Abstract
Clustered regularly interspaced short palindromic repeat/Cas9 (CRISPR/Cas9) system has emerged in recent years as a highly efficient RNA-guided gene manipulation platform. Simultaneous editing or transcriptional activation/suppression of different genes becomes feasible with the co-delivery of multiple guide RNAs (gRNAs). Here, we report that multiple gRNAs linked with self-cleaving ribozymes and/or tRNA could be simultaneously expressed from a single U6 promoter to exert genome editing of dystrophin and myosin binding protein C3 in human and mouse cells. Moreover, this strategy allows the expression of multiple gRNAs for synergistic transcription activation of follistatin when used with catalytically inactive dCas9-VP64 or dCas9-p300core fusions. Finally, the gRNAs linked by the self-cleaving ribozymes and tRNA could be expressed from RNA polymerase type II (pol II) promoters such as generic CMV and muscle/heart-specific MHCK7. This is particularly useful for in vivo applications when the packaging capacity of recombinant adeno-associated virus is limited while tissue-specific delivery of gRNAs and Cas9 is desired. Taken together, this study provides a novel strategy to enable tissue-specific expression of more than one gRNAs for multiplex gene editing from a single pol II promoter.
INTRODUCTION
The Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) Type II system is a bacterial immune system (1) which has gained an influx of scientific interest as a highly efficient molecular scissors to edit the cellular genome (2,3). The endonuclease Cas9 (the most commonly used Cas9 is from Streptococcus pyogenes, SpCas9) is guided by the crRNA and tracrRNA to make a site specific double-strand break (DSB) at the target genome, which is then repaired by either homology-directed repair (HDR) or non-homologous end-joining (NHEJ) so that the target genome is modified. Moreover, catalytically inactive Cas9 (dCas9) and various fusions with transcriptional activator, repressor and recruitment domains have been used to modulate gene expression at targeted loci without introducing irreversible mutations to the genome (4–13).
The crRNA and tracrRNA can be combined into a single guide RNA (gRNA) consisting of a ∼20 nt targeting sequence and a scaffold sequence (1–3). The gRNA is typically expressed in cells under the control of the U6 or U3 snRNA promoters. When multiple gRNAs are expressed, the CRISPR/Cas9 can be guided to simultaneously manipulate multiple genomic loci, which can be achieved by co-transfection of multiple gRNAs in separate constructs (2,3,14–18). Although this approach is highly effective, it would be a challenge for certain applications where the vector capacity and/or vector numbers are limited for simultaneous production of multiple gRNAs. Recently, several strategies have been developed to express multiple gRNAs from a single transcript. One is to use Csy4 endoribonuclease, which can process a transcript containing gRNAs fused with Csy4-cleavable RNA (19,20). The highly conserved tRNA-processing mechanism has also been harnessed for expressing multiple gRNAs from a single transcript (21–25). These strategies use the pol III promoters, which lack tissue specificity, to drive gRNA expression. Ribozymes are a class of enzymatic RNA molecules that adopt a specific tertiary structure allowing for cleavage of themselves at a defined position (26), and have recently been used to express gRNAs from pol II promoters in yeast (27), Leishmania donovani (28) and human cells (19,29). This is particularly important for applications such as gene therapy where tissue specificity is essential to prevent from off-target effects associated with systemic delivery. Recently, we, and several other research groups demonstrated that CRISPR/Cas9 could be used to correct the reading frame of dystrophin in mdx mice, a mouse model of Duchenne muscular dystrophy (DMD), by using two gRNAs to remove the mutant exon 23 (30–33). These studies were pivotal in highlighting the promise of CRISPR/Cas9 in the gene therapy arena, especially pertaining to the orphan disease DMD for which there is no cure and limited treatment options. Overall, this highlights the urgent need to achieve tissue specificity for these in vivo genome editing applications.
In the present study, we explored the feasibility of expressing multiple gRNAs using the self-cleaving ribozymes from a single expression cassette under the control of the U6 promoter. Moreover, we further modified the system so that the gRNAs could be expressed from pol II promoters such as generic CMV and muscle-specific MHCK7.
MATERIALS AND METHODS
Mice
All animal studies were authorized by the Animal Care, Use, and Review Committee of The Ohio State University. SpCas9 knock-in mice with constitutive SpCas9 expression (stock number: 024858) were obtained from the Jackson Laboratory and crossed with mdx mice (C57BL/10ScSn-Dmdmdx/J) to derive SpCas9/mdx mice. Mice including wild-type C57BL/10ScSn were maintained at The Ohio State University Laboratory Animal Resources in accordance with animal use guidelines.
Plasmid construction
Individual gRNAs or minigenes, including ribozymes- or tRNA-linked Dmd, MYBPC3, FST and MYBPC3-DMD gRNAs, were synthesized by Integrated DNA technologies (IDT) and inserted into pLKO-GFP-2A-SpCas9, pLKO-mCherry-2A-SpCas9 or pLKO puro-2A-SpCas9 vectors containing two BsmBI restriction sites as previously described (30). The FST gRNAs were cloned into the Lenti_sgRNA(MS2)_zeo plasmid (Addgene, #61427) with modified cloning site. In order to assemble the HH-i20-tRNA-i23-HDV constructs driven by CMV promoter, two minigenes were used as template to produce fusion PCR products and then inserted into pAAV-MCS. The HH-i20-tRNA-i23-HDV cassette was also subcloned into pLKO-gRNA under the U6 promoter or pRNAT-H1.1 with MHCK7 promoter. All gRNA sequences and primers are listed in Supplementary Data. All minigenes are listed in Supplementary Data.
Cell culture and transfection
Human Embryonic Kidney 293 cells (HEK293) and RAW 264.7 cells were cultured in DMEM (Gibco, Life Sciences, MA, USA) supplemented with 10% FBS (Gibco, Life Sciences) and seeded in six-well plates. When the confluency reached 70%, two μg total plasmids were transfected into each well of HEK293 using 8 μl polyethylenimine (PEI, 25 kDa). RAW 264.7 cells were cultured in RPMI1640 medium (ATCC, Manassas, VA, USA) and transfected with plasmids using Lipofectamine 3000 reagent (Thermo Fisher Scientific, Waltham, MA, USA). C2C12 cells were cultured in DMEM with 20% FBS and electroporated with plasmids as previously described (30,34). After 48 h, cells were collected for genomic DNA and RT-PCR analyses.
