The Notch transmembrane domain exhibits significant structural differences from that of the amyloid precursor protein.
Keywords: Notch, receptor, transmembrane domain, C99, amyloid precursor protein, NMR, structure, bicelles, cholesterol, Membrane
Abstract
γ-Secretase cleavage of the Notch receptor transmembrane domain is a critical signaling event for various cellular processes. Efforts to develop inhibitors of γ-secretase cleavage of the amyloid-β precursor C99 protein as potential Alzheimer’s disease therapeutics have been confounded by toxicity resulting from the inhibition of normal cleavage of Notch. We present biochemical and structural data for the combined transmembrane and juxtamembrane Notch domains (Notch-TMD) that illuminate Notch signaling and that can be compared and contrasted with the corresponding traits of C99. The Notch-TMD and C99 have very different conformations, adapt differently to changes in model membrane hydrophobic span, and exhibit different cholesterol-binding properties. These differences may be exploited in the design of agents that inhibit cleavage of C99 while allowing Notch cleavage.
INTRODUCTION
The Notch receptor initiates one of the most important signaling pathways of mammalian cells. Notch signaling occurs in all vertebrate organisms and has been implicated in an array of developmental patterning choices (1–4). In the nervous system alone, Notch signaling plays key roles in neurogenesis, axon and dendrite growth, synapse plasticity, and neuronal death (5–10). The human Notch receptor is a type I membrane protein with more than 2500 amino acids and a modular domain composition (Fig. 1). Like the amyloid precursor protein (APP), Notch is subjected to a series of proteolytic steps that mature the protein, prime it for signaling, and trigger signal activation. The penultimate step of this process involves cleavage of Notch within its transmembrane domain (TMD) by γ-secretase to release a short peptide (Nβ) into the extracellular milieu and the large Notch intracellular domain (NICD) into the cytosol (11–14). The NICD translocates to the nucleus, where it forms transcriptional activator complexes (15–17) that control expression of many different genes (18, 19). The mechanism of Notch cleavage by γ-secretase cleavage is not well established.
Fig. 1. Domain organization of full-length Notch 1.
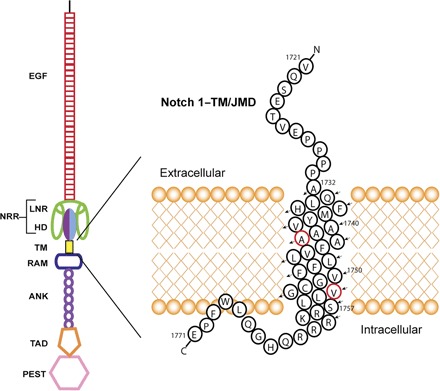
The ligand-binding N-terminal extracellular domain includes the repeat EGF domain followed by three Lin-12/Notch repeats (LNR) and the heterodimerization domain (HD). The LNR segments and the HD comprise the NRR. The transmembrane segment follows. The cytosolic domain contains the RAM domain, followed by ankyrin (ANK) repeats, the transcriptional activation domain (TAD), and the C-terminal terminal proline-, glutamic acid–, serine-, and threonine-rich (PEST) domain. The construct used in this study (the Notch-TMD) is the transmembrane segment plus the immediate juxtamembrane regions, highlighted on the right. Major γ-secretase cut sites are labeled red. The S2 metalloprotease cut site precedes Val1721.
There are experimentally determined structures for various soluble domains from Notch receptors. In previous studies, structures of the ligand-binding epidermal growth factor (EGF) repeats, the negative regulatory region (NRR), the ankyrin repeat domain, and the complete NICD–CSL (CBF-1/suppressor of hairless/Lag-1)–MAML (Mastermind-like) transcription factor complex were determined (20, 21). More recent structural studies have focused on the NRR, the complex of the EGF repeats with a Notch ligand, and a variety of other studies focusing on posttranslationally modified EGF repeats (22–25). Despite this remarkable body of work, there remains no structural information about the important Notch-TMD and its flanking juxtamembrane segments.
The Notch receptor is also related to Alzheimer’s disease (AD), a devastating neurodegenerative disorder that affects tens of millions of people worldwide. The most prominent model for the etiology of AD holds that the production and oligomerization/aggregation of the amyloid-β (Aβ) polypeptide in the brain is the root cause of AD pathogenesis (26). Aβ is the end product of the amyloidogenic pathway, which is initiated by β-secretase release of the large ectodomain of the full-length APP. The remaining 99-residue transmembrane C-terminal domain of APP (C99) is then cleaved by γ-secretase to release the Aβ polypeptide into the ectoplasm and the APP intracellular domain (AICD) into the cytosol (27). It is generally believed that long-term reduction of Aβ production would reduce the risk of AD. One potential way to lower Aβ production is inhibition of γ-secretase cleavage of C99, but this strategy has thus far proven clinically problematic because of the toxicity of γ-secretase inhibitors (28). One γ-secretase inhibitor, semagacestat, made it to phase 3 clinical trials before cancellation (29). Participants in these trials experienced a variety of physical issues, including skin cancers and reduced cognitive capabilities. Inhibition of Notch receptor processing was determined to be the underlying cause of these adverse effects. The inhibition of Notch cleavage by compounds intended to inhibit Aβ release from C99 is therefore a major obstacle to the exploration of γ-secretase inhibitors as potential Alzheimer’s therapeutic agents (28, 29). Given this problem and the broad significance of Notch signaling, a close examination of the structure and biochemistry of the Notch-TMD seems well merited. These studies are also timely in light of the fact that the structure and ligand-binding properties of C99 have recently been characterized (30–38). The stage is thereby set for a compare-and-contrast study of the structures, membrane interactions, and biochemical properties of the Notch-TMD and C99 that may provide the basis for development of improved inhibitors of Aβ production.
RESULTS
Structure of the Notch-TMD
There are significant challenges to assigning side-chain resonances in membrane proteins, including (i) rapid T2 relaxation due to the relatively large size of the complexes formed with the protein and its membrane mimetic (that is, bicelles) and (ii) the huge overlapping background 1H nuclear magnetic resonance (NMR) signals from the bicelles that will be present in spectra unless costly perdeuterated detergents/lipids are used (39). Here, using fully protonated bicelles as the model membrane system for our studies of the Notch-TMD, we were able to assign 95% of the side-chain resonances, complementing previous backbone assignments (40). Sensitivity increases from the use of a cryogenic probe and 3-mm NMR tubes, employment of pulse sequences optimized for a slow tumbling system, and effective data processing were key factors. With these approaches, lipid and detergent signals were minimized in the spectra used to complete NOTCH-TMD side-chain assignments.
The structure of the Notch-TMD in bicelles was determined using a variety of NMR-derived structural restraints combined with XPLOR-NIH calculations [see table S1 for summary of restraints and PROCHECK analysis (41)]. The 10 lowest energy structures were deposited in the Protein Data Bank (PDB; code 5KZO) and exhibited a fairly straight transmembrane helix that spans residues 1732 to 1757. The N-terminal juxtamembrane domain is largely disordered and is connected to the TMD by a tetraproline motif, which forms a type II polyproline helix (Fig. 2A) (42). On the cytosolic C-terminal side, the TMD is flanked by a short water-exposed loop, leading to a membrane reentrant segment centered at a LWF motif, followed by the water-exposed C terminus. A representative structure is shown in Fig. 2A.
Fig. 2. Structures of the Notch-TMD.
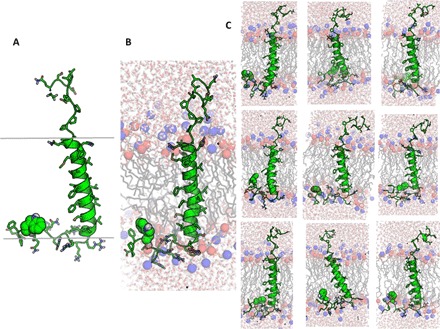
(A) Representative low-energy NMR-determined XPLOR-NIH structure. The approximate location of the membrane span is shown by the gray bars. The residue represented in green van der Waals mode is Trp1768 of the LWF motif. (B) Representative frame from one of the 10 × 60–ns restrained MD trajectories in a hydrated DMPC bilayer. The blue and red balls indicate the positions of the DMPC head group phosphodiester and tertiary amino groups, respectively. (C) Representative frames from the other nine restrained MD trajectories, illustrating variations in TMD tilt, conformational disorder in the JMDs, and the presence of a membrane reentrant segment (LWF) in the cytosolic C-terminal domain.
