Abstract
High-throughput sequencing has greatly facilitated the discovery of long and short non-coding RNAs (ncRNAs), which frequently guide ribonucleoprotein complexes to RNA targets, to modulate their metabolism and expression. However, for many ncRNAs, the targets remain to be discovered. In this study, we developed computational methods to map C/D box snoRNA target sites using data from core small nucleolar ribonucleoprotein crosslinking and immunoprecipitation and from transcriptome-wide mapping of 2΄-O-ribose methylation sites. We thereby assigned the snoRNA guide to a known methylation site in the 18S rRNA, we uncovered a novel partially methylated site in the 28S ribosomal RNA, and we captured a site in the 28S rRNA in interaction with multiple snoRNAs. Although we also captured mRNAs in interaction with snoRNAs, we did not detect 2΄-O-methylation of these targets. Our study provides an integrated approach to the comprehensive characterization of 2΄-O-methylation targets of snoRNAs in species beyond those in which these interactions have been traditionally studied and contributes to the rapidly developing field of ‘epitranscriptomics’.
INTRODUCTION
RNAs are extensively modified in all living organisms (1). Recently, high-throughput approaches have been developed to map 2΄-O-methylated riboses (2΄-O-Me, (2)) and nucleobases carrying the most frequent modifications, including N6-methyladenosine (m6A, (3)), pseudouridine (ψ, (4)) and 5-methylcytosine (m5C, (5)), transcriptome-wide. These studies have catalyzed the birth of ‘epitranscriptomics’ (6) and have rekindled the interest in the functions of RNA modifications and their relevance for human diseases (7,8). Whereas 2΄-O-ribose methylation has long been implicated in the stability and structure of ribosomal RNAs (reviewed in (9)) and m6A appears to modulate the rate of mRNA translation (10–13), the role of most RNA modifications remains to be characterized.
The 2΄-O-methylation of riboses in ribosomal RNAs (rRNAs), small nuclear RNAs (snRNAs) and in Archaea, transfer RNAs (tRNAs) (14–16), is catalyzed by the protein fibrillarin. Fibrillarin is part of a larger ribonucleoprotein (snoRNP) complex whose protein components in mammals and yeast are: FBL (fibrillarin)/Nop1 (17), SNU13/Snu13 (18), NOP56/Nop56 and NOP58/Nop58 (19). As summarized in (20), it is generally accepted that the snoRNP complex assembles sequentially. SNU13/Snu13 initially binds the guide RNA, leading to the folding of the K-turn motif, and the subsequent binding of the NOP56/Nop56:NOP58/Nop58 heterodimer. This complex helps position the guide RNA in its active conformation and is completed by the binding of FBL/Nop1, the snoRNP component responsible for the 2΄-O-methylation enzymatic activity. As we here focus on human snoRNA, to simplify reading we use hereafter the corresponding nomenclature. The guiding C/D-box small nucleolar RNAs (snoRNAs) (in Archaea small RNAs) take their names from conserved C/C’ (RUGAUGA, R = A or G) and D/D’ (CUGA) boxes. Molecules with more complex structure, which can include additional H/ACA boxes and signals that direct their localization to Cajal bodies (therefore called small Cajal body-associated RNAs or scaRNAs (21)) have also been identified and are essential for the modification and proper functioning of snRNAs. The C/C’ and D/D’ boxes are important for snoRNA biogenesis and for the interaction with RNA binding proteins (22). ‘Anti-sense’ elements located upstream of the D and/or D’ boxes, base-pair with the targets. The target nucleotide that pairs with the fifth nucleotide of the anti-sense element acquires the 2΄-O-Me mark. Base-pairing adjacent to the target site can further enhance 2΄-O-methylation (23). Many studies have investigated snoRNA-guided modifications, particularly in yeast (24–27). As a result, features that define snoRNA target sites have been identified and incorporated into computational methods for snoRNA target prediction (28,29). They include a high complementarity to the 3΄ end of the anti-sense box, with no more than one mismatch over at least seven nucleotides, and no bulges (29). A few snoRNAs including U3, U8, U13 have been found to be essential for the processing of rRNA precursors in multiple species, whereas U14 functions in both guiding 2΄-O-methylation as well as rRNA precursor processing (30–33).
Until the introduction of the crosslinking, ligation and sequencing of hybrids (CLASH) (34), experimental characterization of snoRNA target sites was laborious and addressed only a few sites at a time (35). Progress on method development was further driven by the need to generalize target identification approaches to other guide RNAs, such as the miRNAs (36). Interestingly, miRNA–target hybrids are produced by the action of endogenous ligases and can be obtained through crosslinking and immunoprecipitation (CLIP) of Argonaute proteins, without a specific ligation step (37). MiRNA targets inferred from the chimeric reads obtained with CLIP seem to behave more as canonical miRNA targets, responding more strongly to miRNA transfection, than CLASH-determined targets (38). Whether snoRNA–target chimeras can also be obtained from the CLIP of core snoRNPs has not been investigated.
In parallel with the capture of snoRNA–target interactions, efforts were undertaken to map 2΄-O-methylated riboses in ribosomal RNAs, also in high-throughput (2). Taking advantage of the resistance of 2΄-O-methylated riboses to alkaline hydrolysis, the RiboMeth-seq method was used to map 54 annotated and 1 predicted 2΄-O-methylated site in Saccharomyces cerevisiae and is now applied to the profiling of rRNA modifications in human cells as well (39).
Studies from various groups have recently expanded the set of human snoRNAs, beyond those that are catalogued in snoRNAbase (https://www-snorna.biotoul.fr/ (40)) (41–44). Taking advantage of the processing pattern that most C/D-box snoRNAs seem to follow (42) and of the small RNA sequencing data sets generated by the ENCODE consortium, we have recently constructed an updated catalog of human snoRNAs (44). Interestingly, in data sets from both small RNA sequencing and from core snoRNP CLIP we reproducibly identified snoRNA-like sequences which contained only a subset of the C/D box snoRNA-specific sequence elements. For most snoRNA-like molecules we could not predict target sites.
Given the surge in data sets pertaining to snoRNA interactions, we here sought to provide relevant computational analysis methods. First, we developed a model to identify chimeric sequences, composed of a C/D box-containing RNA and a corresponding target part, among the reads obtained by CLIP of core C/D-box snoRNPs. To further enable the functional characterization of the chimera-documented interactions, we developed a model to identify sites of 2΄-O-Me from RiboMeth-seq data (2). Our data supports the concept that some rRNA sites are only partially methylated (39) and indicates that some of the snoRNAs which are not known to guide 2΄-O-methylation interact with sites whose methylation is guided by other snoRNAs. Interactions with strong chimeric read support outside of the canonical snoRNA targets, do not seem to lead to 2΄-O-ribose methylation that can be detected with RiboMeth-seq. This suggests that the sensitivity of RiboMeth-seq is low or that C/D box snoRNA interaction with non-canonical targets may serve yet uncharacterized functions.
MATERIALS AND METHODS
CLIP of snoRNP core proteins
To identify chimeric snoRNA–target reads, we analyzed five CLIP data sets that were published before (42): two NOP58-CLIP (Gene Expression Omnibus (GEO) accession numbers GSM1067861 and GSM1067862), 1 NOP56-CLIP (GEO accession # GSM1067863) and 2 FBL-CLIP (GEO accession # GSM1067864 and GSM1067865). We also generated an additional FBL-CLIP data set with the protocol described in (45) (GEO accession GSE77027).
Identification of snoRNA–target chimera
SnoRNA and target sets
We obtained the most comprehensive annotation of human snoRNA sequences, genome coordinates and known or predicted targets from the human snoRNA atlas that was recently published (44). We downloaded the sequences of known snoRNA targets (rRNA and snRNA) from the snoRNA database (40) and we further obtained tRNA sequences from GtRNAdb (46). We added one tRNA sequence per codon to the set of putative snoRNA targets. The database of putative snoRNA targets thus consisted of the GRCh37 version of the human genome assembly, augmented with rRNA, snRNA and tRNA sequences.
Computational analysis of chimeric reads
Analogous to a previous study that developed a strategy to uncover chimeric miRNA–target reads from Argonaute-CLIP data (37), we here developed a method that uses snoRNA-specific information to identify snoRNA–target chimera in core snoRNP CLIP data sets. The challenge is that the very low frequency of chimeric reads in CLIP data sets and the short length of the snoRNA and target parts in the typically short reads obtained from CLIP can lead to a high rate of false positive chimeras, making it necessary to use additional information, such as the specific pattern of hybridization of the guide RNA to the target.
Read selection
We carried out an initial annotation of CLIP data sets with the CLIPZ web server (47), which provides as output genome-mapped reads with their respective annotations, as well as the unmapped reads. Because we look for snoRNA–target interactions that take place within the snoRNP complex, we expect that target sites are also captured on their own in the core snoRNP CLIP, just as miRNA targets are captured in Argonaute-CLIP (48). Thus, to reduce the search space, we used clusters of at least two overlapping genome-mapped reads as putative target regions. To have sufficiently long snoRNA and target parts in the chimeric reads, we only used unmapped reads longer than 24 nucleotides.
