Abstract
The current classification of cells in an organism is largely based on their anatomic and developmental origin. Cells types and tissues are traditionally classified into those that arise from the three embryonic germ layers, the ectoderm, mesoderm and endoderm, but this model does not take into account the organization of cell type-specific patterns of gene expression. Here, we present computational models for cell type and tissue specification derived from a collection of 921 RNA-sequencing samples from 272 distinct mouse cell types or tissues. In an unbiased fashion, this analysis accurately predicts the three known germ layers. Unexpectedly, this analysis also suggests that in total there are eight major domains of cell type-specification, corresponding to the neurectoderm, neural crest, surface ectoderm, endoderm, mesoderm, blood mesoderm, germ cells and the embryonic domain. Further, we identify putative genes responsible for specifying the domain and the cell type. This model has implications for understanding trans-lineage differentiation for stem cells, developmental cell biology and regenerative medicine.
INTRODUCTION
Careful morphological studies of animal embryos indicate that as the embryo generates a complete body plan, the cells segregate into three germ layers or lineages of cells, composed of the endoderm (liver, lungs, etc.), mesoderm (heart, circulatory system, etc.) and ectoderm (skin, neural tissue, etc.), supplemented with embryonic tissues, extraembryonic tissues (placenta, primitive endoderm) and germ cells (sperm, oocytes) (1–4). Embryonic development has been extensively explored in animals where embryogenesis can proceed in vitro, particularly Xenopus, Zebrafish and Caenorhabditis elegans (5,6), and this work culminated in a description of the complete deterministic cell lineage for C. elegans (7). However, development is less well understood in mouse and other higher mammals, due to the inability to monitor development in vitro for extended periods of time. Instead, elaborate lineage tracing, morphology and genetic studies must tease apart the developmental processes that occur in mammalian embryos as they develop a complete body plan. However, there are several aspects of embryogenesis that are difficult to explain. The three germ lineages form during gastrulation of the embryo, yet cells show surprising plasticity even late in development, as lineage tracing studies indicate cross-lineage seeding can occur much later than gastrulation (1,7–9). In adult tissues, there are no known examples of natural trans-lineage differentiation, suggesting potent barriers blocking these conversions. Similarly, the three germ lineage model of cell type has limitations, as, for example, the neural crest has long been argued as a fourth germ lineage (10).
We reasoned that development leaves an ‘imprint’ upon later cell types, and that this imprint would manifest as lineage-specific gene expression programs that are maintained in adult tissues. By building models of gene expression organization we can then reconstruct developmental patterns from adult tissues. We became interested in using RNA-sequencing (RNA-seq) to understand the systematic organization of cell type by understanding gene expression programs in a global manner. RNA-seq is a powerful technique for the integration of diverse datasets as raw data is stored at an early stage of analysis, permitting the reanalysis of old data as novel computational techniques are developed. Critically, it is possible to uniformly compare data across labs and experimental platforms in a way that is challenging for microarray technologies (11,12), albeit microarray studies can contain many thousands of samples, a scale difficult to achieve with RNA-seq (13,14).
Using a dataset consisting of 921 RNA-seq samples, representing 272 normal mouse cell types or tissues, we built computational models of the global organization of gene expression patterns with the aim to understand how cell types and tissues are related and organized. Our results indicate the existence of new ‘domains’ of cell types, which are distinct from the existing three germ layers. We propose the division of the ectoderm into three domains (neurectoderm, surface ectoderm and the neural crest), and the division of the mesoderm into two new domains (the mesoderm proper and the immune system/blood mesoderm). This analysis resulted in the identification of a set of domain-specific master regulator genes and a topological map of developmental potential. This work constitutes a useful resource of uniformly analyzed RNA-seq data that covers a wide spectrum of mouse cell types and tissues, and the domain-specific genes described here will be of interest for developmental biologists and for researchers interested in cell fate conversions for regenerative medicine.
MATERIALS AND METHODS
RNA-seq working dataset and analysis pipeline
In total, the RNA-seq dataset used in this study consists of 921 biological samples, which resulted in 272 distinct C/Ts, collated from 113 publications (Supplementary Table S1, Supplementary Figure S1A and B). Raw RNA-seq data was downloaded from the short read archive (SRA) (15) and uniformly reanalyzed using RSEM (v1.2.31) (16) and bowtie2 (v2.2.8) and then normalized for GC content using EDASeq (v2.4.1) (17) (Supplementary Figure S1C), as previously described (11,18), except a threshold of 40 GC-normalized tags in any two samples were required to keep a gene. The Ensembl (mm10, v79) transcriptome was used for the RSEM alignment (see also Supplementary Results for a detailed description of the RNA-seq analysis pipeline). Samples in which <10% of the sequence library mapped to the transcriptome or with <0.4 million mapped reads were discarded. The mean expression of biological replicates was taken where biological replicates were available. Sequence depths ranged from 0.4 million mapped sequence tags (NK cells replicate 1), to 166 million mapped sequence tags (HSC MPP1 replicate 2), with a mean of 21 million mapped sequence tags (Supplementary Figure S1D). The typical Pearson correlation between biological replicates was >0.8 (Supplementary Figure S1E). 192 C/Ts have at least one replicate, whilst 80 samples are from a single unreplicated experiment. For the complete 29 267 genes in our selected annotation we could robustly detect 25 075 genes (Supplementary Figure S1F). Of the remaining genes, most were unannotated or predicted gene (Supplementary Figure S1G). The normalized RNA-seq data table, which includes all 25 075 genes (rows) and 272 C/Ts (columns) is included as a Supplemental Data File, and the entire analysis pipeline, starting from the RSEM output is available at https://bitbucket.org/oaxiom/big_tree_pub.
Construction of C/T co-regulatory networks, principal component analysis (PCA) and self-organizing maps (SOMs)
C/T networks were constructed by taking the pair-wise R2 correlation coefficient between all C/Ts and a network was constructed by building edges between nodes (C/Ts), based on weak (R2 > 0.55) or strong (R2 > 0.8) correlations. All networks were laid out using ‘neato’ (http://graphviz.org/) and the analysis was performed using the ‘network’ module of glbase (19). PCA was performed using the ‘pca’ module of glbase (19), which relies on sklearn PCA function. SOMs and PCA were constructed using a filtered set of genes (Supplementary Figure S2), the MDS to seed the SOM training network was generated from the first 13 principal components using the MDS sklearn function. SOMs were generated using the ‘som’ module of glbase.
RESULTS
RNA-seq data collection and properties
To explore the organization of gene expression and its relationship to cell type-specification we set out to collect a dataset that could comprehensively cover the major cell types and tissues in the mouse. We collected publically available RNA-seq data, based on three criteria: (i) normal wild-type cells and tissues, (ii) non-cancerous, non-transformed and (iii) not in vitro-derived. The exceptions were CD4+ T helper cells, bone marrow-derived macrophages, eosinophils, mast cells and dendritic cells, all of which are derived from an already committed hematopoietic precursor cell (Supplementary Table S1). The dataset also included some non-transformed cell types that can be maintained in culture and are thought to be related to their in vivo counterparts: specifically, embryonic stem cells (ESCs), epiblast stem cells (EpiSCs), trophoblast stem cells, fibroblasts, adipocytes and keratinocytes (Supplementary Table S1). Cancerous cells were excluded due to the common observation of exaggerated transcriptomes and distorted responses to stimuli (14,20), and ESC-derived in vitro differentiated cells were excluded, as they typically do not mature into their in vivo equivalents (21). Using these criteria, the final dataset consists of 921 individual RNA-seq experiments, which, after taking the mean of the available biological replicates, resulted in 272 distinct cell types/tissues/treatments (hereafter: ‘C/T’, as previously used in the CellNet studies (20)) (Supplementary Table S1, Supplementary Figure S1A). It would be preferable to use only purified individual cell types instead of mixed heterogeneous tissues. However, this data is not currently available, particularly in less well-studied tissues that cannot be easily dissociated into individual cell types (i.e. those cell types not in the early embryo or in the immune system). For example, the neural crest is only represented by samples derived from tissues, whilst the endoderm, mesoderm and ectoderm are substantially represented by tissue-derived RNA-seq samples (Supplementary Figure S1B). Moreover, although tissues are composed of a mix of cell types, evidence from the C. elegans indicates that tissues and organs are mostly composed of cells derived from a single germ lineage (6,7), and hence tissues may still be a useful dataset to extract global patterns of organization from.
