Abstract
Primary graft dysfunction (PGD) is a principal cause of early morbidity and mortality after lung transplantation, but its pathogenic mechanisms are not fully clarified. Thus far, studies using standard clinical assays have not linked microbial factors to PGD. We previously used comprehensive metagenomic methods to characterize viruses in lung allografts >1 month post-transplant and found that levels of Anellovirus, mainly Torque teno viruses (TTV), were significantly higher than in non-transplant healthy controls. Here we used quantitative PCR to analyze TTV and shotgun metagenomics to characterize full viral communities in acellular bronchoalveolar lavage from donor organs and post-reperfusion allografts in PGD and non-PGD lung transplant recipient pairs. Unexpectedly, TTV DNA levels were 100-fold elevated in donor lungs compared with healthy adults (p=0.0026). Although absolute TTV levels did not differ by PGD status, PGD cases showed a smaller increase in TTV levels from pre- to post-transplant than did control recipients (p=0.041). Metagenomic sequencing revealed mainly TTV and bacteriophages of respiratory tract bacteria, but no viral taxa distinguished PGD cases from controls. These findings suggest that conditions associated with brain death promote TTV replication, and that greater immune activation or tissue injury associated with PGD may restrict TTV abundance in the lung.
Introduction
Lung transplantation is the only long-term option for many patients with end-stage lung diseases. Median survival for lung transplant recipients (LTxRs) is only 5.7 years (1), the lowest among solid-organ transplants (2). Factors limiting survival include primary graft dysfunction (PGD), infection, cellular and antibody mediated rejection, and chronic lung allograft dysfunction (1).
PGD is a form of acute lung injury characterized by hypoxemia, pulmonary edema and alveolar damage that occurs within 72 hours of transplantation (3). Approximately 10–25% of LTxRs experience severe PGD, which is a leading cause of death early post-transplantation (3), and survivors of severe PGD are more likely to develop donor-specific antibodies and chronic rejection (4, 5).
The pathogenesis of PGD is thought to involve both inflammatory and ischemia-reperfusion-related mechanisms. An inflammatory environment develops within the lung after donor brain death, which is perpetuated after reperfusion by recruitment of lymphocytes, macrophages, and neutrophils (3, 6–12). Activated immune cells and reactive oxygen species generated during ischemia-reperfusion are thought to injure lung endothelium and epithelium (3). Host genetic variation in genes involved in oxidant stress responses and innate immunity correlate with risk of PGD (13, 14). PGD has also been associated with inflammasome activation, pattern recognition receptor signaling, and complement activation within the allograft (15–17). These studies raise the possibility of a microbial contribution to pathogenesis.
Culture-based and molecular clinical assays have associated specific bacteria, fungi and community-acquired respiratory viruses with lung transplantation outcomes, such as the development of bronchiolitis obliterans syndrome (BOS) (18–24). Studies using high throughput metagenomic sequencing have demonstrated that lung bacterial, fungal and viral communities are aberrant in lungs of LTxRs compared to healthy controls (22, 25, 26). LTxRs have decreased bacterial diversity, increased bacterial load, outgrowths of pathogens and presence of atypical species (22, 25, 27, 28). One study linked metagenomic bacterial community features and BOS (29).
Our group recently reported high levels of diverse TTVs in acellular bronchoalveolar lavage (BAL) fluid from LTxRs sampled from one month to >10 years following transplant (26). TTVs are small, non-enveloped, circular, negative sense, single-stranded DNA, eukaryotic-cell viruses belonging to the Anellovirus family. These viruses are ubiquitous in the human population (30), with initial infection occurring during childhood (31–33). Changes in blood TTV levels correlate with altered immune states. Plasma TTV DNA levels increase in HIV+ individuals that progress to AIDS and decrease following immune reconstitution with antiretroviral therapy (34–36). Expansion of TTV is seen in blood of solid-organ transplant recipients receiving immunosuppressive therapy (37–39). Thus far, TTV has not been identified as the etiological agent of any disease (30, 40).
In this study we investigated the hypothesis that lung viruses, including TTV, might be associated with PGD. To do this, we analyzed extracellular viruses present in acellular BAL sampled from lung donors immediately prior to organ recovery and from the allograft one hour after transplant and reperfusion. Two complementary approaches were used: 1) a TTV quantitative polymerase chain reaction (qPCR) assay and 2) shotgun metagenomic sequence analysis of all viruses. These assays were also performed on recipient serum samples obtained at the time of reperfusion. We also investigated whether TTV levels were correlated with host cell gene expression, which was previously reported to correlate with PGD in these patients (15).
Methods
Study population and data collected
Stored samples and clinical data were obtained from subjects who had previously been prospectively enrolled in the multicenter Clinical Trials in Organ Transplantation-03 (CTOT-03) (NCT00531921) study. A previous report described the selection of 23 grade 3 PGD cases and controls matched on donor age and pre-transplant recipient diagnosis (15). For one PGD case no specimens remained from their matched control, and one additional PGD case was included for which there was no matching control. Thus, samples were available from 22 case-control pairs and 2 unmatched PGD cases (Table 1). For 15 pairs, all perioperative BAL specimens were available for virome analysis, while for 7 pairs there was insufficient sample remaining from one or more lung samples (Table 1 and S1). Acellular BAL samples were obtained from healthy adult volunteers, as previously reported (Table S2) (41, 42). Serum from a separate group of 11 healthy adult volunteers was also collected. All subjects provided written informed consent under protocols approved by Institutional Review Boards at their respective institutions.
Table 1.
Clinical Features of Lung Transplant Recipients
| Subject ID |
Pair | Group | Samples | Age | Sex | Preoperative Diagnosis |
Donor BAL Bacterial and Fungal Culture |
|---|---|---|---|---|---|---|---|
| 11037 | 1 | Control | DB, RB, RS | 50 | M | CF/Bronch | Negative |
| 11012 | 1 | PGD | DB, RB, RS | 52 | F | CF/Bronch | Negative |
| 13017 | 2 | Control | DB, RB, RS | 36 | F | CF/Bronch | Staphylococcus |
| 12009 | 2 | PGD | DB, RB, RS | 53 | F | CF/Bronch | MRSA |
| 12004 | 3 | Control | RB, RS | 29 | M | CF/Bronch | Negative |
| 13026 | 3 | PGD | DB, RS | 29 | M | CF/Bronch | Negative |
| 12016 | 4 | Control | DB, RB, RS | 47 | F | CF/Bronch | ND |
| 13032 | 4 | PGD | DB, RS | 53 | M | CF/Bronch | Staphylococcus |
| 11005 | 5 | Control | DB, RB, RS | 63 | M | COPD | Staphylococcus, Candida albicans |
| 11004 | 5 | PGD | DB, RB, RS | 64 | F | COPD | Negative |
| 11016 | 6 | Control | DB, RB, RS | 61 | F | COPD | Negative |
| 11024 | 6 | PGD | DB, RB, RS | 61 | M | COPD | Negative |
| 13005 | 7 | PGD | DB, RB, RS | 64 | F | COPD | Staphylococcus |
| 12002 | 8 | Control | DB, RS | 62 | F | COPD | ND |
| 13024 | 8 | PGD | DB, RS | 65 | M | COPD | ND |
| 12001 | 9 | Control | RB, RS | 63 | F | COPD | ND |
| 13035 | 9 | PGD | DB, RB, RS | 63 | M | COPD | Streptococcus |
| 13021 | 10 | Control | DB, RB, RS | 39 | F | ILD | Streptococcus, Candida albicans |
| 11028 | 10 | PGD | DB, RB, RS | 49 | M | ILD | Negative |
| 11036 | 11 | Control | DB, RB, RS | 66 | M | ILD | ND |
| 11030 | 11 | PGD | DB, RB, RS | 56 | M | ILD | ND |
| 11010 | 12 | Control | DB, RB, RS | 49 | F | ILD | Negative |
| 11007 | 12 | PGD | DB, RB, RS | 51 | M | ILD | Negative |
| 11002 | 13 | Control | DB, RB, RS | 62 | F | ILD | Negative |
| 11008 | 13 | PGD | DB, RB, RS | 61 | M | ILD | Staphylococcus |
| 11013 | 14 | Control | DB, RB, RS | 64 | M | ILD | Pseudomonas |
| 11022 | 14 | PGD | DB, RB, RS | 64 | M | ILD | Negative |
| 13003 | 15 | Control | RB, RS | 57 | F | ILD | Negative |
| 11035 | 15 | PGD | DB, RS | 57 | M | ILD | Negative |
| 11032 | 16 | Control | DB, RB, RS | 53 | M | ILD | Streptococcus, Staphylococcus |
| 11042 | 16 | PGD | DB, RB, RS | 54 | M | ILD | Negative |
| 11006 | 17 | Control | DB, RB, RS | 59 | M | ILD | Negative |
| 11046 | 17 | PGD | DB, RB, RS | 60 | M | ILD | Negative |
| 13029 | 18 | Control | DB, RB, RS | 68 | F | ILD | ND |
| 12007 | 18 | PGD | DB, RB, RS | 66 | M | ILD | ND |
| 11040 | 19 | Control | DB, RB, RS | 63 | M | ILD | Negative |
| 12011 | 19 | PGD | DB, RB, RS | 61 | M | ILD | Klebsiella |
| 12015 | 20 | Control | DB, RB, RS | 62 | M | ILD | ND |
| 12013 | 20 | PGD | DB, RB, RS | 61 | M | ILD | ND |
| 11050 | 21 | Control | RS | 58 | M | ILD | Negative |
| 13030 | 21 | PGD | DB, RB, RS | 49 | M | ILD | Negative |
| 13011 | 22 | Control | RB, RS | 38 | M | ILD | Negative |
| 13037 | 22 | PGD | DB, RB, RS | 41 | F | ILD | Staphylococcus |
| 13027 | 23 | Control | DB, RB, RS | 64 | F | ILD | ND |
| 13039 | 23 | PGD | DB, RB, RS | 63 | M | ILD | Negative |
| 11011 | NA | PGD | DB, RS | 60 | M | PPH | Negative |
Subjects were selected from the CTOT-03 study, and patients with PGD were matched with controls as previously described (15). PGD Grade 3 occurring within the first 72 hours following lung transplantation was used as the primary case definition (1).