Detection of snoRNA subsequences in unmapped reads
To speed up the identification of snoRNA subsequences within unmapped reads we first generated all possible sub-sequences of 12 nucleotides in length (‘anchors’) from all snoRNAs. We then searched the unmapped reads for exact matches to any of these anchors and, when a match was found, we carried out the local alignment of the respective snoRNA to the unmapped read with the swalign python package (https://pypi.python.org/pypi/swalign) (parameters for a match = 2, mismatch = −5, gap opening = −6, gap extension = −4). For each chimeric read, we retained only the snoRNA(s) with the best local alignment score. To evaluate the significance of the alignment scores, we applied the same procedure to shuffled reads. For most of the reads, the score of the alignment with the snoRNA presumed to be contained in the read was much higher compared to the score of aligning the snoRNA to a shuffled version of the read (Supplementary Data). Thus, as it appears that many unmapped reads indeed contain snoRNA subsequences, we split chimeric reads into the part that could be aligned to a snoRNA (the ‘snoRNA fragment’) and the rest of the read (‘putative target fragment’). All reads with a putative target fragment of at least 15 nucleotides were considered candidate chimeras which we analyzed further as described below.
Annotation of putative target fragments extracted from chimeric reads
The search space for putative target fragments consisted of CLIPed sites as well as rRNA, snRNA and tRNA sequences, which we explicitly included because the reference genome assembly may not contain all of the repetitive loci of these RNAs. As the PAR-CLIP protocol yields reads in which C nucleotides are incorporated at the sites of crosslinked U's, before carrying out the mapping of the putative target fragments we generated single-point variants of the reads, with one C nucleotide changed to a U (37). For the mapping, we used Bowtie2 (49) in the local alignment mode with the following command line parameters: -f -D100 -L 13 -i C, 1 –score-min C, 30 –local -k 10. For reads that mapped to multiple genomic loci, we checked whether at least one of these loci corresponded to a canonical snoRNA target, rRNA or snRNA. If so, we kept only the canonical locus. Otherwise, we kept all putative target loci. The statistics for each experimental data set can be viewed in Supplementary Data.
Training a model of snoRNA–target interaction
To better distinguishing bona fide snoRNA–target interactions captured in chimeras from false positives, we developed an additional model as follows. We extracted putative target sites that were captured in multiple chimeras with the same snoRNA and had a PLEXY-predicted energy of interaction (28) lower than -12 kcal/mol. From the combined CLIP experiments we identified 362 such sites in the 28S and 18S ribosomal rRNAs. 67 of these are known to undergo 2΄-O-ribose methylation (we called these ‘positives’), whereas for the remaining 295 sites a modification is not so far known to occur (‘negatives’). For each site, we calculated the features described below and trained a model to predict the class (‘positive’ or ‘negative’) of sites in the 28S rRNA. We evaluated the performance of the model using the the known modification sites on the 18S rRNA as true positives and all other sites in the 18S rRNA as true negatives. As the performance was high, we combined the two data sets and retrained a model for the comprehensive identification of snoRNA–target interactions.
Feature definition and computation
Predicted energy of snoRNA–target interaction
PLEXY is a tool for the transcriptome-wide prediction of C/D box snoRNA targets. It uses nearest-neighbor energy parameters to compute thermodynamically stable C/D-box snoRNA - target RNA interactions (28,50), but applies additional rules to further reduce the false positive rate. For each putative target fragment that mapped to the database of putative targets (see section SnoRNA and target sets) we extracted a 50 nucleotides long sequence centered on the target part of the chimeric read, and calculated its interaction energy with the snoRNA also identified from the chimeric read. PLEXY also assigns the position of the snoRNA-induced modification and we kept this information for further analyses. To assess the value of the PLEXY score in identifying bona fide interactions, we shuffled the snoRNA associated with each target part in a chimeric read and repeated the calculation.
Target site accessibility
Known snoRNA–target site interactions involve perfect base-pairing of the nucleotides at the 3΄ end of the anti-sense box, which is anchored at the D box. This interaction region defines the 5΄ end of the target site. Therefore, we defined the accessibility of the target region as the probability that the 5΄-anchored 21 nts-long region in the target is in single stranded conformation within an extended region of 30 nucleotides upstream and 37 nucleotides downstream of 5΄ end of the putative site. We computed this value with CONTRAfold (51).
Nucleotide content of flanking regions
We defined the ‘Flanks A content’ as the proportion of adenines within the 67 nts-long region defined above. We similarly computed frequencies of other nucleotides. Because the frequency of adenines was most predictive of true interaction sites (Supplementary Data) we only used this feature in the model.
Model training
The histograms constructed separately for the positive and negative sites in the 28S and 18S rRNAs indicated that the features described above are informative for distinguishing positive from negative sites (Figure 1) and we therefore trained a generalized linear model (GLM) with the logit link function (logistic regression) using these features, with the Statsmodels python library (52). We built the model based on all 18S rRNA and 28S rRNA sites. The code that we used to extract putative snoRNA–target interactions from CLIP data can be obtained from the github (https://github.com/guma44/snoRNAHybridSearchPipeline) and additional information is available on the accompanying web site (http://www.clipz.unibas.ch/snoRNAchimeras).
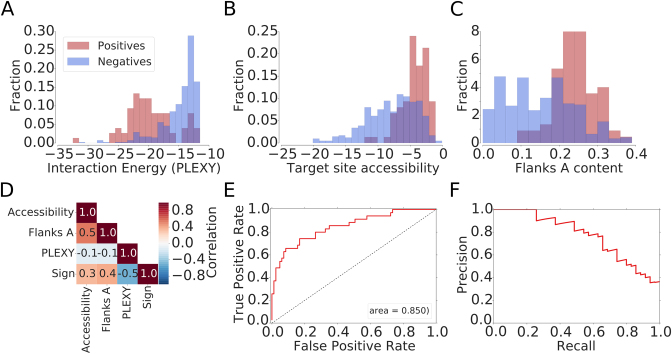
Figure 1.
Features that are relevant for the identification snoRNA–target interactions based on chimeric reads. Distributions of (A) the interaction energy calculated with PLEXY (28), (B) the target site accessibility calculated with CONTRAfold (51) and (C) the A nucleotide composition of the neighborhood of positive (known) and negative (captured in chimeras but unknown) snoRNA interaction sites. (D) Correlation between features used for model training and the indicator function, taking the value of −1 for negative and 1 for positive sites. (E) Receiver operating characteristic (ROC) curve and (F) Precision-Recall (PR) curve constructed based on snoRNA target predictions in 18S rRNA with the model trained on 28S rRNA target sites.
Annotation of modification sites
We annotated the biotypes of the targets in which predicted modification sites resided based on the ENSEMBL version 75 (53) and the RMSK table from University of California Santa Cruz genome browser (54), for the repeat elements. From the known interactions that we retrieved with our model from chimeric reads, we separately extracted those that involve the anti-sense elements at the D and D’ boxes and constructed profiles of coverage of the corresponding snoRNAs by fragments from chimeric reads, relative to the position of the D box. As shown in Supplementary Data, the appropriate anti-sense elements were captured preferentially in chimeric reads, although other parts of the snoRNAs have also been ligated with some frequency to the targets.
