Abstract
We introduce a new concept that utilizes cognate nucleic acid nanoparticles which are fully complementary and functionally-interdependent to each other. In the described approach, the physical interaction between sets of designed nanoparticles initiates a rapid isothermal shape change which triggers the activation of multiple functionalities and biological pathways including transcription, energy transfer, functional aptamers and RNA interference. The individual nanoparticles are not active and have controllable kinetics of re-association and fine-tunable chemical and thermodynamic stabilities. Computational algorithms were developed to accurately predict melting temperatures of nanoparticles of various compositions and trace the process of their re-association in silico. Additionally, tunable immunostimulatory properties of described nanoparticles suggest that the particles that do not induce pro-inflammatory cytokines and high levels of interferons can be used as scaffolds to carry therapeutic oligonucleotides, while particles with strong interferon and mild pro-inflammatory cytokine induction may qualify as vaccine adjuvants. The presented concept provides a simple, cost-effective and straightforward model for the development of combinatorial regulation of biological processes in nucleic acid nanotechnology.
INTRODUCTION
Nucleic acids (RNA and DNA) have been acknowledged as an important building material for nanotechnology due to their biocompatibility and programmability (1-13). The option of both canonical and non-canonical base pairings tremendously expands the diverse set of RNA structural motifs available as building blocks. Programmable multi-tasking as well as the ability to dynamically respond to the environment make nucleic acids an attractive material for tailor-made applications in both biotechnology and personalized therapy.
In one decade, a wide array of artificially designed dynamic DNA assemblies have been shown to respond to a broader spectrum of physicochemical stimuli or ligands. Rationally designed DNA nanomachines can carry out a rotary motion by switching from B- to Z-DNA at high ionic strength (14), sense the pH (15,16) and respond to changes from visible to UV light (17). DNA ‘walkers’ are capable of directional movement based on strand displacement (18,19), enzymatic activity (20,21) or in accordance with the prescriptive DNA origami landscapes (22). DNA boxes with a programmable lid (23) and DNA ‘nanorobots’(24) can be used for delivery and release of different cargos. Recently, a DNA cube that selectively forms a flat 2D structure after hybridization to a specific fusion gene that is characteristic for a prostate cancer cell line was engineered. While authors have demonstrated that the cubes are taken up by a number of cell lines, the dynamic response has only been shown in vitro (25). Although numerous creative innovations of dynamic DNA nanoassemblies have been described, the majority are only functional in vitro and their immediate practical applications in living systems remain unclear.
In addition to being carriers of genetic information, RNAs are now recognized to function as natural scaffolds, enzymes, switches, aptamers and regulators of gene expression and editing. The emerging field of RNA nanotechnology applies the current knowledge related to the structure and function of natural and artificial RNAs to further address specific biomedical challenges by engineering nanodevices that can interact with cellular machinery (2,26). Building dynamic RNA nanoparticles that can communicate with one another will further improve the operation of functional systems. In fact, metabolite and cofactor responsive riboswitches and ribozymes as well as temperature-sensing RNA thermometers are some examples of dynamic RNAs autogenic in nature (27–30). Recently, we reported two approaches of dynamic RNA (31) and RNA-DNA hybrid (32) nanostructures that conditionally activate gene silencing in diseased cells in vitro and in vivo. The first approach is based on computer-generated two-stranded RNA switches that are activated only in the presence of specific mRNAs through interaction with a single-stranded (ss) RNA toehold of the switch (31). The second approach is based on RNA-DNA hybrids with split-functionalities activated only when two complementary copies are introduced into the same cell. Strand exchange, with subsequent intracellular activation of functionalities, is promoted by the interaction of complementary ssDNA (32) or ssRNA (33) toeholds. This concept was further used by other research groups for various applications (34,35). The simultaneous delivery and release of multiple functionalities was achieved by including them all into the longer hybrids (36). Alternatively, RNA and/or DNA nanoscaffolds can be decorated with multiple hybrids and activated by adding individual cognate DNA/RNA hybrids (37,38). This approach, however, requires the simultaneous presence of the nanoparticle and six individual cognate hybrids in the same cell to activate six functionalities. While efficient, previously described nanodevices typically demand intensive computer-assisted design and the use of specifically programmed toeholds.
Here, we set out to design a series of interdependent complementary nucleic acid nanoparticles that take advantage of dynamic interaction and shape-switching to activate multiple functionalities. As opposed to previously described work, this new approach does not require any toeholds to initiate the interactions and their design principles are simplified. Additionally, only two particles are required to simultaneously activate multiple functionalities. The novel interrelated nanoparticles are designed by simply taking the reverse complements of the existing RNA scaffolds and assembling them into the ‘anti-scaffolds,’ as schematically explained in Supplementary Figure S1. As a proof of concept, nucleic acid cubes (1) and their reverse complements, anti-cubes, are extensively characterized in this work (Figure 1A). The interaction of cubes with anti-cubes at physiological conditions leads to conformational changes and to the swift formation of multiple duplexes or fibers that can further activate transcription, Förster resonance energy transfer (FRET), aptamers, and specific gene silencing. We show that the immunostimulatory activity, thermodynamic stability, resistance to nuclease degradation, re-association rate and cost of production for complementary nanoparticles vary tremendously depending on their composition (e.g. DNA versus RNA). In addition, RNA hexameric rings (39) and recently engineered RNA and DNA triangle (40) scaffolds and their corresponding anti-scaffolds are explored.
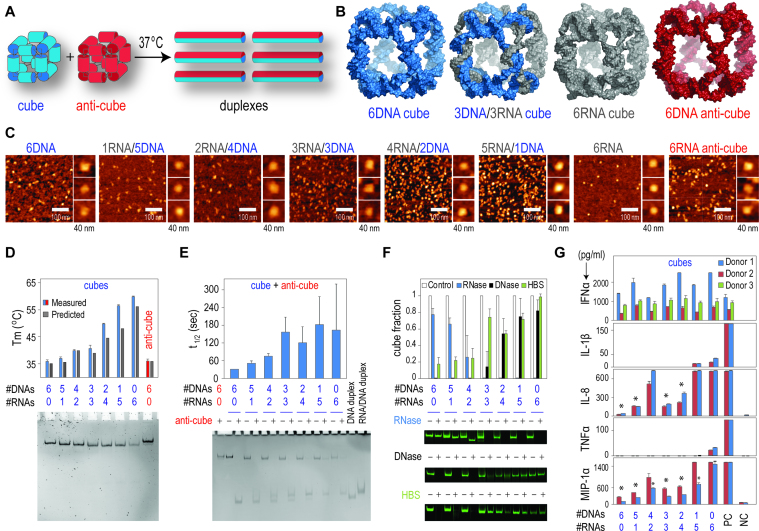
Figure 1.
