Abstract
Most of our knowledge about translation regulatory mechanisms comes from studies on lower organisms. However, the translation control system of higher organisms is less understood. Here we find that in 5΄ untranslated region (5΄UTR) of human Annexin II receptor (AXIIR) mRNA, there are two upstream open reading frames (uORFs) acting in a fail-safe manner to inhibit the translation from the main AUG. These uORFs are unfavorable for re-initiation after termination of uORF translation. Heterogeneous nuclear ribonucleoprotein A2B1 (hnRNPA2B1), hnRNPA0 and ELAV like RNA binding protein 1 (ELAVL1) bind to the 5΄UTR of AXIIR mRNA. They focus the translation of uORFs on uORF1 and attenuate leaky scanning that bypasses uORFs. The cooperation between the two uORFs and the three proteins formed a multiple fail-safe system that tightly inhibits the translation of downstream AXIIR. Such cooperation between multiple molecules and elements reflects that higher organism develops a complex translation regulatory system to achieve accurate and flexible gene expression control.
INTRODUCTION
Appropriate expression level is very important for a gene to exert its functions. AXIIR gene exists solely in human genome, not in other organisms (1). It acts as a receptor to mediate Annexin II (AXII) signal (1–3). Recently, we found that high amount of AXIIR protein could induce cell apoptosis in AXII-independent manner (4). In most cell types, AXIIR protein level is very low and even hardly detectable. It indicates that the expression of AXIIR protein is under tight and accurate control, to ensure its proper functions while avoid adverse effects on cells. However, nothing is known yet about the expression regulation of AXIIR.
Upstream open reading frame (uORF) is one of the major elements that regulate protein translation. It is estimated that ∼50% of mammalian mRNAs contain one or more uORFs (5). Three known mechanisms determine the translation efficiency of the downstream main ORF in mRNAs containing uORFs: leaky scanning (6), re-initiation (7,8) and ribosome stalling (9). Leaky scanning and re-initiation facilitate, while ribosome stalling inhibits the translation of downstream main ORF. Re-initiation requires translation and termination of uORF. Many uORFs exert effect through re-initiation which is believed to be independent of the amino acids the uORFs encode. There are also some uORFs that function through the peptides they encode (10–12). Some of these peptides can stall the ribosome (13), or contribute to mRNA decay (14). Little is known about the factors other than standard cap-dependent initiation factors that work with uORF-dependent translation regulation (15). So far, there is only one paper reporting that uORF and protein cooperate to control translation (16), which is found in Drosophila. It remains unclear whether such translation regulation mechanism involving the cooperation between uORF and protein also exists in higher organisms.
hnRNPA2B1 and hnRNPA0 are RNA binding proteins that belong to the family of ubiquitously expressed heterogeneous nuclear ribonucleoproteins (hnRNPs). hnRNPA2B1 exerts diverse regulatory effects through interacting with different partners (17,18). Little is known about the function of hnRNPA0, except for one paper showing that it stabilizes Gadd45α mRNA (19). ELAVL1, also known as HuR, is a member of ELAVL family of RNA-binding proteins and involves in a variety of important biological activities and diseases (20,21). It selectively binds to AU-rich elements (AREs) in the 3΄ untranslated region (3΄UTR) of mRNAs, and plays roles in stabilizing mRNA (22) and promoting translation (23,24). There are few papers reporting the inhibition effect of ELAVL1 on protein translation. One paper reports that ELAVL1 can bind to 5΄UTR and repress internal ribosome entry site (IRES)-mediated translation (25). Another one also finds that ELAVL1 can bind to 5΄UTR and repress translation, but the mechanism remains unknown (26). ELAVL1 interacts with different molecules to exert its functions (27,28). All the above mentioned three proteins can shuttle between nucleus and cytoplasm (19,29).
In this study, we find that hnRNPA2B1, hnRNPA0 and ELAVL1 bind to the 5΄UTR of AXIIR mRNA and cooperate with uORFs, forming a multiple fail-safe system to tightly inhibit the translation of human AXIIR. The results reveal a complex translation regulatory system in higher organism, in which multiple trans-acting proteins and uORFs cooperatively inhibit translation from downstream main start codon.
MATERIALS AND METHODS
Cell lines
The cell lines used in this study were: HEK293T cell (ATCC), K-562 cell (ATCC), MM.1S cell (ATCC), U266B1 cell (ATCC), RPMI8226 cell and HeLa cell were from Cell Bank of Chinese Academy of Medical Sciences and Peking Union Medical College.
Plasmids
AXIIR 5΄UTR and coding region sequences (CDS) were amplified from human K-562 cell. Fusion genes were spliced by polymerase chain reaction (PCR). Point mutation was achieved using QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent). All the sequences were cloned into pcDNA3.1-Myc-His(-)B vector (Invitrogen). The CDS sequence together with the three adjacent nucleotides upstream of the main start codon was cloned between EcoR I and BamH I sites. All the other 5΄UTR containing sequences were cloned between Xho I and BamH I sites. AXIIR 5΄UTR and enhanced green fluorescent protein (EGFP) fusion gene was cloned between Xho I and EcoR I sites. The coding sequence of mouse TNFAIP3 interacting protein 2 (Tnip2) with C-terminal 6xHis tag was cloned between EcoR I and BamH I sites.
RNA interference
EGFP was firstly cloned into pcDNA3.1-Myc-His(-)B vector (Invitrogen). shRNA coding sequences against human ELAVL1, hnRNPA0 and hnRNPA2B1 were then cloned into it downstream of EGFP stop codon (termed pc-EGFP-shRNA). The amount of transfected shRNA expressing plasmids can be evaluated by measuring the EGFP intensity. Sequences of shRNA (sense strand, 5΄→3΄): ELAVL1 shRNA: GCGTTTATCCGGTTTGACAAA hnRNPA0 shRNA: GGCGGTCGCAGTAATAGTGGA hnRNPA2B1 shRNA: GGAACAGTTCCGTAAGCTCTT negative control shRNA (termed NC shRNA): TCGTATAGAGCTTAAGGGCGG. The inhibition efficiency of shRNAs were measured by SYBR quantitative reverse transcription-PCR (qRT-PCR) analysis of AXIIR mRNA. Human β-actin served as the internal control gene.
mRNA stability analysis
Forty-eight hours after plasmid transfection, Actinomycin D (Sigma) was added to the culture medium at a concentration of 10 µg/ml. Cells were then collected at certain time points after Actinomycin D addition and total RNA was extracted with RNAiso Plus (Takara). For each sample, 3 µg of RNA was used for reverse-transcription with GoScript™ Reverse Transcriptase (Promega). From the 20 µl of reverse-transcription reaction volume, 1 µl was subjected to the following SYBR qRT-PCR analysis of mRNA. Human GAPDH served as the internal control gene. The value of AXIIR was normalized to that of GAPDH and the value of each time point was further normalized to that of the start point, which was designated as 100%.
qRT-PCR
SYBR qRT-PCR reaction was performed in CFX real-time PCR machine (BioRad). Primers were designed by Primer Express 3.0 (ABI). The primer sequences are (5΄→3΄): GAPDH primers: TGGAAGGACTCATGACCACAGT and GCCATCACGCCACAGTTTC AXIIR primers: GTGATTTGGGACTGCTTTCCA and GAGAGTCCGTTTTGCCAGTAGAC β-actin primers: CCAACCGCGAGAAGATGA and CCAGAGGCGTACAGGGATAG. Primers were synthesized by Invitrogen. qRT-PCR was performed using GoTaq® Probe qPCR Master Mix (Promega), according to the manufacturer's instructions.
Transfection
Plasmids were transfected into HEK293T or HeLa cells by Lipomax (Sudgen), following the manufacturer's instructions. Mouse Tnip2 with C-terminal 6xHis tag serves as transfection efficiency control that was co-transfected with AXIIR expressing plasmid. The Tnip2-His fusion protein was blotted by anti-His tag antibody in western blot. EGFP intensity was read by FLUOstar OPTIMA (BMG Lab).
Antibodies
The following antibodies were used for western blot, electrophoretic mobility gel shift assay (EMSA) and Co-IP: anti-β-actin (Abcam), anti-AXIIR (Abcam), anti-ELAVL1 (Abcam), anti-hnRNPA0 (Abcam), anti-hnRNPA2B1 (GeneTex), anti-hnRNP G (Abcam), anti-His tag (Cell Signaling), normal rabbit IgG (Cell Signaling). The secondary antibody was anti-rabbit IgG (Abcam). The density of each band was measured by AlphaEaseFC software (Alpha Innotech).
Photography
Cell images were taken with IX70-SIF2 microscope (OLYMPUS).
RNA pull-down
Biotinylated RNA probe was chemically synthesized (Takara). The sequence of the probe is (5΄→3΄): CUCUCUAAAAUGUUAUCGUUUUUCAUUUGUCUACUAAUUUUC. This probe was used to pull down putative RNA binding proteins from K-526 cells with Magnetic RNA–Protein Pull-Down Kit (Pierce), according to the manufacturer's instructions.
Protein mass spectrometry
It was carried out by the Core Instrument Facility in CAMS/PUMC. Briefly, the gel band was in-gel digested by trypsin and analyzed by nano-LC/MS/MS (TripleTOF 5600, ABsciex). The raw file was searched against human Uniprot database (www.uniprot.org) by MASCOT (V2.3.02). The results were processed by Scaffold (V4.0).
