Abstract
Alternative lengthening of telomeres (ALT) is a telomerase independent telomere maintenance mechanism that occurs in ∼15% of cancers. The potential mechanism of ALT is homology-directed telomere synthesis, but molecular mechanisms of how ALT maintains telomere length in human cancer is poorly understood. Here, we generated TERC (telomerase RNA) gene knockouts in telomerase positive cell lines that resulted in long-term surviving clones acquiring the ALT pathway but at a very low frequency. By comparing these ALT cells with parental telomerase positive cells, we observed that ALT cells possess excessively long telomeric overhangs derived from telomere elongation processes that mostly occur during S phase. ALT cells exhibited preferential elongation of the telomeric lagging strands, whereas telomerase positive cells exhibited similar elongation between leading and lagging strands. We propose that the ALT pathway preferentially occurs at telomeric lagging strands leading to heterogeneous telomere lengths observed in most ALT cancers.
INTRODUCTION
Telomeres are composed of TTAGGG repeated sequences located at the ends of linear chromosomes. In normal human somatic cells each division is accompanied by progressive telomere length shortening due to lack of, or insufficient, telomerase activity. Cancer cells need to acquire a telomere maintenance mechanism during tumorigenesis to proliferate indefinitely. The vast majority of human cancer cells maintain their telomere length via telomerase reactivation (1–3). Therefore anti-telomerase cancer therapy is considered an almost universal cancer target and one that should not affect somatic cells that are telomerase silent (4). One concern of effective anti-telomerase therapeutic approaches is the potential acquired resistance by engagement of the Alternative Lengthening of Telomeres (ALT) pathway (5–7).
ALT is a telomerase-independent telomere maintenance mechanism that occurs in a small subset of cancers (8). Genetic screenings for telomerase mutants demonstrate that such telomerase mutants can survive by acquiring various ALT mechanisms (9–11). In mice, telomerase-expressing tumors exhibit ALT phenotypes in response to abolishing telomerase activity (7,12). Nevertheless, an understanding of ALT engagement in telomerase-positive human cells treated with telomerase inhibitors is not only exceptionally rare but mechanistically not understood (6).
How ALT is activated and extends the telomere is one of the most important unresolved questions in telomere biology. It has been reported that loss of the ATRX gene expression is common, but not universal, in ALT tumors and cell lines (13–15). ATRX knockdown in normal fibroblasts increases the proportion of cells activating ALT and accelerates the occurrence of immortalization (16). Restoration of ATRX expression in ATRX-negative ALT cell lines can result in the loss of ALT activity (17). Therefore, elucidating the recombination-mediated telomere elongation processes may provide a more complete understanding of the ALT mechanism.
In this study, we generated ALT cells, which were derived from TERC (TR) gene knockouts in telomerase-positive human cell lines that resulted in long-term rare surviving clones acquiring the ALT pathway. We then analyzed these telomerase-positive cells with their isogenically derived ALT cells to further dissect the mechanism for maintenance of the ALT phenotype. We observed that ALT cells have longer overhang lengths compared to their parental telomerase-positive cells. Moreover, lagging strand telomeric overhangs are preferentially elongated in ALT cells during S phase. This results in the ALT mechanism preferentially elongating telomeric lagging strands leading to a heterogeneous (both long and short) telomere lengths in ALT cancers.
MATERIALS AND METHODS
Cell culture and TERC gene knockout cell generation
Cells were cultured at 37°C in 5% CO2 in Media-X with 10% cosmic calf-serum (Hyclone). Cell lines were tested for mycoplasma contamination. To generate the TERC KO cell lines, px458 plasmids (Addgene #48138) (18) containing TERC gRNA (5΄-AGCGAGAAAAACAGCGCGCG-(PAM)-3΄) were transfected into SW39, HeLa LT, HAP1, HT1080 (ATCC) or H1299 (ATCC) cells, and GFP-positive cells were sorted in 96-well plates at 48 h post-transfection. We selected the TERC KO clones using digital droplet TRAP and PCR. Cell morphology changes were captured by EVOS FL Cell imaging system (Thermo Scientific). For cell cycle analysis, U2OS (ATCC), HeLa LT or HeLa LT TERC KO cells were synchronized at the G1/S boundary with double thymidine blocks. Cells were incubated with 2 mM thymidine for 20 h, washed 4 times with PBS, and then released into fresh medium for 8 h. Thymidine was re-added for 18 h, and then the cells were washed four times with PBS and released into fresh medium with IdU (5-Iodo-2΄-deoxyuridine) for CsCl separation. U2OS cells were harvested at 6 h for S phase, 9 h for G2 phase, and 15 h for G1 phase. For HeLa LT and HeLa LT TERC KO cells, cells were harvested at 4 h for S phase, 8 h for G2 phase and 13 h for G1 phase. Flow cytometric analysis was performed to determine cell cycle profiles. For RAD51 inhibition, the RAD51 inhibitor (RI-1 Calbiochem) was used.
Viral infection
ATRX shRNA (Sigma-Aldrich TRCN0000013590) was used as previously reported (15). To generate lentivirus, packaging vectors—pMD2.G (Addgene #12259) and psPAX2 (Addgene #12260) were used. pBabe puro U6_hTR (Addgene #27666) (19) and pBabe hygro_loxp-hTERT plasmids were used for the generation of TERC or TERT gene encoding retrovirus. To generate retrovirus, packaging vectors—gag/pol and VSV.G were used. Cells were infected with 3 mg/ml polybrene, and then 3 μg/ml of puromycin or 300 μg/ml of hygromycin were added at least 14 days for selection.
Western blotting
Cells were lysed with NETN lysis buffer (100 mM NaCl, 0.5 mM EDTA, 50 mM Tris–HCl, pH 8.0 and 0.5% NP-40) with protease inhibitor cocktail (Roche cOmplete, EDTA-free). The protein samples were separated by SDS-PAGE gel, and then transferred to nitrocellulose membrane. The following antibodies were used; anti-ATRX antibody (H-300 Santa Cruz Biotechnology, A301-045A Bethyl Laboratories), DAXX (M-112 Santa Cruz Biotechnology), Anti-Lamin A/C (4C11 Cell Signaling), and Actin (A1978 Sigma-Aldrich).
Telomere restriction fragment assay
Telomere restriction fragment assay (TRF) was performed as previously described (20). Briefly, genomic DNA was purified using Puregene Core Kit A (Qiagen) following the manufacturer's instructions. DNA samples were digested with HinfI/RsaI/AluI (NEB, 10 unit each for 1 μg DNA). Digested genomic DNAs were run into 0.85% or 0.7% agarose gels in 1× TAE buffer (20 h, 1 V/cm), The gels were then denatured (0.5 M NaOH and 1.5 M NaCl), neutralized (0.5 M Tris–HCl, pH 8.0 and 2 M NaCl) and then hybridized with a 32P-labeled G-probe to detect the telomere signal.
C-circle assay
The C-circle assay was performed as previously described (21). Briefly, the digested DNAs were incubated with the phi29 polymerase reaction (0.75 unit of phi29 polymerase, 0.1 mg/ml BSA, 0.2 mM dNTP mix and 1× phi29 buffer (NEB)) at 30°C for 12 h and then incubated at 65°C for 20 min. Samples were loaded into a slot-blot, and hybridized with 32P-labeled C-probe in native condition to detect and quantify the amplified C-circle level.
Duplex-specific nuclease method
DSN (duplex-specific nuclease) method was performed as previously described (22) with minor modifications. In brief, genomic DNA was purified using the Blood and Cell culture Midi kit (Qiagen). Precipitated DNA was washed twice with 70% ethanol and suspended in 10 mM Tris–HCl, pH 8.0. Fifteen micrograms of DNA was digested with 4 U DSN (Evrogen) for 2 h at 37°C. Digested DNAs were loaded in alkaline agarose gels (1% or 1.2% agarose made with 50 mM NaOH, 1 mM EDTA), and then run in 50 mM NaOH and 1 mM EDTA at 4°C (17 h, 1 V/cm). DNA was transferred to Hybond-XL (GE Healthcare), and then hybridized with a 32P-labeled C-probe to detect the overhang signal.
In-gel hybridization analysis
In gel hybridization was performed as previously described (23) with minor modifications. Digested genomic DNAs were briefly run into a 0.7% agarose gel (1× TAE, 1 h, 1–2 V/cm) so the telomeric DNAs remained in a tight band. The gels were dried and hybridized with 32P-labeled C-probe (CCCTAA)3 or G-probe (TTAGGG)3 (20) using Rapid-hyb buffer (GE Healthcare) to detect the G-overhang or C-overhang signal. Gels were denatured and neutralized and then hybridized with 32P-labeled G-probe (TTAGGG)3 for total telomere DNA input. G-probe was used for detecting total telomere DNA in leading and lagging strand samples by hybridizing with C-rich sequences (22).