RiboMeth-seq
Preparation and sequencing of RiboMeth-seq libraries
The principle behind RiboMeth-seq is that nucleotides with a 2΄-O-Me ribose are resistant to alkaline hydrolysis. Thus, products of partial alkaline hydrolysis should not start or end at 2΄-O-Me sites, leading to an underrepresentation of these positions among read starts and ends. The read starts and ends thus provide a negative image of the methylation landscape (2). We carried out RiboMeth-seq experiments in HEK 293 cells, using either total RNA or poly(A)-enriched RNA from either the nucleus or cytoplasmic fractions. We also carried out the alkaline hydrolysis for different time intervals of 8, 14 or 20 min. The samples that we prepared were as follows:
RiboMethSeq_HEK_totalRNA_8min
RiboMethSeq_HEK_totalRNA_14min
RiboMethSeq_HEK_totalRNA_20min
RiboMethSeq_HEK_polyARNA_8min
RibomethSeq_HEK_cytoplasmic1_14min
RibomethSeq_HEK_cytoplasmic2_14min
RibomethSeq_HEK_nuclear1_14min
RibomethSeq_HEK_nuclear2_14min
We extracted total RNA with TRI Reagent (Sigma) and prepared the mRNA with the Dynabeads mRNA DIRECT Kit (Life Technologies), from HEK293 cells according to the manufacturer's instructions. For mapping of 2΄-O-methyl sites in rRNA we used 1 μg of total RNA as starting material. To explore the existence of 2΄-O-methyl sites in mRNAs, we used poly(A)-selected RNA (200 ng). In both protocols, the RNA was partially degraded under alkaline conditions in a sodium carbonate/bicarbonate buffer at pH 9.2 for 14 min and then put on ice. Samples were separated parallel to a low molecular weight marker ladder (10–100 nt) on a 15% denaturing polyacrylamide gel for 1 h at 1400 V and 20 W. The gel was stained with GR Green nucleic acid stain (Excellgen) for 3 min and fragmented RNA ranging from 20 to 40 nt was cut out from the gel and extracted overnight in 0.4 M NaCl. The RNA was precipitated with 1 μl of co-precipitant (GlycoBlue) in 75% ethanol at −20°C for 2 h and then centrifuged at maximum speed for 10 min at 4°C. The RNA pellet was washed twice with 70% ethanol and air-dried. The pellet was dissolved in water, the RNA was dephosphorylated with FastAP alkaline phosphatase (Thermo Scientific) at 37°C for 30 min and the enzyme was heat-inactivated at 75°C for 10 min. Subsequently, the RNA was phosphorylated with polynucleotide kinase (Thermo Scientific) in the presence of 1 mM ATP at 37°C for 1 h and then extracted with phenol-chloroform and precipitated in 80% ethanol, washed with 70% ethanol twice and air-dried. The pellet was dissolved in 8 μl mix (4 μl H2O, 1 μl 10× truncated T4 RNA Ligase 2 buffer, 1 μl 100 uM 3΄ rApp-adapter (5΄ adenylated 3΄ adapter, 5΄-App-TGGAATTCTCG GGTGCCAAGG-amino-3΄), 2 μl 50% DMSO), denatured at 90°C for 30 s and chilled on ice. Next, RNasin Plus RNase inhibitor (Promega) and T4 RNA Ligase 2 truncated were added to a final concentration of 2 U/μl and 30 U/μl, respectively, and the reaction was incubated at 4°C for 20 h over night. The next day, 1 μl of RT primer (100 μM; 5΄-GCCTTGGCAC CCAGAGAATTCCA-3΄) was added (for quenching of remaining 3΄ adapter molecules, preventing adapter dimers ligation in the next step), the samples were heated at 90°C for 30 s, at 65°C for 5 min, then placed on ice. A 5΄-adapter ligation mix was then directly added to the sample (1.5 μl 10 mM ATP, 1 μl 100 uM 5΄ RNA Adapter RA5 (Illumina TruSeq RNA sample prep kit), 1 μl T4 RNA Ligase 1 (20 U/μl), 0.5 μl RNasin Plus RNase inhibitor (40 U/μl) and reactions were incubated at 20°C for 1 h and 37°C for 30 min. The RNA was then directly reverse transcribed in a 30 μl reaction by adding dNTPs to 0.5 mM, DTT to 5 mM, 1× SSIV buffer, RNAsin to 2 U/μl and 1 μl Superscript IV reverse transcriptase (Life Technologies). The sample was incubated at 50°C for 30 min and inactivated at 80°C for 10 min. 5 μl of the resulting cDNA was then used in a pilot polymerase chain reaction (PCR) reaction. To this end, aliquots were taken from reactions at every second cycle between 12 and 22 cycles and analyzed on a 2.5% agarose gel. The number of cycles causing a first visible amplification was chosen for a large scale PCR (10 μl cDNA in a 100 μl reaction). The PCR product was ethanol precipitated and run along a 20 bp marker on a 9% non-denaturing polyacrylamide gel in TBE for 1 h at 250 V, 20 W. The gel was dismantled and stained for 3 min with GR Green. PCR products between 125 and 175 bp were cut out, the gel piece was mashed and DNA was eluted overnight into 400 μl of H2O. The supernatant was separated from the gel particles in a SpinX filter column (Costar), NaCl was added to 0.4 M, DNA was ethanol precipitated, the pellet washed in 75% ethanol and dissolved in 20 μl H2O. Libraries were sequenced on an Illumina HiSeq-2500 deep sequencer (GEO accession: GSE77024). Their summary can be found in Supplementary Data.
Mapping of RiboMeth-seq reads
We obtained ∼50 million reads for each of the RiboMeth-seq samples. We removed adaptors with Cutadapt (–minimum-length 15, other parameters left with default values) (55) and mapped the reads with STAR (parameters: –outFilterMultimapNmax 20 –outFilterMismatchNoverLmax 0.05 –scoreGenomicLengthLog2scale 0 –outSAMattributes All) (56) to a human GRCh37 assembly version-based transcriptome composed of rRNAs, snRNAs, tRNAs and snoRNAs (see SnoRNA and target sets section) as well as to lincRNAs, miscRNAs, and all unspliced protein coding genes (obtained from GRCh37 version of ENSEMBL, http://grch37.ensembl.org/index.html (53)).
Computation of the RiboMeth-seq score
For each target of interest such as the 18S rRNA, we calculated the log2 normalized (to a total library size of 106 reads) profile of cleavage positions. We used separately the 5΄ and 3΄ ends of the reads, as both ends are determined by alkaline hydrolysis. We then calculated the angle defined by the log2 coverage values at positions −1, 0 and +1 for each position along the RNA. An angle of 180° indicates that the frequency of cleavage at the three adjacent positions is identical, 0° indicates that the central position has very high coverage compared to the neighboring positions (and is therefore not protected from cleavage) and 360° indicates that the central position has no coverage (and therefore it is protected from cleavage) compared to the neighboring positions. As a RiboMeth-seq score we took the average angle computed based on 5΄ and 3΄ read ends. We used a score threshold of 290° for predicting sites in individual RiboMeth-seq experiments, favoring slightly recall over precision. Detailed statistics for individual experiments can be found in Supplementary Data. Finally, we used putative 2΄-O-Me sites that had a score above the threshold in at least one experiment and calculated their average score across the seven experiments. To determine a threshold for this average score and then compute the PR curve and Matthews correlation coefficient, we included among the positives the 19 sites that were did not score above the threshold in any individual experiment, but are known to undergo methylation. This resulted in a set of 105 known sites in the 18S and 28S rRNAs.
Validation of 2΄-O-methylation sites with RTL-P
Similar to the classic primer extension assays (57), the ‘Reverse Transcription at Low deoxy-ribonucleoside triphosphate (dNTP) followed by polymerase chain reaction’ method (RTL-P, (58)) takes advantage of the observation that cDNA synthesis through a 2΄-O-Me nucleotide is impaired when dNTPs are limiting. However, RTL-P is simpler and more sensitive than primer extension assays. RTL-P consists of a site-specific primer extension by reverse transcriptase at a low dNTP concentration and a semi-quantitative PCR amplification step, followed by agarose gel electrophoresis to obtain ratios of PCR signal intensities. To increase sensitivity and reproducibility, we implemented a real-time PCR (qPCR) step to facilitate the analysis of the signal intensities (qPCR parameters and primer sequences are shown in Supplementary Data).
Validation of 2΄-O-methylation at G2435 in 28S with mass spectrometry
The rRNA fragment isolation for mass spectrometry analysis (MS) was adapted from (59). The isolated fragment was treated with RNase T1 to yield a specific digestion pattern and dephosphorylated prior to LC–MS/MS analysis. As reference we used 11-nts long synthetic RNA oligonucleotides identical in sequence to the 28S rRNA around the G2435 site. 20 pmol of the unmodified synthetic UCCUGAGAGAU as well as the 2΄-O-methylated synthetic variant UCCUG*AGAGAU (the methylated G is indicated by *) were subjected to RNase T1 digestion and dephosphorylation.
Samples were analyzed on a LTQ-Orbitrap Elite mass spectrometer (Thermo Fisher Scientific) using a targeted LC-MS/MS workflow as described recently (60). UCCUG and UCCUG* specific MS assays were generated from the synthetic RNA oligonucleotides and applied to all samples. Data analysis was carried out using the Qual Browser tool of the Xcalibur software (version: 3.0.63). Full details of the sample preparation and LC-MS/MS experiment are described in Supplementary Data.
