Abstract
Telomere repeat binding factor 2 (TRF2) has been increasingly recognized to be involved in telomere maintenance and DNA damage response. Here, we show that TRF2 directly binds SIRT6 in a DNA independent manner and that this interaction is increased upon replication stress. Knockdown of SIRT6 up-regulates TRF2 protein levels and counteracts its down-regulation during DNA damage response, leading to cell survival. Moreover, we report that SIRT6 deactetylates in vivo the TRFH domain of TRF2, which in turn, is ubiquitylated in vivo activating the ubiquitin-dependent proteolysis. Notably, overexpression of the TRF2cT mutant failed to be stabilized by SIRT6 depletion, demonstrating that the TRFH domain is required for its post-transcriptional modification. Finally, we report an inverse correlation between SIRT6 and TRF2 protein expression levels in a cohort of colon rectal cancer patients. Taken together our findings describe TRF2 as a novel SIRT6 substrate and demonstrate that acetylation of TRF2 plays a crucial role in the regulation of TRF2 protein stability, thus providing a new route for modulating its expression level during oncogenesis and damage response.
INTRODUCTION
The telomere repeat binding factor 2 (TRF2) is a key regulator of telomere integrity by blocking ATM signaling and non-homologous end joining (NHEJ) as well as by favoring telomere replication (1–4). In addition to confer telomeric binding specificity of the shelterin complex, TRF2 performs telomeric protective functions through multiple activities, including a direct control of several DDR factors involved in the activation and the propagation of ATM signaling (5–7), the folding of the 3΄ single-stranded G overhang into T-loops (8–12), the regulation of telomeric DNA topology (12) and a restriction of resolvase activity at telomeres (13,14). There are also increasing pieces of evidence showing that TRF2 is also involved in extra-telomeric functions (15). By combining chromatin immunoprecipitation with high-throughput DNA sequencing (ChIP-Seq), TRF2 was shown to occupy a set of interstitial telomeric sequences (ITSs), where it can act as a transcriptional activator (16–19). Another transcriptional activity of TRF2 relies on its binding to the Repressor Element 1-Silencing Transcription factor (REST) involved in the regulation of neural differentiation (20–22).
TRF2 also plays a role in general DNA damage response. It rapidly associates with non-telomeric double strand break sites (DSBs; (23)) where its transient phosphorylation by ATM (24) is required for the fast pathway of DSB repair (25). While depletion of TRF2 impairs homologous recombination (HR) repair and has no effects on NHEJ, overexpression of TRF2 stimulates HR and inhibits NHEJ (26).
The various biological activities of TRF2 rely on its specific protein domains: an N-terminal basic domain rich in glycine and arginine residues (GAR or basic domain), which can bind the non-coding telomeric RNA (TERRA) and DNA junctions in a telomere sequence-independent manner (27,13); a TRFH domain, which behaves as a hub for several proteins involved in DNA repair (28) and which harbors a set of lysine residues implicated in the telomere DNA wrapping ability of TRF2 (12); a flexible hinge domain, which contains the interacting sites of TRF2 with other shelterin proteins, such as RAP1 and TIN2 (29); and a C-terminal Myb/homeodomain-like telobox DNA-binding domain, which has specificity for telomeric TTAGGG repeats (30–32).
The expression of TRF2 is downregulated during aging since its stability decreases during replicative senescence upon p53 activation through a ubiquitin-mediated proteosomal degradation pathway (33,34). In contrast, TRF2 is up-regulated in many cancers (18–19,35–39) where it appears to be directly regulated by the canonical Wnt/b-catenin and WT1 pathways (19,40). In cancer cells, TRF2 can promote oncogenesis by a cell extrinsic mechanism involving Natural Killer cell inhibition through the binding and the activation of the ITS-containing HS3ST4 gene encoding for the heparan sulphate (glucosamine) 3-O-sulphotransferase (18,41). Overall, it emerges that TRF2 plays a key role during development, aging and cancer by controlling cell proliferation through both chromosome maintenance and genome-wide transcriptional regulation (15). In agreement with this view, TRF2-compromised zebrafishes show a premature neuroaging phenotype (42).
Another rate-of-aging regulator of telomere stability, DNA repair and transcriptional regulation is SIRT6, a member of the sirtuin family consisting of conserved proteins with deacylase activities that require the cellular metabolite NAD+ (nicotinamide adenine dinucleotide), thus linking them to cellular metabolism. Loss of SIRT6 leads to the formation of dysfunctional telomeres precipitating cells into cellular senescence (43). SIRT6 also regulates transcriptional silencing at telomeres and subtelomere regions (44). Moreoveer, following DNA damage, SIRT6 is recruited to DSBs ensuring the proper activation of downstream DDR factors leading to an efficient DNA repair. At chromatin level, SIRT6 deacetylates the histone H3 on acetylated K9, K56 (43,45) and the more recently identified K18 residue (46), causing the repression of many genes differently involved in inflammation, aging, genome stability, metabolic pathways and telomere integrity (47–51). Notably, many functions of SIRT6 are linked to its ability to deacetylate and catalyze mono-ADP-ribosylation of nonhistone proteins (52–54), and deacetylate long-chain fatty acil groups (55).
In this study, we identify SIRT6 as a new player among the TRF2-interacting partners. We demonstrate that the TRF2/SIRT6 association does not require DNA and is increased upon replication stress-inducing agents. Moreover, we provide insight into the post-transcriptional regulation of TRF2 whose stability is affected by DNA damage in a SIRT6-dependent manner. Consistent with the data describing SIRT6 as a tumor suppressor (56), an inverse correlation between SIRT6 and TRF2 protein expression levels has been found in a cohort of colorectal cancer (CRC) patients.
MATERIALS AND METHODS
Cells, culture condition and transfection
Human cervix carcinoma HeLa cells were purchased by the ATCC repository and maintained according to the manufacturer's instructions. Human immortilized BJ-hTERT and transformed BJ-EHLT/RasV12 fibroblasts were obtained and maintained as described (57,58). The wild-type and p53-deficient colon carcinoma HCT116 cells were obtained by Dr Vogelstein, Johns Hopkins University. All the cell lines were grown in Dulbecco modified eagle medium (DMEM; Invitrogen, Carlsbad, CA, USA) containing 10% fetal calf serum.
Empty-, TRF2wt- or TRF2cT-overexpressing cells were obtained by infecting with amphotrophic retroviruses generated by transient transfection of retroviral vectors (pBabe-puro-Empty, pBabe-puro-mycTRF2 and pLPC-Myc-TRFcT; the last one was a gift from Eros Lazzerini Denchi, Addgene plasmid #44573; 7) into Phoenix amphotropic packaging cells with JetPEI (Polyplus, New York, NY, USA), according to the manufacturer's instructions. For transient RNA interference experiments, siTRF2 and siPARP1 were purchased from Dharmacon Inc. (Chicago, USA), siSIRT6, siSIRT1 and siGFP were purchased from Santa Cruz Biotechnology Inc. (Santa Cruz, CA, USA) and transfected into HCT116 or HeLa cells with Interferin (Polyplus) according to the manufacturer's instructions.
Drugs and treatments
In the experiments of immunoprecipitation or western blot, the following drugs were used: (S)-(+)-camptothecin at 0.2 or 2 μM for 2 h (CPT), hydroxyurea at 5 mM for 3 h (HU; Sigma Chemicals, Milano, Italy), cisplatin at 5 μg/ml for 2 h (DDP; Prontoplatamine; Pharmacia), paclitaxel at 10 nM for 24 h (Taxol; Bristol-Myers Squibb), bleomycin at 2 μM for 3 h (Bleo; Euro Nippon Kayaku), PARP1 inhibitor NU1025 at 200 μM for 16 h (Santa Cruz Biotechnology, Santa Cruz, CA, USA), Cycloheximide 100 μg/ml for 24 h (CHX; Sigma).
For colony formation assay, cells were seeded at a density of 5 × 104 cells/plate and exposed 24 h later to the following drugs: CPT (0.05–1 μM for 2 h); Bleo (0.1–0.5 μM for 2 h); Taxol (0.1–2 nM for 24 h). At the end of every treatment, 500 cells for each condition were seeded into 60-mm plates and, after 10 days, colonies were stained with 2% methylene blue in a 10% ethanol solution and counted.
Real-time quantitative PCR
RNA was extracted with Trizol reagent (Invitrogen) and converted to complementary DNA with the Tetro Reverse Transcriptase (Bioline, London, UK). Real-time quantitative PCR (qPCR) was performed in triplicate using the 7500 Real Time PCR System (Applied Biosystems, Foster City, CA, USA). The following primers were used: TRF2-FWR 5΄-CTTCTGATGCAAATGCAAAGG-3΄; TRF2-REV 5΄-AGACAGCAAGCACAACAC-3΄; GAPDH-FWR 5΄-AGCCTCCCGCTTCGCTCTCT-3΄; GAPDH-REV 5΄-GCCAGCATCGCCCCACTTGA-3΄.
The specificity of each PCR products was controlled using the melting curve. The relative gene expression levels were calculated using the 2∧(-ΔΔCt) method, where Ct represents the threshold cycle, and GAPDH was used as a reference gene.
Immunofluorescence
Cells fixed with 2% formaldehyde and permeabilized in 0.25% Triton X100 in phosphate buffered saline (PBS) for 5 min at room temperature (RT) were incubated with the following primary antibodies: mAb anti-TRF2 (clone 4A794; Millipore, Billerica, MA, USA); pAb anti-TRF2 (Novus Biologicals, Littleton, CO, USA); pAb anti-SIRT6 (ab62738; Abcam, Cambridge, UK); mAb anti-phospho-Histone H2AX (Ser139, clone JBW301; Millipore). Then, samples were incubated with the secondary antibodies (goat anti-mouse FITC, donkey anti-goat Cy™5, or goat anti-rabbit FITC; Jackson Immunoresearch, Suffolk, UK; 1:250) and nuclei were counterstained with DAPI. Fluorescence signals were analyzed in stained samples recorded using either a Leica DMIRE2 microscope equipped with a Leica DFC 350FX camera and elaborated by Leica FW4000 deconvolution software (Leica, Solms, Germany) or a Zeiss Laser Scanning Microscope 510 Meta and elaborated by Zen2009 software (Zeiss, Oberkochen, Germany).