Cesium chloride (CsCl) separation of leading and lagging telomere DNA
We performed the CsCl separation as previously described (24) with modifications. Briefly, cells were incorporated with 100 μM IdU for 20 h. Five hundred micrograms of genomic DNA was purified using Puregene Core Kit A (Qiagen) and then digested with HinfI/RsaI/AluI. DNAs were mixed with CsCl solution to obtain a final density of 1.770 g/ml. Samples were centrifuged at 55,000 rpm for 20 h using a VTi-90 vertical rotor (Beckman) and then fractions collected. Slot blot for each fraction was performed to collect the leading and lagging strand fractions. Leading DNA was located at a density of 1.790–1.800 g/ml; lagging DNAs was located at a density of 1.760–1.770 g/ml; and unreplicated DNAs were located at a density of 1.740–1.750 g/ml.
Droplet digital telomerase repeat amplification protocol (ddTRAP)
ddTRAP were performed as previously described (25). Briefly, 500 000 cells were harvested then resuspended in 400 μl of TRAP lysis buffer (10 mM Tris–HCl, pH 8.0, 1 mM MgCl2, 1 mM EDTA, 1% NP-40, 0.25 mM sodium deoxycholate, 10% glycerol, 150 mM NaCl, 5 mM beta-mercaptoethanol and 0.1 mM AEBSF) for 30 min. Telomerase extension reactions were performed in extension mix (20 mM Tris–HCl pH 8.3, 1.5 mM MgCl2, 63 mM KCl, 0.05% Tween 20, 1 mM EGTA, 0.05 mM dNTP, 0.2 μM TS primer, 20 μg/ml BSA and 1 μl of cell lysates (1250 cells/μl)) for 40 min at 25°C followed by 95°C for 5 min. ddTRAP reactions were followed in the reaction mix (1× Evagreen ddPCR super mix v2.0 (Biorad), 50 nM TS primer, 50 nM ACX primer and extension product of 50 cells). PCR reaction condition was 95°C, 5 min; then 40 cycles of 95°C, 30 s, 54°C, 30 s, 72°C, 30 s. ddPCR products were measured and analyzed using a droplet reader (Biorad).
Statistical analysis
The student's two-tailed unpaired t test was used to determine statistical significance. The graphs and statistics were generated with Prism (GraphPad software).
RESULTS
Telomerase RNA (TERC) knockout cells engage the alternative lengthening of telomeres (ALT) pathway during crisis, a stage of massive cell death
To test whether the ALT pathway can be activated by telomerase inhibition in human telomerase positive transformed cells, we generated telomerase functional RNA (TERC) knockouts (KO) in the SW39 cell line using the CRISPR/Cas9 genome editing system (Figure 1A). SW39 cells are immortal telomerase positive cells derived from human lung fibroblasts (IMR90 cells) which were stably transfected with SV40 large-T antigen (26). SW39 cells were transformed with a Cas9/TERC targeting guide RNA. Single clones were screened by PCR and a droplet digital PCR-based telomere repeat amplification protocol (ddTRAP) (25) to verify the deletion of the TERC locus and the loss of telomerase activity. SW39 TERC KO cells proliferated ∼21–24 population doublings (PDs) before entering crisis, a period that is accompanied by a balance between cell growth and massive cell death (Supplementary Figure S1A). During crisis, cells acquire many genetic alterations in addition to the edited loss of the TERC gene. We predicted that only a small fraction of cells would escape from crisis by acquiring a telomerase-independent telomere maintenance mechanism. A modification of a fluctuation analysis using SW39 TERC KO cells resulted in emergence of three survival clones out of 12 million cells in crisis resulting in a frequency of immortalization = 2.5 × 10−7 (Supplementary Figure S1B), confirming that overcoming the crisis event occurs at a low frequency.
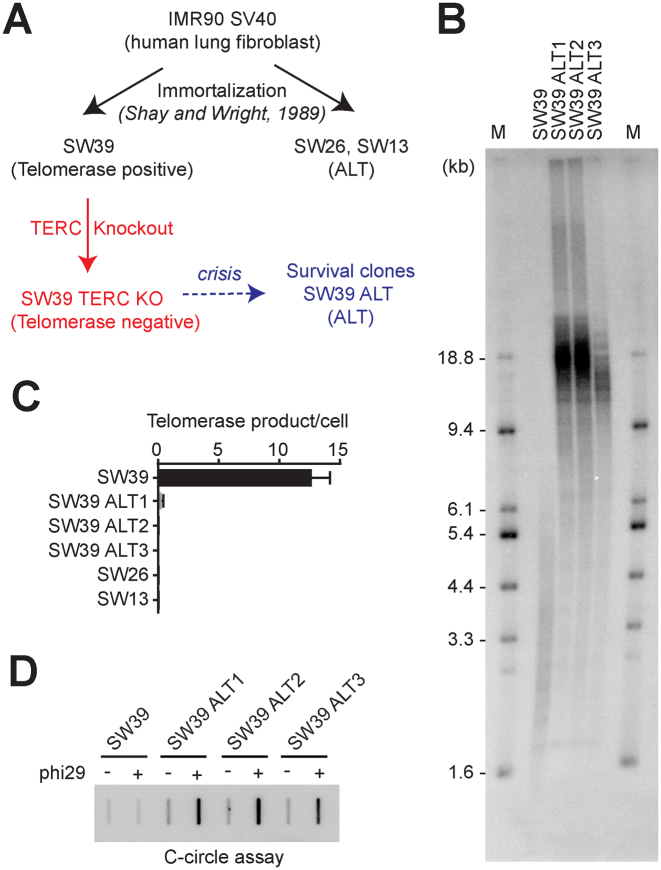
Figure 1.
Telomerase inhibition in telomerase-positive cells onsets the alternative lengthening of telomeres to overcome the crisis. (A) Overview of ALT cell line generation from telomerase positive transformed cells (SW39). SW39 cells were introduced with Cas9/TERC targeting guide RNA and single cell clones were isolated and analyzed. (B) Terminal Restriction Fragment (TRF) analysis in SW39 (parental cell) and SW39 TERC KO survival clones (post-crisis cells). Digested DNA was run on 0.85% agarose gels. M indicates for the marker. (C) Droplet digital PCR based telomere repeat amplification (ddTRAP) analysis in SW39, SW39 TERC KO survival clones (SW39 ALT1, ALT2, and ALT3), SW26 and SW13 cells (mean ± SD; n = 3). (D) C-circle analysis in SW39 and SW39 ALT survival clones. One hundred nanograms of digested DNA from SW39 or SW39 ALT survival clones were used in the phi29 polymerase reaction.
To understand how these surviving clones escaped from crisis, we first measured telomere length using TRF analysis to check whether these clones escaped from crisis by elongating their telomere length. All three clones (herein after termed SW39 ALT1, 2 and 3) exhibited elongated and heterogeneous (long and short) telomere lengths compared to the parental cells (SW39) (Figure 1B). We performed ddTRAP analysis to assess telomerase activity in SW39 ALT cells, SW39, and its sister spontaneously occurring ALT cell lines, SW26 and SW13 as controls (26). As expected, SW39 showed robust telomerase activity, but no telomerase activity was detected in SW26 and SW13 (Figure 1C). In addition, all TERC KO surviving clones of SW39 had no detectable telomerase activity, suggesting that SW39 TERC KO cell lines acquired a telomerase-independent telomere maintenance mechanism.
C-rich circular extra-chromosomal DNAs (C-circles) and ALT-associated PML bodies (APBs) are accepted biomarkers of ALT cells (21). To determine if SW39 ALT cells have these ALT biomarkers, we first performed the C-circle assay in SW39 ALT cells and compared them to SW39 (Figure 1D). We detected a significant increase in C-circle levels in all SW39 ALT clones but not in the parental telomerase positive cell line SW39. Another biomarker of the ALT phenotype is the presence of ALT associated PML bodies (APBs). APB-positive cells were observed in SW39 ALT cells whereas the parental cells were APB-negative (Supplementary Figure S1C). In contrast, we did not detect any alteration of telomeric repeat-containing RNA (TERRA) levels in SW39 ALT cells (Supplementary Figure S1D), which has previously been suggested as another ALT pathway-associated factor (27). The phenotypes in SW39 ALT cells, including the heterogeneous and elongated telomere length, and presence of C-circle and APBs resemble those of ALT cells. These results indicate that telomerase inhibition in human telomerase positive transformed cells can result in engagement of ALT but at a low frequency.
ALT cells have excessively long overhangs
Telomeric overhangs are the binding site for telomerase and are elongated throughout S-phase in telomerase positive human cells (28). Moreover, telomere overhangs are thought to be important as the elongation site for ALT cells as well as telomerase positive cells (29). The exposed 3΄ telomeric end overhang is believed to invade into the sister chromatid, other chromosome ends or extra-chromosomal telomeric DNA repeats that can be used as templates to then elongate the ends via recombination mediated DNA replication (30). Therefore, we tested whether the alterations in telomere maintenance mechanism resulted in overhang length changes. Duplex-specific nuclease (DSN) digests double-stranded DNA into <10 bp fragments while leaving the single stranded telomeric overhangs intact (Figure 2A) (22,31). Using the DSN method, we measured the overhang length in SW39 and SW39 ALT1 cells. Overhang length in SW39 ALT1 cells are variable from 40 nt to 400 nt, whereas parental SW39 cells have 65 nt to 140 nt overhang lengths consistent with previous reports (overhang length in telomerase positive cells: 60–150 nt) (22,24,28) (Figure 2B). These results show that ALT cells can have excessively long overhang populations, possibly due to its less well controlled elongation mechanism compared to telomerase-mediated elongation as previously speculated (29,32,33).
