Abstract
The incidence of whooping cough, a contagious respiratory disease caused by Bordetella pertussis, is on the rise despite existing vaccination programmes. Similar, though usually milder, respiratory symptoms may be caused by other members of the Bordetella genus: B. parapertussis, B. holmesii, and B. bronchiseptica. Pertussis diagnosis is mostly done using PCR, but the use of multiple targets is necessary in order to differentiate the different Bordetella spp. with sufficient sensitivity and specificity. In this study we evaluate a multiplex PCR assay for the differentiation of B. pertussis from other Bordetella spp., using the targets IS481, IS1001, IS1002, and recA. Moreover, we retrospectively explore the epidemiology of Bordetella spp. infections in Belgium, using the aforementioned assay over a three-year period, from 2013 until 2015.
Introduction
Whooping cough or pertussis is a contagious, acute respiratory illness caused by the Gram-negative coccobacillus Bordetella pertussis [1]. It is characterised by a paroxysmal cough, although symptoms can be atypical in infants younger than five months, and even fatal [2]. Pertussis in adolescents and adults is typically milder and presents with less severe symptoms [3], but is still of particular concern due to the associated risk of contagion [4]. Despite existing vaccination programmes, the number of whooping cough cases is still rising in Europe [5, 6] and North America [7].
Besides B. pertussis, three other members of the Bordetella genus are associated with respiratory infection in humans: B. parapertussis, B. holmesii, and to a lesser degree B. bronchiseptica [8]. These Bordetella species can cause respiratory disease with nonspecific symptoms as well as symptoms similar to those caused by B. pertussis, but milder [9]. However, pertussis vaccines do not provide cross-protection against these related Bordetella spp. [10, 11] Therefore, the differentiation of B. pertussis from other Bordetella spp. in respiratory illness is important for the evaluation of the vaccines’ efficacy, as misdiagnosis can lead to misinterpretation of the vaccines’ failure rate. Moreover, accurate identification of B. pertussis is needed to determine the necessity of prophylactic antibiotic treatment for the index cases and their contacts, as its utility in B. parapertussis and B. holmesii cases is still unclear [12, 13, 14].
As culture is difficult and lacks sensitivity, the diagnosis of Bordetella spp. infections often involves real-time polymerase chain reaction (PCR) of nasopharyngeal specimens, which is much more sensitive than bacterial culturing [15, 16, 17]. Segments of insertion sequences (IS) are often used as targets, particularly IS481 and IS1001 for B. pertussis and B. parapertussis respectively. Tests using these targets are sensitive because the IS are present in numerous copies in the bacterial genomes, but they are not species-specific. IS481 is found in B. pertussis, B. holmesii, and some B. bronchiseptica [16]. IS1001 is found in B. parapertussis and some B. bronchiseptica [17]. While hIS1001, which is found in B. holmesii, is highly homologous to IS1001, there are some sequence differences [17]. Thus, IS481 and IS1001 can be used as targets for screening, but confirmation with other targets will improve specificity.
IS1002 and recA have been used previously as secondary targets [18, 19]. IS1002 is found in B. pertussis, B. parapertussis, and some B. bronchiseptica, while recA is found in B. holmesii. Other Bordetella spp. also carry recA, but several polymorphisms allow for specific targeting [18]. When combined, the targets IS481, IS1001, IS1002, and recA can be used to differentiate Bordetella spp. in respiratory illness. IS481 and IS1001 can be used to screen for B. pertussis and B. parapertussis respectively, while IS1002 can confirm a positive screen, or alternatively, absence of IS1002 and presence of recA confirms a case of B. holmesii.
Most commercially available diagnostic PCR assays for pertussis use fewer targets and do not differentiate between B. pertussis and other Bordetella spp. such as B. holmesii. In order to improve specificity, we evaluate the use of the four PCR targets IS481, IS1001, IS1002, and recA, in the differentiation of B. pertussis from other Bordetella spp. that cause respiratory illness. Furthermore, we describe the incidence of respiratory infections caused by Bordetella spp. in Belgium from 2013 until 2015 using the described assay.
Materials and methods
Clinical sample preparation
Prior to nucleic acid (NA) extraction, viscous samples were pre-treated with Sputasol (Oxoid Ltd., Basingstoke, England) in a 1:1 dilution. 100 μl of each sample was used for NA extraction using NucliSENS easyMag (bioMérieux, Grenoble, France) according to the manufacturer’s instructions. Phocine herpesvirus (PhHV) was added as an internal control to monitor the extraction and the amplification’s efficiency [20].