Definition of abbreviations: DB = Donor BAL, RB= Recipient post-reperfusion BAL, RS = Recipient post-transplant serum, CF/Bronch= Cystic fibrosis or bronchiectasis, COPD = Chronic obstructive pulmonary disease including emphysema or alpha-1 antitrypsin deficiency, ILD = Interstitial lung disease including pulmonary fibrosis, PPH = Primary pulmonary hypertension, ND = Not Done
Collection and processing of biological samples
BAL was collected in the operating room from donors immediately before organ procurement (donor BAL) and from allografts in recipients 1 hour after reperfusion (recipient BAL) as previously described (15). Serum was also obtained from recipients 1 hour after reperfusion. A total of 80 perioperative acellular BAL and 46 post-transplantation recipient serum samples were available (Table 1). BAL was processed to minimize host cells and extracellular viral particles were concentrated and nuclease-treated to digest non-encapsidated DNA and RNA, in order to enrich for viral nucleic acids. The full procedure is described in Supplemental Methods. As a positive control, BAL obtained for other indications was spiked with known quantities of the RNA bacteriophage Pseudomonas phi-6 to confirm detection by metagenomic sequencing (Table S3).
Quantitative PCR
TTV qPCR was performed as previously described (26).
Shotgun metagenomic sequencing and bioinformatics pipeline
To study DNA viruses, nucleic acid isolated from virus particle preparations was subjected to DNA whole genome amplification using the GenomiPhi V2 Amplification kit (GE Healthcare; Pittsburg, Pa). To detect RNA viruses, extracted nucleic acids were treated with DNase, reverse transcribed (43) and PCR-amplified as described in Supplemental Methods. Library preparation, Illumina sequencing, and the bioinformatics pipeline employed in sequence analysis are described in Supplemental Methods.
Differential gene expression and gene set variation analysis
Affymetrix Human Gene 1.0 ST Array (Affymetrix; Santa Clara, CA) data published previously on the same cohort of paired PGD cases and controls (15) was used in testing correlations between differential gene expression and TTV levels in the lung (Table S4–S6). Details are provided in Supplemental Methods.
Statistical analyses
Wilcoxon Rank Sum test, Wilcoxon Signed-Rank test, Kruskal-Wallis test, paired two-tailed Student’s T-test, Spearman rank correlation tests, and calculation of gene set enrichment scores were performed in R version 3 (44). Analysis of differences in TTV levels was performed on log10 normalized values. False discovery rate (FDR) using the Benjamini-Hochberg method was applied for multiple testing correction (45).
Results
Study subjects
Viral preparations from lung and blood specimens from 22 pairs of grade 3 PGD patients and matched controls, plus 2 additional PGD subjects, were analyzed (Table 1 and S1). Baseline demographics of study subjects have been previously reported (15) and characteristics relevant to this study are summarized in Table 1. Most subjects received Cytomegalovirus (CMV) prophylaxis, and all but two (Subjects 12013 and 12016) received Basiliximab for induction immunosuppression. Common bacteria and fungi identified by routine clinical culture included Staphylococcus sp., Streptococcus sp., and Candida albicans.
Analysis of TTV genome copy numbers by quantitative PCR
Previously, high levels of TTVs were reported in the respiratory tract of LTxRs in samples collected >1 month post-transplantation (26), and in post-transplant blood (37–39). Here we investigated lung TTV levels at time of transplantation by quantifying TTV genomes in donor BAL, and in recipient post-reperfusion BAL and serum (Fig. 1).
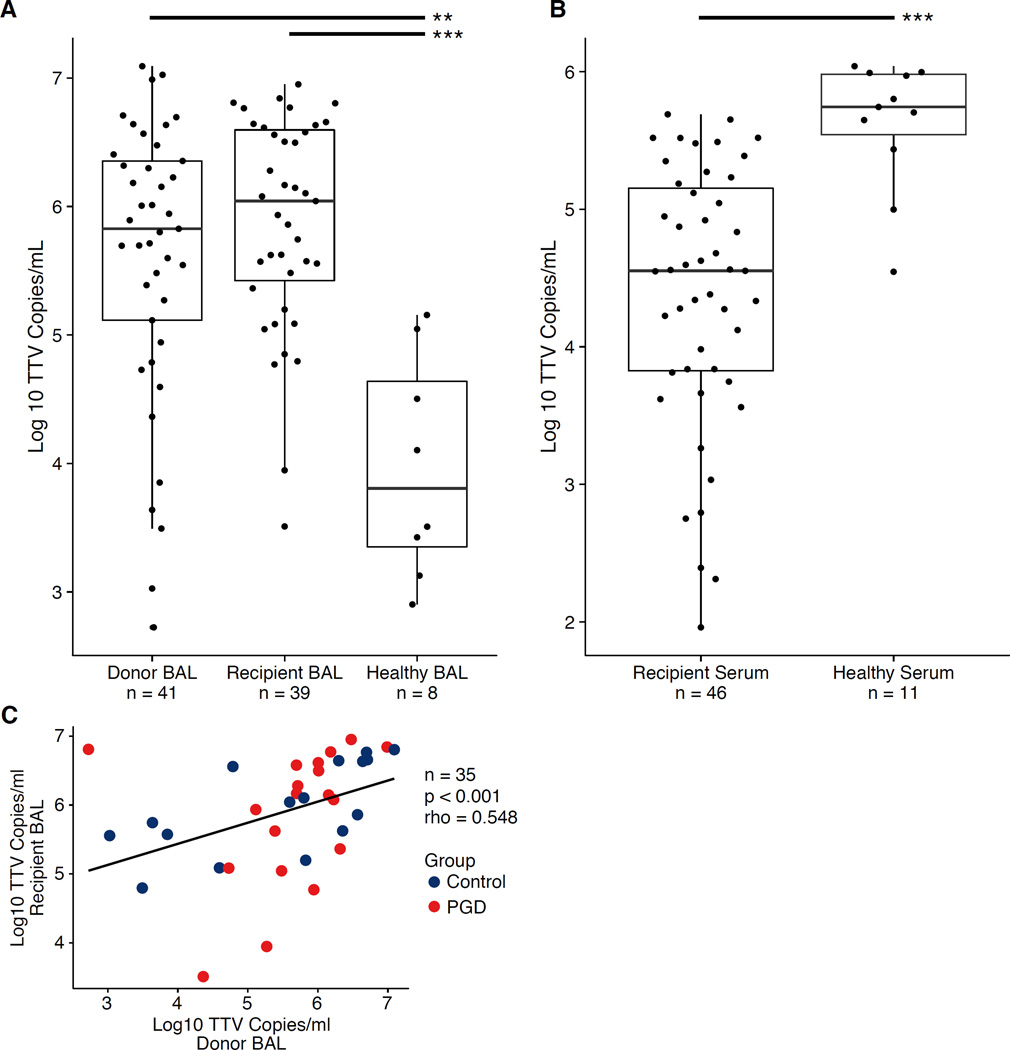
Figure 1. Torque Teno Virus Levels in Lung Transplant Donors, Recipients and Healthy Adults.
