413, bottom panels). (C) Epimerization of amine adduct of t6A. Synthetic ct6A epimers and E. coli total RNA were incubated with Tris buffer (pH 8.5) for 3 h. Nucleosides were analyzed by ODS/ESI-MS to detect Tris-adduct of t6A (m/z 516).
Next we examined epimerization of natural ct6A in E. coli tRNAs. As reported previously (20), when E. coli total RNA was incubated with a buffer at pH 9 for 15 min, we detected d-allo-ct6A along with d-allo-t6A (Figure 6B). d-allo-ct6A in E. coli total RNA was confirmed by coinjection with synthetic specimen. Moreover, we examined epimerization of t6A upon adduct formation with amine. When the synthetic or natural ct6A was incubated with Tris buffer at pH 8.5, both l-type and d-allo-type Tris adducts of t6A were generated (Figure 6C).
Collectively, these findings demonstrate that epimerization of ct6A takes place under alkaline conditions, even at pH 8.5, followed by epimer formation of t6A and its amine-adducts.
DISCUSSION
The crystal structure of the ct6A nucleoside clearly demonstrates that ct6A adopts a hydantoin isoform (Figure 2A), rather than the previously predicted oxazolone isoform (20). We also confirmed that the hydantoin ct6A is actually present in E. coli tRNAs (Figure 4). As shown here, it was impossible to differentiate the two isoforms by a series of NMR and MS analyses, explaining the reason why the chemical structure of ct6A was once assigned to be the oxazolone isoform. For instance, the product ions of CID spectrum of ct6A base (Figure 5A and B) could be assigned to the hydantoin isoform as well as to oxazolone isoform (Supplementary Figure S17) (20). Additionally, a pattern of the scalar couplings observed in 1H–1H-COSY spectra of both isoforms is very similar. In 1H–1H-COSY spectrum of ct6A, we observed cross peaks between NH11–H12, H12–H14, H14–OH14 and H14–H15 attributed to the threonine side chain (Supplementary Figure S18), however, these couplings are present in both hydantoin and oxazolone isoforms. No coupling was found between amino acid side chain and adenine base. In this study, however, we have found three spectroscopic features implying the presence of a hydantoin group in ct6A. First, the unique chemical shift of N6 atom (163 ppm) of ct6A in 15N NMR implies the existence of hydantoin structure (Table 2). Second, the chemical shift of NH11 proton (9.72 ppm) of t6A moved toward high magnetic field by 1 ppm (8.72 ppm) in ct6A (Supplementary Figure S2), supporting the fact that hydrogen bond of NH11–N1 is impossible in the hydantoin isoform (Figure 1A). Third, two characteristic absorption bands at 1780 and 1730 cm−1 in the IR spectrum of ct6A nucleoside (Supplementary Figure S1) also indicate carbonyl bond stretching in the hydantoin ring.
TcdA catalyzes ATP-dependent dehydration of t6A to form ct6A on tRNA (20). Given that TcdA is a ubiquitin-activating E1-like protein with ATPase activity, it is likely that it first adenylates the carboxyl group of t6A to form an activated ester intermediate, and subsequently allows the N6 nitrogen of t6A to attack the C13 carbon to cyclize the side chain with release of AMP. This reaction mechanism resembles the chemical synthesis of ct6A using a carbodiimide-type activating reagent such as EDC. Intriguingly, a previous study reported hydantoin ring formation of the t6A base using N,N’-dicyclohexylcarbodiimide (DCC) (40). The activated ester intermediate of t6A with EDC or DCC is analogous to the adenylate intermediate of t6A generated by TcdA.
