Abstract
We describe a quantitative, high-precision, high-throughput method for measuring the mechanical properties of cells in suspension with a microfluidic device, and for relating cell mechanical responses to protein expression levels. Using a high-speed (750 fps) charge-coupled device camera, we measure the driving pressure Δp, maximum cell deformation εmax, and entry time tentry of cells in an array of microconstrictions. From these measurements, we estimate population averages of elastic modulus E and fluidity β (the power-law exponent of the cell deformation in response to a step change in pressure). We find that cell elasticity increases with increasing strain εmax according to E ∼ εmax, and with increasing pressure according to E ∼ Δp. Variable cell stress due to driving pressure fluctuations and variable cell strain due to cell size fluctuations therefore cause significant variability between measurements. To reduce measurement variability, we use a histogram matching method that selects and analyzes only those cells from different measurements that have experienced the same pressure and strain. With this method, we investigate the influence of measurement parameters on the resulting cell elastic modulus and fluidity. We find a small but significant softening of cells with increasing time after cell harvesting. Cells harvested from confluent cultures are softer compared to cells harvested from subconfluent cultures. Moreover, cell elastic modulus increases with decreasing concentration of the adhesion-reducing surfactant pluronic. Lastly, we simultaneously measure cell mechanics and fluorescence signals of cells that overexpress the GFP-tagged nuclear envelope protein lamin A. We find a dose-dependent increase in cell elastic modulus and decrease in cell fluidity with increasing lamin A levels. Together, our findings demonstrate that histogram matching of pressure, strain, and protein expression levels greatly reduces the variability between measurements and enables us to reproducibly detect small differences in cell mechanics.
Introduction
Cell mechanical properties have been implicated in a wide range of physiological processes and diseases. For example, cell mechanical properties change systematically in malaria (1, 2), asthma (3), inflammation (4), and cancer (1, 5). The need to measure cell mechanical properties in a high-throughput, quantitative way led to the recent development of various microfluidic techniques, such as the optical stretcher (6), hydrodynamic stretching (7), real-time deformability cytometry (8), microfiltration (9), and microconstriction arrays (10, 11, 12). Compared to established low-throughput methods such as atomic force microscopy (13) or optical tweezers (14), these microfluidic systems are easy to implement and, apart from a simple microfluidic chip, do not require expensive specialized equipment other than a microscope, a high-speed camera, and customized image analysis software.
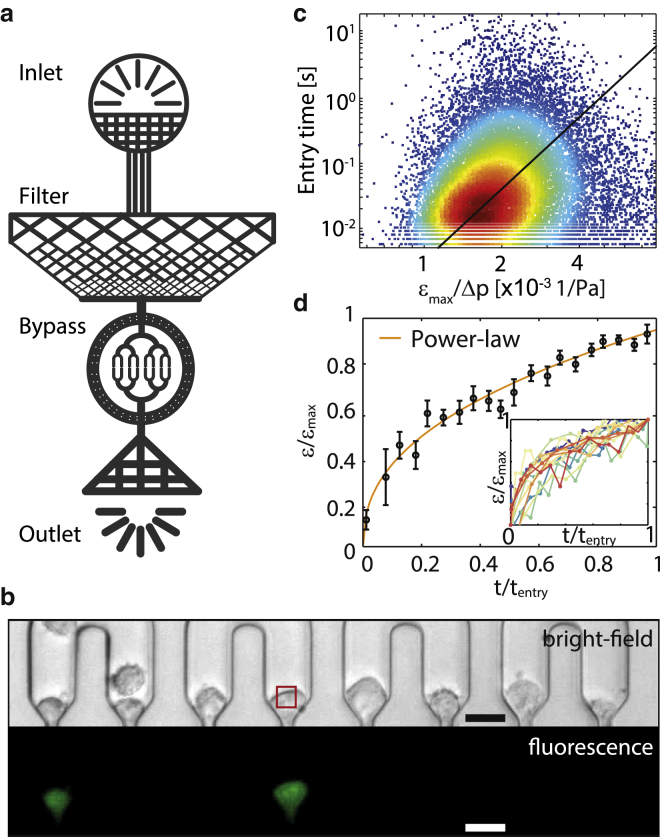
We have recently described a microfluidic method for high-throughput quantitative measurement of cell mechanical properties of suspended cells (15). In this method, we pump cells through an array of eight parallel microconstrictions and measure the pressure drop Δp across the constriction, the cell’s maximum strain εmax, and the entry time tentry of the cells into the constriction. The entry time is the time from the moment the cell is lodged in front of a constriction to the moment the cell has been deformed to the width of the constriction. This is also the moment at which the cell exits the constriction, because the length of the constriction channel in our setup is short compared to the cell diameter, and the travel time through the constriction channel is therefore negligible and below the time resolution of our camera (<1.4 ms).
We have established (15) that pressure, strain, and entry time are related through a power law according to . The parameter E describes the elastic modulus of the cells, the power-law exponent β describes the cells’ fluidity, and the reference time t0 is set to 1 s (16). Rearranging and considering that tentry is the time when the cell strain ε(t) has reached εmax gives
| (1) |
A power-law exponent of β = 0 indicates a purely elastic solid, described by Hooke’s law, in which case there is no time dependency of the creep response, i.e., ε(t) = Δp/E. If Δp/E > εmax, the cell entry time is zero, otherwise it is infinite. For β = 1, Eq. 1 resembles Newton’s law of viscous deformations. Typical values of β for cells are in the range of 0.1–0.4, indicating viscoelastic behavior (16, 17). Both E and β are strongly influenced by the cytoskeleton (actin, microtubules), but also by cell nuclear properties including chromatin condensation and expression levels of nuclear lamina intermediate filaments (15). Moreover, E and β in cells after pharmacological treatment are not independent from each other but scale according to predictions from the theory of soft glassy rheology (13, 15, 16, 17).
Equation 1 assumes that the elastic and dissipative cell mechanical properties are independent of the applied pressure and the maximum strain. However, previous reports have established that cell mechanical properties can be stress- and strain-sensitive (18, 19, 20, 21). Because the applied pressure drop across the microconstrictions in our device can vary during a measurement due to changes in the occupancy of the channel array, the accumulation of cell debris in the filter system, and user adjustments—and because the maximum cell strain also varies from cell to cell due to variable cell diameters—the measured cell mechanical parameters E and β can be subject to a high degree of variability.
