Abstract
The glial scar formation remains a significant barrier to the long term success of neural probes. Micromotion coupled with mechanical mismatch between the probe and tissue is believed to be a key driver of the inflammatory response. In vitro glial scar models present an intermediate step prior to conventional in vivo histology experiments as they enable the cell-device interactions to be tested on a shorter timescale, with the ability to conduct broader biochemical assays. No established in vitro models have incorporated methods to assess device performance with respect to mechanical factors. In this study, we describe an in vitro glial scar model that combines high-precision linear actuators to simulate axial micromotion around neural implants with a 3D primary neural cell culture in a collagen gel. Strain field measurements were conducted to visualize the local displacement within the gel in response to micromotion. Primary brain cell cultures were found to be mechanically responsive to micromotion after one week in culture. Astrocytes, as determined by immunohistochemical staining, were found have a significant increase in cell area and perimeter in response to micromotion compared to static control wells. These results demonstrate the importance of micromotion when considering the chronic response to neural implants. Going forward, this model provides advantages over existing in vitro models as it will enable critical mechanical design factors of neural implants to be evaluated prior to in vivo testing.
Graphical Abstract
A novel 3D in vitro model to probe the mechanical effects of micromotion induced strain around neural implants.
Introduction
Implantable neural implants are being developed for many applications including the treatment of neurological disorders, control of brain machine interfaces, and to understand the connections within neural circuits (1). A universal barrier to the long term success of these implants is the formation of a glial scar around the implant, in which reactive astrocytes surround and isolate the device from viable neurons, negatively impacting device function. Many studies have been conducted to understand the glial scar formation process in vivo and are reviewed elsewhere (2–4).
A large contributor to the glial scar response is thought to be mechanical factors related to micromotion around the implant. Small displacements of the brain occur due to respiration, vascular pulses, and rotational accelerations and create strain fields around the device (5–7). The repetitive motion surrounding the implant is thought to be mostly axial, as it arises from respiration and vascular pulsations when devices are implanted perpendicular to the skull (5). Gilletti et al. measured the magnitude of micromotions present in the rodent skull by placing a displacement sensor on the brain; respiration caused surface displacements up to 30 μm in magnitude, while vascular pulsatility caused displacements up to 4 μm in magnitude. The constant relative motion is thought to drive the chronic response through the constant aggravation of local inflammatory cells. Furthermore, most implants used in experimental animal models are fixed to the skull, thus potentially increasing the magnitude of displacement relative to the brain surface (8). Micromotion may also damage local vasculature which can further drive the inflammatory response through activation of microglia and extravasation of circulating inflammatory cells from the blood, negatively affecting probe function (9).
Finite element analyses have modeled local strain fields around neural implants which result from micromotion (5, 6). Subbaroyan et al. conducted simulations which showed that the local strain fields, and thus the chronic tissue response, could be drastically reduced by incorporating soft materials into neural implants (7). Several studies have been conducted which have aimed to mediate this response by incorporating implant design elements such that the effects of micromotion are reduced. Approaches include incorporating flexible leads (10), constructing implants out of flexible materials (11, 12), and using novel mechanically adaptive composites which soften when implanted in the brain (13, 14).
The performance of these implants is typically evaluated in rodent implantation models. These experiments involve implantation of the devices in the skull and sectioning the brain at various time points post-implantation. Immunohistochemical staining of the sections is used to analyze for inflammatory markers implicated in the glial scar process. These experiments are an essential part of the development process, but are time consuming and costly. The complexity of in vivo systems also makes it difficult to resolve inflammatory pathways on a molecular scale and visualize the dynamics of the cellular response. In vivo studies also introduce confounding variables such as surgical technique, device placement and large blood vessel damage, and tissue heterogeneity.
In vitro models have been developed over the past decade which have made high-throughput methods possible for assessing neural implant biocompatibility. Initial models involved culturing glial cells on a two dimensional (2D) substrate. Polikov et al. demonstrated that primary glial cells grown in culture exhibit many of the hallmarks of glial scar formation following mechanical insult (15). Astrocytes were shown to increase production of glial fibrillary acidic protein (GFAP), and microglia were found in proximity to a microwire device placed on top of the cell layer or a scrape would. Models such as these are planar in nature and are thus not fully representative of the native environment in the brain.
Recent studies have improved upon this model by constructing three dimensional (3D) growth environments for the neural cells (16–18). East et al. demonstrated that astrocytes can be grown in a collagen matrix, and exhibit glial scar characteristics when exposed to inflammatory cytokines (19). Another advantage of 3D models, as highlighted by this study, is that glial cells are less reactive in 3D environments, compared to being grown on a 2D, rigid substrate (19, 20). This enables a more dynamic range to be investigated within the model, as the glial cells are less reactive at the start of the experiment. 3D architecture supports more representative cell-cell communication and cells show less signs of cellular stress (21).
Significant progress has been made towards establishing improved in vitro glial scar models. Few have, however, directly investigated interactions between the implant and glial cells. Those that do incorporate neural implants are predominately static in nature, in which the devices are placed in the culture and then analyzed a set time later (22). Mechanical stresses in existing models are typically present throughout the whole culture and therefore do not assess device-tissue mechanical interactions (23, 24). Any potential mechanical design benefits that the device has to minimize micromotion effects are not captured within such models. Mechanical stimuli have long known to be critical elements tissue engineering applications including vascular tissue engineering (25), bone tissue engineering (26), and muscle tissue engineering (27). The forces present in these systems seek to replicate the in vivo environment and have profound impacts on tissue development and underlying biology in all of these cases. Our objective was to bridge the gap between neural engineering strategies which seek to modulate scar formation through device mechanical factors and existing in vitro glial scar models.
In this study, we have developed a novel 3D in vitro culture model which produces micromotion around the device. Devices inserted into a 3D collagen matrix are cyclically displaced for one week in culture to simulate axial micromotion. As mentioned above, axial micromotion from respiration and vascular pulsations is thought to be the most chronic contributor to the relative motion of the devices. This culture system will enable researchers to rapidly optimize mechanical design constraints for neural implants prior to beginning in vivo studies.
