Abstract
The seven APOBEC3 (A3) enzymes in primates restrict HIV/SIV replication to differing degrees by deaminating cytosine in viral (−)DNA, which forms promutagenic uracils that inactivate the virus. A polymorphism in human APOBEC3C (A3C) that encodes an S188I mutation increases the enzymatic activity of the protein and its ability to restrict HIV-1, and correlates with increased propensity to form dimers. However, other hominid A3C proteins only have an S188, suggesting they should be less active like the common form of human A3C. Nonetheless, here we demonstrate that chimpanzee and gorilla A3C have approximately equivalent activity to human A3C I188 and that chimpanzee and gorilla A3C form dimers at the same interface as human A3C S188I, but through different amino acids. For each of these hominid A3C enzymes, dimerization enables processivity on single-stranded DNA and results in higher levels of mutagenesis during reverse transcription in vitro and in cells. For increased mutagenic activity, formation of a dimer was more important than specific amino acids and the dimer interface is unique from other A3 enzymes. We propose that dimerization is a predictor of A3C enzyme activity.
INTRODUCTION
The human APOBEC3 (A3) family of single-stranded (ss) DNA cytidine deaminases has seven members that act as host restriction factors against retroelements, retroviruses, and other DNA viruses that contain ssDNA intermediates (1). For A3 enzymes to restrict HIV-1 in CD4+ T cells, they must first be encapsidated into the budding virion in order to facilitate cytosine to uracil deaminations on the (−)DNA synthesized by reverse transcriptase (2). When the (−) DNA is copied to form the (+) DNA the uracils template the addition of adenine, which results in C/G→T/A transition mutations that reduce the infectivity of HIV-1 (3–5). The A3 enzymes A3D, A3F, A3G and A3H (haplotypes II, V and VII) are able to restrict HIV-1 infection in this manner to varying degrees (6–12). However, lentiviruses such as HIV-1 have evolved a protein, Vif, which antagonizes the A3 proteins by inducing their degradation (6,12–20). Vif physically interacts with A3 enzymes and functions as a substrate receptor for a Cullin 5 E3 ubiquitin ligase complex inducing the polyubiqitination of the A3 proteins followed by degradation in the proteasome (14). Vif is stabilized in host cells by interacting with the transcriptional cofactor CBFβ and Elongin C (21–23).
For A3 enzymes that fortuitously escape Vif-mediated degradation or in the absence of Vif, the encapsidated A3s must be able to induce enough mutations to inactivate the proviral DNA. To achieve high levels of mutations, the enzymes must efficiently find cytosines for deamination within their preferred target motif within a limited amount of time (1). HIV-1 replication is a dynamic process with (−)DNA synthesis, RNA degradation and (+)DNA synthesis occurring at the same time. A3 enzymes A3F, A3G and A3H have been characterized to locate these targets on ssDNA by facilitated diffusion (1). Facilitated diffusion is Brownian motion driven diffusion of enzymes on DNA that occurs in the absence of an energy source to drive the enzyme's motion (24,25). Facilitated diffusion can involve 1D sliding of the enzyme along the DNA phosphate backbone and 3D translocations that are described as jumping or intersegmental transfer (24,25). Jumping is used to describe the movement of the enzyme as it diffuses within the charged domain of the DNA without directly interacting with the DNA phosphate backbone (24,25). Intersegmental transfer involves a doubly-bound state where the enzyme leaves the charged domain of the DNA and enters into the bulk solution to bind another DNA segment before releasing the first bound DNA (24,25). Sliding allows for deamination of cytosines that are closely spaced (<20 nt) whereas the jumping or intersegmental transfer movements allow for deamination of more distantly spaced targets (26–28). The A3 enzymes that most efficiently induce mutagenesis in HIV-1 proviral DNA, such as A3G and A3H, use a combination of both 1D short-range sliding and 3D long-range scanning movements which enables a rapid sampling of DNA for the preferred target motif (26,29–31). In contrast, A3F that is limited to using only long-range movements induces less mutagenesis than A3G and A3H (32). However, A3G, A3H and A3F are all processive enzymes, meaning they can deaminate multiple cytosines in a single enzyme–substrate encounter, but their level of processivity differs as a result of their scanning movements and this influences their mutagenic efficiency.
The processive mechanism of A3C has not been characterized previous to this study, however, A3C has been found in the majority of studies to be weakly restrictive or not restrictive for HIV-1 replication, yet it is still highly expressed in CD4+ lymphocytes and can be encapsidated (33–39). Recently, a human (h) A3C polymorphism, S188I, which exists in ∼10% of people of African descent was found to enable hA3C S188I to restrict HIV-1 replication 5- to 10-fold more than the common hA3C (38,40). The hA3C S188I was able to dimerize in vitro, unlike the common hA3C (38,41). While this is usually required for encapsidation for other A3s, for hA3C both the common and hA3C S188I were able to encapsidate relatively equally (38). These data suggested that the higher restriction levels induced by hA3C S188I in comparison to the common hA3C were due to differences in the enzymes inherent biochemical characteristics (38).
Here, we investigated the mechanism by which hA3C S188I has greater activity than the common hA3C. Our biochemical analysis demonstrates that the hA3C S188I is more processive and more mutagenic during reverse transcription than the common hA3C. Since we had previously shown that the hA3C S188I mutation correlates with dimerization of the protein (38), we also analyzed the closely related chimpanzee (cA3C) and gorilla A3C (gA3C) enzymes since these encode an S188, like the common hA3C version. Surprisingly, we found that cA3C and gA3C were efficient at inducing mutagenesis during reverse transcription despite the presence of an S188, suggesting another determinant for activity. We found that cA3C and gA3C have achieved this increased activity through dimerization that is mediated by a unique amino acid. These observations are consolidated by the finding that dimerization is required for A3C processivity and although the hominid A3Cs share a common dimer interface, they use different amino acids to form the dimer. Identifying these determinants enables prediction of A3C enzymatic activity.
MATERIALS AND METHODS
Cloning and site-directed mutagenesis
The sequence for hA3C, cA3C and gA3C have been previously described (38,40). The A3C sequences were cloned into a baculovirus transfer vector (pAcG2T or pFAST-bac1) containing an N-terminal GST tag or pcDNA3.1 with an N-terminal HA or V5 tag. Mutants were made by site-directed mutagenesis of the wild-type sequences to create hA3C S188I, hA3C S188I/N115K hA3C N115K, cA3C S188I, cA3C K115N, gA3C S188I and gA3C K115N. All constructed plasmids were verified by DNA sequencing.
Protein expression and purification
Recombinant baculovirus production for expression of wild type and mutant hA3C, cA3C and gA3C in Sf9 cells was carried out using the pACG2T or pFast-bac1 transfer vector as previously described (29,42). Sf9 cells were infected with recombinant GST-A3C virus at an MOI of 1 (hA3C, cA3C, gA3C, cA3C S188I, hA3C S188I/N115K, gA3C S188I and gA3C K115N) or an MOI of 5 (hA3C S188I, hA3C N115K and cA3C K115N). Recombinant baculovirus infected Sf9 cells were harvested after 72 h of infection. Cells lysates treated with RNaseA were incubated with glutathione-Sepharose 4B resin (GE Healthcare) at 4°C and were subjected to a series of salt washes, as previously described (43). The enzymes were eluted with the GST tag in elution buffer (100 mM Tris, pH 8.8, 150 mM NaCl, 10% (v/v) glycerol and 50 mM reduced glutathione). The A3C enzymes were then cleaved from the GST tag in solution at 21°C for 6 h with Thrombin (GE Healthcare) before being dialyzed against 100 mM Tris pH 7.5, 250 mM NaCl, 10% (v/v) glycerol and 1 mM DTT. To purify A3C from the free GST and thrombin, the enzyme stock was diluted to achieve a solution of 50 mM Tris pH 7.5, 50 mM NaCl, 10% (v/v) glycerol and 1 mM DTT for loading onto a DEAE FF column (GE Healthcare). A3C was eluted with a linear gradient of NaCl. The A3C eluted at ∼350 mM NaCl and was >95% pure, as determined by SDS-PAGE (Supplementary Figure S1).
In vitro deamination assay
All ssDNA substrates were obtained from Tri-Link Biotechnologies and have been published previously (32). Reactions were carried out under single-hit conditions (i.e. <15% substrate usage) to ensure that each ssDNA reacted with at most a single enzyme (44). Under these conditions, a processivity factor can be determined by comparing the quantified total amount of deaminations occurring at two sites on the same ssDNA with a calculated theoretical value of deaminations at these two sites if the deamination events were uncorrelated (not processive) (31). A ssDNA substrate containing two 5΄TTC motifs (100 nM) was incubated with 350 nM (gA3C, hA3C S188I, gA3C S188I, cA3C S188I, hA3C S188I/N115K and hA3C N115K) or 700 nM (hA3C, cA3C, cA3C K115N, gA3C K115N) of A3C for 2.5 to 30 min at 37°C in RT buffer (50 mM Tris, pH 7.5, 40 mM KCl, 10 mM MgCl2 and 1 mM DTT). The reaction time was varied on each ssDNA according to the specific activity of the enzymes to ensure ∼10% substrate usage. For intersegmental transfer assays, the A3C:ssDNA ratio (3:1 or 7:1) was kept the same, but increasing concentrations of enzyme and substrate was titrated in (ssDNA: 100–500 nM, A3C: 350–1750 nM or 700–3500 nM). Reactions were started by the addition of the ssDNA substrate. A3C-catalyzed deaminations were detected by treating the ssDNA with uracil DNA glycosylase (New England Biolabs) and heating under alkaline conditions before resolving the fluorescein-labeled ssDNA on 10 or 20% (v/v) denaturing polyacrylamide gels, depending on the sizes of the ssDNA fragments. Gel photos were obtained using a Typhoon Trio multipurpose scanner (GE Healthcare) and integrated gel band intensities were analyzed using ImageQuant (GE Healthcare). The specific activity was calculated from single-hit condition reactions by determining the picomoles of substrate used per minute for a microgram of enzyme.
In vitro reverse transcription assay
Mutagenesis of ssDNA by A3 enzymes during reverse transcription of an RNA template was assessed using a previously established in vitro assay, which models reverse transcription of an RNA template and second-strand synthesis (26). In brief, a synthetic (+)RNA that contains the poly-purine tract (PPT), the catalytic region of the protease gene (120 nt), and lacZα (248 nt) is used. In this construct the PPT is used as a primer for (+)DNA synthesis and enables synthesis of dsDNA. The lacZα serves as a reporter gene for mutations by blue/white screening. To start the assay, the 368 nt RNA template (50 nM) is annealed to a DNA primer (24 nt) and incubated with 1.5 μM of nucleocapsid, 1.2 μM of reverse transcriptase and 500 μM of dNTPs in RT buffer in the presence or absence of 350 nM of each A3C enzyme. The resulting dsDNA that was synthesized from this in vitro system was PCR amplified using Pfu Cx Turbo Hotstart (Agilent Technologies) that can use uracils as a template with high fidelity. These amplicons were then cloned into a pET-Blue vector backbone that allows for blue-white screening of the synthesized lacZα. At least 25 mutated clones for each condition were sequenced.