In this study, we investigate the influence of stress and strain stiffening and explore how Eq. 1 can be extended to account for these effects. We then describe a method for canceling stress or strain stiffening effects when comparing different cell populations. We achieve this by histogram matching, whereby only those cells from two (or more) measurements are included in the analysis that have experienced the same pressure and the same maximum strain. Moreover, we investigate how cell mechanics is influenced by subtle details of measurement and cell culture conditions, such as cell confluency before harvesting, the time since cell harvesting, the choice of the cell suspension medium, or device coating with adhesion-preventing pluronic surfactant. Finally, we explore the effect of protein expression levels in a mixed cell population on the measurement results. Specifically, we transfect cells with a lamin A-green fluorescent protein (GFP) construct and observe them with combined bright-field and fluorescence imaging in our microfluidic device. We then correlate differences in the mechanical properties of individual cells with differences in lamin A-GFP expression levels.
Our results establish that histogram matching of pressure, strain, and protein expression levels greatly reduces the variability between measurements and enables us to reproducibly measure small differences in cell mechanical properties between different groups of cells.
Materials and Methods
Cell culture
K562 leukemia cells (No. CCL-243; American Type Culture Collection, Manassas, VA) are cultured at 37°C and 5% CO2 in Iscove’s Modified Dulbecco’s Medium (IMDM, Cat. No. 12440053; Gibco/Thermo Fisher Scientific, Waltham, MA) containing 10% fetal calf serum (FCS, Cat. No. 16000036; Gibco/Thermo Fisher Scientific) and 1% Penicillin-Streptomycin-Glutamine (PSG, Cat. No. 10378016; Gibco/Thermo Fisher Scientific). K562 lamin A-overexpressing cells are transfected as described in Lange et al. (15). DLD-1 pMCV colon carcinoma cells are a kind gift of Michael Stürzl (Division of Molecular and Experimental Surgery, University Clinics Erlangen) and are cultured in RPMI Medium (Cat. No. 11875093; Gibco/Thermo Fisher Scientific), containing 10% FCS, 1% PSG, and 1% G418 (Cat. No. 11811098; Gibco/Thermo Fisher Scientific). NIH 3T3 mouse embryonic fibroblast cells (No. CRL-1658; American Type Culture Collection) are cultured in Dulbecco’s Modified Eagle Medium (DMEM, Cat. No. 11885084; Gibco/Thermo Fisher Scientific), containing 10% FCS and 1% PSG. Cells are passaged every third day. Actin depolymerization is performed with cytochalasin D (cytoD, Cat. No. C8273; Sigma-Aldrich, St. Louis, MO) at a concentration of 10 μM for a duration of 30 min before measurements. Whole cell fluorescent staining is performed with calcein (Cat. No. C0875; Sigma-Aldrich) at a concentration of 250 nM for a duration of 30 min before measurements.
Design and setup of the device
Each microconstriction device consists of eight parallel microconstrictions that are surrounded by a pressure-equalizing bypass (Fig. 1 a). The pressure across the constriction area is thus clamped by the bypass. The cells enter the device by a punched inlet and first traverse a filter system that holds back debris. After transit through the constriction array, the cells leave the device through a punched outlet. The height and width of the constrictions are chosen to be smaller than the cell diameter and are therefore adapted to the mean size of the measured cell population. The height of the whole device, including the constrictions, is on the order of the cell diameter (K562, 13.6–16.6 μm; DLD-1, 16.6 μm; NIH 3T3, 19 μm). For K562 cells, the width of the constrictions is 5.1–6 μm. For DLD-1 cells, a width of 5.1 μm is used. For NIH 3T3 cells, a constriction width of 7.5 μm is used. All device geometries are confirmed with a μ-surf interferometric confocal microscope (NanoFocus, Oberhausen, Germany) (see Fig. S1 in the Supporting Material).
Figure 1.
(a) Schematic of the microfluidic device with inlet (top), debris filter, constriction area surrounded by a bypass, and outlet (bottom). (b) Bright-field (top) and fluorescence (bottom) image of the constriction region with K562 leukemia cells stained with calcein. Images are contrast-enhanced. Scale bars, 20 μm. The SD of the brightness within each region of interest (indicated by a red square in front of a constriction) is used to calculate cell entry time. (c) tentry versus εmax/Δp of 19,991 K562 leukemia cells. Colors indicate the bivariate kernel density estimate of the data points. (Black line) Orthogonal least-squares fit of Eq. 1 to the data. (d) Strain evolution ε(t) of NIH 3T3 cells (black circles, mean ± SE of 59 cells) during entry into a constriction can be fitted by a power law (orange line). Time t and strain ε are normalized by tentry and εmax before averaging. The parameter t = 0 corresponds to the time point when the cell first encounters the constriction, and t = 1 corresponds to the time point when the cell has reached its maximum strain ε = εmax and leaves the constriction. (Inset) Normalized strain versus normalized time of individual cells. To see this figure in color, go online.
The microconstriction array is made out of polydimethylsiloxane (PDMS, Sylgard 184; Dow Corning, Midland, MI) and is produced with standard soft lithography as described in (15) and (22). After molding and baking the PDMS, the devices are sealed to a glass coverslip through plasma bonding (Zepto; Diener Electronics, Nagold, Germany). Devices are used after a waiting time of at least 48 h after plasma treatment to ensure stable hydrophobic channel wall properties (12). Before measurements, the device is coated with 1 wt % pluronic (BASF, Ludwigshafen, Germany; Cat. No. P2443, Sigma-Aldrich) in PBS for 30 min to decrease nonspecific cell binding to the channel walls (23, 24).
For measurements, we either use naturally suspended cells (K562), or normally adherent cells brought into suspension after trypsinization (DLD-1 and NIH 3T3). Five-million cells are suspended in 1 mL PBS with 1 wt % pluronic. Rigid polyether-ether-ketone tubing (VWR, Radnor, PA) is used to connect the cell reservoirs to the microfluidic device. Cells are then pumped through the channels of the microfluidic device by a hydraulic pump (Bellofram, Newell, WV) using an external pressure range of 1–50 kPa. During experiments, the constriction area of all eight constrictions (Fig. 1 b) is continuously imaged with a 10×, 0.3 NA objective in bright-field mode at a frame rate of 750 fps using a high-speed charge-coupled device (CCD) camera (G680; Allied Vision Technologies, Stadtroda, Germany) mounted to an inverted microscope (DM-IL; Leica Microsystems, Wetzlar, Germany). Videos are recorded with a custom written C-program for further analysis with MATLAB (The MathWorks, Natick, MA). Typically, cell transits of 5000 cells are recorded during a measurement time of 30 min. All measurements are performed at room temperature (22°C).