ct6A is easily hydrolyzed to convert to t6A under mild alkaline conditions (20). In addition, primary amines such as tris(hydroxymethyl)aminomethane or ethanolamine react efficiently with ct6A to generate amides of t6A (20,43,44). The hydantoin ct6A (Figure 1A) has two carbonyl carbons, C10 and C13, both of which could potentially be attacked by a water molecule or an amine. According to the general hydantoin chemistry, the non-ureido carbonyl carbon of a hydantoin compound is more susceptible to the reaction with nucleophiles, yielding the hydantoic acid and its derivatives (45–47). Thus, in the case of ct6A, C13 carbonyl carbon is naturally targeted by hydrolysis (20,40) and amine adduct formation (20,43,44), whereas the ureido C10 is comparatively non-reactive. In addition, we here showed that hydantoin ct6A is susceptible to epimerization under mild alkaline conditions, indicating that H12 in hydantoin ring is acidic enough for such process (48,49). It was shown that hydantoin derivatives are racemized as the result of tautomeric change (50). Just for 5 min incubation of l-ct6A at pH 9, 20–30% of l-ct6A is epimerized to yield d-allo-ct6A, and then it is hydrolyzed to form d-allo-t6A. The epimerization of ct6A and t6A also takes place at pH 8.2 under conventional nucleoside preparation (20). For tRNA preparation used for in vitro translation or other biochemical experiments, aminoacyl-moieties attached to 3΄ termini of aa-tRNAs isolated from the cell are frequently removed by mild alkaline treatment. According to the original methods, tRNA fractions extracted from the cell are incubated with 500 mM Tris-HCL (pH 8.8) (51) or 200 mM glycine buffer (pH 10.3) (52) for their deacylation. In these conditions, ct6A in tRNAs should be epimerized and converted to l-t6A and d-allo-t6A as well as Tris- and glycine-adducts of t6A, respectively. If d-allo-ct6A and d-allo-t6A have different activity in protein synthesis from their l-isomers, translational activity of tRNAs having ct6A should be reconsidered carefully. From these findings, we have learned that usage of primary amines and mild alkaline conditions should be avoided to handle and prepare tRNAs bearing ct6A.
As shown in the crystal structure of ct6A nucleoside, the hydantoin ring adopts a twisted position with a torsion angle of −52.7° against the adenine base (Figure 2A). From the ab initio energy calculation of hydantoin ring rotation, we identified four energy minimum rotamers with different η dihedral angles (127.3°, 67.3°, −52.7° and −122.7°) between the hydantoin and adenine rings (Figure 3). They all have ‘twisted conformation’ due to repulsive interaction between carbonyl oxygens of the hydantoin ring and the N1 and N7 nitrogens of the adenine moiety. In fact, actual conformation of ct6A in the crystal structure adopts one of these energy minimum rotamers. Although we do not have any experimental evidence to determine the orientation of the rings in solution, it is quite natural to assume that ct6A adopts ‘twisted conformation’ in solution, because repulsive interaction between the two rings should come from very basic chemical nature of ct6A. Strong rotational barriers (23 and 17.7 kcal) of the planar arrangement of the two rings of ct6A (Figure 3) restrict free rotation of C6–N6 bond. However, given that these barriers can be overcome by heat energy at normal growth temperatures, the hydantoin ring might be able to rotate to some extent under physiological conditions.
As observed in the crystal structure of the t6A37-containing anticodon stem–loop recognizing the AAG codon at the ribosomal A site (16), the N1–NH11 hydrogen bond extends a planar adenine base that stabilizes codon–anticodon pairing as well as the anticodon stem–loop structure. One possible structural model of the oxazolone ct6A (20) indicated that the oxazolone ring favors being fixed at the planar position with the adenine base to extend its π-conjugated system, enabling a strong stacking interaction with the codon–anticodon helix. This model provides a potential molecular basis for the contribution of ct6A to recognition of the adenine base of the ANN codon. However, the hydantoin ring of ct6A cannot adopt a planar position with the adenine base due to strong coulombic repulsion between the two rings. If there is no conformational restriction in the decoding center, the hydantoin ct6A may adopt one of the four twisted conformations on the ribosome. Taking account of the twisted conformation and low probability of π–π stacking of the hydantoin ring, it is an enigmatic issue how the twisted hydantoin ring stabilizes codon–anticodon interaction at the ribosomal A-site. Structural studies of 70S ribosome complexed with tRNA with ct6A37 and its cognate codon will be required to reveal the actual conformation of the hydantoin ring at the decoding center.
We here confirmed that the hydantoin isoform of ct6A is present in E. coli tRNAs. Presumably, this isoform is present in tRNAs from other organisms. However, we cannot rule out the possibility that the oxazolone isoform of ct6A may serve as an intermediate of the hydantoin isoform during biogenesis of ct6A.
AVAILABILITY
Crystallographic data (excluding structure factors) for the structures reported herein, have been deposited with the Cambridge Crystallographic Data Centre under accession number CCDC 1458975. Copies of the data can be obtained free of charge at http://www.ccdc.cam.ac.uk/.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to all members of the Sochacka and Suzuki laboratories for their technical support and many insightful discussions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Centre in Poland [UMO-2014/13/N/ST5/01591 to M.M.]; Grants-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports and Culture of Japan (to T.S.). Funding for open access charge: Grants-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports, and Culture of Japan.
Conflict of interest statement. None declared.