Size exclusion chromatography
The oligomerization states of the A3C enzymes were determined by loading 10 μg of purified enzyme on a 10 ml Superdex 200 (GE Healthcare) size exclusion column. The column was prepared by pouring the resin bed in a column with 16 cm height and 0.5 cm diameter. The running buffer contained 50 mM Tris pH 8.0, 200 mM NaCl and 1 mM DTT. The chromatograms from the 10 ml Superdex 200 column were constructed by analyzing the integrated gel band intensities of the protein in each fraction after resolution by SDS-PAGE. The gels for each panel were resolved, stained with Oriole fluorescent gel stain, and scanned in parallel. The Bio-Rad standard set was used to generate a standard curve from which molecular weight and oligomerization states of the enzymes were determined.
Protein crosslinking
A3C enzymes (0.5 μM) were incubated with 20 μM BS3 (bis (sulfosuccinimidyl) suberate), an amine to amine crosslinker, in 20 mM HEPES (pH 7.5), 150 mM NaCl and 1 mM DTT for 1 h at 21°C. Crosslinked proteins were resolved by SDS-PAGE, transferred to a nitrocellulose membrane for immunoblotting, and visualized using primary antibody for native A3C (GeneTex) and secondary IRdye labeled goat anti-rabbit antibody compatible with the LI-COR/Odyssey system.
Steady state rotational anisotropy
Steady state fluorescence depolarization (rotational anisotropy) was used to measure the binding affinity of A3C-ssDNA and a 118 nt fluorescein-labeled ssDNA. Reactions were 60 μl and contained fluorescein-labeled ssDNA (10 nM) in RT buffer. The enzyme was titrated into the solution until saturation. For saturation the required titrations used were: hA3C (0–1600 nM); hA3C S188I, cA3C S188I, gA3C S188I, cA3C and gA3C (0–1200 nM); hA3C S188I/N115K, hA3C N115K and cA3C N115K (0–1100 nM); and gA3C K115N (0–1800 nM). A QuantaMaster QM-4 spectrofluorometer (Photon Technology International) with a dual emission channel was used to collect data and calculate anisotropy. Samples were excited with vertically polarized light at 495 nm (6 nm band pass) and vertical and horizontal emissions were measured at 520 nm (6 nm band pass). Apparent dissociation constants (Kd) were obtained by fitting to a rectangular hyperbola or sigmoidal curve using Sigma Plot 11.2 software.
Co-immunoprecipitation
Co-immunoprecipitation (Co-IP) assays were conducted as described previously (45). Briefly, 293T cells (2.5 × 106 cells per 75 cm2 flask) were transfected with 1μg total DNA. Equal amounts of each plasmid for hA3C-HA and hA3C-V5, hA3C S188I-HA and hA3C S188I-V5 or hA3C S188I/N115K-HA and hA3C S188I/N115K-V5 were used to transfect the cells. Genejuice transfection reagent (EMD Millipore) was used according to the manufacturer's instructions. At 72 h post transfection, the cells were washed with PBS and lysed in IP buffer (50 mM Tris–Cl pH 7.4, 1% Nonidet-P40, 0.1% sodium deoxycholate, 10% glycerol, 150 mM NaCl) supplemented with RNaseA (20 μg/ml; Roche) and EDTA-free protease inhibitor (Roche). One half of the precleared supernatant was then incubated with Protein A-agarose conjugated polyclonal rabbit anti-HA antibody (2 μg, Santa Cruz Biotechnology) and the other half (mock) was incubated with Protein A–agarose-conjugated normal rabbit IgG (2 μg, Santa Cruz Biotechnology) at 4°C for 2 h. Resin was washed and the samples were then resolved by SDS-PAGE and transferred to a nitrocellulose membrane. For detection of A3C-HA and A3C-V5 in cell lysates, the membrane was probed with polyclonal Rabbit HA (1:1000, Sigma) and polyclonal Rabbit V5 (1:200, Santa Cruz Biotechnology), respectively. For the loading control, monoclonal mouse anti-α-tubulin (1:1000, Sigma) was used. HA- and rabbit IgG-immunoprecipitated lysates were probed with anti-V5 mouse monoclonal antibodies. After incubation with Horse Radish Peroxidase (HRP) conjugated secondary antibodies, the blots were visualized with X-ray film using Super Signal West Pico chemiluminescence substrate (Thermo-Scientific).
Single-cycle infectivity assay
The 293T cells were plated at a density of 5 × 104 cells per well of a 12-well plate. The next day, the cells were transfected with 0.50 μg pLAI HIV Δvif, 0.15 μg pVSV-G and 0.30 μg pCDNA3.1 A3C-HA or 0.10 μg pCDNA3.1 A3G-HA. Different levels of transfected DNA were used to achieve the same steady state expression levels in cells for A3C-HA and A3G-HA. All transfection amounts of DNA were equalized with empty pCDNA3.1 plasmid. Forty-eight hours after transfection, culture supernatants containing virus were collected, filtered, and used to infect HeLa CD4+ HIV-1 LTR-β-gal cells (MAGI) cells (46). To detect A3 by immunoblotting, harvested supernatants were centrifuged at 16 000 × g for 60 min to recover virions and cells from the same well were washed with PBS and lysed with Laemmli sample buffer. Forty-eight hours after infection of HeLa CD4+ HIV-1 LTR-β-gal cells, Pierce β-galactosidase assay reagent was used to lyse cells and detect β-galactosidase activity through colorimetric detection using a spectrophotometer. Infectivity of each virus was compared by setting infectivity of the ‘No A3’ to 100%. Statistical significance of results was determined using an unpaired t-test.
Quantitative immunoblotting
The 293T cells expressing A3C-HA and A3G-HA from the single-cycle infectivity assays were detected using anti-HA (1:1000; Sigma) in cell lysates (30 μg total protein) and virions (20 μl of concentrated virus). Loading controls for cell lysates (α-tubulin, Sigma) and virions (p24, Cat #3537, NIH AIDS Reagent Program) were detected using mouse monoclonal antibodies. Odyssey software was used to quantitate the amount of p24 in each lane and ensure that similar amounts of virus lysate were loaded. Proteins of interest and loading controls were detected in parallel by using the Licor/Odyssey system (IRDye 680-labeled goat anti-rabbit secondary and IRDye 800-labeled goat anti-mouse secondary antibody). Using Odyssey software, the relative amount of A3 on the blot was calculated by first normalizing each sample lane to the corresponding control. Normalized values were then converted to relative amounts of A3 by setting the hA3C band at 1.0 and calculating the relative amounts of A3 in other lanes.
Sequencing of integrated proviral DNA
Total DNA from infected 293T cells was extracted 48 h after infection using DNAzol reagent (Life Technologies). DNA was treated with DpnI (New England Biolabs) for 1 h at 37°C to remove possible contaminating plasmid DNA and the protease (nt 2280–2631) sequences were amplified by PCR using Q5 polymerase (New England Biolabs). Primers have been published previously (32). The PCR products were purified and cloned with the CloneJET PCR cloning kit (Thermo Fisher). DNA was sequenced with kit-specific primers and carried out at the National Research Council of Canada (Saskatoon, Canada).
Structural models of A3C
The dimer model used was present in the asymmetric unit of the crystal structure (PDB: 3VOW). The model mutations created were done by manual mutagenesis in Coot (Crystallographic Object-Oriented Toolkit), selecting rotamers that minimized clashes to the surrounding residues.
RESULTS
Human A3C S188I is a processive enzyme
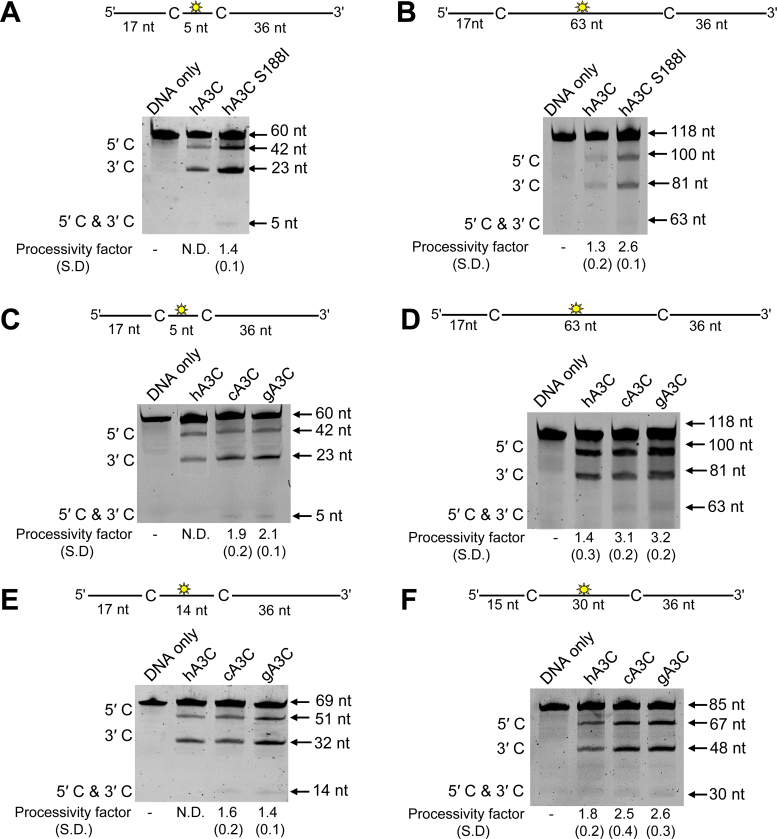
We were interested in understanding why the hA3C S188I polymorphism encodes a more active enzyme than the more common hA3C S188. Since the ssDNA scanning mechanisms used by an A3 enzyme contributes to the efficiency of HIV-1 restriction we first examined the mechanisms by which hA3C scanned ssDNA (1). By characterizing the ssDNA scanning mechanisms we are able to measure enzyme processivity, the ability of the enzyme to deaminate more than one cytosine in a single enzyme-substrate encounter. An in vitro deamination assay was used in which A3C processivity was tested on different synthetic ssDNA substrates that contained two 5΄TTC deamination motifs spaced varying distances apart. Processive deaminations are measured under single-hit conditions (<15% substrate usage) to ensure that each ssDNA substrate was acted upon by only one enzyme during the course of the reaction (44). Under these conditions, a processivity factor is calculated as the ratio of the processive deaminations occurring during the experiment, i.e. deamination of both 5΄TTC motifs in a single enzyme–substrate encounter, in comparison to the calculated theoretical number of deaminations that would occur independently at both 5΄TTC motifs if the enzyme were non-processive (see Materials and Methods) (31). As a reference for the range of processivity, a highly processive deaminase, such as A3G, has processivity factors ranging from 4 to 8, depending on the ssDNA substrate (Supplementary Figure S2) (26,32). An enzyme that is non-processive, such as A3A, would have a processivity factor of 1.0 or an undetectable level of deamination of both motifs under the single-hit reaction kinetics of the experiment (Supplementary Figure S2) (47).
To measure processive sliding movements we used an ssDNA substrate with the deamination targets spaced 5 nt apart. For the common hA3C (S188, referred to as hA3C), a processive deamination band (5΄ and 3΄) was not detected under single-hit reaction kinetics (Figure 1A). Thus, hA3C was unable to processively slide to deaminate multiple cytosines. In contrast, on this substrate the hA3C S188I had a processivity factor of 1.4 (Figure 1A). This processivity factor meant that hA3C S188I was 1.4-fold more likely to catalyze processive deaminations of both the cytosine motifs in a single enzyme–ssDNA encounter than to catalyze both deaminations in separate enzyme–ssDNA encounters. To investigate if the A3Cs had differences in their ability to scan ssDNA by three-dimensional translocations, we utilized ssDNA substrates with more distantly spaced deamination targets (63 nt apart). The processivity factors indicated that hA3C S188I was 2-fold more processive than the more common form of hA3C (Figure 1B). Thus, differences in enzyme processivity could explain why hA3C S188I is able to restrict HIV-1 replication more than hA3C (38).