Fluorescence imaging
For recording the fluorescence signals from transfected cells, a diode-pumped solid-state laser (wavelength 473 nm, VA-I-N-473; Viasho, Beijing, China) with a maximum power of 100 mW is coupled to the epifluorescence port of the microscope. Through a series of excitation and emission filters (Thorlabs, Newton, NJ), light for bright-field imaging (wavelength >590 nm) is separated from the emission wavelength of GFP (∼515–550 nm) (Fig. S2). The CCD camera (G680; Allied Vision Technologies) in the bright-field channel runs at a frame rate of 750 fps with an exposure time of 100 μs. For every third image, the camera triggers a second, synchronized camera (G680; Allied Vision Technologies) in the GFP-channel at a frame rate of 250 fps with an exposure time of 4 ms, which records the GFP-emission light (Fig. 1 b). Only fluorescence images of cells that are immobilized at the constriction entrance are evaluated to avoid motion blurring. For computing the fluorescence intensity of a cell, pixel intensities are background-subtracted and summed over a region of interest corresponding to the channel area in front of the constriction. The intensity sum of all pixels is then normalized by the cell area extracted from bright-field images. Thereby, we obtain a cell-size-independent average fluorescence intensity of the cell in units of counts per cell pixel.
Calculation of cell mechanical properties
We calculate the average cell mechanical parameters E and β from an orthogonal least-squares fit of Eq. 1 to the measured entry times tentry of typically 2000 cells, the maximum cell deformation εmax of each cell, and the pressure drop Δp that each cell experienced during transit. From the recorded images, the cell size is detected from bright-field images of the undeformed cell before it enters the constriction (Fig. 1 b). From the cell size, the maximum deformation εmax of the cell is calculated according to , with rcell being the radius of the undeformed cell, and rcon being half the constriction width.
From the image series of the cell approaching the constriction, the cell speed is measured by tracking the leading cell edge. From the cell speed vcell, the average flow speed vavg is estimated using an experimentally measured relationship according to vcell/vavg = 2.64–1.59 (rcell/rhyd) (Fig. S3), with rhyd being the hydrodynamic radius of the channel, which is calculated from the channel width w and height h according to rhyd = h × w/(h+w) (15). The flow resistance of each constriction and each segment of the channel network is calculated using Hagen-Poiseuille’s law. The pressure drop across each channel is then solved using Kirchhoff’s circuit laws, whereby we take into account the occupancy of all constrictions. Thus, we continuously update the pressure drop across a constriction during a cell transit, depending on the occupancy of all other constrictions. In addition, every time a new cell enters the constriction region, we measure the local flow speed, and the pressure drop over all constriction regions is updated accordingly (15).
Cell entry times are measured from changes in the brightness standard deviation (SD) of regions of interest in front of the constrictions (15), as indicated by the red square in Fig. 1 b.
Statistical analysis
To calculate an average elastic modulus E and fluidity β of a cell population, Eq. 1 is fitted to the measured Δp, εmax, and tentry data from typically 2000 cells using an orthogonal linear least-squares fit implemented in MATLAB, which minimizes the perpendicular distances between the data points and the regression line (Fig. 1 c). Before fitting, tentry and the ratio of εmax/Δp are logarithmically transformed to obtain a linear relationship between log(tentry) and log(εmax/Δp). Standard errors (SEs) for E and β are calculated by bootstrapping, where we repeat the fit 100 times on ensembles of randomly selected cells. This SE corresponds to 1 SD between the fitted values. For testing significant differences when comparing pairs of conditions or cell populations, we compute the area of overlap between the probability density distributions of the fitted parameters. Differences are considered statistically significant for an overlap of <5% (p < 0.05).
Results and Discussion
Time dependence of cell deformations
Equation 1 assumes a power-law creep response of the cells inside the constrictions, ε(t) ∼ tβ. To test the validity of this assumption, we observe the cell shape and measure the time-evolution of the long axis l of NIH 3T3 cells during passage through constrictions (Fig. S4; Movie S1). For this measurement, we use a device that only contains a single constriction to avoid short-time pressure fluctuations during the cells’ transit caused by changing clogging configurations in the device. The device also contains an enlarged channel cavity after the constriction to observe cell relaxations. We approximate the strain as ε(t) = (l(t) − l0)/l0, with l0 being the cell diameter in flow direction before the cell has entered the constriction. We confirm that the time evolution of the strain ε(t) follows a power-law in time (R2 = 0.98) (Fig. 1 d). By contrast, the response of a Kelvin-Voigt model, which is also commonly used to describe visco-elastic cell mechanical properties, fits the creep response with a considerably lower fidelity of R2 = 0.90 (25, 26).