Flow Cytometric Analysis
Cell cycle analysis was performed by flow cytometry (BD Biosciences, Heidelberg, Germany). Adherent cells (2 × 105) were fixed and resuspended in a solution containing propidium iodide at a concentration of 50 μg/ml. Cell percentages in the Sub-G1 phase of the cell cycle were measured using CELLQuest software (BD Biosciences).
Western blotting (WB)
Western blot and detection were performed as previously reported (59). For WB application, the following antibodies were used: mAb anti-TRF2 (Millipore); mAb anti-β-actin (Sigma), mAb anti-p53 DO-1(Cell Signaling, Beverly, MA, USA); pAb anti-SIRT6 (Novus Biologicals, Littleton, CO, USA); mAb anti-c-Myc (clone 9E10; Santa Cruz Biotechnology); mAb anti-PARP1 (clone C2-10) and mAb anti-PAR (clone 10H; Alexis, Lausen, Switzerland); mAb anti-HA (clone 12CA5; Roche, IN, USA); mAb anti-SIRT1 (ab32441); pAb anti-TRF1 (Abcam); mAb anti-phospho-Histone H2AX (Ser139, clone JBW301); pAb anti-phospho-RPA (S4/S8; Bethhyl Laboratories, Montgomery, TX, USA).
Purification of Histidine and GST fusion proteins
Histidine-tagged SIRT6 (SIRT6.27), a gift from John Denu (Addgene plasmid #13739; 54), was transformed and bacteria grown until OD600 0.6λ. The expression of His-SIRT6 was induced with the addition of 0.1 mM isopropyl-D-thiogalactopyranoside (IPTG) for 4 h at 25°C. Cells were harvested and lysed by sonication in 50 mM sodium phosphate and 300 mM NaCl (PBS) with 10 mM imidazole pH 8.0, 0.1 mM phenylmethylsulfonyl fluoride, 10 mg/ml leupeptin, and 5 mg/ml aprotinin. After 30 min of incubation on ice, cell debris was removed by centrifugation at 12 000 × g for 20 min at +4°C. The supernatant was agitated with nickel-nitrilotriacetic acid resin for 1 h at RT. The resin was then washed in PBS with 20 mM imidazole pH 8.0 and protease inhibitors for five times. The protein was eluted agitating nickel-nitrilotriacetic acid resin in PBS with 250 mM imidazole pH 8.0 for 1 h at RT. Eluted protein was pooled, concentrated and dialyzed in 50 mM Tris pH 7.5, 150 mM NaCl, 10% glycerol (wt/vol), and 5 mM DTT and stored at −20°C before use.
GST-tagged proteins were expressed in Escherichia coli (bacteria expression vectors pGEX2T-GST, -TRF2wt, -TRF2basic, -TRF2ΔBΔM and -TRF2myb were produced and obtained by Paul M. Lieberman's lab. as reported in (60)) and purified by using sepharose-coated resin beads, according to the manufactory's instructions. Briefly, the bacteria, transformed with the construct of interest, were grown until OD600 0.6λ and the expression of GST-tagged proteins was induced with the addition of 0.5 mM IPTG for 4 h at 37°C. Bacteria pellets were resuspended and lysed in GST lysis buffer (20 mM Tris–HCl pH 8.0, 100 mM NaCl, 0.5 mM EDTA, 0.5% Triton X-100 and protease inhibitors), sonicated on ice 10 times for 15 s and centrifuged at 20 000 × g for 20 min at 4°C. The supernatants were recovered and added to 2 ml Glutathione Sepharose 4B matrix (Amersham Pharmacia Biotech, NJ, USA). Each matrix was recovered and washed five times with a buffer containing PBS 1×, 1 mM DTT and protease inhibitors (centrifuging as above). The final concentration of matrix bound proteins was evaluated by Comassie Blue Staining. The recombinant GST fusion proteins were eluted from the beads with 10 mM glutathione in a buffer containing 50 mM Tris (pH 7.5), 10 mM DTT and protease inhibitors, dialyzed against PBS using Slide-A-Lyzer Dialysis cassette (Thermo scientific Pierce, Waltham, MA, USA) and then concentrated using a Centricon YM-50 column (Amicon from Millipore).
Pull-down assay and immunoprecipitation (IP)
In the vitro-vitro pulldown assay, 8 pmol of GST-TRF2 recombinant protein and 8 pmol of His-SIRT6 recombinant protein were incubated in 1 ml of GST incubation buffer (20 mM Tris–HCl pH 8, 100 mM KCl, 1 mM EDTA and 0.2% Triton) in agitation at 4°C ON. Successively, in order to precipitate GST recombinant proteins, the buffer was added to 60 μl of Glutathione Sepharose 4B matrix and incubated in agitation at RT for 2 h. After five washes in GST incubation buffer, the precipitated proteins were eluted from Glutathione Sepharose 4B matrix, by adding reducing protein loading buffer and incubating the samples at 95°C for 5 min, and run in a denaturating SDS page.
Regarding the vitro-vivo pull-down or IP experiments, nuclear cell extracts of HeLa or HCT116 cells were obtained by a sequential lysis with buffer A (10 mM Hepes pH 7.9, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 0.6% NP-40, 1 mM DTT and 1 mM PMSF) and buffer C (20 mM Hepes pH 7.9, 400 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM DTT and 1 mM PMSF), which resulted respectively in cytosolic and nuclear fraction isolation. Protein concentration was determined and 700 μg of nuclear fraction was incubated Over Night (ON) at 4°C with 30 μl of recombinant protein-conjugated resin in a buffer containing 50 mM Tris–HCl pH 7.5, 150 mM NaCl, 5 mM EDTA, 0.1% NP-40, 1 mM DTT and 1 mM PMSF. After five washes with a buffer containing 50 mM Tris–HCl pH8, 200 mM NaCl, 0.25% NP-40 and 0.5 mM PMSF, beads were resuspended in 20 μl of reducing protein loading buffer and incubated at 95°C for 5 min. Supernatant was run on a denaturating SDS page. For IP experiments 1 mg of lysate was precleared with protein A/G-Dynabeads (Dynal) in the IP buffer (50 mM Tris pH 7.5, 150 mM NaCl, 5 mM EDTA, 0.1% NP-40, 1 mM DTT) supplemented with protease and phosphatase inhibitors and immunoprecipitated by standard procedures. Myc-tagged proteins were captured directly by anti-Myc magnetic beads (Thermo scientific Pierce). Complexes were washed 5 times with the Wash buffer (50 mM Tris–HCl pH 8, 200 mM NaCl and 0.25% NP40). IP with rabbit-serum, mouse-serum or with nuclear protein extracts from cells that do not express epitope-tagged protein were used as negative controls.
In vivo ubiquitylation assay
To determine whether TRF2 is ubiquitylated, TRF2 IP was performed as above described from TRF2wt-overexpressing HCT116 cells which were transfected with siSIRT6 or control siGFP and after 48 h with pRK5-HA-Ub. 24 h later, the cells were treated with 2 μM CPT for 2 h and then the medium was replaced in absence or presence of 10 μM MG132 for 6 h. Total lysates were obtained by using a buffer containing 50 mM Tris–HCl pH 7.4, 330 mM NaCl, 0.1% (v/v) Triton X-100, 5 mM EDTA, 1% SDS and complete protease inhibitors.
Protein Footprinting
Acetylation of lysines of TRF2 protein was performed by adding 0.5 mM of sulfosuccinimidyl acetate (Thermo scientific) to 8 pmol of the purified recombinant TRF2 protein incubated in 10 mM Tris–HCl pH 8, 150 mM NaCl, 0.5 mM DTT, and 5% glycerol for 30 min at 30°C. The samples were desiccated, resuspended in 50 mM Tris–HCl pH 7.5, 150 mM NaCl, 10 mM NAD+, and 2 mM DTT and incubated with 8 pmol of purified recombinant Sirt6 (Euromedex, Souffelweyersheim, France) for 2 h at 37°C. Samples were resupended in Laemmli loading buffer and boiled for 5 min. Proteins were resolved by SDS-PAGE and submitted to trypsin proteolysis, and profiles of lysines acetylation were analyzed by mass spectrometry (Isabelle Zanella-Cleon IBCP, France, Lyon). We determined the probability of disappearance of lysines acetylation upon SIRT6 addition.
TRF2 acetylation in cell by proteomic analysis
To determine whether TRF2 is acetylated, TRF2 IP was performed as above described and the samples were run on a 10% SDS-polyacrylamide gel by electrophoresis (SDS-PAGE). The SDS-PAGE lanes were cut in the region around 60 kDa and digested separately. Each piece was washed with ultrapure water and CH3CN and subjected to in situ protein digestion as described by Shevchenko (61). Briefly, each slice was reduced with 10 mM 1,4-dithiothreitol (DTT) and alkylated with 54 mM iodoacetamide, then washed and rehydrated in trypsin solution (12 ng/mL) on ice for 1 h. After the addition of ammonium bicarbonate (30 μL, 50 mM, pH 7.5), proteins digestion was allowed to proceed overnight at 37°C. The supernatant was collected and peptides were extracted from the slice using 100% CH3CN and both supernatants were combined. The peptide samples were dried and dissolved in formic acid (FA, 10%) before MS analysis. The peptide mixture (5 μL) was injected into a nano-ACQUITY UPLC system (Waters). Peptides were separated on a 1.7 mm BEH C18 column (Waters) at a flow rate of 400 nl/min. Peptide elution was achieved with a linear gradient (solution A: 95% H2O, 5% CH3CN, 0.1% FA; solution B: 95% CH3CN, 5% H2O, 0.1% FA); 15–50% B over 55 min). MS and MS/MS data were acquired on a LTQ XL high-performance liquid chromatography mass spectrometry system (Thermo-Scientific). The five most intense doubly and triply charged peptide ions were chosen and fragmented. The resulting MS data were processed by Xcalibur software to generate peak lists for protein identifications. Database searches were carried out on the Mascot server. The SwissProt database (release 2016_08, 7 September 2016, 551 987 entries) was employed (settings: two missed cleavages; carbamidomethyl (C) as fixed modification and oxidation (M), phosphorylation (ST) and acetylation (K) as variable modifications; peptide tolerance 80 ppm; MS/MS tolerance 0.8 Da). The experiment was repeated two times. Each run has also been investigated using Xcalibur software (Thermo-Scientific) to integrate the area of all the ion peaks relative to the peptide 174–192 acetylated.
