Abstract
RNA G-quadruplexes (G4s) are formed by G-rich RNA sequences in protein-coding (mRNA) and non-coding (ncRNA) transcripts that fold into a four-stranded conformation. Experimental studies and bioinformatic predictions support the view that these structures are involved in different cellular functions associated to both DNA processes (telomere elongation, recombination and transcription) and RNA post-transcriptional mechanisms (including pre-mRNA processing, mRNA turnover, targeting and translation). An increasing number of different diseases have been associated with the inappropriate regulation of RNA G4s exemplifying the potential importance of these structures on human health. Here, we review the different molecular mechanisms underlying the link between RNA G4s and human diseases by proposing several overlapping models of deregulation emerging from recent research, including (i) sequestration of RNA-binding proteins, (ii) aberrant expression or localization of RNA G4-binding proteins, (iii) repeat associated non-AUG (RAN) translation, (iv) mRNA translational blockade and (v) disabling of protein–RNA G4 complexes. This review also provides a comprehensive survey of the functional RNA G4 and their mechanisms of action. Finally, we highlight future directions for research aimed at improving our understanding on RNA G4-mediated regulatory mechanisms linked to diseases.
G-quadruplexes (G4s) formed by G-rich DNA or RNA sequences are non-canonical structures organized in stacks of tetrads or G-quartets, in which four guanines are assembled in a planar arrangement by Hoogsteen hydrogen bonding. Bioinformatic predictions using a specific search algorithm (Gx-N1-7-Gx-N1-7-Gx-N1-7-Gx, where x ≥ 3 and N = A, U, G or C (1,2)) indicated the presence of 376 000 DNA G4 forming sequences in the human genome that are specifically enriched in telomeres, gene promoters, ribosomal DNA and recombination hotspots. G4s are also frequently located at the very 5΄ end of the first intron (3), and both the 5΄ and 3΄ untranslated regions (UTRs) (4) of mRNAs indicating an important role in mRNA synthesis, expression and function.
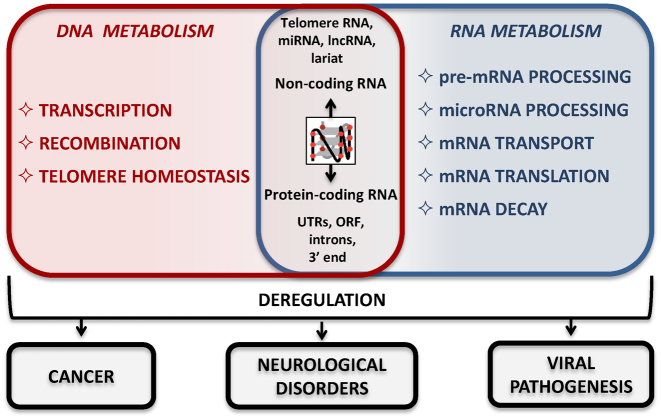
Over the last few years, increasing evidence has emerged supporting the view that RNA G4s are important regulators of key cellular functions (recently reviewed in (5–7)), including telomere homeostasis and gene expression mechanisms (see Figure 1 for an overview of RNA G4 localization, function and impact on disease). Cis-acting G4s associated to mRNAs are now widely accepted as critical regulators of pre-mRNA processing (splicing and polyadenylation), mRNA turnover, mRNA targeting and translation. Consistent with the role of G4s in post-transcriptional events, genome-wide sequencing of DNA G4s in the human genome revealed a high density of G4s in the 5΄ UTRs and splicing sites (8). Recently, G4 motifs have been mapped to non-coding RNAs (ncRNAs), such as long non-coding RNAs (lncRNAs) (9) and precursor microRNAs (pre-miRNAs) (10), indicating the potential of RNA G4s to regulate post-transcriptional gene expression in trans and to control miRNA biogenesis. In addition to post-transcriptional mechanisms, it has been suggested that G4s in ncRNAs affect DNA processes, as demonstrated for G4s at telomeric repeat-containing RNA (TERRA) molecules (11) regulating telomere elongation (12) or transcription (13) and those found in intron lariats which regulate immunoglobulin class switch recombination (14). RNA G4s regulate gene expression not only quantitatively but also qualitatively. Indeed, these structures have been shown to impact processes that change the coding capacity of the genome, such as alternative polyadenylation (15), alternative splicing (16,17) and translational recoding (18). Several independent lines of evidence support the importance of RNA G4s and their biological function both in vitro and in vivo (recently reviewed in (5–7)) (Table 1), including: (i) bioinformatic predictions (1,2,19), (ii) structural analysis (e.g. using nuclear magnetic resonance, crystallography (20) or circular dichroism (21)), (iii) in vitro determination of G4 formation (e.g. using in-line probing (22)), (iv) in cellulo visualization (e.g. using a conformation-specific antibody (23) or a fluorogenic dye (24)), (v) functional in vitro /in cellulo analysis of endogenous (e.g. see (25–27)) or reporter (e.g. see (28–30)) gene expression upon G4 ligand addition. Importantly, widespread formation of RNA G4s in vitro (31,32) has been recently demonstrated by combining reverse transcriptase stalling with next-generation sequencing.
Figure 1.
Overview of RNA G4s: position, proposed function and link with disease.
Table 1. Tools for predicting and analyzing G4 formation.
| Evidence for the occurrence of G-quadruplexes |
|---|
| Bioinformatic prediction and databases |
| Several tools predicting quadruplex-forming propensity are based on the fact that runs of Gs are a requirement for G4 formation. A simple, regular motif (G3+N1–7G3+N1–7G3+N1–7G3+) based on DNA G4 folding studies was originally proposed to describe G4-forming sequences (1,2). Several tools are based on this algorithm, including, QuadDB (128), QGRS mapper (129), Quadfinder (130), QuadBase (131), Greglist (132). More recent algorithms and scoring systems take into account both the neighboring sequences (including G4 Hunter (19), cG/cC score (133)) and the observation that G4s are highly polymorphic in vivo (3,134). Tools. QuadDB (1,128), defines G4 folding rules based on strand stoichiometry, number of tetrads, discontinuities in G-tracts, loop length and composition. Quadfinder (130), prediction of G4 sequences with the flexibility of defining variants of the motif. QGRS mapper (129), prediction of G4 sequences with different parameter settings and possibility to look at sequences with few Gs and long loops; includes a scoring parameter. G4P Calculator (134), orthogonal approach focused on the density of sequences likely to lead to G4s. GRSDB2 and GRS_UTRdb (135,136), list G4s in pre-mRNAs and UTRs. QuadBase (131), ortholog analysis for finding conserved G4s. Greglist (132), list of genes that contain G4 motifs in promoters. G4RNA (137), data retrieval on experimentally tested sequences. G4Hunter (19) takes into account G-richness and G-skewness of a given sequence; provides a G4 propensity score. |
| G4 structure |
| Topologies of G4s depends on the glycosidic conformation (syn or anti), the number of molecules of the nucleic acid involved in their formation (intramolecular, bimolecular or tetramolecular) and the relative orientation of the strands (parallel, antiparallel or mixed). G4 formation depends on several parameters: the number of stacking G-quartets, the length and the nucleotide sequence of the loops, the occurrence of bulges within G-tracks, cation availability and concentration, the presence of consecutive cytosine residues in the surrounding sequence. RNA G4s are more thermodynamically stable, compact and less hydrated than DNA G4s. The 2΄-OH group in the ribose exerts conformational constraints on RNA G4s resulting in more intramolecular interactions, anti conformation and parallel topology. Biophysical techniques: Ultraviolet spectroscopy, Circular dichroism, UV melting, NMR spectroscopy, Crystallography. Drawback: the length of the G4 sequence required for these techniques that does not reflect the in cellulo/in vivo global context. |
| In vitro determination |
| The capability of putative quadruplex sequences to fold into G4 could be assessed experimentally with techniques that use the characteristic of G4s to be stabilized by the presence of a cation (K+>Na+>Li+), and to be modified by G4 small-molecule ligands and trans-acting factors. G4 formation is supported by studies on candidate RNA sequences and, more recently, by transcriptome-wide analysis in vitro (31,32) and in vivo (32). G4 RNA candidate approach: Polyacrylamide gel electrophoresis, reverse transcription pausing assay, DMS (dimethyl sulfate)/footprinting analysis, in-line probing (22), the nucleotide resolutive approach SHALiPE (selective 2΄-hydroxyl acylation with lithium ion-based primer extension) (138), and FOLDeR, a method using 7-deaza-G RNA modifications in combination with secondary structure probing allowing to demonstrate the presence of G4s in long RNAs (139). Transcriptome-wide approaches: RNA G4 (rG4) profiling method that couples rG4-mediated reverse transcriptase pausing with sequencing and generates a global in vitro G4 map (31). More recently, widespread formation of RNA G4s in vitro and in vivo was inferred using DMS treatment before profiling of reverse-transcriptase stops (32). |
| G4 small-molecule ligands |
| Several ligands have been shown to be specific for DNA G4s over other types of DNA structures, including porphyrin, acridine, pentacridium, quinacridine, telomestatin, naphtalene diiamide, bisquinolium derivates. Some of these ligands have been shown to also bind RNA G4s with high affinity and specificity. To date, only two molecules have been demonstrated to exhibit selectivity towards RNA G4s over DNA G4s. RNA/DNA G4 binders: i) TMPyP4 exhibits low affinity for both DNA and RNA G4s, poor selectivity for G4s versus duplex DNA and has opposite effects on RNA G4 formation (50,81,83), ii) Bisquinolium derivates (including Phen-DC3, Phen-DC6, 360A or RR82/R110) are potent binders of DNA and RNA G4s and modulate RNA G4-depedent gene expression (16,25,28,52,82). RNA G4 binders: i) CarboxyPDS (carboxy pyridostatin) triggers selectively RNA G4s within a cellular context (23,140), ii) RGB1, a polyaromatic molecule that binds selectively to RNA G4 structures as compared to DNA G4s or other RNA structures (141). |
| In cellulo probing |
| Facing an urgent need for efficient RNA G4 detection in cellulo, molecular probes that can specifically recognize RNA G4 structures in a simple and reliable way have been recently developed. Structure-specific antibody: BG4 binds both DNA and RNA G4s (23,64). Drawback of immunodetection: fixation and permeabilization of the cells could modify G4 formation; no information on the specific sequences involved in G4 folding. RNA G4 fluorescent probes: CyT (fluorogenic cyanine dye) (24), N-TASQ (naphthoTASQ) (142,143); PyroTASQ (pyrene template-assembled synthetic G-quartet) (144); GTFH (G4-triggered fluorogenic hybridization proble), hybrid probe containing a fluorescent light-up moiety specific to a G4 and an oligonucleotide that hybridize with the specific RNA sequence (145); ThT (thioflavin fluorogenic dye) (146). Advantages: direct detection of RNA G4s in untreated cells (no fixation and no permeabilization). |
The mechanisms underlying the function of RNA G4s in the cell involve in most cases the binding of protein factors (i.e. RNA-binding proteins or RBPs) that modulate G4 conformation and/or serve as a bridge to recruit additional protein regulators (reviewed in (33)) (Figure 2A). Importantly, recent in vivo RNA G4 mapping suggested that RBPs lie at the center of the mechanism that unfolds most eukaryotic RNA G4s (32). In some cases, the biological function of RNA G4s bound to RBPs can involve the interaction with DNA (Figure 2B) to regulate DNA-related processes such as recombination (e.g. (14)) or telomere elongation (e.g. (12,34)). Other examples suggest that the interplay between RNA and DNA can involve the formation of hybrid G4 structures that recruit G-tracts from both the DNA and RNA molecules (Figure 2C) to modulate transcription regulation (35) or telomere homeostasis (36,37). RNA G4s can also exist in equilibrium with hairpin structures (Figure 2D) and play a role in telomere homeostasis (38) or gene expression mechanisms (10,39–41). Finally, sequences with the potential to form G4s might play an active role in the formation or dissolution of stable RNA/DNA hybrids (R-loops) (42–45) (Figure 2E).
