Abstract
In all free-living organisms a late-stage checkpoint in the biogenesis of the small ribosomal subunit involves rRNA modification by an RsmA/Dim1 methyltransferase. The hyperthermophilic archaeon Nanoarchaeum equitans, whose existence is confined to the surface of a second archaeon, Ignicoccus hospitalis, lacks an RsmA/Dim1 homolog. We demonstrate here that the I. hospitalis host possesses the homolog Igni_1059, which dimethylates the N6-positions of two invariant adenosines within helix 45 of 16S rRNA in a manner identical to other RsmA/Dim1 enzymes. However, Igni_1059 is not transferred from I. hospitalis to N. equitans across their fused cell membrane structures and the corresponding nucleotides in N. equitans 16S rRNA remain unmethylated. An alternative mechanism for ribosomal subunit maturation in N. equitans is suggested by sRNA interactions that span the redundant RsmA/Dim1 site to introduce 2΄-O-ribose methylations within helices 44 and 45 of the rRNA.
INTRODUCTION
The rRNAs of all living organisms contain a variety of nucleotide modifications that are added post-transcriptionally and are collectively essential for the process of protein synthesis (1,2). Isomerization of uridines and 2΄-O-ribose methylations are catalyzed by enzyme complexes that are guided to rRNA nucleotides by archaeal sRNAs (3,4) and eukaryotic snoRNAs (5–7). Modifications on the nucleobases, however, are added by enzymes that require no such assistance from small RNA molecules to find their specific nucleotide targets, and this holds true for the enzymes that modify bacterial rRNAs (1,8). The bacterial methyltransferase RsmA (formerly KsgA) and its eukaryotic counterpart Dim1 catalyze N6,N6-dimethylation at two invariant adenosines (A1518 and A1519 in Escherichia coli, Figure 1A) in the loop of helix 45 at the 3΄-end of the small ribosomal subunit RNA (9). The phylogenetic conservation of RsmA indicates that it belongs to the core proteins necessary for cellular life (10,11) and is a requisite part of the minimal translation apparatus (12).
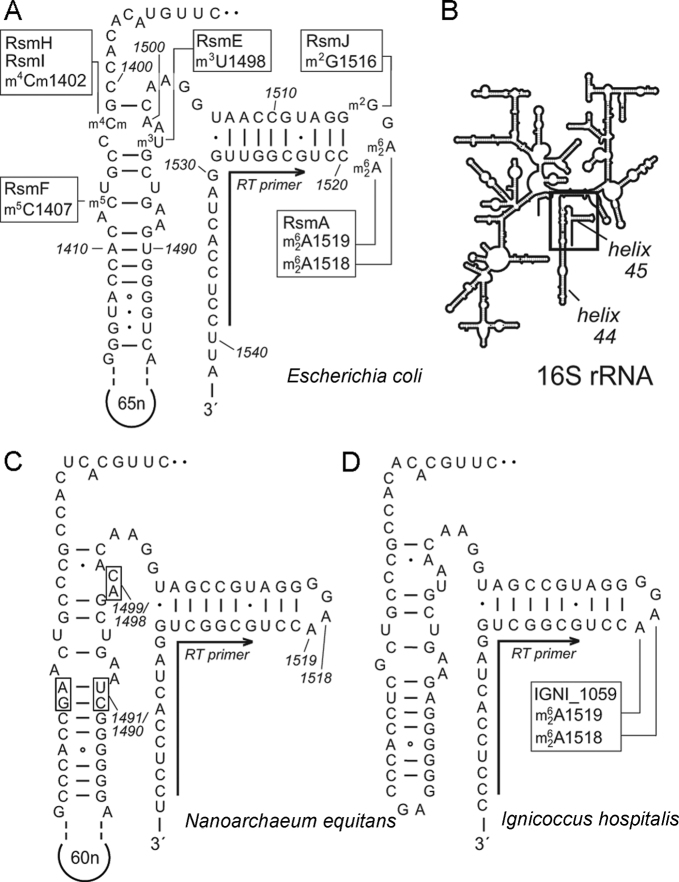
Figure 1.
(A) Secondary structures of helices 44 and 45 in 16S rRNA from Escherichia coli showing the sites of nucleotide methylation and the enzymes involved. These include RsmA, which N6, N6-dimethylates A1518 and A1519. All nucleotide annotations here use the E. coli numbering system. (B) The relative positions of helices 44 and 45 depicted on the 16S rRNA secondary structure schematic. (C) The corresponding structures in Nanoarchaeum equitans and (D) Ignicoccus hospitalis 16S rRNAs show a high degree of conservation in and around helix 45. Nucleotide variations that were used to distinguish and isolate the archaeal sequences are shown on the N. equitans structure (boxes and the 60 nucleotide extension in helix 44). A single oligonucleotide was used for reverse transcriptase (RT) primer extension reactions on all three rRNA, and hybridizes as indicated (arrows) to the conserved sequence at the 3΄-side of helix 45.
Despite its high conservation, RsmA can be inactivated and loss of this function confers resistance to the antibiotic kasugamycin (13). As a side effect of gaining resistance, E. coli rsmA-null mutants are prone to decoding errors (14), exhibit impaired growth, a cold-sensitive phenotype and an overall loss of fitness (15). Some of these deleterious traits are connected with the removal of a late-stage checkpoint in biogenesis of the 30S ribosomal subunit (15,16), and similar effects have been seen in a range of Gram-positive and Gram-negative pathogens (17–19) where RsmA inactivation is accompanied by loss of virulence (20,21). RsmA is also conserved in plant chloroplasts, and loss of this function in Arabidopsis hinders growth at low temperature (22). In eukaryotes as phylogenetically distant as Saccharomyces cerevisiae (23) and human cell lines (24), inactivation of the RsmA homolog Dim1 causes build-up of unprocessed 18S rRNAs in precursors of cytoplasmic ribosomes and is generally a lethal event. In mitochondria, where homologs of RsmA/Dim1 are nuclear-encoded (25,26), adenosine dimethylation by the human homolog h-mtTFB1 is essential for assembly of the small mitoribosomal subunit (27).
Although RsmA/Dim1 homologs play important roles in all free-living organisms and most organelles (28), at the time of writing there are eight instances in the data banks of organelles/organisms that lack a gene for RsmA. Three of these cases, Paranosema grylli (29), Carsonella ruddii (30) and Nasuia deltocephalinicola (31), are bacteria with highly reduced genomes and are obligate intracellular parasites of insects; in two further cases, organellar RsmA is absent in S. cerevisiae (32) and in Euglena gracilis (11); and the three remaining cases are the current members of the hyperthermophilic archaeal phylum of Nanoarchaeota comprising Nanobsidianus stetteri (Nst1) (33), Nanopusillus acidilobi (34) and Nanoarchaeum equitans (35). It is presently unclear how such organisms/organelles compensate for the lack of RsmA/Dim1 during the assembly and activation of their ribosomes.
We have addressed this question in N. equitans, which grows as an obligate commensalist/parasite on its archaeal host Ignicoccus hospitalis. The I. hospitalis host grows at more or less the same rate as a pure culture or in a co-culture with N. equitans, and propagates within the temperatures range 73–98°C (35). The I. hospitalis genome contains almost 1500 open reading frames (ORFs) (36) and includes the RsmA homolog, Igni_1059. The genome of N. equitans is considerably smaller, encoding only 552 ORFs (37) without any obvious RsmA homolog. Due to its severely reduced genetic content, the continuance of N. equitans necessitates the transfer of multiple metabolic components from I. hospitalis through their system of fused membranes (35,38). Molecules larger than metabolites are probably not transferred between the cells (39), although N. equitans cells retain some contamination from I. hospitalis components after isolation making it less than straightforward to draw unambiguous conclusions about protein transfer. Thus at the inception of this study, it was unclear whether N. equitans borrows an RsmA-homolog from I. hospitalis or whether a fundamentally different mechanism of ribosome maturation might be used.