Tissue microarray analysis
Colorectal cancers (CRCs) were obtained from 185 patients who were surgically treated at Regina Elena Cancer Institute between 2000 and 2013. All CRCs were histopathologically re-evaluated on haematoxylin and eosin stained slides and representative areas were marked prior to tissue microarray (TMA) construction. Two core cylinders (1 mm diameter) were taken from selected CRCs and deposited into two separate recipient paraffin blocks using a specific arraying device (Alphelys, Euroclone, Milan, Italy). In cases where informative results on TMA were absent due to missing tissue, no tumor tissue, or unsuccessful staining, we re-analyzed the correspondent routine tissue section. In addition to tumor tissues, the recipient block also received normal colon tissue as negative controls. Two-mμ sections of the resulting microarray block were made and used for immunohistochemical (IHC) analysis after transferring them to SuperFrost Plus slides (Menzel-Gläser, Braunschweig, Germany).
IHC staining on TMA was performed using Monoclonal Antibody (Ab) anti-TRF2 (clone 4A794, Upstate Chemicon, Millipore, USA) and polyclonal Ab anti-SIRT6 (Novusbio) in an automated immunostainer (Bond-III, Leica, Italy). A pH 6 buffer was used as antigen retrieval for the two antibodies according to the manufacturer's protocol.
The levels of TRF2 and SIRT6 were evaluated in terms of intensity of nuclear staining (0 = negative, 1+ = weak, 2+ = moderate, 3+ = strong; Supplementary Figure S7).
Images were obtained at 20x magnification by using a light microscope equipped with a software able to capture images (DM2000 LED, Leica). Scale bar: 100 μm.
Case selection
One hundred and eighty five CRC patients, including 135 colon and 50 rectal carcinomas, with a median follow-up of 66 months (95% CI 61.8–71.5) were retrospectively evaluated. Tumors were staged according to the Unione Internationale Contre le Cancer tumor-node-metastasis system criteria (TNM 7th Edition, L.H. Sobin, M.K. Gospodarowicz, Ch. Wittekind, 2009, UICC). The study was reviewed and approved by the ethics committee of the Regina Elena National Cancer Institute.
Statistical analysis
The experiments have been repeated from three to five times and the results obtained are presented as means ± SD. Significant changes were assessed by using Student's t test two tails for unpaired data, and P values <0.05 were considered significant.
In IHC experiments, levels of both TRF2 and SIRT6 expression were scored semi-quantitatively based on IHC staining intensity. Low intensity cases displayed a 0/1+ IHC score and were considered negative and high intensity cases presented a 2+/3+ IHC score and were considered positive. Relationship between parameters were assed using Chi square test. Significance was defined at the p≤0.05 level. The SPSS® (21.0) statistical program was used for analysis.
RESULTS
TRF2 degradation upon camptothecin treatment
Although DNA damage induced by drugs used in conventional chemotherapy are usually believed to occur throughout genome, a wealth of evidence indicates that telomeres are preferred targets of several genotoxic molecules (62). Here, we investigated the role of the telomere protein TRF2 in the response to camptothecin (CPT), a Topoisomerase I inhibitor that triggers both single- and double-stranded DNA breaks (DSBs). First, we found that overexpression of TRF2 could promote resistance to the drug (Figure 1A and Supplementary Figure S1A and B). Conversely, inhibition of TRF2 by RNA interference made cells more sensitive to CPT (Supplementary Figure S1B). Of note, TRF2 overexpression also conferred resistance to Bleomycin, another replication-dependent DNA-damaging drug, but not to Taxol, an anti-microtubule agent (Supplementary Figure S1C and D).
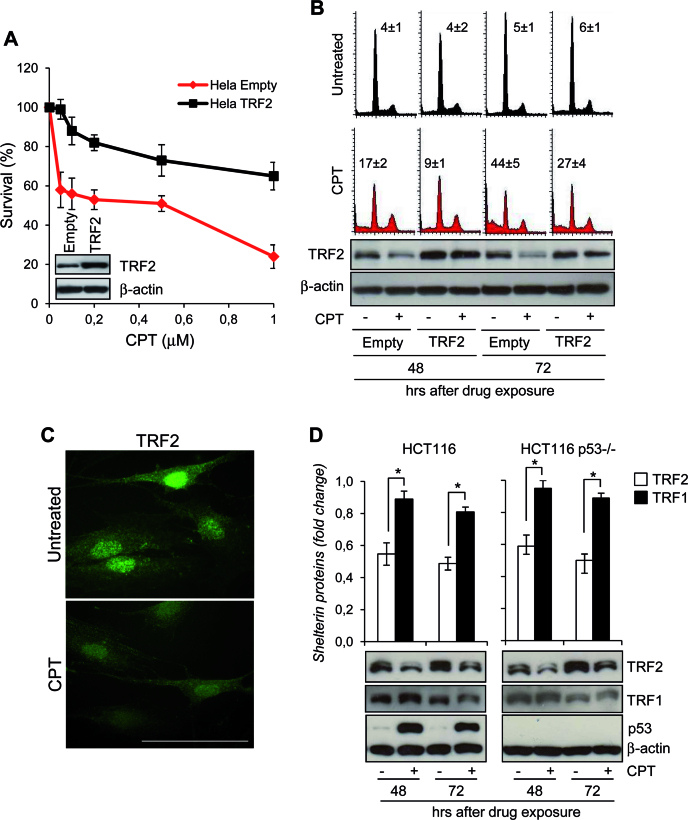
Figure 1.
TRF2 degradation upon CPT treatment. (A) HeLa cells overexpressing TRF2 or Empty vector were treated with Camptothecin (CPT) for 2 h at the indicated doses and colony-forming ability was evaluated. The graph shows the surviving fractions calculated as the ratio of absolute survival of the treated sample/absolute survival of the control sample. Representative images of the cell lines evaluated by western blot are shown. (B) Empty- and TRF2-overexpressing HeLa cells were treated with 0.2 μM CPT for 2 h and then processed at 48 and 72 h after drug exposure for PI staining to evaluate cell cycle analysis at flow cytometry (upper panel), and western blot (WB) with the indicated antibodies to monitor the TRF2 expression levels (lower panel). The percentages of the cells at the Sub-G1 fraction of the cell cycle were reported inside the relative histograms. Representative out of three independent experiments is shown. (C) Representative immunofluorescence (IF) images of TRF2 from BJ-hTERT fibroblasts untreated or treated with 0.2 μM CPT for 2 h and fixed at 48 h after drug exposure. Original magnification, 63x. Scale bar 50 μm. (D) Expression of TRF2, TRF1 and p53 were examined by western blot in p53wt- and p53-null HCT116 cells treated as HeLa cells in A. TRF2 or TRF1 expression levels were expressed in the histograms (upper panel) as fold changes in treated versus untreated samples, after β-actin normalization. All histograms show the mean values of three independent experiments. Bars indicate means ± SD. *P <0.05.
In an attempt to gain insight into the role of TRF2 in drug response, we found a time-dependent decrease of TRF2 protein expression in viable adherent human cervix adenocarcinoma HeLa cells exposed to CPT and this effect was associated with cell death (Figure 1B and C). The CPT-mediated reduction of TRF2 was compensated by the ectopic expression of TRF2 leading to an increased resistance to the drug (Figure 1B). Overall, these results suggest that the sensitivity to CPT is determined by the extent of TRF2 expression and that the beneficial effect of TRF2 overexpression is likely to result from the maintenance of enough TRF2 to protect cells from apoptosis.
The reduction of TRF2 upon CPT treatment was observed in multiple independent cell lines (Figure 1 and Supplementary Figure 2A and B) and at different post-repair time points, depending on the drug dose used (Supplementary Figure S3). Moreover, TRF2 reduction occurred in cell lines possessing either wild type (human colon carcinoma HCT116 cells and immortalized foreskin BJ-hTERT fibroblasts) or inactivated (human transformed foreskin fibroblasts BJ-EHLT/Ras) p53 protein (Figure 1D and Supplementary Figure S2A and B). Interestingly, no changes in TERF2 mRNA were observed (Supplementary Figure S2C) and treatment of cells with the proteasome inhibitor MG132 increased the amount of TRF2 protein and failed to cause the decrease of TRF2 levels upon CPT exposure (Supplementary Figure S2D). Since it was previously shown that p53 activation reduces the stability of TRF2 through a proteasome-dependent mechanism (33), we asked whether p53 was required for the CPT-induced TRF2 destabilization. A similar level of TRF2 down-expression was observed in HCT116 cells carrying wild-type or null alleles of TP53 (Figure 1D) and, of note, CPT was not able to significantly reduce the amount of TRF1, the other telomeric double-strand binding protein which shares with TRF2 both localization and structural organization (Figure 1D). All together these results indicate that TRF2 is specifically degraded upon CPT treatment through a proteasome-mediated p53-independent mechanism.
TRF2 interacts with SIRT6
Consistently with its role in the response to DNA damage (63), endogenous amounts of SIRT6 increased in cells exposed to CPT both in normal and in transformed cells (Figure 2A–B, Supplementary Figure S3A) and this event was associated to an almost complete resolution of DNA damage (Supplementary Figure S3B). Remarkably, SIRT6 appeared to occasionally colocalize with TRF2 (Figure 2C).