Bacterial strain preparation
Reference strains as well as previously well-defined strains from clinical isolates were stored at -80°C prior to use, then grown on laboratory-prepared Regan-Lowe agar plates (Oxoid Ltd., Basingstoke, England) at 35°C. One colony of each strain was suspended in 200 μl Tris-EDTA (pH 8) and boiled to obtain the template NA.
Real-time PCR
Primers and probes for IS481, IS1001, and IS1002 were based on Roorda et al. (2011) [19], those for recA were based on Guthrie et al. (2010) [18], and those used for internal control were based on van Doornum et al. (2003) [21] (sequences in Table 1). Primers and probes were manufactured by Eurogentec (Liège, Belgium), MGB probes were manufactured by Applied Biosystems (Warrington, UK).
Table 1. Sequences and labelling of real-time PCR primers and probes.
| Target | Primer/probe | Sequence (5’-3’) and labels |
|---|---|---|
| IS481 | Forward primer | GCCGGATGAACACCCATAAG [19] |
| Reverse primer | GCGATCAATTGCTGGACCAT [19] | |
| Probe | 6FAM-CGATTGACCTTCCTACGTC-MGBNFQ [19] | |
| IS1001 | Forward primer | AATTGCTGCAAGCCAACCA [19] |
| Reverse primer | CCAGAGCCGTTTGAGTTCGT [19] | |
| Probe | VIC-ACATAGACCGTCAGCAG-MGBNFQ [19] | |
| IS1002 | Forward primer | CTAGGTCGAGCCCTTCTTGTTAAC [19] |
| Reverse primer | GCGGGCAAGCCACTTGTA [19] | |
| Probe | HEX-CATCGTCCAGTTCTGTTGCATCACCC-BHQ1 [19] | |
| recA | Forward primer | CGGTTCGCTGGGTCTCG [18] |
| Reverse primer | CCCGCGGCAGACCAC [18] | |
| Probe | 6FAM-CATCGCATTGGGCG-MGBNFQ [18] | |
| PhHV | Forward primer | GGGCGAATCACAGATTGAATC [21] |
| Reverse primer | GCGGTTCCAAACGTACCAA [21] | |
| Probe | CY5-TTTTTATGTGTCCGCCACCATCTGGATC-BHQ2 [21] |
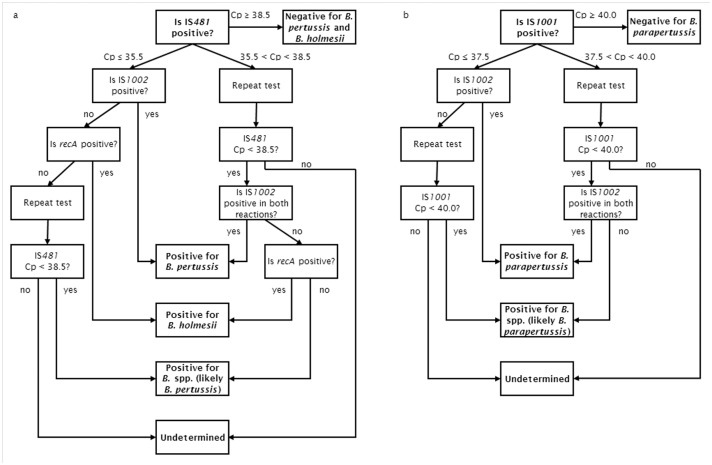
Two different multiplex PCR’s were performed, one for the detection of IS481 and IS1001, and one for the detection of recA and IS1002. A 50 μl reaction mixture consisting of 0.3 μM of each of the primers, 0.2 μM of each of the probes, iQ Multiplex Powermix (Bio-Rad Laboratories, Temse, Belgium), and 5.0 μl of the extracted NA was used in each PCR. Amplification was performed on the LightCycler 480 II PCR system (Roche Diagnostics, Mannheim, Germany). The PCR thermal profile consisted of a 3-minute cycle at 95°C, followed by 45 cycles of 15 seconds at 95°C and 1 minute at 60°C. Acquisition of the fluorescence signal was set during each cycle. A final cooling step of 30 seconds at 40°C was included. The crossing point (Cp) values were determined automatically using the “second derivative maximum” method present in the Roche LightCycler 480 Software, version 1.5. An algorithm with cut-off values was developed and used for the interpretation of the real-time PCR results (Fig 1).