The Igni_1059 enzyme is shown here to act as a true RsmA homolog that N6, N6-dimethylates the two conserved adenosine in helix 45 of 16S rRNA. Igni_1059 retains this function when transferred to heterologous cells, as demonstrated here by the enzyme's ability to complement an rsmA-null mutant of E. coli. However, the helix 45 adenosines in the N. equitans rRNA remain unmodified and thus transfer of Igni_1059 from I. hospitalis could be unequivocally ruled out. A comprehensive map of the other types of modification at the 3΄-end of the 16S rRNAs indicates how sRNA interactions that direct 2΄-O-ribose methylation in this region could represent an alternative means of guiding maturation of the small ribosomal subunit in N. equitans and other nanoarchaea.
MATERIALS AND METHODS
In silico analysis
The KgsA/RsmA protein from E. coli K12 with UniProt accession number (acc. nr.) P06992 and homologs from Pyrococcus yayanosii (WP_013904865.1) and Methanocaldococcus sp. FS406-22 (WP_012981219.1) were used as BLAST (40) queries against ORFs in I. hospitalis and N. equitans. In each case, Igni_1059 (acc. nr. A8ABD5) was identified in I. hospitalis as having the lowest E-value (8 × 10−25 against the E. coli RsmA). No comparable ORF was evident in N. equitans, where the closest hit was NEQ337 (acc. nr. Q74MB4) with a significantly higher E-value of 8 × 10−2. E. coli rRNA methyltransferases (8) were used as queries to screen the N. equitans genome for homologs catalyzing 5-methylpyrimidine modification.
Growth, cloning and RNA purification
Ignicoccus hospitalis was grown in pure culture and in co-culture with N. equitans as previously described (35) and cells were harvested at late log phase. Strains of E. coli were grown in LB medium (41) at 37°C. The wild-type E. coli strain BW25113 was grown without antibiotic, and the Keio collection ΔrsmA knockout strain JW0050 (42) was cultured with kanamycin at 50 μg/ml. E. coli strains transformed with derivatives of plasmid pLJ102 (43) were grown in medium containing 1mM IPTG and ampicillin at 50 μg/ml.
The archaeal genes NEQ337 and Igni_1059 were amplified using the PCR forward primer 5΄-TCATCATTTCATATGGTATTACCTTT and reverse primer 5΄AATAGCTGATCATAAGAAGGTTTAG for NEQ337, and the forward primer 5΄-AGGGGTTCATATGCTGAAGTGGA and reverse primer 5΄-GAATAAAGTGATCACGCTCCTCCC for Igni_1059. Both the forward and both reverse primers contain restriction sites for NdeI and BclI, respectively, for inserting into the expression plasmid pLJ102 (44). RNA from E. coli was purified as described previously (45). Archaeal RNA was extracted by resuspending 0.5 g of pelleted cells in 2.5 ml of 50 mM Tris-Cl, pH 7.5, 10 mM MgCl2, 100 mM NH4Cl. The cells were run twice through a French press, followed by addition of 4 ml of TRIzol® (Life Technologies). After 5 min incubation at room temperature, 0.8 ml chloroform was added. The samples were centrifuged at 12,000 g for 5 min and the aqueous phase was transferred to a new tube for extraction with phenol/chloroform.
Primer extension
A total of 4 pmol Cy5-labeled primer 5΄-(Cy5)GGAGGTGATCCAACCGC (Figure 1) was hybridized to 4 pmol RNA by incubating at 80°C for 2 min in 4.5 μl of 56 mM HEPES pH 7.0 and 112 mM KCl followed by slow cooling to 42°C. Extension and sequencing reactions were performed with 1.5 U avian myelobalstosis virus (AMV) reverse transcriptase (Thermo Scientific) per reaction (46) and analysed on 13% polyacrylamide-urea gels. Bands were visualized on a Typhoon FLA 9500 (GE Healthcare).
Isolation of defined rRNA sequence and analysis by matrix assisted laser desorption-ionization time-of-flight (MALDI-ToF) mass spectrometry
DNA oligodeoxynucleotides were designed to hybridize to the E. coli and two archaeal 16S rRNAs encompassing helix 45 and surrounding nucleotides. About 1 nmol of the oligo complementary to the E. coli sequence (5΄-AAGGAGGTGATCCAGCCGCAGGTTCCCCTACGGTTACCTTGTTACGACTTCACCC), the N. equitans sequence (5΄-GGAGGTGATCCAGCCGCAGGTTCCCCTACGGCTACCTTGTGTCGACTT) or the I. hospitalis sequence (5΄-GGAGGTGATCCAGCCGCAGGTTCCCCTACGGCTACCTTGTTACGACTTCTCCCCCCTC) were mixed with 300 pmol of the respective rRNAs in 62.5 mM HEPES, pH 7 and 125 mM KCl and heated at 90°C for 5 min and slowly cooled to 37°C. After hybridization, rRNAs were digested with RNase A and mung bean nuclease and protected rRNA fragments of approximately the same length as the oligos were isolated on denaturing gels (47,48). Nuclease digestion at the mismatched sequences (underlined bold above) facilitated separation of I. hospitalis molecules from the I. hospitalis/N. equitans rRNA mixture. An additional oligo complimentary to nucleotides 1390–1446 on the 5΄-side of N. equitans helix 44 (Figure 5) was used to isolate this rRNA sequence which differs dramatically from the corresponding structure in I. hospitalis (Figure 1). The rRNA samples were fragmented further with RNase A or T1, and loaded onto MALDI target plates precooled to 4°C. After crystallization of the samples, the plates were gently heated to evaporate any excess water. MALDI-TOF mass spectrometry data was collected in positive mode (UltrafleXtreme, Bruker) (47,48).
Figure 5.
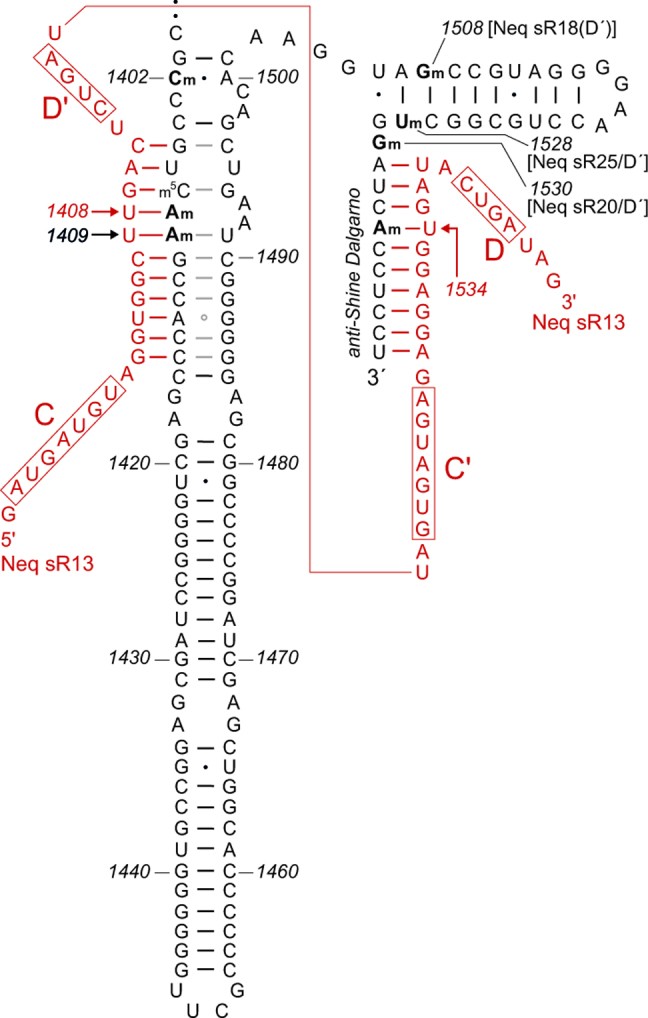
Sites of 2΄O-ribose methylation within helices 44 and 45 of Nanoarchaeum equitans 16S rRNA, showing the complete sequence of this region and the sRNA Neq sR13 (red). Methylation within the decoding site at Am1408 (and presumably also at neighboring Am1409) is directed by the D΄ box of Neq sR13, while methylation at Am1534 within the anti-Shine Dalgarno sequence is directed by the D box of the same sRNA. The Gm1508 methylation is guided by the D΄ box of Neq sR18; Um1528 by the D΄ box of Neq sR25; and Gm1530 by the D΄ box of Neq sR20 with the D box of the same sRNA guiding the methylation at Cm1920 in 23S rRNA. No obvious sRNA candidate for the Cm1402 methylation was apparent in the N. equitans genome or RNA-seq data. The sRNA sequences have previously been reported by Dennis et al. (55).
