Abstract
Higher-order plants and mammals use similar mechanisms to repair and tolerate oxidative DNA damage. Most studies on the DNA repair process have focused on yeast and mammals, in which histone chaperone-mediated nucleosome disassembly/reassembly is essential for DNA to be accessible to repair machinery. However, little is known about the specific role and modulation of histone chaperones in the context of DNA damage in plants. Here, the histone chaperone NRP1, which is closely related to human SET/TAF-Iβ, was found to exhibit nucleosome assembly activity in vitro and to accumulate in the chromatin of Arabidopsis thaliana after DNA breaks. In addition, this work establishes that NRP1 binds to cytochrome c, thereby preventing the former from binding to histones. Since NRP1 interacts with cytochrome c at its earmuff domain, that is, its histone-binding domain, cytochrome c thus competes with core histones and hampers the activity of NRP1 as a histone chaperone. Altogether, the results obtained indicate that the underlying molecular mechanisms in nucleosome disassembly/reassembly are highly conserved throughout evolution, as inferred from the similar inhibition of plant NRP1 and human SET/TAF-Iβ by cytochrome c during DNA damage response.
INTRODUCTION
Plants are constantly exposed to a wide range of biotic and abiotic threats (e.g. UV light, ionising radiation, hydrogen peroxide, pathogens, heat and drought) to which their species have developed responses as they have evolved (1,2). These responses, such as the rapid synthesis of stress-response proteins, are particularly important in plants due to their inherent sessility (1). In plants, as in animals, environmental changes can induce differential gene expression, in which chromatin-remodeling factors play a crucial role (3–5).
Genotoxic agents, such as reactive oxygen species (ROS), ionizing radiation or chemicals, can cause extensive damage to proteins, DNA and lipids, and thereby affect normal cellular functions (6,7). More specifically, high ROS levels cause DNA damage, such as single-strand breaks (SSBs), double-strand breaks (DSBs) and base damage (8–10). The accumulation of such damage may produce lethal mutations, leading to plant genome instability (6,11).
The process of DNA break detection and repair is highly dependent on chromatin, for which histones are crucial (12). Histone dynamics render DNA accessible and provide a target site for the DNA repair machinery (13–18). Although the importance of chromatin dynamics in mammalian and yeast DNA damage repair has been established, little is known about the specific role of histone chaperones in DNA damage response in plants. Furthermore, most analyses of DNA repair pathways have focused on bacteria, yeast and mammals (19). However, recent advances in the study of plant DNA repair demonstrate the use of mechanisms similar to those present in other eukaryotes for the repair or toleration of oxidative DNA damage (19). Indeed, although yeast and mammalian chromatin-remodeling factors present distinctive properties, the mechanisms involved in nucleosome alteration during DNA repair are highly conserved throughout evolution (17,20–22). Strikingly, most of the chromatin-remodeling factors described in yeast and mammals have orthologues in plants, which could be expected to play similar roles in DNA repair response (17,23).
Among these factors, the involvement of histone chaperones in the DNA repair process is essential (14,24). The role of histone chaperones as key regulators of transcriptional activity in chromatin regions with DNA damage has become increasingly clear in recent years (25–29). In this regard, members of the nucleosome assembly protein 1 (NAP1) family of histone chaperones have been reported to be involved in transient chromatin disassembly following DNA damage (18). Specifically, the NAP1 family acts in the nucleosome disassembly/reassembly required during homologous recombination, which is essential for maintenance of genome integrity of Arabidopsis thaliana under stress conditions (30).
NAP1 histone chaperones, which are conserved in species ranging from yeast to humans, have been proposed as facilitating the assembly of newly-synthesized histone H2A and H2B into DNA (31). The NAP1 family in A. thaliana comprises four NAP1 genes (NAP1;1, NAP1;2, NAP1;3 and NAP1;4) and two NAP1-related protein (NRP) genes (NRP1 and NRP2) (30). In A. thaliana, NRPs are multitasking proteins necessary for the binding of histones (32) and histone phosphatase inhibitory properties (33), regulating cell cycle and root meristem formation (32). More specifically, NRP1 plays a crucial role in heat tolerance (34) and the maintenance of genome integrity under genotoxic stresses (30,32).
Recently, we identified NRP1 as a target protein of cytochrome c (Cc) upon the release of the latter from mitochondria into the nucleus of A. thaliana cells submitted to oxidative stress (35). Cc is an electron carrier in the mitochondrial electron transport chain; and its function is tightly regulated by post-translational modifications (36–38). Interestingly, SET/TAF-Iβ, the protein analogous to NRP1 in humans, is also targeted by Cc in human cell nuclei following DNA damage (39,40). However, the role of Cc nuclear translocation in plants remains unknown.
Based on the structural and functional homology of A. thaliana NRP1 and human SET/TAF-Iβ, the present work explores NRP1 histone-binding properties and activity as histone chaperone. Here, we show that NRP1 accumulates in the chromatin of A. thaliana cells soon after DNA breaking. It is shown not only that Cc binds to NRP1, but that this binding results in the inhibition of the latter's ability to act as a histone chaperone. More specifically, Cc is found to bind with NRP1 between the latter's histone-binding (earmuffs) domains and to prevent the latter's binding to histones. Based on these findings, a role is proposed for NRP1 that is quite similar to that played by human SET/TAF-Iβ in chromatin dynamics following DNA damage, thereby shedding light on the significance of NRP1 inhibition by Cc.
MATERIALS AND METHODS
Antibodies
Mouse monoclonal anti-α-Tubulin (-α-Tub, catalog number T8328), secondary horseradish peroxidase (HRP)-conjugated anti-mouse IgG (catalog number A4416) and anti-rabbit IgG (catalog number A0545) were obtained from Sigma-Aldrich. Rabbit polyclonal antibody to histone H3 was from Abcam (catalog number ab1791). Primary rabbit anti-A. thaliana NRP1 was a kind gift from Dr Attila Fehér (Hungarian Academy of Sciences) (34). Rabbit anti-A. thaliana γ-H2AX antibody was kindly donated by Dr Anne B. Britt (University of California) (41).
A. thaliana cell cultures and DNA damage induction
Arabidopsis thaliana MM2d cell suspension cultures (Bayer CropScience) were grown in 500 ml flasks containing 50 ml Murashige and Skoog (MS) medium (Duchefa Biochemie) supplemented with 30 g/l sucrose (Sigma-Aldrich), 0.5 mg/l napthaleneacetic acid (NAA, Sigma-Aldrich), 0.05 mg/ml kinetin (Sigma-Aldrich), 200 mg/l cefotaxime (Duchefa Biochemie) and 200 mg/l penicillin (Duchefa Biochemie) at 110 rpm and 25°C. DNA damage was induced by adding 20 μM camptothecin (CPT) to cell cultures.
Total cell lysates preparation
Cell lysates of 25 ml MM2d A. thaliana cultures containing either untreated or 20 μM CPT-treated cells were prepared. In both cases, cells were harvested following centrifugation at 3000 × g for 15 min, washed twice in phosphate-buffered saline (PBS), pelleted and resuspended in 0.5 ml buffer containing 20 mM HEPES (pH 7.5), 10 mM KCl, 1.5 mM MgCl2, 1 mM ethylenediaminetetraacetic acid (EDTA), 1 mM ethylene glycol tetraacetic acid (EGTA) and 1 mM dithiothreitol (DTT), supplemented with 1 mM phenylmethylsulfonyl fluoride (PMSF), 1× protease inhibitors (Complete, Roche), 1 mM sodium orthovanadate and 1 mM sodium pyrophosphate. Cells were further lysed by sonication (3 cycles 10-s ON, 30-s OFF cycles at 20% amplitude). Cellular debris was then removed through centrifugation at 16 000 × g for 5 min at 4°C.
Crosslinking and chromatin fractionation
Crosslinking and nuclei extraction was performed as described in (42). For this, 25 ml MM2d A. thaliana cells were harvested by centrifuging at 3000 × g for 15 min and rinsed twice thoroughly with PBS. Cultures were submerged in 40 ml of 10 mM dimethyl adipimate (DMA) and vacuum-infiltrated on ice for 15 min. After filtering through a layer of fine nylon mesh, liquid was poured out and washed twice with distilled water. Then, cultures were submerged in 37 ml of 1% formaldehyde and vacuum-infiltrated on ice for 20 min. Crosslinking was stopped by adding 2.5 ml of 2 M glycine under vacuum-infiltration for 5 min. The plant material was filtered again, washed twice with distilled water and dried. The dry plant material was placed in a mortar precooled with liquid nitrogen and then grinded with a pestle to a fine powder. The grounded tissue was dipped in liquid nitrogen and used for chromatin preparation.
Cytosolic fractions were extracted by adding 10 ml of extraction buffer 1 (0.4 M sucrose, 10 mM Tris–HCl pH 8.0, 10 mM MgCl2, 0.1 mM PMSF and 1× protease inhibitors [Complete, Roche]) to the frozen material and mixing thoroughly with a vortex. The solution was filtered twice through a double layer of fine nylon mesh and, then, centrifuged at 3000 × g for 20 min. The pellets were resuspended in 1 ml of extraction buffer 2 (0.25 M sucrose, 10 mM Tris–HCl pH 8.0, 10 mM MgCl2,1% Triton X-100, 0.1 mM PMSF and 1× protease inhibitors) by pipetting up and down. The supernatants were centrifuged at 12 000 × g for 10 min and the pellets were washed in 400 μl of extraction buffer 3 (1.7 M sucrose, 10 mM Tris–HCl pH 8.0, 0.15% Triton X-100, 0.1 mM PMSF and 1× protease inhibitors). An equal volume (400 μl) of extraction buffer 3 was layered to the pellet and centrifuged for 1 h at 16 000 × g. Nuclei were resuspended in 300 μl of nuclei lysis buffer (50 mM Tris–HCl (pH 8.0), 10 mM EDTA, 1% SDS and 1× protease inhibitors) by pipetting up and down and vortexing. The nuclear solutions were sonicated applying 8-s ON, 60-s OFF cycles (10% amplitude) and then centrifuged at 16 000 × g for 5 min.
Chromatin fractionation was performed as described in ref. 43, with some modifications. A. thaliana cells were subjected to subcellular fractionation as above described until obtaining the nuclei. Then, nuclei were washed in 3 mM EDTA, 0.2 mM EGTA, 1 mM DTT and protease inhibitors for 30 min at 4°C. Insoluble chromatin was separated from soluble nuclear fraction by centrifugation at 1700 × g for 4 min at 4°C. Chromatin was washed twice in the same solution and collected by centrifugation at 10 000 × g for 1 min at 4°C. The chromatin pellet was suspended in SDS sample buffer.
Western blot analysis
For the immunoblot detection of A. thaliana γ-H2AX and α-tub in total cell lysates or NRP1 and histone H3 in the subcellular fractions, proteins were resolved by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) in 12% gels and, then, transferred onto polyvinylidene fluoride (PVDF) membranes (EMD Millipore) using a Mini Trans-Blot electrophoretic transfer cell (Bio-Rad). Membranes were blocked using 5% non-fat dry milk solution mixed with Tris-buffered saline (TBS) buffer containing Tween-20 (TBS-T). The immunoblot was performed with primary antibodies incubation overnight (at a dilution of 1:5000 in 5% non-fat dry milk in TBS-T). HRP-conjugated secondary antibodies were used for detection (1:10 000 in 5% non-fat dry milk in TBS-T). The immunoreactive bands were detected using Amersham ECL Plus Western Blotting Detection Reagents (GE Healthcare Life Sciences).