Figure 1.
Analysis of A3C processivity on ssDNA oligonucleotides. Processivity of A3C was tested on ssDNA substrates that contain a fluorescein-labeled deoxythymidine (yellow star) between two 5΄TTC deamination motifs separated by different distances. (A and B) hA3C S188I is more processive than hA3C. (A) Deamination of a 60 nt ssDNA substrate with deamination targets spaced 5 nt apart. Single deaminations of the 5΄C and 3΄C are detected as the appearance of labeled 42- and 23-nt fragments, respectively; double deamination of both C residues on the same molecule results in a 5 nt labeled fragment. (B) Deamination of a 118 nt ssDNA substrate with deaminated cytosines spaced 63 nt apart. Single deaminations of the 5΄C and 3΄C are detected as the appearance of labeled 100- and 81-nt fragments, respectively; double deamination of both C residues on the same molecule results in a 63 nt labeled fragment. (C–F) cA3C and gA3C are more processive than hA3C. (C) Deamination of a 60 nt ssDNA substrate as for panel (A). (D) Deamination of a 118 nt ssDNA as for panel (B). (E) Deamination of a 69 nt ssDNA substrate with deamination targets spaced 14 nt apart. Single deaminations of the 5΄C and 3΄C are detected as the appearance of labeled 51- and 32-nt fragments, respectively; double deamination of both C residues on the same molecule results in a 14 nt labeled fragment. (F) Deamination of an 85 nt ssDNA substrate with deaminated cytosines spaced 30 nt apart. Single deaminations of the 5΄C and 3΄C are detected as the appearance of labeled 67- and 48-nt fragments, respectively; double deamination of both C residues on the same molecule results in a 30 nt labeled fragment. If no 5΄C and 3΄C band was detected, the processivity was denoted with N.D. (not detected). The measurements of enzyme processivity (processivity factor) and the S.D. are shown below the gels. All values are calculated from at least three independent experiments.
Chimpanzee and gorilla A3C processivity is distinct from human A3C processivity
We previously showed the cA3C and gA3C encode a serine at position 188 (38), suggesting that they should be less active than hA3C S188I if the S188I mutation is the only mutation able to confer increased processivity for A3C. Surprisingly, however, on the substrate with deamination targets spaced 5 nt apart, cA3C and gA3C had processivity factors of 1.9 and 2.1, respectively (Figure 1C). Similarly, on ssDNA substrates with deamination targets spaced 63 nt apart, the processivity factors of cA3C and gA3C demonstrated that these enzymes were ∼3-fold more likely to catalyze processive deaminations, but hA3C had a processivity factor of 1.4 (Figure 1D). To extend these results we tested the processivity of hA3C, cA3C and gA3C on substrates with deamination targets spaced 14- and 30-nt apart. Consistently, cA3C and gA3C were more processive than hA3C (Figure 1E and F).
Due to the higher processivity factors of all A3Cs on the ssDNAs with distantly spaced deamination targets, these data suggest that the A3C enzymes were primarily using 3D translocations in order to locate the target cytosines. However, hA3C S188I, cA3C and gA3C were more processive than hA3C. We also determined that the specific activity of cA3C and gA3C were similar to hA3C S188I and 5- to 8-fold higher than hA3C (Table 1). Altogether the data suggested that for cA3C and gA3C, increases in deamination activity did not require an I188, as for hA3C.
Table 1. Specific activities of A3C enzymes.
| Enzyme | Specific activity (pmol/μg/min) |
|---|---|
| hA3C | 0.020 ± 0.005 |
| cA3C | 0.100 ± 0.010 |
| gA3C | 0.150 ± 0.010 |
| hA3C S188I | 0.160 ± 0.050 |
| hA3C N115K | 0.085 ± 0.020 |
| hA3C S188I/N115K | 0.220 ± 0.020 |
| cA3C S188I | 0.210 ± 0.030 |
| cA3C K115N | 0.050 ± 0.007 |
| gA3C S188I | 0.280 ± 0.030 |
| gA3C K115N | 0.080 ± 0.002 |
Dimerization of hominid A3C is mediated through α-helix 6 or β-strand 4
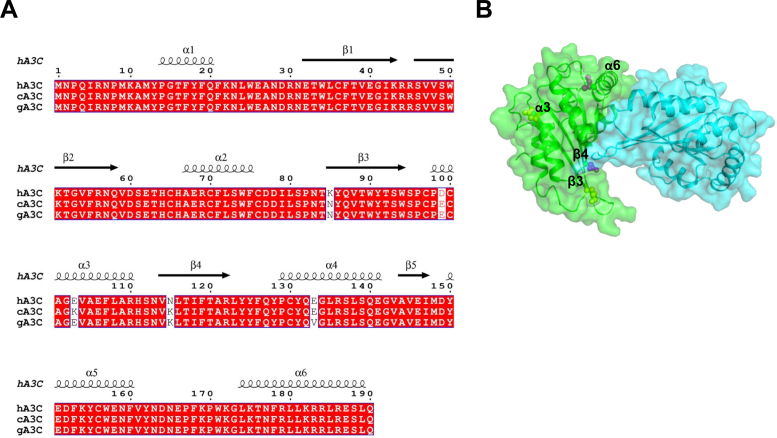
We reasoned that by examining differences in hA3C, cA3C, and gA3C sequences we could identify amino acids that were unique to cA3C and gA3C and investigate if these amino acids enabled increased processivity and specific activity. From this analysis, only the amino acids at positions 85 (β-strand 3), 99 (α-helix 3) and 115 (β-strand 4) that were different in both cA3C and gA3C from hA3C were candidates (Figure 2A). To aid in making mutations we examined the existing crystal structure of hA3C (Figure 2B) (41). Although hA3C is a monomer in solution, the enzyme crystallized as a dimer (41). Since dimerization was important for increased hA3C S188I activity (38), we hypothesized that cA3C and gA3C were more active than hA3C due the ability to dimerize. We thus began by mutating amino acids closest to the predicted dimer interface. The amino acid at position 115 was closer to the predicted dimer interface in the hA3C crystal structure than amino acid 85 or 99 (Figure 2B, β-strand 4). As a result, we converted the cA3C and gA3C amino acid at position 115 to the hA3C amino acid at that position, making a K115N mutant (Figure 2A).
Figure 2.
Sequence alignment and structural analysis of A3C. (A) Sequence alignment of hA3C, cA3C and gA3C with amino acid differences shown in white. The sequence alignment was performed by a Clustal Omega multiple sequence alignment (75) and plotted using the program ESPript (76). (B) Surface representation of a hA3C dimer from the crystal structure (PDB: 3VOW). Amino acids unique to hA3C that are potentially involved in the dimer interface are shown in purple (α-helix 6, S188; β-strand 4, N115) and other amino acids unique to hA3C are shown in yellow (β-strand 3, K85; α-helix 3, D99).
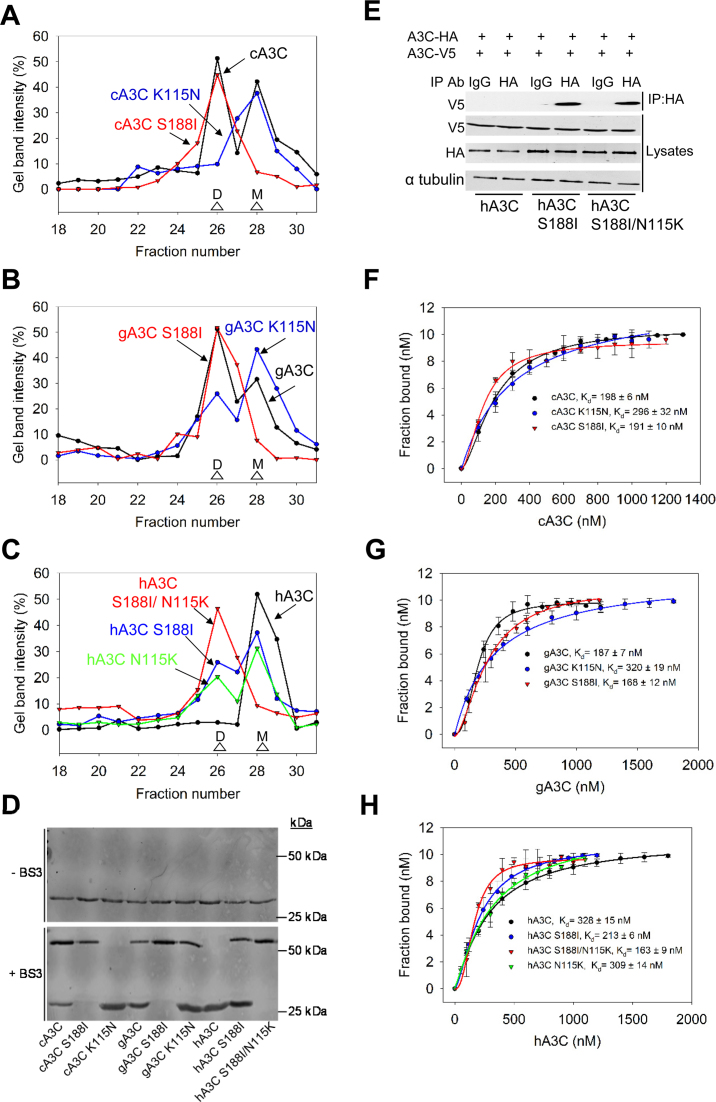
Using size exclusion chromatography (SEC) and a calibration curve, we determined the oligomerization states of cA3C, gA3C and their K115N mutants. Consistent with increased processivity and specific activity, we found that cA3C and gA3C were able to form dimers, similar to hA3C S188I, but in contrast to hA3C (Figure 3A–C and Supplementary Figure S3, apparent molecular weights 45 kDa (dimer) and 19 kDa (monomer)). The cA3C and gA3C SEC showed the presence of both monomer and dimer peaks indicating that the dimer and monomer forms were in equilibrium (Figure 3A and B). Interestingly, the hA3C crystal structure correctly predicted the key dimerization amino acid for cA3C and gA3C (41). The cA3C K115N existed solely as a monomer population, similar to hA3C (Figure 3A and C). The gA3C K115N formed a majority of monomers, however there were still a small proportion of dimers in solution (Figure 3B). This suggested that an amino acid unique to gA3C further stabilized dimerization. Only V133 is unique to gA3C, suggesting that the dimer interface in gA3C may directly or indirectly involve α-helix 4 (Figure 2A). Further, in hA3C a K115 alone can mediate dimerization since a hA3C N115K mutant formed an equilibrium of monomers and dimers (Figure 3C). That the wild type cA3C and gA3C dimerization was similar to hA3C S188I and hA3C N115K (Figure 3A–C) was consistent with the conclusion that dimerization was enabling A3C to be more processive and have a higher specific activity (Figure 1 and Table 1).
Figure 3.
