Abstract
Modular DNA assembly simplifies multigene engineering in Plant Synthetic Biology. Furthermore, the recent adoption of a common syntax to facilitate the exchange of plant DNA parts (phytobricks) is a promising strategy to speed up genetic engineering. Following this lead, here, we present a platform for plant biodesign that incorporates functional descriptions of phytobricks obtained under pre-defined experimental conditions, and systematically registers the resulting information as metadata for documentation. To facilitate the handling of functional descriptions, we developed a new version (v3.0) of the GoldenBraid (GB) webtool that integrates the experimental data and displays it in the form of datasheets. We report the use of the Luciferase/Renilla (Luc/Ren) transient agroinfiltration assay in Nicotiana benthamiana as a standard to estimate relative transcriptional activities conferred by regulatory phytobricks, and show the consistency and reproducibility of this method in the characterization of a synthetic phytobrick based on the CaMV35S promoter. Furthermore, we illustrate the potential for combinatorial optimization and incremental innovation of the GB3.0 platform in two separate examples, (i) the development of a collection of orthogonal transcriptional regulators based on phiC31 integrase and (ii) the design of a small genetic circuit that connects a glucocorticoid switch to a MYB/bHLH transcriptional activation module.
INTRODUCTION
Synthetic Biology aims to apply the engineering principles of Standardization, Modularity and Abstraction of function to Biotechnology. Synthetic Biology is influencing Plant Biotechnology primarily with the adoption of new cloning methods, now renamed as DNA assembly methods. A panoply of new assembly strategies have been developed based either on site-specific recombination (1), PCR-overlap (2,3) or Type IIS enzymes (4–6), which bring the efficiency required to facilitate complex multigene engineering. Type IIS systems based on the original Goldengate strategy (7) are particularly interesting in the context of Synthetic Biology, as they open the way for the definition of assembly standards that, if widely adopted, will facilitate the exchange of DNA parts. In this respect, a common syntax for Goldengate-based methods as MoClo (8) and GoldenBraid (9) has been recently proposed, supported and adopted by developers and users of those technologies (10). This Plant Syntax establishes the physical composition rules that define the way in which individual DNA components (hereafter referred to as phytobricks) (11) are to be connected together to create higher order modules and devices, as for instance how to clone a promoter next to a coding sequence (CDS) and a terminator to create a transcriptional unit (TU).
The definition and adoption of standard rules for physical assembly of genetic elements is a first step forward in Plant Synthetic Biology. A second immediate requirement is the generation of comprehensive collections of parts. These collections should cover a wide range of genetic functions and, for a better use, must be organized in databases that associate DNA parts with biological data. In this way, functional specifications will facilitate standard biological components (i.e. parts, modules or other devices) to be reliably and predictably assembled into higher order functional devices (12). A few collections of standardized parts for Plant Biotechnology have been recently created and deposited in repository databases (13), but to date very little has been advanced in the integration of experimental specifications in those databases. On the other hand, functional documentation can be of very little use unless certain uniformity in the experimental conditions is established beforehand. The definition of standard experimental conditions to be used in the description of parts within a given category is a strategy to partially circumvent this problem. Performing quantitative characterization of biological parts and then summarizing their properties in the form of standard datasheets has been previously proposed as a way to maximize their usability (14). Datasheets physically and functionally describe each element in a collection. Ideally, standard descriptions contained in datasheets should facilitate the creation of new assemblies and the anticipation of their performance (functionality) under different circumstances. This is conceivable specially with those collections whose elements are modular and reusable in biological sense, meaning that once created can be reassembled or replicated without changes (e.g. without introducing assembly seams or PCR-born errors).
In this paper we describe the development of GoldenBraid 3.0 (GB3.0), an assembly platform of reusable genetic elements for Plant Synthetic Biology that incorporates functional descriptions of its synthetic parts. We have built GB3.0 database on top of the previously described GB2.0 assembly system. In its previous version, the GoldenBraid database stored only the sequence information of each DNA element. New genetic devices were assembled using software-assisted tools instructed with the so-called GB physical composition rules (15). The GB3.0 assembly software adopts the new plant standard syntax (PSS) (10) and registers the assembly history of each composite part. Most notably, GB3.0 enables the definition of standard experiments and the introduction of experimental results in the database. Therefore, GB3.0 DNA elements are described by standard datasheets displaying their genealogy, their physical sequence information and their behaviour under standard experimental conditions. We illustrate how the new platform facilitates combinatorial optimization and incremental innovation with two biological examples: first, by assaying new combinations of un-related elements in the database, we created a set of novel orthogonal transcriptional regulators based on phiC31 recombinase. Second we assembled a small genetic circuit where a chemically-inducible regulatory module was connected to a second module controlling in series the activity of the dihydroflavonol 4-reductase (DFR) promoter, resulting in glucocorticoid-dependent modulation of the DFR promoter.
MATERIALS AND METHODS
Nicotiana benthamiana transient expression
For transient expression, plasmids were transferred to Agrobacterium tumefaciens strain GV3101 by electroporation. N. benthamiana plants were grown for 5 to 6 weeks before agroinfiltration in a growing chamber with 24°C (light)/20°C (darkness) in a 16-h-light/8-h-dark photoperiod. Agroinfiltration was carried out as previously described by Orzaez et al. (16). Briefly, overnight Agrobacterium cultures were pelleted and resuspended in agroinfiltration solution (10 mM MES, pH 5.6, 10 mM MgCl2 and 200 μM acetosyringone) to an optical density of 0.1 at 600 nm (OD600). Bacterial suspensions were incubated for 2 h at room temperature on a horizontal rolling mixer and they were mixed for experiments in which more than one GB element was used. Agroinfiltrations were carried out through the abaxial surface of the three youngest leaves of each plant with a 1 ml needle-free syringe.
Luciferase/Renilla assays
Leaves were coinfiltrated with standard GBelements listed in Supplementary Table S3. Leaf samples were collected at 3dpi for SE_001and at 4dpi for SE_002. All GB elements were assembled following GB standard procedures, following the protocols produced by GB software tools in GBcloning webpage. For the determination of the Luc/Ren activity one disc per leaf (d = 0.8 cm, ∼18–19 mg) was excised. For SE_001 leaf discs were kept in plates with or without inducer and samples were frozen in liquid nitrogen at the standard time points. Inductions were performed with D1756-dexamethasone (Sigma Aldrich) or E8875-β-estradiol (Sigma Aldrich) diluted to the final concentrations listed on each experiment in 0.02% Tween-80. For SE_002 excised leaf discs were directly freeze in liquid nitrogen after excision.
Leaf discs were homogenized and extracted with 150 μl of ‘Passive Lysis Buffer’, followed by 15 min of centrifugation (14000 ×g) at 4°C. Then, the supernatant was diluted 2:3 in Passive Lysis Buffer resulting in the working plant extract. Luciferase and Renilla activities were determined following the Dual-Glo® Luciferase Assay System (Promega) manufacturer's protocol with minor modifications: 10 μl of working plant extract, 40 μl of LARII and 40 μl of Stop&Glo Reagent were used. Measurements were made using a GloMax 96 Microplate Luminometer (Promega) with a 2-s delay and a 10-s measurement. Luc/Ren ratios were determined as the mean value of three samples coming from three independent agroinfiltrated leaves of the same plant and were normalized to the Luc/Ren ratio obtained for the internal references GB0166 or GB1116. Elements GB0166 and GB1116 are equivalent constructs featuring a weak pNOS promoter either in a pDGB1-series (GB0166) or a pDGB3-series (GB1116) vector backbone. The election of the internal reference was done according to the backbone of the tested GBelement.
Protoplasts isolation and MOT calculation
N. benthamiana protoplasts were isolated from leaves co-infiltrated 5 days earlier with two Agrobacterium strains carrying GB1287 and GB1288 at 1:1v/v proportion at seven different OD600 (0.1, 0.05 and five 1:3 dilutions ranging from 0.05 to 0.00021). Protoplast isolation was performed as previously described by Sang-Dong Yoo et al. (17) with minor modifications. Vacuum infiltration of cut leaves in enzyme solution was performed for 10 instead of 30 min. After filtration, intact protoplasts were further purified from dead protoplast and remaining cellular debris by the sucrose flotation method on 20% (w/v) sucrose. After the washing steps protoplasts were kept in WI solution (4 mM MES pH 5.7 containing 0.5 M mannitol and 20 mM KCl).
Expression of Yellow and/or the DsRed Fluorescent Proteins on the isolated protoplasts was detected with the photomultiplier of the LSM 780 (Zeiss) confocal microscope. Fluorescence images processing was performed with ImageJ (18) followed by manual counting of the no-fluorescent (untransformed), yellow fluorescent, red fluorescent and yellow and red fluorescent (co-transformed) protoplasts.
