Abstract
Vibrational spectroscopy is increasingly used for the rapid and non-destructive imaging of environmental and medical samples. Both Raman and Fourier-transform infrared (FT-IR) imaging have been applied to obtain detailed information on the chemical composition of biological materials, ranging from single microbial cells to tissues. Due to its compatibility with methods such as stable isotope labeling for the monitoring of cellular activities, vibrational spectroscopy also holds considerable power as a tool in microbial ecology. Chemical imaging of undisturbed biological systems (such as live cells in their native habitats) presents unique challenges due to the physical and chemical complexity of the samples, potential for spectral interference, and frequent need for real-time measurements. This Mini Review provides a critical synthesis of recent applications of Raman and FT-IR spectroscopy for characterizing complex biological samples, with a focus on developments in single-cell imaging. We also discuss how new spectroscopic methods could be used to overcome current limitations of single-cell analyses. Given the inherent complementarity of Raman and FT-IR spectroscopic methods, we discuss how combining these approaches could enable us to obtain new insights into biological activities either in situ or under conditions that simulate selected properties of the natural environment.
Keywords: imaging, isotope labeling, single-cell analysis, vibrational spectroscopy
Introduction
Natural habitats are often physically and chemically complex, which has far-reaching consequences for the spatial distribution of microbial taxa and the processes they mediate (Resat et al., 2012; Vos et al., 2013; Pande et al., 2016; Ratzke and Gore, 2016). Because controlled laboratory experiments rarely capture the heterogeneity present within natural environments, our knowledge of microbial activities is often based on indirect observation. To address this source of uncertainty, there is a need for methods that facilitate the in situ profiling of microorganisms and their activities in complex environments. A full understanding of these topics also requires an ability to study these processes at the level of single cells (Fike et al., 2008; Resat et al., 2012; Roose et al., 2016). Due to its ability to rapidly and non-destructively probe the physiology and activities of microorganisms, vibrational (Raman and FT-IR) microspectroscopy (a combination of microscopy and spectroscopy) shows considerable promise in this respect (Escoriza et al., 2006; Wagner, 2009; Lu et al., 2011). In particular, Raman and infrared imaging have emerged as useful methods for the spatially resolved analysis of biological samples. In this Mini Review, we highlight recent studies that have used these techniques to image single microbial cells within spatially and chemically complex environments. These include pure cultures incubated in contact with physical substrata, multi-species assemblages within their native habitats, as well as other challenging sample types. State-of-the-art approaches for spectral imaging are critically evaluated in order to identify guidelines for future applications of single-cell analyses in microbial ecology.
Raman Imaging
While several types of Raman spectroscopic instrumentation and analytical approaches have been developed, each of these relies on measuring the scattering of monochromatic light as it interacts with a sample. Most photons are elastically scattered and possess the same energy as the incident light beam (also termed Rayleigh scattering). However, a small fraction is inelastically scattered, involving a decrease or an increase in energy compared with the excitation wavelength (Stokes and anti-Stokes Raman scattering, respectively). By providing information on vibrational and other low-frequency transitions in a molecule, both types of inelastically scattered light can be used to determine and differentiate between the chemical composition of solids, liquids and gases. For a more detailed introduction to this technique (as well as infrared spectroscopy), the reader is referred to Skoog et al. (2007) and Lu et al. (2011). Recent advances in the design of high-speed Raman imaging instrumentation have been summarized by Ando et al. (2016). Moreover, developments concerning techniques including surface- and tip-enhanced Raman scattering (SERS and TERS), as well as resonance Raman and coherent anti-Stokes Raman spectroscopy (CARS), are discussed in several reviews (Opilik et al., 2013; Camp and Cicerone, 2015; Cicerone, 2016; Kano et al., 2016).
Two features that make Raman microspectroscopy an ideal technique for single-cell analyses include its direct compatibility with aqueous samples (due to water exhibiting only weak Raman scattering) and its high spatial resolution (Skoog et al., 2007). While a resolution of ∼1 μm is possible using conventional Raman instrumentation, measurements at the nanometer scale are achievable by TERS (Mariani et al., 2010; Opilik et al., 2013; Rusciano et al., 2014). Raman measurements are also well-suited for analyzing motile cells using optical tweezers, as well as monitoring microbial activities by stable isotope probing (SIP) (Chan et al., 2004; Wagner, 2009; Huang et al., 2010; Berry et al., 2015; Wang et al., 2016). Although the real-time Raman imaging of microorganisms remains non-trivial due to issues including background autofluorescence (Polisetti et al., 2016) and weak signal intensities (partly due to a need for low laser excitation power to avoid photodamage), significant progress in this field has already been made. For example, Li et al. (2012) used resonance Raman imaging combined with 13C labeling to identify cells that fixed carbon dioxide in culture and in field-collected seawater samples. By reducing spectral acquisition times to milliseconds, resonance Raman spectroscopy – a method in which the excitation wavelength matches the electronic transition of a selected molecule – was key to enabling the rapid imaging of these samples. It is also possible to visualize selected strains and their locations within habitats including human endothelial cells (Große et al., 2015), macrophages (Silge et al., 2015) and other environments, even when the taxa of interest are present at low abundances (Kalasinsky et al., 2007). Through combining imaging of Staphylococcus aureus cells with a multivariate classification model [based on principal component analysis (PCA) and linear discriminant analysis (LDA)], Große et al. (2015) were further able to detect small differences in the spectral profiles that allowed the authors to discern between intra- and extracellular cells, due to shifts in the physiological state of the bacteria that occur upon host invasion.