This RNA-seq data collection was analyzed using a uniform analysis pipeline beginning with the raw unmapped FASTQ files and ending with normalized tag counts for gene expression (Supplementary Figure S1C–E). In total, we could detect 25 075 out of 29 267 annotated genes (Supplementary Figure S1F). Of the genes we did not detect most were predicted genes (43%), olfactory genes (22%) or lincRNAs (11%) (Supplementary Figure S1G). The FANTOM5 dataset (22) is a similar large-scale atlas of cell type and tissue expression, but we could not adequately merge our dataset with the FANTOM5 dataset, most likely due to differences in the technology used to generate gene expression measurements (RNA-seq versus deepCAGE) and ambiguities in combining gene versus transcript quantitation (Supplementary Figure S3).
Computational models of C/T gene expression organization
We next set out to organize C/Ts into ‘domains’ of related C/Ts, analogous to germ lineages, except the domain organization is derived from the global organization of C/T gene expression. This is based on the idea that development leaves an ‘imprint’ of the preceding developmental process that can be detected in somatic tissues. Two computational approaches were used to model the domains: tree cutting of a clustered co-correlation network (Figure 1) and principal component analysis (PCA) (Figure 2). For the PCA genes were filtered to remove very low expressed genes and genes with low variance (Supplementary Figure S2 and Supplementary Results). We will use these two complementary methods to argue for the division of C/Ts into eight major domains of gene expression: the neurectoderm, neural crest, surface ectoderm, endoderm, mesoderm, blood mesoderm, germ cells and the embryonic domains.
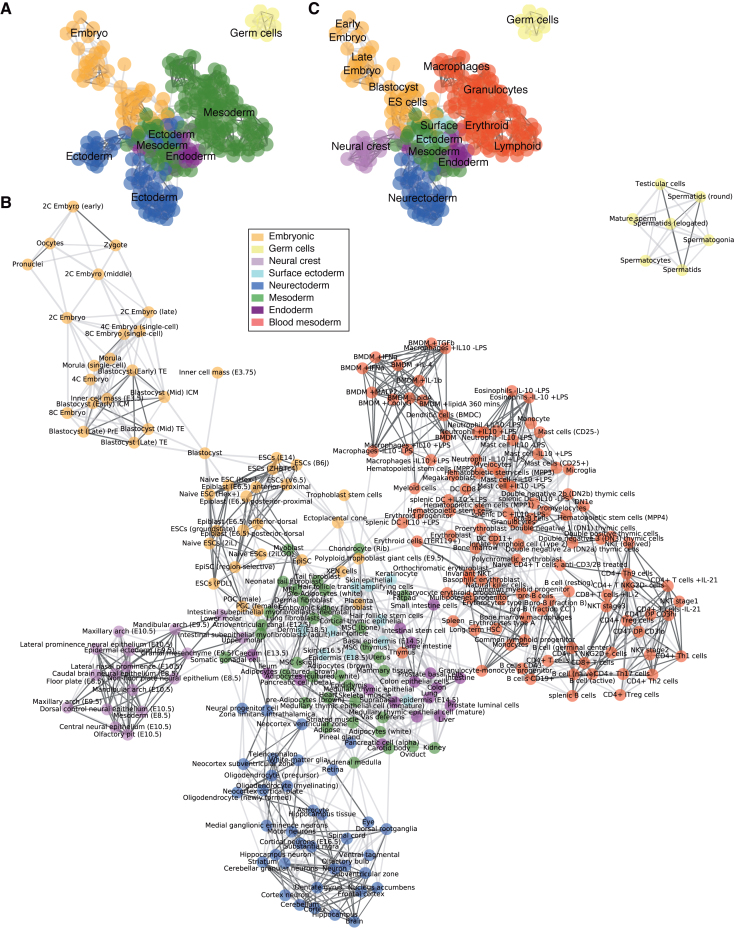
Figure 1.
A relational network of mouse C/Ts. Co-correlation network of mouse C/Ts. C/Ts were correlated (R2) and then clustered together in a network. Strong links (bold lines) are C/Ts with a correlation >0.8 and weak links (dotted lines) are C/Ts with a correlation >0.55. Each C/T in the network could only have a maximum of 10 edges to the best scoring other C/T. All expressed genes were used to generate the co-correlated network. (A) Colors indicate the annotated lineage, using the traditional three-germ lineage model. (B) Same as in panel A, but annotated with the individual C/T name and using a proposed 8 domain model of development. (C) Schematic layout of the network, same as in panel B.
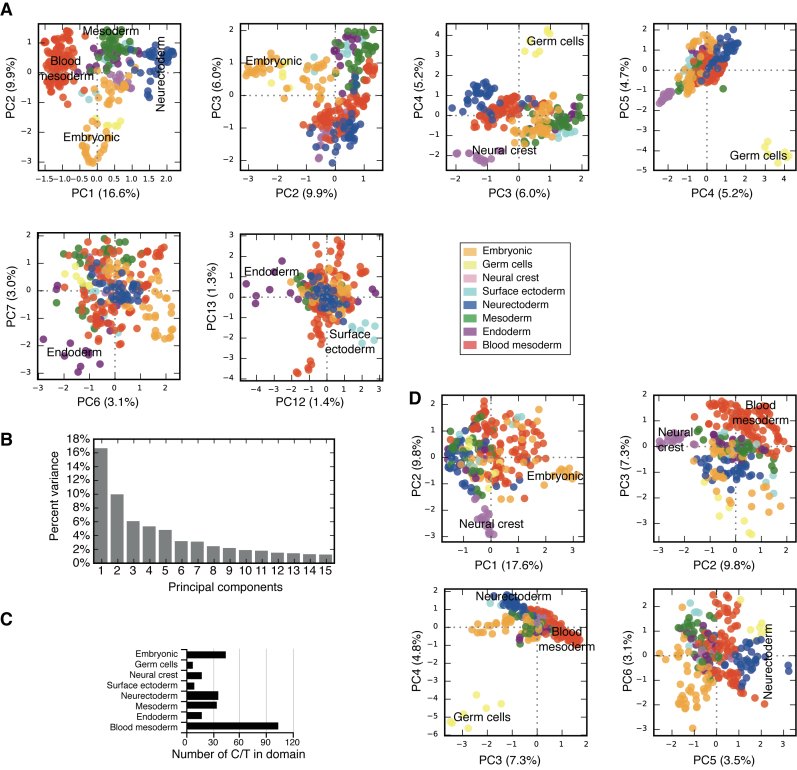
Figure 2.
Principal component analysis (PCA) of C/Ts. A filtered set of genes was used to perfrom PCA (Supplementary Figure S2A). (A) Selected principal component (PC) scatter plots indicating the emergence of domains of C/T identity. Blood mesoderm and neurectoderm emerge at PC1, embryonic and mesoderm at PC2, germ cells at PC4 and PC5, neural crest at PC4, the endoderm at PC7 and the surface ectoderm emerges at PC12. Percentages in brackets after the PC labels are the percent variance explained by each PC. (B) Barchart of the percent explained variance for the first 15 principal components. (C) Barchart of the number of C/Ts in each domain. (D) Selected PCs generated using only lncRNA annotated genes. The embryonic domain emerges at PC1, neural crest at PC2, the blood mesoderm at PC3, germ cells and the neurectoderm at PC4.