RESULTS
Crosslinking and immunoprecipitation of core snoRNPs captures snoRNA–target site chimeras
Although miRNAs and snoRNAs differ entirely in their function, they share the ability to guide ribonucleoprotein complexes to target RNAs. Thus, by analogy with miRNAs (37), we hypothesized that chimeric molecules, composed of snoRNAs and their targets, are captured in CLIP experiments that target one of the core snoRNP proteins. Therefore, we designed a method to identify snoRNA–target chimeric reads from among the unmapped (to genome or transcriptome) reads obtained in six photoreactive nucleoside-enhanced (PAR)-CLIP experiments that targeted one of the NOP58, NOP56 and FBL proteins. We found that on average, ∼10% of the reads that were not mapped to the genome or transcriptome had at least a 12-nt match to a snoRNA. However, only for ∼half of these reads was the remaining, putative target part of the read, longer than 15 nucleotides. Because multi-family snoRNAs have very low expression in the HEK 293 cells, most of the putatively chimeric reads yielded a high-scoring alignment to a single snoRNA, and only ∼20% aligned to multiple snoRNAs. A summary of the data obtained in all of these experiments is shown in Supplementary Data. To determine whether the apparent snoRNA–target chimera do reflect real interactions, we randomized the snoRNA assigned to each target fragment in the chimeras and calculated the predicted energies of interaction of the real and randomized pairs of molecules with PLEXY (28). Although the interaction energy predicted for the presumed chimeras was significantly lower compared to randomized sequence pairs, the difference between the average PLEXY energies was relatively low (∼1.2 kcal/mol, Figure 2A). This indicated that that a more sophisticated approach is needed to reliably identify snoRNA–target interactions from these data, which likely contain a large number of false positives.
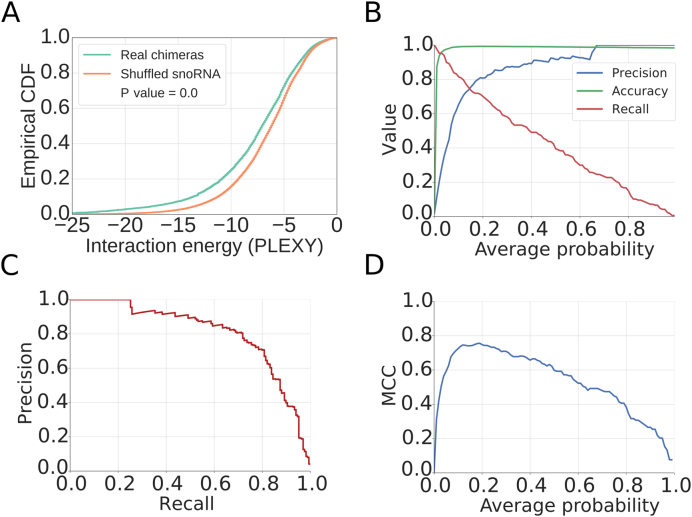
Figure 2.
Characterization of the model for inferring snoRNA–target interactions from chimeric reads. (A) Empirical cumulative distribution function of the interaction energy estimated with PLEXY between target fragment and snoRNA found in the chimera ('Real chimeras') or between target fragment and a randomly assigned snoRNA ('Shuffled snoRNA'). P-value from the Mann–Whitney U test is also shown. (B) Metrics illustrating the performance of the method, as a function of the minimum average probability of the considered sites from the 18S and 28S rRNAs. (C) Precision-Recall curve for the method. (D) Matthews correlation coefficient (MCC) as a function of the minimum average probability of the considered sites.
A model to identify high-confidence snoRNA–target chimeras
For training a model to predict snoRNA–target interactions, we selected presumed snoRNA–rRNA chimeras with low predicted energy of interaction (<−12 kcal/mol), separated them into those containing ‘positive’ target sites (known from previous studies) and those containing ‘negative’ target sites (not known to undergo snoRNA-guided methylation) and compared the distributions of features that have been found to play a role in other small RNA-guided interactions (61) between the two sets. The PLEXY interaction score (28) discriminated best these two data sets (as shown in Figure 1A and D). However, known snoRNA target sites also reside in structurally accessible regions (Figure 1B), rich in adenines (Figure 1C). We used chimeric reads involving the 28S rRNA to train a generalized linear model (GLM) based on these features and then tested the model on chimeric reads involving the 18S rRNA. The area under the receiver operating characteristic (ROC) curve was ∼85%, the model being able to recall 70% of the known interaction sites with 65% precision (Figure 1E and F). We then combined the sites in the 28S and 18S rRNAs, retrained the model, and found that at a score threshold of 0.15 we obtained good performance in predicting rRNA modification sites, with a Matthews correlation coefficient (MCC) of ∼0.75, precision of 0.75 and recall value of 0.74 (Figure 2B–D). Our predictions finally consisted of putative interactions that were supported by chimeric reads from at least two experiments and had a minimum score of 0.15. For completeness, we have also predicted interactions in individual data sets and show the overlap of sites obtained in pairs of experiments in Supplementary Data.
Chimeric reads reveal novel C/D box snoRNA target sites within structural RNAs
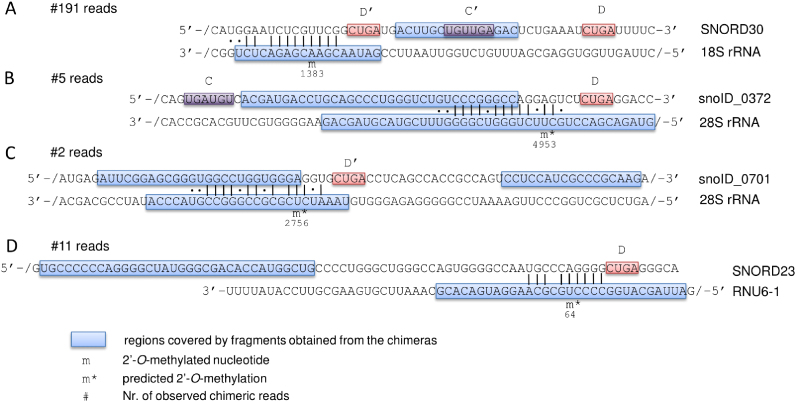
We applied the derived model to the full chimeric read data and identified 980 putative interactions, involving 852 unique target sites. We focused on the snoRNA interactions with structural RNAs, including not only the rRNAs, but also snRNAs, tRNAs and the snoRNAs themselves. Only one of the 2΄-O-Me sites in rRNAs that have been mapped so far is is ‘orphan’, meaning that its guide snoRNA is unknown. Our data indicates that this modification, located at position A1383 in the 18S rRNA (62), is guided by SNORD30 (Figure 3A), a snoRNA which was reported to guide the 2΄-O-methylation at position A3804 in 28S rRNA (63). The chimeric reads also revealed 35 potentially novel 2΄-O-Me sites in rRNAs (13 in 18S rRNA, 21 in 28S rRNA and 1 in 5.8S rRNA), some of which were found in interaction with multiple snoRNAs, thus corresponding to 40 novel interactions. Eleven of the 40 interactions involve snoRNAs that have been so far classified as ‘orphan’ (Supplementary Data). As an example, a snoRNA of unknown family (snoID_372) was found in three experiments in interaction with the 28S rRNA (predicted energy of interaction of −24.8 kcal/mol), in which it may guide the modification at position 4953 (Figure 3B). Similarly, in two experiments we found the recently uncovered snoID_0701 (family unknown) orphan snoRNA, which has low but broad expression across tissues (44), in a very stable (−28.2 kcal/mol) interaction with the 28S rRNA. This snoRNA is predicted to guide the 2΄-O-methylation at position U2756 (Figure 3C).
Figure 3.
Schematic representation of snoRNA–target interactions that are predicted based on chimeric reads from CLIP experiments. For each interaction, the snoRNA sequence is shown at the top and the target sequence at the bottom of the panel. ‘/’ indicates that only part of the sequence is shown, for readability. Regions of both snoRNAs and targets that are represented in the chimeric reads are encompassed in blue boxes. Indicated are also the presumed C/C’ and D/D’ boxes as well as the number of chimeric reads supporting each of the interactions. PLEXY-predicted sites of 2΄-O-methylation are marked by ‘m*’ and the previously mapped site is labeled with ‘m’.
SnRNAs are also known targets of scaRNA-guided 2΄-O-methylation. Of the nine such sites that are known, we were able to recover four over our prediction threshold. Additionally, we identified chimeric reads of the SNORD23, a snoRNA that is currently considered orphan, with the U6 snRNA (Figure 3D). In previous work (42), we have studied the methylation pattern of this snRNA by primer extension. We found evidence of 2΄-O-methylation at positions 60, 62 and 63 of U6, but not at position 64, which is predicted to be modified as a result of the interaction with SNORD23. Thus, the significance of this interaction, supported by 11 reads in our data, remains to be determined.
Additionally, we identified three apparent interactions of snoRNAs with other snoRNAs (SNORD5 with SNORD56, SNORD50 with SNORD57 and SNORD34 with SNORD38A), as well as an intra-molecular chimera of SNORD4B. The predictions are summarized in Supplementary Data and all alignments of putative chimeric reads to putative target sites and snoRNAs can be viewed at http://www.clipz.unibas.ch/snoRNAchimeras/.