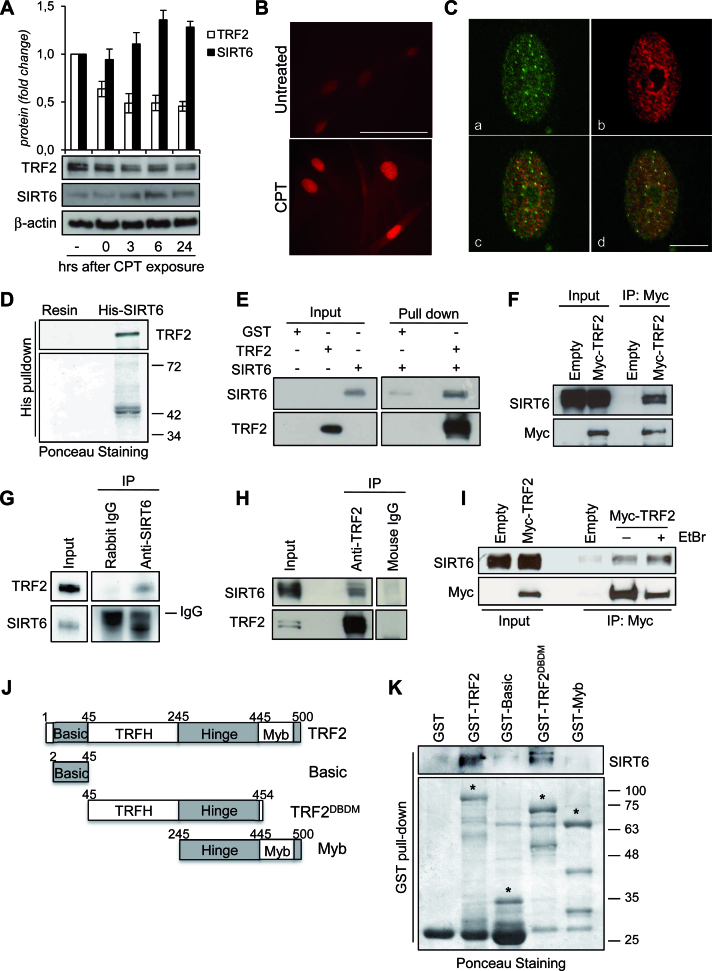
Figure 2.
TRF2 interacts with SIRT6. (A) BJ-hTERT were treated with 0.2 μM CPT for 2 h and then processed for the WB analysis of the expression levels of SIRT6 and TRF2. The expression levels of the two proteins were reported in the histograms (upper panel) as fold changes in treated versus untreated samples, after β-actin normalization. All histograms show the mean values of three independent experiments. Bars indicate means ± SD. (B) Representative IF images of SIRT6 from BJ-hTERT fibroblasts treated as in (A). Original magnification, 63x. Scale bar 50 μm. (C) Representative confocal IF images of untreated BJ-hTERT showing the colocalization between TRF2 and SIRT6. a, TRF2 staining; b, SIRT6 staining; c, merge; d, SIRT6/TRF2 colocalizations showed as white spots after software analysis. Scale bar 10 μm. (D) Pull-down assay of HeLa nuclear extracts with His-tagged SIRT6 protein conjugated with Ni-NTA magnetic beads. The eluted product was analyzed by WB using an antibody against TRF2. The ponceau staining is shown as beads loading control. (E) In vitro GST pull-down assay with GST alone or GST-TRF2 in the presence or absence of His-SIRT6. Following the pull-down, samples were analyzed by WB using antibodies against TRF2 or SIRT6. (F) Anti-Myc immunoprecipitation (IP) followed by WB with anti-SIRT6 or -Myc antibody in nuclear extracts of HCT116 cells infected with Myc-TRF2 or Empty retroviral vectors. (G) Anti-SIRT6 IP in nuclear extracts of HCT116 cells expressing Myc-TRF2. Anti-rabbit IgG IP was used as a negative control. WB was carried out with anti-TRF2 or anti-SIRT6 antibody. (H) Anti-TRF2 IP in nuclear extracts of HCT116 cells. Anti-mouse IgG IP was used as a negative control. WB was carried out with anti-TRF2 or anti-SIRT6 antibody. (I) Nuclear extracts of HCT116 cells infected with Myc-TRF2wt or Empty retroviral vectors were immunoprecipitated with anti-Myc antibody. The IP was performed in presence or in absence of 50 μg/ml EtBr, followed by WB with anti-SIRT6 or -Myc antibody. (J) Schematic representation of TRF2 and its deletion variants used in this study. (K) The various GST-TRF2 proteins or GST alone were affinity-purified and incubated with lysates prepared from HeLa cells, followed by detection with anti-SIRT6 antibody. The purified GST fusion proteins (indicated by the asterisks) were visualized by ponceau staining. Molecular mass markers are expressed in kilodaltons (KDa).
To examine a possible interaction between TRF2 and SIRT6, pull-down experiments were performed. Histidine-tagged recombinant SIRT6 (His-SIRT6) precipitated endogenous TRF2 from nuclear extracts of HeLa cells, indicating that TRF2 interacts with SIRT6 in vitro (Figure 2D). Then, to further analyze whether TRF2 establishes a direct physical association with SIRT6, we performed a co-immunoprecipitation assay using recombinant proteins (His-SIRT6 and GST-TRF2) in vitro. As shown in Figure 2E, GST-TRF2 interacts with SIRT6 in vitro. To determine whether TRF2 and SIRT6 associate also in vivo, myc-TRF2 was ectopically expressed in HeLa cells and immunoprecipitated by an anti-myc antibody. As shown in Figure 2F, endogenous SIRT6 was observed in the anti-myc immunoprecipitates. Reciprocally, TRF2 was recovered in the anti-SIRT6 immunoprecipitates from nuclear extracts of TRF2-overexpressing HeLa cells (Figure 2G). Of note, TRF2/SIRT6 interaction was also detected by immunprecipitating the endogenous TRF2 protein in HCT116 cells (Figure 2H). Next, we examined whether the TRF2/SIRT6 interaction could be mediated by DNA. Therefore, EtBr was added to the cell lysates and throughout the pull-down experiments to disrupt protein-DNA interactions. In the presence of 50 μg/ml EtBr the interaction between TRF2 and SIRT6 persisted, indicating that their reciprocal association is independent of DNA (Figure 2I).
To map the region in TRF2 that is responsible of SIRT6 binding, a series of TRF2 fragments fused to GST (Figure 2J) were used in pull-down experiments. GST-TRF2ΔBΔM maintained the ability to bind to SIRT6, although with lower affinity than GST-full length TRF2. By contrast, GST-Basic and GST-Myb had no detectable binding activity (Figure 2K).
TRF2 and SIRT6 interaction is increased by DNA damage in a PARP-dependent manner
Next, we investigated if TRF2/SIRT6 interaction was influenced by DNA damage. We found that the treatment of HCT116 cells with CPT, Hydroxyurea (HU), Bleomycin (Bleo) or Cisplatin (DDP), but not with Taxol (Tx), led to an increase in the amount of SIRT6 co-precipitated with anti-myc or -TRF2 antibody respect to untreated samples (Figure 3A and Supplementary Figure S4A). Moreover, in immunofluorescence experiments, CPT exposure enhanced the average number of TRF2/SIRT6 colocalizations (Figure 3B and C), which frequently appeared at the same γH2AX foci (Figure 3D and Supplementary Figure S4B), thus indicating that DNA injury would favor the association between SIRT6 and TRF2 at damaged sites.
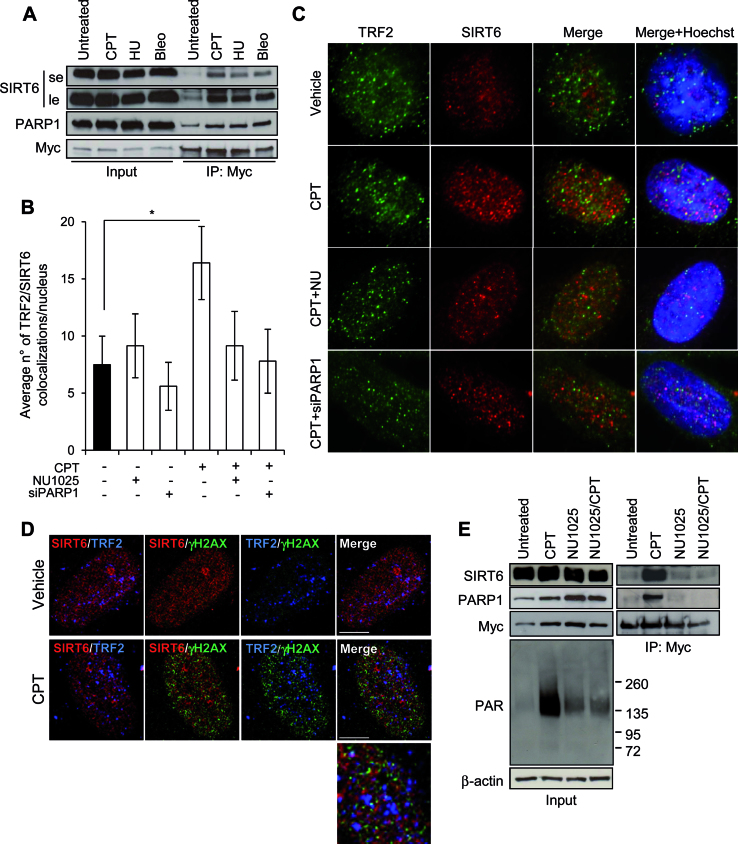
Figure 3.
TRF2 and SIRT6 interaction is increased by DNA damage in a PARP-dependent manner. (A) The IP was conducted on Myc-TRF2-overexpressing HCT116 cells untreated or exposed to the indicated drugs as reported in the Materials and Methods section. Input and immunoprecipitates were then analyzed by WB for the expression of the indicated proteins. le, long exposure; se, short exposure. (B and C) BJ-hTERT fibroblasts were treated with 2 μM CPT for 2 h alone or in combination with NU1025 or siPARP1, fixed and processed for co-IF against TRF2/SIRT6. Histograms report the average number of TRF2/SIRT6 colocalizations per nucleus, expressed as mean values of three independent experiments. Bars indicate means ± SD. *P < 0.05. (C) Representative images of colocalizations are shown (Leica deconvolution microscope 100× magnification). (D) Representative confocal IF images of an untreated or 2 μM CPT exposed sample of BJ-hTERT fibroblast. The single staining for TRF2, SIRT6 and γH2AX are reported in Supplementary Figure S4B. Here the double and triple (merge) staining are showed. In the enlarged panel the triple colocalizations appear as white spots. Scale bar 10 μm. (E) The IP was conducted on Myc-TRF2-overexpressing HCT116 cells untreated or exposed to 2 μM CPT for 2 h, alone or in combination with the PARP1 inhibitor, NU1025. Input and IP were then analyzed by WB for the expression of the indicated proteins.