If we assume that T-DNA distribution of both fluorescent proteins occurs randomly and independently over the cells of agroinfiltrated leaves, it can be considered that the number of T-DNAs of each type entering a cell follows a Poisson distribution. With this consideration, we calculated the multiplicity of transformation (MOT) based on the frequency of co-transformed protoplasts for each tested OD600 following the same approach that Gutiérrrez et al. used for multiplicity of infection (MOI) calculation (19). At least 1000 protoplasts per OD600 were used for calculation.
Software tools development
The website https://gbcloning.upv.es/ was implemented using Django, a Python web framework that supports rapid design and the development of Web-based applications (version 1.8; Django Software Foundation; http://djangoproject.com). The database management system PostgreSQL was chosen to host the schemas of the GBelements and the experiments databases and all software tools accessible on the website were developed using Python. The software-package contains flexible modular blocks that are interconnected and can be classified in five main categories: (i) adaptation of raw DNA sequences to the GB standard, (ii) creation of gene-cassettes from standard parts, (iii) binary assembly of pre-made gene-cassettes, (iv) generation of GBelement datasheets and (v) generation of experiment views. All tools run all functions behind the screen. For the cloning tools, the submitted data are directly passed by Python functions for sequence checking and for output generating by creating a Genbank file and a protocol to each assembly step that is sent back to web server for user download. For generation of experiment views, data submitted are passed by Python functions for plotting a graph with the quantitative values and for incorporating links to the experiments on datasheets of involved GBelements. The source code of all tools is available on the Github repository at https://github.com/pziarsolo/goldenbraid.
Website functionality
The gbcloning website is organized in four different modules (Design, Collection, Experiments and Genome Engineering) as described on the Results section. The user-action workflow between the different cloning tools for the design of either multigene constructs or constructs for genome engineering is explained in detailed on Vazquez-Vilar et al. (20,21) respectively. Access and search to the Collection of GBelements is described at Vazquez-Vilar et al. (20) and also at Sarrion-Perdigones et al. (15). On the ‘Experiments’ section, standard and non-standard experimental information can be incorporated to the user-database (user account is required) and associated to either user or public GBelements at https://gbcloning.upv.es/add/experiment/ following the guidelines specified on the same URL. There are two options to users to access the experiments performed with a GBelement. First, on each GBelement datasheet a list of all public and user-associated experiments with links to them can be found. Second, experiments can be searched at https://gbcloning.upv.es/search/experiment/ following different criteria (experiment ID, keywords or words included on the experiment description and GBelements used on the experiment). Additionally, for standard experiments the search can be filtered by the values of the quantitative outputs.
RESULTS
Refinement of the GB assembly tools and rules: The GB3.0 software package
A renovated website at https://gbcloning.upv.es/ was created to host the new features of GB3.0. As displayed in its front-page, the new GB3.0 web comprises four major sections, namely Design, Collection, Experiment and Genome Engineering.
The Design section contains improved software tools that facilitate in silico assembly of multigenic constructs. Briefly, the GB software comprises three webtools, namely the domesticator, the multipartite assembler and binary assembler. The domesticator tool serves as entry point, converting raw DNA sequences in standard level 0 phytobricks, typically promoters, coding regions, protein domains, terminators, etc. For domestication, raw DNA sequences are cloned into standard entry vectors while internal restriction sites are removed by PCR-mutagenesis. The multipartite assembler takes individual level 0 parts and clones them together to create level 1 GBelements, typically a full transcriptional unit. Later, level 1 elements can be assembled binary (Binary assembler tool) in an iterative way to create level >1 elements, typically multigenic constructs. Although the basis for in silico assembly were developed in GB2.0, the new version incorporates a number of updates and improvements listed below (see Supplementary Figure S1):
Two new part domestication tools (the ‘Phylogeny Search’ and the ‘Synthetic Strategy’) were introduced. As the domestication of coding DNA parts is often hampered by the presence of internal restriction enzyme sites (RES), the first tool searches plant genome databases to find homologous sequences from related species that contain minimal internal restriction sites as potential substitutes. Alternatively, with gene synthesis becoming increasingly affordable, it is often convenient to chemically synthesize RES-free DNA fragments and clone them directly in the Universal entry vector instead of removing internal RES by mutagenesis. The Synthesis Domesticator tool was introduced to facilitate this domestication option.
Domestication tools allow the removal of additional RES (i.e. BpiI) to enhance compatibility with the alternative Type IIS plant assembly method MoClo.
In response to user's requests, a domestication option for intron-containing coding regions was introduced, which facilitates the cloning of ORF from genomic DNA in these specific cases.
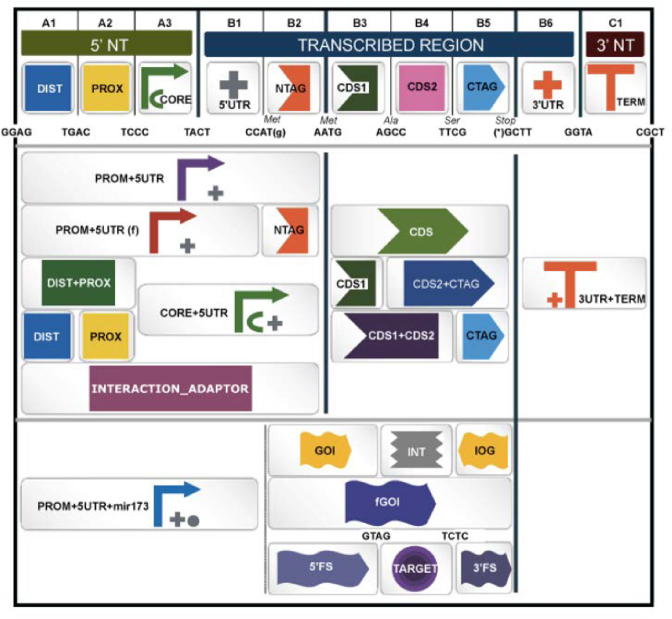
The Collection section serves as interface for the GBdatabase. The GB3.0 database has been reshaped to adapt to the new PSS. Accordingly, all new GB parts introduced in the database shall conform to PSS. A specific synthetic biology open language (SBOL)-inspired symbol was assigned to each level 0 part, and to each relevant combination of parts. An interactive page was designed to browse GB elements in the database using GB symbols (Figure 1). By creating a personal profile with a username and a password, users can introduce their own phytobricks, host them in their private section of the GB3.0 database and use them to create new assemblies. Private parts can be made publicly available under request to the webmaster (as indicated in the webpage). Furthermore, the public database was updated with new structural elements adapted to PSS:
A new set of destination pCAMBIA-based vectors (pDGB3 series) was introduced with increased in planta transformation efficiency.
The AmpR Universal Part Domesticator vector (pUPD) of GB2.0 was substituted by a new CamR pUPD2 as the standard vector where all GB3.0 parts are stored. Adoption of pUPD2-hosted GBparts enhances compatibility with MoClo and with iGEM Registry of Standard Biological Parts. Accordingly, all new GB parts are domesticated using pUDP2. Parts in old pUPD version can be used in new assemblies, but the use of pUPD as entry vector is no longer supported by GB software tools.
Besides GBparts, the extended GB3.0 database harbors now experiments performed with standard GBparts. Experiments are used to feed quantitative descriptions of parts and modules, which are displayed in standard datasheets (see below) that can be searched according to different strings (experiment type, parts involved, etc.). Experiments need to conform to one of the defined standard experimental types. This allows quantitative descriptions to be integrated in the database as structured data. Consequently, the standard experimental data can be used for design purposes (e.g. to optimize circuit design by selecting weak/strong promoters/transcriptional factors). The tool ‘Search GB_experiment’ (https://gbcloning.upv.es/search/experiment/) assists in construct design by retrieving GBelements that conform a given search criteria, e.g. phytobricks showing a certain range of transcriptional activities.
Figure 1.
Browser page displaying the Standard Plant Syntax for Parts database. The figure shows a screenshot of the parts database search browser including specific synthetic biology open language (SBOL)-based images as shortlinks to the different part categories.