Materials and Methods
Materials
Poly-d-lysine (PDL), Hoechst 33258, sodium hydroxide, 10x minimum essential media, and lipopolysaccharides (LPS) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Cell culture media, B27 supplement, fetal bovine serum (FBS), antibiotic/antimycotic (anti/anti), Glutamax, normal donkey serum, trypsin-EDTA, live/dead stain for mammalian cells, and transforming growth factor – beta (TGF-β) were purchased from ThermoFisher/Life Technologies (Waltham, MA, USA). Basic fibroblast growth factor (bFGF) was purchased from R&D systems (Minneapolis, MN, USA). Bovine serum albumin was purchased from Rockland Inc. (Limerick, PA, USA). Fluorescent polymer microspheres (6.0 μm polystyrene particles, yellow green) were purchased from Polysciences Inc. (Warrington, PA, USA). Mouse-anti GFAP antibody was purchased from Millipore Inc. (Billerica, MA, USA).
PDL Coating
T-175 flasks were coated with PDL prior to seeding with primary neural cells. A 50 μg/ml stock solution of poly-D-lysine hydrobromide (MW 70000–150000) was prepared in sterile tissue culture grade water. Approximately 25 ml of solution was placed in the flask and incubated overnight at room temperature. The PDL solution was removed and the plates were rinsed once with tissue culture water. The plates were then allowed to dry before plating cells.
Glial Cell Isolation
Mixed primary embryonic neural cell cultures were established according to a protocol adapted from Liu et al. (28). All procedures were approved by the MIT Committee on Animal Care prior to beginning any experiments. Timed pregnant F344 rats (Charles River Laboratory, Wilmington, MA, USA) were euthanized via CO2 asphyxiation followed by cervical dislocation at days E14–E15. The cell isolation procedure was performed in a designated biosafety hood to maintain sterility throughout the process. The dam’s abdomen was sterilized with 70% ethanol, and the embryos were removed. Each embryo was quickly decapitated and placed in ice cold sterile hanks buffered salt solution. The brain of each embryo was removed under microscope, and placed in dissection media (DMEM cell culture media mixed supplemented with anti/anti). The midbrain section of the brain was isolated. Large blood vessels and connective tissues were removed with a fine pair of forceps. The dissection media was removed and 0.25% trypsin-EDTA was added to the brains for 5–10 minutes at 37°C once all of the midbrain sections were collected. FBS containing cell culture media was then added to neutralize the trypsin. The brain sections were centrifuged at 1000 rpm for 5 minutes and resuspended in cell culture media (Neurobasal media supplemented with B-27, glutamax, anti-anti, 10% FBS). Tissue sections were triturated with 10 ml, 1ml, and 0.2 ml pipette tips (3–5 times each). The cell count was then measured using the Countess II Automated Counter (Invitrogen, Carlsbad, CA, USA) and the cell density was adjusted to 1x106 cells/ml. Cells were seeded in T-175 flasks and maintained for approximately 1 week before seeding in 3D culture.
3D Culture Formation
Three dimensional mixed neural cell cultures were formed using a protocol adapted from East et al. (19). Cells were detached from the T-175 flasks via trypsin-EDTA exposure for 5–10 minutes, and suspended at a density of 3.75 x 107 cells/ml (10x of final concentration). This density was chosen, as it was previously shown to result in maximal cellular viability in 3D neural cell cultures (18). The gel mixture consisted of 10% part concentrated cell mixture, 80% parts type-I rat tail collagen (2mg/ml in .01 N acetic acid), and 10% 10x minimum essential medium. The mixture was neutralized with 1 M sodium hydroxide, until the phenol red pH indicator changed to red. The cell mixture was plated into 24 well plates at a volume of 0.6 ml/well. Final gel dimensions were 16.3 mm in diameter, and 2.9 mm thick. The gels typically adhered to the side walls of the plate throughout the course of the experiment. Samples were excluded if delamination from the side wall due to cellular compaction was observed before or during the micromotion experiment.
The gels were allowed to set at 37 °C for 30 minutes, and were then topped with 0.75 ml of media (Neurobasal media supplemented with B-27, glutamax, anti-anti, 10% FBS). Cultures were maintained with 50% media change every 2–3 days. TC multiwell plates were modified with a hole drilled in the side of the plate to facilitate insertion of the neural implants at the start of the experiment. Plates were resterilized by soaking in 70% ethanol and UV light exposure for 1 hour, followed by 3 washes with sterile DI water. Plates were allowed to dry before seeding with cells. The hole was temporarily plugged shut with a polyimide tube sealed with UV medical grade epoxy prior to beginning the micromotion experiment. The polyimide guide tube was replaced with a sterile polyimide tube prior to beginning an experiment to enable probe insertion into the collagen gel.
Live Dead Stain
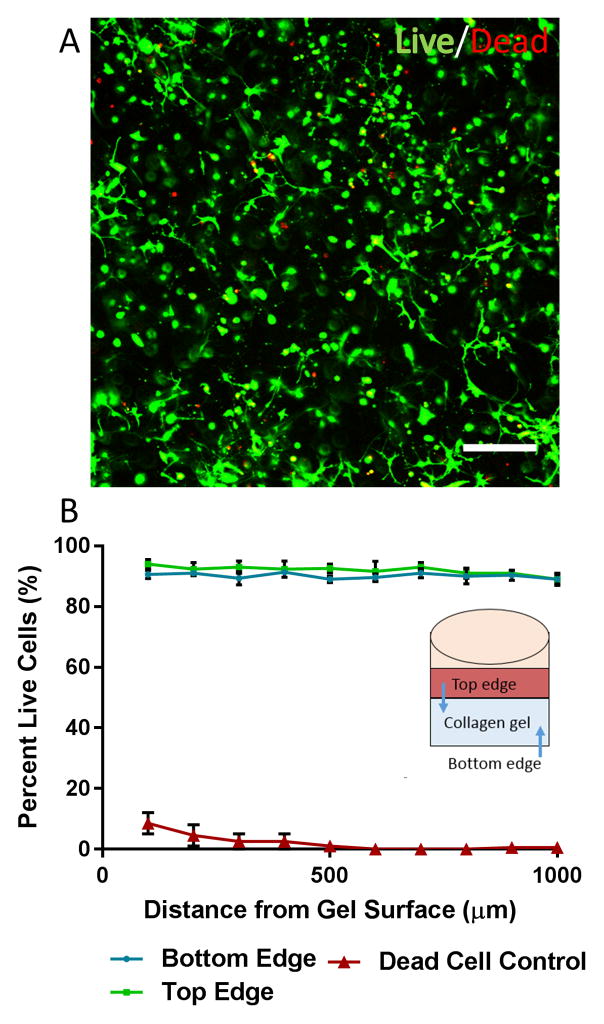
Cell viability within the 3D collagen cell culture construct was assessed by using the Live/Dead stain for Mammalian cells. Briefly, calcein-AM (2.5 μM) and eithidum homodimer-1 (2.5 μM) in sterile D-PBS without calcium and magnesium were added to cultures after 1 week in culture, and incubated for 60 minutes at 37°C. A dead cell control was included by wells to methanol for 30 minutes prior to staining. Stained cultures were imaged via Nikon A1R confocal microscope in the Koch Institute Microscopy Core. Images at 100 μm steps progressing inward towards the center of the gel were obtained at both the top and bottom surface of the gel. The top surface was imaged by flipping the gel after staining. Live/dead cells were manually counted as a function of depth into the gel via ImageJ.