To gain further insight into the structure and membrane interactions of the Notch-TMD, each of the 10 NMR-determined XPLOR-NIH structures was solvated in a disordered phase dimyristoylphosphatidylcholine (DMPC) bilayer and subjected to restrained molecular dynamics (rMD) simulations. The resulting 10 representative structures from these trajectories are shown in Fig. 2 (B and C), with corresponding PROCHECK results in table S1. The key features of the XPLOR-NIH structures were maintained during the rMD simulations, but additional insight was gleaned. Although the rMD structures retain the α-helicity of the TMD, it is observed that the helix does have a tendency to be transiently underwound at Leu1747, shifting the helical register for the entire lower end of the TMD. The reentrant loop in the C-terminal JMD was maintained throughout the simulations. It is also observed that the TMD has a tendency to tilt with respect to the DMPC bilayer normal. The rMD results also provide insight into residue-specific access by lipid chains and water (Fig. 3). There is, of course, a high level of contact between TMD (residues 1732 to 1756) sites and lipid chains in the TMD (Fig. 3A). A slight shoulder in acyl chain exposure is seen on the N-terminal side of the TMD, extending from residues Pro1731 to Phe1737, which indicates that these residues remain close to the membrane surface during the course of the simulation. As expected, this plot also shows increased contact with lipid acyl chains for the cytosolic JMD reentrant loop (Leu-Trp-Phe; residues 1767 to 1769). The results for water contact show that the TM segment (residues 1732 to 1754) is nearly totally devoid of water (Fig. 3B), with the exceptions of sites 1750 and 1753, which are near the initial γ-secretase cleavage site at 1754.
Fig. 3. Water and lipid contacts for the Notch-TMD.
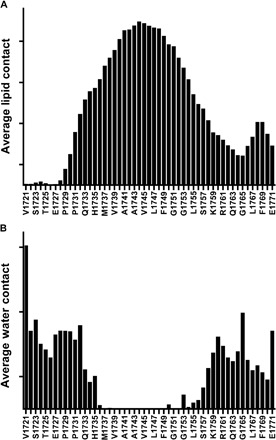
Analysis of the observed frequency of lipid side-chain (A) and water (B) contacts with Notch-TMD side chains in rMD simulations of the Notch-TMD in DMPC bilayers (compilation of results for the final 40 ns of all 10 simulations).
Impact of varying membrane thickness on the Notch-TMD structure and topology
Topological changes induced by changes in membrane thickness as a consequence of varying lipid composition may regulate γ-secretase cleavage of both C99 and Notch. We previously used bicelles containing lipids with different chain lengths and chemical structures (glycerolipids versus sphingolipids) to probe how the conformation and topology of C99 depend on bicelle composition (43). This led us to conduct analogous studies of the Notch-TMD. We examined the Notch-TMD in three sets of bicelles. The first condition was DMPC bicelles in which the lipid component has two relatively short 14-carbon chains; the second was egg sphingomyelin (ESM) bicelles, dominated by sphingomyelin that contains two 16-carbon chains; and the third was milk sphingomyelin (MSM) bicelles, in which the sphingomyelin has the usual C16 ceramide chain plus a fatty amide chain that is usually 22 to 24 carbons long. Transverse relaxation-optimized spectroscopy (TROSY) NMR spectra of the Notch-TMD were collected in these three bicelle types in the presence and absence of water- or lipid-soluble paramagnetic probes, where peak intensity ratios were measured relative to the corresponding diamagnetic reference sample. We also measured backbone 15N chemical shifts for the protein in these three bicelle types.
The site-specific backbone amide 15N chemical shifts for the Notch-TMD (Fig. 4) are nearly identical under all three sets of bicelle conditions, indicating that the Notch-TMD is conformationally tolerant of bicelles containing either different classes of phospholipids (phosphatidylcholine versus sphingomyelin) or lipid components with tail lengths ranging from C14 to C24. Similar conformational robustness was previously observed for C99 (43).
Fig. 4. Comparison of site-specific amide backbone 15N NMR chemical shifts for the Notch-TMD as a function of the lipid used to form the bicelles in each of the three samples.
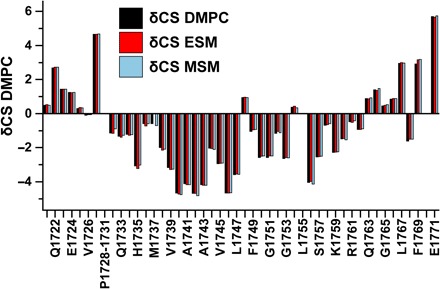
This plot shows the differences between the observed chemical shifts for each site minus the random coil chemical shift value for that amino acid. DMPC has C14 chains. ESM has one C16 chain as part of ceramide and a second usually C16 chain at the fatty amide position. MSM has the ceramide C16 and a usually C22-to-C24 chain at the fatty amide position.
The paramagnetic probe experiments revealed modest differences in model membrane interactions for the Notch-TMD under the three sets of conditions (Fig. 5). There are no significant accessibility pattern differences at the C-terminal (intracellular) end of the TM helix in all three types of bicelles tested. As with C99, a cluster of basic residues (that is, Arg-Lys-Arg-Arg-Arg) terminates the C terminus of the Notch transmembrane segment. These residues serve as an abrupt TM-stop topological motif. Notably, only slight topological differences are seen under the three bicelle conditions for the N-terminal end of the TMD and the N-terminal juxtamembrane domain. This is in stark contrast to C99 (43), where the N-terminal end of the TMD exhibited significant variations in probe accessibility because the lipid component of the bicelles was changed from having relatively short (C14) acyl chains (DMPC) to having long (C22 to C24) chains (MSM).
Fig. 5. Access of the Notch-TMD backbone amide protons to water-soluble (Gd-DTPA) and lipophilic (16-DSA) paramagnetic probes in bicelles containing lipids of different types and chain lengths.
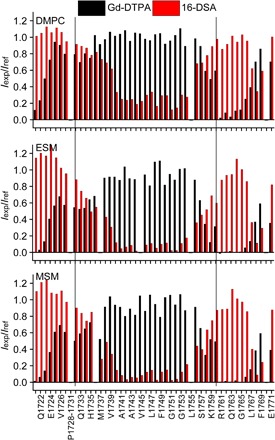
Short chain, DMPC; medium chain, ESM; long chain, MSM. Iexp/Iref is the ratio of the NMR peak intensities for each site in the presence of the probe versus the intensity for that same site under matched diamagnetic conditions. The solid vertical lines indicate the length of the α helix that spans the TMD. The DMPC accessibility plots represent the average of two matched trials, one of which was previously reported (40).
The q = 0.33 dihexanoylphosphatidylcholine (DHPC)–DMPC, DHPC-ESM, and DHPC-MSM bicelles used in this work are rather detergent-rich (three DHPC detergent molecules for each lipid). The q = 0.33 DHPC-DMPC bicelles have been extensively characterized (44–46), particularly with respect to the question of whether they maintain an ideal “bicelle” morphology (DHPC edge-stabilized DMPC bilayer discs) or whether, at this high DHPC content, the assemblies are better described as mixed micelles in which there is little segregation of the lipid and detergent components. Compelling recent experimental evidence favors the bilayered disc morphology (47). For the DHPC-ESM and DHPC-MSM bicelles, there is little data to support one model or the other, although the longer lipid chains of ESM and MSM relative to DMPC might be expected to confer an even higher energetic preference for formation of bilayered discs rather than mixed micelles. Regardless of the bicelle morphologies, the paramagnetic probe results for the Notch-TMD suggest that, when the hydrophobic span of the model membrane is varied, the identities of the residues located at its water-hydrophobic interfaces remain unchanged. This suggests that the Notch-TMD most likely adapts its topology in a real membrane to varying bilayer width by varying its tilt angle with respect to the bilayer normal to maintain the same transmembrane span of residues. This is consistent with the tilting seen for the Notch-TMD in most frames of the rMD DMPC bilayer simulations (Fig. 2, B and C). We postulate that the degree of TMD tilt is determined by the energetic imperative to maintain a roughly constant balance of membrane versus water exposure for N-terminal interfacial residues regardless of bilayer width.