Extraction of DNA, RNA and real time PCR analysis
Genomic DNA was extracted from mouse tissues, C2C12, HEK293 and RAW 264.7 cells. Total RNA was extracted from C2C12 and HEK293 cells by using Zymo Quick-RNA™ MiniPrep kit (Zymo Research Corporation, CA, USA). Real-time PCR were performed as previously described (30,34,35). Briefly, 20 ng genomic DNA was used as template and RT-PCR products were analyzed for gene editing efficiency. Real-time PCR reactions were performed using Radiant™ SYBR Green Hi-ROX qPCR Kits (Alkali Scientific, Pompano Beach, FL) in LightCycler® 480 System and normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Regular RT-PCR was performed using GoTaq® Green Master (Promega, Madison, WI, USA).
Adenovirus production
Ribozymes-linked Dmd gRNAs with U6 promoter (U6-mdx-r-gRNAs) were subcloned into pShuttle-CMV (Clontech, Mountain View, CA, USA). Recombinant adenoviral DNA carrying U6-mdx-r-gRNAs was generated with the AdEasy-1 Adenovirus system (Agilent Technologies, La Jolla, CA, USA) according to the manufacturer's instructions. The adenoviral particles were packaged and amplified in AD293 cells and purified by cesium chloride gradient ultracentrifugation followed by dialysis in storage buffer (10 mM Tris–HCl pH 8.0, 2 mM MgCl2, 4% sucrose) as described previously (30). The titers of the adenovirus preparations were quantified by measuring the OD260 (36).
Adenovirus transduction in vivo
To transduce mouse skeletal muscle in vivo, the quadriceps and gastrocnemius muscles of mdx pups (day 1–4) were injected with ∼2.5 × 1010 viral particles/muscle as previously described (30). One month after transduction, tissues were collected for genomic DNA analysis, western blotting and immunofluorescence staining experiments.
In vivo transfection into mouse skeletal muscle and liver
Vectors for in vivo transfection were prepared using the EndoFreeMaxi Kit (Qiagen). The mouse FDB muscles were electroporated with 40 μg pLKO mCherry-2A-cas9 with i20- and i23- gRNA plasmids or pLKO EGFP-2A-cas9 plus pRNAT-H1.1_MHCK7-HH-i20-tRNA-i23-HDV as previously described (30). For hydrodynamic tail vein injection, plasmid DNA suspended in 2 ml saline was injected via the tail vein in 7–10 s into 8-week-old female mdx mice. Four to seven days after electroporation/hydrodynamic injection, the FDB muscles and livers were dissected out and examined under a Nikon Ti-E inverted fluorescence microscope equipped with an Andor Zyla sCMOS camera a Nikon Super Fluor 20 × 0.75 NA objective lens. Images were recorded using NIS-Elements Advanced Research software package (Nikon) and processed using Photoshop CS5 (Adobe) software package. Genomic DNA was extracted from the tissues as described above for gene editing analysis.
Immunohistochemistry
For immunofluorescence staining, ten μm frozen sections were fixed with 4% paraformaldehyde for 15 min at room temperature. The samples were washed twice with PBS and incubated with blocking solution (PBS, 2% BSA, 0.5% Triton X-100, 0.1% Tween 20, 20% goat serum) for one hour before overnight incubation at 4°C with primary antibodies (rabbit polyclonal anti-dystrophin antibody, Ab15277, Abcam, 1:200). The slides were extensively washed with PBS and incubated with secondary antibodies (Invitrogen 1:500). Finally, the slides were mounted using VECTASHIELD® Mounting Medium with DAPI (Vector Laboratories, Inc.). Slides were analyzed using a Nikon Ti-E inverted fluorescence microscope equipped with an Andor Zyla sCMOS camera a Nikon Super Fluor 20 × 0.75 NA objective lens. Images were recorded using NIS-Elements Advanced Research software package (Nikon) and processed using Photoshop CS5 (Adobe) software package.
Statistical analysis
Data are expressed as mean ± standard error (SEM). Statistical differences were determined by unpaired Student's t test for two groups and one-way ANOVA with Bonferroni's post tests for multiple group comparisons using Prism 5.02 (Graphpad Software, La Jolla, CA, USA). A P-value less than 0.05 was considered to be significant.
RESULTS
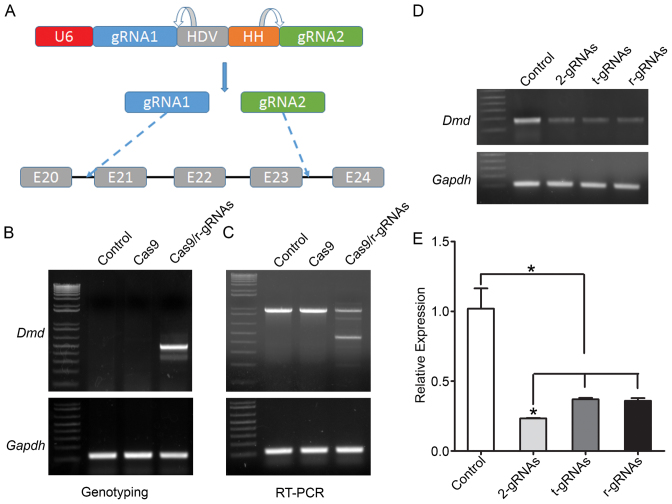
Editing of mouse Dmd gene with ribozyme-linked double gRNAs in C2C12 cells and mdx muscles.