Micromotion Equipment Construction
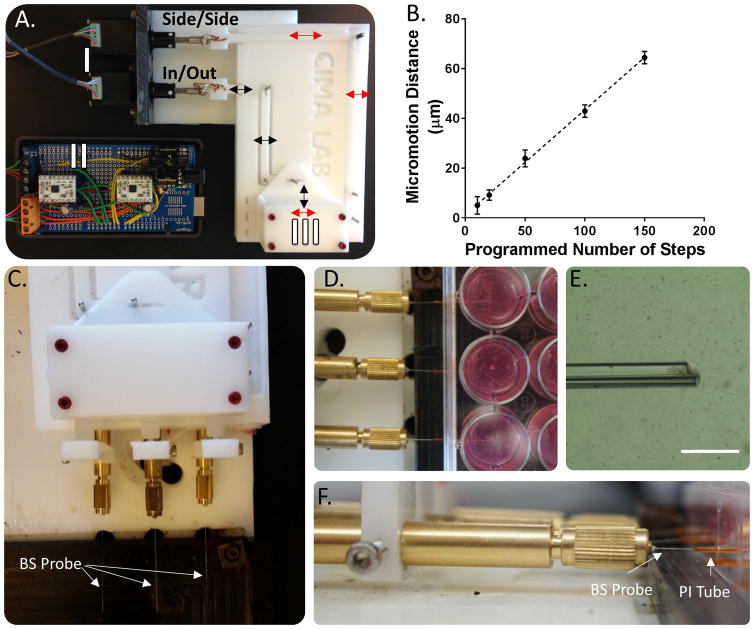
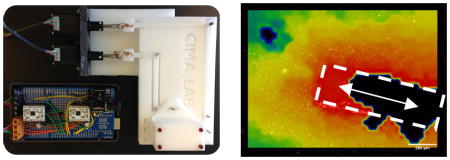
The micromotion apparatus was constructed to simulate micromotion within the tissue culture model (figure 1). Custom designed Delrin components were machined using a computer numerical control mill (Cameron Micromaching Center, Sonora, CA, USA) in our lab. The parts served to interface the devices, held in place with a Teflon lined fiber check (Newport Corporation, Irvine, CA, USA), with two high precision linear actuators (Haydon Kirk Motion Solutions, Milford, NH, USA). The linear actuators were controlled by an Arduino microcontroller (see supplementary information for electrical configuration and the open source code used for this system). A calibration curve was obtained characterizing the relationship between the programmed number of motor steps and the distance travelled by obtaining images under bright field microscopy.
Figure 1. micromotion simulation device.
(a) a custom device was built to replicate the effects of micromotion in a 3d glial scar model. the device consists of two high precision linear actuators (i) that are driven by an arduino microcontroller and ti drv8834 stepper motor drivers (ii). the electronic controller was designed to drive the linear actuators using 1/32nd microstepping accuracy, corresponding to a theoretical .05 micron precision. mechanical coupling components link the device to the actuators and produce displacements both along the device axis and perpendicular to the device holder (iii). arrows are included to indicate the motion of the mechanical coupling components for displacements along the device axis (in/out – black), and perpendicular to the device axis (side/side – red). (b) the device was calibrated by measuring the distance traveled for various numbers of steps ranging from 10 to 150. data shown represents the mean +/− s.d. (c,d) top view of the teflon fiber chucks used to interface the borosilicate probes with the mechanical actuators to produce micromotion within the 3d collagen gel culture. (e) bright field image of a glass probe within the cell culture. scale bar – 400 μm. (f) side view of the fiber chuck and borosilicate probe. a pi guide tube inserted into a drilled hole was used to insert the probe into the collagen gel.