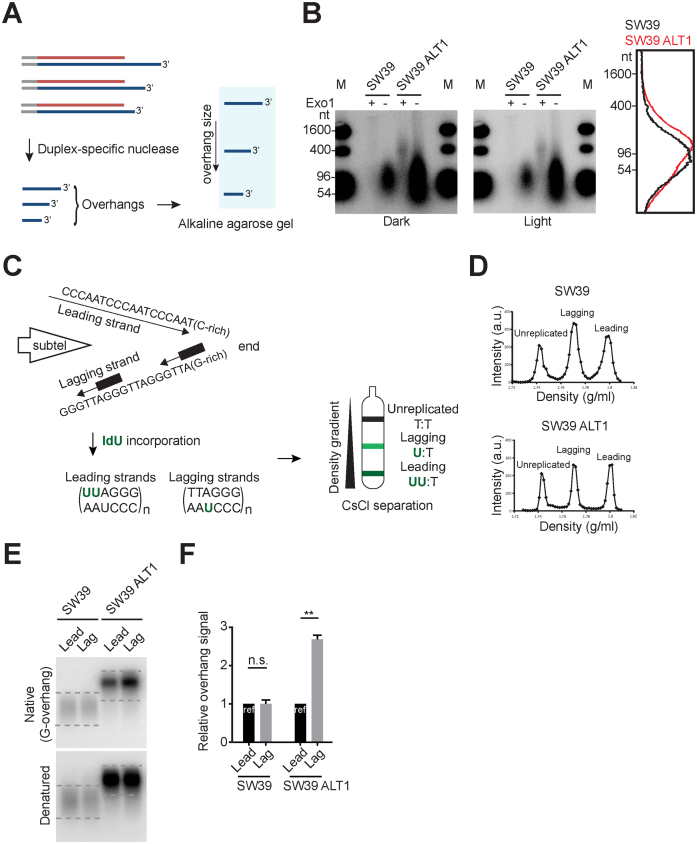
Figure 2.
Excessively elongated lagging overhangs in ALT cells. (A) Illustration demonstrating Duplex-specific nuclease (DSN) overhang assay. DSN enzyme digests double stranded DNA and leaves the single stranded TTAGGG sequences present in the telomeric G-overhangs (3΄-overhangs). (B) DSN overhang assay in SW39 and SW39 ALT1 cells. Digested DNA (15 μg each) was run on 1.2% alkaline agarose gels. C-rich sequence probes were used for detecting G-overhangs. Right: Lines traces of SW39 (black) and SW39 ALT1 (red) gel lanes. (C) Illustration demonstrating CsCl separation of telomeric leading and lagging strands. Cells were usually incubated with IdU (5-Iodo-2΄-deoxyuridine) for 20 h (asynchronous condition) and then leading and lagging strands were separated using CsCl centrifugation. (D) CsCl separation of leading versus lagging strands in SW39 and SW39 ALT1 cells. a.u. represents arbitrary unit. (E) In-gel hybridization of telomeres in SW39 and SW39 ALT1 cells after leading/lagging strand separation. Native gels were hybridized with a C-rich probe to detect the amount of G-overhangs. Denatured gels were hybridized with a G-rich probe to measure the amount of total telomeric DNAs. Dashed lines indicate the quantified area. (F) Quantification of G-overhang intensity in E. Relative ratio of native signal to denatured signal was calculated (mean ± SEM; n = 2) ** indicates P < 0.01 and n.s. indicates non-significant (unpaired Student's t test).
Lagging strands are preferentially extended in ALT cells
In telomerase positive cells, most leading and lagging strand overhangs are extended by telomerase after telomere replication, therefore their leading and lagging overhang lengths are similar (24). However, overhang lengths are totally different in telomerase negative cells [lagging strand overhang (∼100 nt in BJ normal fibroblast cells) > leading strand overhang (∼30 nt in BJ cells)]. Lagging overhangs are determined shortly after completion of DNA replication, whereas leading overhangs are processed throughout S/G2 in telomerase negative cells (23). We tested if these differences in leading versus lagging strands affected the ALT-mediated telomere extension process. Using cesium chloride (CsCl) centrifugation (Figure 2C and D), we separated leading and lagging strands in SW39 ALT1 cells and then measured the overhang signals using in-gel hybridization analysis (Figure 2E). We found that the overhang signal intensity ratio between leading versus lagging overhangs in SW39 ALT1 cell was 1:2.5 while the ratio in the SW39 cell line was 1:1. These results show that SW39 ALT1 cells have a different leading versus lagging strand overhang composition. One possible interpretation is that the lagging overhangs in SW39 ALT1 cells are significantly longer than leading overhangs, whereas SW39 cells have no differences in overhang lengths (Figure 2E). Collectively, these data support the idea that excessive telomere extensions in ALT cells occur more at lagging overhangs.
Telomerase reactivation reverses ALT-mediated telomere extension
We reintroduced the human TERC (TR) gene into SW39 ALT1 cells to test whether reacquiring telomerase activity abrogated the ALT phenotypes in SW39 ALT cells. For this, we introduced TR only (SW39 ALT1 + TR) or both TR and TERT (SW39 ALT1 + TT) into SW39 ALT1 cells. TR and TERT overexpressed SW39 (SW39 + TT) were used as a control. First, we determined that introduction of TR resulted in telomerase reactivation in SW39 ALT1 (Supplementary Figure S2A). Next, we compared the overhang signals. The overhang signals in SW39 ALT1 cells were much stronger compared to SW39+TT cells (Figure 3A and B). Thus, telomerase reactivation in SW39 ALT1 cells significantly reduced the overhang signals. The DSN method confirmed overhang length shortening by reactivation of telomerase. The excessively extended overhangs in SW39 ALT1 cells were reduced in cells reacquiring telomerase activity (Figure 3C). Moreover, the shortest telomere populations (<5 kb in TRF gels) in SW39 ALT1 cells were diminished upon telomerase reactivation (Supplementary Figure S2B). These results are consistent with the previous report that telomerase overexpression dramatically elongates the telomeres, rather than just maintaining the shorter telomere length (34). Lastly, we compared the overhang length after leading versus lagging separation. Telomerase reactivation (SW39 ALT1 + TT) completely diminished preferred lagging overhang elongation in SW39 ALT1 cells. The overhang intensity in leading and lagging strands in SW39 ALT1 + TT cells was almost identical to telomerase positive cells (Figure 3D and E). Collectively these results indicate that telomerase reactivation diminishes the ALT-mediated telomere extension processes.
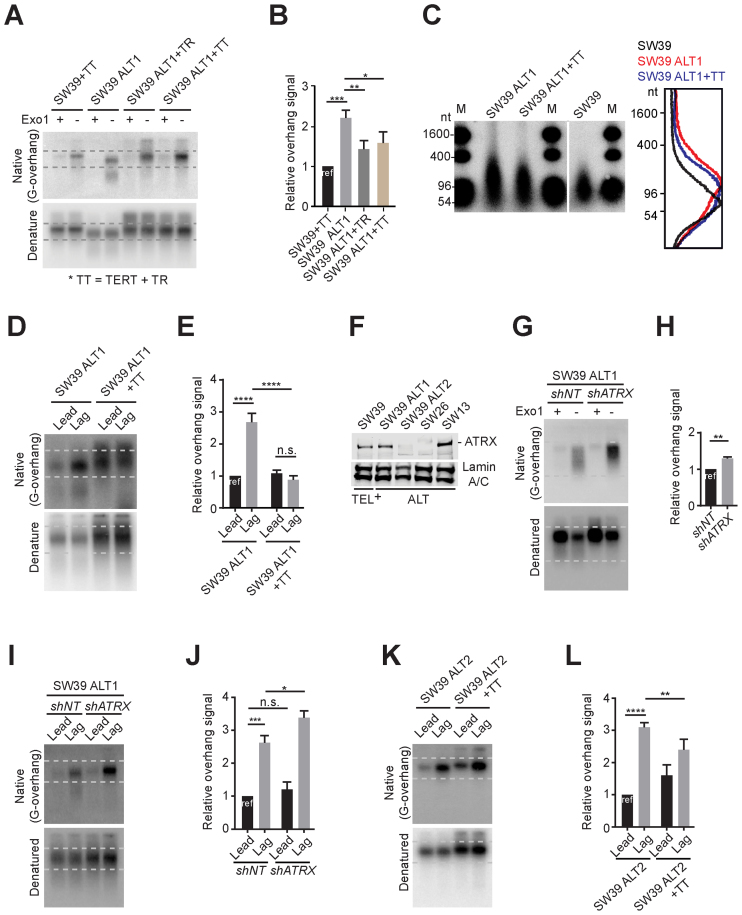
Figure 3.