The blood mesoderm is distinct from the mesoderm proper
In the first approach to understand the organization of gene expression and its relationship to C/T-specification, a network was generated from the pair-wise correlation between all samples based on the strength of the correlation (Figure 1A). Each C/T was annotated with the presumed germ lineage, starting with the endoderm, mesoderm, ectoderm, germ cells and embryonic (oocyte through to ESCs) (Figure 1A). To rule out the influence of alternative, non-biological technical factors that could erroneously influence the clustering of the C/Ts, we collected metadata about the C/Ts, specifically, C/T derivation method, number of studies and replicates per C/T, the read lengths and read type, the sequencing machine and the number of mapped sequence tags (Supplementary Table S1). These metadata were then projected onto the correlation networks and none of the metadata could cluster the C/Ts in a meaningful way (Supplementary Figure S4A–H). We used tree-cutting of the clustered co-correlation matrix to guide the discrimination of specific domains of C/Ts and build major domains of C/T identity (Supplementary Figure S5A and B). Although, tree-cutting does not always show good performance, particularly when compared to gene regulation perturbation networks (20), to date there is not sufficient RNA-seq data to build these perturbation models. Tree-cutting indicated that at two clusters early embryonic and germ cells were distinct and at three clusters the blood mesoderm (hematopoietic system) was distinct (Supplementary Figure S5A and B). An alternative approach using PCA, similarly indicated that the blood mesoderm and neurectoderm split at PC1 and the remaining mesoderm C/Ts and embryonic C/Ts split at PC2 (Figure 2A and B). Additionally, based on the use of E-Cadherin (Cdh1) and N-Cadherin (Cdh2), as a rough proxy for epithelial/mesenchymal C/Ts respectively (23), the blood mesoderm could be classified as neither epithelial nor mesenchymal (Supplementary Figure S5C and D). A similar separation of the blood mesoderm C/Ts from other C/Ts was previously observed in a systematic analysis of microarray data (14,24). Based on these arguments we designated the blood mesoderm as a distinct domain, separate from other mesoderm C/Ts (Figure 1B and C).
Neurectoderm, neural crest and surface ectoderm are separate domains from the ectoderm
Visual inspection of the co-correlation network revealed three areas of the network that were composed of ectoderm C/Ts (Figure 1B). The C/Ts within the areas were related, and suggested three names for these domains: the neurectoderm (brain and spinal cord), surface ectoderm (skin, keratinocytes) and neural crest (molars, mandibular arch). Tree-cutting indicated a division between the neurectoderm and the surface ectoderm as the neurectoderm split from the surface ectoderm at seven clusters (Supplementary Figure S5B). PCA separated the neurectoderm on PC1, the neural crest on PC4 and the surface ectoderm on PC12 (Figure 2A). Based on these arguments we divide cells into eight major domains: Neurectoderm, blood mesoderm, mesoderm, embryonic, neural crest, endoderm and the surface ectoderm, supplemented by germ cells. The number of C/Ts in each domain ranges from the smallest (7, germ cells) to the largest (105, blood mesoderm) (Figure 2C).
The expression pattern of long non-coding RNAs alone can recover the embryonic, neurectoderm, blood mesoderm, germ cell, neural crest and mesoderm domains
The analysis presented above uses combinations of either all expressed genes or filtered gene sets. Potentially other important regulatory genes may contain the pattern of domain-specific gene expression. Long non-coding RNAs (lncRNAs) lack an obvious protein coding-sequence, instead they function as RNA molecules and are implicated in a wide array of developmental and biological processes (25–27). The transcriptome annotation used in this analysis (Ensembl version 79), contained 1789 expressed lncRNA genes (Figure 2D). Intriguingly, when we subjected just these 1789 lncRNAs to PCA, we could recover the embryonic domain (PC1), neural crest (PC2), blood mesoderm (PC3), germ cells (PC4), and neurectoderm (PC4/5) (Figure 2D). At no PC did we observe separation of the endoderm or the surface ectoderm. This result suggests that the domain-specific gene expression pattern is encoded not just within the pattern of expression of coding genes, but also within the expression pattern of lncRNAs.
SOMs support the division of C/Ts into eight domains and suggest further possible subdivisions
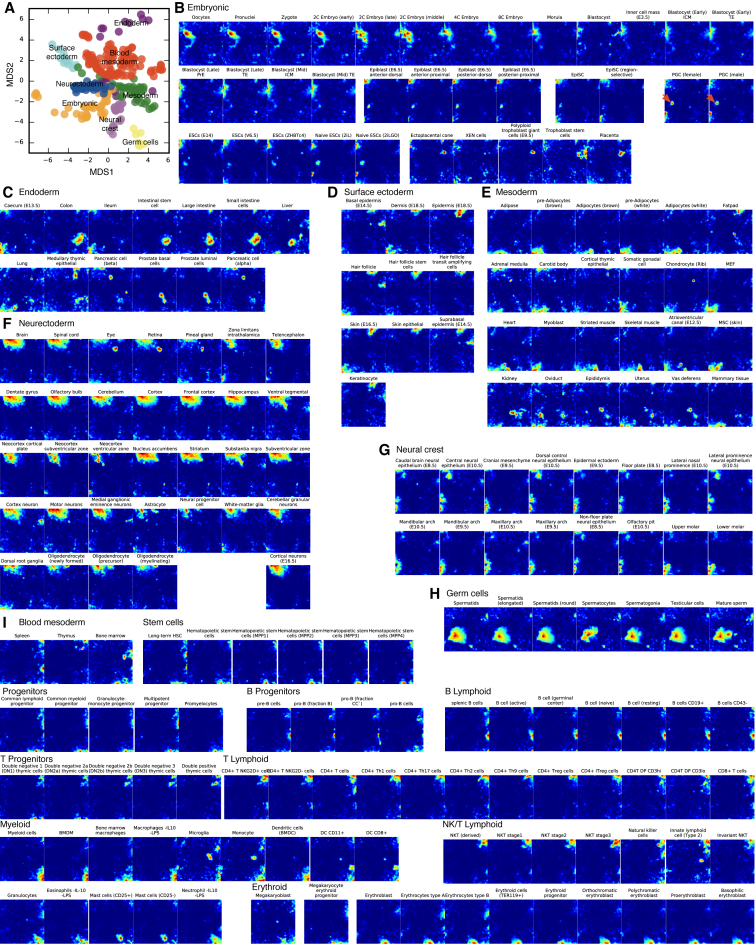
To explore the gene expression pattern of the C/Ts within each domain and explore the domain-specific gene expression programs self-organizing maps (SOMs) were generated for all 272 C/Ts. SOMs are a computational technique that can be used to intuitively represent a C/Ts total gene expression in a single small image (28–30). The SOMs contain all genes, arranged in a co-correlated set of active genes that appear as yellow-red ‘hotspots’. We trained the SOMs using an initial seed generated from a multi-dimensional scale (MDS) plot of the first 13 principle components (Figure 3A). Similar to the correlation networks, projection of metadata onto the trained MDS did not reveal a strong influence of non-biological factors on the arrangement of the C/Ts (Supplementary Figures S6 and S7). Once trained on the MDS, the SOMs also supported nine major divisions, consisting of the embryonic (Figure 3B), endoderm (Figure 3C), surface ectoderm (Figure 3D), mesoderm (Figure 3E), neurectoderm (Figure 3F), neural crest (Figure 3G), germ cells (Figure 3H) and blood mesoderm (Figure 3I). Certain partially committed cells show signs of their eventual fate, for example, primordial germ cells show germ cell domain character (Figure 3B, red arrow). Also there is fine grained structure between different C/Ts within each domain. For example, early versus late embryonic cells show both shared and distinct gene expression profiles (Figure 3B), and myeloid and lymphoid cells of the blood mesoderm can also be discriminated by their SOMs (Figure 3I). However, overall it is remarkable how the SOMs can accurately represent the domains of gene expression in the specific C/Ts.
Figure 3.
Self-organizing maps (SOMs) reveal domain-specific gene expression profiles and C/T identity. (A) The initial seed for the SOMs was generated from an MDS of the first 13 principal components Genes were filtered before being subjected to PCA/MDS/SOM (Supplementary Figure S2). SOMs were generated for all 231 C/Ts, although some SOMs were removed here for clarity. The SOMs are arranged into their respective domains: embryonic (panel B), endoderm (panel C), surface ectoderm (panel D), mesoderm (panel E), neurectoderm (panel F), neural crest (panel G), germ cells (panel H) and the blood mesoderm (panel I). Potential subdivisions are indicated for the blood mesoderm. Putative germ cell character in the primordial germ cells (PGC) is indicated with red arrows.