A3C dimerization is mediated through α-helix 6 or β-strand 4. (A–C) SEC profile for 10 μg of (A) cA3C, cA3C K115N and cA3C S188I; (B) gA3C, gA3C K115N and gA3C S188I and (C) hA3C, hA3C S188I, hA3C N115K and hA3C S188I/N115K from a 10 ml Superdex 200 column was used to calculate the oligomerization state of the enzyme from a standard calibration curve. An M denotes a monomer fraction and a D denotes a dimer fraction. (A) cA3C formed monomers and dimers (apparent molecular weights 19 kDa and 45 kDa, respectively), cA3C S188I formed a stable dimer (apparent molecular weight 45 kDa), and cA3C K115N formed monomers (apparent molecular weight 19 kDa). (B) The gA3C SEC profiles were similar to cA3C, except for gA3C K115N that was mainly monomers (apparent molecular weight 19 kDa), but also retained a small proportion of dimers (apparent molecular weight 45 kDa). (C) hA3C formed monomers in solution (apparent molecular weight 19 kDa), hA3C S188I and hA3C N115K were an equilibrium of monomers and dimers (apparent molecular weights 19 kDa and 45 kDa, respectively) and hA3C S188I/N115K was a stable dimer (apparent molecular weight 45 kDa). The chromatograms were constructed by analyzing the integrated gel-band intensities of each protein in each fraction after resolution by SDS-PAGE (Supplementary Figure S3). (D) A3C enzymes were incubated in the absence or presence of 20 μM BS3 crosslinker and subsequently visualized with SDS-PAGE and immunoblotting. Monomeric A3C enzymes remained as monomers in the presence of crosslinker (cA3C K115N, hA3C). A3C enzymes that were able to form dimers according to SEC, were also present as monomers/dimers (cA3C, gA3C, gA3C K115N, hA3C S188I) or as dimers (cA3C S188I, gA3C S188I, hA3C S188I/N115K) in the presence of the crosslinker. Molecular weight standards are indicated. (E) Coimmunoprecipitation of A3C-V5 with A3C-HA. The A3C-HA and A3C-V5 were transfected in combination and the immunoprecipitation of cell lysates used either anti-HA antibody or Rabbit IgG (mock) and was immunoblotted with antibodies against α-tubulin, HA and V5. Cell lysates show the expression of α-tubulin, HA and V5. (F–H) The apparent Kd of A3C enzymes from a 118 nt ssDNA was analyzed by steady-state rotational anisotropy for (E) cA3C, cA3C S188I and cA3C K115N; (F) gA3C, gA3C S188I, and gA3C K115N and (G) hA3C, hA3C S188I, hA3C N115K and hA3C S188I/N115K. Apparent Kd values are shown in the figure. Hill coefficients for cooperative binding curves are (E) cA3C, 1.6; cA3C S188I, 1.7; (F) gA3C, 1.8; gA3C S188I, 2.1; (G) hA3C S188I, 1.6; hA3C N115K, 1.5; hA3C S188I/N115K, 1.9. Error bars represent the S.D. from three independent experiments.
In order to more rigorously test this conclusion using deamination assays, we first created a panel of mutants at positions 115 and 188 in each of the three hominid A3C enzymes. We introduced the hA3C S188I mutation on α-helix 6 in cA3C and gA3C. Both cA3C S188I and gA3C S188I shifted from an equilibrium of monomer and dimer populations to a stable dimer (Figure 3A and B). We also mutated hA3C to form the hA3C S188I/N115K double mutant. The hA3C S188I/N115K formed a stable dimer (Figure 3C). Analysis of these mutants suggested that the hA3C, cA3C and gA3C dimerization interfaces were the same, but were stabilized by different amino acids.
To confirm the differences in oligomerization as determined by SEC, we used chemical crosslinking and co-IP. For crosslinking, the BS3 amine to amine chemical crosslinker enabled the resolution of A3C complexes by SDS-PAGE, which were then visualized by immunoblotting. Although the amount of protein added to each crosslinking reaction was the same (Figure 3D, −BS3), the total intensities of crosslinked proteins as determined by immunoblotting appeared to be unequal, which may have been due to amino acids that reacted with the BS3 preventing the antibody from binding (Figure 3D, +BS3). Nonetheless, the crosslinked A3C data were consistent with the SEC and identified the same monomeric (cA3C K115N, hA3C), monomeric/dimeric (cA3C, gA3C, gA3C K115N, hA3C S188I) and dimeric (cA3C S188I, gA3C S188I, hA3C S188I/N115K) forms (Figure 3D). We also confirmed that the dimerization was physiological by demonstrating that A3C-HA could co-IP A3C-V5. The co-IP experiment was conducted in the presence of RNaseA to ensure that we were detecting protein-protein interactions. This was tested for hA3C, hA3C S188I, and hA3C S188I/N115K. We found that hA3C S188I and hA3C S188I/N115K, but not hA3C, could co-IP from the lysates of 293T cells, consistent with the SEC and crosslinking data (Figure 3E).
To determine if these differences in dimerization also occurred on ssDNA we used fluorescence depolarization to measure the rotational anisotropy of fluorescently labeled ssDNA when A3C was titrated into the solution. The binding of A3C to the fluorescently labeled ssDNA will result in a change in rotation speed (anisotropy) until the fluorescently labeled ssDNA is saturated with A3C. From these data, an apparent dissociation constant (Kd) was determined. We observed that each A3C was able to bind ssDNA in the nanomolar range, with cA3C, gA3C, and hA3C S188I having <2-fold higher affinity for ssDNA than hA3C (Figure 3F–H). This indicated that the minimal processivity observed for hA3C was not due to the enzyme having a weak interaction with the ssDNA (Figure 1). However, we observed that the nature of the interaction of the A3C enzymes with ssDNA was different. The cA3C, gA3C, and hA3C S188I bound ssDNA cooperatively (Figure 3F–H, Hill coefficients of 1.5–2.1), while hA3C bound ssDNA non-cooperatively (Figure 3H, data fit a rectangular hyperbola). This indicated that the cA3C, gA3C, and hA3C S188I monomers in solution were able to oligomerize by binding ssDNA and the dimers in solution were stabilized or oligomerized further upon binding ssDNA. This was not observed for hA3C that bound ssDNA non-cooperatively and indicated that hA3C remained a monomer even when bound to ssDNA (Figure 3H). With the mutants, we found that the A3C enzymes that were able to form dimers in solution bound to ssDNA cooperatively and the A3C enzymes that lacked the ability to dimerize bound ssDNA non-cooperatively with saturation curves that best fit a rectangular hyperbola by least squares regression analysis (Figure 3F–H). The exception was gA3C K115N that had a minor dimer population, but bound ssDNA non-cooperatively (Figure 3B and G). These data demonstrate that the V133 amino acid is not sufficient to promote dimerization on ssDNA and either I188 or K115 are required for this function. Altogether, the SEC and binding data indicated that the oligomerization state and not the binding affinity for ssDNA differed between hA3C and hA3C S188I, cA3C and gA3C. Thus, the higher processivity of hA3C S188I, cA3C and gA3C correlated with their ability to form dimers.
Dimerization correlates with efficient ssDNA scanning
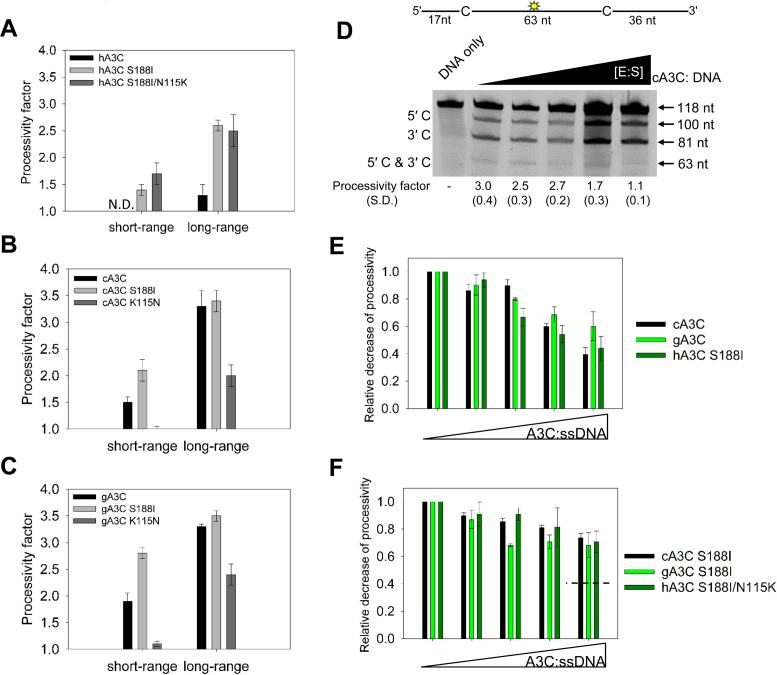
Our analysis of A3C orthologs indicated that dimerization was required for processive ssDNA scanning. However, due to other amino acid differences between the A3C orthologs (Figure 2A) we analyzed the monomer and dimer forms of each A3C ortholog individually to test that improved dimerization would lead to improved processivity. To test for short-range processivity due to sliding we used the substrate with deamination targets separated by 5 nt. We observed that for hA3C, dimerization through the S188I mutation enabled processive sliding and this increased further when the hA3C S188I dimer was stabilized by the N115K mutation (Figure 4A, short-range). For the cA3C S188I stable dimer we observed improved sliding compared to the wild type enzyme (Figure 4B, short-range). In contrast, the monomeric cA3C K115N was not processive (Figure 4B, short-range). The results with the gA3C mutants were similar to cA3C with the gA3C S188I having improved sliding from the wild type enzyme and the gA3C K115N having ∼3-fold reduced sliding from the wild type enzyme (Figure 4C, short-range).
Figure 4.
Dimerization influences processive ssDNA scanning. Processivity of A3C mutants was tested on ssDNA substrates and compared to the wild type enzyme. (A–C) Processivity factor values are shown for short-range movements based on deamination of a 60 nt ssDNA substrate with deamination targets spaced 5 nt apart and long-range movements based on deamination of a 118 nt substrate with deaminated cytosines spaced 63 nt apart for (A) hA3C, hA3C S188I and hA3C S188I/N115K, (B) cA3C, cA3C S188I and cA3C K115N and (C) gA3C, gA3C S188I and gA3C K115N. See Supplementary Figure S4 for a representative gel. (D) Intersegmental transfer ability of cA3C was determined by keeping an A3C/ssDNA ratio of 7:1 constant, but increasing the total reaction components. If the enzyme is able to undergo intersegmental transfer, the assay will result in an apparent decrease in the processivity factor with increasing concentrations of reaction components. The ssDNA substrate contained a fluorescein-labeled deoxythymidine (yellow star) between two deamination targets separated by 63 nt. The measurements of enzyme processivity (processivity factor) and the S.D. are shown below the gel. (E and F) Summary of intersegmental transfer assays shown in Supplementary Figure S5. (E) The monomer/dimer forms of A3C (cA3C, gA3C, hA3C S188I) are better able to undergo intersegmental transfer than the (F) stable dimer forms of A3C (cA3C S188I, gA3C S188I, hA3C S188I/N115K). For comparison, the hatched line in (F) denotes the decrease in processivity observed for monomer/dimer forms of A3C in (E). All values are calculated from at least three independent experiments.
To test for long-range processivity due to jumping or intersegmental transfer we used the substrate with deamination targets separated by 63 nt. For hA3C, the S188I mutation improved the processivity of the enzyme ∼3-fold (Figure 4A, long-range). The formation of a stable dimer did not further improve processivity (Figure 4A–C, long range). However, the cA3C K115N and gA3C K115N mutants were approximately 1.5- to 2- fold less processive than the wild-type enzymes (Figure 4B and C, long-range). The changes in oligomerization also resulted in changes to the specific activity of the enzymes. With the introduction of the hA3C S188I or hA3C S188I/N115K mutation, the hA3C specific activity was increased 8- to 11-fold from the wild-type (Table 1). Stabilization of the cA3C or gA3C dimer with the S188I mutation resulted in a 2-fold increase in specific activity from the wild type enzymes (Table 1). The K115N mutation in cA3C and gA3C resulted in an ∼2-fold lower specific activity than the wild type enzymes (Table 1). Since the amino acid differences were not in the active site, the data suggested that differences in catalytic activity were due to the different efficiencies in the ssDNA scanning mechanisms employed.