Figure 2.
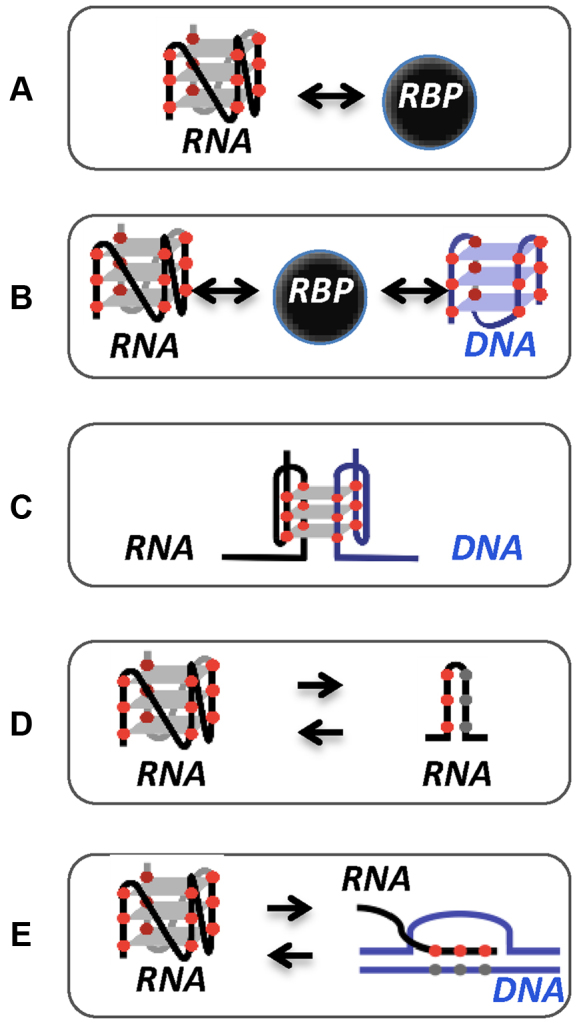
Mechanisms of action underlying the function of RNA G4s. RNA G4 binding to RBP (A), RBP binding to both DNA and RNA G4 (B), intermolecular G4 formed by DNA and RNA strands (C), equilibrium between RNA G4 and hairpin conformation (D), RNA G-rich sequence that can fold into a G4 or hybridize with the C-rich template DNA strand in the R-loop structure (E).
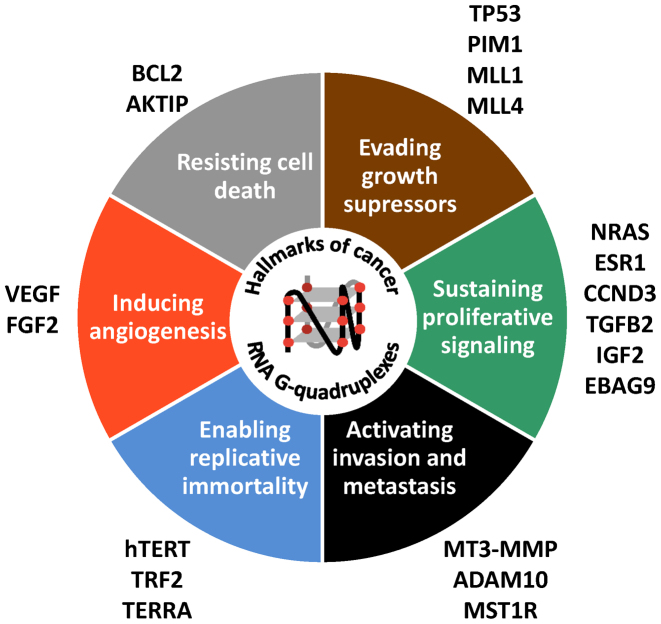
Accumulating evidences suggest that the proposed physiological role of G4s is altered in disease states (for recent reviews see (5,46); (Figure 1)), including neurological disorders and cancer. RNA G4s have been found to regulate the expression of many genes that are associated with the hallmarks of cancer (Figure 3). The ability of RNA G4s to regulate DNA-related processes suggests their possible involvement in the maintenance of genomic stability that is essential to prevent development of diseases including cancer, development defects, immune deficiency and neurodegenerative disorders. They also may play a role in virulence processes in microbial pathogens of humans (reviewed in (47,48)). The connections between RNA G4s and human diseases first emerged from studies showing that these structures act as cis-regulators critical for the mRNA expression of several disease-relevant genes (including the angiogenic factor VEGFA (25,49), the oncogene NRAS (29), the tumor suppressor TP53 (16,50) and the EBV maintenance protein, EBNA1 (26)). G4s formed by repeated G-rich RNA sequences are also targets of disease in repeat expansion disorders through different cis- and trans-acting mechanisms (5). Recently, the expression of several RNA G4-binding proteins have been found to be deregulated in disease contexts (including eIF4A (51), Aven (27), hnRNP A1 (52) or FMRP (53,54)) with important consequences on gene expression, providing further support for a role of RNA G4s in cellular pathology.
Figure 3.
RNA G4s and cancer hallmarks. Examples of RNA G4-containing genes that have been implicated in regulation of each of the hallmarks of cancer.
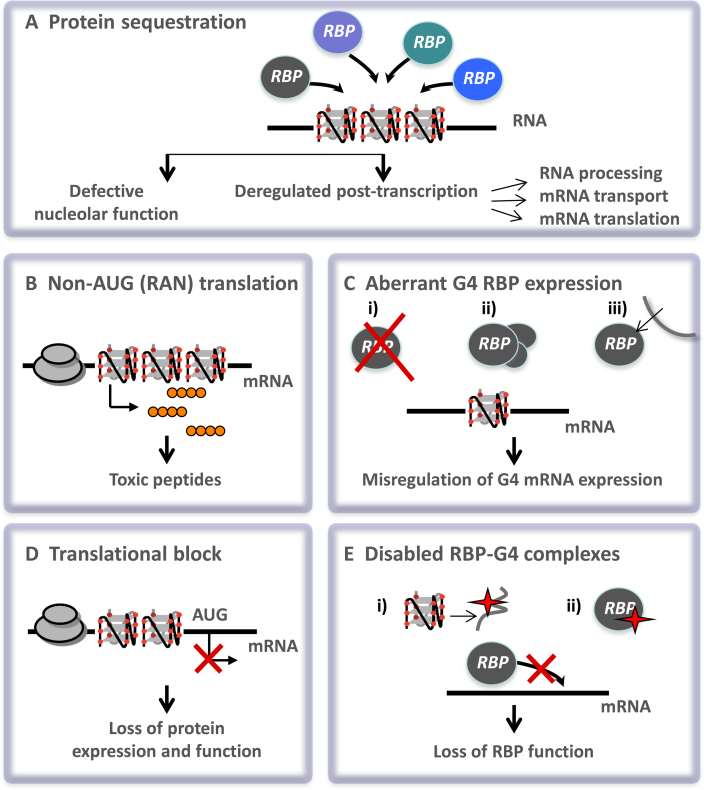
The purpose of our review is to summarize the current knowledge on the proposed molecular mechanisms by which RNA G4s may impact human diseases with particular emphasis on the different models of deregulation supported by a comprehensive account of examples known at present. We also provide a comprehensive list of functional RNA G4s, their regulatory mechanisms and association with diseases (Supplementary Table 1). Based on recent literature investigating the link between RNA G4s and disease entities, several mechanisms of action can be proposed, as listed below and depicted in Figure 4.
Figure 4.
Mechanistic models supporting the proposed link between RNA G4s and human diseases. Based on different examples of RNA G4 associated with disease, five mechanisms of action can be delineated: sequestration of RNA G4-binding proteins impacting the nucleolar function or the regulation of post-transcriptional processes (A), non-AUG (RAN) translation giving raise to toxic peptides (B), altered expression of RNA G4-binding proteins linked to RBP loss (i), RBP overexpression (ii) or RBP mislocalization (iii) (C), translational block by runs of adjacent G-repeats (D) and disabled RNA G4-protein complexes due to mutations (depicted by a red star) in RNA G4s (i) or in RBPs (ii) (E).
