Abstract
L1 elements represent the only currently active, autonomous retrotransposon in the human genome, and they make major contributions to human genetic instability. The vast majority of the 500 000 L1 elements in the genome are defective, and only a relatively few can contribute to the retrotransposition process. However, there is currently no comprehensive approach to identify the specific loci that are actively transcribed separate from the excess of L1-related sequences that are co-transcribed within genes. We have developed RNA-Seq procedures, as well as a 1200 bp 5΄ RACE product coupled with PACBio sequencing that can identify the specific L1 loci that contribute most of the L1-related RNA reads. At least 99% of L1-related sequences found in RNA do not arise from the L1 promoter, instead representing pieces of L1 incorporated in other cellular RNAs. In any given cell type a relatively few active L1 loci contribute to the ‘authentic’ L1 transcripts that arise from the L1 promoter, with significantly different loci seen expressed in different tissues.
INTRODUCTION
Mobile genetic elements make up approximately half of the human genome (1). Long Interspersed Element-1 (L1) retroelements are the only currently active, autonomous family of elements in humans. They make up approximately 17% of the mass of the genome and also drive amplification of non-autonomous elements, such as Alu and SVA (2–5), through an RNA-mediated mechanism. L1 elements continue to insert new copies in the human genome and to generate germ line genetic diseases (6). Recent studies have suggested that not only are L1 elements expressed in many somatic tissues (7) but they are also likely to retrotranspose in somatic tissues throughout the life of an individual (8). This would suggest that they can contribute to genetic instability in somatic tissues that may have implications for human diseases such as cancer and potentially various forms of age-related degeneration (9). Although some tumors support only very low levels of de novo L1 mobilization, a broad range of epithelial tumors have high levels of de novo L1 insertions that are likely to contribute to tumor progression (10–14).
Most of the 500 000 L1 elements are 5΄ truncated at the time of insertion, leaving approximately 5000 full-length elements that contain the internal promoter that is present within the L1 5΄UTR (15). Of those loci that are full length, less than 100 have the capability of coding for retrotranspositionally competent L1 elements and only 5–20 L1 elements in a genome are thought to be potentially responsible for most of the ongoing L1 activity (15,16). These ‘hot’ L1 elements are almost all polymorphic in the human population, meaning that different individuals have different numbers and composition of the ‘hot’ L1 elements. Thus, there is likely to be variable L1 activity in different individuals (9,15). This is further supported by recent analyses of de novo L1 inserts in human tumors that suggest that only a very few L1 loci contribute a large portion of the de novo L1 inserts in a given tumor and that the subset of these contributing loci differs among different types of tumors (10,14,17,18). Thus, an assessment of the expression and activity of these ‘hot’ L1 loci is critical to understanding their impact on genomic instability.
L1 element amplification requires an mRNA and the expression of two proteins encoded in this bicistronic RNA. One protein, ORF1p, is an RNA binding protein with RNA chaperone activity (19). The second protein, ORF2, contains both endonuclease and reverse transcriptase enzymatic activities necessary for the process of L1 integration into genomic DNA (20). Both proteins show a cis preference for their parental RNA, i.e. they preferentially incorporate the specific RNA molecule from which they were translated into a new genomic site (21). In addition to being critical to L1 integration into a new genomic location, the endonuclease activity of ORF2p is capable of generating DNA double-strand breaks that may further contribute to various forms of genomic instability (22).
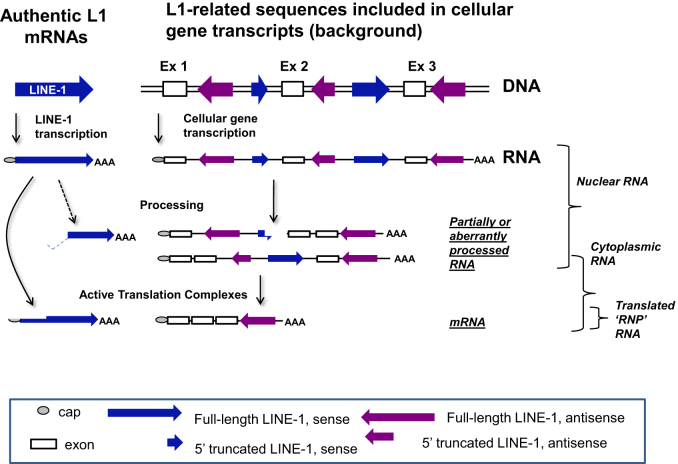
Because L1 elements utilize an RNA intermediate in their amplification process, their promoter is critical to the formation of the full-length transcripts. These authentic, full-length L1 RNAs are essential for L1 amplification. Even if an L1 locus is potentially active as defined using in vitro retrotransposition testing (15), it will not have any impact if it is transcriptionally silent. There is also a vast excess of promoterless fragments of L1 elements spread throughout the genome that can be incorporated into other cellular RNAs during transcription (see Figure 1). It has been shown that any RNA sample containing the nuclear component is heavily contaminated with introns and that cytoplasmic preparations largely, but not completely, remove the intron containing RNAs (23). In particular, there are both truncated and full-length L1-related sequences located in both orientations within the introns of many genes, as well as some 3΄ non-coding exons (24). Thus, we would expect whole-cell RNAs to include many of these L1-related sequences within their primary transcripts (25).
Figure 1.
Sources of L1-related sequences in RNA-Seq studies. L1 elements may be transcribed from their own promoter present in ∼5000–6000 full-length L1 loci in the genome (shown on the left as ‘authentic L1 elements and their mRNAs). In addition, many more truncated L1 sequences existing in and around cellular genes and these can also be incorporated into mRNAs, but not in a way that is relevant to the retrotransposition process. In the nucleus (and therefore whole-cell RNA) there are transcripts of both types. The authentic L1 mRNAs are essentially all in the sense orientation, while L1-related sequences in other genes can be represented in either orientation in the RNA. A small portion of the L1 mRNAs are processed (30,32), but the majority are full length and unprocessed other than polyadenylation and capping. Many fragments of L1 that are present in introns of genes, are eliminated from the RNA as it is spliced, resulting in a decrease of L1-related RNAs in the cytoplasm. However, even in the cytoplasm there is the potential for partially processed mRNAs or mRNAs that include L1-related sequences in the mature mRNA.
Human L1 expression has been analyzed by looking at ORF1p expression (14,26,27), as well as mRNA expression detected by northern blots (7,25). Neither of those approaches, however, allows an assessment of which L1 loci are expressed and whether the expressed L1 loci have potential for activity. Another study utilized the observation that some L1 transcripts extend into downstream flanking sequences (28), allowing for the utilization of quantitative PCR to test expression of individual L1 loci (29). This approach has recently been adapted to RNA-Seq studies to measure downstream transcription from the L1 Ta1 subfamily members that make a 3΄ extended RNA product (14). However, none of the existing methods allow a comprehensive study of actively transcribed L1 loci. The bioinformatic approaches developed here utilize strand-specific RNA-Seq analyses, and a high throughput 5΄ RACE analysis to differentiate between transcripts related to authentic L1 transcription from L1-related sequences that are not produced by L1 promoters (see Figure 1). We show that these complementary methods have different strengths, but together comprehensively map authentic L1 transcripts. Our results generated from applying these approaches demonstrate that a significant subset of L1 elements express at very low levels, but that most of the expression comes from a few dozen loci. Our methods detect expression from larger numbers of active L1 loci than previously reported (14), critically finding that many L1 loci do not make stable 3΄ extended RNAs and that the spectrum of expressed L1 loci varies between cell types.
MATERIALS AND METHODS
RNA preparation and RNA-Seq data generation
Cell lines
Transfection conditions
A total of 4–5 million HeLa or NIH 3T3 cells were seeded in a T75 flask. Transfection was carried out 18–20 h after seeding with 6 µg of plasmid expressing a full-length L1 element driven by a CMV promoter (21) or by its own promoter (30,31) using 24 µl of lipofectamine (ThermoFisher)(in a total of 100 µl of serum free media) and 12 µl of plus reagent (in a total of 200 µl of serum free media). The transfection solution was replaced by culture media 3–4 h after transfection.
RNA preparation
Total RNA was harvested using Trizol extraction 24 h posttransfection as previously described (32). Specifically, a T75 flask of transfected (or a confluent flask of untransfected cells) was scraped into 7.5 ml of Trizol reagent, combined with 4.5 ml of chloroform in a 15 ml conical tube and centrifuged for 30 min at 4C at 4000 rpms. The collected supernatant was combined with 4 ml of chloroform, and centrifuged for 10 min at 4C at 4000 rpms. The resulting supernatant is precipitated with 4 ml of isopropanol overnight in −80°C, centrifuged for 30 min at 4C at 4000 rpms, washed with ethanol and used for further RNA analysis.
