Abstract
Vascular endothelial growth factor (VEGF) plays a pivotal role in angiogenesis. Previous studies focused on transcriptional regulation modulated by proximal upstream cis-regulatory elements (CREs) of the human vegfa promoter. However, we hypothesized that distal upstream CREs may also be involved in controlling vegfa transcription. In this study, we found that the catalytic domain of Seryl-tRNA synthetase (SerRS) interacted with transcription factor Yin Yang 1 (YY1) to form a SerRS/YY1 complex that negatively controls vegfa promoter activity through binding distal CREs at −4654 to −4623 of vegfa. Particularly, we demonstrated that the −4654 to −4623 segment, which predominantly controls vegfa promoter activity, is involved in competitive binding between SerRS/YY1 complex and NFKB1. We further showed that VEGFA protein and blood vessel development were reduced by overexpression of either SerRS or YY1, but enhanced by the knockdown of either SerRS or yy1. In contrast, these same parameters were enhanced by overexpression of NFKB1, but reduced by knockdown of nfkb1. Therefore, we suggested that SerRS does not bind DNA directly but form a SerRS/YY1 complex, which functions as a negative effector to regulate vegfa transcription through binding at the distal CREs; while NFKB1 serves as a positive effector through competing with SerRS/YY1 binding at the overlapping CREs.
INTRODUCTION
Human vascular endothelial growth factor (VEGF) is an endothelial cell-specific mitogen and represents a larger family of growth factors, including VEGFA, VEGFB, VEGFC, VEGFD and placental growth factor, all of which differ in terms of expression pattern, receptor specificity and biological functions (1). In particular, VEGFA has been extensively studied because it is a multifunctional cytokine that localizes in many organs and plays a pivotal role in angiogenesis (2). Human vegfa, located on Chromosome 6 at 6p21.3 (3), is composed of eight exons separated by seven introns (4). Yet, it has several distinct variants, such as VEGFA121, VEGFA145, VEGFA148, VEGFA165, VEGFA183, VEGFA189 and VEGFA206, which occur as a consequence of alternative splicing (1). Many reports demonstrated that each isoform is differentially regulated, depending on its physiological situation (5–7).
The promoter of vegfa has been studied in many species, including mouse, rat, and human. The upstream 1.2 kb region has been studied in the mouse and rat vegfa (8), while the upstream 2.362 kb was investigated in the human vegfa (9). Surprisingly, the human vegfa promoter does not contain a consensus TATA box, but it does share considerable homology, including consensus sites for Sp1/Sp3, AP-2, Egr-1, STAT-3 and HIF-1 at the proximal region of promoters from various species (10). Some studies have reported the interaction of specific trans-acting factors bound at the proximal cis-regulatory elements (CREs) of vegfa. Other studies demonstrated Seryl-tRNA synthetase (SerRS) to be a noncanonical key regulator of angiogenesis based on a report showing that aminoacylation-defective SerRS (SerRS(T429A)) mRNA could restore the vasculature phenotype (11). SerRS contains a nuclear localization signal that directs cellular SerRS into the nucleus (12), suggesting that the effect of SerRS on vasculature development is independent of aminoacylation. Recently, Shi et al. (13) found that SerRS is an antagonist of c-Myc for regulation of vegfa expression through direct competition between SerRS and c-Myc in the proximal CREs of human vegfa.
Thus, most previous studies have focused on the proximal upstream CREs around 2 kb of the human vegfa promoter. Only a few studies have reported on the distal CREs of vegfa, e.g. Ford and D'Amore (14) who demonstrated that microphthalmia-associated transcription factor regulates vegfa promoter activity at −5 to −9 kb of vegfa in ARPE-19, a human retinal pigment epithelium cell line. This evidence supports the hypothesis that the distal upstream CREs of vegfa may also play a role in regulating the vegfa transcription.
Therefore, this study aimed to determine if SerRS and its interacting factors binding at distal upstream CREs such as at -2∼6kb lead to the control vegfa promoter activity. Indeed, we found that Yin Yang 1 (YY1), a transcriptional repressor, interacts with SerRS to form a SerRS/YY1 complex which functions as a negative effector to regulate vegfa transcription through binding distal CREs at −4654 to −4623, while NFKB1 serves as a positive effector through outcompeting SerRS/YY1 for binding at the same distal CREs.
MATERIALS AND METHODS
Gene cloning
Human HEK293T cells were homogenized with TRIzol Reagent (Bio-Rad) to extract total RNAs according to the manufacturer's instructions. First strand of cDNA was synthesized from 2 ng of total RNAs using SuperScript III (Invitrogen). RT-PCR was then performed to clone the coding regions of human SerRS, YY1, NFKB1, GATAD2A and HOXB9.
Plasmid constructs
The coding region of SerRS cDNA was inserted into pCMV-Flag to generate pCMV-Flag-SerRS. The mutated SerRS, SerRS(T429A), was obtained by PCR and inserted into pGEX-5X-1, pCMV-Flag and pCS2+ to generate pGEX-5X-1-SerRS(T429A), pCMV-Flag-SerRS(T429A) and pCS2-SerRS(T429A), respectively. SerRS cDNA without containing a unique domain, UNE-S, was obtained by PCR (SerRS-TBD-CD) and pGEX-5X-1-SerRS-TBD-CD was generated. Similarly, several plasmids were also generated, including pGEX-5X-1-YY1, pCS2-YY1, pCMV-Myc-YY1, pCMV-Flag-YY1, pGEX-5X-1-NFKB1, pCS2-NFKB1, pCMV-Myc-GATAD2A and pCMV-Myc-HOXB9. Plasmid pGL3-vegfa6k, in which the luciferase (luc) reporter was driven by −5868 to +55 of vegfa (15), was provided by the Imaizumi Lab. The −4654 to −4623 and −62 to −36 fragments were deleted from −5868 to +55 to generate pGL3-vegfa6k(△−4654 to −4623), pGL3-vegfa6k(△−62 to −36), and pGL3-vegfa6k(△−4654 to −4623)(△−62 to −36). For luc assay of DNA constructs driven by vegfa(−4654 to −4623) segment, we cloned −4654 to −4623 from pGL3-vegfa6k by PCR and generated three mutated sequences (Supplementary Table S2): (i) vegfa(−4654 to −4623)-mt1, mutation at the binding site for both YY1 and NFKB1, (ii) vegfa(−4654 to −4623)-mt2, mutation at YY1-specific binding site and (iii) vegfa(−4654 to −4623)-mt3, mutation at NFKB1-specific binding site. These fragments were then inserted into pGL3-SV40 to generate their derivative plasmids. Additionally, five copies of vegfa(−4654 to −4623) were synthesized and pGL3-SV-5X-vegfa(−4654 to −4623) was generated. The −4423 to −4392 segment was cloned from pGL3-vegfa6k by PCR and generated pGL3-SV-vegfa(−4423 to −4392) to serve as a negative control (Supplementary Table S2).
Protein expression and purification
Plasmids pGEX-5X-1, pGEX-5X-1- SerRS(T429A), pGEX-5X-1-YY1, pGEX-5X-1-NFKB1, pGEX-6P-1, pGEX-6P-1- SerRS-FL, pGEX-6P-1-SerRS-TBD, pGEX-6P-1-SerRS-CD-UNE-S, pGEX-6P-1-SerRS-CD and pGEX-5X-1-SerRS-TBD-CD were used to express recombinant proteins using 0.1 mM Isopropyl β-d-1-thiogalactopyranoside induction for 1 h at 37°C in an Escherichia coli BL21 expression system and purified by Glutathione resin (Clontech).
GST pull-down assay
Recombinant protein GST-SerRS(T429A) was purified by Glutathione resin (Clontech). GST pull-down assay of GST and GST-SerRS(T429A) with nuclear protein extracts isolated from HEK293T followed the protocol described in the NE-PER™ Nuclear Cytoplasmic Extraction Reagents Kit and Pierce™ Crosslink IP Kit (Thermo Fisher Scientific; TFS). The resultant GST pull-down products were analyzed by SDS-PAGE, followed by silver staining and in-gel digestion. For domain mapping of SerRS, equal amounts of GST and each GST-SerRS recombinant protein were separately incubated with recombinant Flag-YY1 overnight and then pulled down by Glutathione resin (Clontech).
Co-immunoprecipitation (Co-IP)
Plasmids pCMV-Flag-SerRS, pCMV-Flag-SerRS(T429A), pCMV-Myc-YY1, pCMV-Myc-GATAD2A and pCMV-Myc-HOXB9 were used for Co-IP, following the protocols described in the Pierce™ Crosslink IP Kit. The resultant immunoprecipitates were analyzed using antiserum against Flag (Abcam; 1 mg/ml; 1:30000) and Myc (Sigma-Aldrich (SA); 0.5 mg/ml; 1:1000).
In-gel digestion and LC–MS/MS analysis
The procedures were described by Chiang et al. (16) and used Mascot Distiller (Matrix Science). The resultant MGF file was searched using the Mascot search engine (v2.2, Matrix Science) with the following conditions: (i) protein database set as Swiss-Prot; (ii) taxonomy set as Homo sapiens; (iii) one trypsin missed cleavage allowed; (iv) peptide mass tolerance set at ±0.5 Da and fragment mass tolerance set at ±0.5 Da; (v) Carbamidomethyl (Cys) chosen as a fixed modification and (vi) oxidation (Met) and deamidation (Asn and Gln) chosen as variable modifications.