Amino acid sequence analysis
A multiple sequence alignment of A. thaliana NRP1; human SET/TAF-Iβ; yeast NAP1; A. thaliana NAP1;1, NAP1;2, NAP1;3, NAP1;4; and Xenopus laevis and A. thaliana protein core histones H2A, H2B, H3 and H4 was performed with Clustal Omega (European Molecular Biology Laboratory [EMBL]—European Bioinformatics Institute [EBI]). Amino acid sequence identity and similarity was obtained from Basic Local Alignment Search Tool (BLAST) analysis (National Centre for Biotechnology Information [NCBI]).
DNA constructs
The NRP1 gene was cloned in the pET-28a (+) bacterial expression vector under the control of the T7 promoter and adding an N-terminal His-tag. DNA for cloning was obtained via polymerase chain reaction (PCR) using cDNAs purchased from Arabidopsis Biological Resource Centre (ABRC). The oligonucleotides used in the PCR to amplify the NRP1 gene were 5΄-agccatatgatggtcgcggacaagagc-3΄ and 5΄-agtgcggccgctcattcctcaccatct-3΄. A. thaliana Cc was cloned in the pBTR1 vector under the control of the lac promoter, as described in a previous study (44), along with yeast heme lyase for proper heme folding. DNAs encoding X. laevis core histones were obtained in the pET-3a vector from Dr Tim Richmond (Institute of Molecular Biology & Biophysics, ETH Zürich).
Protein expression and purification
A construct containing NRP1 was used to transform the BL21(DE3) competent Escherichia coli strain. Transformed cells were harvested in fresh plates with 50 μg/ml kanamycin at 37°C. 250-ml precultures in Luria–Bertani (LB) medium supplemented with kanamycin were grown overnight and then used to inoculate 2.5-l cultures in 5-l flasks. Following the induction of cultures with 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) and growth at 30°C for 24 h, the cells were harvested at 6000 rpm for 10 min and resuspended in 40 ml lysis buffer composed of 20 mM Tris–HCl buffer (pH 8), 0.8 M NaCl, 10 mM imidazole, 0.01% PMSF, 0.2 mg/ml lysozyme, 5 mM DTT and 0.02 mg/ml DNase. They were then sonicated for 4 min and centrifuged at 20 000 rpm for 20 min. Protein purification was assessed by affinity chromatography using Ni Sepharose 6 Fast Flow medium (GE Healthcare). Proteins were then eluted by applying an imidazole gradient from 0 to 300 mM. The fractions containing protein were concentrated in an Amicon Centrifugal Filter (with MWCO of 10 kDa) until a proper protein concentration was reached and dialyzed against 10 mM sodium phosphate (pH 7.4) for isothermal titration calorimetry (ITC), or against 5 mM sodium phosphate (pH 6.5) for nuclear magnetic resonance (NMR) titrations.
Arabidopsis thaliana Cc was expressed in BL21(DE3) competent E. coli strains. To do so, 25 ml of preculture was shaken at 37°C in LB medium, supplemented with 100 μg/ml ampicillin. 2.5 ml of preculture was used to inoculate 2.5 l of the same media in a 5 l Erlenmeyer flask. The culture was shaken at 30°C for 24 h, after which the cells were harvested at 6000 rpm for 10 min using an Avanti J-25 refrigerated centrifuge (Beckman Coulter). Cells were then resuspended in 1.5 mM borate buffer solution (pH 8.5), sonicated for 4 min and centrifuged at 20 000 rpm for 20 min. For NMR measurements, 15N-labeled Cc was produced in minimal media with 15NH4Cl as nitrogen source. Further purification of Cc was carried out as indicated in previous studies (44). The fractions containing Cc were concentrated in an Amicon Centrifugal Filter (with MWCO of 3 kDa) until reaching the appropriate Cc concentration and dialyzed against 10 mM sodium phosphate (pH 7.4) for ITC. For NMR titrations, Cc was dialyzed against 5 mM sodium phosphate (pH 6.5).
Xenopus laevis core histones H2B, H3 and H4 were expressed in BL21(DE3) competent E. coli strains. Constructs containing core histones were used to transform cells, which were plated in LB medium, supplemented with 100 μg/ml ampicillin. Single colonies were employed to inoculate 25 ml preculture and incubated for 12 h at 37°C. Following induction with 0.5 mM IPTG, cells were grown for 4 h at 25°C in 5 l LB medium. Purification of core histones was performed as indicated previously (45). Chicken erythrocytes H2A–H2B and H3–H4 histone dimers were kindly provided by Dr Arturo Muga (University of the Basque Country, Spain).
All protein solutions contained 1 mM PMSF. Purity of proteins was verified by SDS-PAGE. Protein concentration was assessed using the Bradford protein assay (46) and NRP1 protein concentration was expressed in dimeric form.
Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS)
Gel protein spots were manually excised from micro-preparative gels using pipette tips. The selected proteins were reduced in-gel, alkylated and digested with trypsin. Briefly, spots were washed twice with water, shrunk for 15 min with 100% acetonitrile and dried in a Savant SpeedVac for 30 min. Then, the samples were reduced with 10 mM DTT in 25 mM ammonium bicarbonate for 30 min at 56°C and subsequently alkylated with 55 mM iodoacetamide in 25 mM ammonium bicarbonate for 15 min in the dark. Finally, samples were digested overnight with 12.5 ng/μl sequencing grade trypsin (Roche Molecular Biochemicals) in 25 mM ammonium bicarbonate (pH 8.5) at 37°C. After digestion, the supernatant was collected and 1 μl was spotted onto a MALDI target plate and later air-dried at room temperature. Subsequently, 0.4 μl of a 3 mg/ml α-cyano-4-hydroxy-transcinnamic acid matrix (Sigma) in 50% acetonitrile was added to the dried peptide digest spots and air-dried again at room temperature.
MALDI-TOF MS analyses were performed in Autoflex (Bruker Daltonic) at the Proteomics Service, cicCartuja. Trypsin self-digestion peptidic products were used for internal calibrations. For protein identification, the UniProtKB-SwissProt_160909 database v. 57.7 (497293 sequences, 175274722 residues) was searched using a local license of MASCOT 2.1 through the Global Protein Server v. 3.6 from Applied Biosystems. In all protein identification, the probability scores were greater than the significance threshold set by mascot for a P-value lower than 0.05.
Circular dichroism
Circular dichroism (CD) spectra were recorded in the far-UV range (190–250 nm) at 25°C on a J-815 CD Spectropolarimeter (Jasco), equipped with Peltier temperature control system, using a 1-mm quartz cuvette. NRP1 and histone protein concentrations were 3 and 15 μM, respectively, in 10 mM sodium phosphate buffer (pH 7.4). For each sample, an average of 20 scans was accumulated.
Nucleosome assembly assay
Histone chaperone activity was performed as described in a previous study (47). 0.1 pmol closed circular form of the pBlueScript II SK(−) plasmid (3 kb) was relaxed by pre-incubation with 2.5 U type 1 topoisomerase (Topo I) from wheat germ (Promega) in 50 μl buffer consisting of 10 mM Tris–HCl (pH 8), 100 mM NaCl, 3.5 mM MgCl2, 0.5 mM DTT, 2 mM adenosine triphosphate (ATP) and 0.1 mg/ml bovine serum albumin (BSA) at 37°C for 3 h. The pBlueScript II SK(−) plasmid was purified by classical plasmid DNA extraction using alkaline lysis, as described in other studies (40). 3 pmol of HeLa core histones (EMD Millipore) were pre-incubated with 4 pmol NRP1 in 50 μl assembly buffer composed of 10 mM Tris–HCl (pH 8), 100 mM NaCl, 2 mM MgCl2, 0.5 mM DTT, 2 mM ATP and 0.1 mg/ml BSA) at 37°C for 3 h. When indicated, Cc (0.5–2 μg) was added to the histone-chaperone mixture. Histone–chaperone samples (with or without Cc) were then mixed with the relaxed form plasmid and further incubated at 37°C for 3 h. The reactions were stopped by adding equal volume of stop buffer composed of 20 mM EDTA (pH 8), 1% SDS and 0.2 mg/ml proteinase K and incubated at 37°C for 30 min. Following the reaction, plasmids were extracted using the phenol–chloroform technique and precipitated with ethanol. Plasmids were subjected to electrophoresis with 1% agarose gel, run in Tris–borate–EDTA buffer and further visualized by staining with ethidium bromide.
Micrococcal nuclease digestion assay
Nucleosome assembly was also tested by micrococcal nuclease (MNase) digestion as described in (47). 0.6 μg of the closed circular form of pBlueScript II SK(−) plasmid (3 kb) was relaxed by pre-incubation with 7.5 U Topo I in 50 μl buffer consisting of 10 mM Tris–HCl (pH 8), 50 mM NaCl, 3.5 mM MgCl2, 1 mM DTT, 2 mM ATP and 0.1 mg/ml BSA at 37°C for 3 h. The pBlueScript II SK(−) plasmid was purified as described in other studies (40). Nine picomoles of HeLa core histones (EMD Millipore) were pre-incubated with 12 pmol NRP1 in 50 μl assembly buffer. This one contained 10 mM Tris–HCl (pH 8), 50 mM NaCl, 2 mM MgCl2, 1 mM DTT, 2 mM ATP and 0.1 mg/ml BSA at 37°C for 3 h. When indicated, Cc (0.5–3 μg) was added to the histone-chaperone mixture. Histone–chaperone samples (with or without Cc) were then mixed with the relaxed plasmid solution and incubated at 37°C for 3 h. 5 mM CaCl2 and 15, 30 or 45 U/ml MNase were added to the reaction mixture, followed by incubation at 37°C for 1 min. The reactions were stopped by adding equal volume of a solution containing 20 mM EDTA (pH 8) and 1% SDS. Then, digested DNA was extracted using the phenol–chloroform technique and precipitated with ethanol. Digested DNA was subjected to electrophoresis in 1.8% agarose gel, run in Tris-borate-EDTA buffer for 1 h at 80 V and visualized by ethidium bromide staining.
Isothermal titration calorimetry
All isothermal titration calorimetry (ITC) experiments were performed using a MicroCal Auto-iTC200 isothermal titration calorimeter (Malvern Instruments) at 25°C by titrating NRP1 with Cc or core histones. The reference cell was filled with distilled water. The experiments consisted of 2 μl injections of 300 μM Cc, H2B, H3 or H4 core histones in 10 mM sodium phosphate buffer (pH 7.4) into the sample cell, which initially contained 20 μM NRP1 in the same buffer. All solutions were degassed before the titrations were performed. Titrant was injected at appropriate time intervals to ensure that thermal power signal returned to baseline prior to the following injection. To achieve homogeneous mixing in the cell, stirring speed was maintained constant at 1000 rpm. The data—namely, heat per injection normalized per mole of injectant vs. molar ratio—were analysed with Origin 7.0 (OriginLab Corporation) employing a model that considered identical and independent binding sites. The fitting function contains an adjustable parameter accounting for background heat due to unspecific phenomena (e.g. mechanical injection and slight mismatches between titrant and titrant solution) which proved to be comparable to the intrinsic heat associated with the protein-protein interaction. Calorimeter calibration and performance tests were performed through CaCl2–EDTA titrations using solutions provided by the manufacturer.