(A) Torque teno viruses quantified by qPCR in BAL. Boxes represent the middle two quartiles for each group, with the bold line representing the median value. Dots represent individual samples. Donor BAL was taken prior to organ procurement. Recipient BAL and Recipient serum were taken one hour after organ reperfusion. Quantities of TTV were higher in Donor and Recipient BAL compared to BAL of healthy adults as determined by Wilcoxon Rank Sum test (p=0.0026 and p<0.001, respectively). (B) Quantities of TTV in lung transplant recipient serum were lower compared to healthy adults as determined by Wilcoxon Rank Sum test (p<0.001). (C) Log10 TTV levels in paired Donor and Recipient BAL from individual samples were correlated (p<0.001, Spearman’s rho=0.548. A linear model was fitted to the data and is shown by the black line. The limit of quantification for the qPCR assay ranged from 11–65 copies/reaction. **p<0.01, ***p<0.001
Copies of TTV DNA per mL of acellular BAL varied up to 10,000-fold in both donor and recipient samples (Fig. 1). Donor BAL obtained prior to lung procurement had a median of 670,600 TTV copies/mL, which was 100-fold higher than TTV levels in healthy adult BAL samples (median=6,631; p=0.0026; Wilcoxon Rank Sum test). Copies of TTV in post-reperfusion BAL were approximately 2-fold greater than in samples taken before organ procurement, with a median of 1,102,000 copies/mL (p=0.89 compared with donor BAL; p<0.001 for comparison to healthy controls). There was a positive correlation between TTV levels in lung samples obtained pre- and post-transplantation (p<0.001, Spearman’s rho=0.548; Spearman’s rank correlation test; Fig. 1C). Thus, donor lungs are markedly abnormal with regards to virome populations, even before transplantation.
Conversely, in serum, quantities of TTV immediately post-reperfusion (median=35,530 copies/mL) were lower than in healthy adults (median of 553,996 copies/mL; p<0.001; Wilcoxon Rank Sum test). There was a positive trend correlating TTV levels in BAL and serum of LTxRs immediately post-transplantation (p=0.061, Spearman’s rho=0.303; Spearman’s rank correlation test; Fig. S1).
The high levels of TTV DNA found in donor lungs prior to transplantation were surprising given that donors are selected because they lack overt lung disease. Thus, we investigated clinical features of donors that might be associated with TTV levels (Table S7). Lung TTV DNA levels were inversely correlated with donor age (p=0.036, Spearman’s rho=0.328; Spearman’s rank correlation test). In contrast, there was no correlation between lung TTV levels and donor cause of death, history of purulent secretions, aspiration, or tobacco exposure. There was a positive trend correlating TTV levels in recipient BAL with organ ischemic time (p=0.065, Spearman’s rho=0.298; Spearman’s rank correlation test). However, no correlation was seen between post-reperfusion TTV levels and use of cardiopulmonary bypass or nitric oxide during surgery, administration of blood products or fluids, transplant type, or recipient preoperative diagnosis (Table S7).
We then investigated whether PGD was associated with TTV levels. When evaluated as absolute levels at a single time point, neither donor pre-transplant nor post-reperfusion BAL TTV levels correlated with PGD (p=0.89 and p=0.82, respectively, Wilcoxon Signed-Rank Test), nor did post-reperfusion serum TTV levels (p=0.71, Wilcoxon Signed-Rank Test).
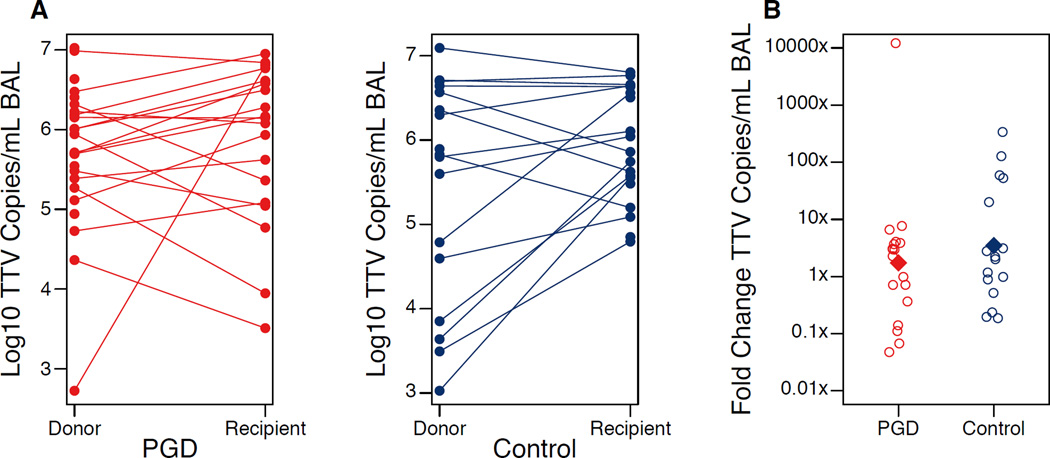
However, TTV dynamics differed significantly between PGD cases and controls (Fig. 2). Non-PGD controls showed an average 3.5-fold increase in lung TTV levels over the perioperative period, whereas the increase was only 1.7-fold in PGD cases. When compared as pairs across the 15 complete sample sets, a 4-fold difference in TTV increase was observed between PGD cases and controls (p=0.041; Wilcoxon Signed Rank Test). Thus, PGD is associated with a significantly lower rise in TTV levels during the peri-transplant period.
Figure 2. Torque Teno Virus Dynamics in Perioperative Period and Association with Primary Graft Dysfunction.
(A) TTV levels from organ pre-procurement (Donor) and post-reperfusion (Recipient) BAL samples are shown for each PGD case and control. Samples from the same organ are connected by a line. (B) The fold change in viral levels in the lung for PGD cases and controls is shown. Empty circles represent individual samples and the filled diamond represents the mean of the group. Both the average and median fold change was lower in PGD cases compared to paired controls (p=0.046 and p=0.041; Paired Student’s T-test and Wilcoxon Signed-Rank test, respectively). The number of paired samples analyzed was 15 due to missing samples for some patients.
Association of TTV levels and differences in mRNA expression in the lung
Gene set enrichment analysis in BAL cells from this cohort of patients previously identified several inflammasome and innate immune pathways for which perioperative change in expression correlated with PGD (15). These pathways included NOD-like receptor signaling, TOLL-like receptor signaling, the IL1 receptor pathway, and NFκB activation by nontypeable Haemophilus influenza.
Therefore, we investigated whether TTV dynamics correlated with gene pathways enriched in PGD, or with the top 9 transcripts linked to these pathways. Neither the PGD-associated pathways nor individual transcripts correlated with TTV dynamics (Table S4 and S5). We then tested whether absolute TTV levels in donor or recipient BAL correlated with transcript expression, but again found no significant relationship (Table S5). Finally, we queried the entire gene expression dataset to determine whether any individual gene (n=16,009) or pathway (n=403) correlated with TTV levels in donor BAL, recipient BAL or perioperative dynamics. Several pathways and gene transcripts demonstrated a nominal correlation with lung TTV levels or dynamics, but none were significant after FDR correction (Table S6). Thus, lung TTV dynamics did not correlate detectably with host cell gene expression patterns.
Metagenomic shotgun sequencing of perioperative samples
To investigate the lung virome further around the time of transplant, and to determine whether other viruses might be linked to PGD, we undertook metagenomic characterization of full viral populations in donor and recipient acellular BAL, as well as in recipient serum. To validate recovery of RNA viruses, 106 plaque forming units of bacteriophage Pseudomonas phage phi-6 were spiked into a non-LTxR BAL sample, which was processed identically to LTxR samples. Of the 369,669 non-human reads generated from this sample’s cDNA library, 135,301 aligned by BLAST to the tripartite phi-6 genome. Coverage of the L, M and S genomic segments was 71, 82 and 40%, respectively (Table S3), confirming viral recovery and detection with our methods.