Dual luc assay
The protocol described by Promega was followed. In HEK293T, we co-transfected 50 ng of phRG-TK (an internal control), and 200 ng of each examined plasmid in the control group. In the experimental group, we co-transfected 50 ng of phRG-TK, 200 ng of each examined plasmid plus 2 μg of plasmid pCS2-SerRS(T429A), 1.5 μg of plasmid pCS2-YY1 or 1.5 μg of plasmid pCS2-NFKB1. In C2C12, we co-transfected 50 ng of pRL-SV40 (an internal control) and 500 ng of each examined plasmid in the control group. In the experimental group, we co-transfected 50 ng of pRL-SV40, 500 ng of each examined plasmid plus 1 μg of plasmid pCS2-SerRS(T429A), 1.5 μg of plasmid pCS2-YY1, 1.5 μg of plasmid pCS2-NFKB1.
Chromatin immunoprecipitation (ChIP) assayf
For analysis of the upstream 6k of vegfa, we used Searching Transcription Factor Binding Sites v 1.3 (TFSEARCH). ChIP assays were performed according to the manufacturer's protocol (EZ-Magna ChIP™ A/G ChIP Kits; Millipore). Nuclear DNAs were sonicated after cells were fixed. IP was performed with 10 μg DNA using one of the following antisera against IgG (Millipore; 1 mg/ml): SerRS (Abnova; 25 mg/ml), YY1 (Santa Cruz (SC); 0.2 mg/ml) and NFKB1 (SC; 2 mg/ml). Elutes were pooled and heated at 62°C with proteinase K (Millipore; 10 mg/ml) at least 2 h to reverse crosslinking. DNA fragments were purified with DNA Spin column (Millipore). The immunoprecipitated DNA was analyzed by PCR and qPCR, which used SYBR Green detection. Starting quantities were determined based on a common standard curve generated using HEK293T genomic DNA. Relative amounts of each chromatin fragment were then extrapolated on the basis of their threshold cycle values and determined by the percentage of the total input DNA. All qPCRs were carried out in triplicate. Primers used for ChIP assay were listed in Supplementary Table S3.
Electrophoretic mobility shift assay (EMSA)
The purified SerRS, YY1 and NFKB1 fusion proteins were used and single-stranded complementary oligonucleotide (co) corresponding to the −4654 to −4623 of vegfa were biotinylated using the Biotin 3΄-end DNA labeling kit (TFS). Biotinylated co pairs were annealed to make double-stranded and biotin-labeled probes (1 μM) by mixing in water, boiling for 5 min, and cooling overnight. Unlabeled co pairs were also annealed to make double-stranded competitor probes (1 μM). EMSA reaction solution was prepared by following the manufacturer's protocol (TFS) except of using 6% polyacrylamide native gel. The biotin-labeled probes were detected using horseradish peroxidase-conjugated streptavidin that binds to biotin and chemiluminescent substrate. For the Super Shift assays, antibodies against YY1 and NFKB1 were added and incubated for 20 min before performing the EMSA.
Western blot analysis
After HEK293T cells were lysed by whole-cell extract buffer (17) containing proteinase inhibitor cocktail (Roche), lysates were separated on SDS-PAGE and then blotted with corresponding antibodies. Antibodies against SerRS (1:1000), YY1 (1:1000), NFKB1 (1:5000), α-tubulin (SA; 1 mg/ml; 1:1000), VEGFA (SC; 0.2 mg/ml; 1:1500), mouse-HRP (SC; 400 mg/ml; 1:5000) and rabbit-HRP (SC; 400 mg/ml; 1:5000), we dechorionized, deyolked and lysed embryos in whole-cell extract buffer containing a proteinase inhibitor cocktail. The total extracted proteins in lysates were separated on SDS-PAGE and blotted with corresponding antibodies. Antibodies against SerRS, YY1 (1:2000), NFKB1 (1:2000), and α-tubulin were used.
qRT-PCR
After HEK293T cells were transfected with plasmid DNA or siRNA, total RNAs were isolated from cells by TRIzol Reagent. 2 μg of total RNA from each sample was transcribed to cDNA by SuperScript ? Reverse Transcriptase. All qPCR were performed using the Single Tube TaqMan® Gene Expression Assay (TFS).
Knockdown by small interfering RNA (siRNA) in cell lines
All siRNAs were designed and synthesized by Qiagen. All siRNAs sequences were listed in Supplementary Table S4. AllStars Negative Control siRNA (Qiagen), a validated non-silencing siRNA, was used as a negative control.
Zebrafish husbandry and microscopy observation
Wild-type and transgenic line Tg(fli1:EGFP)y1 (18) of zebrafish (Danio rerio) was used and their fluorescent signals were visualized by a fluorescent stereomicroscope (MZFLIII, Leica).
Knockdown by antisense morpholino oligonucleotides (MOs)
MOs were purchased from Gene Tools (USA). Their sequences were listed in Supplementary Table S5.
VEGF inhibitor treatment
Dechorionated embryos from 10 to 30 hpf were immersed in E3 medium containing either dimethylsulfoxide (served as control group) or 2.5 μM SU5416 (SA) (served as VEGF-inhibitor-treated group).
Statistical analysis
Data were obtained by the average value from three independent experiments and presented as mean ± SD. Difference levels were analyzed using Student's t-test.
RESULTS
YY1 protein preferentially binds SerRS protein
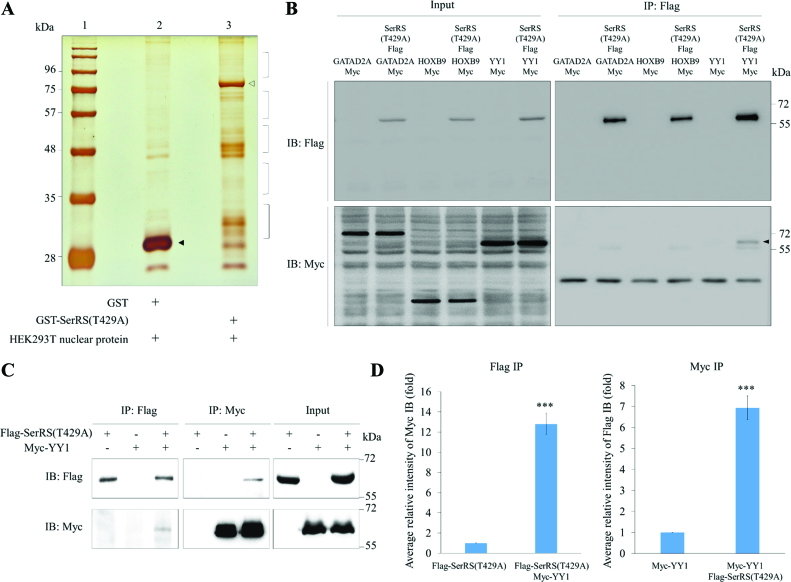
To search for proteins that participate in noncanonical functions of SerRS, we first generated recombinant protein SerRS(T429A) fused with GST. GST-SerRS(T429A) was then used for GST pull-down with nuclear proteins extracted from HEK293T cells. GST proteins pulled down were then separated by SDS-PAGE (Figure 1A). LC–MS/MS was used to analyze the nuclear proteins bound by GST-SerRS(T429A) (Supplementary Table S1). Finally, according to LC–MS/MS scores and numbers of peptides detected per candidate protein, three putative proteins interacting with SerRS(T429A) were selected: GATAD2A, HOXB9 and YY1. Next, using Co-IP, we found that YY1, but not GATAD2A or HOXB9, interacted with SerRS(T429A) (Figure 1B), a result which strongly suggests that YY1 might be a protein bound by SerRS(T429A).
Figure 1.
Screening and identification the putative proteins interacting with SerRS. (A) The GST pull-down assay. Nuclear proteins extracted from HEK293T were pulled down with the recombinant GST-SerRS(T429A). Lane 1, protein markers; lane 2, protein profiles of GST pull-down; lane 3, protein profiles of GST-SerRS(T429A) pull-down. Protein bands marked with brackets on lane 3 were excised for LC–MS/MS. GST and GST-SerRS(T429A) were marked with ▴ and △, respectively. (B) Screening of three putative proteins interacting with SerRS(T429A) by Co-IP. HEK293T cells were co-transfected with a plasmid expressing Flag-tagged SerRS(T429A) and a plasmid expressing Myc-tagged GATAD2A, HOXB9 or YY1. Cell lysate was immunoprecipitated with anti-Flag (IP: Flag), followed by Western blot (IB) using either anti-Flag to detect Flag-SerRS(T429A) (IB: Flag) or anti-Myc to detect Myc-GATAD2A, Myc-HOXB9 and Myc-YY1 (IB: Myc). A positive band was detected only when co-transfecting Flag-SerRS(T429A) and Myc-YY1 (marked with ▴). Input represents 10% of the total cell extract used for each immunoprecipitation. (C) Co-IP demonstrated the direct interaction between SerRS(T429A) and YY1. HEK293T cells were co-transfected with Myc-YY1 and SerRS(T429A). Afterwards, cell lysate was immunoprecipitated with either anti-Flag (IP: Flag) or anti-Myc (IP: Myc), followed by Western blot using anti-Flag to detect Flag-SerRS(T429A) (IB:Flag) and anti-Myc to detect Myc-YY1 (IB:Myc). Myc-YY1 fusion protein was immunoprecipitated with Flag-SerRS(T429A). Input represents 10% of the total cell extract used for each immunoprecipitation. (D) Quantification of the intensities of Flag and Myc, as shown on Co-IP, when the IP intensities of Flag-SerRS(T429A) and Myc-YY1 were individually normalized as 1. Data are calculated from three independent experiments and presented as mean ± SD (n = 3). Student's t-test was used to determine significant differences between each group (*P < 0.05, **P < 0.01 and ***P < 0.005).
