Abstract
The conformational flexibility of a biomolecule may play a crucial role in its biological function. Small-angle x-ray scattering (SAXS) is a very popular technique for characterizing biomolecule flexibility. It can be used to determine a possible structural ensemble of the biomolecule in solution with the aid of a computer simulation. In this article, we present a tool written in Python, which iteratively runs multiple independent enhanced sampling simulations such as amplified collective motions and accelerated molecular dynamics, and an ensemble optimization method to drive the biomolecule toward an ensemble that fits the SAXS data well. The tool has been validated with a protein and an RNA system, i.e., the tandem WW domains of formin-binding protein 21 and the aptamer domain of SAM-1 riboswitch, respectively. These Python scripts are user-friendly and can be easily modified if a different simulation engine is preferred.
Introduction
Small-angle x-ray scattering (SAXS) has been widely used to obtain low-resolution structural information for large biomolecules in solution (1). Compared to other experimental techniques, SAXS is particularly useful for characterizing the flexibility of biomolecules (2). Many recent studies show the possibility of combining experimental SAXS data and computational simulation to interpret biomolecule dynamics in solution (3, 4).
Two strategies, refining-while-sampling and screening-after-sampling, are generally applied to integrate low-resolution SAXS data into the structural modeling of biomolecules (4). For a refining-while-sampling method such as found in Zheng and Tekpinar (5) and Björling et al. (6), a pseudo energy term based on the SAXS data is designed, then a conformation or an ensemble is simulated in optimizing the energy. Such an approach is efficient, but source code must be modified to change the energy function, which may not be an easy job for someone with little programming skill and knowledge about simulation algorithms; furthermore, different research groups prefer different simulation engines. In screening-after-sampling methods such as those found in the literature (7, 8, 9, 10, 11, 12, 13, 14), simulations without SAXS restraint are first carried out to generate a large pool of structures that covers the conformational space of the biomolecule. Then, an ensemble containing a small number of conformations selected from the pool is determined to fit the SAXS data. This strategy is particularly suitable for flexible systems. Although there is no need to change the simulation code, adequate sampling of the conformational space is crucial for such an approach—which is a nontrivial issue, especially for very large biomolecules.
To our knowledge, this article presents a new computational tool for the SAXS-oriented ensemble refinement (SAXS-ER) of flexible biomolecules, which has the advantages of both strategies, but not the disadvantages. Our tool consists of cycles of 1) multiple independent enhanced sampling simulation, and 2) selection of an ensemble that contains a certain number of conformations from the combined trajectories, which best reproduce the SAXS data at this stage, to start the next simulation cycle. We designated this approach as an iterative screening-after-sampling strategy, and it may drive the ensemble of the biomolecule to fit the SAXS data better and better until it converges.
In the Materials and Methods, we introduce the biomolecular systems of interest, the SAXS data acquisition and the computational details of SAXS-ER. In the Results and Discussion, we apply this tool to two biomolecules, the tandem WW domains of formin-binding protein 21 (FBP21-WWs) and the ligand-free SAM-1 riboswitch aptamer domain (free SAM-1 aptamer). Finally, we provide concluding remarks.
Materials and Methods
Biomolecular systems and SAXS data
The formin binding protein 21 (FBP21) is a structural component of the mammalian spliceosomal A/B complex, which plays an important role in pre-RNA splicing (15). The C-terminus of the protein consists of two group-III WW domains (designated “FBP21-WWs”), and a NMR ensemble containing 20 structural models (i.e., PDB: 2JXW) has been resolved (16). Each model has 75 amino acid residues. The individual domains are denoted as “WW1” (residues 6–32) and “WW2” (residues 47–73), respectively, and are structurally well converged. However, a flexible linker (residues 33–46) allows various orientations between the two domains. Because long-range NMR restraints are lacking, we have collected SAXS data for FBP21-WWs (17) to determine the structural ensemble of the protein in solution.
The SAM-1 riboswitch is a RNA element that binds to S-Adenosyl Methionine (SAM) and controls expression of genes for Met and Cys biosynthesis in Gram-positive bacteria (18). The aptamer domain contains 94 nucleotides of the SAM-1 riboswitch in the presence of SAM, and magnesium can form a stable structure (19). However, Stoddard et al. (20) have found that the solution scattering data of the ligand-free SAM-1 aptamer shows an obvious discrepancy with the theoretical curve of the ligand-bound crystal structure, and the former may exist as multiple states in solution. Starting from the crystal structure with the ligand removed, we wanted to construct an ensemble of the free SAM-1 aptamer to reproduce the SAXS data. The SAXS data of the free SAM-1 aptamer was taken from www.bioisis.net with the Bioisis ID: 1SAMRR, and more details can be found in Stoddard et al. (20).