Applying our pipeline to LTxR samples generated a median of 924,291 filtered, paired reads per sample in the BAL DNA library (range 2,668–5,676,284), 150,150 in the BAL cDNA samples (range 268–3,082,400), and 69,342 in serum DNA samples (range 5,241–815,079). On average, human sequences comprised 54.6% of all high quality reads in BAL DNA samples (range 0.20–88.9), 73% in BAL cDNA (range 1.18–98.6) and 80.4% (range 13.2–95.8) in serum DNA.
High quality non-human reads were queried by BLAST against the NCBI viral database and viral taxonomic assignments generated using BROCC (46) as described in Supplemental Methods. To control for contaminating sequences during sample and library preparation (47), buffer blanks were analyzed using the same workflow. On average, less than 1% of high quality, non-human sequences could be assigned to known viral species. The remaining reads likely correspond to bacteria, poorly annotated bacteriophages, human reads that eluded the human filtering step, or potentially novel eukaryotic viruses.
We first inspected read alignments to identify those that were spurious, i.e. attributable to environmental contamination or unfiltered human sequences. For example, alignments to Shamonda and Simbu virus were confined to 90 and 40 base pair regions of the genomes, respectively, and had 92–100% identity to human 45S ribosomal RNA, and were therefore removed. The number of reads annotated as Human endogenous retrovirus K (HERVK) tracked with the extent of human DNA in samples based on the number of reads aligning to the human genome and β-tubulin qPCR (data not shown), and were also removed.
Most subjects had many reads aligning to Human herpesviruses (HHVs) HHV-7, HHV-6A and HHV-6B. However, sequence alignments involved regions of short direct repeats, and herpesvirus genome coverage never exceeded 1%. BLAST queries of these sequences against the human genome revealed high identity to human repetitive regions. Because these reads likely do not represent authentic Herpesvirus detection, they were also removed. Finally, when very few reads align to a particular viral genome, low coverage makes it difficult to discriminate between coincidental alignment (such as to a short and/or non-unique region) and authentic virus detection. Therefore, an empirical threshold was set at 20 reads in a sample aligning to a reference virus in order to confidently call virus detection. Assignments below this threshold are shown in Table S7 and S8.
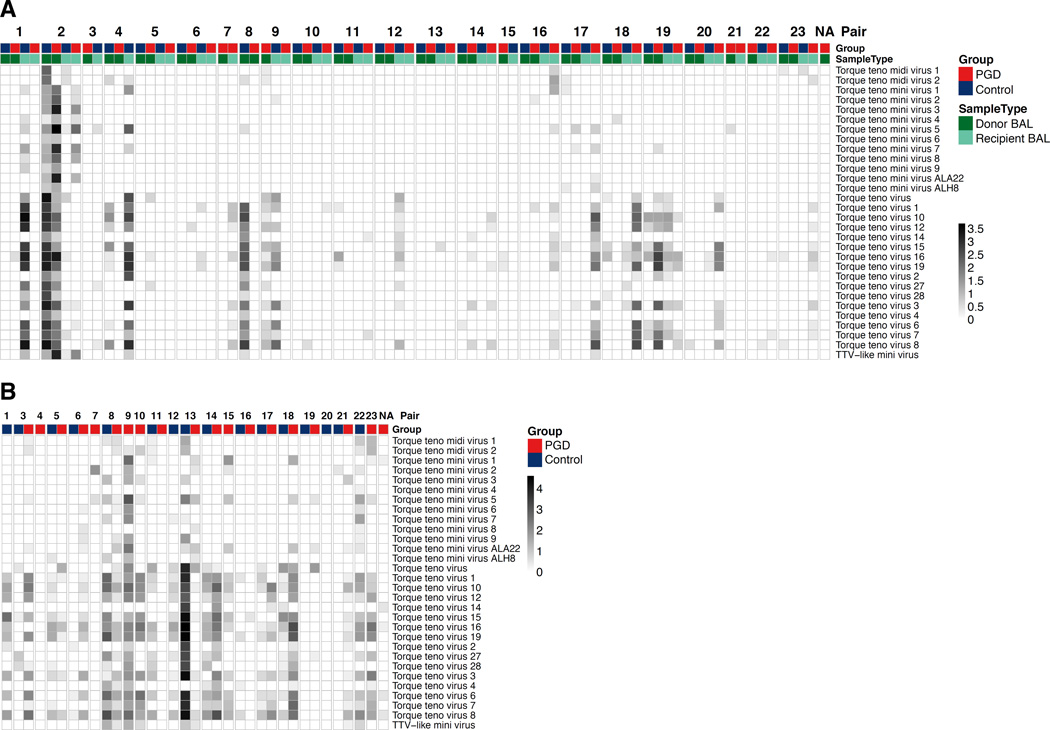
After these filtering steps, a total of 105 viral species from 15 family-level groups were identified in BAL (2 eukaryotic viruses, 1 plant virus and 12 bacteriophage families/unclassified species; Fig. 3 and Fig. S2). Anelloviruses, comprising multiple TTV species, were the most abundant eukaryotic virus in BAL. In serum, we identified reads whose best alignment were to 29 Anellovirus species. In both lung and serum, populations of TTVs appeared highly diverse, both within and between individuals (Fig. 4). The high levels of genome diversity within an individual sample could reflect multiple viral species, or a few novel TTV viruses with low levels of identity to database reference strains.
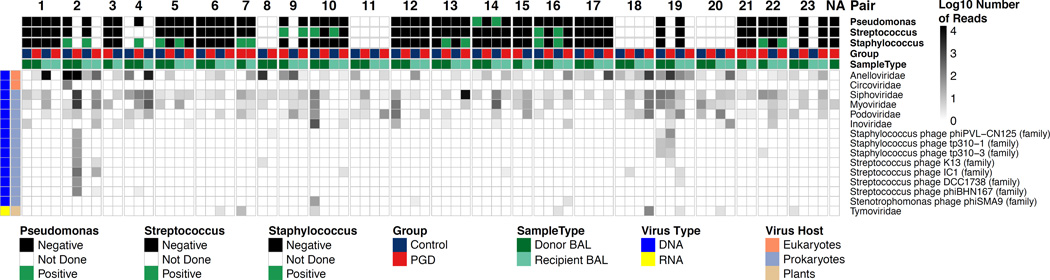
Figure 3. The Perioperative Lung Viral Microbiome.
Displayed is the number of shotgun metagenomic reads of DNA and cDNA libraries from each sample matching known viruses. Each column represents a different BAL sample. Each row represents a viral taxonomy at the family level, or at the species level for those hits that could not be classified into established families (species-level assignments for all viruses are shown in Fig. S2). Sequencing, processing of reads, alignments to viral genomes and removal of spurious hits was carried out as described in Methods. Columns are grouped by PGD-control pairs and labeled according to subject group, pair number and sample type as shown by the color coding across the top of the figure. Results of standard bacterial culture in each sample for the most commonly identified bacteria are also shown annotated on top. Further information on viral type and host is given by the column at the left. The intensity in each block represents the number of reads of each viral family in each sample on a log10 scale.
Figure 4. Diversity of Anellovirus Assignments in Lung and Serum During the Perioperative Period.
(A) Displayed are filtered metagenomic reads from perioperative BAL samples aligning to annotated human Anelloviruses. Each column corresponds to an individual sample and each row corresponds to the top scoring reference Anellovirus genome in the NCBI Viral Database. The intensity of each block represents the number of reads from that sample aligning to that reference species. Columns are grouped by PGD-control pairs and labeled according to subject group, pair number and sample type. (B) Displayed are filtered metagenomic reads from post-transplant recipient serum that aligned to human Anellovirus entries in the NCBI Viral Database. In serum, all viral hits remaining after stringent filtering steps were to Anellovirus family members. Samples that had insufficient DNA after library preparation for Illumina sequencing are omitted.
In addition to TTV, the most abundant viruses seen in lung samples were DNA phages, including Siphoviridae, Myoviridae and Podoviridae, which infect a broad range of oral and respiratory tract bacteria such as Streptococcus pneumoniae, Staphylococcus aureus, Stenotrophomonas maltophilia and Pseudomonas aeruginosa (Fig. 3 and Fig. S2).
