Abstract
Antibody responses against proteins located on the surface or in the apical organelles of merozoites are presumed to be important components of naturally acquired protective immune responses against the malaria parasite Plasmodium falciparum. However, many merozoite antigens are highly polymorphic, and antibodies induced against one particular allelic form might not be effective in controlling growth of parasites expressing alternative forms. The apical membrane antigen 1 (AMA1) is a polymorphic merozoite protein that is a target of naturally acquired invasion-inhibitory antibodies and is a leading asexual-stage vaccine candidate. We characterized the antibody responses against AMA1 in 262 individuals from Papua New Guinea exposed to malaria by using different allelic forms of the full AMA1 ectodomain and some individual subdomains. The majority of individuals had very high levels of antibodies against AMA1. The prevalence and titer of these antibodies increased with age. Although antibodies against conserved regions of the molecule were predominant in the majority of individuals, most plasma samples also contained antibodies directed against polymorphic regions of the antigen. In a few individuals, predominantly from younger age groups, the majority of antibodies against AMA1 were directed against polymorphic epitopes. The D10 allelic form of AMA1 apparently contains most if not all of the epitopes present in the other allelic forms tested, which might argue for its inclusion in future AMA1-based vaccines to be tested. Some important epitopes in AMA1 involved residues located in domain II or III but depended on more than one domain.
Asexual blood stages of the malaria parasite Plasmodium falciparum are responsible for all the pathology associated with the disease and consequently are a major target for disease control strategies including the development of an effective vaccine (22). The merozoite is a particularly attractive target of vaccine interventions because it is the only extracellular stage of the asexual cycle, which makes it accessible to humoral immune responses. Furthermore, the merozoite must invade a new host red blood cell in a short period of time, and blocking this central step in the biology of the parasite would lead to control of a malaria infection. Passive transfer experiments showed many years ago that antibodies are effective in controlling parasite growth in vivo in humans (4, 30), and in vitro they can efficiently block invasion of red blood cells by merozoites (3). It is generally accepted that antibodies are a major component of naturally acquired protective immune responses which, after multiple exposures to the parasite, confer nonsterile immunity to individuals living in areas where malaria is endemic. Thus, induction of appropriate antibody responses should be a main component of any vaccine strategy that aims to mimic and accelerate the development of natural protective immunity.
Some of the antigens located on the surface or in the apical organelles of the merozoite are highly polymorphic. For some of these antigens, it has been established that diversity is the consequence of natural selection, which indicates that immune responses against these antigens are effective in controlling parasite growth (6). However, antigenic diversity can also be a major obstacle for the development of effective vaccines based on these antigens. The immune responses induced might be effective in controlling growth of parasites expressing the same form of the antigen used for the immunization but would affect the growth of parasites expressing alternative forms to a much lesser extent (9, 13, 29).
The apical membrane antigen 1 (AMA1) (27) is one of the best-studied merozoite antigens and one of the most promising malaria vaccine candidates (11, 19, 33). The 83-kDa type I integral membrane protein AMA1 has an N-terminal prosequence followed by three subdomains defined by their disulfide bonds (14). Expressed late in the asexual cycle, at about the time of merozoite release and erythrocyte invasion, AMA1 is processed to a 66-kDa form by cleavage of the prosequence (16, 26). This cleaved form relocates from the membrane of micronemes (2) to the surface of the merozoite (26), and further processing results in the shedding of fragments of 44 and 48 kDa (15, 16). Despite many years of extensive investigation, the precise function of AMA1 is not known, but its location and time of expression suggest a role in the process of merozoite invasion of red blood cells. Furthermore, antibodies against AMA1 efficiently inhibit the process of invasion (13, 18, 19). Recent results suggest that AMA1 might be essential for reorientation of merozoites and formation of the tight junction, which are essential steps for invasion (23). Whatever the precise role of AMA1, the conservation of this protein in all Plasmodium species examined and in other apicomplexan parasites together with the inability to disrupt the P. falciparum ama1 gene (35) indicate that it might be an essential protein for invasion of host red blood cells. More recently, it has been shown that AMA1 is also expressed in sporozoites and has a role in the process of invasion of hepatocytes (32).
Individuals living in areas where malaria is endemic have antibodies against AMA1 (17, 34), and these antibodies inhibit merozoite invasion in vitro (13). Immunization with correctly folded AMA1 conferred high levels of protection in murine and simian models (1, 5, 9, 10, 25, 33). This protection was at least in part mediated by antibodies, as indicated by passive immunization studies conducted with mice (1, 9) and by the correlation between antibody titers and protection (1, 33). A recent trial conducted with Aotus vociferans monkeys challenged with a highly virulent strain of P. falciparum showed that AMA1 is the most efficacious vaccine ever tested in this rigorous system (33). The correct folding of the molecule is immunologically crucial for efficacy, since immunization with reduced and alkylated AMA1 did not induce protection and the antibodies elicited did not inhibit invasion (1, 9, 13). The epitopes for the majority of antibodies against AMA1, especially for those that inhibit invasion, seem to involve more than one subdomain (20).
Although AMA1 lacks the low-complexity regions that are common in other merozoite proteins and does not show any polymorphism in size, a relatively large number of point mutations occur in AMA1. Population genetics analyses provided strong evidence that these polymorphisms are under balancing selection, especially for domains I and III (8, 28), suggesting that polymorphic regions of AMA1 are targets of protective immune responses. Furthermore, polymorphisms in AMA1 are immunologically significant, as immunization with AMA1 induced protection against homologous but not heterologous challenge in murine models (9). Also, antibodies against AMA1 exhibited a marked strain specificity in their ability to inhibit invasion (12, 13, 18, 19), demonstrating that polymorphisms in AMA1 result in evasion of invasion-inhibitory immune responses. However, recent results suggest that immunization with a combination of several forms of AMA1 can induce protection against a broader spectrum of challenging parasites (18), thus overcoming the presumed limitation that strain specificity poses to the efficacy of AMA1 as a component of a vaccine against malaria.
Although previous data indicate that some of the human antibodies against AMA1 are directed against polymorphic epitopes (13), little is known about the relative abundance of antibodies against conserved and polymorphic regions of AMA1 in naturally acquired immune responses. Here, we used different forms of the full AMA1 ectodomain as well as individual domains to characterize the immune responses against this protein in naturally exposed individuals of different ages. We found that in most individuals, antibodies were predominantly directed against conserved regions of the antigen, but a fraction of the antibodies were directed against polymorphic regions. In a small proportion of individuals, the majority of antibodies were directed against polymorphic epitopes.