Following up this experiment, we also employed Co-IP to demonstrate that YY1 is able to interact with mutated SerRS(T429A) (Figure 1C). Quantification data of SerRS and YY1 interaction revealed the following results. (i) When the intensity ratio of Myc level relative to Flag level for the Flag-SerRS(T429A) group was normalized as 1, that of the Flag-SerRS(T429A) plus Myc-YY1 group was 12.8. (ii) When the intensity ratio of Flag level relative to Myc level for the Myc-YY1 group was normalized as 1, that of the Myc-YY1 plus Flag-SerRS(T429A) group was 6.9 (Figure 1D). We then employed Co-IP to demonstrate that YY1 is also able to interact with wild-type SerRS (Supplementary Figure S1A). Quantification of SerRS and YY1 interaction revealed the following results. (i) When the intensity ratio of Myc level relative to Flag level for Flag-SerRS was normalized as 1, that of the Flag-SerRS plus Myc-YY1 was 18.9. (ii) When the intensity ratio of Flag level relative to Myc level for the Myc-YY1 was normalized as 1, that of the Myc-YY1 plus Flag-SerRS was 12.8 (Supplementary Figure S1B), also suggesting that YY1 interacts with SerRS.
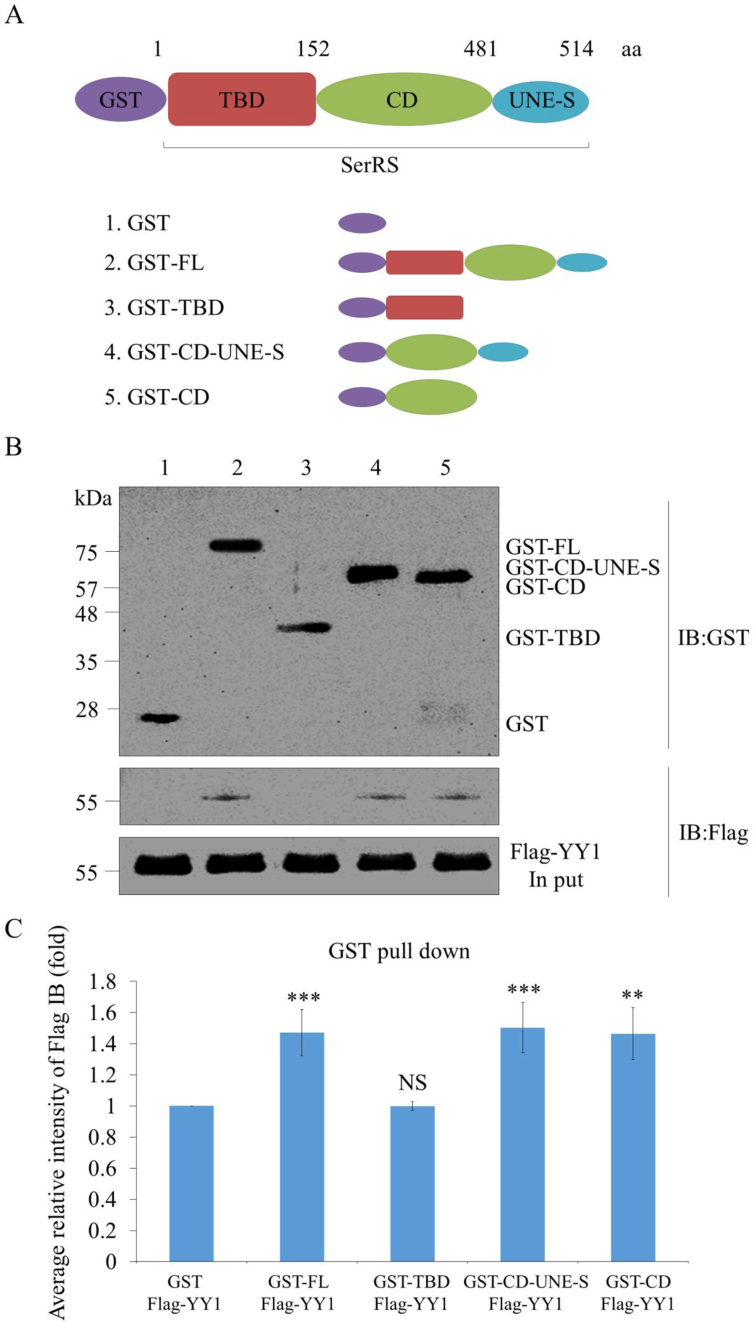
The catalytic domain of SerRS interacts with YY1
We performed SerRS domain mapping to elucidate the domain involved in SerRS-YY1 interaction. Plasmids pGEX-6P-1, pGEX-6P-1-SerRS-FL, pGEX-6P-1-SerRS-TBD, pGEX-6P-1-SerRS-CD-UNE-S and pGEX-6P-1-SerRS-CD were used to generate full-length (FL) recombinant SerRS (rSerRS) and rSerRS with various deleted domains fused with GST, including (i) tRNA-binding domain (TBD), (ii) catalytic domain (CD) combined with UNE-S and (iii) CD only (Figure 2A). After GST pull-down assay, GST-FL could pull down Flag-YY1, suggesting that SerRS and YY1 interact directly (Figure 2B). Domain mapping analysis revealed that the catalytic domain of SerRS is the key domain that interacts with YY1 (Figure 2B). Furthermore, SerRS-YY1 interaction revealed the following quantitative results. When the intensity ratio of Flag level relative to GST level for the GST plus Flag-YY1 was normalized as 1, that of the GST-FL plus Flag-YY1 was 1.32, GST-TBD plus Flag-YY1 was 1.01, GST-CD-UNE-S plus Flag-YY1 was 1.37, and GST-CD plus Flag-YY1 was 1.32 (Figure 2C), indicating that the catalytic domain of SerRS is the motif interacting with YY1.
Figure 2.
Characterization of the biochemical properties of SerRS/YY1 interaction. (A) Schematic illustration of SerRS domains and different deletions. FL: full length; TBD: tRNA-binding domain; CD: catalytic domain; UNE-S: C-terminal appended domain. (B) GST pull-down assay. SerRS/YY1 interaction is direct and mediated by the catalytic domain of SerRS. Different of SerRS domains fused with GST at N-termini were pulled down with purified Flag-YY1. GST-SerRS and Flag-YY1 were detected by Western blot using anti-GST and anti-Flag antibody, respectively. (C) Quantification of GST pull-down assay. GST immunoprecipitation intensities of GST-SerRS were quantified based on Flag-YY1 intensity normalized 1. Data are calculated from three independent experiments and presented as mean ± SD (n = 3). Student's t-test was used to determine significant differences between each group (*P < 0.05, **P < 0.01 and ***P < 0.005). NS: non-significant.
To clarify whether the UNE-S domain of SerRS is also responsible for the formation of SerRS/YY1 complex, we performed SerRS domain mapping to elucidate this question. Plasmids pGEX-6P-1, pGEX-6P-1-SerRS-FL, and pGEX-5X-1-SerRS-TBD-CD were used to generate the full-length (FL) of recombinant SerRS (rSerRS) and the UNE-S-deleted rSerRS fused with GST (Supplementary Figure S2A). After GST pull-down assay, GST-FL could pull down Flag-YY1 (Supplementary Figure S2B). Domain mapping analysis revealed that the UNE-S-deleted SerRS still interacted with YY1 (Supplementary Figure S2B). Furthermore, the interaction between SerRS and YY1 was quantified. As shown in Supplementary Figure S2C, when the intensity ratio of Flag level versus GST level in the GST plus Flag-YY1 group was normalized as 1, the intensity ratio of Flag level versus GST level in the GST-FL plus Flag-YY1 group was 1.45, while that of the GST-TBD-CD plus Flag-YY1 group was 1.41, indicating that the UNE-S domain of SerRS is not required for SerRS/YY1complex formation.
SerRS and YY1 are bound at the distal upstream sequences of vegfa
We performed a ChIP PCR with three primer-pairs designed to scan the distal upstream sequences of vegfa. The site of the three amplicons is located at the distal upstream sequences of vegfa (Figure 3A). The three amplicons immunoprecipitated by anti-SerRS, anti-YY1 and IgG (as a control) from HEK293T cell lysates were amplified by PCR. Results showed that only amplicon 2 had positive bands for SerRS and YY1 ChIPs (Figure 3B). We performed ChIP-qPCR to quantify the total input of chromatin DNA at SerRS and YY1 binding sites. The amounts of DNA immunoprecipitated by anti-SerRS, anti-YY1 and IgG from HEK293T cell lysates were quantified at amplicons 1, 2 and 3. The result showed that the highest expression level was located at amplicon site 2 (Figure 3C), suggesting that SerRS and YY1 might be involved in the activity of vegfa promoter through binding at the distal upstream CREs.
Figure 3.
Identification of the binding sites for SerRS and YY1 located at the upstream distal region of human vegfa. (A) Schematic illustrating the binding sites for SerRS and YY1 from −5868 to +55. The arbitrary amplicon sites were indicated on the bottom. (B) ChIP assay combined with PCR to analyze SerRS and YY1 binding sites. Immunoprecipitated DNA by anti-SerRS, anti-YY1 or control IgG from HEK293T cell lysates was amplified by PCR. A positive band was revealed for both SerRS ChIP and YY1 ChIP shown on amplicon 2. Input represents genomic DNA extract from HEK293T used for PCR analysis with 3 amplicon site primers. (C) ChIP-qPCR was used to quantify the total input of chromatin DNA at SerRS and binding sites. The amount of DNA immunoprecipitated by anti-SerRS, anti-YY1 antibodies and control IgG from HEK293T cell lysates was measured by qPCR on amplicons 1, 2 and 3. The amplicon 2 site located at the distal region of vegfa exhibited the highest expression. Results represent the percentages of total input of chromatin DNA. Data represent mean ± SD (n = 3).