A monomer-dimer equilibrium is required for scanning ssDNA by intersegmental transfer
The long-range processivity movements can be of two types, either jumping or intersegmental transfer. We determined whether the A3C orthologs and their mutants were scanning ssDNA using jumping, intersegmental transfer, or both in order to determine if the stability of the dimer correlated with either of these types of movements.
For A3C that has a single DNA binding domain per polypeptide chain, intersegmental transfer movements would require dimerization to achieve a doubly bound state. We conducted an intersegmental transfer assay by increasing the concentration of the A3C and ssDNA substrate while keeping the ratio of the components the same. In doing this, the reaction environment becomes crowded which makes the enzyme more likely to translocate to a different ssDNA than to translocate within the same ssDNA substrate (29,48). If intersegmental transfer is occurring this results in an apparent decrease in the enzyme's processivity as the enzyme cycles between substrates (29,48). For example, for cA3C we observed that as the concentration of the reaction components increased, the processivity decreased (Figure 4D, cA3C processivity of 3.0 decreased to 1.1). For cA3C the processivity decreased to 1.1, which is essentially non-processive. This meant that the majority of the 3D translocations for cA3C were due to intersegmental transfer and not jumping (Figure 1D and F). Since jumping occurs within the localized charged domain of the DNA it is insensitive to crowding of the reaction (1,29,48). To compare the A3Cs we summarized the intersegmental transfer assay data by calculating the relative decrease of processivity with increasing A3C and ssDNA concentration for the A3Cs that are monomer/dimer (Figure 4E) or dimer (Figure 4F). For the A3Cs with a monomer/dimer equilibrium, all could undergo intersegmental transfer relatively equally as demonstrated by the characteristic decrease of the processivity factor with increasing reaction components (Figure 4E). This suggested that for A3C, dimerization and not specific amino acid motifs was the main determinant for this ssDNA scanning mechanism. However, the stable dimer forms were not able to undergo intersegmental transfer as well (Figure 4F). The processivity factors did not decrease throughout the titration and plateaued (Figure 4F). However, the stable dimer forms of A3C did remain processive and had processivity factors that were not significantly different than the corresponding monomer/dimer A3C form at the initial A3C: ssDNA concentration used in the experiments (gA3C, compare Supplementary Figure S5 panels A and C; hA3C, compare Supplementary Figure S5 panels B and E; cA3C, compare Figure 4D and Supplementary Figure S5D). These data indicate that jumping was used instead of intersegmental transfer (Supplementary Figure S5). Thus, these forms of ssDNA scanning for A3C were interchangeable and did not depend on specific amino acid motifs, but were mediated by dimerization.
Ability to processively scan ssDNA correlates with mutagenesis ability
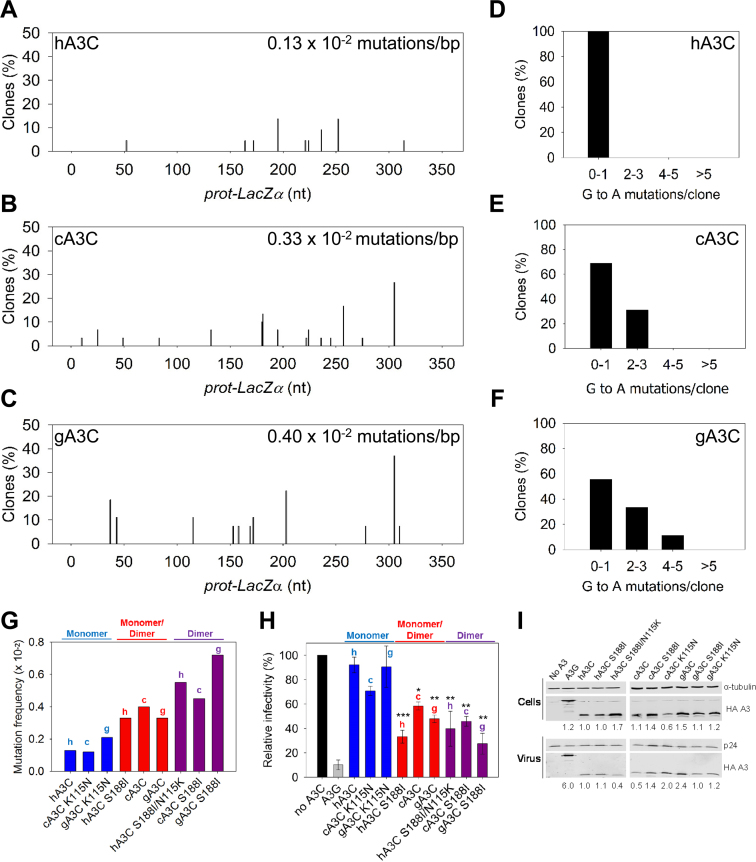
Our results showing that cA3C and gA3C were more processive in comparison to hA3C would predict that they are also able to induce a higher frequency of mutations than hA3C (26,32). To investigate this we used an in vitro assay to test the mutagenic potential of the A3C enzymes in which we could add the same molar amounts of each enzyme. This assay recapitulates reverse transcription of (−) DNA and synthesis of (+) DNA in vitro (see Materials and Methods) (26,32). The mutational spectra showed that hA3C induced at least 2.5-fold less mutations than cA3C and gA3C (Figure 5A–C, hA3C 0.13 × 10−2 mutations/bp; cA3C 0.33 × 10−2 mutations/bp; gA3C 0.40 × 10−2 mutations/bp). We also examined the difference in the mutational load per clone sequenced for each of the enzymes. For hA3C, all of the clones sequenced had at most one G→A mutation per clone (Figure 5D). Since clones were chosen for sequencing based on mutagenesis of the lacZα region (blue/white screening), when hA3C clones contained zero G→A mutations, they were recovered due to other mutations induced by reverse transcriptase. This was in contrast to cA3C and gA3C that were able to cause a greater number of G→A mutations per clone (Figure 5E and F). We next tested the A3C mutants to determine if their mutagenic efficiency would correlate with processivity and dimerization. We summarized the mutation frequency of the A3C enzymes according to their oligomeric state (Figure 5G, monomer, monomer/dimer, dimer). In contrast to other A3C orthologs that formed dimers, the gA3C K115N could not oligomerize on ssDNA, despite forming a small amount of dimers in solution (Figure 3B and G). As a result, for the experiment, we considered gA3C K115N to be a monomer. The results show that the in vitro mutation frequency increases with dimer formation. In comparison to the monomeric A3C forms, the increases were ∼2-fold for monomer/dimer and 3- to 4- fold for dimer and were independent of the A3C ortholog (Figure 5G). The improvement in overall mutations induced was also observed on a per clone basis (Supplementary Figure S6).
Figure 5.
Monomeric A3C induces lower levels of mutagenesis than dimeric A3C. (A–G) An in vitro HIV replication assay was utilized to determine the enzymes abilities to catalyze deaminations during proviral DNA synthesis. (A–C) Spectra of mutations are plotted as the percentage of clones containing a G→A mutation at a particular location (nt) in the 368 nt prot-lacZα construct for (A) hA3C, (B) cA3C or (C) gA3C. (D–F) Histograms illustrate the number of mutations that can be induced by (D) hA3C, (E) cA3C or (F) gA3C within individual clones. (G) Summarized G→A mutation frequency for A3C monomers (hA3C, cA3C K115N, gA3C K115N), monomers/dimers (hA3C S188I, cA3C, gA3C), and dimers (hA3C S188I/N115K, cA3C S188I, gA3C S188I). The graph denotes whether the A3C is from human (h), chimpanzee (c) or gorilla (g). Individual spectra and clonal mutation frequencies not included in Figure 5 are in Supplementary Figure S6. (H) HIV Δvif infectivity was measured by β-galactosidase expression driven by the HIV-1 5΄LTR from HeLa CD4+ HIV-1 LTR-β-gal cells infected with HIV Δvif that was produced in the absence or presence of A3G or A3C orthologs. Relative decrease in virus infectivity is shown for A3G, A3C monomers (hA3C, cA3C K115N, gA3C K115N), A3C monomers/dimers (hA3C S188I, cA3C, gA3C) and A3C dimers (hA3C S188I/N115K, cA3C S188I, gA3C S188I). The graph denotes whether the A3C is from human (h), chimpanzee (c) or gorilla (g). Results normalized to the no A3 condition are shown with the Standard Deviation of the mean calculated from at least three independent experiments. Statistical significance of HIV Δvif restriction for each A3C ortholog was determined in comparison to the monomer form (hA3C, cA3C K115N or gA3C K115N). Designations for significant difference of values were ***P ≤ 0.001, **P ≤ 0.01 or *P ≤ 0.05. (I) Immunoblotting for the HA tag was used to detect A3 enzymes expressed in cells and encapsidated into HIV Δvif virions. The cell lysate and virion loading controls were α-tubulin and p24, respectively. Quantification of the relative amount of A3 was normalized to hA3C.
We also confirmed that these increases in mutagenic efficiency were relevant to virus restriction by conducting single-cycle infectivity assays with virus derived from a LAI Δvif construct. In this assay, an increase in A3-induced mutagenesis results in a decrease in virus infectivity. Consistent with previous studies using HIV-1 Δvif, hA3C does not restrict the virus, A3G highly restricts the virus, and hA3C S188I restricts the virus more than hA3C (∼3-fold more) (Figure 5H) (38). The single-cycle infectivity assays were in agreement with the in vitro mutagenesis assay and demonstrated that monomer/dimer and dimer A3C forms were more able decrease virus infectivity than monomeric A3C, regardless of the A3C ortholog. We confirmed that decreases in viral infectivity were the result of A3-induced mutations by PCR amplifying and sequencing a 351 nt portion of the protease gene from integrated proviral DNA (Table 2). It should be noted that the mutation frequencies recovered from the proviral DNA are not directly comparable to the in vitro assay since the in vitro assay uses a selection process to recover mutated clones and the proviral DNA is sequenced without selection, thus, the latter mutation frequencies are lower. Nonetheless, consistent with the in vitro data (Figure 5A–F and Supplementary Figure S6), with increased dimerization we observed an increase in the total number of mutations (Table 2) and an increase in the number of mutations per clone (Supplementary Figure S7). Specifically, formation of a monomer/dimer resulted in a 2-fold increase in mutation frequency, stable dimers resulted in a 3- to 5-fold increase in mutation frequency, and formation of a monomer resulted in a 2-fold decrease in mutation frequency (Table 2). The only exception is that the cA3C mutation frequency is not as high as the other monomer/dimer forms (hA3C S188I and gA3C) (Table 2). This is also reflected in the similar decreases of infectivity induced by the monomer (cA3C K115N) and monomer/dimer (cA3C) forms (Figure 5H). This is likely due to cA3C encapisdating 2- to 4-fold less than other monomer/dimer A3C forms (hA3C S188I and gA3C) (Figure 5I). Notably, A3G encapsidation is at least 2.5-fold more efficient than A3C, which provides a reasoning for why dimer forms of A3C are still not as effective as A3G in decreasing viral infectivity, although A3G is also more processive than A3C (Figure 5I and Supplementary Figure S2). Despite the level of encapsidation being an additional determining factor to processivity in virus restriction between A3G and A3C, within the A3C orthologs, the decreases in infectivity in the majority of conditions correlates better with the processivity rather than encapsidation. For example, gA3C S188I is encapsidated 2-fold less than gA3C, but is able to restrict virus infectivity 2-fold more (Figure 5H), is the more processive A3C (Figure 4C), is able to induce more mutations (Table 2), and more mutations per clone (Supplementary Figure S7). Similarly, hA3C S188I/N115K is encapsidated 2-fold less than hA3C, but is able to restrict virus infectivity ∼2.5-fold more (Figure 5H), is the more processive A3C (Figure 4A), is able to induce more mutations (Table 2), and more mutations per clone (Supplementary Figure S7). Altogether, these data demonstrate the importance of an enzyme's processive ssDNA scanning mechanisms for inducing mutagenesis in a dynamic system where there is a limited amount of time that the ssDNA is available.