Differential effects of telomerase reactivation on ATRX expression. (A) In-gel hybridization of telomeres in SW39 and SW39 ALT1 cells after introducing TERC or TERT gene expressing retrovirus. Cells were analyzed at 30 days post-infection. (B) Quantification of G-overhang intensity in A. Relative ratio of native signal to denatured signal was calculated (mean ± SD; n = 3). (C) DSN overhang assay in SW39 ALT1, SW39 ALT1 expressing TERC and TERT, and SW39 cells. Digested DNA (10 μg each) was run on 1% alkaline agarose gels (M: marker). Right: Lines traces of SW39 (black), SW39 ALT1 (red) and SW39 ALT1 + TT (blue). (D) In-gel hybridization of leading and lagging strand telomeres in SW39 ALT1 and SW39 ALT1 expressing TERC and TERT cells. (E) Quantification of G-overhang intensity in D. Relative ratio of native signal to denatured signal was calculated (mean ± SD; n = 3). (F) Western-blot for ATRX level from SW39, SW39 ALT1, SW39 ALT2, SW26 and SW13 cell lines. SW39 are telomerase positive and SW39 ALT1, SW39 ALT2, SW26 and SW13 are ALT cells. Lamin A/C was used as a loading control. (G) In-gel hybridization of telomeres in SW39 ALT1 after ATRX shRNA infection. Cells were analyzed 30 days post-infection. (H) Quantification of G-overhang intensity in G. Relative ratio of native signal to denatured signal was calculated (mean ± SD; n = 3). (I) In-gel hybridization of leading and lagging strand telomeres in SW39 ALT1 cells after ATRX shRNA infection. Cells were analyzed 60 days post-infection. (J) Quantification of G-overhang intensity in I. Relative ratio of native signal to denatured signal in leading or lagging strands was calculated (mean ± SD; n = 3). (K) In-gel hybridization of leading and lagging strand telomeres in SW39 ALT2 and SW39 ALT2 expressing TERC and TERT cells. Cells were analyzed 30 days post-infection. (L) Quantification of G-overhang intensity in K. Relative ratio of native signal to denatured signal was calculated (mean ± SD; n = 4). * indicates P < 0.05, ** indicates P < 0.01, *** indicates P < 0.001, **** indicates P < 0.0001 and n.s. indicates non-significant (unpaired Student's t test).
Loss of ATRX facilitates lagging overhangs elongation
Loss of the ATRX gene and mutations in ATRX are hallmarks of ALT-immortalized cells (13–15). To test whether the alteration in ATRX expression correlates with ALT onset in SW39 ALT cells, we first measured the level of ATRX in SW39 ALT cells and SW39 (Figure 3F and Supplementary Figure S3A). As expected, SW39 cells expressed ATRX. The level of ATRX in SW39 ALT1 and SW39 ALT3 were similar to SW39, but ATRX levels in SW39 ALT2 were markedly reduced. We found that spontaneously derived ALT cell lines, SW26 and SW13 exhibited different ATRX expression levels; SW26 was ATRX-negative, while SW13 was ATRX-positive (Figure 3F). These results support the idea that loss of ATRX expression is insufficient to promote ALT (15,16). To directly test the effect of ATRX in ALT, we infected ATRX shRNA in ATRX-positive ALT cells (SW39 ALT1) and analyzed cells 60 days post-infection. We first performed the C-circle assay (Supplementary Figures S3B and C). Depletion of ATRX in SW39 ALT1 cells produced a 3-fold increase of C-circle levels indicating that ATRX depletion in ALT cells may promote ALT activity but is insufficient for engagement of ALT.
We next investigated the mechanism by which ATRX ablation promotes ALT phenotypes. We measured the overhang intensity in SW39 ALT1 after ATRX depletion. Overhang intensity was significantly (∼30%) increased in ATRX-depleted SW39 ALT1 cells (Figure 3G and H), whereas it was not altered in ATRX-depleted SW39 cells (Supplementary Figure S3D and E). Exonuclease I treatment completely eliminated the overhang signal in ATRX-depleted SW39 ALT1 (Figure 3G Exo1+ lanes). This suggests that the distinctly enhanced overhang signal does not originate from an increase of exposed internal single stranded telomeric DNA induced by ATRX depletion. We tested whether ATRX status specifically affected leading or lagging strand extension (Figure 3I and J). We measured the overhang intensity of leading and lagging strand in control and ATRX-depleted SW39 ALT1 cells. The lagging overhangs in SW39 ALT1 cells were further increased after ATRX-depletion despite having similar amounts of leading overhangs. Moreover, the effect of telomerase reactivation in ATRX-negative SW39 ALT2 cells was less dramatic compared to those in ATRX-positive SW39 ALT1 cells (Figure 3K and L). Collectively these data implicate that the ALT pathway can be mediated by preferential elongation of lagging overhangs that is partially suppressed by ATRX.
TERC KO in telomerase-positive cancer cells results in engagement of the ALT pathway
Next, we generated TERC KO in telomerase positive cancer cell lines to test whether the ALT pathway can be activated by telomerase inhibition in a variety of telomerase positive cancer cells. H1299 is a telomerase positive cancer cell line derived from a non-small cell lung cancer patient and after TERC KO H1299 cells divided ∼40 PDs before entering crisis (Figure 4A). We only obtained a single survival clone out of more than one hundred million cells in crisis resulting in a very low frequency of immortalization = 0.9 × 10−8 (Supplementary Figure S1B). Using another cancer cell line HT1080 (fibrosarcoma; mesenchymal origin) we could not obtain any ALT survival clones even though 100 million cells were maintained in crisis (Supplementary Figure S4). Since HT1080 cells have intact p53, HT1080 TERC KO cells entered into a senescence-like stage in late PDs which is one explanation of why we did not obtain any long-term survival clones (Supplementary Figure S4B). Using the HAP1 (derived from chronic myelogenous leukemia) cell line again no ALT survival clones were obtained. Survival clone (H1299 ALT) had no detectable telomerase activity but possessed C-circles (Figure 4B), suggesting that H1299 ALT cells acquired a telomerase-independent telomere maintenance mechanism. Indeed, H1299 ALT cells exhibited elongated and heterogeneous telomere lengths compared to parental cells (H1299 and H1299 TERC KO) (Figure 4C).
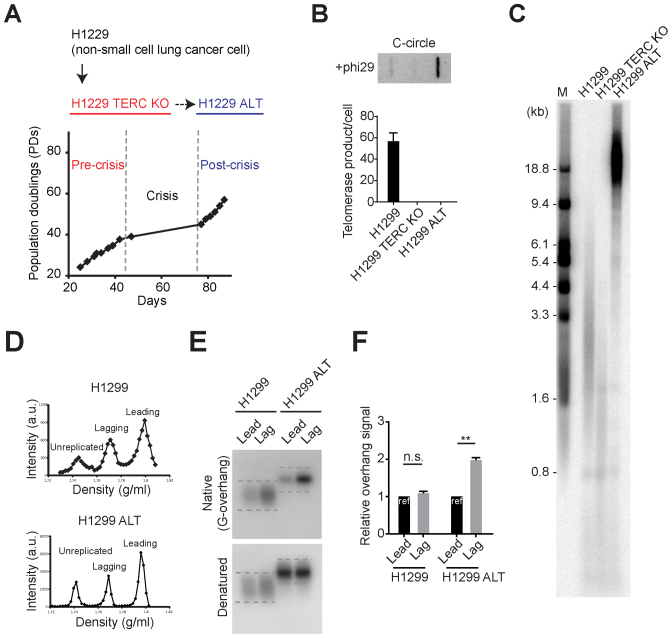
Figure 4.
TERC KO in H1299 cells results in engaging ALT pathway. (A) Overview of ALT cell line generation from telomerase positive cancer cells (H1299). H1299 TERC KO cells were generated by introducing with Cas9/TERC targeting guide RNA. H1299 TERC KO cells divided 40 population doublings before entering crisis. Single survival clone (H1299 ALT) was acquired. (B) DdTRAP analysis (bottom) and C-circle assay (upper) in H1299, H1299 TERC KO and H1299 ALT cells. (C) TRF analysis in H1299, H1299 TERC KO (pre-crisis, PD = 37) and H1299 ALT (post-crisis) cells. Digested DNA was run on 0.8% agarose gels. M indicates for the marker. (D) CsCl separation of leading and lagging strands in H1299 and H1299 ALT cells. a.u. represents arbitrary unit. (E) In-gel hybridization of leading and lagging strand telomeres in H1299 and H1299 ALT cells. (F) Quantification of G-overhang intensity in E. Relative ratio of native signal to denatured signal in leading or lagging strands was calculated (mean ± SEM; n = 2) ** indicates P < 0.01 and n.s. indicates non-significant (unpaired Student's t test).