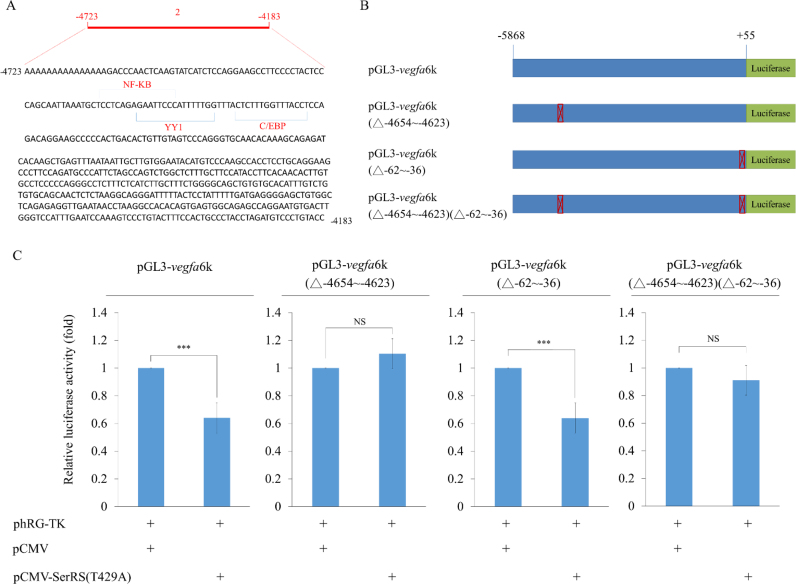
SerRS regulation of vegfa promoter activity depends on factors bound at -4654∼-4623
Using TFSEARCH, we revealed that YY1, NFKB and C/EBP were potentially bound at −4654 to −4623 (Figure 4A). To determine if SerRS was also involved in controlling vegfa promoter activity through binding at distal upstream sequences, we designed several constructs (Figure 4B), including (i) pGL3-vegfa6k, in which the luc reporter was driven by −5868 to +55 (6k) of vegfa; (ii) pGL3-vegfa6k(△−4654 to −4623), in which −4654 to −4623 was deleted from −5868 to +55; (iii) pGL3-vegfa6k(△−62 to −36), in which the −62 to −36 was deleted; (iv) pGL3-vegfa6k(△−4654 to −4623)(△−62 to −36), in which −4654 to −4623 and −62 to −36 were simultaneously deleted and (v) pCMV-SerRS(T429A), which was used to perform luc assay. Compared to the luc activity of HEK293T cells transfected with pGL3-vegfa6k, which was normalized as 100%, the luc activity of cells transfected with pGL3-vegfa6k plus pCMV-SerRS(T429A) was reduced to 63% (Figure 4C), suggesting that the addition of SerRS caused the decrease of vegfa promoter activity. In contrast, compared to the 100% normalized luc activity of pGL3-vegfa6k(△−4654 to −4623)-transfected cells, the luc of pGL3-vegfa6k(△−4654 to −4623) plus pCMV-SerRS(T429A)-transfected cells showed no difference (Figure 4C), suggesting that the distal CREs of −4654 to −4623 was a critical segment for SerRS to regulate the vegfa promoter activity driven by the −5868 to +55. Interestingly, compared to the 100% normalized luc activity of pGL3-vegfa6k(△−62 to −36)-transfected cells, the luc of pGL3-vegfa6k(△−62 to −36) plus pCMV-SerRS(T429A)-transfected cells decreased to 63% (Figure 4C), indicating that SerRS-induced reduction of vegfa promoter activity could not be driven by the −62 to −36 segment alone since promoter activity remained reduced, even when −62 to −36 was deleted. In the case of the 100% normalized luc activity of pGL3-vegfa6k(△−4654 to −4623)(△−62 to −36)-transfected cells, the luc of pGL3-vegfa 6k(△−4654 to −4623) (△−62 to −36) plus pCMV-SerRS(T429A)-transfected cells was only decreased to 90% (Figure 4C), indicating that SerRS primarily affects vegfa promoter activity at the distal region, i.e. the −4654 to −4623 segment. Next, we used the above constructs to perform the luc assay in non-oncogenic cell line C2C12. As shown in Supplementary Figure S3, the results obtained from C2C12 cells were similar to those of HEK293T cells. Thus, we concluded that a distal upstream CREs of −4654 to −4623 predominantly regulates the promoter activity of vegfa.
Figure 4.
The distal −4654 to −4623 is a crucial motif bound by SerRS to control the transcriptional activity of human vegfa. (A) The nucleotide sequences of −4654 to −4623 of vegfa and the putative binding sites for transcription factors YY1, NFKB and C/EBP on −4654 to −4623. (B) Schematic drawing of constructs used for luc assay. The luc activity driven by vegfa -6k (−5868 to +55) (pGL3-vegfa6k), pGL3-vegfa6k with a deletion of −4654 to −4623 from −6k (pGL3-vegfa6k(△−4654 to −4623)), a deletion of −62 to −35 (pGL3-vegfa6k(△−62 to −35)), and a deletion of both −4654 to −4623 and −62 to −35 (pGL3-vegfa6k(△−4654 to −4623)(△−62 to −35)). (C) The luc activity assay. The luc reporter plasmid and pCMV-SerRS(T429A) were co-transfected with each above construct into HEK293T cells. Luc activity was measured in three independent experiments. Data were presented as the mean ± SD (n = 3). *** indicates a significant difference at P < 0.005.
SerRS affects the binding affinity of YY1 and NFKB1 on vegfa at −4654 to −4623
To understand whether SerRS, YY1 and NFKB1 are competitively bound at −4654 to −4623 of vegfa, we employed EMSA and showed that the −4654 to −4623 of vegfa specifically interacted with YY1 (Supplementary Figure S4A) and NFKB1 (Supplementary Figure S4B). The Super-shift assay indicated that the shifted bands were specific (Figure 5A and B). The NFKB1 band-shift competed with that of YY1 (Figure 5C). Unexpectedly, we found that SerRS did not bind to −4654 to −4623 (Supplementary Figure S4C), indicating that SerRS is involved in regulating vegfa promoter activity at −4654 to −4623 in an indirect manner. To understand the impact of SerRS on YY1 binding, we added SerRS as a competitor within the YY1 band-shift experiment. The result showed that SerRS interacted with YY1 to form a SerRS/YY1 complex. This SerRS/YY1 complex bound at the −4654 to −4623 segment at higher affinity than YY1 alone (Figure 5D).
Figure 5.
The presence of SerRS affects the binding affinity of YY1 and NFKB1 on human vegfa at −4654 to −4623. (A) EMSA of DNA fragment −4654 to −4623 with YY1 protein. Anti-YY1 was used to perform the Super-shift assay. (B) EMSA of −4654 to −4623 with NFKB1. Anti-NFKB1 was used to perform the Super-shift assay. (C) EMSA of −4654 to −4623 that was bound competitively between YY1 and NFKB1. (D) The binding preference between −4654 and −4623 with SerRS/YY1 complex and −4654 and −4623 with YY1 alone. (E) ChIP-qPCR to quantify the total input of chromatin DNA bound by YY1 in SerRS-overexpressing (pCMV-SerRS(T429A)) and SerRS-knockdown (SerRS siRNA) cells. The relative percentage of chromatin DNA input of each group was measured when the pCMV-transfected group was set as 100%. Data represented mean ± SD (n = 3). (F) ChIP-qPCR to quantify the total amounts of chromatin DNA bound by NFKB1 in the SerRS-overexpressing (pCMV-SerRS(T429A)) and SerRS-knockdown (SerRS siRNA) cells. The relative percentage of chromatin DNA input of each group was measured when the pCMV-transfected group was set as 100%. Data represented mean ± SD (n = 3). ** and *** indicate the significant differences of values at P < 0.01 and P < 0.005 levels, respectively.
Based on the above evidence, we proposed that SerRS affects the binding affinity of YY1, which, in turn, competes with NFKB1 binding at −4654 to −4623. To further confirm this hypothesis, we quantified the total input of chromatin DNA based on YY1-ChIP in SerRS-overexpression (pCMV-SerRS(T429A)) and SerRS-knockdown (SerRS siRNA) constructs at −4654 to −4623 by qPCR. Results revealed that SerRS-overexpression increased the chromatin occupancy of YY1 at −4654 to −4623, while SerRS-knockdown decreased it (Figure 5E). In contrast, SerRS-overexpression decreased the chromatin occupancy of NFKB1 at −4654 to −4623, while SerRS-knockdown increased it (Figure 5F). Therefore, we concluded that SerRS affects the binding affinity of both YY1 and NFKB1 on vegfa at −4654 to −4623.