Table 2. Analysis of A3-induced mutagenesis of protease DNA from integrated HIVΔvif.
| A3C | Base pairs sequenced | Total G→A mutations | Mutations per kb |
|---|---|---|---|
| hA3C | 9477 | 3 | 0.03 |
| cA3C K115N | 9477 | 2 | 0.02 |
| gA3C K115N | 10179 | 4 | 0.03 |
| hA3C S188I | 10179 | 8 | 0.07 |
| cA3C | 8073 | 3 | 0.04 |
| gA3C | 9126 | 6 | 0.06 |
| hA3C S188I/N115K | 9126 | 15 | 0.16 |
| cA3C S188I | 9126 | 11 | 0.13 |
| gA3C S188I | 10179 | 17 | 0.17 |
DISCUSSION
Our biochemical analysis has established that A3C dimerization correlates with processive DNA scanning. The hA3C S188I variant dimerizes, in contrast to the common hA3C (Figure 3). The cA3C and gA3C were able to dimerize despite containing an S188 by using an amino acid unique to cA3C and gA3C, K115 (Figure 3). These data support the model that dimerization of A3C is primarily mediated through α-helix 6 (human S188I) or β-strand 4 (chimpanzee and gorilla). The importance of these biochemical characteristics is that higher levels of A3C-mediated mutagenesis during reverse transcription requires a dimeric and processive enzyme. Altogether, these data provide a biochemical explanation for why hA3C S188I is able to restrict HIV-1 more than hA3C (38).
Modulation of catalytic activity in the AID/APOBEC family
The A3C data on processivity and specific activity demonstrate that differences in catalytic activity were due to the different efficiencies in the ssDNA scanning mechanisms employed and that this was dependent on the oligomerization state (Figure 1 and Table 1). In the AID/APOBEC family, there have been several common observations that amino acids outside of the active site can mediate catalytic activity. A3F specific activity can be increased by introducing sliding activity through mutations in α-helix 6, a structure that is located away from the active site (32,49,50). A3G and A3A deamination activity can be enhanced by a secondary Zinc ion that binds loop 3, outside of the active site (51). The Zinc ion is not used directly for catalysis, but stabilizes loop 3 in a conformation that promotes the correct orientation of substrate binding for catalysis (51). A3A activity is also enhanced by cooperative dimerization (52). Further, the related family member Activation Induced cytidine Deaminase (AID) that deaminates within specific regions of immunoglobulin genes to promote somatic hypermutation and class switching has been characterized as being a catalytically inefficient enzyme (53–55). The low efficiency of catalysis is despite AID's high processivity and ability to remain bound to ssDNA for an average of 5 min (56). Data indicates that the low efficiency is due to an inaccessibility of the catalytic pocket to DNA (54). It was proposed that this causes a high propensity of catalytically unfavorable ssDNA–AID binding conformations, which may afford some protection against off-target mutations in genomic DNA (53,54). This may also be why the majority of humans carry an A3H allele for a thermodynamically unstable enzyme (haplotypes I, III, IV or VI) although this does not completely protect from the ability of A3H to contribute to mutations that arise during cancer (8,57,58). The relative inactivity of A3C in the majority of humans due the loss of oligomerization may also be a mechanism to decrease off-target mutations, despite its ability to access genomic DNA in cells (38,59).
Processivity of A3C
We observed that the primary mechanism used by the A3C enzymes to locate target cytosines on ssDNA was long-range translocations involving jumping or intersegmental transfer. Even though hA3C S188I, cA3C and gA3C were able to slide on ssDNA, in contrast to hA3C, the processivity factors indicated that they were only 1.5- to 2-fold more likely to catalyze processive deaminations than non-processive deaminations on closely spaced cytosines (Figure 1). Thus, the sliding was a minor component of their processive scanning mechanism. The long-range movements, mediated by jumping or intersegmental transfer, were more significant and enabled hA3C S188I, cA3C and gA3C to undergo processive long-range movements ∼3-fold more than non-processive interactions with ssDNA (Figure 1). Notably, all the A3C enzymes we tested were 2- to 4-fold less processive and 1.7- to 20-fold less mutagenic during in vitro reverse transcription than other A3 enzymes that restrict HIV-1 characterized to date (Figures 4 and 5) (29,31,32). This may explain why although A3C mutations during proviral DNA synthesis increased with dimerization, there was still a portion of proviral clones that only had one mutation even in the presence of a processive and dimeric A3C (Table 2 and Supplementary Figure S7). In agreement with another study by Byeon et al., we found A3C to have less specific activity than other A3s (29,30,32,47,60–62). However, the A3C in the study from Byeon et al. was produced from Escherichia coli and is less active than our A3C produced from Sf9 cells, similar to what has been found for AID produced from Sf9 and E. coli cells (60,63). Based on these data, we expect that hA3C S188I would contribute to mutagenesis along with other A3 enzymes rather than be able to fully suppress the virus independently of other A3s (38).
The ability to oligomerize has been correlated with improved processivity and specific activity for A3G and A3H, although monomers of the enzymes are still catalytically active (26,29,30,43,64). Oligomerization of A3 enzymes allows for multiple binding and interaction domains, which imparts a larger selection of ssDNA scanning mechanisms to efficiently locate the target cytosine. Consistent with previous literature, the current data with A3C enzymes also found that those enzymes that were able to form dimers had higher processivity factors than the enzymes that were unable to dimerize (26,29). Further, the high processivity factors were due to the enzyme not only improving in processivity, but also gaining a processive mechanism, e.g. hA3C S188I improved long-range processivity, but also gained the ability to slide (Figure 4A). The A3C enzymes that formed stable dimers had the most improvement in their ability to catalyze processive deaminations (Figure 4A–C, hA3C S188I/N115K, cA3C S188I, gA3C S188I).
For A3C, a single Zinc coordinating domain (Z-domain) enzyme, dimerization is required for intersegmental transfer to occur. We initially expected that the stable dimer forms of A3C would have enhanced intersegmental transfer since the dissociation or reassociation of dimers on ssDNA would not be rate limiting. Rather, the data supports the conclusion that for A3C, intersegmental transfer required an unstable dimer (Figure 4D–F). The data suggest that the stable dimer is in a conformation where it is not favorable to bind two different ssDNA segments simultaneously. That stable dimer forms of A3C were unable to undergo intersegmental transfer may also explain why enzymes like A3G and A3F that have two DNA binding domains within a single polypeptide chain are unable to utilize intersegmental transfer, under the same reaction conditions tested with A3C (32), although under low salt buffer reaction conditions A3G may be able to undergo intersegmental transfer (30). The improved processivity of the stable A3C dimer forms was through sliding and long-range movements, but the long-range movements were largely comprised of jumping (Figure 4 and Supplementary Figure S5). We found A3C to use long-range movements similarly to A3H, in which jumping and intersegmental transfer are redundant processive mechanisms and for A3C either one can improve the mutagenic efficiency of the enzyme (29).
Our study with A3C raises the possibility that the structures that mediate processivity are different for single and double Z-domain A3 enzymes. In contrast to A3G and A3F that contain two Z-domains, α-helix 6 was not directly involved in mediating A3C sliding (26,32). The hA3C, cA3C and gA3C all had identical amino acids in the α-helix 6 despite differing abilities to slide along ssDNA (Figure 1). In A3C, the ability of α-helix 6 to mediate dimerization or be part of the dimer interface was the only requirement for sliding movements on ssDNA. In contrast, a monomer of A3G can still retain sliding ability and an oligomer of A3F is unable to slide unless specific mutations are made to the loop region extending from α-helix 6 (26,32). In a study of a single Z-domain enzyme, A3H, specific amino acid changes on α-helix 6 could influence sliding of the dimeric A3H (29). Nonetheless, a β2-strand dimerization mutant had compromised sliding ability, despite a wild type α-helix 6 (29). These results suggest that the dimerization interface in single Z-domain A3s creates a groove along the enzyme that promotes an interaction with ssDNA and facilitates sliding (65). The double Z-domain enzymes A3G and A3F may inherently have an extended interaction interface with ssDNA due to possessing two ssDNA binding domains in a single polypeptide chain. This appears to facilitate extensive ssDNA interactions that may also involve bending of the ssDNA around the enzyme (27,66,67).
Dimerization of A3C
Consistent with previous studies in vitro and in cells, we determined that hA3C was a monomer (38,41,68). This is in contrast to Stauch et al. that determined hA3C to be a dimer by co-IP (35). Although Stauch et al. and our study used a similar co-IP strategy, the transfected plasmid amounts were different and Stauch et al. did not use RNaseA in their procedure. These factors can lead to nonspecific interactions in the co-IP (69,70). A surprising feature of our results was that the biochemical data we obtained support a model in which the hA3C dimer formed in the crystal structure did form on a physiologically relevant interface for hA3C S188I, cA3C and gA3C (Figure 2B) (41). However, it should be noted that multiple additional interfaces did form when hA3C was crystallized and these do not appear to represent the dimer interface in hA3C S188I, cA3C or gA3C (41).
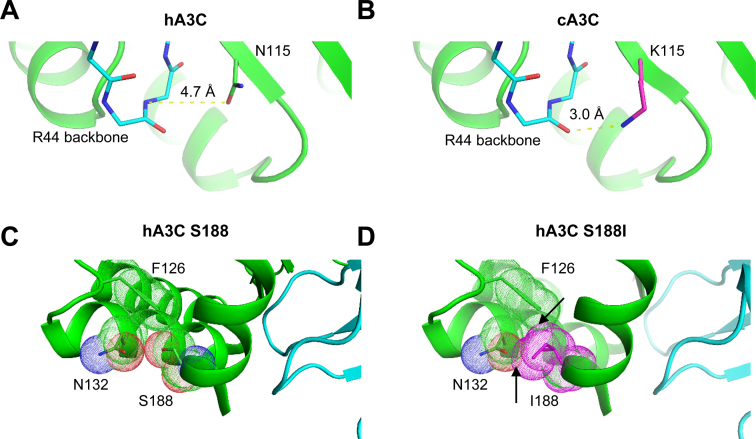
Although the biochemical and co-IP data definitively show that amino acids 115 and 188 mediate A3C dimerization, there is a caveat in our model that relies on the hA3C crystal structure. Namely, visual inspection of amino acids 115 and 188 on the structure of hA3C suggest that amino acid 115, but not amino acid 188, is directly involved in the dimer interface (Figure 2B). For hA3C the N115 is 4.7 Å away from the backbone of R44. In contrast, a model of cA3C K115 positions the backbone of R44 only 3.0 Å away (Figure 6A and B). Thus, the mechanism by which the K115 in cA3C promotes dimerization appears to be due to enabling a new hydrogen bond with R44 on the other subunit (Figure 6A and B). The interface between subunits is already surface complementary and this complementarity is unlikely to be disrupted by the change in the 115 side chain identity. Therefore, the new hydrogen bond formed by the K115 side chain and R44 backbone would mainly serve to strengthen the interface with minimal tradeoffs.