Fine-tunable isothermal re-association of complementary shape-switching nanoparticles. (A) Schematic representation of re-association between the complementary cube and anti-cube nanoparticles. (B) 3D models of nanoparticles. (C) AFM images of RNA, RNA/DNA, DNA cubes and RNA anti-cube. (D) Experimental and predicted melting temperatures of nanoparticles controlled by their compositions and native-PAGE with corresponding assemblies. Error bars indicate s.d.; N = 3 (E) Relative re-association rates of RNA, RNA/DNA and DNA cubes with DNA anti-cubes measured at 25°C and native-PAGE with corresponding re-associations visualized after 30 min of incubation. Error bars indicate s.d.; N = 3. (F) Relative stabilities of nanoparticles in the presence of DNase, RNase and human blood serum. Results are normalized to corresponding non-treated samples. Error bars indicate s.d.; N = 3. (G) Immunostimulatory properties of RNA, RNA/DNA and DNA shape-switching nanoparticles delivered using Lipofectamine 2000. Error bars indicate s.d.; N = 2. Statistically significant results (compared to a positive control, PC) are indicated with asterisks (P-value < 0.05). For IL-1β and TNFα all results are statistically significant. In (D and E), note that the higher number of RNA strands per nanoparticle weakens the extent of total staining.
MATERIALS AND METHODS
All methods are detailed in Supplementary Data.
Nanoparticle assembly and purification
All nanoparticles were assembled by combining individual monomer components in equimolar concentrations following different assembly protocols. For purification and electrophoretic mobility shift assays, 8% non-denaturing native-polyacrylamide gel electrophoresis (PAGE) (37.5:1) was used. All gels were visualized with a Bio-Rad ChemiDoc MP System.
UV-melting experiments
Temperature-dependent absorption measurements were recorded at 260 nm on an Agilent 8453 spectrophotometer coupled with the Agilent 89090 Peltier Temperature Controller.
Kinetics of re-association determination
To determine the kinetics, different compositions of cubes and Cy5-tagged anti-cubes were mixed together and aliquoted at set time points to assess the extent of re-association.
Primary human peripheral blood mononuclear cell and whole blood culture for analysis of interferon and cytokine secretion
The experiments were performed according to the standardized protocol NCL-ITA-10 (http://ncl.cancer.gov/NCL_Method_ITA-10.pdf).
Nuclease digestion assays
Different Alexa 488-labeled nanocubes (1 μM final) were incubated with RNAse, DNase and human blood serum at 37°C. The reaction was stopped by snap cooling samples on dry ice prior to loading on native-PAGE and further analysis.
Computational predictions and 3D modeling
Computational predictions of melting temperatures (Tms) were performed for the different combinations of RNA and DNA strands using new HyperFold. The RNA, RNA/DNA and DNA cube models were built using PyMol. The triangle models were built with the aid of NanoTiler (https://binkley2.ncifcrf.gov/users/bshapiro/software.html).
Fluorescence studies
Activation of FRET
To determine the re-association of nanoparticles in vitro, FRET measurements were performed using a FluoroMax3 (Jobin-Yvon, Horiba).
Activation of Broc-Coli aptamers
All assemblies and re-associations were confirmed on native-PAGE stained with (Z)-4-(3,5-difluoro-4-hydroxybenzylidene)-2-methyl-1-(2,2,2-trifluoroethyl)-1 H-imidazol-5(4 H)-one (DFHBI-1T).
Atomic Force Microscopy (AFM) imaging and sample preparation
AFM imaging was performed on MultiMode AFM Nanoscope IV system (Bruker Instruments, Santa Barbara, CA, USA) in tapping mode. The images were recorded with a 1.5 Hz scanning rate using a TESPA-300 probe from Bruker with a resonance frequency of 320 kHz and a spring constant of about 40 N/m. Images were processed using the FemtoScan Online software package (Advanced Technologies Center, Moscow, Russia) (41,42).
Transfection of human cell lines
For cell culture experiments with nanoparticles, human breast cancer (MDA-MB-231 with or without GFP), prostate cancer (PC-3) and cervical cancer (HeLa) cell lines were used. Lipofectamine 2000 was used for all transfections.
Fluorescent microscopy
To assess the intracellular interaction and shape of nanoparticles, FRET measurements were performed using a LSM 710 confocal microscope (Carl Zeiss) with a 63×, 1.4 NA magnification lens.
Flow Cytometry
BD Accuri C6 or FACSCalibur flow cytometer was used for all experiments. CellQuest or the CFlow Sampler software were used to retrieve the geometric mean fluorescence intensity (gMFI) and the standard error of the mean.
Cell proliferation assay
The viability of cells was assessed through the CellTiter 96® Aqueous One assay (Promega, Madison) following manufacturer's protocol.
Statistics
Statistical significance was determined using a Student's two-tailed t-test conducted with the GraphPad Prism Software. A p-value of less than 0.05 was considered to be statistically significant.
RESULTS AND DISCUSSION
Complementary nanoparticles have controlled rates of re-association and fine-tunable thermodynamic, chemical and immunological properties
An important feature of rationally designed cubes is their ability to efficiently assemble with different ratios of RNA and DNA strands entering their composition (Figure 1B). This design flexibility together with the difference in physicochemical properties between RNA and DNA allows for fine-tuning of the thermodynamic, kinetic, and chemical properties of the interacting nanoparticles (Figure 1D–F). To prove this concept, seven cubes with different ratios of RNA and DNA strands in their compositions were examined. It is worth mentioning that the costs of tested nanoparticles increase by approximately a factor of two with the introduction of each RNA strand into the assembly. The assemblies of all cubes were confirmed by non-denaturing polyacrylamide gel electrophoresis (native-PAGE) and visualized by AFM (Figure 1C). The resolution of AFM does not allow for analysis of structural details of our assemblies, but all the cube nanoparticles appear to be uniform in size and shape and monodisperse. As expected, the relative thermodynamic stability of nanoparticles increased with the higher number of RNA strands introduced into assembly, with the Tms ranging from ∼36°C for DNA cubes to ∼60°C for RNA cubes. Computational predictions using the new version of Hyperfold (31) accurately confirmed the experimental results (Figure 1D and Supplementary Figures S2 and 3).