The new Experiment section contains templates for the introduction of experimental data. In the GB3.0 database the structural information available for each DNA part is enriched with the introduction of functional information derived from experimental results. This is achieved with a new type of element in the GB schema, the GBexperiment. A GBexperiment is defined as a set of data produced as the result of the genetic transformation of one or more GB elements in a plant chassis. The experimental data comprises both the experimental conditions in which the experiment has been performed, as well as any quantitative or qualitative observation (results), obtained as a consequence of the presence of the GB element(s) within the plant chassis. New experiments are introduced in the database by filling a standard questionnaire, where either qualitative (images, text descriptions) or numeric results (in the form of a standard datasheet) can be uploaded. An important feature of the GBexperiment is that it is always associated to a GB element, which corresponds to the complete GB composite part that was employed to perform the test. As the GBdatabase keeps record of individual parts that were combined to build the GB element, every new experiment enriches the description not only of its main GB element, but also of its individual components. Tentatively, we have defined five standard experiment types (SE_001 to SE_005), plus one non-standard template (NS_000). SE_001 and SE_002 templates were designed to accommodate transcriptional activity data resulting from experiments conducted with the normalized Luc/Ren reporter pair in agroinfiltrated Nicotiana benthamiana leaves. SE_002 is reserved to experiments with intact leaves, whereas SE_001 accepts leaf disks incubated with chemical inducers (described in more detail in subsequent sections). SE_003 template describes stable transformation efficiencies, SE_004 is used to collect standardized levels of recombinant protein production, and SE_005 was defined to collect mutagenesis efficiency in CRISPR/Cas9 experiments. Each experiment type, except NS_000, is defined by a number of compulsory experimental conditions, accepts additional (declared) specific conditions and it is expected to produce a limited number of predefined quantitative outputs. SE_001, for instance, has strict rules as for the plant chassis, plant growing conditions, disks size, harvesting and incubation timing, enzyme reaction conditions, use of internal references, etc., but it is agnostic towards the use of chemical inducers, optogenetic signals, temperature, etc. Detailed experimental conditions for each experiment type are shown in https://gbcloning.upv.es/add/experiment/ or in Supplementary Table S1. Examples of all experiments can be consulted in GBdatabase (https://gbcloning.upv.es/search/experiment/) or representative ones in Supplementary Table S2.
Finally, GB3.0 incorporates a new section devoted to genome engineering. This section integrates tools for construction of gRNA and CRISPR/Cas9 assemblies for multiplexing gene editing. The adaptation of CRISPR/Cas9 system to the GB3.0 standard is described elsewhere (21).
The GB3.0 standard datasheet: integrating experimental data
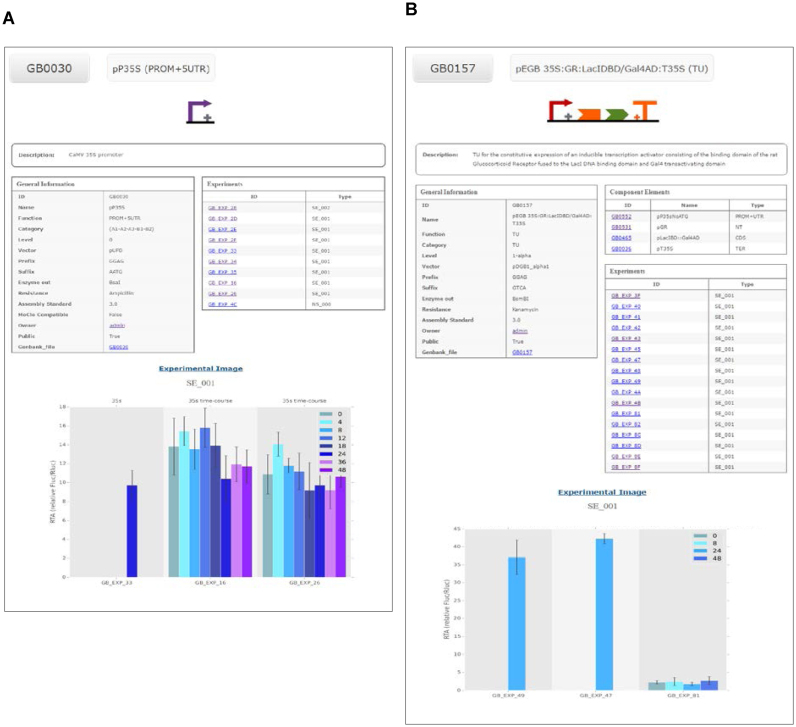
A consequence of the introduction of GB experiments to the GB database is that the GB elements can now incorporate functional information to their descriptions. To handle this information we have designed a basic GB datasheet, a page that displays the most important information of each GB element. The basic datasheet information includes name, type of element (level 0, 1, >1; and the standard position in case of level 0), a short text description of the element, the GB plasmid where it is hosted, a graphical description adapted from the SBOL standard (22) and, when available, the graphical output of representative experiments. The objective of the basic GBdatasheet is to provide the most important information about the GBelement in a single screenshot. In addition, more detailed information is linked to the basic page that comprises, among others, the nucleotide sequence in Genbank and SBOL formats and a clickable list with all the experiments performed with that element.
An example of a GB3.0 datasheet is depicted in Figure 2A, which shows the standard specifications of the level 0 GB0030 phytobrick (see online entry at https://gbcloning.upv.es/feature/GB0030/). GB0030 was constructed based on the constitutive CaMV35S promoter, as indicated in the Description section.This part is flanked by GGAG and AATG sequences and, consistently with the GB specifications and PSS, comprises basic parts A1-A2-A3-B1-B2 (see Figure 1). The datasheet includes, next to the SBOL Visual symbol representative of its standard position, sequence-related information, the plasmid backbone, the presence of internal sites, selection marker and a link to the complete DNA sequence in Genbank and SBOL formats. Level 0 phytobricks are introduced in the system directly via the Domesticator tool and therefore, do not generate genealogy information. Most remarkably, the datasheet contains a clickable list of experiments describing GB0030 activity. Representative experiments are depicted in the lower part of the datasheet, illustrating the functionality of GB0030 under standard conditions. In Figure 2A, three SE_001 experiments involving GB0030 are depicted. Details of each experiment can be obtained by clicking in the corresponding link.
Figure 2.
Examples of two GB3.0 datasheets. (A) Datasheet of the level 0 phytobrick derived from CaMV35S promoter (GB0030). The datasheet includes a SBOL-inspired symbol (top), assembly information such as part category, vector or information of compatibility with the MoClo standard (general information table) and experimental information with links to all experiments performed with devices including this phytobrick (experiments table) and graphs of three experiments (bottom). (B) Datasheet of the composite part GB0157 including a compilation of the SBOL-inspired symbols describing each of its components (top) a list of these components with links to each of them facilitating the traceability of the assembly (component elements table), assembly information such as the vector and phytobrick category (general information table) and experimental information with links to all experiments performed with devices including this element (experiments table) and the graphs of three representative experiments (bottom).
An example of a datasheet describing a composite GBelement is shown in Figure 2B. In this case, GB0157 entry corresponds to a Transcriptional Unit comprising four standard basic parts, GB0552 (the A1-A2-B1 CaMV35S promoter), GB0531 (a B2 N-Terminal fragment) encoding the rat glucocorticoid receptor domain), GB0465 (a B3-B4-B5 CDS encoding a chimeric transcriptional factor comprising the binding domain of LacI and the transcription activation domain (AD) of Gal4) and GB0036 (a B6-C1 phytobrick derived from the transcriptional terminator of the CaMV35S). As shown in Figure 2B, GB0157 datasheet contains a ‘Components Elements’ section describing its genealogy, which is automatically generated during in silico assembly within the GB3.0 frame. All the genealogy elements are clickable and linked to the standard datasheets of the GBelements of the previous assembly level. As in the previous example, GB0157 datasheet contains a list of standard experiments describing this composite part, and displays a representative chart showing the transcriptional activation that GB0157 confers to a minimal promoter containing a LacI operator in presence of dexamethasone, as evaluated using the SE_001 standard (see https://gbcloning.upv.es/feature/GB0157/ for an online version of GB0157 datasheet).
Refinement of the standard measurements of transcriptional activity in plant cells
Our group focuses in the design of transcriptionally-regulated genetic circuits in plants, and therefore we paid particular attention to the establishment of quantitative measurements estimating transcriptional activity, as represented by the proposed standard SE_001. Relative transcriptional activity (RTA) in SE_001 are measured in the N. benthamiana leaves making use of Agrobacterium-mediated transient expression and the Luc/Ren system as indirect measurement of the steady-state transcript levels. Furthermore, the relative luciferase measurements are normalized with an internal reference (GB0166 or GB1116) which is always agroinfiltrated in the same experiment. This reference corresponds to a Luc/Ren reporter construct where Luciferase is driven by the weak promoter of the Nopaline synthase gene. Reference Luc/Ren readings are arbitrarily set as 1.0 rpu (relative promoter units). The addition of this reference was found essential to compare results among different experiments.