SEQUESTRATION OF RBPs
RNA G4s play a role in disease pathogenesis through sequestering RBPs (Figure 4A). This mode of action has been mainly described in non-coding repeat expansion diseases, including frontotemporal dementia and/or amyotrophic lateral sclerosis (FTD/ALS) caused by a large expansion of a non-coding GGGGCC hexanucleotide repeat (HRE) in the first intron of the C9orf72 gene. Using in vitro structural analysis, these G-rich RNA sequences have been shown to form secondary structures, including hairpins and G4s (40,55–57). The mechanism by which G4-forming HRE repeats cause FTD/ALS pathology has been recently investigated. One proposed model predicts that DNA and RNA G4s as well as R-loops formed in the GGGGCC repeat region impair transcription, leading to the accumulation of abortive transcripts that sequester G4-binding proteins (recently reviewed in (58)). Among these RBPs, nucleolin directly and preferentially binds the G4-containing abortive transcripts, resulting in nucleolin mislocalization and nucleolar stress, as evidenced by decreased ribosomal maturation and increased abundance of processing bodies (or P bodies, cytoplasmic foci of mRNA degradation and translational repression). These findings establish a direct link between the G4s of the C9orf72 HRE and the resulting pathological defects in FTD/ALS patients (57).
Another important cause of pathogenesis associated to repeats expansion in C9orf72 is the impaired ability of G4-sequestered RBPs to regulate their targets resulting in defects in RNA processing. This notion is supported by recent findings reporting extensive misregulated RNA processing events (i.e. alternative splicing and polyadenylation) in ALS carrying this expansion (59). Among the RBPs sequestered by the G-rich repeats, hnRNP H/F, a known splicing and polyadenylation regulator previously found to be associated with the C9orf72 G-rich repeats (57,59–61), has been predicted to be a potential regulator of these RNA processing defects. Since this factor can regulate RNA processing by binding to G-rich sequences (50,62) and G-runs forming G4 structures are critical regulators of pre-mRNA processing (15,62,63), it is plausible that sequestration of hnRNP H/F might impact the expression of G4-containing mRNAs in ALS. Recent results support this possibility by showing that hnRNP H associates with G4-forming C9orf72 GGGGCC repeats and colocalizes with G4 foci (visualized using the BG4 antibody (23,64)) in patient derived cells but not in control non-ALS cells (65). The formation of these aggregates correlates with dysregulated splicing of known targets of this factor in C9orf72 patient brains. These findings provide an explanation for global alternative splicing observed in C9orf72 cells (59). While there is no conclusive data establishing that hnRNP H sequestration plays a causal role in ALS, these results further support the involvement of RNA G4 formation and sequestration of this RBP in FTD/ALS pathogenesis. Although this study claims that the interaction between hnRNP H/F and G-rich sequences is linked to G4 formation (in agreement with (50,66)), other sets of data indicate that hnRNP H/F family members can bind G-rich sequences in single-stranded form (67), suggesting that further investigations and structural studies will be required to determine precisely the mode of binding of this RBP.
Overproduced G4-containing transcripts can also directly impact mRNA fate in the cytoplasm. Indeed, it has been proposed that the G4-forming C9orf72 repeats may interfere with the role of endogenous G4-containing tiRNAs (tRNA-derived stress-induced RNAs) in translational regulation that is required for motor neuron survival and confering cytoprotection against stress (69). TiRNA fragments derive from angiogenin (ANG)-mediated cleavage of tRNAs that displace components of the translational initiation machinery (i.e. the cap-binding complex eIF4F), resulting in translational repression and formation of RNA granules (named stress granules, SG). These processes in turn contribute to enhance cell survival subjected to stress conditions (70). The observation that extended C9orf72 repeats inhibit tiRNA-induced SG assembly led to propose that HRE expansions interfere with the function of the endogenous tiRNAs in motor neuron maintenance. Molecular details of such interference are not clear but may involve sequestration of G4 RBPs involved in motor neuron survival, including YB1, nucleolin and hnRNP A3 (69). While it is well established that G4s are formed in both C9orf72 repeats and tiRNAs and that these structures are bound by several RBPs, the role of G4s in this hijacking mechanism remains to be firmly demonstrated. It would be also important to define whether G4-containing tiRNAs could also suppress the toxicity of the pathological HREs.
Finally, RNAs containing the C9orf72 HRE expansion have been recently shown to compromise nucleocytoplasmic transport (71,72) through a mechanism involving the binding of the RBP RanGAP1, a key regulator of the nucleocytoplasmic transport, to the RNA GGGGCC repeat. The G4 ligand TMPyP4 reduces the affinity of RanGAP1 for the repeat RNA G4s (72) and rescues nucleocytoplasmic transport defects (72,73), suggesting that HRE G4s play a role in nucleocytoplasmic transport defects associated to FTD/ALS.
Besides RNA–protein interactions, the sequestration mechanism may also involve RNA–RNA interactions facilitated by intermolecular G4 formation (55). However, it is not known whether these multimolecular RNA G4s occur within cells and whether they might play a toxic role in FTD/ALS by contributing to RNA foci formation.
Based on these data, the G4s formed by the GGGGCC repeats have been proposed as fundamental determinants of the pathogenic mechanisms of C9orf72 repeat expansion linked to FTD/ALS. Collectively, these studies suggest that RNA G4s formed in expanded G-rich repeats may cause disease by sequestrating RBPs, resulting in disruption of RNA homeostasis and thus leading to cell dysfunction. Many RBPs colocalize with RNA foci (including several hnRNPs) but their biological relevance has not been validated. It is important to note that although the formation of these ribonucleoprotein complexes has been proposed to be largely dependent on G4 formation, hairpin structures may be also involved. Indeed, both nucleolin (57) and RanGAP1 (72) may bind hairpins (although they display preference for G4s). While these studies involve RBPs and G4s in the mechanistic model for repeat-associated neurodegenerative diseases, the role of RBP–G4 interactions in pathological conditions awaits further investigation. Functional analysis of RBPs defective in G4-binding may provide more direct evidence of a link between abnormal RBP–G4 interactions and disease pathology. Finally, these studies point to the targeting of these toxic G4-dependent ribonucleoprotein complexes as a possible intervention to prevent the molecular cascade leading to repeat-associated neurodegenerative diseases.
NOVEL TYPE OF TRANSLATION
G4-forming RNA sequences have been proposed to be involved in an atypical form of translation (Figure 4B) that occurs in the absence of an initiating AUG in all possible reading frames, generating homopolymeric proteins with glutamine, serine or alanine repeats (recently reviewed in (74)). This phenomenon, called repeat-associated non-AUG (RAN) translation, has been recently shown to occur on C9orf72 GGGGCC repeats (75,76) and on CGG repeats in the 5΄ UTR of FMR1 (fragile X mental retardation 1) mediating neurodegeneration in fragile X tremor ataxia syndrome (FXTAS) (77). RNA structures (including hairpins (78,79) and G4s (40,55–57)) have been suggested to behave as possible triggers of this noncanonical translation mechanism. It is also possible that the equilibrium between hairpin and G4 conformations may mediate RAN translation (40). How RAN translation occurs has recently begun to emerge. One proposed model of RAN translation at CGG repeats in FMR1 transcripts is that, after cap-dependent initiation of translation (80) and subsequent scanning through the 5΄ UTR, ribosome stalling at secondary structures formed at CGG repeats leads to aberrant translation initiation at non-AUG codons, resulting in the production of polyglycine and polyalanine peptides. While several evidences support the possibility that these G-rich repeats fold into G4 structures (e.g. for the C9orf72 GGGGCC repeats (40,55–57)), the question of whether G4s and G4-binding proteins play a role in the initiation or inhibition of RAN translation awaits further investigation. However, the observation that small molecules binding G4s, such as TMPyP4, can distort the secondary structure of the C9orf72 repeats and disrupt protein interactions (81), suggest that G4 formation may interfere with protein sequestration and/or RAN translation into potentially toxic dipeptides. Although the effect of these molecules has not been tested in cellulo, this study provides the proof of principle that small-molecule ligands can be used to target C9orf72 GGGGCC repeats. Since TMPyP4 does not exhibit RNA G4 selectivity (82) and appears to exert opposite effects on G4 formation (83), it would be important to strengthen these findings by using more selective RNA G4 binders. In support of this notion, small molecules binding to C9orf72 HRE adopting both hairpin and G4 conformations can inhibit RAN translation and foci formation in neurons (40). Overall, these findings highlight G-rich repeats binders as possible FTD/ALS therapeutic.
In conclusion, RNA G4s formed in expanded repeats containing G-rich elements could play a role in disease pathogenesis at the level of mRNA translation through either facilitating or hindering RAN translation.
ALTERED EXPRESSION OF G4 RBPs
Cellular pathology linked to RNA G4s can be ascribed in part to the alteration in the expression of G4 RBPs leading to gene expression deregulation (Figure 4C). One well-documented example of this mechanism of deregulation has been recently provided for eIF4A, a translation initiation factor that plays a key role in removing secondary and tertiary structures during 5΄ UTR ribosome scanning. eIF4A is the molecular target for natural compounds showing promising anticancer activity and its overexpression promotes T-cell acute lymphoblastic leukaemia development in vivo and is required for leukaemia maintenance. Ribosome profiling revealed that eIF4A regulates mRNA translation of transcripts with 5΄ UTRs containing G4-forming motifs composed of CGG motifs. Among the most eIF4A-sensitive mRNAs there are several oncogenes and transcription factors (including MYC, MYB, NOTCH, CDK6, BCL2) (51). Importantly, transcriptome-wide studies revealed the presence of RNA G4s in one third of the CGG-containing transcripts identified in this study (31). The observation that eIF4A-associated transcripts contain guanine quartets composed of CGG motifs has been reported also in breast cancer cells (84,85). Since G4s are general inhibitors of mRNA translation (86,87), a possible model predicts that, in normal conditions, G4s at the 5΄ UTR of these mRNAs would restrain the expression of cancer-prone factors by inhibiting the initiation step of translation. In contrast, overexpression of eIF4A in cancer cells promotes G4 unwinding leading to increased protein synthesis of oncoproteins. Given the importance of cofactors regulating eIF4A RNA-binding and catalytic activity, the question is raised as to whether the eIF4A-dependent mechanism of translational control of G4-containing mRNAs involves additional factors helping eIF4A to target G4s. These cofactors may provide a molecular explanation for the specific recognition of the 12 nucleotides (CGG)4 signature by eIF4A.
