Abstract
The discovery that expansion of a hexanucleotide repeat within a non-coding region of the C9orf72 gene causes amyotrophic lateral sclerosis and frontotemporal dementia raised questions about C9orf72 protein function and potential disease relevance. The major predicted structural feature of the C9orf72 protein is a DENN domain. As DENN domains are best characterized for regulation of specific Rab GTPases, it has been proposed that C9orf72 may also act through regulation of a GTPase target. Recent genetic and cell biological studies furthermore indicate that the C9orf72 protein functions at lysosomes as part of a larger complex that also contains the Smith-Magenis Chromosome Region 8 (SMCR8) and WD repeat-containing protein 41 (WDR41) proteins. An important role for C9orf72 at lysosomes is supported by defects in lysosome morphology and mTORC1 signaling arising from C9orf72 KO in diverse model systems. Collectively, these new findings define a C9orf72-containing protein complex and a lysosomal site of action as central to C9orf72 function and provide a foundation for the elucidation of direct physiological targets for C9orf72. Further elucidation of mechanisms whereby C9orf72 regulates lysosome function will help to determine how the reductions in C9orf72 expression levels that accompany hexanucleotide repeat expansions contribute to both normal physiology and disease pathology.
Keywords: amyotrophic lateral sclerosis, frontotemporal dementia, lysosome, mTORC1, neurodegeneration, C9orf72, SMCR8, Birt-Hogg-Dubé syndrome, FLCN, autophagy
Introduction
In this review, we will first provide a brief overview of amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD). This will be followed by a discussion of the C9orf72 mutations that cause ALS-FTD and the major questions that have arisen concerning disease mechanisms. We will then focus in more depth on new insights into the function of the C9orf72 protein. We will focus in particular on the predicted structural similarity between C9orf72 and folliculin (FLCN), the Birt-Hogg-Dubé syndrome tumor suppressor that has suggested a role for C9orf72 as a GTPase regulating protein. We will furthermore summarize recent insights into the function of the C9orf72 protein with particular focus on the identification of C9orf72 interacting proteins and alterations in lysosome cell biology arising from C9orf72 knockout studies in both mice and human cells. Finally, we will conclude with an overview of additional genes related to lysosomal function that also contribute to ALS-FTD risk.
ALS-FTD disease spectrum
ALS and FTD are lethal adult onset neurodegenerative diseases that predominantly affect the survival of motor neurons and distinct populations of cortical neurons, respectively. While these diseases were long thought to be distinct based on the major motor versus cognitive deficits that they elicit, it is now appreciated that ALS and FTD frequently share overlapping patient symptoms, cellular pathology and genetic origins1, 2. At the level of pathology, a significant subset of ALS and FTD patients have ubiquitinated cytoplasmic inclusions within affected neurons that contain the TAR DNA binding protein of 43 kDa (TARDBP, also known as TDP-43)3. Meanwhile, while there are genes whose mutations strictly promote either ALS or FTD, it is now well known that mutations in other genes can cause ALS and FTD. This overlap between pathology and genetics supports the idea that a major subset ALS and FTD cases can be considered as part of a common disease spectrum1, 2. This concept of an ALS-FTD spectrum is exemplified by disease arising from mutations in the C9orf72 gene.
C9orf72 hexanucleotide expansion is a major cause of ALS-FTD
Analysis of extended families with autosomal dominant inheritance of ALS and FTD with TDP-43 pathology resulted in the discovery of expansions of a GGGGCC hexanucleotide repeat in the first intron of the C9orf72 gene as a cause of both diseases4-6. Subsequent analysis revealed that this genetic perturbation represents the most common known cause of both familial and sporadic forms of ALS and FTD that feature TDP-43 pathology1, 2. It is striking to note that C9orf72 mutations can cause either ALS or FTD within members of the same family7. While specific genetic or environmental factors that underlie such neuronal subtype specific variability in disease manifestation remain uncertain, this genetic overlap in ALS and FTD risk suggests common mechanisms at the subcellular level.
C9orf72 hexanucleotide repeat expansion: Putative disease mechanisms
While questions concerning how expansion of a hexanucleotide repeat within a non-coding region of the C9orf72 gene cause disease have not been conclusively addressed, three major consequences of C9orf72 hexanucleotide repeat expansion have been identified that could contribute either alone or in combination to cause disease. First, sense and antisense RNA foci transcribed from the C9orf72 repeat expansion have been observed in patients carrying the expansion6, 8-11. Furthermore, both DNA and RNA containing the hexanucleotide expansions have been proposed to fold into G quadruplex structures that sequester RNA binding proteins10, 12-17. Second, RNA transcribed from the hexanucleotide expansions can also serve as a template for translation of dipeptide repeat proteins in a non-ATG dependent manner18. Aggregates of such dipeptide repeat proteins are found in the brains and spinal cords of ALS-FTD patients18-22 and have been shown to exert deleterious effects following their transgenic over-expression in various model systems23-25. Third, while C9orf72 hexanucleotide repeat expansions do not alter the coding sequence of the C9orf72 protein, they are accompanied by a reduction in the levels of the C9orf72 mRNA5, 6, 11, 26, 27 that has been attributed to methylation of the locus26-29. Given the autosomal pattern of inheritance, suppression of C9orf72 expression only occurs on one allele and expression from the wild type allele remains intact in repeat expansion carriers. This results in a modest change (∼50% decrease) in C9orf72 levels that might not seem like a major perturbation. However, as progranulin haploinsufficiency is a well-established cause of autosomal dominant FTD with TDP-43 pathology30, 31, there is precedent for the development of a similar neurodegenerative disease through a comparably modest change in gene dosage.
C9orf72 is part of a larger family of DENN domain containing proteins
Does loss of C9orf72 expression contribute to neurodegenerative disease risk? With the knowledge that C9orf72 expression levels are negatively impacted by hexanucleotide expansion, questions naturally arise concerning the function of the C9orf72 protein and whether loss of its function could contribute to disease. Answering this question has long been difficult due to lack of knowledge concerning the normal function of the C9orf72 protein.
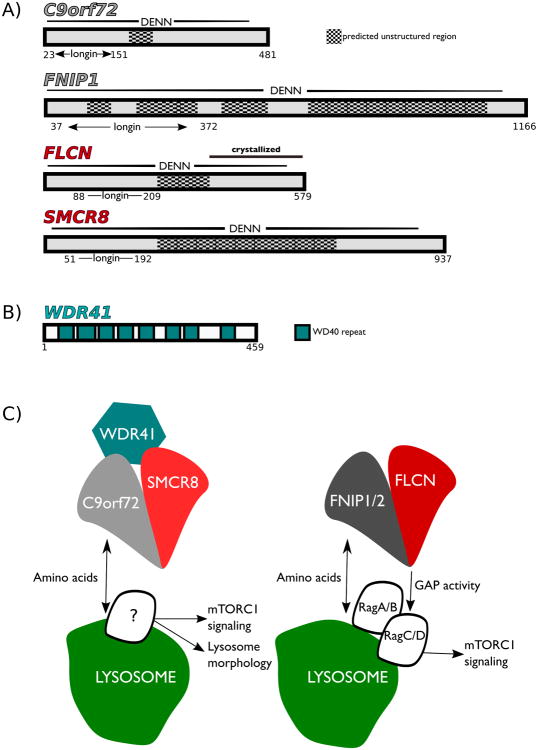
A major clue into the possible functions of C9orf72 arose from bioinformatic studies that predicted the presence of a DENN (differentially expressed in normal and neoplastic cells32) domain as the major structural feature of the C9orf72 protein33, 34. As C9orf72 lacks obvious similarity to other DENN domains at the primary amino acid sequence level, the identification of its putative DENN domain required several important insights into DENN domain structural biology that enabled bioinformatics analyses that predicted the presence of a DENN domain in C9orf72. First was the solution of a high-resolution crystal structure for DENND1B in complex with its target, Rab35, the first such structure for the DENN domain family35. Second was a crystal structure for a C-terminal fragment of folliculin (FLCN), the Birt-Hogg-Dubé syndrome tumor suppressor, which led to the realization that FLCN contained a DENN domain that was highly similar to DENND1B in spite of a lack of strong similarity in their primary amino acid sequences36. This identification of an unanticipated DENN domain in FLCN revealed that the DENN domain family of proteins was larger than previously expected and motivated the search for additional DENN domain containing proteins. Highly sensitive, structure-based homology searches then identified a putative DENN domain in C9orf72 that placed it in the FLCN branch of the DENN domain family (Figure 1A)33, 34. In particular, C9orf72 was predicted to have closest structural similarity to key FLCN binding partners known as FLCN Interacting Proteins 1 and 2 (FNIP1/2), two highly similar proteins that were originally discovered for their ability to robustly interact with FLCN, but whose structural similarity to FLCN was only brought to light by the same analysis that introduced C9orf72 to the DENN domain family. A crystal structure of the N-terminal region of Lst4, the yeast orthologue of FNIP1/2, has since been solved that supports the prediction that FNIP proteins are indeed members of the DENN family37. Of additional interest, another protein of previously unknown function, SMCR8 was predicted to be most structurally similar to FLCN 33, 34. Collectively, the C9orf72, SMCR8, FLCN and FNIP1/2 proteins define a previously unappreciated branch of the DENN domain family tree (Figure 1A).