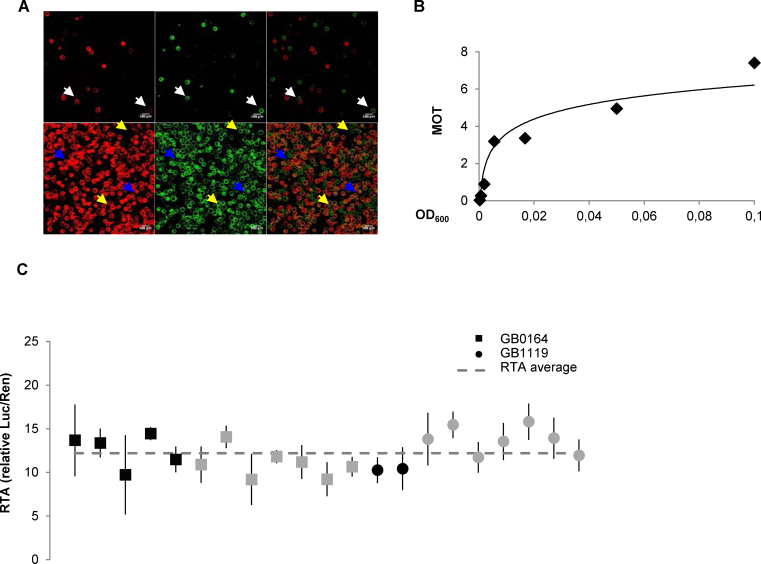
The remaining SE_001 experimental conditions were carefully selected based on detailed observations. Among others, an important parameter is the average number of active T-DNA copies that enter a leaf cell upon agroinfiltration, which depends on the cell density of the Agrobacterium cultures. To estimate this parameter, a strategy similar to that used to estimate the multiplicity of infection (MOI) in viral infections was employed, assuming that the rate of co-transformation of two independent Agrobacterium-transformed constructs (GB1287 and GB1288, carrying Yellow and Red fluorescent proteins respectively, Figure 3A) follows a Poisson distribution. As can be observed in Figure 3B, the number of transcriptionally-active T-DNA copies (defined here as multiplicity of transformation, MOT) adjusts well to a logarithmic function of the optical density of the Agrobacterium culture, with a MOT = 1 obtained at OD600 = 0.002. MOT dependence with OD600 decreases as OD600 increases. We therefore decided to set a standard OD600 = 0.1 (estimated MOT = 7.4) for SE_001 and SE_002, as it balances maximum MOT stability (low dependence to OD600 variations) with low Agrobacterium input and acceptable copy numbers for transactivation studies.
Figure 3.
Refinement of experimental conditions for Plant DNA part characterization in transient expression assays and behaviour of the CaMV35s promoter among experiments. (A) Images of protoplasts co-transformed with GB1287 and GB1288 at OD600 = 0.000617 (top) and OD600 = 0.05 (down). From left to right Red channel, YFP channel and overlay of the two previous pictures. White arrows point to co-transformed protoplasts, blue arrows to protoplasts showing only red fluorescence and yellow arrows to protoplasts showing only yellow fluorescence. (B) Relationship between the OD600 and the multiplicity of transformation (MOT). MOT values were estimated based on the percentage of co-transformed protoplasts (see Materials and Methods for details). (C) Reproducibility of relative transcriptional activity (RTA) measurements in standard experiments. Relative transcriptional activity of the CaMV35S promoter (GB0030), tested as part of devices GB0164 and GB1119 over different experiments. GB0164 has a pGreen backbone while GB1119 has a pCAMBIA backbone. Black squares and circles correspond to measurements at 4dpi while grey squares and circles are measurements determined at different time points. Error bars represent standard deviations of three different leaves expressing the same GBelement on the same experiment.
The experimental data deposited in the GBdatabase was then used as an internal test of the reproducibility of SE_001 and SE_002. We took advantage of the repeated use of GB0030 as internal control in several independent experiments to test the variability of the transcriptional activity of our synthetic version of the CaMV35S constitutive promoter under standard conditions. The GB0030 datasheet contains links to several SE_001 and SE_002 experiments, and the relative transcriptional activity in each experiment was plotted and depicted in Figure 3C. As observed in the Figure, average GB0030-driven transcriptional activity was maintained in a range between 9 and 13 relative units, despite the use of different binary plasmid backbones, plant batches, growth chambers and researchers.
The remaining standard experiments were defined to illustrate the generation of an increasingly informative GBdatabase. We assembled and experimentally tested a number of new GB devices using the five pre-established experiment types and uploaded the results into the database. Supplementary Table S3 shows a non-exhaustive list of characterized devices ranging from protein–protein interactors, constructs for metabolic engineering, recombinant protein production, tomato stable transformation, CRISPR/Cas 9-based mutagenesis and transcriptional regulation, among others.
New orthogonal parts in GB3.0: phiC31 integrase-based transcriptional regulation toolbox
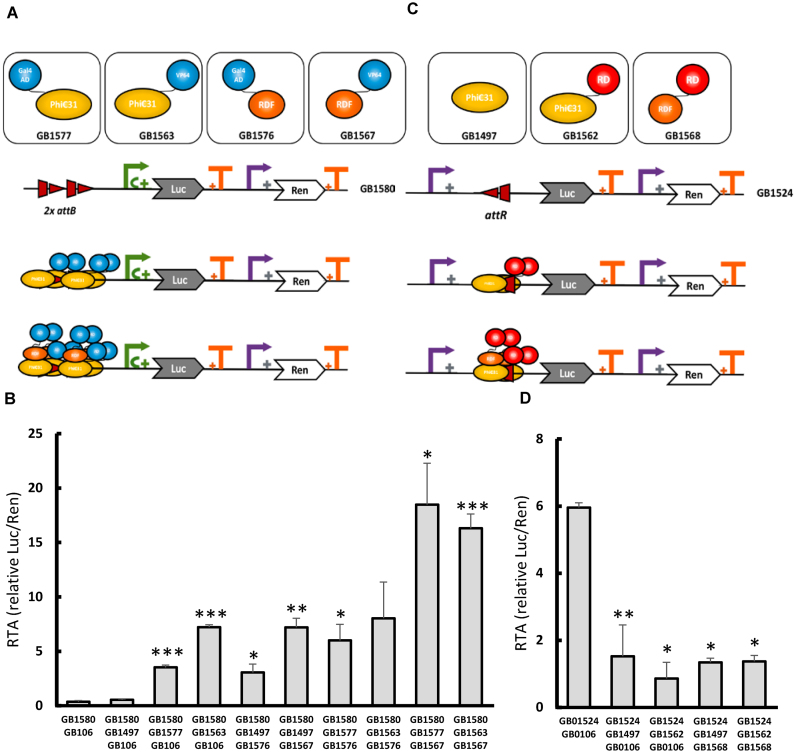
One of the key concepts in Synthetic Biology is orthogonality. Ideally, synthetic genetic devices must function independently of endogenous regulation by the host organism. The GB3.0 platform facilitates the physical combination of orthogonal phytobricks in the collection, and the functional information stored in their datasheets serves to anticipate the outcome of their combination and to harness new biological functions. To illustrate this, we made use of heterologous modular parts present in GB3.0 and designed, assembled and tested a toolbox of novel orthogonal transcriptional regulators for plant synthetic biology. We made use of the following pre-existing genetic elements in the collection (i) level 0 parts with the coding sequence of the phage phiC31 integrase for N-terminal and C-terminal fusions (GB1496 and GB1561), (ii) level 0 parts with the coding sequence of the phage phiC31 recombination directionality factor (RDF) (GB1498 and GB1566), (iii) eukaryotic transcriptional activator domains (GB1186, GB0900) and a eukaryotic repressor domain (GB1175). PhiC31 integrase can bind with high affinity to specific DNA attachment sites, namely attB, attP, attL, attR. In order to perform site-specific recombination, phiC31 integrase requires the presence of both attB and attP sites (regardless of their respective order and orientation) (23). Additionally, phiC31 RDF can bind to Integrase in solution. Integrase:RDF complex can also bind to either attB, attP, attL, attR DNA sequences, but only catalyses recombination in the presence of both attR and attL (regardless of order and orientation) (24). We decided to take advantage of these interactions to build an orthogonal toolbox for transcriptional modulation, combining phiC31 and RDF as binding domains with modular transcriptional activator domains or repressor domains. Additionally, by making use of GB3.0 design and assembly tools, new synthetic promoters incorporating att binding sites as enhancer elements were built to achieve either activation or repression of the reporter Luc/Ren system in response to aforementioned transcription regulators. Transcriptional activators were constructed in four configurations, as illustrated in Figure 4A, by fusing an AD (GAL4 or VP64) either to phiC31 or RDF. All activation configurations were tested under SE_002, using a synthetic operator comprising 2 copies of the same attB phiC31 recognition site (to avoid recombination), next to a minimal 35S as a target promoter followed by Luc/Ren reporter (GB1580) (Figure 4A). The synthetic attB operator is located upstream of the TSS to avoid steric clash between phiC31 integrase and the transcription initiation machinery, and to favour recruitment of the latter by the AD. As can be observed, the orthogonal phiC31 system was very efficient in activating transcription, reaching RTA levels well above GB0030 and induction ratios ranging from 8-fold to 50-fold induction (Figure 4B). The most active combinations comprised both phiC31 and RDF, each carrying different ADs. Transcriptional repression using phiC31 recombination-based orthogonal elements was also tested. In this case, the reporter construct GB1524 comprised a 35S promoter with an attR site inserted immediately downstream of the TSS, followed by Luc/Ren reporter system (Figure 4C). This modification of the DNA sequence reduced RTAs of the 35S promoter from 10 to 6 rpu. PhiC31 was able to reduce reporter expression levels, and this repression was shown to be OD600-dependent (Supplementary Figure S2). The presence of phiC31 fused to a RD (BRD) achieved further repression of this promoter. The observed repression values ranged from 4 to 7-fold (Figure 4D and Supplementary Figure S2). All new orthogonal elements are now part of the GB3.0 collection and provide a new complete toolbox for transcriptional regulation in plants.