Enhanced sampling techniques
Although an MD simulation is popularly used to generate the conformation of a biomolecule (21), a sampling issue may often arise (22). A flexible biomolecule usually exists in multiple conformational states in solution. In a standard MD simulation on a limited timescale, the biomolecule may be trapped in few local states, so that global conformational transitions are rarely sampled due to the complicated energy landscape of the biomolecule (23). Such inefficient sampling in a MD simulation may not be able to properly interpret the experimental SAXS data. Enhanced sampling techniques (24) can be used to resolve this problem.
We previously developed a sampling method known as amplified collective motions (ACM) (25), which calculates a few low-frequency normal modes for a biomolecule in an elastic network model (26), and couples these modes in a high temperature bath in atomic MD simulations to adequately explore the collective motion of the biomolecule. Accelerated molecular dynamics (aMD) (27) improves the sampling efficiency in reducing the energy barrier separating adjacent conformational states of the biomolecule. The method modifies the potential energy and raises these energy wells below a predetermined threshold value, which may allow the biomolecule to sample its conformational space extensively.
The ACM method implemented in the GROMACS-4.5.5 package (28) was used for FBP21-WWs, and aMD encoded in AMBER14 (29) was used for the free SAM-1 aptamer. All of the simulation details are described in the Supporting Material. The sampling efficiency of ACM and aMD compared to the standard MD are shown in Figs. S1 and S2, respectively, in both the root mean square deviation (RMSD) to the starting structures and the principal component analysis (PCA) (30). A description of how to carry out PCA is presented in the Supporting Material.
Ensemble optimization method
The ensemble optimization method (EOM) (7) was used to select an ensemble containing a small number of conformations of the biomolecule from a large structural pool generated in enhanced sampling to fit the SAXS data. Several programs in the ATSAS-2.4.3-1 package (31) were run sequentially. A theoretical scattering profile was calculated in CRYSOL (32) for each conformation in the pool. A master file combining all of the scattering intensities was created in ONEFILE. The program GAJOE was run twice. The first run created the file listing sizes (Rg and Dmax) for all of the conformations, and the second run produced the final EOM. The search procedure for the EOM is to minimize the fitting residual between the experimental and calculated SAXS profiles using the genetic algorithm:
| (1) |
where K is the number of data points in Iexp(q), and σ(q) are experimental errors, Ical(q) is the average of the theoretical scattering profiles of these conformations in the ensemble, and μ is a scaling factor. An automatic subtraction of a constant from the experimental data is allowed to facilitate the fitting. The momentum transfer q equals to 4π sinθ/λ, in which 2θ is the scattering angle and λ is the wavelength. After a certain number of cycles of independent search (the default number is 50), the ensemble with the minimal χ was chosen. The ensemble size was set to a default value of 20 and no repetition was allowed.
Recently, an advanced EOM 2.0 was developed (33), in which the ensemble size can be optimized during the search procedure. We also used this new version of EOM (as implemented in the ATSAS-2.7.2 package) in our SAXS-ER.
SAXS-ER procedure
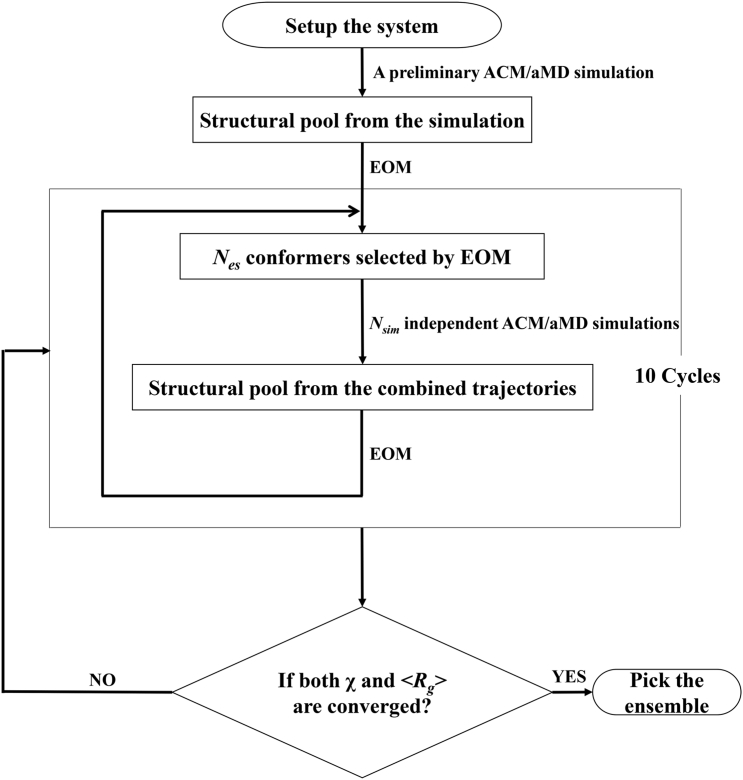
This tool uses several steps (Fig. 1). Python scripts for running the entire process using either ACM or aMD and corresponding tutorials are freely available under a GNU public license from the website https://github.com/pcheng27/SAXS-ER/tree/v1.1, and the DOI is http://doi.org/10.5281/zenodo.243155.