Identification of ribose methylations by RiboMeth-seq
Archaeal RNAs from co-cultures of I. hospitalis/N. equitans were extracted as above and subjected to RiboMeth-seq analysis based on high-throughput sequencing (49) to establish the positions and approximate stoichiometries of 2΄-O-methylations. Briefly, 20–30 μg of RNA were partially degraded in 50 mM NaHCO3/Na2CO3 (pH 9.9) at 90°C for 6 min and fragments of 20–40 nucleotides were purified on 8% denaturing polyacrylamide gels. Adapters were affixed to the fragments by means of a co-transcribed HDV ribozyme and a modified Arabidopsis thaliana tRNA ligase specific for 2΄,3΄-cyclic phosphates to facilitate reverse transcription and sequenced on the Ion Proton sequencing platform. The adapters were trimmed from the reads using Cutadapt v1.2.1, and the remaining sequences were mapped to the I. hospitalis/N. equitans 16S and 23S rRNA sequences using Bowtie 2 v2.1.0. The 5΄- and 3΄-read ends were counted for each position and were scored on the basis of the read counts (49).
RESULTS
Heterologous expression of N. equitans and I. hospitalis methyltransferases in E. coli
In silico analyses of the genome structures indicated that the Igni_1059 gene in I. hospitalis was a probable rsmA homolog. There was no obvious homolog of this gene in N. equitans, and the closest, albeit distantly related, candidate was NEQ337 where similarity to rsmA was limited to the binding region for the methyl donor, S-adenosyl methionine. These genes and an active copy of the E. coli rsmA were inserted into plasmids and expressed in a wild-type and an rsmA-null mutant of E. coli. Dimethylation at the N6-position of adenosine halts the progress of AMV reverse transcriptase (50), and results in gel bands corresponding to the modified A1518 and A1519 nucleotides (Figure 2). Quantification of the gel bands indicated that the RsmA enzyme almost completely dimethylates its adenosine targets in the E. coli wild-type strain, and no further increase in modification was achieved by over-expressing the enzyme from an extra copy of the rsmA gene. The dimethylation stop bands were absent for rRNA from the rsmA-null strain, and were fully re-established by complementation with an active copy of rsmA (Figure 2).
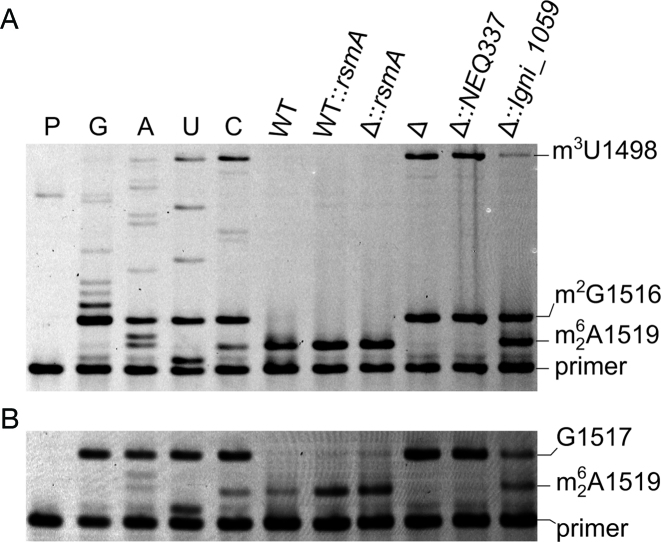
Figure 2.
Primer extension on rRNAs from Escherichia coli strains. (A) The dideoxy sequencing reactions (C, U, A and G) were carried out on rRNA from the rsmA null-mutant (Δ). Extension reactions were on rRNA from the wild-type (WT) strain; the wild-type strain with an extra, plasmid-encoded copy of the E. coli rsmA gene (WT::rsmA); the rsmA null-mutant with plasmid-encoded rsmA (Δ::rsmA); the uncomplemented rsmA null-mutant (Δ); the rsmA null-mutant with plasmid-encoded NEQ337 (Δ::NEQ337); and the rsmA null-mutant with plasmid-encoded Igni_1059 (Δ::Igni_1059). Adenosine dimethylation at A1518/1519 halts reverse transcription; the m2G1516 modification is in all the E. coli strains and causes pausing; the m3U1498 modification (also present in all strains) causes a complete stop. P, primer only. (B) Primer extension on the same rRNA samples replacing dCTP with ddCTP in all reactions to obtain a complete stop at G1517 against which the bands for dimethylated A1518 and A1519 were quantified. Under the growth conditions used here, Igni_1059 dimethylated approximately 50% of the E. coli nucleotides A1519 and/or A1518. These nucleotides were dimethylated to >95% by E. coli's own RsmA enzyme (WT), and no further increase in modification was achieved by over-expressing an extra copy of the rsmA gene (WT::rsmA).
Expression of the recombinant Igni_1059 gene in the rsmA-null strain revealed that this was indeed an rsmA homolog encoding an enzyme that dimethylates the same E. coli 16S rRNA adenosines. Dimethylation at A1518 and A1519 was somewhat less (∼50%) than that achieved by the authentic E. coli enzyme (Figure 2B), although it should be noted that the Igni_1059 enzyme was operating at more than 50°C below its customary temperature. The recombinant Nanoarchaeum enzyme NEQ337 did not generate a stop at A1518 or at A1519 consistent with its low sequence similarity to RsmA.
The primer extension method has certain limitations, and the lack of read-through past dimethylated A1519 prevents quantification of the methylation status of A1518. Furthermore, this method cannot be used for accurate estimations of N6-adenosine monomethylation (50). These gaps in the data were filled by analysis of the modified rRNA region using MALDI-ToF mass spectrometry. The 16S rRNA fragment AGGGGAACp encompassing the loop of helix 45 was found to have an m/z of 2761.7 (mass in Daltons plus one proton) in the wild-type E. coli strain (Figure 3A). The unmodified RNA fragment with this sequence has an m/z of 2691.5, and the 70 Da difference in mass corresponds to five methyl groups, one of which is at m2G1516 (Figure 1A) with the remaining four on the N6-positions of A1518 and A1519. The wild-type E. coli rRNA was thus almost fully methylated with only a small component of unmodified rRNA. The rsmA-null mutant produced a minor unmethylated rRNA peak at m/z 2691 and a second, larger peak at m/z 2705 that contained the m2G1516 modification. This pattern remained unchanged upon expression of NEQ337 in the E. coli null mutant (Figure 3B), consistent with the sequence comparisons indicating that NEQ337 does not belong to the RsmA/Dim1 group of methyltransferases. BLAST searches show that NEQ337 is closely related to tRNA m1A methyltransferases, most notably that from Methanolobus tindarius (W9DVZ5, with an E-value of 10−20).