Upon re-association of two complementary nanoparticles, the formation of duplexes consisting of cube and anti-cube strands was observed (Figure 1E). The re-association of the equimolar concentrations of the cognate cubes after 30 min of incubation was demonstrated via native-PAGE. The intact cubes had lower mobility compared to corresponding duplexes. The re-association of cognate cubes was thermodynamically driven and did not require any toehold interactions (Supplementary Data, Thermodynamics of re-associating cubes). As described in the ‘Materials and Methods’ section, secondary structures for different cubes and the products of their re-associations can now be automatically predicted and generated using HyperFold (Supplementary Figure S5). The kinetics experiments (Figure 1E and Supplementary Figure S4) demonstrated the capability to directly alter the rates of re-association by changes in the nucleic acid makeup. The higher number of RNA strands in cube composition delayed the re-association. As expected, the re-association of RNA cubes and RNA anti-cubes was the most retarded (t1/2 ∼16 min, Supplementary Figure S4). This notion was supported by the measured Tms. The relative chemical stability and the resistance to nuclease degradation can also be tuned by changing the composition of cube (Figure 1F).
Although synthetic RNA and DNA nanoparticles are biodegradable and commonly considered highly biocompatible, various therapeutic RNA and DNA motifs have been found to trigger the human innate immune system, leading to the induction of pro-inflammatory cytokines and type I interferons, thus complicating the translation of these novel therapeutics from bench to clinic (43). Consequences of such immune reaction may be severe and lead to the patient's death. For example, the Phase I clinical trial of MRX34, a nanoparticle formulated miRNA, was halted in September 2016 due to the severe cytokine storm reaction in five patients participating in the study (http://investor.mirnarx.com/releasedetail.cfm?ReleaseID=990204). To estimate immunological compatibility of DNA and RNA cubes with varying ratios of RNA to DNA strands, we assessed the immune response to these particles. This was done in primary human peripheral blood mononuclear cell cultures by measuring the activation and secretion of type I interferon (IFNα) as well as various pro-inflammatory cytokines and chemokines including IL-1β, TNFα, IL-8 and MIP-1α (Figure 1G). IFNα, IL-1β and TNFα are common biomarkers used to estimate the pro-inflammatory potential of nucleic acids during both normal immune responses to viral pathogens and during autoimmune responses to host nucleic acids (44,45). The same markers were used in (pre)clinical studies to estimate the safety of RNA therapeutics (46). We expanded this commonly used test panel by adding pro-inflammatory chemokines IL-8 and MIP-1α (47). To deliver nanoparticles to cells, we used Lipofectamine 2000. Such complexation was used for consistency with other cellular experiments presented in this study, and because expression of the immune receptors sensing nucleic acids predominantly present in the intracellular compartments (e.g. endosomes, cytosol). Although all tested constructs induced the expression of IFNα, IL-8 and MIP-1α, cubes containing six RNA strands were more potent immune stimulants compared to other tested particles. The level of type I interferon induced by RNA cubes was comparable to positive control, a synthetic CpG oligonucleotide (ODN 2216) used as a potent vaccine adjuvant. Therefore, if interferon induction was used as a marker of immunogenicity, RNA cubes could be qualified as promising adjuvants. A similar trend was observed in the cases of pro-inflammatory cytokines IL-1β and TNFα, which were induced only by cubes containing five or six RNA strands. The response, however, was much lower compared to the positive control, a bacterial lipopolysaccharide. High levels of these cytokines are responsible for septic shock syndrome, while their low level is typically induced by vaccine adjuvants. The levels observed with RNA cubes were insignificant in terms of the cytokine storm or septic shock, but were at levels which may contribute to immunogenicity, thus additionally qualifying RNA cubes for a potential vaccine adjuvant application. The levels of chemokines IL-8 and MIP-1α were raised proportionally with an increasing number of RNA strands, except for the 2DNA/4RNA cube, which was as potent as cubes containing a higher number of RNA strands. This data suggested greater immunostimulatory potential of RNA cubes and is consistent with our earlier report (37). This finding also demonstrated that the reduction in number of RNA strands is a viable strategy for reducing any undesirable immunostimulation of these nanoparticles if they are used for systemic delivery. It further suggested that by simply optimizing the ratio between RNA and DNA strands, resulting assemblies that did not induce pro-inflammatory cytokines (e.g. DNA cubes) could be used for drug delivery while RNA cubes with optimal immunomodulatory properties could be used for vaccines and immunotherapy. The precise mechanism of the immune recognition of the particles reported here is a subject for a separate mechanistic study.
Re-association of complementary DNA nanoparticles triggers co-transcriptional formation of RNA nanoparticles
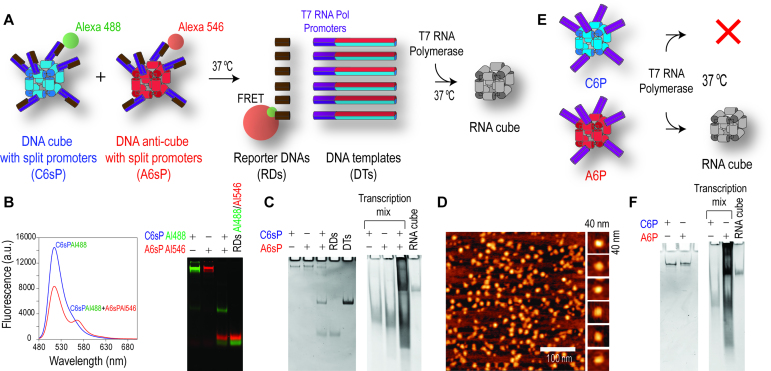
The ability to activate the simultaneous transcription of multiple RNAs and further co-transcriptional assembly of RNA nanostructures is an important step towards the intracellular production of RNA nanoparticles. The endogenous production of functional RNA nanoparticles in mammalian cells will substantially increase their yields while eliminating the complexity of assembly protocols and reducing possible endotoxin contamination. In the attempt to control the transcription with complementary nanoparticles, all six DNA strands of cube and anti-cube were modified with split 20 bps T7 RNA polymerase promoters (Figure 2A and Supplementary Figure S6). T7 RNA polymerase is a single-subunit enzyme that can be expressed in mammalian cells (48) and does not require any additional factors for accurate transcription. In the current design, the presence of both complementary DNA cubes in a transcription mixture was required to undergo shape-switching and formation of dsDNA templates with active T7 RNA polymerase promoters whose further transcription leads to the assembly of RNA cubes. Short reporter DNAs were used to provide an optical response upon re-association (Figure 2B). The co-transcriptionally assembled RNA cubes can be gel purified (Figure 2C) and visualized by AFM (Figure 2D). Even though the co-transcriptional assembly of RNA cubes was efficiently triggered by the re-association of two parent DNA cubes, the run-off transcription of multiple individual DNA templates may not be ideal for co-transcriptional assembly of RNA nanoparticles in the intracellular environment due to potential degradation or compartmentalization of some DNAs, causing stoichiometry problems with transcribed RNA units. To overcome this possible problem, the DNA anti-cube decorated with six complete T7 promoters (Figure 2E) can be used. We then have an assembly of individually potent ssDNA templates (Supplementary Figure S7) required for the co-transcriptional production of RNA cubes (Figure 2F). The advantage of this approach is the precise control over the stoichiometry of the DNA templates and their local availability for transcription. These results paved the way for further development of the intracellular co-transcriptional production of RNA nanoparticles.