Besides SIRT6, PARP1, a major DDR signaling factor (64,65) and a well known interactor of both TRF2 and SIRT6 (52,65,66) was immunoprecipitated by anti-myc antibody more efficiently from nuclear extracts of myc-TRF2 overexpressing cells exposed to CPT, HU or Bleo than from samples of untreated cells (Figure 3A and E). The increased association between SIRT6 and TRF2 cells was abolished in presence of the PARP inhibitor NU1025, both alone or in combination with CPT (Figure 3E). PARP1, which is early activated by CPT treatment in a SIRT6-dependent manner (Supplementary Figure S4C), has a crucial role in promoting the SIRT6/TRF2 interaction. Indeed, when PARP1 was inhibited by pharmacological or genetic approaches, CPT exposure was no longer able to enhance the number of TRF2/SIRT6 colocalizations (Figure 3B and C, Supplementary Figure S4D).
SIRT6 affects TRF2 stability
To examine whether SIRT6 modulates TRF2 levels, siRNA-mediated knockdown of SIRT6 was performed. Increased amounts of the protein but not of TERF2 mRNA were observed in SIRT6-compromised cells (Figure 4A and Supplementary Figure S5A), while under the same experimental conditions TRF1 expression remained unchanged (Supplementary Figure S5B). Of note, inhibition of SIRT1, which is closely related to SIRT6, did not affect TRF2 levels (Supplementary Figure S5C), thus indicating a specific interplay between SIRT6 and the shelterin protein TRF2. The inhibition of protein synthesis by cycloheximide (CHX), analyzed by western blot at different times, showed that SIRT6 knockdown markedly extended the half-life of TRF2 protein (Figure 4B). Down-regulation of SIRT6 was also able to abolish the CPT-dependent impairment of TRF2 both in HCT116 and HeLa cells (Figure 4C and Supplementary Figure S5D).
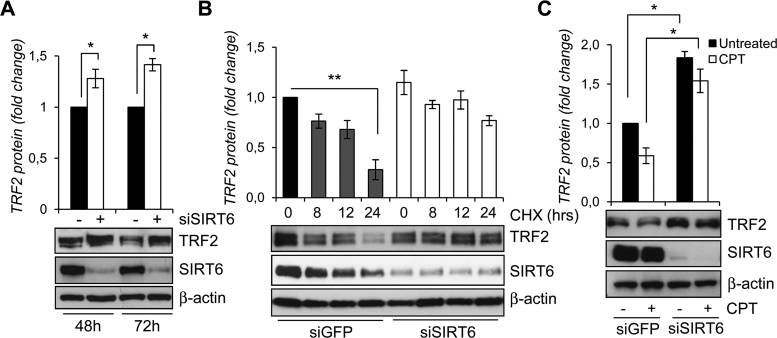
Figure 4.
SIRT6 affects TRF2 stability. (A) WB analysis of TRF2 and SIRT6 levels in siGFP- or siSIRT6-transfected HCT116 cells after 48 and 72 h of siRNA delivery (lower panel). TRF2 expression levels were expressed in the histograms (upper panel) as fold changes in siSIRT6 versus siGFP samples, after β-actin normalization. (B) WB analysis of TRF2 and SIRT6 levels in siGFP- or siSIRT6-transfected HCT116 cells treated with 100 μg/ml cycloheximide for the indicated times (lower panel). TRF2 expression levels were expressed in the histograms (upper panel) as fold changes in treated versus untreated samples, after β-actin normalization. (C) WB analysis of TRF2 and SIRT6 levels in siGFP- or siSIRT6-transfected HCT116 cells after 6 h by the end of 2 μM CPT exposure (lower panel). TRF2 expression levels were expressed in the histograms (upper panel) as fold changes in treated versus untreated samples, after β-actin normalization. All histograms show the mean values of three independent experiments. Bars indicate means ± SD. *P < 0.05; **P < 0.01.
These results clearly demonstrate that SIRT6 promotes TRF2 degradation in response to DNA damage.
TRF2 is deacetylated by SIRT6
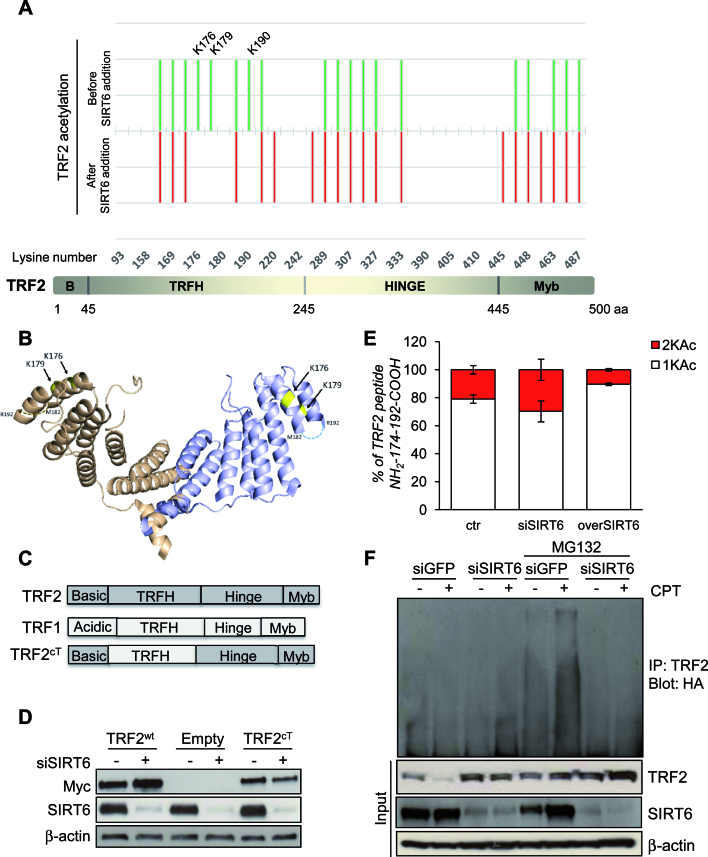
Given that SIRT6 specifically interacts with TRF2 and that SIRT6 protein acts as a deacetylase of various proteins (48–51), we hypothesized that TRF2 is deacetylated by SIRT6 and that this deacetylation targets TRF2 to proteosomal degradation. In order to identify the TRF2 residues that can be deacetylated by SIRT6, purified recombinant protein TRF2 was first chemically acetylated by sulfosuccinimidyl acetate, a compound that specifically acetylates lysines located at the surface of the protein (12) and then incubated with purified SIRT6 protein. Proteins were resolved by SDS-PAGE, submitted to trypsin proteolysis, and profiles of lysines acetylation of TRF2 in presence or in absence of SIRT6 were analyzed by mass spectrometry. The lysines, acetylated before and after SIRT6 addition, are shown in green and red respectively in Figure 5A. Three lysines (K176, K179 and K190) appeared deacetylated upon incubation with SIRT6. Interestingly, they are all located on the TRFH domain of TRF2 as shown in yellow on the 3D structure of the protein (Figure 5B). Of note, K190 is located on a disordered region as indicated with broken lines. Lysines not present in the TRF2 acetylation profile, due to a partial coverage in mass spectrometry were not analyzed (K220, K245, K445 and K458).
Figure 5.
TRF2 is deacetylated by SIRT6. (A) Lower panel: Representation of TRF2 organization in four domains. Upper panel: Footprinting graph showing profile of in vitro acetylation of TRF2 lysines by sulfosuccinimidyl acetate (in green) and TRF2 lysines still acetylated upon SIRT6 addition (in red). The mass spectrometry analysis was done using a nanoLC-MSMS giving both the peptide mass and sequence allowing the determination of the specific acetylated aminoacids. Mass spectrometry analysis gives acetylation profiles of the protein before and after incubation with SIRT6 showing TRF2 lysines deacetylated by SIRT6. (B) Positions of lysines deacetylated upon SIRT6 addition (in yellow), on the 3D structure of the TRFH domain of TRF2 (PDB:3BUA). The third lysine identified, K190 is located on a region of structural disorder, represented as broken lines. (C) Schematic representation of the TRF2cT variant obtained through the domain swapping approach between TRF1 and TRF2. (D) HCT116 cells infected with Myc-TRF2wt, Myc-TRF2cT or Empty retroviral vectors were analyzed by western blot for the expression of Myc-tagged TRF2 variants or SIRT6, after 72 h of siGFP or siSIRT6 transfection. (E) TRF2-overexpressing HCT116 cells were subjected to IP with anti-TRF2 antibody after silencing (siSIRT6) or overexpression (overSIRT6) of SIRT6. Immunoprecipitated TRF2 proteins were loaded on SDS-PAGE and the regions around 60 kDa were cut and, separately, digested as described in material and methods section, to be then analyzed by LC–MS/MS. The relative abundance of the ions of peptide 174–192 mono- (+1KAc) and diacetylated (+2KAc) species was calculated integrating the peak area in each LC–MSMS run and reported in the histograms. (F) After transient transfection with ubiquitin-HA vector, TRF2-overexpressing HCT116 cells were either transfected with siSIRT6 siRNA or control siGFP. Protein lysates prepared from cells after 6 h by the end of treatment with 2 μM CPT, as well as those from untreated cells as negative controls, in presence or not of MG132, were used in immunoprecipitation with anti-TRF2 antibody, followed by western blot with anti-HA. The effectiveness of SIRT6 siRNA and the expression of TRF2 was confirmed by immunoblot using input samples. The levels of β-actin were used as a loading control.
To confirm that the TRFH domain of TRF2 is a specific target of SIRT6 deacetylation, we tested whether a chimeric form of TRF2 (TRF2cT), in which the TRFH domain of TRF2 was replaced by the analogous TRF1 domain (Figure 5C), can be increased upon SIRT6 depletion. Our results indicate that TRF2cT, differently from TRF2wt, failed to be stabilized by SIRT6 down-regulation (Figure 5D).
Moreover, in vivo TRF2 acetylation pattern was investigated by proteomic analysis on HCT116 cells upon SIRT6 depletion and overexpression, focusing on the peptide 174–192 above recognized as a SIRT6-sensible TRF2 domain. In all samples, TRF2 was identified with high confidence (Mascot score always ≥500) and the peptide 174–192 was always found with one or two acetylated lysines. The relative abundance of the ions of 174–192 + 1KAc and 174–192 + 2KAc species was calculated integrating the peak area in each LC–MSMS run (Supplementary Figure S6). As reported in Figure 5E, a clear increase in the percentage of the diacetylated peptide was measured upon in vivo SIRT6 depletion, whereas SIRT6 overexpression favored the presence of the monoacetylated species.