Redundant targeting of known sites of 2΄-O-ribose methylation by multiple snoRNAs
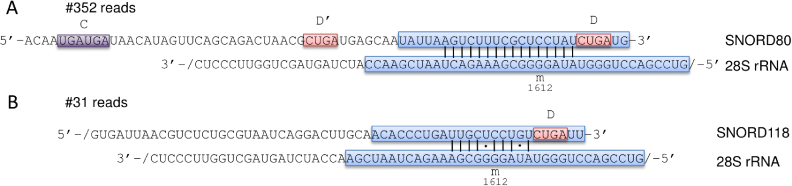
One of the main open questions in the snoRNA field concerns the targets and functions of the 330 orphan snoRNAs, which belong to 219 families (44). As mentioned in the introduction, some of these snoRNAs are involved in pre-rRNA processing. Interestingly however, the chimeric read data shows that SNORD118, also known as U8, a snoRNA which is necessary for the proper maturation of 5.8S and 28S rRNAs (31), interacts with the region of the 28S rRNA where the SNORD80 is known to guide the modification of G1612. The evidence for this interaction is very strong, chimeras having been captured in six distinct experiments (Figure 4). Although the base-pairing between SNORD118 and the putative target site is not as extensive as that of the SNORD80 snoRNA, it still includes 10 consecutive base-pairs, two of which are G-U base pairs. This example suggests that some snoRNAs are capable of interacting with sites whose 2΄-O-methylation is guided by other snoRNAs. We detected fragments from 66 orphan snoRNAs in chimeric reads.
Figure 4.
Similar to Figure 3, representation of the data supporting the interaction of both SNORD80 and SNORD118 with the 28S rRNA, around the known position of 2΄-O-methylation at G1612.
Identification of snoRNA-guided 2΄-O-Me sites with RiboMeth-seq
Surprisingly, ∼300 predicted interaction sites mapped to loci encoding protein-coding genes. To evaluate whether these sites could undergo 2΄-O-methylation, we implemented a high-throughput version of the recently developed RiboMeth-seq method (2). To be able to capture non-canonical targets, we carried out seven experiments, six using total RNA, which contained both the canonical rRNAs targets as well as other RNA species, and one using poly(A)+ RNAs, which was thereby strongly enriched in mRNAs. Two of the total RNA samples were prepared from total cell lysate, two from the nuclear fraction and two from the cytoplasmic fraction.
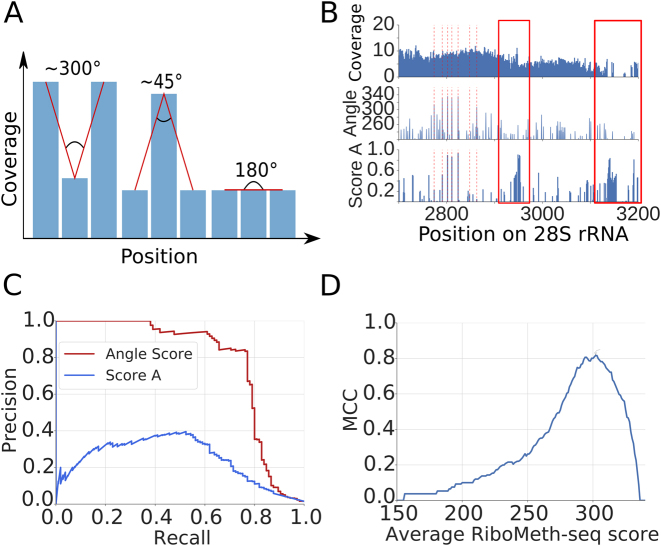
2΄-O-Me sites were previously identified from RiboMeth-seq by comparing the number of reads ending at a particular position in the target with the average number of reads ending at the flanking regions (‘score A’ in (2)). Reasoning that 2΄-O-methylation of adjacent nucleotides is very rare and that 2΄-O-Me positions should yield much fewer cleavage events compared to the unmethylated adjacent nucleotides, we here tested additionally another score. Specifically, at each position of a target of interest (e.g. 18S rRNA), we evaluated the shape (angle) of the trough defined by the log2 normalized read coverage at the specific position and the immediately adjacent positions (Figure 5A). We found that this score yields a higher precision compared to the ‘score A’ proposed before (2) (Figure 5B and C) and a very high Matthews correlation coefficient in classifying the sites (Figure 5D).
Figure 5.
Analysis of RiboMeth-seq data. (A) Strategy for evaluating the RiboMeth-seq data. The score was calculated based on the normalized log2 coverage of a position and of its immediately adjacent neighbors by RiboMeth-seq reads. A large score indicates stronger depletion of the position by 3/5΄ ends of reads and thus resistance to alkaline hydrolysis. (B) Example of a normalized log2 coverage profile along 28S rRNA and calculated scores (Angle and Score A). With red dashed lines positions of known 2΄-O-methylation sites are indicated. The red rectangles indicates regions where no 2΄-O-methylation has been mapped, which is also predicted by the angle score but not by score A. (C) Example of Precision-Recall curves obtained for the two scoring methods applied to rRNAs from the RiboMethSeq_HEK_totalRNA_8min experiment. (D) Matthews correlation coefficient (MCC) plot of average RiboMeth-seq score indicating the optimal angle score.
Applying this method to the combined RiboMeth-seq data, we identified 168 2΄-O-Me sites, 80 of which were known. These included 32 out of the 45 known 2΄-O-Me sites in 18S rRNA (71%), 44 out of the 60 in 28S rRNA (73%), the known site at position 75 in 5.8S rRNA, 2 sites in the U6 snRNA and one site in U1 snRNA. Figure 6 shows the location of previously known 2΄-O-methylation sites in the 18S and 28S rRNAs, as well as the corresponding chimeric read and RiboMeth-seq evidence that we obtained here for these rRNAs. The 88 novel sites were mostly located in canonical snoRNA/scaRNA targets—snRNA, rRNAs and tRNAs—34 being located in other RNA species. Although both the chimeric read method and RiboMeth-seq identified the majority of known 2΄-O-Me sites, with comparable sensitivity (∼70%), none of the 34 novel target sites in structural RNAs that were found in chimeric reads had a RiboMeth-seq score above the threshold.
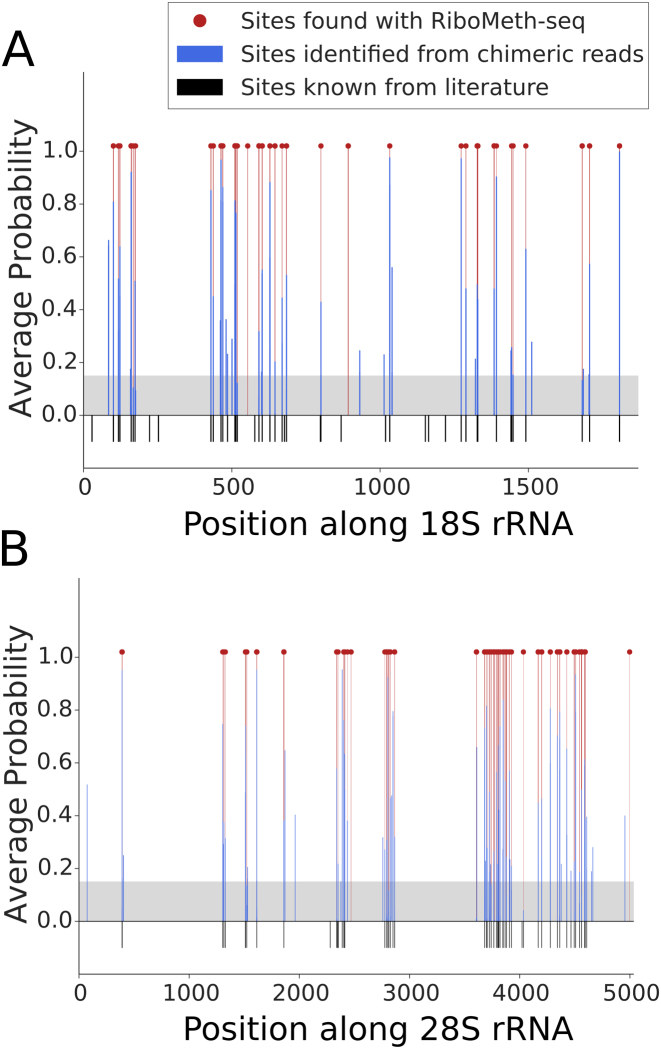
Figure 6.
Location of snoRNA interaction sites and 2΄-O-ribose methylation in the (A) 18S and (B) 28S ribosomal subunits. 2΄-O-Me positions that are known from literature are shown as black bars. Interaction sites identified from chimeric reads are shown as blue bars, with their associated probabilities. The gray area indicates the score threshold that we used to extract the high-confidence sites from chimeric reads. The locations of 2΄-O-Me sites identified with RiboMeth-seq are shown with red lines and dots.