-
1)
Set up the system starting from an initial structure of the biomolecule, and perform a preliminary enhanced sampling (ACM or aMD) simulation.
-
2)
Run EOM on the structural pool generated in the simulation, and pick the ensemble with the minimal χ (Eq. 1) to the SAXS data. The ensemble size is Nes.
-
3)
Starting from the Nes conformers selected in EOM, Nsim (≥Nes) independent ACM/aMD simulations are carried out. Multiple independent short-time simulations may achieve a better sampling than a single long-time simulation (34).
-
4)
Run EOM on the structural pool in combining the Nsim trajectories.
-
5)
Repeat steps 3 and 4 for 10 cycles. If both χ and the average Rg (〈Rg〉) of the ensemble converge, stop the simulation and pick the final ensemble. Otherwise, run the simulation for another 10 cycles.
Figure 1.
General workflow of SAXS-ER.
Results and Discussion
FBP21-WWs
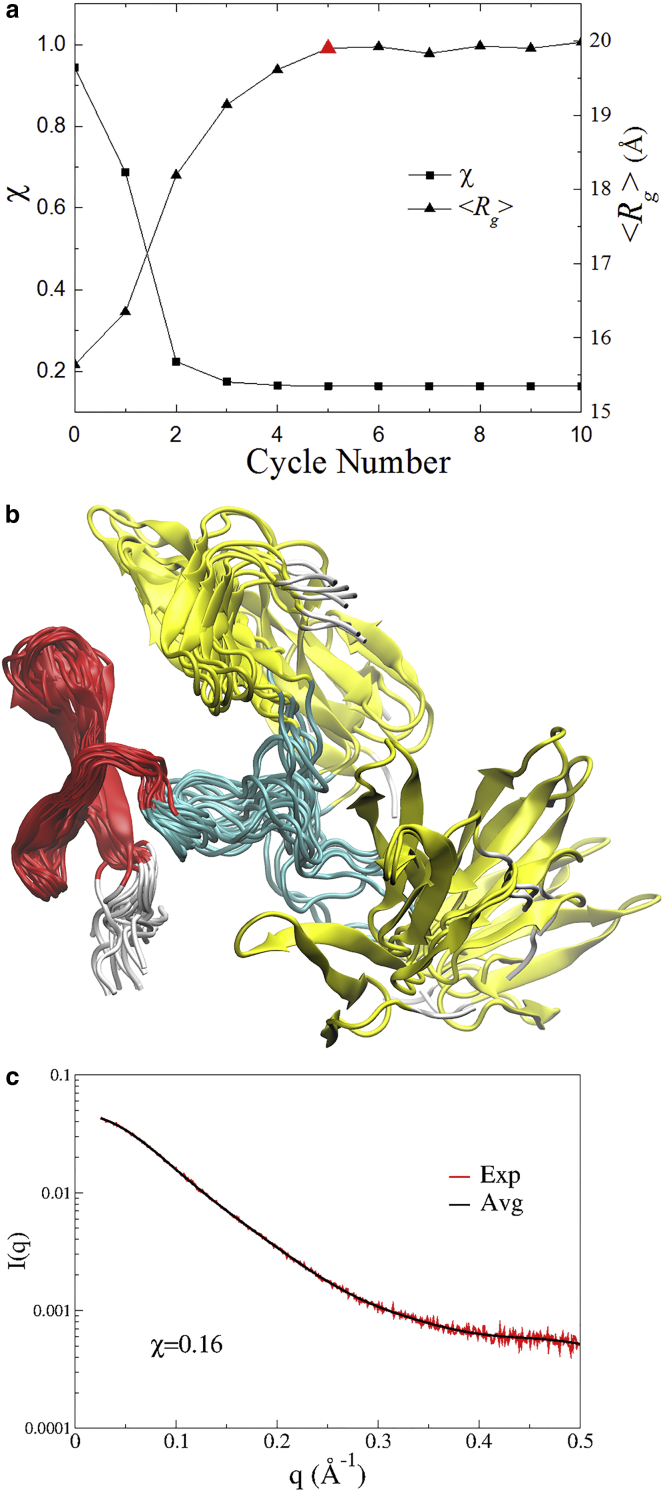
We ran a SAXS-ER starting from model 1 in the NMR structures of FBP21-WWs (16). The ensemble size of EOM was Nes = 20. Each cycle consisted of Nsim = 20 independent 100-ps ACM simulations, in which the low-frequency collective motions were coupled at 500 K. The evolution of the sampled conformational space of FBP21-WWs with the iteration cycles are shown in projections onto the PCA modes (Fig. S3). The SAXS profiles of the ensembles are plotted against cycle number in Fig. S4 to show how they evolve to fit the experimental data. It is found that χ converges very fast in the first three cycles (Fig. 2 a, squares) while 〈Rg〉 of the corresponding ensemble (Fig. 2 a, triangles) starts to saturate at approximately the fifth cycle (Fig. 2 a, red triangle). Considering the low-resolution nature of SAXS data, we used both metrics as criteria to select the final ensemble, to avoid overfitting. Therefore, the ensemble of 20 conformations at the fifth cycle (Fig. 2 b, created by VMD (35)) was selected to reproduce the SAXS profile. The χ-values of individual SAXS curves in the ensemble range from 0.48 to 0.87 (Fig. S5 a), but their average profile gives a χ of 0.16 to the experimental data (Fig. 2 c), which is significantly less than the value of the initial ensemble (Fig. S5 b). This result suggests that the protein should be represented in an ensemble of different conformers. The Rg values of these conformations in