We previously demonstrated that CRISPR/Cas9 system could be used to restore dystrophin expression in mdx mice using two gRNAs targeting the intron 20 and 23 respectively (30). To test whether these two gRNAs could be expressed from a single U6 promoter, we linked the two gRNAs with Hepatitis delta virus (HDV) and Hammerhead (HH) ribozymes (37,38) (Figure 1A). After electroporation of the SpCas9-expressing plasmid into C2C12 cells, these ribozyme-linked gRNAs showed robust gene editing activities as demonstrated by PCR analysis of the genomic DNA (Figure 1B). A 510-bp PCR product was readily detectable in the cells co-transfected with SpCas9 and ribozyme-linked gRNAs, but not in the control cells or the cells transfected with SpCas9 only. RT-PCR analysis also confirmed the expression of the truncated dystrophin as expected in ribozyme-linked gRNAs/SpCas9 transfected cells (Figure 1C). Previously, it was shown that tRNA could also mediate the expression of multiple gRNAs from a single expression cassette (39). We also constructed a plasmid to express the pre-tRNAGly-linked gRNAs. To compare the performance of tRNA-linked gRNAs, ribozyme-linked gRNAs and two separate gRNAs, we used a primer set which detects WT dystrophin alleles but not deleted ones. Transfection of these reagents all induced significant reduction of WT dystrophin allele (Figure 1D). Quantitative real-time PCR analysis showed that all three reagents reduced the WT dystrophin allele by >50%, and that there were no significant difference between tRNA-linked gRNAs or ribozyme-linked gRNAs, although transfection of two separate gRNAs performed slightly better (Figure 1E). This may be due to the incomplete processing and maturation of gRNAs from the ribozyme- and tRNA-linked gRNA transcripts as shown by Northern blotting analysis (Supplementary Data).
Figure 1.
Ribozymes-linked double gRNAs targeting mouse Dmd gene. (A) Diagram showing the two gRNAs targeting the introns 20 and 23, respectively, under the control of human U6 promoter via self-cleaving HDV and HH ribozymes. (B) PCR analysis of genomic DNA extracted from C2C12 cells treated with or without the gRNAs and SpCas9 constructs. (C) RT-PCR analysis of the dystrophin transcript expression in C2C12 cells. (D, E) Regular (D) and quantitative real-time (E) PCR analysis of the WT allele in C2C12 cells treated with or without different versions of gRNAs and SpCas9 constructs. 2-gRNAs, two gRNAs in separate plasmids; t-gRNAs, two gRNAs linked with tRNA; r-gRNAs, two gRNAs linked with ribozymes. Gapdh was used as a reference control. *P < 0.05. All data are representative of a minimum of three experiments.
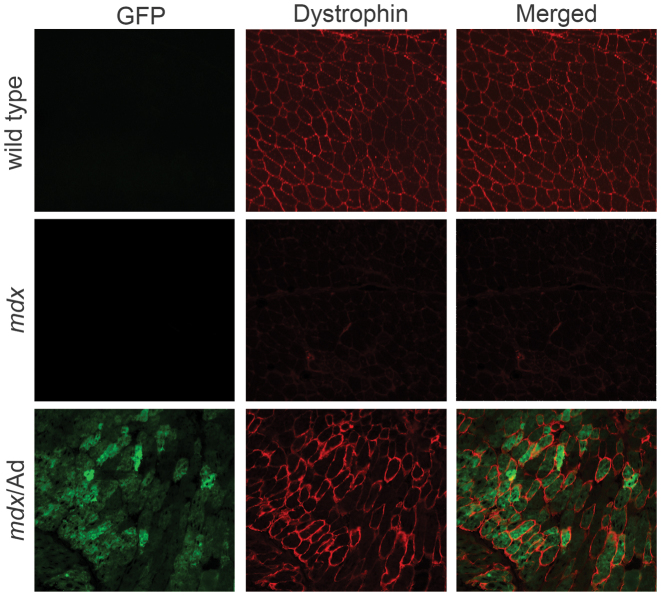
To further investigate whether CRISPR-mediated gene editing could restore dystrophin and its associated protein complex, we injected adenoviral vectors carrying ribozyme-liked i20/i23 gRNAs into the gastrocnemius (GA) muscles of newborn mdx/SpCas9-transgenic pups. Genotyping to detect dystrophin expression showed a very weak band, indicating that the gene editing efficiency was low (data not shown). Western blotting analysis showed that the expression of SpCas9 in the skeletal muscle of the mdx/SpCas9-transgenic mice was much lower than that in the brain and heart (Supplementary Data). Based on these observations, we reasoned that the low gene editing efficiency might be due to the low SpCas9 expression in the skeletal muscle of the transgenic mice. Therefore, we co-injected adenoviral vectors carrying GFP-2A-SpCas9 and ribozyme-gRNAs into the GA muscles of newborn mdx pups. Green fluorescent protein (GFP) signals were readily detectable in the injected muscles, indicating that the adenovirus transduction was successful. Truncated dystrophin gene fragment could be detected by genotyping PCR (Supplementary Data). To study the expression and localization of dystrophin, we performed immunofluorescence staining of the muscle sections. Three weeks after adenovirus transduction, dystrophin expression was restored in the muscle fibers that were positive for GFP, and in GFP-negative (non-treated) mdx muscle fibers, no dystrophin-positive clusters were observed (Figure 2).
Figure 2.
Adenovirus-mediated delivery of ribozymes-linked gRNAs and SpCas9 restored dystrophin expression in mdx muscles. Confocal immunofluorescence images of dystrophin (red) and GFP (green) in muscle cryosections treated with or without EGFP-2A-SpCas9/gRNA adenovirus. The images are representative of three experiments. Scale bar: 100 μm.
Editing of human MYBPC3 gene with ribozyme-linked double gRNAs in HEK293 cells
To test whether the self-cleaving ribozyme could be generally applicable to link other gRNAs, we selected two gRNAs targeting human MYBPC3 gene which encodes myosin binding protein C3 (40) (Figure 3A). After transfection with the SpCas9-encoding plasmid, the ribozyme-linked gRNAs also exerted significant gene editing activities at the human MYBPC3 locus (Figure 3B). Again, the tRNA-linked gRNAs and ribozymes-linked gRNAs performed similarly in editing MYBPC3 (Figure 3B). In this case, they achieved similar levels of gene editing efficiency as that with co-transfection of two individual gRNAs. Quantitative real-time PCR analysis showed that these reagents all reduced the WT MYBPC3 allele by >50% (Figure 3C). Therefore, the ribozymes can be used to link two gRNAs from a single U6 promoter.
Figure 3.
Editing of MYBPC3 gene using ribozymes-linked gRNAs in HEK293 cells. (A) Diagram showing the genomic locus of MYBPC3 with the gRNA targeting sites and primer locations. (B) PCR analysis of genomic DNA extracted from HEK293 cells transfected with the gRNA constructs (2-gRNAs, t-gRNAs or r-gRNAs) and SpCas9 using two sets of primer pairs (F1/R1 and F2/R2). (C) Quantitative real-time PCR analysis of the WT allele in HEK293 cells transfected with or without different gRNAs and SpCas9 constructs. *P < 0.05. All data are representative of a minimum of three experiments.