Surface plasmon resonance
Binding analyses involving NRP1 and its partners, either Cc or histones, were performed by Surface Plasmon Resonance (SPR) using CO SPRi-Biochips and a SPRi-Plex II (Horiba). Cc, H2B, H3 and H4 histones, as well as H2A–H2B and H3–H4 histone dimers, were covalently attached to the matrix using CO direct amine-coupling chemistry. Proteins were immobilized at 1, 5 and 10 μM for Cc, H2B, H3 and H4 histones, whereas for the H2A–H2B and H3–H4 histone dimers concentrations were 0.1, 1 and 10 μM. Bovine serum albumin was attached to the chip surface at reference spots used as control. The binding measurements were performed at 25°C in 10 mM sodium phosphate buffer, pH 7.4. NRP1 at various concentrations (from 0.1 to 1 μM) were flowed on the surface at a 50 μl/min rate. Each solution was injected three times at least. The signals from the reference spot surface were subtracted in every sensogram. The data were analysed with Origin 2016 (OriginLab Corporation).
Nuclear magnetic resonance spectroscopy
One-dimensional (1D) 1H nuclear magnetic resonance (NMR) spectra were recorded to monitor the resonance signal of Met88 of unlabelled reduced Cc (13 μM) in the presence of 6.5 μM unlabelled NRP1 and calf thymus histones (80–300 μg). As a control, 1D 1H NMR measurements of reduced Cc (13 μM) were also taken after adding 6.5 μM BSA or 300 μg calf thymus histones. Titration measurements were prepared in 3 mm NMR containing 0.25 ml volume tubes. All measurements were taken in 5 mM sodium phosphate buffer (pH 6.3) in the presence of 0.1 M sodium ascorbate to ensure the Cc redox state and adding 10% D2O to adjust the lock signal.
To monitor the interaction of reduced Cc with NRP1, 2D [1H, 15N] heteronuclear single-quantum correlation (HSQC) spectra were acquired during the titration of 50 μM 15N-labeled Cc solutions. In these solutions, increasing quantities of NRP1 were added up to the final Cc:NRP1 molar ratio of 1:0.25. Titration measurements were prepared in NMR tubes (Shigemi Inc.) of up to a volume of 0.28 ml and pH values of the samples were verified after each step in the titration. Data were processed using TopSpin NRM 2.0 software (Bruker) and chemical shift perturbation (CSP) analyses were performed with Sparky 3 NMR Assignment and Integration Software (T.D. Goddard and D.G. Kneller, University of California-San Francisco). All measurements were performed on a 700 MHz AVANCE III spectrometer (Bruker) at 25°C. Signals for water were suppressed according to the water suppression by gradient-tailored excitation (WATERGATE) method (48). The NMR signal for 15N and 1H nuclei of reduced plant Cc was already available (Biological Magnetic Resonance Bank [BMRB] accession number: 18828) (49). In order to discriminate between specific instances of line broadening, threshold values were set at ΔΔν1/2 Binding ≥ 14.6 Hz for 15N, as reported previously (50).
Molecular docking
A soft docking algorithm in the Biomolecular Complex Generation with Global Evaluation and Ranking (BiGGER) software package (51) was used to generate in silico models of the Cc:NRP1 and histone:NRP1 complexes. For each run, 500 solutions were generated using a 15° angle step soft dock and a distance of 7 Å. Geometric docking solutions were generated based on protein surface complementarity. The solutions were evaluated and ranked according to their overall score, having taken into account different interaction criteria including hydrophobicity, electrostatics, geometry and side-chain parameters. The centre of mass for all structures was also represented. CSP values were taken from the NMR measurements of the Cc:NRP1 complex and introduced as restraints in the docking calculations. The coordinates file of plant Cc was obtained using a homology model as described previously (44), whereas NRP1 and plant H2A, H2B, H3 and H4 were modelled with MODELLER (52). All molecular graphics of complexes were generated using the UCSF Chimera package (53).
RESULTS
A. thaliana NRP1 is located at chromatin upon DNA damage induction
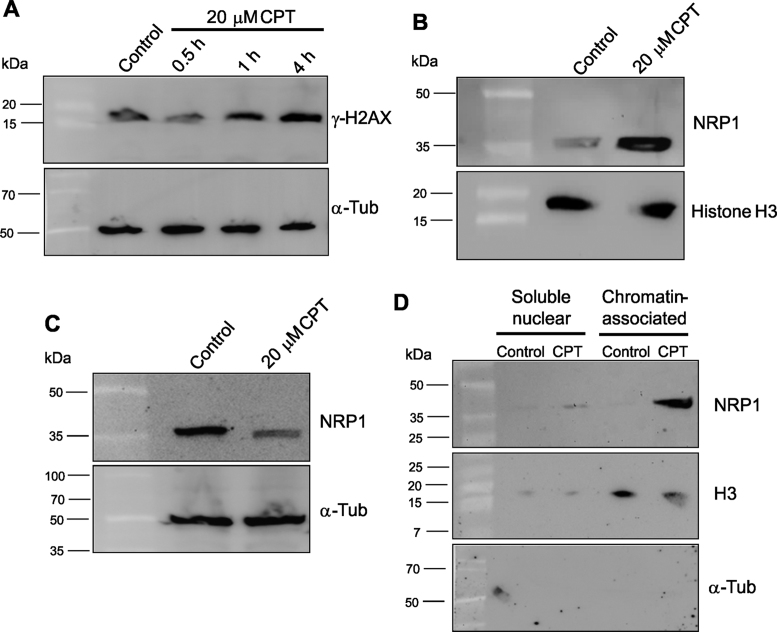
In mammals and yeasts, cellular responses to DNA DSBs are linked to the formation of repair foci, which are multiproteic nuclear complexes formed in the vicinity of DNA breaks, responsible for damage detection and signalling (54–56). Phosphorylated histones H2AX (γH2AX) foci are considered sensitive markers of DNA damage, since γH2AX accumulation occurs in response to DSBs (57). In Arabidopsis, γH2AX foci were detected and connected to an efficient DSBs repair response (56). In order to explore the role of NRP1 in the DNA damage process, A. thaliana cells were treated with the DSBs inducer CPT, and NRP1 cellular location was checked. Western-Blot analysis of total lysates of A. thaliana MM2d cells revealed an accumulation of γH2AX after CPT-treatment (20 μM), which was maximum at 4 h (Figure 1A). Upon treatment with 20 μM CPT for 4 h, nuclei were extracted from A. thaliana cells. Strikingly, NRP1 accumulated in the nuclear fraction from cells undergoing CPT treatment, as compared to non-treated ones (Figure 1B). Co-detection in the nuclear fraction with nuclear-specific histone H3 confirmed the NRP1 location within the nucleus (Figure 1B). In untreated cells, however, NRP1 mainly appears in the cytosolic fractions (Figure 1C). Upon chromatin fractionation, NRP1 was mostly detected in the chromatin-associated fraction after CPT treatment (Figure 1D). This suggests that NRP1 shuttles from the cytosol to the nucleus in response to DNA breaks, subsequently joining the chromatin. These findings are in agreement with a previous report regarding the nuclear NRP1 translocation in response to the DNA damage induced by heat shock (34).
Figure 1.
NRP1 accumulation at the chromatin from CPT-treated A. thaliana cells. (A) Immunodetection of γ-H2AX (16 kDa) in total lysates of MM2d A. thaliana cells treated with 20 μM CPT for 0.5, 1 and 4 h. α-Tub (50 kDa) antibody was used as loading control. (B and C) Detection of NRP1 (38 kDa) in nuclei (B) or in the cytosolic fractions (C) of non-treated or CPT-treated-cells (20 μM CPT) for 4 h. (D) Chromatin fractionation and immunoblotting detection of NRP1 in the soluble nuclear and chromatin-associated fractions of non-treated and CPT-treated cells (20 μM) for 4 h. Specific antibodies against A. thaliana NRP1 and γ-H2AX were used in Western blotting. Antibodies against histone H3 (15 kDa) and α-Tub were used as loading nuclear and cytosolic controls, respectively.
A. thaliana NRP1 is closely related to human SET/TAF-Iβ and exhibits activity characteristic of histone chaperones
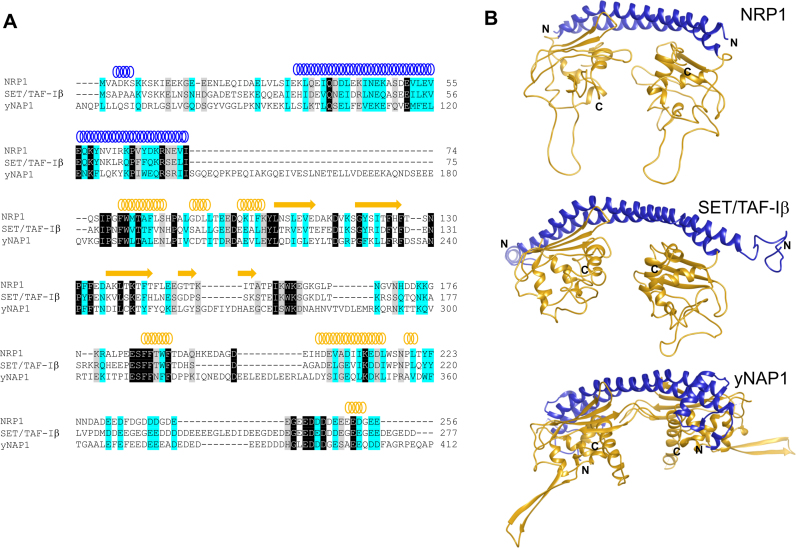
Despite the fact that four genes encoding close NAP1 homologues are present in the genome of A. thaliana, NRPs are more closely related to human SET/TAF-Iβ than to NAP1 proteins (58). In fact, NRP1 demonstrates <30% identity and about 50% similarity with the four members of the A. thaliana NAP1 family (AtNAP1;1, AtNAP1;2, AtNAP1;3 and AtNAP1;4) (Supplementary Figure S1). By contrast, sequence homology between A. thaliana NRP1 and human SET/TAF-Iβ is 45% identity and 66% similarity, while between A. thaliana NRP1 and yeast NAP1 (yNAP1) it is 33% identity and 58% similarity (Figure 2A).
Figure 2.
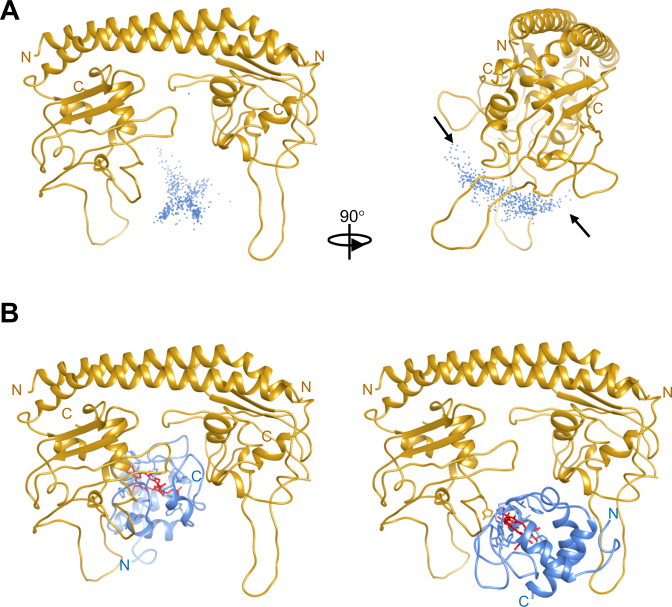
Domain organization of histone chaperone proteins. (A) Multiple amino acid sequence alignment of A. thaliana NRP1 (UniProt entry: Q9CA59-1), human SET/TAF-Iβ (Q01105-2) and yNAP1 (P25293-1) proteins (Clustal Omega). Sequences are colored by similarity as fully conserved residue (black), residues with strongly similar properties (light blue) and residues with weakly similar properties (gray). Secondary structure elements of A. thaliana NRP1 are shown. (B) Ribbon representation of a homology model of NRP1, a model of SET/TAF-Iβ that includes non-observable loops in the crystallographic structure (PDB code: 2e50) and the crystallographic structure of yNAP1 (2z2r). N-terminal and histone-binding domains are depicted in blue and gold, respectively.