Electrophoretic mobility gel shift assay (EMSA)
Nuclear and cytoplasmic proteins of K-562 cells were separated with NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Scientific). The probe used in RNA pull down experiment was also used in EMSA. EMSA was performed with LightShift Chemiluminescent RNA EMSA Kit (Thermo Scientific), according to the manufacturer's instructions. Briefly, 4 µg of proteins were incubated with 5 nM of RNA probe and then subjected to non-denaturing polyacrylamide gel electrophoresis (PAGE).
For the method of adding antibody later in super-shift EMSA, 4 µg of nuclear proteins were incubated with 5 nM of RNA probe, at room temperature for 30 min. A total of 2 µg of each antibody was added to 1/10 of the above mixture, incubated at room temperature for 20 min and then subjected to non-denaturing PAGE. For the method of adding antibody first in super-shift EMSA, 0.91 µg of nuclear proteins were incubated with 3 µg of each antibody, at room temperature for 30 min. The above mixture was then incubated with 2.5 nM of RNA probe at room temperature for 30 min, followed by non-denaturing PAGE.
Co-immuoprecipitation (Co-IP)
One T75 flask of sub-confluent HEK293T cells were collected, and washed with 1× phosphate buffered saline (PBS). Cell pellets were lysed in Pierce IP Lysis Buffer (Pierce) containing Protease Inhibitor Cocktail Set III (Merck) and Phosphatase Inhibitor Cocktail Set IV (Merck), and placed on ice for 10 min. The suspension was centrifuged at 12 000 g for 15 min. Supernatant was transferred to new tube and a portion was reserved for input loading. Protein G Plus/Protein A Agarose Suspension (Millipore) was pre-treated by being suspended in Pierce IP Lysis Buffer, and then centrifuged at 2500 rpm for 3 min, 4°C. This washing step was repeated for four times. The cell lysate was cleared by adding 1 µg of normal rabbit IgG and 20 µl of pre-treated Protein G Plus/Protein A Agarose Suspension to 0.5 ml of cell lysate, and incubated with soft shaking at 4°C for 2 h, followed by centrifugation at 10 000 g for 2 min, 4°C. Supernatant was transferred to new tube. Immunoprecipitation was performed by mixing 0.1 ml of the cleared cell lysate with 2 µg of each of the antibodies or normal rabbit IgG as negative control, and soft shaking overnight. To pull down the Immunoprecipitate, each group was mixed with 40 µl of pre-treated Protein G Plus/Protein A Agarose Suspension and softly shook for 4 h. After centrifugation at 10 000 g for 2 min, 4°C, supernatant was discarded and the pellets were washed with 1 ml of 1× PBS for four–six times. Finally, each sample was mixed with 20 µl of 2× loading buffer, boiled for 5 min and subjected to western blotting.
Statistical analysis
All the experiments were independently repeated in at least triplicate. Data are presented as mean ± standard deviation. Student's t-test was performed based on the data from three experiments. A difference was determined to be statistically significant when P-value was <0.05.
RESULTS
5΄UTR inhibits AXIIR translation
We have previously found that in most human cell types, the protein level of AXIIR was very low, although the mRNA level of AXIIR in these cells was not low. And protein degradation did not account for the low protein level of AXIIR (4). This implied that the expression of AXIIR is regulated at translational level.
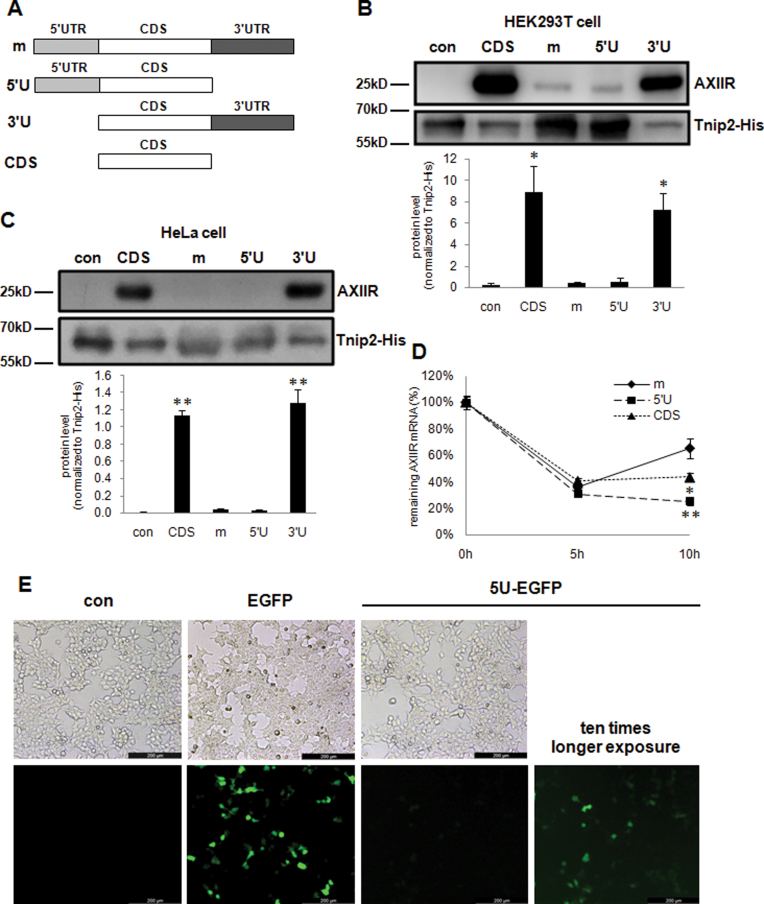
To explore which part of AXIIR mRNA is involved in AXIIR translation, different parts of AXIIR mRNA were cloned into pcDNA3.1-Myc-His(-)B vector (Figure 1A). For the construct of AXIIR coding region alone (termed CDS), the three adjacent nucleotides upstream of the main start codon was also included, to keep the Kozak context for the main start codon. Constructs were transfected into human HEK293T and HeLa cells. Endogenous AXIIR protein in these two cell lines was not detected (4). In both cell lines, construct of intact AXIIR mRNA (termed m) expressed very low level of AXIIR protein, which was hardly detectable. Construct of AXIIR mRNA lacking 3΄UTR (termed 5΄U) expressed similar low level of AXIIR as intact AXIIR mRNA, whereas construct of CDS and of AXIIR mRNA lacking 5΄UTR (termed 3΄U) exhibited similar high level of AXIIR protein (Figure 1B and C). This demonstrated that 5΄UTR of AXIIR mRNA harbored elements that strongly inhibited AXIIR translation.
Figure 1.
Examination of the roles of 5΄UTR and 3΄UTR in AXIIR translation. con: pcDNA3.1-Myc-His(-)B vector control. (A) Illustration of the UTR-deleted constructs. (B and C) Western blot of AXIIR and Tnip2-His protein levels for UTR-deleted constructs in HEK293T and HeLa cells. Different constructs and Tnip2-His expressing plasmid were co-transfected into cells. Western blot was performed 40 h after transfection. Experiments were repeated in triplicates. Quantification of AXIIR level normalized to Tnip2-His was below the blot. Data represent mean±s.d. *P < 0.05, **P < 0.01 (t-test as compared with con) (D) mRNA stability analysis of the UTR-deleted constructs in HEK293T cell. Experiments were repeated in triplicates. Data represent mean ± s.d. *P < 0.05, **P < 0.01 (t-test as compared with m). (E) Photography of EGFP fluorescence, 24 h after plasmid transfection into HEK293T cells. For each column, the upper and lower photographs were taken for the same view.
To exclude the possibility that mRNA stability affects the protein expression of the above constructs, we checked the stability of AXIIR mRNA generated by these constructs. The remaining AXIIR mRNA levels generated by the constructs were similar at 5 h after Actinomycin D treatment. At 10 h, the remaining AXIIR mRNA levels of both 5΄U and CDS were lower than that of the intact AXIIR mRNA and AXIIR mRNA level of CDS was higher than that of 5΄U (Figure 1D). However, the difference between the AXIIR mRNA stability curves of 5΄U and CDS did not match the dramatic difference between their AXIIR protein levels. Moreover, construct m that had relatively stable AXIIR mRNA expressed very low level of AXIIR protein. It demonstrated that the difference in AXIIR protein level between the above constructs was mainly due to regulation at translational level.
To test whether the inhibition effect of 5΄UTR on AXIIR translation was specific to AXIIR, the AXIIR coding region downstream of 5΄UTR was replaced with EGFP coding region which served as a reporter (termed 5U-EGFP). Similar to the above results, EGFP coding region alone without AXIIR 5΄UTR expressed high level of EGFP. By contrast, no EGFP fluorescence was visible in 5U-EGFP (Figure 1E). This demonstrated that the inhibition effect of AXIIR 5΄UTR on translation was not specific for AXIIR. Notably, when the exposure time in photography was extended 10× longer, faint EGFP fluorescence appeared for 5U-EGFP construct. This indicated that AXIIR 5΄UTR strongly inhibited, rather than totally abrogated the downstream protein translation.
uORFs contribute to the effect of 5΄UTR
AXIIR mRNA has a 5΄UTR of 299 nucleotides. There are four uORFs in AXIIR 5΄UTR (Figure 2A and B). Among them, uORF1 is the longest with 165 nucleotides. uORF1, uORF2 and uORF3 overlap. Remarkably, the reading frame of uORF1 and uORF3 are the same, and they share the same double stop codons with an interval of 3 nucleotides. uORF4 does not overlap with other uORFs, and there is merely 3 nucleotides between the stop codon of uORF4 and the downstream main AUG. Additionally, there are two potential non-canonical start codons CUG and AUU upstream of uAUG1. The first non-canonical start codon (termed uStartnc1) has an adjacent double stop codons with an interval of 3 nucleotides. The second non-canonical start codon (termed uStartnc2) is in-frame with uORF1 and uORF3.