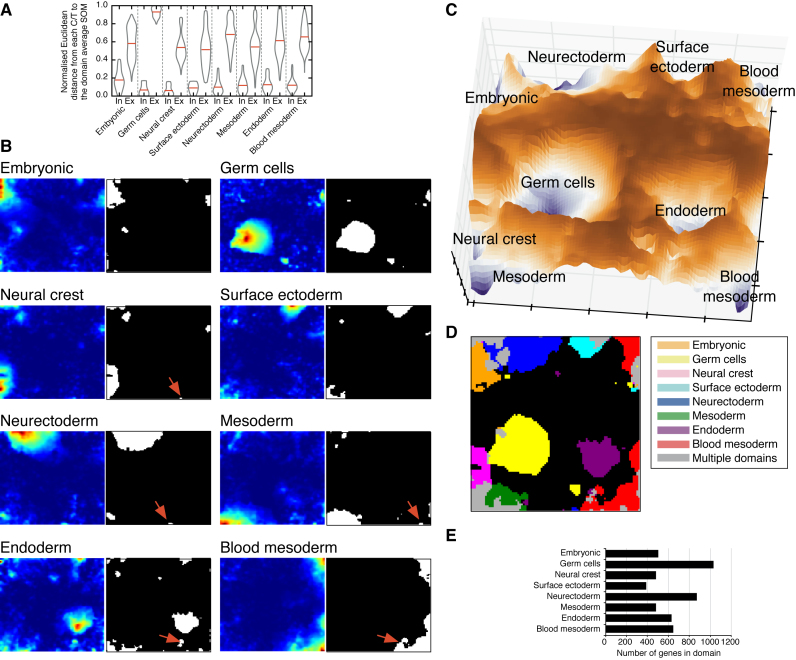
The Euclidean distances between SOMs helps refine C/T domain membership
SOMs can also be used as a measure of similarity to determine if an individual C/T is allocated to the correct domain. For each domain the average SOM was generated using all C/T SOMs within the domain, and the Euclidean distance of each C/T to the domain-average SOM was measured (Supplementary Table S2). The Euclidean distance between each SOM and the domain-average SOM can be used as a proxy to determine C/Ts closer to alternate domains (Supplementary Table S2). We plotted the distributions of inter-domain and extra-domain Euclidean distances as support for genuine separation between the C/Ts that make up the domains (Figure 4A). Overall, the domains were distinct and showed good within-group membership, however, there remained fifteen potential C/Ts for which the SOM was closer to an alternate domain, rather than their presumed developmental domain (Supplementary Figure S8). The distinction between the endoderm and mesoderm is not ideally clear, and nine of the fifteen cell types were either annotated as endoderm when they are presumed to come from the mesoderm or vice versa (Supplementary Figure S8A and B). For the remaining misannotated C/Ts, the pineal gland is derived from the neurectoderm, but was annotated to the mesoderm (Supplementary Figure S8C). Close inspection of the pineal gland SOM indicated a mixed neurectoderm/mesoderm signature (Supplementary Figure S8C) and may reflect difficulties in accurately dissecting this tissue. Similarly, the Dermis (E18.5) C/T showed a mixed surface ectoderm/mesoderm SOM (Supplementary Figure S8D) and may reflect a mixing of the surface ectoderm and mesoderm in the lower layers of the skin. EpiSCs (region-selective) are an embryonic stem cell type that has restricted potential compared to normal EpiSCs (31). Intriguingly, instead of the embryonic domain, these cells were annotated as closer to the neural crest (Supplementary Figure S8E), perhaps suggesting a bias in their developmental potential. Finally, several extraembryonic C/Ts were annotated as closer to the endoderm than the embryonic domains (Supplementary Figure S8F), this ambiguity perhaps reflects similarities in gene expression programs between extraembryonic and embryo-proper tissues. Overall, the SOMs are capable of accurately distinguishing between the different domains.
Figure 4.
SOMs as topological surfaces for domain-specific gene expression programs. (A) Violin plots of the distribution of Euclidean pair-wise distances between all C/Ts within the same domain (Inter-domain ‘In’), or against all C/Ts not within the same domain (Extra-domain ‘Ex’). (B) The mean of all the SOMs in each domain was calculated to generate ‘domain-average SOMs’. Hotspots indicate regions of domain-specific sets of genes. The SOMs were then thresholded and all nodes/genes scoring >0.2 (the top 80%) were collected as specific to a particular domain SOM (white regions). This threshold was chosen empirically to include known domain-specific genes. Red arrows indicate small amounts of blood mesoderm expression shared between multiple domains. (C) The domain-specific SOMs were used to construct a topological surface by taking the maximum depth value from each of the domain-average SOMs. (D) A two-dimensional representation of the combined SOM showing the regions of the SOM specific to a particular domain. Grey areas are shared between two or more domains. (E) The total number of domain-specific genes in each domain and the domain-specific transcription factors detected in the SOMs.
Topological surfaces reveal domain-specific gene expression programs
We next set out to utilize the SOMs as a topological map of the differentiation potential of the cells, and to utilize this map to discover the underlying organization of gene expression that explains the domains. Regions of high co-regulated genes appear as ‘hotspots’ in the SOM maps (yellow-red; Figure 3B–I), and the C/Ts that make up each domain can be combined into a domain-average SOM (Figure 4B). The SOMs also indicate potential contaminating C/Ts in alternate domains, a phenomenon that manifests as small hotspots of genes in common between two or more domains. For example, small amounts of blood mesoderm character is present in most SOMs (Figure 4B, red arrows), likely reflecting circulating immune cells in almost all tissues of the body.
The maximum depth of all of the domain-average SOMs was used to construct a topological map of the C/T domains (Figure 4C). This surface contains hills and valleys that represent particular domains of gene expression (Figure 4D), somewhat like the islands in the ‘ocean expanses’ between cell fates (32,33), or as ‘attractors’ in the state space of dynamical systems (34), although unlike the Waddington epigenetic model it does not contain any downward slope (35). In this topological map, the attractors are sets of domain-specific genes, and the genes can be extracted by taking all of the domain-specific nodes in the bottom 80% of the SOM (the white regions in Figure 4B, and the colored regions in Figure 4D). The domains had between 393–1040 domain-specific genes (Figure 4E, Supplementary Table S3). In total, there were 5093 domain-specific genes and as genes can be in more than one domain, the total set of unique genes was 4870 (Supplementary Table S3).
Extracting domain-specific gene expression programs from the domain average SOMs
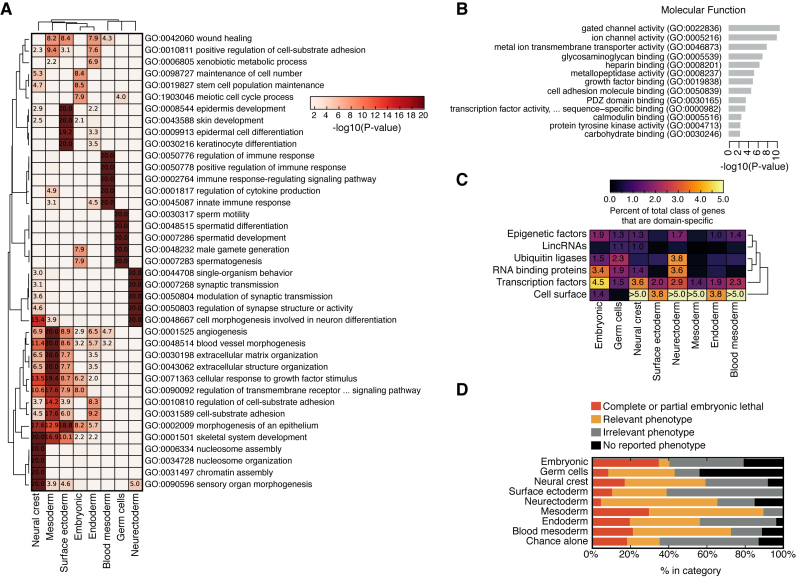
To confirm the role of the domain-specific genes in their respective domains we analyzed the domain-specific genes defined by the SOMs for over-enriched GO terms (Figure 5A). For seven of eight domains we discovered domain-relevant significantly enriched GO terms, specifically, ‘stem cell population maintenance’ (embryonic), ‘spermatogenesis’ (germ cells), ‘skin development’ (surface ectoderm), ‘synaptic transmission’ (neurectoderm), ‘angiogenesis’ (mesoderm), ‘xenobiotic metabolic process’, (endoderm, i.e. liver/intestine function), and ‘regulation of immune response’ (blood mesoderm). For the remaining neural crest domain a relevant term was less obvious and only ‘morphogenesis of an epithelium’ is suggestive of the epithelial to mesenchymal transitions that are critical for the formation of this domain during development (36).