A similar deregulatory mechanism involving RNA helicases on G4-containing mRNAs has been recently proposed for the regulation of mixed lineage leukemia (MLL) proto-oncogene by an Aven-centered complex (27). Aven increases translation of MLL1 and MLL4 leukemic genes by interacting with G4s within the open reading frames (ORFs) and with protein factors associated with the translational machinery (i.e. TDRD3 and SMN). Importantly, optimal translation of MLL1 and MLL4 requires DHX36, a helicase previously reported to unwind RNA G4 structures (88–90). The arginine-glycine-glycine (RGG) domain of Aven plays a central role in this mode of translational regulation since it mediates both Aven–RNA G4 and Aven–protein interactions (27). Since Aven is a pro-survival protein overexpressed in acute leukemia, it has been proposed that this G4-dependent regulatory mechanism might promote survival of leukemic cells and that drugs disrupting this pathway, including G4 ligands, could have a therapeutic potential. This translational mechanism, together with previous findings (26) and recent bioinformatics analysis predicting the presence of 1600 G4s in human ORFs (27), strengthens the notion that in addition to deregulate gene expression via their location within the 5΄ and 3΄ UTRs, G4s might be relevant for the expression of disease-related genes when located within the ORFs.
Another important example of a disease resulting from the altered expression of a G4-binding proteins is the fragile X syndrome (FXS), the most frequent form of intellectual disability caused by FMR1 gene silencing and the lack of the encoded protein, FMRP. The function of this protein in mRNA translation and RNA localization is mediated by its association with specific secondary structures in its target mRNAs, the most well documented being the G4 structure ((91,92) and recently reviewed in (53)). FMRP might inhibit mRNA translation initiation or elongation by binding G4s and by either recruiting trans-acting factors (e.g. CYFIP1 (93)) or binding directly the ribosome to stall translation (94). Notably, as for Aven, FMRP binding to G4s (and surrounding sequences) requires the RGG motif (95,96). Additionally, FMRP is suggested to be a good candidate for the G4-dependent transport of dendritic mRNAs such as PSD-95, Shank-1 or NR2B (see Supplementary Table 1). Indeed, FMRP has been shown to interact with its dendritic mRNA targets by recognizing G4 structures present in their 3΄ UTR and regulating their local translation in neurons (97–99). Therefore, G4–FMRP interactions are thought to regulate hundreds of mRNAs in neuronal cells at different levels and the deregulation involving loss of these interactions are considered as one major pathological mechanism in FXS. The observation that FMRP is overexpressed in cancer and it plays a role in tumor progression (54) raises the possibility that FMRP-dependent G4 mRNA expression regulation may be altered in cancer cells. A similar model of deregulation has been recently suggested also for FXR1, another member of the fragile X-related (FXR) protein family (100). It has been proposed that overexpressed FXR1 in oral squamous cell carcinoma contributed to promote oral cancer progression by deregulating both p21 and the telomerase RNA hTR/TERC turnover and expression. This might occur through a mechanism involving binding of FXR1 to RNA G4 structures, as suggested by luciferase reporter assays after FXR1 silencing.
Deregulated expression of mRNAs with G4 motifs might also result from a modification of the subcellular localization of RNA G4-binding proteins. An example is provided by hnRNP A1, a nuclear pre-mRNA processing regulator that presents high expression and aberrant cytoplasm localization associated with metastatic relapse in patients with invasive breast cancer (52). The observation that cytoplasmic hnRNP A1 increases the translation of the mRNA encoding the tyrosine kinase receptor RON/MST1R through RNA G4 secondary structures in the RON 5΄ UTR suggests that aberrant relocalization of hnRNP A1 in the cytoplasm activates protein synthesis of G4-containing mRNAs. Collectively, these studies suggest that deregulation in the expression of G4 RBPs -including RNA helicases- modulate the expression of transcripts under the control of RNA G4s.
Overall, these studies suggest that deregulation of G4-containing mRNAs due to aberrant expression and regulation of RBPs may be linked to pathological situations.
TRANSLATIONAL BLOCK BY RUNS OF ADJACENT G-REPEATS
Disease-causing expansion of G-rich repeats with the potential to form G4 can directly modify mRNA translation by acting as cis-regulatory elements (Figure 4D). This deregulatory mechanism has been proposed for FXTAS in the FMR1 5΄ UTR. Although methylation of the CGG repeats has been shown to contribute to FMR1 transcriptional downregulation (101), several evidences reveal that silencing of FMR1 transcription does not sufficiently explain all clinical situations and that reduced translation efficiency contribute to the decreased levels of FMRP (102). Indeed, it has been proposed that both reduced polysome formation (i.e. clusters of ribosomes bound to mRNAs during active translation) and stalled ribosome progression on premutation FMR1 mRNAs result in decreased efficiency of its translation in vivo (102,103). The mechanism underlying CGG expansion-mediated FXTAS pathology involves folding of the 5΄ UTR (CGG)n premutation RNA in secondary structures, including G4s (104,105). Translational efficiency of the FMR1 mRNA is decreased by G4s formed by the CGG repeats and can be modulated by G4 RBPs (including hnRNP A2 and CBF-A (105)) and G4 ligands (106).
DISABLING THE FORMATION OF REGULATORY G4-PROTEIN INTERACTIONS
A bioinformatic search of single nucleotide polymorphisms (SNP) within G4-forming sequences in the human 5΄ UTRs revealed that 5% of all predicted 5΄ UTR G4 sequences included at least one SNP. Several identified genes are implicated in various diseases, including cancer (e.g. the RAD51 and CAV2 genes (107)). The observation that a SNP co-localizing with a G4 abolishes quartet structure formation and increases mRNA translation (107), indicates the potential of G4 variants to be involved either in the predisposition, or in the appearance of, various diseases and cancers by altering the gene expression background of a specific individual.
Consistent with this hypothesis, two polymorphisms associated with a G4 in the third intron (PIN3) (16) and a predicted G4 in the 3΄ end region of the TP53 pre-mRNA have been shown to modulate age of tumor onset in TP53 mutation carriers in Brazilian Li-Fraumeni families (108). Recently, the PIN3 polymorphism has been shown to alter the balance between the fully spliced TP53 transcript, encoding TP53 and an incompletely spliced TP53 isoform retaining intron 2, encoding Δ40TP53, an N-terminally deleted TP53 isoform that can act in a dominant-negative manner toward TP53. The alternative splicing associated with this variant is probably the consequence of a modification of the G4 position with respect to the intron/exon boundaries, and is dependent on the presence of G4 ligands or exposure to ionizing radiation (109). Sequence analysis of TP53 in breast tumor samples revealed that the PIN3 polymorphism is associated with a low Δ40TP53:TP53 ratio and better outcome (110). These results suggest that G4s could impact the responses to radiation exposure. In agreement with this hypothesis, we previously showed that a G4 at the TP53 pre-mRNA 3΄ end allows TP53 expression and function after DNA damage induced by UV irradiation. The underlying mechanism involves the interaction of this G4 with hnRNP H/F that, in turn, recruits the pre-mRNA processing machinery at the TP53 polyadenylation signal. Our observation that artificially introduced mutations within the G4 region of TP53 pre-mRNA abrogated its polyadenylation (50) raises the intriguing possibility that natural sequence variants such as tumor-associated mutations or SNPs affecting G4 formation and/or the binding of processing factors could modulate TP53 polyadenylation and function following stress-mediated DNA damage (Figure 4E). A number of sequence variants have been documented surrounding the TP53 polyadenylation site region (i.e. SNPs in dbSNP) some of which, based on bioinformatic approaches, might be expected to impact G4 formation.
Disease-associated mutations in RBPs can also disable the formation of regulatory RBP–G4 complexes (Figure 4E). An example of this deregulatory mechanism has been recently provided for the activation-induced cytidine deaminase (AID) (14), an enzyme essential for immunoglobulin class-switch recombination (CSR). AID binds directly to G4s formed by the intronic lariat RNAs (also called switch RNAs) derived from splicing of the primary transcripts arising from transcription of the recombining S regions. The AID-switch RNA complexes are targeted to the complementary S region DNA in a sequence-specific manner. A point mutation in AID associated with CSR defects (111) impairs binding of AID to both RNA and DNA G4s resulting in defective CSR. Overall, these studies demonstrate that mutations in the RNA G4s or their binding partners may induces disassembly of regulatory complexes and impact both RNA and DNA processes leading to diseases.
PERSPECTIVE
Studies over the past 50 years contributed to characterize the structure and the function of G4s in RNA biology and their role in disease (Figures 1, 2 and 3; Tables 1 and Supplementary Table 1). Accumulating evidences support the existence of overlapping models of deregulation involving G4 structures and leading to different disease entities (Figure 4 and Supplementary Table 1), including neurodegenerative and neurodevelopmental disorders and cancer.
Recently, G4s have been implicated in the molecular mechanisms controlling viral pathogenesis. Indeed, the presence of G4 motifs have been identified in some of the most pathogenic virus (47,48), including the EBV involved in mononucleosis infection and cancer. EBV encodes EBNA1, a genome maintenance protein that is translated through a G4-dependent mechanism linked to viral latency and immune evasion (26,112). RNA G4s have mapped to several mRNAs encoding gammaherpesviral maintenance proteins (26), suggesting that these structures may be responsible for the cis-acting regulation of viral mRNA translation and can be targeted to reduce the burden of gammaherpesvirus-associated malignancies. Moreover, since EBNA1 binds to RNA G4s (113), it might be important to define whether it is engaged in an autoregulatory feedback loop to tightly control its expression during the virus cycle and/or whether it impacts the expression of other viral proteins.
Overall, the common mechanisms of regulation described in this review highlight the importance of trans-acting factors associated with these RNA structures in mediating different pathogenic mechanisms. However, a number of questions remain to be addressed to better understand how these factors mediate the link between RNA G4s and disease.