We compared the overhang composition in H1299 and H1299 ALT cells after leading versus lagging separation (Figure 4D). Consistent with SW39 ALT cells, H1299 ALT cells possess longer lagging overhangs, whereas H1299 telomerase positive cells possess similar leading and lagging overhang composition (Figure 4E and F). Collectively, these results indicate that telomerase inhibition in human telomerase positive cancer cells can also result in engagement of ALT but at a very low frequency.
ALT cancer cells exhibit preferential elongation of lagging overhangs
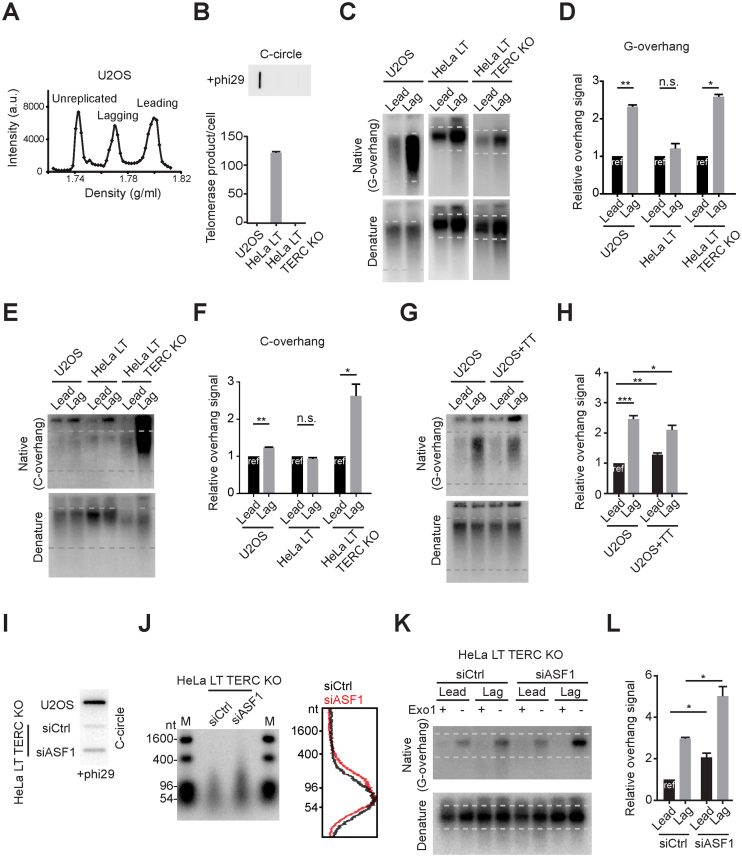
We tested whether the preferential elongation of lagging overhangs in SW39 ALT and H1299 ALT cells is also present in established ALT cancer cells. We analyzed the U2OS cell line, an ALT cancer cell line derived from an osteosarcoma patient. U2OS cells contain canonical telomeric repeat sequences (86.5% of telomeres sequences are TTAGGG), whereas telomere sequences in other ALT cells consist of variant repeats (35). This enabled us to separate leading and lagging strand using CsCl centrifugation in U2OS cells (Figure 5A). CsCl separation enabled us to collect leading and lagging strands that have been fully replicated (Supplementary Figure S5). The lagging overhang signal intensity in U2OS cells was longer than leading overhangs similar to SW39 ALT cells. To check the overhang ratio in cells without a telomere maintenance mechanism, we generated TERC KO cells derived from HeLa LT (with long telomeres in pre-crisis but without a telomere maintenance mechanism; Figure 5B). Interestingly, we found that the overhang ratio in HeLa LT TERC KO cells was 1:2.8 similar to normal BJ cells (23) (Figure 5C and D). We next measured the C-overhangs in this panel of cells. C-overhangs may be due to telomere trimming, a telomere-loop excision process resulting from over-elongated telomere length or telomere uncapping problems (36–38). Telomere trimming has been observed in ALT cells possibly induced by excessive telomere elongation (30,39). We found that C-overhangs in U2OS cells are more abundant in lagging strands, whereas HeLa LT cells have a similar ratio between leading and lagging strands (Figure 5E and F). Interestingly, HeLa LT TERC KO cells have excessive amounts of C-overhangs in lagging strands, possibly due to longer G-overhangs in lagging strands. These data can be interpreted to suggest that telomere trimming processes in ALT cells more frequently occur in lagging strands compared to leading strands, whereas in telomerase positive cells telomere trimming events occur similarly in leading and lagging strands.
Figure 5.
Lagging strand overhangs are preferentially elongated in U2OS and ASF1-depleted HeLa cells. (A) CsCl separation of leading and lagging strands in U2OS cells. a.u. represents arbitrary unit. (B) DdTRAP analysis (bottom) and C-circle assay (upper) in U2OS, HeLa LT and HeLa LT TERC KO cells. (C) In-gel hybridization of leading and lagging strand G-overhangs in U2OS, HeLa LT and HeLa LT TERC KO cells. (D) Quantification of G-overhang intensity in C. Relative ratio of native signal to denatured signal in leading or lagging strands was calculated (mean ± SD; n = 3). (E) In-gel hybridization of leading and lagging strand C-overhangs in U2OS, HeLa LT and HeLa LT TERC KO cells. (F) Quantification of C-overhang intensity in E. Relative ratio of native signal to denatured signal in leading or lagging strands was calculated (mean ± SEM; n = 2). (G) In-gel hybridization of leading and lagging strand telomeres in U2OS and U2OS expressing TERC and TERT cells. (H) Quantification of G-overhang intensity in G. Relative ratio of native signal to denatured signal in leading or lagging strands was calculated (mean ± SD; n = 4). (I) C-circle assay in U2OS and HeLa LT TERC KO cells after control or ASF1 siRNA (siCtrl, siASF1) transfection. Cells were analyzed 3 days post-transfection. (J) DSN overhang assay in HeLa LT TERC KO cells after control or ASF1 siRNA transfection. Right: Lines traces of control siRNA (black) and ASF1 siRNA (red). (K) In-gel hybridization of leading and lagging strand telomeres in HeLa LT TERC KO cells after control or ASF1 siRNA transfection. (L) Quantification of G-overhang intensity in K. Relative ratio of native signal to denatured signal in leading or lagging strands was calculated (mean ± SEM; n = 2). * indicates P < 0.05, ** indicates P < 0.01 and *** indicates P < 0.001 (unpaired Student's t test).
We found that telomerase reactivation significantly decreased the lagging overhang in SW39 ALT1, whereas lagging overhangs were partially reduced in SW39 ALT2 (Figure 3D, E, K and L). This is similar to IMRB+TT cells reported previously (40). We further showed that the effect of telomerase reactivation was related to ATRX status. Consistent with SW39 ALT2 cells, the lagging overhangs were partially reduced when the telomerase (TERT and TERC) genes were ectopically introduced back into ATRX-negative U2OS cells (Figure 5G and H) (15).
A recent study showed that depletion of the histone chaperone ASF1 induced ALT phenotypes mediated by replication stress and the ATR-dependent checkpoint machinery (32). This led us to test whether the ASF1 depletion-induced ALT pathway was mediated by preferential elongation of lagging overhangs. HeLa LT TERC KO cells were transfected with siRNAs targeting ASF1a and ASF1b genes. Consistent with the previous study, ASF1 depletion moderately increased C-circle levels in HeLa LT TERC KO cells (Figure 5I). The DSN method revealed that the overhang lengths were also increased in ASF1-depleted HeLa LT TERC KO cells (Figure 5J). Moreover, we found that ASF1 depletion elongated lagging overhangs more than leading overhangs in HeLa LT TERC KO cells (Figure 5K and L), consistent with the phenotype observed in ALT cells.