MATERIALS AND METHODS
AMA1 constructs.
All proteins were expressed in Escherichia coli with an N-terminal His tag. Antigens were solubilized from inclusion bodies in guanidine-HCl and then affinity purified by using a Ni-nitrilotriacetic acid resin and refolded as previously described (13).
The expressed domain II comprised amino acids 309 to 436, domain III comprised amino acids 436 to 545, and the full ectodomain comprised amino acids 25 to 545, following the previously described subdomain structure of AMA1 (14). The sequences of 3D7, D10, and HB3 AMA1 were described previously (21), but the sequence of 3D7 contained two differences from the published sequence, one in the prodomain and one in domain III (13).
Plasma.
The 262 plasma samples used in this study were collected in cross-sectional surveys conducted in the Wosera region of Papua New Guinea in November 2000 (196 plasma samples) or from patients attending a health center in the same area with a high-density P. falciparum infection (66 plasma samples). The blood samples used for this study have been described before (7, 8).
ELISA.
Enzyme-linked immunosorbent assay (ELISA) experiments were performed essentially as previously described (13) in polyvinyl chloride microtiter plates (Dynatech) coated overnight at 4°C with antigens at 2 μg/ml in phosphate-buffered saline. Plates were blocked with BLOTTO (5% skimmed milk in phosphate-buffered saline [pH 7.3]), washed, and incubated with the relevant plasma sample diluted in BLOTTO for 2 h at room temperature. After washing, the plates were incubated with a horseradish peroxidase-conjugated anti-human immunoglobulin G antibody (Silenus) diluted 1:1,000 in BLOTTO. Bound peroxidase was detected with a citric acid-ABTS [2,2′-azinobis(3-ethylbenzthiazolinesulfonic acid)]-hydrogen peroxide substrate, and the level of antibody binding was determined colorimetrically (405 nm) with a Molecular Dynamics microplate reader.
All plasma samples were tested in duplicate at two different dilutions (1:500 and 1:1,500 for domain II and domain III and 1:1,000 and 1:9,000 for the full ectodomain). Plasma samples were tested in parallel in nine plates coated with the different antigens. Each plate included a standard curve prepared by using a serum of known titer (the titer was defined as the dilution that gave an optical density at 405 nm [OD405] of 0.1). For each plasma sample, the average result from the dilution that gave the OD405 that fell more within the linear part of the standard curve was used for determining the titer by interpolating the standard curve. Each plate also included a blank, a negative control, and a positive control of known titer. If any of these gave aberrant values, the results of the full plate and of other plates containing different allelic forms of the same antigen were discarded and the experiment was repeated.
Data analysis.
A plasma sample was considered positive against a particular construct if the OD405 was higher than the average OD405 plus two standard deviations of 27 sera from Australian donors that had not been exposed to malaria. The threshold for positivity was an OD405 of 0.160 for domain II, 0.128 for domain III, and 0.083 for the full ectodomain.
A plasma sample was considered to exhibit marked allele specificity if it fulfilled one of two criteria: (i) the titer for one particular allelic forms was more than double the titer for another form and (ii) the plasma sample was negative for one allelic form but positive for an alternative form and the positive titer was higher than 1,500 (domain III) or 3,000 (full ectodomain). All plasma samples that exhibited marked allele specificity were retested to confirm the observation, which permitted assessment of the statistical significance of the different recognition of alternative forms by the paired t test.
Inhibition ELISAs.
For selected plasma samples, the dilution necessary to obtain an OD405 of approximately 2.5 in the absence of any inhibitor was determined by endpoint titration. These plasma samples were tested in inhibition ELISA experiments in nine plates in parallel, three of them coated with each alternative form. After the plates were blocked, homologous or heterologous antigen was added at 40 μg/ml in BLOTTO in duplicate wells and titrated by a serial twofold dilution to a concentration of 0.039 μg/ml. The last wells were left without inhibitor. Subsequently, plasma samples were added at the appropriate dilutions with the rest of the ELISA procedure as described above.
RESULTS
Age dependence of the prevalence and titer of antibodies against AMA1.
The majority of individuals from the Wosera region of Papua New Guinea, where malaria is endemic, had antibodies against the AMA1 full ectodomain (Table 1). In contrast, only about 30% of individuals had antibodies against domain II or domain III of this protein (Table 1). The median antibody titer against full-length ectodomain constructs was more than 1 order of magnitude higher than the median for domain II or domain III constructs (Table 1). On the other hand, the titer and prevalence of antibodies against different allelic forms of the full ectodomain were very similar, and the same was true for different allelic forms of domain III. Although similar, titers against the D10 full-length ectodomain were significantly higher than titers against other forms of the same construct (P < 0.05 using Wilcoxon signed-rank test). No differences in the prevalence or titer of antibodies between samples collected from asymptomatic patients and samples from clinical malaria patients with high-density P. falciparum infection were observed. Thus, the results for the two sets of samples are presented together. In asymptomatic patients less than 10 years old, the prevalence of antibodies against full-length ectodomain constructs was significantly higher (P < 0.05 using Fisher's exact test) among individuals with a positive P. falciparum slide (data not shown). No significant differences in hemoglobin levels were observed in any age group between individuals with or without antibodies to the different constructs.
TABLE 1.
Titers of antibodies against different AMA1 constructs in Wosera individuals
| Age group (yr) or total no. of individuals | % Positive serum samples (median antibody titer)b
|
||||||
|---|---|---|---|---|---|---|---|
| Domain II, 3D7 | Domain III
|
Full ectodomain
|
|||||
| 3D7 | D10 | HB3 | 3D7 | D10 | HB3 | ||
| Total (n = 262) | 32 (3,696) | 31 (1,735) | 31 (1,676) | 30 (2,034) | 84 (46,738) | 85 (57,551)a | 84 (51,807) |
| Age group | |||||||
| 0-2 (n = 31) | 10 (1,752) | 6 (2,032) | 3 (3,901) | 0 (0) | 52 (11,221) | 52 (13,807) | 55 (11,564) |
| 3-9 (n = 109) | 24 (3,746) | 11 (1,624) | 13 (1,405) | 13 (1,643) | 77 (23,520) | 78 (24,706)a | 77 (23,916) |
| 10-19 (n = 66) | 39 (5,087) | 38 (1,281) | 38 (1,482) | 36 (2,087) | 98 (78,326) | 98 (83,953)a | 97 (71,955) |
| >20 (n = 56) | 50 (3,466) | 75 (1,798) | 71 (1,793) | 71 (2,047) | 100 (103,860) | 100 (108,664) | 100 (110,703) |
Titer significantly higher (P < 0.05) than the titer against an alternative form of the same construct (using Wilcoxon signed-rank test).