Given that some of these factors bind both DNA and RNA G4s (e.g. the helicases RHAU and DHX9 (114), or the RBP TLS/FUS (12)), the question that remains is whether these factors affect diseases by altering the ‘G4ome’ (including DNA and RNA G4 sequences) through deregulation of the DNA and RNA metabolism. From a more fundamental point of view, do DNA/RNA G4-binding factors provide a functional link between DNA and RNA processes to regulate cellular pathways? Such a functional interplay could have important function in preserving genomic stability. Indeed, recent studies show that protein factors with a dual function in both biochemical processes play a critical role in maintaining cellular genomic integrity (115,116) and that DNA/RNA G4s contribute to genome instability by regulating both DNA-related processes (46) and RNA-based gene expression mechanisms (50).
RNA G4s and factors that modulate their formation/unfolding may also impact genome instability or induce gene expression deregulation through the formation of R-loops. Indeed, these DNA/RNA hybrid structures are formed preferentially when the non-template strand is G-rich (42–44) and their stabilization may depend on the formation of a G4 on the single-stranded exposed DNA strand and a RNA/DNA duplex between the G-rich RNA and the C-rich DNA strand (45). R-loops have important biological functions (including roles in mitochondrial DNA replication, immunoglobulin gene CSR and transcription) but at the same time they are source of genome instability associated to diseases (recently reviewed in (117)). Further work is needed to understand whether cis- or trans-acting (118,119) (including lncRNAs (120)) RNAs with the potential to form G4s play a role in the mechanistic connection between R-loops and pathological processes.
Recent studies identify a role for a subset of RGG motif-containing proteins (including Aven (27), FMRP (95,96), EWS (121), EBNA1 (113) and TLS/FUS (122)) in post-transcriptional control through their ability to bind RNA G4s. These and other proteins harboring these motifs have been implicated in several of the major classes of human diseases, such as cancer and neurological disorders. The observation that RGG motifs are also used for protein–protein interactions and are known substrates for arginine methylation (123) leads to speculate that these protein motifs may facilitate the formation of ribonucleoprotein complexes at RNA G4 structures and that methylation might play a role in modulating these specific interactions.
Another important issue is whether RNA conformational equilibrium involving G4s may represent an additional mechanism underlying the link between G4s and disease. Indeed, a number of studies provided evidence of the formation of G4s in competition with stem-loop structures (10,38–41,124) or with alternate G4 conformations (125). These structural transitions may play a role in telomere homeostasis (38), translational regulation (39,40) and miRNA biogenesis (10,41) as well as seed binding site accessibility (125), and can be modulated by ions (10,41,124), G4 ligands (41) and trans-acting factors, such as ncRNAs (39). Noteworthy, transcriptome-wide analysis of in vitro G4 formation supports the notion that hairpin-G4 transitions may be prevalent in human transcripts and play a role in gene expression regulation (31). The observation that tRNAs influence the equilibrium between hairpin and G4s in the 5΄ UTR of oncogenes suggests that these hairpin-G4 transitions can modulate the expression of disease-relevant genes (39).
Moreover, as human lncRNAs have been predicted to harbour a number of G4s (9) and sequestration of RBPs by RNA G4s appears to be an important pathogenic mechanism, do lncRNAs impact on human diseases by titrating proteins binding functional G4s? Finally, given that RBPs contribute directly or indirectly to remodel these structures in mRNAs and that G4s colocalize with miRNAs (126,127), do the alteration in RBPs expression deregulate miRNA-mediated gene expression mechanisms?
The development of techniques to identify RNA G4s and characterize their partners and function at a genome-wide scale will be essential to understand how diseases can be caused by deregulation of RNA G4 structure and function. The insights we provide here into the link between RNA G4 and disease suggest possible therapeutic interventions that specifically target the deregulated G4s and /or the binding of trans-acting factors to these structures. Many G4 binders (including small-molecule ligands and antisense oligonucleotides) exist that have the potential to modulate G4 conformations and protein–G4 interactions. A better understanding of the interplay between RNA G4s and regulatory protein factors may lead to approaches with improved specificity and selectivity targeting protein–G4 RNA interaction interfaces.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Stéphan Vagner, Gaelle Bougeard, Sergio Di Marco and Morgane Le Bras for critical reading of the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
INSERM; Ligue Nationale Contre le Cancer (to S.M.); Association pour la Recherche contre le Cancer (to A.C., S.M); Emergence GSO (to A.C, to S.M); Lab Excellence Labex TOUCAN. Funding for open access charge: INSERM.
Conflict of interest statement. None declared.
REFERENCES
- 1.Huppert J.L., Balasubramanian S.. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005; 33:2908–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Todd A.K., Johnston M., Neidle S.. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005; 33:2901–2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eddy J., Maizels N.. Conserved elements with potential to form polymorphic G-quadruplex structures in the first intron of human genes. Nucleic Acids Res. 2008; 36:1321–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huppert J.L., Bugaut A., Kumari S., Balasubramanian S.. G-quadruplexes: the beginning and end of UTRs. Nucleic Acids Res. 2008; 36:6260–6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simone R., Fratta P., Neidle S., Parkinson G.N., Isaacs A.M.. G-quadruplexes: emerging roles in neurodegenerative diseases and the non-coding transcriptome. FEBS Lett. 2015; 589:1653–1668. [DOI] [PubMed] [Google Scholar]
- 6.Rhodes D., Lipps H.J.. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015; 43:8627–8637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Agarwala P., Pandey S., Maiti S.. The tale of RNA G-quadruplex. Org. Biomol. Chem. 2015; 13:5570–5585. [DOI] [PubMed] [Google Scholar]
- 8.Chambers V.S., Marsico G., Boutell J.M., Di Antonio M., Smith G.P., Balasubramanian S.. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat. Biotechnol. 2015; 33:877–881. [DOI] [PubMed] [Google Scholar]
- 9.Jayaraj G.G., Pandey S., Scaria V., Maiti S.. Potential G-quadruplexes in the human long non-coding transcriptome. RNA Biol. 2012; 9:81–86. [DOI] [PubMed] [Google Scholar]
- 10.Mirihana Arachchilage G., Dassanayake A.C., Basu S.. A potassium ion-dependent RNA structural switch regulates human pre-miRNA 92b maturation. Chem. Biol. 2015; 22:262–272. [DOI] [PubMed] [Google Scholar]
- 11.Xu Y., Komiyama M.. Structure, function and targeting of human telomere RNA. Methods. 2012; 57:100–105. [DOI] [PubMed] [Google Scholar]
- 12.Takahama K., Takada A., Tada S., Shimizu M., Sayama K., Kurokawa R., Oyoshi T.. Regulation of telomere length by G-quadruplex telomere DNA- and TERRA-binding protein TLS/FUS. Chem. Biol. 2013; 20:341–350. [DOI] [PubMed] [Google Scholar]
- 13.Hirashima K., Seimiya H.. Telomeric repeat-containing RNA/G-quadruplex-forming sequences cause genome-wide alteration of gene expression in human cancer cells in vivo. Nucleic Acids Res. 2015; 43:2022–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng S., Vuong B.Q., Vaidyanathan B., Lin J.Y., Huang F.T., Chaudhuri J.. Non-coding RNA generated following lariat debranching mediates targeting of AID to DNA. Cell. 2015; 161:762–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beaudoin J.D., Perreault J.P.. Exploring mRNA 3΄-UTR G-quadruplexes: evidence of roles in both alternative polyadenylation and mRNA shortening. Nucleic Acids Res. 2013; 41:5898–5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marcel V., Tran P.L., Sagne C., Martel-Planche G., Vaslin L., Teulade-Fichou M.P., Hall J., Mergny J.L., Hainaut P., Van Dyck E.. G-quadruplex structures in TP53 intron 3: role in alternative splicing and in production of p53 mRNA isoforms. Carcinogenesis. 2011; 32:271–278. [DOI] [PubMed] [Google Scholar]
- 17.Gomez D., Lemarteleur T., Lacroix L., Mailliet P., Mergny J.L., Riou J.F.. Telomerase downregulation induced by the G-quadruplex ligand 12459 in A549 cells is mediated by hTERT RNA alternative splicing. Nucleic Acids Res. 2004; 32:371–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu C.H., Teulade-Fichou M.P., Olsthoorn R.C.. Stimulation of ribosomal frameshifting by RNA G-quadruplex structures. Nucleic Acids Res. 2014; 42:1887–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bedrat A., Lacroix L., Mergny J.L.. Re-evaluation of G-quadruplex propensity with G4Hunter. Nucleic Acids Res. 2016; 44:1746–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Campbell N., Collie G.W., Neidle S.. Crystallography of DNA and RNA G-quadruplex nucleic acids and their ligand complexes. Curr. Protoc. Nucleic Acid Chem. 2012; doi:10.1002/0471142700.nc1706s50. [DOI] [PubMed] [Google Scholar]
- 21.Randazzo A., Spada G.P., da Silva M.W.. Circular dichroism of quadruplex structures. Top. Curr. Chem. 2013; 330:67–86. [DOI] [PubMed] [Google Scholar]
- 22.Beaudoin J.D., Jodoin R., Perreault J.P.. In-line probing of RNA G-quadruplexes. Methods. 2013; 64:79–87. [DOI] [PubMed] [Google Scholar]