C-circles are more abundant in lagging strand telomeres compared to leading strand telomeres
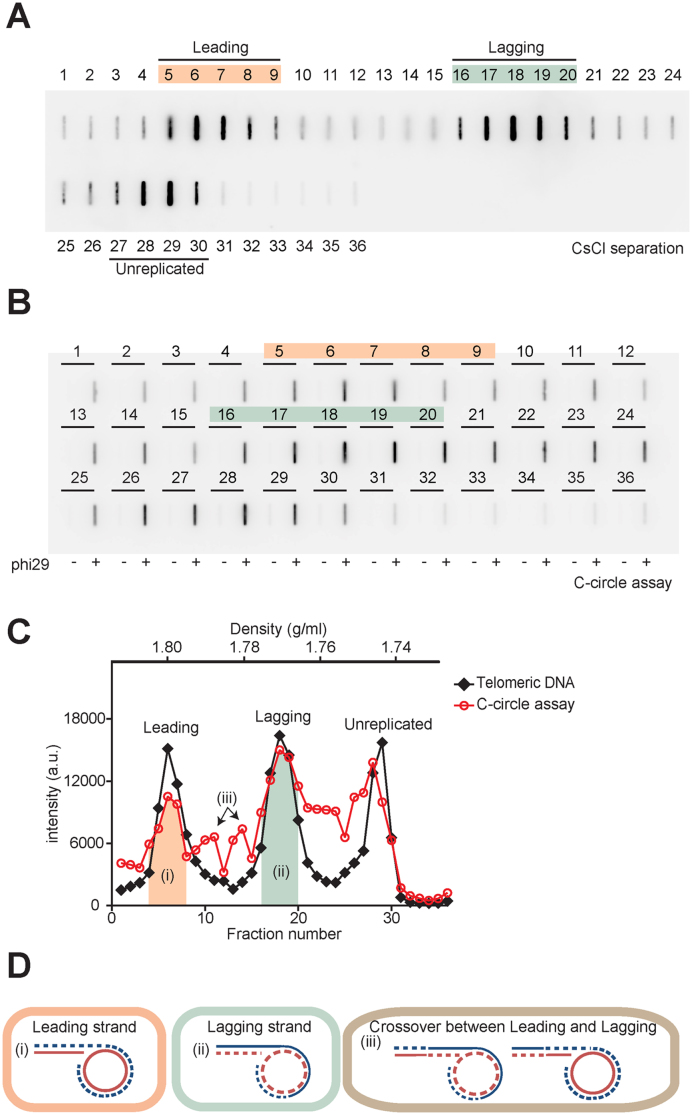
The excessive amounts of C-circles that exist in ALT cells may be important in ALT activity (15,21,32,41,42). To test whether the presence of C-circles has strand preference, we measured C-circle levels after CsCl separation. We performed a phi29 polymerase reaction for each fraction after CsCl centrifugation in U2OS cells (Figure 6A and B). C-circle levels peaked in leading and lagging fractions and mostly overlapped with the telomere DNA peaks as a singlet ((i) and (ii) in Figure 6C). When C-circle levels were quantified in leading strand versus lagging strand fractions, we found that C-circle levels from lagging strand fractions were more abundant compared to the leading strand fractions (leading:lagging = 1:1.63).
Figure 6.
Lagging strand telomeres have more C-circle levels compare to leading strand telomeres. (A) Slot-blot image for CsCl separation. U2OS cells were incubated with IdU 20 h, then separated leading and lagging strand using CsCl centrifugation. Each fraction was dialyzed, and loaded in slot-blots to represent the amount of telomeric DNA. (B) Slot-blot image for C-circle assay for each fraction after CsCl separation. Each fraction was subjected to the phi29 polymerase reaction to measure C-circle levels. (C) Quantification of Telomeric DNA-A and C-circle assay-B. Black Diamond: telomeric DNA, Red circle: C-circle assay. a.u. represents arbitrary unit. (D) Predicted model for the strand-dependent existence of C-circles. (i) C-circle level in the leading strand fraction, (ii) C-circle level in the lagging strand fraction, (iii) C-circle level in the strands after crossover between sister-chromatids. Dashed lines indicate nascent or daughter strands, Solid lines indicate parental strands, Blue lines indicate G-rich sequences and Red lines indicate C-rich sequences.
It has been proposed that C-circles mainly exist in the form of single stranded C-rich repeats with a sub-fraction existing as double stranded (21). If C-circles are a single stranded C-rich circular DNA without major G-rich sequences, it's density in IdU incorporated conditions should be two singlets; ∼1.800 g/ml (CCC-IdU-AA) derived from lagging strand and ∼1.750 g/ml (CCCTAA) derived from leading strand and unreplicated DNA. However, we observed that C-circles peaks mostly overlapped with total telomere DNA peaks in leading, lagging and unreplicated fractions. One possible explanation is that C-circles may present in chromatid fractions possibly annealed with G-overhangs ((i) and (ii) in Figure 6D). The doublet peak between the leading and lagging fraction was also observed and may represent C-circles in crossover strands ((iii) in Figure 6C and D). These results show that C-circles are more abundant in lagging strands which may be used in the telomere elongation mechanism, such as via rolling circle amplification (21).
Telomere elongation in ALT cells mostly occurs during S phase
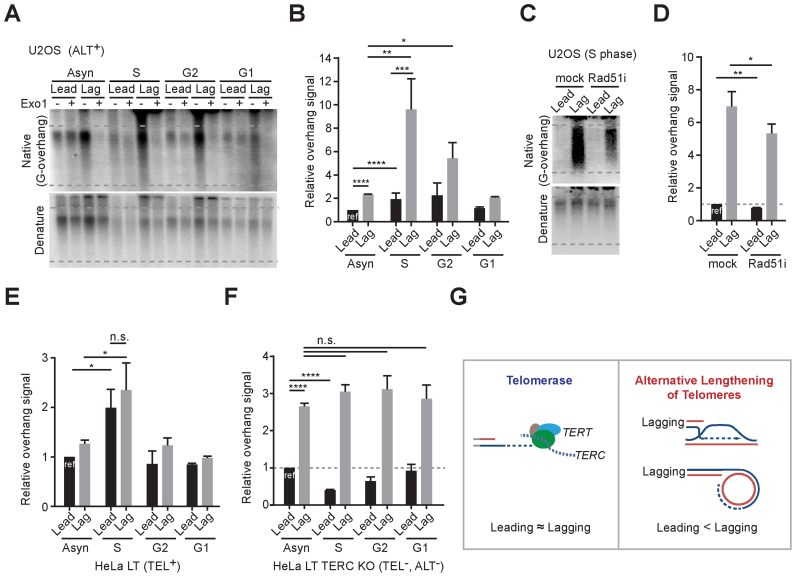
We next investigated the overhang length changes in a cell cycle dependent manner to identify which cell cycle phases are responsible for the ALT-mediated telomere elongation. Cells were synchronized at the G1/S phase by using double thymidine block, then released, and harvested at each cell cycle phase (S, G2 and G1 phase). The overhang intensity in U2OS cells peaked during S phase and gradually declined at the G2 phase (Figure 7A and B). The intensity of both leading and lagging overhangs peaked during S phase, but the increase of lagging overhang intensity was much higher than that of leading overhangs in U2OS cells (Figure 7A and B). These results demonstrate that ALT-mediated telomere elongation processes mostly occur during S phase and that lagging overhangs are preferentially and excessively elongated.
Figure 7.
ALT pathways occur during S phase. (A) In-gel hybridization of leading and lagging strand telomeres in U2OS cells for each cell cycle phases. (B) Quantification of G-overhang intensity in relative ratio of native signal to denatured signal in A (mean ± SD; n = 4). (C) In-gel hybridization of leading and lagging strand telomeres in U2OS cells for S phase with or without 30 μM of RAD51 inhibitor (RI-1) treatment. Cells were synchronized at G1/S phase by using double thymidine block and releases with or without RAD51 inhibitor, and then harvested at 6 hours post release. (D) Quantification of G-overhang intensity in relative ratio of native signal to denatured signal in C (mean ± SD; n = 3). (E) Quantification of G-overhang intensity in relative ratio of native signal to denatured signal in HeLa LT cells (Supplementary Figure S6A) (mean ± SDs; n = 3). (F) Quantification of G-overhang intensity in relative ratio of native signal to denatured signal in HeLa LT TERC KO cells (Supplementary Figure S6B) (mean ± SEM; n = 2). (G) Schematic models for telomere elongation processes in telomerase positive and ALT cells * indicates P < 0.05, ** indicates P < 0.01, *** indicates P < 0.001, **** indicates P < 0.0001 and n.s. indicates non-significant (unpaired Student's t test).
Previous studies showed that RAD51 (mitotic recombinase) depletion or inhibition results in reduction of G-overhangs (30) and interchromosomal telomeric recombination (43,44) in ALT cells. To check whether RAD51-mediated recombination processes affect overhang generation during S phase, we treated U2OS cells with a small molecule RAD51 inhibitor (45). The RAD51 inhibitor treatment reduced the overhang signals in both leading and lagging strands during S phase (Figure 7C and D). These results demonstrate that recombination processes mediate the ALT pathway.
Next, we asked whether the preferential elongation in lagging strands is an ALT-specific phenotype. We compared these observations with telomerase positive cells by analyzing overhang intensity in HeLa LT cells at each cell cycle phase. Telomerase positive HeLa LT cells exhibited increased overhang intensity in both leading and lagging strands at S phase that gradually declined at G2 phase. The overhang intensity were increased up to 2-fold during S phase in both leading and lagging strands (Figure 7E and Supplementary Figure S6A). These results are consistent with previous studies showing that telomerase extends telomere overhangs during S phase, and the CST-pol alpha complex fills in the C-strand during the G2 phase (28,46). In contrast to ALT cells, the extension rate was similar for both leading and lagging strands in telomerase positive cells (Figure 7E and Supplementary Figure S6A) (24). We also measured the cell cycle dependent overhang intensity changes in HeLa LT TERC KO cells to determine the overhang generation processes in cells without a telomere maintenance mechanism. HeLa LT TERC KO cells initially are both telomerase-negative and ALT-negative when derived from telomerase positive HeLa LT cells (Figure 5B). As shown in BJ fibroblasts (23), HeLa LT TERC KO cells exhibited the same lagging overhang intensity throughout the cell cycle, and that leading strand overhangs were processed during S phase (Figure 7F and Supplementary Figure S6B). Collectively these results show that ALT telomeres are preferentially elongated at the lagging overhangs during S phase (Figure 7G).