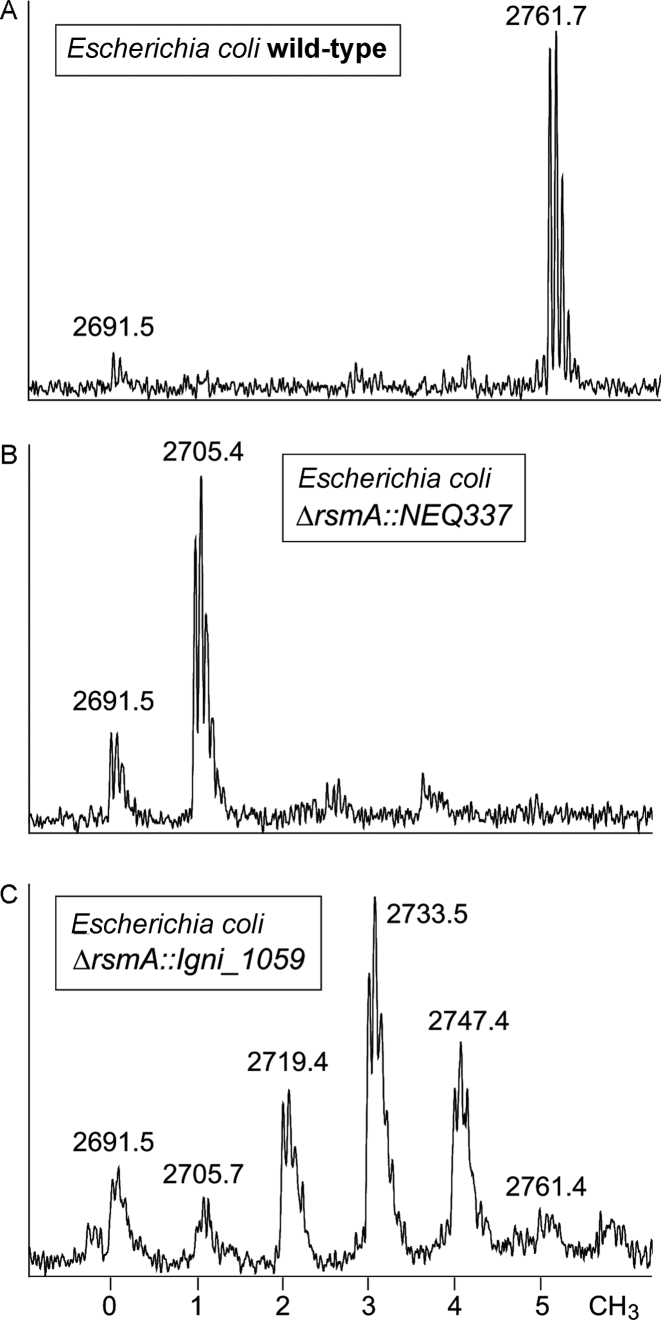
Figure 3.
Mass spectral region for the RNase A fragment AGGGGAACp from the loop of 16S rRNA helix 45 (nucleotides 1513–1520) in the E. coli derivatives. The number of methyl groups added to this sequence is indicated at the bottom of the figure. (A) The wild-type E. coli rRNA fragment at m/z 2761 is stoichiometrically modified with five methyl groups. (B) The spectrum from the E. coli rsmA-null mutant contains a single methyl group at m2G1516 in the m/z 2705 peak, and this pattern remained unchanged upon expression of NEQ337. (C) The E. coli rsmA-null mutant expressing Igni_1059 gave rise to a series of peaks from mono- and dimethylation intermediates at A1518 and A1519.
After complementation of the E. coli null mutant with the Igni_1059 gene, a more complex spectral pattern was evident with an array of peaks at m/z 2691, 2705, 2719, 2733, 2747 and 2761 (Figure 3C). These represent a small amount of the unmethylated AGGGGAACp fragment, and fragments with one methyl group at m2G1516 and one to four extra methyl groups added at A1518/A1519. The data are consistent with the primer extension assays (Figure 2) showing that Igni_1059 is a functional RsmA homolog, whose catalytic reaction had not gone to completion under the sub-optimal temperature conditions.
Analysis of I. hospitalis and N. equitans 16S rRNAs for adenosine dimethylation
Having established that I. hospitalis has a functional RsmA homolog and that N. equitans has none, we then addressed the questions of the extent to which Igni_1059 is expressed in its natural setting within I. hospitalis and whether the enzyme is passed over to N. equitans. Primer extension on rRNA from pure cultures of I. hospitalis cells halted almost completely at positions A1518/A1519 indicating that these adenosines were close to fully dimethylated (E. coli rRNA numbering is used throughout for the archaeal sequences). However, there was considerable reverse transcriptase read-through past these nucleotides using rRNA template mixtures obtained from co-cultures of N. equitans and I. hospitalis (Supplementary Figure S1). The sequence of helix 45 is identical in N. equitans and I. hospitalis (Figure 1), but diverges in helix 44. The rRNA species could therefore be readily differentiated, showing that read-though past A1518/A1519 was solely due to extension on N. equitans rRNA (Supplementary Figure S1).
This result, while demonstrating that N. equitans contains 16S rRNA lacking dimethylation at A1518/A1519, did not rule out the possible existence of a subset of N. equitans 16S rRNA molecules that possessed these modifications. Therefore, to sample the entire complement of 16S rRNA molecules from each organism, DNA oligonucleotides specific for N. equitans or I. hospitalis rRNAs were hybridized to 50–60 nucleotides extending across helices 44 and 45 and any unprotected mismatched sequences were clipped with nucleases. This approach enabled us to fish out fragments of specific lengths for each of the two organisms. The mass spectrum of the N. equitans rRNA fragment showed that it was virtually free from contaminating I. hospitalis rRNA, and vice versa (Figure 4). The helix 45 loop in N. equitans 16S rRNA contained in the AGGGGAACp fragment was devoid of modification (Figure 4C), whereas the corresponding rRNA region from I. hospitalis was almost fully dimethylated at both adenosines (Figure 4F). Thus, Igni_1059 functions effectively in I. hospitalis and is not transferred to substitute for the lack of a homologous enzyme in N. equitans. From this observation it can be inferred that trafficking from I. hospitalis to N. equitans of other molecules as big as or larger than the 27 kDa Igni_1059 is highly unlikely.