Position G2435 in the 28S rRNA, captured in interaction with SNORD2, is partially methylated
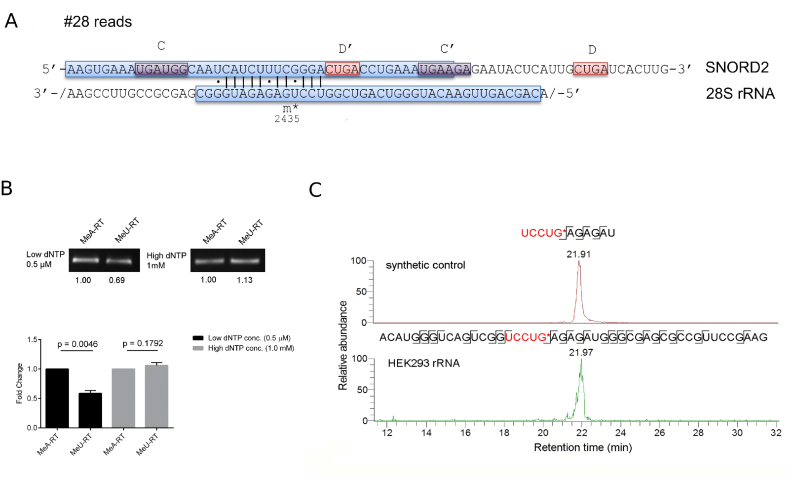
To assess whether the limited sensitivity of RiboMeth-seq could be a reason for the limited validation of sites that are reproducibly captured in chimeric reads, we investigated in depth the predicted SNORD2-guided 2΄-O-methylation of position G2435 in the 28S rRNA. This interaction was captured in four CLIP experiments (Figure 7A).
Figure 7.
SNORD2-guided 2΄-O-methylation of G2435 in the 28S rRNA (A) Schematic representation of the predicted interaction, which is supported by 28 chimeric reads (see also legend of Figure 3). (B) Confirmation of the G2435 2΄-O-methylation by RTL-P followed by agarose gel analysis and followed by qPCR analysis. Error bars represent the standard deviation of the mean, and the P -value of the t-test computed over three replicate experiments, each with three technical replicates is indicated. (C) Targeted LC-MS/MS analysis of UCCUG*, confirming the 2΄-O-methylation at G2435. A synthetic RNA oligonucleotide control (on top) and fragment A2416-G2461 from 28S rRNA (at the bottom) were digested with RNase T1 and specific transitions measured by targeted mass spectrometry.
We applied the recently published method ‘Reverse Transcription at Low deoxy-ribonucleoside triphosphate concentrations followed by polymerase chain reaction’ (RTL-P) (58), which we then followed with qPCR, to improve quantification. After showing that the method yields the expected results on a positive (position A1031 in the human 18S rRNA) and a negative control (U1991 in 28S rRNA) (Supplementary Data), we tested position G2435 in 28S rRNA. We found that the unanchored MeU-RT primer yielded significantly less cDNA and hence PCR product than the anchored MeA-RT primer at low dNTP concentrations (Figure 7B), indicating that the site indeed carries a 2΄-O-Me modification.
To unambiguously show that the RT stoppage at G2435 is due to 2΄-O-methylation, we applied targeted mass spectrometry (60). Figure 7C shows the extracted ion chromatograms of specific UCCUG* fragments that were measured in 28S rRNA as well as in a control sample. We manually checked the identities of the employed fragments using the control sample (Supplementary Data) and found that they matched those obtained from the HEK rRNA (Supplementary Data), confirming the presence of UCCUG* in the HEK sample. The LC-MS analysis also identified the unmodified fragment UCCUG from HEK rRNA (Supplementary Data), albeit at a lower level than UCCUG* (Supplementary Data). These results show that the G2435 28S rRNA site identified among the chimeric reads is predominantly 2΄-O-methylated.
mRNAs captured in chimeras with snoRNAs do not show evidence of 2΄-O-methylation
Finally, we wondered whether some of the chimera-supported interactions that did not reside in the typical snoRNA targets, particularly those annotated as being located in mRNAs, were also below the sensitivity of RiboMeth-seq. We therefore applied RTL-P to four mRNA-annotated sites, located in APP, CCDC93, DHFR and ZC3H12C transcripts, but did not find evidence of 2΄-O-methylation (data not shown).
DISCUSSION
High-throughput sequencing of samples prepared from cells that underwent various treatments have enabled the characterization of transcriptomes at ever increasing depth and resolution. This lead to the realization that the non-coding transcriptome is as large as the protein-coding fraction (64). New members of all classes of RNAs, including miRNAs and snoRNA have also been discovered (65, 66). The large number of novel molecular species increased the need for functional characterization methods, ideally in high-throughput. The aim of our study was to provide such methods for a specific class of non-coding RNAs, the C/D-box snoRNAs.
We have combined two high-throughput approaches, the first aiming to identify direct interactions between snoRNAs and targets and the second to map sites of 2΄-O-methylation transcriptome-wide. The first approach is based on the observation that chimeric reads, resulting from the ligation of a guide RNA to its target by endogenous ligases, are generated during CLIP (37). Whether CLIP of core snoRNP proteins can be used to identify snoRNA targets has not been investigated so far. Due to the low frequency of chimeric sequences (less than a percent of the reads (37)), the large ‘background’ of CLIP (48), and the short length of the snoRNA and target fragments that are captured, a snoRNA-centric analysis, taking into account the specific base-pairing pattern of snoRNAs with targets, is necessary. We found that a model that uses the predicted energy of interaction between the snoRNA and target, the accessibility of the target site and the A nucleotide context of the regions flanking the putative site, can identify over 70% of the known 2΄-O-Me sites in rRNAs, with similar specificity. The model assigns SNORD30 as guide for the ‘orphan’ A1383 site in the 18S rRNA, and identifies an interaction between the SNORD118 snoRNA, so far known to be involved in pre-rRNA processing (31), with G1612 in the 28S rRNA, whose methylation is guided by SNORD80. The multi-copy nature of many of the ‘orphan’ snoRNAs, other homologies that they have in the genome, and the presence of crosslinking-induced mutations in the CLIP data pose substantial challenges to the identification of their targets and will benefit from an increase in the length of the reads generated with CLIP.
The model also predicted 40 novel interactions with rRNAs as well as many outside of structural RNAs. To evaluate 2΄-O-methylation at these sites we implemented the RiboMeth-seq method (67). Although with this method we were able to recover the majority of known methylation sites, we did not find support for 2΄-O-methylation of any novel sites in rRNAs. To determine whether these results are partly due to the limited sensitivity of RiboMeth-seq, we used low-throughput methods to evaluate 2΄-O-methylation at position G2435 site in the 28S rRNA, which was supported by chimeric read data from four experiments. Both RTL-P and mass spectrometry provided evidence for 2΄-O-methylation at this site. These data, as well as a closer inspection of the RiboMeth-seq scores of this site in individual experiments, indicate that the site is only partially methylated. The cause and consequences of partial methylation at rRNA sites will be fascinating topics for future studies, as the evidence for partial and cell type-specific methylation of rRNAs is mounting (39,44). Of note, the interaction of SNORD48 with C1868 in the 28S rRNA, presumed to lead to the observed partial methylation of this site (39) was also captured in our chimeric read data. Another possibility to consider is that the CLIP-derived chimera provide evidence for snoRNA-rRNA interactions that are relevant for rRNA processing but not 2΄-O-methylation. Indeed, it has been proposed that the ancestral function of snoRNAs was in rRNA processing, a function that is still preserved in the U3, U8, U13, and U14 snoRNAs (26,30–32,68,69). Because the corresponding snoRNA-interacting sites may also need to be structurally accessible and have low-energy interaction with the snoRNAs, and because the D/D’ box sequences are short and not perfectly conserved in sequence, our method may misclassify these sites as 2΄-O-methylation sites. Because PLEXY enforces the snoRNA interaction with the target to take place close to already annotated D boxes, we do not expect such cases in our final list of candidates. However, a careful inspection of the hybrids and chimeric read alignments that we provide on the accompanying web site should help identify these cases.
Although the chimeric read data suggested some interactions of snoRNAs with mRNAs, we were not able to validate these with RiboMeth-seq. This could be due to the much lower expression of the mRNAs compared to rRNAs, which makes the reliable detection of troughs in read coverage difficult. However, the RTL-P method also failed to provide evidence of 2΄-O-methylation at mRNA sites (not shown). Thus, these sites may be the result of spurious ligation events. Alternatively, the snoRNA interaction with these sites may have other outcomes than 2΄-O-methylation. Consistent with this hypothesis, a recent study that analyzed globally RNA–RNA interactions also found many interactions of snoRNAs with mRNAs and further demonstrated a function of SNORD83B in controlling the level of its target mRNAs (70).