Figure 1. Parallels between C9orf72:SMCR8 and FLCN:FNIP complexes.
A) Summary of the predicted domain organization of C9orf72, FNIP1, SMCR8 and C9orf72 that illustrates that the DENN domain represents the major folded element in each of these proteins. Also noted at the N-terminal region of each of these DENN domains is a region that has similarity to the Longin domain that is found in many GTPase regulating proteins. B) WDR41 is predicted to contain 8 WD40 repeats62. C) Schematic diagram summarizing functions and lysosomal localization of the C9orf72:SMCR8:WDR41 and FLCN:FNIP complexes.
Regulation of Rab GTPase nucleotide binding status by DENN domain proteins
At least twenty-six proteins are predicted to contain a DENN domain38. The best-characterized function for DENN domains is to act as a guanine nucleotide exchange factors (GEF) for members of the Rab family of GTPases. The human genome is thought to code for 66 Rab GTPases39, many of which have roles in the regulation of intracellular membrane traffic40, 41. Such GTPases cycle between GTP and GDP bound states in parallel with their recruitment on and off intracellular membranes. The spatial and temporal control of these transitions is controlled by the coordinated actions of multiple regulatory proteins that determine both the nucleotide status of the Rabs as well as Rab interactions with specific intracellular membranes. Important Rab regulatory proteins include: GEFs, GDP dissociation inhibitor (GDI) and GTPase activating proteins (GAPs). GEFs preferentially interact with the nucleotide-free form of their target GTPase42, 43 and induce conformational changes that result in the exchange of GDP for GTP44. This GTP-bound form is resistant to extraction from the membrane by GDI45; in this way, GEFs promote the enrichment of GTPases such as Rabs to membranes on which the GEF resides. The membrane localized, GTP-bound, “activated” Rab then recruits diverse effectors related to processes such as membrane traffic and cytoskeletal regulation46. For example, activation of Rab35 at clathrin coats by DENND1 (also known as connecdenn) family members allows for the recruitment of lipid metabolizing and actin cytoskeleton regulating proteins that are critical for early steps in the endocytic pathway47-49. GAPs inactivate Rabs by accelerating the hydrolysis of GTP to GDP, which allows for membrane extraction of the Rab by GDI and its redistribution back into the cytosol44, 45. The predicted presence of a DENN domain in C9orf72 thus suggests a role for C9orf72 in regulating the recruitment of a Rab protein to a specific subcellular membrane where it could in turn recruit diverse effectors.
However, while DENN domain containing proteins are best characterized as Rab GEFs, recent studies into FLCN revealed properties that are not shared by other previously characterized DENN domain proteins. Firstly, rather than acting as a monomer, it forms a strong heterodimer with either of the FNIP1 or FNIP2 proteins that are also predicted to contain a DENN domain as their major folded element33, 34, 37, 50. Secondly, although in vitro assays originally pointed to a role for FLCN as a GEF for Rab3536, subsequent studies provided strong evidence that the major physiological target of FLCN:FNIP heterodimers is not a Rab GTPase but is instead RagA/B:RagC/D GTPase heterodimers51, 52 and that such a function is evolutionarily conserved from yeast to man53. The Rag heterodimer consists of either RagA or RagB bound to RagC or RagD and localizes to the cytoplasmic surface of lysosomes where it plays a major role in regulating the subcellular localization and signaling of the mTOR complex 1 (mTORC1) kinase54. Thirdly, while the FLCN:FNIP heterodimer binds selectively to the nucleotide-free or GDP-bound form of RagA/B (as expected for a RagA/B GEF), its biochemical function is to exert GTPase activating protein (GAP) activity towards RagC/D 51, 52. Such coordination between FLCN:FNIP recruitment to lysosomes by RagA/B interactions coupled with activity towards the RagC/D half of the RagA/B:RagC/D heterodimer may provide a mechanism to help coordinate nucleotide status between subunits of the RagA/B:RagC/D heterodimer. Lastly, the regulation of Rag GTPases by FLCN:FNIP takes place at the surface of lysosomes and is tightly regulated by intracellular amino acid availability51, 52. Such regulation is physiologically important as it ensures that Rag GTPase-dependent activation of mTORC1 (a major positive regulator of cell growth) only occurs in cells that have sufficient amino acid supplies.
Collectively, the structural predictions that C9orf72 contains a DENN domain that is similar to the FLCN and FNIP proteins along with emerging knowledge that FLCN and FNIP DENN proteins function as heterodimers that bind to and exhibit GAP activity towards Rag GTPase heterodimers generated the following specific predictions about C9orf72: 1) C9orf72 might function as part of a larger protein complex; 2) The GTPase target of C9orf72 might not necessarily be a member of the Rab family; 3) C9orf72 (or any complex containing it) might have complex functions relating to GTPase binding, GEF or GAP activity; and 4) Such activities of C9orf72 might be tightly regulated by nutrient availability. Interestingly, recently published studies have provided evidence that strongly supports many of these predictions.
A protein complex containing C9orf72, SMCR8 and WDR41
The predicted structural similarity between C9orf72 and the FLCN:FNIP heterodimer suggested that C9orf72 might form a complex with a second DENN domain containing protein. The analyses that built on the FLCN crystal structure to predict DENN domains in C9orf72 and the FNIPs also identified SMCR8 as having a DENN domain that had previously escaped detection due to low primary sequence similarity with other DENN proteins (Figure 1A). However, even while nothing was known about SMCR8, the predictions that it had structural similarity to C9orf72, FLCN and FNIPs turned out to be prescient; both unbiased proteomics approaches to identify C9orf72 interacting proteins55-58, as well as a targeted investigation of its relationship to FLCN and FNIPs revealed that SMCR8 interacts robustly and directly with C9orf7259. Furthermore, this interaction with SMCR8 is critical for the stability of C9orf72 as C9orf72 levels plummeted when expression of SMCR8 was suppressed through either siRNA of CRISPR KO approaches59, 60. Phylogenetic analysis further supported a close relationship between C9orf72 and SMCR8 as both of these proteins showed a strongly correlated evolutionary pattern of gain and loss59. Additionally, no interactions were detected for C9orf72 with either FLCN and FNIP159 indicating that there is specificity for the formation of FLCN:FNIP and C9orf72:SMCR8 complexes within this sub-family of DENN domain proteins. While SMCR8 was originally named due to its chromosomal location close to the gene responsible for Smith-Magenis syndrome, it was ultimately excluded from having any role in this disease61 and had received little attention until these recent studies identified it as the major binding partner of C9orf72.
The C9orf72:SMCR8 complex was furthermore found to contain a third protein, WD repeat containing protein 41 (WDR41), a WD40 repeat containing protein with a predicted beta-propeller structure (Figure 1B)55-58, 62. The presence of WDR41 (as well as SMCR8) on lysosomal membranes was detected in a previous proteomics study63 but its function had not been previously investigated. Importantly, these interactions between C9orf72, SMCR8 and WDR41 are not transient as the proteins co-purified at close to 1:1:1 stoichiometry55. While C9orf72 and SMCR8 interact strongly even in the absence of WDR41 and depend on one another for their stability59, 60, structural insights into the interactions between C9orf72, SMCR8 and WDR41 remain to be determined.