Figure 4.
New orthogonal TFs toolbox based on phage phiC31 site-specific recombination elements. (A) Schematic representation of orthogonal elements employed in recombinase-based transcriptional activation and their interactions. PhiC31 integrase can bind to attB sites. After binding, activation domains (ADs) (blue) close to the minimal 35S promoter (green arrow) perform transcriptional activation. Interaction between PhiC31 integrase and recombination directionality factor (RDF) increase the number of activator domains (ADs) available, resulting in higher activation. (B) SE_002 assays involving different combinations of the transcriptional activators shown in (A) GB0106 represents a stuffer fragment used to equalize Agrobacterium culture concentrations. GB1497 contains a constitutive PhiC31 TU without AD. (C) Schematic representation of the orthogonal elements employed in recombinase-based transcriptional repression. PhiC31 integrase binds to attR resulting in repression of 35S promoter (purple arrow). Interaction between PhiC31 integrase and RDF increase the number of effective repression domains. (D) SE_002 assays involving different combinations of the transcriptional repressors shown in C. GB0106 represents a stuffer fragment used to equalize Agrobacterium culture concentrations. All relative transcriptional activities are expressed as relative promoter units calculated normalizing Luc/Ren ratios to GB1116. Error bars represent standard deviation of Luc/Ren ratios determined on 3 independent samples. Asterisks indicate significant differences in a T-Test with a *P-value < 0.05, **P-value < 0.01, ***P-value < 0.005.
New gene circuits assembled with GB3.0
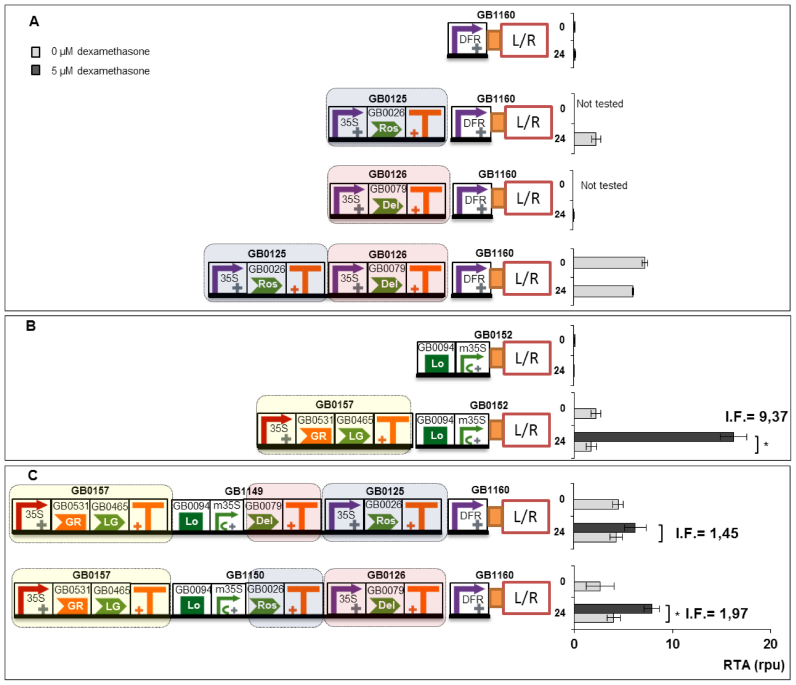
To illustrate the use of GB3.0 in the construction of transcriptional gene circuits, we built and tested two independent genetic devices, and connected one to another, creating a new circuit. The first device represented a simplified version of the endogenous genetic module controlling flavonoid biosynthesis. This device comprised as key elements the transcriptional regulators Rosea1 (GB0026) and Delila (GB0079) and the dihydrofavonol-4-reductase (DFR) promoter (GB0606) operating upstream of the Luc/Ren reporter system (GB1160). Rosea1 and Delila are known to bind a WD40 factor forming quaternary activation complexes with DNA at the promoter regions of several enzymes of the flavonoid pathway, including DFR gene. In the absence of additional transcriptional factors, GB1160 showed negligible transcriptional activity in SE_001 conditions (Figure 5A) as assessed with the Luc/Ren reporter system. When TUs expressing Delila (GB0126) or Rosea1 (GB0125) factors were put next to the DFR construct, transcription activity driven by this promoter increased to low (0.07 rpu) or moderate (2.3 rpu) levels, respectively. The simultaneous co-expression of (GB0126) and (GB0125) resulted in a device with higher (7.2 rpu) activation values for the DFR promoter (see Figure 5A). As it can be deduced, DFR functions as an imperfect AND gate, with Ros1 and Del acting in series in the activation of DFR, but with Ros1 having a stronger influence in DFR expression. Partial AND gate activation illustrates the problem of non-orthogonal systems and probably reflects the ability of Ros1 to recruit endogenous bHLH factors that compensate the absence of Del in the activation of DFR promoter.
Figure 5.
Connection of two transcriptional modules for conferring dexamethasone regulation to the DFR promoter. (A) Relative transcriptional activity of the DFR promoter by Ros1 and Del, either alone or in combination at 0 h or 24 h post-induction (corresponding to 72 h and 96 h post-infiltration, respectively). (B) Dexamethasone-dependent transcriptional activity conferred by GB0157 to a reporter device (GB1052) containing a LacI binding domain (GB0094) next to minimal CaMV35S promoter. (C) Effect of the combination of gene modules in A and B, with Ros1 and Del turning the DFR promoter responsive to dexamethasone. All relative transcriptional activities are expressed as rpu calculated normalizing Luc/Ren ratios to GB0166. Error bars represent standard deviation of Luc/Ren ratios determined on at least two independent leaves. I.F. represent the induction factors calculated by dividing RTA values obtained with and without chemical inducer. Asterisks indicate significant differences in a T-Test with a P-value < 0.05.
The second device (shown in Figure 5B), was a glucocorticoid sensor, also coupled to the Luc/Ren reporter (GB1052). In this device, the sensing of dexamethasone is processed by a constitutively-expressed, chimeric TF (GB0157) comprising a Glucocorticoid Responsive domain (GR) fused to a Gal4 transcription AD and the LacI DNA binding domain. The chimeric TF binds the LacI operator upstream of a Luc/Ren reporter in a dexamethasone-dependent manner. In the absence of chemical inducer, this device shows low transcriptional activity when coupled to Luc/Ren reporter (average 1.7 rpu at t = 24 h). In the presence of 5 μM dexamethasone GB1349 is activated to levels up to 16.0 rpu at t = 24 h resulting in an induction factor of 9.37 (see Figure 5B).
Once both modules were independently characterized, we anticipated that their connection in a single construct would produce a dexamethasone-responsive DFR promoter. This exercise wanted to simulate the ‘hacking’ of an endogenous regulatory module (flavonoid biosynthesis), by making it responsive to a new stimulus. The new circuit admits at least two possible configurations, as both Ros1 and Del TFs can each be directly connected to the dexamethasone sensor. However, attending to the transcriptional activity reported for Ros1 and Del modules separately, it could be easily anticipated that connecting GB0157 to Ros1 would result in a better induction factor (defined as the RTA ratio at t = 24 h with and without dexamethasone) than with Del. To test this model, we constructed the new circuit in both configurations (Figure 5C), and we operated it in presence and absence of inducer. As expected, both circuits resulted in dexamethasone-responsive devices, with the configuration that connected Ros1 to Dexamethasone resulting in a higher induction factor (1.97 and 1.45, respectively).
DISCUSSION
A new wave of innovative crop traits will be needed in an immediate future to respond with sustainable bio-production solutions to a rapidly changing environment (25). Plant Synthetic Biology proposes the rational and systematic design of genetic systems (i.e. regulatory networks, biosynthetic pathways, etc.) as a new breeding strategy for obtaining radically new traits, especially those that are plausibly beyond the repertoire offered by natural variation. GB3.0 proposes a multigene design platform that associates functional DNA elements with biological data. This is to our knowledge the first attempt to create such an integrated platform for Plant Synthetic Biology. Previously, other initiatives such as the BioBricks Foundation in the frame of the iGEM competition (26), and the International Open Facility Advancing Biotechnology (BioFab) in a more-research-oriented scope (27), have produced, characterized and validated large collections of standard biological parts including catalogues of promoters (28), terminators (29), Ribosomal Binding Sites (30), as well as small regulatory devices to support bioengineering but mainly in E. coli and Saccharomyces cerevisiae. To manage and reuse parts and devices, the SBOL standard aims to facilitate the exchange of information and to communicate designs in SynBio (31). SBOL is an extensible standard created to encode additional information beyond an annotated sequence as required by synthetic biology, including measurements of performance characteristics, experimental context information, computational models of behaviour, etc.