Fig 1. Algorithm for the interpretation of the four-target real-time PCR assay for the detection of Bordetella for samples positive for IS481 (a) and IS1001 (b).
Bacterial culture
For culture, clinical specimens were inoculated onto laboratory-prepared Regan-Lowe agar upon arrival at the facilities. Plates were incubated at 35°C in a humidified aerobic atmosphere and examined daily for suspect colonies for up to twelve days.
Suspect colonies were identified using the microflex LT MALDI-TOF platform and MALDI Biotyper 3.0 software (Bruker Daltonics GmbH, Leipzig, Germany) [22, 23]. Identification to the species level was accepted when all matches with a log score above 1.7 belonged to the same species, or when the best match had a log score both above 2.0 and more than 0.200 higher than that of the other matched species.
When MALDI-TOF results were inconclusive, the species was determined based on biochemical characteristics: growth or absence of growth on charcoal agar, Haemophilus agar and MacConkey agar, presence of oxidase and presence of urease. Positive results for all characteristics indicated Bordetella bronchiseptica. Growth on charcoal agar and Haemophilus agar and presence of urease but not oxidase indicated Bordetella parapertussis. Growth on charcoal agar only, combined with the presence of oxidase but not urease indicated Bordetella pertussis [24].
Accuracy of real-time PCR
Accuracy was verified using a group of selected, previously well characterised strains derived from clinical samples, as well as reference strains (ATCC9340, ATCC9797, LMG15945, LMG15946), samples from Quality Control for Molecular Diagnostics (QCMD) 2011, and the extracted NA of different strains from a EUpert-labnet network panel (http://www.ecdc.europa.eu/en/publications/Publications/20120906-TER-EQA-pertussis.pdf). These samples included twenty-seven B. pertussis samples, nineteen B. parapertussis samples, seven B. holmesii samples, four samples of other Bordetella spp. and five non-Bordetella samples.
Analytical specificity of real-time PCR
The specificity of the primers and probes was first checked by performing a BLAST search on their sequences (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Specificity was further tested by performing the previously described real-time PCR on twelve well-defined B. bronchiseptica strains and seven B. holmesii strains, as well as other Bordetella spp. (B. avium (n = 2), B. petrii (n = 2), B. hinzii (n = 4), B. trematum (n = 2), B. ansorpii (n = 2)). The following bacteria of different genera were also tested: Acinetobacter baumani (n = 1), A. iwofii (n = 1), different Burkholderia spp. (n = 20), Enterobacter aerogenes (n = 2), Haemophilus influenzae (n = 2), Klebsiella pneumoniae (n = 1), Moraxella catarrhalis (n = 1), Neisseria meningitides (n = 1), Pseudomonas aeruginosa (n = 2), P. putida (n = 2), Staphylococcus aureus (n = 2), Stenotrophomonas maltophilia (n = 1), and different Streptococcus spp. (n = 8).
Analytical sensitivity of real-time PCR
Analytical sensitivity was determined using three reference strains: ATCC9797 (B. pertussis), ATCC15311 (B. parapertussis), and LMG15945 (B. holmesii). They were suspended in physiological saline at a theoretical concentration of 1.5 × 108 CFU/ml (0.5 McFarland). Serial dilutions were made from these suspensions, and 100-μl aliquots of each dilution were extracted and tested with real-time PCR. Tests were performed ten to twelve times for each strain. The analytical sensitivity was determined by calculating the limit of detection (LOD 95%), i.e. the lowest concentration that is detected 95% of the time, using probit regression analysis in SPSS Statistics 20.0 (IBM Corp., Armonk, NY, USA).
Precision of real-time PCR
Replication experiments were performed to determine precision, using three dilutions (strong positive, positive and weak positive) of the reference strains in physiological water. These samples were tested at least ten times in five different runs over the course of five days. The intra-run and inter-run coefficients of variance (CV) were calculated based on the Cp value of the assay using SPSS Statistics 20.0.
Laboratory-based surveillance of pertussis
Surveillance data on circulating B. pertussis, B. parapertussis and B. holmesii in Belgium, based on the results of the real-time PCR for IS481, IS1001, recA, and IS1002, were gathered retrospectively from 11919 respiratory samples collected from January 2013 until December 2015.
Results
Accuracy of real-time PCR
All clinical strains and reference strains were identified correctly: all B. pertussis strains were positive for IS481 and IS1002, and negative for recA and IS1001. All B. parapertussis strains were positive for IS1001 and IS1002, and negative for recA and IS481. All B. holmesii strains were positive for IS481 and recA, and negative for IS1001 and IS1002.