Figure 2.
Illustration of AXIIR 5΄UTR. (A) Position and length of the uORFs in 5΄UTR. Numbers at both ends of each bar for uORF indicate the start and end nucleotide position of the uORF. The length of each uORF is in the bracket. (B) Sequence of AXIIR 5΄UTR and the uORFs. Non-canonical upstream start codons were marked in purple. The main AUG was in bold and red. The CDS sequence was marked in orange. Stop codons were marked in the same color with their corresponding start codons.
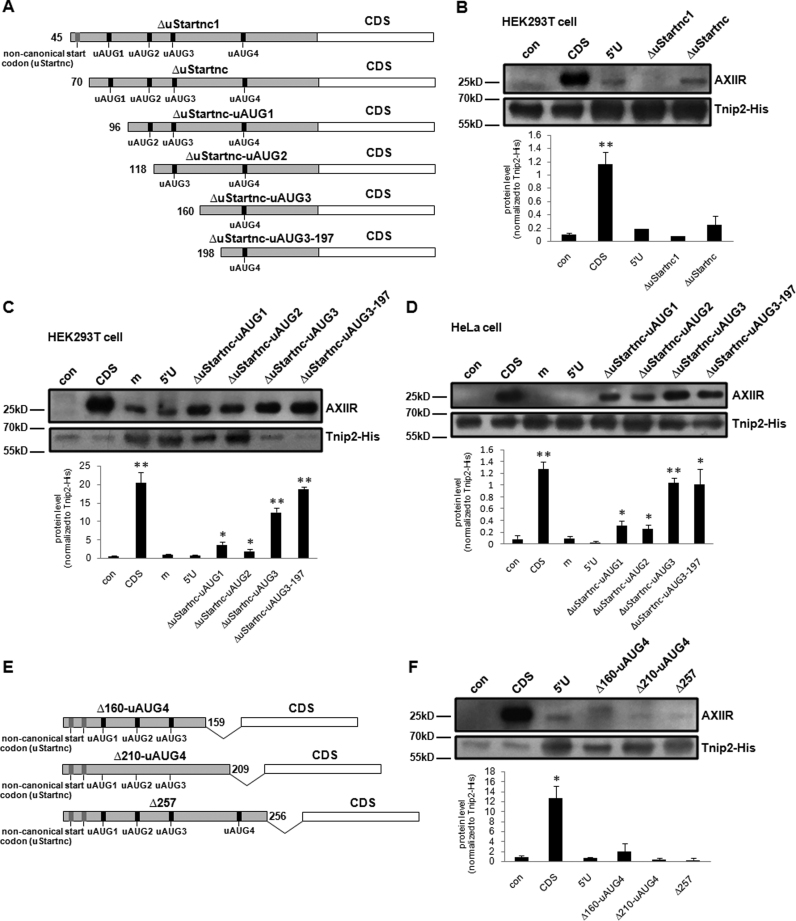
To determine which part accounted for the effect of 5΄UTR, different 5΄-terminal truncated 5΄U were constructed (Figure 3A). Construct ΔuStartnc1 lacked the first non-canonical start codon by deleting 5΄-terminal sequence of 5΄UTR containing uStartnc1. Similarly, ΔuStartnc lacked the two non-canonical start codons. Construct ΔuStartnc-uAUG1 lacked the two non-canonical start codons and uAUG1. Construct ΔuStartnc-uAUG2 lacked the two non-canonical start codons, uAUG1 and uAUG2. Construct ΔuStartnc-uAUG3 lacked the two non-canonical start codons, uAUG1, uAUG2 and uAUG3. Construct ΔuStartnc-uAUG3-197 lacked the 5΄-terminal 197 nucleotides which contained the same start codons as ΔuStartnc-uAUG3, but its 5΄-terminus was even shorter than ΔuStartnc-uAUG3. In HEK293T cell, deletion of the two non-canonical start codons did not significantly affect AXIIR protein level (Figure 3B). Compared with 5΄U, AXIIR protein levels expressed by ΔuStartnc-uAUG1 and ΔuStartnc-uAUG2 were significantly elevated, but still much lower than that by CDS. ΔuStartnc-uAUG1 and ΔuStartnc-uAUG2 exhibited equivalent effect, indicating that deletion of uAUG2 did not improve the effect of ΔuStartnc-uAUG1. That meant uORF2 might not involve in AXIIR translation. AXIIR protein levels expressed by ΔuStartnc-uAUG3 and ΔuStartnc-uAUG3-197 were dramatically increased to 61 and 91% of that by CDS respectively (Figure 3C), suggesting that loss of uAUG1 and uAUG3 reversed much of the inhibition. In HeLa cell, the result was similar, except that AXIIR protein levels expressed by ΔuStartnc-uAUG3 and ΔuStartnc-uAUG3-197 were increased to 82 and 79% respectively of that by CDS (Figure 3D). Significant changes in AXIIR protein level appeared when uAUG1 or both uAUG1 and uAUG3 were deleted. These data implied that among the uORFs, uORF1 and uORF3 might play more important roles in AXIIR translation, uORF4 might have minor effect, while uORF2 and the two non-canonical start codons might not involve. Besides HEK293T and HeLa cells, we also obtained similar results in other cell lines such as K562 cell (data not shown). Therefore, the following experiments were carried out in one cell line.
Figure 3.
Investigation of the roles of different 5΄UTR regions in AXIIR translation. con: pcDNA3.1-Myc-His(-)B vector control. (A) Illustration of AXIIR 5΄UTR 5΄-terminal truncated constructs. Gray bars represent 5΄UTR. White bars represent CDS. Numbers at the beginning of each bar indicate the start nucleotide position of the truncated 5΄UTR. (B–D) Western blot of AXIIR and Tnip2-His protein levels for AXIIR 5΄UTR 5΄-terminal truncated constructs in HEK293T and HeLa cells. Different constructs and Tnip2-His expressing plasmid were co-transfected into cells. Western blot was performed 40 h after transfection. Experiments were repeated in triplicates. Quantification of AXIIR level normalized to Tnip2-His was below the blot. Data represent mean±s.d. *P < 0.05, **P < 0.01 (t-test as compared with 5΄U) (E) Illustration of AXIIR 5΄UTR 3΄-terminal truncated constructs. Gray bars represent 5΄UTR. Numbers at the end of each bar indicate the end nucleotide position of the truncated 5΄UTR. (F) Western blot of AXIIR and Tnip2-His protein levels for AXIIR 5΄UTR 3΄-terminal truncated constructs in HEK293T cell. Legends were the same as those for (B–D).
We went further to construct different 3΄-terminal truncated 5΄U (Figure 3E and Supplementary Figure S1). Construct Δ160-uAUG4 lacked the 3΄-terminal 137 nucleotides of 5΄U, thus lacked the stop codon of uORF1, uORF3 and the whole uORF4. Similarly, Construct Δ210-uAUG4 lacked the 3΄-terminal 87 nucleotides of 5΄U and lacked the stop codon of uORF1, uORF3 and the whole uORF4. Construct Δ257 lacked the 3΄-terminal 40 nucleotides of 5΄U, retained intact uORF1, uORF3 and uAUG4, but lacked the stop codon of uORF4. In these truncations, the last three nucleotides GGG in 5΄UTR which are adjacent upstream of the main AUG were retained to keep the original Kozak sequence context of the main AUG. The above truncated 5΄UTR sequences were ligated to downstream AXIIR coding region. AXIIR protein level expressed by all the three above constructs was not improved, compared with that by 5΄U (Figure 3F). This result showed that lacking of uORF4 or the nucleotides 160–296 in 3΄-terminus of 5΄U did not affect AXIIR translation. It suggested that uORF4, stop codon of uORF1 and uORF3, and the 3΄-terminal sequence of 5΄U played minor role in AXIIR translation. It is notable that 3΄-truncated uORF1 and uORF3 in construct Δ160-uAUG4 were in frame with the main AUG, but were not in construct Δ210-uAUG4. The 3΄-truncated uORF1 and uORF3 in construct Δ210-uAUG4 encountered a stop codon 23 nucleotides downstream of the main AUG (Supplementary Figure S1). Consistently, for Δ160-uAUG4, there was a band slightly bigger than AXIIR. It corresponded to the size of the fusion protein of truncated uORF1 and AXIIR coding region. This suggested that translation could be started from uAUG1.