To analyse the structure of A. thaliana NRP1 and its putative role as a histone chaperone, a structural homology model of NRP1 was designed. As the 3D structure of NRP1 is not currently available, the homology model was constructed using X-ray diffraction data from the NRP1 analogues in human SET/TAF-Iβ (Protein Data Bank [PDB] code: 2E50). Comparison of the overall 3D model of NRP1 with the structure of human SET/TAF-Iβ and yNAP1 (PDB code: 2z2r) revealed similar folding for the three histone chaperones (Figure 2B). Interestingly, resembling the structure of human SET/TAF-Iβ and yNAP1 (59,60), the homology model of NRP1 showed a headphone-shaped homodimer (Figure 2B), composed of a long backbone helix responsible for dimerization with N-terminal domain (Figure 2B: blue region) and an earmuff domain (Figure 2B: gold region) responsible for histone binding.
To dig into the function of NRP1, its ability to bind to histones was also explored. To do so, thermodynamic parameters of the binding of NRP1 to core histones were quantitatively assessed (Table 1). Core histones H2B, H3 and H4 from X. laevis were isolated, rather than used in a mixture, in order to permit comparison of their respective binding affinities with regard to the chaperone. Identity and purity of recombinant proteins were confirmed by MALDI-TOF MS and SDS-PAGE analyses (Supplementary Figures S2 and S3). CD spectra of A. thaliana NRP1 and core histones H2B, H3 and H4 from X. laevis corroborated the proper secondary structure of these proteins (Supplementary Figure S4). Unfortunately, the protein recombinant expression of H2A was unsuccessful.
Table 1. Thermodynamic values inferred from ITC measurements.
| Protein complex | ΔG (kcal mol−1) | ΔH (kcal mol−1) | −TΔS (kcal mol−1) | KD (μM) | n |
|---|---|---|---|---|---|
| H2B:NRP1 | −6.8 | −4.6 | −2.2 | 9.6 | 1.8 |
| H3:NRP1 | −6.4 | −7.1 | 0.7 | 20 | 1.8 |
| H4:NRP1 | −6.8 | −10.4 | 3.6 | 11 | 2.1 |
| H2A–H2B:NRP1 | −7.1 | 58 | −65.1 | 0.1 | 0.9 |
| H3–H4:NRP1 | −2.7 | 52 | −54.7 | 0.4 | 0.9 |
| Cc:NRP1 | −7.2 | −2.2 | −5.0 | 5.0 | 2.0 |
Relative error in KD is 20%.
Error in ΔG is 0.1 kcal/mol, and in ΔH and −TΔS is 0.4 kcal/mol.
Error in n is 0.2.
Thermodynamic equilibrium parameters for interaction of NRP1 with X. laevis H2B, H3 or H4 core histones, H2A–H2B or H3–H4 dimers and A. thaliana Cc. Equilibrium dissociation constant (KD), association enthalpy (ΔH), entropy (−TΔS), Gibbs free energy (ΔG) and stoichiometry of the reaction (n) are shown. The affinity of a protein–protein interaction is defined by the Gibbs energy of the binding (ΔG = −RT ln KA = RT ln KD). ΔG has two different contributions, ΔH and −TΔS, according to the equation ΔG = ΔH − TΔS. NRP1 is expressed as a dimer, whereas core histones and Cc are monomers.
ITC experiments provided direct evidence for the interaction of NRP1 and histones H2B, H3 and H4 (Supplementary Figure S5; Table 1). Because the dissociation constants (KD) are similar to the NRP1 concentration in the calorimetric cell (20 μM), the sigmoidicity of the titration curves is small; that is, the inflection point is not located at the equivalence point (molar ratio of 2). Actually, to exhibit an inflection point, the protein concentration in the cell [P]T must be larger than the dissociation constant (a dimensionless parameter c = n [P]T/KD, where n is the number of binding sites, must satisfy the condition c ≥ 1, or [P]T ≥ KD/n) (61). However, both the dissociation constant and the binding stoichiometry can be estimated in a reliable manner. ITC measurements (Supplementary Figure S5; Table 1) showed binding affinity to be higher for NRP1 and H2B (KD = 9.6 μM) and H4 (KD = 11μM) than for NRP1 and H3 (KD = 20 μM). In all cases, the stoichiometry of histone:NRP1 interactions was found to be 2:1 (Table 1). As NRP1 is a homodimer and core histones were expressed as monomers, it can be inferred from the stoichiometry data that a dimer of histones is bound to one NRP1 dimer. These findings are in agreement with those previously reported; namely, that NRP1 preferentially binds to H2B, rather than to H3 (32). Since histones can form dimers and tetramers, we also examined the NRP1 binding to H2A–H2B and H3–H4 dimers by ITC. As observed in Supplementary Figure S5 and Table 1, NRP1 bound to H2A–H2B and H3–H4 with binding affinity values of 0.1 and 0.4 μM, respectively. As expected, the stoichiometry of histone dimers:NRP1 interactions was found to be 1:1 (Table 1).
To confirm the binding properties of NRP1 interactions with histones, we resorted to SPR measurements. In agreement with ITC data, SPR curves of NRP1 interaction with isolated H2B, H3 or H4 histones were consistently fitted to a multiple-site binding model (Supplementary Figure S6, blue lines). By the contrary, these binding curves poorly fitted a one-site binding model, as shown in Supplementary Figure S6 (red lines). The SPR sensograms corresponding to NRP1 binding to H2A–H2B and H3–H4 dimers were firmly fitted to a 1:1 binding model (Supplementary Figure S6). Noticeably, KD values obtained for NRP1 interactions with isolated or heterodimeric histones were similar to those found for ITC measurements (Supplementary Table S1).
However, as indicated above, NRP1 binds not only to H3, but also to H4, though binding affinity of the chaperone for the former was in fact lower than for the latter. These results contrast with the proposal from an earlier study that NRP1 is a chaperone for H2A/H2B (32). In that study, which did not test with H4, pull-down assays showed NRP1 as binding to A. thaliana H2A and H2B, but hardly at all to H3. However, another study found NRP1 to coimmunoprecipitate with H3 in A. thaliana cell extracts (33). These findings seem to support the results reported here, particularly considering the high degree of sequence identity between A. thaliana and X. laevis core histones H4 (98%), H3 (93%) and H2B (80%) (Supplementary Figure S7). Furthermore, binding affinity values for NRP1 and X. laevis core histones are similar to those previously reported for the former's human analogue, SET/TAF-Iβ, and the same histones (40).
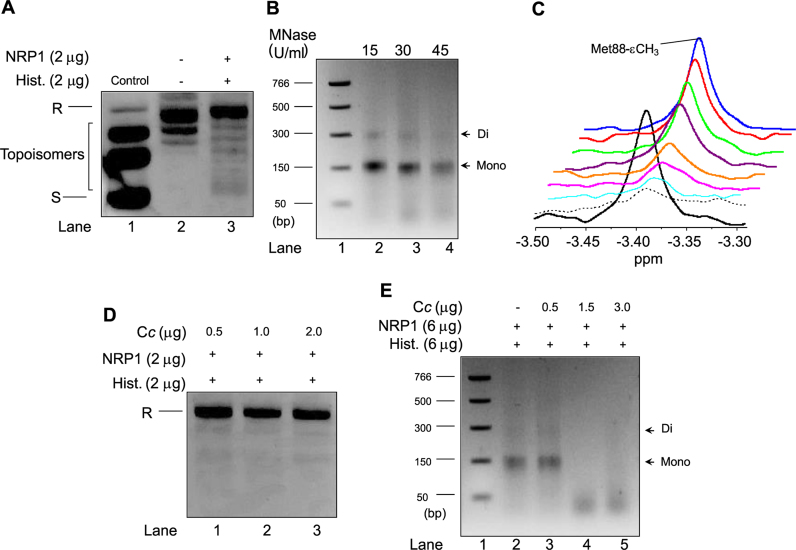
While the NRP1 gene has been described as encoding a histone chaperone (32), no report on the nucleosome assembly activity of NRP1 has been offered to date. To elucidate the function of NRP1 and whether NRP1 performs such histone chaperone activity, a plasmid super-coiling assay—as described in a previous study (47) and measuring the ability of a histone chaperone to introduce negative supercoils on a relaxed plasmid in the presence of HeLa core histones (Supplementary Figure S8A)—was performed. Plasmids were previously relaxed with Topo I (Figure 3A: lane 2). The NRP1-mediated formation of nucleosomes, coupled with the introduction of negative supercoils in the plasmids, was examined in an agarose gel after deproteinization of the samples (Supplementary Figure S8A). As the negatively supercoiled plasmid ran faster than the relaxed ones, they could be easily separated in an agarose gel. NRP1 was shown to promote an increase in the number of negative supercoils of relaxed plasmids (Figure 3A: lane 3), whereas incubation with NRP1 or HeLa core histones separately (Supplementary Figure S8B: lanes 3 and 4) were not found to do the same. The results confirmed NRP1 as demonstrating histone chaperone activity and as stimulating nucleosome assembly in vitro.
Figure 3.
Nucleosome assembly activity of NRP1 and its inhibition by Cc. (A) Plasmid supercoiling analysis was carried out by mixing 2 μg of NRP1 with 200 ng of relaxed plasmid after being treated with Topo I and incubated with 2 μg of HeLa core histones. Relaxed and supercoiled forms of circular DNA plasmid are indicated as R and S, respectively. Lane 1 (control) shows a supercoiled, untreated DNA plasmid, whereas lane 2 corresponds to the DNA plasmid relaxed by Topo I. (B) MNase assay was performed by mixing 6 μg of NRP1 with 600 ng of relaxed plasmid after being treated with Topo I and incubated with 6 μg of HeLa core histones. Plasmid DNA was digested with MNase (15, 30 or 45 U/ml; lanes 2–4). Lane 1 indicates a DNA ladder marker and the size of each band is represented on the left. DNA fragments that correspond to mono- and di-nucleosomes are indicated by arrows on the right. (C) 1D 1H NMR spectra monitoring the Met-88-CH3 signal of reduced Cc in the presence of NRP1 and histones. Details of superimposed 1D 1H NMR spectra of 13 μM reduced Cc either free (black) or bound to 6.5 μM NRP1 (dashed) following addition of increasing concentrations of calf thymus histones (80 μg [light blue], 100 μg [pink], 112 μg [orange], 125 μg [purple], 135 μg [green], 150 μg [red] and 300 μg [blue]). (D) Plasmid supercoiling analysis was carried out as in A, but in the presence of Cc at increasing concentrations (0.5, 1 and 2 μg [lanes 1–3]). Relaxed form of circular DNA plasmid is indicated as R. (E) MNase assay was performed as in B, in the absence (lane 2) and in the presence of Cc at increasing concentrations (0.5, 1.5 and 3 μg [lanes 3–5]). Plasmid DNA was digested with 30 U/ml. Lane 1 indicates a DNA ladder marker and the size of each band is represented on the left. DNA fragments that correspond to mono- and di-nucleosomes are indicated by arrows on the right.