the ensemble range from 16.0 to 26.6 Å, with 〈Rg〉 of 19.9 Å, which is close to the value (19.6 ± 0.4 Å) estimated from the experimental data. This result indicates that the protein may take either a compact or extended conformation in solution, which is consistent with the EOM result for long MD trajectories with 2 μs (36). However, it should be noted that the total timescale of the SAXS-ER is up to 20 ns (100 ps × 20 conformers × 10 cycles). Our tool is efficient because it needs only a relatively short simulation time to obtain an appropriate structural ensemble to fit the SAXS data.
Figure 2.
The SAXS-ER results for FBP21-WWs. (a) The minimal χ and the corresponding 〈Rg〉 at each cycle. The final selected ensemble at the fifth cycle is indicated by a red triangle. (b) The conformers in the final selected ensemble, which are superimposed on the WW1 domain (colored in red). The WW2 domain is colored in yellow, the linker is colored in cyan, and the N- (residues 1–5) and C-terminus (residues 74–75) are both colored in gray. (c) The back-calculated SAXS profile of the final selected ensemble (black) is fitted to the experimental data (red). To see this figure in color, go online.
Starting from different conformations, multiple SAXS-ER runs were carried out with different simulation times for each cycle, or different ACM parameters. These ensembles (Fig. S6) are rather similar to the one shown in Fig. 2 b in the relative orientation between the two WW domains. We also tried a SAXS-ER using EOM 2.0 (33), in which the ensemble size Nes can be optimized (Fig. S7). After the 18th cycle, both χ and 〈Rg〉 became saturated (Fig. S7 a), so we selected the ensemble at this cycle as final. Nes varies from 5 to 20 with the iterative cycles (Fig. S7 b). The final selected ensemble contains 14 conformers (Fig. S7 c), and its back-calculated SAXS profile is in good agreement with the experimental data (Fig. S7 d). Despite its different size, the ensemble shows a similar domain orientation with those with Nes = 20 (Figs. 2 b and S6). The latter include more conformers with similar shapes than the former. Overall, the results indicate that the ensembles generated in SAXS-ER are reliable. The two WW domains can be either close or distant to each other in solution, which may facilitate their cooperative binding with different ligands.