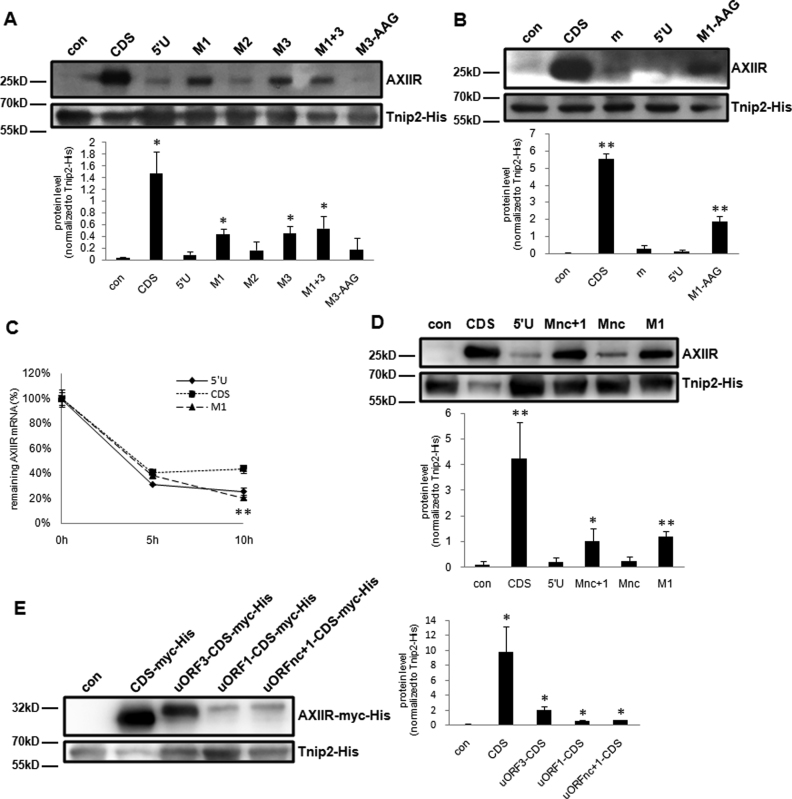
To further confirm the roles of the uORFs on AXIIR translation, start codon of the first three uORFs was individually mutated. Compared with 5΄U, mutation of uAUG1 to stop codon UAG (termed M1) increased AXIIR protein level to 30% of the amount expressed by CDS. Mutation of uAUG2 to stop codon UAG (termed M2) did not improve AXIIR protein level (Figure 4A). Mutation of uAUG1 to AAG (termed M1-AAG) increased AXIIR protein level to 34% of the amount expressed by CDS (Figure 4B), similar to the level of 30% produced by M1. Surprisingly, mutation of uAUG3 to AAG (termed M3-AAG) did not improve AXIIR protein level, whereas mutation of uAUG3 to stop codon UAG (termed M3) increased AXIIR protein level to 31% of the amount expressed by CDS. Double mutation of uAUG1 and uAUG3 (termed M1+3) increased AXIIR protein to an equivalent level as individually mutated M1 and M3 did (Figure 4A). As uORF1 and uORF3 share the same reading frame, this made us suspect that for M3, truncated uORF1 rather than deletion of uORF3 improved AXIIR translation. These data implied that uORF1 might play major role among uORFs. We also checked the stability of AXIIR mRNA generated by construct M1 (Figure 4C). Construct 5΄U and M1 exhibited similar half-life curves, demonstrating that the improvement of AXIIR protein level by M1 was irrelevant to AXIIR mRNA stability. Considering that the remaining AXIIR mRNA level of M1 was 47% of that of CDS at 10 h after Actinomycin D treatment, the effect of uORF1 on AXIIR translation inhibition might be even stronger. Mutation of both two non-canonical start codons to stop codons (termed Mnc) did not improve AXIIR protein level. Double mutation of both two non-canonical start codons and uAUG1 to stop codons (termed Mnc+1) increased AXIIR protein to the same level as M1 did (Figure 4D). Consistent with the truncation result in Figure 3B, this demonstrated that the two non-canonical start codons upstream of uAUG1 might not involve in regulation of AXIIR translation.
Figure 4.
Investigation of the effect of uORFs in AXIIR translation. con: pcDNA3.1-Myc-His(-)B vector control. (A and B) Western blot of AXIIR and Tnip2-His protein levels for uORF1-mutated constructs in HEK293T cell. Different constructs and Tnip2-His expressing plasmid were co-transfected into cells. Western blot was performed 40 h after transfection. Experiments were repeated in triplicates. Quantification of AXIIR level normalized to Tnip2-His was below the blot. Data represent mean ± s.d. *P < 0.05, **P < 0.01 (t-test as compared with 5΄U) (C) mRNA stability analysis of the uAUG-mutated construct in HEK293T cell. Experiments were repeated in triplicates. Data represent mean ± s.d. *P < 0.05, **P < 0.01 (t-test as compared with CDS) (D) western blot of AXIIR and Tnip2-His protein levels for uStartnc-mutated constructs in HEK293T cell. Legends were the same as those for (A and B). (E) Western blot of AXIIR-myc-His and Tnip2-His protein levels for uORF-CDS fusion constructs in HEK293T cell. Different constructs and Tnip2-His expressing plasmid were co-transfected into cells. Western blot was performed 40 h after transfection. AXIIR-myc-His was blotted with anti-His tag antibody. Experiments were repeated in triplicates. Quantification of AXIIR-myc-His level normalized to Tnip2-His was at the right of the blot. Data represent mean±s.d. *P < 0.05, (t-test as compared with con).
Kozak sequence is important for ribosome to recognize translation start codon (30). The main AUG of AXIIR is in strong favorable Kozak sequence context. By contrast, both uAUG1 and uAUG3 are in Kozak sequence context of the same moderate intensity. What's more, uORF1 and uORF3 share the same reading frame and stop codon. Although results in Figure 4A showed that mutation of uORF3 did not affect AXIIR protein level, it could not reach the conclusion that uORF3 did not involve in AXIIR translation. It is possible that uORF1 and uORF3 could substitute for each other.
We then checked whether uORF1 and uORF3 were translated. If uORF is translated, its encoded peptide should exist. However, it is difficult to directly measure the product translated by uORF. Generally, uORF-encoded peptides are short and unstable, which are hard to be detected (31). Therefore, most studies involved in uORF translation fuse uORF with a long protein coding sequence so that the fusion protein containing uORF-encoded peptide can be easily detected (32). So we fused uORFs with AXIIR CDS without main AUG (Supplementary Figure S2), to see whether the uORFs can be translated. All of the CDS were 3΄-tagged with myc and 6xHis tag, to avoid possible interference from endogenous AXIIR. Construct CDS-myc-His is CDS that 3΄-tagged with myc and 6xHis tag. The construct fusing uORF3 with the AXIIR CDS (termed uORF3-CDS-myc-His) also included Kozak sequence context for uAUG3. The construct fusing uORF1 with the AXIIR CDS (termed uORF1-CDS-myc-His) actually contains intact uORF1, uORF2 and uORF3. Construct uORFnc+1-CDS-myc-His contains AXIIR 5΄UTR that covers the two non-canonical uORFs, uORF1 and uORF3. As expected, construct uORF3-CDS-myc-His produced fusion protein bigger than CDS-myc-His. Construct uORF1-CDS-myc-His and uORFnc+1-CDS-myc-His produced the same single fusion protein band slightly bigger than uORF3-CDS-myc-His (Figure 4E). This indicated that in AXIIR 5΄UTR, mainly uORF1 was translated. When upstream sequence including uAUG1 was absent, uORF3 could act as a backup for uORF1.
Collectively, the above results showed that among the uORFs, uORF1 played major role in inhibiting AXIIR translation.
Effect of uORFs depends on their length
In Figure 4E, it was worth noting that the intensity of AXIIR-myc-His band of all the fusion constructs was much lower than that of CDS-myc-His. The intensity of AXIIR-myc-His band of uORF3-CDS-myc-His, uORF1-CDS-myc-His and uORFnc+1-CDS-myc-His was 21, 6 and 7% of that of CDS-myc-His respectively. There are three possible explanations for such difference. Firstly, translation from uAUG1 or uAUG3 was weaker than that from the main AUG, consisting with the difference between Kozak context intensity of uAUG1, uAUG3 and the main AUG. Secondly, elongation of uORF1 or uORF3 was inefficient that intact uORF-CDS fusion protein was very little. Thirdly, when the upstream sequence containing uAUG1 was absent, translation was more easily to be started from uAUG3.
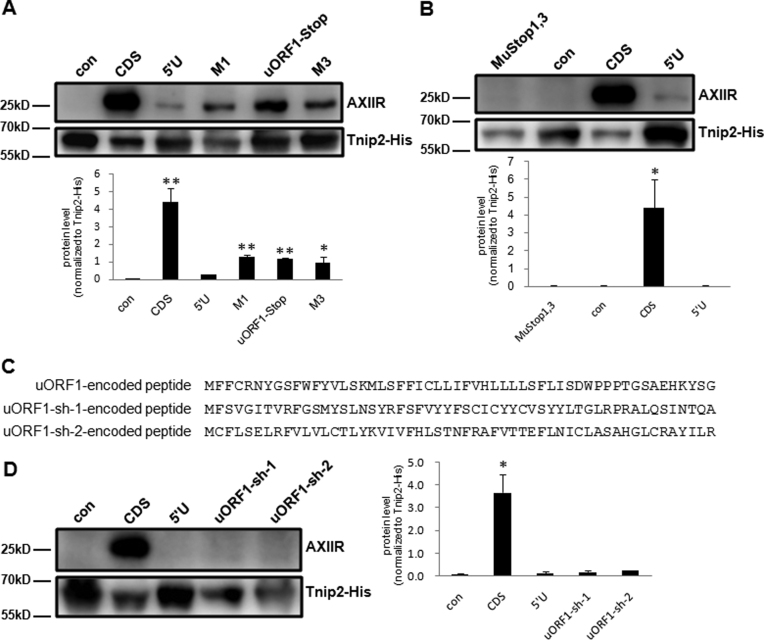
As uORF1 is the main uORF that involved in AXIIR translation inhibition, we then tested whether the length of uORF1 could affect its effect. A stop codon UAG was inserted as the first three nucleotides immediately downstream of uAUG1 to make an early termination of uORF1 (termed uORF1-Stop). Construct M3 in which uAUG3 was mutated to stop codon UAG served as a premature termination of uORF1 whose reading frame length was longer than uORF1-Stop. Compared with 5΄U, both uORF1-Stop and M3 increased AXIIR protein to 27 and 22% of the amount expressed by CDS respectively, equivalent to the level of 29% by M1 (Figure 5A). The common double stop codons shared by uORF1 and uORF3 were also mutated to CGA and CAG (termed MuStop1,3) (Supplementary Figure S3). MuStop1,3 was not in frame with the main AUG and encountered a stop codon downstream of the main AUG. Compared with 5΄U, MuStop1,3 did not improve AXIIR protein level (Figure 5B). These data suggested that the length of the uORFs may be an important factor affecting translation of the AXIIR mRNA.