Median antibody titer in positive samples only.
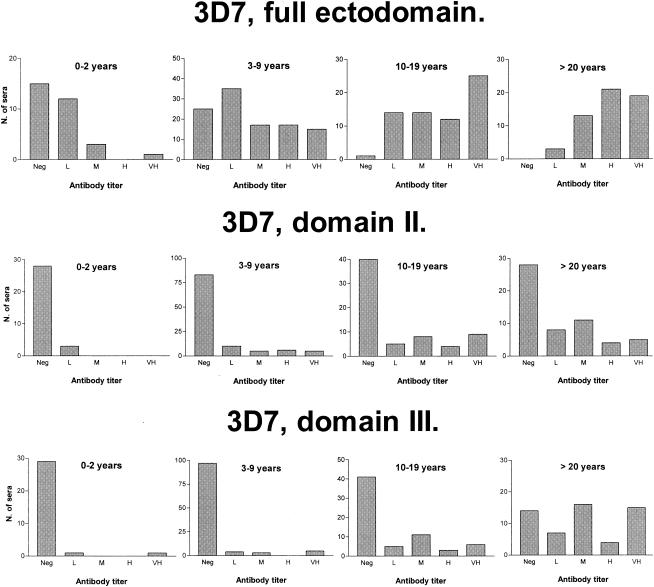
As expected in an area where malaria is endemic, prevalence and titer of antibodies against AMA1 exhibited strong age dependence, which paralleled the development of protective immunity. Approximately half of the individuals less than 3 years of age had antibodies against all three AMA1 full-length ectodomain constructs, whereas essentially all individuals older than 10 years had antibodies against each antigen (Table 1). The majority of positive plasma samples from the youngest age group (less than 3 years old) had antibody titers below 15,000 (Fig. 1), whereas titers above 15,000 were common in children 3 to 9 years old, and much higher titers were also frequently observed. Among older age groups, low antibody titers were relatively rare, and most individuals had high titers (Fig. 1). The difference in the antibody titers between age groups was significant (P < 0.05 using the Mann-Whitney U test) between the first and second and between the second and third age groups but not between the third and fourth age groups.
FIG. 1.
Age dependence of antibody responses to AMA1 constructs. Values are the number of plasma samples (N. of sera) with negative (Neg), low (L), medium (M), high (H), or very high (VH) antibody titers in the age groups indicated. The categories were defined as follows: low, 1 to 15,000 (full ectodomain), 1 to 2,000 (domain II), or 1 to 1,000 (domain III); medium, 15,000 to 50,000 (full ectodomain), 2,000 to 5,000 (domain II), or 1,000 to 2,000 (domain III); high, 50,000 to 150,000 (full ectodomain), 5,000 to 10,000 (domain II), or 2,000 to 3,000 (domain III); very high, more than 150,000 (full ectodomain), more than 10,000 (domain II), or more than 3,000 (domain III).
A similar tendency was observed for domain II and domain III, although for these domains, the increase in prevalence with age was more marked than the increase in antibody titers (Table 1 and Fig. 1). For domain II, the difference in antibody titers was significant only between the second and third age groups, whereas for domain III constructs, the difference was significant between the second and third and between the third an fourth age groups.
Only a small proportion of individuals had very different antibody titers against alternative forms of AMA1.
Most individuals had similar antibody titers against the 3D7, D10, or HB3 allelic form of the full AMA1 ectodomain, with all three correlation coefficients above 0.9 (Table 2). Similarly, the correlation between antibody titers against alternative forms of domain III was high (Table 2). In contrast, the correlation between antibody titers against different AMA1 constructs (domain II, domain III, or full ectodomain) was very low (Table 2), indicating that recognition of one of the individual domains was unrelated to recognition of the other domain or the full ectodomain.
TABLE 2.
Correlation of antibody titers against different AMA1 constructs
| Domain | Correlation coefficient
|
||||||
|---|---|---|---|---|---|---|---|
| Dom. II | Dom. III
|
Full ectodomain
|
|||||
| 3D7 | 3D7 | D10 | HB3 | 3D7 | D10 | HB3 | |
| Dom. II, 3D7 | 1.000 | ||||||
| Dom. III, 3D7 | 0.047 | 1.000 | |||||
| Dom. III, D10 | 0.244 | 0.905 | 1.000 | ||||
| Dom. III, HB3 | 0.212 | 0.929 | 0.988 | 1.000 | |||
| Full ect., 3D7 | 0.682 | 0.236 | 0.390 | 0.380 | 1.000 | ||
| Full ect., D10 | 0.568 | 0.253 | 0.358 | 0.347 | 0.954 | 1.000 | |
| Full ect., HB3 | 0.564 | 0.209 | 0.316 | 0.303 | 0.917 | 0.938 | 1.000 |
Dom., domain; ect., ectodomain.
Marked differences in the binding to different allelic forms of the same construct were observed in a small number of samples (Table 3). Using a stringent criterion for the classification of plasma as showing strong allele specificity, we found that 19 of the plasma samples fell within this category for the full-length ectodomain. These plasma samples were 17.6% of the positive plasma samples from individuals less than 3 years of age, 14% of the samples from individuals 3 to 9 years of age, 4.6% of the samples from individuals 10 to 19 years old, and 1.8% of the samples from individuals more than 20 years old, indicating that the frequency of differential recognition of alternative forms of the AMA1 ectodomain decreased significantly with age (P < 0.02 using Fisher's test). A similar situation occurred with domain III constructs, which were recognized differently by 50, 20, 17.2, and 13.6% of the positive plasma samples in the respective age groups, but in this case, the frequency of differential recognition between different age groups was not significant (P > 0.1).
TABLE 3.