Stress and strain stiffening
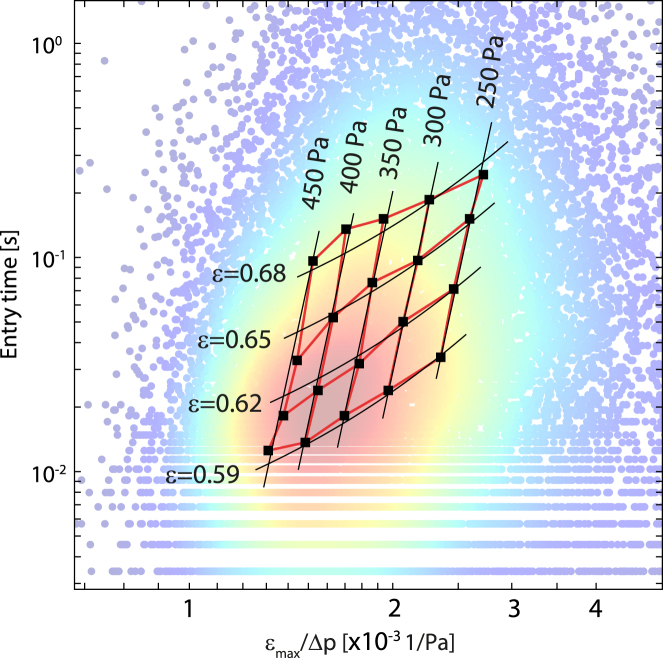
We fit Eq. 1 to the data (tentry, εmax, Δp) from several thousands of cells and compute a population average for the cell elastic modulus and fluidity. This fit corresponds to fitting a straight line to a scatter plot of log(tentry) versus log(εmax/Δp) (15). Data points from individual cells, however, can deviate substantially from the fit line and form a widely distributed, seemingly randomly scattered cloud (Figs. 2 and 3, a and d). Yet a surprisingly large part of the data scatter is not random but occurs systematically: When we sort the individual data points into groups that have experienced similar pressure and strain, and then plot the average entry time versus the average ratio of εmax/Δp for each group separately, we find that the averaged data also form a cloud but with a highly regular, gridlike structure (Fig. 2, black squares). Lines that connect data points of equal strain are aligned more horizontally and are shifted toward larger entry times as the strain increases, implying stain stiffening of the cells (15). Lines that connect points of equal pressure are aligned more vertically but are also shifted toward larger entry times as the pressure increases, implying stress stiffening of the cells (15).
Figure 2.
Stress and strain stiffening: Scatter plot of tentry versus εmax/Δp for 19,991 K562 cells as in Fig. 1c. (Black squares connected by red lines) Median values of tentry versus εmax/Δp for different pressure and strain bins with at least 250 cells in each bin, with a strain bin width of 0.03, a pressure bin width of 50 Pa, and median pressure and strain values as indicated by the numbers next to the grid lines. (Black grid) Fit of Eq. 1 to the binned data, with stress stiffening according to E = E0 + c1 Δp. Fit parameters are E0 = 58 Pa, c1 = 1.11 Pa−1, and β = 0.07. To see this figure in color, go online.
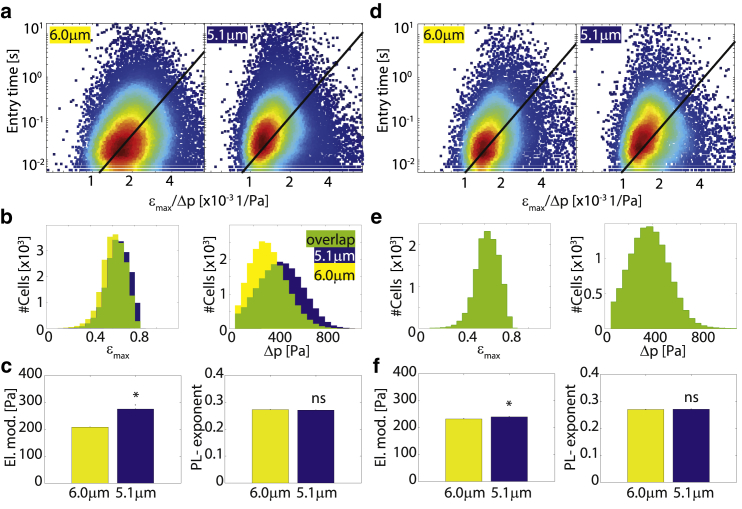
Figure 3.
Histogram matching: (a) Scatter plot tentry versus εmax/Δp for two K562 leukemia cell populations measured with wide (6 μm) and narrower (5.1 μm) constrictions. (Black lines) Fit of Eq. 1 to the data. (b) Strain histograms (left) and stress histograms (right) are shifted to higher values for cells in narrow constrictions (blue) compared to cells in wider constrictions (yellow). Areas where histograms overlap are marked (green). (c) Without histogram matching, cells measured in the narrower constrictions (blue) appear significantly (p < 0.05) stiffer (left) but have a similar fluidity (right) compared to cells measured in the wider constrictions (yellow). (d) Exemplary scatter plots and fit lines of the same measurements as (a) after Δp and εmax histogram matching. (e) Exemplary strain histograms (left) and pressure histograms (right) after histogram matching show perfect overlap for both cell populations. (f) After histogram matching, cells measured in the narrow (blue) and wider (yellow) constrictions both have similar elastic modulus (left) and fluidity (right) values. To see this figure in color, go online.
To a good approximation, the cell elastic modulus E in Eq. 1 increases linearly with increasing stress Δp and strain εmax according to E = E0 + c1Δp + c2εmax. With the additional fit parameters c1 and c2, we can nearly fully account for the scatter of the binned and averaged data points shown in Fig. 2. The fitted stress-stiffening coefficient c1 is on the order of unity, indicating that for a pressure increase of 1 Pa, the cell elastic modulus also increases by 1 Pa. The fitted strain-stiffening coefficient c2 is ∼200 Pa, indicating that for a strain increase of 100%, the cell elastic modulus increases by 200 Pa.
There is a considerable covariance between the fit parameters c1 and c2, however, and the gridlike systematic scatter of the binned data points can be accounted for by considering only stress-stiffening according to E = E0 + c1Δp (Fig. 2, black lines), or only strain stiffening according to E = E0 + c2εmax (data not shown). We therefore opted to fit Eq. 1 to the data with only two constant fit parameters, E and β. These parameters must be interpreted as an effective cell elastic modulus and fluidity that are valid only in the range of pressure and strain values of that particular measurement. To prevent stress or strain stiffening from introducing bias, e.g., when comparing cell populations with different cell sizes or when comparing two measurements obtained with different constriction sizes, it is crucial to control for the strain and stress that the cells have experienced. This can be achieved with a histogram matching method.
Histogram matching
This method selects only those cells from two or more different cell populations for comparison that have experienced both the same range of pressure drop Δp and the same maximum strain εmax. In brief, we compute two-dimensional histograms of distributions for εmax with a bin width of 0.05, and for Δp with a bin width of 50 Pa, similar to one grid tile in Fig. 2. For every εmax/Δp bin, we then randomly exclude cells from the population with the larger number of cells in that particular bin until the number of cells in each histogram-bin match. This ensures that also the histograms of the ratios of εmax/Δp for each of the cell populations match approximately. The random cell exclusion process is repeated for every bootstrap repetition (i.e., 100 times) to ensure that no data point within the overlap region of the histogram is thrown away.