Simultaneous editing of human DMD and MYBPC3 genes using ribozymes-linked four gRNAs
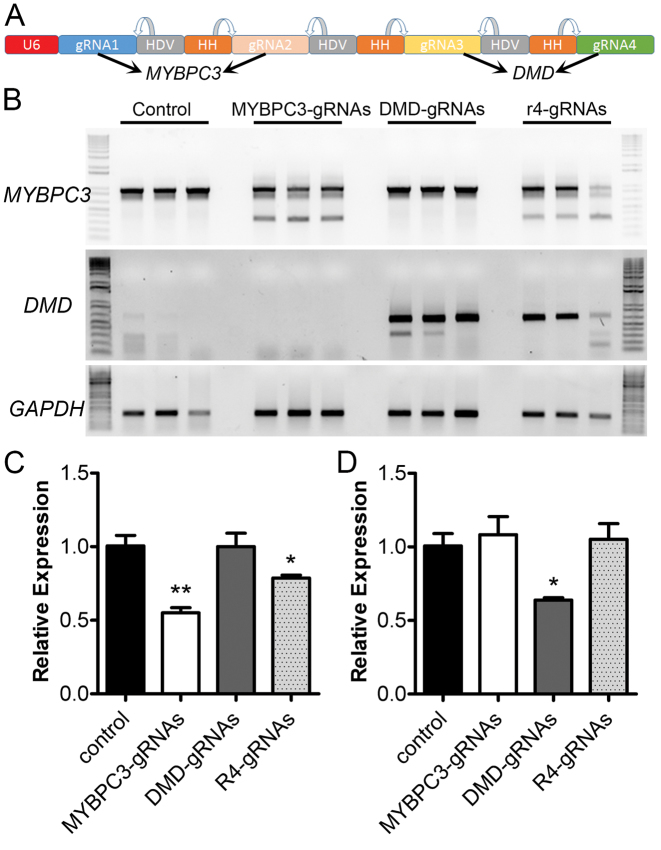
Having demonstrated successful gene editing with two gRNAs linked by ribozymes, we hypothesized that multiple gRNAs can be expressed from a single U6 promoter using the self-cleaving ribozymes. To test this, we assembled four gRNAs together by using the ribozymes (Figure 4A), with the first two targeting human MYBPC3 as in Figure 3 and the latter two targeting the intron 50 and 51 of human dystrophin (DMD). PCR analysis confirmed the successful editing of human MYBPC3 and DMD genes in the transfected cells (Figure 4B). To compare the genome editing efficiency, we performed quantitative PCR to examine the reduction of the WT MYBPC3 or DMD allele. Transfection of separate plasmids encoding the two MYBPC3 gRNAs decreased the WT MYBPC3 allele by ∼45%, while the four gRNAs linked with ribozymes reduced the WT MYBPC3 allele by ∼20% (Figure 4C). In the case of DMD, transfection of two separate DMD gRNAs resulted in 35% reduction of WT DMD allele, and the four gRNAs from a single U6 promoter did not significantly decrease the WT DMD allele (Figure 4D), although the amplicon with correct deletion can be amplified (Figure 4B). In summary, these data suggest that the ribozymes can link multiple gRNAs to a single U6 promoter for gene editing, but there is a reduction in gRNA processing efficiency with an increased number of gRNAs.
Figure 4.
Simultaneous genome editing of human DMD and MYBPC3 genes using a single expression cassette which expresses four gRNAs linked with ribozymes. (A) Diagram showing the gRNA expression cassette. The first two gRNAs are designed to target human MYBPC3 as in Figure 3 and the latter two are to target human DMD in the introns 50 and 51, respectively. (B) PCR analysis of the gene editing activity induced by the four gRNAs linked with ribozymes. All data are representative of a minimum of three experiments. (C, D) Quantitative PCR analysis of the reduction of WT MYBPC3 (C) and DMD (D) alleles after transfection with the specified constructs. **P < 0.01; *P < 0.05.
Synergistic gene activation of FST using ribozymes-linked gRNAs
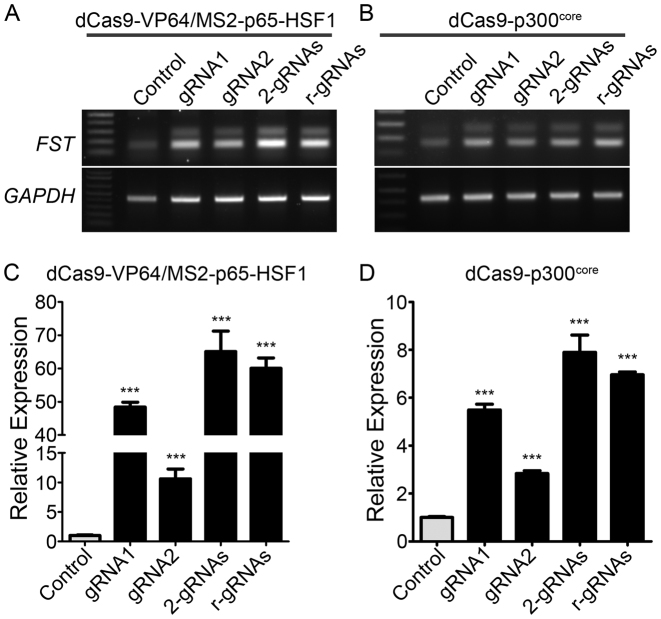
The CRISPR/Cas9 system has been modified to activate gene expression by fusing the catalytic inactive (dCas9) with a transactivation domain such as VP64. However, it was found that a single gRNA was inefficient in activating gene expression by dCas9-VP64 (4–11). Recently, it was found that the gene activation efficiency can be dramatically enhanced by recruiting multiple distinct transcriptional activators together with the dCas9-VP64 (12,41). The modified gene activation systems can robustly activate the gene expression with gRNA targeted to within 200 bp upstream of the transcription start site (13). Therefore, we tested the feasibility of using ribozymes-linked gRNAs to enhance FST gene expression (which encodes a natural muscle mass enhancer follistatin (42)). We assembled two gRNAs linked by ribozymes targeting the human FST promoter region in the lentiviral vector pLKO-MS2-zeocin. Human embryonic kidney cells 293 (HEK293) were transfected with dCas9-VP64, MS2-p65-HSF1 and individual MS2-modified gRNAs or in combination. Both gRNAs could significantly enhance the expression of FST by ∼10- and 48-fold, respectively (Figure 5A and C). Transfection of two gRNAs either in two separate constructs or in ribozymes-linked format produced a higher fold increase (about 65 and 60 fold, respectively) than individual gRNAs, consistent with the previously reported synergistic effect of dCas9-based transcriptional activation.