RNA from cytoplasmic ribonucleoprotein particles (RNPs)
Cells were washed 3x with ice-cold phosphate buffered saline (PBS) and scraped in 5 ml of ice-cold PBS. Cells from two confluent T75 flask were combined in a 15 ml conical tube. Following centrifugation (5 min 3000 ×g at 4°C), supernatant was removed, pelleted cells were resuspended in 500 µl of lysis buffer (1.5 mM KCl, 2.5 mM MgCl2, 5 mM Tris-Cl pH7.5, 1% deoxycholic acid, 1% Triton X-100, 10 µl/ml protease inhibitor cocktail (Sigma), 20 µl/ml of RNAsin (Promega) and incubated on ice for 5 min. Lysed cells were centrifuged (5 min, 3000 ×g at 4°C) and the supernatant was layered on top of a sucrose step gradient, 8.5% top and 17% bottom (8.5%/17% sucrose, 80 mM NaCl, 5 mM MgCl2, 20 mM Tris-Cl, 1 mM DTT, 10 µl/ml protease inhibitor cocktail and 5 µl of RNAsin was added to each tube) followed by ultra-centrifugation for 2 h at 36 500 ×g at 4°C. The resulting pellet was resuspended in 7.5 ml of Trizol. Phenol/chloroform RNA extraction was performed as previously described (7). RNA pellets were resuspended in 50–100 µl of RNAse-free water. RNA samples (10–15 µg of each) were DNAse treated twice using RNase-Free DNase (Qiagen). RNA quality was analyzed by fractionation using agarose gel electrophoresis and an Agilent Bioanalyzer.
RNA sequencing
RNA samples were submitted to the University of Wisconsin Genomics Core for selection of polyadenylated RNAs and TruSeq stranded mRNA library preparation. Samples were pooled in groups of 3–5, and applied to a single lane of an Illumina HiSeq 2000 instrument. Data were sorted based on barcodes attached to each individual sample and analyzed in the various RNA-Seq strategies. Fastq data from all of our RNA-Seq studies has been submitted to the NCBI SRA database under the SRA accession #SRP083758.
We also searched existing databases for RNA-Seq data sets that were generated in a strand-specific manner from polyadenylated cytoplasmic RNAs. The only sample we found that met our criteria involved a HEK293 cell sample in which the cytoplasmic RNA was generated following digitonin treatment of the cells to release cytoplasmic RNAs (33). These data were downloaded as fastq files as SRR1275413 from the Gene Expression Omnibus (GEO) traces website and subjected to the same bioinformatic analyses.
Correlation analysis
Correlation analysis as a statistical method was used to examine the association between different variables. The study hypothesis of interest was tested at the 5% level of significance. All analyses, summaries and listing was performed using SAS software.
Bioinformatic analysis
RNA-seq analysis
To test the nature of L1 transcripts from cells transfected with an L1.3 transfection vector, RNA samples were initially aligned to the L1.3 sequence utilizing STAR (34). STAR provides rapid alignment and is excellent for detecting splicing events. Default conditions were utilized for the alignments of RNA from L1.3 transfected cells because near perfect matches are expected, but we also compared the HeLa cell endogenous RNA where mismatches up to 25% were allowed in the alignments.
Our alignment strategy for RNA-Seq data to the genome for endogenous expression studies was designed to keep in mind the limitations of short-read sequencing technologies in alignments to repetitive sequences. Thus, we have aimed to eliminate any alignments where the best match of a paired-end sequence maps equally well to two or more genomic regions. We have utilized our paired-end, stranded RNA-Seq reads from various cell lines in the BOWTIE alignment program paired with Samtools to create a sorted bam file for output with the command line described in the Supplementary Methods. The resulting bam alignment is then separated into those reads that were transcribed from the top strand of the genome versus the bottom strand using a command line shown in the Supplementary Methods. The strand separation is arbitrary in the sense that it is relative to the genome and not relative to the individual orientation of the L1 elements in the genome.
The bedtools INTERSECT command was then used to map the overlap between the strand-separated files and the oriented annotation .gtf file for either the full-length L1 elements in the reference genome, or for the sequences flanking the polymorphic full-length L1 insertions (see below for annotations) and to count the number of reads mapping to each location. This provides a table of the number of reads that map to each of the annotated L1 loci along with the orientation of the RNA relative to the L1 element.
5΄ Race
We utilized the SMARTer® PCR cDNA Synthesis Kit (Clontech) that relies on the ability of the reverse transcriptase to add several untemplated C residues preferentially at the site of the RNA cap (35). The 1237a primer (5΄-GGCTCCTGAGGCTTCTGCAT-3΄) used for first-strand cDNA synthesis was chosen because it is an excellent match to subfamilies PA1 through PA6, although it would work less well on the very old L1 subfamilies. We then primed second strand synthesis with the SMARTer II A Oligonucleotide (proprietary primer in the kit which primes on the untemplated C residues). This was followed with polymerase chain reaction (PCR) using the 1237a primer and 5΄ PCR primer II (primer to the proprietary second strand primer). In this case, the samples were fractionated on a 1% agarose gel and the approximately 1200 bp band for full-length L1 transcripts was isolated and submitted to the Arizona Genome Institute (http://www.genome.arizona.edu/modules/publisher/item.php?itemid=29) for PacBio sequencing.
PacBio analysis
Because Bowtie cannot handle the long reads from PacBio, these reads were aligned with the GMAP aligner using a command line described in more detail in the Supplementary Methods. The key elements include switches to eliminate the ability to look for RNA splicing, and a switch to only allow a single best alignment. The output from this preliminary alignment was then sorted to obtain the alignments that matched a genomic locus with at least a 99% identity. Those reads were then remapped to the genome and the alignment handled similarly to the RNA-Seq approach with Bedtools Intersect to identify the reads that map to individual full-length L1 loci.
Annotations
RepeatMasker annotations for LINE elements were downloaded from UCSC as a .gtf file. This includes all LINE-related sequences. However, our primary focus was to identify transcription from the small percentage of elements that represent full-length elements that can transcribe a potentially active L1 RNA. In order to identify the most likely group of full-length elements in the hg19 reference genome, we utilized the first 300 bp of the L1.3 full-length L1 element that encompasses the primary region of the L1 promoter to carry out a BLAST search of the human genome. This promoter region is required to generate the authentic, retrotransposition-competent L1 RNAs. We detected approximately 6000 L1 promoter regions in the human genome. We then assumed that ∼6000 bp downstream of that promoter would be associated with the full-length elements. We also reasoned, however, that older L1 elements may have been disrupted or rearranged and therefore would not have a full L1 sequence encompassed in that 6000 bp. Therefore, we carried out a bedtools INTERSECT comparison of the RepeatMasker LINE annotation and our promoter-based annotation and found ∼5000 full-length elements present in the HG19 reference genome that were relatively contiguous with an L1 element. This subset was utilized as a .gtf file in the bioinformatics analyses to identify full-length L1 loci.
We also created annotations for polymorphic L1 elements. These elements were collected from two public databases dbRip (http://dbrip.org) and eul1db (http://eul1db.unice.fr/) (accessed December, 2014). For each element, the two nucleotides flanking the insertion point were obtained from database entries, and 1 kb flanking sequences upstream and downstream of the insertion point were annotated as flanking polymorphic L1 insertions. For those elements that were present in the hg19 reference sequence and determined to be absent among some members of the population, full sequence detail was available to infer full length status. However, due to incomplete sequence information for many L1 polymorphism database entries (i.e. one junction was not sequenced) the full length status of many reported polymorphic elements could not be established; only the position at which the insertion occurred.
RESULTS
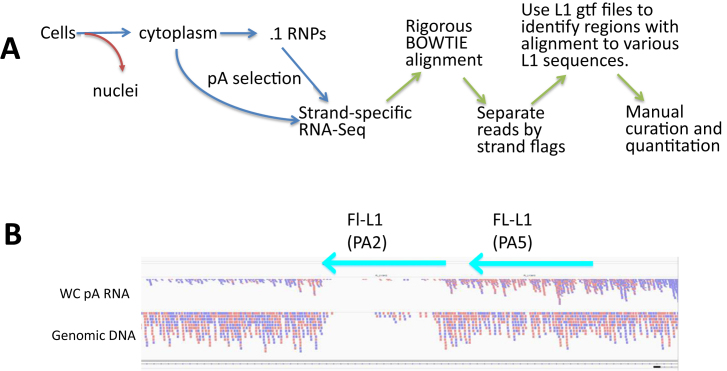
Because most L1 insertional activity is dominated by a relatively few active L1 loci (10,36), we wished to develop RNA-Seq approaches that would allow high throughput analysis of expression from the specific L1 loci. These analyses are complicated by the background from the 99% of L1 fragments in the genome co-transcribed within other genes (24) that cannot contribute to authentic L1 expression or activity (Figure 1). Our primary RNA-Seq approach (described in the RNA-Seq mapping to Individual Full-Length L1 Loci, below) uses BOWTIE in a rigorous alignment that allows us to separate RNA-Seq reads that align preferentially to a unique site in the genome to fragments of L1 versus the full-length L1 elements (Figure 2A and Materials and Methods). We complemented this global RNA-Seq approach with a long 5΄-RACE procedure (see section below) to allow an analysis of transcripts initiating at the beginning of L1 elements to provide the most inclusive analysis of L1-specific RNA expression from individual L1 loci.
Figure 2.