We demonstrate the validity and importance of the histogram matching method by comparing two measurements conducted with two constrictions of differing sizes (6.0 × 13.6 μm vs. 5.1 ± 16.6 μm) but on the same cell type (K562 leukemia cells). From the scatter plots of log(tentry) versus log(εmax/Δp) data (Fig. 3 a) and from the histograms of measured εmax and Δp (Fig. 3 b, left and right), systematic differences between the two measurements are noticeable: cells measured with 5.1 μm constrictions experience a higher maximum deformation than cells measured with 6.0 μm constrictions (Fig. 3 b, left). Cells measured with the smaller constrictions also experience a higher pressure drop Δp, as the experimenter tends to increase the pressure across the narrower device to compensate for the greatly increased entry time (Fig. 3 b, right). The combined effect of an increased strain and pressure cause the elastic modulus to increase due to strain and stress stiffening, whereas the power-law exponent remains similar (Fig. 3 c, left and right). After Δp and εmax histogram matching (exemplary bootstrap fit shown in Fig. 3 d and e), the cell elastic moduli for the two measurement conditions match nearly perfectly (Fig. 3 f). The remaining mismatch of the elastic modulus of 3% (7 Pa) is within the systematic margin of error caused by measurement inaccuracies of the channel geometries (±0.25 μm) between the different devices.
Influence of culture conditions and measurement parameters
We next test to what degree cell mechanical properties are further influenced by measurement parameters such as cell confluency before harvesting, time between cell trypsinization and measurements, choice of cell suspension medium, or device preparation with pluronic. All measurements for a given parameter or measurement condition are conducted on the same day with DLD-1 colon carcinoma cells harvested from identically prepared flasks. Between two and three independent measurements (i.e., with cells from two to three different flasks), each with at least 5000 cells, are performed for each condition. The measurement data for each condition are pooled and histogram matched for comparison. Note, however, that absolute cell elastic modulus values between different types of parameters cannot be compared, as slightly different deformation and pressure histograms have been chosen for different parameter types to allow for an optimal histogram overlap within the parameter group.
Reproducibility
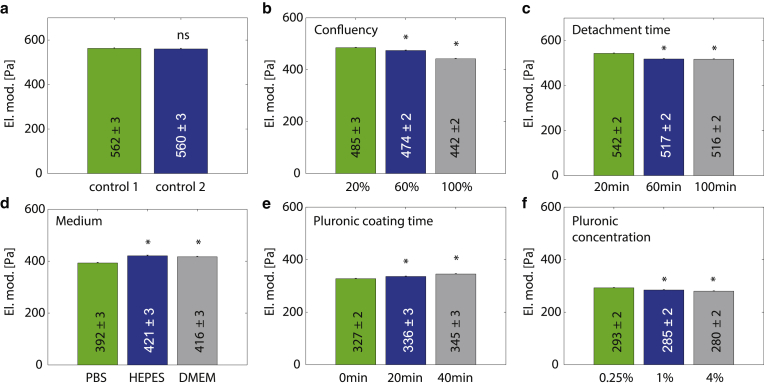
To investigate the measurement reproducibility of our system, we conduct four independent measurements of DLD-1 cells on one day under identical conditions. We then randomly pool two of the measurements into one group (control 1) and compare the resulting mechanical properties with the pooled data from the remaining two measurements (control 2). We find only negligible differences between the elastic modulus (Figs. 4 a and S5 a) or the power-law exponent (Fig. S5 b) between the two groups, demonstrating the reproducibility of the method.
Figure 4.
Influence of culture and measurement conditions on cell mechanical properties of DLD-1 colon carcinoma cells: (a) Elastic moduli of cells from four independent measurements, arbitrarily split into two groups (control 1 and control 2). (b) Cell elastic moduli measured for cell populations with different culture confluency (20, 60, and 100%) before harvesting. (c) Cell elastic moduli in response to different detachment times before measurement (20, 60, and 100 min). (d) Cell elastic moduli measured for three different measurement media: PBS, HEPES-buffer, and DMEM cell culture medium. (e) Cell elastic moduli measured after coating the devices with 1 wt % pluronic for 0, 20, and 40 min. (f) Cell elastic moduli measured with devices coated for 30 min with pluronic concentrations of 0.25, 1, and 4 wt %. n > 1200 for each condition. εmax and Δp histogram matching was performed separately for each (a)–(f). Numbers show mean ± SE. Significant (p < 0.05) differences to first parameter values (green bars) are indicated by asterisks. To see this figure in color, go online.
Confluency
Cells are split 24 h before measurements and are seeded at different densities such that they reach different confluencies (20, 60, and 100%) before harvesting. We find a significant decrease in cell elastic modulus between 20 and 60% confluency, and between 60 and 100% confluency (Figs. 4 b and S5 c). The power-law exponent β also shows a slight decrease from 20 to 100% confluency (Fig. S5 d). Based on this result, we recommend that cell confluency should be tightly controlled in all experiments.
Time between cell trypsinization and measurements
Typically, measurements start ∼15–20 min after harvesting the cells by trypsinization. This time span includes 4–5 min trypsinization, a 4-min centrifugation, a 5-min transport to the microscope and setup, and a 5-min flushing time in which the cells travel through the tubes to the microconstrictions. Measurements typically last for another 30 min. Here, we find that an increase in the waiting time from 20 to 60 min between harvesting and start of the measurement (for which cells are stored in a Falcon tube (Thermo Fisher Scientific) in the incubator) results in a significant decrease of the cell elastic modulus and a slight but significant increase of the power-law exponent (Figs. 4 c and S5, e and f). There is no further decrease in the cell elastic modulus for a waiting time of 100 min. Based on this result, we recommend that measurements should be terminated 1 h after trypsinization at the latest.
Cell suspension medium
After trypsinization and centrifugation, cells are usually resuspended in PBS (pH = 7.25) and kept therein for the time of the measurement. Resuspending cells in HEPES-buffer (50 mM HEPES, 10 mM NaCl, 10 mM glucose, pH = 7.0) instead of PBS significantly increases the cell elastic modulus and decreases the power-law exponent (Figs. 4 d and S5, g and h). Resuspending cells in DMEM (1 g glucose, pH = 8.13) significantly increases the cell elastic modulus, but not the power-law exponent, compared to PBS. It is unclear whether differences in the pH or in the composition of the cell suspension medium have caused these changes. Phenol-Red-containing DMEM is a reasonable choice for a cell suspension medium as it is also the medium in which the cells are cultured. However, PBS improves the image quality and reduces cell clumping compared to DMEM, which justifies its use in our study.