Finally, RiboMeth-seq revealed a few high-confidence sites for which we did not find any corresponding chimeric reads. The low rate of capture of interactions in the chimeric reads may account for this observation. Alternatively, the RiboMeth-seq-documented sites may be resistant to alkaline hydrolysis for reasons other than 2΄-O-Me. Supporting this latter hypothesis, these sites are generally located in rRNAs or snRNAs, molecules that are extensively modified and highly structured. In contrast to the known modification sites in rRNAs, which do not exhibit any nucleotide bias, the new sites recovered by RiboMeth-seq show a strong G-bias (not shown). This could again indicate that these sites are spurious or that modifications are introduced at these sites by specific enzymes such as the transfer RNA methyltransferase 7 protein (71). Interestingly, a recent study reported that G3771 in the 28S rRNA is 2΄-O-methylated, guided by SNORD15A (39). Although we also find strong evidence for the methylation of this site in our RiboMeth-seq data, we did not find chimeric read evidence for SNORD15A acting as guide at this site. Rather, our chimeric read data supports a previous prediction (72) that SNORD15A guides the methylation at A3764 in the 28S rRNA.
Our study thereby provides computational methods that enable the mapping of snoRNA–target interactions in high-throughput. We believe that the application of these two complementary and high-throughput approaches, namely interaction capture via CLIP-seq and RiboMeth-seq will accelerate the accurate assignment of snoRNA guides to already mapped as well as newly discovered sites of 2΄-O-methylation across cell types. This is especially relevant for studying the landscape of rRNA modification, which seems to be much more dynamic than anticipated, and for extending the study of snoRNA-guided methylation beyond species such as yeast and human.
Supplementary Material
ACKNOWLEDGEMENTS
R.G. and D.J.J. would like to thank the members of the Zavolan lab for input on the project.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Marie Curie Initial Training Network RNPnet project [289007 to R.G.] from the European Commission; Marie Curie Initial Training Network RNAtrain project [607720 to F.G.] from the European Commission. Funding for open access charge: Core funding.
Conflict of interest statement. None declared.
REFERENCES
- 1. Li S., Mason C.E.. The pivotal regulatory landscape of RNA modifications. Annu. Rev. Genomics Hum. Genet. 2014; 15:127–150. [DOI] [PubMed] [Google Scholar]
- 2. Birkedal U., Christensen-Dalsgaard M., Krogh N., Sabarinathan R., Gorodkin J., Nielsen H.. Profiling of ribose methylations in RNA by high-throughput sequencing. Angew. Chem. Int. Ed. Engl. 2014; 10.1002/anie.201408362. [DOI] [PubMed] [Google Scholar]
- 3. Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S., Cesarkas K., Jacob-Hirsch J., Amariglio N., Kupiec M. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012; 485:201–206. [DOI] [PubMed] [Google Scholar]
- 4. Schwartz S., Bernstein D.A., Mumbach M.R., Jovanovic M., Herbst R.H., León-Ricardo B.X., Engreitz J.M., Guttman M., Satija R., Lander E.S. et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell. 2014; 159:148–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schaefer M. RNA 5-methylcytosine analysis by bisulfite sequencing. Methods Enzymol. 2015; 560:297–329. [DOI] [PubMed] [Google Scholar]
- 6. Saletore Y., Meyer K., Korlach J., Vilfan I.D., Jaffrey S., Mason C.E.. The birth of the Epitranscriptome: deciphering the function of RNA modifications. Genome Biol. 2012; 13:175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lee M., Kim B., Kim V.N.. Emerging roles of RNA modification: m (6)A and U-tail. Cell. 2014; 158:980–987. [DOI] [PubMed] [Google Scholar]
- 8. Sun W.-J., Li J.-H., Liu S., Wu J., Zhou H., Qu L.-H., Yang J.-H.. RMBase: a resource for decoding the landscape of RNA modifications from high-throughput sequencing data. Nucleic Acids Res. 2015; 10.1093/nar/gkv1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Karijolich J., Kantartzis A., Yu Y.-T.. RNA modifications: a mechanism that modulates gene expression. Methods Mol. Biol. 2010; 629:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Heilman K.L., Leach R.A., Tuck M.T.. Internal 6-methyladenine residues increase the in vitro translation efficiency of dihydrofolate reductase messenger RNA. Int. J. Biochem. Cell Biol. 1996; 28:823–829. [DOI] [PubMed] [Google Scholar]
- 11. Tuck M.T., Wiehl P.E., Pan T.. Inhibition of 6-methyladenine formation decreases the translation efficiency of dihydrofolate reductase transcripts. Int. J. Biochem. Cell Biol. 1999; 31:837–851. [DOI] [PubMed] [Google Scholar]
- 12. Wang X., Lu Z., Gomez A., Hon G.C., Yue Y., Han D., Fu Y., Parisien M., Dai Q., Jia G. et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014; 505:117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang X., Zhao B.S., Roundtree I.A., Lu Z., Han D., Ma H., Weng X., Chen K., Shi H., He C.. N (6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015; 161:1388–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tycowski K.T., You Z.H., Graham P.J., Steitz J.A.. Modification of U6 spliceosomal RNA is guided by other small RNAs. Mol. Cell. 1998; 2:629–638. [DOI] [PubMed] [Google Scholar]
- 15. Dennis P.P., Omer A., Lowe T.. A guided tour: small RNA function in Archaea. Mol. Microbiol. 2001; 40:509–519. [DOI] [PubMed] [Google Scholar]
- 16. Zemann A., op de Bekke A., Kiefmann M., Brosius J., Schmitz J.. Evolution of small nucleolar RNAs in nematodes. Nucleic Acids Res. 2006; 34:2676–2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tollervey D., Lehtonen H., Jansen R., Kern H., Hurt E.C.. Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell. 1993; 72:443–457. [DOI] [PubMed] [Google Scholar]
- 18. Watkins N.J., Véronique S., Bruno C., Stephanie N., Patrizia F., Angela B., Matthias W., Michael R., Christiane B., Reinhard L.. A common core RNP structure shared between the small nucleoar box C/D RNPs and the Spliceosomal U4 snRNP. Cell. 2000; 103:457–466. [DOI] [PubMed] [Google Scholar]
- 19. Gautier T., Bergès T., Tollervey D., Hurt E.. Nucleolar KKE/D repeat proteins Nop56p and Nop58p interact with Nop1p and are required for ribosome biogenesis. Mol. Cell. Biol. 1997; 17:7088–7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Quinternet M., Marc Q., Marie-Eve C., Benjamin R., Decebal T., Bruno C., Xavier M.. Structural features of the box C/D snoRNP Pre-assembly process are conserved through species. Structure. 2016; 24:1693–1706. [DOI] [PubMed] [Google Scholar]
- 21. Jády B.E., Kiss T.. A small nucleolar guide RNA functions both in 2΄-O-ribose methylation and pseudouridylation of the U5 spliceosomal RNA. EMBO J. 2001; 20:541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Caffarelli E., Fatica A., Prislei S., De Gregorio E., Fragapane P., Bozzoni I.. Processing of the intron-encoded U16 and U18 snoRNAs: the conserved C and D boxes control both the processing reaction and the stability of the mature snoRNA. EMBO J. 1996; 15:1121–1131. [PMC free article] [PubMed] [Google Scholar]
- 23. van Nues R.W., Sander G., Grzegorz K., Sloan K.E., Matthew C., David T., Watkins N.J.. Box C/D snoRNP catalysed methylation is aided by additional pre-rRNA base-pairing. EMBO J. 2011; 30:2420–2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tollervey D., Lehtonen H., Carmo-Fonseca M., Hurt E.C.. The small nucleolar RNP protein NOP1 (fibrillarin) is required for pre-rRNA processing in yeast. EMBO J. 1991; 10:573–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tollervey D., Kiss T.. Function and synthesis of small nucleolar RNAs. Curr. Opin. Cell Biol. 1997; 9:337–342. [DOI] [PubMed] [Google Scholar]
- 26. Lafontaine D.L., Tollervey D.. Birth of the snoRNPs: the evolution of the modification-guide snoRNAs. Trends Biochem. Sci. 1998; 23:383–388. [DOI] [PubMed] [Google Scholar]
- 27. Fatica A., Tollervey D.. Insights into the structure and function of a guide RNP. Nat. Struct. Biol. 2003; 10:237–239. [DOI] [PubMed] [Google Scholar]
- 28. Kehr S., Bartschat S., Stadler P.F., Tafer H.. PLEXY: efficient target prediction for box C/D snoRNAs. Bioinformatics. 2011; 27:279–280. [DOI] [PubMed] [Google Scholar]
- 29. Chen C.-L., Perasso R., Qu L.-H., Amar L.. Exploration of pairing constraints identifies a 9 base-pair core within box C/D snoRNA–rRNA duplexes. J. Mol. Biol. 2007; 369:771–783. [DOI] [PubMed] [Google Scholar]
- 30. Kass S., Tyc K., Steitz J.A., Sollner-Webb B.. The U3 small nucleolar ribonucleoprotein functions in the first step of preribosomal RNA processing. Cell. 1990; 60:897–908. [DOI] [PubMed] [Google Scholar]