Cholesterol titration of the Notch-TMD in bicelles
There is much evidence that cholesterol promotes AD. Whereas this phenomenon likely involves multiple mechanisms (48, 49), one seems to be the direct promotion of the amyloidogenic pathway (50, 51). The immediate precursor of Aβ, the transmembrane C99 domain of the APP, has been shown to form a 1:1 complex with cholesterol at physiologically relevant cholesterol concentrations (30, 52). The formation of this complex has been proposed to promote γ-secretase release of Aβ from C99, possibly by promoting partitioning of C99 into lipid rafts, where γ-secretase is enriched (53, 54). The role cholesterol plays in Notch biology/biochemistry has received little attention, possibly because any such role is limited. To test whether the Notch-TMD binds cholesterol, we monitored a cholesterol titration of the protein. Five bicelle samples were prepared, spanning a range of cholesterol concentrations ranging from 0 mole percent (mol %) to 15 mol % (mol % protein relative to moles of lipid). NMR spectra were collected at each titration point and compared to see whether there is any evidence for specific cholesterol binding. The spectral overlay is shown in Fig. 6. In contrast to previous results for C99 (30), only very small Notch-TMD resonance shifts were seen. After plotting the chemical shift changes relative to mole percent cholesterol, plots for the peaks undergoing minor shifts were usually linear, consistent only with very weak binding or completely nonspecific interaction of the Notch-TMD with cholesterol.
Fig. 6. Superimposed 900-MHz TROSY spectra from a cholesterol titration of the Notch-TMD demonstrate an absence of specific association of cholesterol with the Notch-TMD.
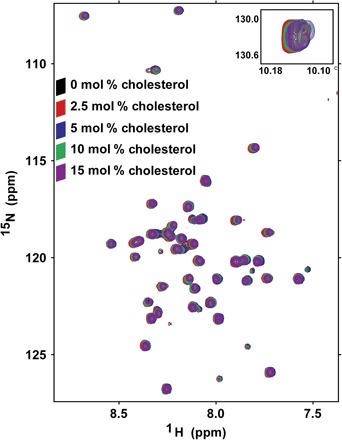
NMR samples were prepared with increasing concentrations of cholesterol (0, 2.5, 5, 10, and 15 mol %). Samples contained ~0.35 mM protein in 15% (w/v) DMPC/DHPC bicelles. There are no significant chemical shift perturbations that suggest direct cholesterol binding [compare to results for C99 (30)]. Moreover, even the modest shifts seen for some residues vary linearly with cholesterol concentration rather than showing any sign of saturation of binding.
DISCUSSION
The structure of bicelle-associated Notch-TMD fills in an important gap in the formidable body of structural data for the Notch 1 receptor (20–25). Given that Notch shares with C99 the property of being subject to intramembrane cleavage by γ-secretase, the results of this work also enable the first compare-and-contrast study of γ-secretase substrates. Accordingly, this study may shed light on the broad substrate specificity of γ-secretase and may also reveal differences between these two proteins that can be exploited in the development of Notch-sparing modulators or inhibitors of C99 cleavage.
Implications of the Notch-TMD structure for Notch signaling and for recognition by γ-secretase
The structural properties of the Notch-TMD domain are likely well adapted for roles in the Notch signaling pathway. The extracellular juxtamembrane region is largely disordered except for the unusual tetraproline motif, leading to the TMD. It seems likely that the disordered linker (Val1721 to Glu1727) and the all-trans tetraproline motif (Pro1728 to Pro1731) serve to help absorb the strain that the TMD might otherwise experience during the forced extension of the extracellular NRR domain (Fig. 1) when the Notch ectodomain engages with a membrane-anchored ligand on an adjacent cell, followed by endocytosis of the bound ligand. The extracellular N terminus of C99 is similar to that of the Notch juxtamembrane ectodomain in its disorder, but the two proteins differ in how the N terminus is linked to the TMD. C99 has a short surface-associated amphipathic helix followed by a water-exposed connecting loop to the TMD (30), whereas the disordered N terminus of Notch is linked to the TMD through a tetraproline segment that adopts a type II polyproline helix. These observations are consistent with the notion that γ-secretase senses the distance of the N termini of potential substrates from the membrane surface, engaging only proteins with short extracellular domains. However, γ-secretase may exhibit little specificity in terms of what substrate juxtamembrane conformations are allowed.
Within the cytosolic juxtamembrane domain of Notch, membrane surface association of the Leu-Trp-Phe1769 segment is interesting in light of its overlap with the Trp-Phe-Pro1770 motif, which is known to be essential for the binding of the RBPJ-associated molecule (RAM) domain (Fig. 1) to a transcriptional activation partner CSL (55, 56). Our results suggest that the Trp-Phe-Pro1770 motif may normally be “tucked away” via membrane surface association, making it unavailable for interaction with CSL or other proteins until the entire NICD is released from the membrane by γ-secretase cleavage. In contrast, C99 lacks any membrane-associated segment immediately following its TMD; instead, it has a disordered 40-residue aqueous loop connecting the TMD to a surface-associated amphipathic helix at the C99 C terminus (30). The very different organization of the intracellular domains of the Notch-TMD and C99 is consistent with the notion that γ-secretase recognition of substrates is tolerant of the nature of the intracellular domains of single-span membrane proteins.
The Notch-TMD is not kinked and does not have a cholesterol-binding site
The TMD of Notch is a straight helix. This is in contrast to C99, for which NMR and electron paramagnetic resonance spectroscopy studies have indicated the presence of a kink associated with a di-Gly motif located near the center of the bilayer (30).
The Notch-TMD in bicelles does not bind cholesterol under the same conditions in which C99 is known to bind cholesterol with physiologically relevant affinity to form a 1:1 complex (30, 52). This biochemical difference highlights key structural features of C99 TMD that are missing in the Notch-TMD. First, as a result of the tandem GXXXG “glycine zipper” sequence, C99 has a flat surface on the extracellular half of its TMD that is well suited for binding the cholesterol ring system. Notch is bereft of GXXXG motifs and does not have a flat surface within its TMD. The glycine zipper also accounts for why the C99 TMD exhibits a modest propensity to dimerize (31, 52, 57–59). Although the possibility that the Notch-TMD dimerizes was suggested by TOXCAT experiments (60), dimerization of the Notch-TMD in the bicelle model membranes used in this work was not detected (40). Also critical for cholesterol binding to C99 are polar residues located in the short amphipathic helix and the connecting loop immediately preceding the TMD; these sites provide hydrogen bonds to the hydroxyl head group of cholesterol. The corresponding juxtamembrane ectodomain of the Notch-TMD is very different, with only the tetraproline motif separating its disordered N terminus and the TMD, a motif ill suited for forming hydrogen bonds with cholesterol.
The interaction and importance of cholesterol in AD remains the topic of intense investigation, with more than 2000 relevant papers already published (48–50, 61). Cholesterol is thought to play important roles in AD progression, with its complexation of C99 likely contributing to these roles. The observed lack of interaction between the Notch-TMD and cholesterol is not surprising because most previous studies of specific roles for lipids in Notch signaling and trafficking imply only indirect effects (62–64).
The fact that C99, but not Notch, has a cholesterol-binding site suggests that it may be possible to develop compounds that target the cholesterol-binding site of C99 to specifically reduce its cleavage by γ-secretase without affecting cleavage of the Notch-TMD. It has been reported that a class of compounds referred to as γ-secretase modulators (GSMs) act by binding to the same glycine zipper face of the C99 TMD that is central to cholesterol binding (65). It is now believed that GSMs do not associate with significant affinity to C99 alone but instead bind only as part of a ternary complex with γ-secretase (51, 66, 67). Our observation in this work of the lack of a flat recess on the Notch-TMD may explain why GSMs do not alter cleavage of Notch by γ-secretase: GSMs likely cannot participate in a ternary complex with Notch and γ-secretase because the Notch-TMD lacks the flat recess formed by the Gly zipper sequence motif at one face of the C99 TMD.
C99 and the Notch-TMD are conformationally robust but adjust topologically in different ways to changes in model membrane hydrophobic thickness
It is known that both the rate of total Aβ production and the ratio of Aβ42 to Aβ40 production are dependent on modulation of γ-secretase activity by membrane thickness (68, 69). Moreover, there are persistent reports that γ-secretase preferentially resides in lipid rafts (53, 54), which are generally thicker than corresponding disordered phase membranes. This work illuminates how two different γ-secretase substrates adapt to changes in membrane thickness and lipid type.
NMR was used to show that the Notch-TMD resembles C99 (43) by adopting a conformational state that is remarkably insensitive to changes in bicelle composition, even when the lipid component was changed from the relatively short C14-based phosphatidylcholine (DMPC) to a form of sphingomyelin (MSM), in which the fatty amide chain is usually 22 to 24 carbons long. This result matches corresponding results for C99 (43). This conformational “robustness” for C99 and the Notch-TMD may reflect a general structural property shared by many membrane proteins, which often must maintain a functional conformational state in the face of marked cellular changes in local membrane lipid compositions (51). We suggest that changes in substrate conformation as a function of lipid composition are unlikely to be a major mechanism regulating the rate by which these substrates are cleaved by γ-secretase.