Next, we asked whether the SIRT6-induced degradation of TRF2 was mediated by ubiquitination. To this end, myc-tagged TRF2 overexpressing HCT116 cells were transiently transfected with HA-ubiquitin and ubiquitin-modified TRF2 was revealed by western blot with anti-HA antibody in the anti-myc immunoprecipitates. To exclude the possibility of detecting ubiquitinated interactors of TRF2, besides to or instead of the target protein itself, immunoprecipitation was carried out on total cellular extracts under denaturing conditions. As showed in Figure 5F, in presence of MG132, CPT-induced ubiquitination of TRF2 was abolished when SIRT6 was reduced.
All together our results indicate that the TRFH domain of TRF2 is required for its post-transcriptional modification.
Inverse correlation between TRF2 and SIRT6 immunohistochemical expression in colorectal cancer
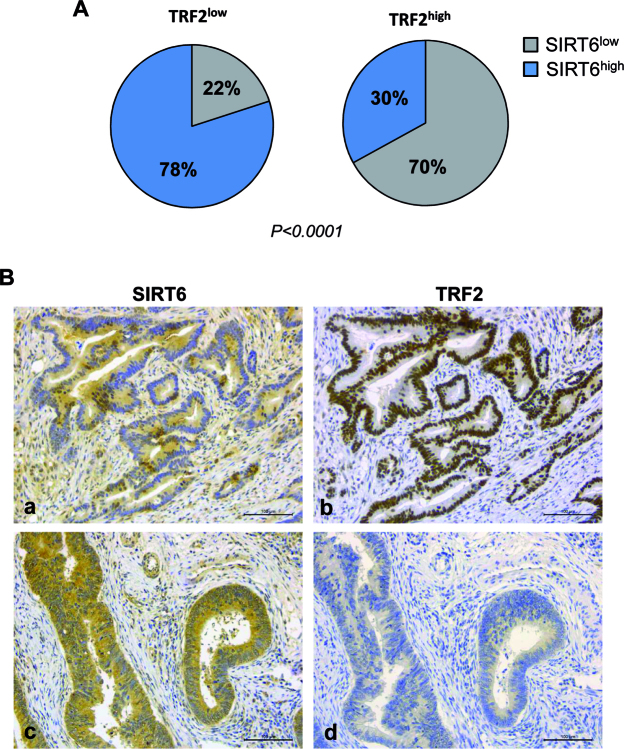
Our results, together with the previously reported low expression and tumor-suppressor functions of SIRT6 in many different types of cancer (67), raised the interesting possibility that the high level of TRF2 observed in many human cancers can be due to a decreased expression of SIRT6. Therefore, we performed immunohistochemical analyses of TRF2 and SIRT6 expression in a cohort of 185 CRC samples (Supplementary Figure S7). The clinical data on the corresponding patients are classified in Supplementary Table SI. Of the entire series of 185 CRCs, 63 (34%) stained negative (score 0/1+, Low) and 122 (66%) positive (score 2+/3+, High) for TRF2, respectively. In contrast, 99 (54%) of the tumors were negative and 86 (46%) positive for SIRT6 showing a significant inverse correlation between the two parameters (P <0.0001; Supplementary Table S2). Indeed, as illustrated in Figure 6A and B, in the subset of TRF2 low CRCs, 49 cases (78%) were SIRT6 high and 14 (22%) SIRT6 low, whereas in the subset of 122 TRF2 high CRCs, 37 (30%) cases were SIRT6 high and 85 (70%) were SIRT6 low (P < 0.0001). Overall, we propose that the oncosuppressive functions of SIRT6 can result at least in part from its ability to destabilize TRF2.
Figure 6.
Inverse correlation between TRF2 and SIRT6 immunohistochemical expression in colorectal cancer. (A) The histograms show the different amount of SIRT6 expression in the 63 TRF2low and in the 122 TRF2high CRC (P < 0.0001). (B) IHC staining of two representative CRC serial sections showing SIRT6low (a) and TRF2high (b), or SIRT6high (c) and TRF2low (d). 3,3΄-Diaminobenzidine (DAB) visualized TRF2 or SIRT6 via a brown precipitate. Magnification 20×. Scale bar: 100 μm.
DISCUSSION
TRF2 emerges as a nodal protein between development, aging and cancer by combining activities involved in telomere protection, global DNA repair and cell type specific transcriptional regulation (15). Accordingly, TRF2 expression integrates various developmentally regulated signals such as those mediated by WNT/β-catenin and WT1 as well as those in response to stress as p53. In addition, several post-translational modifications of TRF2, including acetylation, ubiquitination, methylation and SUMOylation have been described to play important roles in telomere function. This study reveals a novel molecular mechanism for the regulation of TRF2 that involves SIRT6, a member of the sirtuin family of NAD+-dependent enzymes. Indeed, we provide compelling evidences supporting a direct physical interaction between TRF2 and SIRT6 proteins. Their binding does not require DNA, but it is strengthen by DNA-damaging agents in a PARP1-dependent manner. In particular, we identified a new complex, including TRF2 and SIRT6, in which PARP1 takes part. Specifically, TRF2/SIRT6 interaction is increased in cells treated with Camptothecin, Hydroxyurea, Bleomycin or Cisplatin, but not with Taxol; of note all drugs, with the exception of Taxol, have been reported to activate PARP1 (68–71). Interestingly, TRF2 and SIRT6 appeared occasionally to associate in the nucleus and after DNA damage the number of co-localizing spots increased and coincided with γH2AX foci, indicating that, in response of DNA injury, SIRT6, PARP1 and TRF2 may be early recruited to damaged sites. PARP1 is crucial for the response to DNA damage, since the enhanced association between TRF2 and SIRT6 is abolished when PARP1 is inhibited. Our data are consistent with previous findings reporting that TRF2 is involved in non-telomeric DNA damage. In particular, TRF2 has been shown to localize to non-telomeric DNA lesions (23,26) where it is rapidly and transiently phosphorylated by ATM (24), and this interaction is dependent on poly-ADP-ribosylase (PAR) activity of PARP1 (65). Notably, under oxidative stress, SIRT6 activates PARP1's PAR activity by mono-ADP-ribosylation (52), and it is recruited to break sites. This could be the case, since we observed a reduced level of drug-induced PARylation when SIRT6 is depleted. Many functions of SIRT6 are linked to its activity in chromatin, where it catalyzes NAD+ dependent deacetylation of histone H3 on acetylated K9, K56 and K18, allowing the proper unfolding of chromatin and the engagement of downstream DNA damage interacting factors (46,49). Our data indicate that SIRT6 deacetylates TRF2 in vivo and identify K176, K179 and K190 as the putative lysine residues involved. That SIRT6, in addition to deacetylating histone H3, can also deacetylate nonhistone proteins is not unprecedented. Indeed, it has been found to deacetylate the C-terminal binding protein (CtBP) interacting protein (CtIP) involved in a crucial step of DNA end resection through HR (53). Deacetylation of TRF2 occurs on lysine residues located on the TRFH domain, as demonstrated by proteomic analysis. Indeed, overexpression of the TRF2CT mutant, in which the TRFH domain of TRF2 is replaced by the analogous one of TRF1, as result of a domain-swapping approach, failed to be stabilized by SIRT6 depletion. Even if the lysine K293 on the Hinge domain of TRF2 is the only lysine described to be acetylated by p300 (72), our results identify the TRFH as the domain of TRF2 required for its post-transcriptional modification by SIRT6, further strengthen the key role of this region in the protein-protein interaction network.
Our paper goes an important step further revealing the functional significance of TRF2/SIRT6 interaction. In particular, we demonstrate that SIRT6 depletion induces an accumulation of TRF2 with extension of its half-life. Furthermore, during recovery from DNA damage, TRF2 amounts gradually decrease, both in normal and in tumor cells, as effect of a p53-independent proteosomal degradation. Nevertheless, we cannot exclude that Siha1, a p53-inducible E3 ubiquitin ligase, which has been shown to limit TRF2 protein level during telomere-damage signaling and cellular senescence (33) is also involved in SIRT6-mediated TRF2 degradation. Interestingly, in the absence of SIRT6, the decrease of TRF2 levels is almost abrogated and it is associated with a global reduction of the ubiquitinated form of TRF2, indicating that SRT6 is critical for TRF2 ubiquitin-dependent proteolysis. Nevertheless, we cannot exclude that SIRT6 may affect the protein stability of TRF2 by means of other mechanisms. As a chromatin remodeler, in response to DNA damage SIRT6 could alter the histone acetylation status next to lesions, thereby reducing the binding affinity to DNA of TRF2, consequently favoring its degradation. In our system, reduction of TRF2 expression levels during the DDR pathway is associated to cell death. When TRF2 is exogenously overexpressed cell death is attenuated, resulting in resistance to treatment with replication stress inducing agents. In line with the well-known ability of TRF2 to inhibit ATM and Chk2 kinases (5,6), TRF2 degradation could be functional to the proper activation of downstream effectors of DNA signaling (i.e. p53) with consequent impact on downstream cell fate decisions, such as apoptotis and/or senescence. On the other hand, since TRF2 degradation is expected to confer further instability to the already damaged telomeres, it may promote by itself apoptotic cell death. Therefore, in tumors, where TRF2 is aberrantly expressed and its reduction is prevented upon DNA damage, cells can be protected by cell death and become resistant to therapy. Nevertheless, the role of TRF2/SIRT6 interaction in the damage repair pathway and apoptosis remains to be fully elucidated. Other authors underlined the ability of TRF2 to affect tumor cell sensitivity to drug exposure, even though the molecular mechanism has never been elucidated. In particular, aberrant levels of TRF2 have been shown to promote the multidrug resistance (MDR) of gastric cancer cells by interfering DNA damage response (73), while TRF2 depletion can sensitize glioblastoma stem cells to temozolomide, a DNA-alkylating agent currently used to treat glioblastoma (74). Consistently with these results, our findings demonstrate that TRF2 protein levels are subjected to a fine regulation whose alteration could significantly impact on cell survival, reinforcing the notion that TRF2 is a general DNA damage factor. Moreover, we can speculate that impairment of TRF2 degradation could be one of the mechanisms unerlying the increased dosages of TRF2 observed in many human malignancies. In agreement with this hypothesis and with the well-established role of SIRT6 as a tumor suppressor, an inverse correlation between SIRT6 and TRF2 levels has been observed in a cohort of colon rectal cancer patients.