In addition, resonance Raman and SERS have been used to directly image rhizosphere bacteria (Pantoea sp. YR343) on Arabidopsis thaliana root surfaces (Polisetti et al., 2016). This is of interest because Raman-based investigations of plant–microbial interactions are often challenging or impossible due to the strong autofluorescence originating from plant materials. In the study by Polisetti et al. (2016), background interference from the roots was reduced by aging them for 5–15 days. Similar to Große et al. (2015), PCA was used to discriminate bacterial spectra from spectra of other materials. Moreover, using SERS allowed the authors to circumvent the need for a photo-bleaching step which is often employed for the analysis of pigmented cells using conventional Raman instrumentation, but which can result in the degradation of cell components and metabolites that are of importance to understanding bacterially mediated processes in the rhizosphere (Polisetti et al., 2016). Taken together, the studies highlighted above illustrate how advanced Raman imaging techniques and multivariate analyses can be used to generate new insights into the distribution and activities of microorganisms within diverse environments, including systems which have previously been difficult to visualize and where the ability to differentiate between cells and other materials is dependent on detecting minor differences in spectral features. By removing the need for sample treatment steps that are likely to introduce analytical biases, such as sample photo-bleaching prior to the collection of Raman spectra (Polisetti et al., 2016), these techniques can also provide increasingly accurate information on metabolic processes occurring at multiple levels of biological organization (from individual cells to communities).
While a limited number of studies have been published on Raman imaging of microbial strains or uncultured cells within their native environments, new instrumentation is likely to lead to an expansion of this field by enabling reduced spectral acquisition times without a loss of signal intensity (Opilik et al., 2013; Ando et al., 2016; Kano et al., 2016). In addition, combining this approach with well-established methods in microbial ecology (including fluorescence in situ hybridization) (Wang et al., 2016) as well as newer techniques such as bioorthogonal chemical imaging (Berry et al., 2015; Wei et al., 2016) and Raman microfluidics (Chrimes et al., 2013) are likely to find increasing use in the analysis of microbiological samples. Several sample types which have not yet been subjected to Raman imaging have already been characterized using single-point measurements, and therefore represent promising targets for future research. For example, while the Raman-based detection of meningitis-causing pathogens in human cerebrospinal fluid has been achieved (Harz et al., 2009), spatially resolved imaging of such samples could facilitate the development of improved diagnostic tests. One imaging modality that is particularly promising from a microbiological perspective, but which is yet to find widespread use in the field of microbial ecology, is CARS (Krafft et al., 2009; Camp and Cicerone, 2015; Cicerone, 2016). This technique can enable the acquisition of Raman spectra at a rate that is approximately 100 times faster than conventional Raman analyses, making it highly suitable for the real-time imaging of biological samples (Cicerone, 2016). CARS has already been used for the rapid profiling of microorganisms at the subcellular level (Okuno et al., 2010; Yue and Cheng, 2016), and a single study has also employed it to image bacteria within complex matrices including milk and urine (Hong et al., 2016). Another technique which has found surprisingly limited use in the field of microbial ecology is TERS (Mariani et al., 2010; Opilik et al., 2013; Rusciano et al., 2014). However, since this method enables Raman measurements at sub-micron spatial scales, it could be used to analyze microorganisms that are under the conventional size detection limit of ∼1 μm, as well as viral particles present within diverse environmental matrices. Indeed, TERS has already been used for the analysis and classification of viral strains (Hermann et al., 2011; Olschewski et al., 2015).
FT-IR Imaging
While Raman spectroscopy relies on irradiating a sample with a monochromatic laser beam, Fourier-transform infrared (FT-IR) spectroscopy is based on measuring the absorption of polychromatic infrared light. The functional groups in a given molecule are identified according to their vibrational modes at different IR frequencies (for detailed information, see Skoog et al., 2007). Raman analyses depend on a shift in the polarizability of a molecule, whereas FT-IR measurements depend on changes in the dipole moment. Indeed, Raman-active vibrational modes often exhibit weak IR signals and vice versa (with symmetric and asymmetric moieties producing strong Raman and IR spectral bands, respectively), and the two methods provide complementary information on the molecular composition of microbial cells (Lu et al., 2011; Ojeda and Dittrich, 2012; Tang et al., 2013; Wang et al., 2016). Infrared imaging could, therefore, provide insights into microbial physiology in samples that are difficult to analyze using Raman spectroscopy alone. Indeed, high-speed imaging of large (centimeter-scale) sample areas can be achieved using a focal plane array (FPA) detector that enables the simultaneous acquisition of tens of thousands of IR spectra (Dorling and Baker, 2013). Studies employing FPA-based FT-IR analysis are common in biomedical science and have, for example, involved chemical imaging of tissues (Kastyak-Ibrahim et al., 2012; Miller et al., 2013) and cancer cells (Kuimova et al., 2009). Chemical mapping by reflectance FT-IR microspectroscopy has also been used to characterize bacteria on opaque steel surfaces, without a need for destructive sampling (Ojeda et al., 2009). In comparison with Raman analyses, however, few studies have used FT-IR microspectroscopy to investigate single microbial cells within their native environments, potentially due to the coarse spatial resolution (∼10 μm) of conventional FT-IR measurements and water being a strong absorber of IR radiation. Even so, several ways to overcome these challenges have been developed. For example, synchrotron radiation sources have enabled FT-IR measurements at the micron scale (Nasse et al., 2011; Jamme et al., 2013; Saulou et al., 2013) and combining this approach with microfluidics can reduce background interference from water by making it possible to culture cells within a thin layer of fluid (Holman et al., 2009; Loutherback et al., 2015, 2016; Birarda et al., 2016).