After filtering out HHV reads deemed spurious, as described above, we specifically queried the presence of authentic HHV sequences by aligning reads to 10 Human herpesvirus genomes, since reactivation of HHVs, especially CMV, is a risk following organ transplantation (48). No sample exceeded our threshold of 20 reads, but one donor BAL from a PGD case had 12 reads that uniquely aligned to CMV (Table S8). Of note, our analysis of extracellular virions would not detect the presence of latent, non-replicating viral genomes within host cells.
One non-PGD donor BAL revealed sequences that aligned with Circoviruses, which are recognized pathogens in birds and swine, but in humans have primarily been detected in stool (49–51). Other small single stranded circular DNA viruses in the proposed Cyclovirus genus within the Circoviridae family have been reported in cerebrospinal fluid and nasopharyngeal aspirates (51, 52). Our sequence reads covered only 9% of the closest Circovirus reference genome, raising the possibility of a novel eukaryotic virus in this sample.
Although not reaching our threshold of 20 reads in any individual sample, we also detected a total of 26 reads across 4 samples that aligned to Mimiviruses (Table S8), which infect amoebae and have recently been reported in human lung samples, but are of uncertain clinical significance (53, 54).
Detection of RNA viruses was limited. The overwhelming majority of viral reads in the cDNA library were annotated as originating from DNA viruses. These reads might represent viral mRNA or incomplete removal of genomic DNA in cDNA preparation. In some samples there were small numbers of short reads (reflecting genome coverage of 0.6%) aligning to a plant virus (Tymoviridae, Physalis mottle virus). The significance of these reads is uncertain. No RNA viruses known to infect humans were detected in the acellular BAL metagenomic analysis, including an absence of community-acquired respiratory viruses. No viral taxa distinguished PGD cases from controls.
Discussion
Here we describe markedly elevated levels of TTV in the lungs of donors prior to organ recovery. While absolute levels of TTV did not distinguish PGD cases from controls, changes in TTV levels during the perioperative period were significantly associated with PGD.
Finding high levels of TTV in donor lungs was unexpected. Brain death is associated with profound hemodynamic, neurohumoral and inflammatory responses, which result in proinflammatory cytokine elevations and leukocyte infiltration of the lung (55, 56). Although its host cell tropism is poorly understood, TTV is reported to reside in mononuclear immune cells (57–61), and to increase in inflammatory states (62, 63). Thus, local inflammation and leukocyte recruitment to the lung following brain death might result in enhanced TTV replication. Additionally, corticosteroids are generally given to organ donors to modulate inflammation after brain death, and corticosteroid-induced immunosuppression could be another reason for the high TTV levels in donors since host immune function is thought to control virus replication (34, 38, 64). Future studies will be needed to determine whether lung TTVs increase in critically ill patients generally, or if high levels are specifically associated with brain death and/or corticosteroid or other donor treatments.
TTV levels increased from pre- to post-transplant lung samples, but the magnitude of increase was lower in PGD cases than in controls. This difference is consistent with the hypothesis that PGD is linked to tissue viability or immune activation that may restrict TTV levels in the lung. PGD is associated with activation of multiple innate immune responses (15–17) which might control TTV but also mediate injury contributing to PGD. Alternatively, in addition to immune cells, TTV has also been reported to replicate in respiratory epithelia and lung tissue (65, 66). Since ischemia-perfusion injury may result in decreased tissue viability (6–12), this may also limit TTV replication in PGD if permissive cell types are affected.
A prior analysis found that PGD was associated with enrichment in several gene pathways in this cohort. However, we did not find significant associations between peri-transplant TTV dynamics or lung TTV levels and host gene expression patterns. This finding suggests that the association between TTV dynamics and PGD reflects a relationship distinct from previously identified mechanisms linked to host gene expression patterns.
The range of TTV levels in serum of our healthy controls was similar to previous reports (67–70), but our post-reperfusion serum levels were significantly lower (Fig. 1). These levels immediately post-transplant contrast with studies showing elevated blood TTV levels of chronic transplant recipients who are immunosuppressed (37–39). However, a recent report described decreased TTV levels at day 7 in kidney/pancreas transplant recipients receiving anti-thymocyte globulin (ATG), which was ascribed to lymphocytolytic properties of ATG on mononuclear cells believed to support TTV replication (61). Only slight decreases were reported at day 7 in those receiving Basiliximab, which is not lymphocytolytic, but prevents T cell proliferation. We studied samples immediately following induction with Basiliximab. Thus, our results could reflect potential earlier effects of Basiliximab on TTV replication due to acute suppression of T cell proliferation. Two subjects who did not receive induction immunosuppression had markedly lower serum TTV levels compared to other LTxRs and healthy adults (Table S7), opposite to the predictions of this idea. However, induction immunosuppression is withheld in recipients with intercurrent processes which themselves might affect TTV levels, precluding definitive conclusions.
Apart from Anelloviruses, eukaryotic DNA viruses detected by metagenomic sequencing of acellular BAL were sparse and none correlated with PGD. We anticipated that HHVs might be prevalent, but only one sample was positive with few reads aligning to CMV. One sample had sequences annotated as a Circovirus, the significance of which is unclear but could reflect a novel virus. Most viral species detected were bacteriophages, and detection of phages known to infect respiratory tract bacteria implies the presence of their bacterial hosts during transplantation.
In contrast to DNA viruses, we did not find evidence for abundant human RNA viruses in these lung samples. Other studies have succeeded in identifying RNA viruses in human specimens using similar methods (43, 71–73), and our internal control spiked sample confirmed viral RNA recovery. Thus we conclude that actively replicating RNA viruses, which would be released as extracellular particles and therefore present in acellular BAL, are not common and abundant in lung allografts at the time of lung transplantation.
Our initial analysis revealed many sequences that aligned to human and other eukaryotic viruses, but which upon close inspection appeared spurious. These findings underscore the need to carefully inspect viral assignments to distinguish authentic sequences from artifacts. Additionally, alignments with very low read counts are difficult to authenticate, highlighting the challenge in interpreting such low abundance sequences. Thus, virome analysis based simply on automated read alignments may result in over-calling viruses that may not be genuinely present.
This study has several limitations. First, we used acellular BAL, which is suitable for analysis of the extracellular virome but cannot detect viral nucleic acids present within cells in non-replicating or latent forms that are not releasing progeny into the alveolar or airway lumen. In addition, use of acellular BAL limits the ability to characterize bacterial and fungal populations in the sample. Second, there is likely geographic heterogeneity within the lung (74) and any individual sample may represent only part of the viral community present. Third, since PGD can occur up to 3 days post-transplant, it is possible that differences in viral communities could emerge later than our perioperative time-points. Lastly, we did not have pre-transplant recipient serum uniformly available to query whether TTV dynamics in the blood changed in the peri-transplant period and if this relates to PGD incidence.
Finally, an important finding was that peri-transplant samples did not contain evidence indicating active replication of RNA viruses or typical community acquired respiratory viruses known to be associated with late transplant complications. However, small numbers of sequences aligning with unexpected nonhuman viruses, as reported here, might reflect novel human viruses. Further studies are warranted to determine whether TTV or novel viruses are linked to long-term outcomes of lung transplantation.
Supplementary Material
Figure S1: Relationship between Torque Teno Virus Levels in Lung and Peripheral Blood within Subjects
The relationship between transplant recipients’ lung and blood TTV burden (given as Log10 copies/mL on each axis) is shown. TTV levels in the lung after organ reperfusion show a positive trend but were not significantly correlated with levels in serum at the same time point (p=0.061, Spearman’s rho=0.303, Spearman’s rank correlation). A linear model was fitted to the data and is shown by the black line.
Figure S2: Viral Species in Perioperative Lungs
(A) Viral species with a minimum of 20 of metagenomic reads per sample aligning from perioperative BAL DNA and cDNA libraries. Each column corresponds to an individual BAL sample and each row corresponds to a reference viral genome. The intensity of each block represents the number of reads from that sample that match the reference species. Columns are grouped according to subject group, pair number and sample type. Data are the same as those shown in Fig. 3 except here individual species level assignments are shown for all viral hits.
Acknowledgments
We are grateful to subjects and volunteers for providing specimens, to the CTOT investigators, the staff participating in the CTOT-03 and Lung HIV Microbiome Project studies, and to members of the Bushman and Collman laboratories for help and suggestions. This work was supported by NIH grants R01-HL113252, U01-HL098957 and R01-HL087115, K24-HL115354, and received assistance from the Penn Center for AIDS Research (P30-AI045008) and the PennCHOP Microbiome Program. AAA was supported by NSF grant DGE-1321851 and JMD was supported by K23-HL121406.