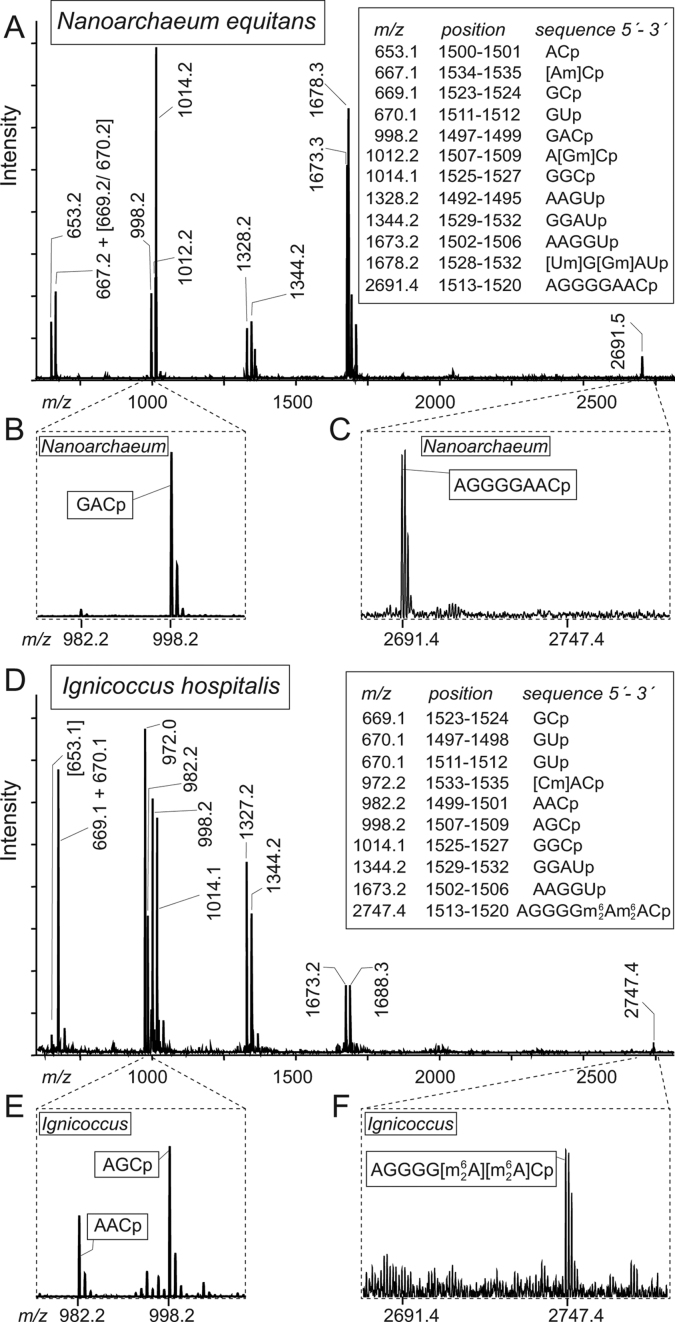
Figure 4.
Analyses of Nanoarchaeum equitans and Ignicoccus hospitalis 16S rRNA fragments containing helix 45. (A) Mass spectrum of RNase A fragments generated from the N. equitans 16S rRNA sequence from nucleotides 1492–1539. The mass/charges (m/z) measured are given above the peaks and matched the theoretical values (box) to within 0.2 Da. (B) Enlargements of the spectrum show that the N. equitans 16S rRNA is virtually clear of I. hospitalis contamination that would give a distinctive AACp fragment at m/z 982.2; and (C) the N. equitans helix 45 loop adenosines are in the fragment AGGGGAACp that flies at m/z 2691.5, indicating that this sequence contains no modification. In addition, several 2΄-O-methylations were evident in this region of the N. equitans 16S rRNA and these are analyzed in detail in Supplementary Figures S2 and S3. (D) The full spectrum from the helix 45 region of 16S rRNA from a pure culture of I. hospitalis. Here, (E) the distinctive AACp fragment is evident, as is (F) the 56 Da increase in mass of the AGGGGAACp fragment indicating that A1518 and A1519 are dimethylated. The additional peaks at m/z 1327 and 1688 arise from modified sequences (Supplementary Figure S3).
Additional methylations within 16S rRNA helices 44 and 45
The MALDI spectra revealed a series of 2΄-O-ribose methylations in the regions neighboring helix 45 (Figure 4). These were evident from the masses of certain RNA fragments and also from the protection against nuclease digestion afforded by 2΄-O-methylation. Such effects can be seen in the N. equitans fragments A(Gm)Cp (nucleotides 1507–1509), (Um)G(Gm)AUp (1528-1532) and (Am)Cp (1534-1535) (Figure 4A, with expanded views of these spectral regions in Supplementary Figure S2). The Gm1508 modification was confirmed in the primer extension analysis of helix 45 (Supplementary Figure S1) by reducing the concentration of dCTP to cause preferential pausing at 2΄-O-methyl guanosine (51).
The neighboring helix 44 has an extended structure in N. equitans compared to I. hospitalis (Figures 1 and 5) and this sequence was isolated and screened by MALDI-MS. Stoichiometric 2΄-O-ribose methylation was observed at N. equitans Am1408 and Am1409 in the RNase A fragment (Am)(Am)GCp from nucleotides 1408–1411 (Supplementary Figure S2B). These modifications were confirmed in the overlapping RNase T1 fragment UC(Am)(Am)Gp (nucleotides 1406–1410), which additionally contained a third methylation localized to the base of U1406 or C1407 (Supplementary Figure S2C). This modification hindered neither RNase cleavage nor primer extension with reverse transcriptase, and is thus consistent with pyrimidine C-5 methylation (48). Some bacteria including E. coli possess the methyltransferase RsmF that modifies m5C1407 (Figure 1A) and N. equitans possesses a homolog of this enzyme, NEQ536 (accession # Q74MV4) with an E-value of 4 × 10−62 compared to E. coli RsmF.
Not all nucleotides were captured in the MALDI-MS analyses with mono- and dinucleotide RNase fragments being particularly elusive (48). We therefore employed an additional analytical procedure involving high-throughput sequencing of the rRNAs (RiboMeth-seq) to check the positions and stoichiometries of 2΄-O-methylations (49). This method confirmed the findings from the MALDI-MS and primer extension experiments for 2΄-O-methylation at N. equitans 16S rRNA nucleotides Am1408, Am1409, Gm1508, Um1528, Gm1530 and Am1534; in addition, 2΄O-methylation was also found on nucleotide Cm1402 (Supplementary Figure S3). Fragments containing nucleotide 1402 were not observed in the MALDI mass spectra suggesting that they had been displaced from the expected mass range by nucleobase modification; sequence ambiguity at G1401 detected in the RiboMeth-seq analyses suggest that this guanine is modified. In the I. hospitalis rRNA, 2΄-methylation was similarly observed at positions 1408, 1409 and 1528, while other sites at Um1380, Cm1397, Gm1505, Gm1527 and Cm1533 revealed a distinctly different modification pattern to that seen in the N. equitans rRNA (Supplementary Figure S3).
DISCUSSION
The bacterial RsmA and eukaryotic Dim1 enzymes are believed to act primarily in quality control mechanisms that prevent immature small ribosomal subunits from entering the translation cycle (15,28). This essential enzymatic function has been maintained in most eukaryotic organelles including mammalian mitochondria (27), and also in mollicutes bacteria with parasitic lifestyles which despite having undergone massive genome reduction have retained their RsmA methyltransferase (12). Further reduction in the size of such genomes using synthetic biology approaches has shown that cells lacking rsmA remain viable albeit with a slower growth rate and a requirement for optimized culturing conditions (52). Taken together these observations suggest that there is a clear selective advantage for organisms to retain rsmA even when undergoing extensive genome reduction.
RsmA function is, however, completely absent in N. equitans despite living under the same extreme physiological conditions as its archaeon host, I. hospitalis, which uses the RsmA homolog Igni_1059 as a conventional means for rRNA methylation and supposedly also for ribosome maturation. Intriguingly, the structures of helices 44 and 45 in N. equitans, with stable G-C rich stems respectively capped with UUCG and GGAA tetraloops (Figure 5) would presumably, if left unhindered, fold rapidly and stably after transcription. It can be envisioned some means of modulating the folding of this rRNA region is required to facilitate the production of functional N. equitans ribosomal subunits.