Out of the twelve QCMD samples, all but one sample tested correctly: four out of five B. pertussis strains were positive for IS481 and IS1002, and negative for recA and IS1001, only one very weak positive (10 CFU/ml) B. pertussis sample was negative for all targets. The B. parapertussis strain was positive for IS1001 and IS1002, and negative for recA and IS481. The B. holmesii strain was positive for IS481 and recA, and negative for IS1001 and IS1002. The B. bronchiseptica, B. hinzii, and non-Bordetella samples tested negative for all targets.
Finally, out of ten NA samples from the EUpert-labnet network panel, eight were identified correctly. One B. pertussis NA sample with a low concentration of 0.02 pg/μl was negative for IS1002, but positive for IS481. One B. holmesii sample was positive for IS1001 as well as for IS481 and recA. Using the algorithm developed for our assay, these results did not lead to an incorrect identification (S1 Table).
Analytical specificity of real-time PCR
BLAST analysis showed that IS481 and IS1001 may cross-react with B. bronchiseptica and B. holmesii. IS1002 was specific for B. pertussis and B. parapertussis. Finally, recA showed cross-reactivity with some Burkholderia spp.
Following BLAST analysis the assay was performed on different strains. Six out of seven B. holmesii strains were negative for both IS1001 and IS1002, one was positive for IS1001. Out of twelve tested B. bronchiseptica strains, four were positive for IS481 and two for IS1001. One IS481-positive strain was also positive for IS1002, and as such could have been misinterpreted as B. pertussis. No cross-reaction was detected in other Bordetella spp.
As BLAST analysis suggested possible cross-reactivity of recA with some Burkholderia spp., this target was tested in twenty Burkholderia strains, belonging to eleven different species. Cross-reaction was detected in one Burkholderia stabilis strain and one Burkholderia multivorans strain. No cross-reaction with any of the other targets was found.
Finally, the assay was tested with various other bacteria, including bacteria that cause respiratory infections as well as commensals. Minor cross-reactivity was found in Stenotrophomonas maltophilia for IS481 and in Enterobacter aerogenes for IS1001, however, the other targets remained negative (S1 Table).
Analytical sensitivity of real-time PCR
The limit of detection (LOD 95%) for IS481 was 145 CFU/ml for B. pertussis and 152 CFU/ml for B. holmesii. For IS1001, the LOD 95% was 242 CFU/ml for B. parapertussis. The recA assay had an LOD 95% of 2577 CFU/ml for B. holmesii. Finally, for IS1002 the LOD 95% was 1954 CFU/ml for B. pertussis and 1725 CFU/ml for B. parapertussis (S1 Table).
Precision of real-time PCR
The coefficients of variance (CV) for the detection of IS481 ranged from 1.1% to 8.4% intra-run, and from 1.8 to 5.4% inter-run. For IS1001, they ranged from 1.7% to 3.7% intra-run, and from 0.8% to 2.0% inter-run. The intra-run CV for recA ranged from 1.8% to 2.4%, and the inter-run CV ranged from 0.8% to 1.2%. Finally, for IS1002, the intra-run and inter-run CV ranged from 1.9% to 4.0% and from 0.3% to 1.9% respectively for B. pertussis, and from 2.3% to 6.2% and from 1.3% to 3.8% respectively for B. parapertussis (S1 Table).
Laboratory-based surveillance of pertussis
The Belgian National Reference Centre (NRC) for B. pertussis performed real-time PCR on 11919 respiratory samples between 2013 and 2015. These data were interpreted using the algorithm presented above (Fig 1). Among these samples, 1189 (10.0%) were found positive for B. pertussis. 872 (73.3%) of these were positive for both IS481 and IS1002 and reported “positive for B. pertussis”, while 317 (26.7%) were positive for IS481 only and reported as “positive for Bordetella spp., probably B. pertussis”. 151 samples (1.3%) were reported positive for B. parapertussis, of which 112 (74.2%) were positive for both IS1001 and IS1002 (reported “positive for B. parapertussis”), and 39 (25.8%) positive for IS1001 only (reported as “positive for Bordetella spp., probably B. parapertussis”). Furthermore, 15 samples (0.1%) were found positive for B. holmesii, and 425 samples (3.6%) were reported as “undetermined”, i.e. they tested positive for IS481 or IS1001 but negative for IS1002 and recA, then negative for all targets upon repeat testing (S2 Table).