Strain Field Measurement
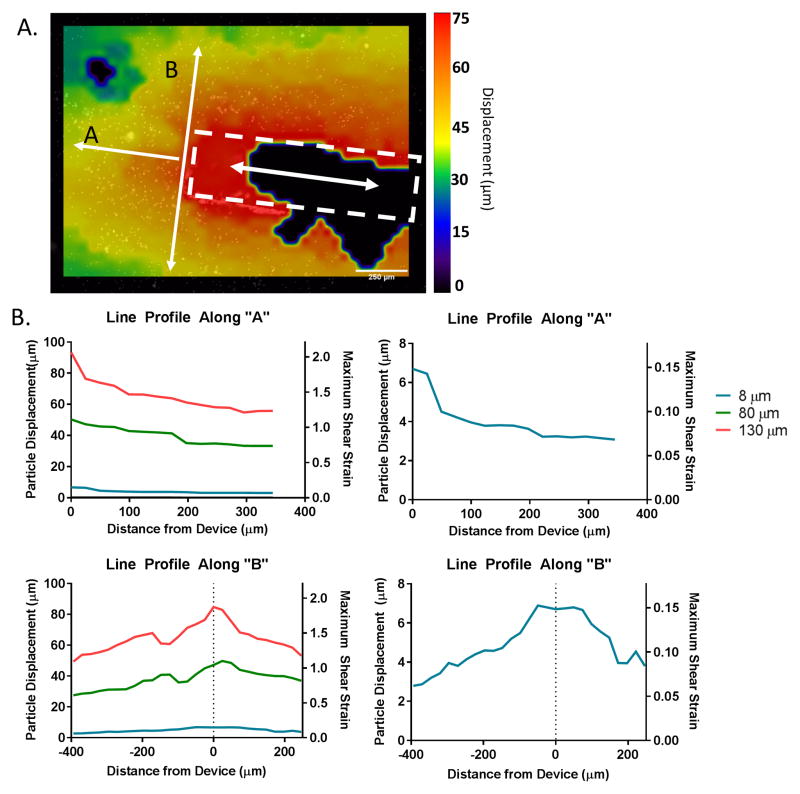
A 2.0 mg/ml collagen gel was embedded with fluorescent latex beads (0.05% w/v, Flouresbrite 6 μm). A probe was loaded into the gel and located under an inverted epifluorescence microscope (Zeiss Axiovert 200, Oberkochen, Germany). The device was coupled to the micromotion simulation device. The device was displaced at a frequency of 2 Hz. A series of fluorescent images were obtained throughout the micromotion process.
The images at the extremes of motion were analyzed via the Particle Image Velocimetry (PIV) plugin for ImageJ to construct the displacement fields around the device. Particle displacements were quantified in the in/out axial direction for multiple steps (8, 80, and 130 μm). Line profiles were obtained both in the perpendicular and axial directions to the device. The corresponding maximum shear strain for these particle displacements was calculated according to the following equations:
Where ux,y corresponds to the measured particle displacements and δx,δy corresponds to the interrogation window used by the PIV ImageJ plugin.
In Vitro Glial Scar Experiment
One week experiments were conducted to demonstrate the utility of our in vitro glial scar model. At the start of the experiment, the sealed polyimide tubes were removed and replaced with new, sterile polyimide tubes. 150 μm borosilicate glass probes were placed approximately 6 mm into the collagen gels. Experimental groups included device with micromotion, device with no motion, positive control, and negative control. Positive control wells were spiked with TGF-β (10 ng/ml) or LPS (10 μg/ml). The distance and frequency travelled for the in vitro experiments were chosen to simulate micromotion effects that have been measured in the brain (30 μm, 2 Hz) (5). The devices were axially oscillated at a total displacement of 30 μm at a frequency of 2 Hz for the entire 168 hour experiment. Experiments in this study were conducted for one week, as this is an intermediate time point similar to those used in previously established in vitro 3D glial scar models (19).
The media was changed to include bFGF (10 ng/ml), which was previously shown to improve the reliability of in vitro glial scar model (29). The cultures were placed in a 37 °C incubator and were maintained with 50% media changes every other day. The motion wells were examined under microscope, at the beginning of the experiment, and during media changes to ensure that devices were moving as intended within the collagen gel throughout the course of the experiment. The 3D collagen gels were fixed after one week in culture with 4% paraformaldehyde in PBS for 45 minutes. The gels were washed 3x in PBS, and stored in PBS prior to immunofluorescent staining.
Immunohistochemical Analysis
3D collagen gels were stained for the expression of GFAP after fixation. Gels were transferred to a glass bottom 24 well plate (#1.5 glass, Cellvis Inc, Mountain View, CA, USA) for improved imaging. Fixed gels were permeabilized in 0.5% triton-x100 in PBS for 30 minutes. Gels were washed three times in 0.05% PBS-tween. Gels were then blocked in 5% donkey serum for 1 hour at room temperature, and washed in PBS-tween. Mouse anti-GFAP-AF488 was diluted in antibody incubation buffer (1:300), and the gels were incubated for 48 hours at 4°C to allow for uniform antibody diffusion into the gels. Antibody incubation buffer consists of 1% bovine serum albumin (BSA), 1% donkey serum, 0.3% triton-x, and 0.01% sodium azide in PBS. Gels were washed in PBS-tween three times for 10 minutes following primary antibody incubation. Gels were then counter stained with Hoechst 33258 (2 μg/ml) for 30 minutes. Gels were then washed in PBS three times, and stored at 4 °C in PBS prior to imaging.
Confocal Microscopy
Confocal microscopy was used to analyze the GFAP expression within the gels after immunofluorescent staining. Z-stacks were obtained using the Nikon A1R confocal microscope in the Koch Institute Microscopy Core. 200 μm z-stacks (7.14 μm optical sections x 28 images per stack) were obtained at three different locations within each well at 10x magnification. Images were obtained in the area directly surrounding the devices, as well as in regions far (≈5–8 mm from the device location) from the device for micromotion device wells. Image exposure settings and resolution were maintained between experimental samples. Images were not manipulated in any way prior to image analysis.
Image Analysis
Maximum intensity projections of the GFAP confocal z-stack images were analyzed via imageJ. A threshold was applied to the image to define the border between cells and background. The particle analysis tool was used to analyze the size and shape of the cells. All objects with an area greater than 40 μm2 (to threshold for cells) were included in the analysis. Metrics including total object area, total object perimeter per image were measured. A minimum of three images per well was analyzed and averaged to get the total average for each experiment. A total of 9 independent experiments were conducted to obtain the final values for each experimental group unless otherwise indicated.
Statistical Analysis
Observed experimental differences were assessed for statistical significance across all experimental groups through a one-way ANOVA test using Prism 6 (In Stat Graphpad, La Jolla, CA, USA). Post hoc analysis (Tukey’s multiple pairwise comparisons test) was performed to compare GFAP area and perimeter size between different groups. Statistical significance was considered p<.05. All data presented represents mean +/− standard error of the mean unless otherwise indicated.