Figure 5.
Validation of the domain-specific genes. (A) Significantly enriched ‘Biological Process (BP)’ gene ontology (GO) terms for all of the genes in each domain. Only the top 10 GO terms are shown for each domain. (B) GO analysis for all genes identified in any domain-specific SOM. GO terms were used from the ‘molecular function’ category. (C) Percentage of the total number of genes annotated as specific to a domain for various functional categories. The percent is indicated if it is ≥1.0% of the total number of genes in its category. (D) Phenotypes for knockout mice for all domain-specific genes were collected from the Mouse Genome Informatics (MGI) website and scored for a relevant or irrelevant knockout phenotype based upon the affected tissues in the mutant mouse (Supplementary Table S4).
Each domain contains many different types of gene with diverse biological function, GO analysis using the ‘molecular function’ category revealed that the cell surface molecules are important components of the domain-specific genes (Figure 5B), in agreement with other reports that gene products localizing to the cell surface are C/T-specific (37,38). The GO analysis also revealed the presence of sequence-specific transcription factors as a major class of domain-specific genes (Figure 5B). To explore this in more detail, we collected various types of regulatory genes and measured what percentage of the total class of genes is specific to a domain (Figure 5C). This analysis again highlighted the cell surface receptors as major determinants of domain-specific expression, along with transcription factors. Other classes of regulatory molecules seemed less important, with the exception of RNA binding proteins in the neurectoderm and embryonic domains and the ubiquitin ligases in the neurectoderm and germ cells (Figure 5C). This domain-specific enrichment of different classes of regulatory molecules hints at differences in developmental control, such as cell autonomous embryonic development, driven by TFs, versus the co-operative action of adult tissues, driven by cell–cell communication. The domain-specific TFs were then scored for a relevant or irrelevant mouse knockout (i.e. does the mouse knockout cause a relevant phenotype in appropriate domain-related tissues?), and, as a control, the genes from one domain were scored as if they were relevant in an alternate domain, to estimate the approximate number of matching phenotypes expected by chance alone (Supplementary Table S4). For all domains, >40% of the TFs showed a relevant domain-specific mouse knockout phenotype (Figure 5D), whilst phenotypes by chance alone were <40%. Interestingly, the TFs in the embryonic domain also showed an increased likelihood of an embryonic lethal mouse knockout phenotype (Figure 5D), supporting their involvement in embryonic development, although as embryonic lethal mouse phenotypes are potentially easier to observe this may bias their increased likelihood. This analysis indicated that these TFs are relevant to their specific domain and so comprise a regulatory module that helps determine the overall domain-specific gene expression program.
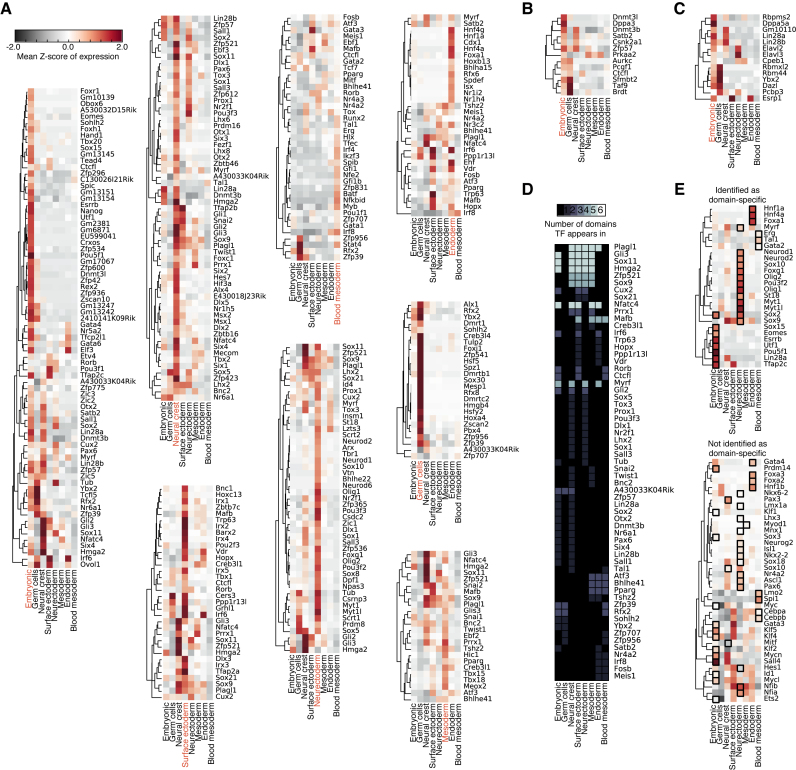
To understand the transcriptional regulatory programs underlying each domain in closer detail, the mean Z-score of expression was measured for all TFs within each domain (Figure 6A, Supplementary Table S5). Within the domains are many well-known C/T-specific regulators (Figure 6A). For example, critical regulators of the embryonic domain included: Pou5f1, Sox2, Nanog, Esrrb, Dnmt3l, Utf1 (34,39,40), along with the primitive endoderm genes, Gata4, Gata6 and Eomes (41), and the trophoblast genes, Hand1 and Tfap2c. Other domains also contained well known C/T-specific regulators: blood mesoderm—Tal1 (42,43), endoderm—Foxa1 and Hnf4a (21), mesoderm—Irx3, Irx5 (44), neural crest—Nr2f1, Msx1, Msx2 (45,46), neurectoderm—Sox10, Olig1, Olig2, Neurod1, Neurod2, Neurod6, Myt1, Myt1l and Pou3f2 (47–49), surface ectoderm—Trp63 (50), and germ cells—Dmrtb1 (51). Other classes of regulatory factors important for specific domains were also identified, for example Dnmt3l and other epigenetic regulatory enzymes were identified as specific to the embryonic domain (Figure 6B, Supplementary Table S6), as were the RNA binding proteins Lin28a/b (Figure 6C, Supplementary Table S7). Additionally the receptors and cell surface molecules emerge as a major class of regulatory molecule (Supplementary Figure S9A–H, Supplementary Table S8) particularly in somatic C/Ts. These non-TF factors may play context-specific roles in their respective domains.
Figure 6.
Transcription factors specific to a domain are important C/T regulators. (A) Heatmaps of the C/T expression of transcription factors identified by the SOMs as domain-specific. The domain is indicated with a red label on the heatmap. The gene expression matrix was first background subtracted to remove the influence of ‘non-expressed genes’ below the threshold of 40 normalized sequence tags, log2 transformed and converted to a gene-wise Z-score. Displayed here is the mean of the Z-scores for all of the C/Ts in the respective domains for all TFs annotated as domain-specific. (B) Heatmap of epigenetic factors in the embryonic domain. Shares the same color-bar scale with panel A. (C) Heatmap of RNA binding proteins in the embryonic domain. Shares the same color-bar scale with panel A. (D) TFs that appear in more than 1 domain. (E) TFs used in trans-domain differentiation protocols, either identified as domain-specific (top) or not identified (bottom). The black squares indicate the target domain the TF was used to differentiate a C/T to.