On the other hand, our results suggest that C99 and the Notch-TMD adjust their membrane topologies differently in response to changes in membrane thickness. Both proteins have a membrane domain stop site composed of consecutive basic residues located just C-terminal to their TMD. However, the Notch-TMD adapts to bicelles containing long-chain lipids in a manner such that the residues comprising its buried transmembrane span remain invariant. Although it is not presently possible to state with certainty that the bicelles used in this work are actual bilayered discs rather than classical mixed micelles, this result suggests that the Notch-TMD may tilt in actual biological membranes so as to maintain a constant span of transmembrane residues in the face of varying bilayer thickness. C99 has very different properties. In bicelles of varying dimensions, the N-terminal start of the C99 TM helix seems to remain fixed in position such that when the span of the model membrane changes, the identity of the residue that starts the TMD adjusts—residues that are within the model membrane when it is thick (-Asn698-Lys-Gly-Ala-Ile-Ile703-) extend into the aqueous phase in thinner model membranes (43). This segment is evidently energetically tolerant of being located at different positions along the bilayer normal in the interfacial region.
The different ways Notch and C99 adjust their topologies in response to changes in membrane thickness may regulate how they initially engage with γ-secretase and the location of the subsequent substrate cleavage site. For C99, it is well established that, although γ-secretase often initially cleaves (“ε-cleavage”) after Thr719, the substrate can also be cleaved at Leu720 (70). Following initial cleavage and release of the AICD, it is believed that the enzyme then processively clips tri- and tetrapeptides from the C-terminal end of the TMD until either Aβ40 or Aβ42 is released into the extracellular space, with Aβ40 being released from C99 that was initially cleaved at Thr719. In contrast, Aβ42 is released from C99 that was originally clipped at Leu720 (71). It has been shown that increasing lipid chain lengths to increase the bilayer width results in an increase in the Aβ40/Aβ42 ratio (68). It has also been shown that adding a pair of nonpolar residues to the C-terminal end of the C99 TMD leads to a decreased Aβ40/Aβ42 ratio, whereas adding nonpolar residues to the N-terminal end of C99 had the opposite effect (72). These results are consistent with the notion that the active site of γ-secretase cleavage is fixed with respect to the center of the membrane. In the case of C99, the fact that its TMD span adjusts asymmetrically with respect to the center of the bilayer as membrane thickness changes implies that the preferred γ-secretase cleavage sites in C99 will vary, consistent with the published results summarized above. In the case of Notch, cleavage is also processive, usually starting at Val1754 and proceeding up to Ala1742, at which point the Nβ polypeptide is released into the extracellular space (11, 12). It is known that there is also an alternate initial TM cut site, Leu1755, and that mutating the C-terminal juxtamembrane lysine to arginine tends to favor cleavage at this site (73). This is consistent with the notion that the initial γ-secretase cut site in both Notch and C99 is determined, at least partly, by site position with respect to the center of the bilayer. A testable prediction of this model is that γ-secretase will tend to cleave the wild-type Notch-TMD at the same sites, regardless of bilayer thickness, if its TMD responds to changes in membrane thickness by altering its tilt—an adjustment that does not change relative residue positions with respect to the center of the bilayer.
CONCLUSIONS
The results of this work help fill in an important gap in the structural biology of the Notch receptor and also highlight differences between the Notch-TMD and C99, which include very different conformations, ligand-binding properties, and modes of topological adaptation to varying model membrane thickness, as summarized in Fig. 7. These differences point to the versatility of γ-secretase in terms of being able to recognize a range of type I single-span membrane proteins with significantly different properties. These differences may also help inform the development of compounds that specifically inhibit or modulate C99 cleavage by γ-secretase without interfering with normal healthy processing of Notch. For example, the cholesterol-binding site of C99 can now be explored as the basis for developing high-affinity compounds that, to some degree, mimic cholesterol and should not bind to Notch. In this regard, it is encouraging to note the success of previous work to design polypeptides that specifically target integrin receptor TMDs (74, 75), in part, by exploiting the unique structural features of the GXXXG motifs found in integrin TMDs and those found in C99 but not in Notch.
Fig. 7. Summary of differences between C99 and the Notch-TMD.
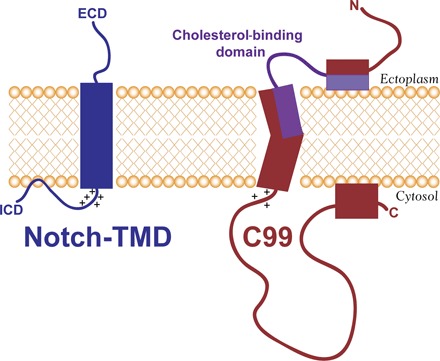
ECD and ICD are the large Notch extracellular and intracellular domains, respectively (see Fig. 1). The general locations of the key residues in the cholesterol binding site of C99 are indicated in purple. Not illustrated here is the propensity of C99, but not the Notch-TMD, to dimerize. Also not illustrated here is the fact that, if the membrane shown in this figure were thinned, the Notch-TMD is predicted by the results of this work to adjust by tilting with respect to the bilayer normal, whereas C99 would remain untilted, with the N-terminal end of its TMD jutting out into the ectoplasm.
MATERIALS AND METHODS
Expression and purification of the N-terminally His6-tagged Notch-TMD for NMR spectroscopy
The Notch-TMD (residues 1721 to 1771; Fig. 1) was expressed and purified into bicelles, as previously described (40), with the solution containing 2% (w/v) DMPC/DHPC bicelles at q = 0.33 (where q equals the lipid-to-detergent mol/mol ratio) in 300 mM imidazole plus 10 mM dithiothreitol (DTT; pH 7.8). The protein-bicelle assemblies were concentrated by centrifugal ultrafiltration, including a 10× buffer exchange cycle, where 15 mM DHPC in 50 mM phosphate and 1 mM EDTA (pH 6.5) was added to a concentrated Notch-TMD solution. The sample was then concentrated 7.5 times to generate approximately 15% bicelles. D2O (10%) was then added, and the pH was reduced to 5.5 using acetic acid, followed by transfer of 180 μl of the solution to a 3-mm NMR tube. Typical final NMR conditions were ~0.5 mM Notch-TMD, 15% DMPC/DHPC bicelles (q = 0.33), 20 mM phosphate, 65 mM imidazole, 2 mM DTT, and 1 mM EDTA (pH 5.5), with 10% D2O. The concentration of the Notch-TMD was spectrophotometrically determined at 280 nm based on its extinction coefficient of 6990 M−1 cm−1.
Side-chain NMR resonance assignments
The U-15N,13C Notch-TMD was prepared for NMR, as previously published (40) and as described above. Backbone resonance assignments from our previous work (40) were also used. Experiments were carried out on a Bruker AV-III 600-MHz spectrometer equipped with inverse detection cryoprobes. Standard Bruker pulse sequences and their hard-coded delays were used for all experiments. Key experiments for the side-chain assignments were the TROSY-(H)C(CO)NH-TOCSY (trhccconhgp3d3) and H(C)(CO)NH (hccconhgp3d2) (76–78). Acquisition for both experiments used a 12-ms DIPSI-2 mixing time for the carbon TOCSY, waltz65 proton decoupling, and garp4 nitrogen decoupling using field strengths of 9.6, 3.6, and 1 kHz, respectively. Relaxation delays were 1.2 s for the TROSY and 1.0 s for the proton-proton experiments. The sweep widths were as follows: 7812 Hz for the direct proton dimension, 1460 Hz for the indirect nitrogen dimensions, and 12074 Hz for the indirect carbon or 5402 Hz for the indirect proton dimension with 2048, 48, 128, and 256 complex points, respectively. Carriers were set to 4.7 parts per million (ppm) (1H), 116.5 ppm (15N), and 40 ppm (13C). Echo/anti-echo detection was used for the nitrogen, and a States-TPPI scheme was used for proton or carbon in the indirect dimension. Water suppression was achieved through gradient coherence selection, with an additional 1-ms sinc-shaped flip-back pulse and a WATERGATE (79) sequence immediately before data acquisition in the carbon TOCSY. To minimize relaxation during acquisition, these elements were omitted in the proton TOCSY, still yielding excellent suppression of all unwanted signal. The carbon TOCSY was acquired with 32 scans per increments, whereas only 16 scans were used for the proton TOCSY, yielding in a total experiment time of 3 days for each experiment.
For the standard Bruker HCCH-TOCSY (hcchdigp3d2) (80, 81), a CPQCI probe was used. The parameters were as follows: Sweep width in the indirect proton dimension was 7812 Hz with 2048 complex data points, and both indirect carbon dimensions were at 9804 Hz and 96 complex points. A DIPSI3 spinlock of 22.6 ms at 9.3-kHz field strength and a 3.9-kHz carbon decoupling field during data acquisition was applied. A States-TPPI scheme was used in both indirect dimensions, using 16 transients with a 1.5-s recycle delay per increment. Water suppression was achieved through gradient coherence selection only. The total experiment time was 70 hours.