Of the samples found positive for B. pertussis with real-time PCR, 28.8% were also positive in culture. For B. parapertussis, this percentage was similar at 29.8%. Only two of the samples reported as “positive for Bordetella spp., probably B. pertussis” and none of the samples reported as “positive for Bordetella spp., probably B. parapertussis” were recovered in culture. These samples were associated with higher Cp values. The association between Cp value and culture recovery is presented in Table 2. Out of the fifteen B. holmesii samples, five were found positive in culture.
Table 2. Association between Cp value and recovery in culture, for samples positive for IS481 (a) and IS1001 (b).
| a | Cp value IS481 | PCR positive | Culture positive (B. pertussis) | Recovery culture/PCR |
| Cp ≤ 20 | 165 | 133 | 80.6% | |
| 20 < Cp ≤ 25 | 122 | 81 | 66.4% | |
| 25 < Cp ≤ 30 | 227 | 89 | 39.2% | |
| Cp > 30 | 675 | 39 | 5.8% | |
| Total | 1189 | 342 | 28.8% | |
| b | Cp value IS1001 | PCR positive | Culture positive (B. parapertussis) | Recovery culture/PCR |
| Cp ≤ 20 | 7 | 7 | 100.0% | |
| 20 < Cp ≤ 25 | 16 | 13 | 81.3% | |
| 25 < Cp ≤ 30 | 22 | 14 | 63.6% | |
| Cp > 30 | 105 | 11 | 10.5% | |
| Total | 151 | 45 | 29.8% |
A few samples were found positive for more than two targets. Seven samples were positive for IS481, IS1001 and IS1002, and were interpreted as co-infection of B. pertussis and B. parapertussis. In culture, two of these were found positive for B. pertussis and three for B. parapertussis. Two other samples were positive for IS481, recA and IS1002 and were interpreted as co-infection of B. pertussis and B. holmesii. These samples were negative in culture.
Discussion
In this study, the use of a multiplex real-time PCR assay for the detection of B. pertussis, B. parapertussis, and B. holmesii was investigated. By combining a sensitive screening method with a confirmation assay, the risk of false positive results is reduced and the specificity increased. The strength of our study lies in the extensive testing of the real-time PCR targets, during validation and during a three-year study with clinical samples.
Our real-time PCR assay uses the targets IS481/IS1001 for screening and recA/IS1002 for confirmation. We showed that this method is accurate, sensitive, fairly specific, and precise. B. pertussis, B. parapertussis, and B. holmesii reference and clinical strains were identified correctly. The detection limit of our method was in line with that of other assays described for IS481 and IS1001, ranging from 145 to 242 CFU/ml for B. pertussis, B. parapertussis and B. holmesii [19, 25, 26, 27]. We also found that the sensitivity of the PCR for recA/IS1002 is around ten times lower than that of the PCR for IS481/IS1001. This lower sensitivity can be explained by the lower copy numbers of recA and IS1002 genes.
The targets used in this study were fairly specific. The targets of the screening assay, IS481 and IS1001, do not show cross-reactivity with other respiratory pathogens. However, they do show cross-reactivity with B. holmesii and sometimes with B. bronchiseptica. IS481, IS1001 and IS1002 have all been previously detected in some B. bronchiseptica strains [19]. Therefore the possibility of cross-reactivity should be taken into account when using these targets. This problem is limited by the use of target combinations: both IS481 (screening) and IS1002 (confirmation) need to be positive to identify B. pertussis, both IS1001 and IS1002 need to be positive to identify B. parapertussis.
Using our method, about a fourth of the cases positive for IS481 remained negative for IS1002 (and recA). These cases are reported as “positive for Bordetella spp., probably B. pertussis”. Samples positive for IS1001 but negative for IS1002 (and recA) are reported as “positive for Bordetella spp., probably B. parapertussis”. Cases such as these occur relatively frequently due to the difference in sensitivity between the screening and the confirmation targets, the latter being present in much lower copy numbers.
For weak positive results (i.e. high Cp values) in the screening assay, cross-reactivity, in particular with B. bronchiseptica, cannot be ruled out. However, B. bronchiseptica is rarely found in respiratory samples from patients displaying symptoms of whooping cough: in over 2000 tested samples only one cross-reacting B. bronchiseptica strain was cultured [28]. Therefore the probability of B. bronchiseptica causing a false positive result may be considered negligible. Because of the low occurrence of B. holmesii and B. bronchiseptica in patients displaying pertussis symptoms, it is acceptable to consider cases with a Cp lower than 38.5 for IS481 (or lower than 40 for IS1001) as probable cases of B. pertussis (or B. parapertussis), even if they are negative for IS1002 and recA.