To compare SAXS-ER with other methods, the program Ranch in the ATSAS package was used to generate 10,000 conformers of FBP21-WWs in rigid-body modeling, and then EOM and EOM 2.0 were run to select an ensemble from the pool to reproduce the experimental SAXS data (Fig. S8). We also used the flexible-meccano statistical coil model (37) to generate a pool containing 10,000 conformers, and again ran EOM and EOM 2.0 to select an ensemble (Fig. S9). These ensembles are different than those generated in SAXS-ER (Figs. 2 b, S6 and S7), which are clearly indicated in their projections onto the 2D plane of the PCA modes (Fig. S10). Ranch or flexible-meccano only consider relatively simple interactions, so the models generated may freely take many different orientations between the two WW domains (Figs. S8 a and S9 a). However, SAXS-ER performs all-atom simulations with a well-defined force field, and the sampled conformations would be physically more reasonable than the simple models. Many conformers in the SAXS-ER ensembles are located at a particular region on the plane (Fig. S10, red circles), whereas the Ranch/flexible-meccano ensembles consist of diverse conformations with a wide distribution (Fig. S10, green and blue circles). Due to the low-resolution nature of the SAXS data and the overfitting problem, the ensembles from SAXS-ER and the other two methods have nearly the same χ-values for fitting the experimental data, but the former may present more realistic protein conformations in solution than the latter. Moreover, additional quantitative information about these conformations, such as the energy and the population, could be extracted from the MD simulation.
Free SAM-1 aptamer
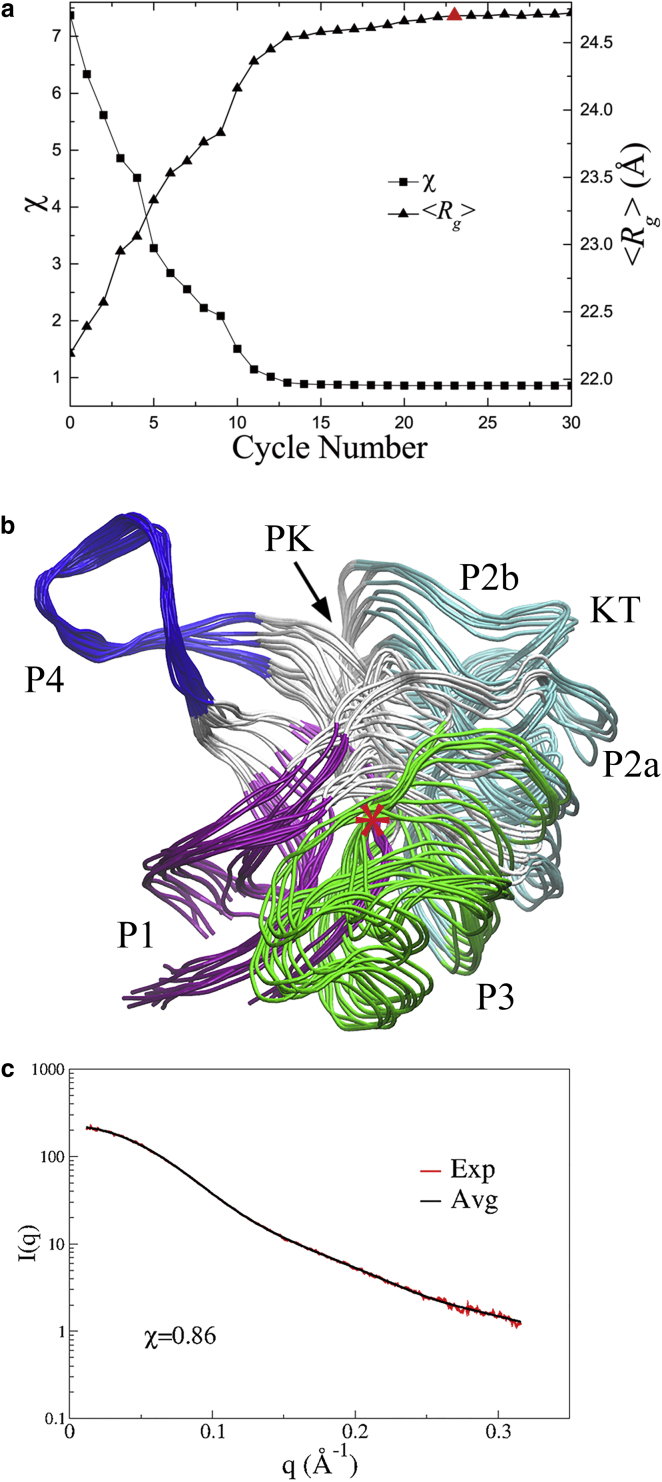
A SAXS-ER of the free SAM-1 aptamer was carried out with up to 30 cycles by using a 200-ns MD trajectory to estimate the aMD parameters. The ensemble size of EOM was Nes = 20. Each cycle consisted of Nsim = 20 independent 100-ps aMD simulations. The evolution of the sampled conformational space with iteration cycle is shown in projections onto the PCA modes (Fig. S11). The SAXS profiles of the ensembles are plotted against the cycle number in Fig. S12, to show how they evolve to fit the experimental data. The value χ (Fig. 3 a, squares) converges more slowly than in the SAXS-ER of FBP21-WWs (Fig. 2 a, squares), from an initial value of 7.37 to a final value of 0.86. The parameter 〈Rg〉 of the corresponding ensemble increases from the initial 22.2 Å to a final value of 24.7 Å (Fig. 3 a, triangles), which is consistent with the value (24.8 ± 0.0 Å) estimated from the experimental SAXS data. We selected the ensemble at the 23rd cycle as final because both χ and 〈Rg〉 were saturated. The 20 conformers in this ensemble were superimposed using the subdomain P4 (Fig. 3 b, blue). The χ-values of individual SAXS curves in the ensemble range from 1.07 to 2.33 (Fig. S13 a), but their average profile gives an χ of 0.86 to the experimental data (Fig. 3 c), which is significantly less than the value of the initial ensemble (Fig. S13 b). The Rg values of these conformers range from 24.1 to 25.2 Å. The result suggests that the aptamer can assume multiple conformations in solution without the ligand. Orientations of the subdomains P1 (Fig. 3 b, violet) and P3 (Fig. 3 b, green), as well as their distances, are variable, which result in the opening/closing of the ligand-binding site. Stoddard et al. (20) generated trajectories of the free aptamer using rigid-body torsion angle MD simulations, and also selected a set of conformations against the SAXS data. Although different simulation algorithms were used, our results show agreement with their ensemble.