While synchrotron-FT-IR analyses require dedicated facilities, advances in the development of high-magnification optics have made it possible to perform FPA-based infrared imaging at a spatial resolution comparable with Raman instruments, even without access to a synchrotron beamline (Findlay et al., 2015). Analyses of cells in aqueous suspensions are additionally possible using attenuated total reflectance (ATR)-FT-IR imaging (Kuimova et al., 2009). Where required, techniques for nano-scale infrared imaging have been developed (Reddy et al., 2013; Centrone, 2015; Amenabar et al., 2017) and even relatively thick aqueous samples can be analyzed by quantum cascade laser-based IR microspectroscopy (Haase et al., 2016). Crucially for the in situ analysis of microbial activities, there is evidence that FT-IR spectroscopy is compatible with SIP and can be used to track the cellular uptake of stable-isotope-labeled carbon (13C) and nitrogen (15N) compounds (Muhamadali et al., 2015). FT-IR microspectropy can detect differences in the spectra of water and heavy water (D2O), due to absorbance peaks corresponding to O–H and O–D bending modes occurring at different wavenumber regions (Miller et al., 2013). While we are unaware of studies that have combined D2O labeling with FT-IR spectroscopy to monitor the activities of individual microbial cells, this has recently been achieved using Raman spectroscopy (Berry et al., 2015), and it is likely that both methods can be used to identify actively metabolizing cells within their native habitats.
Further to the studies discussed above, Muhamadali et al. (2016) evaluated the applicability of three vibrational spectroscopy techniques (FT-IR, conventional Raman and SERS) for differentiating between several clinically relevant taxa including Escherichia coli, Pseudomonas spp., Bacillus spp. and Enterococcus faecium. Of these techniques, infrared spectroscopy was found to provide the most consistent results for the entire sample set (in terms of spectral quality and reproducibility), which led the authors to suggest that FT-IR analyses could be particularly useful for characterizing mixed cultures (also see Wenning et al., 2005). Indeed, FT-IR microspectroscopy has already been used to quantify compare the abundances of bacteria and archaea within subsurface aquifer samples, based on domain-specific CH3:CH2 absorbance ratios (Igisu et al., 2012). In comparison with Raman spectroscopy, there is evidence to suggest that FT-IR analyses can additionally give a higher degree of confidence when there is a need to discriminate between strains belonging to the same species (69 and 89% strain-level prediction accuracies for Raman and FT-IR, respectively, based on chemometric analysis; AlMasoud et al., 2016). Given these results, we anticipate infrared imaging to become an increasingly common technique in the field of microbial ecology, particularly when there is a need for quantitatively analyzing multi-species assemblages and/or in-depth physiological profiling of selected isolates.
Recommendations and Outlook
The spectroscopic imaging of microbial cells in physically and chemically complex samples involves diverse analytical challenges. While addressing these will often require sample-specific optimization steps (such as identifying an appropriate laser wavelength; Edwards et al., 2003; Chan et al., 2004; Jorge Villar et al., 2005), many of them could be overcome by carefully selecting between Raman- and FT-IR-based measurements or a combination of both. Based on the case studies discussed in this Mini Review, it is possible to identify several general guidelines for achieving this (Table 1). The suggestions provided in Table 1 additionally highlight the promising role that live-cell FT-IR imaging could play in environmental microbiological research, further to Raman measurements which have traditionally been more common in this field. The future development of vibrational spectroscopy instrumentation and analytical methods may serve to further enhance the cross-compatibility of Raman and FT-IR techniques (e.g., via improved access to advanced Raman imaging equipment and validation of new protocols for FT-IR-SIP).
Table 1.
Experimental goals associated with the Raman and FT-IR imaging of single microbial cells in complex biological samples.