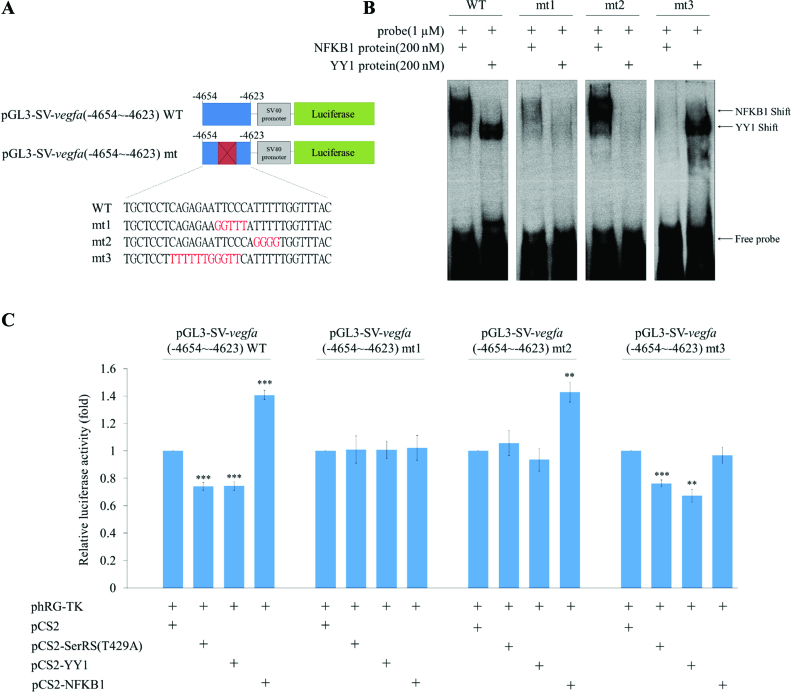
The promoter activity driven by −4654 to −4623 is also dependent on the binding of SerRS/YY1 complex or NFKB1
To know whether the promoter activity driven by a 32-nt (−4654 to −4623) upstream sequences of vegfa without including the complete upstream 6k sequences is also negatively regulated by SerRS/YY1 complex, but positively regulated by NFKB1, we constructed pGL3-SV-vegfa(−4654 to −4623), in which the SV40 promoter combined with −4654 to −4623 (Figure 6A), pGL3-SV-5X-vegfa(−4654 to −4623), in which the SV40 promoter combined with five copies of −4654 to −4623 (Supplementary Figure S5A) and plasmid pGL3-SV-vegfa(−4423 to −4392), in which the SV40 promoter combined with −4423 to −4392, served as a negative control (Supplementary Figure S6A). Compared to the 100% normalized luc activity of pGL3-SV-vegfa(−4654 to −4623)-transfected cells, the luc activity of pGL3-SV-vegfa(−4654 to −4623) plus pCS2-SerRS(T429A)- and pGL3-SV-vegfa(−4654 to −4623) plus pCS2-YY1-transfected cells was, in both cases, reduced to 74% (Figure 6C). In contrast, the luc activity of pGL3-SV-vegfa(−4654 to −4623) plus pCS2-NFKB1-transfected cells was increased to 140% (Figure 6C). Furthermore, as shown in Supplementary Figure S5C, when pGL3-SV-5X-vegfa(−4654 to −4623) containing five copies of −4654 to −4623 plus pCS2-YY1 were employed, a conclusion similar to that obtained from one copy of −4654 to −4623 could be drawn, except that the impact of promoter activity is more effective. In contrast to the results from pGL3-SV-5X-vegfa(−4654 to −4623), when pGL3-SV-vegfa(−4423 to −4392) was employed, its luc activity was not affected by SerRS(T429A), YY1 or NFKB1 (Supplementary Figure S6C). This line of evidence strongly suggested that the luc activity driven by the −4654 to −4623 segment is also negatively regulated by SerRS/YY1, but positively regulated by NFKB1.
Figure 6.
Transcriptional activity driven by the −4654 to −4623 segment of human vegfa is regulated by overexpression of either SerRS/YY1 or NFKB1. (A) Schematic illustration of the luc reporter driven by SV40 promoter (pGL3-SV40), SV40 combined with −4654 to −4623 of vegfa (pGL3-SV-vegfa(−4654 to −4623)) and SV40 combined with mutated −4654 to −4623 (pGL3-SV-vegfa(−4654 to −4623)-mt), as indicated. Three mutated sequences were labeled in red, including mt1, in which the binding sites for both YY1 and NFKB1 were mutated; mt2, in which the binding site for YY1 was mutated; and mt3, in which the binding site for NFKB1 was mutated. (B) EMSA of −4654 to −4623, mutated −4654 to −4623, and NFKB1 and YY1 proteins. (C) The luc activity was obtained from HEK293T cells when transfected with plasmids as indicated. The pGL3-SV-vegfa(−4654 to −4623) served as a control group, and its luc activity was set as 1. The luc activity was measured in three independent experiments. Data were presented as the mean ± SD (n = 3). Student's t-test was used to determine significant differences between each group (*P < 0.05, **P < 0.01 and ***P < 0.005).
Next, we designed three mutant plasmids, including pGL3-SV-vegfa(−4654 to −4623)-mt1, pGL3-SV-vegfa(−4654 to −4623)-mt2 and pGL3-SV-vegfa(−4654 to −4623)-mt3, which respectively reflect mutations at the binding site for both YY1 and NFKB1 (mt1), YY1-specific binding site (mt2), and NFKB1-specific binding site (mt3) (Figure 6A). EMSA results showed that the YY1 band-shift was neither observed in vegfa(−4654 to −4623)-mt1 nor in vegfa (−4654 to −4623)-mt2, whereas the NFKB1 band-shift was neither observed in vegfa(−4654 to −4623)-mt1 nor vegfa(−4654 to −4623)-mt3 (Figure 6B), indicating that these mutated sequences were specific and functional. Compared to the 100% normalized luc of pGL3-SV-vegfa(−4654 to −4623)-mt1-transfected cells, the luc of pGL3-SV-vegfa(−4654 to −4623)-mt1 plus pCS2-SerRS(T429A)-, pGL3-SV-vegfa(−4654 to −4623)-mt1 plus pCS2-YY1- and pGL3-SV-vegfa(−4654 to −4623)-mt1 plus pCS2-NFKB1-transfected cells showed no significant difference (Figure 6C). The 100% normalized 100% activity of pGL3-SV-vegfa(−4654 to −4623)-mt2-transfected cells was compared to the luc activities of cells transfected with pGL3-SV-vegfa(−4654 to −4623)-mt2 plus pCS2-SerRS(T429A) and pGL3-SV-vegfa (−4654 to −4623)-mt2 plus pCS2-YY1, and we also found no significant difference (Figure 6C). However, the luc of pGL3-SV-vegfa(−4654 to −4623)-mt2 plus pCS2-NFKB1-transfected cells increased to 147% (Figure 6C). Finally, compared to the 100% normalized luc activity of pGL3-SV-vegfa(−4654 to −4623)-mt3-transfected cells, the luc activity of pGL3-SV-vegfa(−4654 to −4623)-mt3 plus pCS2-SerRS(T429A)- and pGL3-SV-vegfa(−4654 to −4623)-mt3 plus pCS2-YY1-transfected cells were reduced to 77% and 70%, respectively (Figure 6C). In contrast, the luc activity of pGL3-SV-vegfa(−4654 to −4623)-mt3 plus pCS2-NFKB1-transfected cells was no different from that of pGL3-SV-vegfa(−4654 to −4623)-mt3-transfected cells (Figure 6C).
On the other hand, luc assays were also performed by the siRNA-knockdown approach. Compared to the 100% normalized luc activity of pGL3-SV-vegfa(−4654 to −4623)-transfected cells, the luc activity of pGL3-SV-vegfa(−4654 to −4623) plus SerRS-siRNA- and pGL3-SV-vegfa(−4654 to −4623) plus YY1-siRNA-transfected cells was increased to135% and 133%, respectively (Supplementary Figure S7). In contrast, the luc activity of pGL3-SV-vegfa(−4654 to −4623) plus NFKB1-siRNA-transfected cells was reduced to 81% (Supplementary Figure S7). Additionally, compared to the 100% normalized luc activity of pGL3-SV-vegfa(−4654 to −4623)-mt1-transfected cells, the luc activities of pGL3-SV-vegfa(−4654 to −4623)-mt1 plus SerRS-siRNA-, pGL3-SV-vegfa(−4654 to −4623)-mt1 plus YY1-siRNA- and pGL3-SV-vegfa(−4654 to −4623)-mt1 plus NFKB1-siRNA-transfected cells were no different (Supplementary Figure S7).
We next compared the 100% normalized luc activity of pGL3-SV-vegfa(−4654 to −4623)-mt2-transfected cells to the luc activities of pGL3-SV-vegfa(−4654 to −4623)-mt2 plus SerRS-siRNA- and pGL3-SV-vegfa(−4654 to −4623)-mt2 plus YY1-siRNA-transfected cells. We also found no difference. However, the luc activity of pGL3-SV-vegfa(−4654 to −4623)-mt2 plus NFKB1-siRNA-transfected cells was reduced to 82% (Supplementary Figure S7). Finally, compared to the 100% normalized luc activity of pGL3-SV-vegfa(−4654 to −4623)-mt3-transfected cells, the luc activity of pGL3-SV-vegfa(−4654 to −4623)-mt3 plus SerRS-siRNA- and pGL3-SV-vegfa(−4654 to −4623)-mt3 plus YY1-siRNA-transfected cells was reduced to 135% and 142%, respectively (Supplementary Figure S7). In contrast, the luc activity of pGL3-SV-vegfa(−4654 to −4623)-mt3 plus NFKB1-siRNA-transfected cells was no different from that of pGL3-SV-vegfa(−4654 to −4623)-mt3-transfected cells (Supplementary Figure S7).
Summing up the above results, we found that (a) the promoter activity driven by a segment of −4654 to −4623 of vegfa is also negatively regulated by the binding of SerRS/YY1 complex, but positively regulated by the binding of NFKB1 and (b) once the YY1- and NFKB1-specific binding sequences within −4654 to −4623 segment are mutated, SerRS/YY1 and NFKB1 are totally lost their ability to influence vegfa promoter activity. Therefore, we can conclude that the −4654 to −4623 segment of human vegfa is an important distal upstream CREs in regulating the promoter activity of vegfa.