The present particularities of Plant Biotechnology impose technical constraints that preclude the practical use of microbial-oriented catalogues as BioFab. In principle there is little chance for large inter-kingdom exchange of bioparts due to differences in codon usage and to the lack of functional conservation in many regulatory elements (32). Besides, plant synthetic devices are very often delivered in the form of T-DNAs, and therefore genetic constructs need to be enclosed in Agrobacterium binary vectors. All in all, it seems more operative for the plant community to start developing platforms and standards that are specially adapted for plants, as reflected by the PSS initiative. Fortunately, there is a growing number of plasmid collections for plant SynBio in the Addgene repository covering hundreds of standard elements (8,13,30). However, the development of information managing tools for those collections, such as software tools, protocols for exchange of information, experimental standards or automatization, etc., is to a large extent still lacking. GB3.0 is an example to fill a gap in this direction. GB3.0 can accommodate level 0 phytobricks from GB or MoClo collections to create level >1 composite parts following GoldenBraid cloning procedure. As a proof of compatibility, we incorporated a limited number of (BsmBI-free) MoClo phytobricks from the Addgene repository in the GB3.0 public collection and showed that they can be used to build virtual parts using GB software tools (see e.g. MoClo phytobrick pICSL80001 uploaded to GB database, and GB_UA_1283 and GB_UA_1284 multipartite and binary assemblies respectively made with this phytobrick). Further integration steps (e.g. incorporation of MoClo assembler tools) could be envisioned in the future as collaborative efforts to enhance compatibility. We favour the creation of public phytobrick databases integrating different assembly methods in parallel with curated (dispersed or centralized) repositories that ensure quality and accessibility. In the same direction to enhance compatibility, GB3.0 adapted available tools (33) to produce SBOL XML output files for GBelements, and adopted visual-SBOL-proposed graphics for representation of bioparts. We have also proposed new symbols to represent plant-specific features.
A novelty of GB3.0 is that experimental data are automatically associated to each genetic element. We propose the definition of prototype experiments as the most straightforward way to populate SynBio databases with functional descriptions. The definition of guidelines for minimum experimental information is a widely adopted strategy in high-throughput analysis, as it enables the unambiguous interpretation and the exchange of data in, e.g. microarray experiments or proteomics (34,35). In principle, experimental guidelines for ‘synthetic’ parts need to be more restrictive than microarray experiments-like guidelines, as the objective in the case of phytobricks is provide highly comparable descriptions under identical conditions. Initially, we have defined five standard experiments in GB3.0, encompassing those measuring transcriptional activity (SE_001, SE_002), recombinant protein production (SE_003), transformation efficiency (SE_004) or mutagenesis efficiency for CRISPR/Cas9 constructs (SE_005). This was not aimed to be an exhaustive list but rather a consequence of the main activities in our and the collaborating labs. In the present version of the platform, the definition of new standards is an administration privilege, and therefore the introduction of new standards need to be requested to the web administrators. We proposed Luciferase/Renilla ratios measured in agroinfiltrated N. benthamiana leaves as a general standard for transcriptional activity specifications. Agroinfiltration is a widely used, rapid and straightforward transient expression method amenable for medium and high throughput analysis (36,37). According to this method, transgene expression takes place in differentiated leaf epidermal and parenchyma cells; however the high co-transformation efficiency facilitates the incorporation of trans-acting elements that could ectopically simulate the molecular environment found in other specialized tissues, and therefore extending its applicability to a wide range of experimental setups. Absolute units for measurement of transcriptional and translational activities in Synthetic Biology have been proposed, as for instance the number of polymerases per second or ribosomes per second (14); however, in practical terms the use of relative units calculated with a constant reference, has turned to be more operative. We found that normalization of Luc/Ren ratios to a Nopaline synthase promoter-derived standard element resulted in a robust and reproducible method for estimation of the relative transcriptional activity in agroinfiltration experiments. This is in agreement with previous results in protoplast transformation where normalization with a parameter that compensate for the distinct compositions of differentiated leaf cells (e.g. mesophyll, palisade parenchyma and bundle sheath) and from plants that experienced microclimatic variations was found crucial to eliminate ‘batch effect’ as main source for variability (38).
An important concern when setting up SE_001 standard conditions was the number of T-DNAs that simultaneously enter a plant cell in a typical agroinfiltration experiment, as this could affect reproducibility and interpretation, but also the ability to reliably perform co-transformation with non-linked T-DNAs. Therefore, we carefully investigated the T-DNA co-transformation levels and its dependence of the cell density of Agrobacterium in the infiltration culture. Although the levels of co-transformation in stable transgenics has been throughout investigated (39–41), to our knowledge this question has not been convincingly addressed for the case of agroinfiltration. We followed a strategy similar to that described for estimation of MOI in viral infections and found that at low OD values the T-DNA copy number showed a strong dependence with Agrobacterium concentration, but this dependence sharply declined when OD was close to 0.1, corresponding to a MOT = 7.4. We therefore set 0.1 as standard OD for agroinfiltration, although for certain analysis it would be advisable a MOT value even lower. In the current work, we have found no evidence that T-DNA size is influencing MOT (no bias has been observed in luciferase activity for larger constructs, data not shown). However, the T-DNA constructs assayed so far are of average size range (∼2 kb) and therefore such dependence cannot be discarded for larger constructs. The characterization of increasingly complex constructs will probably require a more comprehensive study of the dependence of MOT with T-DNA size.
We also show that the GB3.0 database itself serves as quality control for the reproducibility of a given experimental standard, as GB3.0 keeps record of all the experiments conducted with the same phytobrick. Thus, we found high consistency and reproducibility within all the experiments involving phytobrick GB0030 (CaMV35S promoter), despite having been conducted by different personnel of our lab over a period of two and a half years. Notwithstanding this, refinements of SE_002 could be proposed to increase throughput as, e.g. whole tissue measurements that skip tissue homogenization steps. In the future, an effort should be made to identify common grounds for experimental comparisons, e.g. defining specific genomic landing paths for each species, agreeing standard growth conditions, etc., so that gene constructs can be reliably characterized in a whole plant genome context. For the time being, the standards proposed here could serve as a first step for functional characterization. In addition, we have defined also NS_000 to serve as repository to those experiments not fitting any current standard. Non-standard experiments provide specific information of each GBelement, but cannot be used as basis for comparisons or for creating functional composition rules.
GB3.0 provides a base for design and assembly of complex genetic devices with novel biological functions. The detailed experimental data associated with individual parts, provides essential information for the prediction of the outcome of a combination of different parts. As an example of GB3.0 versatility, we have described here in more detail the development of a group of orthogonal TFs based on phage phiC31 site-specific recombination system, and the composition of a small circuit that results from the connection of a MYB/bHLH regulatory module to a conditional activation module triggered by Dexamethasone. Streptomyces phage phiC31 integrase has strong affinity and specificity toward its target att recombination sites. We exploited phiC31 as DNA binding domain in chimeric binding domain (BD)-activator or BD-repressor domain fusions with previously characterized heterologous genetic elements. Additionally, we designed two synthetic promoters to assay transcription modulation of Luciferase reporter gene. The relative position of the att sites to the TSS was shown crucial to mediate activation or repression. Assays comprising BD-AD fusions using a promoter with two copies of the attB site upstream of the TSS were shown a successful strategy for transcriptional activation, yielding RTA in the high range. It has been also shown before using Cas9 genome-wide activation that the arrangement of different ADs in close proximity has a cooperative effect (42). The presence of a DNA binding activity close to TSS seems also a general mechanism to induce modest repression levels, as recently reported for Cas9-based gene inactivation (CRISPRi) (43). No significant changes in repression were achieved by co-expression of phiC31 and RDF, both fused to the same RD, suggesting that phiC31:RD binding was sufficient to perform steric hindrance of transcription elongation. Further optimization of this new orthogonal TFs toolbox will be possible by incorporating additional activation/repressor domains, tuning the number of copies and relative position of att sites to TSS and through combination with chemically inducible switches (44). The adaptation of phiC31 recombinase system is, to our knowledge, the first example of a recombinase reconverted into a transcriptional factor, with attB sites reconverted in enhancer elements. This exemplifies a common trend in Synthetic Biology, i.e. the recycling of functional elements for purposes different from those in nature.