| Goal | Recommended technique | Notes | Reference |
|---|---|---|---|
| Analysis of motile cells and/or cell sorting | Raman microspectroscopy | Optical tweezers can be used to trap or move individual cells | Huang et al., 2010; Berry et al., 2015 |
| Detection of cells on autofluorescent and opaque surfaces | Both | Autofluorescence does not interfere with IR measurements; Raman measurements possible using resonance Raman, SERS, sample photobleaching or aging | Ojeda et al., 2009; Polisetti et al., 2016 |
| Addressing other sources of background interference | Both | Water is a strong IR absorber; using microfluidics or an ATR accessory can reduce signal interference | Kuimova et al., 2009; Birarda et al., 2016; Loutherback et al., 2016 |
| Stable isotope probing | Both | Approaches currently better-established for Raman analyses | Wang et al., 2016 |
| Imaging of large (cm-scale) surface areas | FT-IR microspectroscopy | FPA detectors readily available for FT-IR instruments; Raman instrumentation also available, but not as widely accessible | Kuimova et al., 2009; Kastyak-Ibrahim et al., 2012; Miller et al., 2013; Ando et al., 2016 |
| Localization of cells in 3D space | Raman microspectroscopy | Imaging of z-stacks possible using confocal Raman measurements | Große et al., 2015; Silge et al., 2015 |
| High-resolution (including subcellular) measurements | Both | Raman analyses (e.g., TERS) better-established; also possible using FT-IR but requires specialist equipment or access to synchrotron beamline | Mariani et al., 2010; Opilik et al., 2013; Saulou et al., 2013; Rusciano et al., 2014; Findlay et al., 2015 |
| Label-free discrimination between individual strains or taxa | FT-IR microspectroscopy | FT-IR analyses can outperform Raman spectroscopy in terms of spectral quality and reproducibility | AlMasoud et al., 2016; Muhamadali et al., 2016 |
Recommendations for analytical techniques are provided for meeting each goal.
Additionally to considering the benefits and pitfalls inherent to Raman vs. FT-IR measurements, experiments focusing on the imaging of single cells in complex habitats can be expected to profit from combining these techniques with other analytical approaches (Figure 1). Synchrotron-FT-IR microspectroscopy has been paired with synchrotron ultraviolet microspectroscopy and time-of-flight-secondary ion mass spectrometry (ToF-SIMS) for the analysis of human liver tissue, with each technique yielding unique information on the chemical composition of the sample (Petit et al., 2010). Raman microspectroscopy has been combined with nanoscale secondary ion mass spectrometry (NanoSIMS) to quantify the bacterial uptake of deuterium during heavy water labeling experiments (Berry et al., 2015). Moreover, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-ToF-MS) is compatible with microbiological analyses and Raman imaging (Bocklitz et al., 2013; Pande et al., 2016; Stasulli and Shank, 2016). Although it has not yet been applied for the imaging of cells within complex environments such as soils, MALDI-ToF-MS been used to characterize individual bacterial colonies (Pande et al., 2016; Stasulli and Shank, 2016). Promisingly, the technique can be used for strain identification (Singhal et al., 2015) and a method for single-cell MALDI analyses has also been developed (Xiong et al., 2016). Vibrational spectroscopic imaging of microbial cells could be further combined with techniques that provide information on the 3D structure of the surrounding environment. X-ray computed tomography, for example, has been used to visualize roots within undisturbed soil (Mooney et al., 2012). The technique has also been used to produce micron-scale 3D representations of soil pore space (Nunan et al., 2006).
FIGURE 1.
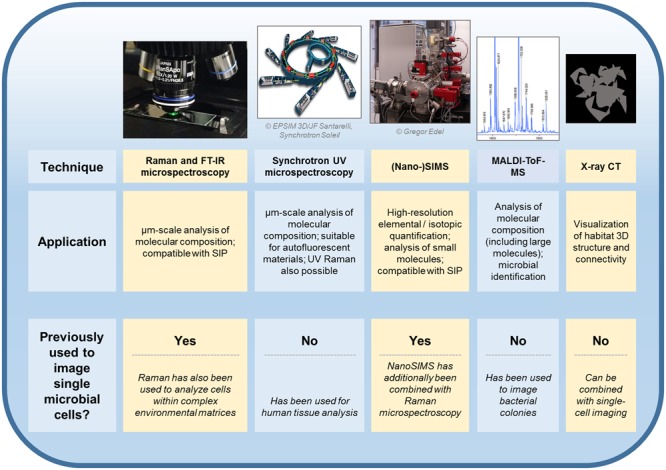
Techniques which have or could be utilized for the in situ imaging of single microbial cells within physically and chemically complex environments. Previously demonstrated applications of each approach are discussed in the main text. FT-IR, Fourier-transform infrared; SIP, stable isotope probing; UV, ultraviolet; (Nano-)SIMS, nanoscale secondary ion mass spectrometry; MALDI-ToF-MS, matrix-assisted laser desorption/ionization mass spectrometry time-of-flight mass spectrometry; CT, computed tomography.
One of the most important challenges involved in the spectral imaging of microorganisms within their native habitats, regardless of the techniques involved, concerns the ability to successfully discriminate between cells and other materials. Additionally, an ability to discern between diverse taxa is required to understand the distribution and activities of microbial cells at the community level. To facilitate research into these topics, we strongly recommend that databases including relevant reference spectra are made available as part of future publications. We also note that using Raman and/or FT-IR spectroscopy alone for the reliable identification of microbial taxa often remains challenging (see FT-IR imaging), and that result using these methods may need to be verified using additional methods. For example, Raman-activated cell sorting has recently been combined with single-cell genomics to identify members of a novel cyanobacterial order within seawater samples (Song et al., 2017). Ultimately, the approaches discussed in this Mini Review could enable us to significantly improve our knowledge of microbial community assembly and the contribution of interspecies interactions to key ecosystem processes, including the cycling of carbon within soils, sediments and other spatially structured habitats.
Author Contributions
All authors listed, have made substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the U.S. Department of Energy (Grant No. BER DE-SC0013887) and the European Research Council (Starting Grant 741623 FunKeyGut).