Figure 5.
Synergistic transcriptional activation of human FST using two gRNAs linked with ribozymes. (A, C) Regular (A) and quantitative (C) RT-PCR analysis of FST expression in HEK293 cells transfected with dCas9-VP64/MS2-p65-HSF1 and different versions of gRNA constructs. (B, D) Regular (B) and quantitative (D) RT-PCR analysis of FST expression in HEK293 cells transfected with dCas9-p300core and different versions of gRNA constructs. GAPDH was used as a reference gene. ***P < 0.001. All data are representative of a minimum of three experiments.
In addition, transcriptional activation of target genes could also be induced by dCas9 fused to the catalytic core of the human acetyltransferase p300 (43). We tested the ribozymes-linked gRNAs and dCas9-p300core in activating FST. Both gRNAs significantly increased FST expression (Figure 5B and D), but the expression level was significantly lower than the dCas9-VP64/MS2-p65-HSF1 system. Similarly, the two gRNAs expressed from two separate constructs or in the ribozymes-linked format produced a synergistic effect in increasing FST expression.
Cell- and tissue-specific gene editing using ribozymes and tRNA-linked gRNAs with pol II promoters
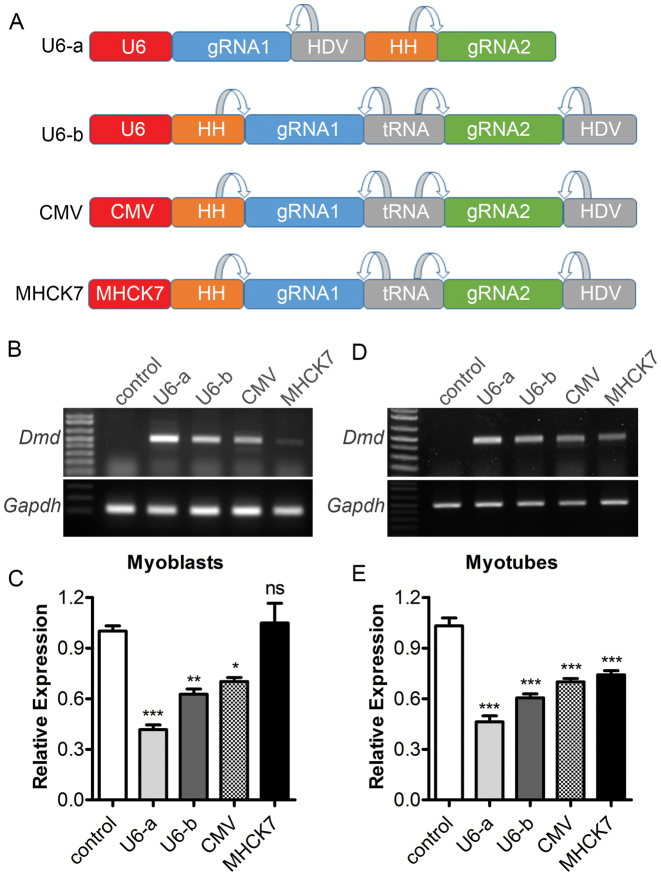
In many applications such as gene editing therapies, it is desirable to limit the expression of CRISPR components in specific tissues to avoid potential off-target activities in non-target tissues. It is possible to restrict the expression of SpCas9 in a particular tissue using tissue-specific promoters. However, expression of gRNA is generally driven by the U6 promoter, a pol III promoter lacking the tissue specificity. We thus tested the feasibility to express the gRNAs from pol II promoters such as generic CMV and muscle-specific MHCK7. The HH ribozyme was placed in front of the first gRNA, which is linked to the second gRNA with a tRNA, followed by the HDV ribozyme (Figure 6A). We placed tRNA between two gRNAs instead of ribozymes because tRNA appeared to be more efficiently processed (Supplementary Data) and repeating sequences are troublesome during the preparation of plasmids or viral vectors. The expression cassette was placed under the control of either the U6, CMV or MHCK7 promoter. Different versions of the gRNA expression cassette were electroporated into C2C12 myoblasts with the SpCas9 plasmid. PCR analysis showed that all constructs, with the exception of the one driven by MHCK7, induced substantial gene editing activities, with U6 producing the strongest effect (Figure 6B and C). However, when the C2C12 cells were differentiated into myotubes, the gene editing activity from the MHCK7 driven plasmid was significantly enhanced (Figure 6D and E). Moreover, transfection of these gRNA constructs into RAW 264.7 cells, a murine macrophage cell line, showed that the MHCK7 driven construct did not induce detectable gene editing activities, while the other constructs did (Supplementary Data). These data are consistent with the muscle specificity of the MHCK7 promoter.
Figure 6.
Expression and activity of two gRNAs linked by ribozymes and tRNA from a single pol II promoter (CMV or MHCK7). (A) Diagram showing different gRNA expression cassette. (B, C) Regular (B) and quantitative (C) RT-PCR analysis of Dmd expression in C2C12 myoblasts transfected with SpCas9 and gRNA constructs driven by different promoters. (D, E) Regular (D) and quantitative. (E) RT-PCR analysis of Dmd expression in C2C12 myotubes transfected with SpCas9 and gRNA constructs driven by different promoters. Gapdh was used as a reference gene. *P < 0.05, **P < 0.01, ***P < 0.001. ns, not significant. All data are representative of a minimum of three experiments.
To test this strategy in vivo, we electroporated the U6 or MHCK7 constructs carrying ribozymes- and tRNA-linked gRNA expression cassette into mouse skeletal muscle (30), or transfected into mouse liver through hydrodynamic injection (44). The U6 construct also carries an independent mCherry expression cassette while the MHCK7 construct has a GFP cassette. Electroporation of these plasmids into mouse flexor digitorum brevis (FDB) muscles resulted in robust fluorescent protein expression by direct epifluorescence observation (Figure 7A). Genome editing activity was readily detected in both U6 and MHCK7 constructs by PCR analysis (Figure 7B). Hydrodynamic injection of the U6 or MHCK7 plasmids resulted in positive liver transfection as shown by expression of mCherry and GFP, respectively (Figure 7A). The MHCK7 construct did not yield any detectable genome editing activity in liver while the U6 construct showed efficient genome editing in this tissue (Figure 7C).