Device coating with pluronic
To prevent the adhesion of cells onto the walls of the microfluidic device, we flush the devices with a 1 wt % pluronic surfactant solution in PBS for 30 min before measurements. Here, we compare noncoated devices with devices coated for 20 or 40 min with 1 wt % pluronic. We find a significant increase in cell elastic modulus for the devices coated for 20 min and for 40 min (Figs. 4 e and S5 i) compared to the noncoated devices. We also observe that a large portion of cells become stuck in the inlet filter and in the constrictions of the uncoated microfluidic devices. Thus, nonspecific adhesion and increased friction between the cells and the walls of the uncoated microfluidic device may have caused a preferential retention of stiffer cells. The power-law exponent β does also show a systematic increase between the three pluronic coating times (Fig. S5 j).
We also investigate the influence of pluronic coating concentration on cell mechanical properties. We compare the usual coating concentration of 1 wt % with a 0.25 wt % and a 4 wt % concentration of pluronic in PBS, each for a 30 min coating time before measurements. We find a significantly higher cell elastic modulus for the 0.25 wt % coating concentration compared to 1 and 4 wt % coating concentration (Figs. 4 f and S5 k), but no changes in the number of cells that become stuck in the inlet filter. Nonetheless, we expect that friction and nonspecific adhesion between the cells and the walls of the device including the inlet filter increases at low pluronic concentrations, and thus we speculate that this may have caused a small but detectable mechanical activation and stiffening of the cells (27). The power-law exponent β shows small and nonsystematic but statistically significant changes for the 1 and 4 wt % compared to the 0.25 wt % coating concentration (Fig. S5 l). Thus, pluronic concentration and coating time both need to be tightly controlled for all measurements.
In summary, although the cell mechanical responses to altered culture and measurement conditions are in most cases relatively small and physiologically irrelevant when considered in isolation, the combined effect of poorly controlled conditions can lead to statistically significant bias.
Influence of protein expression levels
We have demonstrated above that histogram matching effectively compensates for systematic changes in cell mechanical properties that are caused by pressure and strain effects. In the following, we extend the histogram matching approach to evaluate systematic changes in cell mechanical properties that are caused by different protein expression levels. This is relevant foremost in the case of transiently transfected cells where only a part of the cell population, typically between 10 and 90%, is likely to be transfected (28), and where also the amount of expressed proteins varies greatly between transfected cells. We have extended our setup with a second high-speed CCD camera that acquires fluorescence images. This camera is synchronized with the CCD camera for the bright-field images, and thus we can measure for each cell not only the entry into the constriction but also the expression level of a fluorescently tagged protein. This allows us to exclude nontransfected cells from the analysis, or to correlate the GFP-signal intensity with altered mechanical properties to measure a dose-response curve.
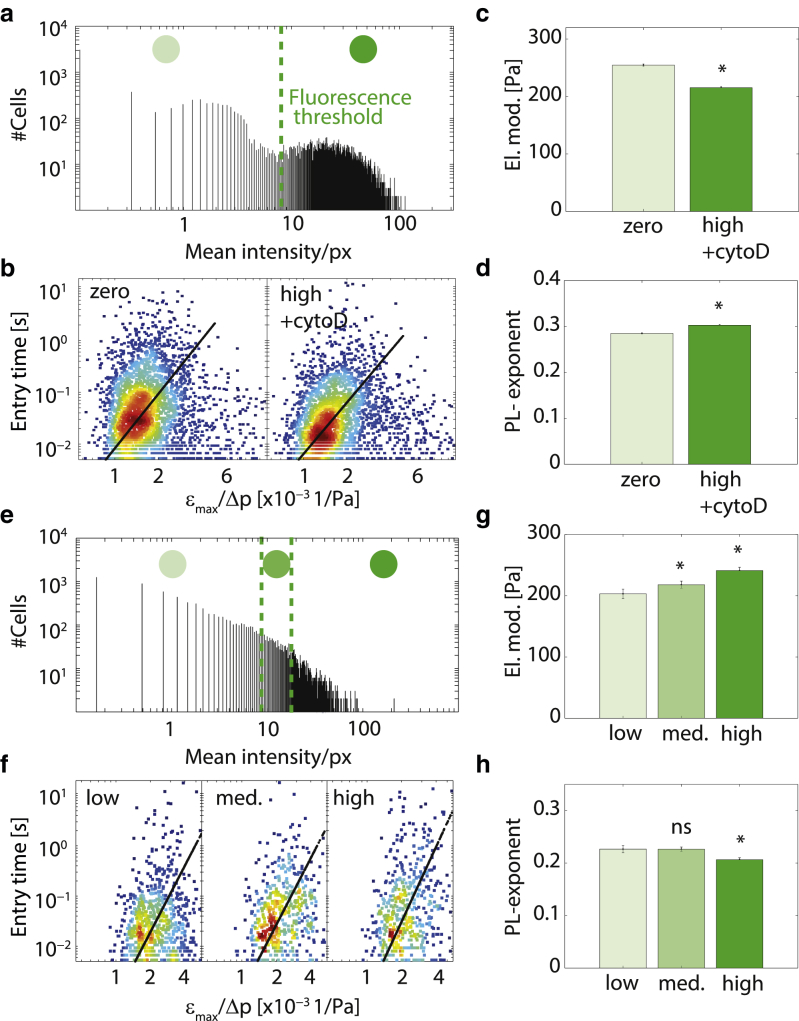
As a proof of principle, we measure a mixed population of K562 leukemia cells. Half of the cells are treated for 30 min with the actin-depolymerizing drug cytochalasin D (10 μM) together with the green fluorescent dye calcein (250 nM). These labeled cells are then mixed with unlabeled, untreated cells and are measured in our microconstriction setup. For the analysis, we split the data into a fluorescent group (>8 count/cell pixel, median = 28 counts/cell pixel) and a nonfluorescent group (≤8 count/cell pixel, median = 0.3 counts/cell pixel), and perform Δp and εmax histogram matching between the two groups (Fig. 5, a and b). As expected, we find a significantly (p < 0.05) decreased cell elastic modulus (Fig. 5 c) and an increased power-law exponent (Fig. 5 d) in the green-fluorescent, cytochalasin D-treated group, which is in accordance with Otto et al. (8) and Bursac et al. (21).