- 23.Biffi G., Di Antonio M., Tannahill D., Balasubramanian S.. Visualization and selective chemical targeting of RNA G-quadruplex structures in the cytoplasm of human cells. Nat. Chem. 2014; 6:75–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu S., Li Q., Xiang J., Yang Q., Sun H., Guan A., Wang L., Liu Y., Yu L., Shi Y. et al. Directly lighting up RNA G-quadruplexes from test tubes to living human cells. Nucleic Acids Res. 2015; 43:9575–9586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cammas A., Dubrac A., Morel B., Lamaa A., Touriol C., Teulade-Fichou M.P., Prats H., Millevoi S.. Stabilization of the G-quadruplex at the VEGF IRES represses cap-independent translation. RNA Biol. 2015; 12:320–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murat P., Zhong J., Lekieffre L., Cowieson N.P., Clancy J.L., Preiss T., Balasubramanian S., Khanna R., Tellam J.. G-quadruplexes regulate Epstein-Barr virus-encoded nuclear antigen 1 mRNA translation. Nat. Chem. Biol. 2014; 10:358–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thandapani P., Song J., Gandin V., Cai Y., Rouleau S.G., Garant J.M., Boisvert F.M., Yu Z., Perreault J.P., Topisirovic I. et al. Aven recognition of RNA G-quadruplexes regulates translation of the mixed lineage leukemia protooncogenes. Elife. 2015; 4:e06234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gomez D., Guedin A., Mergny J.L., Salles B., Riou J.F., Teulade-Fichou M.P., Calsou P.. A G-quadruplex structure within the 5΄-UTR of TRF2 mRNA represses translation in human cells. Nucleic Acids Res. 2010; 38:7187–7198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumari S., Bugaut A., Huppert J.L., Balasubramanian S.. An RNA G-quadruplex in the 5΄ UTR of the NRAS proto-oncogene modulates translation. Nat. Chem. Biol. 2007; 3:218–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Halder K., Benzler M., Hartig J.S.. Reporter assays for studying quadruplex nucleic acids. Methods. 2012; 57:115–121. [DOI] [PubMed] [Google Scholar]
- 31.Kwok C.K., Marsico G., Sahakyan A.B., Chambers V.S., Balasubramanian S.. rG4-seq reveals widespread formation of G-quadruplex structures in the human transcriptome. Nat. Methods. 2016; doi:10.1038/nmeth.3965. [DOI] [PubMed] [Google Scholar]
- 32.Guo J.U., Bartel D.P.. RNA G-quadruplexes are globally unfolded in eukaryotic cells and depleted in bacteria. Science. 2016; 353, doi:10.1126/science.aaf5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brazda V., Haronikova L., Liao J.C., Fojta M.. DNA and RNA quadruplex-binding proteins. Int. J. Mol. Sci. 2014; 15:17493–17517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Biffi G., Tannahill D., Balasubramanian S.. An intramolecular G-quadruplex structure is required for binding of telomeric repeat-containing RNA to the telomeric protein TRF2. J. Am. Chem. Soc. 2012; 134:11974–11976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng K.W., Wu R.Y., He Y.D., Xiao S., Zhang J.Y., Liu J.Q., Hao Y.H., Tan Z.. A competitive formation of DNA:RNA hybrid G-quadruplex is responsible to the mitochondrial transcription termination at the DNA replication priming site. Nucleic Acids Res. 2014; 42:10832–10844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu Y., Kimura T., Komiyama M.. Human telomere RNA and DNA form an intermolecular G-quadruplex. Nucleic Acids Symp. Ser. (Oxf). 2008; 52:169–170. [DOI] [PubMed] [Google Scholar]
- 37.Xu Y., Ishizuka T., Yang J., Ito K., Katada H., Komiyama M., Hayashi T.. Oligonucleotide models of telomeric DNA and RNA form a Hybrid G-quadruplex structure as a potential component of telomeres. J. Biol. Chem. 2012; 287:41787–41796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lacroix L., Seosse A., Mergny J.L.. Fluorescence-based duplex-quadruplex competition test to screen for telomerase RNA quadruplex ligands. Nucleic Acids Res. 2011; 39:e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rode A.B., Endoh T., Sugimoto N.. tRNA shifts the G-quadruplex-hairpin conformational equilibrium in RNA towards the hairpin conformer. Angew. Chem. Int. Ed. Engl. 2016; 55:14315–14319. [DOI] [PubMed] [Google Scholar]
- 40.Su Z., Zhang Y., Gendron T.F., Bauer P.O., Chew J., Yang W.Y., Fostvedt E., Jansen-West K., Belzil V.V., Desaro P. et al. Discovery of a biomarker and lead small molecules to target r(GGGGCC)-associated defects in c9FTD/ALS. Neuron. 2014; 83:1043–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pandey S., Agarwala P., Jayaraj G.G., Gargallo R., Maiti S.. The RNA stem-loop to G-quadruplex equilibrium controls mature microRNA production inside the cell. Biochemistry. 2015; 54:7067–7078. [DOI] [PubMed] [Google Scholar]
- 42.Li X., Manley J.L.. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005; 122:365–378. [DOI] [PubMed] [Google Scholar]
- 43.Ginno P.A., Lim Y.W., Lott P.L., Korf I., Chedin F.. GC skew at the 5΄ and 3΄ ends of human genes links R-loop formation to epigenetic regulation and transcription termination. Genome Res. 2013; 23:1590–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Skourti-Stathaki K., Proudfoot N.J., Gromak N.. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol. Cell. 2011; 42:794–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Duquette M.L., Handa P., Vincent J.A., Taylor A.F., Maizels N.. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev. 2004; 18:1618–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maizels N. G4-associated human diseases. EMBO Rep. 2015; 16:910–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Metifiot M., Amrane S., Litvak S., Andreola M.L.. G-quadruplexes in viruses: function and potential therapeutic applications. Nucleic Acids Res. 2014; 42:12352–12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harris L.M., Merrick C.J.. G-quadruplexes in pathogens: a common route to virulence control. PLoS Pathog. 2015; 11:e1004562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morris M.J., Negishi Y., Pazsint C., Schonhoft J.D., Basu S.. An RNA G-quadruplex is essential for cap-independent translation initiation in human VEGF IRES. J. Am. Chem. Soc. 2010; 132:17831–17839. [DOI] [PubMed] [Google Scholar]
- 50.Decorsiere A., Cayrel A., Vagner S., Millevoi S.. Essential role for the interaction between hnRNP H/F and a G quadruplex in maintaining p53 pre-mRNA 3΄-end processing and function during DNA damage. Genes Dev. 2011; 25:220–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wolfe A.L., Singh K., Zhong Y., Drewe P., Rajasekhar V.K., Sanghvi V.R., Mavrakis K.J., Jiang M., Roderick J.E., Van der Meulen J. et al. RNA G-quadruplexes cause eIF4A-dependent oncogene translation in cancer. Nature. 2014; 513:65–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cammas A., Lacroix-Triki M., Pierredon S., Le Bras M., Iacovoni J.S., Teulade-Fichou M.P., Favre G., Roche H., Filleron T., Millevoi S. et al. hnRNP A1-mediated translational regulation of the G quadruplex-containing RON receptor tyrosine kinase mRNA linked to tumor progression. Oncotarget. 2016; 7:16793–16805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen E., Joseph S.. Fragile X mental retardation protein: a paradigm for translational control by RNA-binding proteins. Biochimie. 2015; 114:147–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Luca R., Averna M., Zalfa F., Vecchi M., Bianchi F., La Fata G., Del Nonno F., Nardacci R., Bianchi M., Nuciforo P. et al. The fragile X protein binds mRNAs involved in cancer progression and modulates metastasis formation. EMBO Mol. Med. 2013; 5:1523–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reddy K., Zamiri B., Stanley S.Y., Macgregor R.B. Jr, Pearson C.E.. The disease-associated r(GGGGCC)n repeat from the C9orf72 gene forms tract length-dependent uni- and multimolecular RNA G-quadruplex structures. J. Biol. Chem. 2013; 288:9860–9866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fratta P., Mizielinska S., Nicoll A.J., Zloh M., Fisher E.M., Parkinson G., Isaacs A.M.. C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms RNA G-quadruplexes. Sci. Rep. 2012; 2:1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Haeusler A.R., Donnelly C.J., Periz G., Simko E.A., Shaw P.G., Kim M.S., Maragakis N.J., Troncoso J.C., Pandey A., Sattler R. et al. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature. 2014; 507:195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang J., Haeusler A.R., Simko E.A.. Emerging role of RNA*DNA hybrids in C9orf72-linked neurodegeneration. Cell Cycle. 2015; 14:526–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Prudencio M., Belzil V.V., Batra R., Ross C.A., Gendron T.F., Pregent L.J., Murray M.E., Overstreet K.K., Piazza-Johnston A.E., Desaro P. et al. Distinct brain transcriptome profiles in C9orf72-associated and sporadic ALS. Nat. Neurosci. 2015; 18:1175–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee Y.B., Chen H.J., Peres J.N., Gomez-Deza J., Attig J., Stalekar M., Troakes C., Nishimura A.L., Scotter E.L., Vance C. et al. Hexanucleotide repeats in ALS/FTD form length-dependent RNA foci, sequester RNA binding proteins, and are neurotoxic. Cell Rep. 2013; 5:1178–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cooper-Knock J., Walsh M.J., Higginbottom A., Robin Highley J., Dickman M.J., Edbauer D., Ince P.G., Wharton S.B., Wilson S.A., Kirby J. et al. Sequestration of multiple RNA recognition motif-containing proteins by C9orf72 repeat expansions. Brain. 2014; 137:2040–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xiao X., Wang Z., Jang M., Nutiu R., Wang E.T., Burge C.B.. Splice site strength-dependent activity and genetic buffering by poly-G runs. Nat. Struct. Mol. Biol. 2009; 16:1094–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Millevoi S., Moine H., Vagner S.. G-quadruplexes in RNA biology. Wiley Interdiscip. Rev. RNA. 2012; 3:495–507. [DOI] [PubMed] [Google Scholar]