All NMR spectra were processed with NMRPipe (82) and analyzed with SPARKY (83). Residual water suppression consisted of a time domain polynomial subtraction. Indirect dimensions were linearly predicted to twice the data size before Gaussian window functions were applied. Polynomial baseline corrections were generally applied in the direct and indirect 15N dimensions. Residue-specific 1H and 13C resonances from the H(CCO)NH- and C(CO)NH-TOCSY experiments were correlated via peaks observed in the HCCH-TOCSY spectrum. Side-chain resonances were assigned by correlations from existing sequential backbone assignments (40). In the few occurrences where 1H-13C–correlated chemical shifts were observed but could not be unambiguously assigned, nuclear Overhauser enhancement spectroscopy (NOESY) data typically resolved ambiguity. The unassigned, although clearly distinguishable, proline chemical shifts in the HCCH-TOCSY spectrum were used to ascertain the presence of all-trans configurations for all prolines based on their Cβ and Cγ chemical shifts (84).
Distance restraints from paramagnetic relaxation enhancement measurements
The wild-type Notch-TMD has one cysteine residue (Cys1752; Fig. 1). For paramagnetic relaxation enhancement (PRE) measurements, samples were prepared in which the native cysteine was spin-labeled. Samples were also prepared where the native Cys1752 was mutated to Ser, and Phe1744 was mutated to Cys and then spin-labeled. Both wild type and the single-Cys mutant were purified into 0.2% n-dodecylphosphocholine (DPC) and spin-labeled using an established method (30, 85). Briefly, each single-Cys form was concentrated to 0.5 mM, and the pH was lowered to 6.5 followed by addition of 2.5 mM DTT to ensure cysteine thiol reduction. The protein was then spin-labeled using the thiol-reactive probe S-(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl methanesulfonothioate (MTSL; Toronto Research Chemicals). A 20-fold excess of MTSL was added relative to ~0.5 mM Notch-TMD [buffer: 65 mM imidazole, 2 mM EDTA, 2.5 mM DTT, and 0.2% DPC (pH 6.5)]. The labeled sample was incubated overnight, followed by buffer exchange with 50 mM sodium phosphate buffer (pH 6.5) to remove the imidazole in preparation for metal ion affinity chromatography. The solution was then mixed overnight with Ni–nitrilotriacetic acid resin, and the resin was washed with 25 column volumes of 50 mM phosphate and 0.2% DPC (pH 7.8) to remove excess MTSL and MTSL-modified DTT. The resin was then rinsed with a 8 × 1 column volume of DHPC in water, followed by 4 × 1 column volume exchange using a 2% (w/v) DMPC/DHPC bicelle mixture (q = 0.33) before the protein was eluted in the same 2% bicelles plus 300 mM imidazole at pH 7.8. The sample was then immediately exchanged into the 20 mM sodium phosphate and 65 mM imidazole NMR buffer, and the pH was reduced to 5.5 using glacial acetic acid.
1H-15N TROSY NMR data were acquired at 900 MHz for the spin-labeled samples. In each case, a spectrum was collected, followed by reduction of the paramagnet and reacquisition of a parameter-matched TROSY spectrum for the now-diamagnetic sample. Reduction of the spin label was carried out by adding ascorbic acid to 20 mM to the sample from a stock solution (pH 5.5). Paired spectra were identically processed using NMRPipe (82) and analyzed in SPARKY (83) to determine the peak intensity ratios from the paramagnetic versus diamagnetic samples. The intensity ratios and the diamagnetic sample linewidths were used to determine distances between the paramagnetic and the backbone amide protons, as previously described (see also below) (30, 85, 86).
Structure calculations
Restraints used in structure calculations included using PRE-derived distances, backbone torsion angles derived from chemical shifts, and NOE distance restraints. Backbone Cα, Cβ, CO, and N chemical shifts (40) were input into TALOS+ to generate dihedral angle restraints (85, 87). Chemical shift index analysis was also used to generate hydrogen bond restraints for the TM segment (88). These backbone dihedral angle and hydrogen bonding restraints were calculated from the backbone chemical shift values (89) and used in structure calculations. The backbone dihedral angles of the all-trans tetraproline motif were initially set to −79° and 149° (42), with generous errors allowed. A three-dimensional (1H,1H,15N)-TROSY-NOESY experiment was carried out on the U-15N Notch-TMD (100-ms mixing time) in DHPC/DMPC bicelles under the standard NMR conditions indicated above to obtain 339 short- and medium-range NOE restraints used in structure calculations. The PRE restraints were very conservatively used, as previously described (39, 85). Briefly, each restraint was classified as “close in space” if the paramagnetic/diamagnetic intensity ratios were less than 0.15, in which case the paramagnetic probe and the target amide proton were assigned as being between 2 and 19 Å apart. Resonances with ratios between 0.15 and 0.85 were converted to explicit distances, as previously described (85, 86), and given generous uncertainties of ±7 Å. Resonances that exhibited little to no PRE effects (intensity ratios greater than 0.85) were loosely restrained to be between 19 and 100 Å apart. The PRE restraints were implemented as NOE-like restraints from the spin label to the backbone amide hydrogen (39, 90).
Structure calculations were conducted using XPLOR-NIH v2.24 (91), similar to previously published works (30, 85). Simulated annealing was carried out with 15,000 steps at 3500 K, with cooling to 100 K. During the temperature-cooling ramp, the van der Waals force constant was varied from 0.004 to 4 kcal mol−1 Å−4. Similarly, force constants were increased for the NOE and PRE restraints from 1.0 to 30.0 kcal mol−1 Å−2. High-temperature simulated annealing was followed by torsion angle and full-atom minimization steps. The lowest energy structures were selected, and the spin-labeled sites associated with the PRE distance restraints were mutated back to their wild-type residue types, followed by energy minimization to allow their side-chain conformations to optimize. Additional details regarding the structure calculation are given in table S1. The 10 XPLOR-NIH models (fig. S1) were deposited in the PDB (code 5KZO).
XPLOR-NIH–based structure determination was followed by AMBER14 rMD to refine the 10 top-scoring Notch-TMD XPLOR-NIH structures in a lipid bilayer. Structures were solvated in an explicit DMPC bilayer, and simulations included the use of NMR-derived distance restraints (85). Simulations used the Lipid14 AMBER lipid force field (92) and the ff12SB AMBER force field. Refinement of each Notch-TMD structure was conducted via a 60-ns production rMD. For each of the 10 trajectories, the average Notch-TMD structure was calculated, and the frame that best matches the average structure was selected as the “representative model” from that trajectory using CPPTRAJ (93). Both the initial X-PLOR structures and the AMBER refinement structures were validated by PROCHECK (see table S1) (41). We also used CPPTRAJ (93) to determine water penetrance and protein-lipid interactions. Additional technical details regarding the rMD calculations are provided in the Supplementary Materials.
Dependence of protein structure and topology on varying bilayer thickness
We have previously described methods to determine paramagnetic probe accessibility to membrane protein sites in bicelles containing lipids of varying chain lengths (43). Briefly, DMPC (C14 chains), ESM (mostly C16), or MSM (mostly C22 to C24) was weighed as a solid into a glass vial. A 2% bicelle solution was generated by adding the appropriate amount of DHPC and water to form a q = 0.33 bicelle mixture that was then used to equilibrate and elute the Notch-TMD from a Ni(II)-resin column. NMR samples of the U-15N Notch-TMD were prepared as described above, concentrated, and split into three parts. The first sample was a paramagnet-free reference sample. The second contained a water-soluble paramagnet, Gd(III)–diethylenetriamine pentaacetic acid [Gd-DTPA; added to 2 mM from a 0.1 M stock solution (pH 5.5)]. The third sample contained the lipophilic paramagnetic probe, 16-doxylstearic acid (16-DSA; 2 mM; Santa Cruz Biotechnology). The 16-DSA sample was prepared by adding an aliquot of 16-DSA stock (2.5 mg/ml) in methanol into a tube and then removing the methanol in a speed vacuum. The protein sample was then added to the dried 16-DSA and gently mixed (to a concentration of 2 mM) before it was transferred into a 3-mm NMR tube. TROSY spectra (900 MHz) were collected for each bicelle condition using a delay time of 4 s between scans to ensure complete relaxation between scans. TROSY peak intensities were measured for the two paramagnet-containing samples and compared to the corresponding peak intensities from the parameter-matched reference sample. The ratios of the peak intensities were plotted as an indicator of site access to the paramagnetic probes.