Antibody titers of individual sera exhibiting different recognition of AMA1 alternative forms
| Plasma sample | Antibody titer
|
|||||
|---|---|---|---|---|---|---|
| 3D7 | D10 | HB3 | ||||
| Full ectodomain
|
||||||
| VGS6 | 111,444 | 101,752 | 9,824a | |||
| VGS23 | 1,110,593 | 1,867,922 | 915,224a | |||
| VGS24 | 22,298 | 21,905 | 10,460a | |||
| VGS35 | 4,054 | 1,975a | 2,235 | |||
| VGS54 | 168,984 | 199,283 | 65,851a | |||
| VGS64 | 720,964 | 762,467 | 279,296a | |||
| VGS80 | 27,950a | 83,953 | 87,930 | |||
| VGS88 | 10,800a | 49,045 | 40,111 | |||
| VGS116 | 4,203a | 20,826 | 23,895 | |||
| VGS128 | 4,278 | 2,586 | 2,000a | |||
| VGS156 | 12,904a | 41,164 | 38,648 | |||
| VGS214 | 1,356a | 5,719 | 6,567 | |||
| VGS217 | 221,779a | 394,171 | 484,749 | |||
| VGS240 | 13,014 | 23,633 | 4,361a | |||
| VGS244 | 315,015 | 416,850 | 191,186a | |||
| RVL19144 | 1,476a | 5,401 | 6,444 | |||
| RVL19161 | 18,238 | 24,706 | 11,533a | |||
| RVL19340 | 5381a | 10,941 | 10,705 | |||
| RVL19342 | 96,186a | 192,398 | 168,993 | |||
| Domain III
|
||||||
| VGS9 | 3,030 | 0a | 0a | |||
| VGS38 | 705a | 2,788 | 2,665 | |||
| VGS47 | 736a | 1,783 | 1,818 | |||
| VGS54 | 0a | 0a | 2,283 | |||
| VGS56 | 3,258 | 3,901 | 0a | |||
| VGS65 | 900a | 842a | 2,053 | |||
| VGS72 | 9,008a | 47,391 | 41,304 | |||
| VGS91 | 1,209a | 2,559 | 3,043 | |||
| VGS92 | 0a | 1,796 | 2,012 | |||
| VGS107 | 5,355 | 1,357a | 1,458a | |||
| VGS164 | 0a | 1,968a | 5,969 | |||
| VGS189 | 3,025a | 11,083 | 10,865 | |||
| VGS253 | 1,864 | 0a | 780a | |||
| RVL19302 | 7,098 | 4,205 | 3,078a | |||
| RVL19564 | 1,236a | 1,122a | 2,581 | |||
Titers that are less than half the titer for an alternative form.
The majority of plasma samples that exhibited strongly different recognition of full-length ectodomain allelic forms had a lower titer of antibodies against either the 3D7 or the HB3 form compared to binding to the two alternative allelic forms (Table 3). Only one plasma sample which had a markedly lower antibody titer against D10 AMA1 was detected, but the difference was not very large (although still significant when tested by paired t test [P < 0.05]), and it was a sample with a low antibody titer. It was noteworthy that some of the plasma samples exhibited very different recognition of 3D7 and D10 AMA1 full-length ectodomains (highly significant when tested by paired t test), which are identical in domain I. For example, plasma sample VGS88 bound D10 AMA1 almost fivefold better than 3D7 AMA1 (Table 3).
Most plasma samples contain a fraction of antibodies directed against allele-specific epitopes.
Inhibition ELISA experiments revealed that plasma samples that exhibited similar recognition of the three different forms of the AMA1 full ectodomain contained a fraction of antibodies directed against allelic form-specific epitopes. The binding of antibodies to the immobilized 3D7 AMA1 full ectodomain was inhibited by soluble HB3 AMA1 to an average of only 84% of the extent of inhibition for soluble 3D7 AMA1 (Table 4). Similarly, the binding to HB3 AMA1 was inhibited by 3D7 AMA1 to an average of only 88% of the extent of inhibition for HB3 (Table 4). These results indicate that a fraction of the antibodies that recognize 3D7 AMA1 bind to epitopes that are not present in HB3 AMA1, and vice versa. On the other hand, D10 AMA1 inhibited binding to the two other allelic forms almost as efficiently as the homologous antigen, but only an average of 91% (3D7) or 83% (HB3) of the binding to D10 AMA1 was competed by the other alternative forms (Table 4). These results indicate that the vast majority of the epitopes present in 3D7 and HB3 AMA1 are present in D10 AMA1 but that some of the epitopes in D10 AMA1 are not present in the other two allelic forms. Also, D10 AMA1 was a more efficient inhibitor of the binding to any of the three antigens, as demonstrated by the lower concentration of this antigen necessary to reduce the OD405 obtained in the absence of inhibitor by 50% (Table 4).
TABLE 4.
Inhibition of antibody binding to AMA1 constructs with homologous and heterologous allelic formsa
| AMA1 construct | Maximum inhibition (%)b | Concn for 50% OD405c | ||
|---|---|---|---|---|
| Full ectodomain
|
||||
| Coating 3D7 | ||||
| 3D7 | 100 | 0.204 | ||
| D10 | 98.1 ± 1.6 | 0.135 | ||
| HB3 | 83.8 ± 3.8 | 0.312 | ||
| Coating D10 | ||||
| 3D7 | 90.7 ± 3.2 | 0.338 | ||
| D10 | 100 | 0.207 | ||
| HB3 | 83.2 ± 4.0 | 0.492 | ||
| Coating HB3 | ||||
| 3D7 | 87.8 ± 4.7 | 0.530 | ||
| D10 | 98.1 ± 2.6 | 0.230 | ||
| HB3 | 100 | 0.381 | ||
| Domain III
|
||||
| Coating 3D7 | ||||
| 3D7 | 100 | 0.255 | ||
| D10 | 100.5 ± 2.0 | 0.223 | ||
| HB3 | 100.2 ± 2.1 | 0.238 | ||
| Coating D10 | ||||
| 3D7 | 98.8 ± 1.5 | 0.288 | ||
| D10 | 100 | 0.287 | ||
| HB3 | 97.8 ± 2.1 | 0.254 | ||
| Coating HB3 | ||||
| 3D7 | 98.7 ± 4.1 | 0.436 | ||
| D10 | 101.5 ± 2.1 | 0.284 | ||
| HB3 | 100 | 0.325 | ||
Values correspond to inhibition ELISA experiments performed with 13 (full ectodomain) or 5 (domain III) different sera, all of which exhibited similar recognition of the different allelic forms in direct ELISA experiments.
Percent inhibition (interpolated from the inhibition curve) obtained with 40 μg of inhibitor/ml relative to inhibition with the same concentration of the homologous form (the allelic form used for the coating). Values are the averages with 95% confidence intervals.