Figure 2.
Isothermal re-association of complementary DNA nanoparticles activates co-transcriptional production of RNA nanoparticles. (A) Schematics of re-association between the complementary DNA cubes and anti-cubes carrying split and therefore inactive T7 RNA polymerase promoters. The re-association of DNA cubes in transcription mixture releases DNA templates with active promoters and allows for co-transcriptional assembly of RNA cubes. (B and C) Native-PAGE and fluorescence experiments visualizing re-association of shape-switching purified DNA cubes and DNA anti-cubes, resulting in formation of DNA templates with activated T7 RNA polymerase promoters and further co-transcriptional assembly of RNA cubes. (D) Co-transcriptionally assembled RNA cubes eluted from native-PAGE and imaged by AFM. (E) RNA cubes can only be formed co-transcriptionally using DNA anti-cubes decorated with six complete T7 RNA Polymerase promoters (A6P) and not from DNA cubes (C6P) due to the directionality of the promoter sequences. (F) Native-PAGE showing the co-transcriptional production of RNA cubes from DNA anti-cubes with six promoters. Following 4 h of incubation, all transcription mixtures were treated with DNase to remove any residual DNA.
Re-association of complementary DNA nanoparticles triggers activation of embedded split RNA aptamers
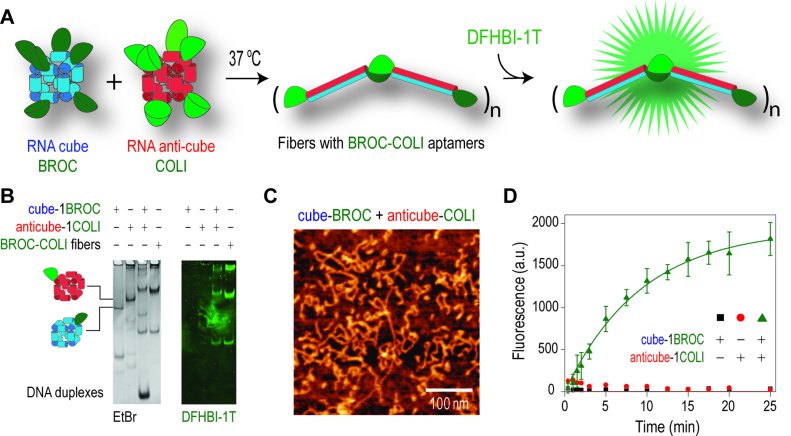
Broccoli is a synthetic RNA aptamer which binds to the fluorophore DFHBI-1T to mimic the fluorescent spectrum of green fluorescent protein (GFP) (49). We hypothesized that the splitting of the Broccoli aptamer sequence into two separate non-functional strands (named Broc and Coli) can be used for monitoring the cube and anti-cube interaction and shape-switching that leads to Broc and Coli re-assembly into an active fluorescent aptamer (Figure 3A). Furthermore, this approach demonstrates the general strategy for conditional re-activation of disconnected functional ssRNAs with complex secondary structure. As predicted, the interaction of Broc-cubes with Coli-anti-cubes leads to the formation of fiber-like structures (Figure 3B and C) containing re-assembled Broc-Coli aptamers, thus providing an alternative optical response of interdependent nanocube interactions. The response was confirmed by native-PAGE (Figure 3B and Supplementary Figure S8) and fluorescent measurements (Figure 3D). Flow cytometry analysis of human cervical cancer cells co-transfected with complementary nanoparticles bearing the split aptamer strands revealed some Broc-Coli re-association into the functional structure, however the difference was not statistically significant compared to just cells treated with the dye (data not shown).
Figure 3.
Activation of functional aptamers with isothermal re-association of shape-switching nanoparticles. (A) Schematics of isothermal re-association and re-assembly of aptamers. (B) Total EtBr and DFHBI-1T stained native-PAGE demonstrates fiber formation and aptamer activation on re-association of cognate cubes. (C) AFM images of the aptamer containing fibers. (D) Re-association and fiber formation can be traced by measuring fluorescence of DFHBI-1T in vitro. Error bars indicate s.d.; N = 3.