Based on the extensive validation of the real-time PCR, an algorithm was developed for its interpretation (Fig 1). Using high copy number targets such as IS481 and IS1001 increases the sensitivity of the assay at the cost of an increased risk of false positives. This risk can be reduced by combining multiple targets, as demonstrated in this study.
The implementation of cut-off values will also reduce false positivity, as shown previously in a quality control study [29], and is recommended by the CDC (http://www.cdc.gov/pertussis/clinical/diagnostic-testing/diagnosis-pcr-bestpractices.html).
In this study, a cut-off value based on the LOD 95%, determined during validation of the assay, was used. Positive results with a Cp value higher than this value should be repeated, and discordant results should be reported to the requesting physician as “undetermined”, to be considered negative or investigated further based on clinical presentation and serology. A second cut-off value to reduce false positivity was implemented as well. Samples with Cp values higher than this cut-off are immediately reported as negative.
During the study period, B. pertussis was diagnosed about ten times as much as B. parapertussis, which was diagnosed about ten times as much as B. holmesii. Less than four percent of the clinical samples could not be reliably identified using our assay (“undetermined”).
By including the target recA in the assay, B. holmesii can be differentiated from B. pertussis. This is important in routine diagnosis as B. holmesii is increasingly reported in Europe, and has often been misidentified as B. pertussis in the past [13, 14, 28]. Using our assay, we found fifteen B. holmesii strains during the study period, suggesting that B. holmesii might indeed occur more frequently than previously reported in Belgium [28].
The algorithm was not developed for the interpretation of co-infections, and they are difficult to identify with the presented method. However, co-infection is likely if the assay is positive for more than two targets. Several possible co-infections were detected during the study that could not be confirmed by culture. Based on the large amount of samples, it seems that the likelihood of co-infections is limited (less than 0.1%).
We demonstrated in this study that there is a high concordance between the Cp value of a sample and the probability of recovery in culture. This finding is consistent with previous results [30]. As culturing of B. pertussis is no longer performed diagnostically, but rather for the acquiring of strains to be used in epidemiologic typing and research, it may be worthwhile to try to culture only samples with low Cp values. For B. pertussis, culturing has a high success rate for Cp values up to 25. For Cp values between 25 and 30, this rate drops to less than 50%. Culturing in these cases should still be considered for laboratories with a particular interest in obtaining as many strains as possible for epidemiological and virulence studies. When Cp values are higher than 30 the success rate becomes negligible (less than 6%) and culturing is not advisable.
Several PCR targets specific to B. pertussis have been described in literature, such as the pertussis toxin promoter (ptxP), recommended by the Pertussis PCR Consensus Group [31]. However, the sensitivity of real-time PCR with ptxP as a target is considerably lower than with IS481 [32]. Other possible targets for B. pertussis, such as the pertactin genes, BP283, BP485, and ptxS1 have also been described, but often show cross-reactivity with other Bordetella spp. [33, 34] Recently an assay with a porin protein as target was described as specific for B. pertussis, but a larger specificity study would be needed to confirm this [32].
In conclusion, multiplex real-time PCR with targets IS481/IS1001 can be used for screening, followed by confirmation with targets recA/IS1002, in order to differentiate B. pertussis from other Bordetella spp. that cause respiratory infection or colonisation.