Figure 5.
Investigation of the effect of uORF1 in AXIIR translation. con: pcDNA3.1-Myc-His(-)B vector control. (A and B) Western blot of AXIIR and Tnip2-His protein levels for 5΄UTR-mutated constructs in HEK293T cell. Different constructs and Tnip2-His expressing plasmid were co-transfected into cells. Western blot was performed 40 h after transfection. Experiments were repeated in triplicates. Quantification of AXIIR level normalized to Tnip2-His was below the blot. Data represent mean ± s.d. *P < 0.05, **P < 0.01 (t-test as compared with 5΄U). (C) Amino acid sequence of wild type and frame-shifted uORF1-encoded peptides. (D) Western blot of AXIIR and Tnip2-His protein levels for uORF1 frame-shifted constructs in HEK293T cell. Legends were the same as those for (A and B) except that quantification of AXIIR level normalized to Tnip2-His was at the right side of the blot.
Re-initiation is one of the important mechanisms involving in uORF-mediated translation regulation. Short uORFs less than 30 codons with long intercistronic region favor downstream re-initiation. In contrast, for RNAs with long uORFs, re-initiation either not occur after termination of long uORF translation or depends on the close proximity between the uORF stop codon and the downstream start codon (33,34). In AXIIR 5΄UTR, uORF1 and uORF3 has 54 and 37 codons, respectively. The distance between the start codon of the main AXIIR ORF and the stop codon of uORF1 and uORF3 is as long as 59 bp. Therefore, we reasoned that re-initiation translation from the main AXIIR AUG was unlikely to occur downstream of uORF1 and uORF3. However, in uORF1-Stop and M3, uORF1 was shortened to one codon and 17 codons, respectively. Re-initiation could occur with such short uORF, resulting in increased translation from the main AUG.
We then asked whether the effect of uORF1 was dependent on the peptide it encoded. Shifting reading frame by inserting or deleting one or two nucleotides in the uORF will change the amino acid sequence of the uORF-encoded peptide. By this way, it can be determined whether the effect of the uORF is peptide-dependent. We made two uORF1 frame-shifted constructs by deleting the first U downstream of uAUG1 (termed uORF1-sh-1), or inserting a G at the second nucleotide downstream of uAUG1 (termed uORF1-sh-2). In both constructs, the Kozak sequence context of uORF1 remained unchanged. uAUG3 was mutated to avoid the interference from possible translation started from uAUG3. And all the stop codons caused by frame-shift were point mutated and one nucleotide was inserted or deleted upstream of uORF1 stop codon, to ensure that the frame-shifted uORF1 stopped at the original position and encoded peptide with the same length as the wild-type (Supplementary Figure S4). Except for the first amino acid Met, uORF1-sh-1, uORF1-sh-2 and wild-type uORF1 shared no other identical amino acids (Figure 5C). Compared with 5΄U, both uORF1-sh-1 and uORF1-sh-2 did not improve AXIIR protein level (Figure 5D), demonstrating that the effect of uORF1 was independent of the peptide it encodes.
hnRNPA2B1, hnRNPA0 and ELAVL1 bind to AXIIR 5΄UTR
Although the above results demonstrated that uORF1 played important role in inhibiting downstream AXIIR translation, and uORF3 acted as a backup for uORF1, uORF alone could not explain all the inhibition of AXIIR translation. As shown in Figure 4A, mutation of both uAUG1 and uAUG3, which completely abrogated uORF1 and uORF3, increased AXIIR protein level to merely 37% of the level produced by construct CDS. This suggested that other mechanisms also involve in the inhibition of AXIIR translation.
The results of truncation experiments showed that the 42-nt sequence between nucleotide 118–159 in AXIIR 5΄UTR recovered 51–62% of AXIIR translation (Figure 3C and D), indicating that this 42-nt sequence played important role. Although this region contained uAUG3 and part of uORF3, uORF3 could not account for all the effect of this 42-nt sequence, as double mutation of both uAUG1 and uAUG3 merely recovered 30% of AXIIR translation (Figure 4A). To examine whether there were any secondary structures within the 42-nt sequence that might account for the translation inhibition effect, we subjected entire AXIIR 5΄UTR to RNA folding form analysis by The Mfold Web Server (http://mfold.rna.albany.edu/?q=mfold). But no stable secondary structure was found. Then we analyzed the nucleotide composition of the 42-nt sequence. Remarkably, within the 42-nt sequence, A and U totally accounted for 73.81% of all the nucleotides and the percentage of U was as high as 52.38%. AREs are the binding sites for many RNA-binding proteins (35,36). This made us speculate that there might be some proteins which could bind to the 42-nt sequence and participated in the inhibition of AXIIR translation.
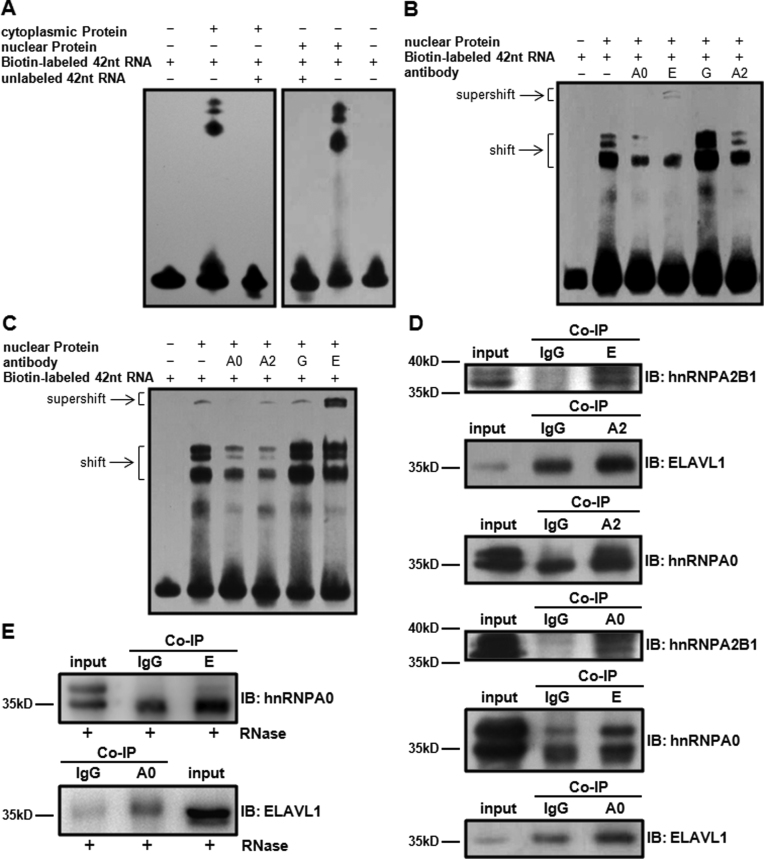
This hypothesis was supported by the result of EMSA. A biotin-labeled 42-nt RNA probe based on the sequence between nucleotide 118–159 in AXIIR 5΄UTR was synthesized. EMSA showed that this probe bound specifically to both nuclear and cytoplasmic proteins of K562 cell (Figure 6A). This 42-nt RNA probe was then used to pull down the putative proteins binding to it. Electrophoresis of the pull-down elute from K562 cell lysate exhibited a band of approximate 36kD, which was not present in control groups (Supplementary Figure S5). Analysis of this band by protein mass spectrometry identified four putative proteins: ELAVL1, hnRNP G, hnRNPA0 and hnRNPA2B1. Considering that more probe bound to nuclear than to cytoplasmic proteins (Figure 6A) and mRNAs firstly encountered nuclear proteins once they were transcribed, nuclear proteins were used to perform super-shift EMSA to verify the binding of the four proteins to the 42-nt RNA probe. When antibodies for the above four proteins were added after incubation of the probe with nuclear proteins, the shift bands dramatically decreased in the groups that ELAVL1 and hnRNPA0 antibody were added. And there were weak super-shift bands when ELAVL1 antibody was added (Figure 6B). Since all the antibodies we used in EMSA were polyclonal, it was reasonable that the shift bands decreased rather than super-shifted. When the above antibodies were incubated with nuclear proteins before adding the probe, the shift bands significantly decreased in the groups with hnRNPA2B1 and hnRNPA0 antibody. In the group when ELAVL1 antibody was added, the shift bands slightly decreased and super-shift bands were observed (Figure 6C). In both super-shift EMSA methods, the shift bands in the group with hnRNP G antibody remained unchanged. The result proved that hnRNPA2B1, hnRNPA0 and ELAVL1 specifically bound to the 42-nt RNA sequence. This was supported by the presence of ELAVL1 binding site 5΄-NNUUNNUUU-3΄ (37) within the 42-nt sequence. The difference between the results acquired by the two super-shift EMSA methods could be due to that different epitopes on the surface of the proteins were accessible to the antibodies in the two methods.
Figure 6.