Re-association of complementary nanoparticles triggers activation of energy transfer and RNA interference in cells
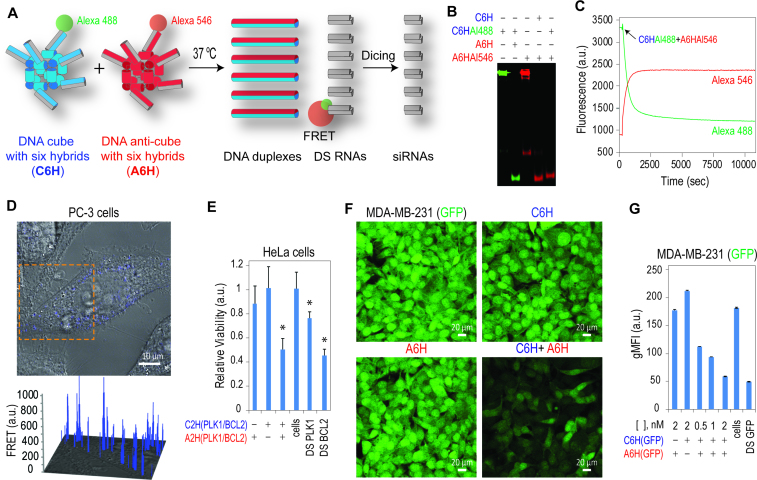
RNAi is a naturally occurring post-transcriptional gene regulation process which represses the expression of specific genes (50). Therefore, exploiting endogenous RNAi mechanisms by externally delivered RNAi inducers is a promising tool in biotechnology and therapy. To have additional control over the initiation of targeted gene silencing is an important step forward leading towards the construction of intracellular logic gates and smart nanoparticles. We decorated two sets of cognate DNA cubes and anti-cubes with split Dicer Substrate (DS) RNAs against either (i) BCL2 and PLK1 (51), well-validated molecular targets whose down-regulation induces apoptosis (52,53) or (ii) GFP (54). The re-association of the cube and anti-cube nanoparticles led to the formation of DS RNAs that could be further activated through dicing by releasing the functional siRNAs (Figure 4A). Additionally, split DS RNAs can be fluorescently labeled with dyes (e.g. Alexa 488 and Alexa 546) chosen to undergo FRET. Thus, the shape-switching of labeled nanoparticles was not only directly visualized by native-PAGE (Figure 4B), but also assessed in real time using FRET (Figure 4C). With the same approach, the intracellular re-association of cubes and anti-cubes in human prostate, breast and cervical cancer cells was traced by FRET (Figure 4D and Supplementary Figure S9). For activation of RNA interference, human cervical cancer cells were treated with nanoparticles releasing BCL2 and PLK1 DS RNAs (Figure 4E and Supplementary Figure S10A). The cell viability was significantly decreased when both cubes were introduced, while individual cubes did not show much effect. To show the generality of the RNAi induction approach, GFP-expressing breast cancer cells were treated with complementary cubes releasing DS RNAs targeting GFP. The extent of GFP silencing was assessed with fluorescent microscopy and quantified by flow cytometry (Figure 4F and G). The results showed an efficient knock-down of GFP production upon the intracellular re-association of cubes and anti-cubes functionalized with six split DS RNAs. By comparison, transfection of cells with individual cubes or anti-cubes did not result in GFP silencing. Re-association occurs intracellularly, as suggested by FRET showing that individual cubes and anti-cubes associated with transfection agent do not re-associate in solution (Supplementary Figure S9B). Efficient GFP silencing was observed at picomolar concentrations of complementary cubes (Supplementary Figure S10B).
Figure 4.
Activation of RNA interference and intracellular FRET with complementary shape-switching nanoparticles. (A) Schematics of isothermal re-association and activation of FRET and RNAi. (B) In vitro re-association of fluorescently labeled cubes and anti-cubes with split DS RNAs was visualized by native-PAGE. (C) FRET time traces during re-association of fluorescently labeled Alexa 488 and Alexa 546 cubes and anti-cubes carrying split Dicer Substrate RNAs (DS RNAs). (D) For intracellular FRET experiments, human prostate cancer (PC-3) cells were co-transfected with fluorescently labeled cubes and anti-cubes and images were taken on the next day. (E) Cell viability assay for HeLa cells transfected with nanoparticles (at 5 nM final) designed to release two DS RNAs against PLK1 and BCL2. Error bars indicate s.d.; N = 3. Statistically significant results (compared to control cells) are indicated with asterisks (P-value < 0.05). (F and G) GFP knockdown assays for human breast cancer cells expressing enhanced GFP (MDA-MB-231/GFP). Prior to transfection, formation of the nanocubes was verified by total EtBr staining of native-PAGE. Three days after the transfection of cells, GFP expression was analyzed with fluorescent microscopy (F) and flow cytometry (G). As the control, pre-formed DS RNAs (at 2 nM final) against PLK1, BCL2 and GFP were used for HeLa and MDA-MB-231 cells respectively. At concentrations higher than 10 nM, some gene silencing was observed for cubes carrying antisense DS RNAs (data not shown). Note that the individual cubes and anti-cubes cause no decrease in GFP production. gMFI corresponds to the geometric mean fluorescence intensity. Error bars denote SEM.
Other examples of complementary nanoparticles
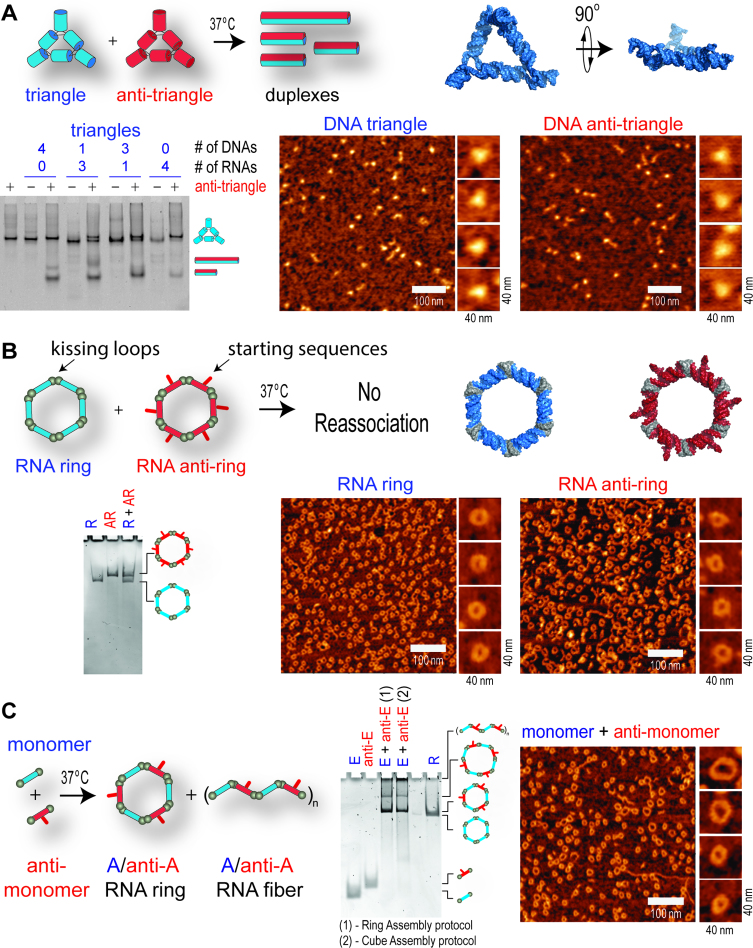