References
- AlMasoud N., Xu Y., Ellis D. I., Rooney P., Turton J. F., Goodacre R. (2016). Rapid discrimination of Enterococcus faecium strains using phenotypic analytical techniques. Anal. Methods 8 7603–7613. 10.1039/C6AY02326F [DOI] [Google Scholar]
- Amenabar I., Poly S., Goikoetxea M., Nuansing W., Lasch P., Hillenbrand R. (2017). Hyperspectral infrared nanoimaging of organic samples based on Fourier transform infrared nanospectroscopy. Nat. Commun. 8:14402 10.1038/ncomms14402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando J., Palonpon A. F., Sodeoka M., Fujita K. (2016). High-speed Raman imaging of cellular processes. Curr. Opin. Chem. Biol. 33 16–24. 10.1016/j.cbpa.2016.04.005 [DOI] [PubMed] [Google Scholar]
- Berry D., Mader E., Lee T. K., Woebken D., Wang Y., Zhu D. (2015). Tracking heavy water (D2O) incorporation for identifying and sorting active microbial cells. Proc. Natl. Acad. Sci. U.S.A. 112 E194–E203. 10.1073/pnas.1420406112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birarda G., Ravasio A., Suryana M., Maniam S., Holman H.-Y. N., Grenci G. (2016). IR-Live: fabrication of a low-cost plastic microfluidic device for infrared spectromicroscopy of living cells. Lab Chip 16 1644–1651. 10.1039/c5lc01460c [DOI] [PubMed] [Google Scholar]
- Bocklitz T. W., Crecelius A. C., Matthäus C., Tarcea N., von Eggeling F., Schmitt M., et al. (2013). Deeper understanding of biological tissue: quantitative correlation of MALDI-ToF and Raman imaging. Anal. Chem. 85 10829–10834. 10.1021/ac402175c [DOI] [PubMed] [Google Scholar]
- Camp C. H., Jr., Cicerone M. T. (2015). Chemically sensitive bioimaging with coherent Raman scattering. Nat. Photonics 9 295–305. 10.1038/nphoton.2015.60 [DOI] [Google Scholar]
- Centrone A. (2015). Infrared imaging and spectroscopy beyond the diffraction limit. Annu. Rev. Anal. Chem. 8 101–126. 10.1146/annurev-anchem-071114-040435 [DOI] [PubMed] [Google Scholar]
- Chan J. W., Esposito A. P., Talley C. E., Hollars C. W., Lane S. M., Huser T. (2004). Reagentless identification of single bacterial spores in aqueous solution by confocal laser tweezers Raman spectroscopy. Anal. Chem. 76 599–603. 10.1021/ac0350155 [DOI] [PubMed] [Google Scholar]
- Chrimes A. F., Khoshmanesh K., Stoddart P. R., Mitchell A., Kalantar-zadeh K. (2013). Microfluidics and Raman microscopy: current applications and future challenges. Chem. Soc. Rev. 42 5880–5906. 10.1039/c3cs35515b [DOI] [PubMed] [Google Scholar]
- Cicerone M. (2016). Molecular imaging with CARS micro-spectroscopy. Curr. Opin. Chem. Biol. 33 179–185. 10.1016/j.cbpa.2016.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorling K. M., Baker M. J. (2013). Rapid FTIR chemical imaging: highlighting FPA detectors. Trends Biotechnol. 31 437–438. 10.1016/j.tibtech.2013.05.008 [DOI] [PubMed] [Google Scholar]
- Edwards H. G. M., Newton E. M., Wynn-Williams D. D., Dickensheets D., Schoen C., Crowder C. (2003). Laser wavelength selection for Raman spectroscopy of microbial pigments in situ in Antarctic desert ecosystem analogues of former habitats on Mars. Int. J. Astrobiol. 1 333–348. 10.1017/S147355040300123X [DOI] [Google Scholar]
- Escoriza M. F., Vanbriesen J. M., Stewart S., Maier J. (2006). Studying bacterial metabolic states using Raman spectroscopy. Appl. Spectrosc. 60 971–976. 10.1366/000370206778397290 [DOI] [PubMed] [Google Scholar]
- Fike D. A., Gammon C. L., Ziebis W., Orphan V. J. (2008). Micron-scale mapping of sulfur cycling across the oxycline of a cyanobacterial mat: a paired nanoSIMS and CARD-FISH approach. ISME J. 2 749–759. 10.1038/ismej.2008.39 [DOI] [PubMed] [Google Scholar]
- Findlay C. R., Wiens R., Rak M., Sedlmair J., Hirschmugl C. J., Morrison J., et al. (2015). Rapid biodiagnostic ex vivo imaging at 1 μm pixel resolution with thermal source FTIR FPA. Analyst 140 2493–2503. 10.1039/c4an01982b [DOI] [PubMed] [Google Scholar]
- Große C., Bergner N., Dellith J., Heller R., Bauer M., Mellmann A., et al. (2015). Label-free imaging and spectroscopic analysis of intracellular bacterial infections. Anal. Chem. 87 2137–2142. 10.1021/ac503316s [DOI] [PubMed] [Google Scholar]
- Haase K., Kröger-Lui N., Pucci A., Schönhals A., Petrich W. (2016). Advancements in quantum cascade laser-based infrared microscopy of aqueous media. Analyst 187 119–134. 10.1039/c5fd00177c [DOI] [PubMed] [Google Scholar]