In addition to interactions with SMCR8 and WDR41, proteomics experiments have also identified an interaction between C9orf72 and the kinases ULK156, 58, 64 and TBK155. Note however that such interactions do not occur at the same high stoichiometry as those that support the stable formation of the C9orf72:SMCR8:WDR41 core components but are still likely to exert regulatory functions on the complex. Along these lines, it is noteworthy that the interaction between C9orf72 and ULK1 is enhanced under starvation conditions and knockdown of SMCR8 results in a modest increase in total ULK1 protein levels58. In addition to the association with these kinases, SMCR8 is also a target of mTORC1-dependent phosphorylation65. These findings also raise interesting parallels with the FLCN:FNIP complex (Figure 1C). FNIP1 and FNIP2 bind the energy sensing kinase AMPK, and FNIP1 is phosphorylated in an AMPK- and mTOR-dependent manner50. In addition, FLCN can also be phosphorylated by ULK166. The functional significance of these similarities concerning interactions and phosphorylation of the C9orf72 and FLCN complexes remains poorly understood. It is likely that new structural insights into the organization of these protein complexes that reveal the locations of interaction interfaces and sites of phosphorylation will be required to fully address these problems.
C9orf72 KO results in lysosome defects in multiple experimental systems
Motivated by interest in the putative connections between C9orf72 depletion and neurodegenerative disease, initial studies of C9orf72 depletion in mice focused on the consequences of its conditional knockout in the nervous system67. However, this strategy did not reveal an overt neurodegenerative phenotype. Such a finding suggested several possible interpretations, ranging from the genuine lack of a role for reduced C9orf72 protein levels as a cause of ALS-FTD, to challenges with modeling such complex, age-dependent, human diseases in mice, or the possibility that the conditional KO strategy did not deplete C9orf72 in the most vulnerable cell types. For example, due to their origins in the monocytic lineage, brain cells such as microglia would not have been affected by the targeted KO of C9orf72 in neuronal progenitors. Indeed, a key role for C9orf72 in macrophages and microglia subsequently emerged from studies of conventional germline C9orf72 KO mice56, 68.
More specifically, an accumulation of enlarged lysosomes was observed in mouse C9orf72 KO macrophages, and microglia56, 68. Mice lacking C9orf72 also exhibited increased levels of SQSTM1/p62 and LC-3 (critical proteins involved in autophagy), potentially indicating an increase in autophagy or a defective clearance of autophagosomes by lysosomes60, 68.
Of additional interest, pathway analysis of genes co-expressed with C9orf72 revealed enrichment for genes involved in lysosome function68. Given the major role played by lysosomes in professional scavenger cells in the degradation and recycling of phagocytosed material, such cells are particularly sensitive to defects in lysosome function69. Key roles for C9orf72 in ensuring the lysosomal function of microglia is of particular interest with respect to neurodegenerative disease given recent insights into potentially important roles played by microglial progranulin (an FTD gene) in the regulation of synaptic pruning70. While there is good reason to focus on C9orf72 functions in macrophages and microglia, in addition to these monocyte lineage phenotypes, defects in both T and B cells have also been reported in C9orf72 KO mice71, 72.
The non-neuronal lysosome phenotypes that were identified in C9orf72 KO mice raise the interesting possibility that reduced C9orf72 expression in cells such as microglia contributes to ALS-FTD disease pathology. However, the cytoplasmic TDP-43 aggregates that robustly arise in human ALS-FTD neurons also suggests that the ability of neurons to clear such aggregates is impaired. Indeed, as autophagy is important for TDP-43 clearance in both mouse and human neurons73, there is a need for more detailed characterization of the autophagy-lysosome pathway in C9orf72 depleted neurons. Collectively, while such findings point to the need to consider how C9orf72 contributes to the physiology of multiple specialized cell types, there is also urgent need for better understanding of the fundamental biochemical functions of the C9orf72 protein whose loss could explain the diverse KO phenotypes as well as disease pathology.
In addition to knockout mouse studies, CRISPR-mediated KO of C9orf72 in human cells also yielded lysosomal phenotypes that included enhanced perinuclear clustering of swollen lysosomes59. Such changes in lysosome morphology were accompanied by impaired activation of mTOR complex 1 (mTORC1) signaling by amino acids59, 60, a process that takes place on the surface of lysosomes and is intimately linked to normal lysosome function74. These findings demonstrate that lysosome morphology and positioning changes following C9orf72 KO are accompanied by important lysosome-related functional consequences. As mTORC1 signaling has pleiotropic functions that collectively promote anabolic processes such as protein translation and suppress catabolic processes such as autophagy, even small changes in mTORC1 signaling have the potential to broadly affect cellular physiology74.
In contrast to the robust complex formed between C9orf72 and SMCR8, further investigation of SMCR8 KO and C9orf72+SMCR8 double KO phenotypes in cultured cells yielded phenotypes that were distinct from those arising from C9orf72 depletion. For example, while both C9orf72 and SMCR8 KO cells have defects in the stimulation of mTORC1 activity by an amino acid stimulus59, 60, SMCR8 KO cells selectively showed an elevated basal level of mTORC1 activity which was accompanied by cellular hypertrophy59. Furthermore, SMCR8 KO cells did not exhibit lysosomal enlargement or perinuclear clustering59. Perhaps most surprisingly, C9orf72+SMCR8 double KO cells had no mTORC1 signaling or lysosome positioning/morphology defects59. These divergent phenotypes arising from C9orf72 and SMCR8 KO have several interesting interpretations including the possibility that C9orf72 and/or SMCR8 have distinct functions that are mediated outside of the C9orf72:SMCR8:WDR41 complex. Alternatively, rather than reflecting loss-of-function, C9orf72 and SMCR8 single KO phenotypes could reflect a dominant, gain-of-function, arising from aberrant function of the remaining binding partner in the respective KOs. Such an interpretation is consistent with the wild type appearance of the double KO cells. Finally, these observations from the analysis of cultured human cells suggest that C9orf72 KO mouse phenotypes relating to lysosomes may arise from mechanisms that are more complex than a simple loss of the normal C9orf72 function. Answers to such questions await the generation and analysis of SMCR8 KO and C9orf72+SMCR8 double KO mice.
C9orf72 localizes to lysosomes
Understanding the subcellular localization of C9orf72 and its regulation is likely to be key to understanding its direct functions. Unfortunately, the combination of limited antibody availability, low protein abundance and lack of KOs to serve as negative controls for staining specificity long presented challenges to addressing the localization of the endogenous C9orf72 protein. Likewise, elucidation of C9orf72 localization via over-expression of tagged proteins is challenging due to the potential need for matching levels of SMCR8 and/or WDR41. This important problem was recently solved through the use of a CRISPR-Cas9 mediated genome editing strategy that inserted an epitope tag into the endogenous C9orf72 gene in cultured human cells59. This strategy enabled the detection of a specific immunofluorescence signal that was robustly enriched on the surface of lysosomes when cells were starved of amino acids but dispersed from the lysosome within minutes of amino acid re-feeding59. This localization of C9orf72 was highly selective for lysosomes and did not strongly overlap with related organelles such as autophagosomes. Localization of C9orf72 to lysosomes nicely parallels the lysosomal phenotypes arising from the C9orf72 KO (see above) and is consistent with the lysosome being an important site of direct functions for C9orf72. In addition to this recent direct demonstration of lysosomal localization for C9orf72, the co-occurrence of both SMCR8 and WDR41 on lysosomes is supported by proteomic analysis of purified lysosomal membranes63.
The negative regulation of C9orf72 lysosome recruitment by intracellular amino acid availability strikingly parallels the behavior of the related FLCN:FNIP protein complex51, 59. However, unlike FLCN:FNIP, which directly binds to and regulates the lysosome-localized Rag GTPases51, 52, the mechanisms that support lysosomal localization and functions of the C9orf72 complex are currently unknown. Related to this topic, unlike FLCN:FNIP, which shows regulated interactions with the Rags and which robustly controls the ability of Rags to recruit mTORC1 to lysosomes, C9orf72 does not co-immunoprecipitate with Rags and mTOR recruitment to lysosomes is intact in the absence of either C9orf72 or SMCR851, 59. Key questions thus remain unanswered about the mechanisms whereby amino acid sensing is coupled to regulation of C9orf72 localization as well as molecular links between C9orf72 localization to lysosomes and the regulation of mTOR signaling.
The localization of C9orf72 to lysosomes may shed light on recent findings relating to C9orf72 and TMEM106b75. Human genetics identified a common TMEM106b variant that protects against FTD risk in both progranulin (also a protein of lysosomes) mutation and C9orf72 repeat expansion carriers76-79. While the specific function of TMEM106b is not clear, it is a lysosome/late endosome-localized type II transmembrane protein whose overexpression induces a pronounced swelling of these organelles along with defects in their acidification and degradative function80, 81. A functional connection between TMEM106b and C9orf72 is supported by the suppression of these TMEM106b over-expression phenotypes in C9orf72-depleted cells80.