With the same design principles, several additional complementary nanoparticles were created by using previously characterized RNA triangles (40) and RNA rings (39) and their reverse complements (Figure 5). We expected that co-incubation of the cognate pairs will lead to the similar collapse of each structure and recombination of smaller subunits as seen in the cube and anti-cube pairs. However, while native-PAGE results showed that the incubation of triangles with anti-triangles led to the shape-switching (Figure 5A), the rings and anti-rings did not interact (Figure 5B). The explanation was offered by comparing ring designing principles with the cube and triangle structures. RNA nanorings did not have exposed single bases like cubes and triangles have in their corners; rather, the rings were formed through interstrand RNA–RNA ‘kissing loops’ (KL). Therefore, due to the absence of exposed bases, the reverse complement strands were unable to elicit shape-switching, and each ring remains intact as is visualized using native-PAGE stained with ethidium bromide (Figure 5B). The use of RNA tertiary interacting motifs (KL) also prevented rings from forming RNA/DNA or DNA/DNA structures (Supplementary Figure S11), as opposed to cubes and triangles. The use of complementary rings provided a simple way of expanding the library of the novel programmable nanoscaffolds based on the existing nanodesigns, thus eliminating any laborious computational design and experimental verification. Although the intact rings would not interact, their individual subunits were complementary and formed both hexameric and octameric rings as well as elongated fiber-like structures as verified by native-PAGE and AFM (Figure 5C and Supplementary Figure S12A). The fabrication of these structures was accomplished using both the established ring and cube assembly protocol, which differ only in the incubation profile. Depending on the kissing loop sequence, the formation of either fibers (e.g. A and anti-A) or rings (e.g. F and anti-F) can be promoted (Supplementary Figure S12A). The formation of functional fibers and rings was achieved by functionalizing one of the monomers (Supplementary Figure S12B).
Figure 5.
Ring and anti-ring nanoparticles do not re-associate but cognate monomers form rings and fibers. (A) Schematics of isothermal re-association of triangles and anti-triangles. AFM of triangles and anti-triangles and native-PAGE of re-associated anti-triangles with triangles after 30 min of incubation. (B) AFM images of RNA rings and anti-rings and native-PAGE showing that they do not interact. (C) The individual monomers of rings and anti-rings form the mixture of hexameric rings and fiber-like structures as shown by native-PAGE and AFM.
CONCLUSION
Diverse examples of mutual relations between interdependent entities exist in nature both at the levels of ecosystems and individual molecular interactions. Herein, we present a new concept of dynamic interdependent nucleic acid nanoparticles. Our approach relies on the physical interaction of two complementary nanoparticles with controllable thermodynamic and chemical properties. Our findings also suggest that by simply optimizing the ratio between RNA and DNA strands entering the composition of assemblies, one can create nanoparticles with optimal immunomodulatory properties when activation of the immune system is desirable (e.g. vaccines and immunotherapy). However, other types of RNA nanoparticles may expose different immunomodulatory properties (55). After interaction of the cognate nanoparticles both in vitro and in human cells, as demonstrated in three different cell lines, constructs undergo isothermal shape-switching resulting in activation of one or more functionalities including RNAi, optical response, transcription and split aptamer re-assembly. Importantly, only two nanoparticles are required to simultaneously activate multiple functionalities and no ssRNA or ssDNA toeholds are needed to initiate the interaction. Moreover, in the case of co-transcriptional assemblies, only one specifically designed DNA nanoparticle is needed to efficiently produce an RNA counterpart. Overall, the presented strategy allows for the use of simple, multifunctional and conditionally activated nanoparticles and provides a promising future for their use in nanobioscience.
Supplementary Material
ACKNOWLEDGEMENTS
Authors would also like to thank Dr Alexander Lushnikov and Dr Alexey Krasnoslobodtsev for performing AFM imaging of the nanoparticles at the Nanoimaging core facility at the University of Nebraska Medical Center and Lauren Lee (UNCC) for her assistance with cell culture experiments.
Author contributions: K.A.A. originated and designed the research project. J.H., E.S., M.V., A.I., M.P., M.C., M.N.B., E.F.K. carried out all gel electrophoresis, fluorescence and cell culture experiments. B.R. measured the Tms. E.F.K. performed AFM imaging. M.A.D. did the immunological studies. E.B., W.K. and B.A.S. contributed with computational studies. J.H., M.P. and K.A.A. co-wrote the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The research was supported by Department of Chemistry UNCC start-up funds and UNCC Faculty Research Grant to KAA. MAD, MV, EB, WKK disclose that this project has been funded in whole or in part with Federal funds from the Frederick National Laboratory for Cancer Research, National Institutes of Health, under contract HHSN26120080001E. This Research was supported [in part] by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the U.S. Government. MP participation on the project was supported [in part] by Slovak Academic Information Agency (SAIA).
Conflict of interest statement. None declared.