REFERENCES
- 1.Machnicka M.A., Milanowska K., Osman Oglou O., Purta E., Kurkowska M., Olchowik A., Januszewski W., Kalinowski S., Dunin-Horkawicz S., Rother K.M. et al. MODOMICS: a database of RNA modification pathways–2013 update. Nucleic Acids Res. 2013; 41:D262–D267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bjork G. Soll D, RajBhandary UL. tRNA: Structure, Biosynthesis, and Function. 1995; Washington, D.C.: American Society for Microbiology; 165–205. [Google Scholar]
- 3.Grosjean H. Grosjean H. DNA and RNA Modification Enzymes: Structure, Mechanism, Function and Evolution. 2009; Austin: Landes Bioscience; 1–18. [Google Scholar]
- 4.Suzuki T. Grosjean H. Fine-Tuning of RNA Functions by Modification and Editing. 2005; 12, Springer-Verlag Berlin and Heidelberg GmbH & Co. KG; 23–69. [Google Scholar]
- 5.Helm M., Alfonzo J.D.. Posttranscriptional RNA modifications: playing metabolic games in a cell's chemical Legoland. Chem. Biol. 2014; 21:174–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Agris P.F., Vendeix F.A., Graham W.D.. tRNA's wobble decoding of the genome: 40 years of modification. J. Mol. Biol. 2007; 366:1–13. [DOI] [PubMed] [Google Scholar]
- 7.Yokoyama S., Nishimura S.. Soll D, RajBhandary UL. tRNA: Structure, Biosynthesis, and Function. 1995; Washington, D.C.: American Society for Microbiology; 207–224. [Google Scholar]
- 8.El Yacoubi B., Bailly M., de Crécy-Lagard V.. Biosynthesis and function of posttranscriptional modifications of transfer RNAs. Annu. Rev. Genet. 2012; 46:69–95. [DOI] [PubMed] [Google Scholar]
- 9.Gustilo E.M., Vendeix F.A., Agris P.F.. tRNA's modifications bring order to gene expression. Curr. Opin. Microbiol. 2008; 11:134–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schweizer M.P., Chheda G.B., Baczynskyj L., Hall R.H.. Aminoacyl nucleosides. VII. N-(Purin-6-ylcarbamoyl)threonine. A new component of transfer ribonucleic acid. Biochemistry. 1969; 8:3283–3289. [DOI] [PubMed] [Google Scholar]
- 11.Niimi T., Nureki O., Yokogawa T., Hayashi N., Nishikawa K., Watanabe K., Yokoyama S.. Recognition of the anticodon loop of tRNA(1)(Ile) by isoleucyl-transfer-RNA synthetase from Escherichia coli. Nucleos. Nucleot. Nucl. 1994; 13:1231–1237. [Google Scholar]
- 12.Yarian C., Townsend H., Czestkowski W., Sochacka E., Malkiewicz A.J., Guenther R., Miskiewicz A., Agris P.F.. Accurate translation of the genetic code depends on tRNA modified nucleosides. J. Biol. Chem. 2002; 277:16391–16395. [DOI] [PubMed] [Google Scholar]
- 13.Phelps S.S., Malkiewicz A., Agris P.F., Joseph S.. Modified nucleotides in tRNA(Lys) and tRNA(Val) are important for translocation. J. Mol. Biol. 2004; 338:439–444. [DOI] [PubMed] [Google Scholar]
- 14.El Yacoubi B., Hatin I., Deutsch C., Kahveci T., Rousset J.P., Iwata-Reuyl D., Murzin A.G., de Crecy-Lagard V.. A role for the universal Kae1/Qri7/YgjD (COG0533) family in tRNA modification. EMBO J. 2011; 30:882–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin C.A., Ellis S.R., True H.L.. The Sua5 protein is essential for normal translational regulation in yeast. Mol. Cell. Biol. 2010; 30:354–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murphy F.V., Ramakrishnan V., Malkiewicz A., Agris P.F. The role of modifications in codon discrimination by tRNA(Lys)UUU. Nat. Struct. Mol. Biol. 2004; 11:1186–1191. [DOI] [PubMed] [Google Scholar]
- 17.Stuart J.W., Gdaniec Z., Guenther R., Marszalek M., Sochacka E., Malkiewicz A., Agris P.F.. Functional anticodon architecture of human tRNALys3 includes disruption of intraloop hydrogen bonding by the naturally occurring amino acid modification, t6A. Biochemistry. 2000; 39:13396–13404. [DOI] [PubMed] [Google Scholar]