RNA-Seq alignment and mappability. (A). The strategy. Our procedure begins with the preparation of the L1 RNA. Although we carry out some analyses on whole-cell RNA preparations, our best results come from isolating either cytoplasmic RNA or mRNA-containing RNPs (51). This eliminates the massive level of intron-related L1 sequences that are present in many primary nuclear transcripts (see Figure 1, (23)). We then carry out polyA selection because the active L1 mRNAs should be polyadenylated and this will remove contaminating RNAs, such as rRNA. The mRNA fraction is then sequenced using a 100 bp, paired-end, strand-specific sequencing protocol that allows us to distinguish transcription in the sense direction through any L1 locus from transcripts in the antisense direction. Our BOWTIE alignment protocol (see Supplementary Methods) is then designed to only keep alignments where each pair of reads (200 bp total) aligns better to one location than any other in the genome. Even perfect alignments are eliminated if there is more than one perfect match in the genome. We then separate the strands of the BAM file that arises from the BOWTIE alignment. We then use SamTools Intersect in conjunction with an annotation file (.gtf) that has the coordinates of all of the full-length L1 elements in the reference genome. This counts the paired-end reads that map specifically to each of the loci in a format that can then be manipulated in excel. These excel outputs are then used to identify the loci with the highest reads which are then curated in the IGV browser relative to the mapped reads in the original BAM file. (B) Mappability. In order to assess just the BOWTIE alignment portion of the protocol, we utilized a paired-end sequence data set from HeLa genomic DNA to align with the same parameters to the reference genome. Note that the pink and blue lines correspond to paired-end reads that were oriented in different orientations during the sequencing. This is expected with random genomic sequencing. Two full-length (>6000 bp) L1 elements are marked with their subfamily designation (PA2 and PA5). The older PA5 element has accumulated sufficient mutations that reads mapped to it almost as well as to the unique flanking regions. The PA2 had much poorer mapping because it is closer to consensus and more of the read pairs multimapped in the genome with equal quality alignments.
One critical feature of all of our studies is that the sequencing is carried out in a strand-specific manner, allowing identification and discrimination of transcripts that are in the sense orientation relative to L1 from those in the antisense orientation. This allows us to estimate the level of background originating from L1-related sequences co-transcribed within other cellular transcripts (Figure 1), because those sequences will be represented in both the sense and antisense directions.
With this in mind we first thought to validate the nature of the L1 transcripts expected to be seen in 100 bp paired-end RNA-Seq performed using different RNA preparations from NIH3T3 and HeLa cells transfected with human L1 expression plasmids (7,21). The L1 expression from these plasmids is supported by use of the endogenous L1 promoter found in the 5΄ UTR of L1 or by a CMV promoter included just upstream of the 5΄ UTR (20). Mouse NIH3T3 cells were chosen to eliminate the background signal from endogenous L1 elements in this initial study, because the mouse L1 sequences are easily differentiated from the human L1 sequences generated from the L1 expression plasmids. Alignment of the RNA-Seq reads to the L1 consensus sequence using STAR (34), demonstrated that L1 elements transfected into mouse NIH 3T3 cells showed expression patterns expected based on previous analysis of L1 expression with northern blots (30,32) (see Supplementary Figure S1). The STAR alignment detected almost exclusive expression of the sense strand L1 transcripts from the plasmid-based expression vectors, with the expected moderate levels of splicing (see Supplementary Table S1) and premature polyadenylation (see Supplementary Figure S1 for discussion of details). Transfection of L1 expression plasmids into HeLa cells showed essentially the same results, but with some background from the endogenous L1 elements as indicated by the presence of antisense reads (Supplementary Figure S1, purple). Supplementary Figure S2 shows the results of RNA-Seq analysis of L1 element-related RNAs extracted from HeLa cells without transfection. This approach confirmed that there is extensive background from L1 sequences unrelated to the L1 replication cycle in the whole-cell RNA preparations as evidenced by the significant presence of antisense reads (purple) and that much, but not all, of this background was eliminated by the use of cytoplasmic RNA preparations (Supplementary Figure S2; compare WC with cyto-RNP). The ability to eliminate these contaminating, irrelevant L1 sequences combined with the development of a rigorous alignment strategy (below) allowed us to proceed with the analysis of endogenous L1 expression.
RNA-Seq mapping to Individual Full-Length L1 loci
Mappability
Each L1-related read can align to many genomic loci. However, due to sequence variation between individual L1 elements a paired-set of RNA reads (DNA or RNA reads) will often align to a single L1 locus in the genome better than any other locus. These uniquely best alignments represent the most likely genomic location responsible for that particular L1 read pair. The only real exception to this ability to obtain a ‘uniquely better’ alignment is for reads that are a perfect match to the L1 consensus and therefore match many loci equally. Because the full-length, ‘authentic’ L1 transcripts that are capable of retrotransposition are expected to be collinear with the genomic DNA (unspliced), we chose a mapping approach that works best with linear, unspliced alignments. For this reason, we used BOWTIE for our alignments (see below) using commands to only accept reads mapped as a concordant pair to one location in the genome better than any other (see Materials and Methods and Supplementary Methods for BOWTIE commands). Younger L1 elements have large regions of identity with the consensus and one another. Therefore, with these stringent alignment conditions fewer (DNA or RNA) reads containing L1 sequences were expected to map ‘uniquely’ to a single young L1 locus, reducing what we term the ‘mappability’ of the locus. This reduction is a direct result of the elimination of the paired-end L1 reads that did not map uniquely to a specific L1 locus (i.e. termed ‘multimapped’) and, therefore, would be excluded under these stringent BOWTIE settings. Without appropriate controls for mappability of individual L1 loci this approach would result in the underestimation of expression from L1 loci with poor ‘mappability’. Thus, an assessment of individual L1 loci mappability is an important control for the correct calling of their expression when using RNA-Seq-based approaches for analysis of endogenous L1 expression.
As a first test to validate the ‘mappability’ (see Materials and Methods for details) we utilized our BOWTIE alignment approach on the HG19 reference genome with Illumina paired-end sequence reads generated from HeLa genomic DNA (Figure 2B and Supplementary Figure S3). If all regions were equally mappable, we would expect a fairly even genomic coverage using this random sequencing approach. In contrast this example shows the results of alignment to a well- and a poorly-mapped full-length L1 locus (PA5 and PA2, respectively) confirming the mappability concept described above (Figure 2B). Almost 35% of the paired-end genomic reads mapped uniquely to genomic regions with an annotation including ‘L1’ in the REPEATMASKER LINE annotations (downloaded from the UCSC browser). Less than 3% of overall DNA reads ‘multimapped’ to multiple L1 loci at identical levels of similarity under these stringent conditions. These multimapped DNA reads represented one-tenth the level of the total LINE-mapped reads. Thus, >90% of L1 genomic reads were able to map to a single location that was better than any other possible genomic locus. Of these uniquely mapping DNA reads, less than one percent of reads mapped to full-length L1 loci present in the reference genome, with the rest mapping to annotated ‘fragments’ of L1 sequences dispersed throughout the genome. This is reasonable given that there are about 5000 loci of 6000 bases in length (3 × 107 bases total) representing about 1% of the human genome. Furthermore, assessment of the number of DNA reads mapped to individual L1 loci showed that most L1 loci have sufficient sequence regions that deviate from the consensus to allow a portion of L1 reads to map uniquely (Supplementary Figure S3B and C). There was an almost linear relationship between L1 subfamily and the number of mapped reads. As expected, the L1s with the most mapped reads were older L1 elements with high levels of sequence divergence relative to the consensus. Similarly, a small number of L1s that had very few reads mapped to them were mostly HS and some PA2 subfamily members with near-consensus sequences (Figure 2B, genomic DNA).
In order to assure that our alignment algorithm was only reporting reads that had a single, best mapping location (termed uniquely mapping in future discussion) in the reference genome we used two assessment criteria. One indication that the method was robust was that out of the 1.5 million L1-mapping reads, only 59 reads aligned to the Y chromosome (Supplementary Figure S3B and C). Because the genetic makeup of HeLa is female, we would expect very little Y chromosome mapping as was seen. Secondly, we carried out manual BLAST alignments of 24 random L1 paired-end mappings chosen from the uniquely mapped paired reads identified by our alignment strategy with stringent settings. All 24 reads mapped to the same location assigned by our BOWTIE mapping. Thus, the best unique genomic match chosen by BOWTIE was also the best location for that pair in the reference genome using BLAST scoring criteria.
RNA-seq mapping
We chose to initiate our analysis of endogenous L1 expression with HeLa cells because they are extensively studied relative to L1 retrotransposition. Analysis of RNA-Seq reads generated using polyA-selected whole-cell RNA from HeLa cells for endogenous L1 expression showed that only 2.7% of the read pairs mapped to L1 annotated sequences (RepeatMasker) in the genome, with as little as 0.03% of reads mapping to the approximately 5000 full-length L1 loci (see Materials and Methods for annotation)(Supplementary Figure S4 and Table 1). This result showed that there is a very strong depletion of L1 sequences in the polyadenylated whole-cell RNA relative to their genomic abundance (17%, (1)). This depletion was even more pronounced in the polyA-selected cytoplasmic RNP fraction with 0.3–1.1% from two repetitions of HeLa (Table 1) of total reads mapping to genomic regions annotated as L1 versus 2.7% for the whole cell. Only 0.002–0.005% (versus 0.03%) of reads mapped to full-length L1 elements in the cytoplasmic preparation (Table 1 HeLa Cyto RNP1 and 2 and Supplementary Figure S4). Even in this cytoplasmic fraction, where the enrichment of authentic L1 transcripts should be the highest, we observe that only 1% of reads mapped to genomic L1 sequences were aligning to the full-length L1 loci and the rest were mapping to truncated L1 fragments (Supplementary Figure S4, graph, red versus black). In all of these analyses, a portion of the L1-related reads could not be mapped uniquely. These multimapped reads discarded by our alignment strategy represented approximately 15% of the number of reads that aligned uniquely, which was determined by alignment of these multimapped reads to the L1 consensus sequence (shown in Table 1 as FL-L1 reads). Consistent with the results observed using the polyA-selected cytoplasmic RNP fraction, we observed a similarly high rate of unique mappability relative to multimapped reads discarded by our approach when analyzing reads generated from the HeLa whole-cell polyA RNA data set. Approaches to deal with genomic loci with poor mappability will be discussed below.