REFERENCES
- 1. Afonin K.A., Bindewald E., Yaghoubian A.J., Voss N., Jacovetty E., Shapiro B.A., Jaeger L.. In vitro assembly of cubic RNA-based scaffolds designed in silico. Nat. Nanotechnol. 2010; 5:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Guo P. The emerging field of RNA nanotechnology. Nat. Nanotechnol. 2010; 5:833–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Guo P., Zhang C., Chen C., Garver K., Trottier M.. Inter-RNA interaction of phage phi29 pRNA to form a hexameric complex for viral DNA transportation. Mol. Cell. 1998; 2:149–155. [DOI] [PubMed] [Google Scholar]
- 4. Haque F., Shu D., Shu Y., Shlyakhtenko L.S., Rychahou P.G., Evers B.M., Guo P.. Ultrastable synergistic tetravalent RNA nanoparticles for targeting to cancers. Nano Today. 2012; 7:245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Khisamutdinov E.F., Jasinski D.L., Guo P.. RNA as a boiling-resistant anionic polymer material to build robust structures with defined shape and stoichiometry. ACS Nano. 2014; 8:4771–4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. He Y., Ye T., Su M., Zhang C., Ribbe A.E., Jiang W., Mao C.. Hierarchical self-assembly of DNA into symmetric supramolecular polyhedra. Nature. 2008; 452:198–201. [DOI] [PubMed] [Google Scholar]
- 7. Yu J., Liu Z., Jiang W., Wang G., Mao C.. De novo design of an RNA tile that self-assembles into a homo-octameric nanoprism. Nat. Commun. 2015; 6:5724. [DOI] [PubMed] [Google Scholar]
- 8. Geary C., Rothemund P.W., Andersen E.S.. RNA nanostructures. A single-stranded architecture for cotranscriptional folding of RNA nanostructures. Science. 2014; 345:799–804. [DOI] [PubMed] [Google Scholar]
- 9. Rothemund P.W.K. Folding DNA to create nanoscale shapes and patterns. Nature. 2006; 440:297–302. [DOI] [PubMed] [Google Scholar]
- 10. Ohno H., Kobayashi T., Kabata R., Endo K., Iwasa T., Yoshimura S.H., Takeyasu K., Inoue T., Saito H.. Synthetic RNA-protein complex shaped like an equilateral triangle. Nat. Nanotechnol. 2011; 6:116–120. [DOI] [PubMed] [Google Scholar]
- 11. Osada E., Suzuki Y., Hidaka K., Ohno H., Sugiyama H., Endo M., Saito H.. Engineering RNA-protein complexes with different shapes for imaging and therapeutic applications. ACS Nano. 2014; 8:8130–8140. [DOI] [PubMed] [Google Scholar]
- 12. Pinheiro A.V., Han D., Shih W.M., Yan H.. Challenges and opportunities for structural DNA nanotechnology. Nat. Nanotechnol. 2011; 6:763–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lee H., Lytton-Jean A.K., Chen Y., Love K.T., Park A.I., Karagiannis E.D., Sehgal A., Querbes W., Zurenko C.S., Jayaraman M. et al. . Molecularly self-assembled nucleic acid nanoparticles for targeted in vivo siRNA delivery. Nat. Nanotechnol. 2012; 7:389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mao C., Sun W., Shen Z., Seeman N.C.. A nanomechanical device based on the B-Z transition of DNA. Nature. 1999; 397:144–146. [DOI] [PubMed] [Google Scholar]
- 15. Modi S., M G.S., Goswami D., Gupta G.D., Mayor S., Krishnan Y.. A DNA nanomachine that maps spatial and temporal pH changes inside living cells. Nat. Nanotechnol. 2009; 4:325–330. [DOI] [PubMed] [Google Scholar]
- 16. Modi S., Nizak C., Surana S., Halder S., Krishnan Y.. Two DNA nanomachines map pH changes along intersecting endocytic pathways inside the same cell. Nat. Nanotechnol. 2013; 8:459–467. [DOI] [PubMed] [Google Scholar]
- 17. Zhou M., Liang X., Mochizuki T., Asanuma H.. A light-driven DNA nanomachine for the efficient photoswitching of RNA digestion. Angew. Chem. Int. Ed. Engl. 2010; 49:2167–2170. [DOI] [PubMed] [Google Scholar]
- 18. Yurke B., Turberfield A.J., Mills A.P., Simmel F.C., Neumann J.L.. A DNA-fuelled molecular machine made of DNA. Nature. 2000; 406:605–608. [DOI] [PubMed] [Google Scholar]
- 19. Shin J.-S., Pierce N.A.. A synthetic DNA walker for molecular transport. J. Am. Chem. Soc. 2004; 126:10834–10835. [DOI] [PubMed] [Google Scholar]
- 20. Bath J., Green S.J., Turberfield A.J.. A free-running DNA motor powered by a nicking enzyme. Angew. Chem. Int. Ed. Engl. 2005; 44:4358–4361. [DOI] [PubMed] [Google Scholar]
- 21. Chen Y., Wang M., Mao C.. An autonomous DNA nanomotor powered by a DNA enzyme. Angew. Chem. Int. Ed. Engl. 2004; 43:3554–3557. [DOI] [PubMed] [Google Scholar]
- 22. Lund K., Manzo A.J., Dabby N., Michelotti N., Johnson-Buck A., Nangreave J., Taylor S., Pei R., Stojanovic M.N., Walter N.G. et al. . Molecular robots guided by prescriptive landscapes. Nature. 2010; 465:206–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Andersen E.S., Dong M., Nielsen M.M., Jahn K., Subramani R., Mamdouh W., Golas M.M., Sander B., Stark H., Oliveira C.L. et al. . Self-assembly of a nanoscale DNA box with a controllable lid. Nature. 2009; 459:73–76. [DOI] [PubMed] [Google Scholar]
- 24. Douglas S.M., Bachelet I., Church G.M.. A logic-gated nanorobot for targeted transport of molecular payloads. Science. 2012; 335:831–834. [DOI] [PubMed] [Google Scholar]
- 25. Bujold K.E., Fakhoury J., Edwardson T.G.W., Carneiro K.M.M., Briard J.N., Godin A.G., Amrein L., Hamblin G.D., Panasci L.C., Wiseman P.W. et al. . Sequence-responsive unzipping DNA cubes with tunable cellular uptake profiles. Chem. Sci. 2014; 5:2449–2455. [Google Scholar]
- 26. Afonin K.A., Lindsay B., Shapiro B.A.. Engineered RNA nanodesigns for applications in RNA nanotechnology. RNA Nanotechnol. 2013; 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jimenez R.M., Polanco J.A., Luptak A.. Chemistry and biology of self-cleaving ribozymes. Trends Biochem. Sci. 2015; 40:648–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mironov A.S., Gusarov I., Rafikov R., Lopez L.E., Shatalin K., Kreneva R.A., Perumov D.A., Nudler E.. Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell. 2002; 111:747–756. [DOI] [PubMed] [Google Scholar]