The amount of VEGFA protein produced in cells is dependent on competitive binding between SerRS/YY1 complex and NFKB1 at −4654 to −4623 in vitro
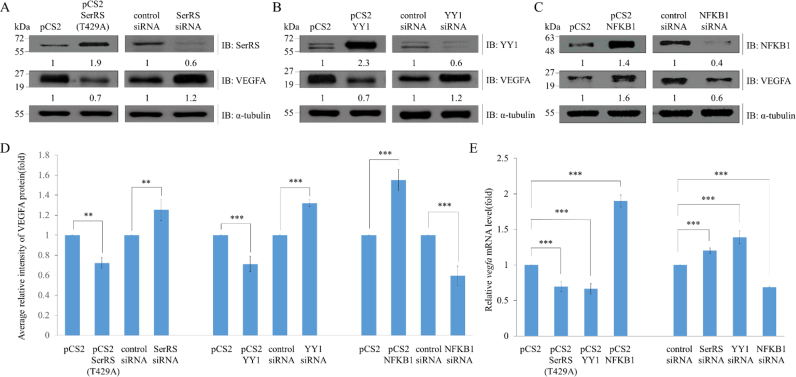
To study whether the amount of VEGFA protein is affected by knockdown and overexpression of SerRS, YY1 or NFKB1, we extracted total proteins from HEK293T cells individually transfected with overexpressed DNA fragments, including plasmids pCS2, pCS2-SerRS(T429A), pCS2-YY1 and pCS2-NFKB1, and DNA knockdown fragments, including control siRNA, SerRS siRNA, YY1 siRNA and NFKB1 siRNA. Total extracted proteins were analyzed by Western blot. The ratio of VEGFA relative to control α-tubulin for pCS2-transfected cells (control group) was normalized as 1, while for the pCS2- SerRS(T429A)-transfected cells, it was 0.7 (Figure 7A and D), indicating that the amount of VEGFA in the pCS2-SerRS(T429A)-transfected cells was reduced, compared to that of control pCS2-transfected cells. On the other hand, VEGFA relative to control α-tubulin level for the siRNA-transfected cells (control group) was normalized as 1, while for SerRS-siRNA-transfected cells, it was 1.2 (Figure 7A and D), indicating that the amount of VEGFA in the SerRS-siRNA-transfected cells was increased, compared to that of the control siRNA-transfected cells.
Figure 7.
The protein level of VEGFA is affected by SerRS, YY1 and NFKB1. (A) Total proteins (20 μg) were extracted from HEK293T cells individually transfected with pCS2, pCS2-SerRS(T429A), control siRNA and SerRS siRNA. Western blot (IB) analysis was performed using antiserum against SerRS, VEGFA, or α-tubulin (served as an internal control). SerRS and VEGFA levels relative to α-tubulin levels are also indicated. (B) Total proteins (20 μg) were extracted from HEK293T cells individually transfected with pCS2, pCS2-YY1, control siRNA and YY1 siRNA. Western blot analysis was performed using a specific antibody against YY1, VEGFA or α-tubulin (served as an internal control). The YY1 and VEGFA levels relative to α-tubulin levels are also indicated. (C) Total proteins (20 μg) were extracted from HEK293T cells individually transfected with pCS2, pCS2-NFKB1, control-siRNA and NFKB1-siRNA. Western blot was performed using a specific antibody against NFKB1, VEGFA or α-tubulin (served as an internal control). NFKB1 and VEGFA levels relative to α-tubulin levels are also indicated. (D) Statistical analysis of the average relative intensity of VEGFA protein level from each group was presented. Data are calculated from three independent experiments and presented as mean±S.D. (n = 3). Student's t-test was used to determine significant differences between each group (*P < 0.05, **P < 0.01 and ***P < 0.005). (E) The level of VEGFA mRNA was detected by qPCR in HEK293T cells transfected with plasmids of control DNA (pCS2), pCS2-SerRS, pCS2-YY1, pCS2-NFKB1, control siRNA, SerRS-siRNA, YY1-siRNA and NFKB1-siRNA. Data are calculated from three independent experiments and presented as mean ± SD (n = 3). Student's t-test was used to determine significant differences between each group (*P < 0.05, **P < 0.01 and ***P < 0.005).
We further evaluated the effect of YY1 on the amount of VEGFA produced in cells. First, VEGFA relative to α-tubulin for pCS2-transfected cells was normalized as 1, while that of pCS2-YY1-transfected cells was 0.7 (Figure 7B and D), indicating that the amount of VEGFA in the pCS2-YY1-transfected cells was reduced, compared to that of control pCS2-transfected cells. Next, VEGFA relative to α-tubulin for control siRNA-transfected cells was normalized as 1, while that of YY1-siRNA-transfected cells was 1.2 (Figure 7B and D), indicating that the amount of VEGFA in the YY1-siRNA-transfected cells was increased, compared to that of control siRNA-transfected cells. We also considered the effect of NFKB1 on the amount of VEGFA produced in cells. First, VEGFA relative to α-tubulin for pCS2-transfected cells was normalized as 1, whereas that of pCS2-NFKB1-transfected cells was 1.6 (Figure 7C and D), indicating that the amount of VEGFA in the pCS2-NFKB1-transfected cells was increased, compared to that of control pCS2-transfected cells. Second, VEGFA relative to α-tubulin for the control siRNA- transfected cells was normalized as 1, while that of NFKB1-siRNA- transfected cells was 0.6 (Figure 7C and D). Here, contrasting results showed that the amount of VEGFA in the NFKB1 siRNA-transfected cells was decreased when compared to the control siRNA-transfected cells.
Additionally, we analyzed the transcriptional level of vegfa by qPCR. Compared to the vegfa mRNA level of cells transfected with pCS2, which was normalized as 100%, the vegfa mRNA levels of cells transfected with pCS2-SerRS(T429A), pCS2-YY1 and pCS2-NFKB1 were 69, 66 and 190%, respectively (Figure 7E). In contrast, compared to the vegfa mRNA level of cells transfected with control siRNA, which was normalized as 100%, the vegfa mRNA levels of cells transfected with SerRS-siRNA, YY1-siRNA and NFKB1-siRNA were 120, 138 and 68%, respectively (Figure 7E). Furthermore, to exclude the possibility that SerRS affects the binding of YY1 or NFKB1 on VEGFA through regulating the expressions of YY1 and NFKB1, we analyzed the protein levels of YY1 and NFKB1 when SerRS was overexpressed or silenced. Compared to the cells transfected with pCS2 and control siRNA, results showed that the protein levels of YY1 and NFKB1 were not significantly different when SerRS was either overexpressed or silenced (Supplementary Figure S8). This line of evidence indicated that the protein level of VEGFA was decreased by adding either SerRS or YY1, but increased by adding NFKB1, suggesting, in turn, a negative effector role of SerRS/YYI complex in the context of human vegfa promoter activity, but a positive effector role for NFKB1.
Change in the amount of SerRS, YY1 and NFKB1 affects trunk angiogenesis in zebrafish embryos
To address whether the binding of either SerRS/YY1 complex or NFKB1 at distal CREs would impact on angiogenesis, we designed MOs to silence SerRS, YY1 and NFKB1, individually. Compared to the zSerRS-control-MO-, zyy1a-MO-control-MO-, and znfkb-control-MO-injected zebrafish embryos, the amounts of zSerRS, zYY1 and zNFKB1 were reduced to 20, 31 and 16% in the zSerRS-MO-, zyy1a-MO- and znfkb-MO-injected zebrafish embryos, respectively (Supplementary Figure S9A–C), indicating that knockdown of MOs is specific and effective.
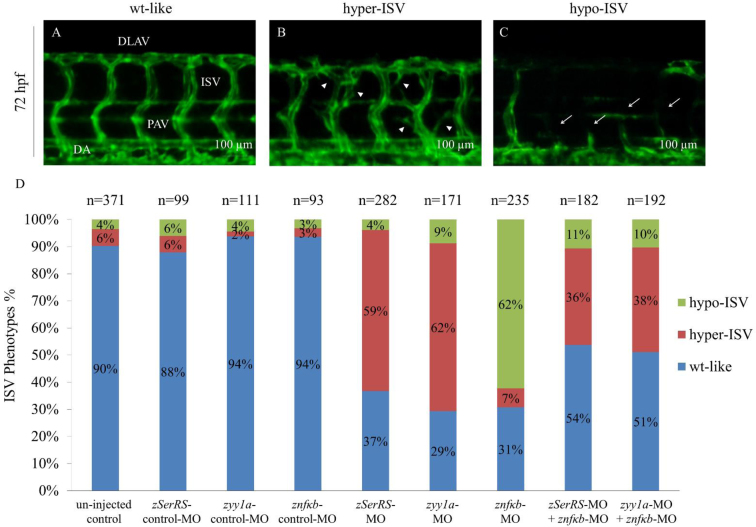
As shown in Figure 8A, in the un-injected control embryos during 72 hpf, we observed that i) blood vessels crossed the transverse myoseptum to form parachordal vessels and ii) most intersegmental vessels (ISVs) grew rostrally from dorsal aorta and then extended caudally, following the chevron-like contours of somites to reach the dorsal-lateral surface where tubes from adjacent ISVs fuse to form dorsal longitudinal anastomotic vessels. Compared to the un-injected embryos and the control-MO-injected embryos, we observed that embryos injected with zSerRS-MO and zyy1a-MO led to 59% and 62% of embryos, respectively, exhibiting abnormally branched blood vessels, i.e. hyper-ISV phenotype, among ISVs in trunk (Figure 8B and D). On the other hand, the znfkb-MO-injected group led to 62% of embryos exhibiting misdirected and less developed ISVs having the appearance of lateral asymmetrical formation, i.e. hypo-ISV phenotype (Figure 8C and D). However, compared to znfkb-MO, the percentages of hypo-ISV phenotype of embryos injected with znfkb-MO plus zSerRS-MO and znfkb-MO plus zyy1a-MO were reduced even more to 11% and 10%, respectively (Figure 8D). Additionally, compared to the percentages of hyper-ISV phenotype of embryos injected with zSerRS-MO and zyy1a-MO, the percentages of embryos injected with znfkb-MO plus zSerRS-MO and znfkb-MO plus zyy1a-MO were reduced to 36 and 38%, respectively (Figure 8D).
Figure 8.