We suggest that an RNA-guided mechanism in N. equitans substitutes for the protein (RsmA/Dim1)-directed checkpoint seen in other organisms (15,24,53). This process would be an integral part of the archaeal mechanism for 2΄-O-ribose methylation where C/D box sRNAs direct a complex of L7Ae/Nop5/fibrillarin proteins to specific rRNA nucleotides (54). The proteins of the fibrillarin complex and sRNAs are abundantly expressed in N. equitans (39,55), and here we draw attention to the sRNA Neq sR13. The 5΄-region of Neq sR13 is complementary to N. equitans 16S rRNA nucleotides 1405–1415 with the D΄ guiding methylation at A1408, while the 3΄-region is complementary to nucleotides 1531–1540 and the D box directs methylation to A1534 (Figure 5). In the comparable, and well-characterized, sRNA complexes of the hyperthermophilic archaeon Pyrococcus furiosus, methylation occurs sequentially with D box-guided modification being dependent on prior modification at the D΄ box (56). This would correspond to modification and release of the N. equitans A1408 region from the sRNA complex prior to modification at A1534. Sequential release of the helix 44/45 regions from Neq sR13, possibly coordinated with the binding of other sRNAs in this region (Figure 5), could create a series of temporal windows enabling the various stages in ribosome assembly to occur in a coordinated fashion.
A similar series of events can be envisaged in a second nanoarchaeon, N. acidilobi, for which the genome sequence has recently been published (34). N. acidilobi also clearly lacks a homolog of rsmA, and the inferred structure its16S rRNA helices 44 and 45 together with a matching sRNA are illustrated in Supplementary Figure S4. The putative sRNA–rRNA interactions indicate that 2΄-O-methylation would be directed via the D΄-box initially to nucleotide A1408 in helix 44, and then to nucleotide G1508 in helix 45 (Supplementary Figure S4) potentially guiding the folding of the 3΄-end of the N. acidilobi 16S rRNA.
While the presence of a functional RsmA homolog in I. hospitalis does not preclude 2΄-O-methylation at some of the equivalent nucleotide positions, the sRNA interactions are distinctly different from those in N. equitans and N. acidilobi. Modification at I. hospitalis position Gm1408 is guided by the D box of the sRNA Iho sR56, while the box D΄ sequence of this sRNA directs methylation to nucleotide U1380. Methylation in the I. hospitalis anti-Shine Dalgarno sequence is shifted one nucleotide to C1533 and involves a presently unknown sRNA. The Gm1508 modification identified in N. equitans (Figure 5) and proposed for N. acidilobi (Supplementary Figure S4) is absent in I. hospitalis (Supplementary Figure S3).
Nucleotide 1408 and its neighboring nucleotide 1409 are of particular functional interest, and both of these are 2΄-O-methylated in N. equitans and I. hospitalis (Supplementary Figure S3). Nucleotides 1408 and 1409 abut the universally conserved adenosines A1492 and A1493 (Figure 1), which are essential components of the ribosomal decoding site (57). Methylation at the 2΄-O-positions of N. equitans Am1408 and Am1409 is stoichiometric, as shown by MALDI-MS (Supplementary Figure S2) and RiboMeth-seq methods (Supplementary Figure S3), and these modifications undoubtedly play a role in modulating the N. equitans decoding site. Methylation at the corresponding nucleotides Gm1408 and Cm1409 in the I. hospitalis 16S rRNA (Supplementary Figure S3) suggests that such modifications play an integral part in the decoding process, and the lack of base conservation at nucleotides 1408 and 1409 indicates that their main contribution to the decoding process is made through their methylated minor groove edges.
The mechanism by which 2΄-O-methylation is directed to nucleotide 1409 remains unclear. The canonical site of sRNA-guided methylation is the fifth nucleotide 5΄ to the D/D΄ box (55,58,59), in this case nucleotide 1408, and we have not uncovered independent sRNAs that might be responsible for the adjacent 1409 modifications. The most straightforward interpretation would be that the D΄ box of Neq sR13 guides methylation at both 1408 and 1409 in N. equitans, and the Iho sR56 D box directs the same pair of modifications in I. hospitalis. In both species, modification at nucleotide 1409 occurs to the same extent as at 1408 (Supplementary Figure S3), which would suggest that both methylations are specific, rather than being caused by random slippage of the modification complex on the rRNA. Determining whether N. acidilobi has the same pattern and mechanism of 1408/1409 methylation awaits empirical analysis of its 16S rRNA (Supplementary Figure S4).
From the recently published sequences (55), sRNAs could be assigned for most of the other sites of 2΄-O-methylation within the N. equitans and I. hospitalis helix 44/45 regions. However, there are additional exceptions including the Cm1402 methylation (Figure 5) for which no suitable sRNA candidate was detected in N. equitans. The D΄ box of Iho sR11 could direct Cm1402 methylation in I. hospitalis, although the level of methylation at this nucleotide (<50%) was below the cut-off for inclusion here (Supplementary Figure S3E).
Interaction of the sRNA components of the 2΄-O-modification complexes have previously been proposed to act as chaperones to facilitate folding of archaeal rRNAs by bringing together regions that are distant from each other in the primary structure (55,60). While such mechanisms are distinct from the role proposed here for the retarding of 16S rRNA folding by Neq sR13, we do see sRNAs that might assist long-range rRNA interaction. For instance, the D΄ box of Neq sR20 directs the Gm1530 methylation adjacent to 16S rRNA helix 45 while the D box of the same sRNA guides methylation at Cm1920 within helix 69 of 23S rRNA. We have confirmed the Cm1920 modification by both mass spectrometry and Ribo-Meth-seq approaches (data not shown). The stage of ribosomal subunit maturation at which these modifications are added is unclear, although it is perhaps worthy of note that these 16S and 23S rRNA helices come into close proximity across the subunit interface upon 30S–50S association to form translationally active ribosomes (61,62).