Table 1. RNA-Seq reads mapped.
| HeLa WC | HeLa cyto RNP 1 | HeLa cyto RNP 2 | XPD WC | XPD cyto RNP | HEK293 cyto RNP | Akata cyto RNP | |
|---|---|---|---|---|---|---|---|
| Total Reads | 55.0 M | 99.5 M | 14.8 M | 51.3 M | 42.9 M | 55.5 M | 44.2 M |
| L1 Reads* | 1.5 M (2.7%) | 300 K (0.3%) | 163 K (1.1%) | 1.14 M (2.3%) | 584 K (1.4%) | 694 K (1.3%) | 1.0 M (2.3%) |
| FL-L1 Reads** | 17.1K (.03%) | 2.2 K (.002%) | 815 (.005%) | 9010 (.018%) | 3359 (.007%) | 7376 (.013%) | 9096 (.02%) |
| FL-L1 sense | 10 039 (.018%) | 1619 (.0016%) | 616 (.004%) | 3740 (.007%) | 2214 (.005%) | 6111 (.011%) | 4657 (.01%) |
| FL-L1 antisense | 7020 (.012%) | 536 (.0005%) | 199 (.001%) | 5270 (.01%) | 1145 (.002%) | 1265 (.002%) | 4439 (.01%) |
| Sense/anti*** | 1.43 | 3.02 | 3.11 | 0.71 | 1.93 | 4.8 | 1.05 |
*Percent of total reads that map to the HG19-LINE annotation uniquely.
**Percent of LINE-mapped reads that map to the full-length L1 elements.
***ratio of sense to antisense reads mapped.
Limiting our analysis only to the uniquely mappable, paired-end reads that align to the full-length L1 loci, we observe a 1.4-fold enrichment for RNAseq reads that were generated from the sense strand of the endogenous L1 element relative to the antisense strand in whole-cell Hela reads (Table 1). The HeLa cytoplasmic RNP fraction demonstrated higher enrichment based on a 3-fold sense/antisense read ratio. The presence of some L1-related reads from the antisense orientation in the cytoplasmic RNP fraction suggested the presence of L1 sequences transcribed as parts of longer transcripts that contain these L1 sequences. As expected, visual analysis of the reads mapping to these loci confirmed that they primarily come from L1-related sequences incorporated in introns of cellular genes (see Supplementary Figure S5 for examples) as well as some 3΄ UTRs.
Because the presence of unprocessed introns is largely due to nuclear RNA species, there is a significant depletion of these L1-related reads in the cytoplasmic RNAs. However, even in the cytoplasmic preparations, some introns seem to be retained in a portion of the RNAs as has been seen before (23). These results demonstrated that limiting our alignments to the cytoplasmic RNP portion and the sense reads uniquely mapping to the full-length L1 loci, only a modest level of manual curation is needed to assure that the alignments are consistent with authentic L1 expression (Supplementary Figure S6).
We carried out similar analyses for other cell lines from other commonly studied cellular lineages, including an SV40-transformed fibroblast (XPD), Akata lymphocytes, and HEK293 human embryonic kidney cells (Table 1). More reads generated using whole-cell RNA extracted from the SV40-transformed fibroblasts corresponded to the antisense than sense strand of the full-length L1. This result suggests that these cells have a very high background from L1-related sequences in other cellular gene transcripts, possibly from some highly expressed genes having intronic L1 elements in the antisense orientation and/or low levels of endogenous L1 expression. This is consistent with low L1 expression levels observed in normal somatic cells using northern blot analysis (7). This is also consistent with the observation in Supplementary Figures S4 and S5 that a limited number of gene regions contribute high levels of background L1-related RNA signals. The same analysis of RNA-Seq reads generated using the cytoplasmic RNP fraction from the XPD-deficient fibroblasts significantly decreased the background resulting in twice as many sense as antisense read pairs mapping to full-length L1 elements. A similar analysis of RNA corresponding to whole cell or RNP fraction from Akata lymphocytes resulted in the sense to antisense full-length L1 transcripts reads ratio of one for the RNP fraction. In contrast, RNA-Seq analysis of endogenous L1 expression using the HEK293 cytoplasmic RNA detected a significantly higher (4.8-fold) sense/antisense ratio of full-length L1 transcripts reads. This result suggests that HEK293 cells may have a significantly higher level of authentic L1 transcription than the other cell types tested.
Sorting the whole-cell HeLa RNA data according to aligned RNA reads mapped in the sense strand of individual full-length loci and comparing them to the same loci in HeLa cytoplasmic RNP preparations demonstrates the profile shown in Supplementary Figure S7. One full-length L1 locus stands out as contributing more than 4% of the sense, full-length L1 reads mapping in the HeLa whole-cell RNA-Seq. Supplementary Figure S5B shows an IGV visualization of all of the HeLa reads to that particular locus (Chr19 FL HeLa sense; L1_FL-5102). The RNA reads are color-coded showing the direction of the RNA from which they were generated. Visual analysis of this locus determined that the L1 element is present in an intron in the same direction as the gene harboring this L1 locus. The read alignments are most consistent with an entire intron of that gene (marked as retained intron) being present in transcripts extracted from the whole-cell RNA, but depleted in the cytoplasmic RNP preparations.
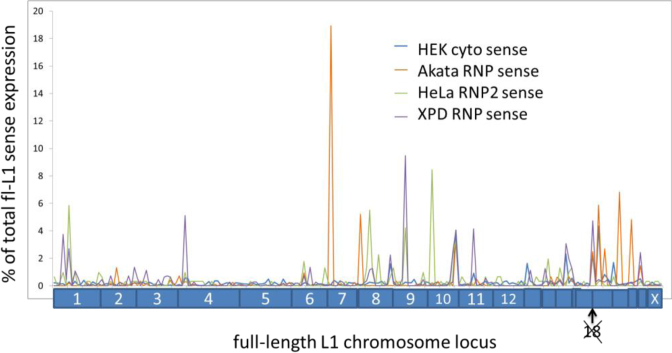
Having evidence from the sense to antisense ratio in Table 1 that HEK 293 cells might have higher levels of authentic L1 expression, we chose to sort the HEK293 data according to the number of reads to each full-length L1 locus and to compare these results collected for each locus to the cytoplasmic RNP preparations from the other cells (Figure 4). In this figure the peaks of L1 expression from individual FL-L1 loci are ordered according to chromosome and the number of mapped reads normalized relative to total FL-L1 expression in those specific cells. Approximately 10–20 out of the 5000 FL-L1 loci show relatively high expression in any given cell type with a tapering group of 50 loci that show very low expression. Thus, 99% of loci remain relatively silent and the majority of the endogenous L1 expression comes from the small group of ten loci. Similar patterns of expression are seen for all analyzed cell types, although the specific expressed L1 loci varied somewhat from one cell line to another (Figure 4). We carried out a Correlation Analysis (Supplementary Table S2) of the expression of individual FL-L1 loci from each cell line and found strong correlations between:
The two independent HeLa RNP preparations as well as with the HeLa whole cell RNA.
The whole-cell RNA preparations versus the cytoplasmic RNP preparations within the specific cell lines for HeLa and the XPD-deficient fibroblasts.
All of the sense full-length L1 expression data.
Figure 4.
Top RNA-Seq alignments to full-length L1 elements from four different cell lineages. Individual loci mapped across all of the chromosomes (labeled on the bottom) of the reference genome for RNP or cytoplasmic RNA data for four different cell lines. A total of 90% of the loci with minimal RNA-Seq mapping are not shown. The loci are ordered according to their chromosomal location, with no loci expressed significantly on chromosome 18. Although there are some loci expressed in multiple different cell types, there are major differences in which loci express in different cells.
We observed no correlation between the sense and antisense reads from full-length L1 loci and highly variable correlation between the antisense reads from the different cell types.