Results
Micromotion Equipment Calibration
The micromotion apparatus was constructed using a combination of machined parts, two high precision linear actuators, Arduino microcontroller (figure 1a), and stepper motor drivers. A 150 μm borosilicate device was attached to the apparatus, and programmed to move a certain amounts of steps in the axial direction. Images captured at the extreme ends of motion were analyzed to produce a calibration curve (figure 1b). It was found there was a linear relationship between distance traveled and the number of steps programmed for both directions of motion. There was minimal backlash in the system, as indicated by the trend line passing near the origin (95% confidence interval for the y-intercept is (−1.1,+3.6 μm). A displacement of approximately 30 μm at a frequency of 2 Hz was chosen for subsequent in vitro glial scar measurements.
In Vitro Strain Field Measurements
The strain field was measured following the displacement of a rigid device implanted in a three dimensional collagen gel (2.0 mg/ml) embedded with fluorescent particles. Images were obtained for three different magnitudes of displacements (8 μm, 80μm, and 130 μm) throughout the course of the displacement. The PIV plugin from imageJ was used to construct the strain field surrounding the device (figure 2a). Line profiles were obtained from these plots (figure 2b). The measured displacements at the surface of the device ranged from 4.23 μm to 84.8 μm. The degree of strain decreased as a function of distance from the device for all three displacement magnitudes investigated. The displacement was 2.8 μm, 28.6 μm, and 51.8 μm at a distance of 400 μm from the implant interface for the 8 μm, 80 μm, and 130 μm displacement respectively.
Figure 2. strain field mapping around the device location.
fluorescent polystyrene mircoparticles (6 μm diameter, yellow green) were embedded in a collagen gel (2.0 mg/ml) to measure the strain fields produced around devices during micromotion (a) imagej software was used to calculate the particle displacement following micromotion using the piv plugin. shown here is the displacement map for an 80 μm displacement. (b) the line profiles were calculated for three different displacements (8 μm, 80 μm, 130 μm) in the micromotion model along and perpendicular to the device axis. the particle displacements and maximum shear strain as a function of distance from the device are shown.
Live/Dead Cell Staining
Live/dead cell staining was conducted to confirm that neural cell viability could be maintained throughout the course of the experiment. The cells were stained with calcein-AM and ethidium homodimer-1 after one week in culture. Confocal microscopy was used to obtain fluorescent images at various depths into the gel from both the top surface and bottom surface (figure 3a). Images were obtained up to 1 mm into the gel from both the top and bottom surface of the gel. Cells could not be effectively imaged beyond this depth due to poor light penetration throughout the gel. The number of live cells (green) and dead cells (red) were counted to produce the total percent viability as a function of depth into the gel (figure 3b). The cell viability was found to be around 90% at both surfaces of the gel. The viability at the top surface was slightly higher than the bottom surface (vs. 93.8 ± 2.7% vs 90.7 ± 2.0%). There was minimal variation in the viability as a function of depth in the gel. At a depth of 1 mm from the surface of the gel, the bottom surface was found to have a viability of 89.4 ± 3.4% and the top surface was found to have a viability of 88.7 ± 2.5%. The dead cell control samples had an average viability of 2.0 ± 3.3%. All values discussed in this section represent the mean ± S.D.
Figure 3. primary neural cell viability in collagen gel after 1 week in culture.
the percentage of live cells was determined using calcein-am/ethidium homodimer-1 cell staining. (a)representative live/dead immunofluorescent image obtained via confocal microscopy. the image was taken at a 300 μm depth in the gel. green=live. red=dead. scale bar 200 μm. (b) calculated cell viability as a function of depth from the gel surface from both the top and bottom edge of the gel (with respect to the media layer). a dead cell control (30 min exposure to methanol) was also included to confirm proper operation of the assay.
Immunohistochemical Analysis
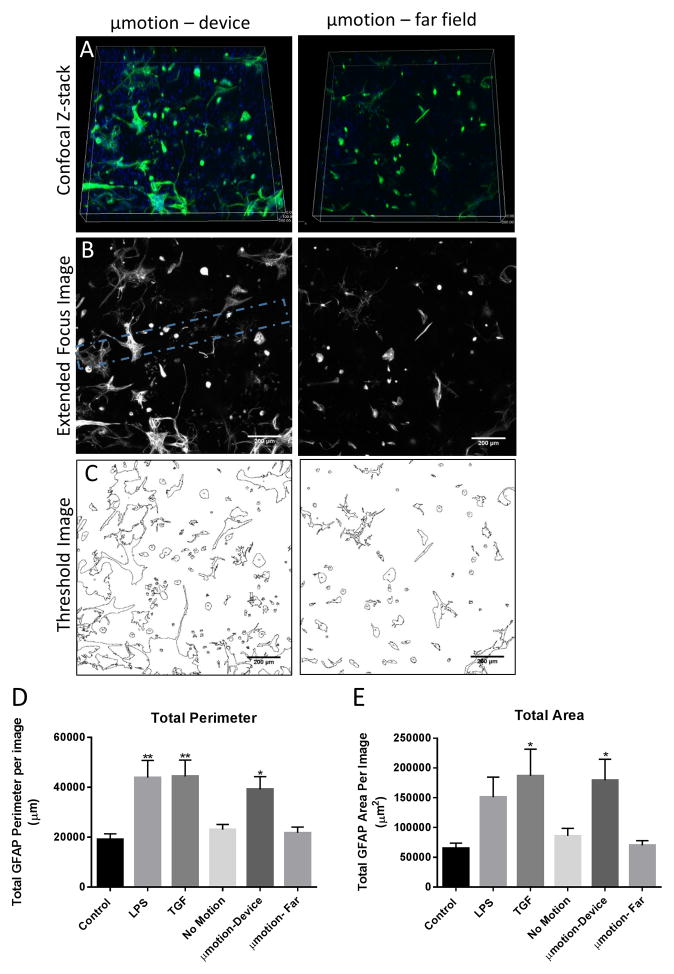
Cells were fixed and stained for the astrocytic marker GFAP to assess changes in the reactivity following 1 week in 3D culture. Antibodies were able to effectively penetrate through the thick collagen gel, and confocal microscopy enabled the region directly surrounding the device to be clearly imaged (figure 4a). Extended focus images were obtained from 200 μm z-stacks and analyzed via imageJ. A threshold was applied to the images to distinguish the cellular boundaries from background (figure 4b).
Figure 4. characteristic images obtained following glial scar experiment and quantified astrocyte morphology.