Identification of AXIIR 5΄UTR binding proteins. A2: anti-hnRNPA2B1 antibody. E: anti-ELAVL1 antibody. A0: anti-hnRNPA0. antibody G: anti-hnRNP G antibody (A) EMSA detection of the binding of the 42-nt sequence between nucleotides 118 and 159 in 5΄UTR with proteins from K-562 cells. (B) Super-shift EMSA verification of the binding of the 42-nt sequence with candidate proteins, antibodies were added after incubation of RNA probe with proteins. (C) Super-shift EMSA verification of the binding of the 42-nt sequence with candidate proteins, antibodies were incubated with the proteins before adding RNA probe. (D) Confirmation of protein interaction by Co-IP. Co-IP was performed 40 h after transfection of construct 5΄U into HEK293T cell. IgG: Co-IP with normal rabbit IgG. E: Co-IP with anti-ELAVL1 antibody. A2: Co-IP with anti-hnRNPA2B1 antibody. A0: Co-IP with anti-hnRNPA0 antibody. (E) Confirmation of direct protein interaction by Co-IP. Legends were the same as those for (D), except that cell lysate was treated with 20 μg/ml of RNase A for 1 h at temperature, prior to Co-IP.
Co-IP was further employed to investigate whether these proteins interacted with each other. hnRNPA2B1 was present in the complex immunoprecipitated by ELAVL1 antibody, and vice versa, hnRNPA0 existed in the complex immunoprecipitated by hnRNPA2B1 antibody, and vice versa, and hnRNPA0 also existed in the complex immunoprecipitated by ELAVL1 antibody and vice versa (Figure 6D). When mRNA was degradated by RNase A prior to Co-IP, hnRNPA0 existed in the complex immunoprecipitated by ELAVL1 antibody and vice versa (Figure 6E). No interaction was observed either between hnRNPA0 and hnRNPA2B1 or between ELAVL1 and hnRNPA2B1 (data not shown). These results showed that hnRNPA0 and ELAVL1 interacted directly. hnRNPA2B1 did not bind to hnRNPA0 and ELAVL1 directly. Instead, hnRNPA2B1 shared a common binding to AXIIR mRNA with hnRNPA0 and ELAVL1.
Collectively, the above data demonstrated that hnRNPA2B1, hnRNPA0 and ELAVL1 bound to the 42-nt sequence between nucleotide 118–159 in AXIIR 5΄UTR.
ELAVL1, hnRNPA0 and hnRNPA2B1 cooperate with uORFs to inhibit AXIIR translation
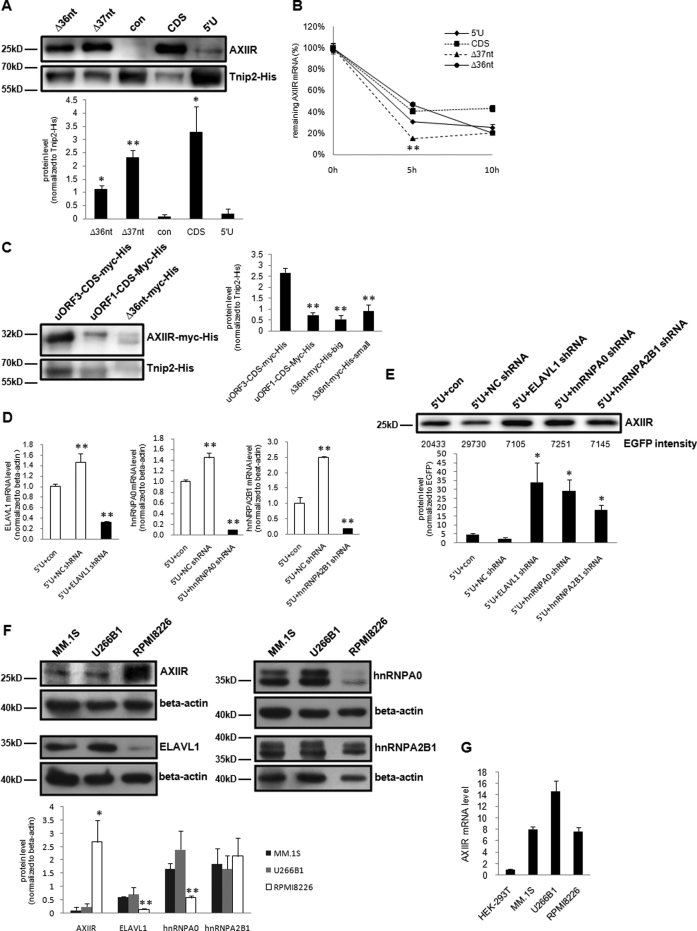
To verify that ELAVL1, hnRNPA0 and hnRNPA2B1 participate in the inhibition of AXIIR translation, we deleted part of the 42-nt sequence to disrupt the binding of the above proteins to AXIIR mRNA. Thirty six and thirty seven nucleotides containing most of the 42-nt sequence were deleted respectively in construct Δ36 and Δ37 nt. In both constructs, uORF3 and its Kozak context were retained to keep the original uAUG structure as much as possible (Supplementary Figure S6). Most of the sequences deleted in the two constructs were the same. However, in Δ37 nt, uORF1 was terminated at a stop codon upstream of the original stop codons, while the stop codon for uORF3 remained unchanged. In Δ36 nt, both uORF1 and uORF3 stopped at the original position. That is, Δ37 nt had shortened uORF1 with 21 codons and shortened uORF3 with 27 codons, Δ36 nt had shortened uORF1 with 42 codons and shortened uORF3 with 26 codons. Compared with 5΄U, Δ36 and Δ37 nt increased AXIIR protein level to 34 and 71% of that CDS did (Figure 7A). It could be inferred that without the binding of ELAVL1, hnRNPA0 and hnRNPA2B1 to AXIIR mRNA, inhibition of AXIIR translation was impaired. In Δ36 nt, shortened uORF1 with 42 codons could still maintain its inhibition on AXIIR translation. However, in Δ37 nt, both uORF1 and uORF3 were shortened to <30 codons and lost their effects, the protein level of AXIIR was greatly increased. The reason that AXIIR translation was not totally recovered in Δ37 nt might be that the 42-nt sequence was not completely deleted. The above results showed that ELAVL1, hnRNPA0 and hnRNPA2B1 work with uORFs to inhibit AXIIR translation.
Figure 7.
Investigation of the roles of ELAVL1, hnRNPA0 and hnRNPA2B1 in AXIIR Translation. (A) Western blot of AXIIR and Tnip2-His protein levels for 42-nt deleted constructs in HEK293T cell. Different constructs and Tnip2-His expressing plasmid were co-transfected into cells. Western blot was performed 40 h after transfection. Experiments were repeated in triplicates. Quantification of AXIIR level normalized to Tnip2-His was below the blot. Data represent mean ± s.d. *P < 0.05, **P < 0.01 (t-test as compared with 5΄U). (B) mRNA stability analysis of 42-nt deleted constructs in HEK293T cell. Experiments were repeated in triplicates. Data represent mean ± s.d. **P < 0.01 (t-test as compared with Δ36 nt). (C) Western blot of AXIIR-myc-His and Tnip2-His protein levels for 42-nt deleted construct in HEK293T cell. Legends were the same as those for (A), except that AXIIR-myc-His was blotted with anti-His tag antibody and quantification of AXIIR-myc-His level normalized to Tnip2-His was at the right side of the blot. **P < 0.01 (t-test as compared with uORF3-CDS-myc-His). (D) Inhibition efficiency of shRNAs against ELAVL1, hnRNPA0 and hnRNPA2B1 in HEK293T cell. Forty-eight hours after co-transfection of 5΄U and pc-EGFP-shRNA, the mRNA levels were measured by SYBR qRT-PCR. NC shRNA, ELAVL1 shRNA, hnRNPA0 shRNA and hnRNPA2B1 shRNA respectively represents their corresponding shRNA-expressing plasmid pc-EGFP-shRNA. (E) Western blot of AXIIR level for shRNA-expressing plasmids in HEK293T cell. 5΄U and shRNA-expressing plasmids were co-transfected into cells. Western blot was performed 48 h after transfection, and the intensity of EGFP was measured simultanously. Experiments were repeated in triplicates. Quantification of AXIIR level normalized to EGFP intensity was below the blot. Data represent mean ± s.d. *P < 0.05, (t-test as compared with 5΄U+con). (F) Western blot of AXIIR, hnRNPA0, ELAVL1, hnRNPA2B1 and β-actin protein level in multiple myeloma cell lines MM.1S, U266B1 and RPMI8226. Experiments were repeated in triplicates. Quantification of AXIIR, hnRNPA0, ELAVL1, hnRNPA2B1 level normalized to β-actin was below the blot. *P < 0.05, **P < 0.01 (t-test as compared with MM.1S) (G) qRT-PCR measurement of AXIIR mRNA level in cell lines.
mRNA stability analysis showed that the mRNA stability curve of Δ36 nt was similar to that of 5΄U (Figure 7B). Although at 5 h after Actinomycin D treatment, the remaining AXIIR mRNA level of Δ37 nt was lower than that of Δ36 nt, their level became the same at 10 h after Actinomycin D treatment. Besides, compared with Δ36, Δ37 nt had higher AXIIR protein level with lower AXIIR mRNA stability. Thus, it could be reasoned that the increasing of AXIIR protein level by Δ36 and Δ37 nt was unrelated to AXIIR mRNA stability.