Figure 3.
The SAXS-ER results for the free SAM-1 aptamer. (a) The minimal χ and the corresponding 〈Rg〉 at each cycle. The final selected ensemble at the 23rd is indicated by a red triangle. (b) The conformers in the final ensemble, which are superimposed on the subdomain P4 (nucleotides 69–82, colored in blue). The location of SAM is approximated by a red star. (c) The back-calculated SAXS profile of the final ensemble (black) is fitted to the experimental data (red). To see this figure in color, go online.
The structural ensemble of another SAXS-ER simulation of the free SAM-1 aptamer is shown in Fig. S14. The RMSDs of all of the P atoms of the conformers in the two ensembles (Figs. 3 b and S14) were measured. For any conformer in one ensemble, at least one conformer in the other ensemble can be found that has a RMSD ≤ 3 Å, which indicates that the two ensembles are similar. A SAXS-ER using EOM 2.0 (33) was also run to optimize the ensemble size (Fig. S15). The ensemble size is five at the most cycles (Fig. S15 b), and the final ensemble at the 28th cycle is shown in Fig. S15 c. Although Nes is significantly smaller, the conformers are still similar to those within the larger (Nes = 20) ensembles (Figs. 3 b and S14) according to the RMSD calculations. The latter contain more conformers with similar RMSD values than the former. It should be noted that it may not be convenient to use the Ranch or flexible-meccano on the free SAM-1 aptamer because it is not as straightforward as for FBP21-WWs to determine which parts are rigid or flexible. However, this is not an issue for our method.
Conclusion
Among the structural modeling methods for biomolecules that integrate SAXS, our SAXS-oriented ensemble refinement tool has the following features: 1) Modification of the complicated simulation code is not required, because this tool uses a screening-after-sampling strategy. 2) Extensive simulations are not necessary to cover the conformational space of the biomolecule. By iteratively running enhanced simulations and EOM, the sampling is efficiently guided by the SAXS data, in a similar manner to refining-while-sampling methods. 3) Although computationally more expensive than other methods that use simple models, SAXS-ER is very suitable for massive parallel computing because it comprises a number of independent simulations. 4) The tool is easy to use and flexible. Any simulation package, sampling method, and ensemble selection algorithm can be chosen by simply changing the Python script.
Author Contributions
Z.Z. and J.P. designed the project; P.C. and J.P. wrote the software; P.C. carried out the simulations, and analyzed the data; and all authors contributed to writing the article.
Acknowledgments
This work is supported by the National Key Basic Research Program of China (under grant No. 2013CB910203), the National Natural Science Foundation of China (under grants No. 31270760 and No. 21573205), the Strategic Priority Research Program of the Chinese Academy of Sciences (under grant No. XDB08030102), and the Supercomputing Center of the University of Science and Technology of China.
Editor: Jill Trewhella.
Footnotes
Peng Cheng and Junhui Peng contributed equally to this work.
Supporting Material and fifteen figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)30238-2.
Supporting Citations
References (38, 39, 40, 41, 42, 43, 44, 45, 46) appear in the Supporting Material.