(a) example 3d rendering of a confocal z-stack of the gfap staining after 1 week of micromotion in a region directly surrounding the implant (left) and a region > 5mm from the device location (right) within the same well. (b) example extended focus confocal images of the gfap staining after 1 week of micromotion in a region scale bar 200 μm. the device location is indicated with a dotted line in the micromotion image. (c) astrocytes were identified after thresholding the image with imagej. these shapes of these images was analyzed using the particle analysis tool to assess differences in cellular morphology (cell perimeter and area). astrocytes were found to exhibit characteristics of glial scar formation after 1 week in culture for positive control wells and micromotion wells. scale bar 200 μm. (d) the total gfap perimeter per image and (e) total gfap area per image as determined by imagej analysis for static control, positive control (tgf-b and lps), device wells (no motion and micromotion), and far field controls. *p<.05, **p<.01 compared to the control. all sample sizes are n=9, besides for no motion which is n=7.
Astrocyte reactivity was compared between groups by assessing the cellular area and perimeter, an approach that has been used in previous in vitro glial scar models (19). The total perimeter and area of GFAP staining was calculated using the imageJ particle analysis tool (figure 4c, d). A statistically significant increase in the total GFAP perimeter was observed in the LPS (43936.5 ± 20543 μm), TGF-β (44360.5 ± 19680 μm), and the micromotion around the device (39151.1 ± 15367 μm) compared to the control wells (19066 ± 6997 μm). There was a slight, but not significant, increase in the wells in which a device was inserted but no motion occurred (23009.6 ± 5640.4 μm), as well as in the regions in the micromotion device wells which are a distance away from the device location (21782.72 ± 6092.98 μm).
Similar trends were observed in the total average GFAP area per image measurements. There was a statistically significant increase in the TGF-β wells (186703 ± 134700 μm2) compared to control wells (65040 ± 26530 μm2) (p<.05). The GFAP area per image was also found to be statistically greater in the regions surrounding the micromotion devices (179200 ± 106720 μm2) compared to the control wells (p<.05). The difference between the LPS positive controls (150838 ± 101495 μm2) and the control wells was not found to be statistically significant in this experiment (p=.14). The no motion well (85600 ± 33400 μm2), and regions far from the device location in the micromotion wells (70100 ± 24024.8 μm2), were slightly increased compared to controls, although this difference was not found to be statistically significant. All values discussed in this section represent the mean ± S.D.
Discussion
There is an ongoing discussion in the field regarding the role that micromotion plays in driving glial scar formation around neural implants (4). Evidence to support this hypothesis has been limited to finite element simulation studies (6, 7) and in vivo studies which rely on histological analysis (8, 30–33). These studies highlight the importance mechanical effects at the tissue-implant interface, but are also confounded by the presence of other factors that are present when using in vivo models including damage from surgical implantation (34), blood-brain barrier breach (9), and the presence of blood-derived macrophages (35). All of these components are likely to play a role in the scar formation process. The results obtained in this in vitro study, however, clearly demonstrate the major contribution that strain fields from micromotion around neural implants have towards driving the glial scar response.
Astrocytes were found to be mechanically responsive in our glial scar model following one week of micromotion. The cells directly within the region of micromotion, as well as the positive control wells exhibited elevated cellular area and perimeter. The total GFAP area increased 2.75-fold in micromotion samples compared to the control wells, and 2.1-fold compared to wells in which devices were present and no motion had occurred. This hypertrophy is typical of the glial scar response (2). This behavior was also observed in previously established glial scar models which have also used TGF-β as a positive control (19). The increase in astrocyte reactivity was primarily isolated to regions directly around the device location in which the local strain was greatest, further highlighting the role that micromotion plays in driving the glial response. No difference in astrocyte reactivity was observed at different locations within the micromotion device confocal z-stacks were observed (supplementary figure 1). This suggests the strain induced from a repetitive 30 μm displacement are sufficient to induce glial activation up to 600 μm away from the implant. (36, 37). Using this model to explore smaller displacements may enable the amount of strain which induces astrocyte hypertrophy to be determined, a result which would greatly improve the neural probe design process.
Areas within the micromotion-strained wells that were far from the device location (>5 mm) were only observed to have marginal increases in area and perimeter compared to controls. This slight increase in reactivity far from the motion devices is likely due to an increased production of inflammatory cytokines within the well in response to the mechanical stimuli (23). Static wells only showed a slight increase in area and perimeter compared to control wells. This response is likely due to the damage from insertion into the well as well as the presence of a foreign body in the culture. Borosilicate glass is generally regarded as biocompatible (38), which potentially explains why the no motion samples did not produce an extensive reaction. The results obtained in this study suggest that strategies which aim to mitigate strain effects from micromotion are essential to improving the long term response to neural implants.
This study was made possible through the creation of a novel in vitro model, in which strain fields resulting from micromotion are replicated using high precision linear actuators. All mechanical and electrical components of the model are relatively inexpensive (total cost <$1000) and are open source. A mixed primary neural culture was used, so that the all of the main cell types involved in the glial scar formation processes were present, and therefore critical inflammatory pathways maintained (2). The 3D nature of the model enables strain fields to be created around the neural implant which resemble those that are observed in vivo. The magnitude and frequency of the displacements were chosen to resemble the micromotion that were measured in the brain due to respiration (5). Devices were chosen to be displaced in the axial direction as this is most representative of repetitive motion which arises from respiration and vascular pulsations, when devices are implanted perpendicular to the skull (5). Future experiments may explore the response to motion perpendicular to the device to resemble rotational acceleration and large scale displacements from concussions The 3D nature of our model also enables devices of more complex geometry, and their effect on glial cells to be quantified in a significantly reduced timeframe compared to conventional in vivo experiments. Future work will also involve expansion of the model so that more devices can be tested simultaneously as well as incorporation of hyaluronic acid components into the culture system to make the cellular environment more representative of the brain extracellular matrix (39).