To explore the underlying mechanism for the effect of the three proteins, the deletion-mutated uORF1 and uORF3 in Δ36 nt was fused with AXIIR CDS without main AUG (termed Δ36 nt–myc-His, Supplementary Figure S6). AXIIR CDS was 3΄-tagged with myc and 6xHis tag, to avoid possible interference from endogenous AXIIR. In construct Δ36 nt–myc-His, binding of the three proteins with AXIIR mRNA was disrupted, while uORF1 still remained its effect. Constructs uORF3-CDS-myc-His and uORF1-CDS-myc-His used in Figure 4E served as control. The result showed that construct Δ36 nt–myc-His mainly produced two bands. The size of the big band (termed Δ36 nt–myc-His-big) was equivalent to that of the fusion protein translated from the second non-canonical start codon AUU. The size of the small band (termed Δ36 nt–myc-His-small) was equivalent to that of the fusion protein translated from uAUG3. The intensity of the two bands was similar to that of the band expressed by uORF1-CDS-myc-His (Figure 7C). This result implied that ELAVL1, hnRNPA0 and hnRNPA2B1 focused the translation of uORFs on uORF1, rather than impeding the elongation of uORF translation. However, even abrogation of both uORF1 and uORF3 could not reverse all the inhibition on AXIIR translation, uORFs only accounted for part of AXIIR translation inhibition. As both uAUG1 and uAUG3 are in Kozak sequence context of moderate intensity, leaky scanning translation bypassing uORFs probably occurred. It could be inferred that ELAVL1, hnRNPA0 and hnRNPA2B1 might also attenuate the leaky scanning bypassing uORFs.
Next we examined whether knockdown of ELAVL1, hnRNPA0 and hnRNPA2B1 would improve AXIIR translation. In most cells, ELAVL1, hnRNPA0 and hnRNPA2B1 proteins are abundantly expressed, whereas AXIIR mRNA is relatively low. When ELAVL1, hnRNPA0 and hnRNPA2B1 were knocked down by shRNA, the remaining protein was still sufficient to inhibit endogenous AXIIR translation (data not shown). Therefore, we chose to co-transfect construct 5΄U and shRNA-expressing plasmids for ELAVL1, hnRNPA0 and hnRNPA2B1 into cells. In this way, the amount of AXIIR mRNA would be high, so that the effect of knockdown of ELAVL1, hnRNPA0 and hnRNPA2B1 would be easily detected. Moreover, EGFP coding sequence was cloned into shRNA-expressing plasmid, so that the intensity of EGFP could serve as transfection efficiency control. As expected, when ELAVL1, hnRNPA0 and hnRNPA2B1 was knocked down (Figure 7D), AXIIR protein level dramatically increased and shRNA against ELAVL1 showed the most significant effect (Figure 7E). This proved that ELAVL1, hnRNPA0 and hnRNPA2B1 involved in inhibition of AXIIR translation.
We went further to check the endogenous AXIIR protein level. Multiple myeloma cell line RPMI8226 expressed much higher level of AXIIR than other two myeloma cell lines MM.1S and U266B1 (Figure 7F), but AXIIR mRNA sequences of the three cell lines were the same and did not have any mutation (data not shown). This again supported that uORFs alone were not sufficient for the remarkable inhibition on AXIIR translation. In RPMI8226 cell expressing high level of AXIIR, the level of hnRNPA0 and ELAVL1 was significantly low, while the level of hnRNPA2B1 was similar to that in the other two cell lines (Figure 7F). However, AXIIR mRNA level in RPMI8226 cell was not higher (Figure 7G). This indicated that reducing the proteins that bind to AXIIR mRNA released AXIIR translation.
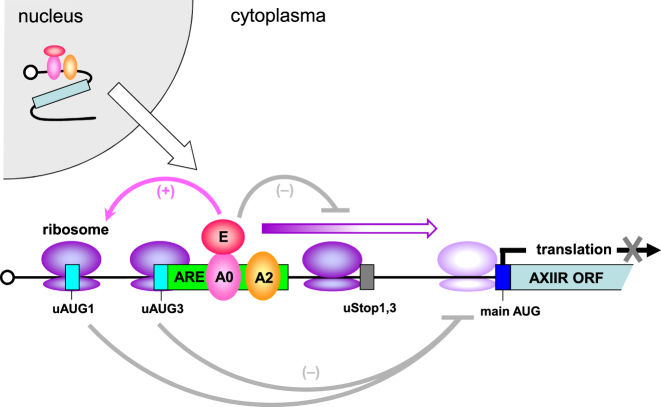
Based on the above results, we proposed a mechanism for the translation regulation of AXIIR (Figure 8). Once AXIIR mRNA was transcribed in the nucleus, it bound trans-acting proteins hnRNPA2B1, hnRNPA0 and ELAVL1. This protein–mRNA complex was then exported to the cytoplasm. Ribosome firstly initiated translation from uAUG1 in AXIIR 5΄UTR. If ribosome bypassed uAUG1, it could still initiate translation from uAUG3. uORF1 and uORF3 acted in such a fail-safe manner to ensure the translation initiation from uORFs. These uORFs are unfavorable for re-initiation after termination of uORF translation. Thus, downstream AXIIR translation was inhibited. However, as both uAUG1 and uAUG3 are in Kozak sequence context of moderate intensity, there was still certain degree of leaky scanning that bypassed uORFs. hnRNPA2B1, hnRNPA0 and ELAVL1 bound ARE that covered uAUG3. They not only focused the translation of uORFs on uORF1, but could also attenuate the leaky scanning bypassing uORFs. The proteins and uORFs acted synergistically and formed a multiple fail-safe system to prevent ribosomes from reaching the downstream main AUG. As a result, AXIIR translation was almost completely prohibited.
Figure 8.
Model of the mechanism underlying the inhibition of AXIIR translation. A2: hnRNPA2B1. E: ELAVL1. A0: hnRNPA0.
DISCUSSION
In summary, we elucidated that human AXIIR translation was inhibited by a multiple fail-safe system consisting two uORFs and trans-acting protein hnRNPA2B1, hnRNPA0 and ELAVL1. This is the first evidence that proteins and uORFs cooperate to regulate translation in higher organism.
So far, there is only one paper describing the cooperation of uORF and protein in translation, and it was found in Drosophila. In that model, one protein SXL regulates one uORF in controlling downstream main ORF translation and the uORF operates via initiation, not elongation or termination (16). Different from this mechanism, our findings show that in human cells, multiple trans-acting proteins work with uORFs to regulate downstream main ORF translation. The proteins not only affect the translation initiation from uORFs, but also attenuate the leaky scanning bypassing uORFs. It revealed that higher organism develops a much more complex translation regulatory system than lower organism.
Our model displays a system composed of multiple fail-safe mechanisms to tightly control AXIIR translation. This reflects the stringency of translation regulation. Such stringent control of gene expression is very important for those genes like AXIIR whose excessive expression is harmful to the host cell. On the other hand, gene expression needs to change with environment. Although the mRNA sequences of most genes generally remain unchanged, trans-acting proteins are readily responsive to environmental changes. Thus, the AXIIR translation regulation system possesses flexibility that can be adjusted. When environment changes, some trans-acting regulatory proteins may be modulated, which in turn adjust the translation of a batch of mRNAs. But different mRNA might respond differently to these regulatory proteins due to different sequence and structure feature of the mRNA. Thus, the AXIIR translation regulation system represents combination of stringency, flexibility, extensive and gene-specific regulation at translational level, making the cells respond to complicated environmental changes more accurately and rapidly.
Little is known about the proteins other than standard cap-dependent initiation factors that involve in translational regulation. hnRNPA2B1, hnRNPA0 and ELAVL1 are important regulatory proteins for gene expression, especially for mRNA stability. But previously they have not been associated with uORF in mechanism. In our model, each protein may exert different function in the protein–AXIIR mRNA complex. The detailed interpretation of their roles in the complex requires further investigations. As hnRNPA2B1, hnRNPA0 and ELAVL1 are ubiquitously expressed in many cells, studies on their roles in translation regulation may reveal the mechanisms underlying more physiological and pathological processes.
Besides hnRNPA2B1, hnRNPA0 and ELAVL1, there may be other RNA-binding proteins that can cooperate with uORFs with similar ways to control protein translation in higher organisms. However, since there are many RNA-binding proteins that recognize different binding site sequences in mRNA (38), it is hard to estimate the scope of such uORF and trans-acting protein cooperative mechanism on translation control. However, since ∼50% of mammalian mRNAs contain one or more uORFs (5), more of such models are expected to be discovered. In-depth investigation of their effects and underlying mechanisms will benefit the efforts both in understanding gene expression regulation and in manipulating recombinant protein expression (39).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Wei Sun in the Core Instrument Facility in CAMS/PUMC for protein mass spectrometry experiment.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Natural Science Foundation of China [31470067 to M.C.]; Major State Basic Research Development Program of China (973 Program) [2011CBA01100 to M.C.]; Program for New Century Excellent Talents in University of China (to M.C.); Xi'an Science and Technology Plan [SF1416 to Q.W.]. Funding for open access charge: by Natural Science Foundation of China [31470067].
Conflict of interest statement. None declared.