Concentration of inhibitor to reduce the OD405 in the absence of inhibitor by 50%. Values are the geometric means.
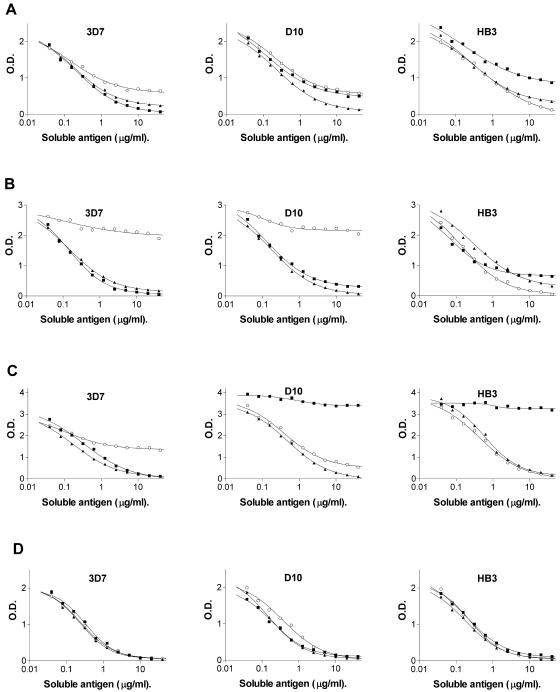
Figure 2 A is a representative example of some of the inhibition curves obtained in experiments performed with plasma samples having similar antibody titers against the different ectodomain forms. However, for some other plasma samples, the allelic form specificity was less marked or the patterns of allelic form specificity were slightly different (see the full set of inhibition curves with full-length ectodomain constructs in the supplemental material [http://www.nimr.mrc.ac.uk/supplementary/cortes]). The degree of allelic form specificity as determined by inhibition ELISA experiments did not appear to depend on age.
FIG. 2.
Inhibition of antibody binding to AMA1 constructs with homologous and heterologous allelic forms. Binding to AMA1 antigens coated onto the ELISA plates (3D7, D10, or HB3 sequence as indicated) was inhibited with soluble antigen 3D7 AMA1 (squares), D10 AMA1 (triangles), or HB3 AMA1 (circles). The AMA1 constructs (coated and inhibitor) were the full ectodomain or domain III as indicated. A) Plasma sample VGS30, full ectodomain; B) plasma sample VGS54, full ectodomain; C) plasma sample VGS88, full ectodomain; D) plasma sample VGS34, domain III.
Inhibition ELISA experiments performed with plasma samples that had a very different antibody titer against different allelic forms of the AMA1 full-length ectodomain confirmed the results obtained with direct ELISA experiments. The HB3 ectodomain failed almost completely to inhibit binding to the 3D7 or to the D10 ectodomain by plasma sample VGS54 (Fig. 2B), which had a much lower titer against the HB3 ectodomain than against the other allelic forms (Table 3). Similarly, the 3D7 ectodomain failed to inhibit binding of VGS88 to the other ectodomain forms (Fig. 2C). This plasma sample had a fivefold-lower titer against 3D7 than against the other forms (Table 3). A parallel situation was observed in inhibition experiments performed with plasma samples VGS6, VGS64, VGS80, and VGS217 (see the supplemental material [http://www.nimr.mrc.ac.uk/supplementary/cortes]).
In contrast to the results with full-length ectodomain constructs, inhibition ELISA experiments performed with domain III constructs provided no evidence for the presence of allelic form-specific antibodies in plasma samples with similar antibody titers against the three different domain III forms (Table 4). Figure 2D is representative of experiments performed with five different plasma samples of this type (see the supplemental material for the full set of inhibition curves with domain III constructs [http://www.nimr.mrc.ac.uk/supplementary/cortes]). However, allelic form-specific recognition occurs in domain III, as revealed by inhibition experiments with samples that had very different titers against the different domain III allelic forms. Inhibition experiments with two such plasma samples, VGS72 and VGS189 (Table 3), clearly showed that the domain III allelic form with lower titer failed to compete binding to the other allelic forms (see the supplemental material [http://www.nimr.mrc.ac.uk/supplementary/cortes]).
As expected from the much lower titers for domain II and domain III compared to the titers for the full ectodomain, these two individual domains failed almost completely to inhibit the binding to the full ectodomain (data not shown).
DISCUSSION
Naturally acquired or vaccine-induced antibodies against AMA1 efficiently inhibit invasion of host red blood cells expressing homologous forms of the antigen but affect parasites expressing alternative forms to a much lesser extent (12, 13, 18, 19). Furthermore, active and passive immunization experiments conducted in murine models showed that antibodies induced upon immunization with AMA1 confer protection against parasites expressing homologous, but not heterologous, forms of the antigen (9). These observations indicate that some of the antibodies against AMA1 target regions of the protein that differ between alternative forms. With the aim of studying the development of antibody responses against different domains of AMA1 in a naturally exposed population with age and to determine whether they are predominantly directed against conserved or allele-specific parts of the protein, we performed serological studies with plasma samples from an age-stratified sample of individuals living in the Wosera region of Papua New Guinea, where malaria is endemic.
The clear age dependence that we observed in the prevalence and levels of antibodies against AMA1, which parallels the development of protective immune responses, is in contrast with the results of previous studies that detected a small age-related change in antibody levels of individuals from Guinea-Bisau and Cameroon and no age dependence in the levels of antibodies in individuals from Senegal (17, 34). Although we cannot rule out the possibility that these results reflect real differences in the antibody responses to AMA1 between these African and Papua New Guinea populations, we consider it more likely that the different results are attributable to the different methodologies in these studies, because the antibody levels detected in these African settings where malaria is highly endemic were much lower than the levels detected in the Papua New Guinea population. It might be of importance that the baculovirus-expressed AMA1 used in the previous studies was highly glycosylated, unlike authentic AMA1 (34). Also, one of those studies (34) used a capture ELISA method instead of the direct ELISA method used here and in the study with samples from Cameroon (17).