Figure 5.
(a–d) Measurement of a mixed K562 leukemia cell population, containing 50% untreated cells and 50% cells treated with cytoD (10 μM, 30 min) and stained with calcein (250 nM, 30 min). (a) Histogram of average fluorescence intensities per cell pixel. (Dashed green line) Fluorescence threshold for separating cells into a zero and a high fluorescence intensity group. (b) Exemplary histogram-matched scatter plots of tentry versus εmax/Δp for cells with zero fluorescence (nontreated) and high fluorescence (cytoD and calcein-treated) after histogram matching. (Black lines) Fit of Eq. 1 to the data. (c) Cell elastic modulus and (d) cell fluidity for the two groups. (e–h) Measurement of lamin A-GFP-transfected K562 leukemia cells. (e) Histogram of average fluorescence intensities per cell pixel. (Dashed green lines) Fluorescence threshold for separating cells into low, medium, and high fluorescence intensity groups. (f) Exemplary histogram-matched scatter plots of tentry versus εmax/Δp for cells with low, medium, and high expression levels of lamin A-GFP. (Black lines) Fit of Eq. 1 to the data. (g) Cell elastic moduli and (h) cell fluidity for the three groups. Significant (p < 0.05) differences compared to the cell group with zero (a–d) and low (e–h) intensity are indicated by asterisks. To see this figure in color, go online.
We next measure the mechanical properties of K562 cells transiently transfected with a lamin A-GFP protein. Lamin A-overexpression has been shown to increase cell elastic modulus (15, 29), which protects the cell nucleus from damage during compression at high deformations (30, 31, 32). After transfection, individual cells show the typical wide distribution of GFP- and thereby lamin A-expression levels (Fig. 5 e). To measure a dose-response curve, we split the data into three subgroups corresponding to low, medium, and high levels of GFP (bins as indicated in Fig. 5 e). The median fluorescence cell pixel intensities for the three groups are 1.2, 11.2, and 25.5 counts/pixel, respectively. We determine E and β separately for the three groups with Δp and εmax histogram matching (Fig. 5 f). With increasing GFP levels, we find a significant (p < 0.05) increase of the cell elastic modulus (Fig. 5 g) and a decrease of the power-law exponent (Fig. 5 h). These data are consistent with a dose-dependent nuclear stiffening for increasing expression levels of lamin A.
Conclusions
In this report, we analyze the influence of stress and strain stiffening on cell mechanical properties measured with a microfluidic microconstriction device. Stress and strain stiffening is a major confounding factor when comparing cell populations that have different mean cell sizes, when comparing measurements from devices with constrictions of different sizes, or when comparing measurements conducted with different mean working pressures. To correct for stress and strain stiffening effects, we introduce the method of stress and strain histogram matching and demonstrate that it greatly reduces the variability between different measurements and ensures high reproducibility. Moreover, we also demonstrate that seemingly minor cell culture details or measurement parameters can confound the resulting data and therefore need to be tightly controlled, such as cell confluency before harvesting, time between harvesting and measurements, choice of cell suspension medium, and details of the device preparation with pluronic. Lastly, we introduce a fluorescence extension to our setup that allows us to measure transiently and incompletely transfected cell populations by excluding untransfected cells from the analysis, or by correlating protein expression levels with cell mechanical properties. We show that we can thereby measure dose-response relationships between the expression of proteins and mechanical cell properties.
Author Contributions
J.R.L., B.F., W.S., and G.W. designed the setup and experiments; J.R.L., S.R., and G.W. developed the data acquisition and analysis software; J.R.L. and M.S. performed the measurements; J.R.L., C.M., and B.F. analyzed the data; T.K. generated the transfected lamin A-GFP cell line; and J.R.L. and B.F. wrote the article.
Acknowledgments
We thank Amy Rowat, Katarina Wolf, Jochen Guck, Yoav Green, and James Butler for helpful discussions, Ingo Thievessen for help with cell culture, and Irina Harder, Bernhard Hensel, Reiner Stadter, and Isabel Gäßner for technical support.
This work was supported by grants from the German Science Foundation (DFG), the Research Training Group 1962 “Dynamic Interactions at Biological Membranes: From Single Molecules to Tissue”, and the Emerging Fields Initiative of the University of Erlangen-Nuremberg.
Editor: Jeffrey Fredberg.
Footnotes
Five figures and one movie are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)30231-X.
Supporting Material
A suspended cell with a diameter larger than the constriction width must deform before passage. This includes both cell narrowing and elongation. After transit, the cell remains deformed for some time and only slowly recovers to its original round shape.