Abbreviations
- ATG
anti-thymocyte globulin
- BAL
bronchoalveolar lavage
- BOS
bronchiolitis obliterans syndrome
- CMV
Cytomegalovirus
- CTOT-03
Clinical Trials in Organ Transplantation-03
- FDR
false discovery rate
- HERVK
Human endogenous retrovirus K
- HHV
Human herpesvirus
- LTxR
lung transplant recipients
- PGD
Primary graft dysfunction
- qPCR
quantitative polymerase chain reaction
- TTV
Torque teno virus
Footnotes
Disclosure
The authors of this manuscript have no conflicts of interest to disclose as described by the American Journal of Transplantation.
Supporting Information
Additional Supporting Information may be found in the online version of this article.
Supplemental Methods
Table S1: BAL Samples Utilized
Table S2: Features of Healthy Adult Lung Samples
Table S3: Recovery of Viral RNA by Unbiased Metagenomic Sequencing of BAL
Table S4: Association of TTV Dynamics with Gene Sets and Pathways Enriched in PGD
Table S5: Association of TTV with Highest Ranked Transcripts Enriched in PGD in Perioperative BAL
Table S6: Association of TTV with Gene Set Variation Analysis of BAL mRNA
Table S7: Correlations between Clinical Variables and Torque Teno Virus
Table S8: Top Scoring Viral Families with Fewer than 20 Reads
Table S9: Top Scoring Viral Species with Fewer than 20 Reads
References
- 1.Yusen RD, Edwards LB, Kucheryavaya AY, Benden C, Dipchand AI, Goldfarb SB, et al. The Registry of the International Society for Heart and Lung Transplantation: Thirty-second Official Adult Lung and Heart-Lung Transplantation Report—2015; Focus Theme: Early Graft Failure. The Journal of Heart and Lung Transplantation. 2015;34(10):1264–1277. doi: 10.1016/j.healun.2015.08.014. [DOI] [PubMed] [Google Scholar]
- 2.Watson CJE, Dark JH. Organ transplantation: historical perspective and current practice. British Journal of Anaesthesia. 2012;108(suppl 1) doi: 10.1093/bja/aer384. [DOI] [PubMed] [Google Scholar]
- 3.Lee JC, Christie JD. Primary Graft Dysfunction. Clinics in Chest Medicine. 2011;32(2):279–293. doi: 10.1016/j.ccm.2011.02.007. [DOI] [PubMed] [Google Scholar]
- 4.Ius F, Sommer W, Tudorache I, Kühn C, Avsar M, Siemeni T, et al. Early donor-specific antibodies in lung transplantation: Risk factors and impact on survival. The Journal of Heart and Lung Transplantation. 2014;33(12):1255–1263. doi: 10.1016/j.healun.2014.06.015. [DOI] [PubMed] [Google Scholar]
- 5.Daud SA, Yusen RD, Meyers BF, Chakinala MM, Walter MJ, Aloush AA, et al. Impact of immediate primary lung allograft dysfunction on bronchiolitis obliterans syndrome. American journal of respiratory and critical care medicine. 2007;175(5):507–513. doi: 10.1164/rccm.200608-1079OC. [DOI] [PubMed] [Google Scholar]
- 6.Eppinger MJ, Deeb GM, Bolling SF, Ward PA. Mediators of ischemia-reperfusion injury of rat lung. The American journal of pathology. 1997;150(5):1773–1784. [PMC free article] [PubMed] [Google Scholar]
- 7.Eppinger MJ, Jones ML, Deeb GM, Bolling SF, Ward PA. Pattern of injury and the role of neutrophils in reperfusion injury of rat lung. The Journal of surgical research. 1995;58(6):713–718. doi: 10.1006/jsre.1995.1112. [DOI] [PubMed] [Google Scholar]
- 8.Fiser, Tribble CG, Long, Kaza AK. Pulmonary macrophages are involved in reperfusion injury after lung transplantation. Pulmonary macrophages are involved in reperfusion injury after lung transplantation. 2001 doi: 10.1016/s0003-4975(01)02407-9. [DOI] [PubMed] [Google Scholar]
- 9.Naidu BV, Krishnadasan B, Farivar AS, Woolley SM, Thomas R, Rooijen N, et al. Early activation of the alveolar macrophage is critical to the development of lung ischemia-reperfusion injury. The Journal of thoracic and cardiovascular surgery. 2003;126(1):200–207. doi: 10.1016/s0022-5223(03)00390-8. [DOI] [PubMed] [Google Scholar]
- 10.Yang Z, Sharma AK, Linden J, Kron IL, Laubach VE. CD4+ T lymphocytes mediate acute pulmonary ischemia–reperfusion injury. The Journal of Thoracic and Cardiovascular Surgery. 2009;137(3):695–702. doi: 10.1016/j.jtcvs.2008.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharma AK, LaPar DJ, Zhao Y, Li L, Lau CL, Kron IL, et al. Natural Killer T Cell–derived IL-17 Mediates Lung Ischemia–Reperfusion Injury. American Journal of Respiratory and Critical Care Medicine. 2011;183(11):1539–1549. doi: 10.1164/rccm.201007-1173OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnston LK, Rims CR, Gill SE, McGuire JK, Manicone AM. Pulmonary Macrophage Subpopulations in the Induction and Resolution of Acute Lung Injury. American journal of respiratory cell and molecular biology. 2012;47(4):417–426. doi: 10.1165/rcmb.2012-0090OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diamond JM, Wigfield CH. Role of innate immunity in primary graft dysfunction after lung transplantation. Current opinion in organ transplantation. 2013;18(5):518–523. doi: 10.1097/MOT.0b013e3283651994. [DOI] [PubMed] [Google Scholar]
- 14.Cantu E, Shah RJ, Lin W, Daye ZJ, Diamond JM, Suzuki Y, et al. Oxidant stress regulatory genetic variation in recipients and donors contributes to risk of primary graft dysfunction after lung transplantation. The Journal of thoracic and cardiovascular surgery. 2015;149(2):596–602. doi: 10.1016/j.jtcvs.2014.09.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cantu E, Lederer DJ, Meyer K, Milewski K, Suzuki Y, Shah RJ, et al. Gene set enrichment analysis identifies key innate immune pathways in primary graft dysfunction after lung transplantation. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2013;13(7):1898–1904. doi: 10.1111/ajt.12283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Naka Y, Marsh HC, Scesney SM, Oz MC, Pinsky DJ. Complement activation as a cause for primary graft failure in an isogeneic rat model of hypothermic lung preservation and transplantation. Transplantation. 1997;64(9):1248–1255. doi: 10.1097/00007890-199711150-00004. [DOI] [PubMed] [Google Scholar]
- 17.Shah RJ, Emtiazjoo AM, Diamond JM, Smith PA, Roe DW, Wille KM, et al. Plasma complement levels are associated with primary graft dysfunction and mortality after lung transplantation. American journal of respiratory and critical care medicine. 2014;189(12):1564–1567. doi: 10.1164/rccm.201312-2121LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khalifah AP, Hachem RR, Chakinala MM, Schechtman KB, Patterson GA, Schuster DP, et al. Respiratory viral infections are a distinct risk for bronchiolitis obliterans syndrome and death. American journal of respiratory and critical care medicine. 2004;170(2):181–187. doi: 10.1164/rccm.200310-1359OC. [DOI] [PubMed] [Google Scholar]
- 19.Botha P, Archer L, Anderson RL, Lordan J, Dark JH, Corris PA, et al. Pseudomonas aeruginosa colonization of the allograft after lung transplantation and the risk of bronchiolitis obliterans syndrome. Transplantation. 2008;85(5):771–774. doi: 10.1097/TP.0b013e31816651de. [DOI] [PubMed] [Google Scholar]
- 20.Weigt SS, Elashoff RM, Huang C, Ardehali A, Gregson AL, Kubak B, et al. Aspergillus colonization of the lung allograft is a risk factor for bronchiolitis obliterans syndrome. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2009;9(8):1903–1911. doi: 10.1111/j.1600-6143.2009.02635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gottlieb J, Schulz TF, Welte T, Fuehner T, Dierich M, Simon AR, et al. Community-acquired respiratory viral infections in lung transplant recipients: a single season cohort study. Transplantation. 2009;87(10):1530–1537. doi: 10.1097/TP.0b013e3181a4857d. [DOI] [PubMed] [Google Scholar]
- 22.Borewicz K, Pragman AA, Kim HB, Hertz M, Wendt C, Isaacson RE. Longitudinal analysis of the lung microbiome in lung transplantation. FEMS microbiology letters. 2013;339(1):57–65. doi: 10.1111/1574-6968.12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakajima T, Palchevsky V, Perkins DL, Belperio JA, Finn PW. Lung transplantation: infection, inflammation, and the microbiome. Seminars in immunopathology. 2011;33(2):135–156. doi: 10.1007/s00281-011-0249-9. [DOI] [PubMed] [Google Scholar]