In conclusion, our study uncovers SIRT6 as a new partner and post-transcriptional regulator of TRF2 that may be part of a higher order complex with functional impacts on DDR, cancer and aging.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Paul M. Lieberman (The Wistar Institute, Philadelphia PA, USA) for providing bacteria expression vectors pGEX2T-GST, -TRF2wt, -TRF2basic, -TRF2ΔBΔM and -TRF2myb. We are indebted to Isabelle Zanella-Cleon for mass spectrometry analysis in vitro (Centre Commun de Micro-analyse des Proteines, IBCP Lyon, France), to Maria Chiara Monti and Agostino Casapullo for mass spectrometry analysis in vivo (Department of Pharmacy, University of Salerno, Italy).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Italian Association for Cancer Research (A.I.R.C.) [16910]; Foundation ARC, Ligue Contre le Cancer (Equipe labellisé), ANR ('Teloloop’ ANR-1582-30020690) and Institut National du Cancer (INCa) (TELOCHROM). Funding for open access charge: Associazione Italiana per la Ricerca sul Cancro [16910].
Conflict of interest statement. None declared.
REFERENCES
- 1.van Steensel B., Smogorzewska A., de Lange T.. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998; 92:401–413. [DOI] [PubMed] [Google Scholar]
- 2.Denchi E.L., de Lange T.. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007; 448:1068–1071. [DOI] [PubMed] [Google Scholar]
- 3.Gilson E., Geli V.. How telomeres are replicated. Nat. Rev. Mol. Cell Biol. 2007; 8:825–38. [DOI] [PubMed] [Google Scholar]
- 4.Ye J., Lenain C., Bauwens S., Rizzo A., Saint-Léger A., Poulet A., Benarroch D., Magdinier F., Morere J., Amiard S. et al. . TRF2 and apollo cooperate with topoisomerase 2alpha to protect human telomeres from replicative damage. Cell. 2010; 142:230–242. [DOI] [PubMed] [Google Scholar]
- 5.Karlseder J., Hoke K., Mirzoeva O.K., Bakkenist C., Kastan M.B., Petrini J.H., de Lange T.. The telomeric protein TRF2 binds the ATM kinase and can inhibit the ATM-dependent DNA damage response. PLoS Biol. 2004; 2:E240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buscemi G., Zannini L., Fontanella E., Lecis D., Lisanti S., Delia D.. The shelterin protein TRF2 inhibits Chk2 activity at telomeres in the absence of DNA damage. Curr. Biol. 2009; 19:874–879. [DOI] [PubMed] [Google Scholar]
- 7.Okamoto K., Bartocci C., Ouzounov I., Diedrich J.K., Yates J.R. 3rd, Denchi E.L.. A two-step mechanism for TRF2-mediated chromosome-end protection. Nature. 2013; 494:502–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Griffith J.D., Comeau L., Rosenfield S., Stansel R.M., Bianchi A., Moss H., de Lange T.. Mammalian telomeres end in a large duplex loop. Cell. 1999; 97:503–514. [DOI] [PubMed] [Google Scholar]
- 9.Amiard S., Doudeau M., Pinte S., Poulet A., Lenain C., Faivre-Moskalenko C., Angelov D., Hug N., Vindigni A., Bouvet P. et al. . A topological mechanism for TRF2-enhanced strand invasion. Nat. Struct. Mol. Biol. 2007; 14:147–154. [DOI] [PubMed] [Google Scholar]
- 10.Doksani Y., Wu J.Y., de Lange T., Zhuang X.. Super-resolution fluorescence imaging of telomeres reveals TRF2-dependent T-loop formation. Cell. 2013; 155:345–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poulet A., Pisano S., Faivre-Moskalenko C., Pei B., Tauran Y., Haftek-Terreau Z., Brunet F., Le Bihan Y.V., Ledu M.H., Montel F. et al. . The N-terminal domains of TRF1 and TRF2 regulate their ability to condense telomeric DNA. Nucleic Acids Res. 2012; 40:2566–2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benarroch-Popivker D., Pisano S., Mendez-Bermudez A., Lototska L., Kaur P., Bauwens S., Djerbi N., Latrick C.M., Fraisier V., Pei B. et al. . TRF2-mediated control of telomere DNA topology as a mechanism for chromosome-end protection. Mol. Cell. 2016; 61:274–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poulet A., Buisson R., Faivre-Moskalenko C., Koelblen M., Amiard S., Montel F., Cuesta-Lopez S., Bornet O., Guerlesquin F., Godet T. et al. . TRF2 promotes, remodels and protects telomeric Holliday junctions. EMBO J. 2009; 28:641–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saint-Leger A., Koelblen M., Civitelli L., Bah A., Djerbi N., Giraud-Panis M.J., Londoño-Vallejo A., Ascenzioni F., Gilson E.. The basic N-terminal domain of TRF2 limits recombination endonuclease action at human telomeres. Cell Cycle. 2014; 13:2469–2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ye J., Renault V.M., Jamet K., Gilson E.. Transcriptional outcome of telomere signalling. Nat. Rev. Genet. 2014; 15:491–503. [DOI] [PubMed] [Google Scholar]
- 16.Simonet T., Zaragosi L.E., Philippe C., Lebrigand K., Schouteden C., Augereau A., Bauwens S., Ye J., Santagostino M., Giulotto E. et al. . The human TTAGGG repeat factors 1 and 2 bind to a subset of interstitial telomeric sequences and satellite repeats. Cell Res. 2011; 21:1028–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang D., Xiong Y., Kim H., He Q., Li Y., Chen R., Songyang Z.. Human telomeric proteins occupy selective interstitial sites. Cell Res. 2011; 21:1013–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Biroccio A., Cherfils-Vicini J., Augereau A., Pinte S., Bauwens S., Ye J., Simonet T., Horard B., Jamet K., Cervera L. et al. . TRF2 inhibits a cell-extrinsic pathway through which natural killer cells eliminate cancer cells. Nat. Cell Biol. 2013; 15:818–828. [DOI] [PubMed] [Google Scholar]
- 19.El Mai M., Wagner K.D., Michiels J.F., Ambrosetti D., Borderie A., Destree S., Renault V., Djerbi N., Giraud-Panis M.J., Gilson E. et al. . The telomeric protein TRF2 regulates angiogenesis by binding and activating the PDGFRbeta promoter. Cell Rep. 2014; 9:1047–1060. [DOI] [PubMed] [Google Scholar]
- 20.Zhang P., Pazin M.J., Schwartz C.M., Becker K.G., Wersto R.P., Dilley C.M., Mattson MP.. Nontelomeric TRF2-REST interaction modulates neuronal gene silencing and fate of tumor and stem cells. Curr. Biol. 2008; 18:1489–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang P., Casaday-Potts R., Precht P., Jiang H., Liu Y., Pazin M.J., Mattson M.P.. Nontelomeric splice variant of telomere repeat-binding factor 2 maintains neuronal traits by sequestering repressor element 1-silencing transcription factor. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:16434–16439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ovando-Roche P., Yu J.S., Testori S., Ho C., Cui W.. TRF2-mediated stabilization of hREST4 is critical for the differentiation and maintenance of neural progenitors. Stem Cells. 2014; 32:2111–2122. [DOI] [PubMed] [Google Scholar]
- 23.Bradshaw P.S., Stavropoulos D.J., Meyn M.S.. Human telomeric protein TRF2 associates with genomic double-strand breaks as an early response to DNA damage. Nat. Genet. 2005; 37:193–197. [DOI] [PubMed] [Google Scholar]
- 24.Tanaka H., Mendonca M.S., Bradshaw P.S., Hoelz D.J., Malkas L.H., Meyn M.S., Gilley D.. DNA damage-induced phosphorylation of the human telomere-associated protein TRF2. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:15539–15544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huda N., Tanaka H., Mendonca M.S., Gilley D.. DNA damage-induced phosphorylation of TRF2 is required for the fast pathway of DNA double-strand break repair. Mol. Cell. Biol. 2009; 29:3597–3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mao Z., Seluanov A., Jiang Y., Gorbunova V.. TRF2 is required for repair of nontelomeric DNA double-strand breaks by homologous recombination. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:13068–13073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deng Z., Norseen J., Wiedmer A., Riethman H., Lieberman P.M.. TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol. Cell. 2009; 35:403–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim H., Lee O.H., Xin H., Chen L.Y., Qin J., Chae H.K., Lin S.Y., Safari A., Liu D., Songyang Z.. TRF2 functions as a protein hub and regulates telomere maintenance by recognizing specific peptide motifs. Nat. Struct. Mol. Biol. 2009; 16:372–379. [DOI] [PubMed] [Google Scholar]
- 29.Giraud-Panis M.J., Pisano S., Benarroch-Popivker D., Pei B., Le Du M.H., Gilson E.. One identity or more for telomeres?. Front. Oncol. 2013; 3:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Broccoli D., Smogorzewska A., Chong L., de Lange T.. Human telomeres contain two distinct Mybrelated proteins, TRF1 and TRF2. Nat. Genet. 1997; 17:231–235. [DOI] [PubMed] [Google Scholar]
- 31.Bilaud T., Koering C.E., Binet-Brasselet E., Ancelin K., Pollice A., Gasser S.M., Gilson E.. The telobox, a Myb-related telomeric DNA binding motif found in proteins from yeast, plants and human. Nucleic Acids Res. 1996; 24:1294–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bilaud T., Brun C., Ancelin K., Koering C.E., Laroche T., Gilson E.. Telomeric localization of TRF2, a novel human telobox protein. Nat. Genet. 1997; 17:236–239. [DOI] [PubMed] [Google Scholar]
- 33.Fujita K., Horikawa I., Mondal A.M., Jenkins L.M., Appella E., Vojtesek B., Bourdon J.C., Lane D.P., Harris C.C.. Positive feedback between p53 and TRF2 during telomere-damage signalling and cellular senescence. Nat. Cell. Biol. 2010; 12:1205–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang J., Uryga A.K., Reinhold J., Figg N., Baker L., Finigan A., Gray K., Kumar S., Clarke M., Bennett M.. Vascular smooth muscle cell senescence promotes atherosclerosis and features of plaque vulnerability. Circulation. 2015; 132:1909–1919. [DOI] [PubMed] [Google Scholar]
- 35.Munoz P., Blanco R., Flores J.M., Blasco M.A.. XPF nuclease-dependent telomere loss and increased DNA damage in mice overexpressing TRF2 result in premature aging and cancer. Nat. Genet. 2005; 37:1063–1071. [DOI] [PubMed] [Google Scholar]