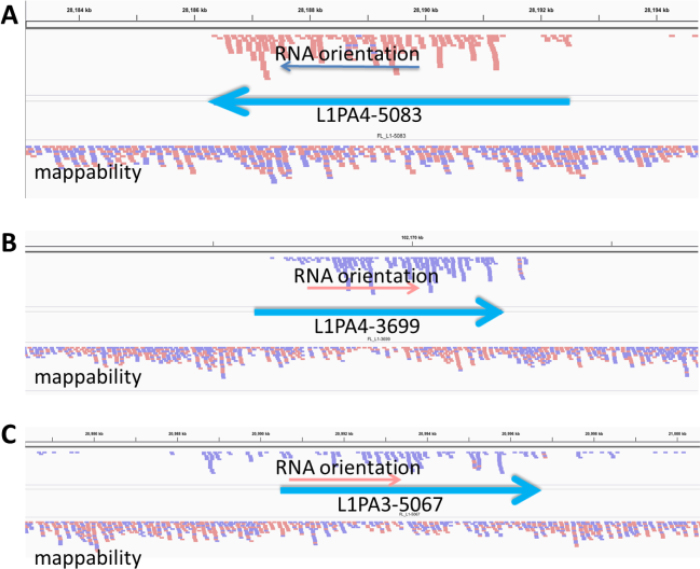
Manual analysis of RNA-Seq reads aligned to full-length L1 loci, particularly those identified in the HEK293 cells, confirmed that the majority of L1 loci showed a pattern of alignments expected to be observed for authentic L1 expression (see Figure 3 and Supplementary Figure S6). In Supplementary Figure S6, panels represent Integrated Genome Viewer (IGV) images for expression of 9 top L1 loci identified by the above described RNA-Seq analysis. Panels A, E, F and H show all reads aligned within the boundaries of the L1 locus, consistent with traditionally predicted L1 expression and with very little 3΄ extension of transcripts beyond the L1. Expression of these L1 loci would not be detected by the previously reported methods relying on readthrough transcripts (Rangwala et al. 2009; Philippe et al. 2016). Panel B, however, shows some read-pairs mapping 3΄ to the L1 locus consistent with the reported read-through of the 3΄ polyA site during L1 transcription for some loci (14,28,29). Panels C, D and G, and to a lesser extent I, all have some reads upstream of the L1 locus that could possibly represent limited 5΄ transduction associated with the L1 promoter initiating transcription upstream of the L1 5΄ UTR (37).
Figure 3.
Top alignments to full-length L1 loci from HEK293 cells. Each full-length L1 locus is marked as a heavy blue arrow with the L1 subfamily and locus number from our annotations listed. The lower alignment labeled mappability represent HeLa genomic DNA reads aligned to the genome to show how well individual loci could align to paired end reads. The upper alignment in each panel shows the cytoplasmic polyA reads aligned uniquely to the genome. Reads in the peach color are oriented as sense to the left. Reads in the lavender color are sense to the right. At the bottom of each panel is the gene annotation if any is shown. The introns are marked with large red arrowheads to show gene orientation and the exons are black boxes. Panels A and B show alignments that are consistent with authentic L1 expression. They have no real alignment upstream and appear to originate from the L1 promoter. Panel A has no reads downstream of the L1, while Panel B shows a few reads extending downstream. Panel C is more uncertain as there are more than expected reads upstream and downstream and show some potential for transcripts coming through this location from another promoter.
Identifying expression from young L1 elements using 3΄ read-through extensions
Using the above described analysis we have identified some expressed L1 loci (L1Hs and older) that can be uniquely mapped in the human genome. Our approach estimated the potential contribution to L1 expression from the youngest (multimapped) L1 loci but did not allow identification of all of these individual expressed loci. Our approach has also established that many expressed L1 loci do not generate 3΄ transductions. To assess which expressed L1 loci can be detected using an approach that relies on generation of 3΄ transductions we have developed an approach similar to that of Philippe et al. (14) that utilizes 3΄ extension of transcripts into unique flanking sequences to identify potential L1 transcripts from elements that are either too close to consensus sequence to allow unique mapping of sequence reads to the L1 element itself, or to map to elements that are not part of the reference genome (see Supplementary Figures S8 and S9). Philippe et al. (14) combine this approach with a PCR-based approach to first identify the polymorphic full-length L1 loci that are specifically found in the genomes of each cell line. This is an excellent approach and previous methods have been published to identify full-length elements with PCR (38) or whole-genome sequencing (39). As a higher throughput but less rigorous alternative, we have proposed that we can utilize the previously annotated L1 polymorphisms (described in more detail in Supplementary Figure S8 and Supplementary description for 3΄ extensions) to identify potential 3΄ extension transcripts that can then be validated by PCR, with the RNA-Seq analysis shown in Supplementary Table S3. This approach will become increasingly robust as the annotations of full-length L1 elements improve. Because of the similarities with Philippe et al. (14), we have described the bioinformatics strategy and bioinformatic command lines more fully in the Supplementary Material. The major finding was to reinforce identification of several loci with few reads in the L1 region due to low mappability (Supplementary Figure S9, panels C and J) as well as provide evidence for expression from several more L1 loci with poor mappability or from the L1 loci that weren't present in the reference genome (Supplementary Figure S9, panels B, D and G).
5΄ Race
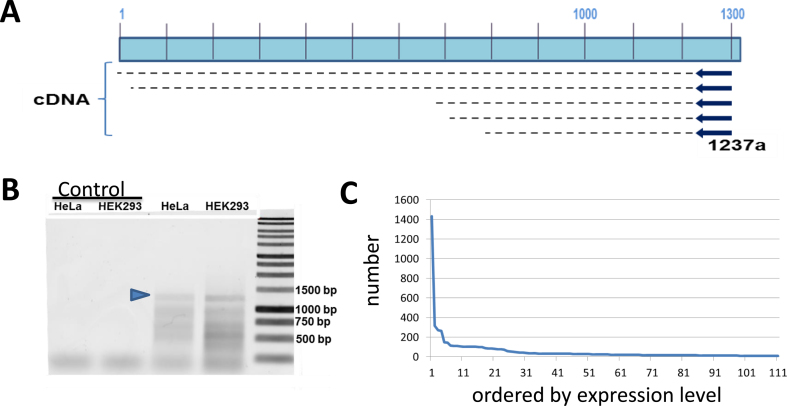
Both our RNA-Seq approach described above, and other previously reported approaches (14,29), have significant limitations in both detection and quantitation of some of the L1 transcripts. Because of these limitations we thought to create an independent approach that overcame some of these shortcomings. One of the key features that distinguish authentic L1 transcripts from cellular transcripts that include some L1 sequences is that the authentic L1 transcript driven by the L1 promoter will start at the beginning of the full-length L1 element. Thus, we carried out 5΄ RACE that strongly enriches for authentic L1 RNAs transcribing from the beginning of the L1 sequence to recover and map the 5΄ ends of authentic L1 RNAs. The 5΄ RACE was designed to generate long (1237 bp) cDNA that when sequenced individually or using NGS approaches it would facilitate mapping to unique L1 loci. PacBio sequencing involves ligation of a ‘dumbbell’ linker on both ends of the fragment, essentially turning it into a single-strand circle. This allows the PacBio sequencing to circle a template of this size multiple times, allowing a consensus sequence to be generated for each fragment (40). This is a standard approach to overcome some of the inherent error rate in PacBio sequencing. We chose to use cytoplasmic RNA extracted from HEK293 cells (see Figure 5) to generate a 5΄ RACE product using a primer complementary to the L1 position 1237. This approach generated both an expected 1200 bp band consistent with full-length transcripts and some smaller bands consistent with the possibility of transcriptional starts around position 700 (Figure 5) as has previously been reported (41). A limited sequencing of these shorter bands by cloning and Sanger sequencing showed that they generally started in the range of 500–700 bp within the L1 promoter and, within the limited sampling, included the same loci as were detected from the 1200 bp band.
Figure 5.
L1 5΄ RACE. In Panel A, we show that we have utilized a primer from position 1237 in the L1.3 sequence to prime reverse transcription on RNA from cytoplasmic RNPs. Note that various cDNA extensions are shown that may occur from authentic L1 RNAs. Products that extend significantly beyond position 1 are likely to represent an L1-related sequence transcribed in a cellular genic mRNA. Panel B shows an agarose gel for the 5΄ RACE reaction. Without the tailing reaction of the initial cDNA, we see only a primer band at the bottom (Control lanes). In the RACE, with HeLa and HEK293 we see a significant band at the expected 1200 bp (marked with a blue arrowhead) and some smaller bands that might be consistent with internal transcription starts. Panel C shows a 1200 bp 5΄ RACE band (blue arrowhead in Panel B) from HEK293 that was fully sequenced with PACBIO and 7500 full-length reads mapped. The individual loci are shown with the number of reads aligned to them. The FL-5219 locus had almost 20% of the total reads in panel C (also see Table 2) with the second highest mapping locus having 5 times fewer reads. Note that 20–30 loci shows modest expression with that trickling off to most loci showing no expression at all out of the ∼5000 loci.
To allow high throughput analysis of the 5΄ RACE, the isolated 1200 bp band was subjected to PacBio sequence analysis (40) resulting in ∼35 000 full-length reads. These 1200 bp reads were aligned to the human genome with GMAP (42). This approach found individual reads aligned to specific L1 loci in the human genome with matches ranging between 92% and 100% match to their best full-length L1 locus. Some of this inaccuracy is probably due to the PacBio error rate as well as the potential for chimeras to form between different L1 loci during the PCR process (43). In order to minimize both of these sources of experimental error, we only accepted reads that mapped to a L1 locus in the human genome with at least 99% accuracy. This left approximately 7500 reads that could be mapped within this accuracy to at least one locus in the human genome. Approximately 10% of the reads still did not map to a single locus better than the others most likely because they were generated from very young L1 HS elements. Manual inspection of a number of the ‘multimapped’ reads from this 5΄-RACE experiment confirmed that they were predominantly L1 HS elements as expected. We verified the quality of the GMAP alignments by showing that 20 randomly chosen reads showed a unique best mapping to the same locus using BLAST. Furthermore, none of the over 7500 reads mapped to the Y chromosome, as HEK293 does not have a Y chromosome.