Cholesterol titration of the Notch-TMD
Samples were prepared essentially as previously described for C99 (30). Briefly, cholesterol-containing bicelles were prepared by dissolving DMPC and cholesterol in chloroform at the desired molar ratio and then removing all solvent by high vacuum, followed by mixing the cholesterol/phospholipid film with DHPC and water to form a 2% (w/v) bicelle solution containing 15 mol % cholesterol (relative to total moles of DHPC + DMPC + cholesterol). This mixture was subjected to freeze-thaw cycles until a clear solution was formed, aliquots of which were then mixed with cholesterol-free bicelles to make a series of q = 0.33 bicelles at 15% (w/v) that contained 0, 2.5, 5, or 10 mol % cholesterol in water. Each of these solutions was used to equilibrate (with no imidazole) and then elute (with 300 mM imidazole) the Notch-TMD from the resin. Samples were than prepared for NMR, as described above, and contained 0.35 mM protein. A 15N-1H TROSY was collected at 318 K for each cholesterol titration point on a Bruker AV-III 900-MHz spectrometer equipped with a CPTCI cryogenic probe.
Supplementary Material
Acknowledgments
Funding: This work was supported by NIH grants RO1 GM106672 and DK069921 as well as by an NIH postdoctoral fellowship to B.M.K. (F32 GM113355). The NMR instrumentation used in this work was supported by NIH S10 RR026677 and NSF DBI-0922862. Author contributions: C.L.D., Z.L., B.M.K., S.M., J.A.S., M.W.V., and R.L.M. conducted the experiments and calculations of this work. All authors participated in data analysis. C.L.D., R.L.M., and C.R.S. wrote the manuscript with input from all authors. C.L.D., B.M.K., R.L.M., and C.R.S. conceived this work and directed the approaches used. Competing interests: The authors declare that they have no competing interests. Data and materials availability: The atomic coordinates of the Notch-TMD structure have been deposited in the PDB under the accession code 5KZO. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors. Correspondence and requests for materials should be addressed to C.R.S. (chuck.sanders@vanderbilt.edu)
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/3/4/e1602794/DC1
Supplementary Methods
fig. S1. Top 20 NMR-determined XPLOR-NIH structures of the Notch-TMD.
table S1. Statistics for Notch-TMD structure determination and structure quality for the 10 final XPLOR-NIH NMR structures and for the 10 representative restrained MD structures.
REFERENCES AND NOTES
- 1.Lanford P. J., Lan Y., Jiang R., Lindsell C., Weinmaster G., Gridley T., Kelley M. W., Notch signalling pathway mediates hair cell development in mammalian cochlea. Nat. Genet. 21, 289–292 (1999). [DOI] [PubMed] [Google Scholar]
- 2.Robey E., Chang D., Itano A., Cado D., Alexander H., Lans D., Weinmaster G., Salmon P., An activated form of Notch influences the choice between CD4 and CD8 T cell lineages. Cell 87, 483–492 (1996). [DOI] [PubMed] [Google Scholar]
- 3.Wahi K., Bochter M. S., Cole S. E., The many roles of Notch signaling during vertebrate somitogenesis. Semin. Cell Dev. Biol. 49, 68–75 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Guruharsha K. G., Kankel M. W., Artavanis-Tsakonas S., The Notch signalling system: Recent insights into the complexity of a conserved pathway. Nat. Rev. Genet. 13, 654–666 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huppert S. S., Le A., Schroeter E. H., Mumm J. S., Saxena M. T., Milner L. A., Kopan R., Embryonic lethality in mice homozygous for a processing-deficient allele of Notch1. Nature 405, 966–970 (2000). [DOI] [PubMed] [Google Scholar]
- 6.Lowell S., Benchoua A., Heavey B., Smith A. G., Notch promotes neural lineage entry by pluripotent embryonic stem cells. PLOS Biol. 4, e121 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bigas A., D’Altri T., Espinosa L., The Notch pathway in hematopoietic stem cells. Curr. Top. Microbiol. Immunol. 360, 1–18 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Kopan R., Ilagan M. X. G., The canonical Notch signaling pathway: Unfolding the activation mechanism. Cell 137, 216–233 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bray S. J., Notch signalling: A simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 7, 678–689 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Ables J. L., Breunig J. J., Eisch A. J., Rakic P., Not(ch) just development: Notch signalling in the adult brain. Nat. Rev. Neurosci. 12, 269–283 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chandu D., Huppert S. S., Kopan R., Analysis of transmembrane domain mutants is consistent with sequential cleavage of Notch by γ-secretase. J. Neurochem. 96, 228–235 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Okochi M., Fukumori A., Jiang J., Itoh N., Kimura R., Steiner H., Haass C., Tagami S., Takeda M., Secretion of the Notch-1 Aβ-like peptide during Notch signaling. J. Biol. Chem. 281, 7890–7898 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Fortini M. E., Notch signaling: The core pathway and its posttranslational regulation. Dev. Cell 16, 633–647 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Jorissen E., De Strooper B., γ-Secretase and the intramembrane proteolysis of Notch. Curr. Top. Dev. Biol. 92, 201–230 (2010). [DOI] [PubMed] [Google Scholar]
- 15.Kovall R. A., More complicated than it looks: Assembly of Notch pathway transcription complexes. Oncogene 27, 5099–5109 (2008). [DOI] [PubMed] [Google Scholar]
- 16.Kitagawa M., Notch signalling in the nucleus: Roles of Mastermind-like (MAML) transcriptional coactivators. J. Biochem. 159, 287–294 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Wu L., Aster J. C., Blacklow S. C., Lake R., Artavanis-Tsakonas S., Griffin J. D., MAML1, a human homologue of Drosophila Mastermind, is a transcriptional co-activator for NOTCH receptors. Nat. Genet. 26, 484–489 (2000). [DOI] [PubMed] [Google Scholar]
- 18.Kageyama R., Ohtsuka T., The Notch-Hes pathway in mammalian neural development. Cell Res. 9, 179–188 (1999). [DOI] [PubMed] [Google Scholar]
- 19.Borggrefe T., Lauthb M., Zwijsenc A., Huylebroeckd D., Oswalde F., Giaimoa B. D., The Notch intracellular domain integrates signals from Wnt, Hedgehog, TGFβ/BMP and hypoxia pathways. Biochim. Biophys. Acta 1863, 303–313 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Gordon W. R., Arnett K. L., Blacklow S. C., The molecular logic of Notch signaling–A structural and biochemical perspective. J. Cell Sci. 121, 3109–3119 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kovall R. A., Blacklow S. C., Mechanistic insights into Notch receptor signaling from structural and biochemical studies. Curr. Top. Dev. Biol. 92, 31–71 (2010). [DOI] [PubMed] [Google Scholar]
- 22.Chillakuri C. R., Sheppard D., Lea S. M., Handford P. A., Notch receptor–ligand binding and activation: Insights from molecular studies. Semin. Cell Dev. Biol. 23, 421–428 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luca V. C., Jude K. M., Pierce N. W., Nachury M. V., Fischer S., Garcia K. C., Structural biology. Structural basis for Notch1 engagement of Delta-like 4. Science 347, 847–853 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu X., Choi S. H., Hu T., Tiyanont K., Habets R., Groot A. J., Vooijs M., Aster J. C., Chopra R., Fryer C., Blacklow S. C., Insights into autoregulation of Notch3 from structural and functional studies of its negative regulatory region. Structure 23, 1227–1235 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weisshuhn P. C., Sheppard D., Taylor P., Whiteman P., Lea S. M., Handford P. A., Redfield C., Non-linear and flexible regions of the human Notch1 extracellular domain revealed by high-resolution structural studies. Structure 24, 555–566 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Selkoe D. J., Hardy J., The amyloid hypothesis of Alzheimer’s disease at 25 years. EMBO Mol. Med. 8, 595–608 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nunan J., Small D. H., Regulation of APP cleavage by α-, β- and γ-secretases. FEBS Lett. 483, 6–10 (2000). [DOI] [PubMed] [Google Scholar]