In conclusion, we show here that the 3΄-ends of the 16S rRNA are heavily modified in the hyperthermophilic archaea N. equitans and I. hospitalis. In addition to the extensive list of 2΄-O-methylations shown in Supplementary Figure S3, base modifications including m5C1407 and at, or adjacent to, C1402 are present in this region. Such modifications are undoubtedly important for maintaining rRNA structures at high temperature (63,64), and we propose here that they also play roles in subunit folding and maturation. Our modification data and the implied involvement of sRNA Neq sR13 suggest that the protein-based checkpoint for small ribosomal subunit maturation found in most organisms could be carried out by an RNA-based mechanism in N. equitans and other Nanoarchaeota.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Claus Asger Lykkebo for help with the figures.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Danish Research Agency [FNU-rammebevilling 10-084554 to S.D.]; Deutsche Forschungsgemeinschaft [Förderkennzeichen HU703/2 to H.H.]; Danish Council for Independent Research/Technology and Production Sciences [FTP-bevilling 10-34611001 to H.N.]. Funding for open access charge: Grant [SDU33086 to S.D.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Decatur W.A., Fournier M.J.. rRNA modifications and ribosome function. Trends Biochem. Sci. 2002; 27:344–351. [DOI] [PubMed] [Google Scholar]
- 2.Grosjean H. Fine-tuning of RNA Functions by Modification and Editing. 2005; NY: Springer Verlag; 442. [Google Scholar]
- 3.Dennis P.P., Omer A., Lowe T.. A guided tour: small RNA function in Archaea. Mol. Microbiol. 2001; 40:509–519. [DOI] [PubMed] [Google Scholar]
- 4.Tran E., Brown J., Maxwell E.S.. Evolutionary origins of the RNA-guided nucleotide-modification complexes: from the primitive translation apparatus. Trends Biochem. Sci. 2004; 29:343–350. [DOI] [PubMed] [Google Scholar]
- 5.Bachellerie J.P., Cavaille J.. Guiding ribose methylation of rRNA. Trends Biochem. Sci. 1997; 22:257–261. [DOI] [PubMed] [Google Scholar]
- 6.Kiss T. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell. 2002; 109:145–148. [DOI] [PubMed] [Google Scholar]
- 7.Lafontaine D.L., Tollervey D.. Birth of the snoRNPs: the evolution of the modification-guide snoRNAs. Trends Biochem. Sci. 1998; 23:383–388. [DOI] [PubMed] [Google Scholar]
- 8.Purta E., O'Connor M., Bujnicki J.M., Douthwaite S.. YgdE is the 2΄-O-ribose methyltransferase RlmM specific for nucleotide C2498 in bacterial 23S rRNA. Mol. Microbiol. 2009; 72:1147–1158. [DOI] [PubMed] [Google Scholar]
- 9.van Buul C.P., van Knippenberg P.H.. Nucleotide sequence of the ksgA gene of Escherichia coli: comparison of methyltransferases effecting dimethylation of adenosine in ribosomal RNA. Gene. 1985; 38:65–72. [DOI] [PubMed] [Google Scholar]
- 10.Harris J.K., Kelley S.T., Spiegelman G.B., Pace N.R.. The genetic core of the universal ancestor. Genome Res. 2003; 13:407–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rife J.P. Grosjean H. Roles of the ultra-conserved ribosomal RNA methyltransferase KsgA in ribosome biogenesis. DNA and RNA Modification Enzymes: Structure, Mechanism, Function and Evolution. 2009; Austin: Landes Bioscience; 512–526. [Google Scholar]
- 12.Grosjean H., Breton M., Sirand-Pugnet P., Tardy F., Thiaucourt F., Citti C., Barre A., Yoshizawa S., Fourmy D., de Crecy-Lagard V. et al. Predicting the minimal translation apparatus: lessons from the reductive evolution of mollicutes. PLoS Genet. 2014; 10:e1004363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Helser T.L., Dahlberg J.E., Davies J.E.. Mechanism of kasugamycin resistance in Escherichia coli. Nat. New Biol. 1972; 235:6–9. [DOI] [PubMed] [Google Scholar]
- 14.O'Connor M., Thomas C.L., Zimmermann R.A., Dahlberg A.E.. Decoding fidelity at the ribosomal A and P sites: Influence of mutations in three different regions of the decoding domain in 16S rRNA. Nucliec Acids Res. 1997; 25:1185–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Connolly K., Rife J.P., Culver G.. Mechanistic insight into the ribosome biogenesis functions of the ancient protein KsgA. Mol. Microbiol. 2008; 70:1062–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siibak T., Remme J.. Subribosomal particle analysis reveals the stages of bacterial ribosome assembly at which rRNA nucleotides are modified. RNA. 2010; 16:2023–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Binet R., Maurelli A.T.. The chlamydial functional homolog of KsgA confers kasugamycin sensitivity to Chlamydia trachomatis and impacts bacterial fitness. BMC Microbiol. 2009; 9:279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ochi K., Kim J.Y., Tanaka Y., Wang G., Masuda K., Nanamiya H., Okamoto S., Tokuyama S., Adachi Y., Kawamura F.. Inactivation of KsgA, a 16S rRNA methyltransferase, causes vigorous emergence of mutants with high-level kasugamycin resistance. Antimicrob. Agents Chemother. 2009; 53:193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tufariello J.M., Jacobs W.R. Jr, Chan J.. Individual Mycobacterium tuberculosis resuscitation-promoting factor homologues are dispensable for growth in vitro and in vivo. Infect. Immun. 2004; 72:515–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McGhee G.C., Sundin G.W.. Evaluation of kasugamycin for fire blight management, effect on nontarget bacteria, and assessment of kasugamycin resistance potential in Erwinia amylovora. Phytopathology. 2011; 101:192–204. [DOI] [PubMed] [Google Scholar]
- 21.Mecsas J., Bilis I., Falkow S.. Identification of attenuated Yersinia pseudotuberculosis strains and characterization of an orogastric infection in BALB/c mice on day 5 postinfection by signature-tagged mutagenesis. Infect. Immun. 2001; 69:2779–2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tokuhisa J.G., Vijayan P., Feldmann K.A., Browse J.A.. Chloroplast development at low temperatures requires a homolog of DIM1, a yeast gene encoding the 18S rRNA dimethylase. Plant Cell. 1998; 10:699–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lafontaine D., Vandenhaute J., Tollervey D.. The 18S rRNA dimethylase Dim1p is required for pre-ribosomal RNA processing in yeast. Genes Dev. 1995; 9:2470–2481. [DOI] [PubMed] [Google Scholar]
- 24.Zorbas C., Nicolas E., Wacheul L., Huvelle E., Heurgue-Hamard V., Lafontaine D.L.. The human 18S rRNA base methyltransferases DIMT1L and WBSCR22-TRMT112 but not rRNA modification are required for ribosome biogenesis. Mol. Biol. Cell. 2015; 26:2080–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Falkenberg M., Gaspari M., Rantanen A., Trifunovic A., Larsson N.G., Gustafsson C.M.. Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nat. Genet. 2002; 31:289–294. [DOI] [PubMed] [Google Scholar]
- 26.Sanyal A., Getz G.S.. Import of transcription factor MTF1 into the yeast mitochondria takes place through an unusual pathway. J. Biol. Chem. 1995; 270:11970–11976. [DOI] [PubMed] [Google Scholar]
- 27.Metodiev M.D., Lesko N., Park C.B., Camara Y., Shi Y., Wibom R., Hultenby K., Gustafsson C.M., Larsson N.G.. Methylation of 12S rRNA is necessary for in vivo stability of the small subunit of the mammalian mitochondrial ribosome. Cell Metab. 2009; 9:386–397. [DOI] [PubMed] [Google Scholar]
- 28.O'Farrell H.C., Pulicherla N., Desai P.M., Rife J.P.. Recognition of a complex substrate by the KsgA/Dim1 family of enzymes has been conserved throughout evolution. RNA. 2006; 12:725–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sokolova Y.Y., Dolgikh V.V., Morzhina E.V., Nassonova E.S., Issi I.V., Terry R.S., Ironside J.E., Smith J.E., Vossbrinck C.R.. Establishment of the new genus Paranosema based on the ultrastructure and molecular phylogeny of the type species Paranosema grylli Gen. Nov., Comb. Nov (Sokolova, Selezniov, Dolgikh, Issi 1994), from the cricket Gryllus bimaculatus Deg. J. Invertebr. Pathol. 2003; 84:159–172. [DOI] [PubMed] [Google Scholar]
- 30.Nakabachi A., Yamashita A., Toh H., Ishikawa H., Dunbar H.E., Moran N.A., Hattori M.. The 160-kilobase genome of the bacterial endosymbiont Carsonella. Science. 2006; 314:267–267. [DOI] [PubMed] [Google Scholar]