We noticed that many TFs were not confined to a single domain but could be found within multiple domains (Figure 6D, Supplementary Table S9), particularly TFs such as Sox9, which was identified as domain-specific for the mesoderm, neural crest, neurectoderm and surface ectoderm domains. This matches closely to Sox9's known biological functions, as Sox9 is known to regulate neural crest development (52), glial cell commitment in the neurectoderm (53), chondrocyte function in the mesoderm (54) and hair stem cell development in the surface ectoderm (55). Other cross-domain TFs may also similarly participate in multiple roles in multiple domains.
Domain-specific genes as transdifferentiation markers
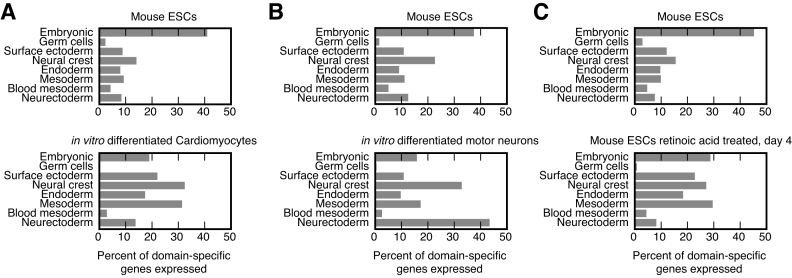
To determine the utility of these TFs in potential trans-differentiation experiments, we analyzed the known TF-mediated trans-differentiation protocols (56). Many of these protocols are within-domain transdifferentiation protocols. For example, in the conversion of fibroblasts to cardiomyocytes, both cell types are mesoderm-derivatives (57), whilst the transdifferentiation of macrophages to B cells, involves two blood mesoderm cell types (58). Of the transdifferentiation protocols that we could identify as crossing a domain, and that were not going from the embryonic domain to a differentiated somatic cell (56), 40% (36/91) of the TFs utilized for transdifferentiation were domain-specific (Figure 6E, Supplementary Table S10). Monte Carlo simulation of the expected number of matching TFs, drawn from all expressed TFs, suggests the expected number of observations by chance alone is 1.2% (1.1/91, 100 000 simulations, P < 0.01 Fisher exact test). Consequently, although we cannot identify all trans-differentiation TFs, the lists of domain-specific TFs is a potentially useful set of genes to extract putative candidate TFs for trans-domain differentiation experiments, and to score the destination of in vitro differentiated cells. As an example of the latter, we analyzed three RNA-seq datasets that profiled the differentiation of mouse ESCs to determine the lineage commitment as the cells are in vitro differentiated. Mouse ESCs differentiated to cardiomyocytes (59) showed a downregulation of embryonic domain genes, and an upregulation of mesoderm and neural crest genes (Figure 7A), and in vitro differentiated motor neurons (60) showed a strong commitment to a neurectoderm cell fate (Figure 7B). Conversely, retinoic acid treatment of ESCs (61) showed a commitment to multiple cell fates (Figure 7C). This analysis shows the utility of these gene sets to identify the direction of in vitro differentiation.
Figure 7.
Domain-specific genes can be used to score in vitro differentiation direction. Domain-specific genes were counted as expressed in mouse ESCs or in their differentiated progeny and are presented as the percent of the total domain-specific genes expressed in those cells. (A) Percent of domain-specific genes from the indicated domains in mouse ESCs (top) or in vitro differentiated cardiomyocytes (bottom). Data was from GSE67868 (59). (B) Percent of domain-specific genes from the indicated domains in mouse ESCs (top) or in vitro differentiated motor neurons (bottom). Data was from GSE60240 (60). (C) Percent of domain-specific genes from the indicated domains in mouse ESCs (top) or mouse ESCs treated with the pro-differentiation agent retinoic acid (bottom). Data was from GSE39523 (61).
DISCUSSION
The three germ layer model of cell type classification is the major model for specification of cell type during development (62). Derived mainly from morphological and anatomical observations it has been wildly successful in defining C/T organization. Here, we reinterpret this model using computational and genomics techniques to understand the global organization of C/Ts and their relationships. Using co-correlation networks, PCA and SOMs these models suggest the existence of eight major domains of C/T identity, corresponding to the neurectoderm, neural crest, surface ectoderm, endoderm, mesoderm, blood mesoderm, germ cells and embryonic domains. Intriguingly we could recover most of this domain-organization using only lncRNA expression, suggesting that the domain-specific pattern of gene expression is embedded within the expression pattern of multiple regulatory molecules.
The analysis performed here is capable of detecting genes required for domain maintenance, but one caveat remains the difficulty of these techniques to detect genes only required during the creation of the domain during development. For example, two TFs critical for the formation of the endoderm and trophectoderm, Sox17 and Cdx2, respectively (63–65), were not detected by the SOMs as specific to the embryonic domain, as would be expected. Similarly, of the known neural crest master regulators, Nr2f1, Nr2f2, Msx1, Msx2 and Tfap2a (45,46), only Nr2f1, Msx1 and Msx2 were detected as domain-specific. Potentially, these missing TFs are only required for initiation and not maintenance of the C/T-domain during development, making their identification difficult when analyzing mature domain-committed C/Ts.
In summary, we propose that mouse C/Ts are composed of eight major domains: the neurectoderm, neural crest, surface ectoderm, endoderm, mesoderm, blood mesoderm, germ cells and embryonic domains. This model has potential implications for understanding development, adult tissue homeostasis and manipulation of cell types in vitro. In combination with other computational approaches such as the perturbation modeling of CellNet (20) or the systematic identification of C/T-specific TFs (13,66), these analyses can be used to optimize in vitro transdifferentiation protocols. In the future it will be desirable to build models of C/T and domain specification in human (67), and also from multiple levels of data, from promoter and enhancer data (22,68,69), to epigenomic data (70), and mass spectrometry-based techniques (71,72), such as in the integrative framework of Mogrify (73), and so build detailed, rationally designed roadmaps for in vitro transdifferentiation of desired cell types. Ultimately, the ability to profile the gene expression of single cells will reshape the concept of cell type identity (43,74,75) and enhance our understanding of the critical concept of cell type.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Ralf Jauch for helpful discussions.
FUNDING
National Natural Science Foundation of China [31471242 and 31550110206 both to A.P.H. 11422108 to Y.W.]. Funding for open access charge: NSFC grants and core Shenzhen funding.
Conflict of interest statement. None declared.