Putative GTPase targets of C9orf72
As summarized above, major functions of C9orf72 are likely to be mediated by the lysosome-localized C9orf72:SMCR8:WDR41 protein complex. The predicted presence of DENN domains in both C9orf72 and SMCR8 as well as similarity to the FLCN:FNIP complex suggest that the direct functions of C9orf72:SMCR8:WDR41 are likely to be mediated through GTPase regulation. Indeed, multiple Rabs have been proposed as putative C9orf72 targets. These include Rab1, Rab5, Rab7, and Rab11 which were found to interact with C9orf72 82. However, a subsequent study failed to recapitulate these interactions but instead found Rabs 8a and 39b were the strongest Rab interactors of C9orf72 while weaker interactions were detected for RAB6a, RAB12, RAB25, RAB33a, and RAB3855. In support of a role for C9orf72 as a GEF for Rab8a and Rab39b, recombinant C9orf72 as well as the whole C9orf72:SMCR8:WDR41 complex stimulated exchange of GDP for GTP on these Rabs in an in vitro assay55. The GEF activity of the complex towards Rab39b was also reported independently58. These observations are consistent with a role for C9orf72 and/or the larger protein complex as a Rab8a and Rab39b GEF. However, there are previously published examples of putative GEF-Rab pairs that fulfilled such binding and in vitro GEF activity criteria but were ultimately found not to be physiologically relevant. For example, FLCN was proposed to act as a Rab35 GEF based on in vitro assays36 but was later shown to function at lysosomes51 as a RagC GAP52. Likewise, in vitro assays initially supported a role for DENN2B as a Rab9 GEF83 but a more in depth analysis later demonstrated a physiological function as a Rab13 GEF84.
With past lessons in mind, several additional experiments would help to rigorously test the model that C9orf72 is a bona fide GEF for Rabs 8a and/or 39b. As it is a broadly conserved property for GEFs to preferentially interact with the nucleotide-free form of their target GTPase42, 43, it is predicted that C9orf72 should preferentially bind to Rab8a or 39b mutants that mimic this state. However, contrary to this expectation, it was reported that the C9orf72 complex did not discriminate between interacting with wildtype versus GDP/nucleotide-free or GTP-locked Rab8a and 39b mutants55. Likewise, as the GTP loading of Rab GTPases is tightly coupled to their recruitment to target membranes, the subcellular localization of their GEFs defines Rab localization85. However, it has yet to be shown that C9orf72, either alone or in complex with SMCR8 and WDR41, is required for the membrane recruitment of either Rab8a or Rab39b. Likewise, as C9orf72 is found on lysosomes59, it is predicted that if it functions as a Rab GEF, that it should promote the recruitment of its target GTPase to lysosomes. However, it has yet to be established that either Rab8a or Rab39b are enriched on C9orf72-positive lysosomes. Instead, Rab8a has been reported to reside on recycling endosomes and Golgi86 while Rab39b localizes to the Golgi87.
Ultimately, the mechanism of any GEF activity for C9orf72 would need to be understood at the structural level as was previously established DENND1B-Rab3535. Thus, while Rab8a and Rab39b represent intriguing candidates as targets for the C9orf72, key additional experiments would greatly help to clarify the physiological relevance and mechanistic basis for any actions of C9orf72, SMCR8 and WDR41 towards these GTPases. Likewise, if other physiologically relevant GTPase targets exist for C9orf72 they remain to be discovered and validated by similar criteria.
C9orf72 and Autophagy
Autophagy serves critical cellular functions through its ability to sequester diverse substrates that include: protein aggregates, damaged organelles and pathogens and deliver them to lysosomes for their degradation. This coupling between autophagosomes and lysosomes protects cells from toxic insults and also serves as an important source of nutrients during starvation. C9orf72 has been linked to autophagy in several ways. C9orf72 is critical for activation of mTORC1 by amino acids59, 60. mTORC1 in turn serves as a major negative regulator of autophagy via its phosphorylation of ULK188-90, a protein that is a critical positive regulator of autophagy induction. mTORC1 also negatively regulates TFEB, TFE3 and MITF91-93, a family of transcription factors regulates the expression of multiple genes related to lysosomes and autophagy94. Impaired mTORC1 activation in C9orf72 KO cells59, 60 would thus be predicted to induce higher levels of autophagy through a release of mTORC1-mediated inhibition on ULK1 and TFEB/TFE3/MITF. However, in contrast to pro-autophagy and lysosome effects arising from relief of mTORC1-dependent inhibition, the lysosome morphology defects that arise in the absence of C9orf7256, 59, 68 are concomitantly predicted to limit the degradation of autophagic cargos. Such an imbalance between autophagosome formation and lysosome-mediated clearance could result in a stress that is toxic to cells. In addition to regulating autophagy through mTORC1-dependent control of ULK1, C9orf72 (likely via the larger SMCR8 and WDR41 protein complex) also interacts with ULK1 and regulates ULK1 levels and activity56, 58. As ULK1 is not currently known to localize to lysosomes, it remains to be determined how such C9orf72-dependent regulation of ULK1 relates to the lysosomal localization of the C9orf72. C9orf72 was also reported to control ULK1 recruitment to sites of autophagosome formation via interactions with the GTP-bound form of Rab1A64.
The net effect of the various mechanisms through which C9orf72 can affect autophagy remains unclear as both increases58, 60 and decreases in autophagy55, 56 have been reported in C9orf72-depleted cells. The challenges in dissecting apart concurrent changes in autophagy induction and lysosome-mediated clearance may help to explain why these recent studies have broadly linked C9orf72 with regulation of autophagy but have come to differing conclusions as to whether C9orf72 promotes or inhibits the autophagic process. Resolution of these discrepancies will likely require a better mechanistic understanding of the direct biochemical and cell biological functions of C9orf72, SMCR8 and WDR41.
Lysosomes and ALS-FTD Genetics
With newfound appreciation of functions for C9orf72 in the autophagy-lysosome pathway, it is natural to ask whether the loss of such functions contributes to ALS-FTD risk in people with C9orf72 hexanucleotide repeat expansions and how such loss of function might synergize with gain-of-function defects that arise in parallel. While a definitive answer to these questions is still lacking, it is interesting that mutations in other autophagy-lysosome pathway genes have been identified as ALS and/or FTD risk factors. These include p62/SQSTM195, optineurin96 and ubiquilin 297, three genes encoding proteins involved in recruiting ubiquitinated cytoplasmic cargos into autophagososmes for their subsequent degradation in lysosomes as well as valosin containing protein98, a protein with regulatory functions related to both autophagosomes and lysosomes99, 100. Meanwhile, mutations in CHMP2B, a component of the ESCRT-III complex that supports the sequestration of ubiquitinated membrane proteins on late endosomes into multivesicular bodies for lysosomal degradation, is also an FTD gene101. Of particular interest, considering that hexanucleotide repeat expansions in C9orf72 only result in a partial reduction in C9orf72 levels, haploinsuffiency for progranulin, a protein of unknown function that resides in the lysosomal lumen102, is a well-established cause of FTD30, 31. Thus, even modest expression changes in key genes related to lysosomes can have important neurodegenerative disease consequences.
Conclusions
Recent studies have shed light onto the basic cellular function of the C9orf72 protein. Important binding partners, including SMCR8 and WDR41 have been found, subcellular localization has been identified and importance for biological processes including mTORC1 signaling, autophagy and lysosome morphology has been established. The role, if any, for C9orf72 function in ALS and FTD remains to be clarified. From a basic cell biological point of view, the similarities of the FLCN:FNIP complex to the C9orf72:SMCR8:WDR41 complex are striking. Future work to clarify the function of each complex and understand the mechanism of action of each complex should yield insights into how each of these two protein complexes achieve their related, but distinct functions in cells. Finally, while the genetic link between C9orf72 and ALS-FTD has focused attention on putative roles for C9orf72 in neurodegenerative disease, connections to lysosome function and mTORC1 signaling suggest the potential for much broader physiological and pathophysiological functions that remain to be investigated.
Synopsis.
Schematic diagram summarizing three distinct mechanisms that have been proposed to explain how hexanucleotide repeat expansion within the C9orf72 gene causes neurodegeneration. A) RNAs transcribed from the hexanucleotide repeat expansion form nuclear foci, which may trap key proteins involved in RNA processing. B) Dipeptide repeat proteins produced by repeat associated non-AUG (RAN) translation form aggregates that may impair the function of membrane-less organelles such as nucleoli and stress granules. Dipeptide repeat proteins may also disrupt transport through nuclear pores. C) The repeat expansion also suppresses expression of the endogenous C9orf72 protein. Based on recent knockout studies, loss of C9orf72 is predicted to impair lysosome function This review focuses on recent progress towards elucidating lysosome-related functions for C9orf72.