Supporting Material
References
- 1.Graewert M.A., Svergun D.I. Impact and progress in small and wide angle x-ray scattering (SAXS and WAXS) Curr. Opin. Struct. Biol. 2013;23:748–754. doi: 10.1016/j.sbi.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 2.Bernadó P., Blackledge M. Structural biology: proteins in dynamic equilibrium. Nature. 2010;468:1046–1048. doi: 10.1038/4681046a. [DOI] [PubMed] [Google Scholar]
- 3.Schneidman-Duhovny D., Kim S.J., Sali A. Integrative structural modeling with small angle x-ray scattering profiles. BMC Struct. Biol. 2012;12:17. doi: 10.1186/1472-6807-12-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Y.H., Peng J.H., Zhang Z.Y. Structural modeling of proteins by integrating small-angle x-ray scattering data. Chin. Phys. B. 2015;24:126101. [Google Scholar]
- 5.Zheng W., Tekpinar M. Accurate flexible fitting of high-resolution protein structures to small-angle x-ray scattering data using a coarse-grained model with implicit hydration shell. Biophys. J. 2011;101:2981–2991. doi: 10.1016/j.bpj.2011.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Björling A., Niebling S., Westenhoff S. Deciphering solution scattering data with experimentally guided molecular dynamics simulations. J. Chem. Theory Comput. 2015;11:780–787. doi: 10.1021/ct5009735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernadó P., Mylonas E., Svergun D.I. Structural characterization of flexible proteins using small-angle x-ray scattering. J. Am. Chem. Soc. 2007;129:5656–5664. doi: 10.1021/ja069124n. [DOI] [PubMed] [Google Scholar]
- 8.Pelikan M., Hura G.L., Hammel M. Structure and flexibility within proteins as identified through small angle x-ray scattering. Gen. Physiol. Biophys. 2009;28:174–189. doi: 10.4149/gpb_2009_02_174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bertini I., Giachetti A., Svergun D.I. Conformational space of flexible biological macromolecules from average data. J. Am. Chem. Soc. 2010;132:13553–13558. doi: 10.1021/ja1063923. [DOI] [PubMed] [Google Scholar]
- 10.Yang S., Blachowicz L., Roux B. Multidomain assembled states of Hck tyrosine kinase in solution. Proc. Natl. Acad. Sci. USA. 2010;107:15757–15762. doi: 10.1073/pnas.1004569107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Różycki B., Kim Y.C., Hummer G. SAXS ensemble refinement of ESCRT-III CHMP3 conformational transitions. Structure. 2011;19:109–116. doi: 10.1016/j.str.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Curtis J.E., Raghunandan S., Krueger S. SASSIE: a program to study intrinsically disordered biological molecules and macromolecular ensembles using experimental scattering restraints. Comput. Phys. Commun. 2012;183:382–389. [Google Scholar]
- 13.Deshmukh L., Schwieters C.D., Clore G.M. Structure and dynamics of full-length HIV-1 capsid protein in solution. J. Am. Chem. Soc. 2013;135:16133–16147. doi: 10.1021/ja406246z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang J.R., Warner L.R., Blackledge M. Transient electrostatic interactions dominate the conformational equilibrium sampled by multidomain splicing factor U2AF65: a combined NMR and SAXS study. J. Am. Chem. Soc. 2014;136:7068–7076. doi: 10.1021/ja502030n. [DOI] [PubMed] [Google Scholar]
- 15.Bedford M.T., Reed R., Leder P. WW domain-mediated interactions reveal a spliceosome-associated protein that binds a third class of proline-rich motif: the proline glycine and methionine-rich motif. Proc. Natl. Acad. Sci. USA. 1998;95:10602–10607. doi: 10.1073/pnas.95.18.10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang X., Beullens M., Shi Y. Structure and function of the two tandem WW domains of the pre-mRNA splicing factor FBP21 (formin-binding protein 21) J. Biol. Chem. 2009;284:25375–25387. doi: 10.1074/jbc.M109.024828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wen B., Peng J., Zhang Z. Characterization of protein flexibility using small-angle x-ray scattering and amplified collective motion simulations. Biophys. J. 2014;107:956–964. doi: 10.1016/j.bpj.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grundy F.J., Henkin T.M. The S box regulon: a new global transcription termination control system for methionine and cysteine biosynthesis genes in gram-positive bacteria. Mol. Microbiol. 1998;30:737–749. doi: 10.1046/j.1365-2958.1998.01105.x. [DOI] [PubMed] [Google Scholar]
- 19.Montange R.K., Batey R.T. Structure of the S-adenosylmethionine riboswitch regulatory mRNA element. Nature. 2006;441:1172–1175. doi: 10.1038/nature04819. [DOI] [PubMed] [Google Scholar]