Figure 7.
In vivo gene editing using ribozymes- and tRNA-linked gRNAs. (A) Epi-fluorescence imaging of isolated skeletal muscle and liver from mice transfected with the MHCK7-driven gRNAs (which carries a GFP expression cassette) or U6-driven gRNAs (which carries a mCherry expression cassette). (B, C) PCR analysis of gene editing activity in skeletal muscle (B) and liver (C) after in vivo transfection. The numbers 1 to 5 indicate different mice tested.
DISCUSSION
The CRISPR/Cas9 system has recently gained a rapid influx of scientific interest as an effective tool for genome editing since its first implementation for such a purpose in 2013 (1–3). CRISPR/Cas9 function to edit the genome by inducing a DSB, which is repaired by the cellular DNA repair mechanisms such as NHEJ and HDR. Such a modification on the genomic DNA is permanent and inheritable. This makes it an attractive platform for therapeutic development, in particular for the treatment of genetic disorders. We and others recently demonstrated that, in DMD mouse model (30–33) and cultured DMD patient-derived cells (45,46), the CRISPR/Cas9-mediated genome editing effectively restored dystrophin expression with two gRNAs targeting the introns flanking the mutant exon, thus highlighting its potential in therapeutic treatment of DMD.
Simultaneously editing two or more sites requires the delivery of two or more gRNAs. Several approaches have been developed to deliver multiple gRNAs. These include co-delivery of multiple gRNAs encoded by each individual constructs (2,3,14–18), Csy4-mediated RNA processing system (19,20), self-cleaving tRNA (21–24) and ribozymes-linked gRNAs (27–29). Although the first method has been shown to be successful, the limitation of this approach is also obvious. For example, manufacturing multiple vectors are expensive and labor intensive. The usage of multiple promoters or performing multiple treatments may not be a clinically viable option as it can increase the risk of adverse side effects occurring through off target effects (47) and immunological complications. Our present study demonstrated that multiple gRNAs could be simultaneously expressed from a single promoter with the use of self-cleaving ribozymes and/or tRNA. This strategy not only saves labor associated with constructing the plasmids, but also reduces the amount of CRISPR/Cas9 components, thus reducing the cost associated with manufacturing the reagents and the potential adverse side effects.
There were no significant differences in terms of gene editing efficiency between ribozymes-linked and tRNA-linked gRNAs from a single construct. However, the co-transfection of separate gRNA-encoding constructs in the case of DMD performed better than expression of the gRNAs from a single transcript, likely due to inefficient processing of these RNA transcripts and/or the reduced expression of longer transcripts from U6 promoter. The Northern blotting analysis showed that tRNAGly cleaved both gRNAs very efficiently (close to 90%). Even though the tRNAgly that we used is plant origin as reported by Yang's group (39), the processing of the tRNA-gRNA transcript was highly efficient in mammalian cells, suggesting the processing of tRNAs is conserved. With ribozymes, cleavage of the first gRNA was ∼95%, but only ∼33% of the second gRNA was in the mature form. Future studies will be needed to optimize the ribozyme design or identify more efficient ribozymes that can cleave the 5΄ of the gRNAs. In addition, the expression of the second gRNA appeared to be much lower than the first gRNA from the ribozyme constructs. This is most likely caused by early termination of transcription due to the presence of four consecutive T residues in the gRNA scaffold (which appeared three times in ribozymes-linked gRNAs) because clusters of four or more Ts in the coding strand serve as termination signals for pol III (48,49). This phenomenon has previously been reported with the use of U6 to transcribe gRNA (50–53). In addition, the pol III promoters such as U6 are less efficient to transcribe long RNAs (54). Interestingly, the expression of the gRNAs from tRNA constructs was not equally reduced. This may be because the tRNA-encoding sequences contain internal promoter elements (box A and B) to recruit the pol III complex (55,56). These small tRNA promoters (only ∼70 bp) are sufficient to drive high expression of tRNA-gRNA fusion transcripts that are efficiently and precisely cleaved by endogenous tRNase Z to release fully functional gRNAs (24).
Another important feature with ribozymes/tRNA strategy to link gRNAs is that it allows the expression of gRNAs from pol II promoters. Lack of tissue specificity using the U6 promoter has been a limiting factor of the CRISPR/Cas9 system. In this study, we demonstrated that the gRNAs linked with ribozymes could be expressed under the control of pol II promoters such as ubiquitous CMV and muscle/heart-specific MHCK7. The MHCK7 promoter was previously engineered to drive gene expression in skeletal and cardiac muscles. Salva et al. demonstrated that the MHCK7 promoter had ∼400-, ∼50- and ∼10-fold higher activity in cardiac, diaphragm, and soleus muscles, respectively when compared to their previous ‘best’ construct creatine kinase 6 (CK6) (57). In addition, they found that the MHCK7 construct showed minimal expression in the liver, lung and spleen after AAV delivery (57). Consistent with the muscle specificity of MHCK7, we showed that the ribozymes- and tRNA-linked gRNAs in the MHCK7 construct induced significant gene editing in C2C12 myotubes, low in C2C12 myoblasts and non-detectable in RAW 264.7 cells. Similarly, the MHCK7 construct induced gene editing in skeletal muscle but not in liver. Overall, these results are of particular importance for the treatment of neuromuscular disorders such as DMD whereby achieving muscle specific gene editing is pivotal for clinical translation. In addition, most DMD patients display and ultimately succumb to cardiomyopathy (58,59) and therefore a promoter such as MHCK7 which displayed specificity to both skeletal and cardiac muscles would be highly desirable. This is particularly useful for in vivo applications when the packaging capacity of rAAV is limited while tissue-specific delivery of gRNAs and SpCas9 is desired.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs Mona El Refaey and Keryn Woodman for fruitful discussions and critical comments.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
U.S. National Institutes of Health [HL116546 and AR064241 to R.H.]. Funding for open access charge: Departmental Fund.
Conflict of interest statement. None declared.