The effects of SerRS, YY1 and NFKB1 on vascular development in zebrafish embryos. The percentages of Tg(fli1:EGFP)y1 embryos exhibiting ISV phenotypes at 72 hpf were counted. Different MOs were injected to specifically knock down the expressions of zebrafish SerRS, yy1a and nfkb, including un-injected control (n = 371), zSerRS-control-MO (4 ng; n = 99), zSerRS-MO (4 ng; n = 282), zyy1a-control-MO (3 ng; n = 111), zyy1a-MO (3 ng; n = 171), znfkb-control-MO (4 ng; n = 93), znfkb-MO (4 ng; n = 235), znfkb-MO (4 ng) plus zSerRS-MO (4 ng) (n = 182), and znfkb-MO (4 ng) plus zyy1a-MO (3 ng) (n = 192). The ISV angiogenesis of Tg(fli1:EGFP)y1embryos was observed under fluorescence microscope at 72 hpf. Three different levels of ISV angiogenesis in embryos were categorized: (A) wild-type-like (wt-like), (B) hypo-phenotype (hypo-ISV, white arrowheads) and (C) hyper-phenotype (hyper-ISV, white arrows). (D) The occurrence of different ISV phenotypes in each group, as indicated, was calculated in percentage of the total number (n) of examined embryos. DLAV: dorsal longitudinal anastomotic vessels; PAV: parachordal vessels; DA: dorsal aorta; ISV: intersegmental vessels. Scale bar: 100 μm.
To exclude the possibility that SerRS/YY1 complex affects blood vessel development through pathways other than VEGF, we employed the VEGF inhibitor SU5416. Compared to 5% of un-injected control embryos which were hyper-ISV, there was 62% of zyy1a-MO-injected embryos that were hyper-ISV (Supplementary Figure S10A–B, E–F and I). However, the SU5416-treated embryos resulted in 0% hyper-ISV and 80% hypo-ISV, while the embryos injected with zyy1a-MO combined with SU5416 led to 2% hyper-ISV and 77% hypo-ISV (Supplementary Figure S10C–D, G–H and I), indicating that injection of zyy1a-MO cannot rescue hypo-ISV defects in embryos treated with the VEGF inhibitor. Thus, we concluded that SerRS/YY1 affects blood vessel development specifically through the VEGF pathway.
DISCUSSION
Based on the luc assay, we demonstrated that SerRS could repress vegfa activity driven by an upstream 6k with a deleted −62 to −36 segment of vegfa. However, using the same assay, SerRS totally lost its ability to repress vegfa activity when driven by an upstream 6k with a deleted −4654 to −4623 segment, suggesting that SerRS requires a distal upstream CRE at −4654 to −4623 to control transcription of human vegfa. Similarly, Ford and D'Amore (14) found that luc activity driven by the upstream 2.1–5 kb of vegfa was significantly decreased, indicating the presence of a potentially important cis-element for repression of vegfa activity within the upstream 2.1–5 kb. Therefore, this author's work supports the above hypothesis because SerRS can reduce vegfa promoter activity through binding the intact −4654 to −4623 segment located at the upstream 2.1–5 kb of vegfa.
YY1 functions as an activator, repressor, or initiator of binding. Many reports demonstrated that YY1 can repress transcriptional activity through competition with transcription factors that bind at overlapping regulatory sequences. The YY1-binding sequences overlaps with that of serum response factor (SRF) in the upstream DNA sequences of α-actin such that YY1 binding with SRF is mutually exclusive (19–21), while YY1 acts as a repressor. Additionally, YY1 represses the transcription of rat serum amyloid Al gene through competing against NFKB which binds at the same regulatory sequences (22). On the other hand, YY1 is also reported to be a positive regulator. De Nigris et al. (23–24) reported that YY1 forms active complex with HIF1α at VEGF promoter to promote VEGF expression.
Moreover, YY1 can modulate transcriptional activity through its interaction with either co-activator or co-repressor. Wang et al. (25) reported that YY1 could increase transcriptional activity in retinoblastoma C33A cells when it interacted with YY1-associated protein (YY1AP), a co-activator, indicating that YY1 plays a role as activator. On the other hand, Yang et al. (26) reported that mRPD3, a mouse homologue of Histone deacetylase RPD3, is able to interact with the glycine-rich domain of YY1. When mRPD3 is overexpressed, transcriptional repressive activity driven by YY1 is increased, indicating that YY1, in this case, plays a repressive role. Here, we demonstrated that YY1 plays a repressive role by its interaction with co-repressor SerRS. Without directly binding DNA sequences, SerRS interacts with YY1 to form a SerRS/YY1 complex, which then binds the −4654 to −4623 of vegfa. SerRS/YY1 complex represses the gene expression of vegfa by successfully competing against NFKB1 which binds the overlapped CREs. Therefore, we suggest that SerRS and YY1 are transcriptional co-repressors that form a complex that competes with NFKB1 for binding at −4654 to −4623, resulting in inhibition of vegfa expression.
We demonstrated that VEGFA was affected by either overexpression or knockdown of NFKB1, SerRS and YY1. Xie et al. (27) reported that suppression of NFKB activity through a phosphorylation mutant, IkBαM, resulted in the decrease of VEGFA protein level, in turn inhibiting angiogenesis in human glioblastoma cells. This finding indicates that NFKB is a positive effector of VEGFA production and support our hypothesis. By either knockdown of vegfa (28) or inhibition of VEGF receptor (29) in Tg(fli1:EGFP)y1 zebrafish embryos, defective blood vessels in somite boundary during angiogenesis could be observed in vivo, indicating that the effect of VEGFA on blood vessel development can be directly monitored. Shi et al. (13) demonstrated that SerRS and c-Myc play opposite function on angiogenesis in zebrafish embryos. More specifically, SerRS decreases VEGFA, while c-Myc increases it. Here we found that blood vessel development in zebrafish embryos proliferates in the presence of SerRS or YY1 knockdown, while it is inhibited by knockdown of NFKB. Importantly, the defects caused by knockdown of either SerRS or YY1 can be rescued by injection of NFKB1, or vice versa. Collectively, we therefore suggest that SerRS/YY1 complex and NFKB1 modulate angiogenesis through controlling the production of VEGFA.
Aminoacyl tRNA synthetase (ARS) plays an essential role in translation. However, multiple ARSs exhibit noncanonical activity, which is manifested in such diverse biological functions as transcription regulation, apoptosis, inflammation and angiogenesis (30–32). Together with different ARS-interacting multifunctional proteins, ARSs can control gene transcription. For example, lysyl-tRNA synthetase associates with transcription factor USF2 and diadenosine polyphosphate Ap4A to form a multiprotein complex that regulates the transcription of USF2-responsive genes (33), which are responsible for tumorigenesis and glucose metabolism. Glutaminyl-tRNA synthetase interacts with apoptosis signal-regulating kinase 1 (ASK1) and inhibits cell death induced by ASK1 in a Glu-dependent manner (34). Thus, these ARSs play important roles in transcription regulation, supporting Park et al. (35) who concluded that ARSs could be considered hub proteins with key roles in protein networks. Additionally, some ARSs are known to be involved in inflammation and angiogenesis. The N-terminus of tyrosyl-tRNA synthetase functions as an angiogenic factor, whereas the C-terminus functions as a cytokine that regulates inflammatory responses (36). Tryptophanyl-tRNA synthetase is induced by interferon-γ stimulation, functioning as an angiostatic factor (37). Glutamyl-prolyl-tRNA synthetase functions as a potent angiostatic factor to suppress angiogenesis through the translational silencing of VEGFA (38). Recently, it has also been reported that SerRS regulates vegfa expression through direct competition between SerRS and c-Myc in the proximal CREs of vegfa (13). Here, we demonstrated that SerRS associates with YY1 to form a complex that competes with NFKB1 for binding at the distal CREs, thereby reducing VEGFA and inhibiting blood vessel formation, thus acting as a potent angiostatic factor in angiogenesis.
SerRS is considered one of the most ancient proteins involved in in protein synthesis. Recent studies demonstrated that a unique SerRS domain, termed UNE-S, exists only in vertebrates and that it harbors a NLS signal directing SerRS into the nucleus to affect vasculature development (12). It is suggested that this noncanonical activity results from the addition of UNE-S to the C-terminus of SerRS through evolution. Therefore, it is plausible that SerRS evolved this new biological function as a potent angiostatic factor for angiogenesis of vertebrates during embryonic development. Such domain additions as eukaryotes become increasingly complex are, correspondingly, progressive and accretive, and this pattern of domain additions is specific to ARS (30–31), which supports the relevance of domain additions for understanding the noncanonical activity of ARS.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the TC5, College of Life Science, NTU, for providing microscope. We also thank Dr Kazunori Imaizumi, Hiroshima University, for providing plasmid pGL3-vegfa6k, Dr Xiang-Lei Yang, Scripps Research Institute, for providing all the SerRS deletion domains plasmids, and the Lawson's Lab, UMASSMED, for giving zebrafish transgenic line Tg(fli1:EGFP)y1.
Huai-Jen Tsai and Chuan-Yang Fu conceived and coordinated the study and wrote the manuscript. Chuan-Yang Fu and Po-Chun Wang designed, performed and analyzed the experiments. All authors reviewed the results and approved the final version of the manuscript.
Footnotes
Present address: Huai-Jen Tsai, No. 46, Sec. 3, Zhongzheng Road, Sanzhi Dist., New Taipei City 252, Taiwan.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry of Science and Technology, Taiwan [105-2313-B-715-001-MY2]. Funding for open access charge: Ministry of Science and Technology, Taiwan.
Conflict of interest statement. None declared.