References
- 1.Suresh S., Spatz J., Seufferlein T. Connections between single-cell biomechanics and human disease states: gastrointestinal cancer and malaria. Acta Biomater. 2005;1:15–30. doi: 10.1016/j.actbio.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 2.Mauritz J.M.A., Esposito A., Kaminski C.F. Biophotonic techniques for the study of malaria-infected red blood cells. Med. Biol. Eng. Comput. 2010;48:1055–1063. doi: 10.1007/s11517-010-0668-0. [DOI] [PubMed] [Google Scholar]
- 3.Fabry B., Maksym G.N., Fredberg J.J. Selected contribution: time course and heterogeneity of contractile responses in cultured human airway smooth muscle cells. J. Appl. Physiol. 2001;91:986–994. doi: 10.1152/jappl.2001.91.2.986. [DOI] [PubMed] [Google Scholar]
- 4.Rosenbluth M.J., Lam W.A., Fletcher D.A. Analyzing cell mechanics in hematologic diseases with microfluidic biophysical flow cytometry. Lab Chip. 2008;8:1062–1070. doi: 10.1039/b802931h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guck J., Schinkinger S., Bilby C. Optical deformability as an inherent cell marker for testing malignant transformation and metastatic competence. Biophys. J. 2005;88:3689–3698. doi: 10.1529/biophysj.104.045476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guck J., Ananthakrishnan R., Käs J. The optical stretcher: a novel laser tool to micromanipulate cells. Biophys. J. 2001;81:767–784. doi: 10.1016/S0006-3495(01)75740-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gossett D.R., Tse H.T.K., Di Carlo D. Hydrodynamic stretching of single cells for large population mechanical phenotyping. Proc. Natl. Acad. Sci. USA. 2012;109:7630–7635. doi: 10.1073/pnas.1200107109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Otto O., Rosendahl P., Guck J. Real-time deformability cytometry: on-the-fly cell mechanical phenotyping. Nat. Methods. 2015;12:199–202. doi: 10.1038/nmeth.3281. [DOI] [PubMed] [Google Scholar]
- 9.Qi D., Kaur Gill N., Rowat A.C. Screening cell mechanotype by parallel microfiltration. Sci. Rep. 2015;5:17595. doi: 10.1038/srep17595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khan Z.S., Vanapalli S.A. Probing the mechanical properties of brain cancer cells using a microfluidic cell squeezer device. Biomicrofluidics. 2013;7:11806. doi: 10.1063/1.4774310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Byun S., Son S., Manalis S.R. Characterizing deformability and surface friction of cancer cells. Proc. Natl. Acad. Sci. USA. 2013;110:7580–7585. doi: 10.1073/pnas.1218806110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nyberg K.D., Scott M.B., Rowat A.C. The physical origins of transit time measurements for rapid, single cell mechanotyping. Lab Chip. 2016;16:3330–3339. doi: 10.1039/c6lc00169f. [DOI] [PubMed] [Google Scholar]
- 13.Cai P., Mizutani Y., Okajima T. Quantifying cell-to-cell variation in power-law rheology. Biophys. J. 2013;105:1093–1102. doi: 10.1016/j.bpj.2013.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dai J., Sheetz M.P. Mechanical properties of neuronal growth cone membranes studied by tether formation with laser optical tweezers. Biophys. J. 1995;68:988–996. doi: 10.1016/S0006-3495(95)80274-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lange J.R., Steinwachs J., Fabry B. Microconstriction arrays for high-throughput quantitative measurements of cell mechanical properties. Biophys. J. 2015;109:26–34. doi: 10.1016/j.bpj.2015.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fabry B., Maksym G.N., Fredberg J.J. Scaling the microrheology of living cells. Phys. Rev. Lett. 2001;87:148102. doi: 10.1103/PhysRevLett.87.148102. [DOI] [PubMed] [Google Scholar]
- 17.Kollmannsberger P., Fabry B. Linear and nonlinear rheology of living cells. Annu. Rev. Mater. Res. 2011;41:75–97. [Google Scholar]
- 18.Levental I., Georges P.C., Janmey P.A. Soft biological materials and their impact on cell function. Soft Matter. 2007;3:299–306. doi: 10.1039/b610522j. [DOI] [PubMed] [Google Scholar]
- 19.Gardel M.L., Nakamura F., Weitz D.A. Stress-dependent elasticity of composite actin networks as a model for cell behavior. Phys. Rev. Lett. 2006;96:088102. doi: 10.1103/PhysRevLett.96.088102. [DOI] [PubMed] [Google Scholar]
- 20.Wang N., Butler J.P., Ingber D.E. Mechanotransduction across the cell surface and through the cytoskeleton. Science. 1993;260:1124–1127. doi: 10.1126/science.7684161. [DOI] [PubMed] [Google Scholar]
- 21.Bursac P., Fabry B., An S.S. Cytoskeleton dynamics: fluctuations within the network. Biochem. Biophys. Res. Commun. 2007;355:324–330. doi: 10.1016/j.bbrc.2007.01.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Madou M.J. CRC Press; Boca Raton, FL: 2002. Fundamentals of Soft Lithography: The Science of Miniaturization. [Google Scholar]
- 23.Sharma S., Johnson R.W., Desai T.A. Evaluation of the stability of nonfouling ultrathin poly(ethylene glycol) films for silicon-based microdevices. Langmuir. 2004;20:348–356. doi: 10.1021/la034753l. [DOI] [PubMed] [Google Scholar]
- 24.Krishnan S., Weinman J., Ober C.K. Advances in polymers for anti-biofouling surfaces. J. Mater. Chem. 2008;18:3405–3413. [Google Scholar]
- 25.Seltmann K., Fritsch A.W., Magin T.M. Keratins significantly contribute to cell stiffness and impact invasive behavior. Proc. Natl. Acad. Sci. USA. 2013;110:18507–18512. doi: 10.1073/pnas.1310493110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tozluoğlu M., Tournier A.L., Sahai E. Matrix geometry determines optimal cancer cell migration strategy and modulates response to interventions. Nat. Cell Biol. 2013;15:751–762. doi: 10.1038/ncb2775. [DOI] [PubMed] [Google Scholar]
- 27.Roca-Cusachs P., Almendros I., Navajas D. Rheology of passive and adhesion-activated neutrophils probed by atomic force microscopy. Biophys. J. 2006;91:3508–3518. doi: 10.1529/biophysj.106.088831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fischer D., Bieber T., Kissel T. A novel non-viral vector for DNA delivery based on low molecular weight, branched polyethylenimine: effect of molecular weight on transfection efficiency and cytotoxicity. Pharm. Res. 1999;16:1273–1279. doi: 10.1023/a:1014861900478. [DOI] [PubMed] [Google Scholar]
- 29.Swift J., Ivanovska I.L., Discher D.E. Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science. 2013;341:1240104. doi: 10.1126/science.1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thiam H.-R., Vargas P., Piel M. Perinuclear Arp2/3-driven actin polymerization enables nuclear deformation to facilitate cell migration through complex environments. Nat. Commun. 2016;7:10997. doi: 10.1038/ncomms10997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Denais C.M., Gilbert R.M., Lammerding J. Nuclear envelope rupture and repair during cancer cell migration. Science. 2016;352:353–358. doi: 10.1126/science.aad7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McGregor A.L., Hsia C.R., Lammerding J. Squish and squeeze-the nucleus as a physical barrier during migration in confined environments. Curr. Opin. Cell Biol. 2016;40:32–40. doi: 10.1016/j.ceb.2016.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A suspended cell with a diameter larger than the constriction width must deform before passage. This includes both cell narrowing and elongation. After transit, the cell remains deformed for some time and only slowly recovers to its original round shape.