Figure 6.
Models of A3C dimerization. (A) For hA3C, N115 (chain A) is 4.7 Å away from the backbone of R44 (chain B). (B) In contrast, a model of cA3C K115 (chain A) positions the backbone of R44 (chain B) only 3.0 Å away. This distance could indicate a new hydrogen bond with potential to stabilize the dimer between chains A and B. (C) In hA3C, S188, shown with van der Waal space filling dots, packs closely to F126 and N132, but does not clash with either. (D) In contrast, a model of hA3C shows how I188 would clash with F126 and N132 (arrows indicating overlap of van der Waal space filling dots). Conformational changes, potentially including a repositioning of the helix to enable formation of an A–B dimer, would be needed to accommodate this amino acid variant.
Although the amino acid at position 188 mutation does not appear to be directly involved in the dimer interface by PISA analysis or in the structural model (41,71), PISA analysis does suggest that amino acids surrounding position 188 on α-helix 6 are involved in dimerization. The most likely explanation for the role of the S188I mutation in altering the dimer affinity is that it causes steric clash with other buried residues that are accommodated by shifting the position of helix 6. This repositioning of the dimer interface could alter the surface complementarity and other interactions, leading to the change in affinity (Figure 6C and D). Based on our studies, the hA3C crystal structure gives a good approximation of the dimer interface, but more mutagenesis studies or crystallization of a dimer form of A3C is required to identify the interface in more detail (41).
A3C forms a dimer interface unique from other A3 dimer interfaces determined thus far (29,43,49,50,52,72–74). Notably, the other dimer interfaces characterized have been from the A3G N-terminal domain in the full length enzyme (Z2-Z1-type Z-domain organization), rhesus macaque A3G N-terminal domain alone (Z2-Z1-type Z-domain organization), A3F C-terminal domain alone (Z2-Z2-type Z-domain organization), A3A (a Z1-type Z-domain), and A3H (a Z3-type Z-domain) (29,43,49,50,52,72–74). A3C is the first Z2-type single Z-domain to have the dimer interface characterized. However, the other Z2-type Z-domains that have been characterized structurally, A3F C-terminal domain and rhesus macaque A3G N-terminal domain further support the involvement of α-helix 6 (49,73). In the rhesus macaque A3G N-terminal domain structure, α-helix 6 and loop 7 are involved in the dimer interface (73). There have been several A3F C-terminal domain (CTD) crystal structures solved and each with a different predicted dimer interface based on crystal contacts (49,50,74). In one A3F crystal structure, the amino acid equivalent to A3C N/K115 in the A3F CTD, N298 is involved in a dimer interface (49). It is not clear why the A3F Z2-type Z-domain CTD structures are not consistent with respect to the dimer contacts made in each of the crystal structures, however, this may be due to different constructs being used between labs to promote crystallization, that the A3F CTD is mutated to improve solubilization, and that it is not the full length A3F enzyme (49,50,74).
CONCLUSIONS
Our results with these three hominid A3C orthologs demonstrates that there is more than one mechanism to maintain enzyme processivity, although the common endpoint requires dimerization of the enzyme. Since A3C is a Z2-type domain, the dimer model may represent how other Z2-type domains such as A3F and A3D dimerize (both have Z2-Z2-type Z-domain organization). That the mechanism for achieving processivity in A3C is based primarily on dimerization rather than specific amino acid motifs that interact with the ssDNA substrate is novel from other A3s and indicates that specific adaptations to maintain activity have been different for individual A3 enzymes. Although we have shown that the dimerization of hA3C through the S188I mutation increases processivity and mutagenic ability, thus providing a reason for why it is able to restrict HIV-1, in contrast to hA3C (38), it remains to be determined if the increased activity naturally found in cA3C and gA3C results in these enzymes restricting SIV. Despite their higher enzymatic activity it has not been demonstrated if cA3C and gA3C are able to encapsidate into SIV, which would be required for them to act as restriction factors. Alternatively, the naturally higher activity of cA3C and gA3C may be used to restrict viruses other than SIV or retroelements. Overall, the data support the model that A3 enzymes must be able to deaminate multiple cytosines processively to be efficient at mutagenesis during reverse transcription and for A3C this requires dimerization.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Canadian Institutes for Health Research [MOP137090 to L.C.; NIH P50 GM082250 to J.S.F., NIH AI30937 to M.E.]; STD & AIDS Research Training Program graduate fellowship [NIH T32 AI07140 to C.J.W.]. Funding for open access charge: Canadian Institutes for Health Research Operating Grant [MOP137090 to L.C.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Feng Y., Baig T.T., Love R.P., Chelico L.. Suppression of APOBEC3-mediated restriction of HIV-1 by Vif. Front. Microbiol. 2014; 5:450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yu Q., Konig R., Pillai S., Chiles K., Kearney M., Palmer S., Richman D., Coffin J.M., Landau N.R.. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat. Struct. Mol. Biol. 2004; 11:435–442. [DOI] [PubMed] [Google Scholar]
- 3. Mangeat B., Turelli P., Caron G., Friedli M., Perrin L., Trono D.. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003; 424:99–103. [DOI] [PubMed] [Google Scholar]
- 4. Harris R.S., Bishop K.N., Sheehy A.M., Craig H.M., Petersen-Mahrt S.K., Watt I.N., Neuberger M.S., Malim M.H.. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003; 113:803–809. [DOI] [PubMed] [Google Scholar]
- 5. Zhang H., Yang B., Pomerantz R.J., Zhang C., Arunachalam S.C., Gao L.. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature. 2003; 424:94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dang Y., Wang X., Esselman W.J., Zheng Y.H.. Identification of APOBEC3DE as another antiretroviral factor from the human APOBEC family. J. Virol. 2006; 80:10522–10533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sheehy A.M., Gaddis N.C., Choi J.D., Malim M.H.. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002; 418:646–650. [DOI] [PubMed] [Google Scholar]
- 8. OhAinle M., Kerns J.A., Li M.M., Malik H.S., Emerman M.. Antiretroelement activity of APOBEC3H was lost twice in recent human evolution. Cell Host Microbe. 2008; 4:249–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liddament M.T., Brown W.L., Schumacher A.J., Harris R.S.. APOBEC3F properties and hypermutation preferences indicate activity against HIV-1 in vivo. Curr. Biol. 2004; 14:1385–1391. [DOI] [PubMed] [Google Scholar]
- 10. Hultquist J.F., Lengyel J.A., Refsland E.W., LaRue R.S., Lackey L., Brown W.L., Harris R.S.. Human and rhesus APOBEC3D, APOBEC3F, APOBEC3G, and APOBEC3H demonstrate a conserved capacity to restrict Vif-deficient HIV-1. J. Virol. 2011; 85:11220–11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Refsland E.W., Hultquist J.F., Luengas E.M., Ikeda T., Shaban N.M., Law E.K., Brown W.L., Reilly C., Emerman M., Harris R.S.. Natural polymorphisms in human APOBEC3H and HIV-1 Vif combine in primary T lymphocytes to affect viral G-to-A mutation levels and infectivity. PLoS Genet. 2014; 10:e1004761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ooms M., Brayton B., Letko M., Maio S.M., Pilcher C.D., Hecht F.M., Barbour J.D., Simon V.. HIV-1 Vif adaptation to human APOBEC3H haplotypes. Cell Host Microbe. 2013; 14:411–421. [DOI] [PubMed] [Google Scholar]
- 13. Etienne L., Hahn B.H., Sharp P.M., Matsen F.A., Emerman M.. Gene loss and adaptation to hominids underlie the ancient origin of HIV-1. Cell Host Microbe. 2013; 14:85–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yu X., Yu Y., Liu B., Luo K., Kong W., Mao P., Yu X.F.. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science. 2003; 302:1056–1060. [DOI] [PubMed] [Google Scholar]
- 15. Mariani R., Chen D., Schrofelbauer B., Navarro F., Konig R., Bollman B., Munk C., Nymark-McMahon H., Landau N.R.. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell. 2003; 114:21–31. [DOI] [PubMed] [Google Scholar]
- 16. Stopak K., de Noronha C., Yonemoto W., Greene W.C.. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol. Cell. 2003; 12:591–601. [DOI] [PubMed] [Google Scholar]
- 17. Sheehy A.M., Gaddis N.C., Malim M.H.. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 2003; 9:1404–1407. [DOI] [PubMed] [Google Scholar]
- 18. Marin M., Rose K.M., Kozak S.L., Kabat D.. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 2003; 9:1398–1403. [DOI] [PubMed] [Google Scholar]
- 19. Kao S., Khan M.A., Miyagi E., Plishka R., Buckler-White A., Strebel K.. The human immunodeficiency virus type 1 Vif protein reduces intracellular expression and inhibits packaging of APOBEC3G (CEM15), a cellular inhibitor of virus infectivity. J. Virol. 2003; 77:11398–11407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wiegand H.L., Doehle B.P., Bogerd H.P., Cullen B.R.. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J. 2004; 23:2451–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jager S., Kim D.Y., Hultquist J.F., Shindo K., LaRue R.S., Kwon E., Li M., Anderson B.D., Yen L., Stanley D. et al. . Vif hijacks CBF-beta to degrade APOBEC3G and promote HIV-1 infection. Nature. 2012; 481:371–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang W., Du J., Evans S.L., Yu Y., Yu X.F.. T-cell differentiation factor CBF-beta regulates HIV-1 Vif-mediated evasion of host restriction. Nature. 2012; 481:376–379. [DOI] [PubMed] [Google Scholar]
- 23. Zhou X., Evans S.L., Han X., Liu Y., Yu X.F.. Characterization of the interaction of full-length HIV-1 Vif protein with its key regulator CBFbeta and CRL5 E3 ubiquitin ligase components. PLoS One. 2012; 7:e33495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Berg O.G., Winter R.B., von Hippel P.H.. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry. 1981; 20:6929–6948. [DOI] [PubMed] [Google Scholar]
- 25. von Hippel P.H., Berg O.G.. Facilitated target location in biological systems. J. Biol. Chem. 1989; 264:675–678. [PubMed] [Google Scholar]
- 26. Feng Y., Chelico L.. Intensity of deoxycytidine deamination of HIV-1 proviral DNA by the retroviral restriction factor APOBEC3G is mediated by the noncatalytic domain. J. Biol. Chem. 2011; 286:11415–11426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Senavirathne G., Jaszczur M., Auerbach P.A., Upton T.G., Chelico L., Goodman M.F., Rueda D.. Single-stranded DNA scanning and deamination by APOBEC3G cytidine deaminase at single molecule resolution. J. Biol. Chem. 2012; 287:15826–15835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stanford N.P., Szczelkun M.D., Marko J.F., Halford S.E.. One- and three-dimensional pathways for proteins to reach specific DNA sites. EMBO J. 2000; 19:6546–6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Feng Y., Love R.P., Ara A., Baig T.T., Adolph M.B., Chelico L.. Natural polymorphisms and oligomerization of human APOBEC3H contribute to single-stranded DNA scanning ability. J. Biol. Chem. 2015; 290:27188–27203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nowarski R., Britan-Rosich E., Shiloach T., Kotler M.. Hypermutation by intersegmental transfer of APOBEC3G cytidine deaminase. Nat. Struct. Mol. Biol. 2008; 15:1059–1066. [DOI] [PubMed] [Google Scholar]