REFERENCES
- 1.Lu G., Maeda H., Reddy S.V., Kurihara N., Leach R., Anderson J.L., Roodman G.D.. Cloning and characterization of the Annexin II receptor on human marrow stromal cells. J. Biol. Chem. 2006; 281:30542–30550. [DOI] [PubMed] [Google Scholar]
- 2.Shiozawa Y., Havens A.M., Jung Y., Ziegler A.M., Pedersen E.A., Wang J., Wang J., Lu G., Roodman G.D., Loberg R.D. et al. Annexin II/Annexin II receptor axis regulates adhesion, migration, homing, and growth of prostate cancer. J. Cell Biochem. 2008; 105:370–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.D'Souza S., Kurihara N., Shiozawa Y., Joseph J., Taichman R., Galson D.L., Roodman G.D.. Annexin II interactions with the annexin II receptor enhance multiple myeloma cell adhesion and growth in the bone marrow microenvironment. Blood. 2012; 119:1888–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiong Y., Fan C., Kong L., Dong L., Zhu N, Zhang J., Wang L., Qin T., Shen Y., Chen M.. Annexin II receptor induces apoptosis independent of Annexin II. Apoptosis. 2013; 18:925–939. [DOI] [PubMed] [Google Scholar]
- 5.Calvo S.E., Pagliarini D.J., Mootha V.K.. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:7507–7512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rossi A., Pisani F., Nicchia G.P., Svelto M., Frigeri A.. Evidences for a leaky scanning mechanism for the synthesis of the shorter M23 protein isoform of aquaporin-4: implication in orthogonal array formation and neuromyelitis optica antibody interaction. J. Biol. Chem. 2010; 285:4562–4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gunišová S., Valášek L.S.. Fail-safe mechanism of GCN4 translational control–uORF2 promotes reinitiation by analogous mechanism to uORF1 and thus secures its key role in GCN4expression. Nucleic Acids Res. 2014; 42:5880–5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schepetilnikov M., Dimitrova M., Mancera-Martínez E., Geldreich A., Keller M., Ryabova L.A.. TOR and S6K1 promote translation reinitiation of uORF-containing mRNAs via phosphorylation of eIF3h. EMBO J. 2013; 32:1087–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ishimura R., Nagy G., Dotu I., Zhou H., Yang X-L., Schimmel P., Senju S., Nishimura Y., Chuang J.H., Ackerman S.L.. Ribosome stalling induced by mutation of a CNS-specific tRNA causes neurodegeneration. Science. 2014; 345:455–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jousse C., Bruhat A., Carraro V., Urano F., Ferrara M., Ron D., Fafournoux P.. Inhibition of CHOP translation by a peptide encoded by an open reading frame localized in the chop 5΄UTR. Nucleic Acids Res. 2001; 29:4341–4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nguyen H.L., Yang X., Omiecinski C.J.. Expression of a novel mRNA transcript for human microsomal epoxide hydrolase (EPHX1) is regulated by short open reading frames within its 5΄-untranslated region. RNA. 2013; 19:752–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Occhi G., Regazzo D., Trivellin G., Boaretto F., Ciato D., Bobisse S., Ferasin S., Cetani F., Pardi E., Korbonits M. et al. A novel mutation in the upstream open reading frame of the CDKN1B gene causes a MEN4 phenotype. PLOS Genet. 2013; 9:e1003350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spevak C.C., Ivanov I.P., Sachs M.S.. Sequence requirements for ribosome stalling by the arginine attenuator peptide. J. Biol. Chem. 2010; 285:40933–40942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaba A., Jacobson A., Sachs M.S.. Ribosome occupancy of the yeast CPA1 upstream open reading frame termination codon modulates nonsense-mediated mRNA decay. Mol. Cell. 2005; 20:449–460. [DOI] [PubMed] [Google Scholar]
- 15.Schleich S., Strassburger K., Janiesch P.C., Koledachkina T., Miller K.K., Haneke K., Cheng Y-S., Chler K.K., Stoecklin G., Duncan K.E. et al. DENR–MCT-1 promotes translation re-initiation downstream of uORFs to control tissue growth. Nature. 2014; 512:208–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Medenbach J., Ivanov Seiler M., Hentze M.W.. Translational control via protein-regulated upstream open reading frames. Cell. 2011; 145:902–913. [DOI] [PubMed] [Google Scholar]
- 17.Barceló C., Etchin J., Mansour M.R., Sanda T., Ginesta M.M., Sanchez-Arévalo Lobo V.J., Real F.X., Capellà G., Estanyol J.M., Jaumot M. et al. Ribonucleoprotein HNRNPA2B1 interacts with and regulates oncogenic KRAS in pancreatic ductal adenocarcinoma cells. Gastroenterology. 2014; 147:882–892. [DOI] [PubMed] [Google Scholar]
- 18.Stockley J., Villasevil M.E., Nixon C., Ahmad I., Leung H.Y., Rajan P.. The RNA-binding protein hnRNPA2 regulates β-catenin protein expression and is overexpressed in prostate cancer. RNA Biol. 2014; 11:755–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reinhardt H.C., Hasskamp P., Schmedding I., Morandell S., van Vugt M.A., Wang X., Linding R., Ong S.E., Weaver D., Carr S.A. et al. DNA damage activates a spatially distinct late cytoplasmic cell-cycle checkpoint network controlled by MK2-mediated RNA stabilization. Mol. Cell. 2010; 40:34–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang S.H., Elemento O., Zhang J., Zhuang Z.W., Simons M., Hla T.. ELAVL1 regulates alternative splicing of eIF4E transporter to promote postnatal angiogenesis. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:18309–18314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masuda K., Abdelmohsen K., Kim M.M., Srikantan S., Lee E.K., Tominaga K., Selimyan R., Martindale J.L., Yang X., Lehrmann E. et al. Global dissociation of HuR-mRNA complexes promotes cell survival after ionizing radiation. EMBO J. 2011; 30:1040–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mukherjee N., Corcoran D.L., Nusbaum J.D., Reid D.W., Georgiev S., Hafner M., Ascano M. Jr, Tuschl T., Ohle U., Keene J.D.. Integrative regulatory mapping indicates that the RNA-binding protein HuR (ELAVL1) couples pre-mRNA processing and mRNA stability. Mol. Cell. 2011; 43:327–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tominaga K., Srikantan S., Lee E.K., Subaran S.S., Martindale J.L., Abdelmohsen K.. Gorospe M Competitive regulation of nucleolin expression by HuR and miR-494. Mol. Cell. Biol. 2011; 31:4219–4231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Durie D., Lewis S.M., Liwak U., Kisilewicz M., Gorospe M., Holcik M.. RNA-binding protein HuR mediates cytoprotection through stimulation of XIAP translation. Oncogene. 2011; 30:1460–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yeh C.H., Hung L.Y., Hsu C., Le S.Y., Lee P.T., Liao W.L., Lin Y.T., Chang W.C., Tseng J.T.. RNA-binding protein HuR interacts with thrombomodulin 5΄-untranslated region and represses internal ribosome entry site–mediated translation under IL-1β Treatment. Mol. Biol. Cell. 2008; 19:3812–3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Winkler C., Doller A., Imre G., Badawi A., Schmid T., Schulz S., Steinmeyer N., Pfeilschifter J., Rajalingam K., Eberhardt W.. Attenuation of the ELAV1-like protein HuR sensitizes adenocarcinoma cells to the intrinsic apoptotic pathway by increasing the translation of caspase-2L. Cell Death Dis. 2014; 5:e1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cao C., Sun J., Zhang D., Guo X., Xie L., Li X., Wu D., Liu L.. The long intergenic noncoding RNA UFC1, a target of microRNA 34a, interacts with the mRNA stabilizing protein HuR to increase levels of β-Catenin in HCC cells. Gestroenterology. 2015; 148:415–426. [DOI] [PubMed] [Google Scholar]
- 28.Wang I.X., So E., Devlin J.L., Zhao Y., Wu M., Cheung V.G.. ADAR regulates RNA editing, transcript stability, and gene expression. Cell Rep. 2013; 5:849–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Erkinheimo T.L., Lassus H., Sivula A., Sengupta S., Furneaux H., Hla T., Haglund C., Butzow R., Ristimäki A.. Cytoplasmic HuR expression correlates with poor outcome and with Cyclooxygenase 2 expression in serous ovarian carcinoma. Cancer Res. 2003; 63:7591–7594. [PubMed] [Google Scholar]
- 30.Kozak M. At least six nucleotides preceding the AUG initiator codon enhance translation in mammalian cells. J. Mol. Biol. 1987; 196:947–950. [DOI] [PubMed] [Google Scholar]
- 31.Slavoff S.A., Mitchell A.J., Schwaid A.G., Cabili M.N., Ma J., Levin J.Z., Karger A.D., Budnik B.A., Rinn J.L., Saghatelian A.. Peptidomic discovery of short open reading frame-encoded peptides in human cells. Nat. Chem. Biol. 2013; 9:59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ebina I., Takemoto-Tsutsumi M., Watanabe S., Koyama H., Endo Y., Kimata K., Igarashi T., Murakami K., Kudo R., Ohsumi A. et al. Identification of novel Arabidopsis thaliana upstream open reading frames that control expression of the main coding sequences in a peptide sequence-dependent manner. Nucleic Acids Res. 2015; 43:1562–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luttermann C., Meyers G.. A bipartite sequence motif induces translation reinitiation in feline calicivirus RNA. J. Biol. Chem. 2007; 282:7056–7065. [DOI] [PubMed] [Google Scholar]
- 34.Meyers G. Characterization of the sequence element directing translation reinitiation in RNA of the calicivirus rabbit hemorrhagic disease virus. J. Virol. 2007; 81:9623–9632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mukherjee N., Jacobs N.C., Hafner M., Kennington E.A., Nusbaum J.D., Tuschl T., Blackshear P.J., Ohler U.. Global target mRNA specification and regulation by the RNA-binding protein ZFP36. Genome Biol. 2014; 15:R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leppek K., Stoecklin G.. An optimized streptavidin-binding RNA aptamer for purification of ribonucleoprotein complexes identifies novel ARE-binding proteins. Nucleic Acids Res. 2014; 42:e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meisner N.C., Hackermüller J., Uhl V., Aszódi A., Jaritz M., Auer M.. mRNA openers and closers: modulating AU-rich element-controlled mRNA stability by a molecular switch in mRNA secondary structure. Chembiochem. 2004; 5:1432–1447. [DOI] [PubMed] [Google Scholar]
- 38.Campbell Z.T., Wickens M.. Probing RNA-protein networks: biochemistry meets genomics. Trends Biochem. Sci. 2015; 40:157–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ferreira J.P., Overton K.W., Wang C.L.. Tuning gene expression with synthetic upstream open reading frames. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:11284–11289. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.