- 64.Biffi G., Tannahill D., McCafferty J., Balasubramanian S.. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013; 5:182–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Conlon E.G., Lu L., Sharma A., Yamazaki T., Tang T., Shneider N.A., Manley J.L.. The C9ORF72 GGGGCC expansion forms RNA G-quadruplex inclusions and sequesters hnRNP H to disrupt splicing in ALS patient brains. Elife. 2016; 5:e17820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.von Hacht A., Seifert O., Menger M., Schutze T., Arora A., Konthur Z., Neubauer P., Wagner A., Weise C., Kurreck J.. Identification and characterization of RNA guanine-quadruplex binding proteins. Nucleic Acids Res. 2014; 42:6630–6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dominguez C., Fisette J.F., Chabot B., Allain F.H.. Structural basis of G-tract recognition and encaging by hnRNP F quasi-RRMs. Nat. Struct. Mol. Biol. 2010; 17:853–861. [DOI] [PubMed] [Google Scholar]
- 69.Ivanov P., O'Day E., Emara M.M., Wagner G., Lieberman J., Anderson P.. G-quadruplex structures contribute to the neuroprotective effects of angiogenin-induced tRNA fragments. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:18201–18206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ivanov P., Emara M.M., Villen J., Gygi S.P., Anderson P.. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol. Cell. 2011; 43:613–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Freibaum B.D., Lu Y., Lopez-Gonzalez R., Kim N.C., Almeida S., Lee K.H., Badders N., Valentine M., Miller B.L., Wong P.C. et al. GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature. 2015; 525:129–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang K., Donnelly C.J., Haeusler A.R., Grima J.C., Machamer J.B., Steinwald P., Daley E.L., Miller S.J., Cunningham K.M., Vidensky S. et al. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature. 2015; 525:56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kwon I., Xiang S., Kato M., Wu L., Theodoropoulos P., Wang T., Kim J., Yun J., Xie Y., McKnight S.L.. Poly-dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science. 2014; 345:1139–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Green K.M., Linsalata A.E., Todd P.K.. RAN translation-What makes it run?. Brain Res. 2016; 1647:30–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zu T., Liu Y., Banez-Coronel M., Reid T., Pletnikova O., Lewis J., Miller T.M., Harms M.B., Falchook A.E., Subramony S.H. et al. RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:E4968–E4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ash P.E., Bieniek K.F., Gendron T.F., Caulfield T., Lin W.L., Dejesus-Hernandez M., van Blitterswijk M.M., Jansen-West K., Paul J.W. 3rd, Rademakers R. et al. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013; 77:639–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Todd P.K., Oh S.Y., Krans A., He F., Sellier C., Frazer M., Renoux A.J., Chen K.C., Scaglione K.M., Basrur V. et al. CGG repeat-associated translation mediates neurodegeneration in fragile X tremor ataxia syndrome. Neuron. 2013; 78:440–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Napierala M., Michalowski D., de Mezer M., Krzyzosiak W.J.. Facile FMR1 mRNA structure regulation by interruptions in CGG repeats. Nucleic Acids Res. 2005; 33:451–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zu T., Gibbens B., Doty N.S., Gomes-Pereira M., Huguet A., Stone M.D., Margolis J., Peterson M., Markowski T.W., Ingram M.A. et al. Non-ATG-initiated translation directed by microsatellite expansions. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:260–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kearse M.G., Green K.M., Krans A., Rodriguez C.M., Linsalata A.E., Goldstrohm A.C., Todd P.K.. CGG repeat-associated non-AUG translation utilizes a cap-dependent scanning mechanism of initiation to produce toxic proteins. Mol. Cell. 2016; 62:314–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zamiri B., Reddy K., Macgregor R.B. Jr, Pearson C.E.. TMPyP4 porphyrin distorts RNA G-quadruplex structures of the disease-associated r(GGGGCC)n repeat of the C9orf72 gene and blocks interaction of RNA-binding proteins. J. Biol. Chem. 2014; 289:4653–4659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bugaut A., Rodriguez R., Kumari S., Hsu S.T., Balasubramanian S.. Small molecule-mediated inhibition of translation by targeting a native RNA G-quadruplex. Org. Biomol. Chem. 2010; 8:2771–2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Morris M.J., Basu S.. An unusually stable G-quadruplex within the 5΄-UTR of the MT3 matrix metalloproteinase mRNA represses translation in eukaryotic cells. Biochemistry. 2009; 48:5313–5319. [DOI] [PubMed] [Google Scholar]
- 84.Modelska A., Turro E., Russell R., Beaton J., Sbarrato T., Spriggs K., Miller J., Graf S., Provenzano E., Blows F. et al. The malignant phenotype in breast cancer is driven by eIF4A1-mediated changes in the translational landscape. Cell Death Dis. 2015; 6:e1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rubio C.A., Weisburd B., Holderfield M., Arias C., Fang E., DeRisi J.L., Fanidi A.. Transcriptome-wide characterization of the eIF4A signature highlights plasticity in translation regulation. Genome Biol. 2014; 15:476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bugaut A., Balasubramanian S.. 5΄-UTR RNA G-quadruplexes: translation regulation and targeting. Nucleic Acids Res. 2012; 40:4727–4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Halder K., Wieland M., Hartig J.S.. Predictable suppression of gene expression by 5΄-UTR-based RNA quadruplexes. Nucleic Acids Res. 2009; 37:6811–6817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Creacy S.D., Routh E.D., Iwamoto F., Nagamine Y., Akman S.A., Vaughn J.P.. G4 resolvase 1 binds both DNA and RNA tetramolecular quadruplex with high affinity and is the major source of tetramolecular quadruplex G4-DNA and G4-RNA resolving activity in HeLa cell lysates. J. Biol. Chem. 2008; 283:34626–34634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lattmann S., Giri B., Vaughn J.P., Akman S.A., Nagamine Y.. Role of the amino terminal RHAU-specific motif in the recognition and resolution of guanine quadruplex-RNA by the DEAH-box RNA helicase RHAU. Nucleic Acids Res. 2010; 38:6219–6233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Booy E.P., Meier M., Okun N., Novakowski S.K., Xiong S., Stetefeld J., McKenna S.A.. The RNA helicase RHAU (DHX36) unwinds a G4-quadruplex in human telomerase RNA and promotes the formation of the P1 helix template boundary. Nucleic Acids Res. 2012; 40:4110–4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Darnell J.C., Jensen K.B., Jin P., Brown V., Warren S.T., Darnell R.B.. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001; 107:489–499. [DOI] [PubMed] [Google Scholar]
- 92.Brown V., Jin P., Ceman S., Darnell J.C., O'Donnell W.T., Tenenbaum S.A., Jin X., Feng Y., Wilkinson K.D., Keene J.D. et al. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001; 107:477–487. [DOI] [PubMed] [Google Scholar]
- 93.Napoli I., Mercaldo V., Boyl P.P., Eleuteri B., Zalfa F., De Rubeis S., Di Marino D., Mohr E., Massimi M., Falconi M. et al. The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell. 2008; 134:1042–1054. [DOI] [PubMed] [Google Scholar]
- 94.Chen E., Sharma M.R., Shi X., Agrawal R.K., Joseph S.. Fragile X mental retardation protein regulates translation by binding directly to the ribosome. Mol. Cell. 2014; 54:407–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Phan A.T., Kuryavyi V., Darnell J.C., Serganov A., Majumdar A., Ilin S., Raslin T., Polonskaia A., Chen C., Clain D. et al. Structure-function studies of FMRP RGG peptide recognition of an RNA duplex-quadruplex junction. Nat. Struct. Mol. Biol. 2011; 18:796–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Vasilyev N., Polonskaia A., Darnell J.C., Darnell R.B., Patel D.J., Serganov A.. Crystal structure reveals specific recognition of a G-quadruplex RNA by a beta-turn in the RGG motif of FMRP. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E5391–E5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Subramanian M., Rage F., Tabet R., Flatter E., Mandel J.L., Moine H.. G-quadruplex RNA structure as a signal for neurite mRNA targeting. EMBO Rep. 2011; 12:697–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang Y., Gaetano C.M., Williams K.R., Bassell G.J., Mihailescu M.R.. FMRP interacts with G-quadruplex structures in the 3΄-UTR of its dendritic target Shank1 mRNA. RNA Biol. 2014; 11:1364–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Stefanovic S., DeMarco B.A., Underwood A., Williams K.R., Bassell G.J., Mihailescu M.R.. Fragile X mental retardation protein interactions with a G quadruplex structure in the 3΄-untranslated region of NR2B mRNA. Mol. Biosyst. 2015; 11:3222–3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Majumder M., House R., Palanisamy N., Qie S., Day T.A., Neskey D., Diehl J.A., Palanisamy V.. RNA-binding protein FXR1 regulates p21 and TERC RNA to bypass p53-mediated cellular senescence in OSCC. PLoS Genet. 2016; 12:e1006306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jin P., Warren S.T.. Understanding the molecular basis of fragile X syndrome. Hum. Mol. Genet. 2000; 9:901–908. [DOI] [PubMed] [Google Scholar]
- 102.Primerano B., Tassone F., Hagerman R.J., Hagerman P., Amaldi F., Bagni C.. Reduced FMR1 mRNA translation efficiency in fragile X patients with premutations. RNA. 2002; 8:1482–1488. [PMC free article] [PubMed] [Google Scholar]
- 103.Feng Y., Zhang F., Lokey L.K., Chastain J.L., Lakkis L., Eberhart D., Warren S.T.. Translational suppression by trinucleotide repeat expansion at FMR1. Science. 1995; 268:731–734. [DOI] [PubMed] [Google Scholar]