Acknowledgments
This work was supported in part by grants from the NIH (GM105718 and AG047270) and The Bluefield Project/Consortium for Frontotemporal Dementia Research to SMF. JA was supported by an NIGMS Fellowship (1F31GM119249-01).
References
- 1.Ling SC, Polymenidou M, Cleveland DW. Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron. 2013;79(3):416–438. doi: 10.1016/j.neuron.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ng AS, Rademakers R, Miller BL. Frontotemporal dementia: a bridge between dementia and neuromuscular disease. Ann N Y Acad Sci. 2015;1338:71–93. doi: 10.1111/nyas.12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, McCluskey LF, Miller BL, Masliah E, Mackenzie IR, Feldman H, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314(5796):130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 4.Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, Kalimo H, Paetau A, Abramzon Y, Remes AM, Kaganovich A, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72(2):257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gijselinck I, Van Langenhove T, van der Zee J, Sleegers K, Philtjens S, Kleinberger G, Janssens J, Bettens K, Van Cauwenberghe C, Pereson S, Engelborghs S, Sieben A, De Jonghe P, Vandenberghe R, Santens P, et al. A C9orf72 promoter repeat expansion in a Flanders-Belgian cohort with disorders of the frontotemporal lobar degeneration-amyotrophic lateral sclerosis spectrum: a gene identification study. Lancet Neurol. 2012;11(1):54–65. doi: 10.1016/S1474-4422(11)70261-7. [DOI] [PubMed] [Google Scholar]
- 6.DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, Kouri N, Wojtas A, Sengdy P, Hsiung GY, Karydas A, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72(2):245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsiung GY, DeJesus-Hernandez M, Feldman HH, Sengdy P, Bouchard-Kerr P, Dwosh E, Butler R, Leung B, Fok A, Rutherford NJ, Baker M, Rademakers R, Mackenzie IR. Clinical and pathological features of familial frontotemporal dementia caused by C9ORF72 mutation on chromosome 9p. Brain. 2012;135(Pt 3):709–722. doi: 10.1093/brain/awr354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mizielinska S, Lashley T, Norona FE, Clayton EL, Ridler CE, Fratta P, Isaacs AM. C9orf72 frontotemporal lobar degeneration is characterised by frequent neuronal sense and antisense RNA foci. Acta Neuropathol. 2013;126(6):845–857. doi: 10.1007/s00401-013-1200-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lagier-Tourenne C, Baughn M, Rigo F, Sun S, Liu P, Li HR, Jiang J, Watt AT, Chun S, Katz M, Qiu J, Sun Y, Ling SC, Zhu Q, Polymenidou M, et al. Targeted degradation of sense and antisense C9orf72 RNA foci as therapy for ALS and frontotemporal degeneration. Proc Natl Acad Sci U S A. 2013;110(47):E4530–4539. doi: 10.1073/pnas.1318835110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooper-Knock J, Walsh MJ, Higginbottom A, Robin Highley J, Dickman MJ, Edbauer D, Ince PG, Wharton SB, Wilson SA, Kirby J, Hautbergue GM, Shaw PJ. Sequestration of multiple RNA recognition motif-containing proteins by C9orf72 repeat expansions. Brain. 2014;137(Pt 7):2040–2051. doi: 10.1093/brain/awu120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donnelly CJ, Zhang PW, Pham JT, Haeusler AR, Mistry NA, Vidensky S, Daley EL, Poth EM, Hoover B, Fines DM, Maragakis N, Tienari PJ, Petrucelli L, Traynor BJ, Wang J, et al. RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron. 2013;80(2):415–428. doi: 10.1016/j.neuron.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reddy K, Zamiri B, Stanley SY, Macgregor RB, Jr, Pearson CE. The disease-associated r(GGGGCC)n repeat from the C9orf72 gene forms tract length-dependent uni- and multimolecular RNA G-quadruplex structures. J Biol Chem. 2013;288(14):9860–9866. doi: 10.1074/jbc.C113.452532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu Z, Poidevin M, Li X, Li Y, Shu L, Nelson DL, Li H, Hales CM, Gearing M, Wingo TS, Jin P. Expanded GGGGCC repeat RNA associated with amyotrophic lateral sclerosis and frontotemporal dementia causes neurodegeneration. Proc Natl Acad Sci U S A. 2013;110(19):7778–7783. doi: 10.1073/pnas.1219643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mori K, Lammich S, Mackenzie IR, Forne I, Zilow S, Kretzschmar H, Edbauer D, Janssens J, Kleinberger G, Cruts M, Herms J, Neumann M, Van Broeckhoven C, Arzberger T, Haass C. hnRNP A3 binds to GGGGCC repeats and is a constituent of p62-positive/TDP43-negative inclusions in the hippocampus of patients with C9orf72 mutations. Acta Neuropathol. 2013;125(3):413–423. doi: 10.1007/s00401-013-1088-7. [DOI] [PubMed] [Google Scholar]
- 15.Lee YB, Chen HJ, Peres JN, Gomez-Deza J, Attig J, Stalekar M, Troakes C, Nishimura AL, Scotter EL, Vance C, Adachi Y, Sardone V, Miller JW, Smith BN, Gallo JM, et al. Hexanucleotide repeats in ALS/FTD form length-dependent RNA foci, sequester RNA binding proteins, and are neurotoxic. Cell Rep. 2013;5(5):1178–1186. doi: 10.1016/j.celrep.2013.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haeusler AR, Donnelly CJ, Periz G, Simko EA, Shaw PG, Kim MS, Maragakis NJ, Troncoso JC, Pandey A, Sattler R, Rothstein JD, Wang J. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature. 2014;507(7491):195–200. doi: 10.1038/nature13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conlon EG, Lu L, Sharma A, Yamazaki T, Tang T, Shneider NA, Manley JL. The C9ORF72 GGGGCC expansion forms RNA G-quadruplex inclusions and sequesters hnRNP H to disrupt splicing in ALS brains. Elife. 2016;5 doi: 10.7554/eLife.17820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zu T, Liu Y, Banez-Coronel M, Reid T, Pletnikova O, Lewis J, Miller TM, Harms MB, Falchook AE, Subramony SH, Ostrow LW, Rothstein JD, Troncoso JC, Ranum LP. RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc Natl Acad Sci U S A. 2013;110(51):E4968–4977. doi: 10.1073/pnas.1315438110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ash PE, Bieniek KF, Gendron TF, Caulfield T, Lin WL, Dejesus-Hernandez M, van Blitterswijk MM, Jansen-West K, Paul JW, 3rd, Rademakers R, Boylan KB, Dickson DW, Petrucelli L. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013;77(4):639–646. doi: 10.1016/j.neuron.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mori K, Weng SM, Arzberger T, May S, Rentzsch K, Kremmer E, Schmid B, Kretzschmar HA, Cruts M, Van Broeckhoven C, Haass C, Edbauer D. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013;339(6125):1335–1338. doi: 10.1126/science.1232927. [DOI] [PubMed] [Google Scholar]
- 21.Schludi MH, May S, Grasser FA, Rentzsch K, Kremmer E, Kupper C, Klopstock T, Arzberger T, Edbauer D German Consortium for Frontotemporal Lobar D, Bavarian Brain Banking A. Distribution of dipeptide repeat proteins in cellular models and C9orf72 mutation cases suggests link to transcriptional silencing. Acta Neuropathol. 2015;130(4):537–555. doi: 10.1007/s00401-015-1450-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mackenzie IR, Arzberger T, Kremmer E, Troost D, Lorenzl S, Mori K, Weng SM, Haass C, Kretzschmar HA, Edbauer D, Neumann M. Dipeptide repeat protein pathology in C9ORF72 mutation cases: clinico-pathological correlations. Acta Neuropathol. 2013;126(6):859–879. doi: 10.1007/s00401-013-1181-y. [DOI] [PubMed] [Google Scholar]