To confirm the nucleosome assembly activity of NRP1, we performed a MNase digestion assay to detect DNA fragments protected by core histones (Figure 3B). After incubation of NRP1 with core histones and relaxed plasmid, MNase was added to digest the linker DNA between nucleosomes. The DNA digested at increasing concentrations of MNase exhibited approximately 150 and 300 bp fragments (Figure 3B, lanes 2–4). This ladder was not detected in the presence of either NRP1 or core histones (Supplementary Figure S8, lanes 2–3). The length of about 150 and 300 bp is the typical size for DNA in a mono- and di-nucleosome (47), thus indicating that NRP1 assembles nucleosomes.
Earmuff domain of NRP1 is engaged in histone binding
To elucidate the functionality of NRP1, further studies were conducted on how NRP1 interacts with core histones. As there is currently no 3D structure available for A. thaliana NRP1 in the PDB, molecular docking simulations for complexes formed by NRP1 and core histones were performed with BiGGER, using homology models of A. thaliana NRP1 and H2A, H2B, H3 and H4 core histones.
The 500 solutions obtained were then evaluated, scored and ranked by BiGGER according to criteria such as geometric and hydrophobic parameters, the electrostatic energy of the interaction and the relative propensity of side chains to interact (Supplementary Figure S9: NRP1 ribbon representations [gold]; histone centres of mass for H2A [green], H2B [purple], H3 [red] and H4 [blue]). Strikingly, the vast majority of histone centres of mass, except for H2B, were positioned between the earmuff domains of NRP1. This is consistent with the recent crystal structure of yNAP1 bound to H2A–H2B heterodimer, in which the concave face of yNAP1 constitutes the binding site for histones (62). In addition, our results agree with previous ones suggesting that core histones position into the acidic cavity of SET/TAF-Iβ (60). Remarkably, the two highest ranked docking solutions are spatially compatible (Supplementary Figure S9: 90° rotated view [centres of mass indicated with arrows]) and consistent with the 2:1 stoichiometry of histone:NRP1 complexes inferred from ITC and SPR measurements (Table 1; Supplementary Table S1; Supplementary Figures S5 and S6). Therefore, the earmuff domain (acidic concave face) of NRP1 may be the binding site for core histones.
NRP1 binds to Cc, blocking the former's capability as histone chaperone
Recently, the authors of the present study described for the first time the interaction of NRP1 with Cc in A. thaliana cells upon induction of oxidative stress (35). Taking into account the close relation between A. thaliana NRP1 and human SET/TAF-Iβ (Figure 2A), the similar histone-binding properties of the two, and the fact that human Cc was recently described as affecting the ability of SET/TAF-Iβ to bind to histones (40), experiments were carried out to determine if Cc affects NRP1 in a similar way.
To detect complex formation between NRP1 and Cc, and to further test the effects on complex formation of the addition of calf thymus histones, 1D 1H NMR spectra of reduced A. thaliana Cc were recorded when free or in the presence of NRP1 and calf thymus histones. Monitoring the signal of the methionine axial ligand of Cc (Met88-ε-CH3) (Figure 3C: solid black line), the resonance was found to broaden beyond the detection limit upon the addition of NRP1 at a Cc:NRP1 ratio of 2:1 (Figure 3C: dashed black line). However, upon titration of the Cc:NRP1 complex with increasing histone concentration, Cc was found to dissociate from NRP1 and the intensity of the former's methionine axial ligand signal was recovered (Figure 3C: coloured lines). Controls of 1D 1H spectra were recorded for reduced Cc free and in the presence of BSA or calf thymus histones (Supplementary Figure S8D), added according to the same ratios as those used for NRP1 and histones. Notably, no change in Met88-ε-CH3 Cc signal intensity was observed upon the separate addition of BSA or histones, suggesting that none of these proteins interacts with Cc (Supplementary Figure S8D).
To further compare the thermodynamic parameters of the complexes formed between NRP1 and Cc and core histones, respectively, ITC measurements of the Cc:NRP1 interaction were performed. Following analysis of the resulting data, it was found that the Cc:NRP1 interaction fit a 2:1 model, with two independent binding sites for Cc on NRP1 (Supplementary Figure S5; Table 1). ITC titrations revealed the interaction between Cc and NRP1 to be exothermic (binding enthalpy [ΔH] = −2.2 kcal mol−1) and primarily entropy-driven (entropy [−TΔS] = −5 kcal mol−1), with a KD of 5 μM. Interestingly, binding affinity of NRP1 with respect to Cc was found to be slightly higher (KD = 5 μM) than that observed with respect to H2B (KD = 9.6 μM), H3 (KD = 20 μM) and H4 (KD = 11 μM) (Table 1). The results show Cc as not only binding to NRP1, but also as a competitor with histones for the NRP1 binding site.
As binding to core histones is a necessary step prior to histone chaperone nucleosome assembly activity, the question was examined of whether this competition between Cc and histones for NRP1 binding sites also compromised the latter's activity as a histone chaperone. Indeed, as evidenced by the absence of negative supercoiling on relaxed plasmids, the activity of NRP1 as a histone chaperone was impaired by addition of increasing amounts of Cc (Figure 3D: lanes 1–3). Such Cc-mediated histone chaperone inhibition was found to occur even at the lowest Cc amount tested (0.5 μg) (Figure 3D: lane 1). Controls were also observed corresponding to the supercoiling assay in the presence of isolated NRP1, HeLa core histones, Cc or a combination (Supplementary Figure S8B: lanes 3–7). We also tested the inhibition of the histone chaperone activity of NRP1 by Cc by means of a MNase digestion assay (Figure 3E). As shown in Figure 3E, adding Cc at 1.5 or 3 μg to reactions resulted in the absence of the 150 bp DNA fragment corresponding to the mono-nucleosome size (Figure 3E, lanes 4–5). These findings demonstrate that Cc competes with core histones for NRP1 binding sites, thereby hampering the latter's ability to function as a histone chaperone.
NRP1 and Cc form a specific, well-defined complex
Structural features of the interaction between NRP1 and Cc were studied to identify the Cc binding site for NRP1 and to analyse interprotein interaction regions at the atomic level. To do this, [1H, 15N] HSQC spectra of fully reduced 15N-labeled Cc, which was either free or bound to unlabelled NRP1, were monitored.
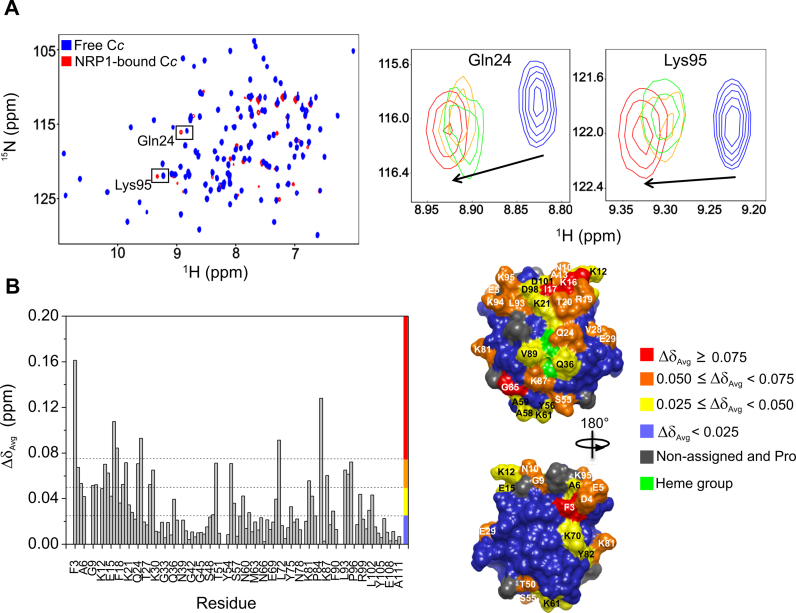
As can be observed, titration of unlabelled NRP1 onto 15N-labeled Cc yielded significant broadening of Cc amide signals, suggesting the formation of a Cc:NRP1 complex (Figure 4A [left]: signals corresponding to Cc free of NRP1 [blue] and Cc bound to NRP1 [red]). It is important to consider that broadening can be caused mainly by one of two effects, namely, slow tumbling or exchange (63). If the unlabelled protein possesses a high molecular weight, as is the case with NRP1 (63.4 kDa), then 15N Cc amide signals will broaden during titration with unlabelled NRP1 due to a slowing down of the tumbling rate. Alternatively, 15N Cc signal broadening can be due to an intermediate or slow exchange between free and bound states (64). This condition is typically associated with dissociation constants in the micromolar range (64), which is consistent with the dissociation constant assessed through ITC analysis (KD = 5 μM).
Figure 4.
NMR titrations of 15N-labeled reduced Cc with NRP1. (A) Left: Superimposed [1H, 15N] 2D HSQC spectra of 15N-labeled Cc, either free or bound to NRP1, at Cc:NRP1 molar ratio of 1:0.5. Right: Details of the superimposed [1H, 15N] HSQC spectra of free Cc (blue) and upon titration with increasing NRP1 concentrations, at differing Cc:NRP1 ratios (1:0.12 [green], 1:0.25 [orange] and 1:0.5 [red]). Gln24 and Lys95 resonances correspond to Cc. Arrows indicate the direction and magnitude of CSPs. (B) Left: Average CSPs (ΔδAvg) experienced by amide NMR signals of Cc in complex with NRP1 (Cc:NRP1 ratio: 1:0.5). Colour bars indicate ΔδAvg categories, as indicated at the right. Right: Mapping of Cc residues perturbed upon binding to NRP1. Cc surfaces are rotated 180° around the vertical axis in each view. Residues are coloured according to their ΔδAvg (ppm).
In addition to general line broadening, enhanced broadening was observed at specific 15N Cc resonances, thereby evidencing their involvement in complex formation. Thus, linewidths (ΔΔν1/2 Binding) of 15N Cc amide signals from [1H, 15N] HSQC spectra of free and NRP1-bound Cc in a Cc:NRP1 ratio of 1:0.5 were analysed (Supplementary Figure S10). Cc resonances displaying enhanced broadening in the 15N dimension corresponded to Lys21, Thr27, Glu29, Gly37, Leu40, Asn41, Leu43, Glu68, Thr71, Tyr75, Lys80, Asp98 and Asp101 (Supplementary Figure S10). Consequently, these residues can be expected to be at or near the NRP1 binding site for Cc. Worth particular mention is that the Cc:NRP1 complex saturation was reached at a ratio of 1:0.5, a finding which matches with the 2:1 stoichiometry previously observed in ITC titrations and is also in agreement with SPR measurements (Table 1; Supplementary Table S1; Supplementary Figures S5 and S6).
Besides enhanced broadening, several amide signals in the [1H, 15N] HSQC spectra of Cc exhibited CSPs upon addition of the histone chaperone. In the two examples depicted in superimposed spectra (Figure 4A: right), signals corresponding to Gln24 and Lys95 gradually shifted following titration at different Cc:NRP1 ratios (1:0.12, 1:0.25 and 1:0.5). It is worth noting that NRP1 is expressed as a homodimer, whereas Cc is a monomer. Since the amide groups of the residues above experienced changes in their chemical environment upon Cc:NRP1 complex formation, residues showing CSPs may belong to the complex interface.
With the aim of identifying the Cc residues most likely to be involved in the complex interface, an analysis of the average CSP values (ΔδAvg) for Cc amide signals was made. As represented (Figure 4B: left), 15N Cc resonances Phe3, Lys16, Ile17, Cys25, Thr71 and Gly85 experienced significant chemical-shift displacements (ΔδAvg ≥ 0.075) upon addition of NRP1. Noticeably, residues at the Cc N-terminal domain (e.g., Asp4, Glu5, Ala6, Gly9, Asn10, Lys12, Ala13, Gly14, Glu15, Phe18, Arg19, Thr20, Lys21, Cys22 and Gln24) displayed moderate (ΔδAvg between 0.075 and 0.050) or slight (ΔδAvg between 0.050 and 0.025) CSPs (Figure 4B: left), evidencing the importance of these Cc residues in the binding to NRP1.