- 31. Chelico L., Pham P., Calabrese P., Goodman M.F.. APOBEC3G DNA deaminase acts processively 3΄ –>5΄ on single-stranded DNA. Nat. Struct. Mol. Biol. 2006; 13:392–399. [DOI] [PubMed] [Google Scholar]
- 32. Ara A., Love R.P., Chelico L.. Different mutagenic potential of HIV-1 restriction factors APOBEC3G and APOBEC3F is determined by distinct single-stranded DNA scanning mechanisms. PLoS Pathog. 2014; 10:e1004024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yu Q., Chen D., Konig R., Mariani R., Unutmaz D., Landau N.R.. APOBEC3B and APOBEC3C are potent inhibitors of simian immunodeficiency virus replication. J. Biol. Chem. 2004; 279:53379–53386. [DOI] [PubMed] [Google Scholar]
- 34. Langlois M.A., Beale R.C., Conticello S.G., Neuberger M.S.. Mutational comparison of the single-domained APOBEC3C and double-domained APOBEC3F/G anti-retroviral cytidine deaminases provides insight into their DNA target site specificities. Nucleic Acids Res. 2005; 33:1913–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stauch B., Hofmann H., Perkovic M., Weisel M., Kopietz F., Cichutek K., Munk C., Schneider G.. Model structure of APOBEC3C reveals a binding pocket modulating ribonucleic acid interaction required for encapsidation. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:12079–12084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Refsland E.W., Stenglein M.D., Shindo K., Albin J.S., Brown W.L., Harris R.S.. Quantitative profiling of the full APOBEC3 mRNA repertoire in lymphocytes and tissues: implications for HIV-1 restriction. Nucleic Acids Res. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Koning F.A., Newman E.N., Kim E.Y., Kunstman K.J., Wolinsky S.M., Malim M.H.. Defining APOBEC3 expression patterns in human tissues and hematopoietic cell subsets. J. Virol. 2009; 83:9474–9485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wittkopp C.J., Adolph M.B., Wu L.I., Chelico L., Emerman M.. A single nucleotide polymorphism in human APOBEC3C enhances restriction of lentiviruses. PLoS Pathog. 2016; 12:e1005865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang T., Zhang W., Tian C., Liu B., Yu Y., Ding L., Spearman P., Yu X.F.. Distinct viral determinants for the packaging of human cytidine deaminases APOBEC3G and APOBEC3C. Virology. 2008; 377:71–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Duggal N.K., Fu W., Akey J.M., Emerman M.. Identification and antiviral activity of common polymorphisms in the APOBEC3 locus in human populations. Virology. 2013; 443:329–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kitamura S., Ode H., Nakashima M., Imahashi M., Naganawa Y., Kurosawa T., Yokomaku Y., Yamane T., Watanabe N., Suzuki A. et al. . The APOBEC3C crystal structure and the interface for HIV-1 Vif binding. Nat. Struct. Mol. Biol. 2012; 19:1005–1010. [DOI] [PubMed] [Google Scholar]
- 42. Bransteitter R., Pham P., Calabrese P., Goodman M.F.. Biochemical analysis of hypermutational targeting by wild type and mutant activation-induced cytidine deaminase. J. Biol. Chem. 2004; 279:51612–51621. [DOI] [PubMed] [Google Scholar]
- 43. Chelico L., Prochnow C., Erie D.A., Chen X.S., Goodman M.F.. Structural model for deoxycytidine deamination mechanisms of the HIV-1 inactivation enzyme APOBEC3G. J. Biol. Chem. 2010; 285:16195–16205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Creighton S., Bloom L.B., Goodman M.F.. Gel fidelity assay measuring nucleotide misinsertion, exonucleolytic proofreading, and lesion bypass efficiencies. Methods Enzymol. 1995; 262:232–256. [DOI] [PubMed] [Google Scholar]
- 45. Baig T.T., Feng Y., Chelico L.. Determinants of efficient degradation of APOBEC3 restriction factors by HIV-1 Vif. J. Virol. 2014; 88:14380–14395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kimpton J., Emerman M.. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated beta-galactosidase gene. J. Virol. 1992; 66:2232–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Love R.P., Xu H., Chelico L.. Biochemical analysis of hypermutation by the deoxycytidine deaminase APOBEC3A. J. Biol. Chem. 2012; 287:30812–30822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lieberman B.A., Nordeen S.K.. DNA intersegment transfer, how steroid receptors search for a target site. J. Biol. Chem. 1997; 272:1061–1068. [DOI] [PubMed] [Google Scholar]
- 49. Bohn M.F., Shandilya S.M., Albin J.S., Kouno T., Anderson B.D., McDougle R.M., Carpenter M.A., Rathore A., Evans L., Davis A.N. et al. . Crystal structure of the DNA cytosine deaminase APOBEC3F: the catalytically active and HIV-1 Vif-binding domain. Structure. 2013; 21:1042–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Siu K.K., Sultana A., Azimi F.C., Lee J.E.. Structural determinants of HIV-1 Vif susceptibility and DNA binding in APOBEC3F. Nat. Commun. 2013; 4:2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Marx A., Galilee M., Alian A.. Zinc enhancement of cytidine deaminase activity highlights a potential allosteric role of loop-3 in regulating APOBEC3 enzymes. Sci. Rep. 2015; 5:18191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bohn M.F., Shandilya S.M., Silvas T.V., Nalivaika E.A., Kouno T., Kelch B.A., Ryder S.P., Kurt-Yilmaz N., Somasundaran M., Schiffer C.A.. The ssDNA mutator APOBEC3A is regulated by cooperative dimerization. Structure. 2015; 23:903–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang M., Yang Z., Rada C., Neuberger M.S.. AID upmutants isolated using a high-throughput screen highlight the immunity/cancer balance limiting DNA deaminase activity. Nat. Struct. Mol. Biol. 2009; 16:769–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. King J.J., Manuel C.A., Barrett C.V., Raber S., Lucas H., Sutter P., Larijani M.. Catalytic pocket inaccessibility of activation-induced cytidine deaminase is a safeguard against excessive mutagenic activity. Structure. 2015; 23:615–627. [DOI] [PubMed] [Google Scholar]
- 55. Mak C.H., Pham P., Afif S.A., Goodman M.F.. A mathematical model for scanning and catalysis on single-stranded DNA, illustrated with activation-induced deoxycytidine deaminase. J. Biol. Chem. 2013; 288:29786–29795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Senavirathne G., Bertram J.G., Jaszczur M., Chaurasiya K.R., Pham P., Mak C.H., Goodman M.F., Rueda D.. Activation-induced deoxycytidine deaminase (AID) co-transcriptional scanning at single-molecule resolution. Nat. Commun. 2015; 6:10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wang X., Abudu A., Son S., Dang Y., Venta P.J., Zheng Y.H.. Analysis of human APOBEC3H haplotypes and anti-human immunodeficiency virus type 1 activity. J. Virol. 2011; 85:3142–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Starrett G.J., Luengas E.M., McCann J.L., Ebrahimi D., Temiz N.A., Love R.P., Feng Y., Adolph M.B., Chelico L., Law E.K. et al. . The DNA cytosine deaminase APOBEC3H haplotype I likely contributes to breast and lung cancer mutagenesis. Nat. Commun. 2016; 7:12918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lackey L., Law E.K., Brown W.L., Harris R.S.. Subcellular localization of the APOBEC3 proteins during mitosis and implications for genomic DNA deamination. Cell Cycle. 2013; 12:762–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Byeon I.J., Byeon C.H., Wu T., Mitra M., Singer D., Levin J.G., Gronenborn A.M.. Nuclear magnetic resonance structure of the APOBEC3B catalytic domain: structural basis for substrate binding and DNA deaminase activity. Biochemistry. 2016; 55:2944–2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Mitra M., Hercik K., Byeon I.J., Ahn J., Hill S., Hinchee-Rodriguez K., Singer D., Byeon C.H., Charlton L.M., Nam G. et al. . Structural determinants of human APOBEC3A enzymatic and nucleic acid binding properties. Nucleic Acids Res. 2014; 42:1095–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Byeon I.J., Ahn J., Mitra M., Byeon C.H., Hercik K., Hritz J., Charlton L.M., Levin J.G., Gronenborn A.M.. NMR structure of human restriction factor APOBEC3A reveals substrate binding and enzyme specificity. Nat. Commun. 2013; 4:1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Pham P., Smolka M.B., Calabrese P., Landolph A., Zhang K., Zhou H., Goodman M.F.. Impact of phosphorylation and phosphorylation-null mutants on the activity and deamination specificity of activation-induced cytidine deaminase. J. Biol. Chem. 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Opi S., Takeuchi H., Kao S., Khan M.A., Miyagi E., Goila-Gaur R., Iwatani Y., Levin J.G., Strebel K.. Monomeric APOBEC3G is catalytically active and has antiviral activity. J. Virol. 2006; 80:4673–4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lu X., Zhang T., Xu Z., Liu S., Zhao B., Lan W., Wang C., Ding J., Cao C.. Crystal structure of DNA cytidine deaminase ABOBEC3G catalytic deamination domain suggests a binding mode of full-length enzyme to single-stranded DNA. J. Biol. Chem. 2015; 290:4010–4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Chelico L., Sacho E.J., Erie D.A., Goodman M.F.. A model for oligomeric regulation of APOBEC3G cytosine deaminase-dependent restriction of HIV. J. Biol. Chem. 2008; 283:13780–13791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Polevoda B., McDougall W.M., Bennett R.P., Salter J.D., Smith H.C.. Structural and functional assessment of APOBEC3G macromolecular complexes. Methods. 2016; 107:10–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Li J., Chen Y., Li M., Carpenter M.A., McDougle R.M., Luengas E.M., Macdonald P.J., Harris R.S., Mueller J.D.. APOBEC3 multimerization correlates with HIV-1 packaging and restriction activity in living cells. J. Mol. Biol. 2014; 426:1296–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ara A., Love R.P., Follack T.B., Ahmed K.A., Adolph M.B., Chelico L.. Mechanism of enhanced HIV restriction by virion coencapsidated cytidine deaminases APOBEC3F and APOBEC3G. J. Virol. 2016; 91, doi:10.1128/JVI.02230-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Letko M., Silvestri G., Hahn B.H., Bibollet-Ruche F., Gokcumen O., Simon V., Ooms M.. Vif proteins from diverse primate lentiviral lineages use the same binding site in APOBEC3G. J. Virol. 2013; 87:11861–11871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Krissinel E., Henrick K.. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007; 372:774–797. [DOI] [PubMed] [Google Scholar]
- 72. Huthoff H., Autore F., Gallois-Montbrun S., Fraternali F., Malim M.H.. RNA-dependent oligomerization of APOBEC3G is required for restriction of HIV-1. PLoS Pathog. 2009; 5:e1000330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Xiao X., Li S.X., Yang H., Chen X.S.. Crystal structures of APOBEC3G N-domain alone and its complex with DNA. Nat. Commun. 2016; 7:12193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Nakashima M., Ode H., Kawamura T., Kitamura S., Naganawa Y., Awazu H., Tsuzuki S., Matsuoka K., Nemoto M., Hachiya A. et al. . Structural Insights into HIV-1 Vif-APOBEC3F Interaction. J. Virol. 2015; 90:1034–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Soding J. et al. . Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011; 7:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Gouet P., Courcelle E., Stuart D.I., Metoz F.. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999; 15:305–308. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.