- 23.Mizielinska S, Gronke S, Niccoli T, Ridler CE, Clayton EL, Devoy A, Moens T, Norona FE, Woollacott IO, Pietrzyk J, Cleverley K, Nicoll AJ, Pickering-Brown S, Dols J, Cabecinha M, et al. C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science. 2014;345(6201):1192–1194. doi: 10.1126/science.1256800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.May S, Hornburg D, Schludi MH, Arzberger T, Rentzsch K, Schwenk BM, Grasser FA, Mori K, Kremmer E, Banzhaf-Strathmann J, Mann M, Meissner F, Edbauer D. C9orf72 FTLD/ALS-associated Gly-Ala dipeptide repeat proteins cause neuronal toxicity and Unc119 sequestration. Acta Neuropathol. 2014;128(4):485–503. doi: 10.1007/s00401-014-1329-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwon I, Xiang S, Kato M, Wu L, Theodoropoulos P, Wang T, Kim J, Yun J, Xie Y, McKnight SL. Poly-dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science. 2014;345(6201):1139–1145. doi: 10.1126/science.1254917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Belzil VV, Bauer PO, Prudencio M, Gendron TF, Stetler CT, Yan IK, Pregent L, Daughrity L, Baker MC, Rademakers R, Boylan K, Patel TC, Dickson DW, Petrucelli L. Reduced C9orf72 gene expression in c9FTD/ALS is caused by histone trimethylation, an epigenetic event detectable in blood. Acta Neuropathol. 2013;126(6):895–905. doi: 10.1007/s00401-013-1199-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gijselinck I, Van Mossevelde S, van der Zee J, Sieben A, Engelborghs S, De Bleecker J, Ivanoiu A, Deryck O, Edbauer D, Zhang M, Heeman B, Baumer V, Van den Broeck M, Mattheijssens M, Peeters K, et al. The C9orf72 repeat size correlates with onset age of disease, DNA methylation and transcriptional downregulation of the promoter. Mol Psychiatry. 2016;21(8):1112–1124. doi: 10.1038/mp.2015.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xi Z, Zinman L, Moreno D, Schymick J, Liang Y, Sato C, Zheng Y, Ghani M, Dib S, Keith J, Robertson J, Rogaeva E. Hypermethylation of the CpG island near the G4C2 repeat in ALS with a C9orf72 expansion. Am J Hum Genet. 2013;92(6):981–989. doi: 10.1016/j.ajhg.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu EY, Russ J, Wu K, Neal D, Suh E, McNally AG, Irwin DJ, Van Deerlin VM, Lee EB. C9orf72 hypermethylation protects against repeat expansion-associated pathology in ALS/FTD. Acta Neuropathol. 2014;128(4):525–541. doi: 10.1007/s00401-014-1286-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baker M, Mackenzie IR, Pickering-Brown SM, Gass J, Rademakers R, Lindholm C, Snowden J, Adamson J, Sadovnick AD, Rollinson S, Cannon A, Dwosh E, Neary D, Melquist S, Richardson A, et al. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature. 2006;442(7105):916–919. doi: 10.1038/nature05016. [DOI] [PubMed] [Google Scholar]
- 31.Cruts M, Gijselinck I, van der Zee J, Engelborghs S, Wils H, Pirici D, Rademakers R, Vandenberghe R, Dermaut B, Martin JJ, van Duijn C, Peeters K, Sciot R, Santens P, De Pooter T, et al. Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature. 2006;442(7105):920–924. doi: 10.1038/nature05017. [DOI] [PubMed] [Google Scholar]
- 32.Chow VT, Lee SS. DENN, a novel human gene differentially expressed in normal and neoplastic cells. DNA Seq. 1996;6(5):263–273. doi: 10.3109/10425179609020873. [DOI] [PubMed] [Google Scholar]
- 33.Zhang D, Iyer LM, He F, Aravind L. Discovery of Novel DENN Proteins: Implications for the Evolution of Eukaryotic Intracellular Membrane Structures and Human Disease. Front Genet. 2012;3:283. doi: 10.3389/fgene.2012.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levine TP, Daniels RD, Gatta AT, Wong LH, Hayes MJ. The product of C9orf72, a gene strongly implicated in neurodegeneration, is structurally related to DENN Rab-GEFs. Bioinformatics. 2013;29(4):499–503. doi: 10.1093/bioinformatics/bts725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu X, Bradley MJ, Cai Y, Kummel D, De La Cruz EM, Barr FA, Reinisch KM. Insights regarding guanine nucleotide exchange from the structure of a DENN-domain protein complexed with its Rab GTPase substrate. Proc Natl Acad Sci U S A. 2011;108(46):18672–18677. doi: 10.1073/pnas.1110415108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nookala RK, Langemeyer L, Pacitto A, Ochoa-Montano B, Donaldson JC, Blaszczyk BK, Chirgadze DY, Barr FA, Bazan JF, Blundell TL. Crystal structure of folliculin reveals a hidDENN function in genetically inherited renal cancer. Open Biol. 2012;2(8):120071. doi: 10.1098/rsob.120071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pacitto A, Ascher DB, Wong LH, Blaszczyk BK, Nookala RK, Zhang N, Dokudovskaya S, Levine TP, Blundell TL. Lst4, the yeast Fnip1/2 orthologue, is a DENN-family protein. Open Biol. 2015;5(12):150174. doi: 10.1098/rsob.150174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chaineau M, Ioannou MS, McPherson PS. Rab35: GEFs, GAPs and effectors. Traffic. 2013;14(11):1109–1117. doi: 10.1111/tra.12096. [DOI] [PubMed] [Google Scholar]
- 39.Klopper TH, Kienle N, Fasshauer D, Munro S. Untangling the evolution of Rab G proteins: implications of a comprehensive genomic analysis. BMC Biol. 2012;10:71. doi: 10.1186/1741-7007-10-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zerial M, McBride H. Rab proteins as membrane organizers. Nat Rev Mol Cell Biol. 2001;2(2):107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- 41.Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009;10(8):513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- 42.Koch D, Rai A, Ali I, Bleimling N, Friese T, Brockmeyer A, Janning P, Goud B, Itzen A, Muller MP, Goody RS. A pull-down procedure for the identification of unknown GEFs for small GTPases. Small GTPases. 2016;7(2):93–106. doi: 10.1080/21541248.2016.1156803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo Z, Ahmadian MR, Goody RS. Guanine nucleotide exchange factors operate by a simple allosteric competitive mechanism. Biochemistry. 2005;44(47):15423–15429. doi: 10.1021/bi0518601. [DOI] [PubMed] [Google Scholar]
- 44.Cherfils J, Zeghouf M. Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol Rev. 2013;93(1):269–309. doi: 10.1152/physrev.00003.2012. [DOI] [PubMed] [Google Scholar]
- 45.Araki S, Kikuchi A, Hata Y, Isomura M, Takai Y. Regulation of reversible binding of smg p25A, a ras p21-like GTP-binding protein, to synaptic plasma membranes and vesicles by its specific regulatory protein, GDP dissociation inhibitor. J Biol Chem. 1990;265(22):13007–13015. [PubMed] [Google Scholar]
- 46.Grosshans BL, Ortiz D, Novick P. Rabs and their effectors: achieving specificity in membrane traffic. Proc Natl Acad Sci U S A. 2006;103(32):11821–11827. doi: 10.1073/pnas.0601617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Allaire PD, Marat AL, Dall'Armi C, Di Paolo G, McPherson PS, Ritter B. The Connecdenn DENN domain: a GEF for Rab35 mediating cargo-specific exit from early endosomes. Mol Cell. 2010;37(3):370–382. doi: 10.1016/j.molcel.2009.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marat AL, McPherson PS. The connecdenn family, Rab35 guanine nucleotide exchange factors interfacing with the clathrin machinery. J Biol Chem. 2010;285(14):10627–10637. doi: 10.1074/jbc.M109.050930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cauvin C, Rosendale M, Gupta-Rossi N, Rocancourt M, Larraufie P, Salomon R, Perrais D, Echard A. Rab35 GTPase Triggers Switch-like Recruitment of the Lowe Syndrome Lipid Phosphatase OCRL on Newborn Endosomes. Curr Biol. 2016;26(1):120–128. doi: 10.1016/j.cub.2015.11.040. [DOI] [PubMed] [Google Scholar]