REFERENCES
- 1. Ferrer-Vaquer A., Viotti M., Hadjantonakis A.K.. Transitions between epithelial and mesenchymal states and the morphogenesis of the early mouse embryo. Cell Adhesion Migration. 2010; 4:447–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tam P.P., Loebel D.A., Tanaka S.S.. Building the mouse gastrula: signals, asymmetry and lineages. Curr. Opin. Genet. Dev. 2006; 16:419–425. [DOI] [PubMed] [Google Scholar]
- 3. Technau U., Scholz C.B.. Origin and evolution of endoderm and mesoderm. Int. J. Dev. Biol. 2003; 47:531–539. [PubMed] [Google Scholar]
- 4. Peng G., Suo S., Chen J., Chen W., Liu C., Yu F., Wang R., Chen S., Sun N., Cui G. et al. Spatial transcriptome for the molecular annotation of lineage fates and cell identity in mid-gastrula mouse embryo. Dev. Cell. 2016; 36:681–697. [DOI] [PubMed] [Google Scholar]
- 5. Aanes H., Winata C.L., Lin C.H., Chen J.P., Srinivasan K.G., Lee S.G., Lim A.Y., Hajan H.S., Collas P., Bourque G. et al. Zebrafish mRNA sequencing deciphers novelties in transcriptome dynamics during maternal to zygotic transition. Genome Res. 2011; 21:1328–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hashimshony T., Feder M., Levin M., Hall B.K., Yanai I.. Spatiotemporal transcriptomics reveals the evolutionary history of the endoderm germ layer. Nature. 2015; 519:219–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sulston J.E., Schierenberg E., White J.G., Thomson J.N.. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 1983; 100:64–119. [DOI] [PubMed] [Google Scholar]
- 8. Tzouanacou E., Wegener A., Wymeersch F.J., Wilson V., Nicolas J.F.. Redefining the progression of lineage segregations during mammalian embryogenesis by clonal analysis. Dev. Cell. 2009; 17:365–376. [DOI] [PubMed] [Google Scholar]
- 9. Viotti M., Nowotschin S., Hadjantonakis A.K.. SOX17 links gut endoderm morphogenesis and germ layer segregation. Nat. Cell Biol. 2014; 16:1146–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hall B.K. The neural crest as a fourth germ layer and vertebrates as quadroblastic not triploblastic. Evol. Dev. 2000; 2:3–5. [DOI] [PubMed] [Google Scholar]
- 11. Hutchins A.P., Takahashi Y., Miranda-Saavedra D.. Genomic analysis of LPS-stimulated myeloid cells identifies a common pro-inflammatory response but divergent IL-10 anti-inflammatory responses. Scientific Rep. 2015; 5:9100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. SEQC/MAQC-III Consortium. A comprehensive assessment of RNA-seq accuracy, reproducibility and information content by the Sequencing Quality Control Consortium. Nat. Biotechnol. 2014; 32:903–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Heinaniemi M., Nykter M., Kramer R., Wienecke-Baldacchino A., Sinkkonen L., Zhou J.X., Kreisberg R., Kauffman S.A., Huang S., Shmulevich I.. Gene-pair expression signatures reveal lineage control. Nat. Methods. 2013; 10:577–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lukk M., Kapushesky M., Nikkila J., Parkinson H., Goncalves A., Huber W., Ukkonen E., Brazma A.. A global map of human gene expression. Nat. Biotechnol. 2010; 28:322–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wheeler D.L., Barrett T., Benson D.A., Bryant S.H., Canese K., Chetvernin V., Church D.M., Dicuccio M., Edgar R., Federhen S. et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2008; 36:D13–D21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li B., Dewey C.N.. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011; 12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Risso D., Schwartz K., Sherlock G., Dudoit S.. GC-content normalization for RNA-Seq data. BMC Bioinformatics. 2011; 12:480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hebenstreit D., Fang M., Gu M., Charoensawan V., van Oudenaarden A., Teichmann S.A.. RNA sequencing reveals two major classes of gene expression levels in metazoan cells. Mol. Syst. Biol. 2011; 7:497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hutchins A.P., Jauch R., Dyla M., Miranda-Saavedra D.. glbase: a framework for combining, analyzing and displaying heterogeneous genomic and high-throughput sequencing data. Cell Regener. 2014; 3:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cahan P., Li H., Morris S.A., Lummertz da Rocha E., Daley G.Q., Collins J.J.. CellNet: network biology applied to stem cell engineering. Cell. 2014; 158:903–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Morris S.A., Cahan P., Li H., Zhao A.M., San Roman A.K., Shivdasani R.A., Collins J.J., Daley G.Q.. Dissecting engineered cell types and enhancing cell fate conversion via CellNet. Cell. 2014; 158:889–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. FANTOM Consortium and the RIKEN PMI and CLST (DGT). Forrest A.R., Kawaji H., Rehli M., Baillie J.K., de Hoon M.J., Haberle V., Lassmann T., Kulakovskiy I.V., Lizio M. et al. A promoter-level mammalian expression atlas. Nature. 2014; 507:462–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zheng H., Hutchins A.P., Pan G., Li Y., Pei D., Pei G.. Where cell fate conversions meet Chinese philosophy. Cell Res. 2014; 24:1162–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mele M., Ferreira P.G., Reverter F., DeLuca D.S., Monlong J., Sammeth M., Young T.R., Goldmann J.M., Pervouchine D.D., Sullivan T.J. et al. The human transcriptome across tissues and individuals. Science. 2015; 348:660–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H., Guernec G., Martin D., Merkel A., Knowles D.G. et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012; 22:1775–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hutchins A.P., Pei D.. Transposable elements at the center of the crossroads between embryogenesis, embryonic stem cells, reprogramming, and long non-coding RNAs. Sci. Bull. 2015; 60:1722–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bao X., Wu H., Zhu X., Guo X., Hutchins A.P., Luo Z., Song H., Chen Y., Lai K., Yin M. et al. The p53-induced lincRNA-p21 derails somatic cell reprogramming by sustaining H3K9me3 and CpG methylation at pluripotency gene promoters. Cell Res. 2015; 25:80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xue J., Schmidt S.V., Sander J., Draffehn A., Krebs W., Quester I., De Nardo D., Gohel T.D., Emde M., Schmidleithner L. et al. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity. 2014; 40:274–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Eichler G.S., Huang S., Ingber D.E.. Gene Expression Dynamics Inspector (GEDI): for integrative analysis of expression profiles. Bioinformatics. 2003; 19:2321–2322. [DOI] [PubMed] [Google Scholar]
- 30. Wirth H., Loffler M., von Bergen M., Binder H.. Expression cartography of human tissues using self organizing maps. BMC Bioinformatics. 2011; 12:306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wu J., Okamura D., Li M., Suzuki K., Luo C., Ma L., He Y., Li Z., Benner C., Tamura I. et al. An alternative pluripotent state confers interspecies chimaeric competency. Nature. 2015; 521:316–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sieweke M.H. Waddington's valleys and Captain Cook's islands. Cell Stem Cell. 2015; 16:7–8. [DOI] [PubMed] [Google Scholar]
- 33. Banerji C.R., Miranda-Saavedra D., Severini S., Widschwendter M., Enver T., Zhou J.X., Teschendorff A.E.. Cellular network entropy as the energy potential in Waddington's differentiation landscape. Scientific Rep. 2013; 3:3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hanna J.H., Saha K., Jaenisch R.. Pluripotency and cellular reprogramming: facts, hypotheses, unresolved issues. Cell. 2010; 143:508–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rajagopal J., Stanger B.Z.. Plasticity in the adult: how should the waddington diagram be applied to regenerating tissues?. Dev. Cell. 2016; 36:133–137. [DOI] [PubMed] [Google Scholar]
- 36. Barrallo-Gimeno A., Nieto M.A.. The Snail genes as inducers of cell movement and survival: implications in development and cancer. Development. 2005; 132:3151–3161. [DOI] [PubMed] [Google Scholar]
- 37. Ramskold D., Wang E.T., Burge C.B., Sandberg R.. An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput. Biol. 2009; 5:e1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ramilowski J.A., Goldberg T., Harshbarger J., Kloppmann E., Lizio M., Satagopam V.P., Itoh M., Kawaji H., Carninci P., Rost B. et al. A draft network of ligand-receptor-mediated multicellular signalling in human. Nat. Commun. 2015; 6:7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chen X., Xu H., Yuan P., Fang F., Huss M., Vega V.B., Wong E., Orlov Y.L., Zhang W., Jiang J. et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008; 133:1106–1117. [DOI] [PubMed] [Google Scholar]
- 40. Hutchins A.P., Choo S.H., Mistri T.K., Rahmani M., Woon C.T., Ng C.K., Jauch R., Robson P.. Co-motif discovery identifies an Esrrb-Sox2-DNA ternary complex as a mediator of transcriptional differences between mouse embryonic and epiblast stem cells. Stem Cells. 2013; 31:269–281. [DOI] [PubMed] [Google Scholar]