- 20.Stoddard C.D., Montange R.K., Batey R.T. Free state conformational sampling of the SAM-I riboswitch aptamer domain. Structure. 2010;18:787–797. doi: 10.1016/j.str.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karplus M., McCammon J.A. Molecular dynamics simulations of biomolecules. Nat. Struct. Biol. 2002;9:646–652. doi: 10.1038/nsb0902-646. [DOI] [PubMed] [Google Scholar]
- 22.Clarage J.B., Romo T., Phillips G.N., Jr. A sampling problem in molecular dynamics simulations of macromolecules. Proc. Natl. Acad. Sci. USA. 1995;92:3288–3292. doi: 10.1073/pnas.92.8.3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Onuchic J.N., Luthey-Schulten Z., Wolynes P.G. Theory of protein folding: the energy landscape perspective. Annu. Rev. Phys. Chem. 1997;48:545–600. doi: 10.1146/annurev.physchem.48.1.545. [DOI] [PubMed] [Google Scholar]
- 24.Mitsutake A., Mori Y., Okamoto Y. Enhanced sampling algorithms. Methods Mol. Biol. 2013;924:153–195. doi: 10.1007/978-1-62703-017-5_7. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Z., Shi Y., Liu H. Molecular dynamics simulations of peptides and proteins with amplified collective motions. Biophys. J. 2003;84:3583–3593. doi: 10.1016/S0006-3495(03)75090-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Atilgan A.R., Durell S.R., Bahar I. Anisotropy of fluctuation dynamics of proteins with an elastic network model. Biophys. J. 2001;80:505–515. doi: 10.1016/S0006-3495(01)76033-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamelberg D., Mongan J., McCammon J.A. Accelerated molecular dynamics: a promising and efficient simulation method for biomolecules. J. Chem. Phys. 2004;120:11919–11929. doi: 10.1063/1.1755656. [DOI] [PubMed] [Google Scholar]
- 28.Hess B., Kutzner C., Lindahl E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 29.Case D.A., Babin V., Kollman P.A. University of California; San Francisco, CA: 2014. AMBER 14. [Google Scholar]
- 30.Amadei A., Linssen A.B., Berendsen H.J.C. Essential dynamics of proteins. Proteins. 1993;17:412–425. doi: 10.1002/prot.340170408. [DOI] [PubMed] [Google Scholar]
- 31.Petoukhov M.V., Franke D., Svergun D.I. New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Cryst. 2012;45:342–350. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Svergun D., Barberato C., Koch M.H.J. CRYSOL—a program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Cryst. 1995;28:768–773. [Google Scholar]
- 33.Tria G., Mertens H.D.T., Svergun D.I. Advanced ensemble modelling of flexible macromolecules using x-ray solution scattering. IUCrJ. 2015;2:207–217. doi: 10.1107/S205225251500202X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caves L.S.D., Evanseck J.D., Karplus M. Locally accessible conformations of proteins: multiple molecular dynamics simulations of crambin. Protein Sci. 1998;7:649–666. doi: 10.1002/pro.5560070314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–28. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Y., Wen B., Zhang Z. Determining structural ensembles of flexible multi-domain proteins using small-angle x-ray scattering and molecular dynamics simulations. Protein Cell. 2015;6:619–623. doi: 10.1007/s13238-015-0162-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ozenne V., Bauer F., Blackledge M. Flexible-meccano: a tool for the generation of explicit ensemble descriptions of intrinsically disordered proteins and their associated experimental observables. Bioinformatics. 2012;28:1463–1470. doi: 10.1093/bioinformatics/bts172. [DOI] [PubMed] [Google Scholar]
- 38.Hockney R.W., Goel S.P., Eastwood J.W. Quiet high-resolution computer models of a plasma. J. Comput. Phys. 1974;14:148–158. [Google Scholar]
- 39.Maier J.A., Martinez C., Simmerling C. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 2015;11:3696–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ryckaert J.P., Ciccotti G., Berendsen H.J.C. Numerical integration of Cartesian equations of motion of a system with constraints—molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 41.Jorgensen W.L., Chandrasekhar J., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 42.Berendsen H.J.C., Postma J.P.M., Haak J.R. Molecular-dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 43.Essmann U., Perera L., Pedersen L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- 44.Duan Y., Wu C., Kollman P. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J. Comput. Chem. 2003;24:1999–2012. doi: 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]
- 45.Bussi G., Donadio D., Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126:014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 46.Hess B. P-LINCS: a parallel linear constraint solver for molecular simulation. J. Chem. Theory Comput. 2008;4:116–122. doi: 10.1021/ct700200b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.