- 24.Verleden SE, Ruttens D, Vandermeulen E, Bellon H, Van Raemdonck DE, Dupont LJ, et al. Restrictive chronic lung allograft dysfunction: Where are we now? The Journal of heart and lung transplantation : the official publication of the International Society for Heart Transplantation. 2015;34(5):625–630. doi: 10.1016/j.healun.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 25.Charlson ES, Diamond JM, Bittinger K, Fitzgerald AS, Yadav A, Haas AR, et al. Lung-enriched organisms and aberrant bacterial and fungal respiratory microbiota after lung transplant. American journal of respiratory and critical care medicine. 2012;186(6):536–545. doi: 10.1164/rccm.201204-0693OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Young JC, Chehoud C, Bittinger K, Bailey A, Diamond JM, Cantu E, et al. Viral metagenomics reveal blooms of anelloviruses in the respiratory tract of lung transplant recipients. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2015;15(1):200–209. doi: 10.1111/ajt.13031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharma P, Diene SM, Thibeaut S, Bittar F, Roux V, Gomez C, et al. Phenotypic and genotypic properties of Microbacterium yannicii, a recently described multidrug resistant bacterium isolated from a lung transplanted patient with cystic fibrosis in France. BMC microbiology. 2013;13:97. doi: 10.1186/1471-2180-13-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dickson RP, Erb-Downward JR, Freeman CM, Walker N, Scales BS, Beck JM, et al. Changes in the lung microbiome following lung transplantation include the emergence of two distinct Pseudomonas species with distinct clinical associations. PloS one. 2014;9(5) doi: 10.1371/journal.pone.0097214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Willner DL, Hugenholtz P, Yerkovich ST, Tan ME, Daly JN, Lachner N, et al. Reestablishment of recipient-associated microbiota in the lung allograft is linked to reduced risk of bronchiolitis obliterans syndrome. American journal of respiratory and critical care medicine. 2013;187(6):640–647. doi: 10.1164/rccm.201209-1680OC. [DOI] [PubMed] [Google Scholar]
- 30.Spandole S, Cimponeriu D, Berca LM, Mihăescu G. Human anelloviruses: an update of molecular, epidemiological and clinical aspects. Archives of virology. 2015;160(4):893–908. doi: 10.1007/s00705-015-2363-9. [DOI] [PubMed] [Google Scholar]
- 31.Ninomiya M, Takahashi M, Nishizawa T, Shimosegawa T, Okamoto H. Development of PCR assays with nested primers specific for differential detection of three human anelloviruses and early acquisition of dual or triple infection during infancy. Journal of clinical microbiology. 2008;46(2):507–514. doi: 10.1128/JCM.01703-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McElvania TeKippe E, Wylie KM, Deych E, Sodergren E, Weinstock G, Storch GA. Increased prevalence of anellovirus in pediatric patients with fever. PloS one. 2012;7(11) doi: 10.1371/journal.pone.0050937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen T, Väisänen E, Mattila PS, Hedman K, Söderlund-Venermo M. Antigenic diversity and seroprevalences of Torque teno viruses in children and adults by ORF2-based immunoassays. The Journal of general virology. 2013;94(Pt 2):409–417. doi: 10.1099/vir.0.046862-0. [DOI] [PubMed] [Google Scholar]
- 34.Thom K, Petrik J. Progression towards AIDS leads to increased torque teno virus and torque teno minivirus titers in tissues of HIV infected individuals. Journal of Medical Virology. 2007;79(1):1–7. doi: 10.1002/jmv.20756. [DOI] [PubMed] [Google Scholar]
- 35.Madsen CD, Eugen-Olsen J, Kirk O, Parner J, Kaae Christensen J, Brasholt MS, et al. TTV viral load as a marker for immune reconstitution after initiation of HAART in HIV-infected patients. HIV clinical trials. 2002;3(4):287–295. doi: 10.1310/8c94-vypq-ng1h-4cnw. [DOI] [PubMed] [Google Scholar]
- 36.Devalle S, Rua F, Morgado MG, Niel C. Variations in the frequencies of torque teno virus subpopulations during HAART treatment in HIV-1-coinfected patients. Archives of Virology. 2009;154(8):1285–1291. doi: 10.1007/s00705-009-0440-7. [DOI] [PubMed] [Google Scholar]
- 37.De Vlaminck I, Khush KK, Strehl C, Kohli B, Luikart H, Neff NF, et al. Temporal response of the human virome to immunosuppression and antiviral therapy. Cell. 2013;155(5):1178–1187. doi: 10.1016/j.cell.2013.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Görzer I, Haloschan M, Jaksch P, Klepetko W, Puchhammer-Stöckl E. Plasma DNA levels of Torque teno virus and immunosuppression after lung transplantation. The Journal of Heart and Lung Transplantation. 2014;33(3):320–323. doi: 10.1016/j.healun.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 39.Görzer I, Jaksch P, Kundi M, Seitz T, Klepetko W, Puchhammer-Stöckl E. Pre-transplant plasma Torque Teno virus load and increase dynamics after lung transplantation. PloS one. 2015;10(3) doi: 10.1371/journal.pone.0122975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hino S, Miyata H. Torque teno virus (TTV): current status. Reviews in medical virology. 2007;17(1):45–57. doi: 10.1002/rmv.524. [DOI] [PubMed] [Google Scholar]
- 41.Charlson ES, Bittinger K, Haas AR, Fitzgerald AS, Frank I, Yadav A, et al. Topographical Continuity of Bacterial Populations in the Healthy Human Respiratory Tract. American Journal of Respiratory and Critical Care Medicine. 2011;184(8):957–963. doi: 10.1164/rccm.201104-0655OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beck JM, Schloss PD, Venkataraman A, Twigg H, Jablonski KA, Bushman FD, et al. Multicenter Comparison of Lung and Oral Microbiomes of HIV-infected and HIV-uninfected Individuals. American journal of respiratory and critical care medicine. 2015;192(11):1335–1344. doi: 10.1164/rccm.201501-0128OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang D, Urisman A, Liu Y-T, Springer M, Ksiazek TG, Erdman DD, et al. Viral Discovery and Sequence Recovery Using DNA Microarrays. PLoS Biology. 2003;1(2) doi: 10.1371/journal.pbio.0000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ihaka R, Gentleman R. R: A Language for Data Analysis and Graphics. Journal of Computational and Graphical Statistics. 1996;5(3):299–314. [Google Scholar]
- 45.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the royal statistical society Series B (Methodological) 1995:289–300. [Google Scholar]
- 46.Dollive S, Peterfreund GL, Sherrill-Mix S, Bittinger K, Sinha R, Hoffmann C, et al. A tool kit for quantifying eukaryotic rRNA gene sequences from human microbiome samples. Genome Biology. 2012;13(7):1–13. doi: 10.1186/gb-2012-13-7-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, Moffatt MF, et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biology. 2014;12(1):1–12. doi: 10.1186/s12915-014-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kotloff RM, Thabut G. Lung transplantation. American journal of respiratory and critical care medicine. 2011;184(2):159–171. doi: 10.1164/rccm.201101-0134CI. [DOI] [PubMed] [Google Scholar]
- 49.Smits SL, Schapendonk CM, van Beek J, Vennema H, Schürch AC, Schipper D, et al. New viruses in idiopathic human diarrhea cases, the Netherlands. Emerging infectious diseases. 2014;20(7):1218–1222. doi: 10.3201/eid2007.140190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Phan TG, da Costa AC, Zhang W, Pothier P, Ambert-Balay K, Deng X, et al. A new gyrovirus in human feces. Virus genes. 2015;51(1):132–135. doi: 10.1007/s11262-015-1210-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Phan TG, Mori D, Deng X, Rajindrajith S, Ranawaka U, Fan Ng TF, et al. Small circular single stranded DNA viral genomes in unexplained cases of human encephalitis, diarrhea, and in untreated sewage. Virology. 2015;482:98–104. doi: 10.1016/j.virol.2015.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Phan TG, Luchsinger V, Avendano LF, Deng X, Delwart E. Cyclovirus in nasopharyngeal aspirates of Chilean children with respiratory infections. Journal of General Virology. 2014;95(Pt 4):922–927. doi: 10.1099/vir.0.061143-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saadi H, Pagnier I, Colson P, Cherif JK, Beji M, Boughalmi M, et al. First isolation of Mimivirus in a patient with pneumonia. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2013;57(4):34. doi: 10.1093/cid/cit354. [DOI] [PubMed] [Google Scholar]