- 31. Peculis B.A., Steitz J.A.. Disruption of U8 nucleolar snRNA inhibits 5.8S and 28S rRNA processing in the Xenopus oocyte. Cell. 1993; 73:1233–1245. [DOI] [PubMed] [Google Scholar]
- 32. Cavaillé J., Hadjiolov A.A., Bachellerie J.P.. Processing of mammalian rRNA precursors at the 3΄ end of 18S rRNA. Identification of cis-acting signals suggests the involvement of U13 small nucleolar RNA. Eur. J. Biochem. 1996; 242:206–213. [DOI] [PubMed] [Google Scholar]
- 33. Li H.D., Zagorski J., Fournier M.J.. Depletion of U14 small nuclear RNA (snR128) disrupts production of 18S rRNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 1990; 10:1145–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kudla G., Granneman S., Hahn D., Beggs J.D., Tollervey D.. Cross-linking, ligation, and sequencing of hybrids reveals RNA-RNA interactions in yeast. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:10010–10015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Maden B.E. Mapping 2΄-O-methyl groups in ribosomal RNA. Methods. 2001; 25:374–382. [DOI] [PubMed] [Google Scholar]
- 36. Helwak A., Kudla G., Dudnakova T., Tollervey D.. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell. 2013; 153:654–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Grosswendt S., Filipchyk A., Manzano M., Klironomos F., Schilling M., Herzog M., Gottwein E., Rajewsky N.. Unambiguous identification of miRNA:target site interactions by different types of ligation reactions. Mol. Cell. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Breda J., Rzepiela A.J., Gumienny R., van Nimwegen E., Zavolan M.. Quantifying the strength of miRNA–target interactions. Methods. 2015; 85:90–99. [DOI] [PubMed] [Google Scholar]
- 39. Krogh N., Jansson M.D., Häfner S.J., Tehler D., Birkedal U., Christensen-Dalsgaard M., Lund A.H., Nielsen H.. Profiling of 2΄-O-Me in human rRNA reveals a subset of fractionally modified positions and provides evidence for ribosome heterogeneity. Nucleic Acids Res. 2016; 10.1093/nar/gkw482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lestrade L., Weber M.J.. snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic Acids Res. 2006; 34:D158–D162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jády B.E., Ketele A., Kiss T.. Human intron-encoded Alu RNAs are processed and packaged into Wdr79-associated nucleoplasmic box H/ACA RNPs. Genes Dev. 2012; 26:1897–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kishore S., Gruber A.R., Jedlinski D.J., Syed A.P., Jorjani H., Zavolan M.. Insights into snoRNA biogenesis and processing from PAR-CLIP of snoRNA core proteins and small RNA sequencing. Genome Biol. 2013; 14:R45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Machyna M., Kehr S., Straube K., Kappei D., Buchholz F., Butter F., Ule J., Hertel J., Stadler P.F., Neugebauer K.M.. The coilin interactome identifies hundreds of small noncoding RNAs that traffic through Cajal bodies. Mol. Cell. 2014; 56:389–399. [DOI] [PubMed] [Google Scholar]
- 44. Jorjani H., Kehr S., Jedlinski D.J., Gumienny R., Hertel J., Stadler P.F., Zavolan M., Gruber A.R.. An updated human snoRNAome. Nucleic Acids Res. 2016; 10.1093/nar/gkw386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Martin G., Gruber A.R., Keller W., Zavolan M.. Genome-wide analysis of pre-mRNA 3΄ end processing reveals a decisive role of human cleavage factor I in the regulation of 3΄ UTR length. Cell Rep. 2012; 1:753–763. [DOI] [PubMed] [Google Scholar]
- 46. Chan P.P., Lowe T.M.. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 2009; 37:D93–D97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Khorshid M., Rodak C., Zavolan M.. CLIPZ: a database and analysis environment for experimentally determined binding sites of RNA-binding proteins. Nucleic Acids Res. 2011; 39:D245–D252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hafner M., Landthaler M., Burger L., Khorshid M., Hausser J., Berninger P., Rothballer A., Ascano M. Jr, Jungkamp A.-C., Munschauer M.. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010; 141:129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tafer H., Hofacker I.L.. RNAplex: a fast tool for RNA–RNA interaction search. Bioinformatics. 2008. [DOI] [PubMed] [Google Scholar]
- 51. Do C.B., Woods D.A., Batzoglou S.. CONTRAfold: RNA secondary structure prediction without physics-based models. Bioinformatics. 2006; 22:e90–e98. [DOI] [PubMed] [Google Scholar]
- 52. Seabold S., Perktold J.. Statsmodels: Econometric and statistical modeling with python. Of the 9th Python in Science Conference. 2010. [Google Scholar]
- 53. Cunningham F., Amode M.R., Barrell D., Beal K., Billis K., Brent S., Carvalho-Silva D., Clapham P., Coates G., Fitzgerald S. et al. Ensembl 2015. Nucleic Acids Res. 2015; 43:D662–D669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rosenbloom K.R., Armstrong J., Barber G.P., Casper J., Clawson H., Diekhans M., Dreszer T.R., Fujita P.A., Guruvadoo L., Haeussler M. et al. The UCSC Genome Browser database: 2015 update. Nucleic Acids Res. 2015; 43:D670–D681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011; 17:10. [Google Scholar]
- 56. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Maden B.E., Corbett M.E., Heeney P.A., Pugh K., Ajuh P.M.. Classical and novel approaches to the detection and localization of the numerous modified nucleotides in eukaryotic ribosomal RNA. Biochimie. 1995; 77:22–29. [DOI] [PubMed] [Google Scholar]
- 58. Dong Z.-W., Shao P., Diao L.-T., Zhou H., Yu C.-H., Qu L.-H.. RTL-P: a sensitive approach for detecting sites of 2΄-O-methylation in RNA molecules. Nucleic Acids Res. 2012; 40:e157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Andersen T.E., Porse B.T., Kirpekar F.. A novel partial modification at C2501 in Escherichia coli 23S ribosomal RNA. RNA. 2004; 10:907–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Bauer M., Ahrné E., Baron A.P., Glatter T., Fava L.L., Santamaria A., Nigg E.A., Schmidt A.. Evaluation of data-dependent and -independent mass spectrometric workflows for sensitive quantification of proteins and phosphorylation sites. J. Proteome Res. 2014; 13:5973–5988. [DOI] [PubMed] [Google Scholar]
- 61. Gumienny R., Zavolan M.. Accurate transcriptome-wide prediction of microRNA targets and small interfering RNA off-targets with MIRZA-G. Nucleic Acids Res. 2015; 10.1093/nar/gkv050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Maden B.E. Identification of the locations of the methyl groups in 18 S ribosomal RNA from Xenopus laevis and man. J. Mol. Biol. 1986; 189:681–699. [DOI] [PubMed] [Google Scholar]
- 63. Tycowski K.T., Shu M.D., Steitz J.A.. A mammalian gene with introns instead of exons generating stable RNA products. Nature. 1996; 379:464–466. [DOI] [PubMed] [Google Scholar]
- 64. Mattick J.S. Non-coding RNAs: the architects of eukaryotic complexity. EMBO Rep. 2001; 2:986–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Morin R.D., O'Connor M.D., Griffith M., Kuchenbauer F., Delaney A., Prabhu A.-L., Zhao Y., McDonald H., Zeng T., Hirst M. et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008; 18:610–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Chen H.-M., Wu S.-H.. Mining small RNA sequencing data: a new approach to identify small nucleolar RNAs in Arabidopsis. Nucleic Acids Res. 2009; 37:e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Birkedal U., Christensen-Dalsgaard M., Krogh N., Sabarinathan R., Gorodkin J., Nielsen H.. Profiling of ribose methylations in RNA by high-throughput sequencing. Angew. Chem. Int. Ed. Engl. 2015; 127:461–465. [DOI] [PubMed] [Google Scholar]
- 68. Tollervey D. A yeast small nuclear RNA is required for normal processing of pre-ribosomal RNA. EMBO J. 1987; 6:4169–4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Enright C.A., Maxwell E.S., Eliceiri G.L., Sollner-Webb B.. 5΄ETS rRNA processing facilitated by four small RNAs: U14, E3, U17, and U3. RNA. 1996; 2:1094–1099. [PMC free article] [PubMed] [Google Scholar]
- 70. Sharma E., Sterne-Weiler T., O'Hanlon D., Blencowe B.J.. Global mapping of human RNA-RNA interactions. Mol. Cell. 2016; 62:618–626. [DOI] [PubMed] [Google Scholar]
- 71. Guy M.P., Shaw M., Weiner C.L., Hobson L., Stark Z., Rose K., Kalscheuer V.M., Gecz J., Phizicky E.M.. Defects in tRNA anticodon loop 2΄-O-methylation are implicated in nonsyndromic X-linked intellectual disability due to mutations in FTSJ1. Hum. Mutat. 2015; 36:1176–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kiss-László Z., Henry Y., Bachellerie J.P., Caizergues-Ferrer M., Kiss T.. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell. 1996; 85:1077–1088. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.