It is widely accepted that CSP magnitude correlates with protein complex dynamics (65,66). When specific complexes yield single well-defined orientation for most of the time, CSPs are considerable. By contrast, highly dynamic complexes exhibit small average CSP values, since the encounter state constitutes a considerable fraction of the complex. For the complex formed between Cc and NRP1, large average CSP values were observed (Figure 4B: left), suggesting the formation of a specific, well-defined complex with predominant relative orientations between the two.
The calculations of ΔδAvg were then used to map the interacting surface of Cc. For this, a previously-reported homology model of the 3D structure of A. thaliana Cc was used (44). Remarkably, residues of 15N Cc demonstrating CSPs were found to be located at the N-terminal and C-terminal domains, as well as at the domain surrounding the heme group (Figure 4B: right). It is also worth noting that Cc also faced the surface pattern surrounding the heme edge to recognize redox counterparts, including Cc oxidase (67), cytochrome bc1 (44,68) and L-galactono-1,4-lactone dehydrogenase (GALDH) (49), which remains in the cytochrome c6-involving photosynthetic complexes (50,69,70).
Moreover, interaction between Cc and NRP1 implicated several lysine residues, such as 12, 16, 21, 61, 70, 81, 87, 94 and 95 (Figure 4B: right), revealing the existence of attractive charge-charge interactions between Cc and NRP1. This latter finding is in agreement with previous works in which lysine residues were found to be involved in the binding of Cc to cytochrome bc1 (44,68,71,72), cytochrome c oxidase (67) and GALDH (49), as well as to partners in apoptosis like Apaf-1 (73) and SET/TAF-Iβ (40).
NRP1 uses its histone-binding domains to recognize Cc
In order to explore the Cc:NRP1 complex interface, NMR-restrained docking was performed with BiGGER, using the previously-mentioned NRP1 model and homology model for A. thaliana Cc (44). As restraints and input for the molecular docking, the CSP data obtained from NMR analysis for the Cc:NRP1 complex at the 1:0.5 ratio were used.
In the 500 NMR-based docking solutions for the Cc:NRP1 complex (Figure 5A: left), Cc geometric centres are depicted around the ribbon representation of NRP1 according to the comprehensive score recorded by the programme. Strikingly, nearly all models were positioned at the same NRP1 domain, that is, between acidic regions at the bottom of the earmuff domain (Figure 5A: left), in agreement with the well-defined Cc:NRP1 complex inferred from NMR data.
Figure 5.
NMR-based BiGGER molecular docking of Cc:NRP1 complex. (A) 500 solutions obtained of NMR-restrained docking for Cc:NRP1 complex. Centres of mass of Cc (blue spheres) and NRP1 ribbon (gold) are represented. Arrows indicate the two highest-score solutions. (B) Ribbon representations of the two best models, according to the overall score. NRP1 in complex shows the same orientation as in A, left.
Although all Cc probe solutions predicted by the restrained docking lied in the same region of NRP1, two differentiated clusters could be observed when NRP1 was rotated 90° (Figure 5A: right), both of which are in agreement with the binding sites observed in ITC and SPR measurements (Table 1; Supplementary Figures S5 and S6) and similar to those found for the complex formed by human Cc and SET/TAF-Iβ (40). Ribbon representations of the highest-scoring models in cluster 1 and 2 revealed heme-driven orientation of Cc and the tight contact between both proteins in the complex (Figure 5B and Supplementary Figure S11). Significantly, the interface of Cc and NRP1 matches those described for histone:NRP1 complexes (Supplementary Figure S11). In fact, with the orientation of the two Cc molecules towards the outer concave surface of NRP1 (Supplementary Figure S11A), their orientation is similar to that of the two H2A, H2B, H3 and H4 monomers at NRP1 histone-binding domains (Supplementary Figure S11B-D). Altogether, these results indicate that the same region of NRP1 recognizes both histones and Cc, thereby explaining the competition among the latter two in binding to NRP1.
DISCUSSION
In this work, we showed that NRP1 is mainly located at the chromatin fraction of A. thaliana cells following DNA breaks. Moreover, the 3D structure of a homology model of the NRP1 histone chaperone is compared with its human and yeast analogue from the NAP1 family. As shown, A. thaliana NRP1, human SET/TAF-Iβ and yeast NAP1 share significant sequence homology and fold in a similar manner. Curiously, despite the fact that four NAP1 genes (AtNAP1;1, AtNAP1;2, AtNAP1;3 and AtNAP1;4) are encoded in the A. thaliana genome, NRP1 demonstrated greater similarity in amino acid sequence and structural homology with human SET/TAF-Iβ. In fact, the two are endowed with similar overall 3D structures and both form headphone-shaped homodimers.
Current understanding of NAP1 proteins is that they bind to newly-synthesized H2A and H2B in the cytoplasm, transport the histones to the nucleus and deposit them on pre-formed H3:H4 complexes (74,75). The results of the present paper, which could not test H2A, show NRP1 to bind to core histones H2B, H3 and H4, while demonstrating a surprisingly greater affinity with respect to H2B and H4 than with respect to H3. These observations are in agreement with previous pull-down experiments in which NRP1 was reported as having binding preference for H2A and H2B, as opposed to H3 (32). However, as the authors of this latter study did not test NRP1 binding to H4, the possibility that NRP1 also acts as a chaperone for H3–H4 in addition to H2A–H2B cannot be ruled out. In fact, in this work NRP1 was found to bind H3–H4 in addition to H2A–H2B heterodimers, although the complex NRP1/H3–H4 was weaker. Moreover, despite the fact that NRP1 had already been proposed as a histone chaperone and its ability to bind to histones was already demonstrated (32), the current work shows for the first time NRP1's role in nucleosome assembly. This, in turn, can explain the role attributed to NRP1 in chromatin remodeling and the regulation of gene expression in A. thaliana (32).
Recently, the authors of the present study described the interaction between NRP1 and Cc in the nucleus of A. thaliana cells upon the induction of programmed cell death (PCD) (35). As this interaction takes place upon DNA damage, the question was investigated of whether NRP1's ability to bind to histones is affected upon binding to Cc. As inferred from NMR measurements, Cc binds to NRP1 and competes with histones for the NRP1 binding site. When comparing the KD value for the NRP1:Cc interaction (5 μM) with those for the interaction between NRP1 and core histones H2B (KD = 9.6 μM), H3 (KD = 20 μM) and H4 (KD = 11 μM), such competition between Cc and core histones to bind to NRP1 seems all the more plausible. Furthermore, such competition explains the suspension of NRP1 nucleosome assembly activity in the presence of Cc, as inferred from the supercoiling and MNase digestion assays. Binding competition and subsequent Cc-mediated hindrance of NRP1 chaperone activity is likely because NRP1 recognizes both histones and Cc at its earmuff domain, as deduced from molecular docking calculations. Notably, Cc exposes its heme crevice when interacting with NRP1, as is also the case in its interactions with partners in the mitochondria cytochrome bc1 (44) cytochrome c oxidase (67) and GALDH (49), as well as with histone chaperone SET/TAF-Iβ (40).
The moderate-to-low affinity observed for NRP1 interacting with histones (KD around 10–20 μM) and Cc (5 μM) is compatible with a regulation of those interactions based on concentration. The main regulatory factor in interaction processes is the binding partner concentration. In constitutive interactions leading to permanent complexes, KD is small (high affinity) but there is little regulation (as long as the two binding partners are present, they will form a complex). However, in transient interactions leading to non-permanent complexes, KD is larger (moderate-to-low affinity) with an intrinsic regulation according to the concentration of the binding partners. Other secondary factors modulating the interaction would be temperature, ionic concentration (for instance pH, salt ions), co-solute, or secondary ligands.
Histone chaperones NAP1L1, NAP1L4, HIRA, FACT, nucleolin, APLF, ASF1, CAF-1, DAXX and p400 have been reported as mediating histone reshaping following DNA lesions (18,25,76–82). Here we show that NRP1 gathers at the chromatin of A. thaliana cells upon DSBs induction. This is in agreement with a previous study showing NRP1 in the nuclear fraction of heat-shocked A. thaliana cells (34). In A. thaliana plants, NRP1 is overexpressed in the nucleus of heat shocked cells, resulting in increased survival (34). Since strong heat shock causes DNA breaks, it seems likely that the overexpression of NRP1 is related to DNA damage response. Taking into account the nucleosome assembly activity exhibited by NRP1 and its accumulation in the damaged chromatin, it is plausible to suggest that the mechanism by which NRP1 contributes to A. thaliana cellular resistance to DNA damage is related to its capability to influence chromatin structure.
As stated above, Cc also interacts with SET/TAF-Iβ in human nuclei when DNA damage occurs (39,40,83). Taking into account that NRP1 and SET/TAF-Iβ are targeted by Cc in both plant and human cells upon DNA damage (35,39), thereby blocking their ability to act as histone chaperones (40), it is tempting to hypothesize a similar role for such inhibition in both organisms.
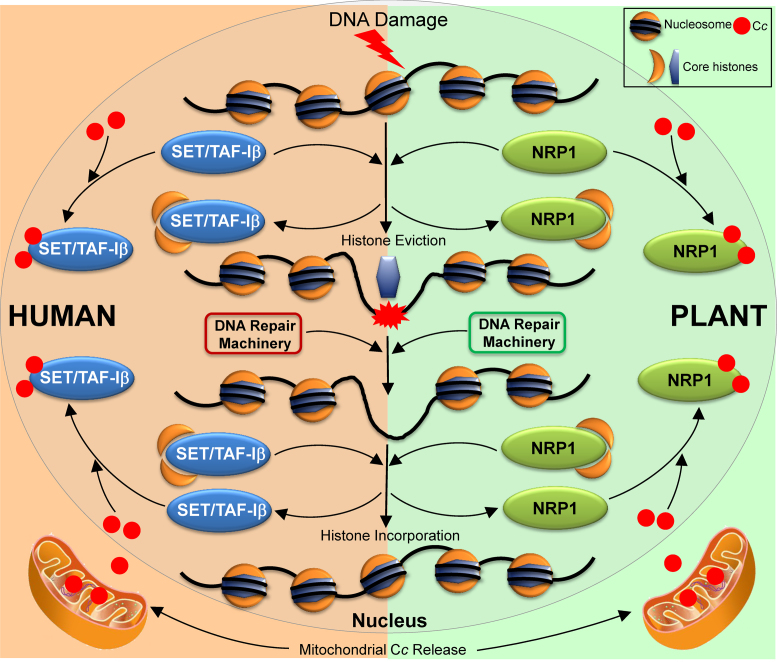
Consequently, it can be proposed that both NRP1 and SET/TAF-Iβ facilitate histone eviction by transiently disassembling nucleosomes at the damaged site, thereby making the damaged DNA accessible to repair factors (Figure 6). Conversely, after a DNA lesion is repaired, NRP1 and SET/TAF-Iβ might restore histones to DNA by reassembling nucleosome structure. In other words, NRP1 and SET/TAF-Iβ may promote localised chromatin remodeling both prior and following repair. However, when Cc is translocated into the plant or human cell nucleus, it targets NRP1 or SET/TAF-Iβ, respectively, and displaces core histones from the binding sites of the histone chaperones, thereby impairing histone eviction after DNA damage and nucleosome assembly following DNA damage response (Figure 6).