The primary objective in this experiment was to explore changes in astrocyte response in the presence of mechanical micromotion, as GFAP is arguably the key factor used in assessing chronic neural implant biocompatibility (2). Future work will investigate the interplay between additional cell types present in the culture such as neurons and microglia. Potential future studies may also examine drug interactions (anti-inflammatory drugs that modulate macrophage (40, 41) or astrocyte behavior (42)), BBB breach (by incorporating IgG or blood borne macrophages in the culture), and incorporation of live cell imaging techniques to further understand the drivers and dynamics of glial scar formation within this model.
Conclusion
Astrocytes grown in a 3D in vitro glial scar model were found to be mechanically responsive to local strain fields. These results clearly illustrate the role that local strain fields have in driving glial scar formation. Previously established in vitro glial scar models have not incorporated local strain fields surrounding the device, either because they are two-dimensional in nature or produce strain throughout the entire culture. The model presented in this study will enable mechanical design factors (such as coatings, mechanically adaptive composites, serpentine connectors etc.) to be tested prior to in vivo work. The tunable nature (adjustable direction, magnitude, and frequency) of our model will enable researches to optimize neural probe design over a wide range of motion conditions.
Supplementary Material
Acknowledgments
This work was supported by funding from the National Institutes of Health Grant R01 EB016101 (MJC) and K99 EB016690 (JCS), the Institute for Soldier Nanotechnologies Grant W911NF-07-D-004, and partially by Cancer Center Support (core) Grant P30-CA145051 from the National Cancer Institute. The authors wish to also thank the Swanson Biotechnology Core at the Koch Institute for Integrative Cancer Research, especially Eliza Vasile for assistance with confocal microscopy.
References
- 1.Kook G, Lee S, Lee H, Cho IJ, Lee H. Neural Probes for Chronic Applications. Micromachines. 2016;7:179. doi: 10.3390/mi7100179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Polikov VS, Tresco PA, Reichert WM. Response of brain tissue to chronically implanted neural electrodes. Journal of Neuroscience Methods. 2005;148:1–18. doi: 10.1016/j.jneumeth.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 3.Mehdi J, John LS, Christoph W, Jeffrey RC. Progress towards biocompatible intracortical microelectrodes for neural interfacing applications. Journal of Neural Engineering. 2015;12:011001. doi: 10.1088/1741-2560/12/1/011001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prodanov D, Delbeke J. Mechanical and Biological Interactions of Implants with the Brain and Their Impact on Implant Design. Frontiers in Neuroscience. 2016;10 doi: 10.3389/fnins.2016.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aaron G, Jit M. Brain micromotion around implants in the rodent somatosensory cortex. Journal of Neural Engineering. 2006;3:189. doi: 10.1088/1741-2560/3/3/001. [DOI] [PubMed] [Google Scholar]
- 6.Hyunjung L, Ravi VB, Wei S, Marc EL. Biomechanical analysis of silicon microelectrode-induced strain in the brain. Journal of Neural Engineering. 2005;2:81. doi: 10.1088/1741-2560/2/4/003. [DOI] [PubMed] [Google Scholar]
- 7.Jeyakumar S, David CM, Daryl RK. A finite-element model of the mechanical effects of implantable microelectrodes in the cerebral cortex. Journal of Neural Engineering. 2005;2:103. doi: 10.1088/1741-2560/2/4/006. [DOI] [PubMed] [Google Scholar]
- 8.Biran R, Martin DC, Tresco PA. The brain tissue response to implanted silicon microelectrode arrays is increased when the device is tethered to the skull. Journal of Biomedical Materials Research Part A. 2007;82A:169–178. doi: 10.1002/jbm.a.31138. [DOI] [PubMed] [Google Scholar]
- 9.Saxena T, et al. The impact of chronic blood–brain barrier breach on intracortical electrode function. Biomaterials. 2013;34:4703–4713. doi: 10.1016/j.biomaterials.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 10.Musallam S, Bak MJ, Troyk PR, Andersen RA. A floating metal microelectrode array for chronic implantation. Journal of Neuroscience Methods. 2007;160:122–127. doi: 10.1016/j.jneumeth.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 11.Sohal HS, et al. The sinusoidal probe: a new approach to improve electrode longevity. Frontiers in Neuroengineering. 2014;7 doi: 10.3389/fneng.2014.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Canales A, et al. Multifunctional fibers for simultaneous optical, electrical and chemical interrogation of neural circuits in vivo. Nat Biotech. 2015 doi: 10.1038/nbt.3093. advance online publication. [DOI] [PubMed] [Google Scholar]
- 13.Simon DM, et al. Design and demonstration of an intracortical probe technology with tunable modulus. Journal of Biomedical Materials Research Part A. 2016 doi: 10.1002/jbm.a.35896. n/a–n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jorfi M, Voirin G, Foster EJ, Weder C. Physiologically responsive, mechanically adaptive polymer optical fibers for optogenetics. Opt Lett. 2014;39:2872–2875. doi: 10.1364/OL.39.002872. [DOI] [PubMed] [Google Scholar]
- 15.Polikov VS, Block ML, Fellous JM, Hong JS, Reichert WM. In vitro model of glial scarring around neuroelectrodes chronically implanted in the CNS. Biomaterials. 2006;27:5368–5376. doi: 10.1016/j.biomaterials.2006.06.018. [DOI] [PubMed] [Google Scholar]
- 16.Hopkins AM, DeSimone E, Chwalek K, Kaplan DL. 3D in vitro modeling of the central nervous system. Progress in Neurobiology. 2015;125:1–25. doi: 10.1016/j.pneurobio.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Puschmann TB, de Pablo Y, Zandén C, Liu J, Pekny M. A Novel Method for Three-Dimensional Culture of Central Nervous System Neurons. Tissue Engineering Part C: Methods. 2013;20:485–492. doi: 10.1089/ten.TEC.2013.0445. [DOI] [PubMed] [Google Scholar]