REFERENCES
- 1. Koch S., Tugues S., Li X., Gualandi L., Claesson-Welsh L.. Signal transduction by vascular endothelial growth factor receptors. Biochem. J. 2012; 437:169–183. [DOI] [PubMed] [Google Scholar]
- 2. Ferrara N. Molecular and biological properties of vascular endothelial growth factor. J. Mol. Med. (Berl.). 1999; 77:527–543. [DOI] [PubMed] [Google Scholar]
- 3. Vincenti V., Cassano C., Rocchi M., Persico G.. Assignment of the vascular endothelial growth factor gene to human chromosome 6p21.3. Circulation. 1996; 93:1493–1495. [DOI] [PubMed] [Google Scholar]
- 4. Tischer E., Mitchell R., Hartman T., Silva M., Gospodarowicz D., Fiddes J.C., Abraham J.A.. The human gene for vascular endothelial growth factor. Multiple protein forms are encoded through alternative exon splicing. J. Biol. Chem. 1991; 266:11947–11954. [PubMed] [Google Scholar]
- 5. Affleck D.G., Bull D.A., Bailey S.H., Albanil A., Connors R., Stringham J.C., Karwande S.V.. PDGF(BB) increases myocardial production of VEGF: shift in VEGF mRNA splice variants after direct injection of bFGF, PDGF(BB), and PDGF(AB). J. Surg. Res. 2002; 107:203–209. [DOI] [PubMed] [Google Scholar]
- 6. Watkins R.H., D'Angio C.T., Ryan R.M., Patel A., Maniscalco W.M.. Differential expression of VEGF mRNA splice variants in newborn and adult hyperoxic lung injury. Am. J. Physiol. 1999; 276:L858–867. [DOI] [PubMed] [Google Scholar]
- 7. Zhang L., Conejo-Garcia J.R., Yang N., Huang W., Mohamed-Hadley A., Yao W., Benencia F., Coukos G.. Different effects of glucose starvation on expression and stability of VEGF mRNA isoforms in murine ovarian cancer cells. Biochem. Biophys. Res. Commun. 2002; 292:860–868. [DOI] [PubMed] [Google Scholar]
- 8. Levy A.P., Levy N.S., Wegner S., Goldberg M.A.. Transcriptional regulation of the rat vascular endothelial growth factor gene by hypoxia. J. Biol. Chem. 1995; 270:13333–13340. [DOI] [PubMed] [Google Scholar]
- 9. Buteau-Lozano H., Ancelin M., Lardeux B., Milanini J., Perrot-Applanat M.. Transcriptional regulation of vascular endothelial growth factor by estradiol and tamoxifen in breast cancer cells: a complex interplay between estrogen receptors alpha and beta. Cancer Res. 2002; 62:4977–4984. [PubMed] [Google Scholar]
- 10. Shima D.T., Kuroki M., Deutsch U., Ng Y.S., Adamis A.P., D'Amore P.A.. The mouse gene for vascular endothelial growth factor. Genomic structure, definition of the transcriptional unit, and characterization of transcriptional and post-transcriptional regulatory sequences. J. Biol. Chem. 1996; 271:3877–3883. [DOI] [PubMed] [Google Scholar]
- 11. Fukui H., Hanaoka R., Kawahara A.. Noncanonical activity of seryl-tRNA synthetase is involved in vascular development. Circ. Res. 2009; 104:1253–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xu X., Shi Y., Zhang H.M., Swindell E.C., Marshall A.G., Guo M., Kishi S., Yang X.L.. Unique domain appended to vertebrate tRNA synthetase is essential for vascular development. Nat. Commun. 2012; 3:681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shi Y., Xu X., Zhang Q., Fu G., Mo Z., Wang G.S., Kishi S., Yang X.L.. tRNA synthetase counteracts c-Myc to develop functional vasculature. Elife. 2014; 3:e02349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ford K.M., D'Amore P.A.. Molecular regulation of vascular endothelial growth factor expression in the retinal pigment epithelium. Mol. Vis. 2012; 18:519–527. [PMC free article] [PubMed] [Google Scholar]
- 15. Miyagi H., Kanemoto S., Saito A., Asada R., Iwamoto H., Izumi S., Kido M., Gomi F., Nishida K., Kiuchi Y., Imaizumi K.. Transcriptional regulation of VEGFA by the endoplasmic reticulum stress transducer OASIS in ARPE-19 cells. PLoS One. 2013; 8:e55155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chiang Y.H., Wu Y.J., Lu Y.T., Chen K.H., Lin T.C., Chen Y.K., Li D.T., Shi F.K., Chen C.C., Hsu J.L.. Simple and specific dual-wavelength excitable dye staining for glycoprotein detection in polyacrylamide gels and its application in glycoproteomics. J. Biomed. Biotechnol. 2011; 2011:780108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lin C.Y., Lee H.C., Fu C.Y., Ding Y.Y., Chen J.S., Lee M.H., Huang W.J., Tsai H.J.. MiR-1 and miR-206 target different genes to have opposing roles during angiogenesis in zebrafish embryos. Nat. Commun. 2014; 4:2829. [DOI] [PubMed] [Google Scholar]
- 18. Lawson N.D., Weinstein B.M.. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev. Biol. 2002; 248:307–318. [DOI] [PubMed] [Google Scholar]
- 19. Gualberto A., LePage D., Pons G., Mader S.L., Park K., Atchison M.L., Walsh K.. Functional antagonism between YY1 and the serum response factor. Mol. Cell. Biol. 1992; 12:4209–4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lee T.C., Chow K.L., Fang P., Schwartz R.J.. Activation of skeletal alpha-actin gene transcription: the cooperative formation of serum response factor-binding complexes over positive cis-acting promoter serum response elements displaces a negative-acting nuclear factor enriched in replicating myoblasts and nonmyogenic cells. Mol. Cell. Biol. 1991; 11:5090–5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee T.C., Shi Y., Schwartz R.J.. Displacement of BrdUrd-induced YY1 by serum response factor activates skeletal alpha-actin transcription in embryonic myoblasts. Proc. Natl. Acad. Sci. U.S.A. 1992; 89:9814–9818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lu S.Y., Rodriguez M., Liao W.S.. YY1 represses rat serum amyloid A1 gene transcription and is antagonized by NF-kappa B during acute-phase response. Mol. Cell. Biol. 1994; 14:6253–6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. de Nigris F., Rossiello R., Schiano C., Arra C., Williams-Ignarro S., Barbieri A., Lanza A., Balestrieri A., Giuliano M. T., Ignarro L. J., Napoli C.. Deletion of Yin Yang 1 protein in osteosarcoma cells on cell invasion and CXCR4/angiogenesis and metastasis. Cancer Res. 2008; 68:1797–1808. [DOI] [PubMed] [Google Scholar]
- 24. de Nigris F., Crudele V., Giovane A., Casamassimi A., Giordano A., Garban H.J., Cacciatore F., Pentimalli F., Marquez-Garban D.C., Petrillo A. et al. . CXCR4/YY1 inhibition impairs VEGF network and angiogenesis during malignancy. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:14484–14489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang C.Y., Liang Y.J., Lin Y.S., Shih H.M., Jou Y.S., Yu W.C.. YY1AP, a novel co-activator of YY1. J. Biol. Chem. 2004; 279:17750–17755. [DOI] [PubMed] [Google Scholar]
- 26. Yang W.M., Inouye C., Zeng Y., Bearss D., Seto E.. Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global regulator RPD3. Proc. Natl. Acad. Sci. U.S.A. 1996; 93:12845–12850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xie T.X., Xia Z., Zhang N., Gong W., Huang S.. Constitutive NF-kappaB activity regulates the expression of VEGF and IL-8 and tumor angiogenesis of human glioblastoma. Oncol. Rep. 2010; 23:725–732. [PubMed] [Google Scholar]
- 28. Li W., Chen J., Deng M., Jing Q.. The zebrafish Tie2 signaling controls tip cell behaviors and acts synergistically with Vegf pathway in developmental angiogenesis. Acta Biochim. Biophys. Sinica. 2014; 46:641–646. [DOI] [PubMed] [Google Scholar]
- 29. Goishi K., Klagsbrun M.. Vascular endothelial growth factor and its receptors in embryonic zebrafish blood vessel development. Curr. Top. Dev. Biol. 2004; 62:127–152. [DOI] [PubMed] [Google Scholar]
- 30. Guo M., Schimmel P.. Essential nontranslational functions of tRNA synthetases. Nat. Chem. Biol. 2013; 9:145–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Guo M., Yang X.L., Schimmel P.. New functions of aminoacyl-tRNA synthetases beyond translation. Nat. Rev. Mol. Cell. Biol. 2010; 11:668–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yang X.L., Schimmel P., Ewalt K.L.. Relationship of two human tRNA synthetases used in cell signaling. Trends Biochem. Sci. 2004; 29:250–256. [DOI] [PubMed] [Google Scholar]
- 33. Lee Y.N., Razin E.. Nonconventional involvement of LysRS in the molecular mechanism of USF2 transcriptional activity in FcepsilonRI-activated mast cells. Mol. Cell. Biol. 2005; 25:8904–8912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ko Y.G., Kim E.Y., Kim T., Park H., Park H.S., Choi E.J., Kim S.. Glutamine-dependent antiapoptotic interaction of human glutaminyl-tRNA synthetase with apoptosis signal-regulating kinase 1. J. Biol. Chem. 2001; 276:6030–6036. [DOI] [PubMed] [Google Scholar]
- 35. Park S.G., Ewalt K.L., Kim S.. Functional expansion of aminoacyl-tRNA synthetases and their interacting factors: new perspectives on housekeepers. Trends Biochem. Sci. 2005; 30:569–574. [DOI] [PubMed] [Google Scholar]
- 36. Wakasugi K., Schimmel P.. Two distinct cytokines released from a human aminoacyl-tRNA synthetase. Science. 1999; 284:147–151. [DOI] [PubMed] [Google Scholar]
- 37. Wakasugi K., Slike B.M., Hood J., Otani A., Ewalt K.L., Friedlander M., Cheresh D.A., Schimmel P.. A human aminoacyl-tRNA synthetase as a regulator of angiogenesis. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:173–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sampath P., Mazumder B., Seshadri V., Gerber C.A., Chavatte L., Kinter M., Ting S.M., Dignam J.D., Kim S., Driscoll D.M., Fox P.L.. Noncanonical function of glutamyl-prolyl-tRNA synthetase: gene-specific silencing of translation. Cell. 2004; 119:195–208. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.