REFERENCES
- 1. Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E.. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012; 337:816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mali P., Yang L.H., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M.. RNA-guided human genome engineering via Cas9. Science. 2013; 339:823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cong L., Ran F.A., Cox D., Lin S.L., Barretto R., Habib N., Hsu P.D., Wu X.B., Jiang W.Y., Marraffini L.A. et al. . Multiplex genome engineering using CRISPR/Cas systems. Science. 2013; 339:819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Konermann S., Brigham M.D., Trevino A.E., Hsu P.D., Heidenreich M., Cong L., Platt R.J., Scott D.A., Church G.M., Zhang F.. Optical control of mammalian endogenous transcription and epigenetic states. Nature. 2013; 500:472–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Maeder M.L., Linder S.J., Cascio V.M., Fu Y.F., Ho Q.H., Joung J.K.. CRISPR RNA-guided activation of endogenous human genes. Nat. Methods. 2013; 10:977–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Perez-Pinera P., Kocak D.D., Vockley C.M., Adler A.F., Kabadi A.M., Polstein L.R., Thakore P.I., Glass K.A., Ousterout D.G., Leong K.W. et al. . RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat. Methods. 2013; 10:973–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cheng A.W., Wang H.Y., Yang H., Shi L.Y., Katz Y., Theunissen T.W., Rangarajan S., Shivalila C.S., Dadon D.B., Jaenisch R.. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 2013; 23:1163–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Qi L.S., Larson M.H., Gilbert L.A., Doudna J.A., Weissman J.S., Arkin A.P., Lim W.A.. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013; 152:1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Larson M.H., Gilbert L.A., Wang X.W., Lim W.A., Weissman J.S., Qi L.S.. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat. Protoc. 2013; 8:2180–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gilbert L.A., Larson M.H., Morsut L., Liu Z.R., Brar G.A., Torres S.E., Stern-Ginossar N., Brandman O., Whitehead E.H., Doudna J.A. et al. . CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013; 154:442–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bikard D., Jiang W.Y., Samai P., Hochschild A., Zhang F., Marraffini L.A.. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 2013; 41:7429–7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zalatan J.G., Lee M.E., Almeida R., Gilbert L.A., Whitehead E.H., La Russa M., Tsai J.C., Weissman J.S., Dueber J.E., Qi L.S. et al. . Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell. 2015; 160:339–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Konermann S., Brigham M.D., Trevino A.E., Joung J., Abudayyeh O.O., Barcena C., Hsu P.D., Habib N., Gootenberg J.S., Nishimasu H. et al. . Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015; 517:583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang H., Yang H., Shivalila C.S., Dawlaty M.M., Cheng A.W., Zhang F., Jaenisch R.. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013; 153:910–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jinek M., East A., Cheng A., Lin S., Ma E., Doudna J.. RNA-programmed genome editing in human cells. Elife. 2013; 2:e00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li J.F., Norville J.E., Aach J., McCormack M., Zhang D.D., Bush J., Church G.M., Sheen J.. Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 2013; 31:688–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhou H.B., Liu B., Weeks D.P., Spalding M.H., Yang B.. Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucleic Acids Res. 2014; 42:10903–10914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shan Q.W., Wang Y.P., Li J., Zhang Y., Chen K.L., Liang Z., Zhang K., Liu J.X., Xi J.J., Qiu J.L. et al. . Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 2013; 31:686–688. [DOI] [PubMed] [Google Scholar]
- 19. Nissim L., Perli S.D., Fridkin A., Perez-Pinera P., Lu T.K.. Multiplexed and programmable regulation of gene networks with an integrated RNA and CRISPR/Cas toolkit in human cells. Mol. Cell. 2014; 54:698–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tsai S.Q., Wyvekens N., Khayter C., Foden J.A., Thapar V., Reyon D., Goodwin M.J., Aryee M.J., Joung J.K.. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat. Biotechnol. 2014; 32:569–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Carter C.W. Jr, Wolfenden R.. tRNA acceptor stem and anticodon bases form independent codes related to protein folding. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:7489–7494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Port F., Bullock S.L.. Augmenting CRISPR applications in Drosophila with tRNA-flanked sgRNAs. Nat. Methods. 2016; 13:852–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Qi W.W., Zhu T., Tian Z.R., Li C.B., Zhang W., Song R.T.. High-efficiency CRISPR/Cas9 multiplex gene editing using the glycine tRNA-processing system-based strategy in maize. BMC Biotechnol. 2016; 16:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mefferd A.L., Kornepati A.V.R., Bogerd H.P., Kennedy E.M., Cullen B.R.. Expression of CRISPR/Cas single guide RNAs using small tRNA promoters. RNA. 2015; 21:1683–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Port F., Bullock S.L.. Expansion of the CRISPR toolbox in an animal with tRNA-flanked Cas9 and Cpf1 gRNAs. Bioarxiv. 2016; doi:10.1101/046417. [Google Scholar]
- 26. Kruger K., Grabowski P.J., Zaug A.J., Sands J., Gottschling D.E., Cech T.R.. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell. 1982; 31:147–157. [DOI] [PubMed] [Google Scholar]
- 27. Gao Y., Zhao Y.. Self-processing of ribozyme-flanked RNAs into guide RNAs in vitro and in vivo for CRISPR-mediated genome editing. J. Integr. Plant Biol. 2014; 56:343–349. [DOI] [PubMed] [Google Scholar]