Antibodies against the AMA1 ectodomain were highly prevalent among individuals exposed to malaria and occurred at very high titers, which may be important for inhibiting merozoite invasion if the on-rate of antibody binding in the short time the merozoite surface is accessible is important (31). In contrast, the prevalence of antibodies against domain II alone or domain III alone was lower. The titers were also lower and did not correlate with titers against the full ectodomain. Structural analyses of domain II (R. F. Anders, personal communication) and domain III (24) indicate that the disulfide bonds in these individually expressed and refolded domains are correct. Thus, the low prevalence and low titers of antibodies against these domains may suggest that the majority of the naturally acquired antibodies against AMA1 are directed against the highly polymorphic domain I. This hypothesis has not been tested because we could not express and correctly refold domain I alone, but a recent report showed that domain I alone, as well as other individual subdomains, is poorly recognized by antibodies against the full-length ectodomain (20). An alternative explanation is that some dominant epitopes in AMA1 depend on more than one domain. Our observation that some plasma samples had dramatically different levels of antibodies against 3D7 or D10, which are identical in domain I, but low levels of antibodies against domain II and domain III favors this latter explanation. The well-documented conformation dependence of anti-AMA1 antibodies (1, 9, 13) is also consistent with the view that some principal epitopes in AMA1 depend on more than one domain. This argues against vaccine development strategies that include only one of the domains of AMA1 if the aim is to mimic the development of naturally acquired protective immune responses. In support of this view, a construct comprising domains I and II induced growth-inhibitory antibodies and competed a significant fraction of the growth inhibition by antibodies against the full ectodomain, but it did not occur with single AMA1 subdomains or other combinations of two subdomains (20).
For experiments with the full ectodomain or domain III, we used three different forms of the antigen corresponding to the sequences of P. falciparum lines 3D7, D10, and HB3. D10 AMA1 differs from 3D7 AMA1 in three positions in the prosequence, one position in domain II, and four positions in domain III, whereas 3D7 AMA1 and HB3 AMA1 differ in 25 positions located in all of the domains (13). The D10 and HB3 sequences differed in 27 positions also distributed among all of the domains. Additionally, the recombinant 3D7 AMA1 used in this study differs from the other two forms in one position located between the prosequence and the beginning of the disulfide-linked core of domain I and one position at the end of domain III (13). For the majority of the plasma samples, we observed very similar antibody titers against the alternative forms. This observation could be explained by essentially all of the antibodies being directed against conserved epitopes. Alternatively, some of the antibodies could be directed against allelic form-specific epitopes, but if this is the case, allele-specific antibodies against the different allelic forms must occur at similar levels. To distinguish between these two possibilities, we performed inhibition ELISA experiments. These experiments revealed that in most individuals, some anti-AMA1 antibodies are directed against epitopes present in some but not other allelic forms of the protein. This fraction of allele-specific antibodies comprised less than 25% of the total anti-AMA1 antibodies in the majority of plasma samples, but this proportion might be underestimated in some cases because we used only three different allelic forms of the ectodomain. Some of the alternative forms used represent rather extreme forms that differ in a large number of polymorphic positions but have the same residue in several other polymorphic sites, and more antigenically distant forms have been reported (12, 18).
Our data suggest that antibodies against conserved epitopes are quantitatively more important than antibodies against polymorphic epitopes but that allele-specific antibodies occur in most, if not all, individuals. The functional role of both types of antibodies is not established, but the observation that polymorphisms in AMA1 are under natural selection indicates that polymorphic sites are targets of protective immunity (8, 28). This observation suggests that antibodies against polymorphic epitopes might constitute the major protective fraction of anti-AMA1 antibodies. Vaccine trials in mice using homologous and heterologous challenge also support this view (9). This is a testable hypothesis that clearly deserves further investigation.
A small subset of plasma samples showed dramatic differences in the recognition of different forms of the complete AMA1 ectodomain, indicating that these samples predominantly contain antibodies against polymorphic epitopes that are present in some of the alternative forms but not others. Different recognition of the different forms of domain III was also detected in a few plasma samples and confirmed by inhibition ELISA experiments. We do not have a satisfactory explanation for why some individuals have very different antibody titers against different allelic forms of AMA1 whereas the majority have very similar titers. It might be the consequence of random repeated exposure to parasites with similar forms of some polymorphic epitopes or, alternatively, it might reflect genetic characteristics of the human host that determine the capacity to mount an antibody response against particular epitopes. The predominance of allele-specific antibodies was significantly more common among younger individuals, suggesting that development of immunity against AMA1 might include an initial phase in which antibodies are mainly directed against polymorphic epitopes followed by a later phase in which antibodies against conserved epitopes are predominant. Our inhibition ELISA experiments, which found no age dependence of the estimated fraction of allele-specific antibodies, did not confirm this hypothesis, but the number of samples that could be tested in these assays was low and far from having any epidemiological significance.
The majority of the plasma samples with very different recognition of the alternative forms of the full ectodomain bound similarly well to D10 AMA1 and to one but not both of the two other allelic forms of the antigen used in this study (3D7 and HB3). Furthermore, in plasma samples with similar titers of antibodies against the three different forms, D10 AMA1 inhibited binding to the two other forms to a greater extent than the other forms inhibited binding to D10. Also, anti-AMA1 antibodies apparently have a higher affinity for D10 AMA1 than for the other forms of the antigen, since lower concentrations of D10 AMA1 were necessary to compete 50% of the binding to any of the three antigens. These results indicate that the majority of the epitopes present in 3D7 or HB3 AMA1 are also present in the D10 antigen, whereas some of the epitopes present in D10 are disrupted by the polymorphisms present in the other alternative forms. If this pattern of reactivity is also a feature of sera from other regions of endemicity, it would argue for the inclusion of the D10 allelic form in a future AMA1-based vaccine. Consistent with these results, antibodies against 3D7 AMA1 inhibited invasion by D10 merozoites more efficiently than they inhibited invasion by homologous 3D7 merozoites but inhibited HB3 merozoite invasion less efficiently (13). In another study in which additional P. falciparum lines were used, the same pattern was observed for the three lines used here (18). Thus, there was a good correlation in the pattern of specificity between ELISA experiments and invasion inhibition experiments.
It is noteworthy that some plasma samples exhibited dramatic differences in their binding to D10 and 3D7 AMA1, which are identical in domain I. This finding indicates that residues outside domain I are part of important allele-specific epitopes in AMA1. In particular, some of the 10 positions that differ between 3D7 and D10 must be part of such epitopes. An analysis of the polymorphisms in AMA1 using transgenic parasite lines also suggested that polymorphic sites located outside domain I are of immunological significance (12). If amino acids in domain II or domain III are part of major polymorphic epitopes in AMA1, these epitopes might also include residues located in another domain, as indicated by the low antibody titers against either of these domains alone. Alternatively, many intradomain conformational epitopes may be present only in the context of the full-length ectodomain. The immunological relevance of polymorphism in domain III had been predicted by population genetics analysis (28), by the clustering of polymorphic sites in solvent-accessible regions of the domain, and by the effect of polymorphism on the invasion-inhibitory activity of affinity-purified human antibodies directed against this domain (24). Thus, it is likely that the four differences in domain III between 3D7 and D10 determine the very different recognition of 3D7 and D10 AMA1 ectodomains by some plasma samples.