Supporting information
(XLSX)
(XLSX)
Acknowledgments
Special acknowledgement goes to all laboratory technicians that contributed to this study, in particular Linda Godau, Steve Jacobs, Elly Roebben, Annick Soetaert, Marina Van Cauwenbergh, Kristof Vandoorslaer, and Anita Wyns.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was performed in the frame of the Belgian National Reference Centre for Bordetella pertussis, supported by the Belgian Ministry of Social Affairs through a fund within the Health Insurance System. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Snyder J, Fisher D. Pertussis in childhood. Pediatr Rev. 2012;33(9):412–20; quiz 20–1. 10.1542/pir.33-9-412 [DOI] [PubMed] [Google Scholar]
- 2.Crowcroft NS, Booy R, Harrison T, Spicer L, Britto J, Mok Q, et al. Severe and unrecognised: pertussis in UK infants. Arch Dis Child. 2003;88(9):802–6. 10.1136/adc.88.9.802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barlow RS, Reynolds LE, Cieslak PR, Sullivan AD. Vaccinated children and adolescents with pertussis infections experience reduced illness severity and duration, Oregon, 2010–2012. Clin Infect Dis. 2014;58(11):1523–9. 10.1093/cid/ciu156 [DOI] [PubMed] [Google Scholar]
- 4.Menard A, Lehours P, Sarlangue J, Bebear C, Megraud F, de Barbeyrac B. Development of a real-time PCR for the identification of Bordetella pertussis and Bordetella parapertussis. Clin Microbiol Infect. 2007;13(4):419–23. 10.1111/j.1469-0691.2006.01659.x [DOI] [PubMed] [Google Scholar]
- 5.de Melker HE, Schellekens JF, Neppelenbroek SE, Mooi FR, Rumke HC, Conyn-van Spaendonck MA. Reemergence of pertussis in the highly vaccinated population of the Netherlands: observations on surveillance data. Emerg Infect Dis. 2000;6(4):348–57. 10.3201/eid0604.000404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sizaire V, Garrido-Estepa M, Masa-Calles J, Martinez de Aragon MV. Increase of pertussis incidence in 2010 to 2012 after 12 years of low circulation in Spain. Euro Surveill. 2014;19(32). [DOI] [PubMed] [Google Scholar]
- 7.Cherry JD. Epidemic pertussis in 2012—the resurgence of a vaccine-preventable disease. N Engl J Med. 2012;367(9):785–7. 10.1056/NEJMp1209051 [DOI] [PubMed] [Google Scholar]
- 8.Mattoo S, Foreman-Wykert AK, Cotter PA, Miller JF. Mechanisms of Bordetella pathogenesis. Front Biosci. 2001;6:E168–86. [DOI] [PubMed] [Google Scholar]
- 9.Cherry JD, Seaton BL. Patterns of Bordetella parapertussis respiratory illnesses: 2008–2010. Clin Infect Dis. 2012;54(4):534–7. 10.1093/cid/cir860 [DOI] [PubMed] [Google Scholar]
- 10.Khelef N, Danve B, Quentin-Millet MJ, Guiso N. Bordetella pertussis and Bordetella parapertussis: two immunologically distinct species. Infect Immun. 1993;61(2):486–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang X, Weyrich LS, Lavine JS, Karanikas AT, Harvill ET. Lack of cross-protection against Bordetella holmesii after pertussis vaccination. Emerg Infect Dis. 2012;18(11):1771–9. 10.3201/eid1811.111544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bolding J, Kamat D. Whooping cough caused by Bordetella parapertussis. Infect Med. 2004;21(6). [Google Scholar]
- 13.Pittet LF, Emonet S, Schrenzel J, Siegrist CA, Posfay-Barbe KM. Bordetella holmesii: an under-recognised Bordetella species. Lancet Infect Dis. 2014;14(6):510–9. 10.1016/S1473-3099(14)70021-0 [DOI] [PubMed] [Google Scholar]
- 14.Pittet LF, Posfay-Barbe KM. Bordetella holmesii: Still Emerging and Elusive 20 Years On. Microbiol Spectr. 2016;4(2). [DOI] [PubMed] [Google Scholar]
- 15.Knorr L, Fox JD, Tilley PA, Ahmed-Bentley J. Evaluation of real-time PCR for diagnosis of Bordetella pertussis infection. BMC Infect Dis. 2006;6:62 10.1186/1471-2334-6-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reischl U, Lehn N, Sanden GN, Loeffelholz MJ. Real-time PCR assay targeting IS481 of Bordetella pertussis and molecular basis for detecting Bordetella holmesii. J Clin Microbiol. 2001;39(5):1963–6. 10.1128/JCM.39.5.1963-1966.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tatti KM, Sparks KN, Boney KO, Tondella ML. Novel multitarget real-time PCR assay for rapid detection of Bordetella species in clinical specimens. J Clin Microbiol. 2011;49(12):4059–66. 10.1128/JCM.00601-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guthrie JL, Robertson AV, Tang P, Jamieson F, Drews SJ. Novel duplex real-time PCR assay detects Bordetella holmesii in specimens from patients with Pertussis-like symptoms in Ontario, Canada. J Clin Microbiol. 2010;48(4):1435–7. 10.1128/JCM.02417-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roorda L, Buitenwerf J, Ossewaarde JM, van der Zee A. A real-time PCR assay with improved specificity for detection and discrimination of all clinically relevant Bordetella species by the presence and distribution of three Insertion Sequence elements. BMC Res Notes. 2011;4:11 10.1186/1756-0500-4-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niesters HG. Quantitation of viral load using real-time amplification techniques. Methods. 2001;25(4):419–29. 10.1006/meth.2001.1264 [DOI] [PubMed] [Google Scholar]