- 28.De Strooper B., Chávez Gutiérrez L., Learning by failing: Ideas and concepts to tackle γ-secretases in Alzheimer’s disease and beyond. Annu. Rev. Pharmacol. Toxicol. 55, 419–437 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Doody R. S., Raman R., Farlow M., Iwatsubo T., Vellas B., Joffe S., Kieburtz K., He F., Sun X., Thomas R. G., Aisen P. S.; Alzheimer’s Disease Cooperative Study Steering Committee, Siemers E., Sethuraman G., Mohs R.; Semagacestat Study Group , A phase 3 trial of semagacestat for treatment of Alzheimer’s disease. N. Engl. J. Med. 369, 341–350 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Barrett P. J., Song Y., Van Horn W. D., Hustedt E. J., Schafer J. M., Hadziselimovic A., Beel A. J., Sanders C. R., The amyloid precursor protein has a flexible transmembrane domain and binds cholesterol. Science 336, 1168–1171 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dominguez L., Foster L., Meredith S. C., Straub J. E., Thirumalai D., Structural heterogeneity in transmembrane amyloid precursor protein homodimer is a consequence of environmental selection. J. Am. Chem. Soc. 136, 9619–9626 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lemmin T., Dimitrov M., Fraering P. C., Dal Peraro M., Perturbations of the straight transmembrane α-helical structure of the amyloid precursor protein affect its processing by γ-secretase. J. Biol. Chem. 289, 6763–6774 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nadezhdin K. D., Bocharova O. V., Bocharov E. V., Arseniev A. S., Structural and dynamic study of the transmembrane domain of the amyloid precursor protein. Acta Nat. 3, 69–76 (2011). [PMC free article] [PubMed] [Google Scholar]
- 34.Nierzwicki Ł., Czub J., Specific binding of cholesterol to the amyloid precursor protein: Structure of the complex and driving forces characterized in molecular detail. J. Phys. Chem. Lett. 6, 784–790 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Pester O., Barrett P. J., Hornburg D., Hornburg P., Pröbstle R., Widmaier S., Kutzner C., Dürrbaum M., Kapurniotu A., Sanders C. R., Scharnagl C., Langosch D., The backbone dynamics of the amyloid precursor protein transmembrane helix provides a rationale for the sequential cleavage mechanism of γ-secretase. J. Am. Chem. Soc. 135, 1317–1329 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sato T., Tang T.-c., Reubins G., Fei J. Z., Fujimoto T., Kienlen-Campard P., Constantinescu S. N., Octave J.-N., Aimoto S., Smith S. O., A helix-to-coil transition at the ε-cut site in the transmembrane dimer of the amyloid precursor protein is required for proteolysis. Proc. Natl. Acad. Sci. U.S.A. 106, 1421–1426 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scharnagl C., Pester O., Hornburg P., Hornburg D., Götz A., Langosch D., Side-chain to main-chain hydrogen bonding controls the intrinsic backbone dynamics of the amyloid precursor protein transmembrane helix. Biophys. J. 106, 1318–1326 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen W., Gamache E., Richardson D., Du Z., Wang C., Expression, purification, and reconstitution of the transmembrane domain of the human amyloid precursor protein for NMR studies. Protein Expr. Purif. 81, 11–17 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Kim H. J., Howell S. C., Van Horn W. D., Jeon Y. H., Sanders C. R., Recent advances in the application of solution NMR spectroscopy to multi-span integral membrane proteins. Prog. Nucl. Magn. Reson. Spectrosc. 55, 335–360 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deatherage C. L., Lu Z., Kim J.-H., Sanders C. R., Notch transmembrane domain: Secondary structure and topology. Biochemistry 54, 3565–3568 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laskowski R. A., Rullmannn J. A. C., MacArthur M. W., Kaptein R., Thornton J. M., AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8, 477–486 (1996). [DOI] [PubMed] [Google Scholar]
- 42.Adzhubei A. A., Sternberg M. J. E., Makarov A. A., Polyproline-II helix in proteins: Structure and function. J. Mol. Biol. 425, 2100–2132 (2013). [DOI] [PubMed] [Google Scholar]
- 43.Song Y., Mittendorf K. F., Lu Z., Sanders C. R., Impact of bilayer lipid composition on the structure and topology of the transmembrane amyloid precursor C99 protein. J. Am. Chem. Soc. 136, 4093–4096 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barbosa-Barros L., Rodríguez G., Barba C., Cócera M., Rubio L., Estelrich J., López-Iglesias C., de la Maza A., López O., Bicelles: Lipid nanostructured platforms with potential dermal applications. Small 8, 807–818 (2012). [DOI] [PubMed] [Google Scholar]
- 45.Dürr U. H. N., Gildenberg M., Ramamoorthy A., The magic of bicelles lights up membrane protein structure. Chem. Rev. 112, 6054–6074 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prosser R. S., Evanics F., Kitevski J. L., Al-Abdul-Wahid M. S., Current applications of bicelles in NMR studies of membrane-associated amphiphiles and proteins. Biochemistry 45, 8453–8465 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Mineev K. S., Nadezhdin K. D., Goncharuk S. A., Arseniev A. S., Characterization of small isotropic bicelles with various compositions. Langmuir 32, 6624–6637 (2016). [DOI] [PubMed] [Google Scholar]
- 48.Carter C. J., Convergence of genes implicated in Alzheimer’s disease on the cerebral cholesterol shuttle: APP, cholesterol, lipoproteins, and atherosclerosis. Neurochem. Int. 50, 12–38 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Giudetti A. M., Romano A., Lavecchia A. M., Gaetani S., The role of brain cholesterol and its oxidized products in Alzheimer’s disease. Curr. Alzheimer Res. 13, 198–205 (2016). [DOI] [PubMed] [Google Scholar]
- 50.Maulik M., Westaway D., Jhamandas J. H., Kar S., Role of cholesterol in APP metabolism and its significance in Alzheimer’s disease pathogenesis. Mol. Neurobiol. 47, 37–63 (2013). [DOI] [PubMed] [Google Scholar]
- 51.Barrett P. J., Sanders C. R., Kaufman S. A., Michelsen K., Jordan J. B., NSAID-based γ-secretase modulators do not bind to the amyloid-β polypeptide. Biochemistry 50, 10328–10342 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Song Y., Hustedt E. J., Brandon S., Sanders C. R., Competition between homodimerization and cholesterol binding to the C99 domain of the amyloid precursor protein. Biochemistry 52, 5051–5064 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Araki W., Tamaoka A., Amyloid beta-protein and lipid rafts: Focused on biogenesis and catabolism. Front. Biosci. 20, 314–324 (2015). [DOI] [PubMed] [Google Scholar]
- 54.Beel A. J., Sakakura M., Barrett P. J., Sanders C. R., Direct binding of cholesterol to the amyloid precursor protein: An important interaction in lipid–Alzheimer’s disease relationships? Biochim. Biophys. Acta 1801, 975–982 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Le Gall M., Giniger E., Identification of two binding regions for the suppressor of hairless protein within the intracellular domain of Drosophila notch. J. Biol. Chem. 279, 29418–29426 (2004). [DOI] [PubMed] [Google Scholar]
- 56.Lubman O. Y., Ilagan M. X. G., Kopan R., Barrick D., Quantitative dissection of the Notch:CSL interaction: Insights into the Notch-mediated transcriptional switch. J. Mol. Biol. 365, 577–589 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nadezhdin K. D., Bocharova O. V., Bocharov E. V., Arseniev A. S., Dimeric structure of transmembrane domain of amyloid precursor protein in micellar environment. FEBS Lett. 586, 1687–1692 (2012). [DOI] [PubMed] [Google Scholar]
- 58.Kienlen-Campard P., Tasiaux B., Van Hees J., Li M., Huysseune S., Sato T., Fei J. Z., Aimoto S., Courtoy P. J., Smith S. O., Constantinescu S. N.; A Research Associate of the National Fund for Scientific Research, Belgium, Octave J.-N., Amyloidogenic processing but not amyloid precursor protein (APP) intracellular C-terminal domain production requires a precisely oriented APP dimer assembled by transmembrane GXXXG motifs. J. Biol. Chem. 283, 7733–7744 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Decock M., El Haylani L., Stanga S., Dewachter I., Octave J. N., Smith S. O., Constantinescu S. N., Kienlen-Campard P., Analysis by a highly sensitive split luciferase assay of the regions involved in APP dimerization and its impact on processing. FEBS Open Bio 5, 763–773 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vooijs M., Schroeter E. H., Pan Y., Blandford M., Kopan R., Ectodomain shedding and intramembrane cleavage of mammalian Notch proteins are not regulated through oligomerization. J. Biol. Chem. 279, 50864–50873 (2004). [DOI] [PubMed] [Google Scholar]
- 61.Martins I. J., Berger T., Sharman M. J., Verdile G., Fuller S. J., Martins R. N., Cholesterol metabolism and transport in the pathogenesis of Alzheimer’s disease. J. Neurochem. 111, 1275–1308 (2009). [DOI] [PubMed] [Google Scholar]