DISCUSSION
Telomerase inhibition can engage the alternative lengthening of telomeres pathway in telomerase positive human cells
The vast majority of human cancer cells maintain their telomere length via telomerase reactivation that permits cancer cells to proliferate indefinitely. Thus, targeting telomerase remains a highly attractive anti-cancer strategy. However, one concern of anti-telomerase therapy is the potential acquired resistance by engagement of ALT. In this study we provide, as far as we could determine, the first direct evidence of activation of ALT in telomerase-positive human cells by knocking out a key telomerase component using genome-editing techniques. The frequency of engaging the ALT mechanism was very low but similar to previous studies for spontaneous engagement of ALT (26).
While we provide evidence for the engagement of ALT in telomerase positive human cells it is important to indicate that we were unsuccessful in obtaining ALT survival clones from TERC KO in two other telomerase positive cancer cell lines; HT1080 and HAP1 cells (no survival clones were acquired from ∼ 1 × 108 cells) (Supplementary Figure S1B). This suggests that engagement of the ALT pathway may be different depending on the background of oncogenic changes within a cell and that some cancer cells may be actually dependent on telomerase for long-term survival. Further analysis recapturing these processes in various cancer cell lines and different genetic background will provide more information to understand the mechanisms for acquiring the ALT telomere maintenance mechanism.
Lagging overhang is preferentially elongated in ALT pathway
The ALT mechanism has been regarded as a recombination-based telomere elongation processes but detailed mechanisms remain to be clarified. It has been suggested that nuclear receptors are involved in these recombination processes (42,47,48), and can be mediated by the meiotic recombinase complex (43). Exposed ssDNA 3΄ telomere overhangs can invade into the sister chromatid or extra-chromosomal telomeric DNA repeats (e.g. C-circles) that can be used as templates, and can then elongate their ends using the DNA replication machinery resulting in net elongation in telomere length. Here, we demonstrated that lagging overhangs are more prone to be involved in telomere elongation processes in ALT pathway. We infer that differences in initial overhang length between leading and lagging strands affect the recombination processes at the telomere, such as filament formation by recombinase, homology search and strand invasion, which rely on its exposed 3΄ end overhang length, leading to more strand invasion via lagging overhangs (Supplementary Figure S7).
We also showed that leading strand overhangs in HeLa LT TERC KO cells (without a telomere maintenance mechanism) are processed during S phase (Figure 7F), which is possibly due to Apollo-mediated C-strand resection (49). We previously showed that C-strand resection processes are rapidly completed in BJ cells (within 1 h after DNA replication completion (23)), but it is delayed in HeLa LT TERC KO cells possibly due to longer telomere length in HeLa LT TERC KO cells (∼18 kb) which may lead to delay for the Apollo recruitment by TRF2 (50). Thus, initial leading strand overhang length in long telomere containing conditions, such as ALT, may be much shorter and may lead to different recombination rates in leading versus lagging overhangs.
Finally, telomeres are fragile sites (51) and especially the lagging strand is more prone to encounter problems during replication processes since it possesses G-rich sequences as a parental strand (52–54). Therefore, these replication problems may also initiate break-induced replication (BIR)-mediated DNA replication processes at the telomere region in ALT cells (55), leading to a preference for lagging strands. We also showed that ATRX depletion partially contributes to preferential elongation in lagging strands (Figure 3). It can be due to loss of its roles in guarding the DNA replication processes upon replication stress or G-quadruplex structure formation (56,57).
In conclusion, our study provides an explanation why most ALT cancer cells contain heterogeneous (short and extremely long telomere lengths) (Supplementary Figure S7). ALT engagement and maintenance is mediated by preferential telomere elongation in lagging strand overhangs.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institute on Aging (NIA) [AG01228 to W.E.W. and J.W.S.]; CPRIT Postdoctoral Cancer Intervention and Prevention Discovery Training Program [RP140110 to J.M.]; Harold Simmons NCI Designated Comprehensive Cancer Center Support Grant [CA142543]; Southland Financial Corporation Distinguished Chair in Geriatric Research (to J.W.S. and W.E.W.); NIH [C06 RR30414]. Funding for open access charge: NIH [C06 RR30414]; NIA [AG01228].
Conflict of interest statement. None declared.
REFERENCES
- 1. Greider C.W., Blackburn E.H.. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985; 43:405–413. [DOI] [PubMed] [Google Scholar]
- 2. Kim N.W., Piatyszek M.A., Prowse K.R., Harley C.B., West M.D., Ho P.L., Coviello G.M., Wright W.E., Weinrich S.L., Shay J.W.. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994; 266:2011–2015. [DOI] [PubMed] [Google Scholar]
- 3. Shay J.W., Bacchetti S.. A survey of telomerase activity in human cancer. Eur. J. Cancer. 1997; 33:787–791. [DOI] [PubMed] [Google Scholar]
- 4. Ouellette M.M., Wright W.E., Shay J.W.. Targeting telomerase-expressing cancer cells. J. Cell. Mol. Med. 2011; 15:1433–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shay J.W., Reddel R.R., Wright W.E.. Cancer. Cancer and telomeres–an ALTernative to telomerase. Science. 2012; 336:1388–1390. [DOI] [PubMed] [Google Scholar]
- 6. Bechter O.E., Zou Y., Walker W., Wright W.E., Shay J.W.. Telomeric recombination in mismatch repair deficient human colon cancer cells after telomerase inhibition. Cancer Res. 2004; 64:3444–3451. [DOI] [PubMed] [Google Scholar]
- 7. Hu J., Hwang S.S., Liesa M., Gan B., Sahin E., Jaskelioff M., Ding Z., Ying H., Boutin A.T., Zhang H. et al. . Antitelomerase therapy provokes ALT and mitochondrial adaptive mechanisms in cancer. Cell. 2012; 148:651–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bryan T.M., Englezou A., Dalla-Pozza L., Dunham M.A., Reddel R.R.. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat. Med. 1997; 3:1271–1274. [DOI] [PubMed] [Google Scholar]
- 9. Lundblad V., Blackburn E.H.. An alternative pathway for yeast telomere maintenance rescues est1- senescence. Cell. 1993; 73:347–360. [DOI] [PubMed] [Google Scholar]
- 10. Cheng C., Shtessel L., Brady M.M., Ahmed S.. Caenorhabditis elegans POT-2 telomere protein represses a mode of alternative lengthening of telomeres with normal telomere lengths. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:7805–7810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Seo B., Kim C., Hills M., Sung S., Kim H., Kim E., Lim D.S., Oh H.S., Choi R.M., Chun J. et al. . Telomere maintenance through recruitment of internal genomic regions. Nat. Commun. 2015; 6:8189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chang S., Khoo C.M., Naylor M.L., Maser R.S., DePinho R.A.. Telomere-based crisis: functional differences between telomerase activation and ALT in tumor progression. Genes Dev. 2003; 17:88–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schwartzentruber J., Korshunov A., Liu X.Y., Jones D.T., Pfaff E., Jacob K., Sturm D., Fontebasso A.M., Quang D.A., Tonjes M. et al. . Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012; 482:226–231. [DOI] [PubMed] [Google Scholar]
- 14. Heaphy C.M., de Wilde R.F., Jiao Y., Klein A.P., Edil B.H., Shi C., Bettegowda C., Rodriguez F.J., Eberhart C.G., Hebbar S. et al. . Altered telomeres in tumors with ATRX and DAXX mutations. Science. 2011; 333:425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lovejoy C.A., Li W., Reisenweber S., Thongthip S., Bruno J., de Lange T., De S., Petrini J.H., Sung P.A., Jasin M. et al. . Loss of ATRX, genome instability, and an altered DNA damage response are hallmarks of the alternative lengthening of telomeres pathway. PLoS Genet. 2012; 8:e1002772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Napier C.E., Huschtscha L.I., Harvey A., Bower K., Noble J.R., Hendrickson E.A., Reddel R.R.. ATRX represses alternative lengthening of telomeres. Oncotarget. 2015; 6:16543–16558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Clynes D., Jelinska C., Xella B., Ayyub H., Scott C., Mitson M., Taylor S., Higgs D.R., Gibbons R.J.. Suppression of the alternative lengthening of telomere pathway by the chromatin remodelling factor ATRX. Nat. Commun. 2015; 6:7538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F.. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013; 8:2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wong J.M., Collins K.. Telomerase RNA level limits telomere maintenance in X-linked dyskeratosis congenita. Genes Dev. 2006; 20:2848–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Herbert B.S., Shay J.W., Wright W.E.. Analysis of telomeres and telomerase. Curr. Protoc. Cell Biol. 2003; doi:10.1002/0471143030.cb1806s20. [DOI] [PubMed] [Google Scholar]