- 104.Khateb S., Weisman-Shomer P., Hershco I., Loeb L.A., Fry M.. Destabilization of tetraplex structures of the fragile X repeat sequence (CGG)n is mediated by homolog-conserved domains in three members of the hnRNP family. Nucleic Acids Res. 2004; 32:4145–4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Khateb S., Weisman-Shomer P., Hershco-Shani I., Ludwig A.L., Fry M.. The tetraplex (CGG)n destabilizing proteins hnRNP A2 and CBF-A enhance the in vivo translation of fragile X premutation mRNA. Nucleic Acids Res. 2007; 35:5775–5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ofer N., Weisman-Shomer P., Shklover J., Fry M.. The quadruplex r(CGG)n destabilizing cationic porphyrin TMPyP4 cooperates with hnRNPs to increase the translation efficiency of fragile X premutation mRNA. Nucleic Acids Res. 2009; 37:2712–2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Beaudoin J.D., Perreault J.P.. 5΄-UTR G-quadruplex structures acting as translational repressors. Nucleic Acids Res. 2010; 38:7022–7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sagne C., Marcel V., Bota M., Martel-Planche G., Nobrega A., Palmero E.I., Perriaud L., Boniol M., Vagner S., Cox D.G. et al. Age at cancer onset in germline TP53 mutation carriers: association with polymorphisms in predicted G-quadruplex structures. Carcinogenesis. 2014; 35:807–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Perriaud L., Marcel V., Sagne C., Favaudon V., Guedin A., De Rache A., Guetta C., Hamon F., Teulade-Fichou M.P., Hainaut P. et al. Impact of G-quadruplex structures and intronic polymorphisms rs17878362 and rs1642785 on basal and ionizing radiation-induced expression of alternative p53 transcripts. Carcinogenesis. 2014; 35:2706–2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Morten B.C., Wong-Brown M.W., Scott R.J., Avery-Kiejda K.A.. The presence of the intron 3 16 bp duplication polymorphism of p53 (rs17878362) in breast cancer is associated with a low Delta40p53:p53 ratio and better outcome. Carcinogenesis. 2016; 37:81–86. [DOI] [PubMed] [Google Scholar]
- 111.Mahdaviani S.A., Hirbod-Mobarakeh A., Wang N., Aghamohammadi A., Hammarstrom L., Masjedi M.R., Pan-Hammarstrom Q., Rezaei N.. Novel mutation of the activation-induced cytidine deaminase gene in a Tajik family: special review on hyper-immunoglobulin M syndrome. Expert Rev. Clin. Immunol. 2012; 8:539–546. [DOI] [PubMed] [Google Scholar]
- 112.Tellam J.T., Zhong J., Lekieffre L., Bhat P., Martinez M., Croft N.P., Kaplan W., Tellam R.L., Khanna R.. mRNA Structural constraints on EBNA1 synthesis impact on in vivo antigen presentation and early priming of CD8+ T cells. PLoS Pathog. 2014; 10:e1004423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Norseen J., Johnson F.B., Lieberman P.M.. Role for G-quadruplex RNA binding by Epstein-Barr virus nuclear antigen 1 in DNA replication and metaphase chromosome attachment. J. Virol. 2009; 83:10336–10346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mendoza O., Bourdoncle A., Boule J.B., Brosh R.M. Jr., Mergny J.L.. G-quadruplexes and helicases. Nucleic Acids Res. 2016; 44:1989–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Vohhodina J., Harkin D.P., Savage K.I.. Dual roles of DNA repair enzymes in RNA biology/post-transcriptional control. Wiley Interdiscip. Rev. RNA. 2016; 7:604–619. [DOI] [PubMed] [Google Scholar]
- 116.Dutertre M., Lambert S., Carreira A., Amor-Gueret M., Vagner S.. DNA damage: RNA-binding proteins protect from near and far. Trends Biochem. Sci. 2014; 39:141–149. [DOI] [PubMed] [Google Scholar]
- 117.Santos-Pereira J.M., Aguilera A.. R loops: new modulators of genome dynamics and function. Nat. Rev. Genet. 2015; 16:583–597. [DOI] [PubMed] [Google Scholar]
- 118.Wahba L., Gore S.K., Koshland D.. The homologous recombination machinery modulates the formation of RNA-DNA hybrids and associated chromosome instability. Elife. 2013; 2:e00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhang J.Y., Zheng K.W., Xiao S., Hao Y.H., Tan Z.. Mechanism and manipulation of DNA:RNA hybrid G-quadruplex formation in transcription of G-rich DNA. J. Am. Chem. Soc. 2014; 136:1381–1390. [DOI] [PubMed] [Google Scholar]
- 120.Cloutier S.C., Wang S., Ma W.K., Al Husini N., Dhoondia Z., Ansari A., Pascuzzi P.E., Tran E.J.. Regulated formation of lncRNA-DNA hybrids enables faster transcriptional induction and environmental adaptation. Mol. Cell. 2016; 61:393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Takahama K., Kino K., Arai S., Kurokawa R., Oyoshi T.. Identification of Ewing's sarcoma protein as a G-quadruplex DNA- and RNA-binding protein. FEBS J. 2011; 278:988–998. [DOI] [PubMed] [Google Scholar]
- 122.Takahama K., Oyoshi T.. Specific binding of modified RGG domain in TLS/FUS to G-quadruplex RNA: tyrosines in RGG domain recognize 2΄-OH of the riboses of loops in G-quadruplex. J. Am. Chem. Soc. 2013; 135:18016–18019. [DOI] [PubMed] [Google Scholar]
- 123.Thandapani P., O'Connor T.R., Bailey T.L., Richard S.. Defining the RGG/RG motif. Mol. Cell. 2013; 50:613–623. [DOI] [PubMed] [Google Scholar]
- 124.Bugaut A., Murat P., Balasubramanian S.. An RNA hairpin to G-quadruplex conformational transition. J. Am. Chem. Soc. 2012; 134:19953–19956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Stefanovic S., Bassell G.J., Mihailescu M.R.. G quadruplex RNA structures in PSD-95 mRNA: potential regulators of miR-125a seed binding site accessibility. RNA. 2015; 21:48–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.O'Day E., Le M.T., Imai S., Tan S.M., Kirchner R., Arthanari H., Hofmann O., Wagner G., Lieberman J.. An RNA-binding protein, Lin28, recognizes and remodels G-quartets in the microRNAs (miRNAs) and mRNAs it regulates. J. Biol. Chem. 2015; 290:17909–17922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mirihana Arachchilage G., Morris M.J., Basu S.. A library screening approach identifies naturally occurring RNA sequences for a G-quadruplex binding ligand. Chem. Commun. (Camb). 2014; 50:1250–1252. [DOI] [PubMed] [Google Scholar]
- 128.Wong H.M., Stegle O., Rodgers S., Huppert J.L.. A toolbox for predicting g-quadruplex formation and stability. J. Nucleic Acids. 2010; 2010:564946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kikin O., D'Antonio L., Bagga P.S.. QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res. 2006; 34:W676–W682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Scaria V., Hariharan M., Arora A., Maiti S.. Quadfinder: server for identification and analysis of quadruplex-forming motifs in nucleotide sequences. Nucleic Acids Res. 2006; 34:W683–W685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yadav V.K., Abraham J.K., Mani P., Kulshrestha R., Chowdhury S.. QuadBase: genome-wide database of G4 DNA–occurrence and conservation in human, chimpanzee, mouse and rat promoters and 146 microbes. Nucleic Acids Res. 2008; 36:D381–D385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhang R., Lin Y., Zhang C.T.. Greglist: a database listing potential G-quadruplex regulated genes. Nucleic Acids Res. 2008; 36:D372–D376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Beaudoin J.D., Jodoin R., Perreault J.P.. New scoring system to identify RNA G-quadruplex folding. Nucleic Acids Res. 2014; 42:1209–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Eddy J., Maizels N.. Gene function correlates with potential for G4 DNA formation in the human genome. Nucleic Acids Res. 2006; 34:3887–3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kikin O., Zappala Z., D'Antonio L., Bagga P.S.. GRSDB2 and GRS_UTRdb: databases of quadruplex forming G-rich sequences in pre-mRNAs and mRNAs. Nucleic Acids Res. 2008; 36:D141–D148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kostadinov R., Malhotra N., Viotti M., Shine R., D'Antonio L., Bagga P.. GRSDB: a database of quadruplex forming G-rich sequences in alternatively processed mammalian pre-mRNA sequences. Nucleic Acids Res. 2006; 34:D119–D124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Garant J.M., Luce M.J., Scott M.S., Perreault J.P.. G4RNA: an RNA G-quadruplex database. Database (Oxford). 2015; bav059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kwok C.K., Sahakyan A.B., Balasubramanian S.. Structural analysis using SHALiPE to reveal RNA G-quadruplex formation in human precursor microRNA. Angew. Chem. Int. Ed. Engl. 2016; 55:8958–8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Weldon C., Behm-Ansmant I., Hurley L.H., Burley G.A., Branlant C., Eperon I.C., Dominguez C.. Identification of G-quadruplexes in long functional RNAs using 7-deazaguanine RNA. Nat. Chem. Biol. 2016; 13:18–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Di Antonio M., Biffi G., Mariani A., Raiber E.A., Rodriguez R., Balasubramanian S.. Selective RNA versus DNA G-quadruplex targeting by in situ click chemistry. Angew. Chem. Int. Ed. Engl. 2012; 51:11073–11078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Katsuda Y., Sato S., Asano L., Morimura Y., Furuta T., Sugiyama H., Hagihara M., Uesugi M.. A small molecule that represses translation of G-Quadruplex-containing mRNA. J. Am. Chem. Soc. 2016; 138:9037–9040. [DOI] [PubMed] [Google Scholar]
- 142.Laguerre A., Hukezalie K., Winckler P., Katranji F., Chanteloup G., Pirrotta M., Perrier-Cornet J.M., Wong J.M., Monchaud D.. Visualization of RNA-quadruplexes in live cells. J. Am. Chem. Soc. 2015; 137:8521–8525. [DOI] [PubMed] [Google Scholar]
- 143.Laguerre A., Wong J.M., Monchaud D.. Direct visualization of both DNA and RNA quadruplexes in human cells via an uncommon spectroscopic method. Sci. Rep. 2016; 6:32141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Laguerre A., Stefan L., Larrouy M., Genest D., Novotna J., Pirrotta M., Monchaud D.. A twice-as-smart synthetic G-quartet: PyroTASQ is both a smart quadruplex ligand and a smart fluorescent probe. J. Am. Chem. Soc. 2014; 136:12406–12414. [DOI] [PubMed] [Google Scholar]
- 145.Chen S.B., Hu M.H., Liu G.C., Wang J., Ou T.M., Gu L.Q., Huang Z.S., Tan J.H.. Visualization of NRAS RNA G-quadruplex structures in cells with an engineered fluorogenic hybridization probe. J. Am. Chem. Soc. 2016; 138:10382–10385. [DOI] [PubMed] [Google Scholar]
- 146.Xu S., Li Q., Xiang J., Yang Q., Sun H., Guan A., Wang L., Liu Y., Yu L., Shi Y. et al. Thioflavin T as an efficient fluorescence sensor for selective recognition of RNA G-quadruplexes. Sci. Rep. 2016; 6:24793. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.