Figure 6.
Proposed model of nucleosome assembly/disassembly impairment by Cc upon DNA damage in human and plant cells. Scheme represents core histone eviction from nucleosomes and histone incorporation by SET/TAF-Iβ or NRP1. The two processes, histone eviction and incorporation, can be impaired by Cc upon nuclear translocation in response to DNA damage.
The comparative analysis of plant and mammalian responses to DNA damage reveals many similarities and some characteristic differences (19). In fact, some plant processes are closer to those in mammals than to those in other well-established eukaryotic models (19). In this work, analogous histone chaperones in plants and humans are shown to be targeted and blocked by Cc upon the induction of DNA damage. The findings presented shed light on the role of histone chaperones in the DNA damage response in plants and contribute to the understanding of the significance of Cc translocation into the nucleus, a process that occurs both in both plants and humans. Altogether, the results highlight the fact that the molecular mechanism regulating DNA damage response is highly conserved throughout evolution and they suggest that the function of Cc with respect to PCD is conserved across plants and humans.
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to thank Dr Tim Richmond (Institute of Molecular Biology & Biophysics, ETH Zürich) for providing plasmids encoding for H2A, H2B, H3 and H4 histones of X. laevis. We also thank Dr Attila Fehér (Hungarian Academy of Sciences) for providing the anti-A. thaliana NRP1 antibody; Dr Anne B. Britt (University of California) for the kind gift of the anti-A. thaliana γ-H2AX antibody; Dr Arturo Muga (University of the Basque Country) for providing the chicken erythrocytes H2A–H2B and H3–H4 histone proteins and Dr Bénédicte Desvoyes and Dr Crisanto Gutiérrez for critical advice.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Spanish Ministry of Economy and Competitiveness [BFU2015-71017/BMC MINECO/FEDER EU, and BFU2013-47064-P]; European Commission (European Regional Development Fund); the Regional Government of Andalusia [BIO198]; Ramon Areces Foundation, the NMR Facility at CITIUS (University of Seville); Biointeractomics Platform (Isla de la Cartuja Scientific Research Centre, Seville); Spanish Ministry of Education, Culture and Sports [FPU013/04373 to F.R.R.]. Funding for open access charge: Spanish Ministry of Economy and Competitiveness [BFU2015-71017/BMC MINECO/FEDER EU].
Conflict of interest statement. None declared.
REFERENCES
- 1.Chen I.P., Haehnel U., Altschmied L., Schubert I., Puchta H.. The transcriptional response of Arabidopsis to genotoxic stress – a high-density colony array study (HDCA). Plant J. 2003; 35:771–786. [DOI] [PubMed] [Google Scholar]
- 2.Rantong G., Gunawardena A.H.L.A.N.. Programmed cell death: Genes involved in signaling, regulation, and execution in plants and animals. Botany. 2015; 93:193–210. [Google Scholar]
- 3.Guyomarc'h S., Bertrand C., Delarue M., Zhou D.-X.. Regulation of meristem activity by chromatin remodeling. Trends Plant Sci. 2005; 10:332–338. [DOI] [PubMed] [Google Scholar]
- 4.Zhao Z., Yu Y., Meyer D., Wu C., Shen W.-H.. Prevention of early flowering by expression of flowering locus c requires methylation of histone H3 K36. Nat. Cell Biol. 2005; 7:1256–1260. [DOI] [PubMed] [Google Scholar]
- 5.Bäurle I., Dean C.. The timing of developmental transitions in plants. Cell. 125:655–664. [DOI] [PubMed] [Google Scholar]
- 6.Anjum N.A., Sofo A., Scopa A., Roychoudhury A., Gill S.S., Iqbal M., Lukatkin A.S., Pereira E., Duarte A.C., Ahmad I.. Lipids and proteins—major targets of oxidative modifications in abiotic stressed plants. Environ. Sci. Pollut. R. 2015; 22:4099–4121. [DOI] [PubMed] [Google Scholar]
- 7.Hossain M.A., Bhattacharjee S., Armin S.-M., Qian P., Xin W., Li H.-Y., Burritt D.J., Fujita M., Tran L.-S.P.. Hydrogen peroxide priming modulates abiotic oxidative stress tolerance: insights from ROS detoxification and scavenging. Front. Plant Sci. 2015; 6:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yakes F.M., Van Houten B.. Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Prco. Natl. Acad. Sci. U.S.A. 1997; 94:514–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Britt A.B. Molecular genetics of DNA repair in higher plants. Trends Plant Sci. 1999; 4:20–25. [DOI] [PubMed] [Google Scholar]
- 10.Bleuyard J.-Y., Gallego M.E., White C.I.. Recent advances in understanding of the DNA double-strand break repair machinery of plants. DNA Repair. 2006; 5:1–12. [DOI] [PubMed] [Google Scholar]
- 11.Gill S.S., Tuteja N.. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010; 48:909–930. [DOI] [PubMed] [Google Scholar]
- 12.Drury G.E., Dowle A.A., Ashford D.A., Waterworth W.M., Thomas J., West C.E.. Dynamics of plant histone modifications in response to DNA damage. Biochem. J. 2012; 445:393–401. [DOI] [PubMed] [Google Scholar]
- 13.Lans H., Marteijn J.A., Vermeulen W.. ATP-dependent chromatin remodeling in the DNA-damage response. Epigenet. Chromatin. 2012; 5:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adam S., Polo S.E.. Blurring the line between the DNA damage response and transcription: the importance of chromatin dynamics. Exp. Cell Res. 2014; 329:148–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lavelle C., Foray N.. Chromatin structure and radiation-induced DNA damage: from structural biology to radiobiology. Int. J. Biochem. Cell Biol. 2014; 49:84–97. [DOI] [PubMed] [Google Scholar]
- 16.Lai W., Li H., Liu S., Tao Y.. Connecting chromatin modifying factors to DNA damage response. Int. J. Mol. Sci. 2013; 14:2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donà M., Mittelsten Scheid O.. DNA damage repair in the context of plant chromatin. Plant Physiol. 2015; 168:1206–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cho I., Tsai P.-F., Lake R.J., Basheer A., Fan H.-Y.. ATP-dependent chromatin remodeling by cockayne syndrome protein b and NAP1-like histone chaperones is required for efficient transcription-coupled DNA repair. PLoS Genet. 2013; 9:e1003407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roldán-Arjona T., Ariza R.R.. Repair and tolerance of oxidative DNA damage in plants. Mut. Res.-Rev. Mutat. 2009; 681:169–179. [DOI] [PubMed] [Google Scholar]
- 20.Jasin M., Rothstein R.. Repair of strand breaks by homologous recombination. Cold Spring Harb. Perspect. Biol. 2013; 5:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seeber A., Hauer M., Gasser S.M.. Nucleosome remodelers in double-strand break repair. Curr. Opin. Genet. Dev. 2013; 23:174–184. [DOI] [PubMed] [Google Scholar]
- 22.Tsabar M., Haber J.E.. Chromatin modifications and chromatin remodeling during DNA repair in budding yeast. Curr. Opin. Genet. Dev. 2013; 23:166–173. [DOI] [PubMed] [Google Scholar]
- 23.Shaked H., Avivi-Ragolsky N., Levy A.A.. Involvement of the Arabidopsis SWI2/SNF2 chromatin remodeling gene family in DNA damage response and recombination. Genetics. 2006; 173:985–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ransom M., Dennehey B.K., Tyler J.K.. Chaperoning histones during DNA replication and repair. Cell. 2010; 140:183–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adam S., Polo S.E., Almouzni G.. Transcription recovery after DNA damage requires chromatin priming by the H3.3 histone chaperone HIRA. Cell. 2013; 155:94–106. [DOI] [PubMed] [Google Scholar]
- 26.Lario L.D., Ramirez-Parra E., Gutierrez C., Spampinato C.P., Casati P.. Anti-silencing function1 proteins are involved in ultraviolet-induced DNA damage repair and are cell cycle regulated by E2F transcription factors in Arabidopsis. Plant Physiol. 2013; 162:1164–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lavelle C., Foray N.. Chromatin structure and radiation-induced DNA damage: From structural biology to radiobiology. Int. J. Biochem. Cell Biol. 2014; 49:84–97. [DOI] [PubMed] [Google Scholar]
- 28.Mandemaker I.K., Vermeulen W., Marteijn J.A.. Gearing up chromatin. Nucleus. 2014; 5:203–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Polo S.E. Reshaping Chromatin after DNA Damage: The choreography of histone proteins. J. Mol. Biol. 2015; 427:626–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao J., Zhu Y., Zhou W., Molinier J., Dong A., Shen W.-H.. NAP1 family histone chaperones are required for somatic homologous recombination in Arabidopsis. Plant Cell. 2012; 24:1437–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dong A., Liu Z., Zhu Y., Yu F., Li Z., Cao K., Shen W.-H.. Interacting proteins and differences in nuclear transport reveal specific functions for the NAP1 family proteins in plants. Plant Physiol. 2005; 138:1446–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu Y., Dong A., Meyer D., Pichon O., Renou J.-P., Cao K., Shen W.-H.. Arabidopsis NRP1 and NRP2 encode histone chaperones and are required for maintaining postembryonic root growth. Plant Cell. 2006; 18:2879–2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bíró J., Farkas I., Domoki M., Ötvös K., Bottka S., Dombrádi V., Fehér A.. The histone phosphatase inhibitory property of plant nucleosome assembly protein-related proteins (NRPs). Plant Physiol. Biochem. 2012; 52:162–168. [DOI] [PubMed] [Google Scholar]
- 34.Bíró J., Domoki M., Fehér A.. NAP-related protein 1 (Atnrp1) overexpression increases the heat tolerance of Arabidopsis cells/plantlets. J. Plant Biochem. Physiol. 2013; 1, doi:10.4172/2329-9029.1000115. [Google Scholar]
- 35.Martínez-Fábregas J., Díaz-Moreno I., González-Arzola K., Janocha S., Navarro J.A., Hervás M., Bernhardt R., Díaz-Quintana A., De la Rosa M.Á.. New Arabidopsis thaliana cytochrome c partners: a look into the elusive role of cytochrome c in programmed cell death in plants. Mol. Cell. Proteomics. 2013; 12:3666–3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Díaz-Moreno I., García-Heredia J.M., Díaz-Quintana A., Teixeira M., De la Rosa M.A.. Nitration of tyrosines 46 and 48 induces the specific degradation of cytochrome c upon change of the heme iron state to high-spin. BBA - Bioenergetics. 2011; 1807:1616–1623. [DOI] [PubMed] [Google Scholar]
- 37.Ly H.K., Utesch T., Díaz-Moreno I., García-Heredia J.M., De La Rosa M.Á., Hildebrandt P.. Perturbation of the redox site structure of cytochrome c variants upon tyrosine nitration. J. Phys. Chem. B. 2012; 116:5694–5702. [DOI] [PubMed] [Google Scholar]
- 38.Guerra-Castellano A., Díaz-Quintana A., Moreno-Beltrán B., López-Prados J., Nieto P.M., Meister W., Staffa J., Teixeira M., Hildebrandt P., De la Rosa M.A. et al. Mimicking tyrosine phosphorylation in human cytochrome c by the evolved tRNA synthetase technique. Chem. Eur. J. 2015; 21:15004–15012. [DOI] [PubMed] [Google Scholar]