- 21. Henson J.D., Cao Y., Huschtscha L.I., Chang A.C., Au A.Y., Pickett H.A., Reddel R.R.. DNA C-circles are specific and quantifiable markers of alternative-lengthening-of-telomeres activity. Nat. Biotechnol. 2009; 27:1181–1185. [DOI] [PubMed] [Google Scholar]
- 22. Zhao Y., Hoshiyama H., Shay J.W., Wright W.E.. Quantitative telomeric overhang determination using a double-strand specific nuclease. Nucleic Acids Res. 2008; 36:e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chow T.T., Zhao Y., Mak S.S., Shay J.W., Wright W.E.. Early and late steps in telomere overhang processing in normal human cells: the position of the final RNA primer drives telomere shortening. Genes Dev. 2012; 26:1167–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chai W., Du Q., Shay J.W., Wright W.E.. Human telomeres have different overhang sizes at leading versus lagging strands. Mol. Cell. 2006; 21:427–435. [DOI] [PubMed] [Google Scholar]
- 25. Ludlow A.T., Robin J.D., Sayed M., Litterst C.M., Shelton D.N., Shay J.W., Wright W.E.. Quantitative telomerase enzyme activity determination using droplet digital PCR with single cell resolution. Nucleic Acids Res. 2014; 42:e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shay J.W., Wright W.E.. Quantitation of the frequency of immortalization of normal human diploid fibroblasts by SV40 large T-antigen. Exp. Cell Res. 1989; 184:109–118. [DOI] [PubMed] [Google Scholar]
- 27. Arora R., Lee Y., Wischnewski H., Brun C.M., Schwarz T., Azzalin C.M.. RNaseH1 regulates TERRA-telomeric DNA hybrids and telomere maintenance in ALT tumour cells. Nat. Commun. 2014; 5:5220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhao Y., Sfeir A.J., Zou Y., Buseman C.M., Chow T.T., Shay J.W., Wright W.E.. Telomere extension occurs at most chromosome ends and is uncoupled from fill-in in human cancer cells. Cell. 2009; 138:463–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cesare A.J., Reddel R.R.. Alternative lengthening of telomeres: models, mechanisms and implications. Nat. Rev. Genet. 2010; 11:319–330. [DOI] [PubMed] [Google Scholar]
- 30. Oganesian L., Karlseder J.. Mammalian 5΄ C-rich telomeric overhangs are a mark of recombination-dependent telomere maintenance. Mol. Cell. 2011; 42:224–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhao Y., Shay J.W., Wright W.E.. Telomere G-overhang length measurement method 1: the DSN method. Methods Mol. Biol. 2011; 735:47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. O'Sullivan R.J., Arnoult N., Lackner D.H., Oganesian L., Haggblom C., Corpet A., Almouzni G., Karlseder J.. Rapid induction of alternative lengthening of telomeres by depletion of the histone chaperone ASF1. Nat. Struct. Mol. Biol. 2014; 21:167–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Draskovic I., Londono Vallejo A.. Telomere recombination and alternative telomere lengthening mechanisms. Front. Biosci. (Landmark Ed.). 2013; 18:1–20. [DOI] [PubMed] [Google Scholar]
- 34. Cristofari G., Lingner J.. Telomere length homeostasis requires that telomerase levels are limiting. EMBO J. 2006; 25:565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lee M., Hills M., Conomos D., Stutz M.D., Dagg R.A., Lau L.M., Reddel R.R., Pickett H.A.. Telomere extension by telomerase and ALT generates variant repeats by mechanistically distinct processes. Nucleic Acids Res. 2014; 42:1733–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pickett H.A., Cesare A.J., Johnston R.L., Neumann A.A., Reddel R.R.. Control of telomere length by a trimming mechanism that involves generation of t-circles. EMBO J. 2009; 28:799–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pickett H.A., Henson J.D., Au A.Y., Neumann A.A., Reddel R.R.. Normal mammalian cells negatively regulate telomere length by telomere trimming. Hum. Mol. Genet. 2011; 20:4684–4692. [DOI] [PubMed] [Google Scholar]
- 38. Oganesian L., Karlseder J.. 5΄ C-rich telomeric overhangs are an outcome of rapid telomere truncation events. DNA Repair (Amst.). 2013; 12:238–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wan B., Yin J., Horvath K., Sarkar J., Chen Y., Wu J., Wan K., Lu J., Gu P., Yu E.Y. et al. . SLX4 assembles a telomere maintenance toolkit by bridging multiple endonucleases with telomeres. Cell Rep. 2013; 4:861–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Episkopou H., Draskovic I., Van Beneden A., Tilman G., Mattiussi M., Gobin M., Arnoult N., Londono-Vallejo A., Decottignies A.. Alternative Lengthening of Telomeres is characterized by reduced compaction of telomeric chromatin. Nucleic Acids Res. 2014; 42:4391–4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cox K.E., Marechal A., Flynn R.L.. SMARCAL1 resolves replication stress at ALT telomeres. Cell Rep. 2016; 14:1032–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Conomos D., Reddel R.R., Pickett H.A.. NuRD-ZNF827 recruitment to telomeres creates a molecular scaffold for homologous recombination. Nat. Struct. Mol. Biol. 2014; 21:760–770. [DOI] [PubMed] [Google Scholar]
- 43. Cho N.W., Dilley R.L., Lampson M.A., Greenberg R.A.. Interchromosomal homology searches drive directional ALT telomere movement and synapsis. Cell. 2014; 159:108–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ramamoorthy M., Smith S.. Loss of ATRX suppresses resolution of telomere cohesion to control recombination in ALT cancer cells. Cancer Cell. 2015; 28:357–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Budke B., Logan H.L., Kalin J.H., Zelivianskaia A.S., Cameron McGuire W., Miller L.L., Stark J.M., Kozikowski A.P., Bishop D.K., Connell P.P.. RI-1: a chemical inhibitor of RAD51 that disrupts homologous recombination in human cells. Nucleic Acids Res. 2012; 40:7347–7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang F., Stewart J.A., Kasbek C., Zhao Y., Wright W.E., Price C.M.. Human CST has independent functions during telomere duplex replication and C-strand fill-in. Cell Rep. 2012; 2:1096–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Conomos D., Stutz M.D., Hills M., Neumann A.A., Bryan T.M., Reddel R.R., Pickett H.A.. Variant repeats are interspersed throughout the telomeres and recruit nuclear receptors in ALT cells. J. Cell Biol. 2012; 199:893–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Marzec P., Armenise C., Perot G., Roumelioti F.M., Basyuk E., Gagos S., Chibon F., Dejardin J.. Nuclear-receptor-mediated telomere insertion leads to genome instability in ALT cancers. Cell. 2015; 160:913–927. [DOI] [PubMed] [Google Scholar]
- 49. Wu P., Takai H., de Lange T.. Telomeric 3΄ overhangs derive from resection by Exo1 and Apollo and fill-in by POT1b-associated CST. Cell. 2012; 150:39–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wu P., van Overbeek M., Rooney S., de Lange T.. Apollo contributes to G overhang maintenance and protects leading-end telomeres. Mol. Cell. 2010; 39:606–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sfeir A., Kosiyatrakul S.T., Hockemeyer D., MacRae S.L., Karlseder J., Schildkraut C.L., de Lange T.. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009; 138:90–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Min J., Choi E.S., Hwang K., Kim J., Sampath S., Venkitaraman A.R., Lee H.. The breast cancer susceptibility gene BRCA2 is required for the maintenance of telomere homeostasis. J. Biol. Chem. 2012; 287:5091–5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zimmermann M., Kibe T., Kabir S., de Lange T.. TRF1 negotiates TTAGGG repeat-associated replication problems by recruiting the BLM helicase and the TPP1/POT1 repressor of ATR signaling. Genes Dev. 2014; 28:2477–2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Arnoult N., Saintome C., Ourliac-Garnier I., Riou J.F., Londono-Vallejo A.. Human POT1 is required for efficient telomere C-rich strand replication in the absence of WRN. Genes Dev. 2009; 23:2915–2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dilley R.L., Verma P., Cho N.W., Winters H.D., Wondisford A.R., Greenberg R.A.. Break-induced telomere synthesis underlies alternative telomere maintenance. Nature. 2016; 539:54–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Law M.J., Lower K.M., Voon H.P., Hughes J.R., Garrick D., Viprakasit V., Mitson M., De Gobbi M., Marra M., Morris A. et al. . ATR-X syndrome protein targets tandem repeats and influences allele-specific expression in a size-dependent manner. Cell. 2010; 143:367–378. [DOI] [PubMed] [Google Scholar]
- 57. Leung J.W., Ghosal G., Wang W., Shen X., Wang J., Li L., Chen J.. Alpha thalassemia/mental retardation syndrome X-linked gene product ATRX is required for proper replication restart and cellular resistance to replication stress. J. Biol. Chem. 2013; 288:6342–6350. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.