- 50.Baba M, Hong SB, Sharma N, Warren MB, Nickerson ML, Iwamatsu A, Esposito D, Gillette WK, Hopkins RF, 3rd, Hartley JL, Furihata M, Oishi S, Zhen W, Burke TR, Jr, Linehan WM, et al. Folliculin encoded by the BHD gene interacts with a binding protein, FNIP1, and AMPK, and is involved in AMPK and mTOR signaling. Proc Natl Acad Sci U S A. 2006;103(42):15552–15557. doi: 10.1073/pnas.0603781103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Petit CS, Roczniak-Ferguson A, Ferguson SM. Recruitment of folliculin to lysosomes supports the amino acid-dependent activation of Rag GTPases. J Cell Biol. 2013;202(7):1107–1122. doi: 10.1083/jcb.201307084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsun ZY, Bar-Peled L, Chantranupong L, Zoncu R, Wang T, Kim C, Spooner E, Sabatini DM. The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Mol Cell. 2013;52(4):495–505. doi: 10.1016/j.molcel.2013.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peli-Gulli MP, Sardu A, Panchaud N, Raucci S, De Virgilio C. Amino Acids Stimulate TORC1 through Lst4-Lst7, a GTPase-Activating Protein Complex for the Rag Family GTPase Gtr2. Cell Rep. 2015;13(1):1–7. doi: 10.1016/j.celrep.2015.08.059. [DOI] [PubMed] [Google Scholar]
- 54.Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141(2):290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sellier C, Campanari ML, Julie Corbier C, Gaucherot A, Kolb-Cheynel I, Oulad-Abdelghani M, Ruffenach F, Page A, Ciura S, Kabashi E, Charlet-Berguerand N. Loss of C9ORF72 impairs autophagy and synergizes with polyQ Ataxin-2 to induce motor neuron dysfunction and cell death. EMBO J. 2016;35(12):1276–1297. doi: 10.15252/embj.201593350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sullivan PM, Zhou X, Robins AM, Paushter DH, Kim D, Smolka MB, Hu F. The ALS/FTLD associated protein C9orf72 associates with SMCR8 and WDR41 to regulate the autophagy-lysosome pathway. Acta Neuropathol Commun. 2016;4(1):51. doi: 10.1186/s40478-016-0324-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xiao S, MacNair L, McLean J, McGoldrick P, McKeever P, Soleimani S, Keith J, Zinman L, Rogaeva E, Robertson J. C9orf72 isoforms in Amyotrophic Lateral Sclerosis and Frontotemporal Lobar Degeneration. Brain Res. 2016;1647:43–49. doi: 10.1016/j.brainres.2016.04.062. [DOI] [PubMed] [Google Scholar]
- 58.Yang M, Liang C, Swaminathan K, Herrlinger S, Lai F, Shiekhattar R, Chen JF. A C9ORF72/SMCR8-containing complex regulates ULK1 and plays a dual role in autophagy. Sci Adv. 2016;2(9):e1601167. doi: 10.1126/sciadv.1601167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Amick J, Roczniak-Ferguson A, Ferguson SM. C9orf72 binds SMCR8, localizes to lysosomes, and regulates mTORC1 signaling. Mol Biol Cell. 2016;27(20):3040–3051. doi: 10.1091/mbc.E16-01-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ugolino J, Ji YJ, Conchina K, Chu J, Nirujogi RS, Pandey A, Brady NR, Hamacher-Brady A, Wang J. Loss of C9orf72 Enhances Autophagic Activity via Deregulated mTOR and TFEB Signaling. PLoS Genet. 2016;12(11):e1006443. doi: 10.1371/journal.pgen.1006443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Slager RE, Newton TL, Vlangos CN, Finucane B, Elsea SH. Mutations in RAI1 associated with Smith-Magenis syndrome. Nat Genet. 2003;33(4):466–468. doi: 10.1038/ng1126. [DOI] [PubMed] [Google Scholar]
- 62.Wang Y, Hu XJ, Zou XD, Wu XH, Ye ZQ, Wu YD. WDSPdb: a database for WD40-repeat proteins. Nucleic Acids Res. 2015;43(Database issue):D339–344. doi: 10.1093/nar/gku1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schroder B, Wrocklage C, Pan C, Jager R, Kosters B, Schafer H, Elsasser HP, Mann M, Hasilik A. Integral and associated lysosomal membrane proteins. Traffic. 2007;8(12):1676–1686. doi: 10.1111/j.1600-0854.2007.00643.x. [DOI] [PubMed] [Google Scholar]
- 64.Webster CP, Smith EF, Bauer CS, Moller A, Hautbergue GM, Ferraiuolo L, Myszczynska MA, Higginbottom A, Walsh MJ, Whitworth AJ, Kaspar BK, Meyer K, Shaw PJ, Grierson AJ, De Vos KJ. The C9orf72 protein interacts with Rab1a and the ULK1 complex to regulate initiation of autophagy. EMBO J. 2016;35(15):1656–1676. doi: 10.15252/embj.201694401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hsu PP, Kang SA, Rameseder J, Zhang Y, Ottina KA, Lim D, Peterson TR, Choi Y, Gray NS, Yaffe MB, Marto JA, Sabatini DM. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science. 2011;332(6035):1317–1322. doi: 10.1126/science.1199498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dunlop EA, Seifan S, Claessens T, Behrends C, Kamps MA, Rozycka E, Kemp AJ, Nookala RK, Blenis J, Coull BJ, Murray JT, van Steensel MA, Wilkinson S, Tee AR. FLCN, a novel autophagy component, interacts with GABARAP and is regulated by ULK1 phosphorylation. Autophagy. 2014;10(10):1749–1760. doi: 10.4161/auto.29640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Koppers M, Blokhuis AM, Westeneng HJ, Terpstra ML, Zundel CA, Vieira de Sa R, Schellevis RD, Waite AJ, Blake DJ, Veldink JH, van den Berg LH, Pasterkamp RJ. C9orf72 ablation in mice does not cause motor neuron degeneration or motor deficits. Ann Neurol. 2015;78(3):426–438. doi: 10.1002/ana.24453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.O'Rourke JG, Bogdanik L, Yanez A, Lall D, Wolf AJ, Muhammad AK, Ho R, Carmona S, Vit JP, Zarrow J, Kim KJ, Bell S, Harms MB, Miller TM, Dangler CA, et al. C9orf72 is required for proper macrophage and microglial function in mice. Science. 2016;351(6279):1324–1329. doi: 10.1126/science.aaf1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Aflaki E, Stubblefield BK, Maniwang E, Lopez G, Moaven N, Goldin E, Marugan J, Patnaik S, Dutra A, Southall N, Zheng W, Tayebi N, Sidransky E. Macrophage models of Gaucher disease for evaluating disease pathogenesis and candidate drugs. Sci Transl Med. 2014;6(240):240ra273. doi: 10.1126/scitranslmed.3008659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lui H, Zhang J, Makinson SR, Cahill MK, Kelley KW, Huang HY, Shang Y, Oldham MC, Martens LH, Gao F, Coppola G, Sloan SA, Hsieh CL, Kim CC, Bigio EH, et al. Progranulin Deficiency Promotes Circuit-Specific Synaptic Pruning by Microglia via Complement Activation. Cell. 2016;165(4):921–935. doi: 10.1016/j.cell.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Atanasio A, Decman V, White D, Ramos M, Ikiz B, Lee HC, Siao CJ, Brydges S, LaRosa E, Bai Y, Fury W, Burfeind P, Zamfirova R, Warshaw G, Orengo J, et al. C9orf72 ablation causes immune dysregulation characterized by leukocyte expansion, autoantibody production, and glomerulonephropathy in mice. Sci Rep. 2016;6:23204. doi: 10.1038/srep23204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sudria-Lopez E, Koppers M, de Wit M, van der Meer C, Westeneng HJ, Zundel CA, Youssef SA, Harkema L, de Bruin A, Veldink JH, van den Berg LH, Pasterkamp RJ. Full ablation of C9orf72 in mice causes immune system-related pathology and neoplastic events but no motor neuron defects. Acta Neuropathol. 2016;132(1):145–147. doi: 10.1007/s00401-016-1581-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Barmada SJ, Serio A, Arjun A, Bilican B, Daub A, Ando DM, Tsvetkov A, Pleiss M, Li X, Peisach D, Shaw C, Chandran S, Finkbeiner S. Autophagy induction enhances TDP43 turnover and survival in neuronal ALS models. Nat Chem Biol. 2014;10(8):677–685. doi: 10.1038/nchembio.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ferguson SM. Beyond indigestion: emerging roles for lysosome-based signaling in human disease. Curr Opin Cell Biol. 2015;35:59–68. doi: 10.1016/j.ceb.2015.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nicholson AM, Rademakers R. What we know about TMEM106B in neurodegeneration. Acta Neuropathol. 2016;132(5):639–651. doi: 10.1007/s00401-016-1610-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gallagher MD, Suh E, Grossman M, Elman L, McCluskey L, Van Swieten JC, Al-Sarraj S, Neumann M, Gelpi E, Ghetti B, Rohrer JD, Halliday G, Van Broeckhoven C, Seilhean D, Shaw PJ, et al. TMEM106B is a genetic modifier of frontotemporal lobar degeneration with C9orf72 hexanucleotide repeat expansions. Acta Neuropathol. 