Acknowledgments
This work was supported by the Australian Agency for International Development and the Australian Cooperative Research Centre for Vaccine Technology.
We are grateful to the Wosera communities that participated in this study for donating blood. We are also grateful to the IMR staff in Maprik for their assistance in the field, to Ariadna Benet and Alice Ura for assistance in the lab, and to Luke Miles for providing the AMA1 domain II recombinant antigen.
Editor: W. A. Petri, Jr.
REFERENCES
- 1.Anders, R. F., P. E. Crewther, S. Edwards, M. Margetts, M. L. Matthew, B. Pollock, and D. Pye. 1998. Immunisation with recombinant AMA-1 protects mice against infection with Plasmodium chabaudi. Vaccine 16:240-247. [DOI] [PubMed] [Google Scholar]
- 2.Bannister, L. H., J. M. Hopkins, A. R. Dluzewski, G. Margos, I. T. Williams, M. J. Blackman, C. H. Kocken, A. W. Thomas, and G. H. Mitchell. 2003. Plasmodium falciparum apical membrane antigen 1 (PfAMA-1) is translocated within micronemes along subpellicular microtubules during merozoite development. J. Cell Sci. 116:3825-3834. [DOI] [PubMed] [Google Scholar]
- 3.Brown, G. V., R. F. Anders, G. F. Mitchell, and P. F. Heywood. 1982. Target antigens of purified human immunoglobulins which inhibit growth of Plasmodium falciparum in vitro. Nature 297:591-593. [DOI] [PubMed] [Google Scholar]
- 4.Cohen, S., G. I. Mc, and S. Carrington. 1961. Gamma-globulin and acquired immunity to human malaria. Nature 192:733-737. [DOI] [PubMed] [Google Scholar]
- 5.Collins, W. E., D. Pye, P. E. Crewther, K. L. Vandenberg, G. G. Galland, A. J. Sulzer, D. J. Kemp, S. J. Edwards, R. L. Coppel, J. S. Sullivan, C. L. Morris, and R. F. Anders. 1994. Protective immunity induced in squirrel monkeys with recombinant apical membrane antigen-1 of Plasmodium fragile. Am. J. Trop. Med. Hyg. 51:711-719. [DOI] [PubMed] [Google Scholar]
- 6.Conway, D. J., and S. D. Polley. 2002. Measuring immune selection. Parasitology 125(Suppl):S3-S16. [DOI] [PubMed] [Google Scholar]
- 7.Cortés, A., M. Mellombo, A. Benet, K. Lorry, L. Rare, and J. C. Reeder. 2004. Plasmodium falciparum: distribution of msp2 genotypes among symptomatic and asymptomatic individuals from the Wosera region of Papua New Guinea. Exp. Parasitol. 106:22-29. [DOI] [PubMed] [Google Scholar]
- 8.Cortés, A., M. Mellombo, I. Mueller, A. Benet, J. C. Reeder, and R. F. Anders. 2003. Geographical structure of diversity and differences between symptomatic and asymptomatic infections for Plasmodium falciparum vaccine candidate AMA1. Infect. Immun. 71:1416-1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crewther, P. E., M. L. S. M. Matthew, R. H. Flegg, and R. F. Anders. 1996. Protective immune responses to apical membrane antigen 1 of Plasmodium chabaudi involve recognition of strain-specific epitopes. Infect. Immun. 64:3310-3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deans, J. A., A. M. Knight, W. C. Jean, A. P. Waters, S. Cohen, and G. H. Mitchell. 1988. Vaccination trials in rhesus monkeys with a minor, invariant, Plasmodium knowlesi 66 kD merozoite antigen. Parasite Immunol. 10:535-552. [DOI] [PubMed] [Google Scholar]
- 11.Dutta, S., P. V. Lalitha, L. A. Ware, A. Barbosa, J. K. Moch, M. A. Vassell, B. B. Fileta, S. Kitov, N. Kolodny, D. G. Heppner, J. D. Haynes, and D. E. Lanar. 2002. Purification, characterization, and immunogenicity of the refolded ectodomain of the Plasmodium falciparum apical membrane antigen 1 expressed in Escherichia coli. Infect. Immun. 70:3101-3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Healer, J., V. Murphy, A. N. Hodder, R. Masciantonio, A. W. Gemmill, R. F. Anders, A. F. Cowman, and A. Batchelor. 2004. Allelic polymorphisms in apical membrane antigen-1 are responsible for evasion of antibody-mediated inhibition in Plasmodium falciparum. Mol. Microbiol. 52:159-168. [DOI] [PubMed] [Google Scholar]
- 13.Hodder, A. N., P. E. Crewther, and R. F. Anders. 2001. Specificity of the protective antibody response to apical membrane antigen 1. Infect. Immun. 69:3286-3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hodder, A. N., P. E. Crewther, M. L. S. M. Matthew, G. E. Reid, R. L. Moritz, R. J. Simpson, and R. F. Anders. 1996. The disulfide bond structure of Plasmodium apical membrane antigen-1. J. Biol. Chem. 271:29446-29452. [DOI] [PubMed] [Google Scholar]
- 15.Howell, S. A., I. Well, S. L. Fleck, C. Kettleborough, C. R. Collins, and M. J. Blackman. 2003. A single malaria merozoite serine protease mediates shedding of multiple surface proteins by juxtamembrane cleavage. J. Biol. Chem. 278:23890-23898. [DOI] [PubMed] [Google Scholar]
- 16.Howell, S. A., C. Withers-Martinez, C. H. Kocken, A. W. Thomas, and M. J. Blackman. 2001. Proteolytic processing and primary structure of Plasmodium falciparum apical membrane antigen-1. J. Biol. Chem. 276:31311-31320. [DOI] [PubMed] [Google Scholar]