- 39.Martínez-Fábregas J., Díaz-Moreno I., González-Arzola K., Janocha S., Navarro J.A., Hervás M., Bernhardt R., Velázquez-Campoy A., Díaz-Quintana A., De la Rosa M.A.. Structural and functional analysis of novel human cytochrome c targets in apoptosis. Mol. Cell. Proteomics. 2014; 13:1439–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.González-Arzola K., Díaz-Moreno I., Cano-González A., Díaz-Quintana A., Velázquez-Campoy A., Moreno-Beltrán B., López-Rivas A., De la Rosa M.A.. Structural basis for inhibition of the histone chaperone activity of SET/TAF-Iβ by cytochrome c. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:9908–9913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Friesner J.D., Liu B., Culligan K., Britt A.B.. Ionizing radiation–dependent γ-H2AX focus formation requires ataxia telangiectasia mutated and ataxia telangiectasia mutated and Rad3-related. Mol. Biol. Cell. 2005; 16:2566–2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Villar C.B.R., Köhler C.. Hennig L, Köhler C. Plant chromatin immunoprecipitation. Plant Developmental Biology, Methods in Molecular Biology. 2010; 655, Springer; 401–411. [DOI] [PubMed] [Google Scholar]
- 43.Li J., Stern D.F.. DNA damage regulates Chk2 association with chromatin. J. Biol. Chem. 2005; 280:37948–37956. [DOI] [PubMed] [Google Scholar]
- 44.Moreno-Beltrán B., Díaz-Quintana A., González-Arzola K., Velázquez-Campoy A., De la Rosa M.A., Díaz-Moreno I.. Cytochrome c1 exhibits two binding sites for cytochrome c in plants. BBA - Bioenergetics. 2014; 1837:1717–1729. [DOI] [PubMed] [Google Scholar]
- 45.Luger K., Rechsteiner T.J., Flaus A.J., Waye M.M.Y., Richmond T.J.. Characterization of nucleosome core particles containing histone proteins made in bacteria. J. Mol. Biol. 1997; 272:301–311. [DOI] [PubMed] [Google Scholar]
- 46.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976; 72:248–254. [DOI] [PubMed] [Google Scholar]
- 47.Umehara T., Chimura T., Ichikawa N., Horikoshi M.. Polyanionic stretch-deleted histone chaperone cia1/Asf1p is functional both in vivo and in vitro. Genes Cells. 2002; 7:59–73. [DOI] [PubMed] [Google Scholar]
- 48.Piotto M., Saudek V., Sklenář V.. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR. 1992; 2:661–665. [DOI] [PubMed] [Google Scholar]
- 49.Hervás M., Bashir Q., Leferink N.G.H., Ferreira P., Moreno-Beltrán B., Westphal A.H., Díaz–Moreno I., Medina M., De la Rosa M.A., Ubbink M. et al. Communication between L–galactono–1,4–lactone dehydrogenase and cytochrome c. FEBS J. 2013; 280:1830–1840. [DOI] [PubMed] [Google Scholar]
- 50.Díaz-Moreno I., Diaz-Quintana A., Molina-Heredia F.P., Nieto P.M., Hansson O., De la Rosa M.A., Karlsson B.G.. NMR analysis of the transient complex between membrane photosystem I and soluble cytochrome c6. J. Biol. Chem. 2005; 280:7925–7931. [DOI] [PubMed] [Google Scholar]
- 51.Palma P.N., Krippahl L., Wampler J.E., Moura J.J.G.. BiGGER: A new (soft) docking algorithm for predicting protein interactions. Proteins: Struct. Funct., Bioinf. 2000; 39:372–384. [PubMed] [Google Scholar]
- 52.Eswar N., Eramian D., Webb B., Shen M.-Y., Sali A.. Structural Proteomics: High-Throughput Methods. 2008; Totowa: Humana Press. [Google Scholar]
- 53.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E.. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004; 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- 54.Niida H., Nakanishi M.. DNA damage checkpoints in mammals. Mutagenesis. 2006; 21:3–9. [DOI] [PubMed] [Google Scholar]
- 55.Lisby M., Barlow J.H., Burgess R.C., Rothstein R.. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004; 118:699–713. [DOI] [PubMed] [Google Scholar]
- 56.Dong Lang J., Smetana O., Sanchez-Calderon L., Lincker F., Genestier J., Schmit A.-C., Houlné G., Chabouté M.-E.. Plant γH2AX foci are required for proper DNA DSB repair responses and colocalize with E2F factors. New Phytol. 2012; 194:353–363. [DOI] [PubMed] [Google Scholar]
- 57.Sharma P., Jha A.B., Dubey R.S., Pessarakli M.. Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. J. Bot. 2012; 2012:26. [Google Scholar]
- 58.Dong A., Zhu Y., Yu Y., Cao K., Sun C., Shen W.H.. Regulation of biosynthesis and intracellular localization of rice and tobacco homologues of nucleosome assembly protein 1. Planta. 2003; 216:561–570. [DOI] [PubMed] [Google Scholar]
- 59.Park Y.-J., Luger K.. The structure of nucleosome assembly protein 1. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:1248–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Muto S., Senda M., Akai Y., Sato L., Suzuki T., Nagai R., Senda T., Horikoshi M.. Relationship between the structure of SET/TAF-I /INHAT and its histone chaperone activity. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:4285–4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Velazquez-Campoy A. Geometric features of the Wiseman isotherm in isothermal titration calorimetry. J. Therm. Anal. Calorim. 2015; 122:1477–1483. [Google Scholar]
- 62.Aguilar-Gurrieri C., Larabi A., Vinayachandran V., Patel N.A., Yen K., Reja R., Ebong I.- O., Schoehn G., Robinson C.V., Pugh B.F. et al. Structural evidence for Nap1-dependent H2A–H2B deposition and nucleosome assembly. EMBO J. 2016; 35:1465–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Williamson M.P. Using chemical shift perturbation to characterise ligand binding. Prog. Nucl. Magn. Reson. Spectrosc. 2013; 73:1–16. [DOI] [PubMed] [Google Scholar]
- 64.Bieri M., Kwan A.H., Mobli M., King G.F., Mackay J.P., Gooley P.R.. Macromolecular NMR spectroscopy for the non-spectroscopist: beyond macromolecular solution structure determination. FEBS J. 2011; 278:704–715. [DOI] [PubMed] [Google Scholar]
- 65.Worrall J.A.R., Liu Y., Crowley P.B., Nocek J.M., Hoffman B.M., Ubbink M.. Myoglobin and cytochrome b5: a nuclear magnetic resonance study of a highly dynamic protein complex. Biochemistry. 2002; 41:11721–11730. [DOI] [PubMed] [Google Scholar]
- 66.Volkov A.N., Ferrari D., Worrall J.A.R., Bonvin A.M.J.J., Ubbink M.. The orientations of cytochrome c in the highly dynamic complex with cytochrome b5 visualized by NMR and docking using HADDOCK. Prot. Sci. 2005; 14:799–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sakamoto K., Kamiya M., Imai M., Shinzawa-Itoh K., Uchida T., Kawano K., Yoshikawa S., Ishimori K.. NMR basis for interprotein electron transfer gating between cytochrome c and cytochrome c oxidase. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:12271–12276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Moreno-Beltrán B., Díaz-Moreno I., González-Arzola K., Guerra-Castellano A., Velázquez-Campoy A., De la Rosa M.A., Díaz-Quintana A.. Respiratory complexes III and IV can each bind two molecules of cytochrome c at low ionic strength. FEBS Lett. 2015; 589:476–483. [DOI] [PubMed] [Google Scholar]
- 69.Díaz-Moreno I., Díaz-Quintana A., Ubbink M., De la Rosa M.A.. An NMR-based docking model for the physiological transient complex between cytochrome f and cytochrome c6. FEBS Lett. 2005; 579:2891–2896. [DOI] [PubMed] [Google Scholar]
- 70.Díaz-Moreno I., Hulsker R., Skubak P., Foerster J.M., Cavazzini D., Finiguerra M.G., Díaz-Quintana A., Moreno-Beltrán B., Rossi G.-L., Ullmann G.M. et al. The dynamic complex of cytochrome c6 and cytochrome f studied with paramagnetic NMR spectroscopy. BBA - Bioenergetics. 2014; 1837:1305–1315. [DOI] [PubMed] [Google Scholar]
- 71.Speck S.H., Ferguson-Miller S., Osheroff N., Margoliash E.. Definition of cytochrome c binding domains by chemical modification: Kinetics of reaction with beef mitochondrial reductase and functional organization of the respiratory chain. Proc. Natl. Acad. Sci. U.S.A. 1979; 76:155–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.König B.W., Osheroff N., Wilms J., Muijsers A.O., Dekker H.L., Margoliash E.. Mapping of the interaction domain for purified cytochrome c1 on cytochrome c. FEBS Lett. 1980; 111:395–398. [DOI] [PubMed] [Google Scholar]
- 73.Yu T., Wang X., Purring-Koch C., Wei Y., McLendon G.L.. A mutational epitope for cytochrome c binding to the apoptosis protease activation factor-1. J Biol. Chem. 2001; 276:13034–13038. [DOI] [PubMed] [Google Scholar]
- 74.Haushalter K.A., Kadonaga J.T.. Chromatin assembly by DNA-translocating motors. Nat. Rev. Mol. Cell Biol. 2003; 4:613–620. [DOI] [PubMed] [Google Scholar]
- 75.Polo S.E., Almouzni G.. Chromatin assembly: a basic recipe with various flavours. Curr. Opin. Genetics Dev. 2006; 16:104–111. [DOI] [PubMed] [Google Scholar]
- 76.Dinant C., Ampatziadis-Michailidis G., Lans H., Tresini M., Lagarou A., Grosbart M., Theil A.F., van Cappellen W.A., Kimura H., Bartek J. et al. Enhanced chromatin dynamics by FACT promotes transcriptional restart after UV-induced DNA damage. Mol. Cell. 2013; 51:469–479. [DOI] [PubMed] [Google Scholar]
- 77.Kobayashi J., Fujimoto H., Sato J., Hayashi I., Burma S., Matsuura S., Chen D.J., Komatsu K.. Nucleolin participates in DNA double-strand break-induced damage response through MDC1-dependent pathway. PLoS ONE. 2012; 7:e49245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mehrotra P.V., Ahel D., Ryan D.P., Weston R., Wiechens N., Kraehenbuehl R., Owen-Hughes T., Ahel I.. DNA repair factor APLF is a histone chaperone. Mol. Cell. 2011; 41:46–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tanae K., Horiuchi T., Matsuo Y., Katayama S., Kawamukai M.. Histone chaperone Asf1 plays an essential role in maintaining genomic stability in fission yeast. PLoS ONE. 2012; 7:e30472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Polo S.E., Roche D., Almouzni G.. New histone incorporation marks sites of UV repair in human cells. Cell. 2006; 127:481–493. [DOI] [PubMed] [Google Scholar]
- 81.Lacoste N., Woolfe A., Tachiwana H., Garea A.V., Barth T., Cantaloube S., Kurumizaka H., Imhof A., Almouzni G.. Mislocalization of the centromeric histone variant CenH3/CENP-A in human cells depends on the chaperone DAXX. Mol. Cell. 2014; 53:631–644. [DOI] [PubMed] [Google Scholar]
- 82.Xu Y., Ayrapetov M.K., Xu C., Gursoy-Yuzugullu O., Hu Y., Price B.D.. Histone H2A.Z controls a critical chromatin remodeling step required for DNA double-strand break repair. Mol. Cell. 2012; 48:723–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Martínez-Fábregas J., Diaz-Moreno I., Gonzalez-Arzola K., Diaz-Quintana A., De la Rosa M.A.. A common signalosome for programmed cell death in humans and plants. Cell Death Dis. 2014; 5:e1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.