In a second example, we employed Rosea1 and Delila MYB/bHLH TFs, which are known to bind a WD40 factor forming quaternary activation complexes with DNA sequences localized in the promoter regions of several enzymes of the anthocyanin biosynthesis pathway, including the DFR gene. In some tissues, like in the tomato fruit engineered to accumulate high levels of hydrosoluble antioxidants, activation of anthocyanin biosynthesis genes strictly requires the simultaneous presence of both Rosea 1 and Delila TFs (45), thus exemplifying a case close to a canonical AND gate with Ros1 and Del acting in series in the activation of DFR. In contrast, we show here that in Nicotiana benthamiana, ectopic expression of Rosea1 and Delila TFs function as an imperfect AND gate in the transcriptional activation of DFR promoter, with Ros1 having a stronger influence in DFR expression. Imperfect AND gate activation illustrates the problem of non-orthogonal systems and probably reflects the ability of Ros1 to recruit endogenous bHLH factors that compensate the absence of Del in the activation of DFR promoter. This lack of orthogonality is a very common situation in Plant SynBio, and it is not devoid of practical relevance, e.g. for food crops, as public opinion polls favour the use of intragenic elements rather than cross-kingdom elements for food crop biotechnology. The constructs shown here prototype this kind of circuits by physically combining a chemical switch and a Ros1/Del (endogenous) module in a single cloning step, and show that this strategy results in a functional combination of both modules. We showed that, by integrating the standardized descriptions of the different elements obtained in separate experiments, it is possible to predict the functional activity of the combined system and to select the configuration that provides higher induction rates to the DFR promoter in Dexamethasone dependent manner. Understandably, as the complexity of the circuit increases, the signal noise ratio decreases, leading to a low induction factor. Additional tuning of the circuits could involve the optimization of the dynamic range of the output with the incorporation of elements with higher dynamic ranges as, e.g. the macrolide-responsive gene expression tool used to engineer optogenetic circuits, which showed induction rates > 750 when assayed in plant protoplasts (46). The activation of entire pathways by ectopic expression/repression of endogenous or homologous TFs has been shown a very successful strategy for metabolic engineering, including biofortification of food crops (45,47,48). Ectopic expression often results from overexpression of the TFs using constitutive promoters or, in a few examples, is made dependent of endogenously regulated factors as, e.g. ripening (47), senescence (49) or tissue specific factors (50–52). A step forward in transcriptional control would involve connecting endogenous pathways (e.g. anthocyanin biosynthesis) with externally operable modules such as optogenetic or chemically inducible switches (44,46,53,54). This would allow to externally operate the biochemical and/or physiological status of the plant, for instance anticipating biotic or abiotic threats or triggering the accumulation of target compounds immediately before harvesting.
In sum, to exercise genetic design with GB3.0 we have constructed more than 350 level ≥1 public elements (single TUs, gene modules), involving more than 100 level 0 phytobricks, and tested them in standard and/or non-standard conditions generating more than 120 experimental entries. GB3.0 proposes standards for experimental data, links new data to genealogy-connected phytobricks and shows how these links provide clues for biodesign of new, increasingly complex and/or orthogonal genetic devices in plants. We think that GB3.0 provides a working solution that accommodates the current needs of Plant Biotechnology, facilitating combinatorial optimization and incremental innovation.
Supplementary Material
ACKNOWLEDGEMENTS
Vazquez-Vilar M. is a recipient of a Junta de Ampliación de Estudios fellowship. The authors also want to thank the COST Action FA1006 for the support in the development of the software tools.
Footnotes
Present address:
Marta Vazquez-Vilar, Laboratory of Systems and Synthetic Biology, Wageningen University, Stippeneng 4, 6708 WE Wageningen, The Netherlands.
Alejandro Sarrion-Perdigones, Department of Biochemistry and Molecular Biology, Verna and Marrs McLean, Houston, TX 77030, USA.
Rocio Ochoa-Fernandez, Institute of Synthetic Biology, University of Düsseldorf, Universitätsstrasse 1, 40225, Düsseldorf, Germany.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Spanish Ministry of Economy and Competitiveness [BIO2013-42193-R and BIO2016-78601-R projects to A.G. and D.O.]. Funding for open access charge: Spanish Ministry of Economy and Competitiveness [BIO2013-42193-R and BIO2016-78601-R projects to A.G. and D.O.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Dalal J., Yalamanchili R., La Hovary C., Ji M., Rodriguez-Welsh M., Aslett D., Ganapathy S., Grunden A., Sederoff H., Qu R.. A novel gateway-compatible binary vector series (PC-GW) for flexible cloning of multiple genes for genetic transformation of plants. Plasmid. 2015; 81:55–62. [DOI] [PubMed] [Google Scholar]
- 2. Cha-aim K., Fukunaga T., Hoshida H., Akada R.. Reliable fusion PCR mediated by GC-rich overlap sequences. Gene. 2009; 434:43–49. [DOI] [PubMed] [Google Scholar]
- 3. Nour-Eldin H.H., Geu-Flores F., Halkier B.A.. USER cloning and USER fusion: the ideal cloning techniques for small and big laboratories. Methods Mol. Biol. 2010; 643:185–200. [DOI] [PubMed] [Google Scholar]
- 4. Engler C., Kandzia R., Marillonnet S.. A one pot, one step, precision cloning method with high throughput capability. PloS One. 2008; 3:e3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Blake W.J., Chapman B.A., Zindal A., Lee M.E., Lippow S.M., Baynes B.M.. Pairwise selection assembly for sequence-independent construction of long-length DNA. Nucleic Acids Res. 2010; 38:2594–2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. De Paoli H.C., Tuskan G.A., Yang X.. An innovative platform for quick and flexible joining of assorted DNA fragments. Sci. Rep. 2016; 6:e19278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Engler C., Gruetzner R., Kandzia R., Marillonnet S.. Golden gate shuffling: a one-pot DNA shuffling method based on type IIs restriction enzymes. PloS One. 2009; 4:e5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Weber E., Engler C., Gruetzner R., Werner S., Marillonnet S.. A modular cloning system for standardized assembly of multigene constructs. PloS One. 2011; 6:e16765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sarrion-Perdigones A., Falconi E.E., Zandalinas S.I., Juarez P., Fernandez-del-Carmen A., Granell A., Orzaez D.. GoldenBraid: an iterative cloning system for standardized assembly of reusable genetic modules. PloS One. 2011; 6:e21622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Patron N.J., Orzaez D., Marillonnet S., Warzecha H., Matthewman C., Youles M., Raitskin O., Leveau A., Farre G., Rogers C. et al. Standards for plant synthetic biology: a common syntax for exchange of DNA parts. New Phytol. 2015; 208:13–19. [DOI] [PubMed] [Google Scholar]
- 11. Liu W., Stewart C.N. Jr. Plant synthetic biology. Trends Plant Sci. 2015; 20:309–317. [DOI] [PubMed] [Google Scholar]
- 12. Wang Y.H., Wei K.Y., Smolke C.D.. Synthetic biology: advancing the design of diverse genetic systems. Annu. Rev. Chem. Biomol. Eng. 2013; 4:69–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Engler C., Youles M., Gruetzner R., Ehnert T.M., Werner S., Jones J.D., Patron N.J., Marillonnet S.. A golden gate modular cloning toolbox for plants. ACS Synth. Biol. 2014; 3:839–843. [DOI] [PubMed] [Google Scholar]
- 14. Canton B., Labno A., Endy D.. Refinement and standardization of synthetic biological parts and devices. Nat. Biotechnol. 2008; 26:787–793. [DOI] [PubMed] [Google Scholar]
- 15. Sarrion-Perdigones A., Vazquez-Vilar M., Palaci J., Castelijns B., Forment J., Ziarsolo P., Blanca J., Granell A., Orzaez D.. GoldenBraid 2.0: a comprehensive DNA assembly framework for plant synthetic biology. Plant Physiol. 2013; 162:1618–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Orzaez D., Medina A., Torre S., Fernandez-Moreno J.P., Rambla J.L., Fernandez-Del-Carmen A., Butelli E., Martin C., Granell A.. A visual reporter system for virus-induced gene silencing in tomato fruit based on anthocyanin accumulation. Plant Physiol. 2009; 150:1122–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yoo S.D., Cho Y.H., Sheen J.. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protocols. 2007; 2:1565–1572. [DOI] [PubMed] [Google Scholar]