The uniquely mapped reads aligned to a number of full-length L1 loci (see Table 2 and Supplementary Table S4), with the frequency at specific loci shown in Figure 5C. Of particular note, locus 5219 on chromosome 22 represents about 20% of the total full-length L1 reads for HEK293 and was found in our other HEK293 RNA-Seq studies. In this study, 85% of the genomic L1 loci did not show a single read mapping to them. Only about 100 loci showed 10 or more reads mapping to them, compared to over 1400 reads mapping to the locus 5219. Table 2 shows the most abundant L1 loci as measured by 5΄ RACE and an excellent overlap between the L1 loci identified by the 5΄ RACE and the RNA-Seq analyses. The only 5΄ RACE-identified L1 loci that did not map in the RNA-Seq studies were the L1HS loci because of their poor mappability. Table 2 also shows that approaches that rely on detection of L1 expression using the reads originating from 3΄ extensions beyond the L1 are only able to detect approximately half of the most highly expressed L1 loci.
Table 2. Read counts to top 5΄ RACE loci and corresponding RNA-Seq counts.
| CHR # | position | position | orient. | L1 identity | subfamily | 5΄ RACE | RNA-Seq | 3΄ reads |
|---|---|---|---|---|---|---|---|---|
| 22 | 29059274 | 29065303 | + | ID = FL_L1-5219 | ‘L1HS’ | 1433 | 24 | 75 |
| 13 | 37724090 | 37730119 | - | ID = FL_L1-4381 | ‘L1PA2’ | 318 | 22 | 4 |
| 7 | 65751841 | 65757868 | - | ID = FL_L1-2762 | ‘L1HS’ | 272 | 24 | 22 |
| 1 | 174812365 | 174818381 | - | ID = FL_L1-0291 | ‘L1PA2’ | 265 | 25 | 31 |
| 2 | 118894577 | 118900596 | - | ID = FL_L1-0602 | ‘L1PA2’ | 148 | 10 | 0 |
| 15 | 64618860 | 64624899 | + | ID = FL_L1-4738 | ‘L1PA2’ | 143 | 4 | 4 |
| 2 | 71638605 | 71644631 | - | ID = FL_L1-0521 | ‘L1HS’ | 114 | 0 | 0 |
| 13 | 39574955 | 39581012 | - | ID = FL_L1-4386 | ‘L1PA2’ | 112 | 100 | 1 |
| 7 | 49719865 | 49725896 | - | ID = FL_L1-2731 | ‘L1HS’ | 110 | 0 | 0 |
| 12 | 76713405 | 76719506 | + | ID = FL_L1-4272 | ‘L1PA3’ | 105 | 22 | 4 |
| 2 | 197770314 | 197776343 | + | ID = FL_L1-0783 | ‘L1HS’ | 104 | 0 | 2 |
| X | 154745678 | 154751709 | - | ID = FL_L1-5789 | ‘L1HS’ | 104 | 0 | 0 |
| 5 | 89958428 | 89964454 | - | ID = FL_L1-2038 | ‘L1PA3’ | 102 | 30 | 0 |
| 11 | 24349498 | 24355547 | + | ID = FL_L1-3806 | ‘L1HS’ | 102 | 2 | 0 |
| 7 | 76357803 | 76363830 | + | ID = FL_L1-2766 | ‘L1PA3’ | 101 | 16 | 4 |
| 4 | 21161014 | 21167044 | + | ID = FL_L1-1333 | ‘L1HS’ | 99 | 0 | 0 |
| 2 | 66618263 | 66624282 | + | ID = FL_L1-0513 | ‘L1PA2’ | 98 | 10 | 0 |
| 3 | 51010569 | 51016603 | - | ID = FL_L1-0946 | ‘L1PA3’ | 88 | 2 | 2 |
| 9 | 89656798 | 89662815 | + | ID = FL_L1-3444 | ‘L1PA3’ | 85 | 29 | 5 |
| 20 | 23406746 | 23412777 | + | ID = FL_L1-5141 | ‘L1HS’ | 83 | 0 | 10 |
| 9 | 85664455 | 85670484 | - | ID = FL_L1-3437 | ‘L1HS’ | 80 | 14 | 8 |
| 12 | 96709723 | 96715749 | + | ID = FL_L1-4311 | ‘L1PA2’ | 78 | 14 | 2 |
| 20 | 8575749 | 8581774 | - | ID = FL_L1-5119 | ‘L1PA2’ | 75 | 13 | 2 |
| 2 | 175269187 | 175275210 | - | ID = FL_L1-0730 | ‘L1PA2’ | 72 | 6 | 0 |
| 5 | 172829800 | 172835828 | - | ID = FL_L1-2242 | ‘L1HS’ | 56 | 0 | 0 |
| 17 | 66404265 | 66410283 | - | ID = FL_L1-4940 | ‘L1PA3’ | 54 | 12 | 2 |
| 1 | 43580199 | 43586164 | - | ID = FL_L1-0029 | ‘L1MB3’ | 50 | 14 | 0 |
DISCUSSION
There is increasing evidence that L1 elements not only cause extensive genetic damage in the germ line leading to disease (6,44), but are also expressed in normal human tissues (7) and tumors (45) where they contribute to genomic instability (8,10–12,17,46). These findings support that L1 activity may contribute to tumorigenesis or aging (9). Although there are ∼500 000 different L1 loci in the human genome (1), the vast majority of them are truncated upon insertion. Of the 5000 full-length copies, as many as 150 have both open reading frames intact (15,47). Of those potentially retrotranspositionally active elements, there is a wide range of retrotransposition capability with only 10–20 representing very active, ‘hot’ L1 elements (15,16). However, there is evidence that even the older L1 elements with incomplete open reading frames could make protein domains that are damaging to the cell (48). Because L1 expression is epigenetically silenced (10,49), the full extent of L1 activity within individual genomes cannot be fully appreciated until we understand the patterns of expression of individual L1 loci.
Since the first discovery of L1 elements, it was recognized that their high copy number and ubiquitous presence throughout the genome made it extremely difficult to differentiate authentic, endogenous full-length L1 transcripts from the presence of L1-related sequences in other RNAs (50). Because of these difficulties, Northern blots have been the primary reliable method for measuring authentic L1 expression from its promoter (7,25,50). Several investigators have now used the tendency of L1 transcripts to extend past their polyadenylation signal into flanking sequences (28,36) to assess expression from individual L1 loci (14,29). Although useful, these methods are limited to detecting stable L1 transcripts including downstream genomic sequences (29) or only the very youngest subfamily (14). In addition, the usefulness of the 3΄ transduction approach is also largely based on the untested assumption that all loci create these extended transcripts. Our data show that a number of relatively highly expressed L1 loci do not show such extensions (Figure 3A, Supplementary Figure S6, Table 2 and Supplementary Table S4) and it is reasonable to assume that the ability to detect L1 loci with these methods is very locus dependent. Our data show that over half of the expressed loci, as measured by 5΄ RACE and RNA-Seq mapping to the body of the L1 do not have reads mapping downstream due to alternative polyA site use (Table 2 and Supplementary Table S4). It is very likely that the specific location and strength of the alternative polyA sites downstream of each L1 locus contribute to the use of alternative polyadenylation or the stability of the alternative transcript.
Our studies involve two totally different approaches than those previously used (14,29) to give a more complete picture of the authentic endogenous L1 site-specific transcriptome. By combining RNA-Seq and large-scale 5΄ RACE studies, we obtain a more complete and quantitative picture of L1 expression. The RNA-Seq analyses in this study are consistent with a modest number of full-length loci dominating most of the authentic L1 expression, although with a much larger number of elements expressed at low levels (Table 2, Supplementary Table S4, Figure 5 and Supplementary Figure S6). Although this result is similar to that reported in another recent paper (14), our approach greatly extends the spectrum of L1 loci that can be analyzed from only a subset of L1-Hs elements to all full-length L1 elements present in the human genome (including L1Hs loci that do not generate 3΄ transductions). 5΄ RACE experiments utilizing longer reads further corroborated our RNA-Seq results, showing that one L1HS locus (L1_5219, Table 2 and Supplementary Table S4) represented 20% of the total authentic L1 expression. This locus was not identified as a highly expressed L1 element by the RNA-Seq approach because of its poor mappability (Supplementary Figure S8A). Our data demonstrate that the top 6 loci identified by the 5΄ RACE analysis contributed over one-third of the expression (Table 2) while the next 35 L1 loci contributed the next third of the expression. Of these expressing loci, about one-third represented the L1 HS subfamily.
Our finding that most authentic L1 expression is limited to a relatively small number of highly expressed full-length L1 loci helps explain the observed trend in L1 amplification within tumors wherein a small number of L1 loci contribute the majority of the new inserts (10,17,18). In particular, our data show that only a very few L1 Hs loci are expressed to high levels in HEK293 cells (Figure 5C). Although there are different biases in our RNA-Seq and 5΄ RACE approaches (RNA-Seq alignments preferentially identify older L1 elements and 5΄ RACE probably shows some bias for younger elements both because of the design of the primer (PA1–PA6 specific) and the accumulation of random mutations at the primer site), our data suggest that there is a significant enrichment for endogenous expression of younger elements and that the older L1 elements are relatively silent, when adjusting for their much higher copy numbers. In addition, the FL-5219 locus that we see dominates expression in HEK293 is also the dominant locus (TTC28) reported as amplifying in colorectal cancer (18).