- 18.Durant P.C., Bajji A.C., Sundaram M., Kumar R.K., Davis D.R.. Structural effects of hypermodified nucleosides in the Escherichia coli and human tRNALys anticodon loop: the effect of nucleosides s2U, mcm5U, mcm5s2U, mnm5s2U, t6A, and ms2t6A. Biochemistry. 2005; 44:8078–8089. [DOI] [PubMed] [Google Scholar]
- 19.Lescrinier E., Nauwelaerts K., Zanier K., Poesen K., Sattler M., Herdewijn P.. The naturally occurring N6-threonyl adenine in anticodon loop of Schizosaccharomyces pombe tRNAi causes formation of a unique U-turn motif. Nucleic Acids Res. 2006; 34:2878–2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyauchi K., Kimura S., Suzuki T.. A cyclic form of N6-threonylcarbamoyladenosine as a widely distributed tRNA hypermodification. Nat. Chem. Biol. 2013; 9:105–111. [DOI] [PubMed] [Google Scholar]
- 21.Crain P.F. Preparation and enzymatic hydrolysis of DNA and RNA for mass spectrometry. Methods Enzymol. 1990; 193:782–790. [DOI] [PubMed] [Google Scholar]
- 22.Kim S., Lee H., Park S.. The structure of Escherichia coli TcdA (also known as CsdL) reveals a novel topology and provides insight into the tRNA binding surface required for N(6)-threonylcarbamoyladenosine dehydratase activity. J. Mol. Biol. 2015; 427:3074–3085. [DOI] [PubMed] [Google Scholar]
- 23.Lopez-Estepa M., Arda A., Savko M., Round A., Shepard W.E., Bruix M., Coll M., Fernandez F.J., Jimenez-Barbero J., Vega M.C.. The crystal structure and small-angle X-ray analysis of CsdL/TcdA reveal a new tRNA binding motif in the MoeB/E1 superfamily. PLoS One. 2015; 10:e0118606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matuszewski M., Sochacka E.. Stability studies on the newly discovered cyclic form of tRNA N(6)-threonylcarbamoyladenosine (ct(6)A). Bioorg. Med. Chem. Lett. 2014; 24:2703–2706. [DOI] [PubMed] [Google Scholar]
- 25.Chheda G., Hong C.I.. Synthesis of naturally occurring 6-ureidopurines and their nucleosides. J. Med. Chem. 1971; 14:748–753. [DOI] [PubMed] [Google Scholar]
- 26.Hong C.I., Chheda G.B.. Towsend LB, Tipson RS. Nucleic Acid Chemistry. 1972; NY: John Wiley & Sons; 661–664. [Google Scholar]
- 27.Hong C.I., Chheda G.B., Dutta S.P., O'Grady-Curtis A., Tritsch G.L.. Synthesis and biological activity of analogs of naturally occurring 6-ureidopurines and their nucleosides. J. Med. Chem. 1973; 16:139–147. [DOI] [PubMed] [Google Scholar]
- 28.Adamiak R.W., Wiewiórowski M.. The modified nucleosides of tRNAs. Synthesis and spectra of some natural ureidonucleosides. Bull. Acad. Pol. Sci., Ser. Sci. Chim. 1975; 23:241–253. [Google Scholar]
- 29.Martin D., Schlimme E.. Preparation of ureidonucleosides of the threonine isomers. Z. Naturforsch. C. 1994; 49:834–842. [Google Scholar]
- 30.Allen F.H. The Cambridge Structural Database: a quarter of a million crystal structures and rising. Acta Crystallogr., Sect. B: Struct. Sci. 2002; 58:380–388. [DOI] [PubMed] [Google Scholar]
- 31.Sakaguchi Y., Miyauchi K., Kang B.I., Suzuki T.. Nucleoside analysis by hydrophilic interaction liquid chromatography coupled with mass spectrometry. Methods Enzymol. 2015; 560:19–28. [DOI] [PubMed] [Google Scholar]
- 32.Suzuki T., Ikeuchi Y., Noma A., Suzuki T., Sakaguchi Y.. Mass spectrometric identification and characterization of RNA-modifying enzymes. Methods Enzymol. 2007; 425:211–229. [DOI] [PubMed] [Google Scholar]
- 33.Arico J.W., Calhoun A.K., Salandria K.J., McLaughlin L.W.. Tetramethylsuccinimide as a directing/protecting group in purine glycosylations. Org. Lett. 2010; 12:120–122. [DOI] [PubMed] [Google Scholar]
- 34.Parthasarathy R., Ohrt J.M., Chheda G.B.. Conformation and possible role of hypermodified nucleosides adjacent to 3΄-end of anticodon in tRNA: N-(purin-6-ylcarbamoyl)-L-threonine riboside. Biochem. Biophys. Res. Commun. 1974; 60:211–218. [DOI] [PubMed] [Google Scholar]