- 41. Fujikura J., Yamato E., Yonemura S., Hosoda K., Masui S., Nakao K., Miyazaki Ji J., Niwa H.. Differentiation of embryonic stem cells is induced by GATA factors. Genes Dev. 2002; 16:784–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wilson N.K., Foster S.D., Wang X., Knezevic K., Schutte J., Kaimakis P., Chilarska P.M., Kinston S., Ouwehand W.H., Dzierzak E. et al. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell. 2010; 7:532–544. [DOI] [PubMed] [Google Scholar]
- 43. Scialdone A., Tanaka Y., Jawaid W., Moignard V., Wilson N.K., Macaulay I.C., Marioni J.C., Gottgens B.. Resolving early mesoderm diversification through single-cell expression profiling. Nature. 2016; 535:289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li D., Sakuma R., Vakili N.A., Mo R., Puviindran V., Deimling S., Zhang X., Hopyan S., Hui C.C.. Formation of proximal and anterior limb skeleton requires early function of Irx3 and Irx5 and is negatively regulated by Shh signaling. Dev. Cell. 2014; 29:233–240. [DOI] [PubMed] [Google Scholar]
- 45. Bronner M.E., LeDouarin N.M.. Development and evolution of the neural crest: an overview. Dev. Biol. 2012; 366:2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rada-Iglesias A., Bajpai R., Prescott S., Brugmann S.A., Swigut T., Wysocka J.. Epigenomic annotation of enhancers predicts transcriptional regulators of human neural crest. Cell Stem Cell. 2012; 11:633–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Najm F.J., Lager A.M., Zaremba A., Wyatt K., Caprariello A.V., Factor D.C., Karl R.T., Maeda T., Miller R.H., Tesar P.J.. Transcription factor-mediated reprogramming of fibroblasts to expandable, myelinogenic oligodendrocyte progenitor cells. Nat. Biotechnol. 2013; 31:426–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yang N., Zuchero J.B., Ahlenius H., Marro S., Ng Y.H., Vierbuchen T., Hawkins J.S., Geissler R., Barres B.A., Wernig M.. Generation of oligodendroglial cells by direct lineage conversion. Nat. Biotechnol. 2013; 31:434–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Caiazzo M., Dell'Anno M.T., Dvoretskova E., Lazarevic D., Taverna S., Leo D., Sotnikova T.D., Menegon A., Roncaglia P., Colciago G. et al. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature. 2011; 476:224–227. [DOI] [PubMed] [Google Scholar]
- 50. Tadeu A.M., Horsley V.. Notch signaling represses p63 expression in the developing surface ectoderm. Development. 2013; 140:3777–3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Raymond C.S., Parker E.D., Kettlewell J.R., Brown L.G., Page D.C., Kusz K., Jaruzelska J., Reinberg Y., Flejter W.L., Bardwell V.J. et al. A region of human chromosome 9p required for testis development contains two genes related to known sexual regulators. Hum. Mol. Genet. 1999; 8:989–996. [DOI] [PubMed] [Google Scholar]
- 52. Cheung M., Briscoe J.. Neural crest development is regulated by the transcription factor Sox9. Development. 2003; 130:5681–5693. [DOI] [PubMed] [Google Scholar]
- 53. Stolt C.C., Lommes P., Sock E., Chaboissier M.C., Schedl A., Wegner M.. The Sox9 transcription factor determines glial fate choice in the developing spinal cord. Genes Dev. 2003; 17:1677–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Akiyama H., Chaboissier M.C., Martin J.F., Schedl A., de Crombrugghe B.. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev. 2002; 16:2813–2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Vidal V.P., Chaboissier M.C., Lutzkendorf S., Cotsarelis G., Mill P., Hui C.C., Ortonne N., Ortonne J.P., Schedl A.. Sox9 is essential for outer root sheath differentiation and the formation of the hair stem cell compartment. Curr. Biol.: CB. 2005; 15:1340–1351. [DOI] [PubMed] [Google Scholar]
- 56. Xu J., Du Y., Deng H.. Direct lineage reprogramming: strategies, mechanisms, and applications. Cell Stem Cell. 2015; 16:119–134. [DOI] [PubMed] [Google Scholar]
- 57. Ieda M., Fu J.D., Delgado-Olguin P., Vedantham V., Hayashi Y., Bruneau B.G., Srivastava D.. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010; 142:375–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Xie H., Ye M., Feng R., Graf T.. Stepwise reprogramming of B cells into macrophages. Cell. 2004; 117:663–676. [DOI] [PubMed] [Google Scholar]
- 59. Morey L., Santanach A., Blanco E., Aloia L., Nora E.P., Bruneau B.G., Di Croce L.. Polycomb regulates mesoderm cell fate-specification in embryonic stem cells through activation and repression mechanisms. Cell Stem Cell. 2015; 17:300–315. [DOI] [PubMed] [Google Scholar]
- 60. Narendra V., Rocha P.P., An D., Raviram R., Skok J.A., Mazzoni E.O., Reinberg D.. CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science. 2015; 347:1017–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Plasschaert R.N., Vigneau S., Tempera I., Gupta R., Maksimoska J., Everett L., Davuluri R., Mamorstein R., Lieberman P.M., Schultz D. et al. CTCF binding site sequence differences are associated with unique regulatory and functional trends during embryonic stem cell differentiation. Nucleic Acids Res. 2014; 42:774–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Edgar R., Mazor Y., Rinon A., Blumenthal J., Golan Y., Buzhor E., Livnat I., Ben-Ari S., Lieder I., Shitrit A. et al. LifeMap discovery: the embryonic development, stem cells, and regenerative medicine research portal. PLoS One. 2013; 8:e66629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Niwa H., Toyooka Y., Shimosato D., Strumpf D., Takahashi K., Yagi R., Rossant J.. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell. 2005; 123:917–929. [DOI] [PubMed] [Google Scholar]
- 64. Niakan K.K., Ji H., Maehr R., Vokes S.A., Rodolfa K.T., Sherwood R.I., Yamaki M., Dimos J.T., Chen A.E., Melton D.A. et al. Sox17 promotes differentiation in mouse embryonic stem cells by directly regulating extraembryonic gene expression and indirectly antagonizing self-renewal. Genes Dev. 2010; 24:312–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Aksoy I., Jauch R., Chen J., Dyla M., Divakar U., Bogu G.K., Teo R., Leng Ng C.K., Herath W., Lili S. et al. Oct4 switches partnering from Sox2 to Sox17 to reinterpret the enhancer code and specify endoderm. EMBO J. 2013; 32:938–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. D'Alessio A.C., Fan Z.P., Wert K.J., Baranov P., Cohen M.A., Saini J.S., Cohick E., Charniga C., Dadon D., Hannett N.M. et al. A systematic approach to identify candidate transcription factors that control cell identity. Stem Cell Rep. 2015; 5:763–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Uhlen M., Hallstrom B.M., Lindskog C., Mardinoglu A., Ponten F., Nielsen J.. Transcriptomics resources of human tissues and organs. Mol. Syst. Biol. 2016; 12:862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Arner E., Daub C.O., Vitting-Seerup K., Andersson R., Lilje B., Drablos F., Lennartsson A., Ronnerblad M., Hrydziuszko O., Vitezic M. et al. Gene regulation. Transcribed enhancers lead waves of coordinated transcription in transitioning mammalian cells. Science. 2015; 347:1010–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Andersson R., Gebhard C., Miguel-Escalada I., Hoof I., Bornholdt J., Boyd M., Chen Y., Zhao X., Schmidl C., Suzuki T. et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014; 507:455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Roadmap Epigenomics Consortium. Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J. et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015; 518:317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Fagerberg L., Hallstrom B.M., Oksvold P., Kampf C., Djureinovic D., Odeberg J., Habuka M., Tahmasebpoor S., Danielsson A., Edlund K. et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteomics: MCP. 2014; 13:397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Yanai I., Peshkin L., Jorgensen P., Kirschner M.W.. Mapping gene expression in two Xenopus species: evolutionary constraints and developmental flexibility. Dev. Cell. 2011; 20:483–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Rackham O.J., Firas J., Fang H., Oates M.E., Holmes M.L., Knaupp A.S., Consortium F., Suzuki H., Nefzger C.M., Daub C.O. et al. A predictive computational framework for direct reprogramming between human cell types. Nat. Genet. 2016; 48:331–335. [DOI] [PubMed] [Google Scholar]
- 74. Shapiro E., Biezuner T., Linnarsson S.. Single-cell sequencing-based technologies will revolutionize whole-organism science. Nat. Rev. Genet. 2013; 14:618–630. [DOI] [PubMed] [Google Scholar]
- 75. Trapnell C., Cacchiarelli D., Grimsby J., Pokharel P., Li S., Morse M., Lennon N.J., Livak K.J., Mikkelsen T.S., Rinn J.L.. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014; 32:381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.