- Harz M., Kiehntopf M., Stöckel S., Rösch P., Straube E., Deufel T., et al. (2009). Direct analysis of clinical relevant single bacterial cells from cerebrospinal fluid during bacterial meningitis by means of micro-Raman spectroscopy. J. Biophotonics 2 70–80. 10.1002/jbio.200810068 [DOI] [PubMed] [Google Scholar]
- Hermann P., Hermelink A., Lausch V., Holland G., Möller L., Bannert N., et al. (2011). Evaluation of tip-enhanced Raman spectroscopy for characterizing different virus strains. Analyst 136 1148–1152. 10.1039/c0an00531b [DOI] [PubMed] [Google Scholar]
- Holman H. Y., Miles R., Hao Z., Wozei E., Anderson L. M., Yang H. (2009). Real-time chemical imaging of bacterial activity in biofilms using open-channel microfluidics and synchrotron FTIR spectromicroscopy. Anal. Chem. 81 8564–8570. 10.1021/ac9015424 [DOI] [PubMed] [Google Scholar]
- Hong W., Liao C.-S., Zhao H., Younis W., Zhang Y., Seleem M. N., et al. (2016). In situ detection of a single bacterium in complex environment by hyperspectral CARS imaging. ChemistrySelect 3 513–517. 10.1002/slct.201600166 [DOI] [Google Scholar]
- Huang W. E., Li M., Jarvis R. M., Goodacre R., Banwart S. A. (2010). Shining light on the microbial world: the application of Raman microspectroscopy. Adv. Appl. Microbiol. 70 153–186. 10.1016/S0065-2164(10)70005-8 [DOI] [PubMed] [Google Scholar]
- Igisu M., Takai K., Ueno Y., Nishizawa M., Nunoura T., Hirai M., et al. (2012). Domain-level identification and quantification of relative prokaryotic cell abundance in microbial communities by micro-FTIR spectroscopy. Environ. Microbiol. Rep. 4 42–49. 10.1111/j.1758-2229.2011.00277.x [DOI] [PubMed] [Google Scholar]
- Jamme F., Vindigni J.-D., Méchin V., Cherifi T., Chardot T., Froissard M. (2013). Single cell synchrotron FT-IR microspectroscopy reveals a link between neutral lipid and storage carbohydrate fluxes in S. cereviciae. PLoS ONE 8:e74421 10.1371/journal.pone.0074421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorge Villar S. E., Edwards H. G. M., Worland M. R. (2005). Comparative evaluation of Raman spectroscopy at different wavelengths for extremophile exemplars. Orig. Life Evol. Biosph. 35 489–506. 10.1007/s11084-005-3528-4 [DOI] [PubMed] [Google Scholar]
- Kalasinsky K. S., Hadfield T., Shea A. A., Kalasinsky V. F., Nelson M. P., Neiss J., et al. (2007). Raman chemical imaging spectroscopy reagentless detection and identification of pathogens: signature development and evaluation. Anal. Chem. 79 2658–2673. 10.1021/ac0700575 [DOI] [PubMed] [Google Scholar]
- Kano H., Segawa H., Okuno M., Leproux P., Couderc V. (2016). Hyperspectral coherent Raman imaging – principle, theory, instrumentation, and applications to life sciences. J. Raman Spectrosc. 47 116–123. 10.1002/jrs.4853 [DOI] [Google Scholar]
- Kastyak-Ibrahim M. Z., Nasse M. J., Rak M., Hirschmugl C., Del Bigio M. R., Albensi B. C., et al. (2012). Biochemical label-free tissue imaging with subcellular-resolution synchrotron FTIR with focal plane array detector. Neuroimage 60 376–383. 10.1016/j.neuroimage.2011.11.069 [DOI] [PubMed] [Google Scholar]
- Krafft C., Dietzek B., Popp J. (2009). Raman and CARS microspectroscopy of cells and tissues. Analyst 134 1046–1057. 10.1039/b822354h [DOI] [PubMed] [Google Scholar]
- Kuimova M. K., Chan K. L. A., Kazarian S. G. (2009). Chemical imaging of live cancer cells in the natural aqueous environment. Appl. Spectrosc. 63 164–171. 10.1366/000370209787391969 [DOI] [PubMed] [Google Scholar]
- Li M., Canniffe D. P., Jackson P. J., Davison P. A., FitzGerald S., Dickman M. J., et al. (2012). Rapid resonance Raman microspectroscopy to probe carbon fixation by single cells in microbial communities. ISME J. 6 875–885. 10.1038/ismej.2011.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loutherback K., Chen L., Holman H.-Y. N. (2015). Open-channel microfluidic membrane device for long-term FT-IR spectromicroscopy of live adherent cells. Anal. Chem. 87 4601–4606. 10.1021/acs.analchem.5b00524 [DOI] [PubMed] [Google Scholar]
- Loutherback K., Giovanni B., Liang C., Holman H.-Y. (2016). Microfluidic approaches to synchrotron radiation-based Fourier transform infrared (SR-FTIR) spectral microscopy of living biosystems. Protein Pept. Lett. 23 273–282. 10.2174/0929866523666160106154035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Al-Qadiri H. M., Lin M., Rasco B. A. (2011). Application of mid-infrared and Raman spectroscopy to the study of bacteria. Food Bioprocess Technol. 4 919–935. 10.1007/s11947-011-0516-8 [DOI] [Google Scholar]