- 28. Zhang W.W., Matlashewski G.. CRISPR-Cas9-mediated genome editing in Leishmania donovani. Mbio. 2015; 6:e00861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yoshioka S., Fujii W., Ogawa T., Sugiura K., Naito K.. Development of a mono-promoter-driven CRISPR/Cas9 system in mammalian cells. Sci. Rep.-UK. 2015; 5:18341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xu L., Park K.H., Zhao L., Xu J., El Refaey M., Gao Y., Zhu H., Ma J., Han R.. CRISPR-mediated genome editing restores dystrophin expression and function in mdx mice. Mol. Ther. 2015; 24:564–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nelson C.E., Hakim C.H., Ousterout D.G., Thakore P.I., Moreb E.A., Castellanos Rivera R.M., Madhavan S., Pan X., Ran F.A., Yan W.X. et al. . In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science. 2016; 351:403–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Long C., Amoasii L., Mireault A.A., McAnally J.R., Li H., Sanchez-Ortiz E., Bhattacharyya S., Shelton J.M., Bassel-Duby R., Olson E.N.. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science. 2016; 351:400–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tabebordbar M., Zhu K., Cheng J.K., Chew W.L., Widrick J.J., Yan W.X., Maesner C., Wu E.Y., Xiao R., Ran F.A. et al. . In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science. 2016; 351:407–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xu L., Zhao P., Mariano A., Han R.. Targeted myostatin gene editing in multiple mammalian species directed by a single pair of TALE nucleases. Mol. Ther. Nucleic Acids. 2013; 2:e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Xu J., El Refaey M., Xu L., Zhao L., Gao Y., Floyd K., Karaze T., Janssen P.M.L., Han R.. Genetic disruption of Ano5 in mice does not recapitulate human ANO5-deficient muscular dystrophy. Skelet. Muscle. 2015; 5:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sweeney J.A., Hennessey J.P. Jr. Evaluation of accuracy and precision of adenovirus absorptivity at 260 nm under conditions of complete DNA disruption. Virology. 2002; 295:284–288. [DOI] [PubMed] [Google Scholar]
- 37. Scott W.G., Murray J.B., Arnold J.R.P., Stoddard B.L., Klug A.. Capturing the structure of a catalytic RNA intermediate: the hammerhead ribozyme. Science. 1996; 274:2065–2069. [DOI] [PubMed] [Google Scholar]
- 38. Nakano S., Chadalavada D.M., Bevilacqua P.C.. General acid-base catalysis in the mechanism of a hepatitis delta virus ribozyme. Science. 2000; 287:1493–1497. [DOI] [PubMed] [Google Scholar]
- 39. Xie K., Minkenberg B., Yang Y.. Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:3570–3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Carrier L., Bonne G., Bahrend E., Yu B., Richard P., Niel F., Hainque B., Cruaud C., Gary F., Labeit S. et al. . Organization and sequence of human cardiac myosin binding protein C gene (MYBPC3) and identification of mutations predicted to produce truncated proteins in familial hypertrophic cardiomyopathy. Circ. Res. 1997; 80:427–434. [PubMed] [Google Scholar]
- 41. Nishimasu H., Ran F.A., Hsu P.D., Konermann S., Shehata S.I., Dohmae N., Ishitani R., Zhang F., Nureki O.. Crystal structure of Cas9 in complex with guide RNA and Target DNA. Cell. 2014; 156:935–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lee S.J., McPherron A.C.. Regulation of myostatin activity and muscle growth. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:9306–9311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hilton I.B., D'Ippolito A.M., Vockley C.M., Thakore P.I., Crawford G.E., Reddy T.E., Gersbach C.A.. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat. Biotechnol. 2015; 33:510–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yin H., Xue W., Chen S., Bogorad R.L., Benedetti E., Grompe M., Koteliansky V., Sharp P.A., Jacks T., Anderson D.G.. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat. Biotechnol. 2014; 32:551–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Young C.S., Hicks M.R., Ermolova N.V., Nakano H., Jan M., Younesi S., Karumbayaram S., Kumagai-Cresse C., Wang D., Zack J.A. et al. . A single CRISPR-Cas9 deletion strategy that targets the majority of DMD patients restores dystrophin function in hiPSC-derived muscle cells. Cell Stem Cell. 2016; 18:533–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Iyombe-Engembe J.P., Ouellet D.L., Barbeau X., Rousseau J., Chapdelaine P., Lague P., Tremblay J.P.. Efficient restoration of the dystrophin gene reading frame and protein structure in DMD myoblasts using the CinDel method. Mol. Ther. Nucleic Acids. 2016; 5:e283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lee C.M., Cradick T.J., Fine E.J., Bao G.. Nuclease target site selection for maximizing on-target activity and minimizing off-target effects in genome editing. Mol. Ther. 2016; 24:475–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Paule M.R., White R.J.. Survey and summary: transcription by RNA polymerases I and III. Nucleic Acids Res. 2000; 28:1283–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nielsen S., Yuzenkova Y., Zenkin N.. Mechanism of eukaryotic RNA polymerase III transcription termination. Science. 2013; 340:1577–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chen B., Hu J., Almeida R., Liu H., Balakrishnan S., Covill-Cooke C., Lim W.A., Huang B.. Expanding the CRISPR imaging toolset with Staphylococcus aureus Cas9 for simultaneous imaging of multiple genomic loci. Nucleic Acids Res. 2016; 44:e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chen B., Gilbert L.A., Cimini B.A., Schnitzbauer J., Zhang W., Li G.W., Park J., Blackburn E.H., Weissman J.S., Qi L.S. et al. . Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013; 155:1479–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Dang Y., Jia G., Choi J., Ma H., Anaya E., Ye C., Shankar P., Wu H.. Optimizing sgRNA structure to improve CRISPR-Cas9 knockout efficiency. Genome Biol. 2015; 16:280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O. et al. . DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013; 31:827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Medina M.F., Joshi S.. RNA-polymerase III-driven expression cassettes in human gene therapy. Curr. Opin. Mol. Ther. 1999; 1:580–594. [PubMed] [Google Scholar]
- 55. White R.J. Transcription by RNA polymerase III: more complex than we thought. Nat. Rev. Genet. 2011; 12:459–463. [DOI] [PubMed] [Google Scholar]
- 56. Dieci G., Fiorino G., Castelnuovo M., Teichmann M., Pagano A.. The expanding RNA polymerase III transcriptome. Trends Genet.: TIG. 2007; 23:614–622. [DOI] [PubMed] [Google Scholar]
- 57. Salva M.Z., Himeda C.L., Tai P.W., Nishiuchi E., Gregorevic P., Allen J.M., Finn E.E., Nguyen Q.G., Blankinship M.J., Meuse L. et al. . Design of tissue-specific regulatory cassettes for high-level rAAV-mediated expression in skeletal and cardiac muscle. Mol. Ther. 2007; 15:320–329. [DOI] [PubMed] [Google Scholar]
- 58. Eagle M., Baudouin S.V., Chandler C., Giddings D.R., Bullock R., Bushby K.. Survival in Duchenne muscular dystrophy: improvements in life expectancy since 1967 and the impact of home nocturnal ventilation. Neuromuscul. Disord. 2002; 12:926–929. [DOI] [PubMed] [Google Scholar]
- 59. Nigro G., Comi L.I., Politano L., Bain R.J.. The incidence and evolution of cardiomyopathy in Duchenne muscular dystrophy. Int. J. Cardiol. 1990; 26:271–277. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.