- 36.Bellon M., Datta A., Brown M., Pouliquen J.F., Couppie P., Kazanji M., Nicot C.. Increased expression of telomere length regulating factors TRF1, TRF2 and TIN2 in patients with adult T-cell leukemia. Int. J. Cancer. 2006; 119:2090–2097. [DOI] [PubMed] [Google Scholar]
- 37.Matsutani N., Yokozaki H., Tahara E., Tahara H., Kuniyasu H., Haruma K., Chayama K., Yasui W., Tahara E.. Expression of telomeric repeat binding factor 1 and 2 and TRF1-interacting nuclear protein 2 in human gastric carcinomas. Int. J. Oncol. 2001; 19:507–512. [PubMed] [Google Scholar]
- 38.Oh B.K., Kim Y.J., Park C., Park Y.N.. Up-regulation of telomere-binding proteins, TRF1, TRF2, and TIN2 is related to telomere shortening during human multistep hepatocarcinogenesis. Am. J. Pathol. 2005; 166:73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakanishi K., Kawai T., Kumaki F., Hiroi S., Mukai M., Ikeda E., Koering C.E., Gilson E.. Expression of mRNAs for telomeric repeat binding factor (TRF)-1 and TRF2 in atypical adenomatous hyperplasia and adenocarcinoma of the lung. Clin. Cancer Res. 2003; 9:1105–1111. [PubMed] [Google Scholar]
- 40.Diala I., Wagner N., Magdinier F., Shkreli M., Sirakov M., Bauwens S., Schluth-Bolard C., Simonet T., Renault V.M., Ye J. et al. . Telomere protection and TRF2 expression are enhanced by the canonical Wnt signalling pathway. EMBO Rep. 2013; 14:356–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cherfils-Vicini J., Zizza P., Gilson E., Biroccio A.. A novel pathway links telomeres to NK-cell activity: Implications for immunotherapy. Oncoimmunology. 2014; 3:e27358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kishi S., Bayliss P.E., Uchiyama J., Koshimizu E., Qi J., Nanjappa P., Imamura S., Islam A., Neuberg D., Amsterdam A. et al. . The identification of zebrafish mutants showing alterations in senescence-associated biomarkers. PLoS Genet. 2008; 4:e1000152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Michishita E., McCord R.A., Berber E., Kioi M., Padilla-Nash H., Damian M., Cheung P., Kusumoto R., Kawahara T.L., Barrett J.C. et al. . SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008; 452:492–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tennen R.I., Bua D.J., Wright W.E., Chua K.F.. SIRT6 is required for maintenance of telomere position effect in human cells. Nat. Commun. 2011; 2:433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang B., Zwaans B.M., Eckersdorff M., Lombard D.B.. The sirtuin SIRT6 deacetylates H3K56Ac in vivo to promote genomic stability. Cell Cycle. 2009; 8:2662–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tasselli L., Xi Y., Zheng W., Tennen R.I., Odrowaz Z., Simeoni F., Li W., Chua K.F.. SIRT6 deacetylates H3K18ac at pericentric chromatin to prevent mitotic errors and cellular senescence. Nat. Struct. Mol. Biol. 2016; 23:434–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Finkel T., Deng C.X., Mostoslavsky R.. Recent progress in the biology and physiology of sirtuins. Nature. 2009; 460:587–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mostoslavsky R., Chua K.F., Lombard D.B., Pang W.W., Fischer M.R., Gellon L., Liu P., Mostoslavsky G., Franco S., Murphy M.M. et al. . Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006; 124:315–329. [DOI] [PubMed] [Google Scholar]
- 49.Toiber D., Erdel F., Bouazoune K., Silberman D.M., Zhong L., Mulligan P., Sebastian C., Cosentino C., Martinez-Pastor B., Giacosa S. et al. . SIRT6 recruits SNF2H to DNA break sites, preventing genomic instability through chromatin remodeling. Mol. Cell. 2013; 51:454–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhong L., D'Urso A., Toiber D., Sebastian C., Henry R.E., Vadysirisack D.D., Guimaraes A., Marinelli B., Wikstrom J.D., Nir T. et al. . The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell. 2010; 140:280–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jia G., Su L., Singhal S., Liu X.. Emerging roles of SIRT6 on telomere maintenance, DNA repair, metabolism and mammalian aging. Mol. Cell. Biochem. 2012; 364:345–350. [DOI] [PubMed] [Google Scholar]
- 52.Mao Z., Hine C., Tian X., Van Meter M., Au M., Vaidya A., Seluanov A., Gorbunova V.. SIRT6 promotes DNA repair under stress by activating PARP1. Science. 2011; 332:1443–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kaidi A., Weinert B.T., Choudhary C., Jackson S.P.. Human SIRT6 promotes DNA end resection through CtIP deacetylation. Science. 2010; 329:1348–1353. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 54.Hallows W.C., Lee S., Denu J.M.. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:10230–10235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Feldman J.L., Baeza J., Denu J.M.. Activation of the protein deacetylase SIRT6 by long-chain fatty acids and widespread deacylation by mammalian sirtuins. J. Biol. Chem. 2013; 288:31350–31356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sebastián C., Zwaans B.M., Silberman D.M., Gymrek M., Goren A., Zhong L., Ram O., Truelove J., Guimaraes A.R., Toiber D. et al. . The histone deacetylase SIRT6 is a tumor suppressor that controls cancer metabolism. Cell. 2012; 151:1185–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Salvati E., Zizza P., Rizzo A., Iachettini S., Cingolani C., D'Angelo C., Porru M., Randazzo A., Pagano B., Novellino E. et al. . Evidence for G-quadruplex in the promoter of vegfr-2 and its targeting to inhibit tumor angiogenesis. Nucleic Acids Res. 2014; 42:2945–2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brunori M., Mathieu N., Ricoul M., Bauwens S., Koering C.E., Roborel de Climens A., Belleville A., Wang Q., Puisieux I., Décimo D. et al. . TRF2 inhibition promotes anchorage-independent growth of telomerase-positive human fibroblasts. Oncogene. 2006; 25:990–997. [DOI] [PubMed] [Google Scholar]
- 59.Biroccio A., Amodei S., Benassi B., Scarsella M., Cianciulli A., Mottolese M., Del Bufalo D., Leonetti C., Zupi G.. Reconstitution of hTERT restores tumorigenicity in melanoma-derived c-Myc low-expressing clones. Oncogene. 2002; 21:3011–3009. [DOI] [PubMed] [Google Scholar]
- 60.Atanasiu C., Deng Z., Wiedmer A., Norseen J., Lieberman P.M.. ORC binding to TRF2 stimulates OriP replication. EMBO Rep. 2006; 7:716–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shevchenko A., Tomas H., Havlis J., Olsen J.V., Mann M.. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Prot. 2006; 1:2856–2860. [DOI] [PubMed] [Google Scholar]
- 62.Lu Y., Leong W., Guérin O., Gilson E., Ye J.. Telomeric impact of conventional chemotherapy. Front. Med. 2013; 7:411–417. [DOI] [PubMed] [Google Scholar]
- 63.Cea M., Cagnetta A., Adamia S., Acharya C., Tai Y.T., Fulciniti M., Ohguchi H., Munshi A., Acharya P., Bhasin M.K. et al. . Evidence for a role of the histone deacetylase SIRT6 in DNA damage response of multiple myeloma cells. Blood. 2016; 127:1138–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Beck C., Robert I., Reina-San-Martin B., Schreiber V., Dantzer F.. Poly(ADP-ribose) polymerases in double-strand break repair: focus on PARP1, PARP2 and PARP3. Exp. Cell Res. 2014; 329:18–25. [DOI] [PubMed] [Google Scholar]
- 65.Saquilabon Cruz G.M., Kong X., Silva B.A., Khatibzadeh N., Thai R., Berns M.W., Yokomori K.. Femtosecond near-infrared laser microirradiation reveals a crucial role for PARP signaling on factor assemblies at DNA damage sites. Nucleic Acids Res. 2016; 44:e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gomez M., Wu J., Schreiber V., Dunlap J., Dantzer F., Wang Y., Liu Y.. PARP1 Is a TRF2-associated poly(ADP-ribose)polymerase and protects eroded telomeres. Mol. Biol. Cell. 2006; 17:1686–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kugel S., Mostoslavsky R.. Chromatin and beyond: the multitasking roles for SIRT6. Trends Biochem. Sci. 2014; 39:72–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Curtin N.J., Szabo C.. Therapeutic applications of PARP inhibitors: anticancer therapy and beyond. Mol. Aspects Med. 2013; 34:1217–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bryant H.E., Petermann E., Schultz N., Jemth A.S., Loseva O., Issaeva N., Johansson F., Fernandez S., McGlynn P., Helleday T.. PARP is activated at stalled forks to mediate Mre11-dependent replication restart and recombination. EMBO J. 2009; 28:2601–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sugimura K., Takebayashi S., Taguchi H., Takeda S., Okumura K.. PARP-1 ensures regulation of replication fork progression by homologous recombination on damaged DNA. J. Cell Biol. 2008; 183:1203–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jones C.B., McIntosh J., Huang H., Graytock A., Hoyt D.G.. Regulation of bleomycin-induced DNA breakage and chromatin structure in lung endothelial cells by integrins and poly(ADP-ribose) polymerase. Mol. Pharmacol. 2001; 59:69–75. [DOI] [PubMed] [Google Scholar]
- 72.Her Y.R., Chung I.K.. p300-mediated acetylation of TRF2 is required for maintaining functional telomeres. Nucleic Acids Res. 2013; 41:2267–2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ning H., Li T., Zhao L., Li T., Li J., Liu J., Liu Z., Fan D.. TRF2 promotes multidrug resistance in gastric cancer cells. Cancer Biol. Ther. 2006; 5:950–956. [DOI] [PubMed] [Google Scholar]
- 74.Bai Y., Lathia J.D., Zhang P., Flavahan W., Rich J.N., Mattson M.P.. Molecular targeting of TRF2 suppresses the growth and tumorigenesis of glioblastoma stem cells. Glia. 2014; 62:1687–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.