- 18. Schneider C.A., Rasband W.S., Eliceiri K.W.. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012; 9:671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gutierrez S., Yvon M., Thebaud G., Monsion B., Michalakis Y., Blanc S.. Dynamics of the multiplicity of cellular infection in a plant virus. PLoS Pathog. 2010; 6:e1001113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vazquez-Vilar M., Sarrion-Perdigones A., Ziarsolo P., Blanca J., Granell A., Orzaez D.. Software-assisted stacking of gene modules using GoldenBraid 2.0 DNA-assembly framework. Methods Mol. Biol. 2015; 1284:399–420. [DOI] [PubMed] [Google Scholar]
- 21. Vazquez-Vilar M., Bernabé-Orts J.M., Fernandez-del-Carmen A., Ziarsolo P., Blanca J., Granell A., Orzaez D.. A modular toolbox for gRNA–Cas9 genome engineering in plants based on the GoldenBraid standard. Plant Methods. 2016; 12:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Quinn J.Y., Cox R.S. 3rd, Adler A., Beal J., Bhatia S., Cai Y., Chen J., Clancy K., Galdzicki M., Hillson N.J.. SBOL Visual: A graphical language for genetic designs. PLoS Biol. 2015; 13:e1002310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Thorpe H.M., Wilson S.E., Smith M.C.. Control of directionality in the site-specific recombination system of the Streptomyces phage phiC31. Mol. Microbiol. 2000; 38:232–241. [DOI] [PubMed] [Google Scholar]
- 24. Khaleel T., Younger E., McEwan A.R., Varghese A.S., Smith M.C.. A phage protein that binds phiC31 integrase to switch its directionality. Mol. Microbiol. 2011; 80:1450–1463. [DOI] [PubMed] [Google Scholar]
- 25. Ort D.R., Merchant S.S., Alric J., Barkan A., Blankenship R.E., Bock R., Croce R., Hanson M.R., Hibberd J.M., Long S.P. et al. Redesigning photosynthesis to sustainably meet global food and bioenergy demand. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:8529–8536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Smolke C.D. Building outside of the box: iGEM and the BioBricks Foundation. Nat. Biotechnol. 2009; 27:1099–1102. [DOI] [PubMed] [Google Scholar]
- 27. Cambray G., Mutalik V.K., Arkin A.P.. Toward rational design of bacterial genomes. Curr. Opin. Microbiol. 2011; 14:624–630. [DOI] [PubMed] [Google Scholar]
- 28. Mutalik V.K., Guimaraes J.C., Cambray G., Mai Q.A., Christoffersen M.J., Martin L., Yu A., Lam C., Rodriguez C., Bennett G. et al. Quantitative estimation of activity and quality for collections of functional genetic elements. Nat. Methods. 2013; 10:347–353. [DOI] [PubMed] [Google Scholar]
- 29. Cambray G., Guimaraes J.C., Mutalik V.K., Lam C., Mai Q.A., Thimmaiah T., Carothers J.M., Arkin A.P., Endy D.. Measurement and modeling of intrinsic transcription terminators. Nucleic Acids Res. 2013; 41:5139–5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mutalik V.K., Guimaraes J.C., Cambray G., Lam C., Christoffersen M.J., Mai Q.A., Tran A.B., Paull M., Keasling J.D., Arkin A.P. et al. Precise and reliable gene expression via standard transcription and translation initiation elements. Nat. Methods. 2013; 10:354–360. [DOI] [PubMed] [Google Scholar]
- 31. Bartley B., Beal J., Clancy K., Misirli G., Roehner N., Oberortner E., Pocock M., Bissell M., Madsen C., Nguyen T. et al. Synthetic biology open language (SBOL) version 2.0.0. J. Integr. Bioinform. 2015; 12:e272. [DOI] [PubMed] [Google Scholar]
- 32. Angov E. Codon usage: nature's roadmap to expression and folding of proteins. Biotechnol. J. 2011; 6:650–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hillson N.J., Rosengarten R.D., Keasling J.D.. j5 DNA assembly design automation software. ACS Synth. Biol. 2012; 1:14–21. [DOI] [PubMed] [Google Scholar]
- 34. Brazma A., Hingamp P., Quackenbush J., Sherlock G., Spellman P., Stoeckert C., Aach J., Ansorge W., Ball C.A., Causton H.C. et al. Minimum information about a microarray experiment (MIAME)[mdash]toward standards for microarray data. Nat. Genet. 2001; 29:365–371. [DOI] [PubMed] [Google Scholar]
- 35. Taylor C.F., Paton N.W., Lilley K.S., Binz P.A., Julian R.K., Jones A.R., Zhu W., Apweiler R., Aebersold R., Deutsch E.W. et al. The minimum information about a proteomics experiment (MIAPE). Nat. Biotechnol. 2007; 25:887–893. [DOI] [PubMed] [Google Scholar]
- 36. Yang Y.N., Li R.G., Qi M.. In vivo analysis of plant promoters and transcription factors by agroinfiltration of tobacco leaves. Plant J. 2000; 22:543–551. [DOI] [PubMed] [Google Scholar]
- 37. Kapila J., DeRycke R., VanMontagu M., Angenon G.. An Agrobacterium-mediated transient gene expression system for intact leaves. Plant Sci. 1997; 122:101–108. [Google Scholar]
- 38. Schaumberg K.A., Antunes M.S., Kassaw T.K., Xu W., Zalewski C.S., Medford J.I., Prasad A.. Quantitative characterization of genetic parts and circuits for plant synthetic biology. Nat. Methods. 2016; 13:94–100. [DOI] [PubMed] [Google Scholar]
- 39. Dafny-Yelin M., Tzfira T.. Delivery of multiple transgenes to plant cells. Plant Physiol. 2007; 145:1118–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tzfira T., Li J., Lacroix B., Citovsky V.. Agrobacterium T-DNA integration: molecules and models. Trends Genet. 2004; 20:375–383. [DOI] [PubMed] [Google Scholar]
- 41. De Buck S., De Wilde C., Van Montagu M., Depicker A.. Determination of the T-DNA transfer and the T-DNA integration frequencies upon cocultivation of Arabidopsis thaliana root explants. Mol. Plant Microbe Interact. 2000; 13:658–665. [DOI] [PubMed] [Google Scholar]
- 42. Konermann S., Brigham M.D., Trevino A.E., Joung J., Abudayyeh O.O., Barcena C., Hsu P.D., Habib N., Gootenberg J.S., Nishimasu H. et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015; 517:583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gilbert L.A., Horlbeck M.A., Adamson B., Villalta J.E., Chen Y., Whitehead E.H., Guimaraes C., Panning B., Ploegh H.L., Bassik M.C. et al. Genome-Scale CRISPR-Mediated control of gene repression and activation. Cell. 2014; 159:647–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Moore I., Samalova M., Kurup S.. Transactivated and chemically inducible gene expression in plants. Plant J. 2006; 45:651–683. [DOI] [PubMed] [Google Scholar]
- 45. Butelli E., Titta L., Giorgio M., Mock H.P., Matros A., Peterek S., Schijlen E.G., Hall R.D., Bovy A.G., Luo J. et al. Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat. Biotechnol. 2008; 26:1301–1308. [DOI] [PubMed] [Google Scholar]
- 46. Muller K., Siegel D., Rodriguez J.F., Gerrer K., Wend S., Decker E.L., Reski R., Weber W., Zurbriggen M.D.. A red light-controlled synthetic gene expression switch for plant systems. Mol. Biosyst. 2014; 10:1679–1688. [DOI] [PubMed] [Google Scholar]
- 47. Davuluri G.R., van Tuinen A., Fraser P.D., Manfredonia A., Newman R., Burgess D., Brummell D.A., King S.R., Palys J., Uhlig J. et al. Fruit-specific RNAi-mediated suppression of DET1 enhances carotenoid and flavonoid content in tomatoes. Nat. Biotechnol. 2005; 23:890–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang Y., Butelli E., Alseekh S., Tohge T., Rallapalli G., Luo J., Kawar P.G., Hill L., Santino A., Fernie A.R. et al. Multi-level engineering facilitates the production of phenylpropanoid compounds in tomato. Nat. Commun. 2015; 6:e8635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ori N., Juarez M.T., Jackson D., Yamaguchi J., Banowetz G.M., Hake S.. Leaf senescence is delayed in tobacco plants expressing the maize homeobox gene knotted1 under the control of a senescence-activated promoter. Plant Cell. 1999; 11:1073–1080. [PMC free article] [PubMed] [Google Scholar]
- 50. Azuma M., Morimoto R., Hirose M., Morita Y., Hoshino A., Iida S., Oshima Y., Mitsuda N., Ohme-Takagi M., Shiratake K.. A petal-specific InMYB1 promoter from Japanese morning glory: a useful tool for molecular breeding of floricultural crops. Plant Biotechnol. J. 2016; 14:354–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Paine J.A., Shipton C.A., Chaggar S., Howells R.M., Kennedy M.J., Vernon G., Wright S.Y., Hinchliffe E., Adams J.L., Silverstone A.L. et al. Improving the nutritional value of Golden Rice through increased pro-vitamin A content. Nat. Biotechnol. 2005; 23:482–487. [DOI] [PubMed] [Google Scholar]
- 52. Houmard N.M., Mainville J.L., Bonin C.P., Huang S., Luethy M.H., Malvar T.M.. High-lysine corn generated by endosperm-specific suppression of lysine catabolism using RNAi. Plant Biotechnol. J. 2007; 5:605–614. [DOI] [PubMed] [Google Scholar]
- 53. Padidam M. Chemically regulated gene expression in plants. Curr. Opin. Plant Biol. 2003; 6:169–177. [DOI] [PubMed] [Google Scholar]
- 54. Kinkema M., Geijskes R.J., Shand K., Coleman H.D., De Lucca P.C., Palupe A., Harrison M.D., Jepson I., Dale J.L., Sainz M.B.. An improved chemically inducible gene switch that functions in the monocotyledonous plant sugar cane. Plant Mol. Biol. 2014; 84:443–454. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.