- 54.Kutikhin AG, Yuzhalin AE, Brusina EB. Mimiviridae, Marseilleviridae, and virophages as emerging human pathogens causing healthcare-associated infections. GMS hygiene and infection control. 2014;9(2) doi: 10.3205/dgkh000236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kutsogiannis DJ, Pagliarello G, Doig C, Ross H, Shemie SD. Medical management to optimize donor organ potential: review of the literature. Canadian journal of anaesthesia = Journal canadien d'anesthésie. 2006;53(8):820–830. doi: 10.1007/BF03022800. [DOI] [PubMed] [Google Scholar]
- 56.Faropoulos K, Apostolakis E. Brain death and its influence on the lungs of the donor: how is it prevented? Transplantation proceedings. 2009;41(10):4114–4119. doi: 10.1016/j.transproceed.2009.09.087. [DOI] [PubMed] [Google Scholar]
- 57.Maggi F, Fornai C, Zaccaro L, Morrica A, Vatteroni ML, Isola P, et al. TT virus (TTV) loads associated with different peripheral blood cell types and evidence for TTV replication in activated mononuclear cells. Journal of medical virology. 2001;64(2):190–194. doi: 10.1002/jmv.1035. [DOI] [PubMed] [Google Scholar]
- 58.Mariscal LF, López-Alcorocho JM, Rodríguez-Iñigo E, Ortiz-Movilla N, de Lucas S, Bartolomé J, et al. TT virus replicates in stimulated but not in nonstimulated peripheral blood mononuclear cells. Virology. 2002;301(1):121–129. doi: 10.1006/viro.2002.1545. [DOI] [PubMed] [Google Scholar]
- 59.Zhong S, Yeo W, Tang M, Liu C, Lin X-r, Ho WM, et al. Frequent detection of the replicative form of TT virus DNA in peripheral blood mononuclear cells and bone marrow cells in cancer patients. Journal of medical virology. 2002;66(3):428–434. doi: 10.1002/jmv.2163. [DOI] [PubMed] [Google Scholar]
- 60.Takahashi M, Asabe S, Gotanda Y, Kishimoto J, Tsuda F, Okamoto H. TT virus is distributed in various leukocyte subpopulations at distinct levels, with the highest viral load in granulocytes. Biochemical and biophysical research communications. 2002;290(1):242–248. doi: 10.1006/bbrc.2001.6183. [DOI] [PubMed] [Google Scholar]
- 61.Focosi D, Macera L, Boggi U, Nelli LC, Maggi F. Short-term kinetics of torque teno virus viraemia after induction immunosuppression confirm T lymphocytes as the main replication-competent cells. The Journal of general virology. 2015;96(Pt 1):115–117. doi: 10.1099/vir.0.070094-0. [DOI] [PubMed] [Google Scholar]
- 62.Maggi F, Pifferi M, Fornai C, Andreoli E, Tempestini E, Vatteroni M, et al. TT virus in the nasal secretions of children with acute respiratory diseases: relations to viremia and disease severity. Journal of virology. 2003;77(4):2418–2425. doi: 10.1128/JVI.77.4.2418-2425.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Walton AH, Muenzer JT, Rasche D, Boomer JS, Sato B, Brownstein BH, et al. Reactivation of multiple viruses in patients with sepsis. PloS one. 2014;9(2) doi: 10.1371/journal.pone.0098819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Focosi D, Maggi F, Albani M, Macera L, Ricci V, Gragnani S, et al. Torquetenovirus viremia kinetics after autologous stem cell transplantation are predictable and may serve as a surrogate marker of functional immune reconstitution. Journal of clinical virology : the official publication of the Pan American Society for Clinical Virology. 2010;47(2):189–192. doi: 10.1016/j.jcv.2009.11.027. [DOI] [PubMed] [Google Scholar]
- 65.Pifferi M, Maggi F, Di Cristofano C, Cangiotti AM, Nelli LC, Bevilacqua G, et al. Torquetenovirus infection and ciliary dysmotility in children with recurrent pneumonia. The Pediatric infectious disease journal. 2008;27(5):413–418. doi: 10.1097/INF.0b013e318162a14f. [DOI] [PubMed] [Google Scholar]
- 66.Okamoto H, Nishizawa T, Takahashi M, Asabe S, Tsuda F, Yoshikawa A. Heterogeneous distribution of TT virus of distinct genotypes in multiple tissues from infected humans. Virology. 2001;288(2):358–368. doi: 10.1006/viro.2001.1097. [DOI] [PubMed] [Google Scholar]
- 67.Haloschan M, Bettesch R, Görzer I, Weseslindtner L, Kundi M, Puchhammer-Stöckl E. TTV DNA plasma load and its association with age, gender, and HCMV IgG serostatus in healthy adults. Age (Dordrecht, Netherlands) 2014;36(5):9716. doi: 10.1007/s11357-014-9716-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Christensen JK, Eugen-Olsen J, SŁrensen M, Ullum H, Gjedde SB, Pedersen BK, et al. Prevalence and prognostic significance of infection with TT virus in patients infected with human immunodeficiency virus. The Journal of infectious diseases. 2000;181(5):1796–1799. doi: 10.1086/315440. [DOI] [PubMed] [Google Scholar]
- 69.Moen EM, Sagedal S, Bjøro K, Degré M, Opstad PK, Grinde B. Effect of immune modulation on TT virus (TTV) and TTV-like-mini-virus (TLMV) viremia. Journal of medical virology. 2003;70(1):177–182. doi: 10.1002/jmv.10356. [DOI] [PubMed] [Google Scholar]
- 70.Mancuso R, Saresella M, Hernis A, Agostini S, Piancone F, Caputo D, et al. Torque teno virus (TTV) in multiple sclerosis patients with different patterns of disease. Journal of medical virology. 2013;85(12):2176–2183. doi: 10.1002/jmv.23707. [DOI] [PubMed] [Google Scholar]
- 71.Wylie KM, Mihindukulasuriya KA, Sodergren E, Weinstock GM, Storch GA. Sequence analysis of the human virome in febrile and afebrile children. PloS one. 2012;7(6) doi: 10.1371/journal.pone.0027735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Holtz LR, Cao S, Zhao G, Bauer IK, Denno DM, Klein EJ, et al. Geographic variation in the eukaryotic virome of human diarrhea. Virology. 2014;468–470:556–564. doi: 10.1016/j.virol.2014.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Handley SA, Thackray LB, Zhao G, Presti R, Miller AD, Droit L, et al. Pathogenic simian immunodeficiency virus infection is associated with expansion of the enteric virome. Cell. 2012;151(2):253–266. doi: 10.1016/j.cell.2012.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Willner D, Haynes MR, Furlan M, Hanson N, Kirby B, Lim YW, et al. Case studies of the spatial heterogeneity of DNA viruses in the cystic fibrosis lung. American journal of respiratory cell and molecular biology. 2012;46(2):127–131. doi: 10.1165/rcmb.2011-0253OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Relationship between Torque Teno Virus Levels in Lung and Peripheral Blood within Subjects
The relationship between transplant recipients’ lung and blood TTV burden (given as Log10 copies/mL on each axis) is shown. TTV levels in the lung after organ reperfusion show a positive trend but were not significantly correlated with levels in serum at the same time point (p=0.061, Spearman’s rho=0.303, Spearman’s rank correlation). A linear model was fitted to the data and is shown by the black line.
Figure S2: Viral Species in Perioperative Lungs
(A) Viral species with a minimum of 20 of metagenomic reads per sample aligning from perioperative BAL DNA and cDNA libraries. Each column corresponds to an individual BAL sample and each row corresponds to a reference viral genome. The intensity of each block represents the number of reads from that sample that match the reference species. Columns are grouped according to subject group, pair number and sample type. Data are the same as those shown in Fig. 3 except here individual species level assignments are shown for all viral hits.