- 21.van Doornum GJ, Guldemeester J, Osterhaus AD, Niesters HG. Diagnosing herpesvirus infections by real-time amplification and rapid culture. J Clin Microbiol. 2003;41(2):576–80. 10.1128/JCM.41.2.576-580.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mellmann A, Cloud J, Maier T, Keckevoet U, Ramminger I, Iwen P, et al. Evaluation of matrix-assisted laser desorption ionization-time-of-flight mass spectrometry in comparison to 16S rRNA gene sequencing for species identification of nonfermenting bacteria. J Clin Microbiol. 2008;46(6):1946–54. 10.1128/JCM.00157-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sauer S, Freiwald A, Maier T, Kube M, Reinhardt R, Kostrzewa M, Geider K. Classification and identification of bacteria by mass spectrometry and computational analysis. PLoS One. 2008;3(7):e2843 10.1371/journal.pone.0002843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Versalovic J. Manual of Clinical Microbiology, 10th edition, chapter 43, ASM Press; 2012. 739–750 [Google Scholar]
- 25.Kosters K, Reischl U, Schmetz J, Riffelmann M, Wirsing von Konig CH. Real-time LightCycler PCR for detection and discrimination of Bordetella pertussis and Bordetella parapertussis. J Clin Microbiol. 2002;40(5):1719–22. 10.1128/JCM.40.5.1719-1722.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Templeton KE, Scheltinga SA, van der Zee A, Diederen BM, van Kruijssen A, Goossens H, et al. Evaluation of real-time PCR for detection of and discrimination between Bordetella pertussis, Bordetella parapertussis, and Bordetella holmesii for clinical diagnosis. J Clin Microbiol. 2003;41(9):4121–6. 10.1128/JCM.41.9.4121-4126.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu Y, Xu Y, Hou Q, Yang R, Zhang S. Triplex real-time PCR assay for detection and differentiation of Bordetella pertussis and Bordetella parapertussis. APMIS. 2010;118(9):685–91. 10.1111/j.1600-0463.2010.02644.x [DOI] [PubMed] [Google Scholar]
- 28.Van den Bossche D, De Bel A, De Smet D, Heylen O, Vekens E, Vandoorslaer K, et al. Prevalence of Bordetella holmesii and Bordetella bronchiseptica in respiratory tract samples from Belgian patients with pertussis-like symptoms by sensitive culture method and mass spectrometry. Acta Clin Belg. 2013;68(5):341–8. 10.2143/ACB.3341 [DOI] [PubMed] [Google Scholar]
- 29.Tatti KM, Martin SW, Boney KO, Brown K, Clark TA, Tondella ML. Qualitative assessment of pertussis diagnostics in United States laboratories. Pediatr Infect Dis J. 2013;32(9):942–5. 10.1097/INF.0b013e3182947ef8 [DOI] [PubMed] [Google Scholar]
- 30.Vestrheim DF, Steinbakk M, Bjornstad ML, Moghaddam A, Reinton N, Dahl ML, et al. Recovery of Bordetella pertussis from PCR-positive nasopharyngeal samples is dependent on bacterial load. J Clin Microbiol. 2012;50(12):4114–5. 10.1128/JCM.01553-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Riffelmann M, Wirsing von Konig CH, Caro V, Guiso N, Pertussis PCRCG. Nucleic Acid amplification tests for diagnosis of Bordetella infections. J Clin Microbiol. 2005;43(10):4925–9. 10.1128/JCM.43.10.4925-4929.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hasan MR, Tan R, Al-Rawahi GN, Thomas E, Tilley P. Evaluation of amplification targets for the specific detection of Bordetella pertussis using real-time polymerase chain reaction. Can J Infect Dis Med Microbiol. 2014;25(4):217–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Probert WS, Ely J, Schrader K, Atwell J, Nossoff A, Kwan S. Identification and evaluation of new target sequences for specific detection of Bordetella pertussis by real-time PCR. J Clin Microbiol. 2008;46(10):3228–31. 10.1128/JCM.00386-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vincart B, De Mendonca R, Rottiers S, Vermeulen F, Struelens MJ, Denis O. A specific real-time PCR assay for the detection of Bordetella pertussis. J Med Microbiol. 2007;56(Pt 7):918–20. 10.1099/jmm.0.46947-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.