- Mariani M. M., Day P. J. R., Deckert V. (2010). Applications of modern micro-Raman spectroscopy for cell analyses. Integr. Biol. 2 94–101. 10.1039/b920572a [DOI] [PubMed] [Google Scholar]
- Miller L. M., Bourassa M. W., Smith R. J. (2013). FTIR spectroscopic imaging of protein aggregation in living cells. BBA Biomembranes 1828 2339–2346. 10.1016/j.bbamem.2013.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney S. J., Pridmore T. P., Helliwell J., Bennett M. J. (2012). Developing X-ray computed tomography to non-invasively image 3-D root systems architecture in soil. Plant Soil 352 1–22. 10.1007/s11104-011-1039-9 [DOI] [Google Scholar]
- Muhamadali H., Chisanga M., Subaihi A., Goodacre M. (2015). Combining Raman and FT-IR spectroscopy with quantitative isotopic labeling for differentiation of E. coli cells at community and single cell levels. Anal. Chem. 87 4578–4586. 10.1021/acs.analchem.5b00892 [DOI] [PubMed] [Google Scholar]
- Muhamadali H., Subaihi A., Mohammadtaheri M., Xu Y., Ellis D. I., Ramanathan R., et al. (2016). Rapid, accurate, and comparative differentiation of clinically and industrially relevant microorganisms via multiple vibrational spectroscopic fingerprinting. Analyst 141 5127–5136. 10.1039/c6an00883f [DOI] [PubMed] [Google Scholar]
- Nasse M. J., Walsh M. J., Mattson E. C., Reininger R., Kajdacsy-Balla A., Macias V., et al. (2011). High-resolution Fourier-transform infrared chemical imaging with multiple synchrotron beams. Nat. Methods 8 413–416. 10.1038/nmeth.1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunan N., Ritz K., Rivers M., Feeney D. S., Young I. M. (2006). Investigating microbial micro-habitat structure using X-ray computed tomography. Geoderma 133 398–407. 10.1016/j.geoderma.2005.08.004 [DOI] [Google Scholar]
- Ojeda J. J., Dittrich M. (2012). “Fourier transform infrared spectroscopy for molecular analysis of microbial cells,” in Microbial Systems Biology: Methods and Protocols (Methods in Molecular Biology) Vol. 881 ed. Navid A. (Berlin: Springer; ) 187–211. [DOI] [PubMed] [Google Scholar]
- Ojeda J. J., Romero-González M. E., Banwart S. A. (2009). Analysis of bacteria on steel surfaces using reflectance micro-Fourier transform infrared spectroscopy. Anal. Chem. 81 6467–6473. 10.1021/ac900841c [DOI] [PubMed] [Google Scholar]
- Okuno M., Kano H., Leproux P., Couderc V., Day J. P. R., Bonn M., et al. (2010). Quantitative CARS molecular fingerprinting of single living cells with the use of the maximum entropy method. Angew. Chem. Int. Ed. Engl. 49 6773–6777. 10.1002/anie.201001560 [DOI] [PubMed] [Google Scholar]
- Olschewski K., Kämmer E., Stöckel S., Bocklitz T., Deckert-Gaudig T., Zell R., et al. (2015). A manual and an automatic TERS based virus discrimination. Nanoscale 7 4545–4552. 10.1039/c4nr07033j [DOI] [PubMed] [Google Scholar]
- Opilik L., Schmid T., Zenobi R. (2013). Modern Raman imaging: vibrational spectroscopy on the micrometer and nanometer scales. Annu. Rev. Anal. Chem. 6 379–398. 10.1146/annurev-anchem-062012-092646 [DOI] [PubMed] [Google Scholar]
- Pande S., Kaftan F., Lang S., Svatoš A., Germerodt S., Kost C. (2016). Privatization of cooperative benefits stabilizes mutualistic cross-feeding interactions in spatially structured environments. ISME J. 10 1413–1423. 10.1038/ismej.2015.212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit V. W., Réfrégiers M., Guettier C., Jamme F., Sebanayakam K., Brunelle A., et al. (2010). Multimodal spectroscopy combining time-of-flight-secondary ion mass spectrometry, synchrotron-FT-IR, and synchrotron-UV microspectroscopies on the same tissue section. Anal. Chem. 82 3963–3968. 10.1021/ac100581y [DOI] [PubMed] [Google Scholar]
- Polisetti S., Bible A. N., Morrell-Falvey J. L., Bohn P. W. (2016). Raman chemical imaging of the rhizosphere bacterium Pantoea sp. YR343 and its co-culture with Arabidopsis thaliana. Analyst 141 2175–2182. 10.1039/c6an00080k [DOI] [PubMed] [Google Scholar]
- Ratzke C., Gore J. (2016). Self-organized patchiness facilitates survival in a cooperatively growing Bacillus subtilis population. Nat. Microbiol. 1 16022 10.1038/nmicrobiol.2016.22 [DOI] [PubMed] [Google Scholar]