- 29. Righetti F., Nuss A.M., Twittenhoff C., Beele S., Urban K., Will S., Bernhart S.H., Stadler P.F., Dersch P., Narberhaus F.. Temperature-responsive in vitro RNA structurome of Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:7237–7242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Winkler W.C., Cohen-Chalamish S., Breaker R.R.. An mRNA structure that controls gene expression by binding FMN. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:15908–15913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bindewald E., Afonin K.A., Viard M., Zakrevsky P., Kim T., Shapiro B.A.. Multistrand structure prediction of nucleic acid assemblies and design of RNA switches. Nano Lett. 2016; 16:1726–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Afonin K.A., Viard M., Martins A.N., Lockett S.J., Maciag A.E., Freed E.O., Heldman E., Jaeger L., Blumenthal R., Shapiro B.A.. Activation of different split functionalities on re-association of RNA-DNA hybrids. Nat. Nanotechnol. 2013; 8:296–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Afonin K.A., Viard M., Tedbury P., Bindewald E., Parlea L., Howington M., Valdman M., Johns-Boehme A., Brainerd C., Freed E.O. et al. . The use of minimal rna toeholds to trigger the activation of multiple functionalities. Nano Lett. 2016; 16:1746–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Groves B., Chen Y.J., Zurla C., Pochekailov S., Kirschman J.L., Santangelo P.J., Seelig G.. Computing in mammalian cells with nucleic acid strand exchange. Nat. Nanotechnol. 2016; 11:287–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rogers T.A., Andrews G.E., Jaeger L., Grabow W.W.. Fluorescent monitoring of RNA assembly and processing using the split-spinach aptamer. ACS Synth. Biol. 2015; 4:162–166. [DOI] [PubMed] [Google Scholar]
- 36. Afonin K.A., Desai R., Viard M., Kireeva M.L., Bindewald E., Case C.L., Maciag A.E., Kasprzak W.K., Kim T., Sappe A. et al. . Co-transcriptional production of RNA-DNA hybrids for simultaneous release of multiple split functionalities. Nucleic Acids Res. 2014; 42:2085–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Afonin K.A., Viard M., Kagiampakis I., Case C.L., Dobrovolskaia M.A., Hofmann J., Vrzak A., Kireeva M., Kasprzak W.K., KewalRamani V.N. et al. . Triggering of RNA Interference with RNA-RNA, RNA-DNA, and DNA-RNA Nanoparticles. ACS Nano. 2015; 9:251–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Afonin K.A., Viard M., Koyfman A.Y., Martins A.N., Kasprzak W.K., Panigaj M., Desai R., Santhanam A., Grabow W.W., Jaeger L. et al. . Multifunctional RNA nanoparticles. Nano Lett. 2014; 14:5662–5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Grabow W.W., Zakrevsky P., Afonin K.A., Chworos A., Shapiro B.A., Jaeger L.. Self-assembling RNA nanorings based on RNAI/II inverse kissing complexes. Nano Lett. 2011; 11:878–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bui M.N., Johnson M.B., Viard M., Satterwhite E., Martins A.N., Ramey H., Marriott I., Li Z., Afonin K.A., Khisamutdinov E.. Versatile RNA tetra-U helix linking motif as a toolkit for nucleic acid nanotechnology. Nanomedicine. 2017; doi:10.1016/j.nano.2016.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shlyakhtenko L.S., Gall A.A., Lyubchenko Y.L.. Mica functionalization for imaging of DNA and protein-DNA complexes with atomic force microscopy. Methods Mol. Biol. 2013; 931:295–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shlyakhtenko L.S., Gall A.A., Filonov A., Cerovac Z., Lushnikov A., Lyubchenko Y.L.. Silatrane-based surface chemistry for immobilization of DNA, protein-DNA complexes and other biological materials. Ultramicroscopy. 2003; 97:279–287. [DOI] [PubMed] [Google Scholar]
- 43. Dobrovolskaia M.A., McNeil S.E.. Immunological and hematological toxicities challenging clinical translation of nucleic acid-based therapeutics. Expert Opin. Biol. Ther. 2015; 15:1023–1048. [DOI] [PubMed] [Google Scholar]
- 44. Blanco P., Palucka A.K., Pascual V., Banchereau J.. Dendritic cells and cytokines in human inflammatory and autoimmune diseases. Cytokine Growth Factor Rev. 2008; 19:41–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Takanohashi A., Prust M., Wang J., Gordish-Dressman H., Bloom M., Rice G.I., Schmidt J.L., Crow Y.J., Lebon P., Kuijpers T.W. et al. . Elevation of proinflammatory cytokines in patients with Aicardi-Goutieres syndrome. Neurology. 2013; 80:997–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zamanian-Daryoush M., Marques J.T., Gantier M.P., Behlke M.A., John M., Rayman P., Finke J., Williams B.R.. Determinants of cytokine induction by small interfering RNA in human peripheral blood mononuclear cells. J. Interferon Cytokine Res. 2008; 28:221–233. [DOI] [PubMed] [Google Scholar]
- 47. Turner M.D., Nedjai B., Hurst T., Pennington D.J.. Cytokines and chemokines: at the crossroads of cell signalling and inflammatory disease. Biochim. Biophys. Acta. 2014; 1843:2563–2582. [DOI] [PubMed] [Google Scholar]
- 48. Elroy-Stein O., Moss B.. Cytoplasmic expression system based on constitutive synthesis of bacteriophage T7 RNA polymerase in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 1990; 87:6743–6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Filonov G.S., Moon J.D., Svensen N., Jaffrey S.R.. Broccoli: rapid selection of an RNA mimic of green fluorescent protein by fluorescence-based selection and directed evolution. J. Am. Chem. Soc. 2014; 136:16299–16308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C.. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998; 391:806–811. [DOI] [PubMed] [Google Scholar]
- 51. Stewart J.M., Viard M., Subramanian H.K., Roark B.K., Afonin K.A., Franco E.. Programmable RNA microstructures for coordinated delivery of siRNAs. Nanoscale. 2016; 8:17542–17550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Reagan-Shaw S., Ahmad N.. Silencing of polo-like kinase (Plk) 1 via siRNA causes induction of apoptosis and impairment of mitosis machinery in human prostate cancer cells: implications for the treatment of prostate cancer. FASEB J. 2005; 19:611–613. [DOI] [PubMed] [Google Scholar]
- 53. Ruckert F., Samm N., Lehner A.K., Saeger H.D., Grutzmann R., Pilarsky C.. Simultaneous gene silencing of Bcl-2, XIAP and Survivin re-sensitizes pancreatic cancer cells towards apoptosis. BMC Cancer. 2010; 10:379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rose S.D., Kim D.H., Amarzguioui M., Heidel J.D., Collingwood M.A., Davis M.E., Rossi J.J., Behlke M.A.. Functional polarity is introduced by Dicer processing of short substrate RNAs. Nucleic Acids Res. 2005; 33:4140–4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Khisamutdinov E.F., Li H., Jasinski D.L., Chen J., Fu J., Guo P.. Enhancing immunomodulation on innate immunity by shape transition among RNA triangle, square and pentagon nanovehicles. Nucleic Acids Res. 2014; 42:9996–10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.