- 35.Parthasarathy R., Ohrt J.M., Chheda G.B.. Modified nucleosides and conformation of anticodon loops: crystal structure of t6A and g6A. Biochemistry. 1977; 16:4999–5008. [DOI] [PubMed] [Google Scholar]
- 36.Altona C., Sundaralingam M.. Conformational analysis of the sugar ring in nucleosides and nucleotides. A new description using the concept of pseudorotation. J. Am. Chem. Soc. 1972; 94:8205–8212. [DOI] [PubMed] [Google Scholar]
- 37.Saenger W. Principles of Nucleic Acids Structure. 1984; NY: Springer. [Google Scholar]
- 38.Buchman R., Komoroski R.A.. A carbon-13 and nitrogen-15 NMR study of some nitrogen heterocycles. J. Heterocyc. Chem. 1980; 17:1089–1092. [Google Scholar]
- 39.Westerman P.W., Botto R.E., Roberts J.D.. Substituent and medium effects on Nitrogen-15 shieldings of compounds with >C=N bonds (imines, oximes and phenylhydrazones). J. Org. Chem. 1978; 43:2590–2596. [Google Scholar]
- 40.Hong C.I., Chheda G.B.. Purinylhydantoins. Facile conversion of the naturally occurring N-(purin-6-ylcarbamoyl)-L-amino acids into 3-purin-6-ylhydantoins and 3-cyclohexyl-1-(purin-6-ylcarbamoyl)hydantoins. J. Med. Chem. 1975; 18:79–84. [DOI] [PubMed] [Google Scholar]
- 41.Rosemeyer H., Toth G., Golankiewicz B., Kazimierczuk Z., Bourgeois W., Kretschmer U., Muth H.P., Seela F.. Syn-anti conformational analysis of regular and modified nucleosides by 1D 1H NOE difference spectroscopy: a simple graphical method based on conformationally rigid molecules. J. Org. Chem. 1990; 55:5784–5790. [Google Scholar]
- 42.Lynch S.R., Tinoco I. Jr. The structure of the L3 loop from the hepatitis delta virus ribozyme: a syn cytidine. Nucleic Acids Res. 1998; 26:980–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nishimura S. Minor components in transfer RNA: their characterization, location, and function. Prog. Nucleic Acid Res. Mol. Biol. 1972; 12:49–85. [PubMed] [Google Scholar]
- 44.Kasai H., Murao K., Nishimura S., Liehr J.G., Crain P.F., Mccloskey J.A.. Structure determination of a modified nucleoside isolated from Escherichia-coli transfer ribonucleic-acid - N-(N-((9-beta-D-ribofuranosylpurin-6-Yl)carbamoyl)threonyl)2-amido-2-hydroxymethylpropane-1,3-diol. Eur. J. Biochem. 1976; 69:435–444. [Google Scholar]
- 45.Blagoeva I.B., Pojarlieff I.G., Dimitrov V.S.. Alkaline-hydrolysis of hydantoin, 3-methylhydantoin, and 1-acetyl-3-methylurea - effect of ring size on cleavage of acylureas. J. Chem. Soc., Perkin Trans. 2. 1978; 887–892. [Google Scholar]
- 46.Meusel M., Gutschow M.. Recent developments in hydantoin chemistry – a review. Org. Prep. Proced. Int. 2004; 36:391–443. [Google Scholar]
- 47.Lopez C.A., Trigo G.G.. The chemistry of hydantoins. Adv. Heterocycl. Chem. 1985; 38:177–228. [Google Scholar]
- 48.Dudley K.H., Bius D.L.. Buffer catalysis of the racemization reaction of some 5-phenylhydantoins and its relation to the in vivo metabolism of ethotoin. Drug Metab. Dispos. 1976; 4:340–348. [PubMed] [Google Scholar]
- 49.Lazarus R.A. Chemical racemization of 5-benzylhydantoin. J. Org. Chem. 1990; 55:4755–4757. [Google Scholar]
- 50.Dakin H.D. The catalytic racemization of optically active hydantoin derivatives and of related substances as the result of tautomeric change. Am. Chem. J. 1910; 44:48–60. [Google Scholar]
- 51.Von Ehrenstein G., Lipmann F.. Experiments on hemoglobin biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 1961; 47:941–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zubay G. The isolation and fractionation of soluble ribonucleic acid. J. Mol. Biol. 1962; 4:347–356. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.