Our RNA-Seq data (Figure 4) also show that different cell types support higher expression levels from a limited number of loci with most loci remaining silent. However, the exact L1 loci being expressed differ significantly from one cell type to another. Thus, the pattern of expression in individuals may be quite different not only due to expression levels from polymorphic ‘hot’ L1 loci (15), but also due to the variation in the pattern of expression of those L1 loci in different tissues within an individual's body. This has significant implications for the potential impact of L1 expression in various somatic tissues during aging (9).
Technical implications
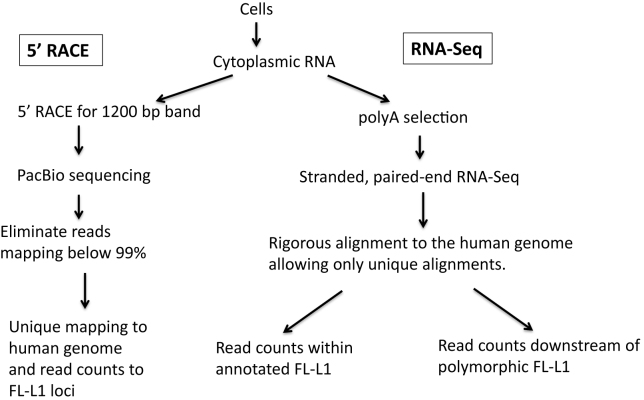
Our findings highlight the difficulties of studying RNA expression from endogenous L1 loci and the rigor that is needed to obtain meaningful results. The high background (>99%) from L1-related sequences incorporated into other RNA species that are unrelated to the L1 retrotransposition process leads to serious and often overlooked issues with interpretation of authentic L1 expression from RNA analyses that do not consider this issue. This background is decreased by (i) isolation of cytoplasmic RNAs, (ii) selection for polyadenylation and (iii) the utilization of methods that eliminate or assess transcripts coming from L1 sequences in the antisense orientation. Figure 6 summarizes our various approaches to identifying transcripts from full-length L1 loci. By isolating cytoplasmic RNA, we eliminate a great deal of background from L1-related reads from introns of genes found in the nucleus. This approach may eliminate some L1 transcripts that never escape the nucleus, or transcripts from the L1 promoter that splice into other genes, either in the sense or antisense orientations. These studies are designed very specifically to identify only full-length transcripts that arise from the L1 promoter and are transported to the cytoplasm for translation as these are expected to be the transcripts that are primarily involved in the retrotransposition process.
Figure 6.
Flowchart for analysis of expression from full-length L1 loci. In all of our approaches we first isolate cytoplasmic RNAs to eliminate a great deal of the background from L1 elements found in the introns of transcripts in the nucleus. Our 5΄ RACE protocol uses the total cytoplasmic RNA for cDNA synthesis with a L1-specific primer that should generate a 1200 base cDNA only from full-length L1 transcripts that initiate at the beginning of the L1 sequence. This 1200 base cDNA is then amplified using the RACE protocol and subjected to PacBio sequencing. Consensus reads from the PacBio for each molecule are aligned to the human genome and those that align throughout their length with an accuracy lower than 99% are eliminated. This not only eliminates fragments with sequence errors, it also eliminates PCR chimeras between different L1 loci. The accurate L1 alignments are then rigorously aligned to the human genome and only reads that align to one full-length L1 locus better than all others are mapped and counted. In the RNA-Seq studies, the same cytoplasmic RNA is subjected to polyA selection to eliminate rRNA and then subjected to a strand-specific, 2 × 100 bp, paired-end RNA-Seq using the Illumina platform. These paired-end reads are aligned rigorously with BOWTIE, accepting only those alignments where both reads align concordantly at one locus better than anywhere else in the human genome. This alignment is used to either count the reads mapping to each individual full-length L1 locus (left branch) or with reads mapping specifically downstream from known polymorphic L1 loci (right branch) as indirect evidence of expression from those loci.
Because of the above factors, most existing databases of RNA-Seq reads are not well suited for analysis of authentic and thus relevant L1 RNA expression. Getting useful information from data sets generated from whole-cell RNAs or non-strand-specific RNA-Seq preparations would require extensive manual curation looking at the pattern of expression around each L1 locus (as in Supplementary Figures S5 and S6), and even then would remain somewhat unreliable. Our results further reiterate that one cannot study mobile element transcripts in the same way one studies transcripts from cellular genes. Utilizing RT-PCR or similar approaches to quantitate L1 transcripts, particularly from whole-cell RNAs would result in products that were almost exclusively generated from background L1 sequences present in contaminating RNAs rather than from those transcribed from the L1 promoter.
Because of the complexities associated with identifying authentic L1 expression, particularly from individual loci, each approach (Figure 6) has specific strengths and weaknesses. Our data demonstrate that the 5΄ RACE approach (Figure 5) has many advantages compared to the existing methods. First, because it generates a PCR product that has been highly enriched for RNAs that are transcribed from the L1 promoter, the 5΄ RACE provides a rapid and relatively quantitative approach to identifying expression from different endogenous L1 loci. It is most robust when cytoplasmic RNA is used, but it will still work with whole-cell RNAs when cytoplasmic RNA is not available (data not shown). Second, the long length of the sequence generated by this approach allows unique identification of most L1 loci and in cases where the specific locus cannot be identified it is typically a member of the L1 Hs subfamily which sequence is too close to consensus. Third, in addition to being a robust and quantitative approach to identify endogenously expressed L1 loci it is also relatively inexpensive because a single PACBIO cell can sequence several multiplexed samples.
The ability to map expression from the vast majority of L1 loci in the human genome now opens up the possibility of doing much more focused and specific studies of both their regulation and impact on the genome. The 5΄ RACE approach provides the most quantitative view of expression, but cannot map uniquely to L1 elements that match the consensus perfectly. The RNA-Seq approach, however, may be able to identify a few loci that are not mapped by 5΄ RACE if they have mutations elsewhere in the L1 element that allow unique mapping, or whose transcription extends into the downstream flanking region (Supplementary Figure S6B, C, D and F). These methods, however, also require some validation that the RNA-Seq reads come from the L1 promoter. In our RNA-Seq approach we look at transcription upstream of the element to eliminate potential inclusion of this L1 sequence into cellular transcripts. Philippe et al. (14) utilized chromatin signals in the Encode database just upstream of an element locus to suggest that it was transcriptionally active in that cell line. This latter method adds an extra reinforcement to the analysis, but also requires either the availability of the appropriate ChIP data or the specific generation of such data.
Supplementary Material
Footnotes
Present addresses:
Travis B. White, Sloan Kettering Institute for Cancer Research, NY, USA.
Dale J. Hedges, St. Jude Children's Hospital, Memphis, TN 38105, USA.