- Reddy R. K., Walsh M. J., Schulmerich M. V., Carney P. S., Bhargava R. (2013). High-definition infrared spectroscopic imaging. Appl. Spectrosc. 67 93–105. 10.1366/11-06568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resat H., Bailey V., McCue L. A., Konopka A. (2012). Modeling microbial dynamics in heterogeneous environments: growth on soil carbon sources. Microb. Ecol. 63 883–897. 10.1007/s00248-011-9965-x [DOI] [PubMed] [Google Scholar]
- Roose T., Keyes S. D., Daly K. R., Carminati A., Otten W., Vetterlein D., et al. (2016). Challenges in imaging and predictive modeling of rhizosphere processes. Plant Soil 407 9–38. 10.1007/s11104-016-2872-7 [DOI] [Google Scholar]
- Rusciano G., Zito G., Isticato R., Sirec T., Ricca E., Bailo E., et al. (2014). Nanoscale chemical imaging of Bacillus subtilis spores by combining tip-enhanced Raman scattering and advanced statistical tools. ACS Nano 8 12300–12309. 10.1021/nn504595k [DOI] [PubMed] [Google Scholar]
- Saulou C., Jamme F., Girbal L., Maranges C., Fourquaux I., Cocaign-Bousquet M., et al. (2013). Synchrotron FTIR microspectroscopy of Escherichia coli at single-cell scale under silver-induced stress conditions. Anal. Bioanal. Chem. 405 2685–2697. 10.1007/s00216-013-6725-4 [DOI] [PubMed] [Google Scholar]
- Silge A., Abdou E., Schneider K., Meisel S., Bocklitz T., Lu-Walther H. W., et al. (2015). Shedding light on host niches: label-free in situ detection of Mycobacterium gordonae via carotenoids in macrophages by Raman microspectroscopy. Cell. Microbiol. 17 832–842. 10.1111/cmi.12404 [DOI] [PubMed] [Google Scholar]
- Singhal N., Kumar M., Kanaujia P. K., Virdi J. S. (2015). MALDI-TOF mass spectrometry: an emerging technology for microbial identification and diagnosis. Front. Microbiol. 6:791 10.3389/fmicb.2015.00791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoog D. A., Holler F. J., Crouch S. R. (2007). Principles of Instrumental Analysis 6th Edn. Belmont: Thomson Brooks / Cole. [Google Scholar]
- Song Y., Kaster A.-K., Vollmers J., Song Y., Davison P. A., Frentrup M., et al. (2017). Single-cell genomics based on Raman sorting reveals novel carotenoid-containing bacteria in the Red Sea. Microb. Biotechnol. 10 125–137. 10.1111/1751-7915.12420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasulli N. M., Shank E. A. (2016). Profiling the metabolic signals involved in chemical communication between microbes using imaging mass spectrometry. FEMS Microbiol. Rev. 40 807–813. 10.1093/femsre/fuw032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M., McEwen G. D., Wu Y., Miller C. D., Zhou A. (2013). Characterization and analysis of mycobacteria and Gram-negative bacteria and co-culture mixtures by Raman microspectroscopy, FTIR, and atomic force microscopy. Anal. Bioanal. Chem. 405 1577–1591. 10.1007/s00216-012-6556-8 [DOI] [PubMed] [Google Scholar]
- Vos M., Wolf A. B., Jennings S. J., Kowalchuk G. A. (2013). Micro-scale determinants of bacterial diversity in soil. FEMS Microbiol. Rev. 37 936–954. 10.1111/1574-6976.12023 [DOI] [PubMed] [Google Scholar]
- Wagner M. (2009). Single-cell ecophysiology of microbes as revealed by Raman microspectroscopy or secondary ion mass spectrometry imaging. Annu. Rev. Microbiol. 63 411–429. 10.1146/annurev.micro.091208.073233 [DOI] [PubMed] [Google Scholar]
- Wang Y., Huang W. E., Cui L., Wagner M. (2016). Single cell stable isotope probing in microbiology using Raman microspectroscopy. Curr. Opin. Biotechnol. 41 34–42. 10.1016/j.copbio.2016.04.018 [DOI] [PubMed] [Google Scholar]
- Wei L., Hu F., Chen Z., Shen Y., Zhang L., Min W. (2016). Live-cell bioorthogonal chemical imaging: stimulated Raman scattering microscopy of vibrational probes. Acc. Chem. Res. 49 1494–1502. 10.1021/acs.accounts.6b00210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenning M., Theilmann V., Scherer S. (2005). Rapid analysis of two food-borne microbial communities at the species level by Fourier-transform infrared microspectroscopy. Environ. Microbiol. 8 848–857. 10.1111/j.1462-2920.2005.00971.x [DOI] [PubMed] [Google Scholar]
- Xiong C., Zhou X., He Q., Huang X., Wang J., Peng W.-P., et al. (2016). Development of visible-wavelength MALDI cell mass spectrometry for high-efficiency single-cell analysis. Anal. Chem. 88 11913–11918. 10.1021/acs.analchem.6b03789 [DOI] [PubMed] [Google Scholar]
- Yue S., Cheng J.-X. (2016). Deciphering single cell metabolism by coherent Raman scattering microscopy. Curr. Opin. Chem. Biol. 33 46–57. 10.1016/j.cbpa.2016.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]