- 62.Chapman G., Major J. A., Iyer K., James A. C., Pursglove S. E., Moreau J. L. M., Dunwoodie S. L., Notch1 endocytosis is induced by ligand and is required for signal transduction. Biochim. Biophys. Acta 1863, 166–177 (2016). [DOI] [PubMed] [Google Scholar]
- 63.Shimizu H., Woodcock S. A., Wilkin M. B., Trubenová B., Monk N. A. M., Baron M., Compensatory flux changes within an endocytic trafficking network maintain thermal robustness of Notch signaling. Cell 157, 1160–1174 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Briot A., Bouloumié A., Iruela-Arispe M. L., Notch, lipids, and endothelial cells. Curr. Opin. Lipidol. 27, 513–520 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kukar T. L., Ladd T. B., Bann M. A., Fraering P. C., Narlawar R., Maharvi G. M., Healy B., Chapman R., Welzel A. T., Price R. W., Moore B., Rangachari V., Cusack B., Eriksen J., Jansen-West K., Verbeeck C., Yager D., Eckman C., Ye W., Sagi S., Cottrell B. A., Torpey J., Rosenberry T. L., Fauq A., Wolfe M. S., Schmidt B., Walsh D. M., Koo E. H., Golde T. E., Substrate-targeting γ-secretase modulators. Nature 453, 925–929 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Crump C. J., Johnson D. S., Li Y.-M., Development and mechanism of γ-secretase modulators for Alzheimer’s disease. Biochemistry 52, 3197–3216 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Beel A. J., Barrett P., Schnier P. D., Hitchcock S. A., Bagal D., Sanders C. R., Jordan J. B., Nonspecificity of binding of γ-secretase modulators to the amyloid precursor protein. Biochemistry 48, 11837–11839 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Holmes O., Paturi S., Ye W., Wolfe M. S., Selkoe D. J., Effects of membrane lipids on the activity and processivity of purified γ-secretase. Biochemistry 51, 3565–3575 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Winkler E., Kamp F., Scheuring J., Ebke A., Fukumori A., Steiner H., Generation of Alzheimer disease-associated amyloid β42/43 peptide by γ-secretase can be inhibited directly by modulation of membrane thickness. J. Biol. Chem. 287, 21326–21334 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Takami M., Funamoto S., γ-Secretase-dependent proteolysis of transmembrane domain of amyloid precursor protein: Successive tri- and tetrapeptide release in amyloid β-protein production. Int. J. Alzheimers Dis. 2012, 591392 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bolduc D. M., Montagna D. R., Seghers M. C., Wolfe M. S., Selkoe D. J., The amyloid-beta forming tripeptide cleavage mechanism of γ-secretase. eLife 5, e17578 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lichtenthaler S. F., Beher D., Grimm H. S., Wang R., Shearman M. S., Masters C. L., Beyreuther K., The intramembrane cleavage site of the amyloid precursor protein depends on the length of its transmembrane domain. Proc. Natl. Acad. Sci. U.S.A. 99, 1365–1370 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tagami S., Okochi M., Yanagida K., Ikuta A., Fukumori A., Matsumoto N., Ishizuka-Katsura Y., Nakayama T., Itoh N., Jiang J., Nishitomi K., Kamino K., Morihara T., Hashimoto R., Tanaka T., Kudo T., Chiba S., Takeda M., Regulation of Notch signaling by dynamic changes in the precision of S3 cleavage of Notch-1. Mol. Cell. Biol. 28, 165–176 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Caputo G. A., Litvinov R. I., Li W., Bennett J. S., DeGrado W. F., Yin H., Computationally designed peptide inhibitors of protein−protein interactions in membranes. Biochemistry 47, 8600–8606 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yin H., Flynn A. D., Drugging membrane protein interactions. Annu. Rev. Biomed. Eng. 18, 51–76 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Grzesiek S., Anglister J., Bax A., Correlation of backbone amide and aliphatic side-chain resonances in 13C/15N-enriched proteins by isotropic mixing of 13C magnetization. J. Magn. Reson. B 101, 114–119 (1993). [Google Scholar]
- 77.Montelione G. T., Lyons B. A., Emerson S. D., Tashiro M., An efficient triple resonance experiment using carbon-13 isotropic mixing for determining sequence-specific resonance assignments of isotopically-enriched proteins. J. Am. Chem. Soc. 114, 10974–10975 (1992). [Google Scholar]
- 78.Logan T. M., Olejniczak E. T., Xu R. X., Fesik S. W., A general method for assigning NMR spectra of denatured proteins using 3D HC(CO)NH-TOCSY triple resonance experiments. J. Biomol. NMR 3, 225–231 (1993). [DOI] [PubMed] [Google Scholar]
- 79.Piotto M., Saudek V., Sklenář V., Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR 2, 661–665 (1992). [DOI] [PubMed] [Google Scholar]
- 80.Bax A., Clore G. M., Gronenborn A. M., 1H-1H correlation via isotropic mixing of 13C magnetization, a new three-dimensional approach for assigning 1H and 13C spectra of 13C-enriched proteins. J. Magn. Reson. 88, 425–431 (1990). [Google Scholar]
- 81.Olejniczak E. T., Xu R. X., Fesik S. W., A 4D HCCH-TOCSY experiment for assigning the side chain 1H and 13C resonances of proteins. J. Biomol. NMR 2, 655–659 (1992). [DOI] [PubMed] [Google Scholar]
- 82.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A., NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995). [DOI] [PubMed] [Google Scholar]
- 83.T. D. Goddard, D. G. Kneller, SPARKY 3 (University of California, 2008).
- 84.Schubert M., Labudde D., Oschkinat H., Schmieder P., A software tool for the prediction of Xaa-Pro peptide bond conformations in proteins based on 13C chemical shift statistics. J. Biomol. NMR 24, 149–154 (2002). [DOI] [PubMed] [Google Scholar]
- 85.Kroncke B. M., Van Horn W. D., Smith J., Kang C., Welch R. C., Song Y., Nannemann D. P., Taylor K. C., Sisco N. J., George A. L. Jr, Meiler J., Vanoye C. G., Sanders C. R., Structural basis for KCNE3 modulation of potassium recycling in epithelia. Sci. Adv. 2, e1501228 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Battiste J. L., Wagner G., Utilization of site-directed spin labeling and high-resolution heteronuclear nuclear magnetic resonance for global fold determination of large proteins with limited nuclear Overhauser effect data. Biochemistry 39, 5355–5365 (2000). [DOI] [PubMed] [Google Scholar]
- 87.Shen Y., Delaglio F., Cornilescu G., Bax A., TALOS+: A hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR 44, 213–223 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wishart D. S., Sykes B. D., Chemical shifts as a tool for structure determination. Methods Enzymol. 239, 363–392 (1994). [DOI] [PubMed] [Google Scholar]
- 89.Shen Y., Bax A., Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. J. Biomol. NMR 56, 227–241 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kang C., Tian C., Sönnichsen F. D., Smith J. A., Meiler J., George A. L. Jr, Vanoye C. G., Kim H. J., Sanders C. R., Structure of KCNE1 and implications for how it modulates the KCNQ1 potassium channel. Biochemistry 47, 7999–8006 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schwieters C. D., Kuszewski J. J., Tjandra N., Clore G. M., The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 160, 65–73 (2003). [DOI] [PubMed] [Google Scholar]
- 92.Dickson C. J., Madej B. D., Skjevik A. A., Betz R. M., Teigen K., Gould I. R., Walker R. C., Lipid14: The Amber lipid force field. J. Chem. Theory Comput. 10, 865–879 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Roe D. R., Cheatham T. E. III, PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 9, 3084–3095 (2013). [DOI] [PubMed] [Google Scholar]
- 94.Wu E. L., Cheng X., Jo S., Rui H., Song K. C., Dávila-Contreras E. M., Qi Y., Lee J., Monje-Galvan V., Venable R. M., Klauda J. B., Im W., CHARMM-GUI Membrane Builder toward realistic biological membrane simulations. J. Comput. Chem. 35, 1997–2004 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jo S., Kim T., Iyer V. G., Im W., CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865 (2008). [DOI] [PubMed] [Google Scholar]
- 96.Case D. A., Cheatham T. E. III, Darden T., Gohlke H., Luo R., Merz K. M. Jr, Onufriev A., Simmerling C., Wang B., Woods R. J., The Amber biomolecular simulation programs. J. Comput. Chem. 26, 1668–1688 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/3/4/e1602794/DC1
Supplementary Methods
fig. S1. Top 20 NMR-determined XPLOR-NIH structures of the Notch-TMD.
table S1. Statistics for Notch-TMD structure determination and structure quality for the 10 final XPLOR-NIH NMR structures and for the 10 representative restrained MD structures.