- 17.Johnson, A. H., R. G. F. Leke, N. R. Mendell, D. Shon, Y. J. Suh, D. Bomba-Nkolo, V. Tchinda, S. Kouontchou, L. W. Thuita, A. M. van der Wel, A. Thomas, A. Stowers, A. Saul, A. Zhou, D. W. Taylor, and I. A. Quakyi. 2004. Human leukocyte antigen class II alleles influence levels of antibodies to the Plasmodium falciparum asexual-stage apical membrane antigen 1 but not to merozoite surface antigen 2 and merozoite surface protein 1. Infect. Immun. 72:2762-2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kennedy, M. C., J. Wang, Y. Zhang, A. P. Miles, F. Chitsaz, A. Saul, C. A. Long, L. H. Miller, and A. W. Stowers. 2002. In vitro studies with recombinant Plasmodium falciparum apical membrane antigen 1 (AMA1): production and activity of an AMA1 vaccine and generation of a multiallelic response. Infect. Immun. 70:6948-6960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kocken, C. H., C. Withers-Martinez, M. A. Dubbeld, A. van der Wel, F. Hackett, A. Valderrama, M. J. Blackman, and A. W. Thomas. 2002. High-level expression of the malaria blood-stage vaccine candidate Plasmodium falciparum apical membrane antigen 1 and induction of antibodies that inhibit erythrocyte invasion. Infect. Immun. 70:4471-4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lalitha, P. V., L. A. Ware, A. Barbosa, S. Dutta, J. K. Moch, J. D. Haynes, B. B. Fileta, C. E. White, and D. E. Lanar. 2004. Production of the subdomains of the Plasmodium falciparum apical membrane antigen 1 ectodomain and analysis of the immune response. Infect. Immun. 72:4464-4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marshall, V. M., L. Zhang, R. F. Anders, and R. L. Coppel. 1996. Diversity of the vaccine candidate AMA-1 of Plasmodium falciparum. Mol. Biochem. Parasitol. 77:109-113. [DOI] [PubMed] [Google Scholar]
- 22.Miller, L. H., D. I. Baruch, K. Marsh, and O. K. Doumbo. 2002. The pathogenic basis of malaria. Nature 415:673-679. [DOI] [PubMed] [Google Scholar]
- 23.Mitchell, G. H., A. W. Thomas, G. Margos, A. R. Dluzewski, and L. H. Bannister. 2004. Apical membrane antigen 1, a major malaria vaccine candidate, mediates the close attachment of invasive merozoites to host red blood cells. Infect. Immun. 72:154-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nair, M., M. G. Hinds, A. M. Coley, A. N. Hodder, M. Foley, R. F. Anders, and R. S. Norton. 2002. Structure of domain III of the blood-stage malaria vaccine candidate, Plasmodium falciparum apical membrane antigen 1 (AMA1). J. Mol. Biol. 322:741-753. [DOI] [PubMed] [Google Scholar]
- 25.Narum, D. L., S. A. Ogun, A. W. Thomas, and A. A. Holder. 2000. Immunization with parasite-derived apical membrane antigen 1 or passive immunization with a specific monoclonal antibody protects BALB/c mice against lethal Plasmodium yoelii yoelii YM blood-stage infection. Infect. Immun. 68:2899-2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Narum, D. L., and A. W. Thomas. 1994. Differential localization of full-length and processed forms of PF83/AMA-1 an apical membrane antigen of Plasmodium falciparum merozoites. Mol. Biochem. Parasitol. 67:59-68. [DOI] [PubMed] [Google Scholar]
- 27.Peterson, M. G., V. M. Marshall, J. A. Smythe, P. E. Crewther, A. Lew, A. Silva, R. F. Anders, and D. J. Kemp. 1989. Integral membrane protein located in the apical complex of Plasmodium falciparum. Mol. Cell. Biol. 9:3151-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Polley, S. D., and D. J. Conway. 2001. Strong diversifying selection on domains of the Plasmodium falciparum apical membrane antigen 1 gene. Genetics 158:1505-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Renia, L., I. T. Ling, M. Marussig, F. Miltgen, A. A. Holder, and D. Mazier. 1997. Immunization with a recombinant C-terminal fragment of Plasmodium yoelii merozoite surface protein 1 protects mice against homologous but not heterologous P. yoelii sporozoite challenge. Infect. Immun. 65:4419-4423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sabchareon, A., T. Burnouf, D. Ouattara, P. Attanath, H. Bouharoun-Tayoun, P. Chantavanich, C. Foucault, T. Chongsuphajaisiddhi, and P. Druilhe. 1991. Parasitologic and clinical human response to immunoglobulin administration in falciparum malaria. Am. J. Trop. Med. Hyg. 45:297-308. [DOI] [PubMed] [Google Scholar]
- 31.Saul, A. 1987. Kinetic constraints on the development of a malaria vaccine. Parasite Immunol. 9:1-9. [DOI] [PubMed] [Google Scholar]
- 32.Silvie, O., J.-F. Franetich, S. Charrin, M. S. Mueller, A. Siau, M. Bodescot, E. Rubinstein, L. Hannoun, Y. Charoenvit, C. H. Kocken, A. W. Thomas, G.-J. van Gemert, R. W. Sauerwein, M. J. Blackman, R. F. Anders, G. Pluschke, and D. Mazier. 2004. A role for apical membrane antigen 1 during invasion of hepatocytes by Plasmodium falciparum sporozoites. J. Biol. Chem. 279:9490-9496. [DOI] [PubMed] [Google Scholar]
- 33.Stowers, A. W., M. C. Kennedy, B. P. Keegan, A. Saul, C. A. Long, and L. H. Miller. 2002. Vaccination of monkeys with recombinant Plasmodium falciparum apical membrane antigen 1 confers protection against blood-stage malaria. Infect. Immun. 70:6961-6967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thomas, A. W., J. F. Trape, C. Rogier, A. Goncalves, V. E. Rosario, and D. L. Narum. 1994. High prevalence of natural antibodies against Plasmodium falciparum 83-kilodalton apical membrane antigen (PF83/AMA-1) as detected by capture-enzyme-linked immunosorbent assay using full-length baculovirus recombinant PF83/AMA-1. Am. J. Trop. Med. Hyg. 51:730-740. [DOI] [PubMed] [Google Scholar]
- 35.Triglia, T., J. Healer, S. R. Caruana, A. N. Hodder, R. F. Anders, B. S. Crabb, and A. F. Cowman. 2000. Apical membrane antigen 1 plays a central role in erythrocyte invasion by Plasmodium species. Mol. Microbiol. 38:706-718. [DOI] [PubMed] [Google Scholar]