- 31.Bennett G.M., Abbà S., Kube M., Marzachì C.. Complete genome sequences of the obligate symbionts Candidatus Sulcia muelleri and Ca. Nasuia deltocephalinicola from the Pestiferous Leafhopper Macrosteles quadripunctulatus (Hemiptera: Cicadellidae). Genome Announc. 2016; 21:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klootwijk J., Klein I., Grivell L.A.. Minimal post-transcriptional modification of yeast mitochondrial ribosomal RNA. J. Mol. Biol. 1975; 97:337–350. [DOI] [PubMed] [Google Scholar]
- 33.Munson-McGee J.H., Field E.K., Bateson M., Rooney C., Stepanaukas R., Young M.J.. Nanoarchaeota, their Sulfolobales Host, and Nanoarchaeota Virus Distribution across Yellowstone National Park Hot Springs. Appl. Environ. Microb. 2015; 81:7860–7868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wurch L., Giannone R.J., Belisle B.S., Swift C., Utturkar S., Hettich R.L., Reysenbach A.L., Podar M.. Genomics-informed isolation and characterization of a symbiotic Nanoarchaeota system from a terrestrial geothermal environment. Nat. Commun. 2016; 7:12115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huber H., Hohn M.J., Rachel R., Fuchs T., Wimmer V.C., Stetter K.O.. A new phylum of Archaea represented by a nanosized hyperthermophilic symbiont. Nature. 2002; 417:63–67. [DOI] [PubMed] [Google Scholar]
- 36.Podar M., Anderson I., Makarova K.S., Elkins J.G., Ivanova N., Wall M.A., Lykidis A., Mavromatis K., Sun H., Hudson M.E. et al. A genomic analysis of the archaeal system Ignicoccus hospitalis-Nanoarchaeum equitans. Genome Biol. 2008; 9:R158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Waters E., Hohn M.J., Ahel I., Graham D.E., Adams M.D., Barnstead M., Beeson K.Y., Bibbs L., Bolanos R., Keller M. et al. The genome of Nanoarchaeum equitans: insights into early archaeal evolution and derived parasitism. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:12984–12988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jahn U., Gallenberger M., Paper W., Junglas B., Eisenreich W., Stetter K.O., Rachel R., Huber H.. Nanoarchaeum equitans and Ignicoccus hospitalis: new insights into a unique, intimate association of two archaea. J. Bacteriol. 2008; 190:1743–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Giannone R.J., Huber H., Karpinets T., Heimerl T., Kuper U., Rachel R., Keller M., Hettich R.L., Podar M.. Proteomic characterization of cellular and molecular processes that enable the Nanoarchaeum equitans–Ignicoccus hospitalis relationship. PLoS One. 2011; 6:e22942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J.. Basic local alignment search tool. J. Mol. Biol. 1990; 215:403–410. [DOI] [PubMed] [Google Scholar]
- 41.Maniatis T., Fritsch E.F., Sambrook J.. Molecular Cloning : a Laboratory Manual. 1982; NY: Cold Spring Harbor Laboratory. [Google Scholar]
- 42.Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K.A., Tomita M., Wanner B.L., Mori H.. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006; 2, doi:10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johansen S.K., Maus C.E., Plikaytis B.B., Douthwaite S.. Capreomycin binds across the ribosomal subunit interface using tlyA-encoded 2΄-O-methylations in 16S and 23S rRNAs. Mol. Cell. 2006; 23:173–182. [DOI] [PubMed] [Google Scholar]
- 44.Andersen N.M., Douthwaite S.. YebU is a m5C methyltransferase specific for 16 S rRNA nucleotide 1407. J. Mol. Biol. 2006; 359:777–786. [DOI] [PubMed] [Google Scholar]
- 45.Desmolaize B., Fabret C., Bregeon D., Rose S., Grosjean H., Douthwaite S.. A single methyltransferase YefA (RlmCD) catalyses both m5U747 and m5U1939 modifications in Bacillus subtilis 23S rRNA. Nucleic Acids Res. 2011; 39:9368–9375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stern S., Moazed D., Noller H.F.. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Meth. Enzymol. 1988; 164:481–489. [DOI] [PubMed] [Google Scholar]
- 47.Andersen T.E., Porse B.T., Kirpekar F.. A novel partial modification at C2501 in Escherichia coli 23S ribosomal RNA. RNA. 2004; 10:907–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Douthwaite S., Kirpekar F.. Identifying modifications in RNA by MALDI mass spectrometry. Meth. Enzymol. 2007; 425:3–20. [DOI] [PubMed] [Google Scholar]
- 49.Birkedal U., Christensen-Dalsgaard M., Krogh N., Sabarinathan R., Gorodkin J., Nielsen H.. Profiling of Ribose Methylations in RNA by High-Throughput Sequencing. Angew. Chem. Int. Edit. 2015; 54:451–455. [DOI] [PubMed] [Google Scholar]
- 50.Madsen C.T., Jakobsen L., Buriankova K., Doucet-Populaire F., Pernodet J.L., Douthwaite S.. Methyltransferase Erm(37) slips on rRNA to confer atypical resistance in Mycobacterium tuberculosis. J. Biol. Chem. 2005; 280:38942–38947. [DOI] [PubMed] [Google Scholar]
- 51.Maden B.E.H., Corbett M.E., Heeney P.A., Pugh K., Ajuh P.M.. Classical and novel approaches to the detection and localization of the numerous modified nucleotides in eukaryotic ribosomal RNA. Biochimie. 1995; 77:22–29. [DOI] [PubMed] [Google Scholar]
- 52.Hutchison C.A. 3rd, Chuang R.Y., Noskov V.N., Assad-Garcia N., Deerinck T.J., Ellisman M.H., Gill J., Kannan K., Karas B.J., Ma L. et al. Design and synthesis of a minimal bacterial genome. Science. 2016; 351:1414. [DOI] [PubMed] [Google Scholar]
- 53.Demirci H., Murphy F., Belardinelli R., Kelley A.C., Ramakrishnan V., Gregory S.T., Dahlberg A.E., Jogl G.. Modification of 16S ribosomal RNA by the KsgA methyltransferase restructures the 30S subunit to optimize ribosome function. RNA. 2010; 16:2319–2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Omer A.D., Ziesche S., Ebhardt H., Dennis P.P.. In vitro reconstitution and activity of a C/D box methylation guide ribonucleoprotein complex. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:5289–5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dennis P.P., Tripp V., Lui L., Lowe T., Randau L.. C/D box sRNA-guided 2΄-O-methylation patterns of archaeal rRNA molecules. BMC Genome. 2015; 16:632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lapinaite A., Simon B., Skjaerven L., Rakwalska-Bange M., Gabel F., Carlomagno T.. The structure of the box C/D enzyme reveals regulation of RNA methylation. Nature. 2013; 502:519–523. [DOI] [PubMed] [Google Scholar]
- 57.Ogle J.M., Ramakrishnan V.. Structural insights into translational fidelity. Annu. Rev. Biochem. 2005; 74:129–177. [DOI] [PubMed] [Google Scholar]
- 58.Kiss-László Z., Henry Y., Bachellerie J.P., Caizergues-Ferrer M., Kiss T.. Site-specific ribose methylation of preribosomal RNA: A novel function for small nucleolar RNAs. Cell. 1996; 85:1077–1088. [DOI] [PubMed] [Google Scholar]
- 59.Reichow S.L., Hamma T., Ferre-D'Amare A.R., Varani G.. The structure and function of small nucleolar ribonucleoproteins. Nucleic Acids Res. 2007; 35:1452–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Watkins N.J., Bohnsack M.T.. The box C/D and H/ACA snoRNPs: key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdis. Rev. RNA. 2012; 3:397–414. [DOI] [PubMed] [Google Scholar]
- 61.Liu Q., Fredrick K.. Intersubunit Bridges of the Bacterial Ribosome. J. Mol. Biol. 2016; 428:2146–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yusupov M.M., Yusupova G.Z., Baucom A., Lieberman K., Earnest T.N., Cate J.H., Noller H.F.. Crystal structure of the ribosome at 5.5 Å resolution. Science. 2001; 292:883–896. [DOI] [PubMed] [Google Scholar]
- 63.Grosjean H., Oshima T.. Gerday C, Glansdorff N. How Nucleic Acids Cope with High Temperature. Physiology and Biochemisty of Extremophiles. 2007; Washington, DC: ASM Press; 39–56. [Google Scholar]
- 64.Kawai G., Yamamoto Y., Kamimura T., Masegi T., Sekine M., Hata T., Iimori T., Watanabe T., Miyazawa T., Yokoyama S.. Conformational rigidity of specific pyrimidine residues in transfer-RNA arises from posttranscriptional modifications that enhance steric interaction between the base and the 2΄-hydroxyl group. Biochemistry. 1992; 31:1040–1046. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.