Monica Concha, Innogenomics, New Orleans, LA 70112, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [P20GM103518 to P.L.D. and V.P.B. and R01GM45668 to P.L.D.; The Life Extension Foundation [to V.P.B.]; Louisiana Board of Regents Support Fund [LEQSF(2015-18)-RD-A-25]; Kay Yow Cancer Foundation. Funding for open access charge: Joe W. and Dorothy Dorsett Brown Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1. Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W. et al. Initial sequencing and analysis of the human genome. Nature. 2001; 409:860–921. [DOI] [PubMed] [Google Scholar]
- 2. Dewannieux M., Esnault C., Heidmann T.. LINE-mediated retrotransposition of marked Alu sequences. Nat. Genet. 2003; 35:41–48. [DOI] [PubMed] [Google Scholar]
- 3. Raiz J., Damert A., Chira S., Held U., Klawitter S., Hamdorf M., Lower J., Stratling W.H., Lower R., Schumann G.G.. The non-autonomous retrotransposon SVA is trans-mobilized by the human LINE-1 protein machinery. Nucleic Acids Res. 2012; 40:1666–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hancks D.C., Goodier J.L., Mandal P.K., Cheung L.E., Kazazian H.H. Jr. Retrotransposition of marked SVA elements by human L1s in cultured cells. Hum. Mol. Genet. 2011; 20:3386–3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boeke J.D. LINEs and Alus–the polyA connection. Nat. Genet. 1997; 16:6–7. [DOI] [PubMed] [Google Scholar]
- 6. Belancio V.P., Deininger P.L., Roy-Engel A.M.. LINE dancing in the human genome: transposable elements and disease. Genome Med. 2009; 1:e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Belancio V.P., Roy-Engel A.M., Pochampally R.R., Deininger P.. Somatic expression of LINE-1 elements in human tissues. Nucleic Acids Res. 2010; 38:3909–3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Evrony G.D., Lee E., Park P.J., Walsh C.A.. Resolving rates of mutation in the brain using single-neuron genomics. Elife. 2016; 5:e12966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Belancio V.P., Blask D.E., Deininger P., Hill S.M., Jazwinski S.M.. The aging clock and circadian control of metabolism and genome stability. Front. Genet. 2014; 5:e455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tubio J.M., Li Y., Ju Y.S., Martincorena I., Cooke S.L., Tojo M., Gundem G., Pipinikas C.P., Zamora J., Raine K. et al. Mobile DNA in cancer. Extensive transduction of nonrepetitive DNA mediated by L1 retrotransposition in cancer genomes. Science. 2014; doi:10.1126/science.1251343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ewing A.D., Gacita A., Wood L.D., Ma F., Xing D., Kim M.S., Manda S.S., Abril G., Pereira G., Makohon-Moore A. et al. Widespread somatic L1 retrotransposition occurs early during gastrointestinal cancer evolution. Genome Res. 2015; 25:1536–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Iskow R.C., McCabe M.T., Mills R.E., Torene S., Pittard W.S., Neuwald A.F., Van Meir E.G., Vertino P.M., Devine S.E.. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell. 2010; 141:1253–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rodic N., Burns K.H.. Long interspersed element-1 (LINE-1): passenger or driver in human neoplasms. PLoS Genet. 2013; 9:e1003402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Philippe C., Vargas-Landin D.B., Doucet A.J., van Essen D., Vera-Otarola J., Kuciak M., Corbin A., Nigumann P., Cristofari G.. Activation of individual L1 retrotransposon instances is restricted to cell-type dependent permissive loci. Elife. 2016; 5:e13926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brouha B., Schustak J., Badge R.M., Lutz-Prigge S., Farley A.H., Moran J.V., Kazazian H.H. Jr. Hot L1s account for the bulk of retrotransposition in the human population. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:5280–5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Beck C.R., Collier P., Macfarlane C., Malig M., Kidd J.M., Eichler E.E., Badge R.M., Moran J.V.. LINE-1 retrotransposition activity in human genomes. Cell. 2010; 141:1159–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Scott E.C., Gardner E.J., Masood A., Chuang N.T., Vertino P.M., Devine S.E.. A hot L1 retrotransposon evades somatic repression and initiates human colorectal cancer. Genome Res. 2016; 26:755–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pitkanen E., Cajuso T., Katainen R., Kaasinen E., Valimaki N., Palin K., Taipale J., Aaltonen L.A., Kilpivaara O.. Frequent L1 retrotranspositions originating from TTC28 in colorectal cancer. Oncotarget. 2014; 5:853–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Martin S.L. Nucleic acid chaperone properties of ORF1p from the non-LTR retrotransposon, LINE-1. RNA Biol. 2010; 7:706–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Moran J.V., Holmes S.E., Naas T.P., DeBerardinis R.J., Boeke J.D., Kazazian H.H. Jr. High frequency retrotransposition in cultured mammalian cells. Cell. 1996; 87:917–927. [DOI] [PubMed] [Google Scholar]
- 21. Wei W., Gilbert N., Ooi S.L., Lawler J.F., Ostertag E.M., Kazazian H.H., Boeke J.D., Moran J.V.. Human L1 retrotransposition: cis preference versus trans complementation. Mol. Cell. Biol. 2001; 21:1429–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gasior S.L., Wakeman T.P., Xu B., Deininger P.L.. The human LINE-1 retrotransposon creates DNA double-strand breaks. J. Mol. Biol. 2006; 357:1383–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zaghlool A., Ameur A., Nyberg L., Halvardson J., Grabherr M., Cavelier L., Feuk L.. Efficient cellular fractionation improves RNA sequencing analysis of mature and nascent transcripts from human tissues. BMC Biotechnol. 2013; doi:10.1186/1472-6750-13-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Medstrand P., van de Lagemaat L.N., Mager D.L.. Retroelement distributions in the human genome: variations associated with age and proximity to genes. Genome Res. 2002; 12:1483–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Deininger P., Belancio V.P.. Detection of LINE-1 RNAs by Northern Blot. Methods Mol. Biol. 2016; 1400:223–236. [DOI] [PubMed] [Google Scholar]
- 26. Sharma R., Rodic N., Burns K.H., Taylor M.S.. Immunodetection of human LINE-1 expression in cultured cells and human tissues. Methods Mol. Biol. 2016; 1400:261–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Harris C.R., Normart R., Yang Q., Stevenson E., Haffty B.G., Ganesan S., Cordon-Cardo C., Levine A.J., Tang L.H.. Association of nuclear localization of a long interspersed nuclear element-1 protein in breast tumors with poor prognostic outcomes. Genes Cancer. 2010; 1:115–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pickeral O.K., Makalowski W., Boguski M.S., Boeke J.D.. Frequent human genomic DNA transduction driven by LINE-1 retrotransposition. Genome Res. 2000; 10:411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rangwala S.H., Zhang L., Kazazian H.H. Jr. Many LINE1 elements contribute to the transcriptome of human somatic cells. Genome Biol. 2009; 10:R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Perepelitsa-Belancio V., Deininger P.. RNA truncation by premature polyadenylation attenuates human mobile element activity. Nat. Genet. 2003; 35:363–366. [DOI] [PubMed] [Google Scholar]
- 31. deHaro D., Kines K.J., Sokolowski M., Dauchy R.T., Streva V.A., Hill S.M., Hanifin J.P., Brainard G.C., Blask D.E., Belancio V.P.. Regulation of L1 expression and retrotransposition by melatonin and its receptor: implications for cancer risk associated with light exposure at night. Nucleic Acids Res. 2014; 42:7694–7707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Belancio V.P., Hedges D.J., Deininger P.. LINE-1 RNA splicing and influences on mammalian gene expression. Nucleic Acids Res. 2006; 34:1512–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lykke-Andersen S., Chen Y., Ardal B.R., Lilje B., Waage J., Sandelin A., Jensen T.H.. Human nonsense-mediated RNA decay initiates widely by endonucleolysis and targets snoRNA host genes. Genes Dev. 2014; 28:2498–2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dobin A., Gingeras T.R.. Mapping RNA-seq reads with STAR. Curr. Protoc. Bioinformatics. 2015; 51, doi:10.1002/0471250953.bi1114s51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Matz M., Shagin D., Bogdanova E., Britanova O., Lukyanov S., Diatchenko L., Chenchik A.. Amplification of cDNA ends based on template-switching effect and step-out PCR. Nucleic Acids Res. 1999; 27:1558–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Macfarlane C.M., Collier P., Rahbari R., Beck C.R., Wagstaff J.F., Igoe S., Moran J.V., Badge R.M.. Transduction-specific ATLAS reveals a cohort of highly active L1 retrotransposons in human populations. Hum. Mutat. 2013; 34:974–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Athanikar J.N., Badge R.M., Moran J.V.. A YY1-binding site is required for accurate human LINE-1 transcription initiation. Nucleic Acids Res. 2004; 32:3846–3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Streva V.A., Jordan V.E., Linker S., Hedges D.J., Batzer M.A., Deininger P.L.. Sequencing, identification and mapping of primed L1 elements (SIMPLE) reveals significant variation in full length L1 elements between individuals. BMC Genomics. 2015; doi:10.1186/s12864-015-1374-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ewing A.D. Transposable element detection from whole genome sequence data. Mob. DNA. 2015; doi:10.1186/s13100-015-0055-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Roberts R.J., Carneiro M.O., Schatz M.C.. The advantages of SMRT sequencing. Genome Biol. 2013; doi:10.1186/gb-2013-14-6-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Alexandrova E.A., Olovnikov I.A., Malakhova G.V., Zabolotneva A.A., Suntsova M.V., Dmitriev S.E., Buzdin A.A.. Sense transcripts originated from an internal part of the human retrotransposon LINE-1 5΄ UTR. Gene. 2012; 511:46–53. [DOI] [PubMed] [Google Scholar]
- 42. Wu T.D., Reeder J., Lawrence M., Becker G., Brauer M.J.. GMAP and GSNAP for Genomic Sequence Alignment: Enhancements to Speed, Accuracy, and Functionality. Methods Mol. Biol. 2016; 1418:283–334. [DOI] [PubMed] [Google Scholar]
- 43. Ji W., Zhang X.Y., Warshamana G.S., Qu G.Z., Ehrlich M.. Effect of internal direct and inverted Alu repeat sequences on PCR. PCR Methods Appl. 1994; 4:109–116. [DOI] [PubMed] [Google Scholar]
- 44. Hancks D.C., Kazazian H.H. Jr. Active human retrotransposons: variation and disease. Curr. Opin. Genet. Dev. 2012; 22:191–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rodic N., Sharma R., Sharma R., Zampella J., Dai L., Taylor M.S., Hruban R.H., Iacobuzio-Donahue C.A., Maitra A., Torbenson M.S. et al. Long interspersed element-1 protein expression is a hallmark of many human cancers. Am. J. Pathol. 2014; 184:1280–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Goodier J.L. Retrotransposition in tumors and brains. Mob. DNA. 2014; doi:10.1186/1759-8753-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Penzkofer T., Dandekar T., Zemojtel T.. L1Base: from functional annotation to prediction of active LINE-1 elements. Nucleic Acids Res. 2005; 33:D498–D500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kines K.J., Sokolowski M., deHaro D.L., Christian C.M., Belancio V.P.. Potential for genomic instability associated with retrotranspositionally-incompetent L1 loci. Nucleic Acids Res. 2014; 42:10488–10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rosser J.M., An W.. L1 expression and regulation in humans and rodents. Front. Biosci. (Elite Ed.). 2012; 4:2203–2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Skowronski J., Singer M.F.. Expression of a cytoplasmic LINE-1 transcript is regulated in a human teratocarcinoma cell line. Proc. Natl. Acad. Sci. U.S.A. 1985; 82:6050–6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kopera H.C., Flasch D.A., Nakamura M., Miyoshi T., Doucet A.J., Moran J.V.. LEAP: L1 element amplification protocol. Methods Mol. Biol. 2016; 1400:339–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.