2014;127(3):407–418. doi: 10.1007/s00401-013-1239-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.van Blitterswijk M, Mullen B, Nicholson AM, Bieniek KF, Heckman MG, Baker MC, DeJesus-Hernandez M, Finch NA, Brown PH, Murray ME, Hsiung GY, Stewart H, Karydas AM, Finger E, Kertesz A, et al. TMEM106B protects C9ORF72 expansion carriers against frontotemporal dementia. Acta Neuropathol. 2014;127(3):397–406. doi: 10.1007/s00401-013-1240-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Finch N, Carrasquillo MM, Baker M, Rutherford NJ, Coppola G, Dejesus-Hernandez M, Crook R, Hunter T, Ghidoni R, Benussi L, Crook J, Finger E, Hantanpaa KJ, Karydas AM, Sengdy P, et al. TMEM106B regulates progranulin levels and the penetrance of FTLD in GRN mutation carriers. Neurology. 2011;76(5):467–474. doi: 10.1212/WNL.0b013e31820a0e3b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Van Deerlin VM, Sleiman PM, Martinez-Lage M, Chen-Plotkin A, Wang LS, Graff-Radford NR, Dickson DW, Rademakers R, Boeve BF, Grossman M, Arnold SE, Mann DM, Pickering-Brown SM, Seelaar H, Heutink P, et al. Common variants at 7p21 are associated with frontotemporal lobar degeneration with TDP-43 inclusions. Nat Genet. 2010;42(3):234–239. doi: 10.1038/ng.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Busch JI, Unger TL, Jain N, Tyler Skrinak R, Charan RA, Chen-Plotkin AS. Increased expression of the frontotemporal dementia risk factor TMEM106B causes C9orf72-dependent alterations in lysosomes. Hum Mol Genet. 2016;25(13):2681–2697. doi: 10.1093/hmg/ddw127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stagi M, Klein ZA, Gould TJ, Bewersdorf J, Strittmatter SM. Lysosome size, motility and stress response regulated by fronto-temporal dementia modifier TMEM106B. Mol Cell Neurosci. 2014;61:226–240. doi: 10.1016/j.mcn.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Farg MA, Sundaramoorthy V, Sultana JM, Yang S, Atkinson RA, Levina V, Halloran MA, Gleeson PA, Blair IP, Soo KY, King AE, Atkin JD. C9ORF72, implicated in amytrophic lateral sclerosis and frontotemporal dementia, regulates endosomal trafficking. Hum Mol Genet. 2014;23(13):3579–3595. doi: 10.1093/hmg/ddu068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yoshimura S, Gerondopoulos A, Linford A, Rigden DJ, Barr FA. Family-wide characterization of the DENN domain Rab GDP-GTP exchange factors. J Cell Biol. 2010;191(2):367–381. doi: 10.1083/jcb.201008051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ioannou MS, Bell ES, Girard M, Chaineau M, Hamlin JN, Daubaras M, Monast A, Park M, Hodgson L, McPherson PS. DENND2B activates Rab13 at the leading edge of migrating cells and promotes metastatic behavior. J Cell Biol. 2015;208(5):629–648. doi: 10.1083/jcb.201407068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Blumer J, Rey J, Dehmelt L, Mazel T, Wu YW, Bastiaens P, Goody RS, Itzen A. RabGEFs are a major determinant for specific Rab membrane targeting. J Cell Biol. 2013;200(3):287–300. doi: 10.1083/jcb.201209113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ang AL, Folsch H, Koivisto UM, Pypaert M, Mellman I. The Rab8 GTPase selectively regulates AP-1B-dependent basolateral transport in polarized Madin-Darby canine kidney cells. J Cell Biol. 2003;163(2):339–350. doi: 10.1083/jcb.200307046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Giannandrea M, Bianchi V, Mignogna ML, Sirri A, Carrabino S, D'Elia E, Vecellio M, Russo S, Cogliati F, Larizza L, Ropers HH, Tzschach A, Kalscheuer V, Oehl-Jaschkowitz B, Skinner C, et al. Mutations in the small GTPase gene RAB39B are responsible for X-linked mental retardation associated with autism, epilepsy, and macrocephaly. Am J Hum Genet. 2010;86(2):185–195. doi: 10.1016/j.ajhg.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, Guan JL, Oshiro N, Mizushima N. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20(7):1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, Kundu M, Kim DH. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20(7):1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13(2):132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Roczniak-Ferguson A, Petit CS, Froehlich F, Qian S, Ky J, Angarola B, Walther TC, Ferguson SM. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal. 2012;5(228):ra42. doi: 10.1126/scisignal.2002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Martina JA, Chen Y, Gucek M, Puertollano R. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy. 2012;8(6):903–914. doi: 10.4161/auto.19653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Erdin S, Huynh T, Ferron M, Karsenty G, Vellard MC, Facchinetti V, Sabatini DM, Ballabio A. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 2012;31(5):1095–1108. doi: 10.1038/emboj.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Napolitano G, Ballabio A. TFEB at a glance. J Cell Sci. 2016;129(13):2475–2481. doi: 10.1242/jcs.146365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fecto F, Yan J, Vemula SP, Liu E, Yang Y, Chen W, Zheng JG, Shi Y, Siddique N, Arrat H, Donkervoort S, Ajroud-Driss S, Sufit RL, Heller SL, Deng HX, et al. SQSTM1 mutations in familial and sporadic amyotrophic lateral sclerosis. Arch Neurol. 2011;68(11):1440–1446. doi: 10.1001/archneurol.2011.250. [DOI] [PubMed] [Google Scholar]
- 96.Maruyama H, Morino H, Ito H, Izumi Y, Kato H, Watanabe Y, Kinoshita Y, Kamada M, Nodera H, Suzuki H, Komure O, Matsuura S, Kobatake K, Morimoto N, Abe K, et al. Mutations of optineurin in amyotrophic lateral sclerosis. Nature. 2010;465(7295):223–226. doi: 10.1038/nature08971. [DOI] [PubMed] [Google Scholar]
- 97.Deng HX, Chen W, Hong ST, Boycott KM, Gorrie GH, Siddique N, Yang Y, Fecto F, Shi Y, Zhai H, Jiang H, Hirano M, Rampersaud E, Jansen GH, Donkervoort S, et al. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature. 2011;477(7363):211–215. doi: 10.1038/nature10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Watts GD, Wymer J, Kovach MJ, Mehta SG, Mumm S, Darvish D, Pestronk A, Whyte MP, Kimonis VE. Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat Genet. 2004;36(4):377–381. doi: 10.1038/ng1332. [DOI] [PubMed] [Google Scholar]
- 99.Papadopoulos C, Kirchner P, Bug M, Grum D, Koerver L, Schulze N, Poehler R, Dressler A, Fengler S, Arhzaouy K, Lux V, Ehrmann M, Weihl CC, Meyer H. VCP/p97 cooperates with YOD1, UBXD1 and PLAA to drive clearance of ruptured lysosomes by autophagy. EMBO J. 2016 doi: 10.15252/embj.201695148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Johnson AE, Shu H, Hauswirth AG, Tong A, Davis GW. VCP-dependent muscle degeneration is linked to defects in a dynamic tubular lysosomal network in vivo. Elife. 2015;4 doi: 10.7554/eLife.07366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cannon A, Baker M, Boeve B, Josephs K, Knopman D, Petersen R, Parisi J, Dickison D, Adamson J, Snowden J, Neary D, Mann D, Hutton M, Pickering-Brown SM. CHMP2B mutations are not a common cause of frontotemporal lobar degeneration. Neurosci Lett. 2006;398(1-2):83–84. doi: 10.1016/j.neulet.2005.12.056. [DOI] [PubMed] [Google Scholar]
- 102.Gowrishankar S, Yuan P, Wu Y, Schrag M, Paradise S, Grutzendler J, De Camilli P, Ferguson SM. Massive accumulation of luminal protease-deficient axonal lysosomes at Alzheimer's disease amyloid plaques. Proc Natl Acad Sci U S A. 2015;112(28):E3699–3708. doi: 10.1073/pnas.1510329112. [DOI] [PMC free article] [PubMed] [Google Scholar]