- 18.LaPlaca MC, Vernekar VN, Shoemaker JT, Cullen DK. Methods in Bioengineering: 3D Tissue Engineering. Artech House Publishers; London: 2010. Three-dimensional neuronal cultures. [Google Scholar]
- 19.East E, Golding JP, Phillips JB. A versatile 3D culture model facilitates monitoring of astrocytes undergoing reactive gliosis. Journal of Tissue Engineering and Regenerative Medicine. 2009;3:634–646. doi: 10.1002/term.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ugbode CI, Hirst WD, Rattray M. Astrocytes Grown in Alvetex® Three Dimensional Scaffolds Retain a Non-reactive Phenotype. Neurochemical Research. 2016;41:1857–1867. doi: 10.1007/s11064-016-1911-3. [DOI] [PubMed] [Google Scholar]
- 21.Puschmann TB, et al. Bioactive 3D cell culture system minimizes cellular stress and maintains the in vivo-like morphological complexity of astroglial cells. Glia. 2013;61:432–440. doi: 10.1002/glia.22446. [DOI] [PubMed] [Google Scholar]
- 22.Jeffery AF, Churchward MA, Mushahwar VK, Todd KG, Elias AL. Hyaluronic Acid-Based 3D Culture Model for In Vitro Testing of Electrode Biocompatibility. Biomacromolecules. 2014;15:2157–2165. doi: 10.1021/bm500318d. [DOI] [PubMed] [Google Scholar]
- 23.Karumbaiah L, et al. The upregulation of specific interleukin (IL) receptor antagonists and paradoxical enhancement of neuronal apoptosis due to electrode induced strain and brain micromotion. Biomaterials. 2012;33:5983–5996. doi: 10.1016/j.biomaterials.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 24.LaPlaca MC, Cullen DK, McLoughlin JJ, Cargill RS., II High rate shear strain of three-dimensional neural cell cultures: a new in vitro traumatic brain injury model. Journal of Biomechanics. 2005;38:1093–1105. doi: 10.1016/j.jbiomech.2004.05.032. [DOI] [PubMed] [Google Scholar]
- 25.Jeong SI, et al. Mechano-active tissue engineering of vascular smooth muscle using pulsatile perfusion bioreactors and elastic PLCL scaffolds. Biomaterials. 2005;26:1405–1411. doi: 10.1016/j.biomaterials.2004.04.036. [DOI] [PubMed] [Google Scholar]
- 26.Sladkova M, de Peppo G. Bioreactor Systems for Human Bone Tissue Engineering. Processes. 2014;2:494. [Google Scholar]
- 27.Riehl BD, Park JH, Kwon IK, Lim JY. Mechanical Stretching for Tissue Engineering: Two-Dimensional and Three-Dimensional Constructs. Tissue Engineering Part B, Reviews. 2012;18:288–300. doi: 10.1089/ten.teb.2011.0465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu B, Hong JS. Primary rat mesencephalic neuron-glia, neuron-enriched, microglia-enriched, and astroglia-enriched cultures. Methods Mol Med. 2003;79:387–395. doi: 10.1385/1-59259-358-5:387. [DOI] [PubMed] [Google Scholar]
- 29.Polikov VS, Su EC, Ball MA, Hong JS, Reichert WM. CONTROL PROTOCOL FOR ROBUST IN VITRO GLIAL SCAR FORMATION AROUND MICROWIRES: ESSENTIAL ROLES OF BFGF AND SERUM IN GLIOSIS. Journal of neuroscience methods. 2009;181:170–177. doi: 10.1016/j.jneumeth.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thelin J, et al. Implant size and fixation mode strongly influence tissue reactions in the CNS. PLoS One. 2011;6:e16267. doi: 10.1371/journal.pone.0016267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Köhler P, et al. Influence of Probe Flexibility and Gelatin Embedding on Neuronal Density and Glial Responses to Brain Implants. PLoS ONE. 2015;10:e0119340. doi: 10.1371/journal.pone.0119340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nguyen JK, et al. Mechanically-compliant intracortical implants reduce the neuroinflammatory response. Journal of Neural Engineering. 2014;11:056014. doi: 10.1088/1741-2560/11/5/056014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schander A, et al. Design and fabrication of novel multi-channel floating neural probes for intracortical chronic recording. Sensors and Actuators A: Physical. 2016;247:125–135. [Google Scholar]
- 34.Kelsey AP, Amy CB, Wade KS, Jeffrey RC. Stab injury and device implantation within the brain results in inversely multiphasic neuroinflammatory and neurodegenerative responses. Journal of Neural Engineering. 2012;9:046020. doi: 10.1088/1741-2560/9/4/046020. [DOI] [PubMed] [Google Scholar]
- 35.Ravikumar M, et al. The roles of blood-derived macrophages and resident microglia in the neuroinflammatory response to implanted Intracortical microelectrodes. Biomaterials. 2014;35:8049–8064. doi: 10.1016/j.biomaterials.2014.05.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fee MS. Active Stabilization of Electrodes for Intracellular Recording in Awake Behaving Animals. Neuron. 2000;27:461–468. doi: 10.1016/s0896-6273(00)00057-x. [DOI] [PubMed] [Google Scholar]
- 37.Bayly PV, et al. Deformation of the Human Brain Induced by Mild Acceleration. Journal of neurotrauma. 2005;22:845–856. doi: 10.1089/neu.2005.22.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parthasarathy KS, et al. Biocompatibilities of sapphire and borosilicate glass as cortical neuroprostheses. Magnetic Resonance Imaging. 2007;25:1333–1340. doi: 10.1016/j.mri.2007.03.020. [DOI] [PubMed] [Google Scholar]
- 39.Wang TW, Spector M. Development of hyaluronic acid-based scaffolds for brain tissue engineering. Acta Biomaterialia. 2009;5:2371–2384. doi: 10.1016/j.actbio.2009.03.033. [DOI] [PubMed] [Google Scholar]
- 40.Hachim D, LoPresti ST, Yates CC, Brown BN. Shifts in macrophage phenotype at the biomaterial interface via IL-4 eluting coatings are associated with improved implant integration. Biomaterials. 2017;112:95–107. doi: 10.1016/j.biomaterials.2016.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rennaker RL, Miller J, Tang H, Wilson DA. Minocycline increases quality and longevity of chronic neural recordings. Journal of Neural Engineering. 2007;4:L1. doi: 10.1088/1741-2560/4/2/L01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spataro L, et al. Dexamethasone treatment reduces astroglia responses to inserted neuroprosthetic devices in rat neocortex. Experimental Neurology. 2005;194:289–300. doi: 10.1016/j.expneurol.2004.08.037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.